Abstract
Background
Pediatric obesity-related asthma is more severe and less responsive to medications than normal-weight asthma. Obese asthmatic children have non-atopic Th1 polarized systemic inflammation that correlates with pulmonary function deficits, but the pathways underlying Th1 polarized inflammation are not well understood.
Objective
We compared the CD4+ T cell transcriptome in obese children with asthma to that in normal-weight children with asthma to identify key differentially expressed genes associated with Th1 polarized inflammation.
Methods
CD4+ T cell transcriptome-wide differential gene expression was compared between 21 obese and 21 normal-weight children using directional RNA-seq. High confidence differentially expressed genes were verified in the first cohort and validated in a second cohort of 20 children (10 obese and 10 normal-weight children) using qRT-PCR.
Results
Transcriptome-wide differential gene expression among obese asthmatic children was enriched for genes including VAV2, DOCK5, PAK3, PLD1, CDC42EP4 and CDC42PBB, which are associated with CDC42, a small GTP protein, that is linked with T cell activation. Upregulation of MLK3 and PLD1, genes downstream of CDC42 in the MAPK and mTOR pathways, and the inverse correlation of CDC42EP4 and DOCK5 transcript counts with FEV1/FVC ratio, together support a role of CDC42 in the Th1 polarization and pulmonary function deficits found in obesity-related asthma.
Conclusions
Our study identifies the CDC42 pathway as a novel target that is upregulated in Th cells of obese asthmatic children, suggesting its role in non-atopic Th1 polarized systemic inflammation and pulmonary function deficits found in pediatric obesity-related asthma.
Keywords: Asthma, obesity, T helper cell transcriptome, children
Introduction
Obesity and asthma are two of the most common pediatric diseases, particularly among African Americans and Hispanics.1, 2 Obesity is a known predictor of childhood asthma3 and obesity-related asthma is distinct from normal-weight asthma.4 Obese children with asthma have higher disease morbidity,5 lower pulmonary function6 and lower responsiveness to bronchodilators as compared to normal-weight children with asthma.7 Although truncal adiposity,8 metabolic abnormalities, including insulin resistance and dyslipidemia,9, 10 and systemic inflammation,6 have been postulated, the precise mechanisms underlying pediatric obesity-related asthma are not well elucidated.
Obese asthmatic children have evidence of non-atopic Th1 polarized systemic inflammation,6, 11 that is associated with pulmonary function deficits,6, 11 and differs from atopic inflammation associated with classic childhood asthma.12 Moreover, insulin resistance mediates the association of Th1 polarization with pulmonary function deficits.11 While extensive investigation of immune pathways in context of atopic asthma13 has led to the development of targeted therapy, including omalizumab,14 the lack of a similar understanding of mechanisms underlying non-atopic immune responses, as observed in the context of obesity-related asthma,6, 11 is associated with limited therapeutic options for obese children with asthma.
To address these gaps in knowledge, we compared the CD4+ (Th) cell transcriptome of obese asthmatic children to that of normal-weight asthmatic children of African American and Hispanic ethnicity to elucidate the mechanistic immune pathways underlying Th1 polarization. We hypothesized that gene expression in CD4+ T cells from obese asthmatics differs from that in normal-weight asthmatics and its investigation will allow for the identification of key molecules underlying the non-atopic Th1 polarized inflammation associated with the obese asthma phenotype.
Methods
Study population
Two study cohorts comprised of African American and Hispanic children, ages 7–11 years, with obese asthma and normal-weight asthma, were recruited from clinics at Children’s Hospital at Montefiore between 7/2013 to 6/2016. The first discovery cohort comprised of 42 children, including 21 obese and 21 normal-weight children with asthma. The second validation cohort comprised of 20 children, including 10 obese and 10 normal-weight children with asthma. Obesity was defined as Body Mass Index (BMI) >95th percentile for age and gender. Asthma was classified based on the clinical diagnosis made by a health care provider that was confirmed on electronic medical records. All participants completed a research study visit at the Clinical Research Center at the Montefiore Medical Center when they underwent anthropometric measurements, skin prick allergy testing to six allergens (tree mix, grass mix, ragweed, dust mite (D. pteronyssinus), cockroach, mouse, cat and mold), and fasting phlebotomy as previously described.6 Pulmonary function testing was abstracted from the medical charts. It was performed as per the American Thoracic guidelines and included spirometry and lung volumes quantification by nitrogen washout as previously described.6 Percent predicted values were calculated for spirometry indices using the National Health and Nutrition Examination Survey (NHANES) prediction equations and for lung volume indices using equations developed by the American Thoracic Society workshop, as previously described.6 The Institutional Review Board at Albert Einstein College of Medicine approved the study.
Study measures
Separation of CD4+ T cells
Given the association of insulin resistance with Th1 polarization,11 we processed fasting blood for cell and serum separation. Peripheral blood mononuclear cells (PBMCs) were separated using the Ficoll Hypaque method. CD4+ T cells were isolated from the PBMCs by negative selection using magnetic beads (Easy Sep, Stem Cell Technologies, Tukwila, WA) to avoid any ex-vivo CD4+ T cell stimulation. The CD4+ T cell purity was 95–98% as confirmed by flow cytometry (see Figure E1 in online repository). T cell proportions did not differ between the obese (26.6±5%) and normal-weight (27.1±7.4%) samples. Unstimulated T cells were used for transcriptome quantification to elucidate differential gene expression more reflective of in-vivo conditions. Serum separated from fasting blood was used for insulin quantification using radioimmunoassay (Millipore Corporation) on a Wizard2 gamma counter (Perkin Elmer Corporation). Lipids were quantified by enzymatic immunoassay and measured on AU400 chemistry autoanalyzer (Beckman-Coulter Corporation).
CD4+ T cell transcriptome quantification by directional RNA-seq
The expression levels of genes in the CD4+ T cell transcriptome were quantified in the discovery cohort of 42 participants. CD4+ T cell RNA from the validation cohort of 20 participants was utilized to validate the transcriptome findings from the discovery cohort. RNA extracted from 2×106 CD4+ T cells using Qiazol (Qiagen Inc, Valencia, CA) underwent quality control testing (2100 Bioanalyzer, Agilent Technologies, Santa Clara, CA) and samples with RNA integrity number of 8 or greater underwent processing for directional RNA-seq library preparation. After removal of ribosomal RNA with the Ribo Zero-rRNA removal kit (Illumina Inc., San Diego, CA), reverse transcription was performed using the SuperScriptIII First-Strand Synthesis system (Thermo Fisher Scientific, Waltham MA) followed by second strand cDNA synthesis using dUTP (Thermo Fisher Scientific, Waltham MA). The double stranded cDNA was fragmented with Covaris (200–300bp target length), end-repaired, dA tailed and adaptors added for the Illumina sequencer to allow multiplexing of 8 samples per lane. To maintain directional information, i.e. transcribing strand-specific information,15 a combination of dUTP incorporation and uracil-DNA glycosylase were used. All libraries underwent Bioanalyzer testing for quality control and were sequenced on Illumina HiSeq 2500 as 100 bp single-end reads.
Alignment and analysis of directional RNA-seq libraries
All bioinformatics analyses were performed on a high performance computing cluster at Albert Einstein College of Medicine. Several quality control steps were taken to ensure accuracy of analyzed results. Picard-tools v 1.11916 was used to demultipex and generate FastQ files, which were then trimmed for poor quality bases and adaptor sequences using Trim Galore! v. 0.3.7.17 Using STAR v. 2.5.1b18 and the gene annotations from Ensembl release 83,19 the trimmed reads were aligned to the UCSC hg38 human genome assembly, and gene counts were generated.
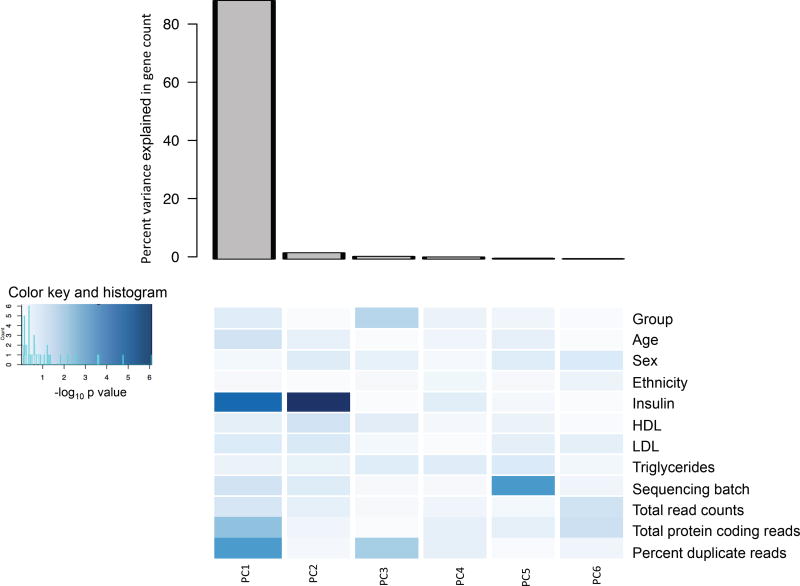
The gene counts were normalized and between-group comparison was performed using DESeq20 on R statistical software, version 3.2.2. Using principal component analysis, we additionally investigated the contribution of biological and technical covariates in the variance of normalized counts between the study groups (Figure 1). While fasting insulin was the only biological variable associated with variance, several technical factors, including total number of reads, protein coding reads, percent duplicate reads, and sequencing batch were determined to be significantly contributing to the variance in the gene counts. There was no association of lipids (HDL, LDL, or triglycerides) with gene count variance. Given the association of gene-count variance with both biological and technical factors, multivariate linear regression analysis was conducted to identify between-group differential gene expression, adjusting for the biologic and each of the technical factors. Age, gender and ethnicity were included in the model for their demographic significance. Genes identified by multivariate analysis to be differentially expressed among obese asthmatics with a between group p-value <0.05 and a false discovery rate (FDR) q-value of <0.05 were retained as high confidence differentially expressed genes for verification and validation studies. The Search Tool for the Retrieval of Interacting Genes/Proteins (STRING) database v.1021 was used to identify relationships between differentially expressed genes. The gene expression data and patient characteristics are available at Gene Expression Omnibus number GSE86430.
Figure 1.
Principal component analysis of biological and technical covariates. Principal component 1 was the most important component as seen in the bar graph. The heatmap depicts the strength of the contribution of the study group, and of the biologic and technical covariates, on the variance of gene count for each of the top 6 principal components, highlighting the individual factors that contribute to principal component 1. The darker the intensity of blue color, the stronger was the contribution of the covariate in the variance of the gene count in each principal component as shown in the color key.
Verification and validation of directional RNA-seq results
A subset of the high confidence differentially expressed genes was verified by qRT-PCR on RNA isolated from CD4+ T cells from 32 children from the discovery cohort (16 children per study group). These genes and additional related genes were further validated in the validation cohort. Verification and validation was done by quantitative PCR using the TaqMan gene expression assay with commercially available qPCR primers (Thermo Fisher Inc, Waltham, MA) and analyzed by the ΔΔCT method.
Protein quantification as additional validation of differential gene expression
CDC42EP4 and DOCK5 proteins were quantified to additionally validate differential gene expression, and phosphorylated and total p38 and S6K1 proteins were quantified as evidence for MAPK and mTOR pathway activation, in T cell lysates from a subset of 5 obese and 4 normal-weight asthmatic samples, and normalized to actin. All antibodies, except for CDC42EP4 (Fisher Scientific), were purchased from Santa Cruz Biotechnology, Inc. Western blot experiments were run as per protocol (Bio-Rad laboratories). Immunoblot band intensity was quantified using the ImageJ software (National Institutes of Health).
Statistical Analysis
Clinical characteristics were compared between obese and normal-weight asthmatics using the Student’s T test for continuous variables and Χ2 or Fisher-exact test for categorical variables. Between-group comparison of the log10 transformed differential gene expression by qRT-PCR was done using Student’s T test. Normalized gene counts of the high confidence differentially expressed genes were log transformed for correlation analysis with pulmonary function indices. Statistical analysis was performed on STATA version 14.
Results
Study population
The clinical characteristics of the discovery and validation cohort are summarized in Table 1 and Table 2 respectively. The obese asthmatics did not differ in age, gender, or ethnicity distribution from the normal-weight asthmatic children in either the discovery or the validation cohort. More normal-weight than obese children had evidence of atopic sensitization in the discovery cohort. In keeping with prior studies, obese children in the discovery cohort had lower percent-predicted FVC, FEV1, RV, and FRC and RV/TLC ratio as compared to normal-weight children with asthma. In the validation cohort, while RV, RV/TLC ratio and FRC were lower among obese children as compared to normal-weight children, only FRC was significantly different between the two groups. The pulmonary function values were consistent between the obese asthmatic groups in the two cohorts, other than percent-predicted RV, but there was greater variability in FVC, FEV1 and RV/TLC ratio between the two normal-weight groups.
Table 1.
Demographic and clinical characteristics of the discovery cohort.
| Obese Asthmatic (n=21) |
Normal-weight Asthmatic (n=21) |
P value | |
|---|---|---|---|
| Age (years) | 9.1±1.5 | 8.9± 0.6 | 0.76 |
| Males (n(%)) | 8 (38.1) | 10 (47.6) | 0.75 |
| Hispanic (n(%)) | 15 (71.4) | 12 (57.4) | 0.52 |
| BMI-z score | 1.9 ± 0.4 | 0.06 ± 0.8 | <0.001 |
| Atopic sensitization# | 17 (81) | 20 (95.2) | 0.01 |
| Insulin | 11.9 ± 7.5 | 8.5 ± 4.7 | 0.08 |
| FVC* | 102 ± 11.1 | 89.7 ± 12.3 | 0.002 |
| FEV1* | 96.2 ± 16.7 | 87.1 ± 16.4 | 0.08 |
| FEV1/FVC | 81.7 ± 7.5 | 84.5 ± 8.4 | 0.27 |
| TLC* | 92.6 ± 13.5 | 92 ± 11 | 0.88 |
| RV* | 86.6 ± 36.2 | 120.4 ± 33.7 | 0.004 |
| RV/TLC* | 22 ± 6.2 | 31.8 ± 6 | <0.001 |
| ERV* | 77.2 ± 17.7 | 81.1 ± 29.7 | 0.62 |
| FRC* | 85.3 ± 19.4 | 105.8 ± 23.4 | 0.005 |
| Medication use | |||
| Inhaled steroids | 11 (52.3) | 9 (42.9) | 0.75 |
| Montelukast | 13 (61.9) | 9 (42.9) | 0.35 |
| Inhaled steroid/ long acting beta agonist combination therapy | 3 (14.3) | 4 (19) | 1 |
All continuous variables are reported as mean±SD.
Categorical variables (gender (males), ethnicity (Hispanic)), atopic sensitization, and medication use are reported as group-specific numerical frequency and percentages.
Pulmonary function variables are reported as percent predicted values, other than FEV1/FVC and RV/TLC ratio, which are reported as percentages.
Atopic sensitization was defined as skin prick test reactivity to one or more allergens.47
Table 2.
Demographic and clinical characteristics of the validation cohort
| Obese Asthmatic (n=10) |
Normal-weight Asthmatic (n=10) |
P value | |
|---|---|---|---|
| Age | 8.8±1 | 8.6± 1.4 | 0.71 |
| Males (n(%)) | 5 (50) | 8 (80) | 0.34 |
| Hispanic (n(%)) | 5 (50) | 5 (50) | 1 |
| BMI-z score | 1.8±0.5 | 0.1 ± 1.01 | <0.001 |
| Atopic sensitization# | 8 (80) | 8 (80) | 1 |
| Insulin | 13.9 ± 5.8 | 12.2 ± 6.2 | 0.6 |
| FVC* | 102 ± 14 | 105 ± 22 | 0.76 |
| FEV1* | 91.5 ± 19 | 100.1 ± 21 | 0.36 |
| FEV1/FVC | 77.1 ± 8 | 82.9 ± 4 | 0.08 |
| TLC* | 94.4 ± 10 | 98 ± 14 | 0.54 |
| RV* | 100.4 ± 32 | 110.1 ± 22 | 0.43 |
| RV/TLC | 25.6 ± 7 | 27.9 ± 4 | 0.39 |
| ERV* | 71.1 ± 24 | 99.8 ± 25 | 0.02 |
| FRC* | 90.1 ± 18 | 112.2 ± 17 | 0.02 |
| Medication use | |||
| Inhaled steroids | 5 (50) | 5 (50) | 1 |
| Montelukast | 9 (90) | 7 (70) | 0.58 |
| Inhaled steroid/ long acting beta agonist combination therapy | 4 (40) | 2 (20) | 0.62 |
All continuous variables are reported as mean±SD.
Categorical variables (gender (males), ethnicity (Hispanic)), atopic sensitization and medication use are reported as group-specific numerical frequency and percentages.
Pulmonary function variables are reported as percent predicted values, other than FEV1/FVC and RV/TLC ratio, which are reported as percentages.
Atopic sensitization was defined as skin prick test reactivity to one or more allergens.
Analysis and validation of differential CD4+ T cell gene expression
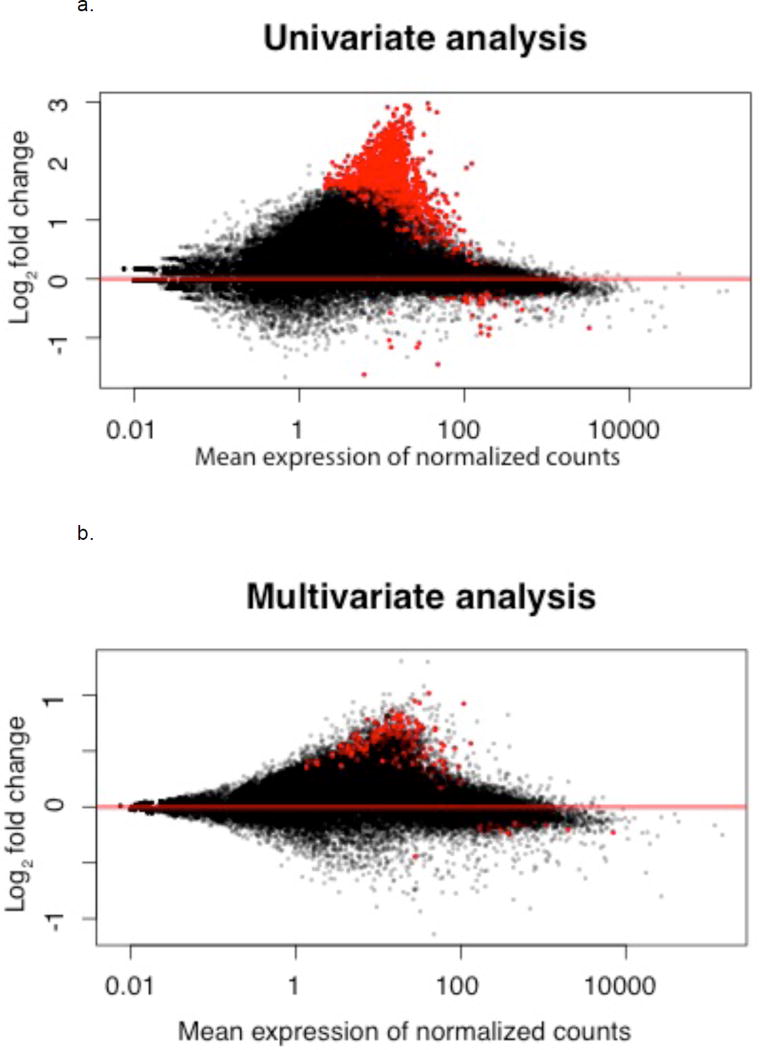
Univariate analysis using DESeq revealed differential gene expression of 1371 genes in obese asthmatic compared to normal-weight asthmatic CD4+ T cells with more genes being upregulated in the obese compared to normal-weight asthmatic cells (Figure 2a). After adjusting in multivariate regression analysis for the biologic and technical factors associated with gene count variance, 319 genes were differentially expressed among obese as compared to normal-weight asthmatic children (between-group p value<0.05, q-value<0.05) (Figure 2b, Table E1 in online repository). Of these, 89 genes overlapped between the initial DESeq analysis and multivariate analysis (see Table E2 in online repository).
Figure 2.
MA plot of fold change and mean gene expression using DESeq analysis, a) before adjustment for biological and technical co-variates (univariate analysis) and b) after adjustment for biological and technical co-variates (multivariate analysis). The red dots denote differentially expressed genes among obese asthmatics at FDR q-value<0.05.
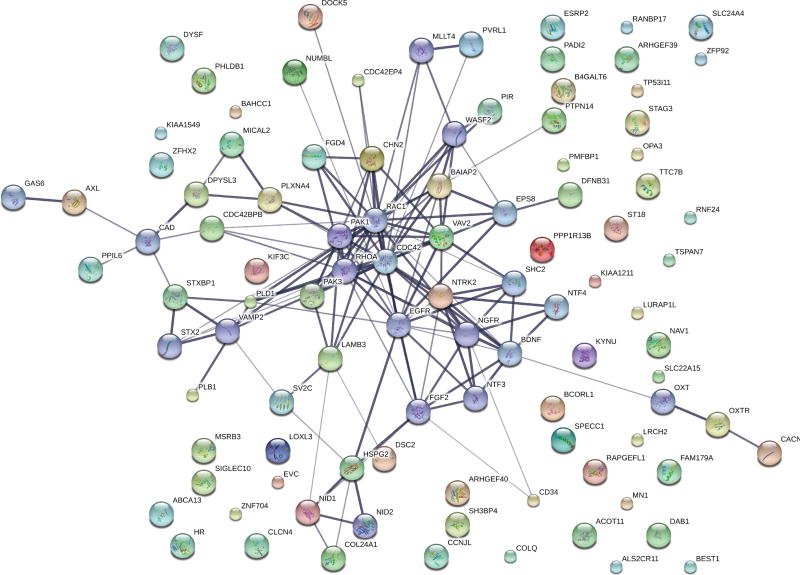
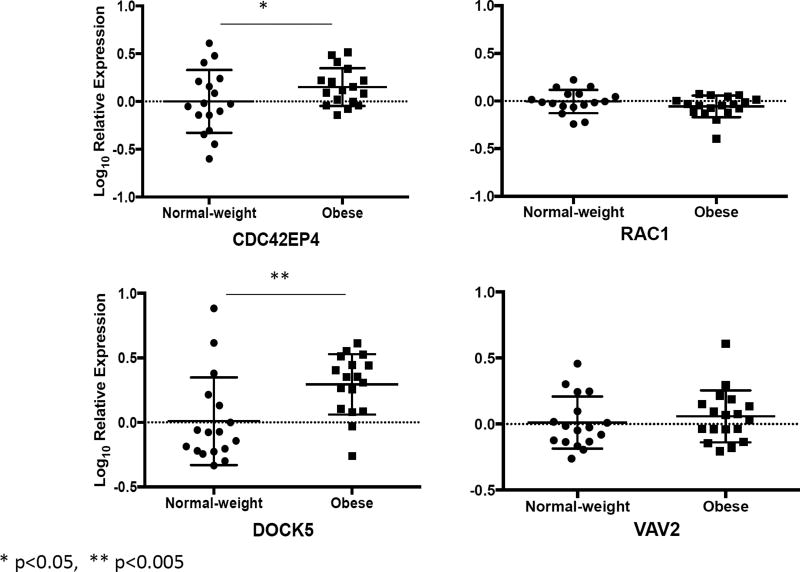
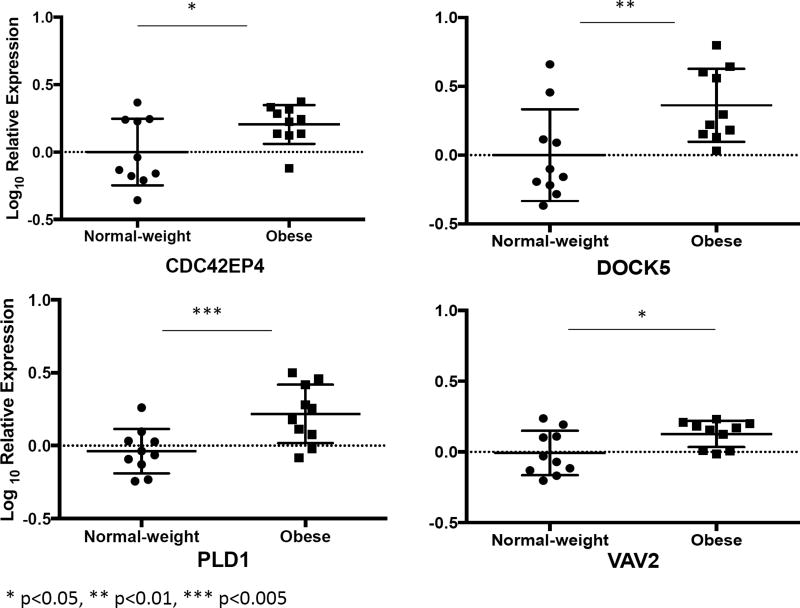
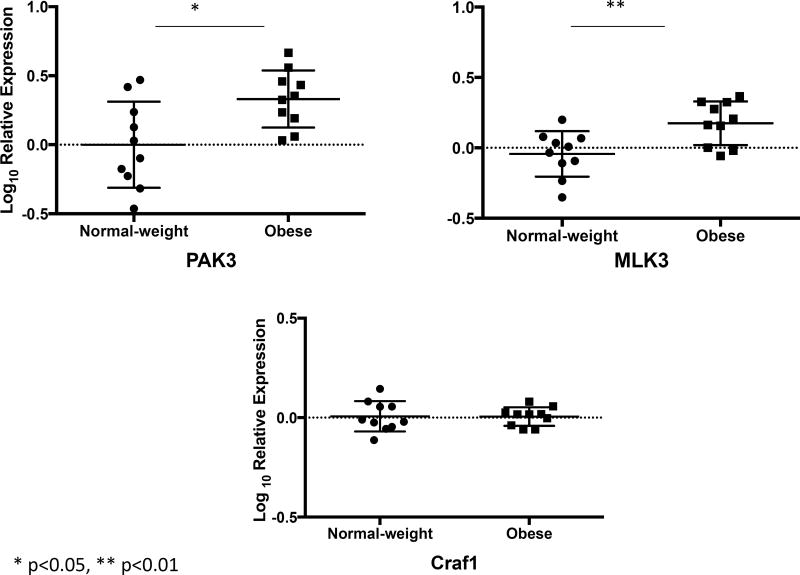
Gene network analysis of these 89 genes revealed CDC42 to be at the hub of the differentially expressed genes (Figure 3). Moreover, 24 of the 89 upregulated genes among obese asthmatics were associated with modulation of small GTP binding proteins, CDC42 and RAC1 (Table 3). Of particular relevance were VAV2, DOCK5, PAK3, PLD1, CDC42BPB and CDC42EP4 that either interact directly with the CDC42 protein (CDC42EP4), are upstream modulators (VAV2 and DOCK5) or are downstream targets of CDC42 (PAK3, PLD1, and CDC42BPB). We verified the expression of CDC42EP4, since it directly interacts with CDC42, and DOCK5 and VAV2, its upstream modulators by qRT-PCR (Figure 4). While CDC42EP4 and DOCK5 were significantly higher among obese asthmatics, there was a non-significant trend of higher VAV2 in obese asthmatics (Figure 4, E2). Since several differentially expressed genes were associated with both CDC42 and RAC1 activation, we quantified RAC1 expression and found no differential expression (Figure 4). We, therefore, focused on validating these differentially expressed genes and additional downstream genes (PAK3, PLD1, MLK3 and cRaf1) in the validation cohort (Figure 5) as confirmation of upregulation of the CDC42 pathway in obese asthmatic CD4+ T cells. While MLK3, PAK3, and PLD1 were upregulated in obese asthmatics, cRaf1 was not differentially expressed (Figure 5).
Figure 3.
Association between differentially expressed high confidence genes. The thickness of the connecting lines reflects the strength of the association.
Table 3.
Genes associated with activation of small GTP proteins, CDC42 and RAC1
| Ensembl ID | Associated Gene Name |
log2 Fold Change |
q-value |
|---|---|---|---|
| ENSG00000168453 | HR | 2.599971402 | 2.32E-05 |
| ENSG00000204764 | RANBP17 | 2.251636516 | 0.000253142 |
| ENSG00000157388 | CACNA1D | 1.8684354 | 0.002357941 |
| ENSG00000137135 | ARHGEF39 | 1.850566818 | 0.001725958 |
| ENSG00000115318 | LOXL3 | 1.663949396 | 0.015247401 |
| ENSG00000163803 | PLB1 | 1.649412493 | 0.0013521 |
| ENSG00000087842 | PIR | 1.61362157 | 0.020860028 |
| ENSG00000173406 | DAB1 | 1.553990563 | 0.016608231 |
| ENSG00000139132 | FGD4 | 1.453418533 | 0.008949179 |
| ENSG00000075651 | PLD1 | 1.361719192 | 0.018931263 |
| ENSG00000160293 | VAV2 | 1.332433107 | 0.012568743 |
| ENSG00000130147 | SH3BP4 | 1.310248419 | 0.031128422 |
| ENSG00000142512 | SIGLEC10 | 1.290791208 | 0.030722074 |
| ENSG00000165801 | ARHGEF40 | 1.285779282 | 0.034584495 |
| ENSG00000105245 | NUMBL | 1.242714423 | 0.014961879 |
| ENSG00000106069 | CHN2 | 1.220243828 | 0.018341102 |
| ENSG00000167601 | AXL | 1.216368793 | 0.049907113 |
| ENSG00000175866 | BAIAP2 | 1.125602885 | 0.021294253 |
| ENSG00000147459 | DOCK5 | 1.101075872 | 0.03490964 |
| ENSG00000198752 | CDC42BPB | 0.87291003 | 0.022874905 |
| ENSG00000088808 | PPP1R13B | 0.642280439 | 0.020469995 |
| ENSG00000101236 | RNF24 | 0.572603051 | 0.028734724 |
| ENSG00000077264 | PAK3 | 0.538432709 | 0.0141306 |
Figure 4.
Verification of a subset of genes in the first cohort of samples by qRT-PCR. Log10 fold differential gene expression is compared between obese and normal-weight children with asthma.
Figure 5.
Validation of the subset of genes in the second cohort of samples by qRT-PCR. Log10 fold differential gene expression was compared between obese asthmatic and normal-weight asthmatic Th cells. In addition to genes investigated in the verification experiment (Figure 4), differential expression of additional genes downstream of CDC42 including MLK3 and cRAF1 were investigated. While expression of MLK3 was higher in obese asthmatic Th cells, that of cRAF1 did not differ between obese and normal-weight asthmatic Th cells.
Immunoblot analysis was done to confirm that higher mRNA expression of CDC42EP4 and DOCK5 translated into higher protein expression. In keeping with the RNA-Seq and qPCR data, we found a trend towards higher levels of these two proteins, although the trend did not reach statistical significance (see Figure E3 in online repository). Similarly, there was a non-significant trend towards higher levels of S6K1 phosphorylation in obese asthmatic T cells, which was supportive of activation of the CDC42-mTOR signaling pathway.
To elucidate clinical significance, we correlated gene transcript counts with pulmonary function variables. We found an inverse association between log-transformed normalized gene counts of CDC42EP4 and DOCK5 and FEV1/FVC ratio, only among obese asthmatics (Figure 6) ascribing clinical relevance to the differential gene expression.
Figure 6.
Association of CDC42EP4 and DOCK5 gene counts with FEV1/FVC ratio. Log10 transformed CDC42EP4 and DOCK5 gene counts were inversely correlated with FEV1/FVC ratio only in obese asthmatic children.
Discussion
Our study provides novel evidence that the transcriptome of CD4+ T cells derived from obese asthmatic children differs from that of CD4+ T cells from children with normal-weight asthma. Specifically, we found that several genes associated with the small GTP-binding protein, CDC42, were upregulated in T cells from obese children with asthma. While VAV2 and DOCK5 are upstream of, and activate CDC42, CDC42EP4 interacts directly with CDC42 and CDC2BPB, PAK3, and PLD1 are downstream effectors, mediating the different effects of this ubiquitous GTP protein. Further, CDC42EP4 and DOCK5 gene transcript counts correlated with lower airway obstruction, suggesting that CDC42 pathway may have clinical implications in childhood obesity-related asthma.
Few of these genes have been previously studied in the context of inflammation in asthma or obesity.22 However, the role of CDC42 and its related proteins has been researched extensively in the context of T cell physiology. VAV2 and DOCK5, upregulated in obese asthmatic T cells, are guanine exchange factors that activate CDC42.23 VAV2 is ubiquitously expressed, and is associated with activation of CDC4224 and its downstream c-JUN transcription factor,24 leading to CDC42-mediated activation of the MAP kinase pathway.25 DOCK proteins are less well studied in T cells. Although DOCK 5, a member of DOCK family A,26 is classically associated with activation of RAC1,27 it influences degranulation response in mast cells through the NCK-AKT pathway, independent of the RAC1 activation28 suggesting that it may have additional effects on immune cells. Since our study is the first to identify upregulation of DOCK5 and its association with CDC42 in T cells, further investigation of its effects and related downstream molecules are needed to identify its role in T cells.
We also confirmed upregulation of downstream targets of CDC42 including PAK3, MLK3, PLD1, but not cRaf1. PAK3, with CDC42, plays a role immunological synapse formation,29–31 and is associated with activation of MAPK and mTOR immune pathways.32–34 Higher MLK3 expression also supports downstream activation of MAPK pathway as a potential mechanism underlying Th1 polarization among obese asthmatic children. Similarly, PLD1 activation by CDC4235 mediates S6K1 activation in the mTOR pathway,33 which regulates Th1 and Th17 differentiation.36 Quantified in resting cells, we found a small non-significant trend for higher p-S6K1 in obese asthmatic T cells. Together, upregulated gene expression of upstream guanine exchange factors and downstream effectors provide evidence of activation of CDC42-related pathways that have been associated with Th cell activation and may explain the differentiation bias towards a Th1 lineage,36 a phenotype observed among obese asthmatic children.6, 11 Moreover, the inverse association of CDC42EP4 and DOCK5 gene counts with FEV1/FVC ratio uniquely in obese asthmatics provides additional evidence of its relevance in the obese asthma phenotype.
In light of the association of non-atopic systemic inflammation with disease burden in obese asthmatic adults37 and children,6, 11 we speculate that CDC42 activation in obese asthmatic T cells may play several roles, including skewing of T cell differentiation to a Th1/Th17 profile, increased T cell tissue recruitment by facilitating transmigration, and T cell activation. Since these pathways are distinct from those activated in atopic asthma,38 our findings begin to identify immune pathways associated with non-atopic immune responses in obese asthma phenotype. Activation of Rho GTPases, including CDC42, has been proposed to underlie T cell activation in Th1 mediated autoimmune diseases like lupus.39 Validation of our findings would support potential extension of therapeutics used for autoimmune diseases to obesity-related asthma. Although the direct mechanism by which circulating T cells influence pulmonary physiology in obese asthma is not known at this time, we hypothesize that T cells may potentially increase neutrophilic airway inflammation40 or may influence the airway smooth muscle contractility and proliferation,41 mechanisms that have been proposed to underlie obese asthma phenotype that warrant further investigation.
We have previously reported an association between insulin resistance and systemic Th1 polarization in urban minority children.11 In the current study, insulin was significantly associated with gene count variance. T cell activation is associated with aerobic glycolysis42 with increased cellular glucose uptake due to translocation of glucose transporter 1 (GLUT1),43 and de-novo expression of the insulin receptor on the cell surface,44 suggesting a metabolic role for insulin in T cell activation. Since the insulin signaling pathway utilizes CDC42 for glucose transport in adipocytes,45 we hypothesize that the same mechanism may be utilized to address the activated T cell’s increased glucose needs.42 Thus, upregulation of the CDC42 pathway identifies a need for further exploration of these immuno-metabolic pathways in chronic diseases such as obesity-related asthma.
Our study has several strengths. It is the first study to investigate the Th cell transcriptome using directional RNA-seq, and identifies CDC42 pathway as a key upregulated pathway in obese asthmatic children that is associated with lower airway obstruction. The Th cell transcriptome findings were validated in a separate cohort of 10 obese and 10 normal-weight children with asthma. We also identified the influence of technical and biological covariates on differential gene expression in a disease state, highlighting the importance of addressing these to allow for the identification of high confidence genes. These findings in negatively selected unstimulated T cells, unmodified by any external mitogenic influences, are more reflective of in-vivo responses, and are novel since prior studies of unstimulated T cell transcriptome did not find differences between cases and controls.46 However, there are certain limitations. Asthma diagnosis in our cohort was based on physician diagnosis rather than objective assessment of airway reactivity. We found small fold differences in the gene expression and non-significant differences in the corresponding and downstream proteins. Since these cells were derived from obese children with well-controlled asthma in the absence of a recent exacerbation, it is unlikely that the cells would be markedly activated with elevated protein levels as may be observed in the setting of an asthma exacerbation. Furthermore, the lack of an obese non-asthmatic control group limited the understanding of the extent to which differential gene expression was driven by obesity alone. In addition, our study is cross sectional in nature so a cause-effect relationship could not be derived. Nonetheless, the upregulation of several genes associated with the CDC42 pathway, upstream of both MAPK and mTOR pathways, suggest that these pathways may be poised for activation in the setting of poor disease control or an exacerbation, and thereby provides direction for further investigation to identify the key molecules that may be associated with non-atopic Th1 systemic inflammation in obesity-related asthma.
Conclusions
In summary, we found upregulation of several genes associated with the small GTP protein CDC42, which is involved in many T cell functions, including antigen recognition via the immunological synapse, intracellular transport and vesicle formation for cytokine release, and T cell activation with activation of the MAPK and the mTOR pathways. These genes and their associated pathways provide direction for further investigation of key regulatory molecules underlying the non-atopic T cell inflammation in obese asthmatics that may potentially serve as therapeutic targets for pediatric obesity-related asthma.
Supplementary Material
Key Messages.
T helper cells from obese asthmatic children have a distinct transcriptome when compared to T helper cells from normal-weight children.
Genes related to CDC42, a GTP protein, which plays a role in T cell activation, were upregulated in T helper cells from obese asthmatic children.
MLK3 and PLD1, genes downstream of CDC42 in the MAPK and mTOR pathways respectively, were upregulated in obese asthmatic T helper cells suggesting that pathways activated distal to CDC42 may play a role in the non-atopic T helper 1 immune responses previously observed in obese asthmatic children.
Transcript abundance of genes in the CDC42 pathway directly correlated with lower airway obstruction in obese asthmatic children.
Acknowledgments
We are grateful to all the participating children and their families for taking the time to complete the study protocol. We also acknowledge the staff at the Epigenomics Shared Facility and the High-Performance Computing Core Facility at Albert Einstein College of Medicine for the sequencing and post sequencing processing of the RNA-Seq experiments.
Grant support: This study was supported in part by the K23 HL118733 (D.R.), Feldstein Medical Foundation, NIH/NCRR CTSA Grant 1 UL1 TR001073-01, 1 TL1 TR001072-01, 1 KL2 TR001071-01 (Albert Einstein Institute of Clinical and Translational Research) from the National Center for Advancing Translational Sciences (NCATS), a component of the National Institutes of Health (NIH). The views expressed in this article do not communicate an official position of the NIH/NCRR.
Abbreviations
- Th
T helper
- BMI
Body Mass Index
- PBMCs
Peripheral blood mononuclear cells
- qRT-PCR
Quantitative reverse transcription-polymerase chain reaction
- rRNA
Ribosomal RNA
- cDNA
complementary DNA
- FDR
False Discovery Rate
- NHANES
National Health and Nutrition Examination Survey
- TNF
Tumor necrosis factor
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Ogden CL, Carroll MD, Kit BK, Flegal KM. Prevalence of obesity and trends in body mass index among US children and adolescents, 1999–2010. JAMA. 2012;307:483–90. doi: 10.1001/jama.2012.40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Akinbami LJ, Simon AE, Rossen LM. Changing Trends in Asthma Prevalence Among Children. Pediatrics. 2016;137 doi: 10.1542/peds.2015-2354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chen YC, Dong GH, Lin KC, Lee YL. Gender difference of childhood overweight and obesity in predicting the risk of incident asthma: a systematic review and meta-analysis. Obes Rev. 2013;14:222–31. doi: 10.1111/j.1467-789X.2012.01055.x. [DOI] [PubMed] [Google Scholar]
- 4.Wenzel SE. Asthma phenotypes: the evolution from clinical to molecular approaches. Nat Med. 2012;18:716–25. doi: 10.1038/nm.2678. [DOI] [PubMed] [Google Scholar]
- 5.Lang JE, Hossain J, Smith K, Lima JJ. Asthma severity, exacerbation risk, and controller treatment burden in underweight and obese children. J Asthma. 2012;49:456–63. doi: 10.3109/02770903.2012.677895. [DOI] [PubMed] [Google Scholar]
- 6.Rastogi D, Canfield S, Andrade A, Hall CB, Isasi CR, Rubinstein A, et al. Obesity-associated asthma in children: A distinct entity. Chest. 2012;141:895–905. doi: 10.1378/chest.11-0930. [DOI] [PubMed] [Google Scholar]
- 7.McGarry ME, Castellanos E, Thakur N, Oh SS, Eng C, Davis A, et al. Obesity and bronchodilator response in black and Hispanic children and adolescents with asthma. Chest. 2015;147:1591–8. doi: 10.1378/chest.14-2689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chen Y, Rennie D, Cormier Y, Dosman JA. Waist circumference associated with pulmonary function in children. Pediatr Pulmonol. 2009;44:216–21. doi: 10.1002/ppul.20854. [DOI] [PubMed] [Google Scholar]
- 9.Arshi M, Cardinal J, Hill RJ, Davies PS, Wainwright C. Asthma and insulin resistance in children. Respirology. 2010;15:779–84. doi: 10.1111/j.1440-1843.2010.01767.x. [DOI] [PubMed] [Google Scholar]
- 10.Cottrell L, Neal WA, Ice C, Perez MK, Piedimonte G. Metabolic abnormalities in children with asthma. Am J Respir Crit Care Med. 2011;183:441–8. doi: 10.1164/rccm.201004-0603OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Rastogi D, Fraser S, Oh J, Huber AM, Schulman Y, Bhagtani RH, et al. Inflammation, Metabolic Dysregulation and Pulmonary Function Among Obese Asthmatic Urban Adolescents. Am J Resp Crit Care Med. 2015;191:149–60. doi: 10.1164/rccm.201409-1587OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Guilbert TW, Mauger DT, Lemanske RF., Jr Childhood asthma-predictive phenotype. J Allergy Clinical Immunol Pract. 2014;2:664–70. doi: 10.1016/j.jaip.2014.09.010. [DOI] [PubMed] [Google Scholar]
- 13.Georas SN, Guo J, De Fanis U, Casolaro V. T-helper cell type-2 regulation in allergic disease. Eur Respir J. 2005;26:1119–37. doi: 10.1183/09031936.05.00006005. [DOI] [PubMed] [Google Scholar]
- 14.Busse WW, Morgan WJ, Gergen PJ, Mitchell HE, Gern JE, Liu AH, et al. Randomized trial of omalizumab (anti-IgE) for asthma in inner-city children. N Engl J Med. 2011;364:1005–15. doi: 10.1056/NEJMoa1009705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Podnar J, Deiderick H, Huerta G, Hunicke-Smith S. Next-Generation Sequencing RNA-Seq Library Construction. Cur Protoc Mol Biol. 2014;106:1–19. doi: 10.1002/0471142727.mb0421s106. [DOI] [PubMed] [Google Scholar]
- 16.http://broadinstitute.github.io/picard/.].
- 17.http://www.bioinformatics.babraham.ac.uk/projects/trim_galore/.].
- 18.Dobin A, Davis CA, Schlesinger F, Drenkow J, Zaleski C, Jha S, et al. STAR: ultrafast universal RNA-Seq aligner. Bioinformatics. 2013;29:15–21. doi: 10.1093/bioinformatics/bts635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Flicek P, Amode MR, Barrell D, Beal K, Billis K, Brent S, et al. Ensembl 2014. Nucleic Acids Res. 2014:D749–55. doi: 10.1093/nar/gkt1196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Anders S, Huber W. Differential expression analysis for sequence count data. Genome Biol. 2010;11:R106. doi: 10.1186/gb-2010-11-10-r106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Szklarczyk D, Franceschini A, Wyder S, Forslund K, Heller D, Huerta-Cepas J, et al. STRING v10: protein-protein interaction networks, integrated over the tree of life. Nucleic Acids Res. 2015;43 doi: 10.1093/nar/gku1003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lee SM, Choi HJ, Oh CH, Oh JW, Han JS. Leptin increases TNF-α expression and production through phospholipase D1 in Raw 264.7 cells. PLoS One. 2014;9:e102373. doi: 10.1371/journal.pone.0102373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Liu BP, Burridge K. Vav2 activates Rac1, Cdc42, and RhoA downstream from growth factor receptors but not beta1 integrins. Mol Cell Biol. 2000;20:7160–9. doi: 10.1128/mcb.20.19.7160-7169.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Abe K, Rossman KL, Liu B, Ritola KD, Chiang D, Campbell SL, et al. Vav2 is an activator of Cdc42, Rac1, and RhoA. J Biol Chem. 2000;275:10141–9. doi: 10.1074/jbc.275.14.10141. [DOI] [PubMed] [Google Scholar]
- 25.Kyriakis JM, Avruch J. Mammalian MAPK signal transduction pathways activated by stress and inflammation: a 10-year update. Physiol Rev. 2012;92:689–737. doi: 10.1152/physrev.00028.2011. [DOI] [PubMed] [Google Scholar]
- 26.Namekata K, Kimura A, Kawamura K, Harada C, Harada T. Dock GEFs and their therapeutic potential: neuroprotection and axon regeneration. Prog Retin Eye Res. 2014;43:1–16. doi: 10.1016/j.preteyeres.2014.06.005. [DOI] [PubMed] [Google Scholar]
- 27.Watanabe M, Terasawa M, Miyano K, Yanagihara T, Uruno T, Sanematsu F, et al. DOCK2 and DOCK5 act additively in neutrophils to regulate chemotaxis, superoxide production, and extracellular trap formation. J Immunol. 2014;193:5660–7. doi: 10.4049/jimmunol.1400885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ogawa K, Tanaka Y, Uruno T, Duan X, Harada Y, Sanematsu F, et al. DOCK5 functions as a key signaling adaptor that links FcεRI signals to microtubule dynamics during mast cell degranulation. J Exp Med. 2014;211:1407–19. doi: 10.1084/jem.20131926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Salazar-Fontana LI, Barr V, Samelson LE, Bierer BE. CD28 engagement promotes actin polymerization through the activation of the small Rho GTPase Cdc42 in human T cells. J Immunol. 2003;171:2225–32. doi: 10.4049/jimmunol.171.5.2225. [DOI] [PubMed] [Google Scholar]
- 30.Vicente-Manzanares M, Sánchez-Madrid F. Role of the cytoskeleton during leukocyte responses. Nature Rev Immunol. 2004;4:110–22. doi: 10.1038/nri1268. [DOI] [PubMed] [Google Scholar]
- 31.Tskvitaria-Fuller I, Seth A, Mistry N, Gu H, Rosen MK, Wülfing C. Specific patterns of Cdc42 activity are related to distinct elements of T cell polarization. J Immunol. 2006;177:1708–20. doi: 10.4049/jimmunol.177.3.1708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zhong B, Jiang K, Gilvary DL, Epling-Burnette PK, Ritchey C, Liu J, et al. Human neutrophils utilize a Rac/Cdc42-dependent MAPK pathway to direct intracellular granule mobilization toward ingested microbial pathogens. Blood. 2003;101:3240–8. doi: 10.1182/blood-2001-12-0180. [DOI] [PubMed] [Google Scholar]
- 33.Fang Y, Park IH, Wu AL, Du G, Huang P, Frohman MA, et al. PLD1 regulates mTOR signaling and mediates Cdc42 activation of S6K1. Curr Biol. 2003;13:2037–44. doi: 10.1016/j.cub.2003.11.021. [DOI] [PubMed] [Google Scholar]
- 34.Kaga S, Ragg S, Rogers KA, Ochi A. Activation of p21-CDC42/Rac-activated kinases by CD28 signaling: p21-activated kinase (PAK) and MEK kinase 1 (MEKK1) may mediate the interplay between CD3 and CD28 signals. J Immunol. 1998;160:4182–9. [PubMed] [Google Scholar]
- 35.Henage LG, Exton JH, Brown HA. Kinetic analysis of a mammalian phospholipase D: allosteric modulation by monomeric GTPases, protein kinase C, and polyphosphoinositides. J Biol Chem. 2006;281:3408–17. doi: 10.1074/jbc.M508800200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Delgoffe GM, Pollizzi KN, Waickman AT, Heikamp E, Meyers DJ, Horton MR, et al. The kinase mTOR regulates the differentiation of helper T cells through the selective activation of signaling by mTORC1 and mTORC2. Nat Immunol. 2011;12:295–303. doi: 10.1038/ni.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Peters MC, McGrath KW, Hawkins GA, Hastie AT, Levy BD, Israel E, et al. Plasma interleukin-6 concentrations, metabolic dysfunction, and asthma severity: a cross-sectional analysis of two cohorts. Lancet Respir Med. 2016;4:574–84. doi: 10.1016/S2213-2600(16)30048-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Holt PG, Sly PD. Interaction between adaptive and innate immune pathways in the pathogenesis of atopic asthma: operation of a lung/bone marrow axis. Chest. 2011;139:1165–71. doi: 10.1378/chest.10-2397. [DOI] [PubMed] [Google Scholar]
- 39.Pernis A. Rho GTPase-mediated pathways in mature CD4+ T cells. Autoimmunity Reviews. 2009;9:199–203. doi: 10.1016/j.autrev.2008.07.044. [DOI] [PubMed] [Google Scholar]
- 40.Scott HA, Gibson PG, Garg ML, Upham JW, Wood LG. Sex hormones and systemic inflammation are modulators of the obese-asthma phenotype. Allergy. 2016;71:1037–47. doi: 10.1111/all.12891. [DOI] [PubMed] [Google Scholar]
- 41.Ramos-Barbón D, Fraga-Iriso R, Brienza NS, Montero-Martínez C, Verea-Hernando H, Olivenstein R, et al. T Cells localize with proliferating smooth muscle alpha-actin+ cell compartments in asthma. Am J Resp Crit Care Med. 2010;182:317–24. doi: 10.1164/rccm.200905-0745OC. [DOI] [PubMed] [Google Scholar]
- 42.Wang R, Dillon CP, Shi LZ, Milasta S, Carter R, Finkelstein D, et al. The transcription factor Myc controls metabolic reprogramming upon T lymphocyte activation. Immunity. 2011;35:871–82. doi: 10.1016/j.immuni.2011.09.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Macintyre AN, Gerriets VA, Nichols AG, Michalek RD, Rudolph MC, Deoliveira D, et al. The glucose transporter Glut1 is selectively essential for CD4 T cell activation and effector function. Cell Metab. 2014;20:61–72. doi: 10.1016/j.cmet.2014.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Stentz FB, Kitabchi AE. Hyperglycemia-induced activation of human T-lymphocytes with de novo emergence of insulin receptors and generation of reactive oxygen species. Biochem Biophys Res Commun. 2005;335:491–5. doi: 10.1016/j.bbrc.2005.07.109. [DOI] [PubMed] [Google Scholar]
- 45.Usui I, Imamura T, Huang J, Satoh H, Olefsky JM. Cdc42 is a Rho GTPase family member that can mediate insulin signaling to glucose transport in 3T3-L1 adipocytes. J Biol Chem. 2003;278:13765–74. doi: 10.1074/jbc.M208904200. [DOI] [PubMed] [Google Scholar]
- 46.Quinn EM, Coleman C, Molloy B, Dominguez Castro P, Cormican P, Trimble V, et al. Transcriptome Analysis of CD4+ T Cells in Coeliac Disease Reveals Imprint of BACH2 and IFNγ Regulation. PloS One. 2015;10:e0140049. doi: 10.1371/journal.pone.0140049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Salo PM, Arbes SJ, Jr, Jaramillo R, Calatroni A, Weir CH, Sever ML, et al. Prevalence of allergic sensitization in the United States: results from the National Health and Nutrition Examination Survey (NHANES) 2005–2006. J Allergy Clin Immunol. 2014;134:350–9. doi: 10.1016/j.jaci.2013.12.1071. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.