Key Points
Question
What are the tumor biological differences in invasive breast cancers from patients of African and European ancestry?
Findings
In the cohort study from The Cancer Genome Atlas, a racial disparity in breast cancer–free interval and distribution of aggressive subtypes of breast cancers was detected. After taking into account differences in prevalence of intrinsic subtypes, modest molecular differences in gene expression, protein expression, somatic mutations, and DNA methylation patterns were observed, and most significantly, higher genetic contribution to estrogen receptor–negative breast cancer was seen in black patients than in white patients.
Meaning
Biological differences between breast cancers in blacks and whites may be linked to differences in the distribution of germline genetic variants, and interventions to improve cancer risk assessment and optimal use of more effective targeted therapies have the potential to close the widening mortality gap between black and white patients with breast cancer.
Abstract
Importance
African Americans have the highest breast cancer mortality rate. Although racial difference in the distribution of intrinsic subtypes of breast cancer is known, it is unclear if there are other inherent genomic differences that contribute to the survival disparities.
Objectives
To investigate racial differences in breast cancer molecular features and survival and to estimate the heritability of breast cancer subtypes.
Design, Setting, and Participants
Among a convenience cohort of patients with invasive breast cancer, breast tumor and matched normal tissue sample data (as of September 18, 2015) were obtained from The Cancer Genome Atlas.
Main Outcomes and Measures
Breast cancer–free interval, tumor molecular features, and genetic variants.
Results
Participants were 930 patients with breast cancer, including 154 black patients of African ancestry (mean [SD] age at diagnosis, 55.66 [13.01] years; 98.1% [n = 151] female) and 776 white patients of European ancestry (mean [SD] age at diagnosis, 59.51 [13.11] years; 99.0% [n = 768] female). Compared with white patients, black patients had a worse breast cancer-free interval (hazard ratio, HR=1.67, 95 CI: 1.02-2.74; P = .043). They had a higher likelihood of basal-like (odds ratio, 3.80; 95% CI, 2.46-5.87; P < .001) and human epidermal growth factor receptor 2 (ERBB2 [formerly HER2])–enriched (odds ratio, 2.22; 95% CI, 1.10-4.47; P = .027) breast cancer subtypes, with the Luminal A subtype as the reference. Blacks had more TP53 mutations and fewer PIK3CA mutations than whites. While most molecular differences were eliminated after adjusting for intrinsic subtype, the study found 16 DNA methylation probes, 4 DNA copy number segments, 1 protein, and 142 genes that were differentially expressed, with the gene-based signature having an excellent capacity for distinguishing breast tumors from black vs white patients (cross-validation C index, 0.878). Using germline genotypes, the heritability of breast cancer subtypes (basal vs nonbasal) was estimated to be 0.436 (P = 1.5 × 10−14). The estrogen receptor–positive polygenic risk score built from 89 known susceptibility variants was higher in blacks than in whites (difference, 0.24; P = 2.3 × 10−5), while the estrogen receptor–negative polygenic risk score was much higher in blacks than in whites (difference, 0.48; P = 2.8 × 10−11).
Conclusions and Relevance
On the molecular level, after adjusting for intrinsic subtype frequency differences, this study found a modest number of genomic differences but a significant clinical survival outcome difference between blacks and whites in The Cancer Genome Atlas data set. Moreover, more than 40% of breast cancer subtype frequency differences could be explained by genetic variants. These data could form the basis for the development of molecular targeted therapies to improve clinical outcomes for the specific subtypes of breast cancers that disproportionately affect black women. Findings also indicate that personalized risk assessment and optimal treatment could reduce deaths from aggressive breast cancers for black women.
This cohort study from The Cancer Genome Atlas investigates racial differences in breast cancer molecular features and survival and estimates the heritability of breast cancer subtypes.
Introduction
Breast cancer is the most common tumor in women. While the incidence among black and white women in the United States has converged at 135 cases per 100 000 women per year in recent years, the mortality gap has continued to increase, with a 42% higher death rate in black patients. The reasons for this survival gap are multifactorial and may include access to care and inherent biological tumor differences. One known biological cause of this racial disparity is the higher frequency of basal-like or triple-negative breast cancers (TNBCs) (ie, negative for estrogen receptor [ER], progesterone receptor, and human epidermal growth factor receptor 2 [ERBB2, formerly HER2]) among black women. However, there is a paucity of data on additional biological or genomic differences that explain the higher mortality rate in blacks. Increased genetic predisposition due to higher risk allele frequencies at the TERT (OMIM 187270) locus and a higher prevalence of specific lifestyle factors, such as lack of breastfeeding, may contribute to intrinsic subtype frequency differences seen across ethnicities.
To address this knowledge gap, we systematically investigated molecular features, including gene expression, protein expression, somatic mutations, somatic DNA copy number alteration (CNA), and DNA methylation patterns, between breast cancers of black and white patients and examined differences in breast cancer recurrence and overall survival in relation to these molecular features. In addition, we assessed germline genetic variants for breast cancer intrinsic subtypes and estimated the heritability of these subtypes.
Methods
Study Cohort
This study used breast cancer data (as of September 18, 2015) from The Cancer Genome Atlas (TCGA), which are publicly available from TCGA Data Portal (https://tcga-data.nci.nih.gov/docs/publications/tcga/). Data fields were processed using established methods unless otherwise specified (eTable 1 in Supplement 1). We estimated genomic race using principal component analysis of germline genotype data from 1062 patients (eMethods in Supplement 2). Patients were grouped into genomic black (≥50% African ancestry) or genomic white (≥90% European ancestry). Herein, we present analytical methods unique to this study, while genomic data generation and processing have been previously described. The samples were collected under institutional review board–approved protocols at each participating institution and analysis was performed by the African American TCGA Breast Cancer Working Group. This study used only deidentified data from TCGA Project.
Gene Expression by RNA Sequencing
After excluding genes not expressed or not variable (zero reads or SD<1), upper quartile normalized RNA sequencing data were log2 transformed. Prediction analysis of microarray 50 (PAM50) intrinsic subtypes were called as previously described. Multinomial logistic regression was used to examine the association between breast cancer subtype and genomic race.
Differentially expressed genes between black patients and white patients were identified by linear regression, fitting a model adjusting for age at diagnosis and batch (36 batches) and further adjusting for intrinsic subtype. False discovery rates (FDRs) by Benjamini-Hochberg and Bonferroni-adjusted P values were calculated to control for multiple testing. Hierarchical clustering analysis with a Spearman rank correlation similarity measure was used for visualization. We used elastic net penalized logistic regression to create a race-differentiated gene signature, with cross-validation to tune the penalty parameters. Using 10-fold cross-validation data sets, area under the receiver operating characteristic curve, ie, C index (AUROC), was calculated to assess the discriminating capacity of the signature.
Protein Expression
Reverse phase protein array data contain expression levels for 215 proteins and phosphorylated proteins for 745 patients (114 black and 631 white). Linear regression models were used to identify differentially expressed proteins, adjusting for age at diagnosis and batches. Proteins that passed the multiple testing correction with FDRs less than 0.05 were included in a multivariable logistic regression. AUROC was calculated using 10-fold cross-validation data sets.
Somatic Mutation and DNA CNAs
We examined genes with mutations in coding regions, integrating information from DNA and RNA sequencing. The numbers of mutated genes and CNA segments per patient were compared between blacks and whites using Wilcoxon rank sum tests. Recurrently mutated genes and recurrent CNAs were identified by MutSigCV2 and GISTIC, respectively. Differences in mutation frequency or CNAs were determined with Fisher exact test and logistic regression to adjust for subtype.
DNA Methylation
DNA methylation (Infinium HumanMethylation450; Illumina) data were available for 124 black cases and 517 white cases. Of 480 721 probes, we identified 13 811 cancer-associated hypermethylation probes located within 1500 base pairs of a transcription start site and 9436 cancer-associated hypomethylation probes that overlapped with putative enhancer regions (eMethods in Supplement 2). We used linear regression models to identify race-specific DNA methylation, with adjustment for age, tumor purity, and subtype.
Survival Analysis
We used a disease-free survival definition that included all breast cancer–related events to define the breast cancer–free interval (BCFI). Events for BCFI were defined as locoregional recurrences, distant metastases, new primary tumors in the breast, or deaths from breast cancer. The BCFI was defined as the time from the date of the initial pathological diagnosis to the date of the events defined above, the date of last contact, or the date of death. For BCFI analysis, 17 patients with stage IV cancer at diagnosis were excluded. Another 17 patients who died with tumor but without an indication of a defined new tumor event were excluded for lack of information, resulting in 896 patients for BCFI analysis. Overall survival was defined as the date of diagnosis to the date of last contact or death (in 930 patients). Because of a limited duration of follow-up, survival analysis may be underpowered to detect racial disparity, especially for ER-positive breast cancer. To compensate for this limitation, we also calculated the PAM50-based risk of recurrence score, which estimates the biological potential of recurrence.
The Kaplan-Meier method was used to generate survival curves. We fit a series of Cox proportional hazards regression models, starting from a model with race only, then a multivariable model adjusting for age and American Joint Committee on Cancer stage, then subtypes, and then further adjusting for molecular features (gene expression, protein expression, or mutation) that were differentially present between blacks and whites. Only molecular features associated with BCFI in Cox proportional hazards regression models were considered potential mediators.
Germline Variants
DNA from germline samples was hybridized to arrays (SNP Array 6.0; Affymetrix) to genotype single-nucleotide polymorphisms (SNPs). Of 93 breast cancer susceptibility loci identified in previous genome-wide association studies, a total of 24 were genotyped. For the remaining SNPs, we conducted imputation analysis using IMPUTE2 software, and 65 SNPs could be imputed reliably (imputation score >0.7; mean, 0.97). Therefore, a total of 89 SNPs were analyzed in this study. First, we compared SNP allele frequencies of these 89 SNPs between racial groups using Fisher exact test, and we determined if they varied by subtype after adjusting for race using logistic regression. Second, because the power for assessing individual SNPs was limited in the TCGA data set, we derived polygenetic risk scores (PRSs) for ER-positive and ER-negative breast cancers to investigate the combined effects of 89 SNPs using the following equation:
| PRS=β1χ1+β2χ2+…+βκχκ+βnχn, |
where βκ is the per-allele log odds ratio for breast cancer associated with risk alleles published previously and where χκ is the number of risk alleles (0, 1, or 2) for the kth SNP. Third, because many breast cancer susceptibility loci are yet to be identified, we tried to extract breast cancer subtype–related information using all genetic variants on Affymetrix arrays. With a mixed-effects model implemented in GCTA, we estimated subtype heritability. The phenotype in the mixed-effects model is a simplified subtype (basal vs nonbasal). To increase statistical power, we also estimated the heritability of ESR1 and ERBB2 gene expression, 2 genes that are important components of the PAM50 subtypes.
Results
Molecular Portraits of Breast Tumors by Race
In the TCGA cohort, there were 154 black patients of African ancestry and 776 white patients of European ancestry. While not population based, many of our findings were in line with previous population-based research. For example, black patients were diagnosed at a younger age and were more likely to have ER-negative or nonluminal subtypes of breast cancers (Table 1). After adjusting for age, black patients had a higher odds of basal-like (odds ratio, 3.80; 95% CI, 2.46-5.87; P < .001) and HER2-enriched (odds ratio, 2.22; 95% CI, 1.10-4.47; P = .027) breast cancer subtypes than white patients, with the luminal A subtype as the reference.
Table 1. Description of Breast Cancer Cohorta.
| Variable | White (n = 776) |
Black (n = 154) |
P Value |
|---|---|---|---|
| Age at diagnosis, mean (SD), y | 59.51 (13.11) | 55.66 (13.01) | .0009b |
| Sex, No. (%) | |||
| Female | 768 (99.0) | 151 (98.1) | .40 |
| Male | 8 (1.0) | 3 (1.9) | |
| PAM50 subtype, No. (%) | |||
| Luminal A | 432 (55.7) | 52 (33.8) | <.0001 |
| Luminal B | 161 (20.7) | 27 (17.5) | |
| HER2 | 44 (5.7) | 12 (7.8) | |
| Basal | 114 (14.7) | 56 (36.4) | |
| Normal | 24 (3.1) | 7 (4.5) | |
| ER, No. (%) | |||
| Positive | 597 (76.9) | 94 (61.0) | <.0001c |
| Negative | 138 (17.8) | 60 (39.0) | |
| Indeterminate or not evaluated | 39 (5.0) | 0 | |
| PR, No. (%) | |||
| Positive | 519 (66.9) | 79 (51.3) | <.0001c |
| Negative | 213 (27.4) | 75 (48.7) | |
| Indeterminate or not evaluated | 44 (5.7) | 0 | |
| HER2, No. (%) | |||
| Positive | 110 (14.2) | 20 (13.0) | .80c |
| Negative | 648 (83.5) | 129 (83.8) | |
| Indeterminate or not available | 18 (2.3) | 5 (3.2) | |
| AJCC stage, No./total No. (%) | |||
| I | 138/762 (18.1) | 28/151 (18.5) | .56 |
| II | 424/762 (55.6) | 91/151 (60.3) | |
| III | 186/762 (24.4) | 29/151 (19.2) | |
| IV | 14/762 (1.8) | 3/151 (2.0) | |
| Overall survival median follow-up time, mo | 29.8 | 31.8 | .57b |
| Breast cancer–free interval median follow-up time, mo | 27.5 | 29.4 | .69b |
Abbreviations: AJCC, American Joint Committee on Cancer; ER, estrogen receptor; PR, progesterone receptor.
Some values do not sum to heading totals because of missing data.
Wilcoxon rank sum test. All other comparisons are by Fisher exact test.
Statistical tests were performed using counts of positive and negative cases only.
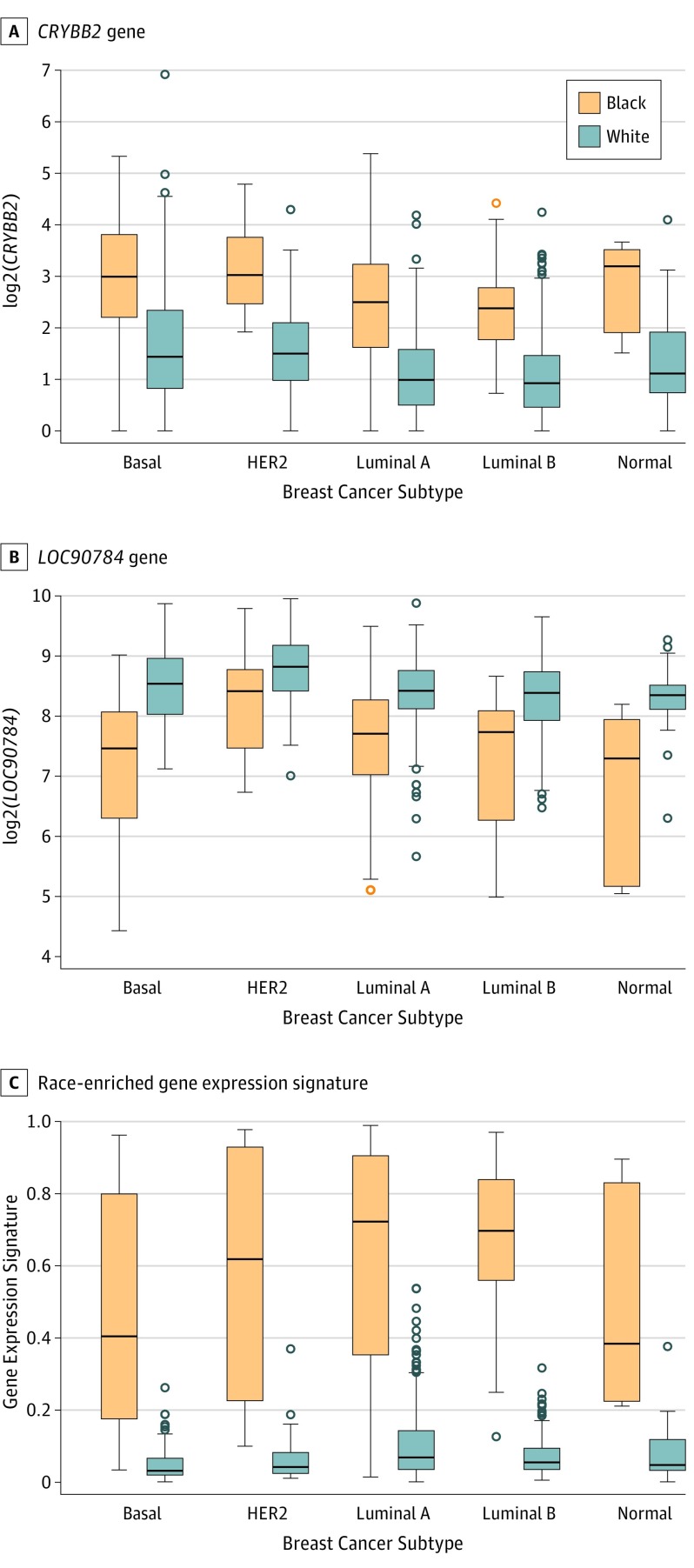
We identified 9232 genes (49.1%) differentially expressed between black and white patients after adjusting for age and batch effects (nominal P < .05). After adjusting for subtype, 142 genes had Bonferroni-adjusted P < .05 (eTable 2 in Supplement 1). The top 2 differentially expressed genes were LOC90784 (no OMIM accession number to date) and CRYBB2 (OMIM 123620) (Figure 1). Unsupervised clustering analysis of the 142 genes identified a branch enriched for black patients in each subtype (eFigure 1 in Supplement 2, right). Using penalized logistic regression, we developed a gene expression signature from the 142 genes distinguishing breast tumor samples from blacks and whites, with a cross-validation AUROC of 0.878.
Figure 1. Plots of the CRYBB2 and LOC90784 Genes and Race-Enriched Gene Expression Signature Estimated Using a Penalized Regression Model, by Subtype and Race.
All comparisons between black patients and white patients within each subtype were statistically significant (P < .001 for all tests) except HER2-enriched subtype for the LOC90784 gene (P = .017).
We found 25 proteins differentially expressed between tumors from blacks and whites after adjusting for age, batch, and subtype (eTable 3 in Supplement 1). Expression of these proteins can moderately distinguish black and white patients (eFigure 2 in Supplement 2). Two proteins (caspase-8 and Src) had FDRs less than 0.05, but only caspase-8 had P < .05 after Bonferroni adjustment. A logistic regression model that included the 2 proteins yielded an AUROC of 0.629 in the cross-validation analysis.
We did not find any difference in the total number of mutations per patient between blacks and whites. However, the total number of CNA segments was higher in black patients compared with white patients within the luminal A subtype (eFigure 3 in Supplement 2). Comparing mutation frequencies of the 68 recurrently mutated genes, 44 regions of gain and 28 regions of loss, we found that 13 of them were significantly different between the 2 races (Table 2 and eTable 4 and eTable 5 in Supplement 1). Compared with white patients, black patients had a higher proportion of TP53 (OMIM 191170) mutations (51.7% vs 31.3%) and 8q24.21/MYC (OMIM 190080) amplification (30.9% vs 20.4%) and a lower proportion of PIK3CA (OMIM 171834) mutations (23.7% vs 35.6%). However, after adjusting for subtype, no mutations and only 4 CNAs (11q23.1 deletion, 21q21.1 deletion, 8p12 amplification, and 12q15 amplification) remained statistically significant, with slightly higher frequencies in blacks.
Table 2. Recurrently Mutated Genes and DNA Copy Number Alterations Differing by Race.
| Gene or Cytobanda | No. (%) | P Valueb | Adjusted P Valuec | |
|---|---|---|---|---|
| Black | White | |||
| Mutation | ||||
| TP53 | 61/118 (51.7) | 234/748 (31.3) | 2.5 × 10−5 | .85 |
| PIK3CA | 28/118 (23.7) | 266/748 (35.6) | .012 | .60 |
| FBXW7 | 5/118 (4.2) | 9/748 (1.2) | .031 | .10 |
| Deletion | ||||
| 8p23/CSMD1 | 22/152 (14.5) | 66/759 (8.7) | .035 | .41 |
| 13q14/RB1 | 13/152 (8.6) | 31/759 (4.1) | .035 | .37 |
| 11q23.1 | 6/152 (3.9) | 10/759 (1.3) | .037 | .023 |
| 21q21.1 | 5/152 (3.3) | 8/759 (1.1) | .050 | .031 |
| Amplification | ||||
| 8q24.21/MYC | 47/152 (30.9) | 155/759 (20.4) | .0072 | .44 |
| 8p12 | 20/152 (13.2) | 43/759 (5.7) | .0024 | 6.4 × 10−4 |
| 8q11 | 16/152 (10.5) | 45/759 (5.9) | .049 | .14 |
| 19q12/CCNE1 | 14/152 (9.2) | 27/759 (3.6) | .0046 | .25 |
| 5p15/TERT | 10/152 (6.6) | 22/759 (2.9) | .049 | .46 |
| 12q15/MDM2 | 10/152 (6.6) | 21/759 (2.8) | .026 | .011 |
Genes after virgules are putative oncogenic drivers or fragile sites located within GISTIC peaks. Accession numbers for the genes are given in the text except for the following: FBXW7 (OMIM 606278), CSMD1 (OMIM 608397), RB1 (OMIM 604041), CCNE1 (OMIM 123837), and MDM2 (OMIM 164785).
Fisher exact test.
Adjusted for PAM50 subtype.
We identified 9 hypermethylated DNA methylation probes significantly associated with race after adjusting for age, tumor purity, and subtype (multiple testing adjusted P < .001). The DNA methylation levels of all 9 probes were higher in blacks (eFigure 4 in Supplement 2). Of the hypomethylated enhancer probes, 7 were significantly associated with race after adjusting for age, tumor purity, and subtype (multiple testing adjusted P < .001). The DNA methylation levels were lower in black patients than in white patients, suggesting that enhancers at these loci may be more active in black patients (eFigure 5 in Supplement 2).
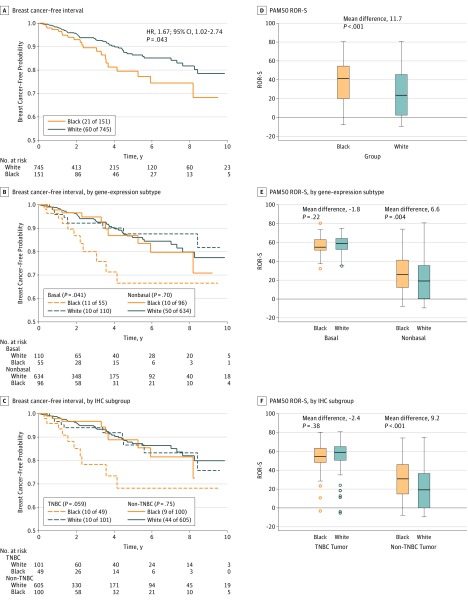
Breast Cancer Recurrence and Survival by Race
During a median follow-up of 29 months, 81 BCFI recurrence events occurred. There was a significant difference in BCFI between blacks and whites (hazard ratio [HR], 1.67; 95 CI, 1.02-2.74; P = .043), with the difference most pronounced in basal-like cancers or TNBCs (Figure 2, A-C). By contrast, blacks had a higher PAM50 risk of recurrence score than whites, with the difference most pronounced in non–basal-like cancers or non-TNBCs (Figure 2, D-F). In the multivariable Cox proportional hazards regression model adjusting for age and American Joint Committee on Cancer stage, blacks exhibited a significantly worse outcome (HR, 1.90; 95% CI, 1.14-3.16; P = .014). Further adjusting for subtype, the HR for race was reduced slightly (HR, 1.73; 95% CI, 1.02-2.95; P = .043). We found more racial differences in BCFI outcomes in basal-like breast cancer (HR, 2.36) than in nonbasal subtypes (HR, 1.28), although none reached statistical significance due to limited events within each subgroup (eTable 6 in Supplement 1).
Figure 2. Kaplan-Meier Curves of Breast Cancer–Free Interval.
The survival analyses were performed using all follow-up time, although we plotted the curves for the first 10 years only. Shown are all patients (A), patients with basal and nonbasal tumors (B), patients with triple-negative breast cancer (TNBC) and non-TNBC tumors (C), PAM50 risk of recurrence score (ROR-S) for all patients (D), patients with basal and nonbasal tumors (E), and patients with TNBC and non-TNBC tumors (F). HR indicates hazard ratio; IHC, immunohistochemistry.
The TP53 mutation was a positive predictor for recurrence (age- and stage–adjusted HR, 2.04; 95% CI, 1.29-3.22; P = .002), and TP53 mutation frequency varied between blacks and whites. The racial difference in BCFI remained statistically significant after adjusting for TP53 mutation status (HR, 2.10; 95% CI, 1.22-3.61; P = .007). The racial-enriched gene expression signature was prognostic across all patients (age and stage–adjusted HR, 2.06; 95% CI, 0.95-4.46; P = .066); after adding it to the model, the HR for race was reduced to 1.56 (95% CI, 0.60-4.02; P = .36). The 2 most significantly differentially expressed proteins (caspase-8 and Src), the PIK3CA mutation, MYC amplification, and the 4 CNAs with different frequencies between racial groups were not prognostic factors for BCFI. Race was not associated with overall survival (eFigure 6 in Supplement 2). However, blacks with basal-like subtype or TNBCs trended toward worse overall survival compared with whites in subgroup analyses.
Germline Variants and the Heritability of Breast Cancer Subtypes
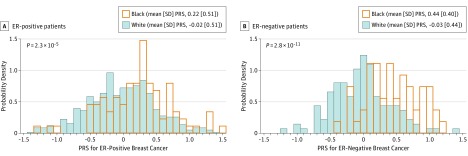
We found significant allele frequency differences between blacks and whites for 76 of 89 breast cancer susceptibility SNPs in case-only analyses (eTable 7 in Supplement 1). After adjusting for race, 4 SNPs were significantly associated with intrinsic subtype (eTable 8 in Supplement 1). The risk allele (T) of SNP rs10069690 in TERT was higher in blacks than whites and was associated with a higher odds of basal-like relative to luminal A subtype breast cancer, which is consistent with studies showing that rs10069690 was associated with ER-negative breast cancer. Two SNPs (rs10069690 and rs7726159) in TERT were associated with a lower odds of HER2-enriched relative to luminal A subtype breast cancer. The risk allele (C) of rs11814448 was a common allele in blacks but was rare in whites, and it was associated with a lower odds of luminal B subtype tumors. In contrast, the risk allele (A) of SNP rs34084277 in BABAM1 (OMIM 612766) was lower in blacks than whites, and it was associated with a higher odds of basal-like compared with luminal A subtype breast cancer, a finding consistent with previous genome-wide association studies showing that variants in the BABAM1 gene were associated with ER-negative breast cancer. To summarize the combined effect of 89 SNPs and understand how allele frequency differences between blacks and whites in these common variants contribute to racial differences in subtype distribution, we calculated breast cancer PRSs. We found that blacks had higher ER-positive and ER-negative PRSs than whites, and the racial difference in ER-negative PRSs was even larger (Figure 3), suggesting that breast cancer (especially ER-negative breast cancer) in blacks has a greater genetic contribution than in whites.
Figure 3. Polygenic Risk Score (PRS), Stratified by Race.
Polygenic risk scores were calculated based on allele frequencies of 89 genetic variants and log odds ratios of 89 genetic variants for estrogen receptor (ER)–positive and ER-negative breast cancers obtained from the literature.
In a heritability analysis of breast cancer subtype, we found that 43.6% (SE, 24.5%; P = 1.5 × 10−14) of subtype variation can be explained by genome-wide germline variants. We estimated the heritability of ESR1 (OMIM 133430) and ERBB2/HER2 (OMIM 164870) gene expression as continuous variables and found the heritability of ESR1 to be 39.8% (SE, 16.0%; P = 1.3 × 10−14) and the heritability of ERBB2/HER2 to be 17.3% (SE, 16.1%; P = .098). The ER-positive PRS built from 89 known susceptibility variants was higher in blacks than in whites (difference, 0.24; P = 2.3 × 10−5), while the ER-negative PRS was much higher in blacks than in whites (difference, 0.48; P = 2.8 × 10−11).
Discussion
In the first ancestry-based comprehensive analysis of multiple platforms of genomic and proteomic data of its kind to date, we found significant molecular differences between invasive breast cancers from genomically defined black and white patients. Black patients were more likely to have basal-like and ERBB2-enriched subtypes and less likely to have the luminal A subtype than white patients. While most tumor genomic differences between races can be captured by subtype, we found a modest number of genomic features, including gene expression (n = 142), protein (n = 1), DNA copy number (n = 4), and DNA methylation (n = 16), that were different between these ancestry groups after adjusting for subtype.
We found a higher risk of recurrence for black patients relative to white patients, in particular for basal-like cancers or TNBCs. Blacks had a higher PAM50 risk of recurrence score than whites, especially for non–basal-like or non-TNBC breast cancer subtypes. These seemingly perplexing findings may reflect heterogeneous tumor biology: basal-like breast cancer has rapid recurrence, so racial disparity in outcome can be captured in TCGA, which has a limited duration of follow-up, while non–basal-like breast cancer has a longer time to recurrence. The higher risk of recurrence for black patients relative to white patients was attenuated after taking into account subtype and was further reduced after adjusting for the race-specific gene expression signature; it should be noted that this signature correlates highly with ancestry, so collinearity existed. Several investigations have examined gene expression by race in breast cancer: all studies except one found race-specific gene expression patterns. Results from these studies are not entirely consistent because of variable sample sizes and differing analytical methods. However, one gene that has been consistently found to be higher in tumors from blacks than from whites is the CRYBB2 gene. We showed that CRYBB2 expression was higher in black patients than in white patients within each breast cancer subtype. Expression of CRYBB2 was also found to be higher in normal breast tissue from blacks than those from whites. Furthermore, CRYBB2 had higher expression in prostate cancer and colorectal cancer from black Americans compared with white Americans. Taken together, these findings indicate that CRYBB2 is likely a true race-specific gene, and its differential expression by race is likely due to genetic background via expression quantitative trait loci regulation. We conducted Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway analysis of the 142 genes differentially expressed between blacks and whites and found an enrichment in ether lipid (P = .023) and alpha-linolenic acid (P = .049) metabolic pathways. Such race-differential expression in lipid-metabolizing genes could be due to expression quantitative trait loci regulation or may reflect differences in the nutritional status or obesity rates between blacks and whites.
We estimated that more than 40% of the variation in intrinsic breast cancer subtype can be explained by inherited germline variants (known and unknown), suggesting that a significant proportion of racial differences in subtype frequencies are due to genetic factors. This finding is supported by previous studies in which predisposing germline variants varied in frequency by race. The heritability of ESR1 was higher than that of ERBB2/HER2, which extends previous observations that the ER-positive proportion varies widely across populations, while the ERBB2-positive proportion does not. For known breast cancer susceptibility loci, most SNPs showed different allele frequencies between blacks and whites. We used PRSs to summarize the combined effect of 89 known germline variants and found that differences in allele frequencies between blacks and whites can explain racial differences in the distribution of subtypes, with blacks having a much higher PRS for ER-negative breast cancer. Taken together, these findings underscore the need for larger genetic epidemiological studies to identify additional biological factors that contribute to racial frequency differences in breast cancer subtypes.
This comprehensive assessment of multiple omics data types, including somatic mutations, CNAs, gene expression, protein expression, DNA methylation, and germline variants, identified important differences between breast cancers from black and white patients. Compared with previous analyses of TCGA breast cancer data, we included 154 black women, updating the data sets presented previously in studies by Keenan et al (n = 105) and Stewart et al (n = 53). The survival analysis in our study provided a more stable assessment of racial disparity because it included more events (81 vs 34) than the previous analysis. We also used genomically determined race to avoid possible misclassification and to eliminate missing data in the self-reported race variable.
Limitations
The present study has some limitations. First, TCGA cohort consists of convenience samples, so it may not represent a general breast cancer patient population. Nonetheless, most of our findings are similar to previous publications with respect to the frequency differences and outcomes of blacks with basal-like cancers or TNBCs. Second, a short follow-up time and unclear definition of cause of death may have affected the investigation of racial disparity in cancer recurrence. Third, socioeconomic and environmental factors were not available in TCGA, so their contribution to racial disparity could not be evaluated. Fourth, TCGA may not have sufficient power to test racial differences in each breast cancer subtype because stringent multiple testing procedures need to be taken.
Conclusions
In summary, this study demonstrated that there are biological differences between breast cancers in black patients and white patients that are linked to genetic ancestry broadly and to specific inherited gene variants. Future studies are warranted to investigate genetic and nongenetic factors that contribute to heterogeneity in the development of distinct breast cancer subtypes, as well as how these factors influence response to therapy and survival outcomes in diverse populations.
eTables
eMethods and eFigures
References
- 1.Torre LA, Bray F, Siegel RL, Ferlay J, Lortet-Tieulent J, Jemal A. Global cancer statistics, 2012. CA Cancer J Clin. 2015;65(2):87-108. [DOI] [PubMed] [Google Scholar]
- 2.Siegel RL, Miller KD, Jemal A. Cancer statistics, 2015. CA Cancer J Clin. 2015;65(1):5-29. [DOI] [PubMed] [Google Scholar]
- 3.DeSantis CE, Fedewa SA, Goding Sauer A, Kramer JL, Smith RA, Jemal A. Breast cancer statistics, 2015: convergence of incidence rates between black and white women. CA Cancer J Clin. 2016;66(1):31-42. [DOI] [PubMed] [Google Scholar]
- 4.Daly B, Olopade OI. A perfect storm: how tumor biology, genomics, and health care delivery patterns collide to create a racial survival disparity in breast cancer and proposed interventions for change. CA Cancer J Clin. 2015;65(3):221-238. [DOI] [PubMed] [Google Scholar]
- 5.Carey LA, Perou CM, Livasy CA, et al. Race, breast cancer subtypes, and survival in the Carolina Breast Cancer Study. JAMA. 2006;295(21):2492-2502. [DOI] [PubMed] [Google Scholar]
- 6.Bauer KR, Brown M, Cress RD, Parise CA, Caggiano V. Descriptive analysis of estrogen receptor (ER)–negative, progesterone receptor (PR)–negative, and HER2-negative invasive breast cancer, the so-called triple-negative phenotype: a population-based study from the California Cancer Registry. Cancer. 2007;109(9):1721-1728. [DOI] [PubMed] [Google Scholar]
- 7.Ihemelandu CU, Leffall LD Jr, Dewitty RL, et al. Molecular breast cancer subtypes in premenopausal and postmenopausal African-American women: age-specific prevalence and survival. J Surg Res. 2007;143(1):109-118. [DOI] [PubMed] [Google Scholar]
- 8.Yang XR, Sherman ME, Rimm DL, et al. Differences in risk factors for breast cancer molecular subtypes in a population-based study. Cancer Epidemiol Biomarkers Prev. 2007;16(3):439-443. [DOI] [PubMed] [Google Scholar]
- 9.Sweeney C, Bernard PS, Factor RE, et al. Intrinsic subtypes from PAM50 gene expression assay in a population-based breast cancer cohort: differences by age, race, and tumor characteristics. Cancer Epidemiol Biomarkers Prev. 2014;23(5):714-724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kohler BA, Sherman RL, Howlader N, et al. Annual report to the nation on the status of cancer, 1975-2011, featuring incidence of breast cancer subtypes by race/ethnicity, poverty, and state. J Natl Cancer Inst. 2015;107(6):djv048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Haiman CA, Chen GK, Vachon CM, et al. ; Gene Environment Interaction and Breast Cancer in Germany (GENICA) Consortium . A common variant at the TERT-CLPTM1L locus is associated with estrogen receptor–negative breast cancer. Nat Genet. 2011;43(12):1210-1214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Millikan RC, Newman B, Tse CK, et al. Epidemiology of basal-like breast cancer. Breast Cancer Res Treat. 2008;109(1):123-139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cancer Genome Atlas Network Comprehensive molecular portraits of human breast tumours. Nature. 2012;490(7418):61-70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ciriello G, Gatza ML, Beck AH, et al. ; TCGA Research Network . Comprehensive molecular portraits of invasive lobular breast cancer. Cell. 2015;163(2):506-519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Parker JS, Mullins M, Cheang MC, et al. Supervised risk predictor of breast cancer based on intrinsic subtypes. J Clin Oncol. 2009;27(8):1160-1167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zou H, Hastie T. Regularization and variable selection via the elastic net. J R Stat Soc Series B Stat Methodol. 2005;67(2):301-320. [Google Scholar]
- 17.Wilkerson MD, Cabanski CR, Sun W, et al. Integrated RNA and DNA sequencing improves mutation detection in low purity tumors. Nucleic Acids Res. 2014;42(13):e107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lawrence MS, Stojanov P, Polak P, et al. Mutational heterogeneity in cancer and the search for new cancer-associated genes. Nature. 2013;499(7457):214-218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mermel CH, Schumacher SE, Hill B, Meyerson ML, Beroukhim R, Getz G. GISTIC2.0 facilitates sensitive and confident localization of the targets of focal somatic copy-number alteration in human cancers. Genome Biol. 2011;12(4):R41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hudis CA, Barlow WE, Costantino JP, et al. Proposal for standardized definitions for efficacy end points in adjuvant breast cancer trials: the STEEP system. J Clin Oncol. 2007;25(15):2127-2132. [DOI] [PubMed] [Google Scholar]
- 21.Michailidou K, Hall P, Gonzalez-Neira A, et al. ; The Breast and Ovarian Cancer Susceptibility Collaboration; Hereditary Breast and Ovarian Cancer Research Group Netherlands (HEBON); and The GENICA (Gene Environment Interaction and Breast Cancer in Germany) Network . Large-scale genotyping identifies 41 new loci associated with breast cancer risk. Nat Genet. 2013;45(4):353-361, 361e1-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Garcia-Closas M, Couch FJ, Lindstrom S, et al. ; The Gene Environmental Interaction Network Breast Cancer (GENICA); kConFab Investigators; Familial Breast Cancer Study (FBCS); and Australian Breast Cancer Tissue Bank (ABCTB) . Genome-wide association studies identify four ER negative–specific breast cancer risk loci. Nat Genet. 2013;45(4):392-398e2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mavaddat N, Pharoah PD, Michailidou K, et al. Prediction of breast cancer risk based on profiling with common genetic variants. J Natl Cancer Inst. 2015;107(5):djv036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Huo D, Feng Y, Haddad S, et al. Genome-wide association studies in women of African ancestry identified 3q26.21 as a novel susceptibility locus for oestrogen receptor negative breast cancer. Hum Mol Genet. 2016;25(21):4835-4846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Michailidou K, Beesley J, Lindstrom S, et al. ; BOCS; kConFab Investigators; AOCS Group; NBCS; The GENICA Network . Genome-wide association analysis of more than 120,000 individuals identifies 15 new susceptibility loci for breast cancer. Nat Genet. 2015;47(4):373-380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Howie BN, Donnelly P, Marchini J. A flexible and accurate genotype imputation method for the next generation of genome-wide association studies. PLoS Genet. 2009;5(6):e1000529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yang J, Lee SH, Goddard ME, Visscher PM. GCTA: a tool for genome-wide complex trait analysis. Am J Hum Genet. 2011;88(1):76-82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Antoniou AC, Wang X, Fredericksen ZS, et al. ; EMBRACE; GEMO Study Collaborators; HEBON; kConFab; SWE-BRCA; MOD SQUAD; GENICA . A locus on 19p13 modifies risk of breast cancer in BRCA1 mutation carriers and is associated with hormone receptor–negative breast cancer in the general population. Nat Genet. 2010;42(10):885-892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Stewart PA, Luks J, Roycik MD, Sang QX, Zhang J. Differentially expressed transcripts and dysregulated signaling pathways and networks in African American breast cancer. PLoS One. 2013;8(12):e82460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Martin DN, Boersma BJ, Yi M, et al. Differences in the tumor microenvironment between African-American and European-American breast cancer patients. PLoS One. 2009;4(2):e4531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Field LA, Love B, Deyarmin B, Hooke JA, Shriver CD, Ellsworth RE. Identification of differentially expressed genes in breast tumors from African American compared with Caucasian women. Cancer. 2012;118(5):1334-1344. [DOI] [PubMed] [Google Scholar]
- 32.Sturtz LA, Melley J, Mamula K, Shriver CD, Ellsworth RE. Outcome disparities in African American women with triple negative breast cancer: a comparison of epidemiological and molecular factors between African American and Caucasian women with triple negative breast cancer. BMC Cancer. 2014;14:62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Grunda JM, Steg AD, He Q, et al. Differential expression of breast cancer–associated genes between stage- and age-matched tumor specimens from African- and Caucasian-American women diagnosed with breast cancer. BMC Res Notes. 2012;5:248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.D’Arcy M, Fleming J, Robinson WR, Kirk EL, Perou CM, Troester MA. Race-associated biological differences among luminal A breast tumors. Breast Cancer Res Treat. 2015;152(2):437-448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Chavez-Macgregor M, Liu S, De Melo-Gagliato D, et al. Differences in gene and protein expression and the effects of race/ethnicity on breast cancer subtypes. Cancer Epidemiol Biomarkers Prev. 2014;23(2):316-323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Jovov B, Araujo-Perez F, Sigel CS, et al. Differential gene expression between African American and European American colorectal cancer patients. PLoS One. 2012;7(1):e30168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wallace TA, Prueitt RL, Yi M, et al. Tumor immunobiological differences in prostate cancer between African-American and European-American men. Cancer Res. 2008;68(3):927-936. [DOI] [PubMed] [Google Scholar]
- 38.Bower KM, Thorpe RJ Jr, Rohde C, Gaskin DJ. The intersection of neighborhood racial segregation, poverty, and urbanicity and its impact on food store availability in the United States. Prev Med. 2014;58:33-39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wang Y, Beydoun MA. The obesity epidemic in the United States: gender, age, socioeconomic, racial/ethnic, and geographic characteristics: a systematic review and meta-regression analysis. Epidemiol Rev. 2007;29:6-28. [DOI] [PubMed] [Google Scholar]
- 40.Garcia-Closas M, Hall P, Nevanlinna H, et al. ; Australian Ovarian Cancer Management Group; Kathleen Cuningham Foundation Consortium for Research Into Familial Breast Cancer . Heterogeneity of breast cancer associations with five susceptibility loci by clinical and pathological characteristics. PLoS Genet. 2008;4(4):e1000054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Howlader N, Altekruse SF, Li CI, et al. US incidence of breast cancer subtypes defined by joint hormone receptor and HER2 status. J Natl Cancer Inst. 2014;106(5):dju055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Keenan T, Moy B, Mroz EA, et al. Comparison of the genomic landscape between primary breast cancer in African American versus white women and the association of racial differences with tumor recurrence. J Clin Oncol. 2015;33(31):3621-3627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kroenke CH, Sweeney C, Kwan ML, et al. Race and breast cancer survival by intrinsic subtype based on PAM50 gene expression. Breast Cancer Res Treat. 2014;144(3):689-699. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
eTables
eMethods and eFigures