Abstract
The rice cultivar (Oryza sativa L.) BRS AG, developed by Embrapa Clima Temperado, is the first cultivar designed for purposes other than human consumption. It may be used in ethanol production and animal feed. Different abiotic stresses negatively affect plant growth. Soil salinity is responsible for a serious reduction in productivity. Therefore, the objective of this study was to evaluate the gene expression and the activity of antioxidant enzymes (SOD, CAT, APX and GR) and identify their functions in controlling ROS levels in rice plants, cultivar BRS AG, after a saline stress period. The plants were grown in vitro with two NaCl concentrations (0 and 136 mM), collected at 10, 15 and 20 days of cultivation. The results indicated that the activity of the enzymes evaluated promotes protection against oxidative stress. Although, there was an increase of reactive oxygen species, there was no increase in MDA levels. Regarding genes encoding isoforms of antioxidant enzymes, it was observed that OsSOD3-CU/Zn, OsSOD2-Cu/Zn, OsSOD-Cu/Zn, OsSOD4-Cu/Zn, OsSODCc1-Cu/Zn, OsSOD-Fe, OsAPX1, OsCATB and OsGR2 were the most responsive. The increase in the transcription of all genes among evaluated isoforms, except for OsAPX6, which remained stable, contributed to the increase or the maintenance of enzyme activity. Thus, it is possible to infer that the cv. BRS AG has defense mechanisms against salt stress.
Electronic supplementary material
The online version of this article (doi:10.1007/s12298-017-0467-2) contains supplementary material, which is available to authorized users.
Keywords: Oryza sativa L., Oxidative stress, Reactive oxygen species, Salinity
Introduction
Soil salinity affects plants by osmotic and ionic effects and oxidative stress (Pitman and Lauchli 2002). With high salt contents in the soil, the absorption of water and nutrients by plants and the permeability of the membranes are hindered. Such changes affect water and nutrient balance, causing changes in the hormone metabolism, gas exchange and production of reactive oxygen species, which will compromise cell expansion and division and consequently plant growth, leading to accelerated senescence and plant death (Prisco and Gomes Filho 2010).
Reactive oxygen species (ROS) are produced by the plant’s normal metabolic processes such as photosynthesis and respiration. However, during stress situations, the metabolic imbalance leads to the formation of ROS in excess, causing oxidative stress. Among the various types of ROS, singlet oxygen (1O2), superoxide radicals (O·−2), hydrogen peroxide (H2O2) and radical hydroxyl (OH·) may be formed (Gupta and Huang 2014). When the production of ROS is much higher than the volume that can be metabolized, they may react with several cellular components. When in excess, it may cause damage to lipids, proteins and DNA, leading to a structural function change and/or inhibition thereof (Ferreira et al. 2007). On the other hand, plants have a complex antioxidant system whose function is to protect cells from damage caused by ROS. The main enzymatic components of the defense system are superoxide dismutase (SOD), catalase (CAT), ascorbate peroxidase (APX), glutathione reductase (GR), monodehydro ascorbate reductase (MDAR) and dehydroascorbate reductase (DAR) (Silveira et al. 2010; Ahmad 2014).
SOD appears to be the first to act in the line of defense regarding the elimination of ROS, performing the dismutation of superoxide radicals into hydrogen peroxide. The conversion of hydrogen peroxide into water and oxygen is performed by the CAT and the APX with the participation of the GR involved in the ASH/GSH cycle.
CAT, APX, SOD and GR enzymes are encoded by different gene families. In rice, eight APx genes encode APX enzymes located in different compartments: two cytosolic isoforms encoded by OsAPx1 and OsAPx2, two peroxisome/glyoxysome isoforms encoded by OsAPx3 and OsAPx4 and chloroplastidic isoforms encoded by OsAPx5, OsAPx6, OsAPx7 and OsAPx8. The OsAPX6 isoform was also located in mitochondria (Teixeira et al. 2004).
All SOD isoforms are nuclear - encoded and directed to their respective sub-cellular compartments, when necessary, through amino-terminal targeting sequences (Gill and Tuteja 2010). SODs are classified according to their metal cofactor and/or its sub-cellular location: Fe-SOD is present in chloroplasts; Mn-SOD is present in mitochondria and peroxisomes, and Cu/Zn-SODs is present in chloroplasts, peroxisomes, cytosol and possibly in the extracellular space (Alscher et al. 2002). According to Bowler et al. (1992), the CAT enzyme is represented by a small family called CatA, CatB and CatC. Three genes encoding GR have also been described for rice: OsGR2 encoding cytosolic isoform of GR (Kaminaka et al. 1998), and OsGR1 and OsGR3 encoding chloroplastic GR isoforms (Bashir et al. 2007). The expression of genes encoding APX, CAT, SOD and GR are modulated by several environmental stimuli, including saline stress.
Rice is a crop of great economic importance and widely cultivated worldwide. The cultivar BRS AG, developed by Embrapa Clima Temperado of Pelotas/RS, is intended for the production of ethanol and/or animal feed, meeting the demand of the southern Rio Grande do Sul (RS) region. Thus, BRS AG is the first rice cultivar developed for purposes other than human consumption because it does not meet the standards required by consumers (Magalhães Júnior et al. 2012).
Due to the lack of physiological information on the stress response in rice plants of the cultivar BRS AG and the importance of this new variety in the production of biofuels, we sought to investigate the gene expression of the enzymes involved in the control of ROS levels in rice plants cultivar BRS AG after a salt stress period.
Materials and methods
Plant material
The experiment was conducted at the Plant Tissue Culture Laboratory of the Department of Botany, Institute of Biology of the Federal University of Pelotas (UFPel), located in the city of Capão do Leão, RS. Rice seeds, cultivar BRS AG, from the Estação Experimental Terras Baixas (Embrapa - Clima Temperado) were used.
The seeds were manually peeled, sterilized and placed to germinate in test tubes containing 15 mL of Murashige and Skoog (1962) MS medium, with half the concentration of salt sources and supplemented with two different concentrations of NaCl (0 and 136 mM), 10 gL−1 of sucrose, 100 mg L−1 of myo-inositol and solidified by adding 7 gL−1 of agar and pH adjusted to 5.8. The cultures were kept in an environmental greenhouse at 28 °C in the dark for 5 days. After this period, the plants were transferred to a growth room with photoperiod of 16 h light and 8 h of darkness and temperature of 25 ± 2 °C.
Experimental design
The experimental design was completely randomized with a 2 × 4 factorial design representing two concentrations of NaCl (0 and 136 mM) and three collection times (10, 15 and 20 days). Three replications were performed, each represented by 15 seedlings. For the analyses of gene expression and enzymatic antioxidant activity, the plant material was collected and stored in an ultrafreezer at −80 °C until analyses.
Data were submitted to analysis of variance (p ≤ 0.05) and the mean values were compared by Tukey test at 5% probability using the statistical software using the statistical program SAS 9.3 (SAS Institute, Cary NC).
Determination of ROS
The superoxide radical (O·−2) was determined according to Li et al. (2010). Leaf tissues (200 mg) were macerated in a 65 mM phosphate buffer (pH 7.8) and centrifuged at 5000 g for 10 min. The supernatant was mixed with 65 mM phosphate buffer (pH 7.8) and 10 mM hydroxylamine hydrochloride, and then incubated at 25 °C for 20 min. Then, 17 mM sulfanilamide and 7 mM α-naphthylamine were added to the mixture. The absorbance of the solution was measured at 530 nm after resting for 20 min at 25 °C. A standard curve with nitrogen dioxide radical (NO2) was used to calculate the O·−2 generation rate.
For the determination of hydrogen peroxide (H2O2) contents, the methodology proposed by Velikova et al. (2000) was used. 200 mg of leaf tissue, macerated in 2.0 mL of 0.1% solution (w/v) of trichloroacetic acid (TCA) were used. The samples were homogenized and centrifuged at 12,000 g for 15 min at 4 °C. Subsequently, 0.25 mL was collected from the supernatant, to which 0.75 mL of buffer solution was added, consisting of 10 mM potassium phosphate (pH 7.0) and 1 mL of 1 M potassium iodide (KI), resulting in a final volume of 2.0 mL in each tube. Absorbance readings were taken in a spectrophotometer at 390 nm and the H2O2 content was calculated by comparing the readings obtained with the standard curve of H2O2 expressed in µmol of H2O2 g−1 of MF.
Lipid peroxidation was determined as described by Heath and Packer (1968). Approximately 200 mg of leaves were macerated in liquid N2 and homogenized in 2.0 mL of trichloroacetic acid (TCA) at 0.1% (w/v). The samples were centrifuged at 12,000g for 15 min at 4 °C. A 0.5 mL aliquot of the supernatant was added to 1.5 mL of thiobarbituric acid (TBA at 0.5% in TCA at 10%), prepared at the time of use. The mixture was incubated at 90 °C for 20 min. Thereafter, the reaction was stopped by cooling on ice and the readings were taken by a spectrophotometer at 535 nm and 600 nm. TBA forms complexes with malondialdehyde (MDA), a byproduct of the peroxidation process. The concentration of the MDA/TBA complex was calculated and the peroxidation was expressed as nmol MDA g−1 of MF.
Extraction of antioxidant enzymes
Leaf samples (200 mg) were soaked in liquid N2 and homogenized in 2.0 mL of extraction buffer: 100 mM potassium phosphate (pH 7.8), 0.1 mM ethylenediaminetetraacetic acid (EDTA) and 10 mM ascorbic acid. The homogenate was centrifuged at 13,000 g for 15 min at 4 °C. The supernatant was collected to determine the activity of superoxide dismutase (SOD), catalase (CAT), ascorbate peroxidase (APX) and glutathione reductase (GR). Total proteins were determined with the same buffer solution and the quantification was made using the Bradford (1976) method. The values regarding total protein contents were expressed as mg protein g−1 of MF.
Determination of enzymatic activities
The SOD activity (EC 1.15.1.1) was evaluated by the ability of the enzyme to inhibit the photoreduction of nitro blue tetrazolium (NBT) in a reaction medium containing 50 mM potassium phosphate (pH 7.8), 14 mM methionine, EDTA 0.1 µM, NBT 75 µM and riboflavin 2 µM (Giannopolitis and Reis 1977). The tubes with the reaction medium and the samples were illuminated for 7 min under 20 W. As a control, the same reaction medium without the sample was illuminated under the same conditions, while the blank solution was kept in the dark. Readings were taken at 560 nm in a spectrophotometer, where one unit of SOD was considered as the amount of enzyme able to inhibit by 50% the photoreduction of NBT under the experimental conditions. The SOD activity was expressed in U mg−1 of protein.
To determine CAT activity (EC 1.11.1.6), the method described by Azevedo et al. (1998) was used with some modifications. Its activity was monitored in a spectrophotometer evaluating the degradation of H2O2 at 240 nm for 2 min in a reaction medium containing 100 mM potassium phosphate buffer (pH 7.0), 12.5 mM H2O2 and 50 µL of plant extract incubated at 28 °C. As blank solution, the same reaction medium free of plant extract was used. The activity of the enzyme CAT was expressed in µmol H2O2 min−1 mg−1 of protein.
The activity of APX (EC 1.11.1.1) was determined according to Nakano and Asada (1981) by monitoring the oxidation rate of ascorbate at 290 nm. The reaction medium was incubated at 28 °C. It consisted of 100 mM potassium phosphate buffer (pH 7.0), 0.5 mM ascorbic acid and 0.1 mM H2O2. The decrease in absorbance was monitored for 2 min from the start of the reaction and the activity of APX was expressed in μmol ASC min-1 mg-1 of protein.
The activity of GR (CE 1.6.4.2) was determined according to Cakmak et al. (1993) by the decrease of absorbance at 340 nm due to the oxidation of NADPH. The reaction consisted of 50 mM phosphate buffer (pH 7.8), 1 mM oxidized glutathione (GSSG), 75 μM NADPH and the aliquot of the enzyme (100 μg L−1).
RT-qPCR analysis
Each macerated plant sample (100 mg) was transferred to a 1.5 mL microtube free of nuclease, to which Link® Pure Kit (Invitrogen™) extraction buffer was immediately added. The quantity of total RNA was measured by spectrophotometer at 260 and 280 nm, whose ratio provides an estimate of RNA purity. The integrity of the nucleic acid was assessed by agarose gel electrophoresis at 1.5%.
First, the samples were treated with DNAse I using the Amplification Grade DNAse I kit (Sigma-Aldrich®) to remove possible residues of genomic DNA following the manufacturer’s recommendations. Total RNA samples treated with DNase I were subjected to RT-PCR (reverse transcription PCR) for complementary DNA synthesis. The single strand cDNA synthesis per sample was made from RNA (1 μg μL−1) using the Superscript First-Strand Synthesis System for RT-PCR kit (Invitrogen).
RT-qPCR reactions were performed in a Bio-Rad CFX Real Time Thermo Cycler using the fluorophore LightCycler® 480 SYBR Green I Master (Roche®). Primers with specific amplicons (single peak with a dissociation of the bands of PCR products) and amplification efficiency close to 100% (E ≅ 1.8−2.2) were used. The primers are listed in the Supplementary Material 1 (Vighi et al. 2017). The rice gene OsUBQ10 (Moraes et al. 2015) was used as a quantitative internal control (reference gene - Tested in our experimental conditions). The relative quantification of the differential expression was performed using the comparative threshold cycle method as described by Livak and Schmittgen (2001).
Results
Determination of superoxide radicals, hydrogen peroxide and lipid peroxidation
For the superoxide content there was no statistical difference between the stress times in the presence of NaCl (Fig. 1a).
Fig. 1.
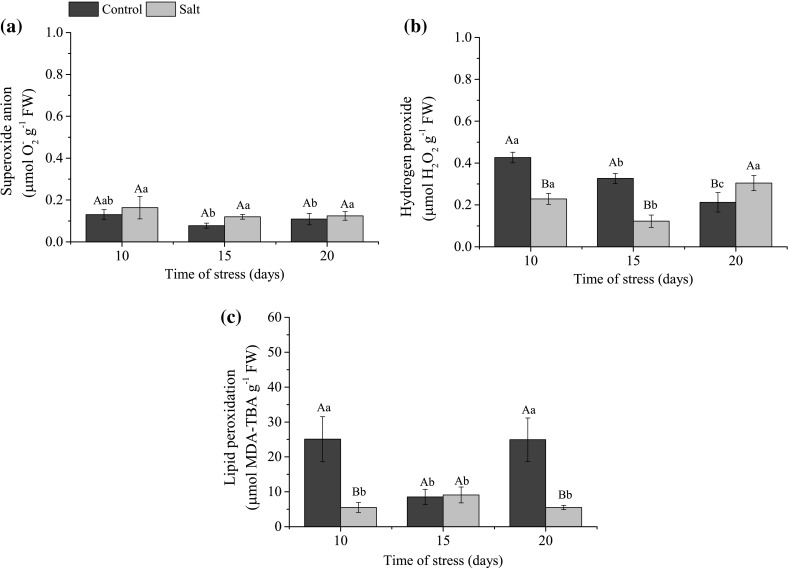
Superoxide (a), hydrogen peroxide (b), lipid peroxidation (c) contents of rice plants, cultivar BRS AG, subjected to saline stress (136 mM). The values are represented by the mean of each treatment ± SD (n = 3). Uppercase letters indicate a significant difference (p ≤ 0.05) between days under stress conditions for the same treatment and lowercase letters indicate a difference between the NaCl concentration and the control treatment at each day
By analyzing the formation of hydrogen peroxide in rice plants under salt stress conditions, there was a higher content in plants treated with NaCl at 10 and 20 days (Fig. 1b). The plants under salt stress (136 mM NaCl) showed decreased levels of H2O2, below control levels at 10 and 15 days.
The results of lipid peroxidation in rice plants, cultivar BRS AG, determined by MDA content are shown in the Fig. 1c. It can be observed that in plants grown in saline medium, MDA level, not differing significantly between stress times. For the control treatment, lipid peroxidation levels fluctuated over stress times. MDA values were higher than stressed plants at 10 and 20 days.
Activity of antioxidant enzymes
The activity of superoxide dismutase (SOD) was increased in rice plants, cultivar BRS AG, grown under salinity conditions (136 mM NaCl) during the initial development phase. Mean values at 10 and 15 days (24.85 and 21.18) were significantly higher when compared to control plants (11.52 and 9.50), respectively. An enzymatic activity was observed during stress time for control plants and under salt stress, noting that there were no significant differences in SOD activity for these plants (Fig. 2a).
Fig. 2.
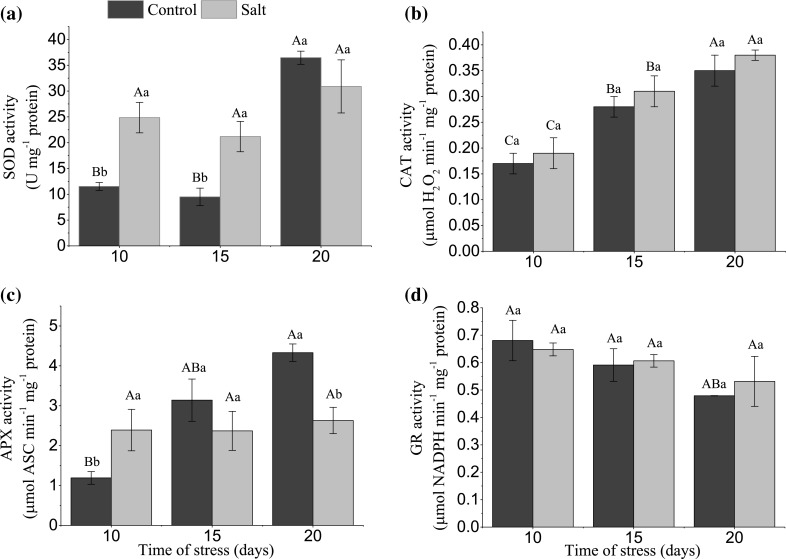
Activity of the enzyme superoxide dismutase (a), catalase (b), ascorbate peroxidase (c), and glutathione reductase (d) in rice plants, cultivar BRS AG, subjected to salinity conditions at different times. Means of activity ± SD (n = 3), where uppercase letters indicate a statistical difference between stress days for the same treatment and lowercase letters indicate differences between the control treatment and salt at each day
For the activity of catalase (CAT), a significant activity increase was observed with increasing stress exposure times. The values were 0.31 and 0.38 for periods of 15 and 20 days of stress, respectively. Control plants showed a higher activity of CAT at 20 days. There were no significant differences between control and stressed plants after 10 days of exposure to stress (Fig. 2b).
The activity of ascorbate peroxidase (APX), in plants under stress, was higher only at 10 days, with an average value of 2.39, when compared to the plants (1.19) that did not grow in a saline environment, as can be seen in Fig. 2c. At 20 days, the opposite was noticed: there was an increased activity of APX in control plants, with 4.33 in relation to the value of 2.63 of plants that were under salt stress. The increase in stress time had no influence on the activity of the APX enzyme in rice plants. In relation to the control, the increased activity occurred at 20 days and did not differ statistically from the values found at 15 days. There was a decrease in activity at 10 days, but it was not statistically different from that observed at 15 days.
According to the results shown in Fig. 2d, it was observed that between control plants and plants subjected to salt stress there were no differences regarding the enzyme activity of glutathione reductase. In plants that germinated in a medium with a concentration of 136 mM NaCl, there was a higher GR activity at 10 days (0.648), 15 (0.606) and 20 (0.532) days. The same behavior was observed for the control plants.
Expression of genes that encode isoforms of enzymes of the antioxidant system
According to the analysis of variance, it was possible to verify that the levels of transcripts of the genes of CAT, SOD, APX and GR enzymes in rice plants subjected to salt stress (136 mM NaCl) significantly changed in function of stress times (Table 1).
Table 1.
Relative quantification (RQ) of the expression of the genes that encode the enzymes of the antioxidant system (superoxide dismutase, ascorbate peroxidase, glutathione reductase and catalase) in rice plants, cultivar BRS AG, subjected to salt stress conditions (136 mM de NaCl)
| Genes | 0 | 10 days | 15 days | 20 days |
|---|---|---|---|---|
| OsAPX1 | 1.00B | 0.46 ± 0.19B | 6.05 ± 0.46A | 6.31 ± 0.56A |
| OsAPX2 | 1.00C | 1.13 ± 0.31C | 3.24 ± 0.63B | 5.62 ± 0.21A |
| OsAPX3 | 1.00C | 1.69 ± 0.42BC | 1.48 ± 0.33BC | 2.75 ± 0.44A |
| OsAPX4 | 1.00B | 2.07 ± 0.74AB | 2.49 ± 0.28A | 1.67 ± 0.17AB |
| OsAPX5 | 1.00C | 2.39 ± 0.35B | 2.07 ± 0.78BC | 5.30 ± 0.29A |
| OsAPX6 | 1.00A | 1.79 ± 0.08A | 1.93 ± 0.17A | 1.97 ± 0.88A |
| OsAPX7 | 1.00BC | 2.64 ± 0.16A | 2.08 ± 0.88AB | 1.93 ± 0.14AB |
| OsAPX8 | 1.00B | 0.77 ± 0.18B | 1.38 ± 0.41AB | 1.93 ± 0.61A |
| OsSOD4-Cu/Zn | 1.00C | 2.44 ± 0.10B | 3.38 ± 0.37A | 2.12 ± 0.12B |
| OsSOD3-CU/Zn | 1.00B | 2.65 ± 0.45A | 2.70 ± 0.44A | 2.37 ± 0.25A |
| OsSOD2-Cu/Zn | 1.00B | 2.57 ± 0.49A | 2.49 ± 0.40A | 1.90 ± 0.30A |
| OsSODCc1-Cu/Zn | 1.00D | 1.48 ± 0.04C | 3.43 ± 0.16A | 2.09 ± 0.06B |
| OsSOD-Cu/Zn | 1.00B | 1.71 ± 0.41A | 1.78 ± 0.05A | 2.02 ± 0.22A |
| OsSOD-Fe | 1.00B | 0.91 ± 0.11A | 0.65 ± 0.18A | 0.68 ± 0.17A |
| OsCATA | 1.00C | 1.14 ± 0.11C | 1.76 ± 0.34B | 3.69 ± 0.27A |
| OsCATB | 1.00B | 1.81 ± 0.54B | 1.90 ± 0.45B | 5.83 ± 0.52A |
| OsCATC | 1.00D | 3.77 ± 0.18A | 2.85 ± 0.25B | 1.79 ± 0.39C |
| OsGR1 | 1.00C | 1.88 ± 0.36AB | 2.03 ± 0.15A | 1.48 ± 0.07B |
| OsGR2 | 1.00B | 4.76 ± 0.20A | 1.71 ± 0.55B | 1.54 ± 0.63B |
| OsGR3 | 1.00B | 0.49 ± 0.16C | 1.19 ± 0.12B | 2.14 ± 0.09A |
Statistical difference between stress times (10, 15 and 20 days) are indicated by uppercase letters
* Significance level: p ≤ 0.05, Tukey test, represented by the mean ± SD (n = 3)
In this study, six of the eight SOD isoforms known for rice were analyzed because there was no amplification with the primers tested for two isoforms. In rice plants germinated in a saline environment, the genes OsSOD3-CU/Zn, OsSOD2-Cu/Zn, OsSOD-Fe and OsSOD-Cu/Zn had a similar expression behavior, as shown in Table 1. There were no differences in the expression of these genes at 10, 15 and 20 days. For the genes OsSOD4-Cu/Zu and OsSODCc1-Cu/Zn, there was an increase in their expression at 15 days of stress, with a decrease in expression at 20 days.
The expression of the gene of the APX enzyme is shown in Table 1. Among the eight genes evaluated, OsAPX1, OsAPX2 and OsAPx8 showed a significantly increased expression (p ≤ 0.05) in the presence of salt stress at 15 and 20 days when compared to their respective control treatments. On the other hand, the genes OsAPX3 and OsAPX5 were more expressed at the end of the stress time (20 days) when compared to their respective control treatments and the other stress times. Regarding the genes OsAPX7 and OsAPX4, there was a significant expression increase at 10 and 15 days of treatment, respectively. The expression values for both genes did not differ between the times 10, 15 and 20 days. The expression of the gene OsAPX6 remained stable over the stress time and did not show a significant difference. Among the eight genes, OsAPX1 presented expression values higher than the other genes.
In rice, the enzyme catalase consists of a small family represented by the genes OsCATA, OsCATB and OsCATC. The cultivar BRS AG presented a change in expression levels after 20 days of exposure to stress related to the genes OsCATA and OsCATB. They were significantly different when compared to 10 and 15 days of stress and the control plants. Also, as can be seen in Table 1, the gene OsCATC showed an increase in expression when compared to the control, especially in plants after 10 days of salt stress. Comparing the mean expression values of the genes of catalase, it was found that OsCATB has a higher expression value (RQ = 5.83) at 20 days.
For the enzyme glutathione reductase (GR), it was observed that the highest average expression value occurred for the isoform OsGR2 at 10 days of exposure to salt, with a RQ = 4.76. For this gene, this value was significantly higher when compared to its respective control and the other stress exposure times. The gene OsGR1 showed an increased expression at 10 and 15 days. For OsGR3, plants grown at 20 days, under salinity conditions, showed higher expression values in relation to the other times (Table 1).
Discussion
The acclimation of plants to salinity conditions is often associated with increased levels of reactive oxygen species (ROS) such as superoxide (O·−2), hydroxyl radical (HO.), hydrogen peroxide (H2O2) and singlet oxygen (1O2), which may be toxic to cells. ROS are produced during the aerobic metabolism of the plant. However, ROS production increases during stress conditions because of the disrupting of electron transport system and also due to the metabolic activities that occur in chloroplasts, mitochondria and peroxisomes (Ahmad et al. 2012). Under normal conditions, the ROS are efficiently eliminated by an enzymatic and non-enzymatic antioxidant system, whereas during salt stress, the excessive production may cause an imbalance in the immune system (Sofo et al. 2015). The superoxide radical is a moderately reactive free radical with a short half-life (2–4 μs). However, it cannot pass through the membrane and should be immediately converted to H2O2. If it is not eliminated, it may interact with other molecules forming a hydroxyl radical (HO.) extremely harmful to the cell (Sharma et al. 2012).
In this study, the concentration of O·−2 in plants grown under saline conditions was higher than the control plants at 5 days after germination. This increase may be closely related to the low activity of SOD on that day. However, there was a decrease in superoxide levels during the stress time. At the same time, the activity of SOD at 10 and 15 days was significantly higher in stressed plants. At 20 days, the activity resembled the control treatment. It is understood that the activity of this enzyme followed superoxide levels produced by rice plants under unfavorable growth conditions because, when its activity was increased, superoxide levels remained low.
In a study related to salinity stress in rice cultivars, Mishra et al. (2013) found that, at a concentration of 140 mM NaCl, leaves and roots of the sensitive cultivar (Malviya-36) showed an increase in the superoxide concentration with the increase in exposure times (5–20 days) to stress conditions. However, the superoxide content decreased over time in plants tolerant to salt stress (CSR-27), which is in agreement with the results found for the cv. BRS AG in this study.
In an experiment with tolerant (CSR-1 and Pokkali) and sensitive (MI-48, IR-28) rice plants, two Triticum aestivum cultivars (tolerant to salt: KRL-19, and sensitive to salt: WH-542) submitted to saline concentrations of 50 and 100 mM showed an increase in the SOD activity in plants tolerant to salt at all stress times and salt concentrations in the medium. This behavior was observed for the cultivar BRS AG only at 10 and 15 days in plants subjected to stress (Chawla et al. 2013; Mandhania et al. 2006). In rye grass plants (Lolium perenne) cultivated in 250 mM NaCl, during 4, 8 and 12 day periods, Hu et al. (2012) observed that the activity of SOD increased only in plants that remained for 4 days under salt stress. This was similar to the results obtained for the cv. BRS AG in the present study.
Rice plants, cultivar BRS AG, showed increased mean expression values of the genes OsSOD3-CU/Zn, OsSOD2-Cu/Zn and OsSOD-Cu/Zn at 10, 15 and 20 days, and OsSOD4-Cu/Zn and OsSODCc1-Cu/Zn at 15 days, suggesting that most of the SOD activity was due to these genes. Similar results were observed by Mishra et al. (2013), who observed an increase in SOD activity directly related to the gene Cu/Zn-SOD for the rice cultivars Malviya (sensitive) and CSR-27 (tolerant) exposed to salinity levels of 70 and 140 mM NaCl, respectively.
Interestingly, Menezes-Benavente et al. (2004) observed that only the gene sodCc2 was transcribed in rice plants cultivated in vitro under salinity conditions (250 mM NaCl). In plants grown in greenhouse, both the genes sodCc1 and sodCc2 were significantly expressed. Morita et al. (2011) showed that salt stress (100 mM NaCl) caused an increase in the transcription of the genes sodCc1 and sodCc2 in the rice cultivar Nipponbare, differing from those found for the cultivar BRS AG, where all Cu/Zn-dependent genes were expressed in at least one stress time.
At 15 and 20 days, the low content of H2O2 at a concentration of 136 mM NaCl indicates that the increased activity of CAT and APX could be responsible for detoxifying the H2O2 produced by SOD. There are contrasting literature data concerning this variable in saline conditions, where the content of H2O2 was reported to increase (Wang et al. 2005) and decrease (Turan and Tripathy 2013) under salt stress. At the same time, this increase was at first not harmful to the cells, probably not occurring the formation of radicals. Unlike the results of this study, Das et al. (2016), studying the rice cv. MTU1010 under salt stress, reported an increase in hydrogen peroxide and MDA levels under salt stress.
Hydrogen peroxide is moderately reactive and has a relatively long half-life (1 ms). It is known that hydrogen peroxide, as well as other ROS, is a metabolite not only toxic to the cells but may also serve as a signaling molecule, being beneficial at low concentrations and toxic when in excess (Sharma et al. 2012). It is understood that the levels found for the cultivar BRS AG were low and did not cause damage. A wide variety of environmental stimuli leads to a temporary increase in H2O2. This phenomenon is a signal that transmits the initial stimuli to a final response in the metabolism, which may explain the increase in these ROS at 20 days of stress. Due to the long life and the high permeability through membranes, hydrogen peroxide is beginning to be accepted by the scientific community as a secondary messenger in the presence of ROS (Gill and Tuteja 2010), and that it acts as a key regulator in a wide variety of physiological processes.
In this study, catalase showed a low activity at 10 days in plants that germinated in a saline medium. The activity of APX remained stable over time, with values higher than the control only at 10 days. Consequently, the increase in the activity of catalase over time and the maintenance of APX levels were effective in protecting and maintaining the balance of ROS in the cell, reducing hydrogen peroxide levels.
According to Mishra et al. (2013), the salt tolerance of rice plants is due to high levels of non-enzymatic antioxidants and antioxidant enzymes during stress. The results for the cultivar BRS AG are in agreement with those obtained by those authors and by Chawla et al. (2013). They reported an increased catalase enzyme activity in rice plants after salt stress at concentrations of 50, 100 and 140 mM with increases in stress exposure times.
According to Khan and Panda (2008), the decrease in the enzymatic activity of catalase in rice roots promotes the accumulation of H2O2, and this, when present in the cell via Haber–Weiss reaction, may form hydroxyl radicals. The hydroxyl radical is known to damage the biological membrane and react with most of the compounds present in biological systems, contributing to lipid peroxidation. The relation between levels of H2O2 and catalase activity was also observed by Chunthaburee et al. (2015) for rice genotypes. Tolerant plants with an increased catalase activity had lower levels of H2O2 when they were cultured in 100 mM NaCl.
In a study using the cultivar Kum Doi Saket, a pigmented rice, under in vitro saline conditions, Umnajkitikorn et al. (2013) obtained similar results to those of this study. They observed a higher activity of APX in stressed plants compared to their respective controls only 4 days after the addition of 150 mM NaCl to the medium. These results are in agreement with those found in this study, where the activity of APX was higher in plants treated with salt compared to their respective controls only at one stress time (10 days). Regarding the activity of catalase, different results were found by these authors: a decrease in the CAT activity during the germination of plants in a saline medium.
Upon analyzing the cultivar BRS AG, it was observed that all APX genes, except for OsAPX6, showed increase in expression values during the stress time. This behavior evidences that the increase in the transcript level may have contributed to the maintenance of the enzyme activity during the stress period. The increase in the expression of the gene OsAPX7 after 10 days, in response to salt stress, for the cultivar BRS AG, may be correlated with the activity of the enzyme. OsAPX7, present in chloroplasts, certainly contributed to the elimination of H2O2 since catalase does not appear to act in this organelle, although there are some studies reporting the opposite (Mhamdi et al. 2010). However, at 20 days of stress, the significant increase in the expression of OsAPX1, OsAPX2, OsAPx8, OsAPX3 and OsAPX5 is not related to the increase in enzyme activity. On this day, an increase in enzymatic activity was observed for plants that were not under stress. This could be because increase in transcription were not being translated into proteins in plants that were under stress. According to Dale and Schantz (2002), the increase in the amount of mRNA does not necessarily correlate with the amount of protein produced.
Another point to be considered is that all APXs uses reduced ascorbate (AsA) as a specific electron donor to reduce H2O2 into H2O and O2. Thus, this low concentration may inhibit the activity of APX. Furthermore, the protection performed by APX, as well as its activity, occurs in conjunction with the enzymes GR, MDAR and DAR, which comprise the ascorbate/glutathione cycle (Silveira et al. 2010). Thus, a high level of endogenous AsA is essential to maintain the antioxidant system, which is essential for the protection of plants against oxidative damage. Under special conditions, in which the ascorbic acid concentration is less than 20 μM, the APX activity is rapidly lost. APXs present in chloroplasts are less stable. Therefore, the APX of the cytosol and the membrane have inactivation half-times of approximately 1 h or more, while it is less than 30 s for mitAPX and chlAPX (Shigeoka et al. 2002). The fact that the enzyme activity does not correspond so significantly with the increase in gene expression can also be explained by the low availability of reduced ascorbate, a necessary substrate for the activity of this enzyme. Thus, ascorbate may also be acting directly on ROS.
Menezes-Benavente et al. (2004) evaluated the expression of genes of the antioxidant system of rice plants exposed for 11 days to salt stress (250 mM NaCl). They found cytosolic APX was positively regulated in plants up to 3 days of stress. This also occurred in this experiment: both genes that encode the cytosolic isoforms increased the expression in the presence of stress, especially APX1, which showed higher expression values compared to the other genes.
Results similar to those of the present study were observed by Yamane et al. (2010) in relation to APX isoforms. The induction was observed for the genes OsAPX1, OsAPX4, OsAPX6 and OsAPx7, while the expression of the cytosolic gene OsAPX2 did not change due to salinity, and the transcription level of OsAPx8 was slightly decreased by salinity in the basal region of rice leaves under stress conditions (200 mM NaCl).
Our results agree with those found by Teixeira et al. (2006) for the rice cultivar Taim, except with respect to the behavior observed for the gene OsAPx8. Similarly, they found that the genes OsAPX2, OsAPx7 and OsAPx8 had changed transcription levels in response to a treatment with NaCl. The expression of OsAPX2 and OsAPx7 was increased, while the accumulation of OsAPx8 transcripts were strongly repressed in plants subjected to salt stress.
In rice plants cv. Nipponbare, the expression of the gene OsAPX4 was induced by abiotic stresses, including salt stress (100 mM NaCl) (Guan et al. 2010). The cultivar BRS AG had an average OsAPX4 expression, being higher in stressed plants at all times.
The increase in the expression during the stress times can be correlated with the increase in enzyme activity and the increase in stress time, although this is not different from control plants. The results of this study indicate that increased levels of transcripts of OsCATA, OsCATB and OsCATC contributed to maintain the activity of the CAT enzyme, and OsCATB was the most responsive to salt stress. Under salinity conditions similar to our study, Kim et al. (2007) found that rice plants had expression values positively regulated for OsCATB, which showed the highest expression in our study, which evaluated the cultivar BRS AG. However, unlike our results, they observed no increase in expression for the genes OsCATA and OsCATC when the plants were exposed to saline conditions.
The enzyme glutathione reductase (GR) is essential for the elimination of ROS and for the maintenance of reduced glutathione (GSH) by the ascorbate/glutathione cycle. For the cultivar BRS AG grown under saline conditions, the activity of GR was not significant among treatments. In rice plants cv. CSR10 and Pusa Basmati1 (PB1), Turan and Tripathy (2013) found that the activity of GR increased in response to salt stress (200 mM) in both cultivars. The increase in GR activity in response to stress was higher in sensitive plants. These results are not in agreement with those found in this study for the cv. BRS AG.
For all times, the activity of the enzyme GR and the transcripts levels appear to be related. Under conditions similar to our study, Kim et al. (2007) observed an increase in the expression of two GR genes in rice plants under salt stress (130 mM NaCl), which were positively regulated at all stress times (3, 5 and 6 days). Turan and Tripathy (2013), studying rice plants under salt stress, reported an increase in the expression of OsGR2 both for the sensitive and the tolerant genotype.
Lipid peroxidation occurs when ROS levels are excessive in the cell, affecting its normal operation due to the oxidative stress caused by the production of radicals derived from lipids. According to Hernández et al. (2001), the growing evidence that there is damage to the membrane under salt stress conditions is related to the increased production of highly toxic reactive oxygen species. The oxidation of fatty membrane acids by reactive oxygen species may result in byproducts that aggravate the damage. Among them, malondialdehyde (MDA) is the main and most studied product of lipid peroxidation (Del Rio et al. 2005). However, the cultivar BRS AG did not show increased MDA levels, which indicates that ROS did not cause cellular damage. This response may be related to the fact that the MS medium contains myo-inositol, an osmoprotector, which regulates cell homeostasis, and influences various physiological processes, such as seed germination. Khan and Panda (2008), studying the response of rice plants under saline stress conditions, showed an increase in lipid peroxidation regarding both cultivars. However, the level was higher for the sensitive cultivar compared to the tolerant cultivar. The authors point out that the protection mechanism was more efficient under salt stress in the tolerant cultivar, consequently with a lower level of lipid peroxidation.
Conclusions
According to the results, it can be concluded that: (1) maintaining the activity of SOD, CAT, APX and GR over stress time is effective in protecting the cv. BRS AG against oxidative stress; (2) the SOD enzyme activity reduces superoxide levels in stressed plants over stress exposure times; (3) the increase in ROS not necessarily increases MDA levels; (4) under salt stress, some genes such as OsSOD3-CU/Zn, OsSOD2-Cu/Zn, OsSOD-Cu/Zn, OsSOD4-Cu/Zn, OsSOD-Fe and OsSODCc1-Cu/Zn may contribute to increase the activity of the enzyme superoxide dismutase; (5) the increase in the transcription of all evaluated isoforms, except for OsAPX6, contributed to the increase or the maintenance of the enzyme activity. Thus, it is possible to infer that the cv. BRS AG has defense mechanisms against salt stress.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Acknowledgements
This work was supported by the Brazilian funding agency Coordenação de Aperfeiçoamento de Pessoal de Nível Superior (CAPES), Fundação de Amparo à Pesquisa do Rio Grande do Sul (FAPERGS) and Conselho Nacional de Desenvolvimento Científico e Tecnológico (CNPq).
Author’s contributions
Conceived and designed the experiments LSP, LCB and AMMJ. Wrote, edited and analyzed the data: TR, LCB, MNA and ILV. Revised the paper: LSP, AMMJ and EJBB. Conducted the experiments: TR, LCB, MNA and ILV. All authors readed the paper and approved the final manuscript.
Compliance with ethical standards
Conflict of interest
The authors declare that they have no competing interests.
Footnotes
Electronic supplementary material
The online version of this article (doi:10.1007/s12298-017-0467-2) contains supplementary material, which is available to authorized users.
References
- Ahmad P. Oxidative damage to plants: antioxidant networks and signaling. Amsterdam: Elsevier Science, Academic Press; 2014. p. 672. [Google Scholar]
- Ahmad P, Bhardwaj R, Tuteja N. Plant signaling under abiotic stress environment. In: Ahmad P, Prasad MNV, editors. Environmental adaptations and stress tolerance of plants in the era of climate change. New York: Springer; 2012. pp. 297–323. [Google Scholar]
- Alscher RG, Erturk N, Heath LS. Role of superoxide dismutases (SODs) in controlling oxidative stress in plants. J Exp Bot. 2002;53:1331–1341. doi: 10.1093/jexbot/53.372.1331. [DOI] [PubMed] [Google Scholar]
- Azevedo RA, Alas RM, Smith RJ, Lea PJ. Responses of antioxidant enzymes to transfer from elevated carbon dioxide to air and ozone fumigation, in the leaves androots of wild type and catalase-deficient mutant of barley. Physiol Plant. 1998;104:280–292. doi: 10.1034/j.1399-3054.1998.1040217.x. [DOI] [Google Scholar]
- Bashir K, Nagasaka S, Itai RN, Kobaysahi T, Takahashi M, Nakanishi H. Expression and enzyme activity of glutathione reductase is upregulated by Fe-deficiency in graminaceo us plants. Plant Mol Biol. 2007;64:277–284. doi: 10.1007/s11103-007-9216-1. [DOI] [PubMed] [Google Scholar]
- Bowler C, Montagu MV, Inze D. Superoxide dismutase and stress tolerance. Ann Rev. 1992;43:83–116. doi: 10.1146/annurev.me.43.020192.000503. [DOI] [Google Scholar]
- Bradford M. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem. 1976;72:248–254. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- Cakmak I, Strbac D, Marschner H. Activities of hydrogen peroxide-scavenging enzymes in germinating wheat seeds. J Exp Bot. 1993;44:127–132. doi: 10.1093/jxb/44.1.127. [DOI] [Google Scholar]
- Chawla S, Jain S, Jain V. Salinity induced oxidative stress and antioxidant system in salt-tolerant and salt-sensitive cultivars of rice (Oryza sativa L.) J Plant Biochem Biotechnol. 2013;22:27–34. doi: 10.1007/s13562-012-0107-4. [DOI] [Google Scholar]
- Chunthaburee S, Dongsansuk S, Sanitchon J, Pattanagul W, Theerakulpisut P. Physiological and biochemical parameters for evaluation and clustering of rice cultivars differing in salt tolerance at seedling stage. Saudi J Biol Sci. 2015;23:467–477. doi: 10.1016/j.sjbs.2015.05.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dale JW, Schantz MV. From genes to genomes. Concptes and applications of DNA technology. Hoboken: Wiley; 2002. [Google Scholar]
- Das P, Seal P, Biswas AK. Regulation of growth, antioxidants and sugar metabolism in rice (Oryza sativa L.) seedlings by NaCl and its reversal by silicon. Am J Plant Sci. 2016;7:623–638. doi: 10.4236/ajps.2016.73055. [DOI] [Google Scholar]
- Del Rio D, Stewart AJ, Pellegrini N. A review of recent studies on malondialdehyde as toxic molecule and biological marker of oxidative stress. Nutr Metab Cardiovasc. 2005;15:316–328. doi: 10.1016/j.numecd.2005.05.003. [DOI] [PubMed] [Google Scholar]
- Ferreira ICFR, Abreu RMV. Stress oxidativo, antioxidantes e fitoquímicos. Bioanálise. 2007;2:32–39. [Google Scholar]
- Giannopolitis CN, Reis SK. Superoxide dismutases: II. Purification and quantitative relationship with water soluble protein in seedlings. Plant Physiol. 1977;59:315–318. doi: 10.1104/pp.59.2.315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gill SS, Tuteja N. Reactive oxygen species and antioxidante machinery in abiotic stress tolerance in crop plants. Plant Physiol Biochem. 2010;48:909–930. doi: 10.1016/j.plaphy.2010.08.016. [DOI] [PubMed] [Google Scholar]
- Guan G, Xia D, Liu S. OsAPX4 gene response to several environmental stresses in rice (Oryza sativa L.) Afr J Biotechnol. 2010;9:5908–5913. [Google Scholar]
- Gupta B, Huang B. Mechanism of salinity tolerance in plants: physiological, biochemical, and molecular characterization. Int J Genomics. 2014;701596:18. doi: 10.1155/2014/701596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heath RL, Packer L. Photoperoxidation in isolated chloroplast. I. Kinetics and stoichiometry of fatty acid peroxidation. Arch Biochem Biophys. 1968;125:189–198. doi: 10.1016/0003-9861(68)90654-1. [DOI] [PubMed] [Google Scholar]
- Hernández JA, Ferrer MA, Jiménez A, Barceló AR, Sevilla F. Antioxidant system and O2/H2O2 production in the apoplast of pea leaves: its relation with salt-induced necrotic lesion in minor veins. Plant Physiol. 2001;127:817–831. doi: 10.1104/pp.010188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu L, Li H, Pang H, Fu J. Responses of antioxidant gene, protein and enzymes to salinity stress in two genotypes of perennial ryegrass (Lolium perenne) differing in salt tolerance. J Plant Physiol. 2012;169:146–156. doi: 10.1016/j.jplph.2011.08.020. [DOI] [PubMed] [Google Scholar]
- Kaminaka H., Morita S., Nakajima M., Masumura T., Tanaka K. Gene Cloning and Expression of Cytosolic Glutathione Reductase in Rice (Oryza Sativa L.) Plant and Cell Physiology. 1998;39:1269–1280. doi: 10.1093/oxfordjournals.pcp.a029330. [DOI] [PubMed] [Google Scholar]
- Khan MH, Panda SK. Alterations in root lipid peroxidation and antioxidative responses in two rice cultivars under NaCl-salinity stress. Acta Physiol Plant. 2008;30:81–89. doi: 10.1007/s11738-007-0093-7. [DOI] [Google Scholar]
- Kim D, Shibato J, Agrawal JK, Fujihara S, Iwahashi H, Kim DH, Shim LS, Rakwal R. Gene transcription in the leaves of rice undergoing salt-induced morphological changes (Oryza sativa L.) Mol Cell. 2007;24:45–59. [PubMed] [Google Scholar]
- Li C, Bai T, Maa F, Hana M. Hypoxia tolerance and adaptation of anaerobic respiration to hypoxia stress in two Malus species. Sci Hortic. 2010;124:274–279. doi: 10.1016/j.scienta.2009.12.029. [DOI] [Google Scholar]
- Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 22DDCT method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- Magalhães Júnior AM, Fagundes P, Franco DF, Andres A, Nunes CD, Petrini JÁ, Martins JF, Severo A, Moraes OP, MouraF, Streck EA, Aguiar G, Facchinello PH, Huber MP, Krüger TK (2012) BRS AG: cultivar de arroz irrigado desenvolvida para produção de álcool de cereais e/ou alimentação animal. Comunicado Técnico—Embrapa Clima Temperado. https://www.embrapa.br/web/mobile/publicacoes/-/publicacao/1021670/brs-ag-cultivar-de-arroz-irrigado-desenvolvida-como-materia-prima-para-producao-dealcool-de-cereais-eou-alimentacao-animal. Accessed 02 Mar 2016
- Mandhania S, Madan S, Sawhney V. Antioxidant defense mechanism under salt stress in wheat seedlings. Biol Plant. 2006;50:227–231. doi: 10.1007/s10535-006-0011-7. [DOI] [Google Scholar]
- Menezes-Benavente L, Teixeira FK, Kamei CLA, Margis-Pinheiro M. Salt stress induces altered expression of genes encoding antioxidant enzymes in seedlings of a Brazilian indica rice (Oryza sativa L.) Plant Sci. 2004;166:323–331. doi: 10.1016/j.plantsci.2003.10.001. [DOI] [Google Scholar]
- Mhamdi A, Queval G, Chaouch S, Vanderauwera S, Van Breusegem F, Noctor G. Catalase function in plants: a focus on Arabidopsis mutants as stress-mimic models. J Exp Bot. 2010;61:4197–4220. doi: 10.1093/jxb/erq282. [DOI] [PubMed] [Google Scholar]
- Mishra P, Bhoomika K, Dubey RS. Differential responses of antioxidative defense system to prolonged salinity stress in salt-tolerant and salt-sensitive Indica rice (Oryza sativa L.) seedlings. Protoplasma. 2013;250:3–19. doi: 10.1007/s00709-011-0365-3. [DOI] [PubMed] [Google Scholar]
- Moraes GP, Benitez LC, Do Amaral MN, Vighi IL, Auler PA, Maia Da LC, Bianchi VJ, Braga EJB. Evaluation of reference genes for RT-qPCR studies in the leaves of rice seedlings under salt stress. Genet Mol Res. 2015;14:2384–2398. doi: 10.4238/2015.March.27.24. [DOI] [PubMed] [Google Scholar]
- Morita S, Nakatani S, Koshib T, Masumura T, Ogihara Y, Tanaka K. Differential expression of two cytosolic ascorbate peroxidases and two superoxide dismutase genes in response to abiotic stress in rice. Rice Sci. 2011;18:157–166. doi: 10.1016/S1672-6308(11)60023-1. [DOI] [Google Scholar]
- Murashige T, Skoog FA. Revised medium for rapid growth and bio essays with tobacco tissue culture. Physiol Plant. 1962;15:473–497. doi: 10.1111/j.1399-3054.1962.tb08052.x. [DOI] [Google Scholar]
- Nakano Y, Asada K. Hydrogen peroxide is scavenged by ascobate-specific peroxidase in spinach chloroplasts. Plant Cell Physiol. 1981;22:867–880. [Google Scholar]
- Pitman MG, Lauchli A (2002) Global impact of salinity and agricultural ecosystems. In: Lauchli A, Luttge V (eds) Salinity: environment-plants molecules, 1st edn. Springer, Dordrecht, pp 3–20
- Prisco JT, Gomes Filho E (2010) Fisiologia e bioquímica do estresse salino em plantas. In: Ghey HR, da Dias NS, de Lacerda CF (eds) Manejo da salinidade na agricultura: Estudos básicos e aplicados 1st edn. Expressão Gráfica e Editora, Fortaleza, pp 143–159
- Sharma P, Bhushan A, Dubey RS, Pessarakli M. Reactive oxygen species, oxidative damage, and antioxidative defense mechanism in plants under stressful conditions. J Bot. 2012;2012:1–26. doi: 10.1155/2012/217037. [DOI] [Google Scholar]
- Shigeoka S, Ishikawa T, Tamoi M, Miyagawa Y, Takeda T, Yabuta Y. Regulation and function of ascorbate peroxidase isoenzymes. J Exp Bot. 2002;53:1305–1319. doi: 10.1093/jexbot/53.372.1305. [DOI] [PubMed] [Google Scholar]
- Silveira JAG, Silva SLF, Silva EN, Viégas RA (2010) Mecanismos biomoleculares envolvidos com a resistência ao estresse salino em plantas. In: Ghey HR, da Dias NS, de Lacerda CF (eds) Manejo da salinidade na agricultura: Estudos básicos e aplicados 1st edn. Expressão Gráfica e Editora, Fortaleza, pp 161–180
- Sofo A, Scopa A, Nuzzaci M, Vitti A. Ascorbate peroxidase and catalase activities and their genetic regulation in plants subjected to drought and salinity stresses. Int J Mol Sci. 2015;16:13561–13578. doi: 10.3390/ijms160613561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teixeira FK, Menezes-Benavente L, Margis R, Margis-Pinheiro M. Analysis of the molecular evolutionary history of the ascorbate peroxidase gene family: inferences from the rice genome. J Mol Evol. 2004;59:761–770. doi: 10.1007/s00239-004-2666-z. [DOI] [PubMed] [Google Scholar]
- Teixeira FK, Menezes-Benavente L, Galvão VC, Margis R, Margis-pinheiro M. Rice ascorbate peroxidase gene family encodes functionally diverse isoforms localized in different subcellular compartments. Planta. 2006;224:300–314. doi: 10.1007/s00425-005-0214-8. [DOI] [PubMed] [Google Scholar]
- Turan S, Tripathy BC. Salt and genotype impact on antioxidative enzymes and lipid peroxidation in two rice cultivars during de-etiolation. Protoplasma. 2013;250:209–222. doi: 10.1007/s00709-012-0395-5. [DOI] [PubMed] [Google Scholar]
- Umnajkitikorn K, Faiyue B, Saengnil K. Enhancing antioxidant properties of germinated Thai rice (Oryza sativa L.) cv. Kum Doi Saket with salinity. Rice Res Open Access. 2013;1:1–8. [Google Scholar]
- Velikova V, Yordanov I, Edreva A. Oxidative stress and some antioxidant systems in acid rain-treated bean plants. Plant Sci. 2000;151:59–66. doi: 10.1016/S0168-9452(99)00197-1. [DOI] [Google Scholar]
- Vighi IL, Benitez LC, Amaral MN, Moraes GP, Auler PA, Rodrigues GS, Deuner S, Maia LC, Braga EJB. Functional characterization of the antioxidant enzymes in rice plants exposed to salinity stress. Biol Plant. 2017;61:1–11. doi: 10.1007/s10535-017-0727-6. [DOI] [Google Scholar]
- Wang Y, Wisniewski M, Meilan R, Cui M, Webb R, Fuchigami L. Overexpression of cytosolic ascorbate peroxidase in tomato confers tolerance to chilling and salt stress. J Am Soc Hortic Sci. 2005;130:167–173. [Google Scholar]
- Yamane K, Mitsuya S, Taniguchi M, Miyake H. Transcription profiles of genes encoding catalase and ascorbate peroxidase in the rice leaf tissues under salinity. Plant Prod Sci. 2010;13:164–168. doi: 10.1626/pps.13.164. [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


