Abstract
Iso-cytoplasmic restorers possess the same male sterile cytoplasm as the cytoplasmic male sterile (CMS) lines, thereby minimizing the potential cyto-nuclear conflict in the hybrids. Restoration of fertility of the wild abortive CMS is governed by two major genes namely, Rf3 and Rf4. Therefore, assessing the allelic status of these restorer genes in the iso-cytoplasmic restorers using molecular markers will not only help in estimating the efficiency of these genes either alone or in combination, in fertility restoration in the hybrids in different environments, but will also be useful in determining the efficacy of these markers. In the present study, the efficiency of molecular markers in identifying genotypes carrying restorer allele of the gene(s) Rf3 and Rf4, restoring male fertility of WA cytoplasm in rice was assessed in a set of 100 iso-cytoplasmic rice restorers using gene linked as well as candidate gene based markers. In order to validate the efficacy of markers in identifying the restorers, a sub-set of selected 25 iso-cytoplasmic rice restorers were crossed with four different cytoplasmic male sterile lines namely, IR 79156A, IR 58025A, Pusa 6A and RTN 12A, and the pollen and spikelet fertility of the F1s were evaluated at three different locations. Marker analysis showed that Rf4 was the predominant fertility restorer gene in the iso-cytoplasmic restorers and Rf3 had a synergistic effect on fertility restoration. The efficiency of gene based markers, DRCG-RF4-14 and DRRM-RF3-10 for Rf4 (87%) and Rf3 (84%) genes was higher than respective gene-linked SSR markers RM6100 (80%) and RM3873 (82%). It is concluded that the gene based markers can be effectively used in identifying fertility restorer lines obviating the need for making crosses and evaluating the F1s. Though gene based markers are more efficient, there is a need to identify functional polymorphisms which can provide 100% efficiency. Three iso-cytoplasmic restorers namely, PRR 300, PRR 363 and PRR 396 possessing both Rf4 and Rf3 genes and good fertility restoration have been identified which could be used further in hybrid rice breeding.
Keywords: Hybrids, Iso-cytoplasmic restorers, Gene based markers, Fertility restorer genes, Pollen fertility
Introduction
Rice is the staple food of more than 1.25 billion people in most parts of India. An increase in rice production is necessary to bring self-sufficiency in the country to feed the burgeoning population. India has the largest acreage under paddy at 43.40 million hectare with a production of 157.20 m tones as compared to China having 30.87 million hectare under rice cultivation with a total production of 208.23 m tones. Productivity of China (6.74 t/ha) is almost twice that of productivity in India (3.62 t/ha) (FAOSTAT 2014). The higher productivity of China has been mainly attributed to the large-scale adoption of hybrid rice with hybrids accounting for more than half of cultivated rice area in the country (17 mha out of 30 mha) (Xie et al. 2014).
Hybrid technology is considered as one of the most potential, efficient, productive and sustainable options for breaking yield levels witnessed in several crops including rice (Kutka 2011; Peng et al. 2004; Sheeba et al. 2009). In spite of its huge potential, the area under hybrid rice during 2013 was merely 2.8 mha in India, which is only 6.5% of the total cultivated area under rice. There is a need to promote hybrid rice through systematic enhancement of heterosis in rice. In indica rice, wild abortive (WA) cytoplasm, derived from Oryza sativa f. sp. spontanea (Lin and Yuan 1980; Li and Yuan 1986) is one of the most widely used cytoplasmic male sterility (CMS) source for hybrid rice production, due to its complete and stable fertility restoration.
For developing heterotic hybrid, there is a need to identify/develop restorers with better restoration efficiency in the hybrids. Evaluation for fertility restoration is mainly done through test crossing with CMS lines and evaluating the fertility (both pollen and spikelet) of the F1 hybrids, which is laborious and time consuming. Evaluation of fertility restoration is based on three indices namely per cent fertile pollen, bagged and open seed-setting of the panicles; of which per cent fertile pollens is one of the reliable criteria for determining fertility (Ghara et al. 2012). Compared to the allo-cytoplamsic restorers, the iso-cytoplasmic restorer lines are unique as they carry the same male sterile cytoplasm which minimizes the potential cyto-nuclear conflict between the cytoplasmic and nuclear genes. Additionally, these iso-cytoplasmic restorers carry male sterile cytoplasm and therefore, there is no need for test crossing to assess their restoration potential. However, restoration of fertility of the WA CMS is governed by two major loci namely, Rf3 located on chromosome 1 and Rf4 on chromosome 10 (Yao et al. 1997) and the information on the status of these restorer genes in the iso-cytoplasmic restorers can help in estimating the efficiency of these genes, either alone or in combination in restoring fertility in the test cross hybrids in different environments.
Molecular markers based on/linked to genes governing fertility restoration greatly help in precise selection, saves time and overcomes the drawbacks of phenotypic screening (Revathi et al. 2013). Thus, the current investigation was undertaken with the objective of characterizing the iso-cytoplasmic restorer lines for Rf3 and Rf4 alleles using gene linked as well as gene based markers, and their effects on pollen and spikelet fertility in test crosses derived from crosses between CMS lines and iso-cytoplasmic restorer lines in different environments.
Materials and methods
Plant material
Iso-cytoplasmic restorer lines of rice derived from the commercial hybrids released across the country were used in this study. The pure F1 seeds of top 25 hybrids were grown at ICAR-IARI farm during 2010, panicles were bagged to self-pollinate, producing selfed seeds resulting in F2 seeds. These second filial generation seeds were grown at off season nursery at RBGRC, ICAR-IARI, Aduthurai, Tamil Nadu. Plants with better panicle exsertion, spikelet fertility and plant yield in F2 generation were selected and advanced to F3, and superior progenies with good spikelet fertility were advanced till F6 generation. In F6 generation, 100 lines showing good spikelet fertility and plant yield were selected. These 100 lines were evaluated in augmented design and one superior iso-cytoplasmic restorer from each of the 25 hybrids was selected and test crosses were attempted with four diverse WA based cytoplasmic male sterile lines namely, IR 79156A, IR 58025A, Pusa 6A and RTN 12A. The 100 test cross hybrids were evaluated at three locations namely Delhi, Karnal and Pusa (Bihar) in augmented design.
Analysis of Rf genes using molecular markers
The 100 iso-cytoplasmic rice restorer lines were grown in portrays with 12 plants in each well. Fresh tissues from green, young and healthy plants were collected 10 days after germination. The plant tissue was chopped into small pieces into 2 mL eppendorf tube, dipped into liquid nitrogen and crushed using Mixer Mill MM400 (Restsch GmbH, Germany). Crushed tissues were further processed using CTAB method to isolate high quality DNA (Stewart and Via 1993). Presence of restorer allele of the Rf genes were screened among 100 iso-cytoplasmic lines using two gene linked markers namely, RM 3873 and RM 6100, and two candidate gene based markers namely, DRRM-RF3-10 and DRCG-RF4-14 (Suresh et al. 2012; Revathi et al. 2013) for Rf3 and Rf4, respectively (Table 1).
Table 1.
Classification of 100 iso-cytoplasmic rice restorer lines based on alleles detected byRf3 and Rf4 gene linked or gene based markers
| Markers | Rf3Rf3 | Rf3rf3 | rf3rf3 | Total | Markers | Rf4Rf4 | Rf4rf4 | rf4rf4 | Total |
|---|---|---|---|---|---|---|---|---|---|
| RM3873 | 33 | 12 | 55 | 100 | RM6100 | 73 | 4 | 18 | 95 |
| DRRM RF3-10 | 27 | 3 | 70 | 100 | DRCG RF4-14 | 77 | 5 | 18 | 100 |
| No. of genotypes in common | 10 | 0 | 41 | 51 | No. of genotypes in common | 63 | 2 | 12 | 77 |
Pollen fertility and spikelet fertility analysis
For the analysis of pollen fertility (PF), panicle from the main tiller of each plant was collected 1 day before the onset of anthesis. The panicles were immediately kept in the fixative solution consisting of ethanol and glacial acetic acid in the ratio of 3:1. Three randomly selected spikelets from different positions in the panicle were split open and the anthers were smeared in a drop of 1% Iodine-Potassium Iodide (I-KI) on a glass slide separately and visualized under compound microscope. The round and stained pollen were considered as fertile, while shriveled and unstained pollen were considered as sterile. Test crosses were classified into different groups based on the proportion of stained-round pollens (per cent PF) as completely sterile (0% PF), partially sterile (1–30% PF), partially fertile (30–60% PF) and fertile (>60% PF), as proposed by Chaudhury et al. (1981). After harvesting the plant, panicle from the main tiller of each plant was kept in envelope and numbers of filled grains as well as chaffy seeds were counted and spikelet fertility percentage was calculated. Data was taken for randomly selected five plants and the average value was used for the calculation. The efficiency of molecular markers in identifying fertility restorers was computed using SPSS software.
Results
Screening of iso-cytoplasmic restorer lines with gene linked and candidate gene based markers for Rf3 and Rf4 genes
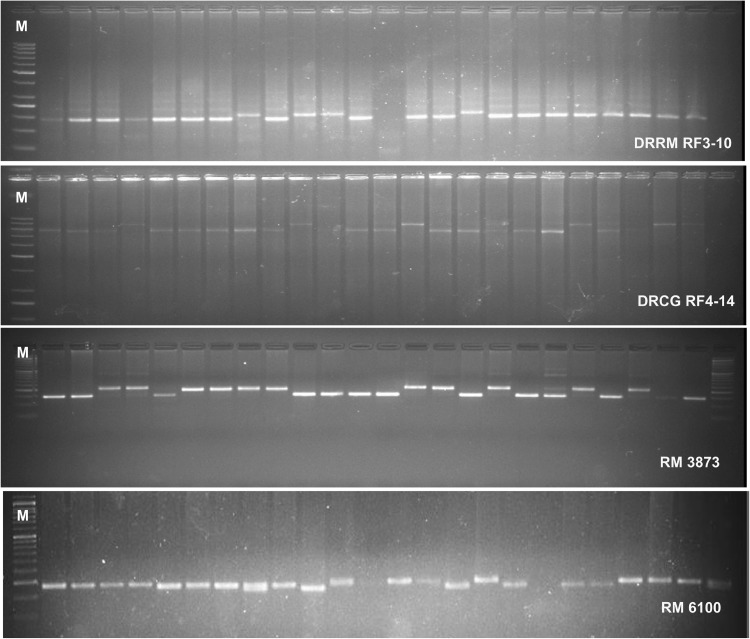
A set of 100 iso-cytoplasmic restorer lines were screened with gene linked markers namely, RM 3873 and RM 6100 as well as candidate gene based markers namely, DRRM-RF3-10 and DRCG-RF4-14 for Rf3 and Rf4, respectively. The fragments amplified using these markers in different rice genotypes were classified into restorer and non-restorer alleles based on the restorer check, PRR78 and maintainer check, Pusa 6B as well as in comparison with previous reports (Suresh et al. 2012; Revathi et al. 2013). Representative amplification profiles of 4 markers are presented in Fig. 1.
Fig. 1.
A representative amplification profile of the gene based and linked markers for fertility restoration in the iso-cytoplasmic restorer lines. Marker: DNA Ladder 100 bp, DRRM-RF3-10: R—210 bp, NR—190 bp, DRCG-RF4-14: R—800 bp, NR—900 bp, RM3873: R—210 bp, NR—155 bp, RM 6100: R—150 bp, NR—140 bp where, R = Restorer allele, NR = Non-restorer allele
For Rf3, 10 out of the 100 genotypes were found to be homozygous for restorer alleles with both linked as well as gene based markers namely, RM 3873 and DRRM-RF3-10 while there were 41 genotypes (41.0%), which were homozygous for the non-restorer allele of Rf3 with these two markers (Table 1). With RM 3873, 33.0% genotypes (33 genotypes) were homozygous for Rf3 restorer allele and 55.0% genotypes (55 genotypes) were homozygous for non-restorer allele rf3, whereas with DRRM-RF3-10, only 27.0% genotypes (27 genotypes) were homozygous for Rf3 allele while 70.0% genotypes (70 genotypes) were homozygous for non-restorer allele, rf3. Further, 12.0% and 3.0% genotypes (12 and 3 genotypes) were found to be heterozygous for Rf3 with RM 3873 and DRRM-RF3-10, respectively.
With respect to Rf4 gene, 63 genotypes (63.0%) were homozygous for restorer allele of Rf4 with both gene linked marker, RM 6100 and gene based marker DRCG-RF4-14, while 12 genotypes were found homozygous for non-restorer allele for Rf4 (Table 1). About 73.0% genotypes (73 genotypes) were homozygous for restorer allele Rf4 for RM 6100 whereas it was 77.0% (77 genotypes) with DRCG-RF4-14. Eighteen percent genotypes (18 genotypes) were found to be homozygous for non-restorer allele (rf4rf4). Further, 4.0% and 5.0% genotypes (4 and 5 genotypes) were found to be heterozygous (Rf4rf4) with RM 6100 and DRCG-RF4-14, respectively.
Taking into consideration all the four markers used for analysis of the fertility restorer genes, the genotypes were grouped into different combinations namely, Rf3Rf3Rf4Rf4, Rf3Rf3rf4rf4, rf3rf3Rf4Rf4, rf3rf3rf4rf4, Rf3rf3Rf4Rf4, Rf3Rf3Rf4rf4, etc., (Table 2). A total of four marker combinations namely, DRRM-RF3-10 + DRCG RF4-14, DRRM-RF3-10 + RM 6100, DRCG-RF4-14 + RM 3873 and RM3873 + RM 6100 were identified using one marker each for Rf3 and Rf4 genes, respectively. Frequency of iso-cytoplasmic restorer lines carrying only Rf4 genes (40%) was highest followed by the frequency of lines carrying both Rf3 and Rf4 genes (>22%) and more than 10% lines were found to carry the non-restorer allele of Rf3 and Rf4 genes.
Table 2.
Classification of hundred iso-cytoplasmic rice restorer lines for both Rf3 and Rf4 gene linked and gene based markers
| Genotype | DRRM-RF3-10 + DRCG RF4-14 | DRRM-RF3-10 + RM 6100 | DRCG-RF4-14 + RM 3873 | RM3873 + RM 6100 |
|---|---|---|---|---|
| Rf3Rf3Rf4Rf4 | 23 | 23 | 23 | 24 |
| Rf3Rf3rf4rf4 | 5 | 4 | 10 | 8 |
| rf3rf3Rf4Rf4 | 49 | 50 | 42 | 42 |
| rf3rf3rf4rf4 | 16 | 17 | 9 | 11 |
| Rf3rf3Rf4Rf4 | 2 | 2 | 9 | 8 |
| Rf3Rf3Rf4rf4 | 0 | 1 | 1 | 1 |
| Rf3rf3Rf4rf4 | 1 | 1 | 1 | 2 |
| Rf3rf3rf4rf4 | 0 | 0 | 2 | 2 |
| rf3rf3Rf4rf4 | 4 | 2 | 3 | 2 |
| Total | 100 | 100 | 100 | 100 |
Evaluation of test crosses for fertility restoration
A total of 100 test crosses derived by crossing twenty-five selected iso-cytoplasmic restorer lines with four male sterile lines namely, IR 79156A, IR 58025A, Pusa 6A and RTN 12A were subjected to pollen and spikelet fertility evaluation. The test crosses were grouped into different categories based on the extent of pollen fertility. Pollen fertility in the all test crosses ranged from 0 to 100% with an average pollen fertility of 78.82% over all the locations (Table 3). The average pollen fertility with IR 79156A, IR 58025A, Pusa 6A and RTN 12A were found to be 80.03, 78.03, 80.10 and 77.11%, respectively. The test crosses derived from IR 79156A showed highest pollen fertility across different locations followed by Pusa 6A, IR 58025A and RTN 12A, respectively. Spikelet fertility was also calculated across different locations, and IR 79156A (70.36%) and RTN 12A (69.75) produced test crosses with highest spikelet fertility. When test crosses were planted in early conditions in Delhi, test crosses derived from IR 79156 A (86.38%) exhibited highest pollen fertility followed by IR 58025A (82.61%) and RTN 12A (81.80%). But when trial was sown in late conditions in Delhi, test crosses derived from Pusa 6A (84.69%) followed by IR 58025A (76.45%) had highest pollen fertility. At Pusa (Bihar), test crosses with RTN 12A (80.54%) showed its superiority whereas in Karnal, test crosses with IR 79156A (80.51%) were found to be better in pollen fertility. The spikelet fertility of the test crosses with four male sterile lines had similar/comparable performances.
Table 3.
An overview of the pollen fertility status in test crosses generated with different CMS lines
| CMS lines | Parameters | Delhi (Early) | Delhi (late) | Bihar | Karnal | ||||
|---|---|---|---|---|---|---|---|---|---|
| PF (%) | SF (%) | PF (%) | SF (%) | PF (%) | SF (%) | PF (%) | SF (%) | ||
| IR 79156A | Mean | 86.38 | 73.54 | 74.82 | 63.52 | 78.41 | 74.28 | 80.51 | 70.10 |
| Maximum | 93.24 | 92.06 | 98.03 | 85.67 | 91.80 | 92.65 | 92.70 | 82.49 | |
| Minimum | 68.00 | 57.76 | 1.86 | 18.46 | 40.70 | 41.78 | 41.20 | 39.70 | |
| IR 58025A | Mean | 82.61 | 71.99 | 76.45 | 61.70 | 76.38 | 73.90 | 76.71 | 68.58 |
| Maximum | 94.93 | 86.07 | 93.23 | 80.78 | 89.29 | 93.39 | 93.13 | 81.18 | |
| Minimum | 29.13 | 28.54 | 0.60 | 30.50 | 6.44 | 11.49 | 0.00 | 44.27 | |
| Pusa 6A | Mean | 79.97 | 65.65 | 84.69 | 62.70 | 77.41 | 74.61 | 78.34 | 73.92 |
| Maximum | 93.14 | 84.70 | 94.90 | 84.43 | 89.32 | 90.62 | 94.18 | 96.31 | |
| Minimum | 7.96 | 32.51 | 13.19 | 43.24 | 33.33 | 40.67 | 0.00 | 53.13 | |
| RTN 12A | Mean | 81.80 | 72.23 | 76.19 | 64.70 | 80.54 | 73.00 | 69.92 | 69.06 |
| Maximum | 93.75 | 85.44 | 95.60 | 86.84 | 92.75 | 90.67 | 89.45 | 83.71 | |
| Minimum | 1.54 | 59.10 | 19.46 | 36.25 | 2.24 | 8.90 | 0.00 | 52.20 | |
Where, PF (%) = Pollen fertility percentage, SF (%) = Spikelet fertility percentage
Efficacy of markers in identifying potential restorers
Based on genotypic data on gene linked/gene based markers and phenotypic data on pollen fertility, the efficiency of the markers in identification of restorers was calculated. The test crosses with more than 60% pollen fertility were considered as complete restorers. Based on the correspondence between the presence of restorer alleles and the pollen fertility of more than 60 per cent in the test crosses, the efficiency of the markers in identification of restorers from the diverse set of rice genotypes were estimated (Table 4).
Table 4.
Per cent efficiency of the gene linked and gene based markers in identification of potential restorers: (a) Spikelet fertility, (b) pollen fertility
| Parameter | Location | RM3873 | RM6100 | DRRM RF3-10 | DRCG RF4-14 |
|---|---|---|---|---|---|
| (a) | |||||
| Genotypes homozygous for Restorer allele | 33 | 73 | 27 | 77 | |
| Genotypes homozygous for non-R allele | 55 | 18 | 70 | 18 | |
| Genotypes used in test crosses | 25 | 25 | 25 | 25 | |
| Test crosses (F1) made with R allele genotypes (all four CMS lines) | 36 | 64 | 40 | 76 | |
| Test crosses (F1) made with R allele genotypes and restorer phenotype (>60% PF) | Delhi (Early) | 34 | 56 | 37 | 68 |
| Delhi (Late) | 29 | 52 | 35 | 66 | |
| Karnal | 33 | 55 | 35 | 68 | |
| Bihar | 31 | 58 | 36 | 69 | |
| Percent efficiency (%) = with R allele (>60% PF) | Delhi (Early) | 89 | 87 | 90 | 92 |
| Delhi (Late) | 76 | 68 | 79 | 81 | |
| Karnal | 83 | 80 | 84 | 84 | |
| Bihar | 82 | 87 | 95 | 89 | |
| (b) | |||||
| Genotypes homozygous for Restorer allele | 33 | 73 | 27 | 77 | |
| Genotypes homozygous for non-R allele | 55 | 18 | 70 | 18 | |
| Genotypes used in test crosses | 25 | 25 | 25 | 25 | |
| Test crosses (F1) made with R allele genotypes (all four CMS lines) | 36 | 64 | 40 | 76 | |
| Test crosses (F1) made with R allele genotypes and restorer phenotype (>60% SF) | Delhi (Early) | 32 | 53 | 37 | 66 |
| Delhi (Late) | 19 | 40 | 30 | 53 | |
| Karnal | 29 | 51 | 32 | 65 | |
| Bihar | 31 | 54 | 33 | 64 | |
| Percent efficiency (%) = with R allele (>60% SF) | Delhi (Early) | 78 | 77 | 79 | 86 |
| Delhi (Late) | 15 | 29 | 54 | 45 | |
| Karnal | 66 | 67 | 68 | 76 | |
| Bihar | 82 | 74 | 78 | 75 | |
SF spikelet fertility, PF pollen fertility
Pollen fertility
The per cent efficiency for gene linked markers namely, RM 3873 and RM 6100 were 82.5 and 80.5%, respectively whereas, for the gene based markers namely, DRRM-RF3-10 and DRCG-RF4-14, the efficiency were observed to be 83.75 and 86.75%, respectively. The results of different locations were compared and it was observed that the markers were more efficient in identifying the restoration potential, when the test crosses were planted in normal planting season. In cases, where planting was delayed, the efficiency of the markers was comparatively lower.
Spikelet fertility
It was observed that the comparative efficiency of molecular markers in identifying potential restorers based on spikelet fertility was lesser than on the basis of pollen fertility. In timely sown conditions, a number of test crosses with IR 79156A had shown maximum pollen (100%) as well as spikelet fertility (92%) followed by IR 58025A. In late planting conditions, test crosses with Pusa 6A and RTN 12A showed better spikelet fertility. Even though, the efficacy of markers for spikelet fertility was lesser than that of pollen fertility, gene based markers were still more efficient as compared to gene linked markers in identifying the restorer lines.
Correlation of hybrids performance with their parental value
Correlation was computed for pooled data of all iso-cytoplasmic restorers used for crossing (male sterile line wise) (Table 5). Comparison was also made between the test crosses with their respective male lines for detailed information on type of correlation for each cross combination (Table 6a, b). No significant correlation was observed between the iso-cytoplasmic restorer lines and their cross combinations as far as pollen as well as spikelet fertility were concerned. This remained constant when restorer genes were absent/present singly or together.
Table 5.
Correlation between iso-cytoplasmic restorers and their hybrid performances for pollen and spikelet fertility
| IR 79156 A | IR 58025 A | Pusa 6 A | RTN 12 A | |||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Del (Early) | Del (Late) | Bihar | Karnal | Del (Early) | Del (Late) | Bihar | Karnal | Del (Early) | Del (Late) | Bihar | Karnal | Del (Early) | Del (Late) | Bihar | Karnal | |
| Pollen fertility | ||||||||||||||||
| PRR-DEL2014 | 0.202 | 0.291 | −0.140 | 0.174 | −0.190 | 0.048 | −0.417** | −0.040 | 0.020 | 0.025 | −0.213 | 0.572** | −0.047 | 0.340 | 0.268 | −0.060 |
| PRR-ADT | 0.120 | 0.127 | 0.064 | −0.020 | −0.004 | 0.025 | −0.001 | −0.326 | 0.197 | 0.198 | 0.099 | −0.089 | 0.012 | 0.191 | −0.188 | −0.144 |
| PRR-DEL2015 | 0.196 | 0.320 | −0.177 | 0.160 | −0.141 | 0.170 | −0.436** | 0.041 | −0.029 | −0.050 | −0.242 | 0.485** | −0.058 | 0.351 | 0.444 | −0.053 |
| Spikelet fertility | ||||||||||||||||
| PRR-DEL2014 | −0.124 | −0.049 | −0.047 | 0.074 | −0.001 | 0.086 | 0.052 | −0.038 | 0.110 | 0.220 | −0.244 | 0.284 | 0.296 | 0.255 | 0.249 | 0.186 |
| PRR-ADT | 0.168 | −0.076 | −0.134 | 0.019 | −0.066 | −0.053 | 0.087 | 0.031 | −0.235 | 0.005 | 0.203 | −0.175 | −0.372 | −0.247 | 0.032 | −0.266 |
| PRR-DEL2015 | 0.094 | 0.227 | −0.055 | 0.315 | 0.639** | 0.150 | 0.381 | 0.400** | 0.451** | 0.210 | 0.218 | −0.034 | 0.171 | 0.315 | 0.084 | −0.210 |
Where PRR-DEL2014 = PRR (Iso-cytoplasmic restorer) lines grown in Delhi (2014), PRR-ADT = PRR (Iso-cytoplasmic restorer) lines grown in Aduthurai, Tamil Nadu (2014–2015) and PRR-DEL2015 = PRR (Iso-cytoplasmic restorer) lines grown in Delhi (2015)
Table 6.
Association between iso-cytoplasmic restorer lines and their derived hybrid performances for (a) pollen fertility, (b) spikelet fertility
| TC1 | TC2 | TC3 | TC4 | TC5 | TC6 | TC7 | TC8 | TC9 | TC10 | TC11 | TC12 | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| (a) | ||||||||||||
| PRR1 | −0.99 | −0.23 | 0.60 | 0.09 | 0.24 | −0.97 | −0.37 | 0.56 | −1.00 | −0.23 | 0.05 | 0.17 |
| PRR2 | −0.99 | −0.24 | 0.62 | 0.07 | 0.25 | −0.97 | −0.39 | 0.54 | −1.00 | −0.25 | 0.07 | 0.19 |
| PRR3 | −0.99 | −0.21 | 0.59 | 0.10 | 0.22 | −0.97 | −0.36 | 0.57 | −1.00 | −0.22 | 0.04 | 0.16 |
| PRR4 | 1.00 | 0.29 | −0.66 | −0.02 | −0.30 | 0.98 | 0.43 | −0.50 | 1.00 | 0.30 | −0.12 | −0.24 |
| PRR5 | −1.00 | −0.29 | 0.66 | 0.02 | 0.31 | −0.98 | −0.44 | 0.50 | −1.00 | −0.30 | 0.12 | 0.24 |
| PRR6 | −0.99 | −0.25 | 0.62 | 0.07 | 0.26 | −0.97 | −0.39 | 0.54 | −1.00 | −0.25 | 0.08 | 0.19 |
| PRR7 | 0.98 | 0.20 | −0.58 | −0.11 | −0.21 | 0.96 | 0.35 | −0.58 | 1.00 | 0.21 | −0.03 | −0.15 |
| PRR8 | −0.99 | −0.51 | 0.82 | −0.22 | 0.53 | −1.00 | −0.64 | 0.27 | −0.95 | −0.52 | 0.36 | 0.47 |
| PRR9 | 0.99 | 0.25 | −0.62 | −0.06 | −0.26 | 0.98 | 0.40 | −0.53 | 1.00 | 0.26 | −0.08 | −0.20 |
| PRR10 | −0.96 | −0.63 | 0.89 | −0.36 | 0.64 | −0.98 | −0.74 | 0.14 | −0.90 | −0.63 | 0.48 | 0.59 |
| PRR11 | −0.99 | −0.47 | 0.79 | −0.17 | 0.48 | −1.00 | −0.60 | 0.32 | −0.97 | −0.48 | 0.31 | 0.42 |
| PRR12 | 0.99 | 0.24 | −0.61 | −0.08 | −0.25 | 0.97 | 0.38 | −0.55 | 1.00 | 0.24 | −0.06 | −0.18 |
| PRR13 | −1.00 | −0.34 | 0.69 | −0.03 | 0.35 | −0.99 | −0.48 | 0.45 | −0.99 | −0.35 | 0.17 | 0.29 |
| PRR14 | 1.00 | 0.29 | −0.65 | −0.02 | −0.30 | 0.98 | 0.43 | −0.50 | 1.00 | 0.30 | −0.12 | −0.24 |
| PRR15 | −1.00 | −0.31 | 0.67 | 0.00 | 0.33 | −0.99 | −0.45 | 0.48 | −1.00 | −0.32 | 0.15 | 0.26 |
| PRR16 | −1.00 | −0.46 | 0.78 | −0.16 | 0.47 | −1.00 | −0.59 | 0.33 | −0.97 | −0.47 | 0.30 | 0.41 |
| PRR17 | −0.99 | −0.50 | 0.81 | −0.21 | 0.51 | −1.00 | −0.63 | 0.29 | −0.96 | −0.51 | 0.34 | 0.45 |
| PRR18 | −0.99 | −0.52 | 0.82 | −0.23 | 0.53 | −1.00 | −0.65 | 0.26 | −0.95 | −0.53 | 0.37 | 0.48 |
| PRR19 | −1.00 | −0.41 | 0.75 | −0.11 | 0.43 | −1.00 | −0.55 | 0.38 | −0.98 | −0.42 | 0.25 | 0.36 |
| PRR20 | −1.00 | −0.43 | 0.76 | −0.13 | 0.44 | −1.00 | −0.57 | 0.36 | −0.98 | −0.44 | 0.27 | 0.38 |
| PRR21 | 0.99 | 0.25 | −0.62 | −0.07 | −0.26 | 0.98 | 0.39 | −0.54 | 1.00 | 0.26 | −0.08 | −0.20 |
| PRR22 | −1.00 | −0.40 | 0.74 | −0.10 | 0.41 | −1.00 | −0.53 | 0.40 | −0.98 | −0.41 | 0.24 | 0.35 |
| PRR23 | −1.00 | −0.42 | 0.76 | −0.12 | 0.44 | −1.00 | −0.56 | 0.37 | −0.98 | −0.43 | 0.26 | 0.38 |
| PRR24 | −1.00 | −0.37 | 0.71 | −0.06 | 0.38 | −1.00 | −0.51 | 0.43 | −0.99 | −0.38 | 0.20 | 0.32 |
| PRR25 | −0.99 | −0.27 | 0.64 | 0.04 | 0.28 | −0.98 | −0.41 | 0.52 | −1.00 | −0.28 | 0.10 | 0.22 |
| (b) | ||||||||||||
| PRR1 | −0.89 | 0.90 | 0.53 | 0.73 | 0.64 | 0.30 | 0.81 | −0.65 | −0.99 | −0.99 | 0.54 | −0.58 |
| PRR2 | 0.89 | −0.17 | 0.38 | −0.98 | 0.25 | 0.60 | −0.95 | 1.00 | 0.71 | 0.69 | −1.00 | 1.00 |
| PRR3 | 0.57 | −1.00 | −0.87 | −0.32 | −0.93 | −0.72 | −0.44 | 0.22 | 0.79 | 0.81 | −0.08 | 0.13 |
| PRR4 | 0.57 | −1.00 | −0.86 | −0.32 | −0.93 | −0.71 | −0.44 | 0.22 | 0.79 | 0.81 | −0.08 | 0.13 |
| PRR5 | 0.64 | 0.24 | 0.72 | −0.82 | 0.61 | 0.87 | −0.74 | 0.88 | 0.38 | 0.35 | −0.94 | 0.92 |
| PRR6 | −0.37 | 0.97 | 0.95 | 0.11 | 0.99 | 0.85 | 0.24 | 0.00 | −0.64 | −0.66 | −0.14 | 0.09 |
| PRR7 | 0.86 | −0.92 | −0.58 | −0.69 | −0.69 | −0.36 | −0.77 | 0.60 | 0.97 | 0.98 | −0.49 | 0.53 |
| PRR8 | −0.57 | 1.00 | 0.86 | 0.33 | 0.92 | 0.71 | 0.45 | −0.23 | −0.79 | −0.81 | 0.09 | −0.14 |
| PRR9 | 0.57 | 0.32 | 0.78 | −0.77 | 0.68 | 0.91 | −0.68 | 0.83 | 0.29 | 0.26 | −0.90 | 0.88 |
| PRR10 | 0.51 | 0.39 | 0.82 | −0.72 | 0.73 | 0.94 | −0.62 | 0.79 | 0.22 | 0.19 | −0.87 | 0.84 |
| PRR11 | 0.12 | 0.72 | 0.98 | −0.39 | 0.94 | 1.00 | −0.26 | 0.48 | −0.18 | −0.21 | −0.60 | 0.56 |
| PRR12 | 0.13 | 0.72 | 0.98 | −0.40 | 0.94 | 1.00 | −0.27 | 0.49 | −0.18 | −0.21 | −0.61 | 0.57 |
| PRR13 | 0.52 | 0.37 | 0.81 | −0.73 | 0.72 | 0.93 | −0.64 | 0.80 | 0.24 | 0.21 | −0.88 | 0.85 |
| PRR14 | 0.09 | 0.74 | 0.99 | −0.36 | 0.95 | 1.00 | −0.23 | 0.45 | −0.21 | −0.24 | −0.58 | 0.53 |
| PRR15 | 0.33 | 0.56 | 0.91 | −0.58 | 0.85 | 0.99 | −0.46 | 0.66 | 0.03 | 0.00 | −0.76 | 0.73 |
| PRR16 | 0.87 | −0.13 | 0.42 | −0.97 | 0.29 | 0.63 | −0.93 | 0.99 | 0.68 | 0.66 | −1.00 | 1.00 |
| PRR17 | 0.37 | −0.97 | −0.95 | −0.10 | −0.99 | −0.85 | −0.23 | 0.00 | 0.63 | 0.66 | 0.14 | −0.09 |
| PRR18 | 0.51 | 0.39 | 0.82 | −0.72 | 0.73 | 0.93 | −0.63 | 0.79 | 0.23 | 0.20 | −0.87 | 0.85 |
| PRR19 | −0.95 | 0.82 | 0.39 | 0.82 | 0.52 | 0.15 | 0.89 | −0.76 | −1.00 | −1.00 | 0.66 | −0.70 |
| PRR20 | −0.43 | −0.47 | −0.87 | 0.66 | −0.79 | −0.96 | 0.55 | −0.73 | −0.13 | −0.10 | 0.82 | −0.79 |
| PRR21 | 0.44 | 0.46 | 0.86 | −0.67 | 0.78 | 0.96 | −0.57 | 0.74 | 0.15 | 0.12 | −0.83 | 0.80 |
| PRR22 | 0.63 | 0.25 | 0.73 | −0.82 | 0.62 | 0.87 | −0.73 | 0.87 | 0.37 | 0.34 | −0.93 | 0.91 |
| PRR23 | −0.91 | 0.88 | 0.49 | 0.76 | 0.60 | 0.26 | 0.84 | −0.69 | −0.99 | −1.00 | 0.58 | −0.62 |
| PRR24 | −0.05 | 0.83 | 1.00 | −0.22 | 0.99 | 0.98 | −0.09 | 0.32 | −0.35 | −0.38 | −0.45 | 0.41 |
| PRR25 | 0.41 | 0.49 | 0.88 | −0.64 | 0.80 | 0.97 | −0.53 | 0.72 | 0.11 | 0.08 | −0.81 | 0.78 |
| TC13 | TC14 | TC15 | TC16 | TC17 | TC18 | TC19 | TC20 | TC21 | TC22 | TC23 | TC24 | TC25 | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| (a) | |||||||||||||
| PRR1 | −0.87 | 0.93 | 0.95 | −0.95 | 0.17 | −0.98 | 0.75 | −0.63 | 0.92 | 0.39 | 0.62 | −0.99 | 0.45 |
| PRR2 | −0.86 | 0.94 | 0.94 | −0.95 | 0.18 | −0.98 | 0.76 | −0.61 | 0.91 | 0.38 | 0.63 | −0.99 | 0.43 |
| PRR3 | −0.87 | 0.93 | 0.95 | −0.96 | 0.15 | −0.99 | 0.74 | −0.64 | 0.92 | 0.41 | 0.61 | −0.99 | 0.46 |
| PRR4 | 0.83 | −0.95 | −0.93 | 0.93 | −0.23 | 0.97 | −0.79 | 0.57 | −0.89 | −0.33 | −0.67 | 0.98 | −0.38 |
| PRR5 | −0.83 | 0.95 | 0.93 | −0.93 | 0.24 | −0.97 | 0.79 | −0.57 | 0.89 | 0.33 | 0.67 | −0.98 | 0.38 |
| PRR6 | −0.86 | 0.94 | 0.94 | −0.95 | 0.19 | −0.98 | 0.76 | −0.61 | 0.91 | 0.37 | 0.64 | −0.99 | 0.43 |
| PRR7 | 0.88 | −0.92 | −0.96 | 0.96 | −0.14 | 0.99 | −0.73 | 0.65 | −0.93 | −0.42 | −0.60 | 0.99 | −0.47 |
| PRR8 | −0.67 | 1.00 | 0.81 | −0.81 | 0.46 | −0.88 | 0.92 | −0.36 | 0.75 | 0.09 | 0.83 | −0.90 | 0.15 |
| PRR9 | 0.85 | −0.94 | −0.94 | 0.94 | −0.19 | 0.98 | −0.76 | 0.61 | −0.91 | −0.37 | −0.64 | 0.99 | −0.42 |
| PRR10 | −0.56 | 1.00 | 0.72 | −0.72 | 0.58 | −0.81 | 0.96 | −0.22 | 0.65 | −0.05 | 0.90 | −0.84 | 0.01 |
| PRR11 | −0.71 | 0.99 | 0.84 | −0.84 | 0.42 | −0.91 | 0.89 | −0.40 | 0.78 | 0.14 | 0.80 | −0.93 | 0.20 |
| PRR12 | 0.86 | −0.94 | −0.95 | 0.95 | −0.18 | 0.98 | −0.75 | 0.62 | −0.91 | −0.38 | −0.63 | 0.99 | −0.44 |
| PRR13 | −0.80 | 0.97 | 0.91 | −0.91 | 0.28 | −0.96 | 0.82 | −0.53 | 0.86 | 0.28 | 0.71 | −0.97 | 0.34 |
| PRR14 | 0.83 | −0.95 | −0.93 | 0.93 | −0.23 | 0.97 | −0.79 | 0.57 | −0.89 | −0.33 | −0.67 | 0.98 | −0.39 |
| PRR15 | −0.82 | 0.96 | 0.92 | −0.92 | 0.26 | −0.96 | 0.80 | −0.55 | 0.88 | 0.31 | 0.69 | −0.98 | 0.36 |
| PRR16 | −0.72 | 0.99 | 0.84 | −0.85 | 0.41 | −0.91 | 0.89 | −0.41 | 0.79 | 0.15 | 0.79 | −0.93 | 0.21 |
| PRR17 | −0.69 | 1.00 | 0.82 | −0.82 | 0.45 | −0.89 | 0.91 | −0.37 | 0.76 | 0.11 | 0.82 | −0.91 | 0.17 |
| PRR18 | −0.67 | 1.00 | 0.80 | −0.81 | 0.47 | −0.88 | 0.92 | −0.35 | 0.74 | 0.08 | 0.84 | −0.90 | 0.14 |
| PRR19 | −0.75 | 0.99 | 0.87 | −0.87 | 0.36 | −0.93 | 0.86 | −0.46 | 0.82 | 0.20 | 0.76 | −0.95 | 0.26 |
| PRR20 | −0.74 | 0.99 | 0.86 | −0.86 | 0.38 | −0.92 | 0.87 | −0.44 | 0.81 | 0.18 | 0.78 | −0.94 | 0.24 |
| PRR21 | 0.86 | −0.94 | −0.94 | 0.95 | −0.19 | 0.98 | −0.76 | 0.61 | −0.91 | −0.37 | −0.64 | 0.99 | −0.43 |
| PRR22 | −0.76 | 0.98 | 0.88 | −0.88 | 0.34 | −0.94 | 0.86 | −0.47 | 0.83 | 0.22 | 0.75 | −0.95 | 0.28 |
| PRR23 | −0.74 | 0.99 | 0.86 | −0.87 | 0.37 | −0.93 | 0.87 | −0.45 | 0.81 | 0.19 | 0.77 | −0.94 | 0.25 |
| PRR24 | −0.78 | 0.98 | 0.89 | −0.90 | 0.31 | −0.95 | 0.84 | −0.50 | 0.85 | 0.25 | 0.73 | −0.96 | 0.31 |
| PRR25 | −0.84 | 0.95 | 0.93 | −0.94 | 0.21 | −0.98 | 0.78 | −0.59 | 0.90 | 0.35 | 0.65 | −0.98 | 0.40 |
| (b) | |||||||||||||
| PRR1 | −0.98 | 0.82 | −0.07 | −0.87 | 0.96 | −0.62 | −0.64 | 0.24 | 0.97 | −0.08 | −0.98 | −0.41 | 0.87 |
| PRR2 | 0.74 | −0.01 | 0.85 | 0.10 | −0.35 | −0.28 | 1.00 | 0.65 | −0.36 | −0.76 | 0.40 | 0.98 | −0.91 |
| PRR3 | 0.76 | −0.99 | −0.41 | 1.00 | −0.97 | 0.92 | 0.21 | −0.67 | −0.97 | 0.54 | 0.96 | −0.07 | −0.53 |
| PRR4 | 0.76 | −0.99 | −0.40 | 1.00 | −0.98 | 0.92 | 0.21 | −0.67 | −0.97 | 0.54 | 0.96 | −0.07 | −0.54 |
| PRR5 | 0.42 | 0.39 | 0.99 | −0.30 | 0.05 | −0.63 | 0.88 | 0.90 | 0.04 | −0.96 | 0.01 | 0.98 | −0.67 |
| PRR6 | −0.60 | 1.00 | 0.59 | −0.98 | 0.90 | −0.98 | 0.01 | 0.82 | 0.90 | −0.71 | −0.88 | 0.29 | 0.34 |
| PRR7 | 0.96 | −0.85 | 0.01 | 0.90 | −0.98 | 0.67 | 0.59 | −0.30 | −0.98 | 0.14 | 0.99 | 0.35 | −0.84 |
| PRR8 | −0.76 | 0.99 | 0.40 | −1.00 | 0.98 | −0.91 | −0.21 | 0.67 | 0.97 | −0.54 | −0.96 | 0.07 | 0.54 |
| PRR9 | 0.34 | 0.46 | 1.00 | −0.38 | 0.14 | −0.70 | 0.84 | 0.93 | 0.12 | −0.98 | −0.08 | 0.96 | −0.60 |
| PRR10 | 0.27 | 0.53 | 1.00 | −0.45 | 0.21 | −0.75 | 0.80 | 0.96 | 0.20 | −0.99 | −0.15 | 0.93 | −0.54 |
| PRR11 | −0.14 | 0.82 | 0.91 | −0.76 | 0.58 | −0.95 | 0.50 | 0.99 | 0.57 | −0.96 | −0.53 | 0.72 | −0.16 |
| PRR12 | −0.13 | 0.82 | 0.91 | −0.76 | 0.58 | −0.95 | 0.50 | 0.99 | 0.56 | −0.97 | −0.52 | 0.72 | −0.17 |
| PRR13 | 0.28 | 0.52 | 1.00 | −0.43 | 0.20 | −0.74 | 0.81 | 0.95 | 0.18 | −0.99 | −0.14 | 0.94 | −0.55 |
| PRR14 | −0.17 | 0.84 | 0.90 | −0.78 | 0.61 | −0.96 | 0.47 | 0.99 | 0.60 | −0.95 | −0.56 | 0.69 | −0.13 |
| PRR15 | 0.08 | 0.68 | 0.98 | −0.61 | 0.39 | −0.86 | 0.67 | 0.99 | 0.38 | −1.00 | −0.34 | 0.85 | −0.37 |
| PRR16 | 0.72 | 0.03 | 0.87 | 0.07 | −0.31 | −0.31 | 0.99 | 0.68 | −0.33 | −0.79 | 0.37 | 0.99 | −0.89 |
| PRR17 | 0.60 | −0.99 | −0.60 | 0.98 | −0.90 | 0.98 | −0.01 | −0.82 | −0.90 | 0.71 | 0.87 | −0.29 | −0.34 |
| PRR18 | 0.27 | 0.53 | 1.00 | −0.44 | 0.21 | −0.75 | 0.80 | 0.95 | 0.19 | −0.99 | −0.15 | 0.94 | −0.54 |
| PRR19 | −1.00 | 0.72 | −0.22 | −0.79 | 0.91 | −0.49 | −0.75 | 0.09 | 0.92 | 0.07 | −0.94 | −0.54 | 0.93 |
| PRR20 | −0.18 | −0.60 | −0.99 | 0.52 | −0.30 | 0.81 | −0.74 | −0.98 | −0.28 | 1.00 | 0.24 | −0.90 | 0.46 |
| PRR21 | 0.20 | 0.59 | 1.00 | −0.51 | 0.28 | −0.80 | 0.75 | 0.97 | 0.27 | −1.00 | −0.22 | 0.91 | −0.48 |
| PRR22 | 0.41 | 0.40 | 0.99 | −0.31 | 0.06 | −0.64 | 0.88 | 0.90 | 0.05 | −0.96 | 0.00 | 0.98 | −0.66 |
| PRR23 | −0.99 | 0.79 | −0.12 | −0.85 | 0.95 | −0.58 | −0.68 | 0.20 | 0.96 | −0.04 | −0.97 | −0.45 | 0.89 |
| PRR24 | −0.31 | 0.91 | 0.82 | −0.87 | 0.72 | −0.99 | 0.33 | 0.96 | 0.70 | −0.90 | −0.67 | 0.58 | 0.02 |
| PRR25 | 0.16 | 0.62 | 0.99 | −0.54 | 0.32 | −0.82 | 0.73 | 0.98 | 0.30 | −1.00 | −0.26 | 0.89 | −0.44 |
Where, PRR1.PRR 300, PRR2.PRR 307, PRR3.PRR 311, PRR4.PRR 314, PRR5.PRR 317, PRR6.PRR 323, PRR7.PRR 326, PRR8.PRR 329, PRR9.PRR 334, PRR10.PRR 337, PRR11.PRR 342, PRR12.PRR 347, PRR13.PRR 348, PRR14.PRR 354, PRR15.PRR 358, PRR16.PRR 363, PRR17.PRR 367, PRR18.PRR 368, PRR19.PRR 372, PRR20.PRR 376, PRR21.PRR 381, PRR22.PRR 386, PRR23.PRR 390, PRR24.PRR 395, PRR25.PRR 396
Identification of promising restorers
Out of 100 iso-cytoplasmic restorer lines, 20 of them were found to possess restorer alleles of both Rf3 and Rf4 genes (Table 7). These lines can be used as potential restorers with various male sterile lines. In the test crosses involving twenty-five iso-cytoplasmic restorers, a set of six restorers possessing restorer alleles of both Rf3 and Rf4 with good per se performance for yield and yield component traits, and pollen fertility of more than 80% in the test crosses have been identified (Table 8). Effects of presence of fertility restorer genes on the extent of pollen and spikelet fertility of test crosses generated from iso-cytoplasmic restorer lines were compared at four locations. Both pollen as well as spikelet fertility were found to be comparable (> 60%) when fertility restorer alleles (Rf3 and Rf4 genes) were present singly or in combination. In few combinations having male parents with/without restorer alleles of the fertility restorer genes, the extent of pollen and spikelet fertility was below 60% (Table 9a, b). Thus, in this case an unknown alleles/genes might be responsible for fertility restoration, which needs further in-depth study.
Table 7.
Identification of iso-cytoplasmic restorer lines carrying both Rf3 and Rf4 genes based on gene based markers
| S. No. | Identified restorer lines | Parental hybrids |
|---|---|---|
| 1 | PRR 300, PRR 302, PRR 303 | DRRH 2 |
| 2 | PRR 304, PRR 305, PRR 306 | DRRH 3 |
| 3 | PRR 308 | PSD 3 |
| 4 | PRR 313 | PRH 10 |
| 5 | PRR 332, PRR 333 | Sahyadri 4 |
| 6 | PRR 360, PRR 361, PRR 363 | PA 6201 |
| 7 | PRR 367 | PA 6444 |
| 8 | PRR 389, PRR 390 | PAC 835 |
| 9 | PRR 394, PRR 395 | PAC 837 |
| 10 | PRR 396, PRR 398 | KRH 2 |
Bold italics—lines also present in the set used (25 selected) for crossing
Table 8.
Per se performance of some promising iso-cytoplasmic restorer lines possessing both Rf3 and Rf4 genes
| CMH | PH | NT | PL | SF | TW | PF | DFF | YPP |
|---|---|---|---|---|---|---|---|---|
| PRR 300 | 96.07 | 10.78 | 28.75 | 82.91 | 22.58 | 91.65 | 89.94 | 27.62 |
| PRR 363 | 111.22 | 11.47 | 25.70 | 75.83 | 16.22 | 91.57 | 99.44 | 20.00 |
| PRR 367 | 98.70 | 9.87 | 24.05 | 84.72 | 21.20 | 86.35 | 93.44 | 14.73 |
| PRR 390 | 112.14 | 9.56 | 27.79 | 87.92 | 22.94 | 89.67 | 94.69 | 21.60 |
| PRR 395 | 94.75 | 10.76 | 23.76 | 86.58 | 28.05 | 88.22 | 95.02 | 23.62 |
| PRR 396 | 119.44 | 9.69 | 25.74 | 78.74 | 22.24 | 86.43 | 100.69 | 25.37 |
Where PH plant height, NT number of tillers, PL panicle length, SF spikelet fertility, TW test weight, PF pollen fertility, DFF days to fifty percent flowering and YPP yield per plant
Table 9.
Performance of test crosses derived from iso-cytoplasmic restorer lines carrying restorer alleles of Rf3 and Rf4 genes for (a) pollen fertility and (b) spikelet fertility across three locations
| Restorers | Performance of hybrids generated | Iso-cytoplasmic restorers (Parents) | |||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Delhi (Early) | Delhi (Late) | Bihar | Karnal | Delhi 2014 | Aduthurai 2015 | Delhi 2015 | |||||||||||||
| IR 79156A | IR 58025A | Pusa 6A | RTN 12A | IR 79156A | IR 58025A | Pusa 6A | RTN 12A | IR 79156A | IR 58025A | Pusa 6A | RTN 12A | IR 79156A | IR 58025A | Pusa 6A | RTN 12A | ||||
| (a) | |||||||||||||||||||
| Restorers with restorer allele of Rf3 gene only | |||||||||||||||||||
| PRR 314 | 89.8 | 91.3 | 92.2 | 54.6 | 80.8 | 89.7 | 86.5 | 93.1 | 84.7 | 86.7 | 90.3 | 76.6 | 88.6 | 94.9 | 80.4 | 92.4 | 81.4 | 79.6 | 81.8 |
| PRR 334 | 75.2 | 28.4 | 80.7 | 91.8 | 3.1 | 1.5 | 76.6 | 95.0 | 84.5 | 4.9 | 37.8 | 83.5 | 89.5 | 54.3 | 87.4 | 75.3 | 94.6 | 91.6 | 95.0 |
| PRR 396 | 83.3 | 86.5 | 86.4 | 77.6 | 81.0 | 91.3 | 54.2 | 92.3 | 80.3 | 69.5 | 85.4 | 85.1 | 88.0 | 85.6 | 55.8 | 91.3 | 83.9 | 93.0 | 82.3 |
| Restorers with restorer allele of Rf4 gene only | |||||||||||||||||||
| PRR 317 | 91.3 | 90.8 | 89.9 | 82.7 | 90.4 | 91.1 | 68.5 | 72.9 | 82.5 | 71.6 | 91.4 | 93.5 | 87.2 | 83.6 | 81.4 | 92.0 | 79.1 | ||
| PRR 323 | 86.9 | 78.6 | 91.1 | 88.7 | 91.5 | 88.2 | 78.4 | 89.0 | 50.9 | 78.2 | 85.3 | 70.5 | 81.7 | 89.2 | 83.5 | 88.4 | 87.3 | 91.0 | 86.7 |
| PRR 326 | 89.3 | 90.1 | 92.6 | 67.5 | 11.1 | 91.2 | 84.0 | 79.7 | 63.0 | 90.4 | 81.6 | 69.4 | 93.0 | 79.4 | 86.2 | 95.8 | 94.6 | 95.9 | |
| PRR 329 | 86.0 | 89.5 | 88.9 | 71.1 | 78.8 | 84.2 | 69.5 | 88.4 | 86.2 | 91.6 | 83.8 | 92.1 | 89.8 | 84.4 | 7.5 | 88.6 | 91.0 | 87.0 | |
| PRR 337 | 75.9 | 78.5 | 82.9 | 78.1 | 5.7 | 2.9 | 88.3 | 53.3 | 83.7 | 93.7 | 5.4 | 87.6 | 81.6 | 66.4 | 66.9 | 63.6 | 93.1 | 31.9 | |
| PRR 342 | 92.5 | 88.1 | 84.2 | 82.1 | 88.9 | 90.0 | 96.1 | 88.2 | 82.1 | 74.8 | 84.9 | 82.9 | 89.0 | 78.2 | 91.2 | 89.9 | 90.8 | 89.4 | |
| PRR 354 | 88.8 | 89.1 | 88.5 | 80.3 | 87.8 | 86.9 | 87.8 | 84.5 | 87.8 | 80.7 | 75.2 | 81.7 | 85.1 | 87.4 | 76.0 | 94.1 | 91.5 | 94.7 | |
| PRR 368 | 90.9 | 84.5 | 90.3 | 86.7 | 83.4 | 86.5 | 75.5 | 87.2 | 87.8 | 87.9 | 84.2 | 11.0 | 14.7 | 73.8 | 85.6 | 77.7 | 97.1 | 64.5 | |
| PRR 372 | 77.4 | 85.9 | 49.6 | 90.5 | 85.8 | 93.1 | 90.7 | 79.4 | 83.9 | 49.2 | 90.0 | 75.0 | 87.0 | 80.0 | 87.1 | 41.5 | 91.2 | 96.7 | 88.8 |
| PRR 376 | 93.2 | 85.1 | 87.1 | 89.6 | 6.2 | 84.8 | 87.9 | 92.9 | 68.5 | 83.2 | 97.2 | 77.7 | 89.6 | 78.2 | 64.0 | 16.6 | 90.0 | 95.7 | 87.4 |
| PRR 381 | 86.5 | 89.2 | 87.3 | 91.5 | 79.7 | 86.3 | 80.9 | 89.6 | 71.1 | 85.8 | 94.1 | 91.2 | 86.1 | 82.4 | 88.0 | 77.7 | 92.2 | 87.7 | 92.8 |
| PRR 386 | 62.6 | 86.2 | 87.0 | 84.0 | 11.4 | 82.1 | 89.6 | 81.8 | 67.5 | 85.3 | 74.9 | 68.8 | 64.0 | 40.8 | 71.7 | 89.5 | 89.8 | 89.4 | |
| Restorers with restorer alleles of both Rf3 and Rf4 genes | |||||||||||||||||||
| PRR 300 | 88.5 | 84.2 | 89.9 | 5.9 | 88.2 | 90.3 | 84.8 | 89.1 | 85.3 | 62.8 | 89.3 | 86.9 | 87.0 | 81.2 | 83.1 | 1.1 | 90.7 | 93.9 | 90.3 |
| PRR 307 | 67.3 | 73.0 | 82.1 | 84.6 | 4.5 | 87.9 | 90.1 | 86.7 | 81.0 | 84.0 | 75.2 | 89.5 | 86.1 | 89.9 | 1.5 | 73.4 | 88.6 | 71.4 | |
| PRR 358 | 84.3 | 94.0 | 86.6 | 91.1 | 83.0 | 87.1 | 89.3 | 96.8 | 91.0 | 91.0 | 85.5 | 88.7 | 92.7 | 1.5 | 90.3 | 58.6 | 93.6 | 50.1 | |
| PRR 363 | 90.6 | 72.3 | 86.5 | 87.5 | 81.5 | 91.1 | 91.4 | 75.4 | 92.3 | 88.6 | 86.3 | 61.6 | 78.7 | 83.3 | 90.5 | 91.2 | 93.6 | 89.9 | |
| PRR 367 | 90.1 | 86.5 | 81.8 | 93.3 | 89.5 | 89.1 | 89.3 | 82.3 | 88.4 | 86.7 | 79.2 | 79.2 | 82.2 | 85.9 | 87.2 | 85.3 | 93.4 | 80.3 | |
| PRR 390 | 87.3 | 87.8 | 78.6 | 81.9 | 90.9 | 82.7 | 93.8 | 90.5 | 74.6 | 92.7 | 84.0 | 89.1 | 78.1 | 87.9 | 84.9 | 90.2 | 88.5 | 94.9 | 85.7 |
| PRR 395 | 88.9 | 79.9 | 88.6 | 8.6 | 78.4 | 89.1 | 86.7 | 79.7 | 78.6 | 88.0 | 90.5 | 6.0 | 3.7 | 87.1 | 68.6 | 86.2 | 95.3 | 83.2 | |
| Restorers lacking restorer alleles of both Rf3 and Rf4 genes | |||||||||||||||||||
| PRR 311 | 84.6 | 87.3 | 7.0 | 82.9 | 3.8 | 52.0 | 73.6 | 17.7 | 81.0 | 77.9 | 84.9 | 86.3 | 11.7 | 71.6 | 82.8 | 75.1 | 85.5 | 74.1 | |
| PRR 347 | 88.7 | 67.3 | 39.0 | 83.2 | 88.1 | 87.5 | 87.4 | 70.9 | 54.9 | 28.2 | 85.4 | 81.8 | 85.1 | 61.5 | 87.0 | 81.3 | 93.8 | 88.8 | 94.4 |
| PRR 348 | 90.6 | 66.7 | 80.7 | 76.5 | 62.9 | 35.4 | 68.5 | 51.4 | 62.5 | 85.1 | 88.6 | 86.9 | 53.0 | 84.9 | 79.7 | 83.4 | 80.5 | 90.9 | 77.6 |
| Mean | 85.2 | 81.6 | 78.0 | 77.6 | 65.3 | 70.6 | 84.4 | 80.8 | 77.7 | 75.2 | 85.4 | 75.9 | 76.1 | 76.0 | 75.9 | 69.2 | 85.0 | 91.7 | 81.7 |
| Min | 62.6 | 28.4 | 7.0 | 5.9 | 3.1 | 1.5 | 54.2 | 17.7 | 50.9 | 4.9 | 37.8 | 5.4 | 11.0 | 3.7 | 1.5 | 1.1 | 58.6 | 79.6 | 31.9 |
| Max | 93.2 | 94.0 | 92.2 | 93.3 | 91.5 | 93.1 | 93.8 | 96.8 | 88.4 | 92.7 | 97.2 | 91.2 | 92.1 | 94.9 | 89.9 | 92.4 | 95.8 | 97.1 | 95.9 |
| (b) | |||||||||||||||||||
| Restorers with restorer allele of Rf3 gene only | |||||||||||||||||||
| PRR 314 | 62.6 | 71.3 | 77.5 | 79.9 | 63.1 | 61.1 | 61.5 | 84.4 | 85.3 | 97.8 | 64.9 | 72.4 | 74.5 | 69.0 | 70.8 | 84.1 | 92.37 | 84.35 | 90.46 |
| PRR 334 | 55.8 | 26.6 | 31.5 | 77.5 | 19.7 | 31.8 | 46.7 | 64.0 | 61.6 | 17.8 | 38.8 | 74.3 | 42.5 | 47.1 | 58.2 | 62.2 | 86.7 | 89.63 | 50.59 |
| PRR 396 | 69.4 | 75.5 | 77.0 | 83.0 | 67.9 | 87.9 | 71.9 | 63.3 | 95.0 | 85.1 | 87.5 | 71.7 | 72.1 | 72.7 | 69.7 | 85.0 | 80.01 | 82.06 | 74.16 |
| Restorers with restorer allele of Rf4 gene only | |||||||||||||||||||
| PRR 317 | 67.2 | 73.0 | 66.0 | 65.2 | 64.6 | 65.9 | 60.2 | 77.0 | 84.5 | 50.5 | 69.7 | 67.8 | 58.6 | 64.2 | 90.51 | 90.45 | 87.8 | ||
| PRR 323 | 63.5 | 75.0 | 51.1 | 73.0 | 63.2 | 63.5 | 55.7 | 64.4 | 92.0 | 79.3 | 75.6 | 12.2 | 66.7 | 84.0 | 49.1 | 66.0 | 78.33 | 85.32 | 78.41 |
| PRR 326 | 77.1 | 79.6 | 80.7 | 71.5 | 82.0 | 87.6 | 60.4 | 97.0 | 61.0 | 72.7 | 68.7 | 77.6 | 70.3 | 71.9 | 75.6 | 93 | 79.09 | 84.44 | |
| PRR 329 | 68.8 | 65.5 | 62.2 | 61.7 | 51.1 | 59.4 | 64.1 | 81.0 | 79.7 | 80.4 | 80.5 | 64.8 | 78.5 | 58.7 | 60.9 | 85.55 | 98.78 | 88.76 | |
| PRR 337 | 66.9 | 61.3 | 62.4 | 61.0 | 69.3 | 57.8 | 57.0 | 33.9 | 85.5 | 81.0 | 6.5 | 71.3 | 73.6 | 66.5 | 68.8 | 84.34 | 89.47 | 55.91 | |
| PRR 342 | 71.9 | 75.1 | 80.5 | 85.8 | 65.2 | 77.3 | 65.4 | 92.5 | 86.3 | 88.7 | 72.3 | 81.6 | 70.9 | 61.6 | 70.9 | 88.88 | 99.05 | 79.57 | |
| PRR 354 | 64.3 | 71.4 | 65.2 | 63.0 | 50.1 | 56.1 | 58.8 | 83.8 | 84.0 | 78.2 | 67.4 | 74.9 | 68.7 | 61.3 | 59.8 | 85.43 | 91.13 | 80.77 | |
| PRR 368 | 71.7 | 70.3 | 72.0 | 73.3 | 60.0 | 58.2 | 61.4 | 98.4 | 68.0 | 79.3 | 66.8 | 75.8 | 57.3 | 68.1 | 88.8 | 80.25 | 83.77 | 60.1 | |
| PRR 372 | 73.3 | 66.7 | 59.5 | 59.6 | 63.5 | 54.3 | 63.5 | 50.1 | 95.8 | 62.1 | 86.5 | 61.5 | 72.6 | 67.3 | 59.7 | 56.7 | 73.91 | 87.84 | 85.25 |
| PRR 376 | 64.0 | 65.6 | 61.9 | 62.9 | 56.8 | 60.5 | 60.7 | 59.5 | 65.1 | 68.6 | 68.7 | 78.9 | 63.3 | 50.1 | 56.6 | 64.8 | 82.03 | 81.3 | 84.35 |
| PRR 381 | 78.0 | 83.3 | 80.4 | 78.3 | 70.4 | 73.8 | 66.9 | 65.9 | 97.2 | 61.7 | 87.9 | 76.8 | 77.7 | 64.8 | 71.0 | 70.7 | 87.7 | 88.63 | 84.47 |
| PRR 386 | 66.9 | 63.0 | 77.1 | 69.9 | 64.9 | 63.5 | 76.9 | 91.2 | 65.1 | 79.6 | 81.7 | 71.6 | 59.9 | 73.0 | 54.0 | 84.51 | 84.34 | 66.86 | |
| Restorers with restorer alleles of both Rf3 and Rf4 genes | |||||||||||||||||||
| PRR 300 | 90.1 | 77.5 | 75.1 | 80.2 | 78.1 | 61.3 | 73.8 | 72.3 | 66.2 | 92.4 | 81.2 | 80.3 | 78.7 | 75.0 | 70.6 | 72.3 | 80.82 | 84.57 | 83.34 |
| PRR 307 | 69.3 | 79.2 | 72.0 | 77.3 | 46.1 | 64.2 | 64.7 | 72.7 | 58.3 | 85.3 | 63.2 | 68.8 | 79.3 | 82.3 | 55.6 | 88.31 | 86.3 | 83.46 | |
| PRR 358 | 70.8 | 73.0 | 56.7 | 73.9 | 69.0 | 57.4 | 59.0 | 54.6 | 66.6 | 85.6 | 86.4 | 73.7 | 64.5 | 71.5 | 73.9 | 88.77 | 90.63 | 85.06 | |
| PRR 363 | 75.6 | 42.6 | 68.6 | 58.0 | 75.4 | 60.6 | 65.0 | 82.5 | 80.7 | 73.8 | 87.2 | 69.8 | 70.5 | 70.7 | 81.8 | 89.97 | 78.27 | 59.27 | |
| PRR 367 | 70.3 | 83.7 | 76.2 | 86.0 | 79.6 | 76.6 | 76.4 | 78.0 | 81.3 | 80.4 | 59.7 | 75.1 | 82.6 | 65.7 | 73.2 | 88.3 | 77.65 | 88.23 | |
| PRR 390 | 84.1 | 68.0 | 76.2 | 72.2 | 69.8 | 71.2 | 69.3 | 73.5 | 64.2 | 84.7 | 75.3 | 76.4 | 81.8 | 74.0 | 77.4 | 75.0 | 80.76 | 93.31 | 89.7 |
| PRR 395 | 83.0 | 78.4 | 62.8 | 52.2 | 62.3 | 80.6 | 82.4 | 65.8 | 87.2 | 87.2 | 88.8 | 81.0 | 76.4 | 58.3 | 74.1 | 85.6 | 91.22 | 82.93 | |
| Restorers lacking restorer alleles of both Rf3 and Rf4 genes | |||||||||||||||||||
| PRR 311 | 57.6 | 64.3 | 54.8 | 75.6 | 70.1 | 32.0 | 57.3 | 66.1 | 85.1 | 64.5 | 51.7 | 82.0 | 69.7 | 63.2 | 80.6 | 87.08 | 78.11 | 84.97 | |
| PRR 347 | 72.9 | 61.8 | 46.1 | 73.4 | 46.1 | 75.0 | 66.4 | 52.2 | 62.8 | 72.7 | 64.6 | 77.4 | 66.9 | 67.5 | 61.3 | 67.3 | 83.91 | 94.88 | 73.61 |
| PRR 348 | 78.0 | 60.9 | 63.0 | 63.6 | 67.1 | 67.7 | 76.2 | 61.6 | 84.9 | 80.1 | 75.1 | 82.4 | 77.0 | 64.3 | 70.0 | 84.5 | 88.04 | 95.9 | 38.39 |
| Mean | 70.9 | 68.5 | 63.9 | 72.2 | 63.4 | 64.2 | 65.5 | 63.7 | 82.0 | 74.8 | 75.4 | 69.0 | 72.0 | 68.6 | 65.8 | 70.8 | 85.4 | 87.4 | 76.8 |
| Min | 55.8 | 26.6 | 31.5 | 52.2 | 19.7 | 31.8 | 46.7 | 33.9 | 58.3 | 17.8 | 38.8 | 6.5 | 42.5 | 47.1 | 49.1 | 54.0 | 73.9 | 77.7 | 38.4 |
| Max | 90.1 | 83.7 | 80.4 | 86.0 | 85.8 | 87.9 | 87.6 | 84.4 | 98.4 | 97.8 | 88.8 | 87.2 | 81.8 | 84.0 | 82.3 | 88.8 | 93.0 | 99.1 | 90.5 |
Pollen fertility
In the test crosses derived from restorer lines with restorer allele of Rf3 gene, one cross combination IR 58025A/PRR 334 in both early and late sowing conditions at Delhi showed lesser pollen fertility of 28.4% and 44.1%, respectively. Another combination of Pusa 6A/PRR 396 was also low performing in Karnal and in late sown conditions of Delhi (Table 9a). As far as cross combinations with parental lines carrying Rf4 gene were concerned, Pusa 6A/PRR 372 (49.6%, Delhi-early sown), IR 58025A/PRR 372 (49.2%, Delhi-late sown), IR 79156 A/PRR 376 (6.2%, Delhi-late sown), IR 58025A/PRR 326 (11.1%, Delhi-late sown), IR 58025A/PRR 386 (11.4%, Delhi-late sown), IR 58025A/PRR 372 (49.2%, Delhi-late sown), Pusa 6A/PRR386 (40.8%, Karnal), IR79156 A/PRR 368 (11.1%, Karnal) and IR 58025A/PRR 368 (14.7%, Karnal) showed a lesser pollen fertility behaviour. One restorer line namely, PRR 337 also showed a lesser pollen fertility in four combinations with different male sterile lines throughout various locations. In cases of combinations derived from parents having restorer alleles of both Rf3 and Rf4 genes, RTN 12/PRR 300 was found to be inferior for pollen fertility at Karnal (1.1%) and in early sown conditions at Delhi (5.9%). Other cross combination namely, RTN 12A/PRR 307 (1.5%, Karnal), IR79156 A/PRR 307 (4.5%, Delhi-late sown), RTN 12A/PRR 395 (8.6%, Delhi-early; 6.0, Bihar) and IR 58025A/PRR 395 (3.7%, Karnal) had low pollen fertility. Three restorer lines lacking restorer alleles of both Rf3 and Rf4 genes showed good pollen fertility except for several combinations. These lines may possess novel source of fertility restoration, which needs to be characterized in detail.
Spikelet fertility
Among the hybrids generated by using three iso-cytoplasmic restorer lines carrying restorer allele of Rf3 gene, all except one (when RTN 12A was used as a female parent) showed a spikelet fertility of > 60% in all combinations. As far as cross combinations with restorer lines carrying restorer allele of Rf4 gene were concerned, 29 out of 192 (48 crosses evaluated at four locations) showed a lesser spikelet fertility (Table 9b). When cross combinations having parental lines carrying restorer alleles of both Rf3 and Rf4 genes were compared, twelve out of 112 cross combinations (over locations) showed less pollen fertility (Tables 5, 6).
Multilocational evaluation of crosses using GGE biplot
100 cross combinations were evaluated at three locations, namely, IARI (New Delhi; early and late sown conditions), IARI Regional station in Karnal (Haryana) and, IARI Regional station at Pusa (Bihar) during Kharif 2015. Based on GGE bi-plot analysis ideal genotypes were identified. The genotype with the highest mean performance and absolute stability is considered an ideal genotype (Yan and Kang 2003). In the bi-plot, an ideal genotype gets placed at the center of the concentric circle. Average environment (tester) coordinate (AEC), passes through the bi-plot origin and the deviation from either the bi-plot origin or the AEC line indicates higher GXE interaction and less stability.
Pollen fertility
GGE biplot analysis of multilocation data
In the polygon, the polygon is formed by connecting the vertex genotypes with straight lines and the rest of the genotypes were placed within the polygon. The polygon vertices are genotype markers that are maximally remote from the bi-plot centre. The lines dividing the bi-plot into sectors represent a set of hypothetical environments. If a line at an angular vertex of the polygon falls within one sector with an environment marker (or with several markers), that means the yield capacity of this genotype was highest in this particular environment. The vertex hybrids for Delhi (early sown condition) were derived from PRR 358 and PRR 348, when IR 79156A was used as a female parent (Fig. 2a). PRR 363 (with IR 58025A), PRR 326 (with Pusa 6A) and PRR 376 (with RTN 12A) were the vertex hybrids with the longest distance from origin, which indicate that these were the best or poorest in some or all environments. For Delhi (late sown conditions), PRR 376 and PRR 368 (with IR 79156A), PRR 334 (with IR 58025A), PRR 390 (with Pusa 6A), PRR363 and PRR 334 (with RTN 12A) were the responsive restorers as far as mean performance of hybrids are concerned. PRR 354 and PRR 395 (with IR 79156A), PRR 367 (with IR 58025A), PRR 390 (with Pusa 6A) had produced the vertex hybrids for Karnal. For Bihar IR 58025A/PRR 329, IR 58025A/PRR 347, Pusa 6A/PRR 372 were the superior cross combinations.
Fig. 2.
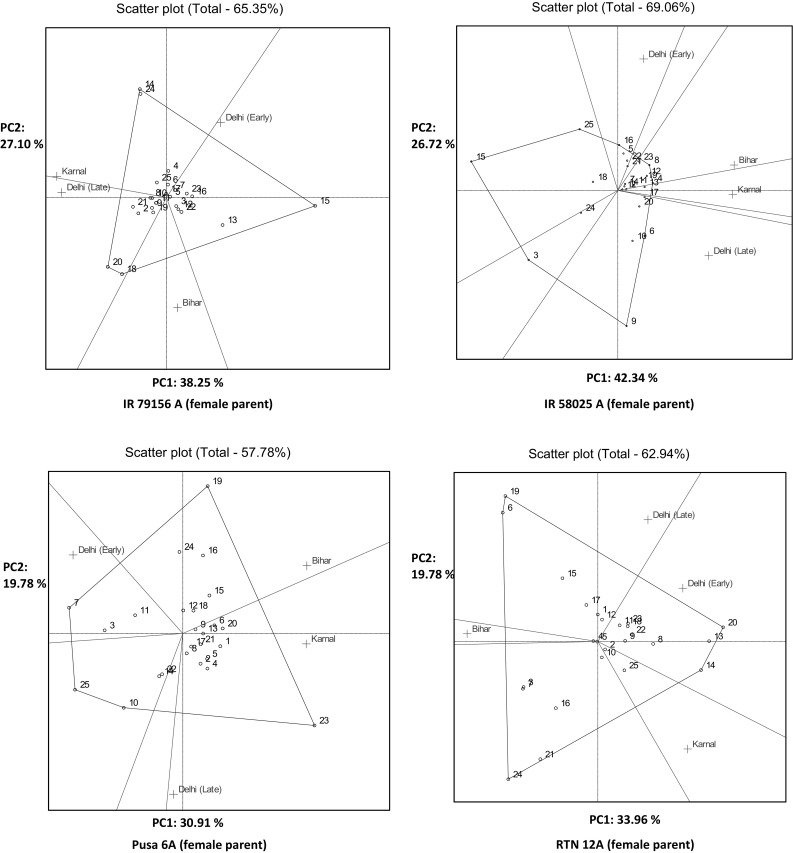
a GGE biplot of rice hybrids for pollen fertility based on which won where pattern. Restorer lines used: 1. PRR 300, 2.PRR 307, 3.PRR 311, 4.PRR 314, 5.PRR 317, 6.PRR 323, 7.PRR 326, 8.PRR 329, 9.PRR 334, 10.PRR 337, 11.PRR 342, 12.PRR 347, 13.PRR 348, 14.PRR 354, 15.PRR 358, 16.PRR 363, 17.PRR 367, 18.PRR 368, 19.PRR 372, 20.PRR 376, 21.PRR 381, 22.PRR 386, 23.PRR 390, 24.PRR 395, 25.PRR 396, b GGE biplot of rice hybrids for spikelet fertility based on which won where pattern. Restorer lines used: 1. PRR 300, 2.PRR 307, 3.PRR 311, 4.PRR 314, 5.PRR 317, 6.PRR 323, 7.PRR 326, 8.PRR 329, 9.PRR 334, 10.PRR 337, 11.PRR 342, 12.PRR 347, 13.PRR 348, 14.PRR 354, 15.PRR 358, 16.PRR 363, 17.PRR 367, 18.PRR 368, 19.PRR 372, 20.PRR 376, 21.PRR 381, 22.PRR 386, 23.PRR 390, 24.PRR 395, 25.PRR 396
Ranking genotypes based on mean yield performance and stability
Among the test crosses generated by using IR 79156A as female parents, cross combinations derived from PRR 381 had shown highest pollen fertility. The other restorers were PRR 307 > PRR 334 > PRR 329 > PRR 337. All these restorers showed less GXE interaction and thus stable as they are projected on AEC line only. Cross combinations derived from PRR 358 had shown less pollen fertility, whereas PRR 354 and PRR 395 had also shown a less stable behavior as depicted by their distance from AEC line. When IR 58025A was used as A line, hybrids derived from PRR 314 was the ideal one followed by PRR 347 > PRR 372 > PRR 348 > PRR 329. Similarly, with Pusa 6A, PRR 300 > PRR 376 > PRR 317 > PRR 314 > PRR 307 > PRR 323 had shown higher pollen fertility (Fig. 3a). As far as RTN 12A was considered PRR 368 followed by PRR 390, PRR 342, PRR 386, PRR 329 and PRR 334 were close to the center of concentric ring and thus high performers. The identified hybrids were also stable with a lesser proportion of GXE interactions.
Fig. 3.

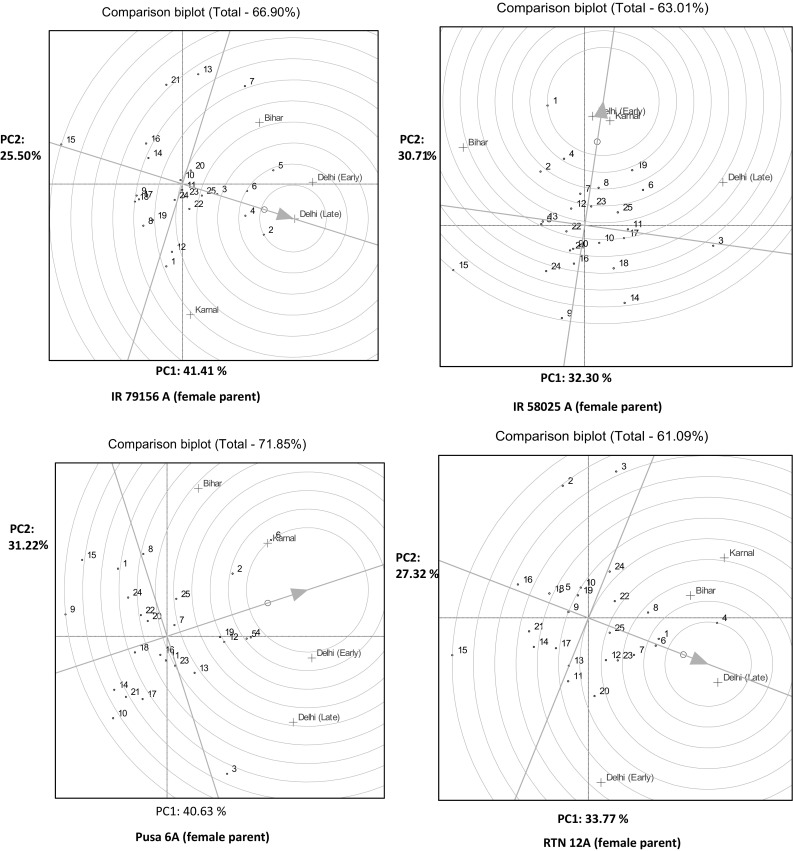
a GGE biplot of ideal genotype and comparison of the genotypes with the ideal genotype (pollen fertility). Restorer lines used: 1. PRR 300, 2.PRR 307, 3.PRR 311, 4.PRR 314, 5.PRR 317, 6.PRR 323, 7.PRR 326, 8.PRR 329, 9.PRR 334, 10.PRR 337, 11.PRR 342, 12.PRR 347, 13.PRR 348, 14.PRR 354, 15.PRR 358, 16.PRR 363, 17.PRR 367, 18.PRR 368, 19.PRR 372, 20.PRR 376, 21.PRR 381, 22.PRR 386, 23.PRR 390, 24.PRR 395, 25.PRR 396, b GGE biplot of ideal genotype and comparison of the genotypes with the ideal genotype (spikelet fertility). Restorer lines used: 1. PRR 300, 2.PRR 307, 3.PRR 311, 4.PRR 314, 5.PRR 317, 6.PRR 323, 7.PRR 326, 8.PRR 329, 9.PRR 334, 10.PRR 337, 11.PRR 342, 12.PRR 347, 13.PRR 348, 14.PRR 354, 15.PRR 358, 16.PRR 363, 17.PRR 367, 18.PRR 368, 19.PRR 372, 20.PRR 376, 21.PRR 381, 22.PRR 386, 23.PRR 390, 24.PRR 395, 25.PRR 396
Spikelet fertility
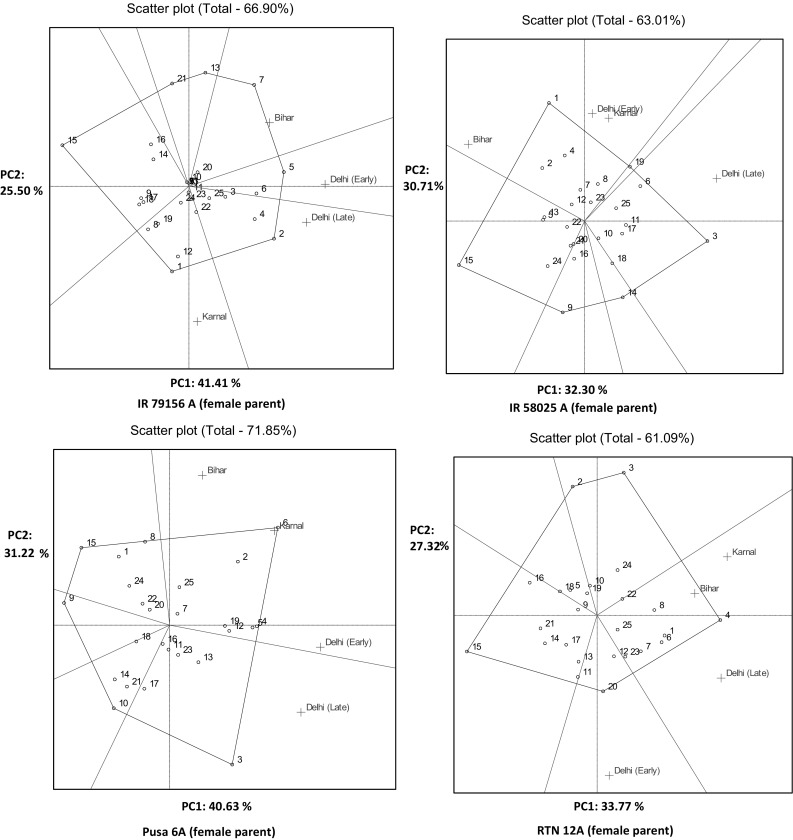
Polygon view of GGE biplot analysis of MET data
PRR 317 for Delhi (early sown condition), PRR 307 for Delhi (late sown condition), PRR 300 (for Karnal) and PRR 326 (for Pusa-Bihar) produced hybrids with the highest spikelet fertility, when IR 79156A was used as a female parent. The vertex hybrids for a mega environment consisting of Delhi (early sown condition), Karnal and Bihar were derived from PRR1 whereas for Delhi (late sown condition), PRR 311 produced the vertex hybrids when IR 58025A was used as a female parent (Fig. 2b). With Pusa 6A, Delhi (early sown condition), Karnal and Bihar again formed a mega environment where PRR 323 followed by PRR 307 has shown its superiority. PRR 311 maintained in superiority in late sown conditions of Delhi. With RTN 12A Bihar, Karnal and Delhi (late sown condition) were grouped together and PRR 314 was found to be promising. In early sown condition of Delhi, PRR 376 was found to be promising.
Ranking of genotypes based on mean and stability performance
Among the test crosses generated by using IR 79156A as female parent, the cross combination derived from PRR 307 was superior for spikelet fertility. The other restorers were PRR 314 > PRR 323 > PRR311 > PRR317. These all have a less GXE interaction and thus were stable as they were projected on AEC line only (Fig. 3b). Using IR 58025A as female parent, hybrids derived from PRR 300 was the ideal one followed by PRR 314 > PRR 372 > PRR 307 > PRR 329. Similarly, with Pusa 6A, PRR 314 > PRR 317 > PRR 307 > PRR 372 > PRR 347 had shown higher spikelet fertility. As far as RTN 12A was considered PRR 323 followed by PRR 300, PRR 314, PRR 326, PRR 329 and PRR 390 were close to the centre of concentric ring and thus were high performer. The identified hybrids were also stable with a lesser proportion of GXE interactions.
Discussion
Identification of fertility restorer lines is one the foremost essential requirements in hybrid rice breeding. Conventionally two crop seasons are required, one season for making test crosses and another season for evaluating the test cross hybrids for pollen and spikelet fertility. Molecular mapping of the genes governing fertility restoration has helped in identification of restorer lines without phenotypic screening involving test crossing. Nas et al. (2003) demonstrated for the first time the use of molecular markers for selecting restorer lines. Further, in case of WA cytoplasm of rice, two genes responsible for fertility restoration namely Rf3 and Rf4 have been mapped (Alavi et al. 2009; Prakash 2003) and candidate gene based markers have also been developed (Suresh et al. 2012) and used for identifying restorer lines. It is now possible to routinely use molecular markers to select putative restorers of WA cytoplasm among a set of rice genotypes. The identified ones can then be test crossed with appropriate CMS lines to confirm their fertility restoration and check the magnitude of heterosis. Molecular markers can also aid in pyramiding of the restorer genes into an elite genotype and identification of plants with both restorer genes without environmental influence and developmental stage.
In our study, both gene based and linked markers showed an efficiency of > 80% in identification of potential restorers. Revathi et al. (2013) observed that the efficacy of RM 6100 and RM 3873 was 80–85 and 90%, respectively, whereas Suresh et al. (2012) reported the efficacy of gene based markers namely, DRCG-RF4-14 and DRRM-RF3-10 around 92% for identification of restorers. In general, gene based markers showed higher efficiency as compared to gene linked markers for identifying the restorers. However, gene based markers are not functional markers i.e., present at the functional domain of fertility restoration genes and thus a 100% efficacy can’t be achieved. In case of iso-cytoplasmic restorer lines, results showed that there was a large variation in pollen fertility among them, indicating that the genomic background also played a crucial role in fertility restoration and possibility of interaction with modifiers cannot be ruled out (Pande et al. 1990; Virmani 1987; Salgotra et al. 2002). It has been observed earlier that the differential restoration behaviour of the genotypes to CMS lines with same WA cytoplasmic source may be due to interaction between the genes of the maintainer and pollinator parents (Virmani 1987).
Our study also establishes the need for appropriate sowing times to assess the restoration ability of the iso-cytoplasmic restorers. The late sown test crosses hybrids had expressed poor pollen fertility restoration indicating that lower temperature during flowering time may affect pollen fertility even in the presence of restorer allele. Low temperature induced spikelet sterility at booting stage in cooler weather has been reported in pureline varieties of rice as well (Alvarado 1999; Shimono et al. 2010), which may be attributed to the improper pollen development, leading to a shortage of viable pollen at flowering (Farrell et al. 2006). The present study also emphasizes the need for considering both pollen fertility as well as spikelet fertility parameters for identifying restorers as spikelet fertility is influenced by several other factors such as anther dehiscence, translocation rate of starch and other polysaccharides to the grains (spikelets), balance between translocation and respiration rate in addition to pollen fertility (Falodun et al. 2012). Even lesser pollen fertility tends to provide higher seed set, due to the ability of single fertile pollen to fertilize a spikelet (Joshi et al. 2007).
Considerable reduction in the need for hybrid evaluation can be achieved possibly by using inbred line information to predict hybrid performance (Betran et al. 2003). In our experiment, association between parental and their hybrid performances were found to be lower at all the environments for both pollen as well as spikelet fertility. This necessitates the need to develop cross combinations for their further evaluation for development of superior hybrids as the performances of crosses cannot be judged based on the performance of their parental lines. Khalique et al. (1977) had also reported no association between parental and their hybrid yield performances in rice, which has been reiterated by Samanci (1996), probably due to new gene/allele combinations in the hybrids. In each generation, gene combination changes based on the allelic contribution of both parental lines and thus it is very hard to predict the performance of hybrids based on their parental breeding value. Any resemblance between these performances might be due to chance. However, Virmani et al. (1982) reported that F1 hybrids correlated significantly with their parental traits and thus selecting parental lines from elite breeding lines based on yield performance may be a fruitful approach.
The present study shows that a marker combination of DRRM-RF3-10 and DRCG-RF4-14 was found to be the best combination to identify potential restorers with restorer alleles of both Rf3 and Rf4 genes. Use of these markers can considerably reduce the amount of work done in making and evaluating test crosses to identify effective restorer lines in a hybrid rice breeding program. Based on multi-location evaluation of the test cross hybrids, five iso-cytoplasmic restorers namely, PRR 381, PRR 307, PRR 334, PRR 329, PRR 337 have been identified which produced stable fertility restoration across locations. These restorers identified in the present study can be further assessed for their combining ability with diverse WA CMS lines and used in producing better rice hybrids.
References
- Alavi M, Ahamadidhan A, Kamkar B, Kalateh M. Mapping Rf3 locus in rice by SSR and CAPS markers. Int J Genet Mol Biol. 2009;7:121–126. [Google Scholar]
- Alvarado R (1999) Influence of air temperature on rice population, length of period from sowing to flowering and spikelet sterility. In: Hill JE, Hardy B (eds) Proceedings of the second temperate rice conference. Sacramento, California, pp 63–68
- Betran FJ, Ribaut JM, Beck D, De Leon DG. Genetic diversity, specific combining ability, heterosis in tropical maize under stress and non-stress environments. Crop Sci. 2003;43:797–806. doi: 10.2135/cropsci2003.7970. [DOI] [Google Scholar]
- Chaudhury RC, Virmani SS, Khush GS. CMS lines in rice. Andhra Agric J. 1981;38:302–303. [Google Scholar]
- Falodun D, Njoku KL, Ogunyebi AL, Akinola MO. Spikelet numbers, filled grains and spikelet fertility potential of NERICA rice (Mecux, Tox and WitA.4) grown in hydrocarbon polluted soils. J Res Environ Sci Toxicol. 2012;1(8):201–206. [Google Scholar]
- FAO (2014) FAO STAT agricultural data (on line), In: http://www.fao.org/available at http://faostat.fao.org, Rome, Italy: Food and Agriculture Organization (FAO)
- Farrell TC, Fox KM, Williams RL, Fukai S. Genotypic variation for cold tolerance during reproductive development in rice: screening with cold air and cold water. Field Crops Res. 2006;98:178–194. doi: 10.1016/j.fcr.2006.01.003. [DOI] [Google Scholar]
- Ghara AG, Nematzadeh G, Bagheri N, Ebrahimi A, Oladi M. Molecular and cytological evaluation of male sterile and restorer lines in hybrids rice. Int Res J Appl Basic Sci. 2012;3(1):183–189. [Google Scholar]
- Govinda Raj K, Virmani SS. Genetics of fertility restoration of ‘WA’ type cytoplasmic genic male sterility in rice. Crop Sci. 1988;28:787–792. doi: 10.2135/cropsci1988.0011183X002800050013x. [DOI] [Google Scholar]
- Joshi BK, Subedi LP, Gurung SB, Sharma RC. Pollen and spikelet analysis in F1 rice hybrids and their parents. Nepal Agric Res J. 2007;8:120–126. [Google Scholar]
- Khalique MA, Joarder OI, Euus AM. Heterosis and combining ability in a diallel cross of rice (Oryzasativa L.) Bangladesh J Agric Sci. 1977;4(2):137–145. [Google Scholar]
- Kutka F. Open-pollinated vs. Hybrid maize cultivars. Sustainability. 2011;3:1531–1554. doi: 10.3390/su3091531. [DOI] [Google Scholar]
- Li YC, Yuan LP. Genetic analysis of fertility restoration in male sterile lines of rice. Manila, Rice Genetics: IRRI; 1986. pp. 617–632. [Google Scholar]
- Lin SC, Yuan LP (1980) Hybrid rice breeding in China. In: Innovative approaches to rice breeding. IRRI, Manila, Philippines, pp. 35–51
- Nas TMS, Casal CL, Li Z, Virmani SS. Application of molecular markers for identification of restorers. Rice Genet News lett. 2003;20:69–71. [Google Scholar]
- Pande K, Ratho SN, Patnaik RN, Jachuk PJ. Fertility restoration in cytoplasmic male sterile lines in rice. Oryza. 1990;27:232–238. [Google Scholar]
- Peng S, Laza RC, Visperas RM, Khush GS, Virk P, Zhu D (2004) Rice: progress in breaking the yield ceiling. “New directions for a diverse planet”. In: Proceedings of the 4th international crop science congress, 26 Sep–1 Oct 2004, Brisbane, Australia Published on CDROM. Web site www.cropscience.org.au
- Prakash P (2003) Molecular mapping of fertility restorer gene(s) and validation of Rf-gene linked markers in rice. M.Sc. Dissertation, Indian Agricultural Research Institute, New Delhi
- Revathi P, Medoju P, Singh AK, Sundaram RM, Sravan R, Senguttuvel P, Kemparaju KB, Hariprasad AS, Ramesha MS, Neeraja CN, Rani NS, Viraktamath BC. Efficiency of molecular markers in identifying fertility restoration trait of WA-CMS system in rice. Indian J Genet Plant Breed. 2013;73(1):89–93. doi: 10.5958/j.0019-5200.73.1.012. [DOI] [Google Scholar]
- Salgotra RK, Katoch PC, Kaushik RP. Identification of restorers and maintainers for cytoplasmic genic male sterile lines of rice. Oryza. 2002;39:55–57. [Google Scholar]
- Samanci B. Phenotypic correlations between maize inbreds and their single cross hybrids in short season areas. Euphytica. 1996;89:291–296. [Google Scholar]
- Sheeba NK, Viraktamath BC, Sivaramakrishnan S, Gangashetti MG, Pawan K, Sundaram RM. Validation of molecular markers linked to fertility restorer gene(s) for WA-CMS lines of rice. Euphytica. 2009;167:217–227. doi: 10.1007/s10681-008-9865-4. [DOI] [Google Scholar]
- Shimono H, Okada M, Inoue M, Nakamura H, Kobayashi K, Hasegawa T. Diurnal and seasonal variations in stomatal conductance of rice at elevated atmospheric CO2 under fully open-air conditions. Plant Cell Environ. 2010;33:322–331. doi: 10.1111/j.1365-3040.2009.02057.x. [DOI] [PubMed] [Google Scholar]
- Stewart CN, Jr, Via LE. A rapid CTAB DNA isolation technique useful for RAPD fingerprinting and other PCR applications. Bio Tech. 1993;14(5):748–750. [PubMed] [Google Scholar]
- Suresh PB, Srikanth B, Hemanth VK, Rao IS, Vemireddy LR, Dharika N, Sundaram RM, Ramesha MS, Rao KRSS, Viraktamath BC, Neeraja CN. Fine mapping of Rf3 and Rf4 fertility restorer loci of WA-CMS of rice (Oryza sativa L.) and validation of the developed marker system for identification of restorer line. Euphytica. 2012;187:421–435. doi: 10.1007/s10681-012-0737-6. [DOI] [Google Scholar]
- Virmani SS (1987) Hybrid rice breeding. Cytogenetic relationship between two cytoplasmic male sterile lines. Int Rice Res Newsl 12:14
- Virmani SS, Aquino RC, Khush GS. Heterosis breeding in rice (Oryza sativa L.) Theor Appl Genet. 1982;63:373–380. doi: 10.1007/BF00303911. [DOI] [PubMed] [Google Scholar]
- Xie F, He Z, Esguerra MQ, Qiu F, Ramanathan V. Determination of heterotic groups for tropical indica hybrid rice germplasm. Theor Appl Genet. 2014;127:407–417. doi: 10.1007/s00122-013-2227-1. [DOI] [PubMed] [Google Scholar]
- Yan W, Kang MS. GGE biplot analysis: a graphical tool for breeders, geneticists and agronomists. 1. Boca Raton: CRC Press LLC.; 2003. p. 271. [Google Scholar]
- Yao FY, Xu CG, Yu SB, Li JX, Gao YJ, Li XH, Zhang Q. Mapping and Genetic analysis of two fertility restorer loci in the wild abortive cytoplasmic male sterility system of rice (Oryza sativa L.) Euphytica. 1997;98:183. doi: 10.1023/A:1003165116059. [DOI] [Google Scholar]
- Yuan LP, Virmani SS (1988) Status of hybrid rice research and development. In: Hybrid rice. International Rice Research Institute, Manila, Philippines, pp 7–24