Abstract
BACKGROUND: Mounting evidence suggests that long noncoding RNAs (lncRNAs) are closely related to pathological complete response (pCR) in neoadjuvant treatment of breast cancer. Here, we construct lncRNA associated models to predict pCR rate. METHODS: LncRNA expression profiles of breast cancer patients treated with neoadjuvant chemotherapy (NAC) were obtained from Gene Expression Omnibus by repurposing existing microarray data. The prediction model was firstly built by analyzing the correlation between pCR and lncRNA expression in the discovery dataset GSE 25066 (n = 488). Another three independent datasets, GSE20194 (n = 278), GSE20271 (n = 178), and GSE22093 (n = 97), were integrated as the validation cohort to assess the prediction efficiency. RESULTS: A novel lncRNA signature (LRS) consisting of 36 lncRNAs was identified. Based on this LRS, patients with NAC treatment were divided into two groups: LRS-high group and LRS-low group, with positive correlation of pCR rate in the discovery dataset. In the validation cohort, univariate and multivariate analyses both demonstrated that high LRS was associated with higher pCR rate. Subgroup analysis confirmed that this model performed well in luminal B [odds ratio (OR) = 5.4; 95% confidence interval (CI) = 2.7-10.8; P = 1.47e-06], HER2-enriched (OR = 2.5; 95% CI = 1.1-5.7; P = .029), and basal-like (OR = 5.5; 95% CI = 2.3-16.2; P = 5.32e-04) subtypes. Compared with other preexisting prediction models, LRS demonstrated better performance with higher area under the curve. Functional annotation analysis suggested that lncRNAs in this signature were mainly involved in cancer proliferation process. CONCLUSION: Our findings indicated that our lncRNA signature was sensitive to predict pCR rate in the neoadjuvant treatment of breast cancer, which deserves further evaluation.
Introduction
Breast cancer, one of the most common cancers, is still a fatal malignant tumor that could lead to nearly half a million of deaths in a year [1]. Neoadjuvant chemotherapy (NAC) has emerged as a new and effective option for locally advanced breast cancer. By shrinking tumor size before surgery, NAC is able to improve the feasibility of conventional breast operation and conservative surgery in locally advanced breast cancer patients [2]. In addition, numerous studies have shown that a pathological complete response (pCR) after NAC is significantly correlated with good further clinical outcome [3]. However, other data also demonstrated that only less than 40% to 50% breast cancer patients can achieve pCR after NAC [4]. Therefore, clinicians usually need to make a decision as to whether NAC is necessary or not for breast cancer patients by evaluating the pros and cons of chemotherapy before the operation according to patients' tumor stage, histologic grade, age, estrogen receptor (ER), and human epidermal growth factor receptor 2 (HER2) status as these clinicopathological characteristics are associated with the probability of pCR [5]. Meanwhile, utilizing different mRNA signatures to predict pCR rate when making a decision is also attracting researchers' attention. Over the past decades, breast cancer has been classified into five different “intrinsic subtypes”, including luminal A, luminal B, HER2 enriched, basal-like, and normal-like according to its mRNA expression pattern [6]. To date, much data have exhibited that different subtypes of breast cancer have distinct biological behaviors as well as responses to NAC [7]. Specifically, basal-like tumors generally achieve much higher pCR rate than luminal A [8]. Oncotype DX, Gene70, and Gene Expression Grade Index (GGI) signature are previously established mRNA signatures employed to predict breast cancer patient survival [9], [10], [11]. It has also been shown that high expression level of these mRNA signatures is associated with higher probability of pCR [12].
As a matter of fact, at least 98% of the human genome is transcribed into noncoding RNAs rather than protein-coding mRNAs, implying that these novel moleculars also play a vital role in biological processes and are potential biomarkers to powerfully predict NAC response in breast cancer in addition to mRNA [13]. Long noncoding RNAs (lncRNAs) are generally non–protein-coding transcripts longer than 200 nucleotides in length [14]. These long RNAs function directly at RNA level rather than being translated into functional proteins. Recent studies have revealed that lncRNAs are involved in varieties of biological processes including tumorigenesis, invasion, metastasis, and drug resistance of breast cancer at both transcriptional and posttranscriptional levels [15], [16], [17]. What's more, a growing number of lncRNAs are validated to be associated with patients' outcomes and pCR in NAC, thus providing a new option for model construction to predict pCR of NAC besides mRNA signature [18]. However, there is no lncRNA signature built based on a large number of breast cancer patients treated with NAC up to now.
In this study, we aimed at identifying an lncRNA signature to fully investigate the relationship between lncRNA expression pattern and pCR rate in breast cancer patients with NAC treatment. By reannotating previously published Affymetrix HG-U133A array profiles from Gene Expression Omnibus (GEO), we established an lncRNA signature comprising of 36 lncRNAs from NAC-treated patients. Then, the prediction efficiency of this signature was further assessed in a validation cohort with three independent datasets. Our final finding indicated that this signature could be potentially utilized as a novel biomarker to predict pCR rate in addition to traditional clinicopathological markers and mRNA signature, which needs further evaluation.
Materials and Methods
Gene Expression Profiles of NAC-Treated Patients Obtained from GEO
We searched for qualified gene expression datasets in regard to breast cancer with NAC treatment in GEO database, which is publically available. Only datasets that met the following criteria were selected: First, gene expression data were assayed using Affymetrix HG-U133 A platform. Second, pCR in the study was defined as the absence of invasive tumor in both breast and lymph nodes [19]. Third, nearly or more than 100 cases were included in the dataset. Finally, 1041 NAC-treated breast cancer patients in total were collected from GSE25066 [20], GSE20194 [21], GSE20271 [22], and GSE22093 [23] datasets. GSE25066, which consists of 488 samples, was used as the discovery dataset for model construction, while another 553 patients from GSE20194, GSE20271, and GSE22093 were combined and used as the validation dataset. Patients' characteristics of both discovery and validation datasets are listed in Table 1.
Table 1.
NAC Patients' Clinicopathological Characteristics Included in the Discovery and Validation Cohort
| Characteristics | Discovery GSE25066 (n = 488) |
Validation |
|||
|---|---|---|---|---|---|
| All Trials (n = 553) |
GSE20194 (n = 278) |
GSE20271 (n = 178) |
GSE22093 (n = 97) |
||
| Age | |||||
| ≤50 | 268 | 287 | 133 | 100 | 54 |
| >50 | 220 | 264 | 144 | 78 | 42 |
| Unknown | 0 | 2 | 1 | 0 | 1 |
| ER | |||||
| Negative | 200 | 249 | 114 | 80 | 55 |
| Positive | 287 | 304 | 164 | 98 | 42 |
| Unknown | 1 | 0 | 0 | 0 | 0 |
| HER2 | |||||
| Negative | 466 | 463 | 219 | 151 | 97 |
| Positive | 5 | 85 | 59 | 26 | 0 |
| Unknown | 17 | 1 | 0 | 1 | 0 |
| cT | |||||
| T0-1 | 29 | 42 | 26 | 13 | 3 |
| T2 | 245 | 274 | 147 | 76 | 51 |
| T3 | 140 | 113 | 50 | 37 | 26 |
| T4 | 74 | 121 | 53 | 51 | 17 |
| Unknown | 0 | 3 | 2 | 1 | 0 |
| cN | |||||
| N0 | 154 | 159 | 79 | 59 | 21 |
| N1 | 231 | 212 | 125 | 71 | 16 |
| N2 | 64 | 79 | 31 | 38 | 10 |
| N3 | 39 | 54 | 42 | 9 | 3 |
| Unknown | 0 | 49 | 1 | 1 | 47 |
| Grade | |||||
| 1-2 | 201 | 225 | 117 | 76 | 32 |
| 3 | 252 | 271 | 152 | 72 | 47 |
| Unknown | 35 | 57 | 9 | 30 | 18 |
| pCR | |||||
| Yes | 99 | 110 | 56 | 26 | 28 |
| No | 389 | 443 | 222 | 152 | 69 |
cT, clinical tumor stage; cN, clinical nodal status.
Interrogate lncRNA Expression by Repurposing Microarray Probes
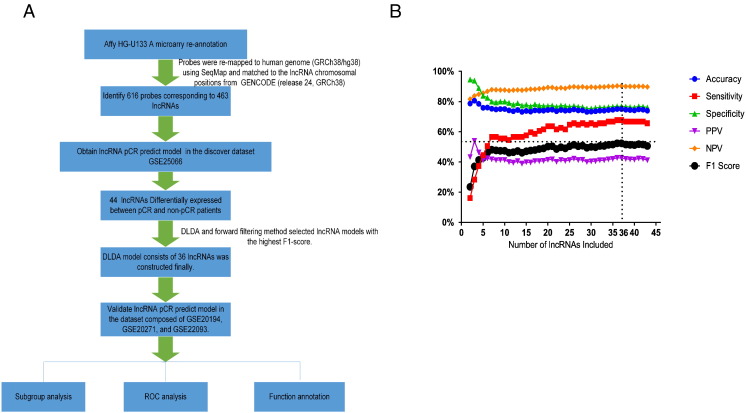
All raw data were normalized by Robust Multichip Average algorithm [24]. ComBat was utilized to remove the potential batch effects when combining batches of gene expression dataset [25]. LncRNA expression of the NAC-treated patients was obtained by remapping the probes of the array to human genome (GRCh38/hg38) using SeqMap [26] and then matching the probes to the lncRNA chromosomal positions from GENCODE (release 24, GRCh38) [27], [28], [29]. A total of 616 probes corresponding to 463 lncRNAs were obtained in the end. LncRNA expression values of multiple probes that target the same lncRNA were averaged arithmetically. Figure 1A demonstrated the order of the analysis and model construction.
Figure 1.
The construction and optimization of the LRS model for pCR prediction. (A) The diagram of the construction and validation of the lncRNA pCR prediction model. (B) The 44 lncRNAs significantly different between non-pCR and pCR cases in the discovery dataset were employed for constructing the predictive DLDA model. The accuracy, sensitivity, specificity, PPVs, NPVs, and F1 score were calculated with each number of lncRNAs. This LRS ranked the highest F1 score when comprising the first 36 lncRNAs.
Diagonal Linear Discriminant Analysis Prediction Model Construction
Of the aforementioned 463 lncRNAs, 44 lncRNAs in the discovery dataset were found to be differentially expressed between the non-pCR and pCR patients by Welch's t test. (P < .001). These 44 lncRNAs were further used to construct the predictive model with Diagonal Linear Discriminant Analysis (DLDA) [30]. According to a previously described method, we employed the forward filtering method to optimize the model [31]. In the end, the DLDA model of 36 lncRNAs ranked the highest F1 score in the discovery dataset and was therefore defined as the lncRNA signature (LRS) for pCR prediction of NAC.
Comparison of the Efficiency of LRS with Other Signatures
Pam50 intrinsic subtypes, Oncotype DX, Gene70, and GGI score were obtained by using “genefu” package in R [32]. The receiver operating characteristic (ROC) curves of LRS and other signatures were plotted and compared by R package ROCR [33]. The area under the curve (AUC) was calculated correspondingly.
Function Annotation of the lncRNA Signature
Integrative analysis of lncRNA-mRNA association was employed to infer the potential function of the lncRNAs included in the LRS. We calculated Pearson correlation between lncRNAs and mRNAs in the discovery cohort to identify mRNAs that positively coexpressed with lncRNAs in LRS (Pearson correlation coefficient > 0.4 and ranked top 0.5%) [29], [34]. Then these mRNAs were further annotated by the Database for Annotation, Visualization, and Integrated Discovery (DAVID) using the functional annotation clustering option (version 6.8) [35], [36]. Clusters with enrichment score higher than 3.0 and functional annotations with P value lower than .001 were considered to be statistically significant. Finally, Cytoscape with the Enrichment Map plugin was used to visualize the significant enrichment results [37].
Statistical Analysis
The microarray datasets downloaded from GEO database were analyzed by R software (version 3.3.1) and Bioconductor. The clinicopathological parameters in the datasets were analyzed using two-sided χ2 test and Fisher's exact test with a P value < .05 as the cutoff. The logistic regression model was employed to perform the univariate and multivariate analyses.
Result
Establishment and Validation of the lncRNA Prediction Model for pCR
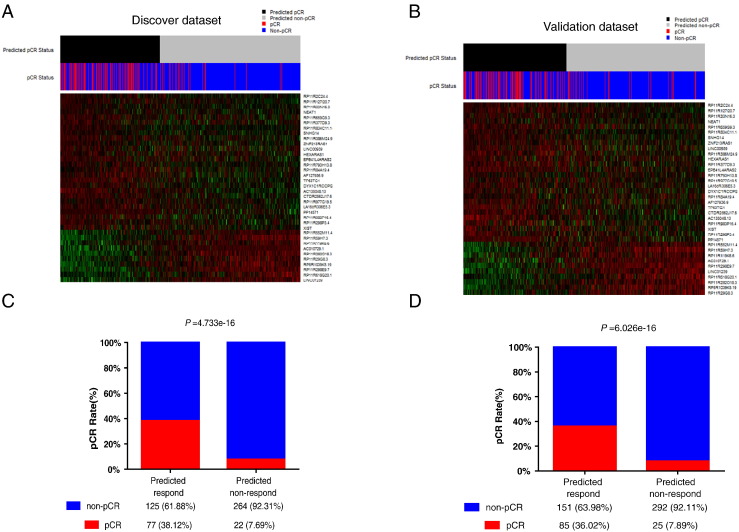
By reannotating microarray probes in the discovery dataset, 616 probes that corresponded to 463 lncRNAs were identified in total. Of the 463 lncRNAs we found, 44 lncRNAs differentially expressed between non-pCR (n = 389) and pCR (n = 99) patients with P < .01, which were further used for model construction. Next, through DLDA with model optimization by leaving one out cross-validation and forward filtering method, we finally observed that the DLDA model of 36 lncRNAs ranked the highest F1 score, which divided patients into two groups: chemotherapy-sensitive group (n = 202) with high LRS score and chemotherapy-insensitive group (n = 286) with low LRS score (Figure 1B). The accuracy, sensitivity, specificity, positive predictive values (PPVs), negative predictive values (NPVs), and F1 score were 69.9%, 77.8%, 67.9%, 38.1%, 92.3%, and 51.2%, respectively. The heat map illustrated that these two groups based on LRS score had distinct lncRNA expression patterns (Figure 2A). More importantly, the high–LRS score group was more likely to achieve a higher pCR rate when compared with the other one (Figure 2C, P = 4.733e-16). Multivariate analysis showed that only tumor grade (P = .04922), LRS score (P = .00373), and intrinsic subtype (P = .02463) were independent factors for pCR in the discovery set (Table 2).
Figure 2.
The lncRNA expression profile and pCR prediction were analyzed in the discovery and validation dataset. NAC patients were classified into LRS-high (predicted pCR) and LRS-low (predicted non-pCR) groups. lncRNA expression profile and pCR distribution are shown in (A) the discovery dataset and (B) the validation dataset. Then, patients' pCR statuses in different LRS groups were further compared in (C) the discovery dataset and (D) the validation dataset.
Table 2.
Univariate and Multivariate Analysis for Parameters Associated with pCR in the Discovery Dataset
| Characteristics | n | pCR | Univariate Analysis |
Multivariate Analysis |
||||
|---|---|---|---|---|---|---|---|---|
| OR | 95% CI | P | OR | 95% CI | P | |||
| Age | ||||||||
| ≤50 | 268 | 60 | 1 | |||||
| >50 | 220 | 39 | 0.75 | 0.47-1.17 | .204 | |||
| Unknown | ||||||||
| ER | ||||||||
| Negative | 200 | 69 | 1 | |||||
| Positive | 287 | 30 | 0.22 | 0.14-0.35 | 6.20e-10 | 0.70 | 0.38-1.28 | .25115 |
| Unknown | 1 | |||||||
| HER2 | ||||||||
| Negative | 466 | 93 | 1 | |||||
| Positive | 5 | 2 | 2.67 | 0.35-16.36 | .285 | |||
| Unknown | 17 | 4 | ||||||
| cT | ||||||||
| T0-2 | 274 | 58 | 1 | |||||
| T3-4 | 214 | 41 | 0.88 | 0.56-1.38 | .584 | |||
| Unknown | 0 | |||||||
| cN | ||||||||
| Negative | 154 | 28 | 1 | |||||
| Positive | 334 | 71 | 1.21 | 0.75-2.00 | .433 | |||
| Unknown | ||||||||
| Grade | ||||||||
| 1-2 | 201 | 13 | 1 | |||||
| 3 | 252 | 77 | 6.36 | 3.53-12.36 | 5.73e-09 | 2.07 | 1.02-4.40 | .04922 |
| Unknown | 35 | |||||||
| lncRNA score | ||||||||
| Low score | 286 | 22 | 1 | |||||
| High score | 202 | 77 | 7.39 | 4.47-12.68 | 4.41e-14 | 2.74 | 1.40-5.51 | .00373 |
| Intrinsic subtype | ||||||||
| Luminal A | 124 | 2 | 1 | |||||
| Others | 364 | 97 | 25.78 | 7.99-157.88 | 6.76e-06 | 6.91 | 1.50-50.6 | .02463 |
| Oncotype DX | ||||||||
| Low & medium score | 80 | 2 | 1 | |||||
| High score | 408 | 97 | 12.16 | 3.74-74.79 | 5.73e-04 | 1.41 | 0.33-9.76 | .67782 |
| Gene70 | ||||||||
| Low score | 82 | 3 | 1 | |||||
| High score | 406 | 96 | 8.15 | 2.96-33.77 | 4.63e-04 | 0.92 | 0.25-4.49 | .91205 |
| GGI | ||||||||
| Low score | 195 | 13 | 1 | |||||
| High score | 293 | 86 | 5.82 | 3.25-11.24 | 2.15e-08 | 0.94 | 0.41-2.23 | .88345 |
To further evaluate the prediction efficiency of pCR with the LRS, we collected a total of 553 breast cancer patients with NAC treatment by integrating three independent datasets (GSE20194, GSE20271, and GSE22093) as the validation cohort. As expected, the results gained by using validation dataset (Figure 2, B and D) shared similar findings with those of the discovery dataset, with distinct lncRNA expression patterns and higher pCR rate in the high–LRS score group (P = 6.026e-16). The accuracy, sensitivity, specificity, PPV, NPV, and F1 score for validation dataset were 68.2%, 77.3%, 65.9%, 36.0%, 92.1%, and 49.1%, respectively. Multivariate analysis demonstrated that only ER status (P = 3.79e-05), HER2 status (P = .01147), and LRS score (P = 2.97e-05) were independent factors for predicting pCR in the validation dataset (Table 3).
Table 3.
Univariate and Multivariate Analysis for Predicting Parameters Associated with pCR in the Validation Dataset
| Characteristics | n | pCR | Univariate Analysis |
Multivariate Analysis |
||||
|---|---|---|---|---|---|---|---|---|
| OR | 95% CI | P | OR | 95% CI | P | |||
| Age | ||||||||
| ≤50 | 287 | 58 | 1 | |||||
| >50 | 264 | 52 | 0.97 | 0.64-1.47 | .881 | |||
| Unknown | 2 | 0 | ||||||
| ER | ||||||||
| Negative | 249 | 83 | 1 | 1 | ||||
| Positive | 304 | 27 | 0.19 | 0.12-0.31 | 1.51e-11 | 0.28 | 0.15-0.50 | 3.79e-05 |
| Unknown | 0 | 0 | ||||||
| HER2 | ||||||||
| Negative | 463 | 81 | 1 | 1 | ||||
| Positive | 85 | 29 | 2.00 | 1.25-3.26 | 4.33e-3 | 2.18 | 1.19-4.00 | .01147 |
| Unknown | 1 | 0 | ||||||
| cT | ||||||||
| T0-2 | 316 | 66 | 1 | |||||
| T3-4 | 234 | 44 | 0.88 | 0.57-1.34 | .546 | |||
| Unknown | 3 | 0 | ||||||
| cN | ||||||||
| Negative | 159 | 21 | 1 | |||||
| Positive | 345 | 66 | 1.56 | 0.93-2.71 | .101 | |||
| Unknown | 49 | 23 | ||||||
| Grade | ||||||||
| 1-2 | 225 | 18 | 1 | 1 | ||||
| 3 | 271 | 80 | 4.84 | 2.86-8.60 | 1.67e-08 | 1.58 | 0.82-3.11 | .17662 |
| Unknown | 57 | 12 | ||||||
| lncRNA score | ||||||||
| Low score | 317 | 25 | 1 | 1 | ||||
| High score | 236 | 85 | 6.57 | 4.10-10.89 | 3.6e-14 | 4.17 | 2.18-8.36 | 2.97e-05 |
| Intrinsic Subtype | ||||||||
| Luminal A | 122 | 5 | 1 | 1 | ||||
| Others | 431 | 105 | 7.54 | 3.31-21.73 | 1.74e-05 | 0.99 | 0.28-3.96 | .98660 |
| Oncotype DX | ||||||||
| Low & medium score | 81 | 3 | 1 | 1 | ||||
| High score | 472 | 107 | 7.62 | 2.77-31.51 | 6.87e-04 | 1.07 | 0.27-5.42 | .92960 |
| Gene70 | ||||||||
| Low score | 101 | 5 | 1 | 1 | ||||
| High score | 452 | 105 | 5.81 | 2.54-16.80 | 1.93e-04 | 0.86 | 0.27-3.04 | .80190 |
| GGI | ||||||||
| Low score | 226 | 21 | 1 | 1 | ||||
| High score | 327 | 89 | 3.65 | 2.23-6.22 | 6.74e-07 | 1.35 | 0.65-2.89 | .43185 |
Relationship between Clinicopathological Features and LRS Model
We uncovered that high LRS score was significantly associated with ER negativity (P < 2.2e-16), T3 and T4 tumors’ size (P < .017), lymph node positivity (P < .026), grade 3 tumor (P < 2.2e-16), and non–luminal A subtype (P < .026) in discovery dataset. In the validation set, high–LRS score patients were more likely to be ER negative (P < 2.2e-16), HER2 positive (P = .047), grade 3 (P < 2.2e-16), and non–luminal A subtype (P < 2.2e-16). (Table 4).
Table 4.
Association between Clinicopathological Parameters and LRS in the Discovery and Validation Dataset
| Characteristics | Discovery Set |
Validation Set |
||||
|---|---|---|---|---|---|---|
| LRS Low | LRS High | P | LRS Low | LRS High | P | |
| n | 286 | 202 | 317 | 236 | ||
| Age | ||||||
| ≤50 | 154 | 114 | 168 | 119 | ||
| >50 | 132 | 88 | .6356 | 148 | 116 | .6165 |
| Unknown | 0 | 0 | 1 | 1 | ||
| ER | ||||||
| Negative | 43 | 157 | 85 | 164 | ||
| Positive | 242 | 45 | <2.2e-16 | 232 | 72 | <2.2e-16 |
| Unknown | 1 | 0 | 0 | 0 | ||
| HER2 | ||||||
| Negative | 275 | 191 | 277 | 190 | ||
| Positive | 1 | 4 | .1651 | 40 | 45 | .0474 |
| Unknown | 10 | 7 | 0 | 1 | ||
| cT | ||||||
| T0-2 | 174 | 100 | 186 | 130 | ||
| T3-4 | 112 | 102 | .01673 | 128 | 106 | .3749 |
| Unknown | 0 | 0 | 3 | 0 | ||
| cN | ||||||
| Negative | 102 | 52 | 104 | 55 | ||
| Positive | 184 | 150 | .02615 | 202 | 143 | .1717 |
| Unknown | 0 | 0 | 10 | 39 | ||
| Grade | ||||||
| 1-2 | 174 | 27 | 176 | 49 | ||
| 3 | 97 | 155 | <2.2e-16 | 101 | 170 | <2.2e-16 |
| Unknown | 15 | 20 | 40 | 17 | ||
| Intrinsic subtype | ||||||
| Luminal A | 135 | 2 | 117 | 5 | ||
| Others | 151 | 200 | <2.2e-16 | 200 | 231 | <2.2e-16 |
| Oncotype DX | ||||||
| Low & medium score | 79 | 1 | 77 | 4 | ||
| High score | 207 | 201 | 4.211e-15 | 240 | 232 | 2.647e-13 |
| Gene70 | ||||||
| Low score | 81 | 1 | 96 | 5 | ||
| High score | 205 | 201 | 1.527e-15 | 221 | 231 | <2.2e-16 |
| GGI | ||||||
| Low score | 180 | 15 | 188 | 38 | ||
| High score | 106 | 187 | <2.2e-16 | 129 | 198 | <2.2e-16 |
| pCR status | ||||||
| pCR | 22 | 77 | 25 | 85 | ||
| Non-pCR | 264 | 125 | <2.2e-16 | 292 | 151 | <2.2e-16 |
Evaluation of the Prediction of LRS in Different Intrinsic Subtypes
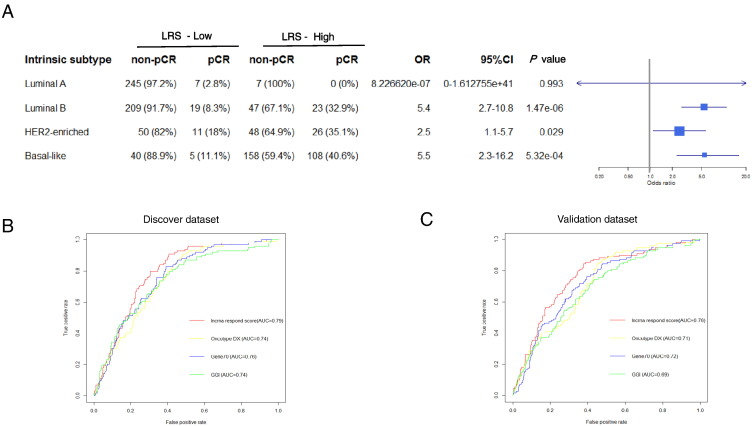
By using PAM50 subtypes classification, 259 cases of luminal A, 298 cases of luminal B, 135 cases of HER2-enriched, 311 cases of basal-like, and 38 cases of normal-like subtype were identified in the whole 1041 NAC-treated patients. The normal-like subtype was excluded in the subgroup analysis because of its small sample size. In luminal A subtype, which is less likely to undergo NAC, LRS failed to predict pCR rate. However, in luminal B, HER2-enriched, and basal-like subtypes, high LRS score was significantly associated with a higher chance to achieve pCR (Figure 3A).
Figure 3.
The evaluation of the predictive power of LRS in the subgroup analysis and comparison of ROCs with other different prediction models. As determined by Pam50, luminal A, luminal B, HER2-enriched, and basal-like subtypes of breast cancer were subjected to subgroup analysis to evaluate the predictive power of LRS in all NAC patients (A). Then, ROCs of the LRS, Oncotype DX, Gene70, and GGI were compared in (B) the discovery and (C) validation datasets. The AUC was also calculated for each curve.
In detail, for luminal B subtype, LRS-high patients achieved a higher pCR rate compared with LRS-low patients [32.9% vs 8.3%; odds ratio (OR) = 5.4; 95% confidence interval (CI) = 2.7-10.8; P = 1.47e-06]. That is quite similar for HER2-enriched and basal-like subtypes, with 35.1% versus 18%; OR = 2.5; 95% CI = 1.1-5.7; P = .029 and 40.6% versus 11.1%; OR = 5.5; 95% CI = 2.3-16.2; P = 5.32e-04, respectively.
Comparison of LRS Predictive Power with Other Preexisting Signatures
We then compared the predictive capability of LRS with other preexiting signatures by ROC curves (Figure 3, B and C). The AUC value of LRS was approximately 0.8, indicating that it can effectively distinguish pCR patients from non-pCR patients. Compared with other preexisting predictive signatures, our LRS performed better with higher AUC values in both discovery and validation datasets when used to predict pCR response of patients undergoing NAC.
Combination of LRS with Other Different pCR Predictors
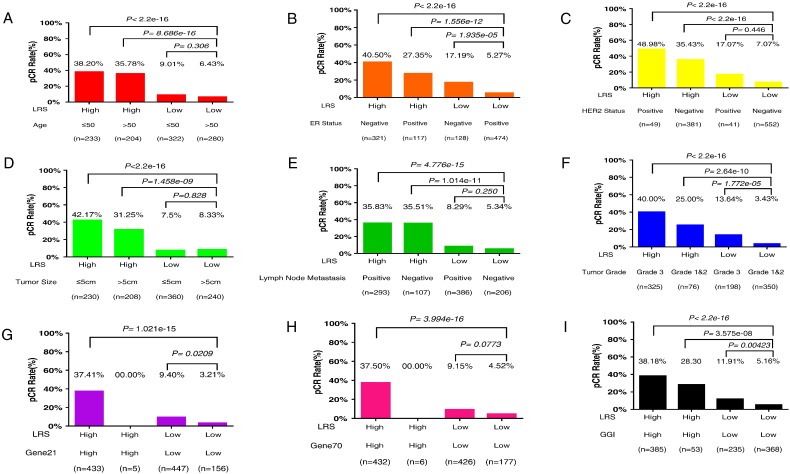
In order to inspect the clinical significance of LRS, we then integrated LRS with age, ER status, HER2 status, tumor size, lymph node metastasis status, tumor grade, Gene21 index, Gene70 index, and GGI which divided patients into four groups respectively. Breast cancer patients with high LRS score and younger age (≤50 years old), negative ER status, positive HER2 status, smaller tumor size (≤5 cm), positive lymph node, tumor of grade 3, Gene21 index high, Gene70 index high, and GGI high were significantly more likely to achieve pCR after NAC (Figure 4), which implied that LRS could be potentially utilized to improve pCR prediction in combination with clinicopathological biomarkers and preexisting predictive models in clinical practice.
Figure 4.
The results of the analysis combining LRS and other different pCR predictors performed for the whole group of patients. In order to inspect the clinical significance of LRS, we combined LRS with other factors which may be related to pCR. Age (A), ER status (B), HER2 status (C), tumor size (D), lymph node metastasis status (E), tumor grade (F), Gene21 index (G), Gene70 index (H), and GGI (I) which were available in our cohort were integrated, respectively, with LRS and divided patients into four groups. The pCR rates were compared among each of the four groups.
Identification of Biological Processes Related to the lncRNA Signature
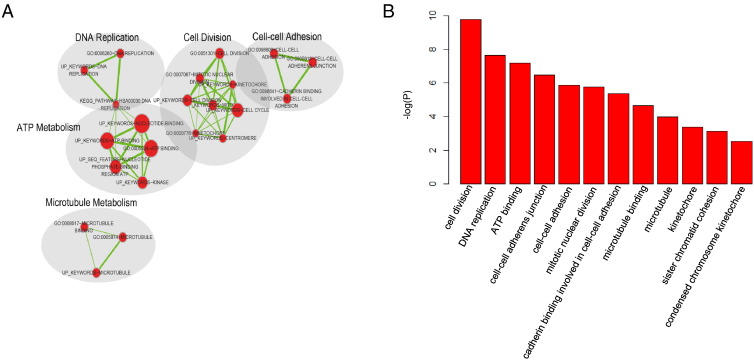
We conducted Gene Ontology (GO) enrichment analysis to determine the biological processes associating with LRS [38]. To this end, we firstly employed Pearson correlation to identify the most correlated mRNAs with each of our lncRNAs (correlation effecient > 0.4 and ranked top 0.5%). Then, using functional annotation clustering option from DAVID (https://david.ncifcrf.gov/, version 6.8), we collected annotation clusters with enrichment score > 3.0 and P value less than .001 (supplementary file). Finally, Cytoscape was employed to visualize the significantly enriched clusters based on similar functions (Figure 5, A), and statistically significant GO processes were also plotted by P value (Figure 5, B). Surprisingly, cancer-related functional clusters such as DNA replication, cell cycle, cell-cell adhesion, and microtubule metabolism were clearly identified, indicating that the LRS signature might mainly be related to cancer proliferation process.
Figure 5.
Enrichment analysis of the mRNAs positively correlated with lncRNAs in LRS. mRNAs that positively correlated with lncRNAs in LRS were analyzed by DAVID functional annotation tool. And results were organized by functional enrichment map in Cytoscape (A). Each node represents a functional term annotated by the DAVID tool. The size of the node represents the number of genes in the terms. (B) Then, the significant GO terms in the functional annotation were illustrated with barplot according to their P value.
Discussion
During the past decades, lots of effort have been devoted by clinicians to develop tools for the prediction of the response to NAC in order that the candidate patients could undergo the most suitable therapy. Since the response to NAC is related to patients' clinicopathological features/characteristics, a pCR prediction nomogram has been built based on clinical stage, ER status, histologic grade, and number of preoperative chemotherapy cycles [39]. Moreover, some researchers also utilize Oncotype DX and Gene70 score (Mamaprint score), which are the most widely accepted mRNA signatures, for prediction of response to NAC for luminal subtype breast cancer. Until now, a number of models have been utilized to predict breast cancer survival and/or pCR in NAC.
In this study, we identified a 36-lncRNA signature to predict pCR with high specificity by using lncRNA remapping approach in 488 breast cancer patients treated with NAC. Furthermore, LRS signature was demonstrated to be an independent factor that highly related to the pCR of NAC-treated patients by univariate and multivariate analysis in both discovery and validation sets. It has also been shown that the combination of LRS with other pCR predictors significantly improves the prediction of pCR in NAC. This LRS was the first lncRNA signature built on a large scale of NAC-treated patients. It could be potentially utilized as a prediction tool with high specificity to assess the possibilities of pCR for patients who undergo NAC, in addition to traditional clinicopathological biomarkers and mRNA signatures.
Oncotype DX score and Gene70 score have been originally built to predict patients' prognosis, while GGI has been developed for better assessing histologic grade firstly. Although these mRNA signatures can also predict the pCR rate despite their original goal, they were not specifically developed for patients treated with NAC. Until now, there is still no gene signature specifically designed for pCR prediction to NAC. It is noteworthy that our LRS was the first gene model developed for pCR rate prediction to NAC patients specifically. With this end of LRS, clinicians could select the most suitable candidates for NAC to better realize individualized therapy.
Luminal B, HER2-enriched, and basal-like are the three major subtypes of breast cancers that are more likely to go through NAC. In these three subgroups, LRS distinguished non-pCR from pCR patients significantly, although it did not work well for HER2-enriched subgroup which might be mainly due to its distinct biological behaviors. More importantly, it should be noted that all patients in our datasets treated with NAC regiments did not receive trastuzumab. HER2, which drives breast cancer progression, invasion, and metastasis, was reported to have an unneglectable influence on NAC in varieties of ways [40], [41], [42]. Without the treatment of trastuzumab, lower pCR rate might be induced by HER2, which led to the relatively inaccuracy of our signature in the HER2-enriched subtype. Meanwhile, we also included luminal A subtype in the subgroup analysis, which usually does not undergo NAC. LRS was not able to predict NAC responses effectively in luminal A subtype, which is also a common issue for many other predictive models with unknown reasons [43].
In both discovery and validation datasets, patients with high LRS score were more likely to be ER negative and grade 3. Previous studies suggested that ER-negative breast cancer especially triple-negative breast cancer is generally more aggressive or grows faster than ER-positive [44]. On the other hand, grade 3 tumors typically also have a higher proliferation rate than lower grades [45]. What's more, gene enrichment analysis demonstrated that these 36 lncRNAs were mainly associated with tumor proliferation. Therefore, we hypothesize that patients with high LRS score were more sensitive to NAC because of the high proliferation rate of their tumors. Chemotherapeutic agents are usually cytotoxic by means of interfering with cell division [46]. Cancer cells are more susceptible to these agents because they have a higher proliferation rate than normal cells. In other words, high cell proliferation rate leads to a high sensitivity of chemotherapy [47]. Patients with high LRS score were more likely to be with high proliferation tumors, which caused their being more sensitive to NAC.
It must be acknowledged that there are some limitations in this study. First, lncRNAs in our signature came from the reannotation of probes in microarray platform. LncRNAs that could not be identified by repurposing microarray data could be omitted and thus might affect the sensitivity and specificity of the analysis results. Second, only a handful of lncRNAs identified in this study have been functionally characterized before. Experimental studies on these lncRNAs are desperately needed to provide important information to understand their functional roles. What's more, further validation of the signature in clinical trials will be a better option to finally turn it into clinical practice.
In conclusion, our study suggests an important role of the lncRNA signature in predicting pCR rate of NAC in addition to traditional clinicopathological markers and mRNA signature, which might deserve further evaluation. What's more, these findings also imply a therapeutic strategy through targeting these lncRNAs to improve patients' clinical outcomes in the future.
The following are the supplementary data related to this article.
Competing Interests
The authors declare that they have no competing interests. All authors read and approved the final manuscript.
Footnotes
Funding: This work was supported by grants from National Natural Science Foundation of China (grant number: 81472462), Medical Guidance Foundation of Shanghai Municipal Science and Technology Commission (grant number: 15411966400), and Technology Innovation Act Plan of Shanghai Municipal Science and Technology Commission (grant numbers: 14411950200, 14411950201, 15411952500, 15411952501).
References
- 1.Harbeck N, Gnant M. Breast cancer. Lancet. 2016 doi: 10.1016/S0140-6736(16)31891-8. [DOI] [PubMed] [Google Scholar]
- 2.Kaufmann M, von Minckwitz G, Bear HD, Buzdar A, McGale P, Bonnefoi H, Colleoni M, Denkert C, Eiermann W, Jackesz R. Recommendations from an international expert panel on the use of neoadjuvant (primary) systemic treatment of operable breast cancer: new perspectives 2006. Ann Oncol. 2007;18(12):1927–1934. doi: 10.1093/annonc/mdm201. [DOI] [PubMed] [Google Scholar]
- 3.Rastogi P, Anderson SJ, Bear HD, Geyer CE, Kahlenberg MS, Robidoux A, Margolese RG, Hoehn JL, Vogel VG, Dakhil SR. Preoperative chemotherapy: updates of National Surgical Adjuvant Breast and Bowel Project Protocols B-18 and B-27. J Clin Oncol. 2008;26(5):778–785. doi: 10.1200/JCO.2007.15.0235. [DOI] [PubMed] [Google Scholar]
- 4.Fisher B, Brown A, Mamounas E, Wieand S, Robidoux A, Margolese RG, Cruz AB, Jr., Fisher ER, Wickerham DL, Wolmark N. Effect of preoperative chemotherapy on local-regional disease in women with operable breast cancer: findings from National Surgical Adjuvant Breast and Bowel Project B-18. J Clin Oncol. 1997;15(7):2483–2493. doi: 10.1200/JCO.1997.15.7.2483. [DOI] [PubMed] [Google Scholar]
- 5.Chang J, Powles TJ, Allred DC, Ashley SE, Clark GM, Makris A, Assersohn L, Gregory RK, Osborne CK, Dowsett M. Biologic markers as predictors of clinical outcome from systemic therapy for primary operable breast cancer. J Clin Oncol. 1999;17(10):3058–3063. doi: 10.1200/JCO.1999.17.10.3058. [DOI] [PubMed] [Google Scholar]
- 6.Eroles P, Bosch A, Perez-Fidalgo JA, Lluch A. Molecular biology in breast cancer: intrinsic subtypes and signaling pathways. Cancer Treat Rev. 2012;38(6):698–707. doi: 10.1016/j.ctrv.2011.11.005. [DOI] [PubMed] [Google Scholar]
- 7.Prat A, Fan C, Fernandez A, Hoadley KA, Martinello R, Vidal M, Viladot M, Pineda E, Arance A, Munoz M. Response and survival of breast cancer intrinsic subtypes following multi-agent neoadjuvant chemotherapy. BMC Med. 2015;13:303. doi: 10.1186/s12916-015-0540-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.von Minckwitz G, Martin M. Neoadjuvant treatments for triple-negative breast cancer (TNBC) Ann Oncol. 2012;23(Suppl. 6):vi35–vi39. doi: 10.1093/annonc/mds193. [DOI] [PubMed] [Google Scholar]
- 9.Harris LN, Ismaila N, McShane LM, Andre F, Collyar DE, Gonzalez-Angulo AM, Hammond EH, Kuderer NM, Liu MC, Mennel RG. Use of Biomarkers to Guide Decisions on Adjuvant Systemic Therapy for Women With Early-Stage Invasive Breast Cancer: American Society of Clinical Oncology Clinical Practice Guideline. J Clin Oncol. 2016;34(10):1134–1150. doi: 10.1200/JCO.2015.65.2289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Naoi Y, Noguchi S. Multi-gene classifiers for prediction of recurrence in breast cancer patients. Breast Cancer. 2016;23(1):12–18. doi: 10.1007/s12282-015-0596-9. [DOI] [PubMed] [Google Scholar]
- 11.Haibe-Kains B, Desmedt C, Piette F, Buyse M, Cardoso F, Van't Veer L, Piccart M, Bontempi G, Sotiriou C. Comparison of prognostic gene expression signatures for breast cancer. BMC Genomics. 2008;9:394. doi: 10.1186/1471-2164-9-394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Liu R, Lv QL, Yu J, Hu L, Zhang LH, Cheng Y, Zhou HH. Correlating transcriptional networks with pathological complete response following neoadjuvant chemotherapy for breast cancer. Breast Cancer Res Treat. 2015;151(3):607–618. doi: 10.1007/s10549-015-3428-x. [DOI] [PubMed] [Google Scholar]
- 13.Mattick JS, Makunin IV. Non-coding RNA. Hum Mol Genet. 2006;15(1):R17–29. doi: 10.1093/hmg/ddl046. [DOI] [PubMed] [Google Scholar]
- 14.Spizzo R, Almeida MI, Colombatti A, Calin GA. Long non-coding RNAs and cancer: a new frontier of translational research? Oncogene. 2012;31(43):4577–4587. doi: 10.1038/onc.2011.621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zhang F, Zhang L, Zhang C. Long noncoding RNAs and tumorigenesis: genetic associations, molecular mechanisms, and therapeutic strategies. Tumour Biol. 2016;37(1):163–175. doi: 10.1007/s13277-015-4445-4. [DOI] [PubMed] [Google Scholar]
- 16.Jiang C, Li X, Zhao H, Liu H. Long non-coding RNAs: potential new biomarkers for predicting tumor invasion and metastasis. Mol Cancer. 2016;15(1):62. doi: 10.1186/s12943-016-0545-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Deng H, Zhang J, Shi J, Guo Z, He C, Ding L, Tang JH, Hou Y. Role of long non-coding RNA in tumor drug resistance. Tumour Biol. 2016;37(9):11623–11631. doi: 10.1007/s13277-016-5125-8. [DOI] [PubMed] [Google Scholar]
- 18.Jiang YZ, Liu YR, Xu XE, Jin X, Hu X, Yu KD, Shao ZM. Transcriptome Analysis of Triple-Negative Breast Cancer Reveals an Integrated mRNA-lncRNA Signature with Predictive and Prognostic Value. Cancer Res. 2016;76(8):2105–2114. doi: 10.1158/0008-5472.CAN-15-3284. [DOI] [PubMed] [Google Scholar]
- 19.Tanioka M, Shimizu C, Yonemori K, Yoshimura K, Tamura K, Kouno T, Ando M, Katsumata N, Tsuda H, Kinoshita T. Predictors of recurrence in breast cancer patients with a pathologic complete response after neoadjuvant chemotherapy. Br J Cancer. 2010;103(3):297–302. doi: 10.1038/sj.bjc.6605769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hatzis C, Pusztai L, Valero V, Booser DJ, Esserman L, Lluch A, Vidaurre T, Holmes F, Souchon E, Wang H. A genomic predictor of response and survival following taxane-anthracycline chemotherapy for invasive breast cancer. JAMA. 2011;305(18):1873–1881. doi: 10.1001/jama.2011.593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Popovici V, Chen W, Gallas BG, Hatzis C, Shi W, Samuelson FW, Nikolsky Y, Tsyganova M, Ishkin A, Nikolskaya T. Effect of training-sample size and classification difficulty on the accuracy of genomic predictors. Breast Cancer Res. 2010;12(1):R5. doi: 10.1186/bcr2468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Tabchy A, Valero V, Vidaurre T, Lluch A, Gomez H, Martin M, Qi Y, Barajas-Figueroa LJ, Souchon E, Coutant C. Evaluation of a 30-gene paclitaxel, fluorouracil, doxorubicin, and cyclophosphamide chemotherapy response predictor in a multicenter randomized trial in breast cancer. Clin Cancer Res. 2010;16(21):5351–5361. doi: 10.1158/1078-0432.CCR-10-1265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Iwamoto T, Bianchini G, Booser D, Qi Y, Coutant C, Shiang CY, Santarpia L, Matsuoka J, Hortobagyi GN, Symmans WF. Gene pathways associated with prognosis and chemotherapy sensitivity in molecular subtypes of breast cancer. J Natl Cancer Inst. 2011;103(3):264–272. doi: 10.1093/jnci/djq524. [DOI] [PubMed] [Google Scholar]
- 24.Gautier L, Cope L, Bolstad BM. RA Irizarry, affy--analysis of Affymetrix GeneChip data at the probe level. Bioinformatics. 2004;20(3):307–315. doi: 10.1093/bioinformatics/btg405. [DOI] [PubMed] [Google Scholar]
- 25.Johnson WE, Li C, Rabinovic A. Adjusting batch effects in microarray expression data using empirical Bayes methods. Biostatistics. 2007;8(1):118–127. doi: 10.1093/biostatistics/kxj037. [DOI] [PubMed] [Google Scholar]
- 26.Jiang H, Wong WH. SeqMap: mapping massive amount of oligonucleotides to the genome. Bioinformatics. 2008;24(20):2395–2396. doi: 10.1093/bioinformatics/btn429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Harrow J, Frankish A, Gonzalez JM, Tapanari E, Diekhans M, Kokocinski F, Aken BL, Barrell D, Zadissa A, Searle S. GENCODE: the reference human genome annotation for The ENCODE Project. Genome Res. 2012;22(9):1760–1774. doi: 10.1101/gr.135350.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Du Z, Fei T, Verhaak RG, Su Z, Zhang Y, Brown M, Chen Y, Liu XS. Integrative genomic analyses reveal clinically relevant long noncoding RNAs in human cancer. Nat Struct Mol Biol. 2013;20(7):908–913. doi: 10.1038/nsmb.2591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sun J, Chen X, Wang Z, Guo M, Shi H, Wang X, Cheng L, Zhou M. A potential prognostic long non-coding RNA signature to predict metastasis-free survival of breast cancer patients. Sci Rep. 2015;5:16553. doi: 10.1038/srep16553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hess KR, Anderson K, Symmans WF, Valero V, Ibrahim N, Mejia JA, Booser D, Theriault RL, Buzdar AU, Dempsey PJ. Pharmacogenomic predictor of sensitivity to preoperative chemotherapy with paclitaxel and fluorouracil, doxorubicin, and cyclophosphamide in breast cancer. J Clin Oncol. 2006;24(26):4236–4244. doi: 10.1200/JCO.2006.05.6861. [DOI] [PubMed] [Google Scholar]
- 31.Sota Y, Naoi Y, Tsunashima R, Kagara N, Shimazu K, Maruyama N, Shimomura A, Shimoda M, Kishi K, Baba Y. Construction of novel immune-related signature for prediction of pathological complete response to neoadjuvant chemotherapy in human breast cancer. Ann Oncol. 2014;25(1):100–106. doi: 10.1093/annonc/mdt427. [DOI] [PubMed] [Google Scholar]
- 32.Gendoo DM, Ratanasirigulchai N, Schroder MS, Pare L, Parker JS, Prat A, Haibe-Kains B. Genefu: an R/Bioconductor package for computation of gene expression-based signatures in breast cancer. Bioinformatics. 2016;32(7):1097–1099. doi: 10.1093/bioinformatics/btv693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sing T, Sander O, Beerenwinkel N, Lengauer T. ROCR: visualizing classifier performance in R. Bioinformatics. 2005;21(20):3940–3941. doi: 10.1093/bioinformatics/bti623. [DOI] [PubMed] [Google Scholar]
- 34.Liao Q, Liu C, Yuan X, Kang S, Miao R, Xiao H, Zhao G, Luo H, Bu D, Zhao H. Large-scale prediction of long non-coding RNA functions in a coding-non-coding gene co-expression network. Nucleic Acids Res. 2011;39(9):3864–3878. doi: 10.1093/nar/gkq1348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Huang da W, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc. 2009;4(1):44–57. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
- 36.Huang da W, Sherman BT, Lempicki RA. Bioinformatics enrichment tools: paths toward the comprehensive functional analysis of large gene lists. Nucleic Acids Res. 2009;37(1):1–13. doi: 10.1093/nar/gkn923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Merico D, Isserlin R, Stueker O, Emili A, Bader GD. Enrichment map: a network-based method for gene-set enrichment visualization and interpretation. PLoS One. 2010;5(11):e13984. doi: 10.1371/journal.pone.0013984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ashburner M, Ball CA, Blake JA, Botstein D, Butler H, Cherry JM, Davis AP, Dolinski K, Dwight SS, Eppig JT. Gene ontology: tool for the unification of biology. The Gene Ontology Consortium. Nat Genet. 2000;25(1):25–29. doi: 10.1038/75556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Rouzier R, Pusztai L, Delaloge S, Gonzalez-Angulo AM, Andre F, Hess KR, Buzdar AU, Garbay JR, Spielmann M, Mathieu MC. Nomograms to predict pathologic complete response and metastasis-free survival after preoperative chemotherapy for breast cancer. J Clin Oncol. 2005;23(33):8331–8339. doi: 10.1200/JCO.2005.01.2898. [DOI] [PubMed] [Google Scholar]
- 40.Arteaga CL, Sliwkowski MX, Osborne CK, Perez EA, Puglisi F, Gianni L. Treatment of HER2-positive breast cancer: current status and future perspectives. Nat Rev Clin Oncol. 2011;9(1):16–32. doi: 10.1038/nrclinonc.2011.177. [DOI] [PubMed] [Google Scholar]
- 41.Buzdar AU, Ibrahim NK, Francis D, Booser DJ, Thomas ES, Theriault RL, Pusztai L, Green MC, Arun BK, Giordano SH. Significantly higher pathologic complete remission rate after neoadjuvant therapy with trastuzumab, paclitaxel, and epirubicin chemotherapy: results of a randomized trial in human epidermal growth factor receptor 2-positive operable breast cancer. J Clin Oncol. 2005;23(16):3676–3685. doi: 10.1200/JCO.2005.07.032. [DOI] [PubMed] [Google Scholar]
- 42.Buzdar AU, Valero V, Ibrahim NK, Francis D, Broglio KR, Theriault RL, Pusztai L, Green MC, Singletary SE, Hunt KK. Neoadjuvant therapy with paclitaxel followed by 5-fluorouracil, epirubicin, and cyclophosphamide chemotherapy and concurrent trastuzumab in human epidermal growth factor receptor 2-positive operable breast cancer: an update of the initial randomized study population and data of additional patients treated with the same regimen. Clin Cancer Res. 2007;13(1):228–233. doi: 10.1158/1078-0432.CCR-06-1345. [DOI] [PubMed] [Google Scholar]
- 43.Ignatiadis M, Singhal SK, Desmedt C, Haibe-Kains B, Criscitiello C, Andre F, Loi S, Piccart M, Michiels S, Sotiriou C. Gene modules and response to neoadjuvant chemotherapy in breast cancer subtypes: a pooled analysis. J Clin Oncol. 2012;30(16):1996–2004. doi: 10.1200/JCO.2011.39.5624. [DOI] [PubMed] [Google Scholar]
- 44.Dent R, Trudeau M, Pritchard KI, Hanna WM, Kahn HK, Sawka CA, Lickley LA, Rawlinson E, Sun P, Narod SA. Triple-negative breast cancer: clinical features and patterns of recurrence. Clin Cancer Res. 2007;13(15 Pt 1):4429–4434. doi: 10.1158/1078-0432.CCR-06-3045. [DOI] [PubMed] [Google Scholar]
- 45.Beresford MJ, Wilson GD, Makris A. Measuring proliferation in breast cancer: practicalities and applications. Breast Cancer Res. 2006;8(6):216. doi: 10.1186/bcr1618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Mitchison TJ. The proliferation rate paradox in antimitotic chemotherapy. Mol Biol Cell. 2012;23(1):1–6. doi: 10.1091/mbc.E10-04-0335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Fasching PA, Heusinger K, Haeberle L, Niklos M, Hein A, Bayer CM, Rauh C, Schulz-Wendtland R, Bani MR, Schrauder M. Ki67, chemotherapy response, and prognosis in breast cancer patients receiving neoadjuvant treatment. BMC Cancer. 2011;11:486. doi: 10.1186/1471-2407-11-486. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.