Abstract
The TCP family is a group of plant-specific transcription factors. TCP genes encode proteins harboring bHLH structure, which is implicated in DNA binding and protein–protein interactions and known as the TCP domain. TCP genes play important roles in plant development and have been evolutionarily and functionally elaborated in various plants, however, no overall phylogenetic analysis or expression profiling of TCP genes in Zea mays has been reported. In the present study, a systematic analysis of molecular evolution and functional prediction of TCP family genes in maize (Z. mays L.) has been conducted. We performed a genome-wide survey of TCP genes in maize, revealing the gene structure, chromosomal location and phylogenetic relationship of family members. Microsynteny between grass species and tissue-specific expression profiles were also investigated. In total, 29 TCP genes were identified in the maize genome, unevenly distributed on the 10 maize chromosomes. Additionally, ZmTCP genes were categorized into nine classes based on phylogeny and purifying selection may largely be responsible for maintaining the functions of maize TCP genes. What’s more, microsynteny analysis suggested that TCP genes have been conserved during evolution. Finally, expression analysis revealed that most TCP genes are expressed in the stem and ear, which suggests that ZmTCP genes influence stem and ear growth. This result is consistent with the previous finding that maize TCP genes represses the growth of axillary organs and enables the formation of female inflorescences. Altogether, this study presents a thorough overview of TCP family in maize and provides a new perspective on the evolution of this gene family. The results also indicate that TCP family genes may be involved in development stage in plant growing conditions. Additionally, our results will be useful for further functional analysis of the TCP gene family in maize.
Electronic supplementary material
The online version of this article (doi:10.1007/s12298-017-0476-1) contains supplementary material, which is available to authorized users.
Keywords: TCP, Transcription factor, Maize, Expression
Introduction
Transcription factors are important regulators for cell proliferation and cell growth, and they also play great roles in various developmental processes. In plants, an increasing number of gene families encoding different transcription factors have been identified, such as CCCH, HD-Zip and TCP transcription factors (Ma et al. 2016; Peng et al. 2012; xue et al. 2014). Herein, TCP genes have vital roles in plant development progress. They encode for a group of plant-specific transcription factors, expressed in different tissues and involved in regulating cell growth and development. The TCP family of transcription factors were named after the first four characterized members, including teosinte branched1 (TB1) from maize (Zea mays), cycloidea (CYC) from snapdragon (Antirrhinum majus) and proliferating cell factors1 and 2 (PCF1 and PCF2) from rice (Oryza sativa). Besides, these proteins are characterized by a highly conserved, approximately 60 residue TCP domain in the N-terminal region, which contains a basic helix-loop-helix (bHLH) structure involved in DNA binding and protein interactions (Kosugi and Ohashi 1997). TCP proteins can be further classified into two subfamilies based on the primary structures of their DNA binding domains. Class I TCP (also known as TCP-P) proteins contain only a conserved TCP domain, whereas class II TCP (also known as TCP-C) proteins contain an R domain which is predicted to form a coiled coil and likely mediates protein interactions and other domains (Cubas 2002; Martín-Trillo and Cubas 2010).
Over the past few decades, TCP genes have been identified in diverse plant species, such as Arabidopsis and rice. These genes encode proteins involved in the processes of cell proliferation, growth and cell differentiation, such as regulating shoot apical meristem (SAM) development, lateral branching (Hubbard 2003), leaf development and flower development. In Arabidopsis, TCP genes in class I such as AtTCP7, AtTCP8, AtTCP22, AtTCP23 play important roles in traits of leaf development and the shape of the flower (Aguilar-Martínez and Sinha 2013; Tomotsugu et al. 2007). Through yeast two-hybrid and direct protein–protein interaction assays, the AtTCP2, -3, -11, -15 were found to interact with different components and then regulate circadian clock activity (Giraud et al. 2010), while AtTCP14 and AtTCP15 regulates the cell division in the young internodes, influencing the height of plant (Kieffer et al. 2011). A recent research indicates that AtTCP14/AtTCP15 interact with ubiquitin receptor DA1, DA1-related proteins (DAR1, DAR2), and subsequently repress endoreduplication by directly regulating the expression of cell-cycle genes (Yuancheng et al. 2015). At the same time, AtTCP14 also regulates embryonic development (Rueda-Romero et al. 2012). In rice, two class I members, PCF1 and PCF2 regulate the transcription of proliferating cell nuclear antigen (PCNA), an auxiliary protein of DNA polymerase (Kosugi and Ohashi 1997). By contrast, in plants, class II genes have greatly contributed to the modification of existing traits and the evolution of novel morphological traits. For example, the AtTCP04 that controls leaf development encodes a TCP family transcription factor containing an miR319-binding site, and it influences the size and shape of leaves (Ori et al. 2007). OsTB1 is a unique factor in control of axillary buds in rice, and functions as a negative regulator of lateral branching (Takeda et al. 2003). Losing of these genes may influence the number of lateral branches. CYC-like genes participate in petal shape formation and may help determine floral bilateral symmetry (Carpenter et al. 1999). Tb1/brc1-type mutants in maize and wheat exhibit different increased numbers of branches, implying that tb-1 suppress the occurrence of lateral branches (Hubbard et al. 2002; Lewis et al. 2008), which may be related to the corresponding gene expression.
Maize is an important cereal crop that serves as a model plant for the study of genetics, evolution and other basic biological processes (Yang et al. 2011). Functionally important TCP proteins has been comprehensively studied in diverse plants, such as Arabidopsis, rice, Prunus mume and so on, but their evolutionary fate or functions in maize still remain unknown. With the high quality maize genome sequences, genome-wide analysis of maize TCP family is plausible. In the present study, we performed a comprehensive analysis of the maize TCP transcription factor family. We identified 29 non-redundant TCP transcription factor genes in maize and subjected them to systematic analysis, revealing their phylogenetic relationships, chromosomal locations, gene duplication status, substitution rates, gene structures, conserved motifs and expression profiles. Furthermore, we performed a thorough comparative analysis of maize TCP genes with those from Arabidopsis and rice. Interestingly, we found that the expansion of the TCP family in maize may have been caused by segmental duplication and is not associated with tandem duplication. The results of our genome-wide analysis of the ZmTCP gene family will contribute to further studies involving the functional characterization of TCP proteins in maize, and it can also help to identify and perform comprehensive analysis of the TCP transcription factor family in other species.
Materials and methods
Identification of TCP genes in the maize genome
The http://www.maizesequence.org/index.html was used to download the maize (Z. mays L.B73) database searched to detect ZmTCP gene sequences (Sheng et al. 2014). DNATOOLS software was then used to build a local database from the latest complete maize genome sequence information, including nucleotide sequences and protein sequences. The conserved TCP DNA-binding domain sequence was obtained from the Pfam protein family database (http://pfam.sanger.ac.uk/). And it was used as a query to search against the maize protein database with the BLASTP program (P value = 0.001). In order to confirm the putative TCP genes in the maize genome, the amino acid sequences of all the proteins were searched for the presence of TCP domains by Pfam and SMART (http://smart.embl-heidelberg.de/). After examining, the redundant genes was removed, and all of the non-redundant TCP genes were used for further analysis. The molecular weight (kDa) and isoelectric point (PI) of each protein were calculated using the ExPASy program (http://www.expasy.org/tools/).
Phylogenetic analysis, gene location and duplication
Multiple sequence alignments were conducted on the amino acid sequences of TCP proteins in the Z. mays, rice and Arabidopsis genomes using Cluster X version 1.83 with default settings (Thompson et al. 1994). Subsequently, MEGA 6.0 software was employed to construct an unrooted tree based on the alignment using neighbor-joining (N-J), bootstrap analysis was performed using 1000 replicates. Additionally, a separate phylogenetic tree was constructed based on all of the TCP protein sequence in Z. mays for further analysis, but bootstrap analysis was performed using 100 replicates. BlastP searches were performed against the Z. mays genome database in Phytozome (http://www.phytozome.net/zeamays.phb) to obtain information about the physical locations of all ZmTCP genes. The diagram of the chromosome locations of TCP genes was generated using MapInspect software (http://mapinspect.software.informer.com/). The duplication of TCP genes were also investigated. The duplication is important to expand, to confirm the duplicate gene, all of the confirmed ZmTCP protein sequences were aligned using ClustalW, their evolutionary distances were computed and all identical sequences were checked manually to remove redundant sequences. Finally, consisting with the criteria on the following, the duplication were admitted: length of aligned sequences cover > 80% of the longer gene; similarity of the aligned regions > 80%.
Gene structure analysis and identification of conserved motifs
The TCP gene sequences and CDS were extracted from the downloaded maize genome sequences. Identification of the exon/intron organization of the ZmTCP genes was performed with Gene Structure Display Server (GSDS; http://gsds.cbi.pku.edu.cn/) by aligning the cDNAs with their corresponding genomic DNA sequences. To identify the conserved motifs, the maize TCP proteins sequences were submitted to the online Multiple Expectation maximization for Motif Elicitation (MEME) program, with the following parameters: any number of repetitions, the optimum width = 6–200 residues, and maximum number of motifs = 20.
Calculation of Ka/Ks values
The number of non-synonymous substitutions per non-synonymous site (Ka) and the number of synonymous substitutions per synonymous site (Ks) of duplicated genes were calculated using DnaSPv5.0. The ratio of non-synonymous to synonymous nucleotide substitutions (Ka/Ks) between paralogs was analyzed to detect the mode of selection. To estimate the timing of the duplication events, the Ks value was translated into duplication time in millions of years based on a rate of λ substitutions per synonymous site per year. The duplication time (T) was calculated as T = Ks/2λ × 10−6 Mya (λ = 6.56 × 10−9 for grasses) (Gaut et al. 1996). Indeed, a sliding window analysis of Ka/Ks ratios was performed with the following parameters: window size, 150 bp; step size, 9 bp.
RNA isolation and real-time quantitative RT-PCR analysis
To detect the expression profiles of TCP genes in Z. mays, total RNA was prepared using RNA Plus, followed by DNase I treatment to remove any genomic DNA contamination. RNA concentrations were determined using a NanoDrop ND-1000 UV–Vis spectrophotometer, and the integrity of the RNA was assessed on a 2% (w/v) agarose gel. Next, 1 µg of total RNA was reverse transcribed into cDNA using a Transcriptor First Strand cDNA Synthesis Kit (Roche Switzerland). Quantitative RT-PCR was carried out using an ABI PRISM 7300 real-time PCR system. Each reaction contained 12.5 µl FastStart Universal SYBR Green Master (ROX), 2 µl cDNA sample and 1 µl primer (10 ng/µl). Each pair of primers was designed using Primer Express 3.0 software, targeting an amplicon size of 90–190 bp. The thermal cycling conditions were as follows: 95 °C for 10 min, followed by 40 cycles at 95 °C for 15 s and 60 °C for 1 min. The specificity of the reactions was verified by melting curve analysis. The relative mRNA level for each gene was calculated as 2−ΔΔCT value in comparison to unstressed seedlings (Livak and Schmittgen 2001). The maize Actin 1 gene was used as an internal control for normalization. At least three replicates of each cDNA sample were performed for quantitative RT-PCR analysis.
Detection of orthologous gene pairs
The OrthoMCL (http://orthomcl.org/orthomcl/) was used to identify the orthologs of TCP genes among maize, rice and sorghum (Li et al. 2003). The relationships of the orthologous pairs among the three species were plotted using Circos (http://circos.ca/) (Krzywinski et al. 2009).
Results
Identification and genomic location of TCP proteins in maize
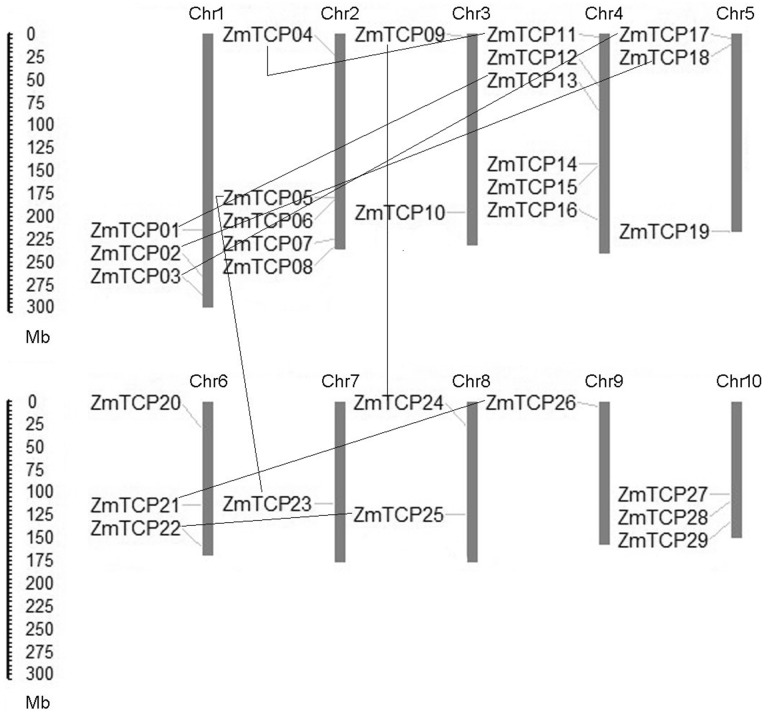
The sequences of TCP domain HMM profile (PF03634) were employed as a query to search against the maize genome database with the BlastP program. Originally, a total of 46 TCP genes were identified. After removing redundant sequences, a total of 29 maize genes were obtained. Subsequently, all these 29 proteins were submitted for Pfam matches to check whether TCP domains exist. The total number of TCP proteins in maize is greater than that in rice and Arabidopsis (Table 1). At last, details about each ZmTCP isoelectric point (pI), molecular weight (Mw), the length of protein and chromosome location are provided in Table 2. The TCP proteins consisted of 93 to 471 amino acids (aa), with an average of 225 aa, thus exhibiting diversity in their lengths. Moreover, the molecular weight ranged from 15.1 to 69.6 kDa and isoelectric point varied from 5.21 to 11.95. All information of ZmTCP protein sequences are summarized in Table 2. Furthermore, based on the starting position of each gene on the chromosome, the 29 maize TCPs were mapped onto chromosomes (Fig. 1). All the 29 putative proteins were found to harbor TCP domains and regarded as TCP family members. We next designated these 29 TCP genes ZmTCP1 to ZmTCP29 according to their physical locations (from top to bottom) on chromosomes 1–10. The distribution of maize TCP genes varies among chromosomes and appears to be uneven. Chromosome 4 possesses as many as six TCP genes (the largest number of TCP genes on a chromosome), while no more than three TCP genes were located on the other chromosomes. Chromosome 7, 8 and 9 contain only one maize TCP gene. Two ZmTCPs are located on chromosome 3 and 8, and three ZmTCPs are located on chromosome 1, 5, 6 and 10.
Table 1.
Number of TCP genes in the maize, rice and Arabidopsis genomes
| Category | Maize | Arabidopsis | Rice |
|---|---|---|---|
| PCF | 7 | 13 | 10 |
| CIN | 12 | 8 | 12 |
| CYC/TB1 | 10 | 3 | 3 |
| Total number | 29 | 24 | 25 |
Table 2.
The list of TCP genes in maize
| Gene name | Sequenced ID | Length (aa) | pI | MW (Da) | Chr. location |
|---|---|---|---|---|---|
| ZmTCP01 | GRMZM2G166687 | 168 | 5.21 | 26,112.37 | 1:214307551..214308535 |
| ZmTCP02 | AC233950.1 | 280 | 7.99 | 39,856.39 | 1:265745979..265747712 |
| ZmTCP03 | GRMZM2G115516 | 286 | 9.30 | 39,395.37 | 1:285309991..285314535 |
| ZmTCP04 | AC234521.1 | 189 | 10.60 | 27,713.53 | 2:22267351..22268478 |
| ZmTCP05 | GRMZM2G110242 | 199 | 5.99 | 29,902.05 | 2:179980551..179982009 |
| ZmTCP06 | GRMZM2G088440 | 129 | 8.55 | 21,139.24 | 2:181657719..181660104 |
| ZmTCP07 | GRMZM2G414114 | 276 | 8.67 | 42,293.04 | 2:224523636..224527847 |
| ZmTCP08 | GRMZM2G020805 | 330 | 9.44 | 46,223.44 | 2:234795896..234802008 |
| ZmTCP09 | AC205574.3 | 410 | 6.78 | 58,828.29 | 3:3475915..3478576 |
| ZmTCP10 | GRMZM2G166946 | 206 | 8.89 | 30,475.09 | 3:194884444..194887065 |
| ZmTCP11 | GRMZM2G055024 | 316 | 5.99 | 29,902.05 | 4:2546850..2550515 |
| ZmTCP12 | GRMZM2G062711 | 127 | 11.95 | 21,582.06 | 4:55919615..55920304 |
| ZmTCP13 | GRMZM2G060319 | 209 | 5.76 | 29,352.33 | 4:82358722..82359791 |
| ZmTCP14 | GRMZM2G135461 | 93 | 6.83 | 15,089.23 | 4:142927564..142929311 |
| ZmTCP15 | GRMZM2G078077 | 161 | 9.99 | 22,969.73 | 4:145614348..145615398 |
| ZmTCP16 | GRMZM2G003944 | 279 | 9.33 | 40,759.97 | 4:204959815..204961840 |
| ZmTCP17 | GRMZM2G089361 | 282 | 9.57 | 39,053.19 | 5:4945066..4946626 |
| ZmTCP18 | AC190734.2 | 268 | 6.72 | 38,894.31 | 5:11847304..11848383 |
| ZmTCP19 | GRMZM2G445944 | 158 | 10.24 | 21,115.65 | 5:216602509..216603638 |
| ZmTCP20 | GRMZM2G031905 | 101 | 9.88 | 16,009.05 | 6:27464014..27465629 |
| ZmTCP21 | GRMZM2G142751 | 274 | 7086 | 39,088.24 | 6:112589573..112591576 |
| ZmTCP22 | GRMZM2G120151 | 219 | 7.92 | 30,902.69 | 6:158367934..158370996 |
| ZmTCP23 | GRMZM2G064628 | 204 | 6.05 | 30,231.37 | 7:110781361..110782852 |
| ZmTCP24 | GRMZM2G015037 | 471 | 6.30 | 69,551.53 | 8:24472981..24476366 |
| ZmTCP25 | GRMZM2G035944 | 195 | 9.04 | 29,585.31 | 8:122903771..122906285 |
| ZmTCP26 | GRMZM2G113888 | 296 | 8.93 | 404,658.95 | 9:5003891..5006082 |
| ZmTCP27 | GRMZM2G096610 | 130 | 6.84 | 18,325.70 | 10:100072291..100073204 |
| ZmTCP28 | GRMZM2G458087 | 117 | 8.44 | 18,829.39 | 10:110203457..110205129 |
| ZmTCP29 | GRMZM2G465091 | 153 | 10.01 | 21,010.86 | 10:132033330..132034374 |
Fig. 1.
Locations of 29 TCP proteins on ten maize chromosomes. a The scale on the left is in megabases. Chromosome numbers are indicated above each bar. The gene names on the left side of each chromosome correspond to the approximate locations of each TCP gene. The line in chromosome was arranged to show duplicated genes
Phylogenetic analysis of TCP proteins
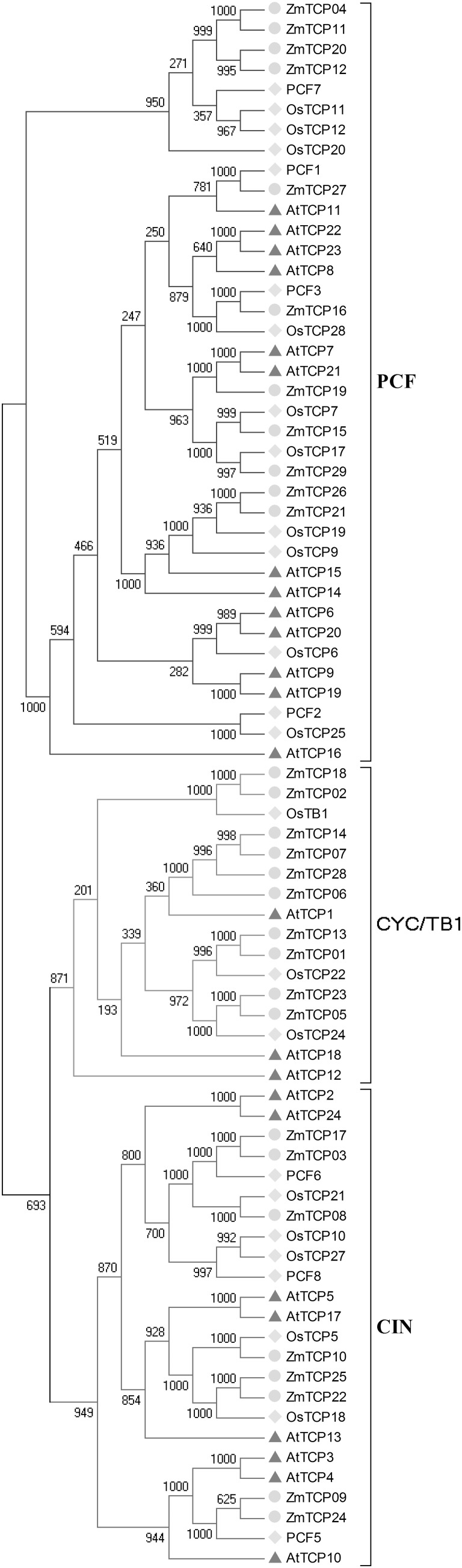
To elucidate the evolutionary history of TCP genes, we constructed an unrooted phylogenetic tree using the neighbor-joining (N-J) method of all maize TCP genes, compared with Arabidopsis and rice TCP genes reported previously, to generate a phylogenetic tree (“Methods” section, Fig. 2) (Aguilar-Martínez et al. 2007; Pagnussat et al. 2005). Based on the subdomain of these TCP genes, the 29 TCP members were divided into two classes. In general, genes clustered in the same group of a phylogenetic tree often share similar functional features. The Arabidopsis TCP genes clustered in group PCF in our phylogenetic tree were previously reported to participate in proliferation, suggesting that these genes may play essential roles in plant growth (Martín-Trillo and Cubas 2010).
Fig. 2.
Phylogenetic analysis. The phylogenetic tree was constructed using previously reported Arabidopsis and rice TCP sequences and maize TCP sequences using the neighbor-joining (N-J) method, with 1000 bootstrap replicates. The protein sequences could be divided into two distinct subfamilies. Class II was further divided into two group (CIN and CYC/TB1). Blue lines represent PCF, and dark blue for CIN, red for CYC/TB1 (colour figure online)
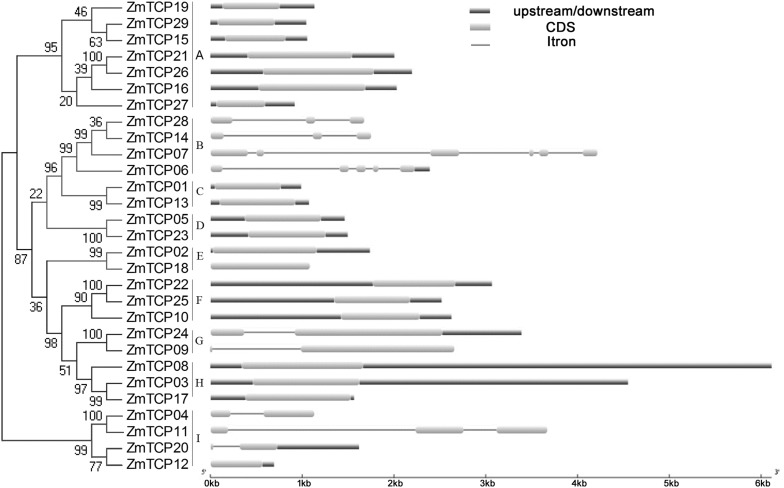
In order to decipher the evolutionary relationships within maize TCP family member, an N-J tree was constructed, only with the sequences of 29 maize TCP proteins. Based on the topological relationship of maize TCP genes (Fig. 3), we identified eight sister pairs, all of which had high bootstrap support. The TCP genes were further categorized into nine major subgroups, including A, B, C, D, E, F, G, H and I. The A and I subfamilies belong to class I, while B, C, D, E, F, G and H belong to class II. Class II can also be divided into two subfamily, CIN and TB1/CYC, with B, C, D and E belonging to TB1/CYC and F, G and H belonging to CIN. The largest subgroup is A, which contains seven members. Some subgroups only contain two genes, such as C, D, E and G.
Fig. 3.
Phylogenetic relationships and exon/intron structures of maize TCP proteins. The unrooted tree was generated with the MEGA4.0 program using the full-length amino acid sequences of the 29 TCP proteins by the N-J method, with 100 bootstrap replicates. Exons and introns are indicated by yellow lines and thin gray lines, respectively. The untranslated regions (UTRs) are indicated by blue lines (colour figure online)
Structure and domain conservation analysis of TCP proteins
To further confirm the phylogenetic relationships and to examine the diversity of ZmTCP gene structure, we compared the exon/intron organizations in the coding sequences of individual ZmTCP genes in Z. mays, revealing that only nine genes are disrupted by introns (ZmTCP04, -06, -07, -09, -11, -14, -20, -24, -28), while the remaining genes lack introns. The introns are present in subgroups B, G and I. Among the genes, ZmTCP07 has five introns and ZmTCP06 has six introns. These results are consistent with the results of phylogenetic analysis, in which genes clustered in the same group display similar exon/intron structures, especially regarding the number of introns. However, in some cases, the number of exons/introns varies among genes clustered together in the phylogenetic tree. For example, the introns in subgroup B are different, ZmTCP14(2), ZmTCP28(2), ZmTCP07(5), ZmTCP06(4).
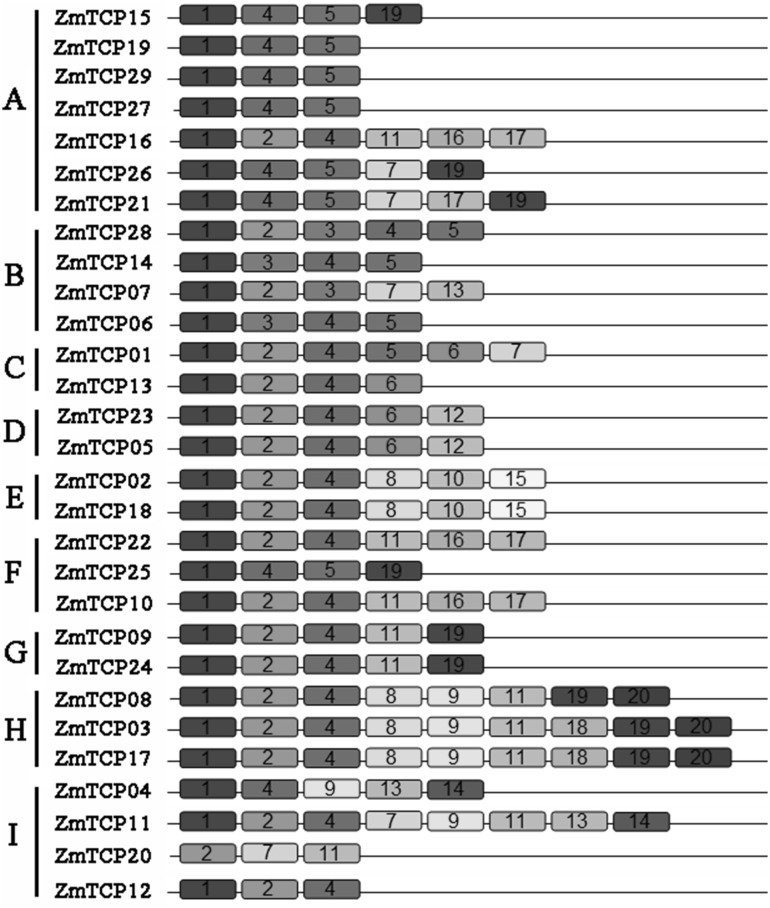
We further searched for conserved motifs in TCP proteins using the MEME web server to examine the diversity of motif compositions among ZmTCPs. As shown in Fig. 4, 20 conserved motifs (designated motif 1–20) were identified in maize TCP proteins. All ZmTCPs have conserved domain, motif 1 and motif 2 represent a highly conserved 60-residue-long DNA-binding structure at the N-terminus, called the TCP conserved domain. The proteins in the same subgroup share similar conserved motifs, but high divergence was observed among subgroups. All the TCP proteins harbor motif 1 or motif 2. All proteins in subgroup A contain motif 1 and motif 4, while all ZmTCPs in subgroup H possess motif 1, 2, 4, 8, 9, 11, 19, 20. Proteins in subgroup E contain motif 10, which does not exist in other subgroups. Generally speaking, the results of phylogenetic analysis are supported by the domain architecture and exon/intron structures.
Fig. 4.
Protein motifs of ZmTCPs. Each color represents a specific motif in the protein identified using the MEME motif search tool. The order of the motifs only represents the motif in the protein sequence and does not represent the actual location and size (colour figure online)
Gene duplication analysis
Gene duplication events are important events implicated in gene family expansion, mainly including tandem and segmental duplications (Leister 2004). To detect tandem or segmental duplications within maize TCP members, we firstly defined genes locating in the same clade of the phylogenetic tree, which exhibits high bootstrap values, as co-paralogs in the same species (Fig. 3). Secondly, these paralogs were further designated as segmental duplication if they are co-paralogs and located on duplicated chromosomal blocks as proposed by Wei et al. (2007). Tandem duplication generates gene copies through chromosome crossing over. Finally, based on phylogenetic analysis and chromosomal locations of ZmTCPs, a total of 16 maize TCP genes was found to be involved in segmental duplication, while no tandem duplication events were detected. All three ZmTCPs on chromosome 1 have taken part in segmental duplication events. Chromosome 10 has three ZmTCP genes, but no duplication events occurred on this chromosome. Whereas chromosome 7 and 9 share two ZmTCP genes, both of them belong to segmental duplication events. Segmental duplication occurred in all subgroups except subgroup B (Table 3).
Table 3.
Types of ZmTCP genes
| Identifier (Zea Mays) | Name | Type |
|---|---|---|
| GRMZM2G166687 | ZmTCP01 | CYC/TB1 |
| AC233950.1 | ZmTCP02 | CYC/TB1 |
| GRMZM2G115516 | ZmTCP03 | CIN |
| AC234521.1 | ZmTCP04 | CIN |
| GRMZM2G110242 | ZmTCP05 | CYC/TB1 |
| GRMZM2G088440 | ZmTCP06 | CYC/TB1 |
| GRMZM2G414114 | ZmTCP07 | CYC/TB1 |
| GRMZM2G020805 | ZmTCP08 | CIN |
| AC205574.3 | ZmTCP09 | CIN |
| GRMZM2G166946 | ZmTCP10 | CIN |
| GRMZM2G055024 | ZmTCP11 | CIN |
| GRMZM2G062711 | ZmTCP12 | CIN |
| GRMZM2G060319 | ZmTCP13 | CYC/TB1 |
| GRMZM2G135461 | ZmTCP14 | CYC/TB1 |
| GRMZM2G078077 | ZmTCP15 | PCF |
| GRMZM2G003944 | ZmTCP16 | PCF |
| GRMZM2G089361 | ZmTCP17 | CIN |
| AC190734.2 | ZmTCP18 | CYC/TB1 |
| GRMZM2G445944 | ZmTCP19 | PCF |
| GRMZM2G031905 | ZmTCP20 | CIN |
| GRMZM2G142751 | ZmTCP21 | PCF |
| GRMZM2G120151 | ZmTCP22 | CIN |
| GRMZM2G064628 | ZmTCP23 | CYC/TB1 |
| GRMZM2G015037 | ZmTCP24 | CIN |
| GRMZM2G035944 | ZmTCP25 | CIN |
| GRMZM2G113888 | ZmTCP26 | PCF |
| GRMZM2G096610 | ZmTCP27 | PCF |
| GRMZM2G458087 | ZmTCP28 | CYC/TB1 |
| GRMZM2G465091 | ZmTCP29 | PCF |
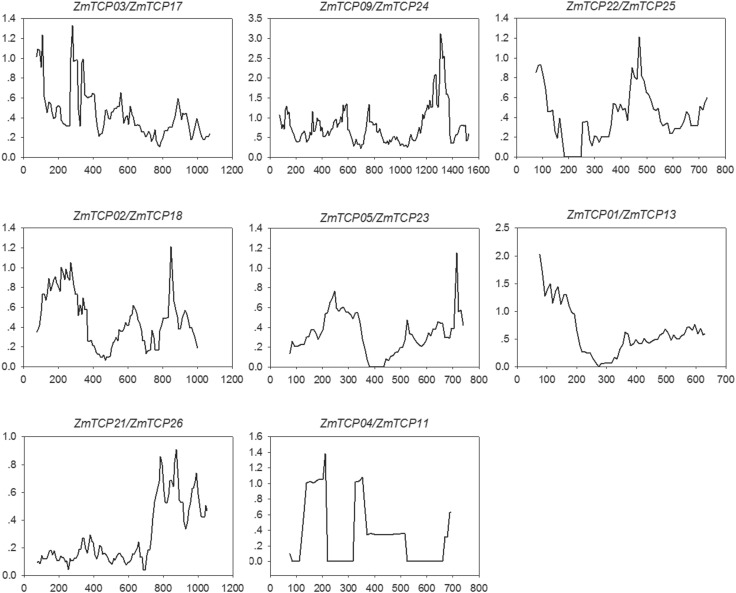
To explore the selective constraints on duplicated TCP genes, we calculated the Ks and Ka/Ks ratio for each duplicated TCP gene pairs (Table 4). The ratio of synonymous mutations (Ka) to non-synonymous mutations (Ks), (Ka/Ks) is widely applied to measure the rate of genetic evolution, which is also referred to as selection pressure. According to the natural selection theory, Ka/Ks > 1 indicates positive selective, while Ka/Ks = 1 indicates neutral selection and a ratio < 1 indicates negative or purifying selection. In the present study, all eight duplicated pairs were investigated. The results show that the Ka/Ks ratios of all eight pairs were < 1, indicating the effect of strong purifying selection and the slow evolution rate in TCP protein sequences. Based on Ka/Ks analysis, we conclude that purifying selection is the major evolutionary force on maize TCP genes. The duplication of the eight paralogous gene pairs was estimated to have occurred 2.43–19.38 million years ago (Mya), according to the substitution rate of 6.5 × 10−9 substitutions per site per year (Gaut et al. 1996; Li et al. 2013). Therefore, we performed sliding window analysis of the Ka/Ks ratios between each pair of TCP genes (Fig. 5), which directly reflects the relationships between paralogous genes.
Table 4.
Ka/Ks analysis and estimated divergence time for ZmTCP gene pairs in maize
| Paralogous pairs | Ks | Ka | Ka/Ks | Duplication date (MY) | Duplication type |
|---|---|---|---|---|---|
| ZmTCP13-ZmTCP03 | 0.1515 | 0.0599 | 0.3954 | 11.65 | Segmental |
| ZmTCP09-ZmTCP24 | 0.1463 | 0.1003 | 0.6860 | 11.25 | Segmental |
| ZmTCP25-ZmTCP22 | 0.1007 | 0.0526 | 0.5223 | 7.75 | Segmental |
| ZmTCP18-ZmTCP02 | 0.2042 | 0.0879 | 0.4305 | 15.71 | Segmental |
| ZmTCP23-ZmTCP05 | 0.2519 | 0.0728 | 0.2890 | 19.38 | Segmental |
| ZmTCP13-ZmTCP01 | 0.2479 | 0.1556 | 0.6277 | 19.07 | Segmental |
| ZmTCP26-ZmTCP21 | 0.2341 | 0.0681 | 0.2909 | 18.01 | Segmental |
| ZmTCP04-ZmTCP11 | 0.0316 | 0.0177 | 0.5601 | 2.43 | Segmental |
Fig. 5.
Sliding window plots of duplicated ZmTCP genes. X-axes indicate nucleotide positions
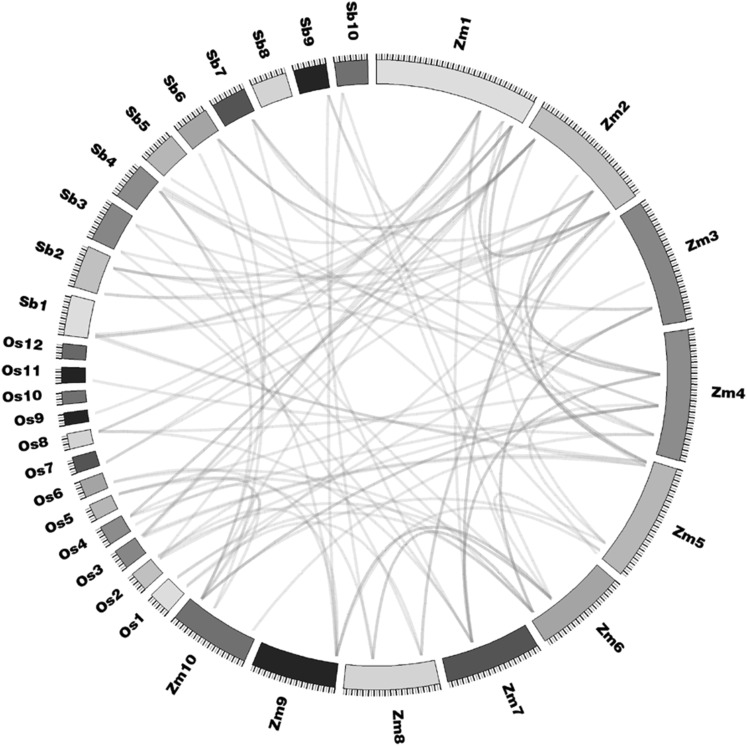
Interspecies microsynteny analysis
Previous study demonstrate that genes originated from a common ancestor may have chromosome collinearity among species, and whole-genome duplications is an important driving force for gene family expansion (Abrouk et al. 2010). To better understand collinearity of ZmTCP members in monocotyledons maize, rice and sorghum, we identified 92 orthologs in maize, rice and sorghum by OrthoMCL, which was used to generate a microsynteny map (Fig. 6, Table S1). Based on these comparisons, 32 collinear gene pairs between maize and rice, 34 collinear gene pairs between maize and sorghum, 22 of the maize gene are present in collinear gene pairs, ZmTCP06, -07, -11, -12, -14, -20 and -28 have paralogs within the maize, which might have resulted from ancient tetraploidy processes during the course of evolution. ZmTCP01 have two collinear gene pair in rice and sorghum respectively (ZmTCP01/LOC_Os08g33530.1, ZmTCP01/LOC_Os09g24480.1, ZmTCP01/Sb07g021140.1, ZmTCP01/Sb02g024450.1), indicating gene loss in maize. Interestingly, some collinear gene pairs identified between maize and sorghum were not identified between maize and rice, such as ZmTCP19/Sb04g038140.1, perhaps due to the progenitor of sorghum and the two progenitors of maize diverged at nearly the same time (approximately 11.9 Mya). The duplication events were evident on every chromosome except for chr8 in sorghum, as well as chr10 and chr12 in rice.
Fig. 6.
Microsynteny map of TCP genes in maize, rice and sorghum. The blue lines show the duplicated genes in the three species. Different colors represent maize, rice and sorghum chromosomes. Sb, sorghum chromosome; Zm, maize chromosome; Os, rice chromosome (colour figure online)
Analysis of the expression of ZmTCP genes in different developmental stages
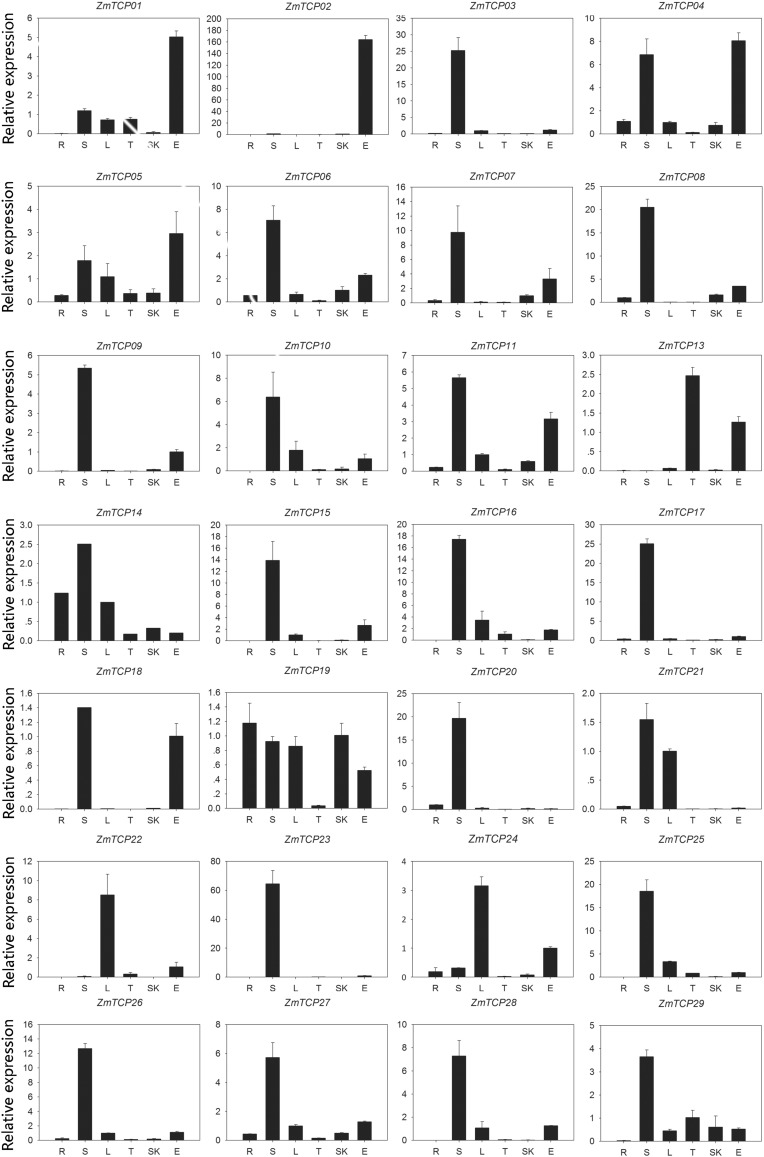
We performed quantitative reverse-transcription PCR (qRT-PCR) to investigate the expression profiles of TCP genes in different organs in maize. The results show that the 29 ZmTCP genes exhibited differential expression patterns in the six tissues examined (Fig. 7). Twenty of the genes (ZmTCP03, -06, -07, -08, -09, -10, -14, -15, -16, -17, -18, -19, -20, -21, -23, -25, -26, -27, -28 and -29) showed high expression in stems. ZmTCP01, -02, -04 and -05 were highly expressed in ears. ZmTCP22 and -24 showed high expression in leaves, with relatively low expression in other tissues. ZmTCP13 was mainly expressed in tassels. ZmTCP14 and ZmTCP19 mainly expressed in roots, stems and leaves. At the same time, the expression of ZmTCP12 was not detected in any of the organs, perhaps this gene is specifically expressed in other organs.
Fig. 7.
Relative expression of ZmTCP genes analyzed by qRT-PCR in different organs. The qRT-PCR data are expressed relative to that of maize Actin1 (set to 1). X-axis labels represent root, stem, leaf, tassel, silk and ear tissue. Y-axis labels indicate relative expression levels
Members possessing similar sequences are clustered in the same subfamilies (Ma et al. 2014), these genes may have similar expression patterns or functions. For example, the genes in subgroup A, subgroup B and subgroup H were all highly expressed in stems, while ZmTCP19 was expressed in all six tissues, suggesting that it may play roles at multiple developmental stages. Some genes clustered in the same phylogenetic subgroup have similar expression profile. For example, ZmTCP04 and ZmTCP11 were highly expressed in stems and ears, and ZmTCP02 and ZmTCP18 were highly expressed in ears.
Discussion
TCP genes are a class of plant-specific transcription factor, which have been identified in diverse plants. TCP proteins can influence cell proliferation and differentiation to regulate corresponding growth and developmental progress. Although the plant-specific TCP genes have been analyzed in Arabidopsis, rice and Gossypium raimondii, this gene family has not previously been examined or annotated in maize. In this study, 29 non-redundant TCP genes were identified in maize, whereas, Arabidopsis and rice contain only 24 and 25 TCP genes, respectively. Segmental duplication plays important role in the evolutionary history of maize TCP genes. The higher number of TCP genes in maize versus Arabidopsis (24) and rice (25) suggests that the expansion of the maize TCP family was caused by plant evolutionary diversification and duplication events such as segmental duplication and tandem duplication. However, only segmental duplication appears to have played an important role in ZmTCP evolution. The plant genome and biological evolutionary processes generate variations in original genetic material, genetic drift and choices, leading to the evolution of new gene functions and gene networks. Duplication is the main driver of gene expansion. In this study, we found that segmental duplication was the primary driver of TCP gene expansion, which is consistent with findings from Arabidopsis and rice TCP families (Yao et al. 2007). Almost every chromosome in maize contains duplicated genes. Therefore, we hypothesized that the maize TCP gene family is a slowly evolving gene family during evolution. The segmental duplications might have played a key role in the expansion of the maize TCP gene family, which was also observed in some other maize gene families, such as CCCH zinc fingers (Peng et al. 2012). The results of Ka/Ks analysis suggest that the duplication of ZmTCP genes occurred in 2.43–19.38 Mya. The prevalence of orthologous pairs and segmental duplication in all three species suggests that the TCP gene family is a conserved and slowly evolving family in plants, which may also explain a little difference numbers of TCP proteins in these three species. Microsynteny analysis also supports the notion that duplication led to gene expansion. 22 maize TCP genes have orthologs in rice and sorghum. Therefore, we hypothesized that the maize TCP gene is a slowly evolving gene family during evolution.
Comparative genomics showed that the euchromatic regions are highly conserved between rice and maize (Wei et al. 2007). The current results also show that maize ZmTCP and rice TCP genes are highly conserved. Many TCP genes in maize have two or more counterparts in rice with high protein sequence similarity (orthologs), implying that maize and rice may have a common ancestor. We also analyzed the gene structures and exon/intron structures of ZmTCP genes. There are eight pairs of paralogous maize TCPs, all of which have conserved structures, especially genes in the same subgroup. These findings suggest that genes in the same subgroup may have similar functions. However, some differences still exists. For example, structural analysis showed that ZmTCP04 and ZmTCP11 have different numbers of introns; ZmTCP04 has one intron, while ZmTCP11 has two introns, which may be due to intron loss or gain during the process of evolution. We further studied the conserved motifs in maize TCP proteins. Class I and class II proteins play different roles, as they compete for various targets or partners. The seven subfamily A members have similar motifs, indicating that the gene structure of the TCP family is highly conserved. Class II members have diverse motifs, which may be due to their functional diversification. Class I family genes are mainly expressed in plant meristems, and they are specifically expressed. Class II genes are extensively expressed in different organs, in addition to meristematic cells, these genes are also expressed in flowers, leaves, roots, seeds and some other vegetative organs (Reeves and Olmstead 2003). We speculate that TCP family genes may function throughout plant development. Furthermore, several motifs were exclusively observed in certain groups, such as motif 14 (I), motif 15 (E), motif 17 (F). These motifs might contribute to the specific functions of the TCP proteins, such as maintaining genome stability, DNA methylation, replication and transcriptional regulation.
The results show that the 29 ZmTCP genes exhibited differential expression patterns in the six surveyed tissues (Fig. 6). Twenty-two of the genes (ZmTCP01, -03, -04, -05, -06, -07, -08, -09, -10, -11, -15, -16, -17, -18, -20, -22, -24, -25, -26, -27, -28 and -29) were highly expressed in stems and ears, implying that these genes may play important roles in the growth of axillary organs and the formation of ears. These results are consistent with the results of a previous study demonstrating that in maize, TB1 represses the growth of axillary organs and promotes the formation of female inflorescences (Doebley et al. 1997); perhaps the ZmTCP genes have a similar function. ZmTCP21, -22 and -24 were highly expressed in leaves and at relatively low levels in other tissues, indicating that they may play important roles in leaf development. ZmTCP13 mainly expressed in tassels, suggesting that it may be related to pollen formation. ZmTCP14 and ZmTCP19 were mainly expressed in roots, stems and leaves, implying that these genes function in these organs. At the same time, ZmTCP12 expression was not detected in the six organs examined, but it may be expressed in other organs. Divergent expression patterns in paralogous pairs may lead to evolutionary diversification of gene functionalization. Most duplicated ZmTCP genes exhibited distinct expression patterns. These results suggest that chromosomal duplication events led to the divergence between duplicated genes and may have contributed to the diversity of gene function over the course of evolution.
Conclusions
In summary, the TCP family has important functions in various developmental processes in plants. This study will prove to be useful in further study of the functions of ZmTCP genes, particularly for members with potentially important functions in plant growth. However, further experiments should be conducted to directly examine the functions of ZmTCP genes.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Acknowledgements
We thank members of the National Engineering Laboratory of Crop Stress Resistance/Key Laboratory of Crop Biology of Anhui Province for their assistance in this study.
Funding
This work was supported by the Key Project of Chinese National Programs for Fundamental Research and Development (2014CB138200), and the National Natural Science Foundation of China (31201217, 31301324).
Author contributions
WC, PJ and XL conceived and designed this research; WC and GH performed the experiment; WC, GH, HJ analyzed the data; WC, PJ, XL and HJ contributed reagents/materials/analysis tools; WC, PJ wrote the manuscript. All authors read and approved the manuscript.
Compliance with ethical standards
Conflict of interest
The authors declare that they have no conflict of interest.
Footnotes
Electronic supplementary material
The online version of this article (doi:10.1007/s12298-017-0476-1) contains supplementary material, which is available to authorized users.
References
- Abrouk M, et al. Palaeogenomics of plants: synteny-based modelling of extinct ancestors. Trends Plant Sci. 2010;15:479–487. doi: 10.1016/j.tplants.2010.06.001. [DOI] [PubMed] [Google Scholar]
- Aguilar-Martínez JA, Sinha N. Analysis of the role of Arabidopsis class I TCP genes AtTCP7, AtTCP8, AtTCP22, and AtTCP23 in leaf development. Plant Evol Dev. 2013;4:406. doi: 10.3389/fpls.2013.00406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aguilar-Martínez JA, Poza-Carrión C, Cubas P. Arabidopsis BRANCHED1 acts as an integrator of branching signals within axillary buds. Plant Cell. 2007;19:458–472. doi: 10.1105/tpc.106.048934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carpenter R, Copsey L, Vincent C, Clark J, Coen E. Control of organ asymmetry in flowers of antirrhinum. Cell. 1999;99:367–376. doi: 10.1016/S0092-8674(00)81523-8. [DOI] [PubMed] [Google Scholar]
- Cubas P. Role of TCP genes in the evolution of morphological characters in angiosperms. Syst Assoc Spec. 2002;65:247–266. [Google Scholar]
- Doebley J, Stec A, Hubbard L. The evolution of apical dominance in maize. Nature. 1997;386:485. doi: 10.1038/386485a0. [DOI] [PubMed] [Google Scholar]
- Gaut BS, Morton BR, McCaig BC, Clegg MT. Substitution rate comparisons between grasses and palms: synonymous rate differences at the nuclear gene Adh parallel rate differences at the plastid gene rbcL. Proc Natl Acad Sci. 1996;93:10274–10279. doi: 10.1073/pnas.93.19.10274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giraud E, et al. TCP transcription factors link the regulation of genes encoding mitochondrial proteins with the circadian clock in Arabidopsis thaliana. Plant Cell. 2010;22:3921–3934. doi: 10.1105/tpc.110.074518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hubbard L. Expression patterns and mutant phenotype of teosinte branched1 correlate with growth suppression in maize and teosinte. Genetics. 2003;162:1927–1935. doi: 10.1093/genetics/162.4.1927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hubbard L, McSteen P, Doebley J, Hake S. Expression patterns and mutant phenotype of teosinte branched1 correlate with growth suppression in maize and teosinte. Genetics. 2002;162:1927–1935. doi: 10.1093/genetics/162.4.1927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kieffer M, Master V, Waites R, Davies B. TCP14 and TCP15 affect internode length and leaf shape in Arabidopsis. Plant J. 2011;68:147–158. doi: 10.1111/j.1365-313X.2011.04674.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kosugi S, Ohashi Y. PCF1 and PCF2 specifically bind to cis elements in the rice proliferating cell nuclear antigen gene. Plant Cell. 1997;9:1607–1619. doi: 10.1105/tpc.9.9.1607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krzywinski M, et al. Circos: an information aesthetic for comparative genomics. Genome Res. 2009;19:1639–1645. doi: 10.1101/gr.092759.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leister D. Tandem and segmental gene duplication and recombination in the evolution of plant disease resistance gene. Trends Genet. 2004;20:116. doi: 10.1016/j.tig.2004.01.007. [DOI] [PubMed] [Google Scholar]
- Lewis JM, et al. Overexpression of the maize teosinte branched1 gene in wheat suppresses tiller development. Plant Cell Rep. 2008;27:1217–1225. doi: 10.1007/s00299-008-0543-8. [DOI] [PubMed] [Google Scholar]
- Li L, Jr, Stoeckert CJ, Roos DS. OrthoMCL: identification of ortholog groups for eukaryotic genomes. Genome Res. 2003;13:2178–2189. doi: 10.1101/gr.1224503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Z, et al. Molecular evolution of the HD-ZIP I gene family in legume genomes. Gene. 2013;533:218–228. doi: 10.1016/j.gene.2013.09.084. [DOI] [PubMed] [Google Scholar]
- Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2−∆∆CT method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- Ma X, Ma J, Fan D, Li C, Jiang Y, Luo K (2016) Genome-wide Identification of TCP Family Transcription Factors from Populus euphratica and Their Involvement in Leaf Shape Regulation. Scientific Reports 6. doi:10.1038/srep32795 [DOI] [PMC free article] [PubMed]
- Ma J, Wang Q, Sun R, Xie F, Jones DC, Zhang B. Genome-wide identification and expression analysis of TCP transcription factors in Gossypium raimondii. Sci Rep. 2014;4:6645. doi: 10.1038/srep06645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martín-Trillo M, Cubas P. TCP genes: a family snapshot ten years later. Trends Plant Sci. 2010;15:31–39. doi: 10.1016/j.tplants.2009.11.003. [DOI] [PubMed] [Google Scholar]
- Ori N, et al. Regulation of lanceolate by miR319 is required for compound-leaf development in tomato. Nat Genet. 2007;39:787–791. doi: 10.1038/ng2036. [DOI] [PubMed] [Google Scholar]
- Pagnussat GC, et al. Genetic and molecular identification of genes required for female gametophyte development and function in Arabidopsis. Development. 2005;132:603–614. doi: 10.1242/dev.01595. [DOI] [PubMed] [Google Scholar]
- Peng X, et al. CCCH-type zinc finger family in maize: genome-wide identification, classification and expression profiling under abscisic acid and drought treatments. PLoS ONE. 2012;7:e40120. doi: 10.1371/journal.pone.0040120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reeves PA, Olmstead RG. Evolution of the TCP gene family in Asteridae: cladistic and network approaches to understanding regulatory gene family diversification and its impact on morphological evolution. Mol Biol Evol. 2003;20:1997–2009. doi: 10.1093/molbev/msg211. [DOI] [PubMed] [Google Scholar]
- Rueda-Romero P, Barrero-Sicilia C, Gómez-Cadenas A, Carbonero P, Oñate-Sánchez L. Arabidopsis thaliana DOF6 negatively affects germination in non-after-ripened seeds and interacts with TCP14. J Exp Bot. 2012;63:1937–1949. doi: 10.1093/jxb/err388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheng L, et al. A genome-wide analysis of the AAAP gene family in maize. J Proteom Bioinform. 2014;7:23–33. [Google Scholar]
- Takeda T, et al. The OsTB1 gene negatively regulates lateral branching in rice. Plant J Cell Mol Biol. 2003;33:513–520. doi: 10.1046/j.1365-313X.2003.01648.x. [DOI] [PubMed] [Google Scholar]
- Thompson JD, Higgins DG, Gibson TJ. CLUSTAL W: improving the sensitiv ity of progressive multiple sequence alignment through sequence weighting, position specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomotsugu K, Masahiko F, Masao T, Masaru OT. TCP transcription factors control the morphology of shoot lateral organs via negative regulation of the expression of boundary-specific genes in Arabidopsis. Plant Cell. 2007;19:473–484. doi: 10.1105/tpc.106.044792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei F, et al. Physical and genetic structure of the maize genome reflects its complex evolutionary history. PLoS Genet. 2007;3:e123. doi: 10.1371/journal.pgen.0030123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xue C, Zhu C, Hualin Z, Yang Z, Beijiu C, Yan X. Genome-wide analysis of soybean HD-zip gene family and expression profiling under salinity and drought treatments. PLoS ONE. 2014;9:e87156. doi: 10.1371/journal.pone.0087156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Z, et al. Systematic analysis of sequences and expression patterns of drought-responsive members of the HD-zip gene family in maize. PLoS ONE. 2011;6:e28488. doi: 10.1371/journal.pone.0028488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yao X, Ma H, Wang J, Zhang D. Genome-wide comparative analysis and expression pattern of TCP gene families in Arabidopsis thaliana and Oryza sativa. J Integr Plant Biol. 2007;49:885–897. doi: 10.1111/j.1744-7909.2007.00509.x. [DOI] [Google Scholar]
- Yuancheng P, et al. The ubiquitin receptors DA1, DAR1, and DAR2 redundantly regulate endoreduplication by modulating the stability of TCP14/15 in Arabidopsis. Plant Cell. 2015;27:649–662. doi: 10.1105/tpc.114.132274. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.