Abstract
Rice being a staple cereal is extremely susceptible towards abiotic stresses. Drought and salinity are two vital factors limiting rice cultivation in Eastern Indo-Gangetic Plains (EIGP). Present study has intended to evaluate the consequences of salinity stress on selected drought tolerant rice genotypes at the most susceptible seedling stage with an aim to identify the potential multi-stress (drought and salt) tolerant rice genotype of this region. Genotypic variation was obvious in all traits related to drought and salt susceptibility. IR84895-B-127-CRA-5-1-1, one of the rice genotypes studied, exhibited exceptional drought and salinity tolerance. IR83373-B-B-25-3-B-B-25-3 also displayed enhanced drought and salt tolerance following IR84895-B-127-CRA-5-1-1. Variations were perceptible in different factors involving photosynthetic performance, proline content, lipid peroxidation, K+/Na+ ratio. Accumulation of reactive oxygen species (ROS) disintegrated cellular and sub-cellular membrane leading to decreased photosynthetic activities. Therefore, accumulation and detoxification of reactive oxygen species was also considered as a major determinant of salt tolerance. IR84895-B-127-CRA-5-1-1 showed improved ROS detoxification mediated by antioxidant enzymes. IR84895-B-127-CRA-5-1-1 seedlings also displayed significant recovery after removal of salt stress. The results established a direct association of ROS scavenging with improved physiological activities and salt tolerance. The study also recommended IR84895-B-127-CRA-5-1-1 for improved crop performance in both drought and saline environments of EIGP. These contrasting rice genotypes may assist in understanding the multiple stress associated factors in concurrent drought and salt tolerant rice genotypes.
Keywords: Drought, Genotype, Rice, ROS, Salinity
Introduction
Global climatic variability and depleting soil conditions are emerging as major threats to rice cultivation. Drought and salinity are the most detrimental abiotic stresses for sustainable rice production in the rainfed ecosystems of the Eastern Indo-Gangetic Plains (EIGP) (Sekar and Pal 2012). In India, almost 13.6 and 6.3 M ha rice growing area is affected by drought and salinity respectively, resulting in severe yield loss and food insecurity (Huke and Huke 1997; Singh et al. 2010). Therefore, efforts to identify rice genotypes for the fragile ecosystems are intensifying to attain food security (Singh et al. 2016). Exploration for potential drought and salinity resilient rice genotypes of EIGP may lead to sustainable rice production in the rainfed environments of India.
Drought and salinity adversely affect rice in terms of growth and productivity (Hazmana et al. 2016). Both of the abiotic stresses decrease water uptake ability of plants that reduce plant growth by inhibiting cell division and accelerating cell death (Munns 2002). Drought results in vigorous root system in plants and leads to prominent modification in root structure by means of increased branching and density. On the other hand, drought also causes leaf rolling and senescence in rice (Chaves et al. 2003). Major physiological processes are hampered due to drought and salinity (Kumar et al. 2009, 2014). Photosynthesis, the foremost metabolic process regulating crop production is severely affected by drought and salinity due to reduced stomatal conductance (Yusuf et al. 2010). Reduction in stomatal conductance also lowers transpiration rate by stomatal closure and increase plant survival ability by restricting water reserves in the root zone (Zhang and Kirkham 1995). Consequently, exchange of water vapour and CO2 through stomata also becomes limited due to stomatal closure resulting in increased leaf-turgor (Chaves et al. 2009; Farooq et al. 2009). Osmotic stress leads to degradation in chlorophyll pigments (Chutia and Borah 2012). Salinity-induced movement of salt into root is associated with transpiration flux, obligatory in maintenance of plant water status. Unregulated transpiration causes ion toxicity in plant aerial parts. High ionic concentration disturbs ion homeostasis, cell membrane function and interferes with internal solute balance. Excessive Na+ accumulated during salt stress, competitively inhibits K+ uptake and disrupts K+/Na+ ratio of cells. Therefore, acclimation of plants to saline atmosphere conjointly depends on their capability to prevent Na+ toxicity and to uphold K+ concentration which is closely related to overcome salt-induced injury (Ma et al. 2014). Two crucial factors by which plants combat with salinity stress involve Na+ efflux and compartmentalization of Na+ in vacuole (Deinlein et al. 2014).
Abiotic stress tolerance of plants is a complex phenomenon and consists of interactions between stress factors and various molecular, biochemical and physiological activities affecting plant growth and development. Accumulation of osmo-protectant is a primary and critical response under osmotic stress and has been positively correlated with osmotic stress tolerance of several crops (Bray et al. 2000). Proline has been reported as the most widely distributed multifunctional osmolyte facilitating osmotic regulation and also protecting the subcellular components in stressed plants (Zhao et al. 2014). Exposure of plants to environmental stresses such as, drought and salt increase the production of reactive oxygen species (ROS) including singlet oxygen (1O2), superoxide (O·−2), hydrogen peroxide (H2O2) and hydroxyl radical (OH·) through enhanced leakage of electrons to molecular oxygen, which triggers aging in plants (Jiang et al. 2016). Excessive ROS production leads to oxidative damages in plants through peroxidation of polyunsaturated fatty acids (DaCosta and Huang 2007). As a consequence, photosynthetic pigments, proteins and nucleic acids also get oxidised (Yordanov et al. 2000). Tolerance towards drought and salinity involves ROS metabolism through a multifaceted antioxidant assembly that counter the deleterious effects of ROS (Miller et al. 2010; Jiang et al. 2016). Enzymatic antioxidant system consisting of superoxide dismutase (SOD), ascorbate peroxidase (APX), catalase (CAT), glutathione peroxidase (GPX) and peroxiredoxin (PrxR) play important role in ROS detoxification (Miller et al. 2010). Peroxidases, present in all subcellular compartments are the key enzymes that catalyse the conversion of H2O2 to water and O2 (Gratao et al. 2005). Peroxidase uses guaiacol or pyrogallol as electron donor. Several classes of peroxidases which act as important antioxidants can decompose H2O2 (Frary et al. 2010). Decline in cytosolic peroxidase activity followed by an accumulation of H2O2 has suggested its role as an indicator for promoting senescence in plants (Ye et al. 2000). Detoxification of ROS with different antioxidant enzymes is another potentially important tolerance trait upon salinity stress (Zhu et al. 2005). Quantity of antioxidant enzymes marks the intensity of stress and stress-tolerant genotypes show significantly high levels of antioxidant enzyme activities under stress conditions (Alscher et al. 2002).
Tolerance in rice varies with genotypes and developmental stages, with early vegetative stage being relatively more susceptible (Krishnamurthy et al. 2011; Platten et al. 2013). Recent studies have also established the crosstalk of drought and salinity stress regulatory genes in rice (Sharma et al. 2013; Hong et al. 2016). The present study aimed to identify the drought tolerant rice genotypes of EIGP. Furthermore, the consequences of salinity stress on the selected drought tolerant Eastern Indo-Gangetic rice genotypes were also analysed. The study included comparative analyses of morpho-physiological and biochemical alterations with special emphasis on ROS generation and detoxification through antioxidant enzyme activity.
Materials and methods
Plant material and growth conditions
Screening for drought tolerance was performed with 10 rice genotypes (IR64, IR83373-B-B-25-3, IR83387-B-B-27-4, IR83929-B-B-291-2-1-1-2, IR84895-B-127-CRA-5-1-1, IR84899-B-179-16-1-1-1, IR88964-24-2-1-4, Rajendra Bhagwati, Rajendra Sweta, Sahbhagi Dhan) recommended for north eastern plains zone (NEPZ) as suggested by the breeder. Rice genotypes were obtained from Division of Crop Research, ICAR Research Complex for Eastern Region (ICAR-RCER), Patna. Details of 10 genotypes and their pedigree information have been presented in Table 1. Experiments were performed in two phases, with a first phase of screening for drought tolerance. On the basis of screening, 2 promising drought tolerant rice genotypes (IR83373-B-B-25-3 and IR84895-B-127-CRA-5-1-1) were selected for further studies. These two genotypes were exposed to salinity stress and the consequences of salinity were elaborately studied. An established salt sensitive genotype IR64 was considered as negative control.
Table 1.
Detailed pedigree information of rice genotypes (n = 10) considered for screening under PEG-6000 induced drought stress
| Name of the considered rice genotypes | Parents | Variety/advanced breeding lines |
|---|---|---|
| IR64 | IR5657-33-2-1/IR2061-465-1-5-5 | Checked variety |
| IR83373-B-B-25-3 | IR71700-247-1-1-2/IR72875-94-3-3-2 | Advanced breeding line |
| IR83387-B-B-27-4 | IR72022-46-2-3-3-2/Sambha Mahsuri | Advanced breeding line |
| IR83929-B-B-291-2-1-1-2 | IR78878-53-2-2-2/CT6510-24-1-2 | Advanced breeding line |
| IR84895-B-127-CRA-5-1-1 | IR77080-B-34-3/IRRI123 | Advanced breeding line |
| IR84899-B-179-16-1-1-1 | IR78877-208-B-1-1/IRRI132 | Advanced breeding line |
| IR88964-24-2-1-4 | IRRI123/IR05N372 | Advanced breeding line |
| Rajendra Bhagwati | RAU1397-18-3-7-9-4-2 | High yielding variety of Bihar (EIGP) |
| Rajendra Sweta | Sita/Pusa Basmati-1//Katarni | High yielding variety of Bihar (EIGP) |
| Sahbhagi Dhan | IR55419-04*2 Way Rarem IR55419-04 (IR12979-24-1 (Brown)/UPRLRi5) | Checked variety |
Seeds of selected rice genotypes were surface sterilized by washing with 0.5% sodium hypochlorite solution for 5 min followed by 3–4 times extensive washing with de-ionized water and were allowed to germinate in dark conditions for 48 h in plant growth chamber. After germination, the seedlings were further grown in hydroponic system supplemented with ½ Yoshida medium (Yoshida et al. 1972) under control conditions (28 ± 2 °C, 12 h light and dark cycle, 60–80% relative humidity) (Kumar et al. 2012).
Drought and salt stress treatments
Drought stress in experimental hydroponics’ media was induced by polyethylene glycol (PEG-6000) solution (Zhang et al. 2017). Salt stress was exerted by 200 mM sodium chloride (NaCl) in the hydroponics’ media (Kumar et al. 2009). 12 days old seedlings were divided into two groups, one group was treated with 20% PEG-6000/200 mM NaCl for 24, 48 and 72 h (stressed) and other group was kept untreated (control). After removal of stress, the seedlings were transferred to fresh media for 7 days recovery.
Morphological studies
Root and shoot length of the stressed (S) and non-stressed (C) seedlings was measured with thread. Relative change of root and shoot length for each genotype was calculated using the following formula (Kumar et al. 2009).
Measurement of gas exchange parameters
Net photosynthesis (PN), stomatal conductance (gs) and transpiration rate (E) were measured in the fully expanded leaf of all rice genotypes under normal and salt stressed conditions using an IRGA (Li-6400XT, LICOR Inc, Lincoln, NE, USA) following the protocol of Li et al. (2011). The instrument was stabilized according to the manufacturer’s guidelines. Steady state levels of reference CO2 was observed before taking the measurements. Photosynthetic parameters were measured at photosynthetically active radiation (PAR) of 1000 μmol m−2 s−1 (photon), CO2 concentration of 370 μmol mol−1 at 30 °C and expressed in per unit leaf area.
Determination of relative water content
Relative water content (RWC) was determined according to the protocol of Weatherley (1950). Fully expanded leaves were collected and weighed quickly omitting the mid-rib to record the fresh weight. The samples were hydrated to full turgidity by floating on de-ionized water for 4 h. After incubation, the surface moisture of samples was removed and immediately weighed to obtain fully turgid weight. The dry weight of the samples was taken after proper drying in a hot air oven at 80 °C for 24 h. RWC was calculated for each sample following the formula.
Quantification of total chlorophyll content
Chlorophyll was estimated according to Arnon (1949). 0.1 g leaf samples of each genotype were frozen in liquid N2 and ground. Total chlorophyll was extracted in 80% acetone at 4 °C and then absorbance in extracts was recorded at 645 and 663 nm on a UV–Vis spectrophotometer (Model BIO 200, Thermo). Total chlorophyll concentration was calculated from the following equation, where, V = volume of solvent and W = weight of leaf (in gram).
Estimation of proline content
Proline content was estimated using the standard protocol (Bates et al. 1973). Proline was extracted from 0.1 g leaf tissue of both stressed and non-stressed seedlings. The absorbance was measured at 520 nm and proline content was calculated from the standard curve.
Measurement of MDA content
MDA content was determined by thiobarbituric acid (TBA) test according to Heath and Packer (1968). After extracting MDA with 0.5% TBA in 20% trichloroacetic acid (TCA), absorbance was measured at 532 nm. Values of non-specific absorption recorded at 600 nm were subtracted from the values recorded at 532 nm. MDA content was calculated according to its extinction coefficient ε = 155 mM−1 cm−1.
Detection of H2O2 accumulation
Hydrogen peroxide was detected by the DAB uptake method (Fryer et al. 2002). Whole seedlings (control and salt stressed) of different rice genotypes under study were infiltrated with 0.5 mg ml−1 freshly prepared DAB staining solution; chlorophyll was removed by incubating seedlings in 90% ethanol. H2O2 was visualized as brown colour due to DAB polymerization.
Assay of peroxidase enzyme (POX) activity
Lyophilized plant tissue was homogenized with ice-cold phosphate buffer (pH 7.0). Total POX activity was determined at 436 nm by its ability to convert guaiacol to tetraguaiacol (Polle et al. 1994). Increase in absorbance was recorded by addition of H2O2 at 436 nm for 2 min.
Activity staining for peroxidase (POX) isozymes
Plant extracts containing equal amounts of protein were subjected to discontinuous polyacrylamide gel electrophoresis (PAGE) under non-denaturing and non-reducing conditions (Laemmli 1970). Peroxidase activity was detected using the method of Graham et al. (1964). Gels were incubated in darkness, till the POX activity-containing band visualized carefully. Band intensity was measured using Adobe Photoshop version 7.
Ionomic analysis
Endogenous Na+ and K+ content was determined according to Kumar et al. (2009), by digesting 0.1 g of whole seedling tissue of four rice genotypes in 0.1% HNO3. Ions were extracted in distilled water by boiling it twice for 30 min each. The filtrate was used to measure specifications with a flame photometer (Toshiba, Japan).
Statistical analysis
The data were analysed statistically with Statistics 8.1, USA software for analysis of variance (ANOVA) using 2 factorial complete randomized design. Treatments were compared by computing the ‘F’ test. All experiments were performed in triplicate (n = 3). Each value is presented in the form of mean ± standard error and the lowest standard deviations of mean with a reading considering at least three samples per experiment for each genotype and each condition. The standard error of mean (± SE) and differences between treatments was compared pair wise by critical difference (CD) at 5% level of probability.
Results
Screening for potential drought tolerant rice genotypes of Eastern Indo-Gangetic Plains
The present study included a screening of 10 rice genotypes of EIGP under 20% PEG-6000 induced drought stress for 24, 48 and 72 h (Fig. 1). The screening (n = 10) was performed at seedling stage, based on ratio of root/shoot, RWC, total chlorophyll and proline content. Average root/shoot ratio in 10 rice genotypes during 0, 24, 48 and 72 h drought stress ranged within 0.2–0.4, 0.3–0.4, 0.4–0.5 and 0.4–0.6, respectively (Fig. 1a). Drought stress resulted in an overall increase in root/shoot ratio of all rice genotypes. IR84899-B-179-16-1-1-1, Rajendra Sweta and Sahbhagi Dhan showed almost similar root/shoot ratio after 72 h of stress (0.4-0.5). IR83373-B-B-25-3-B-B-25-3 and IR84895-B-127-CRA-5-1-1 exhibited the highest root/shoot ratio after 72 h of stress (0.5 and 0.6, respectively) among the considered genotypes, whereas, IR64 showed the lowest value for root/shoot ratio (0.4 after 72 h stress). Relative water content under control condition was within the range of 85.2–96.4% in the considered genotypes, which decreased with successive intensification of stress and ranged within 55.5–79.1, 20.8–67.2 and 15.4–51.4%, respectively after 24, 48 and 72 h stress (Fig. 1b). IR88964-24-2-1-4, Rajendra Bhagwati and Rajendra Sweta exhibited higher percentage of loss in RWC (71.3, 75.1 and 80.7% decrease after 72 h of stress). The minimum decrease in RWC among the 10 genotypes was observed in IR83373-B-B-25-3-B-B-25-3 and IR84895-B-127-CRA-5-1-1 after 72 h of drought treatment (58.3 and 46.6% respective decrease), whereas, IR64 showed the maximum decrease (83.6% relative decrease after 72 h stress). Total chlorophyll content in all rice genotypes ranged between 6.7 and 8.5 μg g−1 FW during non-stressed condition (Fig. 1c). 20% PEG treatment caused massive reduction of total chlorophyll content in all rice genotypes, which ranged within 5.1–7.0, 2.7–5.6 and 1.0–4.2 μg g−1 FW, respectively after 24, 48 and 72 h stress. IR83373-B-B-25-3-B-B-25-3 and IR84895-B-127-CRA-5-1-1 exhibited minimum decrease in total chlorophyll content even after 72 h of treatment (58.3 and 46.6% decrease, respectively). Proline content in all rice genotypes was in the range of 0.18–0.28 μg g−1 DW under non-stressed condition, which increased in all genotypes with sequential increase in duration of stress and ranged between 0.3–0.5, 0.4–0.7 and 0.5–0.9 μg g−1 DW, respectively after 24, 48 and 72 h of stress (Fig. 1d). IR83373-B-B-25-3-B-B-25-3 and IR84895-B-127-CRA-5-1-1 exhibited considerably high proline content after 72 h of stress (0.8 and 0.9 μg g−1 DW respectively), among all the genotypes studied.
Fig. 1.
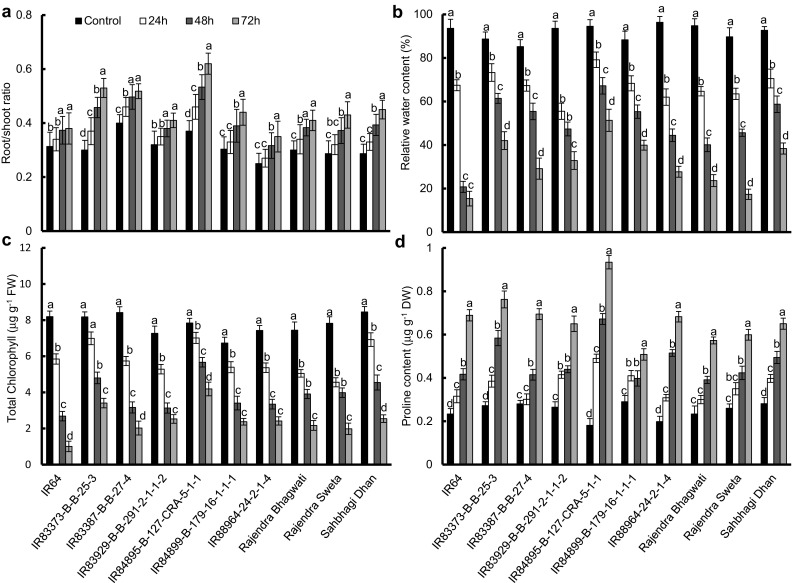
Screening of drought tolerant rice genotypes of Eastern Indo-Gangetic Plains. 12 days old seedlings of 10 Eastern Indo-Gangetic rice genotypes were exposed to 20% PEG-6000 induced drought stress for 24, 48 and 72 h. On the basis of morpho-physiological and biochemical performances enhanced drought tolerant genotypes were selected. a Root/shoot ratio, b Relative water content, c Chlorophyll content, d Proline content. Error bars represent mean standard deviation (n = 3). Different letters above the error bars indicate significantly different values (ANOVA, P ≤ 0.05)
Effect of salinity stress on morpho-physiological activities of promising drought tolerant rice genotypes
All of the morphological parameters were influenced by genotypic variation but exhibited the similar trend under stress condition. The selected rice genotypes (n = 3) showed significant decrease in root and shoot length after 72 h of 200 mM NaCl-induced salt stress (P ≤ 0.01) (Fig. 2a, b). IR84895-B-127-CRA-5-1-1 seedlings displayed minimum decrease in root length after 24 h (17.7%) as compared to IR83373-B-B-25-3 and IR64 (21.5 and 67.4%, respectively) (Fig. 2a). Overall, root length decreased with increasing duration of stress and after 72 h of salt treatment, decline in root length was the lowest in IR84895-B-127-CRA-5-1-1 (33.2%) as compared to IR83373-B-B-25-3 and IR64 (59.7 and 87.8%, respectively). Shoot length also showed the similar pattern of relative decrease as that of root in the considered genotypes (Fig. 2b). IR64, IR83373-B-B-25-3 and IR84895-B-127-CRA-5-1-1 exhibited respective 31.7, 23.9 and 10.5% relative decrease in shoot length after 24 h of stress. With increasing duration of stress, the condition deteriorated, eventually initiating desiccation of the shoot. There were visual variations between the three genotypes after their exposure to NaCl stress, with IR64 being severely damaged. After 72 h stress IR64, IR83373-B-B-25-3 and IR84895-B-127-CRA-5-1-1 showed respective 57.4, 42.6 and 21.7% relative decrease in shoot length. After elimination of stress, a recovery period of 7d was provided to the seedlings. After recovery period, the considered genotypes showed different ability to recover from the stress (Fig. 2d); maximum IR84892 seedlings recovered from the stress, with a reduction in root length equivalent to 24 h of stress (21.2 and 15.0% decrease in root and shoot length, respectively). IR83373-B-B-25-3 showed very poor recovery (51.1 and 32.2% decrease in root and shoot length, respectively) whereas IR64 was almost unable to recover (86.2 and 60.8% decrease in root and shoot length, respectively).
Fig. 2.
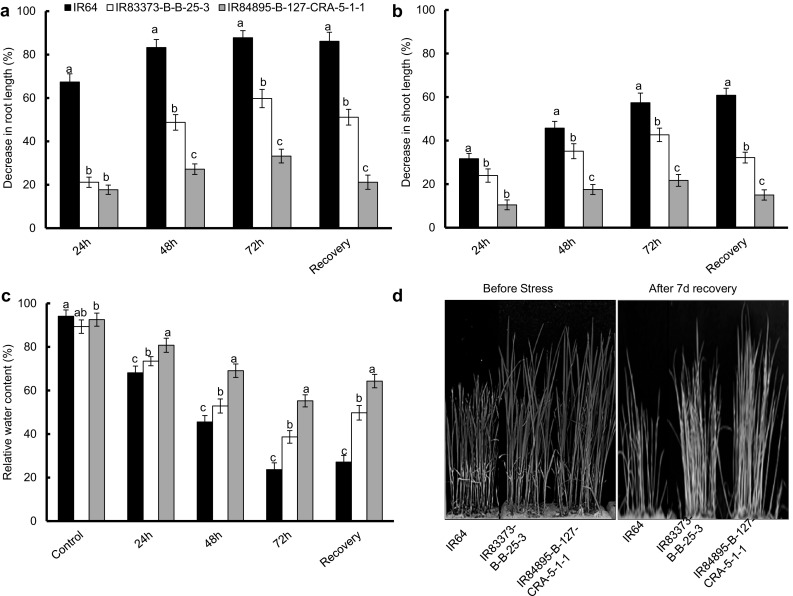
Evaluation of salt tolerance in selected Eastern Indo-Gangetic rice genotypes based on morphological performances. a Relative decrease in root length, b Relative decrease in shoot length, c Relative water content, d Representative images of rice seedlings before exposure to salt stress and during post-stress recovery treatment of 7 days. Error bars represent mean standard deviation (n = 3). Different letters above the error bars indicate significantly different values (ANOVA, P ≤ 0.05)
Relative water content was observed to be severely decreased with duration of stress, but the amount of water loss varied significantly in the selected genotypes (P ≤ 0.01) (Fig. 2c). Among the three genotypes, IR64 showed maximum water loss at very early phase of stress (25.9% loss after 24 h), which increased with extent of stress and the seedlings were unable to revive after the recovery treatment. In IR84895-B-127-CRA-5-1-1 seedlings, cells did not completely loss their water content even after 72 h of stress (55.2%) and started to recover after removal of stress (64.3%). IR83373-B-B-25-3 showed an intermediary response to the stress with respective 15.9, 36.4 and 50.7% loss in water content after respective 24, 48 and 72 h of stress. After recovery, IR83373-B-B-25-3 reached almost the condition similar to 48 h stress (49.7% water loss). Salt-exposed seedlings exhibited decrease in total chlorophyll content as compared to the non-stressed seedlings (Fig. 4a). IR84895-B-127-CRA-5-1-1 showed the minimal decrease in chlorophyll content during initial phase of stress, which intensified with duration of stress (23.1 and 55.1% decrease, respectively). IR84895-B-127-CRA-5-1-1 had the lowest chlorophyll degradation, even after 72 h of stress, contrasting with IR64 (86.9% decrease). IR83373-B-B-25-3 showed huge amount of chlorophyll degradation (77.7% decrease) after 72 h of stress as in IR64. After recovery period, IR64 seedlings were unable to revive degraded chlorophyll content (0.4 μg g−1 FW), whereas IR84895-B-127-CRA-5-1-1 seedlings exhibited an outstanding recovery and its chlorophyll content reached almost that of the 24 h stress condition (6.1 μg g−1 FW).
Fig. 4.
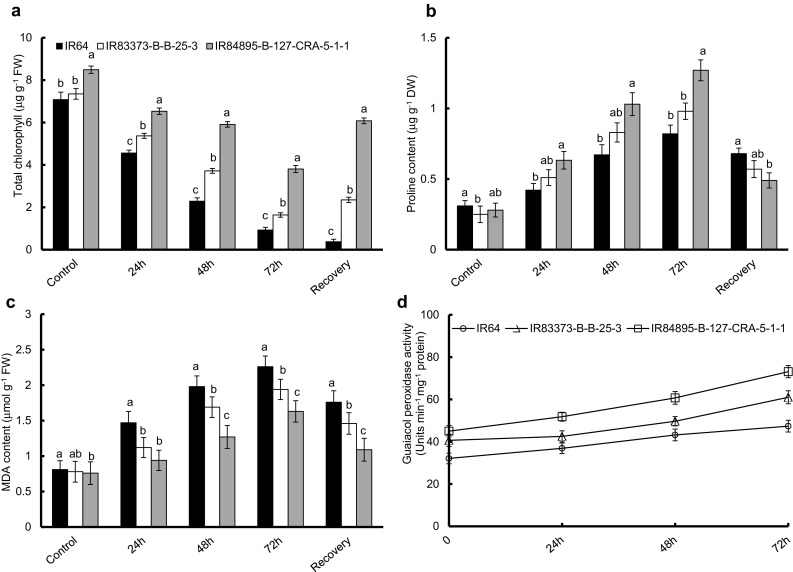
Assessment of salt tolerance in selected rice genotypes based on biochemical studies. a Chlorophyll content, b MDA content, c Proline content, d POX activity. Error bars represent mean standard deviation (n = 3). Different letters above the error bars indicate significantly different values (ANOVA, P ≤ 0.05)
Photosynthetic rate (PN) showed the same trend as that of total chlorophyll content (Fig. 3a). IR84895-B-127-CRA-5-1-1 seedlings exposed to various degree of salt stress showed significant difference in PN as compared to the other two genotypes (P ≤ 0.01). IR84895-B-127-CRA-5-1-1 exhibited the lowest decrease in PN after 48 and 72 h of stress as compared to the other two genotypes (58.2 and 65.2% decrease respectively). IR64 displayed a sharp drop in PN rate when exposed to stress for 24 h (70.8% decrease), which further decreased with successive increase in stress (94.7% decrease) showing lowest PN of 0.43 μmol m−2 s−1 after 72 h of stress. After recovery, PN differed to great extent among the three genotypes. IR83373-B-B-25-3 had very low PN (2.9 μmol m−2 s−1) as compared to IR84895-B-127-CRA-5-1-1, which was successfully revived attaining the PN same as that of the 24 h stress (5.1 μmol m−2 s−1). IR64 was unable to withstand the severity of 72 h NaCl stress and PN tended toward zero (0.2 μmol m−2 s−1) and could not recover after elimination of stress. Salinity stress resulted in decreased stomatal conductance and transpiration rate (Fig. 3b, c). After 24 h of stress, IR64 seedlings showed 84 and 65% respective decrease in stomatal conductance and transpiration rate, contrasting with, IR84895-B-127-CRA-5-1-1 (58 and 12% decrease in stomatal conductance and transpiration rate, respectively). With successive increase in duration of stress, stomatal conductance and transpiration rate in IR64 declined so much that it was unable to recover (0.01 and 0.5 mol H2O m−2 s−1 respectively). IR84895-B-127-CRA-5-1-1 seedlings recovered their stomatal conductance and transpiration rate in such a way that they reached almost that of the 24 h-stressed seedlings (0.25 and 6.19 mol H2O m−2 s−1 respectively). Correlation matrix among different physiological and biochemical parameters showed PN to be significantly correlated with chlorophyll content, MDA, gs, E and RWC (P ≤ 0.001). Lipid peroxidation highly correlated with K+/Na+ ratio, RWC and proline content. Leaf chlorophyll content showed high correlation with K+/Na+ ratio, RWC, MDA and proline content. Moreover, transpiration rate highly correlated with stomatal conductance (P ≤ 0.001) (Table 2).
Fig. 3.
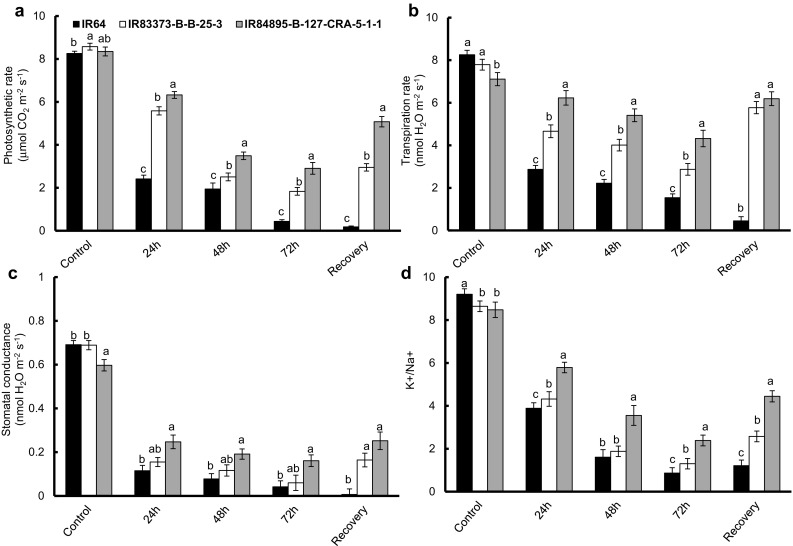
Effect of salinity stress on physiological performances and ionomic profile of selected rice genotypes. a Photosynthetic rate, b Transpiration rate, c Stomatal conductance, d Ratio of K+/Na+. Error bars represent mean standard deviation (n = 3). Different letters above the error bars indicate significantly different values (ANOVA, P ≤ 0.05)
Table 2.
Pearson correlation among different traits of selected rice genotypes under salt stress
| Chl | ||||||
| 0.0004*** | E | |||||
| 0.0537* | 0.0003** | K+/Na+ | ||||
| 0.0005*** | 0.0001*** | 0.0002*** | MDA | |||
| 0.0012*** | 0.0004*** | 0.0001*** | 0.0009*** | PN | ||
| 0.0049** | 0.0064** | 0.0025** | 0.0036** | 0.0057** | Proline | |
| 0.0055** | 0.0005*** | 0.0007*** | 0.0003*** | 0.0001*** | 0.0092*** | RWC |
| 0.0021** | 0.0002*** | 0.0035** | 0.0004*** | 0.0001*** | 0.0011*** | 0.0001*** |
Chl chlorophyll content, E transpiration rate, K+/Na+–K+/Na+ ratio, MDA malondialdehyde content, P N photosynthetic rate, RWC relative water content. *P ≤ 0.05, **P ≤ 0.01, ***P ≤ 0.001
Salinity stress intensified proline content and membrane lipid peroxidation
Proline content among non-stressed genotypes ranged between 0.2 and 0.3 μg g−1 DW (Fig. 4b). Salinity stress for varying period resulted in significant elevation in proline in all of the seedlings (P ≤ 0.01), which was remarkably higher in IR84895-B-127-CRA-5-1-1. After 24 h of stress, proline content amplified up to 2.3-fold in IR84895-B-127-CRA-5-1-1. After 24 h of stress, proline content elevated up to 1.4-fold and twofold in IR64 and IR83373-B-B-25-3 respectively. Proline level increased gradually with duration of stress and maximum proline content was estimated after 72 h of stress. In case of IR84895-B-127-CRA-5-1-1, 72 h of NaCl stress led to 4.5-fold increase in proline level. Recovery treatment facilitated the reduction in proline content by 0.4-fold than that of 72 h stress in IR84895-B-127-CRA-5-1-1 (1.27 μg g−1 DW), which was depicted by a better recovery as compared to the other two genotypes. IR64 and IR83373-B-B-25-3 seedlings did not show such massive decline in proline level after recovery period (0.68 and 0.57 μg g−1 DW). MDA content was estimated to measure lipid peroxidation in all seedlings after NaCl treatment (Fig. 4c). MDA content in unstressed seedlings was within a range of 0.7–0.8 μmol g−1 FW for the three genotypes. IR64 seedlings showed 1.8, 2.4 and 2.8-fold respective increase in MDA content after perceiving 24, 48 and 72 h of salinity stress which did not recover even after removal of stress (1.8 μmol g−1 FW). IR83373-BB-25-3 also persisted a high MDA content throughout the stress (1.1, 1.7 and 1.9 μmol g−1 FW after 24, 48 and 72 h of stress respectively) and were unable to recover (1.5 μmol g−1 FW). However, IR84895-B-127-CRA-5-1-1 seedlings retained considerably low MDA content after 24 and 48 h of stress (0.9 and 1.3 μmol g−1 FW respectively), which increased after 72 h of salt stress (1.6 μmol g−1 FW). After recovery period, MDA content in IR84895-B-127-CRA-5-1-1 declined to same as that of the 24 h stressed condition (1.1 μmol g−1 FW).
Elevated Na+ accumulation disturbed ionic homeostasis under salinity stress
Salinity stress resulted in significant accumulation of Na+ among three genotypes with increased duration of stress (data not shown). IR64 seedlings maintained higher level Na+ throughout the stress. Na+ content in IR 64 seedlings subjected to salt stress for 24 and 48 h were much higher (6.2 and 9.1 μg g−1 FW respectively) than that of the non-stressed seedlings (3.8 μg g−1 FW). IR84895-B-127-CRA-5-1-1 showed the lowest increase in Na+ content after 48 h of stress (4.9 and 6.9 μg g−1 FW after 24 and 48 h) than that observed in IR83373-B-B-25-3 (5.1 and 8.6 μg g−1 FW after 24 and 48 h respectively). After 72 h of stress, all genotypes were saturated with Na+ and lost their competence to check more Na+ entry. Hence a marginal increase in Na+ accumulation was observed. Interestingly, recovered IR84895-B-127-CRA-5-1-1 seedlings had a lower accumulation of Na+ (4.4 μg g−1 FW) contrasting to the other two genotypes. During non-stressed condition, the K+/Na+ ratio was between 8.5 and 9.2 for the three genotypes. With introduction of stress, IR64 displayed a sharp drop in the ratio (57.8% decrease) as compared to that of IR84895-B-127-CRA-5-1-1 (31.7% decrease) after 24 h of stress. IR83373-BB-25-3 showed an average response after 24 h stress (50.0% decrease). After 72 h of stress IR64 and IR83373-B-B-25-3 showed huge decline in the ratio (90.6 and 84.9% decrease). After recovery period, IR84895-B-127-CRA-5-1-1 and IR83373-B-B-25-3 revived the K+/Na+ ratio. IR84895-B-127-CRA-5-1-1 showed the K+/Na+ ratio to reach that of the 24 h stress condition (4.4), while IR 64 and IR83373-B-B-25-3 were unable to improve their respective ratio (1.2 and 2.6 respectively).
ROS-induced oxidative damage and antioxidant enzyme mediated detoxification of ROS
Rice seedlings exposed to salt stress exhibited extreme oxidative damage induced by increased H2O2 levels, as observed from accumulation of brown product after staining with DAB (Fig. 5a). IR64 seedlings were severely affected by salt after initial stress period; whereas, IR83373-B-B-25-3 was conspicuously affected after 48 h of stress. IR84895-B-127-CRA-5-1-1 seedlings resisted the severity of stress even after 72 h as evidenced by lesser accumulation of brown products; suggesting their better ability to eliminate salt induced damage among the selected genotypes.
Fig. 5.
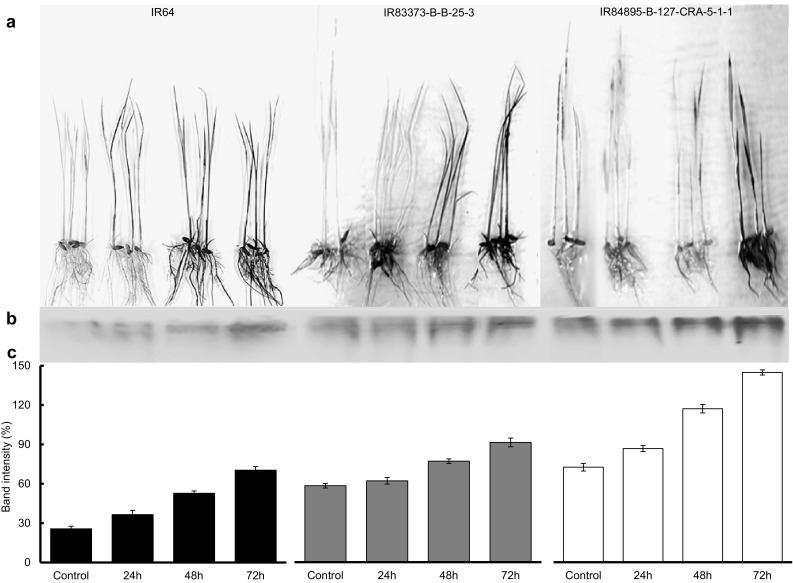
Study of ROS accumulation and detoxification in selected rice genotypes and evaluation of their salinity tolerance. a DAB staining of whole rice seedlings, b Activity staining for POX isozymes, c Relative intensity of bands in the POX gel. Error bars represent mean standard deviation (n = 3)
IR84895-B-127-CRA-5-1-1 exhibited a sharp rise in POX activity with increasing intensity of salt stress and maximum activity was observed after 72 h of stress (51.9, 60.7 and 73.1 Units min−1 mg−1 protein after 24, 48 and 72 h of stress respectively) (Fig. 4d). IR64 showed almost similar activity throughout the stress with marginal increase (36.9, 43.2 and 47.4 Units min−1 mg−1 protein respectively). In IR83373-B-B-25-3, POX activity was almost unaffected at the initial phase of stress up to 48 h (42.5 and 49.7 Units min−1 mg−1 protein respectively) which increased abruptly after 72 h (61.0 Units min−1 mg−1 protein). POX activity obtained from biochemical assay was further validated through in gel analysis. As shown in Fig. 5b and c, IR 64 showed bands of minimum intensity after 24 h stress (36.5%) but with gradual increase in stress, the band became prominent and maximum band intensity was obtained after 72 h of stress (70.4%). IR83373-B-B-25-3 showed almost similar POX activity after 0 and 24 h of stress (40.7 and 42.5 Units min−1 mg−1 protein respectively) which was represented by bands of almost same intensity (58.2 and 61.9%). In IR84895-B-127-CRA-5-1-1, intense band appeared after the initial phase of stress, which was maximum after 72 h stress, indicating better ROS detoxification activity.
Discussion
Rice cultivation has been found to be severely interrupted by abiotic stresses including drought and salinity (Singh et al. 2016). Drought stress is considered as a massive hindrance in Eastern Indo-Gangetic Kharif cropping system (Kumar et al. 2014). Salt stress is a major constraint for rice cultivation in coastal saline belt of eastern India (Ali et al. 2014). Therefore, exploration of potential drought and salt tolerant Indo-Gangetic rice genotypes has become a serious concern to feed the world’s growing population. Furthermore, comprehensive analysis of combined drought and salinity tolerance in single rice genotype may help in understanding the dual mechanism in an innovative way. Present study highlighted the variability in salt tolerance among drought tolerant rice genotypes of EIGP during most sensitive early vegetative stage with an emphasis on morphological, physiological and biochemical aspects.
Evaluating differential responses of rice genotypes under salinity stress
Salt stress interrupted growth and physiological activities of all rice genotypes, but the degree of fatality was diverse depending upon their relative adaptability and tolerance mechanism. Salt stress treatment inhibited overall plant growth, which might be due to the restriction of cell division and enhanced cell death (Wu et al. 2012). Maximum decrease in root and shoot length was observed in IR64, whereas IR84895-B-127-CRA-5-1-1 seemed to be acclimatized itself with salinity and sustained its growth throughout the treatment, establishing its better salt tolerance than the other two genotypes (Fig. 2a, b). NaCl stress resulted in maximum loss of water in IR64, contrasting with IR84895-B-127-CRA-5-1-1, and thus could not be revived after removal of stress (Fig. 2c). Salinity had adverse effect on relative water content which might be due to high osmotic pressure generated restriction in water uptake of plant cells (Summart et al. 2010). Experimental evidences from recovery treatment supported IR 84895-B-127-CRA-5-1-1 to be the most tolerant rice genotype among the three. Stomatal conductance was correlated with photosynthetic rate and coordinated with CO2 requirement of mesophyll cells (Yusuf et al. 2010). Decrease in stomatal conductance resulted in declined photosynthetic rate as confirmed by Brugnoli and Lauteri (1991) (Fig. 3c). Although chlorophyll content and consequently photosynthesis rate decreased in all three genotypes under NaCl stress (Figs. 3a, 4a), but IR84895-B-127-CRA-5-1-1 always maintained a higher chlorophyll content and photosynthetic rate as compared to IR83373-B-B-25-3 and IR64. Chaves et al. (2009) has already described loss of leaf water content as probable cause for reduced photosynthetic rate. Salinity stress also resulted in decreased transpiration rate and stomatal conductance in all rice genotypes. Minimum decrease in transpiration rate and stomatal conductance was exhibited by IR84895-B-127-CRA-5-1-1, which may cause reduced uptake of salt, as most of the salt uptake in rice occurs passively through the transpiration stream (Flowers and Yeo 1981). Thus IR84895-B-127-CRA-5-1-1 showed a better tolerance towards the stress exerted by 200 mM NaCl.
Osmotic adjustment is considered as the initial response to osmotic stress and can be measured by means of osmoprotectants like proline (Liu et al. 2014). Proline accumulation in plant tissue has been correlated with osmoregulation and may be considered as an indicator of the adaptive response under salt stress (Zhao et al. 2014). Moreover, proline has also been described to stabilize RUBISCO during NaCl stress (Ma et al. 2014). Among the considered genotypes, IR84895-B-127-CRA-5-1-1 exhibited the highest increase in free proline content with successive intensification of salinity, possibly for enhanced osmo-protective activity (Fig. 4b).
Salinity tolerance in plants is stimulated by the conservation of ion homeostasis with high K+/Na+ ratio in cell cytoplasm (Kumar et al. 2009). Ionic analysis indicated IR84895-B-127-CRA-5-1-1 seedlings had better ability to maintain ion homeostasis with a higher K+/Na+ ratio by detaining Na+ concentration inside the cells. This can be a reflection of several advanced strategies acquired by the tolerant genotype to balance Na+ level either by restricting entrance of Na+ ions into cells or withdrawal of excess Na+ (Ma et al. 2014).
Improved ROS detoxification contributes to better salt tolerance
Salinity stress is also associated with higher accumulation of ROS (Asada 2006). Among the three genotypes, IR64 and IR83373-B-B-25-3 had the increased ROS accumulation indicating its susceptibility towards salinity. IR84895-B-127-CRA-5-1-1 showed the minimum accumulation of ROS, representing its better salinity tolerance. Accumulation of ROS leads to cellular and sub-cellular membrane injury (Aspinall and Paleg 1981). Lipid peroxidation (MDA formation) might be indicative of the membrane injury (Riazi et al. 1985). IR84895-B-127-CRA-5-1-1 seedlings showed the minimum MDA content even after 72 h stress contrasting with IR64 and IR83373-B-B-25-3, which may be attributed to antioxidant enzyme mediated improved ROS scavenging (Jiang et al. 2016). Deleterious effect of ROS on cell is determined by the equilibrium between the production and detoxification of ROS through antioxidant enzyme system (Xiong et al. 2002). Reactive oxygen species-induced cell toxicity could be eliminated in IR84895-B-127-CRA-5-1-1 through strong antioxidant defence mechanism against ROS built-up (Fig. 5). Furthermore, increased activity of POX in IR84895-B-127-CRA-5-1-1 seedlings might result in enhanced ROS scavenging, providing better protection under salinity (Fig. 6).
Fig. 6.
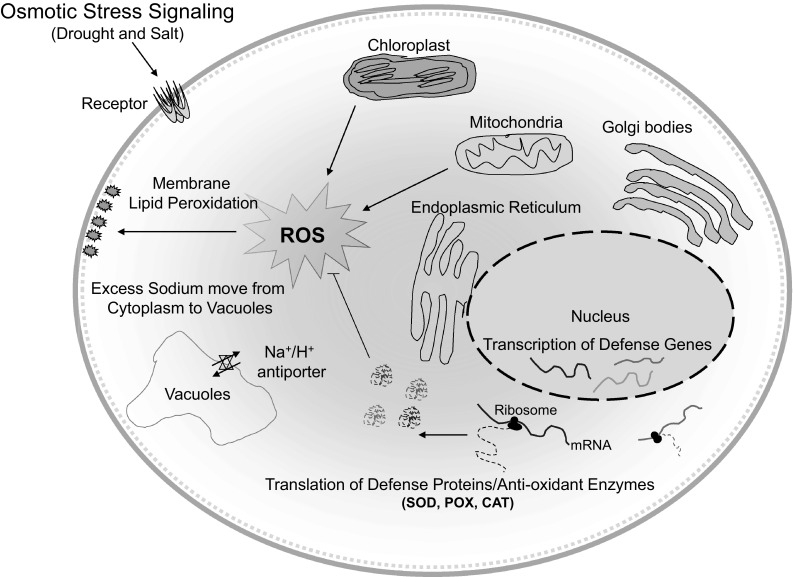
Schematic presentation elucidating the ROS signaling mechanism during plant response to different osmotic stresses (drought and salt). Osmotic stress in plants leads to the accumulation of ROS triggering severe oxidative damage. Excessive ROS generation causes disintegration of plasma membrane and different cell organelles involved in vital metabolic processes of cell. ROS-mediated signaling pathway stimulates gene expression for antioxidant enzymes within the cell that provides osmotic stress tolerance in plants through detoxification of ROS
Establishment of promising multiple stress tolerant rice genotype of Eastern Indo-Gangetic Plains
Establishment of multiple stress tolerant rice genotype of Eastern Indo-Gangetic Plains was based on elaborated studies carried out with the potential drought tolerant rice genotypes under high salinity stress. A comprehensive study was performed employing morpho-physiological and biochemical characterisation. Accumulation and detoxification of ROS under salinity stress was the vital part of the exploration that supported the former observations.
Morpho-physiological studies were executed to recognize the potential traits may aid in breeding for multiple stress tolerance. Drought and salt exposed seedlings exhibited an overall decrease in chlorophyll content and thereby photosynthetic rate which may be caused by the over accumulation of ROS due to osmotic stress (Chaves et al. 2009). Alternatively, reduction in photosynthetic rate can also be attributed to reduced stomatal conductance under drought and salt stress (Brugnoli and Lauteri 1991). Reduced degradation of chlorophyll and higher photosynthetic rate in IR84895-B-127-CRA-5-1-1 supported the hypothesis (Figs. 3a, 4a). Transpiration rate may be associated with stomatal conductance and unavailability of enough water due to lower conductance might be a reason for diminished transpiration as evident in IR83373-B-B-25-3-B-B-25-3 and IR64. However, upholding higher rate of conductance in IR84895-B-127-CRA-5-1-1 after 72 h of stress has facilitated this genotype to maintain the photosynthetic rate. Moreover, maintaining higher RWC in IR84895-B-127-CRA-5-1-1 also supported this assumption. Then again, excessive accumulation of Na+ in plant cells decreased the K+/Na+ ratio and disrupted the ion homeostasis. But IR84895-B-127-CRA-5-1-1 protected the cells from Na+ toxicity and maintained higher K+/Na+ ratio which may be related to the better osmotic tolerance of this genotype. As estimated from the membrane lipid peroxidation (MDA content), conductance may be restricted as a result of the ROS-induced membrane damage (Shao et al. 2008). IR84895-B-127-CRA-5-1-1 exhibited lower accumulation of ROS, due to its improved detoxification mechanism mediated by increased POX activity throughout the stress and thus had the lowest membrane lipid peroxidation (Fig. 6).
Correlation among different characters was evaluated to identify the salinity-tolerant genotype (Table 2). Photosynthetic rate of the genotypes exhibited significant correlation with several traits, including leaf chlorophyll content, RWC, stomatal conductance, transpiration rate and lipid peroxidation P ≤ 0.001 (***). Furthermore, lipid peroxidation was found to be significantly correlated with K+/Na+ ratio, proline content and RWC P ≤ 0.001 (***). Considering all the evaluations, IR84895-B-127-CRA-5-1-1 has displayed sustainable morpho-physiological activities and ROS detoxification among the considered genotypes. Therefore, IR84895-B-127-CRA-5-1-1 may contribute as a donor line in breeding for multiple stress tolerant varieties of EIGP.
Conclusions
This study highlighted the search for a rice genotype with better tolerance towards drought and salinity based on morphological, physiological, ionomic and biochemical analyses. In summary, IR84895IR84895-B-127-CRA-5-1-1 exhibited considerably enhanced drought and salinity tolerance at seedling stage. Improved salinity tolerance in IR84895-B-127-CRA-5-1-1 was associated with improved ROS detoxification mediated by up-regulated antioxidant enzyme activity and greater recovery ability after removal of stress, as compared to IR83373-B-B-25-3-B-B-25-3 and IR64. Thus, IR84895IR84895-B-127-CRA-5-1-1 may be considered as the most potential drought and salt tolerant rice genotype in EIGP for fragile ecosystems. On the whole, this study established complexity and plasticity of the rice genotypes under stress conditions and the outcomes may draw a new insight in exploiting multiple stress environments.
Acknowledgements
The work was supported by research grants to GK from the University Grants Commission and Science and Engineering Research Board-Department of Science and Technology, Government of India.
Abbreviations
- DAB
Diaminobenzidine
- DW
Dry weight
- FW
Fresh weight
- MDA
Malondialdehyde
- NaCl
Sodium chloride
- PEG
Polyethylene glycol
- POX
Peroxidase
- PN
Photosynthetic rate
- ROS
Reactive oxygen species
- RWC
Relative water content
- TBA
Thiobarbituric acid
- TCA
Trichloroacetic acid
References
- Ali NM, Yeasmin L, Gantait S, Goswami R, Chakraborty S. Screening of rice landraces for salinity tolerance at seedling stage through morphological and molecular markers. Physiol Mol Biol Plants. 2014;20(4):411–423. doi: 10.1007/s12298-014-0250-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alscher RG, Erturk N, Heath LS. Role of superoxide dismutases in controlling oxidative stress in plants. J Exp Bot. 2002;53:1331–1341. doi: 10.1093/jexbot/53.372.1331. [DOI] [PubMed] [Google Scholar]
- Arnon DI. Copper enzymes in isolated chloroplasts, polyphenoxidase in Beta vulgaris. Plant Physiol. 1949;24:1–15. doi: 10.1104/pp.24.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Asada K. Production and scavenging of reactive oxygen species in chloroplasts and their functions. Plant Physiol. 2006;141:391–396. doi: 10.1104/pp.106.082040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aspinall D, Paleg LG. The physiology and biochemistry of drought resistance in plants. Australia: Academic Press; 1981. Proline accumulation: physiological aspects; pp. 205–241. [Google Scholar]
- Bates LS, Waldren RP, Teare ID. Rapid determination of free proline for water stress studies. Plant Soil. 1973;39:205–207. doi: 10.1007/BF00018060. [DOI] [Google Scholar]
- Bray EA, Bailey-Serres J, Weretilnyk E. Gruissem W, Buchannan B, Jones R (2000) Responses to abiotic stresses, biochemistry and molecular biology of plants, Rockville, MD American Society of Plant Physiologists, pp 1158–1203
- Brugnoli E, Lauteri M. Effects of salinity on stomatal conductance, photosynthetic capacity and carbon isotope discrimination of salt-tolerant (Gossypium hirsutum L.) and salt-sensitive (Phaseolus vulgaris L.) C3 non-halophytes. Plant Physiol. 1991;95:628–635. doi: 10.1104/pp.95.2.628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaves MM, Maroco JP, Pereira JS. Understanding plant responses to drought—from genes to the whole plant. Funct Plant Biol. 2003;30:239–264. doi: 10.1071/FP02076. [DOI] [PubMed] [Google Scholar]
- Chaves MM, Flexas J, Pinheiro C. Photosynthesis under drought and salt stress: regulation mechanisms from whole plant to cell. Ann Bot. 2009;103:551–560. doi: 10.1093/aob/mcn125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chutia J, Borah SP. Water stress effects on leaf growth and chlorophyll content but not the grain yield in traditional rice (Oryza sativa Linn.) genotypes of Assam. India: II. Protein and proline status in seedlings under PEG induced water stress. Am J Plant Sci. 2012;3(7):971–980. doi: 10.4236/ajps.2012.37115. [DOI] [Google Scholar]
- DaCosta M, Huang B. Changes in antioxidant enzyme activities and lipid peroxidation for Bentgrass Species in response to drought stress. J Am Soc Hortic Sci. 2007;132(3):319–326. [Google Scholar]
- Deinlein U, Stephan AB, Horie T, Luo W, Xu G, Schroeder JI. Plant salt-tolerance mechanisms. Trends Plant Sci. 2014;19(6):371–379. doi: 10.1016/j.tplants.2014.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farooq M, Wahid A, Lee DJ. Exogenously applied polyamines increase drought tolerance of rice by improving leaf water status, photosynthesis and membrane properties. Acta Physiol Plant. 2009;31:937–945. doi: 10.1007/s11738-009-0307-2. [DOI] [Google Scholar]
- Flowers TJ, Yeo AR. Variability in the resistance of sodium chloride salinity within rice (Oryza sativa L.) varieties. New Phytol. 1981;88:363–373. doi: 10.1111/j.1469-8137.1981.tb01731.x. [DOI] [Google Scholar]
- Frary A, Gol D, Keles D, Okmen B, Pinar H, Sigva HO, Yemenicioglu A, Doganlar S. Salt tolerance in Solanum pennellii: antioxidant response and related QTL. BMC Plant Biol. 2010;10:58–74. doi: 10.1186/1471-2229-10-58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fryer MJ, Oxborough K, Mullineaux PM, Baker NR. Imaging of photo-oxidative stress responses in leaves. J Exp Bot. 2002;53:1249–1254. [PubMed] [Google Scholar]
- Graham RC, Lundholm U, Arnovsky MJK. Cytochemical demonstration of peroxidase activity with 3-amino-9-ethylcarbazole. J Histochem Cytochem. 1964;13:150–159. doi: 10.1177/13.2.150. [DOI] [PubMed] [Google Scholar]
- Gratao PL, Polle A, Lea PJ, Azevedo RA. Making the life of heavy metal stressed plants a little easier. Funct Plant Biol. 2005;32:481–494. doi: 10.1071/FP05016. [DOI] [PubMed] [Google Scholar]
- Hazmana M, Hause B, Eiche E, Riemann M, Nick P. Different forms of osmotic stress evoke qualitatively different responses in rice. J Plant Physiol. 2016;202:45–56. doi: 10.1016/j.jplph.2016.05.027. [DOI] [PubMed] [Google Scholar]
- Heath RL, Packer L. Photoperoxidation in isolated chloroplast. I. Kinetics and stoichiometry of fatty acid peroxidation. Arch Biochem Biophys. 1968;125:189–198. doi: 10.1016/0003-9861(68)90654-1. [DOI] [PubMed] [Google Scholar]
- Hong Y, Zhang H, Huang L, Li D, Song F. Overexpression of a stress-responsive NAC transcription factor gene ONAC022 improves drought and salt tolerance in rice. Front Plant Sci. 2016;7(4):1–19. doi: 10.3389/fpls.2016.00004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huke RE, Huke EH. Rice area by type of culture, South, Southeast, and East Asia. Los Banos: IRRI; 1997. [Google Scholar]
- Jiang Y, Qiu Y, Hu Y, Yu D (2016) Heterologous expression of AtWRKY57 confers drought tolerance in Oryza sativa. Front Plant Sci. 7(145). doi:10.3389/fpls.2016.00145 [DOI] [PMC free article] [PubMed]
- Krishnamurthy P, Ranathunge K, Nayak S, Schreiber L, Mathew MK. Root apoplastic barriers block Na+ transport to shoots in rice (Oryza sativa L.) J Exp Bot. 2011;62(12):4215–4228. doi: 10.1093/jxb/err135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar G, Purty RS, Sharma MP, Singla-Pareek SL, Pareek A. Physiological responses among Brassica species under salinity stress show strong correlation with transcript abundance for SOS pathway-related genes. J Plant Physiol. 2009;166:507–520. doi: 10.1016/j.jplph.2008.08.001. [DOI] [PubMed] [Google Scholar]
- Kumar G, Kushwaha HR, Purty RS, Kumari S, Singla-Pareek SL, Pareek A. Cloning, structural and expression analysis of OsSOS2 in contrasting cultivars of rice under salinity stress. Genes Genomes Genomics. 2012;6(1):34–41. [Google Scholar]
- Kumar S, Dwivedi SK, Singh SS, Bhatt BP, Mehta P, Elanchezhian R, Singh VP, Singh ON. Morphophysiological traits associated with reproductive stage drought tolerance of rice (Oryza sativa L.) genotypes under rain-fed condition of eastern Indo-Gangetic Plain. Indian J Plant Physiol. 2014;19(2):87–93. doi: 10.1007/s40502-014-0075-x. [DOI] [Google Scholar]
- Laemmli UK. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- Li X, Zhang L, Ma L. Effects of preconditioning on photosynthesis of rice seedlings under water stress. Procedia Environ Sci. 2011;11:1339–1345. doi: 10.1016/j.proenv.2011.12.201. [DOI] [Google Scholar]
- Liu G, Li X, Jin S, Liu X, Zhu L. Overexpression of rice NAC gene SNAC1 improves drought and salt tolerance by enhancing root development and reducing transpiration rate in transgenic cotton. PLoS ONE. 2014;9(1):e86895. doi: 10.1371/journal.pone.0086895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma DM, Xu WR, Li HW, Jin FX, Guo LN, Wang J, Dai HJ, Xu X. Co-expression of the Arabidopsis SOS genes enhances salt tolerance in transgenic tall fescue (Festuca arundinacea Schreb.) Protoplasma. 2014;251:219–231. doi: 10.1007/s00709-013-0540-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller G, Suzuki N, Ciftci-Yilmaz S, Mittler R. Reactive oxygen species homeostasis and signalling during drought and salinity stresses. Plant Cell Environ. 2010;33:453–467. doi: 10.1111/j.1365-3040.2009.02041.x. [DOI] [PubMed] [Google Scholar]
- Munns R. Comparative physiology of salt and water stress. Plant, Cell Environ. 2002;25(2):239–250. doi: 10.1046/j.0016-8025.2001.00808.x. [DOI] [PubMed] [Google Scholar]
- Platten JD, Egdane JA, Ismail AM. Salinity tolerance, Na+ exclusion and allele mining of HKT1; 5 in Oryza sativa and O. glaberrima: many sources, many genes, one mechanism? BMC Plant Biol. 2013;13:32–47. doi: 10.1186/1471-2229-13-32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Polle A, Otter T, Seifert F. Apoplastic peroxidases and lignification in needles of Norway Spruce Picea abies L. Plant Physiol. 1994;106:53–60. doi: 10.1104/pp.106.1.53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riazi A, Matsuda K, Arslan A. Water-stress induced changes in concentrations of proline and other solutes in growing regions of young barley leaves. J Exp Bot. 1985;36:1716–1725. doi: 10.1093/jxb/36.11.1716. [DOI] [Google Scholar]
- Sekar I, Pal S. Rice and wheat crop productivity in the Indo-Gangetic plains of india: changing pattern of growth and future strategies. Ind J Agric Eco. 2012;67(2):238–252. [Google Scholar]
- Shao H, Chu L, Shao M, Li S, Yao J. Bioengineering plant resistance to abiotic stresses by the global calcium signal system. Biotech Adv. 2008;26:503–510. doi: 10.1016/j.biotechadv.2008.04.004. [DOI] [PubMed] [Google Scholar]
- Sharma R, Vleesschauwer DD, Sharma MK, Ronald PC. Recent advances in dissecting stress-regulatory crosstalk in rice. Mol Plant. 2013;6(2):250–260. doi: 10.1093/mp/sss147. [DOI] [PubMed] [Google Scholar]
- Singh G, Bundela DS, Sethi M, Lal K, Kamra SK. Remote sensing and GIS for appraisal of salt-affected soils in India. J Environ Qual. 2010;39:5–15. doi: 10.2134/jeq2009.0032. [DOI] [PubMed] [Google Scholar]
- Singh R, Singh Y, Xalaxo S, et al. From QTL to variety-harnessing the benefits of QTLs for drought, flood and salt tolerance in mega rice varieties of India through a multi-institutional network. Plant Sci. 2016;242:278–287. doi: 10.1016/j.plantsci.2015.08.008. [DOI] [PubMed] [Google Scholar]
- Summart J, Thanonkeo P, Panichajakul S, Prathepha P, McManus MT. Effect of salt stress on growth, inorganic ion and proline accumulation in Thai aromatic rice, Khao Dawk Mali 105, callus culture. Afr J Biotechnol. 2010;9(2):145–152. [Google Scholar]
- Weatherley PE. Studies in the water relations of the cotton plant I. The field measurement of water deficits in leaves. New Phytol. 1950;49:81–97. doi: 10.1111/j.1469-8137.1950.tb05146.x. [DOI] [Google Scholar]
- Wu A, Allu AD, Garapati P, Siddiqui H, Dortay H, Zanor MI, Asensi-Fabado MA, Munne-Bosch S, Antonio C, Tohge T, Fernie AR, Kaufmann K, Xue GP, Mueller-Roeber B, Balazadeha S. JUNGBRUNNEN1, a reactive oxygen species-responsive NAC transcription factor, regulates longevity in Arabidopsis. Plant Cell. 2012;24:482–506. doi: 10.1105/tpc.111.090894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiong L, Schumaker KS, Zhu JK. Cell signaling during cold, drought and salt stress. Plant Cell. 2002;14:165–183. doi: 10.1105/tpc.010278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye Z, Rodriguez R, Tran A, Hoang H, de los Santos D, Brown S, Vellanoweth RL. The developmental transition to flowering represses ascorbate peroxidase activity and induces enzymatic lipid peroxidation in leaf tissue in Arabidopsis thaliana. Plant Sci. 2000;158:115–127. doi: 10.1016/S0168-9452(00)00316-2. [DOI] [PubMed] [Google Scholar]
- Yordanov I, Velikova V, Tsonev T. Plant responses to drought, acclimation and stress tolerance. Photosynthetica. 2000;38:171–186. doi: 10.1023/A:1007201411474. [DOI] [Google Scholar]
- Yoshida S, Forno DA, Cock JH, Gomez KA. Laboratory manual for physiological studies of rice, 3. Manila: International Rice Research Institute; 1972. pp. 1–83. [Google Scholar]
- Yusuf MA, Kumar D, Rajwanshi R, Strasser RJ, Tsimilli-Michael M, Govindjee Sarin NB. Overexpression of c-tocopherol methyl transferase gene in transgenic Brassica juncea plants alleviates abiotic stress: physiological and chlorophyll a fluorescence measurements. Biochim Biophys Acta. 2010;1797:1428–1438. doi: 10.1016/j.bbabio.2010.02.002. [DOI] [PubMed] [Google Scholar]
- Zhang J, Kirkham MB. Water relations of water-stressed, split-root C4 (Sorghum bicotor; Poaceae) and C3 (Helianthus annuus; Asteraceae) plants. Am J Bot. 1995;82:1220–1229. doi: 10.2307/2446244. [DOI] [Google Scholar]
- Zhang J, Long Y, Xue M, Xiao X, Pei X. Identification of microRNAs in response to drought in common wild rice (Oryza rufipogon Griff.) shoots and roots. PLoS ONE. 2017;12(1):e0170330. doi: 10.1371/journal.pone.0170330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao X, Wang W, Zhang F, Deng J, Li Z, et al. Comparative metabolite profiling of two rice genotypes with contrasting salt stress tolerance at the seedling stage. PLoS ONE. 2014;9(9):e108020. doi: 10.1371/journal.pone.0108020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu JK, Chinnusamy V, Jagendorf A. Understanding and improving salt tolerance in plants. Crop Sci. 2005;45:437–448. doi: 10.2135/cropsci2005.0437. [DOI] [Google Scholar]


