Abstract
As asthma is a disease that results from host x environment interactions, an approach which allows assessment of the impact of the environment on the host is needed to understand disease. Metabolomics has appealing potential as an application to study pathways to childhood asthma development. The objective of this review is to provide an overview of metabolomics methods, and their application to understanding host x environment pathways in asthma development. We reviewed recent literature on advances in metabolomics and their application to study pathways to childhood asthma development. We highlighted 1) the potential of metabolomics in understanding the pathogenesis of disease and the discovery of biomarkers, 2) choice of metabolomics techniques, biospecimen handling, and data analysis, 3) the application to studying the role of environment on asthma development, 4) review of metabolomics applied to the outcome of asthma, 5) recommendations for application of metabolomics based –omics data integration in understanding disease pathogenesis, and 6) limitations. In conclusion metabolomics allows use of biospecimens to identify useful biomarkers and pathways involved in disease development, and subsequently to inform a greater understanding of the disease pathogenesis and endotypes, and predicting the clinical course of childhood asthma phenotypes.
Keywords: metabolomics, asthma, respiratory infections, systems approach
Introduction - What is Metabolomics?
Metabolomics is the study of the metabolite composition, the metabolome, of a cell type, tissue, organ, or organism.1 The metabolome is the collection of endogenous small molecules that mark specific fingerprints of cellular biochemistry.2 Metabolomics measures global sets of low molecular weight metabolites (including amino acids, organic acids, sugars, fatty acids, lipids, steroids, small peptides, vitamins, etc.), thus providing a “snapshot” of relevant biological processes. It provides a readout of metabolic activity status in relation to genetic variations, gene expression, or external stimuli.3 Such external stimuli include infections4 and allergens5, where specific metabolome profile marks the interaction between the environmental agent and host molecules, i.e. gene x environment (Deoxyribonucleic acid (DNA), ribonucleic acid (RNA), proteins, lipids, and other enzymes). Metabolites, in addition to being produced directly by the host organism, can be derived by host microbiota, as well as transformed from xenobiotic, dietary, and other exogenous sources.6
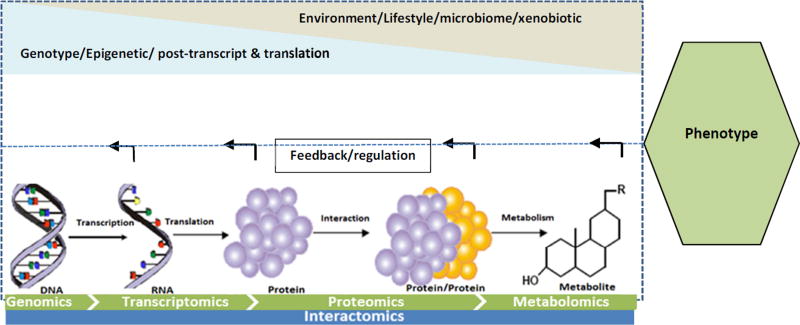
Situated at the end of the -omics tetralogy (Figure 1), metabolomics provides an opportunity to answer questions that are not possible with other -omics technologies. The spectrum from genomics to proteomics can provide information on what might be happening in a cell (probable cause of phenotype), processes that are subject to epigenetic regulation and post-translational modifications. Metabolomics on the other hand provides a snapshot of the entire physiology of the host and its response to the environment, which can be associated with the outcome phenotype (for example healthy vs disease) and endotypes.3,7 Metabolomics combines high-throughput analytical techniques with bioinformatics tools to provide information on a large number of metabolites simultaneously.2,8
Figure 1.
Tetralogy of -omics. The metabolome is at the end of the -omics spectrum and is influenced both by the changes to the upstream -omics (epigenome, post transcription and translation modifications) and environmental stimuli. The metabolites also play a role in regulation of the upstream -omics functions through feedback loops.
Why apply metabolomics approaches to studying host x environment pathways?
Metabolomics is an appealing application to monitor environment-host interaction since measured metabolites reflect alterations/dysfunctions of metabolic fluxes of various organs and cells.4 This method can also be used to trace back upstream molecular pathways, and as such reveal gene-environment interactions in disease development pathways.9 Thus, metabolomics offers the potential to identify biomarkers for host susceptibility, to assess response to environmental risk factors, to monitor for subsequent development of persistent wheeze and asthma, and elucidate biologic pathways.
How metabolomics may address the challenges of studying human infant environmental exposure and overcome limitations of animal models
Studying early life respiratory morbidity in infants and children is challenging. In vivo experiments with environmental exposures such as viral infection in infants are not possible due to obvious ethical issues.25 Thus, investigations to understand the pathogenesis are commonly conducted in either animal models26 or adult volunteers.27 However, experiments in adult human populations cannot be extrapolated to infant populations because of the difference in immune systems, lung development, prior exposures, and the impact of and response to environment in different age hosts.
As an example, the use of murine models to study the pathogenesis of respiratory syncytial virus (RSV) as an early life environmental asthma risk factor has not fulfilled its promise largely due to differences in immune system28 and anatomy29 between humans and animals. In addition, animal models of human RSV infection are semi-permissive and the most permissive primates like chimpanzees, African green monkeys, and baboons still require a large virus inoculum to establish a significant infection.30 Thus these viruses are not natural pathogens in animals, and these experimental models do not replicate what occurs in human disease.
With recent development of -omics methods, previously unrecognized molecular level host-environment interactions can be investigated with repeated collection of less invasive bio-specimens and integration of high-throughput metabolomics and other -omics data using bioinformatics tools.31 Metabolomics presents a chance to augment in vivo studies in primates and monitor the natural course of response to risk factor exposure. Metabolomics is also intrinsically suitable for in vitro study of primary airway cells and for ex vivo experimental models on explanted airway tissues, although ex vivo methods are limited by ethical issues of tissue collection and short-term viability of the tissue.32
Despite the promise of discovery of novel molecular biomarkers and new insights into disease pathobiology, there are important limitations and design issues that must be considered. The metabolic profile of an organism is dynamic, thus it is important to recognize that metabolites only represent a snapshot of current bodily or cellular activity. While a strength of this approach is that the measure of the metabolome represents the reaction to an exposure or what is happening at a single point in time in a dynamic system, the dynamic flux of metabolites must be considered. Metabolomics experiments are also hampered by methodological difficulties including very low molecular concentrations, variability and lack of standardized sampling, as well as complex analytical and data processing methods.33,34
Metabolomics experimental approaches
Metabolomics experiments are roughly categorized into two approaches: untargeted (global) and targeted. These strategies differ in many aspects, first and foremost in study objective (discovery vs hypothesis testing), but also in the level of quantitation, complexity of sample preparation, experimental accuracy and precision, and number of metabolites detected.
Untargeted metabolomics is an unbiased analysis of the metabolite composition of the biological entity in a specific physiological state under given environmental conditions.35 We should note that with limitations of current analytical platforms, as well as the conditions under which the samples are collected and processed, it is impossible to cover all metabolites in an unbiased manner. However, an untargeted approach can be still regarded as unbiased because no metabolite is identified prior to sample analysis. The hypothesis tested in this scenario is not associated with a particular metabolite or group of metabolites (metabolome), though knowledge of metabolite classes of biological interest helps in selecting an appropriate analytical platform and the sample preparation method to enhance detection of metabolites of interest.36 The global nature of untargeted metabolomics experiments enables novel areas of metabolism to be identified, but it is currently technically infeasible to positively identify all detected molecules.36 Despite the technological progress, the main disadvantage of untargeted metabolomics is the time required to process the extensive amounts of raw data, the difficulties in identifying and characterizing unknown small molecules, the reliance on the intrinsic analytical coverage of the platform employed, and the bias towards detection of high-abundance molecules.37 In general, epidemiological studies using untargeted approaches pose challenges as to platform selection and metabolite identification, and further studies are needed to explore replication, synthesis, and impact of current results.38 Nevertheless, untargeted metabolomics has been used to discriminate severity and phenotypes of asthma. 39, 40
Targeted metabolite profiling is an approach aimed at quantification of a prior known subset of metabolites that usually are of related chemical structure and/or biological activity.38,33 The targeted approach takes advantage of the understanding of a vast array of metabolic enzymes, their kinetics, end products, and the known biochemical pathways in which the set of metabolites takes part.37 Bias toward low abundance molecules is reduced with methods such as triple quadrupole mass spectrometer (TQMS) that are quantitatively reliable and allow for quantification of low-concentration metabolites that are difficult to detect.1 In addition, sample preparation can be optimized to reduce the dominance of high-abundance molecules.38 Targeted metabolomics have been applied to distinguish asthma patients from controls without disease. The metabolites measured in these targeted studies include adenosine, Adenosine monophosphate, purine, alkanes, aldehydes, ketones, and volatile organic compounds (VOCs) in exhaled breath condensate from children.42,43
Metabolomics analytical technologies
The two most common analytical approaches for the generation of metabolomics data are nuclear magnetic resonance (NMR) spectroscopy and mass spectrometry (MS). NMR is a spectroscopic technique based on the principle of energy absorption and re-emission of the atom nuclei due to variations in an external magnetic field.44 NMR produces spectral data that allow for quantification of the concentration and for characterization of chemical structure of metabolites.
Mass spectrometry acquires spectral data in the form of a mass-to-charge ratio (m/z) and a relative intensity of the ionized compound.45 MS based metabolomics is generally preceded by a separation step, which reduces the complexity of the biological sample and allows the MS analysis of different sets of molecules at different times.46 The most common separation techniques in MS technology are liquid chromatography (LC) and gas chromatography (GC) columns, termed as LC-MS and GC-MS techniques.47
The choice of metabolomics techniques should be based on the objective of the study, type of sample, and resource availability. NMR is highly selective, highly reproducible, requires less sample preparation, and it produces spectra that correlate directly and linearly with compound concentration.48 However, NMR has relatively low sensitivity, and accordingly only the most abundant species can generally be detected.49 On the other hand, mass spectrometry, when combined with effective sample preparation and chromatographic separation, has high sensitivity and specificity, as well as good dynamic range, which makes it advantageous especially for targeted metabolomics.49 Among major weaknesses of MS in metabolomics is quantification; the type of sample preparation used and its molecular environment affects signal intensity.49
While NMR and MS based analytical platforms are currently time-consuming for clinical and translational application, there are new advances in measuring VOCs in real-time, specifically for exhaled breath components. Semiconductor metal oxide (SMO)-based chemiresistive sensors are being used widely due to their amenability to miniaturization, for example development of electronic nose (E-nose), which enhances mobility and cost effectiveness.50 SMO measures the electrical resistance signal from the buildup of VOC at the surface of the polymer inducing swelling of the polymer film.51 However, low sensitivity and less selectivity of SMO-based sensors are limitations for precise phenotype and endotype identification of diseases.52
The choice of biospecimen for the study of the metabolomics of respiratory disease
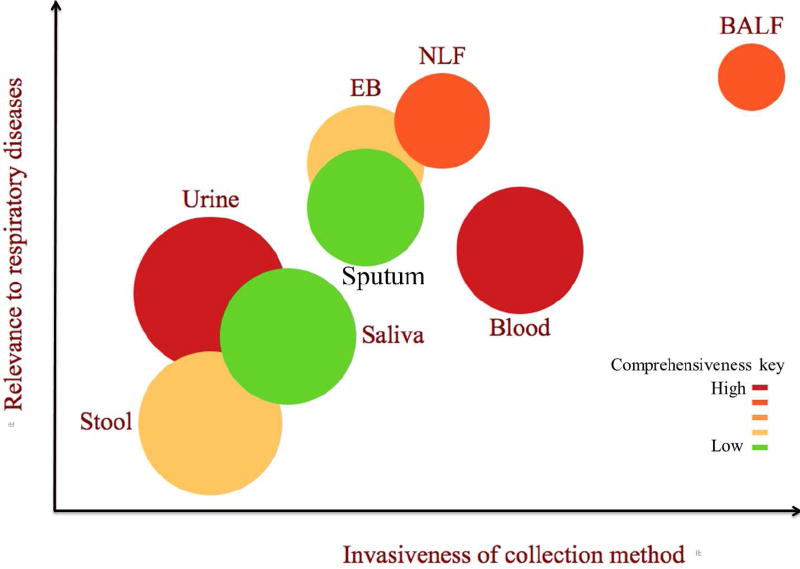
Biospecimens that can be used in metabolomics experiments for studying asthma development, response to environmental exposures, and asthma morbidity, include urine, stool, blood (plasma, serum, and whole blood), sputum, saliva, exhaled breath (condensate (EBC) and direct (EB)), nasal lavage fluid, and bronchoaveolar lavage fluid. The choice of biospecimen should be based on relevance and suitability to the research question and ease of sample collection procedures. Figure 2 provides a visual of rank of the biospecimen with regard to ease of collection (access, abundance, and invasiveness of collection method), specificity to the pathophysiology, and comprehensiveness of metabolite composition. The detail of biospecimen properties along with advantages and disadvantages for metabolomics experiment to study the development of asthma and response to environmental exposure are described in Table 1.
Figure 2.
Schematic depiction of the position of the biospecimen with regard to the invasiveness of collection (x-axis), relevance to airway environmental exposure (y-axis), and comprehensiveness of metabolites composition (represented by color gradient) and abundance of the biospecimen for collection (represented by size of the bubbles). The quantification of the biospecimen properties is subjective judgment of the authors on the scale of 1 to 10. EB=Exhaled breath, NLF= Nasal lavage fluid, and BALF=Bronchoalveolar lavage fluid.
Table 1.
Characteristics and physiological relevance of common biospecimen used to study metabolomics of environmental response, asthma pathogenesis, and asthma phenotypes and endotypes.
| Biospecimen | Characteristics and physiological relevance to airway infection and diseases | Advantage | Disadvantage |
|---|---|---|---|
| Urine |
|
|
|
| Blood |
|
|
|
| Induced Sputum |
|
|
|
| Saliva |
|
|
|
| Exhaled breath (condensates and non-condensates) |
|
|
|
| Nasal lavage fluid (NLF) |
|
|
|
| Broncho aveolar lavage fluid (BALF) |
|
|
|
| Stool |
|
|
|
Study design
Metabolomics epidemiological studies face unique challenges of bias starting with study design and selection of target population. Like other clinical-translational investigations, metabolomics studies should first have a well-defined question and include a study population of representative subjects from a well-defined population in whom unrelated factors that impact the measure of the metabolome can be held constant (e.g. medications, diet, timing, etc.).71 As an example, if a study is designed to include both control and disease subjects, the subjects need to be similar at baseline with regard to such characteristics as hospitalization, medication, nutrition, genetic variants, and physical activity.71 Heterogeneity in severity of the disease among selected subjects should also be similar to the target population. If the study is designed as a time series (repeated measure within subjects), the effect of age on metabolites should additionally be considered. However, the initial biomarkers that are identified under the minimum difference between case and control populations need to be verified in heterogeneous populations to ascertain real world clinical application of the biomarkers.
Determining the sample size
Like all experiments, metabolomics studies require a certain number of samples to be able to effectively estimate the significance of the difference between study groups. Currently there is no standard power calculation approach for metabolomics, in part because the effect size of the metabolites on phenotype and number of metabolites to be identified are unknown a priori.72 While an adaptive design can help in overcoming the challenges of a priori unknown effect size in determining sample size, challenges stemming from the large dimensional nature of the data remain, especially in the case of untargeted metabolomics where the number of metabolites detected exceeds the number of samples. Furthermore, if metabolites belong to the same metabolic pathways, their abundances are highly correlated.73,72 Recently, Blaise and colleagues73 introduced a new approach, based on multivariate simulation, which deals with the highly correlated structure and high-dimensionality of metabolic phenotyping data.
Sample collection, handling, and analysis
For metabolomics studies, the sample collection, sample processing, and appropriate sample storage procedures should ensure that all samples are treated in the same manner.74,75,76 Uniformity in type and brand of collection tubes, aliquot tubes, and pipette tips is crucial. It is important to freeze the samples immediately and minimize factors such as, freeze-thaw cycles, contamination, and differing preservation methods to reduce small changes to the metabolic profiles.77,78,79 Freezing immediately is also critical as some metabolites transform very rapidly if enzymatic activity is not stopped completely.80,81
Studies using biospecimen repositories should ensure that samples have identical histories in terms of collection, storage, and processing, which require careful planning especially for samples that are collected as part of large multipurpose studies. Like most biological molecules, metabolites degrade overtime even when frozen. Thus the earliest possible analysis and avoiding samples with freeze-thaw history prevents potential loss of biomarker analytes.81,82
Biological samples undergoing metabolomics analyses require a standardized metabolite extraction protocol.48,83 The protocol needs to be optimized and validated to specific biospecimen type and metabolite species of interest. The intrinsic characteristics of the biospecimen such as presence of water, salt, and protein can pose challenges depending on the analytical platform, thus additional steps are required to suppress any undesired noise.84 Pooled quality control and blank samples must be included with each filtration/extraction batch of samples to allow for the monitoring of variability in sample processing and acquisition, especially for studies which include longitudinal and recurrent sampling.85,86
Metabolomics data preprocessing and analysis
Analysis of metabolomics data poses challenges common to other –omics and some challenges unique to metabolomics, including high dimensionality, use of multiple analytical methods (NMR, LC/GC-MS, etc.), high degree of collinearity between features, non-random missing data, nonlinearity, and non-normality. High-throughput NMR and MS spectral data need to undergo preprocessing in order to ensure the quality and replicability.87 Once the metabolite features are filtered, detected, identified, quantified, normalized, and scaled, data analysis can be performed either on the spectra itself (normalized peak intensity) or on the resultant concentration table. Parametric univariate tests such as t-test and analysis of variance (ANOVA), and nonparametric univariate tests such as Mann Whitney U and Kruskal Wallis one way analysis88 can be applied to identify biomarkers associated with the outcome of interest. It is important to recognize that univariate tests suffer from the multiple test problem and need correction.89 Unsupervised multivariate statistics such as principal component analysis (PCA), self-organizing map (SOM), and hierarchical cluster analysis (HCA) are useful to explore pattern and clusters and to assess data quality problems such outliers and batch effects.90,91 Supervised methods such as partial least square discriminant analysis (PL-SDA) and orthogonal PLS-DA92,93 can be applied both to concentration tables and spectra for discriminating between disease phenotypes and endotypes, prediction, and biomarker identification. Machine learning tools such as hidden Markov, Bayesian, support vector machine (SVM), random forest, and neural network are useful for prediction and biomarker identification in large samples.94,95
The use of metabolomics data to identify biomarkers using supervised multivariate and machine learning methods has limitations, including overfitting, which denotes the over specification of a model on the basis of irrelevant variables that separate the groups, but are not related to the classification criterion itself. To remedy the overfitting problem, adequate sample size are needed, the model should be calibrated with the application of internal cross-validation or bootstrapping, and the results should be validated in an independent external sample. Ideally, an independent team conducts the external validation step on independent subjects and on an independent analytical platform, but it is sufficient if blinded validation finds correlation between biomarker and the clinically important outcome.
Knowledge based methods for biological data interpretation
Following robust statistical analysis, the selected metabolites should be interpreted within the context of relevant metabolic pathways. Metabolic pathways are groups of metabolites and associated genes and proteins that are related to the same biological process and directly or indirectly related by one or multiple enzymatic reactions.96,97 Several curated biological databases have been constructed and referenced by metabolomics investigators over the last decade.98,99,100 Open source and commercial bioinformatics tools are available to access, draw, edit, and visualize the pathway networks related to the metabolites identified.101,102
In addition, metabolites representation in metabolic pathways can be objectively assessed with metabolite set enrichment analysis (MSEA) and quantitative enrichment analysis (QEA).103 MSEA and QEA evaluate the probability that metabolites are represented in the pathway more than expected by chance or due to phenotype. MSEA uses a preselected set of metabolites while QEA is based on the concentration of metabolites from experiments. The latter accounts for correlation between metabolites, whether a few metabolites significantly change, or whether there is a pattern of change in which a large number of metabolites change slightly but consistently.104
Application of metabolomics to study the impact of environment on the host
Using the example of infant respiratory viral infection as an early life exposure and asthma risk factor it is easy to demonstrate how metabolomics sheds light on the impact of an environmental exposure on the host, on elucidating disease pathways, and on identifying biomarkers of disease.
Metabolic profiling has been utilized in infectious disease models to enhance prognostic or diagnostic methods, and to gain insight into disease pathogenesis. 105,106,107 Evidence shows that viruses takeover and reprogram metabolic pathways, demonstrating modified metabolism or new metabolic pathways that enhance virulence. 107.108,109 Although to date, very few studies have applied metabolomics methods to investigate infant RSV and human rhinovirus (HRV) infections, it is one area in which metabolomics is beginning to be applied to characterize disease pathogenesis and endotypes. Atzei and colleagues110 studied a group of preterm neonates hospitalized due to RSV bronchiolitis comparing two 1H-NMR urine spectra from two bronchiolitis patients, which revealed alterations in several compounds like creatinine, betaine and glycine.110 Our group has developed the only other data to date profiling and contrasting the urinary metabolome of healthy and RSV infected infants, demonstrating that five metabolites were significantly decreased during acute RSV infection, represented in essential and nonessential amino acids, leukotriene, and urea cycle, and vitamin B metabolism pathways.111 Schee and colleagues profiled VOC in exhaled breath of preschool children who wheezed (with and without HRV infection) and did not wheeze demonstrating that the VOC profile between preschool children with and without acute respiratory wheeze differ, and a sustained and distinct VOC profile was observed in children with HRV-induced wheeze after resolution of symptoms.112 Fowler and colleagues have also demonstrated that VOC can be used to distinguish those with high and low pathogen load in lower respiratory tract among intensive care unit intubated and ventilated patients.113
Review of metabolomics applied to the outcome of childhood asthma
The first pediatric asthma metabolomics investigation was carried out by Carraro and colleagues114 less than a decade ago. They applied untargeted NMR metabolomics on EBC samples of 25 asthma patients and 11 controls between the ages of 7 and 15 years. Metabolomics profiles clearly discriminated children with and without asthma, with a 95% success rate in their classification, and identified acetylated and oxidized compounds that distinguished the children. In subsequent studies, researchers have applied untargeted NMR115, targeted LC-MS (targeting adenosine monophosphate and purine116 and leukotriene117), untargeted LC-MS118 and targeted GC-MS (targeting VOC119,120,121,43 and alkanes, alkenes, aldehydes, and ketones42) metabolomics technologies in EB120,121,42,43 and EBC116,117,118 samples to distinguish between asthmatic and healthy children,116,117,120,121,43 phenotypes of asthma,118 wheezers and non wheezers,43 and transient wheezers and asthmatics42. In addition, E-nose VOC pattern recognition in EB has been successfully used to discriminate between asthmatic and control, as well as between mild and severe asthma in children.122
In plasma sample based targeted LC-MS lipid metabolomics application, McGeachie and colleagues123 found that monoHETE_0863 and sphingosine-1-phosphate could predict asthma control. In addition, integration of metabolomics and genetics data revealed that sphingolipid metabolism and immune response pathways were associated with asthma control. Similarly, Fitzpatrick and colleagues39 used untargeted LC-MS in plasma samples from children to discriminate mild and severe asthma. Fitzpatrick and colleagues39 identified two metabolic pathways (glycine, serine and threonine metabolism and N-acylethanolamine and N-acyltransferase pathways) associated with asthma severity. In addition, Saude and colleagues124 and Mattarucchi and colleagues40 applied targeted NMR and LC-MS to urine samples. Saude and colleagues showed that 23 and 28 metabolites separated stable asthmatic versus healthy controls, and stable asthmatics versus acute asthmatic patients, respectively. Mattarucchi and colleagues additionally found decreased excretion of methyl-imidazole acetic acid, urocanic acid and a metabolite similar to the structure of an isolusine–proline fragment in the asthmatics. These studies demonstrate that metabolomics may be used to identify biomarkers of disease and healthy states, as well as progression of disease states.
Limitations of metabolomics
Metabolomics provides a unique opportunity to study the interaction between environment and host because the metabolites represent the response to both environmental stimulation and upstream genetic and regulatory modification (epigenetics, transcription, post-translational modification). However, metabolomics by itself may not capture the range of environmental agent characteristics or the range of host responses during the interaction. First, an environmental stimulus unleashes a cascade of host molecular pathways, and it is difficult to pinpoint which pathway is precisely associated with the specific stimuli. Second, metabolic profiles are subject to random fluctuations, and can be influenced by diet, sleep patterns, age, smoking, and many other variables that mask the effects of disease or toxicity. Third, many of the changes in metabolites are subtle and below detection limits of commonly applied analytical techniques. Furthermore, there are a variety of factors that influence the type and amount of metabolites detected: timing of sample collection, the sample collection procedure, sample processing, stabilization, stability and storage, extraction procedures, dilution of sample, type and number of analytical methods used, and preferences of analytical assays for metabolites with certain physico-chemical properties. Identifying biomarkers of disease from this background noise is a complex analytical and statistical challenge.
Integration with other -omics data and future directions
Disease development occurs in complex biological systems, where genetic, regulatory, and environmental stimuli trigger a broad range of input-output cascades in the biological system. The pathogenesis of disease development needs to be examined within the context of this broad biological system. An emerging approach is to integrate data from a range of -omics studies (genome, epigenome, transcriptome, proteome, and metabolome), which may provide greater opportunity to understand and model the process of disease development in the context of complex biological systems.125,126 Vertical and longitudinal integration of multi-omics data may reveal not only what will happen following exposure to an environmental risk factor, but also the long-term imprint of the exposure on the immune response and airway function leading to an understanding of the mechanisms through which the risk factor contributes to asthma development, and ultimately how asthma, or the disease under study, might be prevented.
Phenotyping and endotyping with multi-omics data can be performed using clustering, latent class models, and projection based methods. Further functional analyses using network and pathway inference can be performed with combining database and bioinformatics tools. Analysis of causal relationships using multi-omics data from experiments under different conditions and/or at different time points can be modeled in probabilistic causal networks. However, multi-omics data integration tools are in their infancy, and integration of data is complicated by many unresolved factors including handling of data with different measurement scales (eg. binary, ordinal, and interval) and how to manage missing data.
An example of progress in bioinformatics for integrating complex data, is the development of a promising method that overcomes the scale issue (integrative phenotyping framework, iPF) by Kim and coworkers and they have demonstrated its use in feature discovery and integrative clustering in high dimensional space.127 They applied their iPF method to a large dataset of lung samples of genomic and clinical data and identified endotypes of Chronic Obstructive Pulmonary Disease and Interstitial Lung Disease. However, the -omics data integrated using this method was limited to messenger (mRNA) and micro RNA (miRNA). There is still a need to extend such an approach to upstream (genome) and downstream (proteome and metabolome) -omics data.
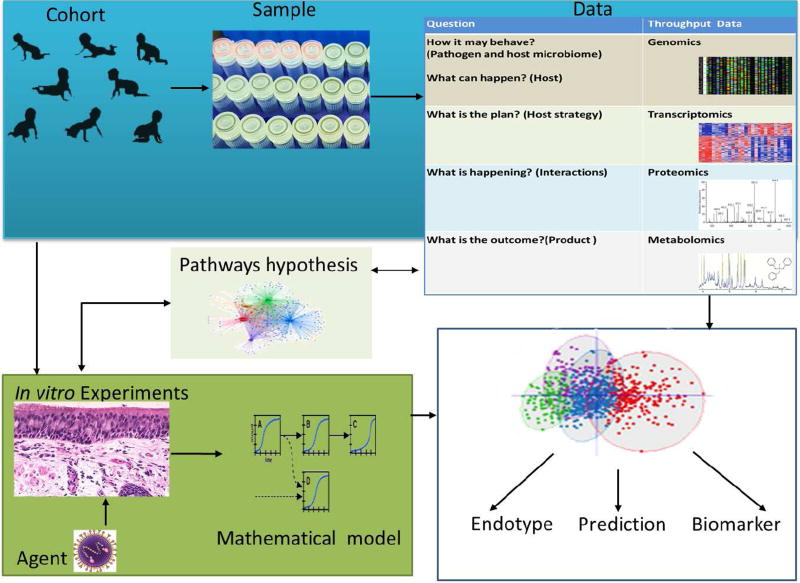
Multi-omics data integration and analysis pipelines for studying the pathogenesis of disease and the influence of environmental risk factors are scarce. de Steenhuijsen Piters and colleagues were able to integrate host blood transcriptome and nasopharyngeal microbiome data to show that RSV infection immune response and disease severity are modulated by dominant colonizing microbiota.128 On a larger scale, the complexity of multi-omics data is challenging for integration because the data is generated not only from the analysis of host biospecimens, but may also include sequencing of the host microbiome, the environmental microbiome, the virus genome, and even –omics data from controlled in vitro and ex vivo experiments in host cell or tissue cultures. The multisource -omics data integration approach can be thought of as a top-down systems approach, the integration of -omics data at a population level; and a bottom-up systems approach, which integrates experimental data from primary cell in vitro experiments (Figure 3). The bottom-up systems approach provides detailed and validating information for the pathways hypotheses generated from the top-down systems approach. The integration of the -omics data from the host and in response to environment is crucial in elucidating disease pathogenesis at a systems level. This is a novel approach, and contrary to traditional approaches where both are studied separately. In this regard, asthma consortiums and cohorts such as the Unbiased Biomarkers for the Prediction of Respiratory Disease Outcomes (U-BIOPRED) project, the Severe Asthma Research Program (SARP), and the NIH Environmental Influences on Child Health Outcomes (ECHO) Children’s Respiratory Research and Environment Workgroup (CREW) have great potential in integrating complex clinical and biomolecular data and identifying and predicting various endotypes and phenotypes of asthma.129,130 Commercial and public collaborative initiative such as European Translational Information and Knowledge Service (eTRIKS) have developed a graph based knowledge integration tool that could help to integrate experimental and clinical knowledge to map disease pathways.131,132
Figure 3.
Illustration of multi-omics systems approach using a birth cohort (top-down systems approach) and in vitro and ex vivo experiments (bottom-up systems approach) to study pathogenesis and biomarker discovery using an example of reparatory viral infection exposure and acute morbidity, and associated subsequent chronic respiratory disease.
In conclusion, metabolomics measures relevant biological processes, providing readout of metabolic activity status in relation to genetic variations, gene expression, or environmental stimuli. Metabolic profiles are unique in providing a snapshot of the interaction between the environmental agent and host molecules, providing an opportunity to answer questions that have not been addressed with other upstream -omics technologies. However, remaining, but an area of active development, is addressing the challenge of integrating and interpreting these complex data in order to address mechanistic questions about disease pathogenesis, disease phenotyping and endotyping, and disease prediction.
Acknowledgments
Financial support: This work was supported in part by National Institute of Health U19AI95227, K24 AI 77930, R21HD087864, and T32HL087738.
Abbreviations
- ANOVA
Analysis of variance
- CREW
Children’s Respiratory Research and Environment Workgroup
- E-nose
Electronic nose
- ECHO
Environmental Influences on Child Health Outcomes
- eTRIKS
European Translational Information and Knowledge Service
- EB
Exhaled Breath
- EBC
Exhaled breath condensate
- HCA
Hierarchical cluster analysis
- HRV
Human rhinovirus
- iPF
Integrative phenotyping framework
- LC
Liquid chromatography
- GC
Gas chromatography
- RNA
Ribonucleic acid
- mRNA
Messenger RNA
- MSEA
Metabolite set enrichment analysis
- miRNA
Micro RNA
- NIH
National Institute of Health
- NMR
Nuclear magnetic resonance
- PL-SDA
Partial least square discriminant analysis
- PCA
Principal component analysis
- QEA
Quantitative enrichment analysis
- RSV
Respiratory syncytial virus
- SOM
Self-organizing map
- SARP
Severe Asthma Research Program
- MS
Spectroscopy and mass spectrometry
- SMO
Semiconductor metal oxide
- TQMS
Triple quadrupole mass spectrometer
- SVM
Support vector machine
- U-BIOPRED
Unbiased Biomarkers for the Prediction of Respiratory Disease Outcomes
- VOC
Volatile organic compound
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Potential Conflict of interest: The authors declare no conflicts of interest.
References
- 1.Patti GJ, Yanes O, Siuzdak G. Innovation: Metabolomics: the apogee of the omics trilogy. Nat Rev Mol Cell Bio. 2012;13(4):263–269. doi: 10.1038/nrm3314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Gowda GN, Zhang S, Gu H, Asiago V, Shanaiah N, Raftery D. Metabolomics-based methods for early disease diagnostics. Expert Rev Mol Diagn. 2008;8(5):617–633. doi: 10.1586/14737159.8.5.617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Idle JR, Gonzalez FJ. Metabolomics. Cell Metab. 2007;6(5):348–351. doi: 10.1016/j.cmet.2007.10.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Pacchiarotta T, Deelder AM, Mayboroda OA. Metabolomic investigations of human infections. Bioanalysis. 2012;4(8):919–925. doi: 10.4155/bio.12.61. [DOI] [PubMed] [Google Scholar]
- 5.Scrivo R, Casadei L, Valerio M, Priori R, Valesini G, Manetti C. Metabolomics approach in allergic and rheumatic diseases. Curr Allergy Asthma Rep. 2014;14(6):1–1. doi: 10.1007/s11882-014-0445-5. [DOI] [PubMed] [Google Scholar]
- 6.Johnson CH, Patterson AD, Idle JR, Gonzalez FJ. Xenobiotic metabolomics: major impact on the metabolome. Annu Rev Pharmacol Toxicol. 2012;52:37–56. doi: 10.1146/annurev-pharmtox-010611-134748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kettunen J, Tukiainen T, Sarin AP, Ortega-Alonso A, Tikkanen E, Lyytikainen LP, et al. Genome-wide association study identifies multiple loci influencing human serum metabolite levels. Nat Genet. 2012;44:269–276. doi: 10.1038/ng.1073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Serkova NJ, Standiford TJ, Stringer KA. The emerging field of quantitative blood metabolomics for biomarker discovery in critical illnesses. Am J Resp Crit Care Med. 2011;184(6):647–655. doi: 10.1164/rccm.201103-0474CI. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gieger C, Geistlinger L, Altmaier E, Hrabe de Angelis M, Kronenberg F, et al. Genetics meets metabolomics: a genome-wide association study of metabolite profiles in human serum. PLoS Genet. 2008;4(11):e1000282. doi: 10.1371/journal.pgen.1000282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.GBD 2013 Mortality and Causes of Death Collaborators. Global, regional, and national age-sex specific all-cause and cause-specific mortality for 240 causes of death, 1990–2013: a systematic analysis for the Global Burden of Disease Study 2013. Lancet. 2015;385:117–171. doi: 10.1016/S0140-6736(14)61682-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Nair H, Nokes DJ, Gessner BD, Dherani M, Madhi SA, Singleton RJ, et al. Global burden of acute lower respiratory infections due to respiratory syncytial virus in young children: a systematic review and meta-analysis. Lancet. 2010;375(9725):1545–55. doi: 10.1016/S0140-6736(10)60206-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Tregoning JS, Schwarze J. Respiratory viral infections in infants: causes, clinical symptoms, virology, and immunology. Clin Microbiol Rev. 2010;23(1):74–98. doi: 10.1128/CMR.00032-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cox DW, Bizzintino J, Ferrari G, Khoo SK, Zhang G, Whelan S, et al. Human rhinovirus species C infection in young children with acute wheeze is associated with increased acute respiratory hospital admissions. Am J Resp Crit Care Med. 2013;188(11):1358–64. doi: 10.1164/rccm.201303-0498OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bacharier LB, Cohen R, Schweiger T, Yin-DeClue H, Christie C, Zheng J, et al. Determinants of asthma after severe respiratory syncytial virus bronchiolitis. J Allergy Clin Immunol. 2012;130:91–100. doi: 10.1016/j.jaci.2012.02.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Beigelman A, Bacharier LB. The role of early life viral bronchiolitis in the inception of asthma. Curr Opin Allergy Clin Immunol. 2013;13:211–216. doi: 10.1097/ACI.0b013e32835eb6ef. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jartti T, Lehtinen P, Vuorinen T, Ruuskanen O. Bronchiolitis: age and previous wheezing episodes are linked to viral etiology and atopic characteristics. Pediatr Infect Dis J. 2009;28:311–31710. doi: 10.1097/INF.0b013e31818ee0c1. [DOI] [PubMed] [Google Scholar]
- 17.Stein RT, Sherrill D, Morgan WJ, Holberg CJ, Halonen M, Taussig LM, et al. Respiratory syncytial virus in early life and risk of wheeze and allergy by age 13 years. The Lancet. 1999;354(9178):541–5. doi: 10.1016/S0140-6736(98)10321-5. [DOI] [PubMed] [Google Scholar]
- 18.Wu P, Dupont WD, Griffin MR, Carroll KN, Mitchel EF, Gebretsadik T, et al. Evidence of a causal role of winter virus infection during infancy in early childhood asthma. Am J Resp Crit Care Med. 2008;178(11):1123–9. doi: 10.1164/rccm.200804-579OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kawasaki Y, Aoyagi Y, Abe Y, Go H, Imamura T, Kaneko M, et al. Serum KL-6 levels as a biomarker of lung injury in respiratory syncytial virus bronchiolitis. J Med Virol. 2009;81(12):2104–2108. doi: 10.1002/jmv.21634. [DOI] [PubMed] [Google Scholar]
- 20.Mejias A, Dimo B, Suarez NM, Garcia C, Suarez-Arrabal MC, Jartti T, et al. Whole blood gene expression profiles to assess pathogenesis and disease severity in infants with respiratory syncytial virus infection. PLoS Med. 2013;10:e1001549. doi: 10.1371/journal.pmed.1001549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Harvala H, McIntyre CL, McLeish NJ, Kondracka J, Palmer J, Molyneaux P, et al. High detection frequency and viral loads of human rhinovirus species A to C in fecal samples; diagnostic and clinical implications. J Med Virol. 2012;84(3):536–42. doi: 10.1002/jmv.23203. [DOI] [PubMed] [Google Scholar]
- 22.Rosas-Salazar C, Gebretsadik T, Carroll KN, Reiss S, Wickersham N, Larkin EK, et al. Urine Club Cell 16-kDa Secretory Protein and Childhood Wheezing Illnesses After Lower Respiratory Tract Infections in Infancy. Pediatr Allergy Immu. 2015;28(3):158–164. doi: 10.1089/ped.2015.0528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Openshaw PJ. A gene expression signature for RSV: clinical implications and limitations. PLoS Med. 2013;10(11):e1001550–e1001550. doi: 10.1371/journal.pmed.1001550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Peng B, Li H, Peng XX. Functional metabolomics: from biomarker discovery to metabolome reprogramming. Protein & Cell. 2015;6(9):628–637. doi: 10.1007/s13238-015-0185-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Thompson WW, Shay DK, Weintraub E, Brammer L, Cox N, Anderson LJ, et al. Mortality associated with influenza and respiratory syncytial virus in the United States. JAMA. 2003;289(2):179–186. doi: 10.1001/jama.289.2.179. [DOI] [PubMed] [Google Scholar]
- 26.Renukaradhya GJ, Narasimhan B, Mallapragada SK. Respiratory nanoparticle-based vaccines and challenges associated with animal models and translation. J Control Release. 2015;219:622–631. doi: 10.1016/j.jconrel.2015.09.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kim YI, DeVincenzo JP, Jones BG, Rudraraju R, Harrison L. Respiratory Syncytial Virus Human Experimental Infection Model: Provenance, Production, and Sequence of Low-Passaged Memphis-37 Challenge Virus. PLoS One. 2014;9(11):e113100. doi: 10.1371/journal.pone.0113100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mestas J, Hughes CC. Of mice and not men: differences between mouse and human immunology. J Immunol. 2004;172:2731–2738. doi: 10.4049/jimmunol.172.5.2731. [DOI] [PubMed] [Google Scholar]
- 29.Irvin CG, Bates JH. Measuring the lung function in the mouse: the challenge of size. Respir Res. 2003;4(4):1–9. doi: 10.1186/rr199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Papin JF, Wolf RF, Kosanke SD, Jenkins JD, Moore SN, Anderson MP, et al. Infant baboons infected with respiratory syncytial virus develop clinical and pathological changes that parallel those of human infants. Am J Physiol Lung Cell Mol Physiol. 2013;304(8):L530–9. doi: 10.1152/ajplung.00173.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.De Chassey B, Meyniel-Schicklin L, Vonderscher J, André P, Lotteau V. Virus-host interactomics: new insights and opportunities for antiviral drug discovery. Genome Med. 2014;6(11):115. doi: 10.1186/s13073-014-0115-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Blume C, Davies DE. In vitro and ex vivo models of human asthma. Eur J Pharm Biopharm. 2013;84(2):394–400. doi: 10.1016/j.ejpb.2012.12.014. [DOI] [PubMed] [Google Scholar]
- 33.Leung TF, Ko FW, Wong GW. Recent advances in asthma biomarker research. Ther Adv Respir Dis. 2013;7(5):297–308. doi: 10.1177/1753465813496863. [DOI] [PubMed] [Google Scholar]
- 34.Moschino L, Zanconato S, Bozzetto S, Baraldi E, Carraro S. Childhood asthma biomarkers: present knowledge and future steps. Paediatr Respir Rev. 2015;16(4):205–12. doi: 10.1016/j.prrv.2015.05.001. [DOI] [PubMed] [Google Scholar]
- 35.Fiehn O. Metabolomics-the link between genotypes and phenotypes. Plant Mol Biol. 2002;48(1):155–171. [PubMed] [Google Scholar]
- 36.Dunn WB, Erban A, Weber RJ, Creek DJ, Brown M, Breitling R, et al. Mass appeal: metabolite identification in mass spectrometry-focused untargeted metabolomics. Metabolomics. 2013;9(1):44–66. [Google Scholar]
- 37.Roberts LD, Souza AL, Gerszten RE, Clish CB. Targeted metabolomics. Curr Protoc Mol Bio. 2012:30–2. doi: 10.1002/0471142727.mb3002s98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ioannidis JP, Khoury MJ. Improving validation practices in “omics” research. Science. 2011;334(6060):1230–1232. doi: 10.1126/science.1211811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Fitzpatrick AM, Park Y, Brown LA, Jones DP. Children with severe asthma have unique oxidative stress-associated metabolomic profiles. J Allergy Clin Immunol. 2014;133(1):258–261. doi: 10.1016/j.jaci.2013.10.012. J. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Mattarucchi E, Baraldi E, Guillou C. Metabolomics applied to urine samples in childhood asthma; differentiation between asthma phenotypes and identification of relevant metabolites. Biomed Chromatogr. 2012;26:89–94. doi: 10.1002/bmc.1631. [DOI] [PubMed] [Google Scholar]
- 41.Dudley E, Yousef M, Wang Y, Griffiths WJ. Targeted metabolomics and mass spectrometry. Adv Protein Chem Struct Biol. 2010;80:45–83. doi: 10.1016/B978-0-12-381264-3.00002-3. [DOI] [PubMed] [Google Scholar]
- 42.Caldeira M, Perestrelo R, Barros AS, Bilelo MJ, Morete A, Camara JS, et al. Allergic asthma exhaled breath metabolome: a challenge for comprehensive two-dimensional gas chromatography. J Chromatogr A. 2012;1254:87–97. doi: 10.1016/j.chroma.2012.07.023. [DOI] [PubMed] [Google Scholar]
- 43.van de Kant KD, van Berkel JJ, Jöbsis Q, Passos VL, Klaassen EM, van der Sande L, et al. Exhaled breath profiling in diagnosing wheezy preschool children. Eur Respir J. 2013;41(1):183–8. doi: 10.1183/09031936.00122411. [DOI] [PubMed] [Google Scholar]
- 44.Ho CS, Lam CW, Chan MH, Cheung RC, Law LK, Lit LC, et al. Electrospray ionisation mass spectrometry: principles and clinical applications. Clin Biochem Rev. 2003;24(1):3–12. [PMC free article] [PubMed] [Google Scholar]
- 45.Draper J, Enot DP, Parker D, Beckmann M, Snowdon S, Lin W, et al. Metabolite signal identification in accurate mass metabolomics data with MZedDB, an interactive m/z annotation tool utilising predicted ionisation behaviour’rules’. BMC Bioinformatics. 2009 Jul 21;10(1):1. doi: 10.1186/1471-2105-10-227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Alonso A, Marsal S, Julia A. Analytical methods in untargeted metabolomics: state of the art in 2015. Front Bioeng Biotechnol. 2015;3(23):1–20. doi: 10.3389/fbioe.2015.00023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Theodoridis G, Gika HG, Wilson ID. Mass spectrometry-based holistic analytical approaches for metabolite profiling in systems biology studies. Mass Spectrom Rev. 2011;30(5):884–906. doi: 10.1002/mas.20306. [DOI] [PubMed] [Google Scholar]
- 48.Beckonert O, Keun HC, Ebbels TM, Bundy J, Holmes E, Lindon JC, et al. Metabolic profiling, metabolomic and metabonomic procedures for NMR spectroscopy of urine, plasma, serum and tissue extracts. Nat Protoc. 2007;2(11):2692–703. doi: 10.1038/nprot.2007.376. [DOI] [PubMed] [Google Scholar]
- 49.Veenstra TD. Metabolomics the final frontier. Genome Med. 2012;4(4):40. doi: 10.1186/gm339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Tisch U, Haick H. Chemical sensors for breath gas analysis: the latest developments at the Breath Analysis Summit 2013. J Breath Res. 2014 Mar 28;8(2) doi: 10.1088/1752-7155/8/2/027103. 027103. [DOI] [PubMed] [Google Scholar]
- 51.Lewis NS. Comparisons between mammalian and artificial olfaction based on arrays of carbon black-polymer composite vapor detectors. Acc Chem Res. 2004;37:663–72. doi: 10.1021/ar030120m. [DOI] [PubMed] [Google Scholar]
- 52.Scarlata S, Pennazza G, Santonico M, Pedone C, Antonelli Incalzi R. Exhaled breath analysis by electronic nose in respiratory diseases. Expert Rev Mol Diagn. 2015;15(7):933–56. doi: 10.1586/14737159.2015.1043895. [DOI] [PubMed] [Google Scholar]
- 53.Magdeldin S, Hirao Y, Elguoshy A, Xu B, Zhang Y, Fujinaka H, et al. A proteomic glimpse into human ureter proteome. Proteomics. 2016;16(1):80–4. doi: 10.1002/pmic.201500214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Wu J, Gao Y. Physiological conditions can be reflected in human urine proteome and metabolome. Exp Rev Proteomics. 2015;12(6):623–36. doi: 10.1586/14789450.2015.1094380. [DOI] [PubMed] [Google Scholar]
- 55.Emwas AH, Roy R, McKay RT, Ryan D, Brennan L, Tenori L, et al. Recommendations and standardization of biomarker quantification using NMR-based metabolomics with particular focus on urinary analysis. J Proteome Res. 2016;15(2):360–73. doi: 10.1021/acs.jproteome.5b00885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Psychogios N, Hau DD, Peng J, Guo AC, Mandal R, Bouatra S, et al. The human serum metabolome. PloS One. 2011;6(2):e16957. doi: 10.1371/journal.pone.0016957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Yin P, Lehmann R, Xu G. Effects of pre-analytical processes on blood samples used in metabolomics studies. Anal Bioanal Chem. 2015;407(17):4879–92. doi: 10.1007/s00216-015-8565-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Bakakos P, Schleich F, Alchanatis M, Louis R. Induced sputum in asthma: from bench to bedside. Curr Medl Chem. 2011;18(10):1415–22. doi: 10.2174/092986711795328337. [DOI] [PubMed] [Google Scholar]
- 59.Nicholas B, Djukanović R. Induced sputum: a window to lung pathology. Biochem SocTrans. 2009;37(4):868–72. doi: 10.1042/BST0370868. [DOI] [PubMed] [Google Scholar]
- 60.Rehak NN, Cecco SA, Csako G. Biochemical composition and electrolyte balance of “unstimulated” whole human saliva. Clin Chem Lab Med. 2000;38:335–343. doi: 10.1515/CCLM.2000.049. [DOI] [PubMed] [Google Scholar]
- 61.Dame ZT, Aziat F, Mandal R, Krishnamurthy R, Bouatra S, Borzouie S, et al. The human saliva metabolome. Metabolomics. 2015;11(6):1864–83. [Google Scholar]
- 62.Lee YH, Wong DT. Saliva: an emerging biofluid for early detection of diseases. Am J Dent. 2009;22(4):241. [PMC free article] [PubMed] [Google Scholar]
- 63.Ahmadzai H, Huang S, Hettiarachchi R, Lin JL, Thomas PS, Zhang Q. Exhaled breath condensate: a comprehensive update. Clin Chem Lab Med. 2013;51(7):1343–61. doi: 10.1515/cclm-2012-0593. [DOI] [PubMed] [Google Scholar]
- 64.Kuban P, Fore F. Exhaled breath condensate: determination of non-volatile compounds and their potential for clinical diagnosis and monitoring. A review. Anal Chim Acta. 2013;805:1–18. doi: 10.1016/j.aca.2013.07.049. [DOI] [PubMed] [Google Scholar]
- 65.Maniscalco JM, Matkin CO, Maldini D, Calkins DG, Atkinson S. Assessing killer whale predation on Steller sea lions from field observations in Kenai Fjords, Alaska. Mar Mamm Sci. 2007;23(2):306–21. [Google Scholar]
- 66.Dodig S, Cepelak I. Exhaled breath condensate-from an analytical point of view. Biochem Med (Zagreb) 2013;23(3):281–95. doi: 10.11613/BM.2013.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Schoenebeck B, May C, Güldner C, Respondek G, Mollenhauer B, Hoeglinger G, et al. Improved preparation of nasal lavage fluid (NLF) as a noninvasive sample for proteomic biomarker discovery. Biochim Biophysica Acta. 2015;1854(7):741–5. doi: 10.1016/j.bbapap.2015.01.015. [DOI] [PubMed] [Google Scholar]
- 68.Mendivil CO, Koziel H, Brain JD. Metabolic hormones, apolipoproteins, adipokines, and cytokines in the alveolar lining fluid of healthy adults: Compartmentalization and physiological correlates. PloS One. 2015;10(4):e0123344. doi: 10.1371/journal.pone.0123344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Fronius M, Clauss WG, Althaus M. Why do we have to move fluid to be able to breathe? Front Physiol. 2012;3:146. doi: 10.3389/fphys.2012.00146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Stone MD, Harvey SB, Nelsestuen GL, Reilly C, Hertz MI, Wendt CH. Elevated peptides in lung lavage fluid associated with bronchiolitis obliterans syndrome. PloS One. 2014;9(1):e84471. doi: 10.1371/journal.pone.0084471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Kohler I, Verhoeven A, Derks RJ, Giera M. Analytical pitfalls and challenges in clinical metabolomics. Bioanalysis. 2016;8(14):1509–32. doi: 10.4155/bio-2016-0090. [DOI] [PubMed] [Google Scholar]
- 72.Vinaixa M, Samino S, Saez I, Duran J, Guinovart JJ, Yanes O. A guideline to univariate statistical analysis for LC/MS-based untargeted metabolomics-derived data. Metabolites. 2012;2(4):775–95. doi: 10.3390/metabo2040775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Blaise BJ, Correia G, Tin A, Young JH, Vergnaud AC, Lewis M, et al. Power Analysis and Sample Size Determination in Metabolic Phenotyping. Anal Chem. 2016;88(10):5179–88. doi: 10.1021/acs.analchem.6b00188. [DOI] [PubMed] [Google Scholar]
- 74.Bernini P, Bertini I, Luchinat C, Nincheri P, Staderini S, Turano P, et al. Standard operating procedures for pre-analytical handling of blood and urine for metabolomic studies and biobanks. J Biomol NMR. 2011;49:231–243. doi: 10.1007/s10858-011-9489-1. [DOI] [PubMed] [Google Scholar]
- 75.Rosenling T, Stoop MP, Smolinska A, Muilwijk B, Coulier L, Shi S, et al. The impact of delayed storage on the measured proteome and metabolome of human cerebrospinal fluid. Clin Chem. 2011;57:1703–1711. doi: 10.1373/clinchem.2011.167601. [DOI] [PubMed] [Google Scholar]
- 76.Tzoulaki I, Ebbels TM, Valdes A, Elliott P, Ioannidis JP. Design and analysis of metabolomics studies in epidemiologic research: a primer on -omic technologies. Am J Epidemiol. 2014;180:129–139. doi: 10.1093/aje/kwu143. [DOI] [PubMed] [Google Scholar]
- 77.Gika HG, Theodoridis GA, Wilson ID. Liquid chromatography and ultra-performance liquid chromatography-mass spectrometry fingerprinting of human urine: sample stability under different handling and storage conditions for metabonomics studies. J Chromatogr A. 2008;1189:314–322. doi: 10.1016/j.chroma.2007.10.066. [DOI] [PubMed] [Google Scholar]
- 78.Gika HG, Theodoridis GA, Wingate JE, Wilson ID. Within-day reproducibility of an HPLC-MS-based method for metabonomic analysis: application to human urine. J Proteome Res. 2007;6:3291–3303. doi: 10.1021/pr070183p. [DOI] [PubMed] [Google Scholar]
- 79.Teahan O, Gamble S, Holmes E, Waxman J, Nicholson JK, Bevan C, et al. Impact of analytical bias in metabonomic studies of human blood serum and plasma. Anal Chem. 2006;78:4307–4318. doi: 10.1021/ac051972y. [DOI] [PubMed] [Google Scholar]
- 80.Yang W, Chen Y, Xi C, Zhang R, Song Y, Zhan Q, et al. Liquid chromatography-tandem mass spectrometry-based plasma metabonomics delineate the effect of metabolites’ stability on reliability of potential biomarkers. Anal Chem. 2013;85(5):2606–10. doi: 10.1021/ac303576b. [DOI] [PubMed] [Google Scholar]
- 81.Cuhadar S, Koseoglu M, Atay A, Dirican A. The effect of storage time and freeze-thaw cycles on the stability of serum samples. Biochem Med (Zagreb) 2013;23(1):70–7. doi: 10.11613/BM.2013.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Bhatnagar BS, Bogner RH, Pikal MJ. Protein stability during freezing: separation of stresses and mechanisms of protein stabilization. Pharm Dev Technol. 2007;12(5):505–23. doi: 10.1080/10837450701481157. [DOI] [PubMed] [Google Scholar]
- 83.Emwas AH, Luchinat C, Turano P, Tenori L, Roy R, Salek RM, et al. Standardizing the experimental conditions for using urine in NMR-based metabolomic studies with a particular focus on diagnostic studies: a review. Metabolomics. 2015;11:872–894. doi: 10.1007/s11306-014-0746-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Yin P, Lehmann R, Xu G. Effects of pre-analytical processes on blood samples used in metabolomics studies. Anal Bioanal Chem. 2015;407(17):4879–92. doi: 10.1007/s00216-015-8565-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Broadhurst DI, Kell DB. Statistical strategies for avoiding false discoveries in metabolomics and related experiments. Metabolomics. 2006;2:171–196. [Google Scholar]
- 86.Dunn WB, Wilson ID, Nicholls AW, Broadhurst D. The importance of experimental design and QC samples in large-scale and MS-driven untargeted metabolomic studies of humans. Bioanalysis. 2012;4:2249–2264. doi: 10.4155/bio.12.204. [DOI] [PubMed] [Google Scholar]
- 87.Berg RA, Hoefsloot HC, Westerhuis JA, Smilde AK, Werf MJ. Centering, scaling, and transformations: improving the biological information content of metabolomics data. BMC Genomics. 2006;7(1):1. doi: 10.1186/1471-2164-7-142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Bartel J, Krumsiek J, Theis FJ. Statistical methods for the analysis of high-throughput metabolomics data. Comput Struct Biotechnol J. 2013;4(5):1–9. doi: 10.5936/csbj.201301009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.van den Oord EJ. Controlling false discoveries in genetic studies. American Journal of Medical Genetics Part B: Neuropsych Gen. 2008;147(5):637–44. doi: 10.1002/ajmg.b.30650. [DOI] [PubMed] [Google Scholar]
- 90.Misra BB, der Hooft JJ. Updates in metabolomics tools and resources: 2014–2015. Electrophoresis. 2016;37(1):86–110. doi: 10.1002/elps.201500417. [DOI] [PubMed] [Google Scholar]
- 91.Worley B, Powers R. Multivariate analysis in metabolomics. Curr Metabolomics. 2013;1(1):92. doi: 10.2174/2213235X11301010092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Barker M, Rayens W. Partial least squares for discrimination. J Chemom. 2003;17(3):166–73. [Google Scholar]
- 93.Trygg J, Wold S. Orthogonal projections to latent structures (O-PLS) J Chemom. 2002;16(3):119–28. [Google Scholar]
- 94.Bickel PJ, Brown JB, Huang H, Li Q. An overview of recent developments in genomics and associated statistical methods. Philos Trans A Math Phys Eng Sci. 2009;367(1906):4313–37. doi: 10.1098/rsta.2009.0164. [DOI] [PubMed] [Google Scholar]
- 95.Trygg J, Holmes E, Lundstedt T. Chemometrics in metabonomics. J Proteome Res. 2007;6(2):469–79. doi: 10.1021/pr060594q. [DOI] [PubMed] [Google Scholar]
- 96.Durek P, Walther D. The integrated analysis of metabolic and protein interaction networks reveals novel molecular organizing principles. BMC Sys Biol. 2008;2(1):100. doi: 10.1186/1752-0509-2-100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Srere PA. Complexes of sequential metabolic enzymes. Annu Rev Biochem. 1987;56:89–124. doi: 10.1146/annurev.bi.56.070187.000513. [DOI] [PubMed] [Google Scholar]
- 98.Kanehisa M, Goto S. KEGG: kyoto encyclopedia of genes and genomes. Nucleic Acids Res. 2000;28(1):27–30. doi: 10.1093/nar/28.1.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Ma H, Sorokin A, Mazein A, Selkov A, Selkov E, Demin O, et al. The Edinburgh human metabolic network reconstruction and its functional analysis. Mol Systems Biol. 2007;3(1) doi: 10.1038/msb4100177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Wishart DS, Tzur D, Knox C, Eisner R, Guo AC, Young N, et al. HMDB: the human metabolome database. Nucleic Acids Res. 2007;35(suppl 1):D521–6. doi: 10.1093/nar/gkl923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Shannon P, Markiel A, Ozier O, Baliga NS, Wang JT, Ramage D, et al. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res. 2003;13(11):2498–504. doi: 10.1101/gr.1239303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Kramer A, Green J, Pollard J, Tugendreich S. Causal analysis approaches in ingenuity pathway analysis. Bioinformatics. 2014;30(4):523–30. doi: 10.1093/bioinformatics/btt703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Xia J, Wishart DS. MSEA: a web-based tool to identify biologically meaningful patterns in quantitative metabolomic data. Nucleic Acids Res. 2010;38(suppl 2):W71–7. doi: 10.1093/nar/gkq329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Booth SC, Weljie AM, Turner RJ. Computational tools for the secondary analysis of metabolomics experiments. Comput Struct Biotechnol J. 2013;4(5):1–3. doi: 10.5936/csbj.201301003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Chandler JD, Hu X, Ko EJ, Park S, Lee YT, Orr M, et al. Metabolic pathways of lung inflammation revealed by high-resolution metabolomics (HRM) of H1N1 influenza virus infection in mice. Am J Physiol Regul Integr Comp Physiol. 2016;311(5):R906–16. doi: 10.1152/ajpregu.00298.2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Lee H. Procalcitonin as a biomarker of infectious diseases. Korean J Inten Med. 2013;28(3):285–91. doi: 10.3904/kjim.2013.28.3.285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Milner JJ, Wang J, Sheridan PA, Ebbels T, Beck MA, Saric J. 1 H NMR-based profiling reveals differential immune-metabolic networks during influenza virus infection in obese mice. PloS One. 2014;9(5):e97238. doi: 10.1371/journal.pone.0097238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Ritter JB, Wahl AS, Freund S, Genzel Y, Reichl U. Metabolic effects of influenza virus infection in cultured animal cells: Intra-and extracellular metabolite profiling. BMC Syst Biol. 2010;4(1):1. doi: 10.1186/1752-0509-4-61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Sanchez EL, Lagunoff M. Viral activation of cellular metabolism. Virology. 2015;479:609–18. doi: 10.1016/j.virol.2015.02.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Atzei A, Atzori L, Moretti C, Barberini L, Noto A, Ottonello G, et al. Metabolomics in paediatric respiratory diseases and bronchiolitis. J Materner Fetal Neonatal Med. 2011;24(2):59–62. doi: 10.3109/14767058.2011.607012. [DOI] [PubMed] [Google Scholar]
- 111.Turi KN, Steinhoff M, Watanabe M, Romick-Rosendale L, Moore ML, Anderson LJ, et al. Metabolomics Approach to Understanding the Pathogenesis of Respiratory Syncytial Virus (RSV) Infection. Am J Respir Crit Care Med. 2016;193:A4613. [Google Scholar]
- 112.van der Schee MP, Hashimoto S, Schuurman AC, van Driel JS, Adriaens N, van Amelsfoort RM, et al. Altered exhaled biomarker profiles in children during and after rhinovirus-induced wheeze. Eur Respir J. 2015;45(2):440–8. doi: 10.1183/09031936.00044414. [DOI] [PubMed] [Google Scholar]
- 113.Fowler SJ, Basanta-Sanchez M, Xu Y, Goodacre R, Dark PM. Surveillance for lower airway pathogens in mechanically ventilated patients by metabolomic analysis of exhaled breath: a case-control study. Thorax. 2015;70(4):320–5. doi: 10.1136/thoraxjnl-2014-206273. [DOI] [PubMed] [Google Scholar]
- 114.Carraro S, Rezzi S, Reniero F, Heberger K, Giordano G, Zanconato S, et al. Metabolomics applied to exhaled breath condensate in childhood asthma. Am J Respir Crit Care Med. 2007;175:986–90. doi: 10.1164/rccm.200606-769OC. [DOI] [PubMed] [Google Scholar]
- 115.Sinha A, Krishnan V, Sethi T, Roy S, Ghosh B, Lodha R, Kabra S, Agrawal A. Metabolomic signatures in nuclear magnetic resonance spectra of exhaled breath condensate identify asthma. Eur Respir J. 2012 Feb 1;39(2):500–2. doi: 10.1183/09031936.00047711. [DOI] [PubMed] [Google Scholar]
- 116.Esther CR, Boysen G, Olsen BM, Collins LB, Ghio AJ, Swenberg JW, et al. Mass spectrometric analysis of biomarkers and dilution markers in exhaled breath condensate reveals elevated purines in asthma and cystic fibrosis. Am J Physiol Lung Cell Mol Physiol. 2009;296(6):L987–93. doi: 10.1152/ajplung.90512.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Montuschi P. LC/MS/MS analysis of leukotriene B 4 and other eicosanoids in exhaled breath condensate for assessing lung inflammation. J. Chromatogr. B Analyt. Technol. Biomed. Life Sci. 2009;877(13):1272–80. doi: 10.1016/j.jchromb.2009.01.036. [DOI] [PubMed] [Google Scholar]
- 118.Carraro S, Giordano G, Reniero F, Carpi D, Stocchero M, Sterk PJ, et al. Asthma severity in childhood and metabolomic profiling of breath condensate. Allergy. 2013;68(1):110–7. doi: 10.1111/all.12063. [DOI] [PubMed] [Google Scholar]
- 119.Dallinga JW, Robroeks CM, Van Berkel JJ, Moonen EJ, Godschalk RW, Jöbsis Q, et al. Volatile organic compounds in exhaled breath as a diagnostic tool for asthma in children. Clin Exp Allergy. 2010;40(1):68–76. doi: 10.1111/j.1365-2222.2009.03343.x. [DOI] [PubMed] [Google Scholar]
- 120.Gahleitner F, Guallar-Hoyas C, Beardsmore CS, Pandya HC, Thomas CP. Metabolomics pilot study to identify volatile organic compound markers of childhood asthma in exhaled breath. Bioanalysis. 2013;5(18):2239–47. doi: 10.4155/bio.13.184. [DOI] [PubMed] [Google Scholar]
- 121.Smolinska A, Klaassen EM, Dallinga JW, van de Kant KD, Jobsis Q, Moonen EJ, et al. Profiling of volatile organic compounds in exhaled breath as a strategy to find early predictive signatures of asthma in children. PLoS One. 2014;9(4):e95668. doi: 10.1371/journal.pone.0095668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Dragonieri S, Schot R, Mertens BJ, Le Cessie S, Gauw SA, Spanevello A, et al. An electronic nose in the discrimination of patients with asthma and controls. J Allergy Clin Immunol. 2007;120(4):856–62. doi: 10.1016/j.jaci.2007.05.043. [DOI] [PubMed] [Google Scholar]
- 123.McGeachie MJ, Dahlin A, Qiu W, Croteau-Chonka DC, Savage J, Wu AC, et al. The metabolomics of asthma control: a promising link between genetics and disease. Immun Inflamm Dis. 2015;3(3):224–38. doi: 10.1002/iid3.61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Saude EJ, Skappak CD, Regush S, Cook K, Ben-Zvi A, Becker A, et al. Metabolomic profiling of asthma: diagnostic utility of urine nuclear magnetic resonance spectroscopy. Journal of Allergy and Clinical Immunology. 2011;127(3):757–64. doi: 10.1016/j.jaci.2010.12.1077. [DOI] [PubMed] [Google Scholar]
- 125.Lock EF, Dunson DB. Bayesian consensus clustering. Bioinformatics. 2013;29(20):2610–6. doi: 10.1093/bioinformatics/btt425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Shen R, Olshen AB, Ladanyi M. Integrative clustering of multiple genomic data types using a joint latent variable model with application to breast and lung cancer subtype analysis. Bioinformatics. 2009;25(22):2906–12. doi: 10.1093/bioinformatics/btp543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Kim S, Herazo-Maya JD, Kang DD, Juan-Guardela BM, Tedrow J, Martinez FJ, et al. Integrative phenotyping framework (iPF): integrative clustering of multiple omics data identifies novel lung disease subphenotypes. BMC Genomics. 2015;16(1):924. doi: 10.1186/s12864-015-2170-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.de Steenhuijsen Piters WA, Heinonen S, Hasrat R, Bunsow E, Smith B, Suarez-Arrabal MC, et al. Nasopharyngeal Microbiota, Host Transcriptome and Disease Severity in Children with Respiratory Syncytial Virus Infection. Am J Respir Crit Care Med. 2016;194(9):1104–15. doi: 10.1164/rccm.201602-0220OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Wu W, Bleecker E, Moore W, Busse WW, Castro M, Chung KF, et al. Unsupervised phenotyping of Severe Asthma Research Program participants using expanded lung data. J Allergy Clin Immunol. 2014;133(5):1280–8. doi: 10.1016/j.jaci.2013.11.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Kupczyk M, Wenzel S. US and European severe asthma cohorts: what can they teach us about severe asthma? J Int Med. 2012;272(2):121–32. doi: 10.1111/j.1365-2796.2012.02558.x. [DOI] [PubMed] [Google Scholar]
- 131.Lysenko A, Roznovaţ IA, Saqi M, Mazein A, Rawlings CJ, Auffray C. Representing and querying disease networks using graph databases. BioData Min. 2016;9(1):23. doi: 10.1186/s13040-016-0102-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Satagopam V, Gu W, Eifes S, Gawron P, Ostaszewski M, Gebel S, et al. Integration and visualization of translational medicine data for better understanding of human diseases. Big Data. 2016;4(2):97–108. doi: 10.1089/big.2015.0057. [DOI] [PMC free article] [PubMed] [Google Scholar]