Abstract
A mutant strain of Vibiro alginolyticus with an in-frame deletion of the toxR gene was constructed to reveal the role of ToxR in the physiology and virulence of V. alginolyticus. The statistical analysis showed no significant difference in the growth ability, swarming motility, activity of extracellular protease and the virulence by injection (the value of LD50) between the wild-type and the toxR mutant. However, the deletion of toxR could decrease the level of biofilm formation. The comparative proteomic analysis demonstrated the deletion mutation of toxR could up-regulate the expression of glutamine synthetase and levansucrase, and down-regulate the expression of 10 proteins such as OmpU, DnaK, etc. These results suggest that ToxR may be involved in the early stages of infection by influencing colonization of the bacteria on the surface of the intestine through enhancing the biofilm information of V. alginolyticus via modulating the expression of glutamine synthetize, levansucrase and OmpU.
Keywords: Vibrio alginolyticus, toxR mutant, Comparative proteomic analysis, Physiology, Virulence
Introduction
Vibrio alginolyticus is a halophilic (salt-tolerant) Gram-negative bacterium found naturally in temperate marine and estuarine environments [1]. This bacterium is one of the important epizootic pathogens causing vibriosis outbreak and high mortality in sea animals with clinical symptoms of bacterial septicaemia and skin ulcer. It is also an opportunistic pathogen commonly associated with ear infections and gastrointestinal diseases in humans [2]. In addition, clinical isolates usually have high lethality in fish and high cytotoxicity to various cultured cells compared with environmental isolates [3]. Several putative virulence factors of V. alginolyticus, such as metalloprotease, alkaline serine protease and iron acquisition factors, have been reported in vivo and in vitro [4–6].
ToxR was first identified in V. cholerae in which it regulates several virulence factors in response to changes in environmental parameters like pH, temperature, amino acid composition and osmolarity of the growth medium [7]. ToxR has also been identified and characterized from a variety of Vibrio species and strains isolated from marine organisms and environments. The regulatory protein ToxR of V. cholerae contains a cytoplasmic DNA-binding-transcriptional activation domain, a trans-membrane domain and a periplasmic domain [8]. ToxR and TcpP of V. cholera could stimulate the expression of some virulence genes by combining to activate the toxT promoter, and then ToxT activates various virulence genes including cholera toxin (CT) and the toxin co-regulated pilus (TCP) directly [9]. Lee et al. [10] reported ToxR could stimulate the expression of the hemolysin gene vvhA of V. vulnificus. ToxR of V. parahaemolyticus could regulate acid, bile salts, and sodium dodecyl sulfate stresses and affect intestinal colonization in an orogastric adult murine model [11]. Similarly, Chen et al. [12] found ToxR of V. alginolyticus might have enhanced the bile resistance and biofilm formation.
Biofilm is microbial communities that are embedded in a matrix comprising polysaccharides, nucleic acids and extracellular proteins. The matrix enables the biofilm to adhere to the surfaces of planktonic cells, forms the scaffold for the construction of the biofilm, and provides mechanical stability once the biofilm is established. When grown in biofilm, bacteria have access to nutrients accumulating at interfaces [13]. Vibrio can survive as free-living or in association with zooplankton and form biofilm on biotic and abiotic surfaces in order to protect themselves with the exopolymer barrier [14]. The biofilm can also provide protection from toxic compounds, such as antibiotics, thermal stress, and predation [15]. Although biofilm formation has been studied extensively in V. cholera and V. harveyi [14, 16], much of the focus has been directed toward the survival of Vibrio in aquatic environment and its role in pathogenicity, and little is known about the related genes that can regulate biofilm formation.
In the current study, a mutant strain of V. alginolyticus with an in-frame deletion of the toxR gene was constructed, and then physiology and virulence of the mutant were observed. Furthermore, we compared the proteomes of the wild-type and toxR mutant to identify the proteins possibly regulated by toxR gene, and to determine whether some of the proteins may be involved in regulating biofilm formation.
Materials and Methods
Bacterial Strains, Plasmids and Media
The bacterial strains and plasmids used in this study are shown in Table 1. V. alginolyticus strain HY9901 was isolated from the diseased Epinephelus coioides and maintained in our laboratory. V. alginolyticus was grown in tryptic soy broth (TSB, Huankai, Guangzhou, China) supplemented with 2% NaCl at 25 °C. Escherichia coli strains were cultured in Luria broth (LB, Huankai, Guangzhou, China) at 37 °C. When required, appropriate antibiotics such as ampicillin (Amp, 100 μg ml−1) or chloramphenicol (Cm, 30 μg ml−1) were added.
Table 1.
Plasmids and bacterial strains used in this study
| Strains and plasmids | Phenotype | Source or references |
|---|---|---|
| E. coli strains | ||
| DH5α | SupE44ΔlacU169 (φ80lacZΔM15) hsdR17 recA1 gyrA96 thi-1 relA1 | Sangon |
| MC1061 λpir | lacY1 galK2 ara-14 xyl-5 supE44 λpir | Rubires et al. [31] |
| MC1061-pRE112-toxR | MC1061 containing plasmid of pRE112-toxR, Cmr | This study |
| S17-1 λpir | Tpr Smr recA thi pro hsdR−M+RP4:2-Tc:Mu:Km Tn7 λpir | |
| S17-1-pRE112-toxR | S17-1 containing plasmid of pRE112-toxR, Cmr | This study |
| Vibiro alginolyticus strain | ||
| HY9901 | Wild-type, isolated from the diseased fish | Cai et al. [5] |
| HY9901-pRE112-toxR | HY9901 containing plasmid of pRE112-toxR, Cmr | This study |
| HY9901ΔtoxR | HY9901 carrying an in-frame deletion of toxR 206-812 | This study |
| C-HY9901ΔtoxR | HY9901ΔtoxR containing plasmid of pACYC184-toxR, Cmr | This study |
| Plasmid | ||
| pMD18-T | Cloning vector; Ampr | TaKaRa |
| pRE112 | pGP704 suicide plasmid, pir dependent, oriT, oriV, sacB, Cmr | Simon et al. [32] |
| pRE112-toxR | pRE112 containing toxR gene in-frame deletion of toxR 206-812, Cmr | This study |
| pACYC184 | Cmr, Tcr | Amersham |
| pACYC184-toxR | pACYC184 containing toxR gene, Cmr | This study |
Construction of a toxR Mutant
PCR-amplified DNA fragments used for constructing the in-frame deletion mutation of toxR were generated by overlap PCR [17]. Briefly, two PCR fragments were obtained from V. alginolyticus strain HY9901 genomic DNA with the primer pairs of ToxR-for/ToxR-int-rev and ToxR-int-for/ToxR-rev (Table 2) using the pfu Taq polymerase (Promega, Madison, WI). The resulting products generated a 216-bp fragment containing the DNA upstream of toxR and a 230-bp fragment containing the DNA downstream of the toxR. A 15 bp overlap in the sequences (Italic) permitted amplification of a 442-bp product containing a deletion from nucleotides 206–812 of toxR during a second PCR with primers of ToxR-for and ToxR-rev, which were introduced at a Kpn I or Sac I restriction site (underlined), respectively. The resulting PCR product was digested with Kpn I or Sac I, then inserted into the same sites of the suicide plasmid pRE112 which carried a sacB sucrose-sensitivity gene and conferred chloramphenicol resistance, generating the recombinant plasmid pRE112-toxR transformed into E. coli MC1061 λpir and subsequently S17-1 λpir. The plasmid pRE112-toxR was sequenced to confirm the correct construction. E. coli S17-1 λpir containing the plasmid pRE112-toxR was conjugated with wild-type V. alginolyticus strain HY9901. Recipient cells were plated on TSA containing the antibiotics chloramphenicol (30 μg ml−1) to select the clone pRE112-toxR that had integrated the vector by a single crossover of allelic exchange. The conjugants were plated on counter-selective TSA containing 10% (w/v) sucrose. Colonies that grew on this medium were tested for chloramphenicol sensitivity to ensure the loss of plasmid by using TSA supplemented with chloramphenicol. The selected strains were confirmed by PCR analysis and sequencing of the chromosome. One of the mutants was named HY9901ΔtoxR and subjected to further study.
Table 2.
Sequences of primers used in this study
| Name | Sequence (5′—3′) | Targeting sequence | Product size (bp) |
|---|---|---|---|
| ToxR-T1 | AAGAACTAAATGACTAACAT | Flanking region and ORF sequence of toxR | 1106 |
| ToxR-T2 | GAGATATTTCACGTTTGA | ||
| ToxR-for | CGGGGTACCATGACTAACATTGGCACC (KpnI) | Upstream of toxR | 216 |
| ToxR-int-rev | AGTCATCCACCTCAAAACCTTG | ||
| ToxR-int-for | TTGAGGTGGATGACTACGTAACACTGCGTAT | Downstream of toxR | 230 |
| ToxR-rev | CGGAGCTCAGTATCGTCAGGCAAGCT (SacI) | ||
| ToxR-F1 | CGCGGATCCTTATTTGCAGATGTCTGT (BamHI) | ORF sequence of toxR | 879 |
| ToxR-F2 | ACATGCATGCATGACTAACATTGGCAC (SphI) |
To create a complementary plasmid for the toxR mutant, the toxR gene was amplified using primers of ToxR-F1 and ToxR-F2 (Table 2), and then the amplified fragment was cloned into BamH I and Sph I sites of the pACYC184 to construct pACYC184-toxR. The plasmid pACYC184-toxR was first transformed into E. coli DH5α and plasmid DNA was isolated, and transformed into the HY9901ΔtoxR mutant to produce the complementary strain C-HY9901ΔtoxR. The presence of the plasmid was confirmed by PCR analysis and sequencing.
Characterization of V. alginolyticus Strain HY9901ΔtoxR
The phenotypes of the mutant were characterized including growth ability, biofilm formation, extracellular protease activities and swarming motility changes as previously described [14]. Briefly, the wild-type strain HY9901, the mutant strain HY9901ΔtoxR and the complementary strain C-HY9901ΔtoxR were each cultured in TSA at 28 °C for 24 h. To measure the growth level of bacteria in TSB, overnight cultures of wild-type, the mutant and the complementary strain were inoculated into TSB with an initial OD600 of 0.01, respectively. Samples were removed every 1 h and OD600 was measured. Biofilm formation, extracellular protease activities, swarming motility were assayed and measured [18–20]. All experiments were performed in triplicate.
The medium lethal dose (LD50) determination was conducted to assess the virulence of wild-type, the mutant and the complementary strain. The grouper fish (E. coioides) approximately weighing 50 g were obtained from a commercial farm in Zhanjiang city of China, cultured in tanks with circulating sea water at 28 °C and fed to satiation daily with commercial dry pellets. After 7 days of acclimatizing in the tank, the LD50 was determined to assess the mortality titration response of the wild-type, the mutant and the complementary strain. Briefly, the different strains of V. alginolyticus were grown to an OD600 of 0.4 in TSB, pelleted by centrifugation, and resuspended in sterilized phosphate-buffered saline (PBS, pH 7.4) to densities of 1 × 108, 1 × 107, 1 × 106, 1 × 105 and 1 × 104 cfu ml−1. For each bacterial density, a group of 20 grouper fish was injected intraperitoneally with 0.1 ml of bacterial suspension, and the control group was injected intraperitoneally with 0.1 ml of PBS. All of the experiments repeated in triplicate. The fish were monitored for mortality for 7 days and the values of LD50 were calculated.
Two-Dimensional Gel Electrophoresis (2-DE) Profiles of Whole-Cell Lysates and the Identification of Protein Spots
Vibiro alginolyticus strain HY9901ΔtoxR and HY9901 were grown in TSB supplemented with 2% NaCl with gentle agitation at 28 °C for 18 h, respectively. Preparation of whole-cell protein extract and 2-DE was carried out essentially [21]. The first-dimensional isoelectric focusing (IEF) and second-dimensional SDS-gel electrophoresis were performed according to the Amersham Biosciences manual. IEF was carried out with IPG strips (Immobiline DryStripTM, pH 4–7, 7 cm, GE Healthcare) by the IPGphor TM system (GE Healthcare, Uppsala, Sweden). After IEF, the IPG strips were equilibrated for two intervals of 15 min in the equilibration solution. Equilibrated strips were placed onto 12.5% SDS–polyacrylamide gels, with a Bio-Rad Cell system (Bio-Rad, Hercules, CA, USA). Electrophoresis was performed with an initial constant current of 80 V for 30 min, followed by 180 V until the tracking dye reached the gel bottom. The 2-DE experiments were repeated in triplicate.
Gel analysis was processed, and then the detection and calculation of standardized abundance of protein spots were carried out [21]. The volume ratio was calculated as the ratio of logarithmic values of the standardized volumes. Student’s t test was used to determine the differential expression of protein spots. The differential spots reported had p values of ≤ 0.01 and volume ratio of ≥ 2.0. The differential protein spots were excised, digested with trypsin and then analyzed by Liquid Chromatography Mass Spectrometry analysis (LC–MS/MS) between wild-type and the mutant [22]. MS/MS fragmentation patterns for each protein spot were searched against the NCBI database using a MASCOT search engine (www.matrixscience.com) and NMPDR database (http://www.nmpdr.org/FIG/wiki/view.cgi). Protein identification was considered significant if at least 2 different peptides were matched to the database. Mixture hits were ignored.
Results
Construction of the toxR Mutant
To understand the possible role of toxR in V. alginolyticus, an unmarked toxR deletion mutant was constructed by allelic exchange. Using the double selection strategy of allelic exchange mutagenesis by means of suicide vector pRE112, we deleted 607-bp (residues 206–812) of the toxR gene in V. alginolyticus strain HY9901, and obtained the toxR mutant. The mutant was confirmed by the mutant’s inability to grow on TSA supplemented with chloramphenicol (30 μg ml−1), and also verified by generating a 442-bp fragment after PCR analysis. Chromosomal sequencing showed that the mutation did not alter the reading frame of the downstream genes of toxR.
Characterization of the toxR Mutant
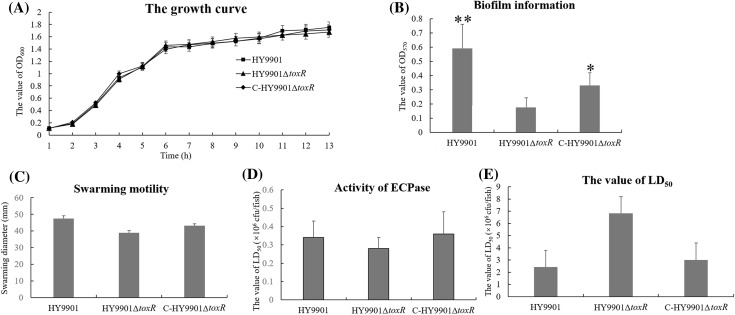
Several physiological phenotypes were compared among the mutant, wild-type and the complementary strain (Fig. 1). The mutant showed no discernible morphological difference from wild-type and the complementary strain when cultured in TSB. The mutant also showed a similar growth level with wild-type and the complementary strain. No significant difference of the activity of ECPase and swarming diameter was found between the mutant and wild-type when cultured in TSA, or TSB medium (p > 0.05). The toxR mutant of V. alginolyticus had suppressed biofilm formation, which was confirmed by the statistical analysis (p < 0.05).
Fig. 1.
The physiological phenotypes of the mutant, wild-type and the complementary strain. a The growth curve; b the biofilm formation; c the swarming motility; d the activity of ECPase; e the value of LD50. *p < 0.05; **p < 0.01
To investigate the virulence of ToxR in vivo, the grouper fish were intraperitoneally injected with the mutant, wild-type and the complementary strain, and the value of LD50 was then determined. The results showed the values of LD50 for the mutant, wild-type and the complementary strain were 6.82 × 106, 2.42 × 106 and 3.01 × 106 cfu per fish, respectively, indicating no significant difference in the virulence among the mutant, wild-type and the complementary strain under the experimental conditions (p > 0.05). The pathologic signs of vibriosis were observed by ulcers on the skin, hemorrhagic and swelling in the liver and kidney in the experimental group. None of fish died or became diseased in the control group.
2-DE Profiles of Whole-Cell Lysates and Protein Identification
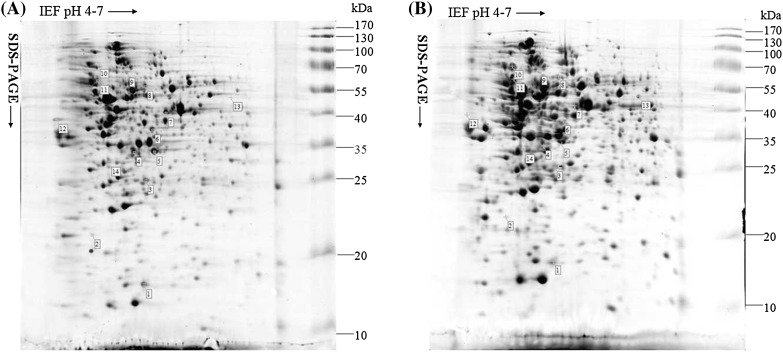
The whole-cell lysates from wild-type and the mutant were resolved by 2-DE, in triplicates for each of the two independent biological samples. A total number of 427 protein spots were detected, out of which 2 (0.05%) proteins increased and 12 (2.81%) decreased in expression levels significantly (p < 0.01) after Coomassie R-250 staining (Fig. 2). Most of the spots were found to be in the pH 4–7 range and had molecular masses of 10–130 kDa. Of the 427 protein spots detected, 14 protein spots of interest were excised to detect by LC–MS/MS analysis (Table 3). The LC–MS/MS results showed only 12 protein spots corresponding to 12 proteins could be identified among 14 different protein spots, and the LC–MS/MS results of spot 2 and 3 were negative due to the very low abundance of the proteins. In the identified 12 proteins, the expression levels of 2 proteins including glutamine synthetase and levansucrase were up-regulated, and the expression levels of 10 proteins including metabolism-related proteins, transport-related proteins, molecular chaperone DnaK and outer membrane protein U (OmpU) were down-regulated in the toxR mutant compared with that in wild-type (Table 3).
Fig. 2.
2D map (pH 4–7) of a whole-cell lysate of Vibrio alginolyticus strain HY9901 and HY9901ΔtoxR. The positions of molecular weight standards are indicated on the right. a The typical 2D gel of Vibrio alginolyticus strain HY9901. b The typical 2D gel of Vibrio alginolyticus strain HY9901ΔtoxR
Table 3.
Identified protein spots with changed expression levels by LC-MALDI
| Protein no. | Protein name | R | Accession no. | Function | Subcellular localization |
|---|---|---|---|---|---|
| T1 | Pterin-4-alpha-carbinolamine dehydratase | − 1.76 | gi|91227037 | Biosynthesis of cofactors, prosthetic groups, and carriers | Cell inner membrane |
| T4 | Electron transfer flavoprotein, alpha-subunit | − 1.57 | gi|91224794 | Iron-binding | Cell inner membrane, cytoplasm |
| T5 | Amino acid ABC transporter, periplasmic amino acid-binding protein | − 1.42 | gi|91227965 | Transport and binding proteins | Periplasm |
| T6 | Putative pyruvate dehydrogenase E1 component, alpha subunit | − 1.29 | gi|91225779 | Unknown | Cytoplasm |
| T7 | Dihydrolipoamide acetyltransferase | − 1.54 | gi|91225777 | Energy metabolism | Cytoplasm |
| T8 | Pyruvate kinase | − 1.27 | gi|91227129 | Energy metabolism | Cell inner membrane, cytoplasm |
| T9 | d-fructose-6-phosphate amidotransferase | − 1.42 | gi|91227131 | Energy metabolism | Cytoplasm |
| T10 | Molecular chaperone DnaK | − 1.31 | gi|91225094 | Promote protein folding, interaction, and translocation, both constitutively and in response to stress, | Cytoplasm |
| T11 | Glutamine synthetase | + 1.36 | gi|91224957 | Amino acid biosynthesis | Cytoplasm |
| T12 | Outer membrane protein U | − 1.88 | gi|237846493 | Cell envelope | Cell outer membrane |
| T13 | Chain A, crystal structure of levansucrase | + 1.53 | gi|38492844 | Hydrolysis of sucrose | Extracell |
| T14 | Hypothetical protein V12G01_14960 | − 1.23 | gi|91227427 | Unknown | Cytoplasm |
R = + (the mutant strain/wild-type strain) for up-regulated proteins, and R = − (wild-type strain/the mutant strain) for down-regulated
Discussion
Vibiro alginolyticus can adopt several survival strategies in aquatic environments. The bacterium can survive as free-living or in association with zooplankton, and penetrate the surface barriers of the host and disseminate effectively in the host tissue as a major cause of fish bacterial septicaemia [12]. Biofilm formation is a multicellular behavior by which bacteria can colonize a surface of the host tissue, leading to resistance to antibiotics and host immune-killing [23]. It was reported that V. alginolyticus invaded the intestine of fish where some adverse factors such as anti-microbial peptides could easily be overcome for successful colonization and initiation of infection with the assistance of biofilm formation [24]. In this study, a mutant strain of V. alginolyticus with an in-frame deletion of toxR gene was constructed to reveal the role of ToxR in the physiology and virulence of V. alginolyticus. ToxR was shown to enhance biofilm formation, which should assist bacterial survival and colonization in the fish intestine. However, we found the deletion mutation of toxR didn’t clearly affect the growth ability, swarming motility, activity of ECPase and the virulence (the value of LD50). These indicate that ToxR may not be needed for the development of disseminated disease. Therefore, ToxR may be only involved in the early stages of infection by influencing colonization of the bacteria on the surface of the intestine through enhancing the biofilm information. The results were in accordance with that of Chen et al. [12] in V. alginolyticus strain ZJ51-O.
Biofilm formation of Vibrio depends on specific structural genes (flagella, pili and exopolysaccharide biosynthesis) and complex regulatory processes (two-component regulators, quorum sensing and c-di-GMP signaling) [25]. In the study, the deletion mutation of toxR could up-regulate the expression of glutamine synthetase and levansucrase, and down-regulate the expression of 10 proteins such as OmpU. It was proved that biofilm information was suppressed in case of glutamine synthetase mutant in Mycobacterium bovis [26]. The glutamine synthetase also overexpressed during biofilm growth in exponential and stationary phases of bacterial growth by DNA array analysis of an Escherichia coli biofilm formation [27]. It was reported that levansucrase could contribute to biofilm formation in Pseudomonas syringae, and its expression occurred mainly in the early stages of bacterial growth and biofilm development [28]. Levansucrase is also involved in tolerance to NaCl, sucrose and desiccation, and in biofilm formation in Gluconacetobacter diazotrophicus [29]. In the investigation of biofilm formation, the ToxR of V. alginolyticus had significantly enhanced biofilm formation. Therefore, there are good theoretical reasons to believe that the expression of glutamine synthetase and levansucrase might have related to biofilm formation of V. alginolyticus.
However, an evidence showed that the expression of OmpU was positively regulated by ToxR, and a toxR mutant showed a thicker biofilm with better surface area coverage compared to that of wild-type in V. anguillarum [30]. Interestingly, our results showed that the expression of OmpU was suppressed and biofilm formation was decreased in the toxR mutant, and were inconsistent with that in V. anguillarum. This indicated OmpU could negatively regulate biofilm formation in V. alginolyticus. Furthermore, it should be further studied that how OmpU negatively regulates biofilm formation of V. alginolyticus.
In conclusion, the results of our study showed that ToxR didn’t clearly affect the growth ability, swarming motility, activity of ECPase and the virulence, and could only enhance biofilm formation of V. alginolyticus probably via modulating the expression of glutamine synthetase, levansucrase and OmpU. The suggests ToxR may be involved in the early stages of infection by influencing colonization of the bacteria on the surface of the intestine through enhancing the biofilm information, and cannot affect directly the later stage of disseminated infection as indicated by the failure of toxR mutation to attenuate the virulence of V. alginolyticus.
Acknowledgements
This work was supported by the National Natural Science Foundation of China (No. 31572656), Fishing Port Construction and Fishery Development Funds of Guangdong Province (Prevention of Fish Disease), Natural Science Foundation of Guangdong Province (No. 2015A030308020, 2017A030307033 and 2017A030313174), and High-level Talents Project of Sailing Plan in Guangdong Province (No. GDOU2017030501).
References
- 1.Cai SH, Lu YS, Jian JC, Wang B, Huang YC, Tang JF, Ding Y, Wu ZH. Protection against Vibrio alginolyticus in crimson snapper Lutjanus erythropterus immunized with a DNA vaccine containing the ompW gene. Dis Aquat Organ. 2013;106:39–47. doi: 10.3354/dao02617. [DOI] [PubMed] [Google Scholar]
- 2.Austin B. Vibrios as causal agents of zoonoses. Vet Microbiol. 2010;140:310–317. doi: 10.1016/j.vetmic.2009.03.015. [DOI] [PubMed] [Google Scholar]
- 3.Lu YS, Feng JM, Wu ZH, Jian JC. Genotype analysis of collagenase gene by PCR-SSCP in Vibrio alginolyticus and its association with virulence to marine fish. Curr Microbiol. 2011;62:1697–16703. doi: 10.1007/s00284-011-9919-z. [DOI] [PubMed] [Google Scholar]
- 4.Wang TY, Chen YC, Kao LW, Chang CY, Wang YK, Liu YH, Feng JM, Wu TK. Expression and characterization of the biofilm-related and carnosine-hydrolyzing aminoacylhistidine dipeptidase from Vibrio alginolyticus. FEBS J. 2008;275:5007–5020. doi: 10.1111/j.1742-4658.2008.06635.x. [DOI] [PubMed] [Google Scholar]
- 5.Cai SH, Wu ZH, Jian JC, Lu YS. Cloning and expression of the gene encoding an extracellular alkaline serine protease from Vibrio alginolyticus strain HY9901, the causative agent of vibriosis in Lutjanus erythopterus (Bloch) J Fish Dis. 2007;30:493–500. doi: 10.1111/j.1365-2761.2007.00835.x. [DOI] [PubMed] [Google Scholar]
- 6.Wang Q, Liu Q, Ma Y, Zhou L, Zhang Y. Isolation, sequencing and characterization of cluster genes involved in the biosynthesis and utilization of the siderophore of marine fish pathogen Vibrio alginolyticus. Arch Microbiol. 2007;188:433–439. doi: 10.1007/s00203-007-0261-6. [DOI] [PubMed] [Google Scholar]
- 7.DiRita VJ, Mekalanos JJ. Periplasmic interaction between two membrane regulatory proteins, ToxR and ToxS, results in signal transduction and transcriptional activation. Cell. 1991;64:29–37. doi: 10.1016/0092-8674(91)90206-E. [DOI] [PubMed] [Google Scholar]
- 8.Reich KA, Schoolnik GK. The light organ symbiont Vibrio fischeri possesses a homolog of the Vibrio cholerae transmembrane transcriptional activator ToxR. J Bacteriol. 1994;176:3085–3088. doi: 10.1128/jb.176.10.3085-3088.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Withey JH, DiRita VJ. The toxbox: specific DNA sequence requirements for activation of Vibrio cholerae virulence genes by ToxT. Mol Microbiol. 2006;59:1779–1789. doi: 10.1111/j.1365-2958.2006.05053.x. [DOI] [PubMed] [Google Scholar]
- 10.Lee SE, Shin SH, Kim SY, Kim YR, Shin DH, Chung SS, Lee ZH, Lee JY, Jeong KC, Choi SH, Rhee JH. Vibrio vulnificus has the transmembrane transcription activator ToxRS stimulating the expression of the hemolysin gene vvhA. J Bacteriol. 2000;182:3405–3415. doi: 10.1128/JB.182.12.3405-3415.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Whitaker WB, Parent MA, Boyd A, Richards GP, Boyd EF. The Vibrio parahaemolyticus ToxRS regulator is required for stress tolerance and colonization in a novel orogastric streptomycin-induced adult murine model. Infect Immun. 2012;80:1834–1845. doi: 10.1128/IAI.06284-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chen C, Wang QB, Liu ZH, Zhao JJ, Jiang X, Sun HY, Ren CH, Hu CQ. Characterization of role of the toxR gene in the physiology and pathogenicity of Vibrio alginolyticus. Antonie Van Leeuwenhoek. 2012;101:281–288. doi: 10.1007/s10482-011-9632-8. [DOI] [PubMed] [Google Scholar]
- 13.Johnson TL, Fong JC, Rule C, Rogers A, Yildiz FH, Sandkvist M. The type II secretion system delivers matrix proteins for biofilm formation by Vibrio cholerae. J Bacteriol. 2014;196:4245–4252. doi: 10.1128/JB.01944-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Valeru SP, Wai SN, Saeed A, Sandström G, Abd H. ToxR of Vibrio cholerae affects biofilm, rugosity and survival with Acanthamoeba castellanii. BMC Res Notes. 2012;5:33. doi: 10.1186/1756-0500-5-33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Islam MS, Jahid MI, Rahman MM, Rahman MZ, Kabir MS, Sack DA, Schoolnik GK. Biofilm acts as a microenvironment for plankton associated Vibrio cholerae in the aquatic environment of Bangladesh. Microbiol Immunol. 2007;51:369–379. doi: 10.1111/j.1348-0421.2007.tb03924.x. [DOI] [PubMed] [Google Scholar]
- 16.Karunasagar I, OttaIndrani SK, Karunasagar I. Biofilm formation by Vibrio harveyi on surfaces. Aquaculture. 1996;140:241–245. doi: 10.1016/0044-8486(95)01180-3. [DOI] [Google Scholar]
- 17.Ho SN, Hunt HD, Horton RM, Pullen JK, Pease LR. Site-directed mutagenesis by overlap extension using the polymerase chain reaction. Gene. 1989;77:51–59. doi: 10.1016/0378-1119(89)90358-2. [DOI] [PubMed] [Google Scholar]
- 18.Zhou ZJ, Pang HY, Ding Y, Cai J, Huang YC, Jian JC, Wu ZH. VscO, a putative T3SS chaperone escort of Vibrio alginolyticus, contributes to virulence in fish and is a target for vaccine development. Fish Shellfish Immunol. 2013;35:1523–1531. doi: 10.1016/j.fsi.2013.08.017. [DOI] [PubMed] [Google Scholar]
- 19.Fletcher M. The effects of culture concentration and age, time, and temperature on bacterial attachment to polystyrene. Can J Microbiol. 1977;23:1–6. doi: 10.1139/m77-001. [DOI] [Google Scholar]
- 20.Mathew J, Tan Y, Rao PSS, Lim T, Leung K. Edwardsiella tarda mutants defective in siderophore production, motility, serum resistance and catalase activity. Microbiology. 2001;147:449–457. doi: 10.1099/00221287-147-2-449. [DOI] [PubMed] [Google Scholar]
- 21.Pang HY, Zhang XZ, Wu ZH, Jian JC, Cai SH, Liang J. Identification of novel immunogenic proteins of Vibrio alginolyticus by immunoproteomic methodologies. Aquac Res. 2013;44:472–484. doi: 10.1111/j.1365-2109.2012.03150.x. [DOI] [Google Scholar]
- 22.Stickland HG, Davenport PW, Lilley KS, Griffin JL, Welch M. Mutation of nfxB causes global changes in the physiology and metabolism of Pseudomonas aeruginosa. J Proteome Res. 2010;9:2957–2967. doi: 10.1021/pr9011415. [DOI] [PubMed] [Google Scholar]
- 23.Verstraeten N, Braeken K, Debkumari B, Fauvart M, Fransaer J, Vermant J, Michiels J. Living on a surface: swarming and biofilm formation. Trends Microbiol. 2008;16:496–506. doi: 10.1016/j.tim.2008.07.004. [DOI] [PubMed] [Google Scholar]
- 24.Chen Q, Yan Q, Wang K, Zhuang Z, Wang X. Portal of entry for pathogenic Vibrio alginolyticus into large yellow croaker Pseudosciaena crocea, and characteristics of bacterial adhesion to mucus. Dis Aquat Organ. 2008;80:181–188. doi: 10.3354/dao01933. [DOI] [PubMed] [Google Scholar]
- 25.Watnick PI, Lauriano CM, Klose KE, Croal L, Kolter R. The absence of a flagellum leads to altered colony morphology, biofilm development and virulence in Vibrio cholerae O139. Mol Microbiol. 2001;39:223–235. doi: 10.1046/j.1365-2958.2001.02195.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chandra H, Basir SF, Gupta M, Banerjee N. Glutamine synthetase encoded by glnA-1 is necessary for cell wall resistance and pathogenicity of Mycobacterium bovis. Microbiology. 2010;156:2677–3669. doi: 10.1099/mic.0.043828-0. [DOI] [PubMed] [Google Scholar]
- 27.Schembri MA, Kjaergaard K, Klemm P. Global gene expression in Escherichia coli biofilms. Mol Microbiol. 2003;48:253–267. doi: 10.1046/j.1365-2958.2003.03432.x. [DOI] [PubMed] [Google Scholar]
- 28.Laue H, Schenk A, Li H, Lambertsen L, Neu TR, Molin S, Ullrich MS. Contribution of alginate and levan production to biofilm formation by Pseudomonas syringae. Microbiology. 2006;152:2909–2918. doi: 10.1099/mic.0.28875-0. [DOI] [PubMed] [Google Scholar]
- 29.Velázquez-Hernández ML, Baizabal-Aguirre VM, Cruz-Vázquez F, Trejo-Contreras MJ, Fuentes-Ramírez LE, Bravo-Patiño A, Cajero-Juárez M, Chávez-Moctezuma MP, Valdez-Alarcón JJ. Gluconacetobacter diazotrophicus levansucrase is involved in tolerance to NaCl, sucrose and desiccation, and in biofilm formation. Arch Microbiol. 2011;193:137–149. doi: 10.1007/s00203-010-0651-z. [DOI] [PubMed] [Google Scholar]
- 30.Wang SY, Lauritz L, Jass J, Milton DL. A ToxR homolog from Vibrio anguillarum serotype O1 regulates its own production, bile resistance, and biofilm formation. J Bacteriol. 2002;184:1630–1639. doi: 10.1128/JB.184.6.1630-1639.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Rubirés X, Saigi F, Piqué N, Climent N, Merino S, Albertí S, Tomás JM, Regué M. A gene (wbbL) from Serratia marcescens N28b (O4) complements the rfb-50 mutation of Escherichia coli K-12 derivatives. J Bacteriol. 1997;179:7581–7586. doi: 10.1128/jb.179.23.7581-7586.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Simon R, Priefer U, Pühler A. A Broad Host Range Mobilization System for In Vivo Genetic Engineering: Transposon Mutagenesis in Gram Negative Bacteria. Nat Biotechnol. 1983;1:784–791. doi: 10.1038/nbt1183-784. [DOI] [Google Scholar]