Abstract
Melanoma is a cancer that exhibits one of the most aggressive and heterogeneous features. The incidence rate escalates. A high number of clones harboring various mutations contribute to an exceptional level of intratumor heterogeneity of melanoma. It also refers to metastases which may originate from different subclones of primary lesion. Such component of the neoplasm biology is termed intertumor and intratumor heterogeneity. These levels of tumor heterogeneity hinder accurate diagnosis and effective treatment. The increasing number of research on the topic reflects the need for understanding limitation or failure of contemporary therapies. Majority of analyses concentrate on mutations in cancer-related genes. Novel high-throughput techniques reveal even higher degree of variations within a lesion. Consolidation of theories and researches indicates new routes for treatment options such as targets for immunotherapy. The demand for personalized approach in melanoma treatment requires extensive knowledge on intratumor and intertumor heterogeneity on the level of genome, transcriptome/proteome, and epigenome. Thus, achievements in exploration of melanoma variety are described in details. Particularly, the issue of tumor heterogeneity or homogeneity given BRAF mutations is discussed.
Introduction
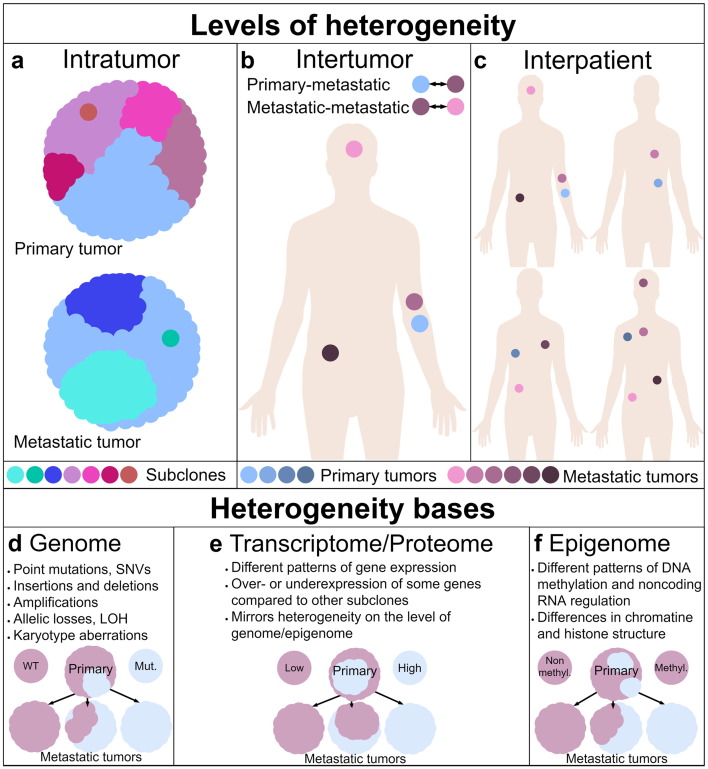
Cancers contain morphologically different cells, and this was observed more than 100 years ago [1]. Further research showed that these cells differ regarding their genomes, epigenomes, transcriptomes, and proteomes, as well as their motility, metabolism, and angiogenic, proliferative, immunogenic, and metastatic potential [2]. The coexistence of cells of distinct phenotypic and molecular features within a tumor is named intratumor heterogeneity (ITH) or intercellular heterogeneity (Figure 1A) [3]. Intratumor heterogeneity is found in most, probably all, solid human tumors [4]. It was documented in a variety of types of solid tumors cancer, including brain cancers [5], [6], [7], [8], [9], [10], [11], [12], breast cancers [4], [13], [14], [15], [16], [17], [18], [19], [20], [21], [22], [23], lung cancers [3], [12], [24], [25], thyroid cancers [12], [26], as well as gastrointestinal cancers [12], [18], [27], [28], [29], [30], [31], [32], [33], [34], urinary cancers [12], [18], [35], [36], [37], [38], [39], and reproductive system cancers [12], [18], [35], [36], [37], [38], [39], [40], [41]. Likewise, heterogeneity of hematopoietic malignancies was found [42], [43], [44], [45], [46]. Moreover, multiple tumors of the same type, found in one patient, may differ significantly. This level of heterogeneity is named intertumor intrapatient heterogeneity (Figure 1B) [47]. The third level of heterogeneity is termed interpatient heterogeneity (Figure 1C) and concerns the differences among tumors within different patients.
Figure 1.
Tumor heterogeneity. Levels of heterogeneity. (A) The differences among tumor cells are termed intratumor or intercellular heterogeneity. The dominant subclone ( ) in the primary tumor (top) was also the founder of metastases (bottom). (B) The differences among the primary tumor (
) in the primary tumor (top) was also the founder of metastases (bottom). (B) The differences among the primary tumor ( ) and metastatic (
) and metastatic ( ) tumors and among metastases constitute intrapatient intertumor heterogeneity. (C) The differences among tumors from different patients are termed interpatient heterogeneity. Heterogeneity bases. (D) Genetic heterogeneity arises from various changes within genome. On this scheme, the primary tumor (top) contains cells wild type given some gene as well as cells harboring a mutated allele. Wild-type subclone was the founder of two metastases (bottom left). However, one of them acquired a mutation in this gene (middle). The mutated subclone was the founder of third metastasis. (E) Heterogeneity of transcriptome and proteome constitute heterogeneity of gene expression. Low-expressing subclone was the founder of one metastasis, which is homogeneous given this gene expression. However, the other two metastatic tumors were formed by a high-expressing subclone, although both metastases heterogeneously express this gene on mRNA/protein marker level. (F) The primary tumor is homogeneous given methylation status of some gene promoter – remain nonmethylated. One metastasis mirrors the status of primary tumor, while the other two metastatic tumors are heterogeneous. One tumor (middle) exhibits epigenetic heterogeneity developed during progression since one of the tumor cells acquired an epimutation. The last metastasis is homogeneous because the founder cell acquired an epimutation. Therefore, methylation of promoter is present in all tumor cells.
) tumors and among metastases constitute intrapatient intertumor heterogeneity. (C) The differences among tumors from different patients are termed interpatient heterogeneity. Heterogeneity bases. (D) Genetic heterogeneity arises from various changes within genome. On this scheme, the primary tumor (top) contains cells wild type given some gene as well as cells harboring a mutated allele. Wild-type subclone was the founder of two metastases (bottom left). However, one of them acquired a mutation in this gene (middle). The mutated subclone was the founder of third metastasis. (E) Heterogeneity of transcriptome and proteome constitute heterogeneity of gene expression. Low-expressing subclone was the founder of one metastasis, which is homogeneous given this gene expression. However, the other two metastatic tumors were formed by a high-expressing subclone, although both metastases heterogeneously express this gene on mRNA/protein marker level. (F) The primary tumor is homogeneous given methylation status of some gene promoter – remain nonmethylated. One metastasis mirrors the status of primary tumor, while the other two metastatic tumors are heterogeneous. One tumor (middle) exhibits epigenetic heterogeneity developed during progression since one of the tumor cells acquired an epimutation. The last metastasis is homogeneous because the founder cell acquired an epimutation. Therefore, methylation of promoter is present in all tumor cells.
Melanoma is one of the most aggressive, complex, and heterogeneous cancers [12]. It is an exceedingly metastatic cancer that is related, among others, with lineage-specific transcription factors characteristic for melanocytes which migrate from neural crest during development [48]. Moreover, melanoma is highly refractory to conventional chemotherapy. It responds well to both targeted and immunotherapy; however, the resistance occurs in a significant percentage of patients, especially in targeted therapy. Progress in melanoma treatment has been discussed in several papers [49], [50]. Importantly, melanoma was the third most prevalent cancer in 2016 in the United States [51], whereas it was the seventh most prevalent in 2012 in Europe [52]. In 2012, it was estimated that there were over 100,000 new cases in Europe and about 22,300 deaths from melanoma [52]. It is becoming more frequent with a three-fold increase in incidence rates over the last four decades [53], [54].
The mutation rate of melanoma is the highest from all cancers [55]. Based on The Cancer Genome Atlas (TCGA) data, the number of mutations per Mb ranged from 0.1 to 100.0 [55] with an average of 16.8 mutations per Mb [56]. The most relevant signaling pathway for melanoma transformation is the Ras signaling pathway, in particular, the mitogen-activated protein kinase (MAPK) pathway [57], which is dysregulated in about 80% of melanomas [58]. Based on molecular features, melanomas are divided into four groups, i.e., BRAF−, NRAS−, NF1− mutant, and triple wild type [56]. About half of the melanomas have a mutation in the serine/threonine-protein kinase B-Raf (v-Raf murine sarcoma viral oncogene homolog B) gene, especially the BRAF V600E mutation [59], [60], [61], [62], [63]. Mutations in the NRAS (neuroblastoma RAS viral oncogene homolog) gene are harbored by 15% to 30% of melanoma tumors [61], [64], [65], [66], [67], whereas mutations in NF-1 gene are present in 12% to 18% of melanomas [68].
Recently, a significant progress in melanoma tumor heterogeneity research has been made. In this review, we summarize the current knowledge about intratumor and intertumor heterogeneity in melanoma with emphasis on intratumor genetic heterogeneity and heterogeneous gene expression within tumors.
Intratumor Heterogeneity in Melanoma
It is known that intratumor heterogeneity (Figure 1A) can arise independently from point mutations, copy number changes, i.e., deletions and duplications, whole chromosomal rearrangements, gene expression, epimutations, or reversible phenotypic changes [69], [70], [71]. The phenomenon of intratumor heterogeneity is commonly explained by Darwinian-like clonal evolution of a single tumor [72].
Genetic Heterogeneity
The main cause of genetic heterogeneity (Figure 1D) is genomic instability [47] which is considered to be one of the hallmarks of cancer [73]. This is due to, among others, inhibited or reduced DNA damage response mechanisms and microenvironmental factors, such as hypoxia, acidosis, and reactive oxygen species [69]. Despite the monoclonal origin of most cancers, new clones arise during tumor progression which is due to the continuous acquisition of mutations, i.e., hypermutability. This promotes division into subclones and therefore causes an increase of intratumor heterogeneity [74]. The coexistence of distinct subclones creates a complex network of interaction which affects cancer progression [2]. Moreover, genetic heterogeneity provides the material with which selection can work [2], [21] allowing Darwinian-like clonal evolution.
Mutations which occur early in tumor evolution are called common or ubiquitous and are present in all regions [3], [27]. Generally, almost all tumor cells harbor them; thereby, they are rather homogeneous within a tumor (e.g., NRAS or BRAF mutations; Table 1). Mutations which occur later are present in only some regions and are called shared or branches, whereas mutations which occur the latest in tumor progression are present in only one compartment (one subclone); therefore, they are called private [3], [27]. The latter are the basis of genetic intratumor heterogeneity. The degree of intratumor genetic heterogeneity is usually defined as the number of alterations, i.e., SNVs, CNVs, LOH, karyotype aberrations, and other mutations, present in some compartments within a tumor but not detectable in others.
Table 1.
Intratumor Heterogeneity or Homogeneity of the BRAF Genotype and Expression Status within Melanoma Tumors
| Ref. | Samples | Methods | Number (%) of Heterogeneous Tumors |
|
|---|---|---|---|---|
| Overall | Among BRAF Mutated Tumors | |||
| [76] | 5 PT | scPCR, Sanger seq | 4/5 (80%) | 4/4 (100%) |
| [85] | 49 PT | IHC | 29/49 (59%) | 29/31 (94%) |
| [86] | 50 PT 139 MT |
HRM, qPCR, IHC | 25/189 (13%) | 25/88 (28%) |
| [87] | 20 PT | IHC | 2/20 (10%) | 2/7 (29%) |
| [88] | 100 PT | PNA qPCR, IHC, NGS, Cap seq | 7/100 (7%) | 7/59 (12%) |
| [89] | 124 PT 76 MT |
IHC | 10/200 (5%) | 10/94 (11%) |
| [90] | 22 MI1S 56 PT 93 MT |
Pyro, IHC | 2/171 (1%) | 2/75 (3%) |
| [91] | 140 PT 171 MT |
IHC | 0/239 (0%) | 0/137 (0%) |
| Overall | 79/973 (8.1%) | 79/495 (16.0%) | ||
| [63] | 104 NT from TCGA | NGS | BRAF-M% ranged from 8% to 97% | |
| [92] | 75 PT 88 MT |
NGS | BRAF-M% ranged from 0% to 92% (median for PT: 28% and for MT: 26%) | |
| [63] | 475 NT | Pyro, qPCR, dPCR | BRAF-M% ranged from 10% to 90%. | |
| [93] | 9 PT | LCM, Direct seq, SNaPshot assay | BRAF-M% ranged from 0% to 81% | |
| [94] | 52 MT | Pyro, Sanger seq, qPCR | BRAF-M% ranged from 3% to 80% (median 41.3%) | |
| [95] | 47 MT | dPCR, HRM, Pyro | BRAF-M% ranged from 1% to 79% | |
| [96] | 40 PT 37 MT |
Sequenom MassARRAY | BRAF-M% ranged from 8% to 53% | |
| Overall | BRAF-M% ranged from 0% to 97% | |||
Abbreviations: PT, primary tumors; MT, metastasis tumors; NT, undefined tumors; MIS, melanoma in situ; scPCR, single-cell PCR; Sanger seq, Sanger sequencing; qPCR, quantitative PCR (also known as real-time PCR); IHC, immunohistochemistry [anti-BRAF V600 (VE1)]; PNA qPCR, peptide nucleic acid quantitative PCR; Cap seq, capillary sequencing; Pyro, pyrosequencing; LCM, laser capture microdissection; Direct seq, direct sequencing; BRAF-M%, BRAF mutant allele frequencies.
The development of advanced research techniques, such as digital polymerase chain reaction (dPCR) or next-generation sequencing (NGS), provides tools for more comprehensive and sophisticated cancer research. These methods enable the detection of mutations even if they are present in only a small subpopulation of tumor cells. Therefore, they are extremely useful for tumor heterogeneity research. It was established that the limit of detection of the mutant allele differs significantly for various methods and ranged from 1% to 2% of mutated alleles detected by high-accuracy methods like high-resolution melting (HRM) PCR, allele-specific real-time PCR, and pyrosequencing (NGS) to as much as 20% of mutated alleles detected by conventional Sanger sequencing [75]. Less precise methods in research do not provide the detection of whole spectrum of mutations, in particular, heterogeneous mutations which are present in only a small percentage of tumor cells, which would be detected by more accurate methods, for instance, NGS [74], [76], [77]. This leads to an underestimation of tumor heterogeneity and may result in a false homogeneity.
Mutations in Cancer-Related Genes
Heterogeneity of genes which are related with signaling pathway is particularly relevant because it directly affects tumor cell fitness. Heterogeneity in this field results in differences in features of subclones within a tumor, for instance, different proliferation rate, different response to treatment, or even different resistance to treatment.
The percentage of heterogeneous tumors, in the context of the BRAF genotype, was very variable and ranged from 0% to 97% in various studies (Table 1). Because of these discrepancies and the fact that BRAF mutation is one of the most common in melanoma, we performed a systematic review which showed that the overall number of heterogeneous tumors, considering the BRAF genotype, was 79 (8.1%) out of 973 (it was 16.0% among BRAF mutated tumors). Moreover, the BRAF mutant allele frequency (MAF) was variable and ranged from 0% to 97%. The relatively low rate of BRAF intratumor heterogeneity is due to the fact that BRAF mutations arise early during melanoma and thus are present in the majority of tumor cells [65], [70]. The variability of results received by various authors results from the use of different methods. Therefore, it is expected that the most appropriate method for BRAF analysis will be established and used in clinical practice [78], [79], [80], [81]. Some authors suggested that it is real-time PCR–based analysis, for instance, Cobas 4800 BRAF V600 Mutation Test, which enables detection of mutation allele at more than 5% frequency [78], [80], [82], whereas others suggested immunohistochemistry [79], [81], [83] and then pyrosequencing (NGS) for VE1-negative or uninterpretable cases because of the highest accuracy and efficiency of this method [79], [82]. In addition to accuracy, consideration should be given to cost and turnaround time to establish the appropriate genotype testing methods for clinical practice. It was shown that NGS-based testing is cheaper and has lower mean turnaround time than conventional methods, i.e., real-time quantitative PCR HRM and Sanger sequencing for BRAF, real-time quantitative PCR and Sanger sequencing for NRAS, and conventional PCR and Sanger sequencing for KIT [84].
Chiappetta et al. [64] used laser capture microdissection to isolate 3 areas from each of 15 melanoma tumors. Discrepancies in BRAF or NRAS mutation status in at least one area compared to the whole tumor mutational analysis were found in 4 (36%) out of 11 mutated tumors. Wilmott et al. [97] observed two distinct subclones within BRAF inhibitor-resistant melanoma metastasis. Subclone A harbored the BRAF V600E mutation but was NRAS wild type, whereas subclone B harbored both the BRAF V600E and NRAS G13R mutations.
Gremel et al. [98] observed two subclones with different response to imatinib (tyrosine kinase inhibitor) after analyses of cell-free circulating tumor DNA (ctDNA) in metastatic vaginal mucosal melanoma. One subclone was KIT mutated and responded to imatinib, whereas the other was KIT wild type and, therefore, did not respond to imatinib.
Vivancos et al. [99] investigated the nevus of Ota, primary melanoma, and recurrent tumor from one patient. Using amplicon sequencing, they established that all tumors contained cells with the GNAQ mutation (MAF was 3%, 46%, and 21%, respectively) as well as cells with wild-type allele. Moreover, they showed that only part of the tumor cells harbored the mutated allele of MMP10, BAP1, COL4A4, FN3K, PLD3, LRRC25, MYH3, and TP53 genes. Furthermore, genetic profiling of the recurrent tumor in four different regions was performed. MAF of all these genes was different among examined compartments. For instance, the MAF of BAP1 ranged from 38% to 65%, whereas it equaled 48% after the bulk tumor analyses. The MAF of TP53 equaled 2% in three regions, whereas it was 53% in the fourth region.
Shain et al. [100] investigated the evolution of melanoma from precursor lesions. They sequenced 293 cancer-relevant genes in 150 areas of 37 primary tumors and their precursors. Significant intratumor genetic heterogeneity was observed. Moreover, they confirmed that neoplastic cells acquire mutations during progression; thereby, tumor cells harbored a substantial number of additional mutations compared with their precursors.
Single Nucleotide Variations (SNVs) and Copy Number Variations (CNVs) within Genome
The most comprehensive analysis of intratumor genetic heterogeneity was published by Andor et al. [12]. They used the exome sequencing data of paired tumor and normal cells of 1165 primary tumor samples derived from 12 cancer types, including cutaneous melanoma. Clones within tumors were detected based on somatic SNVs and CNVs. At the time of biopsy or surgical resection, four clones coexisted in a tumor on average. Furthermore, 86% of tumors contained at least two clones. Among all cancers, melanoma tumors were the most heterogeneous. All of them contained more than one clone. Moreover, about half of the melanomas contained at least nine distinct clones within the tumors. The number of clones was investigated by Ding et al. [101] as well, who used whole genome sequencing (WGS) to characterize 15 metastatic melanoma tumors. They found that 11 (73%) tumors contained multiple distinct subclonal populations of tumor cells. Regions of the genome exhibiting highly genomic stability were selected based on the loss of heterozygosity (LOH) and CNV analyses. Mutations from these regions were used for clustering. The frequency of founding clone cluster ranged among tumors from 22.24 to 46.80 (mean = 33.55), whereas the frequency of subclones clusters ranged from 0.00 to 45.60 (mean = 16.35). Sanborn et al. [102] investigated primary tumors and corresponding metastases from eight patients. Whole exome sequencing (WES) was performed to detect SNVs and CNVs. They reported that each patient had at least one tumor with subclonal CNVs. The number of such subclonal CNVs ranged from 1 to 10 per patient, with a maximum of 6 per tumor. Moreover, they detected identical subclonal CNV in two locoregional metastases within one patient. They concluded that each of these tumors were formed by at least two distinct tumor cell populations. Thereby, the current view of the mechanisms of metastasis was confirmed. Gremel et al. [98] observed two clones with eight and six exclusive SNVs, respectively, after analyses of cell-free ctDNA in metastatic vaginal mucosal melanoma, as mentioned above.
Harbst et al. [70] sequenced the whole exome of six metastases and two primary melanoma tumors. The number of nonsynonymous mutations per tumor ranged from 45 to 814 (mean = 489). On average, 13.0% (ranged from 2.7% to 37.8%) of them were heterogeneous, i.e., were not detected within all samples obtained from one tumor. Moreover, they established that 12% of cancer driver mutations, i.e., mutations that increase tumor cells fitness, confer a growth advantage, and thereby promote cancer progression, were heterogeneous. The most common driver mutations in melanoma, i.e., BRAF, NRAS, and KIT mutations, if present, were observed within all regions at similar allele frequencies. This is consistent with our findings (the BRAF mutation; Table 1). In their research, heterogenous mutations were principally passenger mutations, i.e., mutations that do not confer a growth advantage and do not contribute to cancer development.
Anaka et al. [103] found significant genetic heterogeneity in melanoma lymph node metastases and derived early-passage cell lines from three patients. Using amplicon sequencing and SNP microarrays, they showed that the number of CNVs in eight regions within one tumor from the first patient varied from 44 to 133. Almost 20% of all aberrations were found in just one compartment, whereas about 35% were found in two or three regions. Only about 20% of the aberrations were detected in all examined compartments. Moreover, significant heterogeneity was observed for genes with impact on melanoma biology, which proves that distinct subpopulations of tumor cells may exhibit different fitness. Likewise, cell lines derived from this tumor were heterogeneous for many aberrations; however, clones were less heterogeneous than parental tumors since the number of harbored CNVs varied between 55 and 69. Similarly, clones obtained from the other two cell lines were heterogeneous as well. Therefore, the usefulness of cell lines in intratumor genetic heterogeneity studies was established. However, possible differences between the results obtained from tumors and cell lines, namely. from in vivo and in vitro conditions, must be taken into consideration in tumor heterogeneity research. This issue is discussed in the context of gene expression below. Moreover, they established that the copy number profile of the parental cell line represented an average from all cells (all clones) in the line. Hence, clones with the closest profile to the parental cell line profile were the dominant clones.
Turajlic et al. [104] examined one paired primary and metastatic acral melanoma. NGS, i.e., WGS and WES, enabled them to estimate the MAF within each tumor. The MAFs of SNVs in primary tumor ranged from 21% to 82% (mean = 45.3%), whereas in metastases, it was from 11% to 84 % (mean = 45.1%) and thus was various for different mutations. The degree of intratumor heterogeneity in primary and metastatic tumors was similar. Gandolfi et al. [67] used molecular inversion probe assay technology to investigate whole-genome CNVs in 41 primary melanomas. They found that tumors exhibited significant intratumor heterogeneity given their chromosomal alterations.
De Lange et al. [105] investigated 64 uveal melanomas (UMs). They used dPCR to count the frequency of aberrations (CNVs) and to determine their sequence. They observed that tumors commonly contained only a subpopulation of cells with GNAQ mutation; a loss of chromosome 1p, 3, or 16q; or a gain of chromosome 6p or 8q. Moreover, 14 tumors were heterogeneous for monosomy 3 and chromosome 16q loss. In one tumor, 71.8% of the cells contained the GNAQ Q209P mutation, whereas 36% of the cells harbored monosomy 3. Therefore, monosomy 3 occurred after GNAQ mutation. Thereby, they confirmed that the different time of the occurrence of mutations is one of the causes of intratumor heterogeneity. If one mutation occurs later than another, then it is absent in some tumor cells, and therefore, it is heterogeneous within the tumor.
LOH
Chromosomal alterations, especially allelic losses, are common in cancer, including melanoma [106]. Losses of the tumor suppressor gene lead to acceleration of cancer progression. Therefore, intratumor heterogeneity of LOH is commonly investigated.
Nakayama et al. [107] used a microsatellite-PCR allelic losses (LOH) analysis of eight markers on chromosomes 1p, 3p, 6p, 9p, 9q, 10q, and 11q to investigate 79 in-transit melanoma metastases from 25 patients. Tumors from six patients who exhibited intertumor heterogeneity (described in the subsection “Intertumor: Genetic Heterogeneity”) were chosen for further research. Six (23%) of the 26 selected tumors displayed intratumor heterogeneity for at least one marker. Likewise, Bogdan et al. [106] studied heterogeneity of LOH on chromosomes 1p, 9p, 9q, and 17p within 29 areas in 11 melanoma metastases. They found that 10 (91%) out of 11 metastases were heterogeneous given LOH in at least two markers. The degree of intratumor heterogeneity for each marker was highly variable. Eight (73%) out of 11 tumors were heterogeneous given LOH on chromosome 9p, whereas 4 (36%) were heterogeneous given 1p, 2 (18%) given 17p, and 1 (9%) given 9q. Rao et al. [108] examined LOH of 9 markers on chromosomes 1p, 5q, 9p, 10q, and 17p in 16 tumors, including primary and metastatic melanoma. They found that 12 (75%) tumors exhibited intratumor heterogeneity. The degree of intratumor heterogeneity for each tumor was different. Two (17%) tumors were heterogeneous given five markers, one (8%) given four markers, one (8%) given three markers, and three (25%) given two markers. Heterogeneity of loss of chromosome 9 was observed in one examined spitzoid melanoma by DiSano et al. [109] who used array-based comparative genomic hybridization (aCGH).
Uguen et al. [110] investigated intratumor heterogeneity of chromosomes 6, 8, 9, and 11 in 12 primary melanomas. Fluorescent in situ hybridization (FISH) was performed to analyze four distinct areas of each tumor. Intratumor heterogeneity of at least one marker was found in all tumors. Likewise, Takata et al. [111] found intratumor heterogeneity of LOH of chromosomes 6q, 9p, 10q, and 18q in 8 out of 10 primary tumors. All heterogeneous tumors contained at least two distinct subclones. Moreover, one tumor contained at least six genetically different subclones.
Dopierala et al. [112] used multiplex ligation-dependent probe amplification to characterize intratumor heterogeneity of chromosomes 1, 3, 6, and 8 in 32 UMs. Intratumor genetic heterogeneity was detected in 24 (75%) tumors. The most heterogeneous loci were CDKN1A (6p21.2), which exhibited heterogeneity in 11 (35%) out of 31 examined tumors; RP1 (8q11.23), which was heterogeneous in 11 (34%) out of 32 tumors; and ROBO1 (3p12.2), which was heterogeneous in 10 (31%) out of 32 tumors. Moreover, 15 (47%) tumors exhibited intratumor heterogeneity in at least 1 locus on chromosome 3. Likewise, Lake et al. [113] used multiplex ligation-dependent probe amplification to investigate heterogeneity of chromosomes 1p, 3, 6, and 8 between the intraocular and extraocular parts of 10 UM tumors. Only in three (30%) tumors were intraocular and extraocular parts homogeneous given these chromosomes. Seven (70%) tumors showed heterogeneity in at least one chromosome arm. Four tumors showed gains of 6p in intraocular parts, whereas they showed disomy in extraocular parts. Three tumors were heterogeneous for chromosome 3. Nevertheless, no heterogeneity for chromosome 8 was observed.
To conclude, 80 (67.8%) out of 118 examined tumors exhibited intratumor heterogeneity given LOH in at least 1 locus. The degree of intratumor heterogeneity differs significantly for each marker. Likewise, the degree was different for each tumor, with both relatively homogeneous and highly heterogeneous tumors.
Wide Karyotype Aberrations
Helmbold et al. [114] analyzed 54 superficial spreading melanomas. DNA image cytometry was used to investigate DNA stem lines, i.e., a group of proliferative cells with the same DNA content, in 196 measuring fields (ranged from 2 to 9 per tumor). They found that 23 out of 54 (43%) tumors contained at least 2 distinct tumor cell populations (on average 1.46 per tumor). In 22 cases (96%), different populations occurred spatially separated. In one case (4%), the populations were mixed in all measuring fields.
Intratumor heterogeneity of monosomy 3 is commonly observed in melanoma tumors, especially in UM. This alteration is a strong predictive factor. It is associated with a high metastatic risk and therefore with poor prognosis. It results from the BAP1 gene (3p21) loss which is essential in the acquisition of metastatic capacity [115]. Therefore, subclones with monosomy 3 may be the founders of metastases more common than subclones with disomy of chromosome 3. Monosomy 3 was found in fine needle aspiration biopsy of choroidal melanoma by Chang et al. [116]. Monosomy 3 was reported in 93 patients. The percentage of positive cells ranged from 4.7% to 100% (mean = 62.9%). All positive cells were detected in 6 (6%). Mensink et al. [117] chose 16 UMs with possible heterogeneity of chromosome 3 after FISH. Six (38%) tumors showed homogeneity of monosomy 3, whereas the remaining 10 (62%) tumors contained at least 2 clones with a different percentage of monosomy 3. Sandinha et al. [118] studied the mixed-cell type of choroidal melanoma. In 7 (32%) out of 22 tumors, they found that epithelioid cells exhibited monosomy 3, whereas spindle cells contained 2 copies of chromosome 3. Maat et al. [119] established that at least 7 (14%) out of 50 UMs were heterogeneous considering monosomy 3. On the contrary, Meir et al. [120] did not find heterogeneity of monosomy 3 in eight UMs. However, such results could be due to the small number of analyzed regions, i.e., only two dissected samples from each tumor.
Summarizing discussed research, 111 (58.7%) out of 189 examined tumors were heterogeneous given monosomy 3.
Heterogeneity of Gene Expression
Heterogeneous gene expression (Figure 1E) is the consequence of genomic and epigenomic alterations, or selection by heritable factors, nonheritable factors, or both [69], [121]. These changes may be observed on the level of transcriptome solely (RNA sequencing, reverse transcription PCR, in situ hybridization) or involve proteome (Western blotting, immunohistochemistry, Orbitrap liquid chromatography mass spectrometry, zymography). Nonheritable heterogeneity may result from different source. It is known that cancer stem cells (CSCs) contribute to heterogeneity of gene expression [2], [122], which is confirmed by the ability to reestablish intratumor heterogeneity of gene expression by CSCs. Moreover, clonal selection and phenotype switching may create and maintain tumor heterogeneity. Interestingly, Chapman et al. [123] observed that distinct tumor subpopulations, i.e., MITF-high and MITF-low, cooperated in tumor invasion. Therefore, preservation of tumor heterogeneity without clonal selection or phenotype switching is also possible. Moreover, all of the aforementioned sources of heterogeneity are influenced by the tumor microenvironment, hypoxia, and inflammation. It creates a network of close links and connections that results in tumor heterogeneity. However, it remains unclear which mechanism is critical for establish and maintenance of heterogeneous gene expression. Currently, phenotype switching and microenvironmental factors seem to take the lead.
In melanoma, two main phenotypes are distinguished regarding gene expression. Proliferative but weakly motile, invasive, and metastatic melanoma cells have transcriptional profile similar to the neural-crest transcriptome [124]. Genes upregulated in this phenotype are related with cell cycle, proliferation, and well-known melanocyte and melanoma markers, i.e., SOX10, PAX3, and MITF [125]. The latter is established as a marker of proliferative phenotype. The second phenotype, i.e., invasive and metastatic, is probably mainly triggered by tumor microenvironmental factors such as glutamine limitation, hypoxia, and inflammation through TGFβ and JNK signaling [124], [126], [127]. This phenotype is characterized by high expression of AXL (tyrosine-protein kinase receptor UFO), and is regulated by AP-1 and TEAD [125]. Both phenotypes coexist within tumors [125], [128]. Moreover, switching from proliferative to invasive phenotype as well as from invasive to proliferative may occur [129]. It is known that Wnt signaling pathway plays relevant role in phenotype switching [130]. At the center of phenotype switching regulation is MITF. It integrates a variety of signals in tumor cell and regulates both proliferation and invasiveness [131]. Importantly, phenotype switching is essential from a clinical point of view. Through phenotype switching from proliferative to invasive, melanoma cells may acquire resistance to treatment. In response to therapy, cells upregulate expression of neural crest and melanocyte markers and growth slowly [132]. Tumor cells with higher expression of MITF are more sensitive to MAPK inhibitor therapy than invasive melanoma cells with lower MITF but higher AXL and NF-κB expression [133], [134]. Therefore, low expression of MITF predicts early resistance to targeted therapies [135]. It has been shown for in vitro condition that slowly cycling state of tumor cells in response to treatment is only temporary. After several days of growth in drug-free medium, melanoma cell population returns to its initial state and reestablishes heterogeneity [132]. Similarly, phenotype switching contributes to resistance to immunotherapy [126]. Moreover, besides switching between proliferative and invasive phenotypes, switching of melanoma tumor cells similar to epithelial-mesenchymal transition (EMT) described for epithelial tumors has been reported [136].
Melanoma cell lines are used in many studies, including tumor heterogeneity research. Therefore, Vincent et al. [137] examined the utility of melanoma cell lines as a tumor model in gene expression research. Transcriptomes of 42 melanoma cell lines were compared with data obtained from tumors (TCGA) and single melanoma cells. They concluded that, in general, cell lines faithfully represent the mutational and transcriptional profiles of melanoma tumors, except immune-associate genes. Moreover, utility of tumor-derived cell lines in tumor heterogeneity research was confirmed by Anaka et al. [103]. Therefore, cell lines can be used as a reliable model in tumor heterogeneity studies but with the awareness of potential differences between studies in vitro and in vivo.
Heterogeneous Patterns of Gene Expression
Harbst et al. [70] used microarray-based gene expression profiling to classify tumors into four groups, i.e., high-immune, MITF-low proliferative, MITF-high pigmentation, and normal-like. They revealed that 50% (four out of eight) of examined tumors exhibited intratumor heterogeneity when considering genes determining classification into molecular groups. Moreover, the levels of mRNA of MITF and its target genes, i.e., MLANA and TYR, as well as immune checkpoint genes, i.e., CTLA4 and PD-1, displayed intratumor heterogeneity.
Tirosh et al. [128] performed single-cell RNA sequencing of 4645 tumor cells isolated from 19 patients with metastatic melanoma. First, they established that cell cycle phase-specific genes were expressed by small subpopulations of tumor cells. Based on the expression of these signatures, tumor cells were classified as cycling or noncycling. The fraction of cycling cells varied among the examined tumors from 1% to 30% (on average 13.5%). This classification was confirmed by Ki-67 staining which was heterogeneous as well. Moreover, higher expression of JARID1B (lysine-specific demethylase 5B), i.e., a putative marker of CSCs, by slow-cycling cells confirmed the results of other research [138]. Furthermore, 468 tumor cells from 4 regions within 1 melanoma were examined. They found that 229 genes were expressed higher in the first region when compared with other regions. These genes were associated with inflammation, response to stress, survival, and melanoma progression. This confirmed that heterogeneity of gene expression concerns genes directly affecting tumor cell fitness. Moreover, a similar pattern of expression was found in T cells from the first region compared with tumor cells from this area. Therefore, spatial effects affecting the pattern of gene expression in various cells were suggested. Moreover, both AXL-high and MITF-high populations of tumor cells were detected in tumors classified after bulk analysis as AXL-high or MITF-high. Likewise, single-cell RNA sequencing was performed by Gerber et al. [139]. Cells from three melanoma biopsies were examined. Based on a self-organizing map, they observed substantial heterogeneous patterns of gene expression. This heterogeneity was driven by genes associated with proliferation, stroma, and MITF/AXL programs, thereby, similar to previously discussed research, by genes directly affecting tumor cell fitness. Furthermore, heterogeneous immunostaining for TOP2A, Ki-67, and ITGA1, i.e., proliferation markers, was observed.
A heterogeneous spatial pattern of angiogenesis-specific gene expression was found by Demou et al. [140]. A higher expression of about half of these genes was observed in cells forming networks when compared to nests, i.e., monolayers of randomly positioned cells.
Heterogeneity of gene expression in freshly isolated melanoma cells, cultured patient-derived melanoma cells, as well as melanoma cell lines was investigated by Mirkina et al. [141]. The percentage of cluster of differentiation (CD)63+ ranged from 39% to 89% (mean = 64%) depending on the studied cell line, while frequency of CD24+ cells ranged from 4% to 48%. Likewise, the fraction of erythropoietin receptor (EPO-R)–positive cells ranged from 4% to 40%. Moreover, EPO-R was expressed in the distinct subpopulation of melanoma cells coexpressing CD24.
As previously mentioned, Anaka et al. [103] found genetic heterogeneity in three melanoma metastases and patient-derived cell lines. They also confirmed heterogeneity on the level of transcriptome. Gene expression profiling revealed that the parental cell line profile represents an average of profiles of all clones. Furthermore, they established that clones with the expression profile most similar to the parental cell line (GSEA), namely, clones which were dominant, expressed genes typical for aggressive metastatic melanoma. Ennen et al. [142] characterized gene expression in two melanoma cell lines, i.e., 501Mel and 1205LU. The first one was MITF-high, rapidly proliferating in vitro but poorly invasive, motile, and tumorigenic in nude mice. Conversely, the second was MITF-negative, slow proliferating in vitro but invasive, motile, and highly tumorigenic in nude mice. First, these cell lines were examined in vitro. Although the 501Mel cell line was classified as MITF-high, cells with low MITF expression were detected by immunostaining and high-throughput single-cell qPCR in monolayer cultures. This heterogeneity was even more substantial in cells grown as melanospheres. Furthermore, they injected subcutaneously 501Mel and 1205Lu cells into nude mice. In 501Mel-derived tumors, three groups of cells were distinguished, i.e., high-MITF, intermediate-MITF, and low-MITF and its target gene expression. Moreover, a small subpopulation of cells expressed markers of invasion, i.e., ZEB1, GLI2, MYOF, or drug resistance, i.e., ABCB5. Immunostaining and immunohistochemistry revealed that tumors exhibited highly heterogeneous expression of MITF, CEACAM1, and POU5F1. A small subpopulation strongly expressing BIRC3 was observed as well. Hence, 501Mel-derived tumors were highly heterogeneous. In 1205Lu-derived tumors, cells were distinguished based on high-ZEB1 or low-ZEB1 expression. Furthermore, small subpopulations with different expression of some genes were observed. In contrast to 501Mel-derived tumors, expression of POU5F1 was strong and homogeneous within whole 1205Lu-derived tumors. Moreover, they established that there was a group of genes differentially expressed in in vitro conditions, i.e., in monolayers or spheres, and in in vivo tumors. Therefore, it has once again been emphasized that different conditions contribute to the differences among the results obtained by researchers from in vitro and in vivo studies.
Heterogeneous Expression of Cell Adhesion– and EMT-Related Genes
Haqq et al. [143] isolated RNA from melanoma and nevus and used microarrays to analyze gene expression. Laser capture microdissection of the primary tumor was performed to isolate radial and vertical growth phases within the tumor. No genes activated in vertical growth compared with radial growth were identified. Instead, loss of expression of many cell adhesion receptors and extracellular matrix molecules, for instance, integrin α2, laminin γ2, MMP10, and CDH3, was reported. Different expression of MMP10 and CDH3 was confirmed by IHC. Therefore, temporal heterogeneity, i.e., loss of expression of many genes during tumor progression, was confirmed. Furthermore, heterogeneous expression of adhesion-related proteins, i.e., ICAM-1/CD54, CEACAM-1/CD66a, L1/CD171, was revealed by Mirkina et al. [141] as well.
Croteau et al. [144] used quantitative reverse transcriptase PCR and Western blot to distinguish clones from VMM5 melanoma cell line considering their MMP-1 expression. Substantial heterogeneity was reported. One clone (C9) expressed about 24 times the amount of MMP-1 expressed by another clone (C4). Furthermore, clones were injected orthotopically into nude mice. They found that the clone with the highest MMP-1 expression (C9) grew more rapidly in vivo than the clone with the lowest expression (C4). Moreover, tissue and explants analysis showed that both tumors expressed MMP-1 mRNA and protein at a high level. Conversely to in vitro studies, C4 tumors produced more MMP-1 than C9 tumors. They concluded that there are mechanisms within the tumor microenvironment which affect the MMP-1 expression.
Kim et al. [145] found heterogeneous expression of EMT markers among melanoma cell lines using Western blotting. Moreover, individual cells with detectable expression of MITF were observed among very low expressing MITF cells. Therefore, they suggested that melanoma tumors may contain a mixture of phenotypes with some cells expressing high MITF and E-cadherin and other cells expressing low MITF but high N-cadherin, Slug, and AXL. The former can be characterized by noninvasive behavior, whereas the latter by more invasive behavior. Also, Demou et al. [140] found that extracellular matrix cell adhesion genes were expressed on a higher level in tumor cells forming nests than vascular-like networks in 3D cell cultures.
Heterogeneous Expression of Members of the Signaling Pathways
Intratumor heterogeneity of the BRAF mutant protein expression was revealed by IHC which is presented in Table 1. Richmond-Sinclair et al. [146] observed heterogeneous expression of P-MAPK, also known as extracellular signal–regulated kinase (ERK), Brn-2, as well as pRb, p53, and p16, i.e., tumor suppressor proteins, in 114 melanoma tumors revealed by IHC. Similarly, Wilmott et al. [97] observed heterogeneous expression of the ERK gene. They distinguished two subclones based on IHC staining. In subclone A, 95% of tumor cells had high p-ERK1/2 expression, whereas in subclone B, only 3% of tumor cells were p-ERK1/2 positive. Moreover, these subclones exhibited different proliferative rates. In subclone A, 10% of tumor cells were Ki-67 positive, whereas in subclone B, it was just 3%. Furthermore, in subclone A, 46% of tumor cells were cyclin D positive, while in subclone B, it was 74% of tumor cells. Likewise, IHC revealed intratumor heterogeneity of c-KIT expression in mucosal melanoma [147]. The percentage of positive cells ranged from 10% to more than 75%. Mirkina et al. [141] noted heterogeneous expression of cytokine and growth factor receptors on melanoma cells obtained from freshly isolated samples. Less than 5% of tumor cells were positive after each anti-ErbB2, anti-c-MET, anti-G-CSF-R, and anti-KIT staining. Similarly, endoglin was weakly expressed by a small subpopulation of tumor cells. The fraction of ErbB4-positive and EPO-R–positive cells varied from 6% to 20%, whereas the fraction of ErbB3-positive and IGFI-R–positive cells ranged from 21% to 60%. Moreover, DiSano et al. [109] found heterogeneous cytoplasmic and nuclear staining after IHC with an anti-p16 antibody in one spitzoid melanoma tumor. Heterogeneous expression of transcription factors, i.e., NF-κB and JunB, was observed as well [128].
The existence of two subpopulations, i.e., Nodal-high and Nodal-low expressing cells, within melanoma metastases was detected by Seftor et al. [148]. Therefore, fluorescence-activated cell sorting (FACS) was used to separate these subpopulations from melanoma cell lines and compare their features. They observed that significantly more tumor colonies were formed in soft agar by Nodal-high expressing cells than Nodal-low. Hence, they suggested the importance of the Nodal pathway in tumor forming.
Intravital imaging of signaling reporter cell lines was performed by Manning et al. [149] to examine intratumor heterogeneity of Notch and serum response factor (SRF) signaling pathways. Only a few Notch-active cells or SRF-active cells were observed within tumors. SRF-active cells were scattered within tumors in small groups or as individual positive cells. Moreover, only an average of 6.6% of cells (ranged from 0% to 22% per field of view) was motile. The authors established that tumor motility is associated with higher transcriptional activity of both Notch and SRF signaling pathways. Furthermore, IHC staining revealed that the expression of EZH2, i.e., regulator of higher Notch and SRF activity, was heterogeneous in 27 human melanomas as well as 2 melanoma mouse models. Expression was the highest in cells with the lowest levels of pigmentation in invasive tumor margins.
Cintra Lopes Carapeto et al. [85] performed IHC to assess heterogeneity of marker expression in four different areas within each of 49 acral lentiginous melanoma tumors and 60 compound nevi. The most heterogeneous protein was MYC which was heterogeneous in 75% of tumors. PTEN expression was heterogeneous in 71% of tumors, SCF in 53%, KIT in 47%, cyclin D1 37%, and BRAF in 53% of tumors (Table 1). Moreover, heterogeneity of expression was observed in nevi, however, in a lower degree than in melanomas.
Recently, monoclonal antibodies inhibiting the interaction of programmed death protein 1/programmed death-ligand 1 (PD-1/PD-L1), such as nivolumab or pembrolizumab, have been considered as promising drugs, especially for melanoma [150]. Because of this, studies conducted by Madore et al. [151] are particularly important. They used IHC to investigate intratumor and intertumor (described in the subsection “Intertumor: Heterogeneity of Gene Expression”) heterogeneity of PD-L1 expression in 43 primary and 96 metastatic melanoma tumors from 58 patients. Of these tumors, 71 (51%) contained PD-L1–positive cells. The percentage of PD-L1–expressing cells as well as the intensity of staining was highly heterogeneous. The median fraction of PD-L1–positive cells among all tumors was 1%, whereas it was 8% considering only samples with PD-L1 expression. Most commonly, small populations of cells expressing PD-L1 within tumors were observed, especially in peripheral compartments and in close association with tumor-infiltrating lymphocytes. Moreover, PD-L1 expression was associated with a higher tumor-infiltrating lymphocytes grade, whereas no correlation with other prognostic factors was established. Similarly, tumor heterogeneity of PD-L1 expression was observed by Sunshine et al. [152].
Kuzbicki et al. [153] examined 126 melanoma tumors. They performed IHC staining to assess whether the level of cyclooxygenase-2 (COX-2, also known as prostaglandin-endoperoxide synthase 2) expression may serve as a melanoma prognostic marker. They found significant correlation between a high level of COX-2 expression in primary lesions and shorter survival as well as other prognostic factors, such as tumor thickness, ulceration, or more invasive histologic subtype. Moreover, considerable intratumor heterogeneity of COX-2 expression was found. The fraction of COX-2–positive cells varied from about 65% in primary tumors to over 95% in metastases. In primary tumors, they observed a slightly higher expression in peripheral areas compared with the central regions (about 70% and 65% of COX-2-positive cells).
Likewise, Botti et al. [154] observed heterogeneous expression of both PD-L1 and COX-2 in primary tumors and lymph node metastases. Furthermore, they reported significant correlation between expression of COX-2 and PD-L1, and observed coexpression of these proteins.
Heterogeneous Expression of Melanoma Markers
Sigalotti et al. [121] used 14 single-cell clones generated from the primary culture of melanoma tumors and performed reverse transcription PCR and electrophoresis to investigate intratumor heterogeneity of cancer/testis antigens (CTAs) expression. They found that MAGE (melanoma-associated antigen)-A1, MAGE-A2, MAGE-A3, MAGE-A4, MAGE-A6, GAGE (G antigen) 1-6, SSX (synovial sarcoma X) 1-5, and PRAME (preferentially expressed antigen in melanoma) were heterogeneously expressed. Of all antigens, MAGE-A3 displayed the most substantial heterogeneity with undetectable expression in five clones (36%), weak in six (43%), and very strong in three clones (21%). Moreover, one clone expressed 130 times the amount of MAGE-A3 mRNA expressed by the other. Likewise, heterogeneity of MAGE-C1/CT7 and MAGE-C2/CT10 expression in 54 mucosal melanoma tumors was found by Curioni-Fontecedro et al. [155]. However, they used IHC and thus investigated heterogeneity on the level of proteome, whereas previously discussed research examined heterogeneity on the level of transcriptome.
Widmer et al. [156] observed heterogeneity of MLANA (protein melan-A, also known as melanoma antigen recognized by T cells 1) expression in primary melanoma. Moreover, GLUT1 (glucose transporter 1) expression was heterogeneous within the tumor, i.e., was upregulated with increasing distance from the blood vessels within the tumor. Furthermore, they established that intratumor heterogeneity may be induced by location and microenvironmental factors, among others, by hypoxia. In this case, switching from proliferative to invasive tumor cell phenotype was triggered in the HIF1a (hypoxia-inducible factor 1a)-dependent mechanism. Lenggenhager et al. [157] observed heterogeneous expression of TRP-2, i.e., membrane-bound melanosomal enzyme connected with melanin biosynthesis and thereby with the melanoma differentiation marker. Interestingly, they established that TRP-2-negative cells were a highly aggressive subpopulation. Moreover, Oiso et al. [158] presented one case of amelanotic vulvar melanoma with highly intratumor heterogeneity of melanoma markers expression revealed by IHC.
CSCs and Heterogeneous Expression of CDs
Intratumor heterogeneity of gene expression has been shown by many scientists who have investigated the melanoma CSC model. This model assumes existence of a small tumor cell subpopulation within tumor with characteristic features, among others, the ability to self-renew and to differentiate into all lineages of tumor cells [159]. The CSC model was first described for hematologic cancers. Further, it was extended for many solid cancers. However, the CSC model in case of melanoma is still controversial [160]. Herein, we focus on tumor heterogeneity described during CSC research and the not CSC model itself. We refer to several papers in which controversies related with CSCs in melanoma are discussed more precisely [160], [161], [162], [163], [164].
The subpopulation of tumor cells expressing ABC transporters, which provide exclusion of Hoechst dye and anticancer drugs, as well as those with ALDH and JARID1B expression was established as putative melanoma stem cells. Kuzbicki et al. [165] used IHC to investigate the expression of JARID1B in 30 nevi, 27 primary melanomas, 4 metastases, and 1 local recurrence. In all primary and metastatic tumors, expression was detected. About 70% and 75% of tumor cells were JARID1B positive in primary and metastatic tumors, respectively. Therefore, no significant differences in the percentage of JARID1B+ cells between primary and metastatic tumors were observed. Furthermore, expression in melanomas was significantly higher compared with nevi.
Boiko et al. [166] used FACS and found that CD271, also known as NGFR (nerve growth factor receptor), was heterogeneously expressed in 9 (90%) out of 10 melanomas analyzed. The percentage of CD271-positive cells ranged from 2.5% to 41.0% (mean = 16.7%). Moreover, they established that CD271 was expressed differently in xenografted tumors (into Rag2−/− γc−/− mice) as well, and the percentage of positive cells ranged from 6.4% to 75.3% (mean = 26.3%). Furthermore, they showed that CD271+ cells were capable of tumor induction (were tumorigenic) and reestablished the original CD271 expression heterogeneity. Therefore, CD271+ cells have stem cells properties. Civenni et al. [167] confirmed that CD271+ cells are tumorigenic and are able to reestablish tumor heterogeneity. However, they noted a lower percentage of CD271+ cells within tumors. They did not detect CD271 expression in 31 (57%) primary melanoma biopsies; in 14 (26%) biopsies, they found that less than 5% of tumor cells were CD271+, whereas in 9 (17%), more than 5% of tumor cells were CD271+. Conversely, Quintana et al. [168] did not found differences in tumorigenicity between CD271+ and CD271− cells. Therefore, it was established that different conclusions obtained by researchers resulted from different methods of tumor cells preparation, especially during enzymatic dissociation of tumor tissue [164]. CD271 was shown to be a good marker of desmoplastic melanoma [169], [170]. High percentage of CD271-positive cells within tumors [169] contradicts the CSC model, according to which CSCs represent a very small percentage of the population. Moreover, due to the fact that expression of CD271 is upregulated by inflammation [171], it undermines the role of a reliable marker of CSCs. Using FACS, Held et al. [172] distinguished three subpopulations in each short-term tumor cell culture, i.e., CD34+p75−, CD34−p75−, and CD34−p75+. Moreover, they reported that the tumor biopsies showed a similar fraction of these subpopulations. Furthermore, reestablishing tumor heterogeneity by CD34−p57− subpopulation was shown. Markers of tumor-initiating cells, i.e., subpopulation with the capability to seed new tumors but unable to reestablish tumor heterogeneity, were sought by Schatton et al. [173]. They identified a subpopulation of cells expressing ABCB5 which were enriched for tumor-initiating cells. The level of ABCB5 expression was assessed by IHC. In seven freshly derived melanoma biopsies, the frequency of ABCB5-positive tumor cells ranged from 1.6% to 20.4% (mean = 10.1%). Moreover, the expression of nestin/NES (on average 28.7% of tumor cells were positive), TIE1 (22.9%), CD31 (0.7%), BMPRCD144 (0.5%), and CD20 (0.4%) was heterogeneous as well. Furthermore, flow cytometry and IHC of ABCB5+ population-derived primary and secondary xenografts in nude mice showed that ABCB5+ tumor cells reestablished parental tumor heterogeneity, i.e., ABCB5+ population-derived tumors contained ABCB5+ cells as well as ABCB5−. Using FACS, Lai et al. [174] assessed that, in primary and metastatic melanoma cell lines, the subset of CD133+ cells ranged from 0.2% to 76.3%. However, Moznani et al. [175] found in fresh surgical biopsies from seven tumors that the percentage of CD133+ cells was less than 1% (mean = 0.43%). Therefore, the percentage of CD133+ cells strictly depends on the conditions, i.e., in vitro or in vivo. Furthermore, the tumor-initiating properties of the CD133+ cells were shown. Similarly to the previous studies, Fang et al. [176] found small subpopulations of tumor cells with stem cell properties in patient-derived metastatic melanoma cultures.
Moreover, heterogeneity of A2B5, ABCB5, CD34, CD44, CD49B, CD49D, CD49f, CD54, CD133, CD166, GD2, gp100, L1CAM, MCAM, Melan-A, HNK-1, Nestin, and Oct3/4 expression in primary melanoma or patient-derived melanoma cell lines was observed [168], [177], [178].
Side Population (SP)
Luo et al. [179] examined SP cells, i.e., cells having stem cell–like properties and expressing ABC transporters, in melanoma. In all three primary and five metastatic melanoma tumors, SPs were detected. The fraction of SP cells was small, ranging from 0.13% to 0.70% (mean = 0.33%). Furthermore, microarray analysis was performed to compare expression levels of all 48 ABC transporters between SP and other tumor cells. SP cells expressed 13 times more ABCB1 and ABCB5 than cells from non-SP main population (MP). Upregulation of expression of these transporters was confirmed by quantitative reverse transcriptase PCR and flow cytometric analysis. Likewise, SP cells in melanoma were investigated by Wouters et al. [138]. They detected SP in all 38 examined melanoma tumors. Similarly to previous research, the fraction of SP cells ranged from 0.1% to 2.2% of tumor cells (mean = 0.4%). Moreover, they established that SP had higher tumorigenic activity than MP and could reestablish parental tumor heterogeneity. Whole-genome expression of SP and MP from four primary melanomas was compared using microarray. They detected 462 different expressing genes between SP and MP (≥1.5 times up or down). Interestingly, expression of antiapoptotic factors was significantly higher in SP compared with MP cells
Epigenetic Heterogeneity
Epigenetic regulation is the highest level of the intracellular regulation system. Its effects on pathogenesis, progression, and resistance to treatment of melanoma are becoming better understood; nonetheless, many issues remain unclear [180], [181]. Heterogeneity of tumor cells may also be present on epigenetic level (Figure 1F). To have a comprehensive view of the differences presented here, the following components of epigenetics need to be considered: chromatin structure and histone modification, in particular acetylation, DNA, especially promoter sequence, methylation, X-chromosome inactivation and noncoding RNA, i.e., microRNA and long noncoding RNAs, regulation. Epigenetic dysregulation results from a primary lesion on a lower level of genetic organization (genomic/transcriptomic) ,or it can be solely a causative change. However, miRNA may diffuse to surrounding cells and tissues and, by doing so, may affect distant locations. This phenomenon transfers the changes from the intracellular level even up to the circulatory level. Nevertheless, epigenetic heterogeneity remains poorly explored and therefore requires further intensive research.
Sigalotti et al. [121] showed that methylation of CTA promoters generates intratumor heterogeneity of CTA expression. DNA methylation analyses were performed for three single-cell clones from metastatic tumor characterized by the highest levels and the lowest levels of MAGE-A3 expression as well as undetectable MAGE-A3 expression. They found that methylation of CpG dinucleotides located in an MAGE-A3 promoter was substantially higher in clones which did not express MAGE-A3 when compared with clones which expressed MAGE-A3. Moreover, CpG methylation at some positions, which was common in clones with low expression, was rare in those with high expression. Furthermore, they showed that DNA hypomethylating agent 5-aza-2′-deoxycytidine (5-AZA-dCyd) induced CTA expression in all negative clones. They believed that using 5-AZA-dCyd may provide the possibility to overcome of the limitations in CTA-based vaccine therapies caused by intratumor heterogeneity. Rastetter et al. [182] investigated the methylation state of tumor suppressor gene promoters in 15 primary and 19 metastatic tumors. Using methylation-specific PCR, they showed that 70% (16 out of 23) of cases exhibited an intratumoral heterogeneous methylation pattern in at least one promoter (17%, 20%, 33%, 40%, and 45% for RASSF1A, MGMT, DAPK, Rb, and p16 respectively). Methylation of tumor suppressor gene promoters led to accelerated progression of tumor cells; therefore, the intratumor heterogeneity of methylation pattern may directly enhance differences among clones within the tumor.
Intertumor Heterogeneity in Melanoma
It is known that the main cause of intertumor (intrapatient) heterogeneity (Figure 1B) is intratumor heterogeneity (Figure 1A) of the primary tumor [102], [183], [184]. To be precise, heterogeneity of circulating tumor cells (CTCs) or CTC clusters. Metastatic tumors may arise from different, i.e., dominant or minor, subpopulations within tumors. Another cause of intertumor heterogeneity is the constant evolution of tumors. The acquisition of novel mutations during tumor progression results in branching of the phylogenetic tree. Therefore, heterogeneity between primary and metastatic tumors, and among multiple metastases, i.e., intermetastases heterogeneity, may be detected.
Genetic Heterogeneity
Different mutations among primary tumor and metastases, or multiple metastatic tumors, are commonly observed. These discrepancies constitute intertumor genetic heterogeneity.
Mutations in Cancer-Related Genes
In a similar manner, as with intratumor heterogeneity, we performed a systematic review of intertumor heterogeneity of BRAF mutation status (Table 2). Considering all tumors examined by various methods, 15.5% (229 of 1,477) exhibited discordant BRAF status. Moreover, as expected, the rate of discordance in the BRAF genotype increases with the number of tumors, i.e., was 8% (2/25) in patients with one primary tumor and one metastasis, 18% (6/33) in patients with two metastases, and 20% (2/10) in patients with three metastases [185]. The meta-analysis showed that intertumor heterogeneity of BRAF mutations exists and has clinical relevance [186]. Nevertheless, intertumor heterogeneity of BRAF mutations still remains controversial, and some researchers claim that the majority of discordances are due to false-positive or false-negative results as well as contamination with non-tumor cells [75].
Table 2.
Intertumor Heterogeneity of BRAF Mutation Status
| Ref. | Samples | Methods | Number (%) of Tumors or Pairs with Different BRAF Genotype |
|---|---|---|---|
| [187] | 2 MT | qPCR | MT-MT 2/2 (100%) |
| [188] | 114 PT 86 MT |
Direct seq | PT-MT 7/24 (29%) MT-MT 2/2 (100%) |
| [89] | 124 PT 76 MT |
IHC | 63/135 (47%) |
| Direct seq | 4/135 (3%) | ||
| [96] | 40 PT 37 MT |
Sequenom MassARRAY | 9/17 (53%) |
| [189] | 59 PT 68 MT |
Direct seq | 5/13 (38%) |
| [93] | 18 PT 94 MT |
Direct seq, MS-PCR | PT-MT 8/18 (44%) |
| MT-MT 5/19 (26%) | |||
| [88] | 100 PT 37 MT |
PNA qPCR, IHC, NGS Cap Seq | 5/15 (33%) |
| [190] | 40 PT 34 MT |
Southern blot | 11/34 (32%) |
| IHC | 11/40 (28%) | ||
| [188] | 24 PT 24 MT |
Direct seq | 5/24 (21%) |
| [191] | 44 PT 194 MT |
Pyro, Cap seq | 10/53 (19%) |
| [192] | 25 PT 25 MT |
qPCR, ARMS | PT-MT 4/25 (16%) MT-MT 2/6 (33%) |
| [91] | 140 PT 171 MT |
qPCR | 23/140 (16%) |
| IHC | 1/140 (1%) | ||
| [61] | 102 PT 165 MT |
Direct seq | 15/99 (15%) |
| [193] | 48 PT 48 MT |
Pyro | 7/48 (15%) |
| IHC | 3/48 (6%) | ||
| [185] | 74 PT 89 MT |
Direct seq | 10/74 (14%) |
| [183] | 25 PT 50 MT |
NGS | 3/25 (12%) |
| [90] | 22 MIS 56 PT 93 MT |
Pyro, IHC | 2/30 (7%) |
| [86] | 50 PT 139 MT |
HRM, qPCR | 3/54 (6%) |
| IHC | 0/54 (0%) | ||
| [92] | 75 PT 88 MT |
NGS | 4/75 (5%) |
| [65] | 70 PT 88 MT |
SSCP | 3/71 (4%) |
| [194] | 39 PT 255 MT |
Pyro | 2/57 (4%) |
| Overall | 229/1477 (15.5%) | ||
Abbreviations: PT, primary tumors; MT, metastasis tumors; NT, undefined tumors; qPCR, quantitative (real-time) PCR; MIS, melanoma in situ; IHC, immunohistochemistry; Direct seq, direct sequencing; MS-PCR, mutant-specific PCR; PNA qPCR, peptide nucleic acid real-time PCR; NGS, next-generation sequencing; Cap seq, capillary sequencing; Pyro, pyrosequencing; HRM, high-resolution melting; ARMS, amplification refractory mutation system; SSCP, single-strand conformation polymorphism.
Egberts et al. [195] investigated the mutational status of the BRAF and NRAS gene as well as TERT-promoter in multiple primary melanomas. Intertumor heterogeneity between first and subsequent primary tumors given all three genes was noted in 59 (61%) out of 96 patients. Only 24% of patients exhibited intertumor homogeneity given all three genes.
Lin et al. [76] compared BRAF MAF in three primary and matched metastatic or recurrence tumors. In two pairs, an increase in BRAF mutant allele frequency in metastatic or recurrence tumor compared to primary tumor was observed. Moreover, a small fraction of BRAF V600K mutant allele was detected after PCR, subcloning, and sequencing in primary tumors which was classified as BRAF wild type by conventional sequencing. In seven (78%) out of nine corresponding metastases, the frequency of BRAF mutant allele was substantially higher. Therefore, this suggests selection of the BRAF mutant allele during melanoma progression. Likewise, intertumor heterogeneity of BRAF mutation was observed by Sakaizawa et al. [196].
Vivancos et al. [99] confirmed that tumor cells acquired mutations during progression. From nine examined mutations which were present in recurrent tumor, six were present in primary tumor, whereas only two were present in nevus of Ota. Moreover, MAFs varied among the tumors. Harbst et al. [197] examined 27 patients with multiple metastatic melanoma tumors. In 22 (81%) patients, private mutations and copy number alterations, i.e., specific to only one tumor within the patient, were detected by NGS. Moreover, they established that BRAF and NRAS mutations, if present in the first metastasis, were preserved in subsequent tumors, whereas some mutations, like PTEN, were acquired during cancer progression. Houben et al. [188] found discrepancies in the NRAS genotype between primary and matched metastatic tumors in 2 (8%) out of 24 patients. Similarly, Colombino et al. [61] noted discordant NRAS status among matched tumors in 7 (7%) out of 99 patients. Moreover, in 11 (69%) out of 16 patients with p16 CDKN2A (cyclin-dependent kinase Inhibitor 2A) mutations, discordancy among tumors was detected. It is worth noting that, in all these patients, only metastatic tumor cells harbored p16 CDKN2A mutations, i.e., tumor suppressor protein mutations, whereas primary tumors were wild type. Additional mutations in TP53 and p16 CDKN2A genes in lymph node metastases when compared with the paired primary tumor in 3 (12%) out of 25 patients were noted by Casula et al. [183] as well.
Goswami et al. [198] investigated 265 pairs of primary and metastatic tumors, including 20 melanomas. Interestingly, they observed mutations present exclusively in primary tumors, whereas other studies showed additional mutations mainly in metastases. For instance, one primary tumor harbored KIT mutation which was absent in paired metastasis, and the other primary tumor harbored KRAS mutation which was absent in metastasis. This was due to the acquisition of mutations by primary tumor after dissemination to metastases.
Hugo et al. [199] found that melanoma tumors acquired MAPKi (mitogen-activated protein kinases inhibitor) resistance in different ways, including genetic alterations. Seven (88%) out of eight patients receiving BRAFi (BRAF inhibitor), who had multiple tumors examined, exhibited at least one discrepancy in genes conferring resistance to MAPKi among tumors. For instance, in one patient, two (22%) out of nine tumors exhibited copy number gain of the BRAF gene, whereas three (33%) exhibited SNVs or indels in the KRAS gene. Likewise, three (50%) out of six patients receiving BRAFi+MEKi (BRAF inhibitor and MEK inhibitor combination) with multiple tumors exhibited at least one discrepancy. For instance, one patient had three (43%) out of seven tumors with BRAF copy number gain and copy number loss of p16 CDKN2A, three (43%) BRAF wild-type tumors while exhibiting however copy number loss of p16 CDKN2A, and one (14%) tumor wild type for both genes.
CTCs enable noninvasive monitoring of tumor heterogeneity and plasticity over time. Therefore, Fusi et al. [200] established a method for the analysis of BRAF mutation in circulating melanoma cells. In 2 (10%) out of 21 patients, discordancy of the BRAF genotype between bulk tumors and CTCs from peripheral blood samples was observed. In one patient, two (40%) out of five CTCs were BRAF mutated, whereas the tissue sample was BRAF wild type. All CTCs from another patient were BRAF mutated, whereas the tissue sample was BRAF wild type. Likewise, heterogeneity of CTCs was observed by Sakaizawa et al. [196].
SNVs and CNVs within Genome
Reuben et al. [201] sequenced whole exome of 33 metastatic tumors from 15 patients. They found that the percentage of intertumor heterogeneous nonsynonymous mutations was 37%, 52%, and 43% for multiple metastases obtained from targeted therapy, checkpoint blockade, and treatment-naïve patients, respectively. Furthermore, they showed that the large percentage of neoantigens were unique for one metastasis, i.e., an average of 29% neoantigens for patients receiving targeted treatment, 40% for patients receiving immune checkpoint blockade, and 37% for treatment-naïve patients. Moreover, they established that intrapatient intertumor heterogeneity was noticeable; however, it was not as pronounced as interpatient heterogeneity (Figure 1C). Sanborn et al. [102] confirmed that the main cause of intertumor heterogeneity is intratumor heterogeneity. WES in three (37.5%) out of eight patients revealed at least two distinct parental subpopulations in primary tumors shared only with some metastases. Therefore, they reported that these metastases arose from different subpopulations within the primary tumors, not evolved from other metastases. In one patient, sequencing revealed that at least two SNVs were present subclonally within the primary tumor, were absent in locoregional metastasis, and were present in two others metastases, i.e., lymph node and distant skin metastasis. Conversely, at least two SNVs were present subclonally within the primary tumor as well as in locoregional metastasis, although they were absent in two other metastases. Therefore, they concluded that locoregional metastasis arose from one subpopulation within primary tumor, whereas lymph node and distant skin metastases arose from another. Furthermore, identical CNV was observed in the subclonal level in two locoregional metastases within one patient. Thus, each of these tumors had to be formed by at least two distinct tumor cell populations.
Ding et al. [101] compared data obtained from WGS from two pairs of metastatic tumors from two patients. One pair exhibited almost identical profiles of rearrangements and CNVs, whereas in the other pair, some mutations were present in one tumor but were absent in the other, i.e., were intertumor heterogeneous. Despite these differences, further analysis revealed that both tumors arose from the same clone in the primary tumor.
Turajlic et al. [104] compared MAF of 44 genes between primary and metastasis acral melanomas. In both tumors, 39 SNVs were confirmed. For 16 (41%) SNVs, the MAF differed by less than 10% between primary and metastatic tumors, although for 8 (21%) SNVs, the MAF differed by more than 20%. Moreover, for 23 (59%) SNVs, the MAF was higher in metastasis compared with primary tumor. Furthermore, 27 and 32 short insertions and deletions (indels) were identified as somatic in the coding regions in primary and metastatic tumors, respectively. Only one (2%) indel was common to both primary and metastatic tumors, whereas others (98%) were exclusive for primary tumors or metastases and thus were heterogeneous. Furthermore, they found 57 and 71 somatic structural variations (SVs) in the primary tumor and metastasis, respectively. Of them, 55 (75%) were present in both tumors, whereas others (25%) exhibited intertumor heterogeneity
LOH
Katona et al. [202] performed PCR amplification of five microsatellite markers and gel electrophoresis to find the heterogeneous loci of LOH. In 3 (23%) out of 13 patients, the primary and all metastatic tumors shared the same LOH pattern. In 10 (77%) patients, there were discrepancies between the primary and corresponding metastatic tumors or among multiple metastases (intermetastases). Moreover, in six (46%) patients, metastases showed LOH at a greater number when compared with the matched primary tumor. Therefore, they suggested that some LOH may be involved in the acquisition of metastatic potential. Nakayama et al. [107] found that 6 (8%) out of 79 in-transit melanomas exhibited different LOH status in at least 1 marker compared with other metastases from the same patient. Moreover, they examined 10 corresponding primary tumors and found that 5 (50%) patients exhibited additional LOH in metastases. Furthermore, Swoboda et al. [184] found deletions exclusively in the primary melanoma cell line or in the matched metastatic melanoma cell line.
Takata et al. [111] investigated 15 metastases from 8 patients apart from 10 primary tumors as described in the subsection “Intratumor: Genetic Heterogeneity.” For five patients, more than one tumor was examined. Four (80%) patients exhibited the same pattern of LOH in all metastatic tumors. Moreover, the pattern of LOH in metastases was identical to those in all tumor cells or in the subclones within the primary tumor. Nevertheless, one (20%) patient showed intertumor heterogeneity between two metastases, and three (60%) patients exhibited a discordant pattern of LOH between the primary and metastatic tumors.
Bahrami et al. [203] analyzed LOH at eight loci, i.e., 1p, 6q, 10q, 11q, 18q, and three loci in 9p, as well as X-chromosome inactivation in nine primary melanoma tumors and corresponding metastases. Discrepancies between primary and metastatic tumors as well as among multiple metastases were observed. The most heterogeneous locus were 9p, i.e., D9S171, [discordant status in seven (78%) out of nine patients] and 6q, i.e., D6S305 [discordant status in six (75%) out of eight patients]. The number of discordant loci among tumors within patients ranged from 0 to 6 (mean = 3).
Uguen et al. [110] used FISH targeting chromosomes 6, 8, 9, and 11 to characterize 10 paired primary and metastatic tumors. They found intertumor heterogeneity in at least 1 marker in 9 (90%) out of 10 patients. One (10%) patient exhibited heterogeneity among primary and metastatic tumor as well as among metastases.
Besides intratumor heterogeneity, Gandolfi et al. [67] investigated intertumor heterogeneity of one primary and two corresponding metastatic tumors. They found that only a subpopulation of cells within the primary tumor exhibited deletion of chromosome 9 (intratumor heterogeneity). Two distant metastases showed the same alterations as the primary tumor, except the chromosome 9 deletion. Therefore, it is probable that these metastases derived from a population of cells with normal chromosome 9 status. Moreover, they did not find any additional alterations compared to the primary tumor. In contrast to the previously discussed studies, they suggested that the majority of genetic alterations are already present within the primary tumor at the time of metastasis spread.
Heterogeneity of Genes’ Expression
Intertumor heterogeneity of BRAF mutant expression was revealed by IHC which is presented in Table 2.
Haqq et al. [143] found that gene expression may vary significantly between the primary tumor and corresponding metastasis. Likewise, Reuben et al. [201] found a significant intermetastases heterogeneity in all 15 patients studied. They observed heterogeneity of expression of immune-related genes, i.e., cytokines, chemokines, human leukocyte antigen (HLA) molecules, adhesion molecules as well as interferon (IFN) pathway genes. Moreover, in more than half of the patients, intertumor heterogeneity of PD-L1 expression was observed [151].
Harbst et al. [197] found that, in half of the examined patients (14 out of 28), multiple metastases belonged to different molecular subtypes of a gene expression. Interestingly, Hugo et al. [199] observed significant intertumor heterogeneity of mechanisms leading to resistance to MAPKi through alterations in gene expression, i.e., underexpression or overexpression.
Kuzbicki et al. [165] found that the expression levels of JARID1B were similar in matched primary and metastatic tumors, and primary and recurrent tumors, though only a small sample was investigated, i.e., two cases with paired primary and metastatic tumors and one case with primary and recurrent tumors. Therefore, their results have little statistical significance. Furthermore, intertumor heterogeneity of COX-2 expression was investigated by Kuzbicki et al. [153]. They showed that for all 39 cases of primary and matched metastatic tumors, the percentage of COX-2–positive cells was similar or higher in the primary tumor.
Epigenetic Heterogeneity
Different methylation patterns between paired cell lines were investigated by Chatterjee et al. [204]. Reduced representation bisulfite sequencing was used to compare genome-wide DNA methylation patterns in three cutaneous primary and metastatic melanoma cell line pairs. They found 22,745, 7220, and 7520 differentially methylated fragments, i.e., fragments with at least 25% mean methylation difference, between the primary and corresponding metastatic melanoma cell lines. Generally, they established that primary cell lines were hypermethylated compared with the matched metastatic melanoma cell lines. Rastetter et al. [182] found that all four cases with multiple metastases exhibited a heterogenous methylation pattern between tumors. They suggested that differences between metastases are due to their origin from a distinct subpopulation within the primary tumor. Therefore, the main cause of intertumor epigenetic heterogeneity is intratumor heterogeneity. Katona et al. [202] found that 4 (31%) out of 13 patients had a discordant pattern of X-chromosome inactivation between the primary and matched metastatic tumors. These results were similar to those obtained by Bahrami et al. [203], who observed intratumor heterogeneity of X-chromosome inactivation in one (25%) out of four patients.
Similarly to previous results, Hugo et al. [199] found that multiple tumors within patient may exhibit different epigenetic alterations leading to acquisition of MAPKi resistance.
Implications for Diagnosis and Treatment
The main consequences of melanoma tumor heterogeneity relevant from a clinical point of view are the obstacles to diagnosis and treatment.
Diagnosis: Difficult Detection of Small Subclones Harboring Rare Mutations
Characterization of an entire spectrum of tumor cells, commonly exhibiting variable, distinct features, is necessary since all tumor cells have to be killed for successful therapy [156]. Tumor heterogeneity is the main obstruction for personalized cancer medicine [103], [104]. Therefore, it is necessary to perform an analysis of multiple biopsies within all tumors with very high accuracy to detect rare mutations present commonly in small subclones. Due to intratumor heterogeneity, a single biopsy may be insufficient for the evaluation of the alterations landscape of the whole tumor [15], [33], [67], [94], [103], [108], [205]. For instance, Lake et al. [113] established that biopsies of an extraocular, thereby more accessible, part of UM may not represent a whole tumor spectrum of mutations.
Bulk analysis leads to the detection of dominant clones, although this may omit minor subpopulations, for instance, SP or CSC populations, which are often crucial for tumor progression and the effectiveness of therapy. Moreover, it is suggested that the quantitative evaluation of mutations, for instance, BRAF mutations, is more reliable and useful than qualitative evaluation and therefore ought to be a standard procedure in mutational status defining [63]. At present, NGS seems to be the most accuracy, the cheapest, the fastest, and therefore the most appropriate method for cancer-relevant genes analyses, including BRAF, NRAS, and KIT. Due to very low frequency of many private mutations which are the basis of intratumor heterogeneity as well as tumor cell dispersal resulting from high motility of some clones [206], the increase of coverage, which enables decrease of detection threshold in NGS analyses, is required to ensure the highest accuracy [84], [206].
One of the implications of intertumor heterogeneity for diagnosis is the necessity to examine multiple biopsies from each tumor because one biopsy alone might not represent the entire mutational spectrum of all tumors. It has been shown for other types of cancer, e.g. clear cell renal cell carcinoma [207], that multisite biopsies are far more accurate and more advantageous than routine. Nonetheless, mutational screening restricted to only one biopsy is common in clinical practice, also in cases of melanoma [187]. Therefore, even though the patient could be qualified for treatment with, for instance, a specific inhibitor, the patient would not be qualified for treatment if the only examined sample would be wild type, i.e., would not harbor a mutant allele [185], [187], [191]. Saint-Jean et al. [185] found that in 5 (7%) out of 74 patients with at least 2 tumors analyzed, the second or third sample was BRAF mutated whereas the first or the first and second samples were BRAF wild type. Therefore, without repeated biopsies, these patients would not have been treated with BRAFi vemurafenib.
Interestingly, Helmbold et al. [114] showed that only 65% of measuring fields (127 out of 196) were found to represent their tumors reliably, i.e., contained all of the tumor cell stem lines. The rate of representative fields was even lower in a group of stem line heterogeneous tumors and equaled 32% (33 out of 102). Therefore, they established that measuring at least three fields per tumor was necessary to identify all stem lines within a tumor with a likelihood of 95%. Moreover, Saint-Jean et al. [185] highlighted the necessity of multiple biopsies during disease progression, especially in BRAF wild-type patients, apart from biopsies of all tumors. It was confirmed by Gray et al. [208] who detected the NRAS Q61 mutations which provide MAPKi resistance in cell-free ctDNA extracted from three (43%) out of seven patients with disease progression after initial response to dabrafenib/trametinib treatment. Therefore, further treatment would be ineffective. In biopsies obtained prior to MAPKi therapy from these patients, the NRAS mutations were not detected. Temporal heterogeneity, especially under intense selection during treatment, makes constant tumor monitoring necessary. However, the choice of therapy is often made on the basis of archived primary tumor material [209]. Our review of the literature shows that this clinical practice has to advance on the basis of novel research results.
Tumor heterogeneity studies are even more important since degree of heterogeneity of melanoma may serve as a prognostic marker. Higher heterogeneity is often associated with a poorer prognosis [12], [210]. Relation between cancer progression and the degree of heterogeneity was first shown for progression of Barrett's esophagus to esophageal adenocarcinoma [31]. Association between tumor heterogeneity and cancer progression as well as prognosis is poorly explored in melanoma. However, it was shown that heterogeneity of protein marker expression is a more powerful indicator of malignancy in acral lentiginous melanomas than protein marker expression alone [85]. Nonetheless, further studies are necessary.
Recently, the characterization of CTCs and cell-free ctDNA, i.e., “liquid biopsy” [211], seems to be a promising approach to noninvasively monitor the progression of cancer at the whole-body level, especially during and after therapy, without neglecting or underestimation of tumor heterogeneity [196], [208], [212], [213], [214]. Moreover, liquid biopsies enable the disease monitoring when biopsies are not available and the earlier prediction of cancer progression, response to treatment, or relapse after its than radiological diagnostics [98]. Further research and development of liquid biopsies analysis methods are required for their improvement and dissemination in clinical practice.
Treatment: Heterogeneity of Treatment Targets and Existence of Drug-Resistant Populations
Tumor heterogeneity becomes even more relevant, especially from a clinical point of view, if it concerns genes or their products, which provide a new advantage and increase tumor cell fitness [103], or if it concerns treatment targets. It is known that genetic heterogeneity as well as nongenetic acquired heterogeneity induced by microenvironmental factors may lead to treatment resistance [156]. Drugs that target one pathway may induce the emergence of another clone harboring a mutation in member of alternative pathway providing further tumor progression [144]. These drugs may also cause changes in the clonal composition of the tumor leading to the dominance of drugs-resistance clones [102]. Intertumor heterogeneity, i.e., tumor-specific mutations and alternating mutant allele frequency, may lead to differences in treatment responses among multiple tumors within a patient [101]. Reuben et al. [201] established that heterogeneous response to targeted therapy and immune checkpoint blockade is observed in the majority of patients. Interestingly, in 13 (22%) patients, the differences in rate of tumor growth between metastatic tumors were greater than 50%.
Since tumor growth probably depends on a small fraction of tumor cells, the elimination of this crucial subpopulation would eradicate tumors more effectively than targeting all tumor cells [215]. Schmidt et al. [215] showed that target elimination of a small subpopulation of tumor cells, i.e., CD20+ cells which represented less than 2% of the tumor cells, causes effective eradication of melanoma tumors. Furthermore, phenotypes present in only a minority of cells common are crucial for survival and regrowth during or after therapy [103]. The existence of multiple drug-resistant subpopulations of cells within tumors remains a major challenge for future therapy approaches. Schatton et al. [173] showed that anti-ABCB5 monoclonal antibody treatment, i.e., targeting one of the CSCs markers, inhibited tumor growth and formation in mice.
Tumor clonal composition is changing during treatment. It is due to heterogeneity of therapy. Moreover, it results from different response to treatment of each clone within tumor [98]. The most pronounced changes occur after therapy with monoclonal antibodies and small molecule inhibitors. After anti-ABCB5 treatment, the percentage of ABCB5+ decreased from, on average, 10% to less than 1% [173]. It was found that after BRAFi treatment, BRAF expression decreases significantly and becomes more heterogeneous [81]. Moreover, in tumors treated with vemurafenib (BRAFi) or dabrafenib (BRAFi) and trametinib (RAF/MEKi), the fraction of AXL-high cells is higher compared to the pretreatment state [128]. Gremel et al. [98] noted that the KIT MAF in cell-free ctDNA was highly variable during treatment with different agents, i.e., increased to about 15% during dacarbazine treatment, further decreased to almost zero after imatinib (tyrosine kinase inhibitor), increased to 30% after ipilimumab (anti–CTLA-4), and finally achieved about 45% after pembrolizumab (anti-PD-1), which results from a different response of two clones to each of the therapy.
Intratumor and intertumor heterogeneity of the BRAF mutation may contribute to the range of initial responses [192] and cause decreased efficiency [200] and resistance to BRAF inhibitors [192]. Albeit, Riveiro-Falkenbach et al. [91] claimed that BRAF heterogeneity cannot cause BRAFi resistance. However, their views may be biased due to observed homogeneity of BRAF gene within all examined tumors examined by them (Table 1). It was shown that quantification of the BRAF mutated allele within tumors is a predictive marker for response to BRAF inhibitors [216]. A large BRAF mutation allele fraction is associated with the best response rate to BRAFi, such as vemurafenib. In this research, progression-free survival after vemurafenib (BRAFi) treatment was significantly higher in patients with high mutation levels (more than 50%) during the first 10 months of treatment, although it was lower thereafter. Helias-Rodzewicz et al. [63] found that two patients with high BRAF mutation levels had prolonged progression-free survival, thereby confirming previous studies. Moreover, it is suggested that generally patients with lower intratumor and intertumor heterogeneity are more likely to respond better to treatment [201].
Hugo et al. [199] observed a diversity of genetic and nongenetic alterations leading to the acquisition of MAPKi resistance. Moreover, they established that tumors within one patient may achieve resistance in different ways. Therefore, heterogeneity of acquired resistance, as well as heterogeneity present at the beginning of treatment, is an issue that must be taken into consideration in choosing the method of treatment and must be further explored. Interestingly, it was shown that the mechanisms of MAPKi resistance may be heterogeneous within one tumor as well as among tumors [217], [218]. Therefore, both intratumor and intertumor heterogeneity of resistance mechanisms is observed.
Conclusions and Future Directions
Melanoma is highly heterogeneous. Despite a considerable progress in tumor heterogeneity research, many issues remain unclear and, therefore, controversial. A comprehension of the mechanisms driving tumor heterogeneity and targeting them in new therapy approaches is required [144]. Moreover, understanding them would allow overcoming difficulties in diagnosis and treatment caused by heterogeneity. Besides investigating tumor heterogeneity, phylogenetic analyses, which enable more comprehensive view, are required to understand tumor biology [219].
In the era of personalized medicine, fast, cheap, accurate, and effective diagnostic methods are necessary. Both intratumor and intertumor heterogeneity must be considered in diagnosis and treatment planning. Moreover, the preclinical stages of therapy development must take into account tumor heterogeneity [123]. Knowing the clonal composition of each tumor is necessary to optimize therapy approaches [101]. In addition, finding a key population of tumor cells and targeting therapy against it would provide effective tumor eradication.
Conflict of Interest
All authors declare no conflict of interest. Research was supported by the grant from I Faculty of Medicine, Medical University of Warsaw: 1M15/NM3/16.
Compliance with Ethical Standards
The authors declare that they have no conflict of interest.
References
- 1.Dick JE. Stem cell concepts renew cancer research. Blood. 2008;112(13):4793–4807. doi: 10.1182/blood-2008-08-077941. [DOI] [PubMed] [Google Scholar]
- 2.Marusyk A, Polyak K. Tumor heterogeneity: causes and consequences. Biochim Biophys Acta. 2010;1805(1):105–117. doi: 10.1016/j.bbcan.2009.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Zhang J, Fujimoto J, Zhang J, Wedge DC, Song X, Zhang J, Seth S, Chow CW, Cao Y, Gumbs C. Intratumor heterogeneity in localized lung adenocarcinomas delineated by multiregion sequencing. Science. 2014;346(6206):256–259. doi: 10.1126/science.1256930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Nassar A, Radhakrishnan A, Cabrero IA, Cotsonis GA, Cohen C. Intratumoral heterogeneity of immunohistochemical marker expression in breast carcinoma: a tissue microarray-based study. Appl Immunohistochem Mol Morphol. 2010;18(5):433–441. doi: 10.1097/PAI.0b013e3181dddb20. [DOI] [PubMed] [Google Scholar]
- 5.Coons SW, Johnson PC, Shapiro JR. Cytogenetic and flow cytometry DNA analysis of regional heterogeneity in a low grade human glioma. Cancer Res. 1995;55(7):1569–1577. [PubMed] [Google Scholar]
- 6.Mora J, Cheung NK, Gerald WL. Genetic heterogeneity and clonal evolution in neuroblastoma. Br J Cancer. 2001;85(2):182–189. doi: 10.1054/bjoc.2001.1849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Okada Y, Hurwitz EE, Esposito JM, Brower MA, Nutt CL, Louis DN. Selection pressures of TP53 mutation and microenvironmental location influence epidermal growth factor receptor gene amplification in human glioblastomas. Cancer Res. 2003;63(2):413–416. [PubMed] [Google Scholar]
- 8.Snuderl M, Fazlollahi L, Le LP, Nitta M, Zhelyazkova BH, Davidson CJ, Akhavanfard S, Cahill DP, Aldape KD, Betensky RA. Mosaic amplification of multiple receptor tyrosine kinase genes in glioblastoma. Cancer Cell. 2011;20(6):810–817. doi: 10.1016/j.ccr.2011.11.005. [DOI] [PubMed] [Google Scholar]
- 9.Sottoriva A, Spiteri I, Piccirillo SG, Touloumis A, Collins VP, Marioni JC, Curtis C, Watts C, Tavare S. Intratumor heterogeneity in human glioblastoma reflects cancer evolutionary dynamics. Proc Natl Acad Sci U S A. 2013;110(10):4009–4014. doi: 10.1073/pnas.1219747110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Szerlip NJ, Pedraza A, Chakravarty D, Azim M, McGuire J, Fang Y, Ozawa T, Holland EC, Huse JT, Jhanwar S. Intratumoral heterogeneity of receptor tyrosine kinases EGFR and PDGFRA amplification in glioblastoma defines subpopulations with distinct growth factor response. Proc Natl Acad Sci U S A. 2012;109(8):3041–3046. doi: 10.1073/pnas.1114033109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Tirosh I, Venteicher AS, Hebert C, Escalante LE, Patel AP, Yizhak K, Fisher JM, Rodman C, Mount C, Filbin MG. Single-cell RNA-seq supports a developmental hierarchy in human oligodendroglioma. Nature. 2016;539(7628):309–313. doi: 10.1038/nature20123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Andor N, Graham TA, Jansen M, Xia LC, Aktipis CA, Petritsch C, Ji HP, Maley CC. Pan-cancer analysis of the extent and consequences of intratumor heterogeneity. Nat Med. 2016;22(1):105–113. doi: 10.1038/nm.3984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Aubele M, Mattis A, Zitzelsberger H, Walch A, Kremer M, Hutzler P, Hofler H, Werner M. Intratumoral heterogeneity in breast carcinoma revealed by laser-microdissection and comparative genomic hybridization. Cancer Genet Cytogenet. 1999;110(2):94–102. doi: 10.1016/s0165-4608(98)00205-2. [DOI] [PubMed] [Google Scholar]
- 14.Benetkiewicz M, Piotrowski A, Diaz De Stahl T, Jankowski M, Bala D, Hoffman J, Srutek E, Laskowski R, Zegarski W, Dumanski JP. Chromosome 22 array-CGH profiling of breast cancer delimited minimal common regions of genomic imbalances and revealed frequent intra-tumoral genetic heterogeneity. Int J Oncol. 2006;29(4):935–945. [PubMed] [Google Scholar]
- 15.Cottu PH, Asselah J, Lae M, Pierga JY, Dieras V, Mignot L, Sigal-Zafrani B, Vincent-Salomon A. Intratumoral heterogeneity of HER2/neu expression and its consequences for the management of advanced breast cancer. Ann Oncol. 2008;19(3):595–597. doi: 10.1093/annonc/mdn021. [DOI] [PubMed] [Google Scholar]
- 16.Fujii H, Marsh C, Cairns P, Sidransky D, Gabrielson E. Genetic divergence in the clonal evolution of breast cancer. Cancer Res. 1996;56(7):1493–1497. [PubMed] [Google Scholar]
- 17.Geyer FC, Weigelt B, Natrajan R, Lambros MB, de Biase D, Vatcheva R, Savage K, Mackay A, Ashworth A, Reis-Filho JS. Molecular analysis reveals a genetic basis for the phenotypic diversity of metaplastic breast carcinomas. J Pathol. 2010;220(5):562–573. doi: 10.1002/path.2675. [DOI] [PubMed] [Google Scholar]
- 18.Klein CA, Blankenstein TJ, Schmidt-Kittler O, Petronio M, Polzer B, Stoecklein NH, Riethmuller G. Genetic heterogeneity of single disseminated tumour cells in minimal residual cancer. Lancet. 2002;360(9334):683–689. doi: 10.1016/S0140-6736(02)09838-0. [DOI] [PubMed] [Google Scholar]
- 19.Navin N, Kendall J, Troge J, Andrews P, Rodgers L, McIndoo J, Cook K, Stepansky A, Levy D, Esposito D. Tumour evolution inferred by single-cell sequencing. Nature. 2011;472(7341):90–94. doi: 10.1038/nature09807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Navin N, Krasnitz A, Rodgers L, Cook K, Meth J, Kendall J, Riggs M, Eberling Y, Troge J, Grubor V. Inferring tumor progression from genomic heterogeneity. Genome Res. 2010;20(1):68–80. doi: 10.1101/gr.099622.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Park SY, Gonen M, Kim HJ, Michor F, Polyak K. Cellular and genetic diversity in the progression of in situ human breast carcinomas to an invasive phenotype. J Clin Invest. 2010;120(2):636–644. doi: 10.1172/JCI40724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Siitonen SM, Isola JJ, Rantala IS, Helin HJ. Intratumor variation in cell proliferation in breast carcinoma as determined by antiproliferating cell nuclear antigen monoclonal antibody and automated image analysis. Am J Clin Pathol. 1993;99(3):226–231. doi: 10.1093/ajcp/99.3.226. [DOI] [PubMed] [Google Scholar]
- 23.Teixeira MR, Pandis N, Bardi G, Andersen JA, Heim S. Karyotypic comparisons of multiple tumorous and macroscopically normal surrounding tissue samples from patients with breast cancer. Cancer Res. 1996;56(4):855–859. [PubMed] [Google Scholar]
- 24.de Bruin EC, McGranahan N, Swanton C. Analysis of intratumor heterogeneity unravels lung cancer evolution. Mol Cell Oncol. 2015;2(3):e985549. doi: 10.4161/23723556.2014.985549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zito Marino F, Liguori G, Aquino G, La Mantia E, Bosari S, Ferrero S, Rosso L, Gaudioso G, De Rosa N, Scrima M. Intratumor Heterogeneity of ALK-Rearrangements and Homogeneity of EGFR-Mutations in Mixed Lung Adenocarcinoma. PLoS One. 2015;10(9):e0139264. doi: 10.1371/journal.pone.0139264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Le Pennec S, Konopka T, Gacquer D, Fimereli D, Tarabichi M, Tomas G, Savagner F, Decaussin-Petrucci M, Tresallet C, Andry G. Intratumor heterogeneity and clonal evolution in an aggressive papillary thyroid cancer and matched metastases. Endocr Relat Cancer. 2015;22(2):205–216. doi: 10.1530/ERC-14-0351. [DOI] [PubMed] [Google Scholar]
- 27.Cao W, Wu W, Yan M, Tian F, Ma C, Zhang Q, Li X, Han P, Liu Z, Gu J. Multiple region whole-exome sequencing reveals dramatically evolving intratumor genomic heterogeneity in esophageal squamous cell carcinoma. Oncogenesis. 2015;4:e175. doi: 10.1038/oncsis.2015.34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Friemel J, Rechsteiner M, Frick L, Bohm F, Struckmann K, Egger M, Moch H, Heikenwalder M, Weber A. Intratumor heterogeneity in hepatocellular carcinoma. Clin Cancer Res. 2015;21(8):1951–1961. doi: 10.1158/1078-0432.CCR-14-0122. [DOI] [PubMed] [Google Scholar]
- 29.Giaretti W, Monaco R, Pujic N, Rapallo A, Nigro S, Geido E. Intratumor heterogeneity of K-ras2 mutations in colorectal adenocarcinomas: association with degree of DNA aneuploidy. Am J Pathol. 1996;149(1):237–245. [PMC free article] [PubMed] [Google Scholar]
- 30.Gonzalez-Garcia I, Sole RV, Costa J. Metapopulation dynamics and spatial heterogeneity in cancer. Proc Natl Acad Sci U S A. 2002;99(20):13085–13089. doi: 10.1073/pnas.202139299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Merlo LM, Shah NA, Li X, Blount PL, Vaughan TL, Reid BJ, Maley CC. A comprehensive survey of clonal diversity measures in Barrett's esophagus as biomarkers of progression to esophageal adenocarcinoma. Cancer Prev Res (Phila) 2010;3(11):1388–1397. doi: 10.1158/1940-6207.CAPR-10-0108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Raimondi C, Nicolazzo C, Gradilone A, Giannini G, De Falco E, Chimenti I, Varriale E, Hauch S, Plappert L, Cortesi E. Circulating tumor cells: exploring intratumor heterogeneity of colorectal cancer. Cancer Biol Ther. 2014;15(5):496–503. doi: 10.4161/cbt.28020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Stoecklein NH, Hosch SB, Bezler M, Stern F, Hartmann CH, Vay C, Siegmund A, Scheunemann P, Schurr P, Knoefel WT. Direct genetic analysis of single disseminated cancer cells for prediction of outcome and therapy selection in esophageal cancer. Cancer Cell. 2008;13(5):441–453. doi: 10.1016/j.ccr.2008.04.005. [DOI] [PubMed] [Google Scholar]
- 34.Zhang XC, Xu C, Mitchell RM, Zhang B, Zhao D, Li Y, Huang X, Fan W, Wang H, Lerma LA. Tumor evolution and intratumor heterogeneity of an oropharyngeal squamous cell carcinoma revealed by whole-genome sequencing. Neoplasia. 2013;15(12):1371–1378. doi: 10.1593/neo.131400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Alvarado C, Beitel LK, Sircar K, Aprikian A, Trifiro M, Gottlieb B. Somatic mosaicism and cancer: a micro-genetic examination into the role of the androgen receptor gene in prostate cancer. Cancer Res. 2005;65(18):8514–8518. doi: 10.1158/0008-5472.CAN-05-0399. [DOI] [PubMed] [Google Scholar]
- 36.Clark J, Attard G, Jhavar S, Flohr P, Reid A, De-Bono J, Eeles R, Scardino P, Cuzick J, Fisher G. Complex patterns of ETS gene alteration arise during cancer development in the human prostate. Oncogene. 2008;27(14):1993–2003. doi: 10.1038/sj.onc.1210843. [DOI] [PubMed] [Google Scholar]
- 37.Konishi N, Hiasa Y, Matsuda H, Tao M, Tsuzuki T, Hayashi I, Kitahori Y, Shiraishi T, Yatani R, Shimazaki J. Intratumor cellular heterogeneity and alterations in ras oncogene and p53 tumor suppressor gene in human prostate carcinoma. Am J Pathol. 1995;147(4):1112–1122. [PMC free article] [PubMed] [Google Scholar]
- 38.Macintosh CA, Stower M, Reid N, Maitland NJ. Precise microdissection of human prostate cancers reveals genotypic heterogeneity. Cancer Res. 1998;58(1):23–28. [PubMed] [Google Scholar]
- 39.Prudnikova TY, Soulitzis N, Kutsenko OS, Mostovich LA, Haraldson K, Ernberg I, Kashuba VI, Spandidos DA, Zabarovsky ER, Grigorieva EV. Heterogeneity of d-glucuronyl C5-epimerase expression and epigenetic regulation in prostate cancer. Cancer Med. 2013;2(5):654–661. doi: 10.1002/cam4.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Fujii H, Yoshida M, Gong ZX, Matsumoto T, Hamano Y, Fukunaga M, Hruban RH, Gabrielson E, Shirai T. Frequent genetic heterogeneity in the clonal evolution of gynecological carcinosarcoma and its influence on phenotypic diversity. Cancer Res. 2000;60(1):114–120. [PubMed] [Google Scholar]
- 41.Varley KE, Mutch DG, Edmonston TB, Goodfellow PJ, Mitra RD. Intra-tumor heterogeneity of MLH1 promoter methylation revealed by deep single molecule bisulfite sequencing. Nucleic Acids Res. 2009;37(14):4603–4612. doi: 10.1093/nar/gkp457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Anderson K, Lutz C, van Delft FW, Bateman CM, Guo Y, Colman SM, Kempski H, Moorman AV, Titley I, Swansbury J. Genetic variegation of clonal architecture and propagating cells in leukaemia. Nature. 2011;469(7330):356–361. doi: 10.1038/nature09650. [DOI] [PubMed] [Google Scholar]
- 43.Campbell PJ, Pleasance ED, Stephens PJ, Dicks E, Rance R, Goodhead I, Follows GA, Green AR, Futreal PA, Stratton MR. Subclonal phylogenetic structures in cancer revealed by ultra-deep sequencing. Proc Natl Acad Sci U S A. 2008;105(35):13081–13086. doi: 10.1073/pnas.0801523105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Landau DA, Carter SL, Stojanov P, McKenna A, Stevenson K, Lawrence MS, Sougnez C, Stewart C, Sivachenko A, Wang L. Evolution and impact of subclonal mutations in chronic lymphocytic leukemia. Cell. 2013;152(4):714–726. doi: 10.1016/j.cell.2013.01.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Notta F, Mullighan CG, Wang JC, Poeppl A, Doulatov S, Phillips LA, Ma J, Minden MD, Downing JR, Dick JE. Evolution of human BCR-ABL1 lymphoblastic leukaemia-initiating cells. Nature. 2011;469(7330):362–367. doi: 10.1038/nature09733. [DOI] [PubMed] [Google Scholar]
- 46.Li S, Garrett-Bakelman FE, Chung SS, Sanders MA, Hricik T, Rapaport F, Patel J, Dillon R, Vijay P, Brown AL. Distinct evolution and dynamics of epigenetic and genetic heterogeneity in acute myeloid leukemia. Nat Med. 2016;22(7):792–799. doi: 10.1038/nm.4125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Burrell RA, McGranahan N, Bartek J, Swanton C. The causes and consequences of genetic heterogeneity in cancer evolution. Nature. 2013;501(7467):338–345. doi: 10.1038/nature12625. [DOI] [PubMed] [Google Scholar]
- 48.Gupta PB, Kuperwasser C, Brunet JP, Ramaswamy S, Kuo WL, Gray JW, Naber SP, Weinberg RA. The melanocyte differentiation program predisposes to metastasis after neoplastic transformation. Nat Genet. 2005;37(10):1047–1054. doi: 10.1038/ng1634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Rodriguez-Cerdeira C, Gregorio MC, Lopez-Barcenas A, Sanchez-Blanco E, Sanchez-Blanco B, Fabbrocini G, Bardhi B, Sinani A, Guzman RA. Advances in Immunotherapy for Melanoma: A Comprehensive Review. Mediat Inflamm. 2017;2017:3264217. doi: 10.1155/2017/3264217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Cosgarea I, Ritter C, Becker JC, Schadendorf D, Ugurel S. Update on the clinical use of kinase inhibitors in melanoma. J Dtsch Dermatol Ges. 2017;15(9):887–893. doi: 10.1111/ddg.13321. [DOI] [PubMed] [Google Scholar]
- 51.Miller KD, Siegel RL, Lin CC, Mariotto AB, Kramer JL, Rowland JH, Stein KD, Alteri R, Jemal A. Cancer treatment and survivorship statistics, 2016. CA Cancer J Clin. 2016;66(4):271–289. doi: 10.3322/caac.21349. [DOI] [PubMed] [Google Scholar]
- 52.Ferlay J, Steliarova-Foucher E, Lortet-Tieulent J, Rosso S, Coebergh JW, Comber H, Forman D, Bray F. Cancer incidence and mortality patterns in Europe: estimates for 40 countries in 2012. Eur J Cancer. 2013;49(6):1374–1403. doi: 10.1016/j.ejca.2012.12.027. [DOI] [PubMed] [Google Scholar]
- 53.Garbe C, Leiter U. Melanoma epidemiology and trends. Clin Dermatol. 2009;27(1):3–9. doi: 10.1016/j.clindermatol.2008.09.001. [DOI] [PubMed] [Google Scholar]
- 54.Siegel RL, Miller KD, Jemal A. Cancer statistics, 2017. CA Cancer J Clin. 2017;67(1):7–30. doi: 10.3322/caac.21387. [DOI] [PubMed] [Google Scholar]
- 55.Lawrence MS, Stojanov P, Polak P, Kryukov GV, Cibulskis K, Sivachenko A, Carter SL, Stewart C, Mermel CH, Roberts SA. Mutational heterogeneity in cancer and the search for new cancer-associated genes. Nature. 2013;499(7457):214–218. doi: 10.1038/nature12213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Genomic classification of cutaneous melanoma. Cell. 2015;161(7):1681–1696. doi: 10.1016/j.cell.2015.05.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Govindarajan B, Bai X, Cohen C, Zhong H, Kilroy S, Louis G, Moses M, Arbiser JL. Malignant transformation of melanocytes to melanoma by constitutive activation of mitogen-activated protein kinase kinase (MAPKK) signaling. J Biol Chem. 2003;278(11):9790–9795. doi: 10.1074/jbc.M212929200. [DOI] [PubMed] [Google Scholar]
- 58.Hachey SJ, Boiko AD. Therapeutic implications of melanoma heterogeneity. Exp Dermatol. 2016;25(7):497–500. doi: 10.1111/exd.13002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Lee JH, Choi JW, Kim YS. Frequencies of BRAF and NRAS mutations are different in histological types and sites of origin of cutaneous melanoma: a meta-analysis. Br J Dermatol. 2011;164(4):776–784. doi: 10.1111/j.1365-2133.2010.10185.x. [DOI] [PubMed] [Google Scholar]
- 60.Boursault L, Haddad V, Vergier B, Cappellen D, Verdon S, Bellocq JP, Jouary T, Merlio JP. Tumor homogeneity between primary and metastatic sites for BRAF status in metastatic melanoma determined by immunohistochemical and molecular testing. PLoS One. 2013;8(8):e70826. doi: 10.1371/journal.pone.0070826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Colombino M, Capone M, Lissia A, Cossu A, Rubino C, De Giorgi V, Massi D, Fonsatti E, Staibano S, Nappi O. BRAF/NRAS mutation frequencies among primary tumors and metastases in patients with melanoma. J Clin Oncol. 2012;30(20):2522–2529. doi: 10.1200/JCO.2011.41.2452. [DOI] [PubMed] [Google Scholar]
- 62.Dong J, Phelps RG, Qiao R, Yao S, Benard O, Ronai Z, Aaronson SA. BRAF oncogenic mutations correlate with progression rather than initiation of human melanoma. Cancer Res. 2003;63(14):3883–3885. [PubMed] [Google Scholar]
- 63.Helias-Rodzewicz Z, Funck-Brentano E, Baudoux L, Jung CK, Zimmermann U, Marin C, Clerici T, Le Gall C, Peschaud F, Taly V. Variations of BRAF mutant allele percentage in melanomas. BMC Cancer. 2015;15:497. doi: 10.1186/s12885-015-1515-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Chiappetta C, Proietti I, Soccodato V, Puggioni C, Zaralli R, Pacini L, Porta N, Skroza N, Petrozza V, Potenza C. BRAF and NRAS mutations are heterogeneous and not mutually exclusive in nodular melanoma. Appl Immunohistochem Mol Morphol. 2015;23(3):172–177. doi: 10.1097/PAI.0000000000000071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Omholt K, Platz A, Kanter L, Ringborg U, Hansson J. NRAS and BRAF mutations arise early during melanoma pathogenesis and are preserved throughout tumor progression. Clin Cancer Res. 2003;9(17):6483–6488. [PubMed] [Google Scholar]
- 66.Sensi M, Nicolini G, Petti C, Bersani I, Lozupone F, Molla A, Vegetti C, Nonaka D, Mortarini R, Parmiani G. Mutually exclusive NRASQ61R and BRAFV600E mutations at the single-cell level in the same human melanoma. Oncogene. 2006;25(24):3357–3364. doi: 10.1038/sj.onc.1209379. [DOI] [PubMed] [Google Scholar]
- 67.Gandolfi G, Longo C, Moscarella E, Zalaudek I, Sancisi V, Raucci M, Manzotti G, Gugnoni M, Piana S, Argenziano G. The extent of whole-genome copy number alterations predicts aggressive features in primary melanomas. Pigment Cell Melanoma Res. 2016;29(2):163–175. doi: 10.1111/pcmr.12436. [DOI] [PubMed] [Google Scholar]
- 68.Kiuru M, Busam KJ. The NF1 gene in tumor syndromes and melanoma. Lab Investig. 2017;97(2):146–157. doi: 10.1038/labinvest.2016.142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Gillies RJ, Verduzco D, Gatenby RA. Evolutionary dynamics of carcinogenesis and why targeted therapy does not work. Nat Rev Cancer. 2012;12(7):487–493. doi: 10.1038/nrc3298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Harbst K, Lauss M, Cirenajwis H, Isaksson K, Rosengren F, Torngren T, Kvist A, Johansson MC, Vallon-Christersson J, Baldetorp B. Multiregion Whole-Exome Sequencing Uncovers the Genetic Evolution and Mutational Heterogeneity of Early-Stage Metastatic Melanoma. Cancer Res. 2016;76(16):4765–4774. doi: 10.1158/0008-5472.CAN-15-3476. [DOI] [PubMed] [Google Scholar]
- 71.Quintana E, Shackleton M, Foster HR, Fullen DR, Sabel MS, Johnson TM, Morrison SJ. Phenotypic heterogeneity among tumorigenic melanoma cells from patients that is reversible and not hierarchically organized. Cancer Cell. 2010;18(5):510–523. doi: 10.1016/j.ccr.2010.10.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Nowell PC. The clonal evolution of tumor cell populations. Science. 1976;194(4260):23–28. doi: 10.1126/science.959840. [DOI] [PubMed] [Google Scholar]
- 73.Hanahan D, Weinberg RA. Hallmarks of cancer: the next generation. Cell. 2011;144(5):646–674. doi: 10.1016/j.cell.2011.02.013. [DOI] [PubMed] [Google Scholar]
- 74.Gerlinger M, Swanton C. How Darwinian models inform therapeutic failure initiated by clonal heterogeneity in cancer medicine. Br J Cancer. 2010;103(8):1139–1143. doi: 10.1038/sj.bjc.6605912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Riveiro-Falkenbach E, Santos-Briz A, Rios-Martin JJ, Rodriguez-Peralto JL. Controversies in intrapatient melanoma BRAFV600E mutation status. Am J Dermatopathol. 2017;39(4):291–295. doi: 10.1097/DAD.0000000000000710. [DOI] [PubMed] [Google Scholar]
- 76.Lin J, Goto Y, Murata H, Sakaizawa K, Uchiyama A, Saida T, Takata M. Polyclonality of BRAF mutations in primary melanoma and the selection of mutant alleles during progression. Br J Cancer. 2011;104(3):464–468. doi: 10.1038/sj.bjc.6606072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Horak P, Frohling S, Glimm H. Integrating next-generation sequencing into clinical oncology: strategies, promises and pitfalls. ESMO Open. 2016;1(5):e000094. doi: 10.1136/esmoopen-2016-000094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Anderson S, Bloom KJ, Vallera DU, Rueschoff J, Meldrum C, Schilling R, Kovach B, Lee JR, Ochoa P, Langland R. Multisite analytic performance studies of a real-time polymerase chain reaction assay for the detection of BRAF V600E mutations in formalin-fixed, paraffin-embedded tissue specimens of malignant melanoma. Arch Pathol Lab Med. 2012;136(11):1385–1391. doi: 10.5858/arpa.2011-0505-OA. [DOI] [PubMed] [Google Scholar]
- 79.Colomba E, Helias-Rodzewicz Z, Von Deimling A, Marin C, Terrones N, Pechaud D, Surel S, Cote JF, Peschaud F, Capper D. Detection of BRAF p.V600E mutations in melanomas: comparison of four methods argues for sequential use of immunohistochemistry and pyrosequencing. J Mol Diagn. 2013;15(1):94–100. doi: 10.1016/j.jmoldx.2012.09.001. [DOI] [PubMed] [Google Scholar]
- 80.Halait H, Demartin K, Shah S, Soviero S, Langland R, Cheng S, Hillman G, Wu L, Lawrence HJ. Analytical performance of a real-time PCR-based assay for V600 mutations in the BRAF gene, used as the companion diagnostic test for the novel BRAF inhibitor vemurafenib in metastatic melanoma. Diagn Mol Pathol. 2012;21(1):1–8. doi: 10.1097/PDM.0b013e31823b216f. [DOI] [PubMed] [Google Scholar]
- 81.Schirosi L, Strippoli S, Gaudio F, Graziano G, Popescu O, Guida M, Simone G, Mangia A. Is immunohistochemistry of BRAF V600E useful as a screening tool and during progression disease of melanoma patients? BMC Cancer. 2016;16(1):905. doi: 10.1186/s12885-016-2951-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Sener E, Yildirim P, Tan A, Gokoz O, Tezel GG. Investigation of BRAF mutation analysis with different technical platforms in metastatic melanoma. Pathol Res Pract. 2017 doi: 10.1016/j.prp.2017.01.010. [DOI] [PubMed] [Google Scholar]
- 83.Anwar MA, Murad F, Dawson E, Abd Elmageed ZY, Tsumagari K, Kandil E. Immunohistochemistry as a reliable method for detection of BRAF-V600E mutation in melanoma: a systematic review and meta-analysis of current published literature. J Surg Res. 2016;203(2):407–415. doi: 10.1016/j.jss.2016.04.029. [DOI] [PubMed] [Google Scholar]
- 84.de Unamuno Bustos B, Murria Estal R, Perez Simo G, de Juan Jimenez I, Escutia Munoz B, Rodriguez Serna M, Alegre de Miquel V, Llavador Ros M, Ballester Sanchez R, Nagore Enguidanos E. Towards Personalized Medicine in Melanoma: Implementation of a Clinical Next-Generation Sequencing Panel. Sci Rep. 2017;7(1):495. doi: 10.1038/s41598-017-00606-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Cintra Lopes Carapeto F, Neves Comodo A, Germano A, Pereira Guimaraes D, Barcelos D, Fernandes M. Marker protein expression combined with expression heterogeneity is a powerful indicator of malignancy in acral lentiginous melanomas. Am J Dermatopathol. 2017;39(2):114–120. doi: 10.1097/DAD.0000000000000635. [DOI] [PubMed] [Google Scholar]
- 86.Manfredi L, Meyer N, Tournier E, Grand D, Uro-Coste E, Rochaix P, Brousset P, Lamant L. Highly Concordant Results Between Immunohistochemistry and Molecular Testing of Mutated V600E BRAF in Primary and Metastatic Melanoma. Acta Derm Venereol. 2016;96(5):630–634. doi: 10.2340/00015555-2326. [DOI] [PubMed] [Google Scholar]
- 87.Busam KJ, Hedvat C, Pulitzer M, von Deimling A, Jungbluth AA. Immunohistochemical analysis of BRAF(V600E) expression of primary and metastatic melanoma and comparison with mutation status and melanocyte differentiation antigens of metastatic lesions. Am J Surg Pathol. 2013;37(3):413–420. doi: 10.1097/PAS.0b013e318271249e. [DOI] [PubMed] [Google Scholar]
- 88.William B, Claudia M, Virginia A, Lorenza P, Francesco S, Bruna D, Francesco C, Marina G, Alberto B, Bianchi-Scarra G. Heterogeneity and frequency of BRAF mutations in primary melanoma: Comparison between molecular methods and immunohistochemistry. Oncotarget. 2016 doi: 10.18632/oncotarget.14094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Eriksson H, Zebary A, Vassilaki I, Omholt K, Ghaderi M, Hansson J. BRAFV600E protein expression in primary cutaneous malignant melanomas and paired metastases. JAMA Dermatol. 2015;151(4):410–416. doi: 10.1001/jamadermatol.2014.3689. [DOI] [PubMed] [Google Scholar]
- 90.Verlinden I, van den Hurk K, Clarijs R, Willig AP, Stallinga CM, Roemen GM, van den Oord JJ, zur Hausen A, Speel EJ, Winnepenninckx VJ. BRAFV600E immunopositive melanomas show low frequency of heterogeneity and association with epithelioid tumor cells: a STROBE-compliant article. Medicine (Baltimore) 2014;93(28):e285. doi: 10.1097/MD.0000000000000285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Riveiro-Falkenbach E, Villanueva CA, Garrido MC, Ruano Y, Garcia-Martin RM, Godoy E, Ortiz-Romero PL, Rios-Martin JJ, Santos-Briz A, Rodriguez-Peralto JL. Intra- and Inter-Tumoral Homogeneity of BRAF(V600E) Mutations in Melanoma Tumors. J Invest Dermatol. 2015;135(12):3078–3085. doi: 10.1038/jid.2015.229. [DOI] [PubMed] [Google Scholar]
- 92.Satzger I, Marks L, Kerick M, Klages S, Berking C, Herbst R, Volker B, Schacht V, Timmermann B, Gutzmer R. Allele frequencies of BRAFV600 mutations in primary melanomas and matched metastases and their relevance for BRAF inhibitor therapy in metastatic melanoma. Oncotarget. 2015;6(35):37895–37905. doi: 10.18632/oncotarget.5634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Yancovitz M, Litterman A, Yoon J, Ng E, Shapiro RL, Berman RS, Pavlick AC, Darvishian F, Christos P, Mazumdar M. Intra- and inter-tumor heterogeneity of BRAF(V600E))mutations in primary and metastatic melanoma. PLoS One. 2012;7(1):e29336. doi: 10.1371/journal.pone.0029336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Mesbah Ardakani N, Leslie C, Grieu-Iacopetta F, Lam WS, Budgeon C, Millward M, Amanuel B. Clinical and therapeutic implications of BRAF mutation heterogeneity in metastatic melanoma. Pigment Cell Melanoma Res. 2016 doi: 10.1111/pcmr.12569. [DOI] [PubMed] [Google Scholar]
- 95.Lamy PJ, Castan F, Lozano N, Montelion C, Audran P, Bibeau F, Roques S, Montels F, Laberenne AC. Next-Generation Genotyping by Digital PCR to Detect and Quantify the BRAF V600E Mutation in Melanoma Biopsies. J Mol Diagn. 2015;17(4):366–373. doi: 10.1016/j.jmoldx.2015.02.004. [DOI] [PubMed] [Google Scholar]
- 96.Kaji T, Yamasaki O, Takata M, Otsuka M, Hamada T, Morizane S, Asagoe K, Yanai H, Hirai Y, Umemura H. Comparative study on driver mutations in primary and metastatic melanomas at a single Japanese institute: A clue for intra- and inter-tumor heterogeneity. J Dermatol Sci. 2016 doi: 10.1016/j.jdermsci.2016.10.006. [DOI] [PubMed] [Google Scholar]
- 97.Wilmott JS, Tembe V, Howle JR, Sharma R, Thompson JF, Rizos H, Lo RS, Kefford RF, Scolyer RA, Long GV. Intratumoral molecular heterogeneity in a BRAF-mutant, BRAF inhibitor-resistant melanoma: a case illustrating the challenges for personalized medicine. Mol Cancer Ther. 2012;11(12):2704–2708. doi: 10.1158/1535-7163.MCT-12-0530. [DOI] [PubMed] [Google Scholar]
- 98.Gremel G, Lee RJ, Girotti MR, Mandal AK, Valpione S, Garner G, Ayub M, Wood S, Rothwell DG, Fusi A. Distinct subclonal tumour responses to therapy revealed by circulating cell-free DNA. Ann Oncol. 2016;27(10):1959–1965. doi: 10.1093/annonc/mdw278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Vivancos A, Caratu G, Matito J, Munoz E, Ferrer B, Hernandez-Losa J, Bodet D, Perez-Alea M, Cortes J, Garcia-Patos V. Genetic evolution of nevus of Ota reveals clonal heterogeneity acquiring BAP1 and TP53 mutations. Pigment Cell Melanoma Res. 2016;29(2):247–253. doi: 10.1111/pcmr.12452. [DOI] [PubMed] [Google Scholar]
- 100.Shain AH, Yeh I, Kovalyshyn I, Sriharan A, Talevich E, Gagnon A, Dummer R, North J, Pincus L, Ruben B. The Genetic Evolution of Melanoma from Precursor Lesions. N Engl J Med. 2015;373(20):1926–1936. doi: 10.1056/NEJMoa1502583. [DOI] [PubMed] [Google Scholar]
- 101.Ding L, Kim M, Kanchi KL, Dees ND, Lu C, Griffith M, Fenstermacher D, Sung H, Miller CA, Goetz B. Clonal architectures and driver mutations in metastatic melanomas. PLoS One. 2014;9(11):e111153. doi: 10.1371/journal.pone.0111153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Sanborn JZ, Chung J, Purdom E, Wang NJ, Kakavand H, Wilmott JS, Butler T, Thompson JF, Mann GJ, Haydu LE. Phylogenetic analyses of melanoma reveal complex patterns of metastatic dissemination. Proc Natl Acad Sci U S A. 2015;112(35):10995–11000. doi: 10.1073/pnas.1508074112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Anaka M, Hudson C, Lo PH, Do H, Caballero OL, Davis ID, Dobrovic A, Cebon J, Behren A. Intratumoral genetic heterogeneity in metastatic melanoma is accompanied by variation in malignant behaviors. BMC Med Genet. 2013;6:40. doi: 10.1186/1755-8794-6-40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Turajlic S, Furney SJ, Lambros MB, Mitsopoulos C, Kozarewa I, Geyer FC, Mackay A, Hakas J, Zvelebil M, Lord CJ. Whole genome sequencing of matched primary and metastatic acral melanomas. Genome Res. 2012;22(2):196–207. doi: 10.1101/gr.125591.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.de Lange MJ, van Pelt SI, Versluis M, Jordanova ES, Kroes WG, Ruivenkamp C, van der Burg SH, Luyten GP, van Hall T, Jager MJ. Heterogeneity revealed by integrated genomic analysis uncovers a molecular switch in malignant uveal melanoma. Oncotarget. 2015;6(35):37824–37835. doi: 10.18632/oncotarget.5637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Bogdan I, Xin H, Burg G, Boni R. Heterogeneity of allelic deletions within melanoma metastases. Melanoma Res. 2001;11(4):349–354. doi: 10.1097/00008390-200108000-00005. [DOI] [PubMed] [Google Scholar]
- 107.Nakayama T, Taback B, Turner R, Morton DL, Hoon DS. Molecular clonality of in-transit melanoma metastasis. Am J Pathol. 2001;158(4):1371–1378. doi: 10.1016/S0002-9440(10)64088-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Rao UN, Jones MW, Finkelstein SD. Genotypic analysis of primary and metastatic cutaneous melanoma. Cancer Genet Cytogenet. 2003;140(1):37–44. doi: 10.1016/s0165-4608(02)00651-9. [DOI] [PubMed] [Google Scholar]
- 109.DiSano K, Tschen JA, Cho-Vega JH. Intratumoral heterogeneity of chromosome 9 loss and CDKN2A (p16) protein expression in a morphologically challenging spitzoid melanoma. Am J Dermatopathol. 2013;35(2):277–280. doi: 10.1097/DAD.0b013e31826b187b. [DOI] [PubMed] [Google Scholar]
- 110.Uguen A, Uguen M, Talagas M, Gobin E, Marcorelles P, De Braekeleer M. Fluorescence in situ hybridization testing of chromosomes 6, 8, 9 and 11 in melanocytic tumors is difficult to automate and reveals tumor heterogeneity in melanomas. Oncol Lett. 2016;12(4):2734–2741. doi: 10.3892/ol.2016.4949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Takata M, Morita R, Takehara K. Clonal heterogeneity in sporadic melanomas as revealed by loss-of-heterozygosity analysis. Int J Cancer. 2000;85(4):492–497. [PubMed] [Google Scholar]
- 112.Dopierala J, Damato BE, Lake SL, Taktak AF, Coupland SE. Genetic heterogeneity in uveal melanoma assessed by multiplex ligation-dependent probe amplification. Invest Ophthalmol Vis Sci. 2010;51(10):4898–4905. doi: 10.1167/iovs.09-5004. [DOI] [PubMed] [Google Scholar]
- 113.Lake SL, Damato BE, Dopierala J, Baudo MM, Taktak AF, Coupland SE. Multiplex ligation-dependent probe amplification analysis of uveal melanoma with extraocular extension demonstrates heterogeneity of gross chromosomal abnormalities. Invest Ophthalmol Vis Sci. 2011;52(8):5559–5564. doi: 10.1167/iovs.10-6845. [DOI] [PubMed] [Google Scholar]
- 114.Helmbold P, Altrichter D, Klapperstuck T, Marsch W. Intratumoral DNA stem-line heterogeneity in superficial spreading melanoma. J Am Acad Dermatol. 2005;52(5):803–809. doi: 10.1016/j.jaad.2004.12.049. [DOI] [PubMed] [Google Scholar]
- 115.Harbour JW, Onken MD, Roberson ED, Duan S, Cao L, Worley LA, Council ML, Matatall KA, Helms C, Bowcock AM. Frequent mutation of BAP1 in metastasizing uveal melanomas. Science. 2010;330(6009):1410–1413. doi: 10.1126/science.1194472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Chang MY, Rao NP, Burgess BL, Johnson L, McCannel TA. Heterogeneity of monosomy 3 in fine needle aspiration biopsy of choroidal melanoma. Mol Vis. 2013;19:1892–1900. [PMC free article] [PubMed] [Google Scholar]
- 117.Mensink HW, Vaarwater J, Kilic E, Naus NC, Mooy N, Luyten G, Bruggenwirth HT, Paridaens D, de Klein A. Chromosome 3 intratumor heterogeneity in uveal melanoma. Invest Ophthalmol Vis Sci. 2009;50(2):500–504. doi: 10.1167/iovs.08-2279. [DOI] [PubMed] [Google Scholar]
- 118.Sandinha T, Farquharson M, McKay I, Roberts F. Correlation of heterogeneity for chromosome 3 copy number with cell type in choroidal melanoma of mixed-cell type. Invest Ophthalmol Vis Sci. 2006;47(12):5177–5180. doi: 10.1167/iovs.06-0332. [DOI] [PubMed] [Google Scholar]
- 119.Maat W, Jordanova ES, van Zelderen-Bhola SL, Barthen ER, Wessels HW, Schalij-Delfos NE, Jager MJ. The heterogeneous distribution of monosomy 3 in uveal melanomas: implications for prognostication based on fine-needle aspiration biopsies. Arch Pathol Lab Med. 2007;131(1):91–96. doi: 10.5858/2007-131-91-THDOMI. [DOI] [PubMed] [Google Scholar]
- 120.Meir T, Zeschnigk M, Masshofer L, Pe'er J, Chowers I. The spatial distribution of monosomy 3 and network vasculogenic mimicry patterns in uveal melanoma. Invest Ophthalmol Vis Sci. 2007;48(5):1918–1922. doi: 10.1167/iovs.06-1308. [DOI] [PubMed] [Google Scholar]
- 121.Sigalotti L, Fratta E, Coral S, Tanzarella S, Danielli R, Colizzi F, Fonsatti E, Traversari C, Altomonte M, Maio M. Intratumor heterogeneity of cancer/testis antigens expression in human cutaneous melanoma is methylation-regulated and functionally reverted by 5-aza-2'-deoxycytidine. Cancer Res. 2004;64(24):9167–9171. doi: 10.1158/0008-5472.CAN-04-1442. [DOI] [PubMed] [Google Scholar]
- 122.Roesch A. Melanoma stem cells. J Dtsch Dermatol Ges. 2015;13(2):118–124. doi: 10.1111/ddg.12584. [DOI] [PubMed] [Google Scholar]
- 123.Chapman A, Fernandez del Ama L, Ferguson J, Kamarashev J, Wellbrock C, Hurlstone A. Heterogeneous tumor subpopulations cooperate to drive invasion. Cell Rep. 2014;8(3):688–695. doi: 10.1016/j.celrep.2014.06.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Hoek KS, Schlegel NC, Brafford P, Sucker A, Ugurel S, Kumar R, Weber BL, Nathanson KL, Phillips DJ, Herlyn M. Metastatic potential of melanomas defined by specific gene expression profiles with no BRAF signature. Pigment Cell Res. 2006;19(4):290–302. doi: 10.1111/j.1600-0749.2006.00322.x. [DOI] [PubMed] [Google Scholar]
- 125.Verfaillie A, Imrichova H, Atak ZK, Dewaele M, Rambow F, Hulselmans G, Christiaens V, Svetlichnyy D, Luciani F, Van den Mooter L. Decoding the regulatory landscape of melanoma reveals TEADS as regulators of the invasive cell state. Nat Commun. 2015;6:6683. doi: 10.1038/ncomms7683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Falletta P, Sanchez-Del-Campo L, Chauhan J, Effern M, Kenyon A, Kershaw CJ, Siddaway R, Lisle R, Freter R, Daniels MJ. Translation reprogramming is an evolutionarily conserved driver of phenotypic plasticity and therapeutic resistance in melanoma. Genes Dev. 2017;31(1):18–33. doi: 10.1101/gad.290940.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Riesenberg S, Groetchen A, Siddaway R, Bald T, Reinhardt J, Smorra D, Kohlmeyer J, Renn M, Phung B, Aymans P. MITF and c-Jun antagonism interconnects melanoma dedifferentiation with pro-inflammatory cytokine responsiveness and myeloid cell recruitment. Nat Commun. 2015;6:8755. doi: 10.1038/ncomms9755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Tirosh I, Izar B, Prakadan SM, Wadsworth MH, II, Treacy D, Trombetta JJ, Rotem A, Rodman C, Lian C, Murphy G. Dissecting the multicellular ecosystem of metastatic melanoma by single-cell RNA-seq. Science. 2016;352(6282):189–196. doi: 10.1126/science.aad0501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Hoek KS, Eichhoff OM, Schlegel NC, Dobbeling U, Kobert N, Schaerer L, Hemmi S, Dummer R. In vivo switching of human melanoma cells between proliferative and invasive states. Cancer Res. 2008;68(3):650–656. doi: 10.1158/0008-5472.CAN-07-2491. [DOI] [PubMed] [Google Scholar]
- 130.Webster MR, Kugel CH, III, Weeraratna AT. The Wnts of change: how Wnts regulate phenotype switching in melanoma. Biochim Biophys Acta. 2015;1856(2):244–251. doi: 10.1016/j.bbcan.2015.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Carreira S, Goodall J, Denat L, Rodriguez M, Nuciforo P, Hoek KS, Testori A, Larue L, Goding CR. Mitf regulation of Dia1 controls melanoma proliferation and invasiveness. Genes Dev. 2006;20(24):3426–3439. doi: 10.1101/gad.406406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Fallahi-Sichani M, Becker V, Izar B, Baker GJ, Lin JR, Boswell SA, Shah P, Rotem A, Garraway LA, Sorger PK. Adaptive resistance of melanoma cells to RAF inhibition via reversible induction of a slowly dividing de-differentiated state. Mol Syst Biol. 2017;13(1):905. doi: 10.15252/msb.20166796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Kemper K, de Goeje PL, Peeper DS, van Amerongen R. Phenotype switching: tumor cell plasticity as a resistance mechanism and target for therapy. Cancer Res. 2014;74(21):5937–5941. doi: 10.1158/0008-5472.CAN-14-1174. [DOI] [PubMed] [Google Scholar]
- 134.Konieczkowski DJ, Johannessen CM, Abudayyeh O, Kim JW, Cooper ZA, Piris A, Frederick DT, Barzily-Rokni M, Straussman R, Haq R. A melanoma cell state distinction influences sensitivity to MAPK pathway inhibitors. Cancer Discov. 2014;4(7):816–827. doi: 10.1158/2159-8290.CD-13-0424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Muller J, Krijgsman O, Tsoi J, Robert L, Hugo W, Song C, Kong X, Possik PA, Cornelissen-Steijger PD, Geukes Foppen MH. Low MITF/AXL ratio predicts early resistance to multiple targeted drugs in melanoma. Nat Commun. 2014;5:5712. doi: 10.1038/ncomms6712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Li FZ, Dhillon AS, Anderson RL, McArthur G, Ferrao PT. Phenotype switching in melanoma: implications for progression and therapy. Front Oncol. 2015;5:31. doi: 10.3389/fonc.2015.00031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Vincent KM, Postovit LM. Investigating the utility of human melanoma cell lines as tumour models. Oncotarget. 2017 doi: 10.18632/oncotarget.14443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Wouters J, Stas M, Gremeaux L, Govaere O, Van den Broeck A, Maes H, Agostinis P, Roskams T, van den Oord JJ, Vankelecom H. The human melanoma side population displays molecular and functional characteristics of enriched chemoresistance and tumorigenesis. PLoS One. 2013;8(10):e76550. doi: 10.1371/journal.pone.0076550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Gerber T, Willscher E, Loeffler-Wirth H, Hopp L, Schadendorf D, Schartl M, Anderegg U, Camp G, Treutlein B, Binder H. Mapping heterogeneity in patient-derived melanoma cultures by single-cell RNA-seq. Oncotarget. 2016 doi: 10.18632/oncotarget.13666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Demou ZN, Hendrix MJ. Microgenomics profile the endogenous angiogenic phenotype in subpopulations of aggressive melanoma. J Cell Biochem. 2008;105(2):562–573. doi: 10.1002/jcb.21855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Mirkina I, Hadzijusufovic E, Krepler C, Mikula M, Mechtcheriakova D, Strommer S, Stella A, Jensen-Jarolim E, Holler C, Wacheck V. Phenotyping of human melanoma cells reveals a unique composition of receptor targets and a subpopulation co-expressing ErbB4, EPO-R and NGF-R. PLoS One. 2014;9(1):e84417. doi: 10.1371/journal.pone.0084417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Ennen M, Keime C, Kobi D, Mengus G, Lipsker D, Thibault-Carpentier C, Davidson I. Single-cell gene expression signatures reveal melanoma cell heterogeneity. Oncogene. 2015;34(25):3251–3263. doi: 10.1038/onc.2014.262. [DOI] [PubMed] [Google Scholar]
- 143.Haqq C, Nosrati M, Sudilovsky D, Crothers J, Khodabakhsh D, Pulliam BL, Federman S, Miller JR, III, Allen RE, Singer MI. The gene expression signatures of melanoma progression. Proc Natl Acad Sci U S A. 2005;102(17):6092–6097. doi: 10.1073/pnas.0501564102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Croteau W, Jenkins MH, Ye S, Mullins DW, Brinckerhoff CE. Differential mechanisms of tumor progression in clones from a single heterogeneous human melanoma. J Cell Physiol. 2013;228(4):773–780. doi: 10.1002/jcp.24225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Kim JE, Leung E, Baguley BC, Finlay GJ. Heterogeneity of expression of epithelial-mesenchymal transition markers in melanocytes and melanoma cell lines. Front Genet. 2013;4:97. doi: 10.3389/fgene.2013.00097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Richmond-Sinclair NM, Lee E, Cummings MC, Williamson R, Muller K, Green AC, Hayward NK, Whiteman DC. Histologic and epidemiologic correlates of P-MAPK, Brn-2, pRb, p53, and p16 immunostaining in cutaneous melanomas. Melanoma Res. 2008;18(5):336–345. doi: 10.1097/CMR.0b013e32830d8329. [DOI] [PubMed] [Google Scholar]
- 147.Satzger I, Schaefer T, Kuettler U, Broecker V, Voelker B, Ostertag H, Kapp A, Gutzmer R. Analysis of c-KIT expression and KIT gene mutation in human mucosal melanomas. Br J Cancer. 2008;99(12):2065–2069. doi: 10.1038/sj.bjc.6604791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Seftor EA, Seftor RE, Weldon DS, Kirsammer GT, Margaryan NV, Gilgur A, Hendrix MJ. Melanoma tumor cell heterogeneity: a molecular approach to study subpopulations expressing the embryonic morphogen nodal. Semin Oncol. 2014;41(2):259–266. doi: 10.1053/j.seminoncol.2014.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Manning CS, Hooper S, Sahai EA. Intravital imaging of SRF and Notch signalling identifies a key role for EZH2 in invasive melanoma cells. Oncogene. 2015;34(33):4320–4332. doi: 10.1038/onc.2014.362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Mahoney KM, Freeman GJ, McDermott DF. The next immune-checkpoint inhibitors: PD-1/PD-L1 blockade in melanoma. Clin Ther. 2015;37(4):764–782. doi: 10.1016/j.clinthera.2015.02.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Madore J, Vilain RE, Menzies AM, Kakavand H, Wilmott JS, Hyman J, Yearley JH, Kefford RF, Thompson JF, Long GV. PD-L1 expression in melanoma shows marked heterogeneity within and between patients: implications for anti-PD-1/PD-L1 clinical trials. Pigment Cell Melanoma Res. 2015;28(3):245–253. doi: 10.1111/pcmr.12340. [DOI] [PubMed] [Google Scholar]
- 152.Sunshine JC, Nguyen PL, Kaunitz GJ, Cottrell TR, Berry S, Esandrio J, Xu H, Ogurtsova A, Bleich KB, Cornish TC. PD-L1 Expression in Melanoma: A Quantitative Immunohistochemical Antibody Comparison. Clin Cancer Res. 2017;23(16):4938–4944. doi: 10.1158/1078-0432.CCR-16-1821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Kuzbicki L, Lange D, Stanek-Widera A, Chwirot BW. Intratumoral expression of cyclooxygenase-2 (COX-2) is a negative prognostic marker for patients with cutaneous melanoma. Melanoma Res. 2016;26(5):448–456. doi: 10.1097/CMR.0000000000000282. [DOI] [PubMed] [Google Scholar]
- 154.Botti G, Fratangelo F, Cerrone M, Liguori G, Cantile M, Anniciello AM, Scala S, D'Alterio C, Trimarco C, Ianaro A. COX-2 expression positively correlates with PD-L1 expression in human melanoma cells. J Transl Med. 2017;15(1):46. doi: 10.1186/s12967-017-1150-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Curioni-Fontecedro A, Pitocco R, Schoenewolf NL, Holzmann D, Soldini D, Dummer R, Calvieri S, Moch H, Mihic-Probst D, Fitsche A. Intratumoral Heterogeneity of MAGE-C1/CT7 and MAGE-C2/CT10 Expression in Mucosal Melanoma. Biomed Res Int. 2015;2015:432479. doi: 10.1155/2015/432479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Widmer DS, Hoek KS, Cheng PF, Eichhoff OM, Biedermann T, Raaijmakers MI, Hemmi S, Dummer R, Levesque MP. Hypoxia contributes to melanoma heterogeneity by triggering HIF1alpha-dependent phenotype switching. J Invest Dermatol. 2013;133(10):2436–2443. doi: 10.1038/jid.2013.115. [DOI] [PubMed] [Google Scholar]
- 157.Lenggenhager D, Curioni-Fontecedro A, Storz M, Shakhova O, Sommer L, Widmer DS, Seifert B, Moch H, Dummer R, Mihic-Probst D. An Aggressive Hypoxia Related Subpopulation of Melanoma Cells is TRP-2 Negative. Transl Oncol. 2014;7(2):206–212. doi: 10.1016/j.tranon.2014.02.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Oiso N, Yoshida M, Kawara S, Kawada A. Amelanotic vulvar melanoma with intratumor histological heterogeneity. J Dermatol. 2010;37(6):537–541. doi: 10.1111/j.1346-8138.2010.00895.x. [DOI] [PubMed] [Google Scholar]
- 159.Clarke MF, Dick JE, Dirks PB, Eaves CJ, Jamieson CH, Jones DL, Visvader J, Weissman IL, Wahl GM. Cancer stem cells--perspectives on current status and future directions: AACR Workshop on cancer stem cells. Cancer Res. 2006;66(19):9339–9344. doi: 10.1158/0008-5472.CAN-06-3126. [DOI] [PubMed] [Google Scholar]
- 160.Brinckerhoff CE. Cancer stem cells (CSCs) in melanoma: there's smoke, but is there fire? J Cell Physiol. 2017;232(10):2674–2678. doi: 10.1002/jcp.25796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Hoek KS, Goding CR. Cancer stem cells versus phenotype-switching in melanoma. Pigment Cell Melanoma Res. 2010;23(6):746–759. doi: 10.1111/j.1755-148X.2010.00757.x. [DOI] [PubMed] [Google Scholar]
- 162.Kumar D, Gorain M, Kundu G, Kundu GC. Therapeutic implications of cellular and molecular biology of cancer stem cells in melanoma. Mol Cancer. 2017;16(1):7. doi: 10.1186/s12943-016-0578-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Schatton T, Frank MH. Cancer stem cells and human malignant melanoma. Pigment Cell Melanoma Res. 2008;21(1):39–55. doi: 10.1111/j.1755-148X.2007.00427.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Shakhova O, Sommer L. Testing the cancer stem cell hypothesis in melanoma: the clinics will tell. Cancer Lett. 2013;338(1):74–81. doi: 10.1016/j.canlet.2012.10.009. [DOI] [PubMed] [Google Scholar]
- 165.Kuzbicki L, Lange D, Straczynska-Niemiec A, Chwirot BW. JARID1B expression in human melanoma and benign melanocytic skin lesions. Melanoma Res. 2013;23(1):8–12. doi: 10.1097/CMR.0b013e32835d5d6f. [DOI] [PubMed] [Google Scholar]
- 166.Boiko AD, Razorenova OV, van de Rijn M, Swetter SM, Johnson DL, Ly DP, Butler PD, Yang GP, Joshua B, Kaplan MJ. Human melanoma-initiating cells express neural crest nerve growth factor receptor CD271. Nature. 2010;466(7302):133–137. doi: 10.1038/nature09161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Civenni G, Walter A, Kobert N, Mihic-Probst D, Zipser M, Belloni B, Seifert B, Moch H, Dummer R, van den Broek M. Human CD271-positive melanoma stem cells associated with metastasis establish tumor heterogeneity and long-term growth. Cancer Res. 2011;71(8):3098–3109. doi: 10.1158/0008-5472.CAN-10-3997. [DOI] [PubMed] [Google Scholar]
- 168.Quintana E, Shackleton M, Sabel MS, Fullen DR, Johnson TM, Morrison SJ. Efficient tumour formation by single human melanoma cells. Nature. 2008;456(7222):593–598. doi: 10.1038/nature07567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Lazova R, Tantcheva-Poor I, Sigal AC. P75 nerve growth factor receptor staining is superior to S100 in identifying spindle cell and desmoplastic melanoma. J Am Acad Dermatol. 2010;63(5):852–858. doi: 10.1016/j.jaad.2009.11.688. [DOI] [PubMed] [Google Scholar]
- 170.Radfar A, Stefanato CM, Ghosn S, Bhawan J. NGFR-positive desmoplastic melanomas with focal or absent S-100 staining: further evidence supporting the use of both NGFR and S-100 as a primary immunohistochemical panel for the diagnosis of desmoplastic melanomas. Am J Dermatopathol. 2006;28(2):162–167. doi: 10.1097/01.dad.0000183696.46573.ee. [DOI] [PubMed] [Google Scholar]
- 171.Landsberg J, Kohlmeyer J, Renn M, Bald T, Rogava M, Cron M, Fatho M, Lennerz V, Wolfel T, Holzel M. Melanomas resist T-cell therapy through inflammation-induced reversible dedifferentiation. Nature. 2012;490(7420):412–416. doi: 10.1038/nature11538. [DOI] [PubMed] [Google Scholar]
- 172.Held MA, Curley DP, Dankort D, McMahon M, Muthusamy V, Bosenberg MW. Characterization of melanoma cells capable of propagating tumors from a single cell. Cancer Res. 2010;70(1):388–397. doi: 10.1158/0008-5472.CAN-09-2153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Schatton T, Murphy GF, Frank NY, Yamaura K, Waaga-Gasser AM, Gasser M, Zhan Q, Jordan S, Duncan LM, Weishaupt C. Identification of cells initiating human melanomas. Nature. 2008;451(7176):345–349. doi: 10.1038/nature06489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Lai CY, Schwartz BE, Hsu MY. CD133+ melanoma subpopulations contribute to perivascular niche morphogenesis and tumorigenicity through vasculogenic mimicry. Cancer Res. 2012;72(19):5111–5118. doi: 10.1158/0008-5472.CAN-12-0624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Monzani E, Facchetti F, Galmozzi E, Corsini E, Benetti A, Cavazzin C, Gritti A, Piccinini A, Porro D, Santinami M. Melanoma contains CD133 and ABCG2 positive cells with enhanced tumourigenic potential. Eur J Cancer. 2007;43(5):935–946. doi: 10.1016/j.ejca.2007.01.017. [DOI] [PubMed] [Google Scholar]
- 176.Fang D, Nguyen TK, Leishear K, Finko R, Kulp AN, Hotz S, Van Belle PA, Xu X, Elder DE, Herlyn M. A tumorigenic subpopulation with stem cell properties in melanomas. Cancer Res. 2005;65(20):9328–9337. doi: 10.1158/0008-5472.CAN-05-1343. [DOI] [PubMed] [Google Scholar]
- 177.Sharma BK, Manglik V, O'Connell M, Weeraratna A, McCarron EC, Broussard JN, Divito KA, Simbulan-Rosenthal CM, Rosenthal DS, Zapas JL. Clonal dominance of CD133+ subset population as risk factor in tumor progression and disease recurrence of human cutaneous melanoma. Int J Oncol. 2012;41(5):1570–1576. doi: 10.3892/ijo.2012.1590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.Sztiller-Sikorska M, Hartman ML, Talar B, Jakubowska J, Zalesna I, Czyz M. Phenotypic diversity of patient-derived melanoma populations in stem cell medium. Lab Investig. 2015;95(6):672–683. doi: 10.1038/labinvest.2015.48. [DOI] [PubMed] [Google Scholar]
- 179.Luo Y, Ellis LZ, Dallaglio K, Takeda M, Robinson WA, Robinson SE, Liu W, Lewis KD, McCarter MD, Gonzalez R. Side population cells from human melanoma tumors reveal diverse mechanisms for chemoresistance. J Invest Dermatol. 2012;132(10):2440–2450. doi: 10.1038/jid.2012.161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.de Unamuno B, Palanca S, Botella R. Update on melanoma epigenetics. Curr Opin Oncol. 2015;27(5):420–426. doi: 10.1097/CCO.0000000000000217. [DOI] [PubMed] [Google Scholar]
- 181.Martinez-Cardus A, Vizoso M, Moran S, Manzano JL. Epigenetic mechanisms involved in melanoma pathogenesis and chemoresistance. Ann Transl Med. 2015;3(15):209. doi: 10.3978/j.issn.2305-5839.2015.06.20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182.Rastetter M, Schagdarsurengin U, Lahtz C, Fiedler E, Marsch W, Dammann R, Helmbold P. Frequent intra-tumoural heterogeneity of promoter hypermethylation in malignant melanoma. Histol Histopathol. 2007;22(9):1005–1015. doi: 10.14670/HH-22.1005. [DOI] [PubMed] [Google Scholar]
- 183.Casula M, Colombino M, Manca A, Caraco C, Botti G, Ascierto PA, Lissia A, Cossu A, Palmieri G. Low Levels of Genetic Heterogeneity in Matched Lymph Node Metastases from Patients with Melanoma. J Invest Dermatol. 2016;136(9):1917–1920. doi: 10.1016/j.jid.2016.05.103. [DOI] [PubMed] [Google Scholar]
- 184.Swoboda A, Rasin-Streden D, Schanab O, Okamoto I, Pehamberger H, Petzelbauer P, Mikula M. Identification of genetic disparity between primary and metastatic melanoma in human patients. Genes Chromosomes Cancer. 2011;50(9):680–688. doi: 10.1002/gcc.20890. [DOI] [PubMed] [Google Scholar]
- 185.Saint-Jean M, Quereux G, Nguyen JM, Peuvrel L, Brocard A, Vallee A, Knol AC, Khammari A, Denis MG, Dreno B. Is a single BRAF wild-type test sufficient to exclude melanoma patients from vemurafenib therapy? J Invest Dermatol. 2014;134(5):1468–1470. doi: 10.1038/jid.2013.378. [DOI] [PubMed] [Google Scholar]
- 186.Valachis A, Ullenhag GJ. Discrepancy in BRAF status among patients with metastatic malignant melanoma: a meta-analysis. Eur J Cancer. 2017;81:106–115. doi: 10.1016/j.ejca.2017.05.015. [DOI] [PubMed] [Google Scholar]
- 187.Richtig E, Schrama D, Ugurel S, Fried I, Niederkorn A, Massone C, Becker JC. BRAF mutation analysis of only one metastatic lesion can restrict the treatment of melanoma: a case report. Br J Dermatol. 2013;168(2):428–430. doi: 10.1111/j.1365-2133.2012.11121.x. [DOI] [PubMed] [Google Scholar]
- 188.Houben R, Becker JC, Kappel A, Terheyden P, Brocker EB, Goetz R, Rapp UR. Constitutive activation of the Ras-Raf signaling pathway in metastatic melanoma is associated with poor prognosis. J Carcinog. 2004;3(1):6. doi: 10.1186/1477-3163-3-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 189.Shinozaki M, Fujimoto A, Morton DL, Hoon DS. Incidence of BRAF oncogene mutation and clinical relevance for primary cutaneous melanomas. Clin Cancer Res. 2004;10(5):1753–1757. doi: 10.1158/1078-0432.ccr-1169-3. [DOI] [PubMed] [Google Scholar]
- 190.Saroufim M, Habib RH, Gerges R, Saab J, Loya A, Amr SS, Sheikh S, Satti M, Oberkanins C, Khalifeh I. Comparing BRAF mutation status in matched primary and metastatic cutaneous melanomas: implications on optimized targeted therapy. Exp Mol Pathol. 2014;97(3):315–320. doi: 10.1016/j.yexmp.2014.09.008. [DOI] [PubMed] [Google Scholar]
- 191.Heinzerling L, Baiter M, Kuhnapfel S, Schuler G, Keikavoussi P, Agaimy A, Kiesewetter F, Hartmann A, Schneider-Stock R. Mutation landscape in melanoma patients clinical implications of heterogeneity of BRAF mutations. Br J Cancer. 2013;109(11):2833–2841. doi: 10.1038/bjc.2013.622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 192.Bradish JR, Richey JD, Post KM, Meehan K, Sen JD, Malek AJ, Katona TM, Warren S, Logan TF, Fecher LA. Discordancy in BRAF mutations among primary and metastatic melanoma lesions: clinical implications for targeted therapy. Mod Pathol. 2015;28(4):480–486. doi: 10.1038/modpathol.2014.136. [DOI] [PubMed] [Google Scholar]
- 193.Yaman B, Kandiloglu G, Akalin T. BRAF-V600 mutation heterogeneity in primary and metastatic melanoma: a study with pyrosequencing and immunohistochemistry. Am J Dermatopathol. 2016;38(2):113–120. doi: 10.1097/DAD.0000000000000404. [DOI] [PubMed] [Google Scholar]
- 194.Edlundh-Rose E, Egyhazi S, Omholt K, Mansson-Brahme E, Platz A, Hansson J, Lundeberg J. NRAS and BRAF mutations in melanoma tumours in relation to clinical characteristics: a study based on mutation screening by pyrosequencing. Melanoma Res. 2006;16(6):471–478. doi: 10.1097/01.cmr.0000232300.22032.86. [DOI] [PubMed] [Google Scholar]
- 195.Egberts F, Bohne AS, Kruger S, Hedderich J, Rompel R, Haag J, Rocken C, Hauschild A. Varying Mutational Alterations in Multiple Primary Melanomas. J Mol Diagn. 2016;18(1):75–83. doi: 10.1016/j.jmoldx.2015.07.010. [DOI] [PubMed] [Google Scholar]
- 196.Sakaizawa K, Goto Y, Kiniwa Y, Uchiyama A, Harada K, Shimada S, Saida T, Ferrone S, Takata M, Uhara H. Mutation analysis of BRAF and KIT in circulating melanoma cells at the single cell level. Br J Cancer. 2012;106(5):939–946. doi: 10.1038/bjc.2012.12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 197.Harbst K, Lauss M, Cirenajwis H, Winter C, Howlin J, Torngren T, Kvist A, Nodin B, Olsson E, Hakkinen J. Molecular and genetic diversity in the metastatic process of melanoma. J Pathol. 2014;233(1):39–50. doi: 10.1002/path.4318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 198.Goswami RS, Patel KP, Singh RR, Meric-Bernstam F, Kopetz ES, Subbiah V, Alvarez RH, Davies MA, Jabbar KJ, Roy-Chowdhuri S. Hotspot mutation panel testing reveals clonal evolution in a study of 265 paired primary and metastatic tumors. Clin Cancer Res. 2015;21(11):2644–2651. doi: 10.1158/1078-0432.CCR-14-2391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 199.Hugo W, Shi H, Sun L, Piva M, Song C, Kong X, Moriceau G, Hong A, Dahlman KB, Johnson DB. Non-genomic and Immune Evolution of Melanoma Acquiring MAPKi Resistance. Cell. 2015;162(6):1271–1285. doi: 10.1016/j.cell.2015.07.061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 200.Fusi A, Berdel R, Havemann S, Nonnenmacher A, Keilholz U. Enhanced detection of BRAF-mutants by pre-PCR cleavage of wild-type sequences revealed circulating melanoma cells heterogeneity. Eur J Cancer. 2011;47(13):1971–1976. doi: 10.1016/j.ejca.2011.04.013. [DOI] [PubMed] [Google Scholar]
- 201.Reuben A, Spencer CN, Prieto PA, Gopalakrishnan V, Reddy SM, Miller JP, Mao X, De Macedo MP, Chen J, Song X. Genomic and immune heterogeneity are associated with differential responses to therapy in melanoma. NPJ Genom Med. 2017;2(1):10. doi: 10.1038/s41525-017-0013-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 202.Katona TM, Jones TD, Wang M, Eble JN, Billings SD, Cheng L. Genetically heterogeneous and clonally unrelated metastases may arise in patients with cutaneous melanoma. Am J Surg Pathol. 2007;31(7):1029–1037. doi: 10.1097/PAS.0b013e31802b3488. [DOI] [PubMed] [Google Scholar]
- 203.Bahrami S, Cheng L, Wang M, Jones TD, Malone JC, Billings SD. Clonal relationships between epidermotropic metastatic melanomas and their primary lesions: a loss of heterozygosity and X-chromosome inactivation-based analysis. Mod Pathol. 2007;20(8):821–827. doi: 10.1038/modpathol.3800833. [DOI] [PubMed] [Google Scholar]
- 204.Chatterjee A, Stockwell PA, Ahn A, Rodger EJ, Leichter AL, Eccles MR. Genome-wide methylation sequencing of paired primary and metastatic cell lines identifies common DNA methylation changes and a role for EBF3 as a candidate epigenetic driver of melanoma metastasis. Oncotarget. 2016 doi: 10.18632/oncotarget.14042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 205.Siegel RL, Miller KD, Jemal A. Cancer statistics, 2016. CA Cancer J Clin. 2016;66(1):7–30. doi: 10.3322/caac.21332. [DOI] [PubMed] [Google Scholar]
- 206.Pongor L, Harami-Papp H, Mehes E, Czirok A, Gyorffy B. Cell dispersal influences tumor heterogeneity and introduces a bias in NGS data interpretation. Sci Rep. 2017;7(1):7358. doi: 10.1038/s41598-017-07487-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 207.Erramuzpe A, Cortes JM, Lopez JI. Multisite tumor sampling enhances the detection of intratumor heterogeneity at all different temporal stages of tumor evolution. Virchows Arch. 2017 doi: 10.1007/s00428-017-2223-y. [DOI] [PubMed] [Google Scholar]
- 208.Gray ES, Rizos H, Reid AL, Boyd SC, Pereira MR, Lo J, Tembe V, Freeman J, Lee JH, Scolyer RA. Circulating tumor DNA to monitor treatment response and detect acquired resistance in patients with metastatic melanoma. Oncotarget. 2015;6(39):42008–42018. doi: 10.18632/oncotarget.5788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 209.Caiado F, Silva-Santos B, Norell H. Intra-tumour heterogeneity—going beyond genetics. FEBS J. 2016;283(12):2245–2258. doi: 10.1111/febs.13705. [DOI] [PubMed] [Google Scholar]
- 210.Ly E, Cardot-Leccia N, Ortonne JP, Benchetrit M, Michiels JF, Manfait M, Piot O. Histopathological characterization of primary cutaneous melanoma using infrared microimaging: a proof-of-concept study. Br J Dermatol. 2010;162(6):1316–1323. doi: 10.1111/j.1365-2133.2010.09762.x. [DOI] [PubMed] [Google Scholar]
- 211.Huynh K, Hoon DS. Liquid biopsies for assessing metastatic melanoma progression. Crit Rev Oncog. 2016;21(1-2):141–154. doi: 10.1615/CritRevOncog.2016016075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 212.De Souza LM, Robertson BM, Robertson GP. Future of circulating tumor cells in the melanoma clinical and research laboratory settings. Cancer Lett. 2017;392:60–70. doi: 10.1016/j.canlet.2017.01.023. [DOI] [PubMed] [Google Scholar]
- 213.Gray ES, Reid AL, Bowyer S, Calapre L, Siew K, Pearce R, Cowell L, Frank MH, Millward M, Ziman M. Circulating Melanoma Cell Subpopulations: Their Heterogeneity and Differential Responses to Treatment. J Invest Dermatol. 2015;135(8):2040–2048. doi: 10.1038/jid.2015.127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 214.Kaisaki PJ, Cutts A, Popitsch N, Camps C, Pentony MM, Wilson G, Page S, Kaur K, Vavoulis D, Henderson S. Targeted Next-Generation Sequencing of Plasma DNA from Cancer Patients: Factors Influencing Consistency with Tumour DNA and Prospective Investigation of Its Utility for Diagnosis. PLoS One. 2016;11(9):e0162809. doi: 10.1371/journal.pone.0162809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 215.Schmidt P, Kopecky C, Hombach A, Zigrino P, Mauch C, Abken H. Eradication of melanomas by targeted elimination of a minor subset of tumor cells. Proc Natl Acad Sci U S A. 2011;108(6):2474–2479. doi: 10.1073/pnas.1009069108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 216.Lebbe C, How-Kit A, Battistella M, Sadoux A, Podgorniak MP, Sidina I, Pages C, Roux J, Porcher R, Tost J. BRAF(V600) mutation levels predict response to vemurafenib in metastatic melanoma. Melanoma Res. 2014;24(4):415–418. doi: 10.1097/CMR.0000000000000088. [DOI] [PubMed] [Google Scholar]
- 217.Kemper K, Krijgsman O, Cornelissen-Steijger P, Shahrabi A, Weeber F, Song JY, Kuilman T, Vis DJ, Wessels LF, Voest EE. Intra- and inter-tumor heterogeneity in a vemurafenib-resistant melanoma patient and derived xenografts. EMBO Mol Med. 2015;7(9):1104–1118. doi: 10.15252/emmm.201404914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 218.Van Allen EM, Wagle N, Sucker A, Treacy DJ, Johannessen CM, Goetz EM, Place CS, Taylor-Weiner A, Whittaker S, Kryukov GV. The genetic landscape of clinical resistance to RAF inhibition in metastatic melanoma. Cancer Discov. 2014;4(1):94–109. doi: 10.1158/2159-8290.CD-13-0617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 219.Schwartz R, Schaffer AA. The evolution of tumour phylogenetics: principles and practice. Nat Rev Genet. 2017;18(4):213–229. doi: 10.1038/nrg.2016.170. [DOI] [PMC free article] [PubMed] [Google Scholar]