Abstract
In agricultural ecosystems, pest insects, pathogens, and reduced soil fertility pose major challenges to crop productivity and are responsible for significant yield losses worldwide. Management of belowground pests and diseases remains particularly challenging due to the complex nature of the soil and the limited reach of conventional agrochemicals. Boosting the presence of beneficial rhizosphere organisms is a potentially sustainable alternative and may help to optimize crop health and productivity. Field application of single beneficial soil organisms has shown satisfactory results under optimal conditions. This might be further enhanced by combining multiple beneficial soil organisms, but this remains poorly investigated. Here, we inoculated wheat plots with combinations of three beneficial soil organisms that have different rhizosphere functions and studied their effects on crop performance. Plant beneficial Pseudomonas bacteria, arbuscular mycorrhizal fungi (AMF), and entomopathogenic nematodes (EPN), were inoculated individually or in combinations at seeding, and their effects on plant performance were evaluated throughout the season. We used traditional and molecular identification tools to monitor their persistence over the cropping season in augmented and control treatments, and to estimate the possible displacement of native populations. In three separate trials, beneficial soil organisms were successfully introduced into the native populations and readily survived the field conditions. Various Pseudomonas, mycorrhiza, and nematode treatments improved plant health and productivity, while their combinations provided no significant additive or synergistic benefits compared to when applied alone. EPN application temporarily displaced some of the native EPN, but had no significant long-term effect on the associated food web. The strongest positive effect on wheat survival was observed for Pseudomonas and AMF during a season with heavy natural infestation by the frit fly, Oscinella frit, a major pest of cereals. Hence, beneficial impacts differed between the beneficial soil organisms and were most evident for plants under biotic stress. Overall, our findings indicate that in wheat production under the test conditions the three beneficial soil organisms can establish nicely and are compatible, but their combined application provides no additional benefits. Further studies are required, also in other cropping systems, to fine-tune the functional interactions among beneficial soil organisms, crops, and the environment.
Keywords: plant-growth promoting rhizobacteria, biofertilizer, Steinernema, Heterorhabditis, wheat, biological control, insect pest, plant growth promotion
Introduction
In addition to poor soil fertility, soil pests and pathogens pose major threats to the health and productivity of crops in agricultural ecosystems resulting in important yield losses every year (Oerke, 2006; Kupferschmied et al., 2013). The use of fertilizers, fungicides, nematicides, and insecticides to counter these problems can have important negative consequences, such as the persistence of these agrochemicals in the soil, water, and food with potential negative impacts on the environment and consumers (Bale et al., 2008; Lichtfouse et al., 2009; Kupferschmied et al., 2013; Johnson et al., 2016). Hence, new and more sustainable pest and disease control strategies need to be explored for a next-generation agriculture and the application of beneficial soil organisms (BeSO) presents a promising alternative for maintaining crop health and productivity (Bommarco et al., 2013; Bender et al., 2016).
Various BeSO are known to enhance plant performance, e.g., by directly promoting plant growth, by stimulating plant defenses, by facilitating nutrient acquisition by the plant, or by protecting the plant from pathogens and pests (Philippot et al., 2013; Rasmann and Turlings, 2016; Venturi and Keel, 2016). The three groups of BeSO investigated in the present study fulfill one or several of these beneficial functions, i.e., plant-growth promoting rhizobacteria (PGPR), arbuscular mycorrhizal fungi (AMF), and entomopathogenic nematodes (EPN). Root-colonizing bacteria belonging to the Pseudomonas fluorescens group are well-characterized PGPR that have the ability to induce systemic plant defenses and ward off soil-borne pathogens, in particular pathogenic fungi and oomycetes, including Gaeumannomyces, Thielaviopsis, Rhizoctonia, Fusarium oxysporum, and Pythium (Haas and Défago, 2005; Mercado-Blanco and Bakker, 2007; Hol et al., 2013; Vacheron et al., 2015). To date, several biocontrol products that are based on PGPR pseudomonads are on the market (Berg, 2009; Kupferschmied et al., 2013). Moreover, certain subgroups, in particular the two species Pseudomonas protegens and Pseudomonas chlororaphis exhibit potent oral insecticidal activity notably against Lepidopteran pests (Kupferschmied et al., 2013; Ruffner et al., 2013; Flury et al., 2016).
AMF are well-known beneficial symbionts that colonize the roots of the majority of land plants (Schueßler et al., 2001; van der Heijden et al., 2015). AMF form extensive hyphal networks that provide water and nutrients to their host plant. AMF are key actors in processes such as the mineralization of phosphorus and nitrogen and enhancing the nutrient up-take by plant roots (Jakobsen et al., 1992; Mäder et al., 2000; Smith et al., 2004; van der Heijden et al., 2006). AMF primarily improve plant nutrition, but they can also contribute to enhance the tolerance of their host plant against biotic and abiotic stresses (van der Heijden et al., 2015). Numerous AMF species, e.g., Rhizoglomus irregulare, are commercialized as inoculum to improve soil fertility (Lekberg and Koide, 2005; Pellegrino et al., 2015) and plant productivity (Hijri, 2016; Köhl et al., 2016). Today, the agronomic use of AMF includes the direct augmentation or inoculation of seedlings in nurseries before transplanting to the field (Jeffries et al., 2003) and seed coating (e.g., Ijdo et al., 2011).
Finally, EPN of the genera Steinernema and Heterorabditis are well-known biocontrol agents that selectively search their insect hosts and kill them within 2–3 days with the aid of mutualistic bacteria of the genera Xenorhabdus and Photorhabdus, respectively (Georgis et al., 2006; Kaya et al., 2006; Dillman et al., 2012; Campos-Herrera, 2015; Lacey et al., 2015). Their wide distribution in soils throughout the world (Adams et al., 2006) and the availability of commercial products (Lacey et al., 2015) make them excellent products in integrated pest management (IPM) programs or in organic production, both for augmentation or restoration of naturally occurring EPN (Campos-Herrera, 2015). However, their performance and activity is affected by biotic and abiotic factors, and hence, their efficacy depends on soil characteristics, agricultural management practices, and competition within the food web (Stuart et al., 2015).
The three groups of organisms—Pseudomonas, AMF, and EPN—occur naturally in most arable soils and commercial formulations are available for agronomic use (Stockwell and Stack, 2007; Berg, 2009; Kupferschmied et al., 2013; Lacey et al., 2015). Previous greenhouse and field studies have reported varying effects on plant health and growth when combining inoculants of these BeSO groups. For example, combinations of certain Pseudomonas strains provided better control of the wheat disease take-all than did the individual strains alone (Pierson and Weller, 1994). Positive effects have been also recorded when combining bacteria, such as Pseudomonas or Azospirillum strains, with fungi, including the AMF Glomus (Frey-Klett et al., 2007; Couillerot et al., 2012; Walker et al., 2012), Fusarium or Trichoderma (Fogliano et al., 2002; Yigit and Dikilitas, 2007). Similarly, EPN have been combined with other BeSO, with differing results. For example, the combination of Steinernema kraussei with the entomopathogenic fungus (EPF) Metarhizium anisopliae resulted in a synergistic effect in the control of Otiorhynchus sulcatus in strawberry (Ansari et al., 2010), while the combination of Steinernema ichnusae with the EPF Beauveria bassiana resulted in clear antagonism and competition for the host under controlled laboratory conditions (Tarasco et al., 2011).
Field applications of single BeSO have shown to greatly enhance plant growth and health in various crops (Jeffries et al., 2003; Berg, 2009; Kupferschmied et al., 2013; Campos-Herrera, 2015; Lacey et al., 2015), but the putative positive effect of combining various BeSO remains poorly predictable. The Swiss National Research Programme 68 (NRP 68) “Sustainable use of soil as a resource” (www.nrp68.ch) provided the framework for our multidisciplinary investigations into BeSO and their possible role in novel strategies for sustainable soil management. As part of this, we evaluated, for the first time, the simultaneous application of Pseudomonas, AMF, and EPN inoculants in field experiments, using wheat as the model crop. We hypothesized that the combined application of these BeSO would show greater benefits for the crop than their individual application.
Materials and methods
Beneficial organisms
Selected species of BeSO, all known to naturally occur in Swiss soils (Campos-Herrera et al., 2015a; Jaffuel et al., 2016; Schlaeppi et al., 2016; Imperiali et al., 2017), were applied depending on the objective and design of each field experiment (Figure S1). The BeSO that were used included two species of the genus Pseudomonas, three AMF species and four EPN species and they were applied as inoculants either individually or in various combinations in the different experiments (Tables 1, 2).
Table 1.
Beneficial soil organisms applied individually or in combinations in the field experiments.
| Beneficial group/species | Strain | Treatment code | Application type | GenBank accession no. | Reference or source |
|---|---|---|---|---|---|
| PSEUDOMONAS BACTERIA | |||||
| Pseudomonas chlororaphis | PCL1391a | B2 | Aqueous | NZ_LFUT01000004 | Chin-A-Woeng et al., 1998 |
| Pseudomonas protegens | CHA0a | B1 | Aqueous | NC_021237 | Stutz et al., 1986 |
| ARBUSCULAR MYCORRHIZAL FUNGI | |||||
| Claroideoglomus claroideum | SAF12c | F4 | Substrate | n.a.d | Swiss collection of arbuscular miychorrizal fungi (SAF) |
| Funneliformis mosseae | SAF11c | F3 | Substrate | n.a.d | SAF |
| Rhizoglomus irregulareb | INOQ Top | F1 | Substrate | n.a.d | Inog GmbH, Schnega |
| Rhizoglomus irregulareb | SAF22c | F2 | Substrate | DQ377990 | Germany, SAF |
| ENTOMOPATHOGENIC NEMATODES | |||||
| Heterorhabditis bacteriophora | Andermatt | N2 | Aqueous | KJ938576 | Andermatt Biocontrol AG, Grossdietwil, Switzerland |
| Heterorhabditis megidis | Andermatt | N1 | Aqueous | KJ938577 | Andermatt Biocontrol AG, Grossdietwil, Switzerland |
| Steinernema carpocapsae | D-83 | N3 | Aqueous | KJ818295 | Jaffuel et al., 2016 |
| Steinernema feltiae | RS-5 | N4 | Aqueous | KJ938569 | Jaffuel et al., 2017 |
Rifampicin-resistant variants of strains CHA0 and PCL1391 were used as inoculants in the field trials (see Materials and Methods).
Rhizoglomus irregulare was previously referred to as Rhizophagus irregularis and earlier as Glomus intraradices (Sieverding et al., 2015).
Strain ID referring to the Agroscope AMF strain collection, http://www.agroscope.ch/saf.
n.a., not available.
Table 2.
Details on the characteristics of the three field experiments used to assess effects of inoculation of beneficial soil organisms (pseudomonads, entomopathogenic nematodes, and arbuscular mycorrhizal fungi) on growth, health, and yield of spring wheat.
| Field trials | COMBINATION | PERFORMANCE-1 | PERFORMANCE-2 |
|---|---|---|---|
| Coordinates SN/EW | 46.397676/6.260763 | 46.397455/6.260166 | 46.39502/6.260444 |
| Sowing/inoculation day | 18th March 2014 | 18th March 2014 | 27th March 2015 |
| Treatments (Treatments codes refer to Table 1) |
Control (no inoculants) B1: P. protegens CHA0-Rif B2: P. chlororaphis PCL1391-Rif N1: H. megidis Andermatt N2: H. bacteriophora Andermatt N3: S. carpocapsae D83 N4: S. feltiae RS5 F1-H: R. irregulare INOQ Top (high dosage, 250 ml/row) F1-L: R. irregulare INOQ Top (low dosage, 50 ml/row) F2: R. irregulare SAF22 F3: F. mosseae SAF11 F4: C. claroideum SAF12 AMF control (substrate only) B1+ N2 B1+ F1 N2 + F1 B1+ N2 + F1 |
Control (no inoculants) B1: P. protegens CHA0-Rif B2: P. chlororaphis PCL1391-Rif N2: H. bacteriophora Andermatt BM: B1 + B2 B1 + N2 B2 + N2 BM + N2 |
Control (no inoculants) BM: B1 + B2 NM: N1 + N2 + N4 F1-L AMF control (substrate only) BM + NM NM + F1-L BM + F1-L BM + NM + F1-L |
| Number of treatments | 17 | 8 | 9 |
| Number of replicates (plots) per treatment | Four replicates in randomized complete block design | Four replicates in randomized complete block design | Nine replicates in randomized complete block design |
| Size of plots | 1.5 m2 | 6.75 m2 | 9 m2 |
| Number of plant rows per plot | 5 | 5 | 5 |
| Wheat seeds per m of row | ~80 | ~140 | ~140 |
| Bacterial inoculum (CFU in 400 ml H2O per meter of row) | B1: 1.19 × 109
B2: 1.21 × 109 |
B1: 1.42 × 109
B2: 3.37 × 109 |
B1: 4.875 × 108
B2: 8.25 × 108 |
| AMF inoculum | F1-H: 80 F1-L: 16 F2: 250 F3: 250 F4: 250 AMF control: 250 |
Not contributed | F1-L: 16 AMF control: 16 |
| Nematode inoculum | 50 infective juveniles/cm2/4 L | 50 infective juveniles/cm2/8 L | 50 infectice juveniles/cm2/8 L |
| Pest insect stress | Heavy natural infestation with Oscinella frit | Heavy natural infestation with Oscinella frit | No relevant Oscinella frit infestation |
| Soil type and texture | Sandy loam (clay, 25.5%; silt, 34.3%; sand; 40.2%) | Sandy loam (clay, 25.5%; silt, 34.3%; sand, 40.2%) | Loam (clay, 14.5%; silt, 26.8%; sand, 58.7%) |
To monitor the bacteria following field application, the bacterial inoculants, i.e., P. protegens strain CHA0 (Stutz et al., 1986) and P. chlororaphis strain PCL1391 (Chin-A-Woeng et al., 1998) were tagged with a spontaneous resistance to rifampicin following previously described protocols (Natsch et al., 1994). Briefly, spontaneous rifampicin-resistant derivatives were obtained following plating concentrated cell suspensions of each parental strain on King's medium B agar (KMB) (King et al., 1954) supplemented with 100 μg/ml of rifampicin and incubated for 3 days. A CHA0-Rif derivative and a PCL1391-Rif derivative (Table 1), which stably maintained rifampicin resistance and displayed wild-type growth and antifungal and insecticidal activities, were selected. For the preparation of the bacterial field inocula, the selected rifampicin resistant strains were grown overnight at 25°C in lysogeny broth (LB) (Bertani, 1951) containing 100 μg/ml of rifampicin. Aliquots of 200 μl of each culture were spread on KMB plates without antibiotics. After incubation at 27°C for 16 h, bacterial cells were harvested and washed in sterile distilled water. The optical density at 600 nm (OD600) of the bacterial cell suspensions was adjusted to 0.15 corresponding to a cell density of 8 × 107 CFU ml−1. These bacterial stock suspensions were maintained on ice until final dilution and use on the field sites.
The AMF strains Claroideoglomas claroideum SAF12, Funneliformis mosseae SAF11 and R. irregulare SAF22 were selected from the Swiss Collection of Arbuscular Mycorrhizal Fungi (SAF) at Agroscope (Reckenholz, Zurich, www.agroscope.ch/saf; Table 1). The inocula were prepared as described by Schlaeppi et al. (2016). Briefly, AMF were propagated over 6 months in the greenhouse in autoclaved sand:soil (85:15%; v/v) as substrate and using Plantago lanceolata as host. The final inoculum contained pieces of plant roots mixed with the substrate containing AMF hyphae and spores (SAF12, Propagation 0510, P. lanceolata roots were colonized by 28% and 763 spores could be washed from 25 g substrate; SAF#11, Propagation 0711, 27% and 29 spores; SAF#22, Propagation 0813; 97% and 475 spores). In addition, a “mock” inoculum consisting of Plantago roots and substrate free of AMF propagules was prepared following the same protocol and this mock treatment was termed “AMF control” (Quality inspection of the mock inoculum, Propagation 0711, roots were not colonized by AMF and no spores could be washed from 25 g of substrate). The COMBINATION experiment also comprised a treatment where nothing was applied, named “control”. AMF inocula as well as the mock-inoculum were mixed in separate plastic bags and stored at room temperature until use. In addition, the commercial AMF inoculum R. irregulare TOP (INOQ GmbH, Schnega, Germany, www.inoq.de) was used as obtained from Otto Hauenstein Samen AG (Rafz, Switzerland, www.hauenstein.ch). For the second trial (PERFORMANCE-2), we utilized the lower dosage of the commercial inoculum based on the COMBINATION experiment results. For the PERFORMANCE-2 trial we utilized autoclaved commercial inoculum as AMF control treatment.
For the EPN, infective juveniles (IJs) of four species were prepared in adjusted suspensions. Heterorhabiditis species were obtained from a commercial source (Andermatt Biocontrol, Grossdietwil, Switzerland, www.andermattbiocontrol.com), whereas Steinernema species were propagated from field collected populations under laboratory conditions following protocols described by Campos-Herrera et al. (2015a) (Table 1). All nematodes were received or harvested within 2 weeks prior to field application. The day before application, the EPN inoculant suspensions were prepared in sterile water. To this end, IJs were counted and their density was adjusted to deliver the required field concentration per experimental unit (Table 2) by using separate containers. Containers were kept at 5°C overnight and transported in coolers to the field. In addition, laboratory infections of Galleria mellonella larvae by the inoculant EPN at field concentrations were used to verify their infectivity for each experiment (Jaffuel et al., 2017).
Experimental designs
From spring 2014 to summer 2015, three field experiments were conducted in wheat plots and the applications of beneficial soil organisms were adapted for each experiment. All the experiments were carried out with the commercial spring wheat variety “Rubli” in the experimental plots, whereas the commercial triticale variety “Trado” was seeded in the buffer zones. Fields were bordered by strips of non-managed grassland. The three experiments were named as follows: COMBINATION (2014), PERFORMANCE-1 (2014), and PERFORMANCE-2 (2015) (Table 2). The selection of the applied organisms and combinations of treatments were adapted on results of the preceding trial. The first experiment (COMBINATION) was set up to test various species of each group of beneficials and first combinatory treatments. The second experiment was designed to evaluate wheat yield effects after combining bacteria and EPN (PERFORMANCE-1). In this experiment, the AMF treatment was not included due to limitations in scaling the production of inoculum for the large plot size. Finally, the PERFORMANCE-2 experiment consisted of the full bacteria-AMF-EPN combinations during the subsequent season (Table 2).
All the experiments were conducted in neighboring experimental field sites located near Prangins, Switzerland (see Table 2 for coordinates). The sites belong to Agroscope, research center of Changins, (Nyon, Switzerland) and have documented crop and management sequences for the last 30 years. The field sites chosen for the experiments had no overlapping areas to avoid cross-contaminations with inoculants. None of the experiments had irrigation systems. The soil type was sandy loam for the COMBINATION and PERFORMANCE-1 trials and loam for the PERFORMANCE-2 trial (Table 2). General agronomic preparations for all the experiments included tillage (15 cm deep) and harrowing about 4 days before seeding. The seeding machine “Hege Seedmatic” (Hege Maschinen, Eging am See, Germany) allowed the customized seeding for each plot size and arrangement (Table 2; Figure S1) and was modified to keep the seed furrows open after placing the seeds. After seeding, the plots were marked for the corresponding treatments (see Figure S1 for the exact field design of each of the three experiments) and inoculated on the same day with the beneficial soil organisms. In combination treatments, the application followed the order bacteria, EPN, and AMF.
Bacteria were applied as a cell suspension to the seed furrows (plant rows) using treatment-specific watering cans. Final cell suspensions were prepared directly on the field from bacterial stock suspensions (OD600 0.15; i.e., 8 × 107 CFU ml−1) by adjusting with water to obtain the required volumes (400 ml per meter of row) and bacterial cells (8 × 108 CFU per meter of row) (Table 2). Similarly, EPN were applied in variable volumes depending on plot size using treatment-specific watering cans. They were applied to entire plots (not just the rows), and in all the cases, the final concentration was 0.5 Mio. IJs/m2 (equivalent to 50 IJs/cm2, Grewal and Peters, 2005). Finally, AMF inocula were applied manually employing 250-ml glass beakers. The material was applied directly over the seeds in the furrows, thereby gently mixing seeds and inoculum with a small hoe. AMF control plots received the same quantity of AMF-free substrate. Control plots were treated with the same volumes of BeSO-free water. Immediately after treatment application, the seeds were covered with soil using hoes to close the seed furrows. All equipment used for inoculant application was thoroughly cleaned and disinfested between manipulations using 70% ethanol.
Weed control included the application of the herbicides Azur (Omya AG, Switzerland) against monocots 2 weeks after seeding and Apell (Syngenta AGRO SA) against dicots shortly before earing (BBCH 45-50). When necessary, some persistent weeds (Galium spp., Setaria spp.) were controlled manually. No fungicides nor nematicides were applied during any of the experiments. The insecticide Karate Zeon (Lambda-Cyhalotryne, Syngenta Agro GmbH) was applied in the PERFORMANCE-2 experiment against cereal mining dipterae such as the frit fly and hessian fly conducted in 2015, but not in the 2014 COMBINATION and PERFORMANCE-1 experiments. Plots were fertilized once by supplementing nitrogen in liquid (Lonza-sol N liquid, Basel, Switzerland) at 62 kg ha−1 of to reach 155 units N and potassium (K2O) at 30.6 kg ha−1. The PERFORMANCE-2 trial was covered with a black hail net during the first 2–3 weeks to protect the seeds and young plants from cold conditions and predation by birds and small mammals.
Sampling of beneficial organisms and measuring of plant traits
Pseudomonas bacteria
The presence of P. protegens CHA0-Rif and P. chlororaphis PCL1391-Rif was evaluated in both inoculated plots and non-inoculated control plots, as well as in the buffer zone around the experimental plots, and in the border zone (grassland) around the field site to control for possible cross contamination. This analysis was conducted four times during the growing season (i.e., at seeding, end of the winter, at earing, and maturity) in selected experiments (Table 3). For this, the root systems from four wheat plants per plot (triticale plants and grass for the buffer and border zones, respectively) were collected, pooled, washed, and gently dried with paper towels. Roots were weighed, cut into pieces (about 15 cm long), placed in 50-ml Falcon tubes (Greiner Bio One, Germany) containing 40 ml of sterile water and kept overnight at 4°C. All sampling equipment was cleaned with 70% ethanol between samples to avoid cross-contaminations. Samples were vigorously agitated on a rotary shaker at 180 rpm for 20 min, and roots were removed and dried at 80°C for 3 days to obtain the dry weight. The remaining suspensions were transferred to fresh sterile Falcon tubes on ice and centrifuged at 8,500 rpm (9,300 g) at 4°C. The supernatant was discarded and the pellet was re-suspended in 1 ml of sterile water. Each sample was then serially diluted and dilutions spread on KMB supplemented with 100 μg/ml of cycloheximide and 100 μg/ml of rifampicin (Scanferlato et al., 1990). The colonies were counted and the results were expressed as colony forming units (CFU) per gram of dry root weight.
Table 3.
Description of the type of measurements and methods employed and timing in each of the field experiments.
| Organisms | Type of measurement | Method | Reference | COMBINATION | PERFORMANCE-1 | PERFORMANCE-2 |
|---|---|---|---|---|---|---|
| Bacteria | Sample type | Composite samples of wheat roots | Authors | Wheat roots | Wheat roots | Wheat roots |
| Quantification at seeding | CFU counting on selective media | Authors | 28.03.2014 | 28.03.2014 | 27.03.2015 | |
| Quantification during wheat growth | CFU counting on selective media | Authors | 03.06.2014 | 03.06.2014 25.06.2014 22.07.2014 |
27.04.2015 18.05.2015 29.06.2015 |
|
| Tracing presence in buffer zones | CFU counting on selective media | Authors | Not done | 03.06.2014 | Not done | |
| Tracing in non-agricultural soil of border zone | CFU counting on selective media | Authors | Not done | 25.06.2014 22.07.2014 |
Not done | |
| AMF | Sample type | Composite samples of wheat roots | Authors | Wheat roots | Not contributed | Wheat roots |
| Quantification of inoculum | Real-time qPCR using primers targeting the inocula | Authors | At harvest | Not contributed | At harvest | |
| Determination of AMF community | AMF community sequencing | Schlaeppi et al., 2016 | At harvest | Not contributed | Not done | |
| EPN | Sample type | Composite soil sample | 12 soil cores/plot | 15 soil cores/plot | 15 soil cores/plot | |
| EPN presence: pre-inoculation (baseline) | Species-specific primers/probes and real time qPCR | Campos-Herrera et al., 2015a | 27.03.2014 (Baseline) | 27.03.2014 (Baseline) | 27.03.2015 (Baseline) | |
| EPN presence: post-augmentation | Species-specific primers/probes and real time qPCR | Campos-Herrera et al., 2015a | 25.06.2014 | 25.06.2014 | 17.06.2015 | |
| EPN activity: insect-baits | Galleria bait | Bedding and Akhurst, 1975 | 25.06.2014 | 25.06.2014 | 17.06.2015 | |
| Soil food web assemblage (nematophagous fungi, free-living nematodes and ectophoretic bacteria) | Species-specific primers/probes and real time qPCR | Atkins et al., 2005; Zhang et al., 2006; Campos-Herrera et al., 2011b, 2012, 2015b; Pathak et al., 2012 | 27.03.2014 (Baseline) 25.06.2014 |
27.03.2014 (Baseline) 25.06.2014 |
27.03.2015 (Baseline) 17.06.2015 |
|
| Plantsa | Height (average per plot) | Measured from shoot base to the upper growth | Authors | – | – | 23.04.2015 05.06.2015 18.05.2015 01.06.2015 08.06.2015 |
| Weight | Eight plants | Authors | – | – | At harvest | |
| Density (% of plot surface covered by plants)/number of plants per linear meter | Visual scoring | Authors | – | 14.05.2014 21.05.2014 11.06.2014 |
06.05.2015 18.05.2015 18.06.2015 29.06.2015 |
|
| Chlorophyll activity | N-tester (YARA) | Authors | – | – | 08.06.2015 | |
| Yield (g seeds/plot); | Weighing wheat seeds at dough developmental stage | Authors | – | At harvest | At harvest | |
| Thousand-seed weight (TSW) | Marvin seed analyzer | Gegas et al., 2010 | – | – | At harvest | |
| Protein content (%) | Near-infrared spectroscopy | Authors | – | – | At harvest | |
| Insect pest and pathogen incidence | Visual counts | Authors | Weekly | Weekly | Weekly |
Measurements were made for the three field experiments, but data are considered not representative due to the small size of the plots (COMBINATION assay) and/or the highly heterogeneous growth of the wheat plants within the plots following heavy frit fly damage in the 2014 COMBINATION and PERFORMANCE-1 assays.
Arbuscular mycorrhizal fungi
The inoculation success of the different AMF inocula was traced by quantitative PCR comparing their abundance in wheat roots sampled from inoculated, non-inoculated or mock-inoculated plots (Table 3). At harvest, the root systems of four plants per plot were pooled to become one sample. The fine roots (deeper than ca. 3 cm in soil) were cut from the root system using scissors, hackled into small pieces (1–2 cm long) with a scalpel and thereby homogenizing all root fragments of the four plants. The root samples were lyophilized and then ground to a fine powder using a Retsch Ball Mill (model MM301; settings 30 s at 30 Hz using one 1-cm steel ball). DNA was extracted from ~200 mg of fine root powder utilizing the NucleoSpin® Plant II kit from Macherey-Nagel following the instructions. DNA concentrations were determined on a Varian Eclipse Fluorescence plate reader using Quant-iT PicoGreen dsDNA Assay Kit (Invitrogen) and Herring Sperm DNA (Invitrogen) as standard solution. The R. irregulare strains INOQ Top and SAF22 were quantified by qPCR utilizing primers developed by Alkan et al. (2006) and Bender et al. (unpublished), respectively. The AMF signals were expressed relative to a plant signal obtained with qPCR primers targeting the wheat ADP-ribosylation factor (Giménez et al., 2011). Triplicate amplifications were performed in 20 μl reactions using the HOT FIREPol® EvaGreen® qPCR Mix Plus (no ROX) from Solis Biodyne (www.sbd.ee, Estonia) on a Bio-Rad CFX96 Touch™ Real-Time PCR Detection System (www.bio-rad.com, USA). Reactions contained 4 μl qPCR mix (5X), 1 μl of each primer (10 μM), 9 μl distilled sterile water, and 5 μl template (5 ng DNA). The cycling program consisted of a 15 min initial denaturation step at 95°C followed by 40 cycles (95°C for 15 s, 63°C for 40 s for both R. irregulare primer sets or 60°C for 10 s for the wheat reference primers, 72°C for 20 s) and a 10 min final extension step at 72°C. Melting curve analysis consisted of a gradient from 65 to 95°C, increasing by half degrees/per 10 s to determine the uniformity of the amplicons. Raw data were imported from the qPCR cycler into the LinRegPCR program to determine the Ct and efficiency (E) values using a common fluorescence threshold for all samples (Ruijter et al., 2009). F. mosseae and Claroideoglomus claroideum were quantified with species-specific TaqMan probes following the protocols developed by Thonar et al. (2012). Template amounts were calculated for each reaction using the individual E, averaged among the replicates of each sample and expressed relative to the plant signal. Of note, for samples of the COMBINATION trial, we determined also the whole AMF community by amplicon sequencing (Schlaeppi et al., 2016).
Entomopathogenic nematodes, soil food web, and post-application activity
A total of 18 soil organisms were identified and quantified before application (baseline) and post-augmentation (Table 3) to detect possible trophic cascade effects due to EPN augmentation (Campos-Herrera et al., 2013). These organisms comprised seven EPN species (all previously described for the area, Campos-Herrera et al., 2015a), four free-living nematodes (FLNs) that compete with EPN for the insect cadaver (Campos-Herrera et al., 2012, 2015b), six nematophagous fungi (NF) (Campos-Herrera et al., 2015a), and one nematode surface-associated bacterium (Enright and Griffin, 2005; Campos-Herrera et al., 2011a) (Table S1). Briefly, a composite soil sample composed of several cores (2.5 cm diameter, 20 cm depth, see Table 3 for exact quantities per experiment) were taken per plot and kept on ice for transportation to the laboratory. The nematode community and other soil organisms were extracted from aliquots of 300–400 g of fresh soil by sucrose-centrifugation (Jenkins, 1964), concentrated in 1.5 ml tubes and stored at −80°C until processed, following Campos-Herrera et al. (2015a,b). Briefly, DNA was extracted from soil samples as well as from pure cultures for the generation of standard curves (when living material was available) with the Power Soil DNA Isolation Kit (MO BIO laboratories, Inc.). If no living material was available for a target organism, we employed plasmids with the whole sequence of interest to establish our positive control (Table S1; Campos-Herrera et al., 2015a). The quality and quantity of each DNA sample was analyzed prior to use (1 μl per duplicate, Nanodrop 1000, Thermo Scientific, Wilmington, DE, USA).
Species-specific primers and probes were employed in real time qPCR assessment of the 18 soil organisms (Atkins et al., 2005; Zhang et al., 2006; Campos-Herrera et al., 2011a,b, 2012, 2015a,b; Pathak et al., 2012), following the MIQE procedures (Bustin et al., 2009). All samples were run in duplicates (unknown, positive, and negative controls) employing optical 100-well gene disc reaction plates (Biolabo, scientific instruments, Switzerland) on a Corbett Research real time PCR machine. Final reactions, concentrations, and protocols were used as previously described (Campos-Herrera et al., 2015a,b). Nematode quantification from the soil samples was done with a 10-fold dilution of the DNA, whereas the identification and quantification of NF and surface-associated bacteria required the use of total DNA without dilutions (see details in Campos-Herrera et al., 2015a). A correction factor was derived from the dilution series to transform qPCR data to numbers of IJs. Finally, a sub-sample of fresh soil was dried to allow quantification per 100 g of dry soil.
In addition to the EPN soil food web, we evaluated the EPN activity at post-application sampling times (Table 3), as previously described by Campos-Herrera et al. (2015a) and Jaffuel et al. (2016). Briefly, two aliquots of 200 g of fresh soil per sample were baited with larvae of Galleria mellonella (Lepidoptera: Pyralidae) to test the suppressive potential of the soil. Following a modified procedure as described by Bedding and Akhurst (1975), each subsample (from augmented or not augmented plots) was baited with five final instar G. mellonella larvae (commercial stock, Au Pêcheur SARL Neuchâtel, Switzerland) in two independent rounds. After exposure for 4 days, the cadavers were recovered from the soil, thoroughly rinsed with tap water, and individually placed in White traps (White, 1927). Under a stereoscope, we checked for nematode emergence every 2–3 days to determine the organisms responsible for larval mortality. We recovered the nematodes in tap water upon emergence. The cadavers for which no obvious cause of death could be determined after 1 month of incubation were discarded after dissection. The DNA of the progeny leaving the cadavers was extracted using the QIAamp DNA mini kit (Qiagen), purity checked (Nanodrop system), adjusted to the range of 0.5–1 ng/μl, and species identity assessed by qPCR as described above (Campos-Herrera et al., 2015a; Jaffuel et al., 2016).
Plant traits
A total of eight measurements recorded the evolution of plant growth, productivity, and health. They were: average plant height per plot, plant density per plot, chlorophyll activity (N-tester), seed yield, thousand-seed weight, plant weight, plant protein content, and presence of pest insects and pathogens (Table 3). Regular monitoring of the experiments ensured the status of development into each phenostage. Most of the agronomical traits presented herein were measured at harvest (Table 3).
Statistical analysis
All experimental field trials presented a Randomized Complete Block design (Figure S1). The data from each group of beneficial organisms were analyzed following standard procedures for their data presentation, transformation, standardization, and normalization whenever necessary. In the case of the EPN activity, data from the G. mellonella baits were expressed as the percentage of larval mortality per plot, averaged by treatment. The activity was determined with respect to the total mortality caused only by nematodes. For the EPN soil food web analysis, all the organisms (EPN, FLN, NF, and bacteria) quantified by using qPCR were expressed per 100 g of dry soil. The parasitism of nematodes by NF was expressed as “infection rate” (IR), which was calculated by dividing the DNA quantity of each species by the total amount of DNA (Campos-Herrera et al., 2012; Duncan et al., 2013). Similarly, to estimate the total FLN and NF, we divided all data within a species by the highest measurement for that species, which allowed the standardization of the units of measurement among species ranging from 0 to 1 (de Rooij-van der Goes et al., 1995).
Unless specified, all significant differences between treatments were assessed by one–way ANOVA, using Tukey's HSD test, considering block as co-variable (V 20.0, IBM SPSS Inc., Chicago, IL). In some cases, t-tests were employed to compare pre- and post-augmentation or control vs. a specific treatment. If necessary, data were transformed to conform the assumptions of normality and equal variances (transformation method is indicated with the respective statistics). The bacterial colonization data were statistically assessed with a non-parametric Kruskal-Wallis test, followed by a post-hoc test (Dunn's test). With the exception of the Pseudomonas root colonization data (presented as log10 of the obtained values ± SEM) all data are presented as mean ± SEM of untransformed values.
Results
Survival and persistence of pseudomonas inoculants
In the COMBINATION trial, P. protegens CHA0-Rif and P. chlororaphis PCL1391 reached similar population densities that surpassed the threshold of ~105 CFU per gram of roots, which is the level needed for a plant-beneficial effect (Haas and Défago, 2005) (Figure 1A; Table 4). However, in the PERFORMANCE-1 trial, the population density of the P. chlororaphis strain on wheat roots was significantly lower (P < 0.05) than the density of the P. protegens strain at all three monitoring times (Figure 1B; Table 4). In this trial, in general, for strain CHAO-Rif, alone or in the combinations, we observed a better progression of the population if compared with the strain PCL1391-Rif. If both strains were present in the same treatment, our agar plates almost only reported P. protegens CHA0-Rif colonies (personal observation). Moreover, in clear contrast to CHA0-Rif, PCL1391-Rif never approached the population threshold for plant-beneficial effects, neither when applied alone nor when combined with other BeSO, (Figure 1B; Table 4). In combination with a commercial population of the EPN H. bacteriophora, the density of CHA0-Rif was significant reduced for the June 2014 sample. July 2014, i.e., about 1 month later, CHA0-Rif still maintained its population density in presence of the nematode inoculant while PCL1391-Rif was no longer detectable (Figure 1B; Table 4). In the 2015 PERFORMANCE-2 trial, bacterial numbers approached or surpassed the threshold for plant-beneficial effects at all three sampling times (Figure 1C; Table 4). In general, no significant differences among treatments were observed, but there was a trend of higher bacterial population densities in the April 2015 and May 2015 samplings when bacterial inoculants where combined with the EPN inoculant mixture. In contrast, an opposite trend was observed for June 2015 samples. Finally, as already observed in the PERFORMANCE-1 trial, strain P. protegens CHA0-Rif dominated the colonization, while P. chlororaphis PCL1391 was hardly detected (Figure S2).
Figure 1.
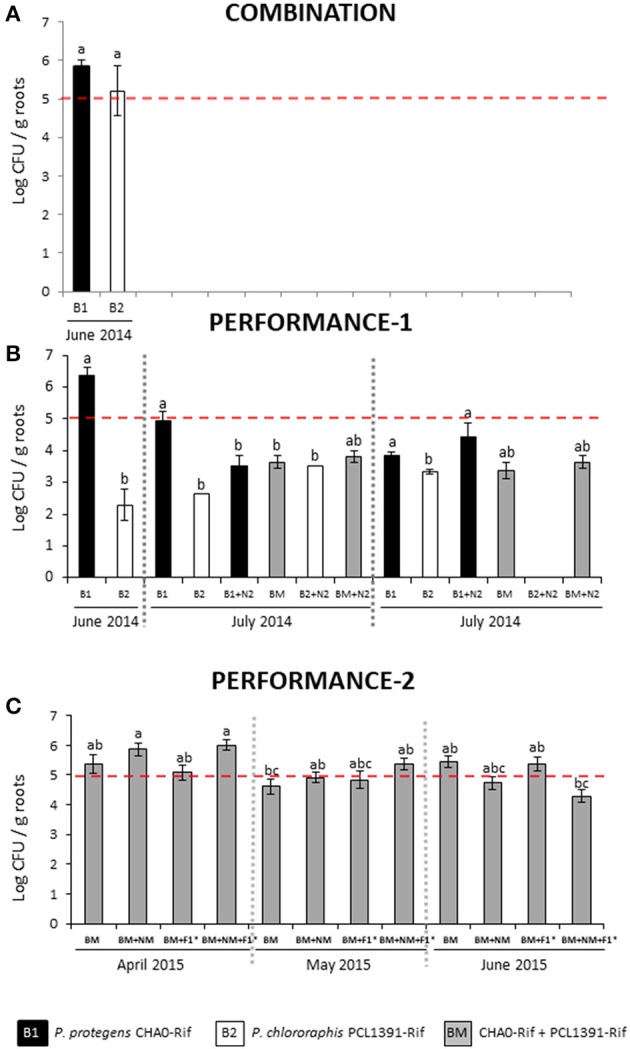
Survival of Pseudomonas protegens strain CHA0-Rif (B1) and Pseudomonas chlororaphis strain PCL1391-Rif (B2) on wheat roots in the COMBINATION (A), PERFORMANCE-1 (B), and PERFORMANCE-2 (C) field trials. Bacterial strains were inoculated individually or in combinations with the entomopathogenic nematode (EPN) Heterorhabditis bacteriophora (N2), an EPN mixture (NM; comprising Heterorhabditis megidis, H. bacteriophora, and Steinernema feltiae) and the arbuscular mycorrhizal fungus Rhizoglomus irregularis (F1*). Inoculants were monitored by selective plating on KMB supplemented with rifampicin (100 μg/ml) and cycloheximide (100 μg/ml) at three different time points following seed furrow inoculation. The dashed red line indicates the generally agreed threshold (~105 CFU per g root) required to provoke beneficial plant effects with plant growth-promoting pseudomonads (Haas and Défago, 2005). Bar graphs show means of log10 transformed CFU values per gram of dry roots weight (± SEM). Significant differences between treatments were calculated with one-way ANOVA (significance level P < 0.05) followed by the Tukey post-hoc test, or with a non-parametric Kruskal-Wallis test (significance level P < 0.05), followed by Dunn's test for post-hoc comparisons. Different letters indicate statistical significance at P < 0.05. Inoculants were not detected in the buffer and border zones of the field assays. No Rifampicin-resistant background population was detected at the field sites.
Table 4.
Statistical analysis for beneficial soil organisms and plant traits in the three field experiments.
| Organisms | Type of measurement | Statistical method | COMBINATIONa | PERFORMANCE-1a | PERFORMANCE-2a |
|---|---|---|---|---|---|
| Bacteria | CFU quantification I | One–way ANOVA (Tukey's HSD test); Kruskal-Wallis (Dunn's test) | F(1, 6) = 0.1526, n.s | F(1, 2) = 2.571, ** | F(3, 15) = 0.991, n.s. |
| CFU quantification II | One–way ANOVA (Tukey's HSD test); Kruskal-Wallis (Dunn's test) | –b | F(5, 12) = 3.675, ** | F(3, 16) = 0.656, n.s. | |
| CFU quantification III | One–way ANOVA (Tukey's HSD test); Kruskal-Wallis (Dunn's test) | – | F(4, 15) = 2.311, n.s. | F(3, 16) = 1.570, n.s. | |
| AMF | Quantification of INOQ Topc | One–way ANOVA | F(2, 9) = 10.42, ** | Not contributed | – |
| Quantification of SAF22c | T-tests | T = 10.377 *** | Not contributed | – | |
| Quantification of INOQ Top in combination samplesc | One–way ANOVA | F(5, 18) = 3.712, * | Not contributed | F(4, 36) = 0.571, n.s. | |
| EPN | EPN presence: pre-inoculation (baseline) | One–way ANOVA (Tukey's HSD test) | F(7, 24) = 1.273, n.s. | F(6, 21) = 0.525, n.s. | F(4, 40) = 0.281, n.s. |
| EPN presence: post-augmentation | One–way ANOVA (Tukey's HSD test) | F(7, 24) = 4.604, ** | F(6, 21) = 2.194, § | F(6, 21) = 2.888, * | |
| EPN activity: insect-baits | One–way ANOVA (Tukey's HSD test) | F(7, 24) = 3.317, * | F(6, 21) = 1.243, n.s. | F(6, 21) = 0.722, n.s. | |
| Free-living nematodes: pre-inoculation | One–way ANOVA (Tukey's HSD test) | F(7, 24) = 1.051, n.s. | F(6, 21) = 0.498, n.s. | F(6, 21) = 0.119, n.s. | |
| Free-living nematodes: post-augmentation | One–way ANOVA (Tukey's HSD test) | F(7, 24) = 0.395, n.s. | F(6, 21) = 1.025, n.s. | F(6, 21) = 0.341, n.s. | |
| Nematophagous fungi: pre-inoculation | One–way ANOVA (Tukey's HSD test) | F(7, 24) = 0.675, n.s. | F(6, 21) = 0.288, n.s. | F(6, 21) = 0.618, n.s. | |
| Nematophagous fungi: post-augmentation | One–way ANOVA (Tukey's HSD test) | F(7, 24) = 5.820, n.s. | F(6, 21) = 1.384, n.s. | F(6, 21) = 0.582, n.s. | |
| Plants | Height (at harvest) | One–way ANOVA (Tukey's HSD test) | – | – | F(8, 72) = 1.009, n.s. |
| Density (% plot covered by plants at harvest) | One–way ANOVA (Tukey's HSD test) Kruskal-Wallis (Dunn's test) | – | F(7, 88) = 17.219, *** | F(8, 72) = 0.756, n.s. | |
| Chlorophyll activity (N-tester) | One–way ANOVA (Tukey's HSD test) | – | – | F(8, 72) = 0.161, n.s. | |
| Yield (g seeds/plot) | One–way ANOVA (Tukey's HSD test) | – | F(7, 23) = 2.069, ** | F(8, 72) = 0.026, n.s. | |
| Thousand-seed weight (TSW) | One–way ANOVA (Tukey's HSD test) | – | – | F(8, 72) = 0.129, n.s. | |
| Protein content | One–way ANOVA (Tukey's HSD test) | – | – | F(8, 72) = 0.300, n.s. |
Data are presented as the statistical values, degree of freedom and probability levels:
P < 0.1,
P < 0.05,
P < 0.01,
P < 0.001, n.s., not significant.
For these variables, obtained data were not representative because of highly heterogeneous growth of the wheat plants within the plots following frit fly damage and thus were not considered for statistical analysis.
Statistics corresponding to data from two sets of primers, i.e., by Alkan et al. (2006) for INOQ Top, and Bender et al. (unpublished) for SAF22.
In all three field trials, no rifampicin-resistant bacteria were detected in the non-inoculated control treatments, in the buffer zones or in the grassland border zones at the experimental sites (data not shown), hence, the reported CFU data for the augmented bacteria required no baseline correction.
In general, the applied bacteria survived under field conditions until the end of the crop season (Figure 1), although the threshold required to provoke beneficial plant effects (~105 CFE per g root) was not always attained in all the trials or treatments. Inoculation was successful in all, with good traceability of the different bacterial inocula without cross-contamination. Consistently, the P. protegens inoculant showed higher presence on wheat roots, but the effects of the combination with other BeSO were not conclusive and depended on the BeSO species, on the time of exposure to the field conditions and differed between trials.
AMF inoculation success
For the COMBINATION trial we mainly used the locally well-adapted AMF R. irregulare (Schlaeppi et al., 2016) (Table 1). We confirmed successful wheat root inoculation for both R. irregulare strains that we tested, as well as our custom strain SAF22 and the commercial inoculum INOQ Top (Figure 2A; Table 4). The higher dosage of the inoculum INOQ Top (80 g per row) corresponded approximately to the amount of SAF22 inoculum and it appeared that both R. irregulare strains colonized the wheat roots to a similar extent. The reduced dosage of the commercial inoculum (16 g per row) was reflected in lower levels of root colonization and only showed a minor tendency of augmentation. We also traced the inoculation of the F. mosseae strain SAF11 and the C. claroideum strain SAF12 using specific qPCR primers (Thonar et al., 2012). We did not detect these AMF species at the field site (data not shown) confirming the findings of a previous AMF community profiling (Schlaeppi et al., 2016). Hence, we concluded that these strains failed to establish at the tested field site in the wheat roots. In summary, R. irregulare could be augmented in wheat AMF communities using the strains SAF22 or INOQ Top, while this was not successful for F. mosseae SAF11 and C. claroideum SAF12.
Figure 2.
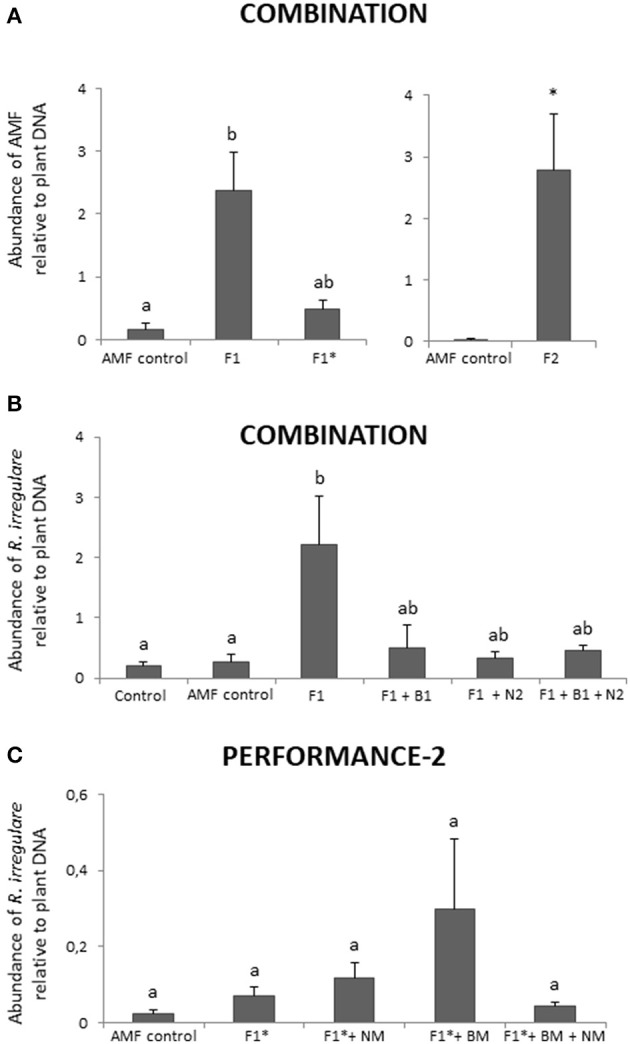
Abundance of Rhizoglomus irregulare in wheat roots in the COMBINATION (A,B) and PERFORMANCE-2 (C) field trials. (A) In the COMBINATION experiment, R. irregulare strain INOQ TOP was inoculated comparing high (F1) vs. low (F1*) dosages, with one of the treatments including the AMF strain SAF22 (F2). (B) In the same experiment, R. irregulare INOQ TOP (F1) was quantified in combination with bacteria, i.e., Pseudomonas protegens CHA0-Rif (B1), and nematodes, i.e., Heterorhabditis bacteriophora Andermatt (N2). (C) In the PERFORMANCE-2 experiment, R. irregulare INOQ TOP at the lower dosage (F1*) was used for the combination treatments with bacterial mixture (BM; i.e., P. protegens + Pseudomonas chlororaphis) and nematode mixture (NM; i.e., Heterorhabditis megidis + H. bacteriophora + Steinernema feltiae; for details see Figure S1). Control, non-inoculated control; AMF control, substrate control for AMF inoculation. R. irregulare was measured with quantitative PCR employing species-specific primers developed by Alkan et al. (2006) for INOQ TOP or their modified variants with enhanced specificity for SAF22 (Bender et al., unpublished). Bar graphs report mean normalized (R. irregulare relative to plant DNA) abundance (± SEM; COMBINATION, n = 4; PERFORMANCE-2, n = 7–9). Statistical analyses were performed on log-transformed data; asterisks and different letters indicate statistical significance at P < 0.05 for t-test and one-way ANOVA followed by the Tukey post-hoc test, respectively.
Plots in which soil beneficial organisms showed low AMF colonization levels were not different from control (nothing applied) and mock (application of carrier substrate without AM fungus) plots (Figure 2B; Table 4). These measured abundances of R. irregulare correspond to the native strain in the field and indicated that the application of the carrier substrate on its own did not affect the root colonization by the AM fungus. Although, the level of root colonization by R. irregulare showed a slight tendency to increase in the combination treatments of the AM fungus with bacteria, nematodes or both, the AM fungus was not augmented to the same extent as in single application. These first insights on combining soil beneficial organisms suggest possible negative effects on the AMF inoculum if combined with bacteria or nematodes.
In the PERFORMANCE-2 experiment, the commercial R. irregulare strain INOQ Top was inoculated using a lower dosage level to larger plots compared to the previous experiments (Table 2). Again, there was a tendency of increased colonization of the wheat roots in the combined treatments, however, high inter-plot variation precluded statistic support for this effect (Figure 2C; Table 4). It remains to be validated whether the colonization by this R. irregulare strain is particularly facilitated if applied in combination with the Pseudomonas bacteria.
In summary, R. irregulare successfully colonized wheat roots if inoculated alone, and in the combination experiments, we found varying augmentation efficiencies for R. irregulare if combined with pseudomonads, EPN or both, indicating that interactions with these beneficial soil organisms are context dependent.
Nematode survival, activity, and interactions with soil food web members
In all plots, very low numbers of background populations were detected as also found by Campos-Herrera et al. (2015a) and Jaffuel et al. (2017) in the same area. Five to seven species naturally occurred at the experimental field sites, and these species included the taxa that we augmented. Prior to inoculations (baseline; Table 3), there were no differences among treatments for any measured variable (EPN, free-living nematodes and nematophagous fungi; data not shown) in any of the three field trials. The evaluation of EPN soil food web members (free-living nematodes and nematophagous fungi) only revealed natural temporal fluctuations between baseline (pre-inoculation) and post EPN augmentation (data not shown), whereas their presence was not significantly affected by the EPN augmentation (alone or in combination) (Figure S3; Table 4). The nematophagous fungi and free-living nematodes species were in agreement with those already described by Campos-Herrera et al. (2015a,b) and Jaffuel et al. (2017). Finally, the ectophoretic bacterium P. nematophilus was not detected in any of plots (control or augmented).
In the COMBINATION trial, the EPN species H. megidis and S. carpocapsae were recovered in only 25% of the plots, 4 months after augmentation. In contrast, the species S. feltiae and H. bacteriphora, of which the latter was also combined with other BeSO, were detected in 100% of the plots, at the end of the season. The augmentation with S. feltiae was the only treatment with a significant increase in total EPN numbers compared with the native populations (Figure 3A; Table 4). The remarkable persistence of S. feltiae, which was the only species detected in the soil in their corresponding plots, was in agreement with the nematode activity measured in the laboratory as % mortality of G. mellonella producing progeny. This was the only treatment with significantly higher activity than the control in the whole trial (Figure 4A; Table 4).
Figure 3.
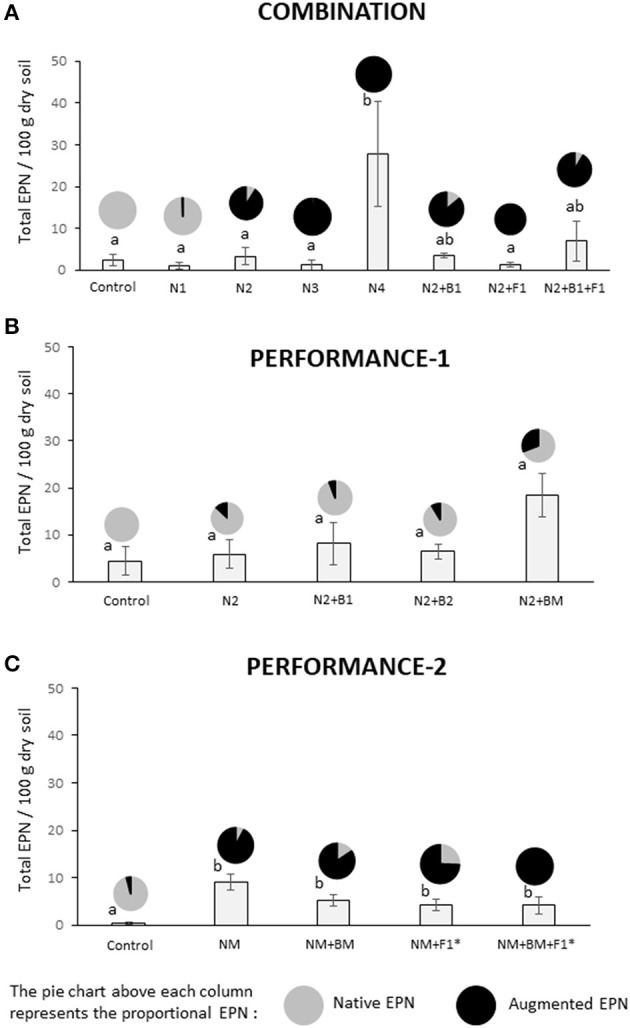
End of the season presence of inoculant and resident entomopathogenic nematodes in the COMBINATION (A), PERFORMANCE-1 (B), and PERFORMANCE-2 (C) field trials. Four different EPN species Heterorhabditis megidis (N1), Heterorhabditis bacteriphora (N2), Steinernema carpocapsae (N3), and Steinernema feltiae (N4) were inoculated individually or in combination with Pseudomonas protegens (B1), Pseudomonas chlororaphis (B2) and Rhizoglomus irregularis at two dosages (F1 and F1*). Mixtures of EPN (N1+N2+N4) or of the two bacteria (B1+B2) are indicated with NM and BM, respectively (for details see Figure S1). To determine the persistence of the EPN in soil of the different nematode inoculants as well as the impact of each treatment on the resident population of entompathogenic nematodes (EPN), a DNA extraction procedure followed by a qPCR approach was performed. Data are expressed as total EPN 100 g−1 of dry soil. Bar graphs report means (± SEM) and pie-charts show the proportion of native EPN vs. augmented EPN. Significant differences between treatments were calculated with one-way ANOVA (significance level P < 0.05) followed by the Tukey post-hoc test. Different letters indicate statistical significance at P < 0.05.
Figure 4.
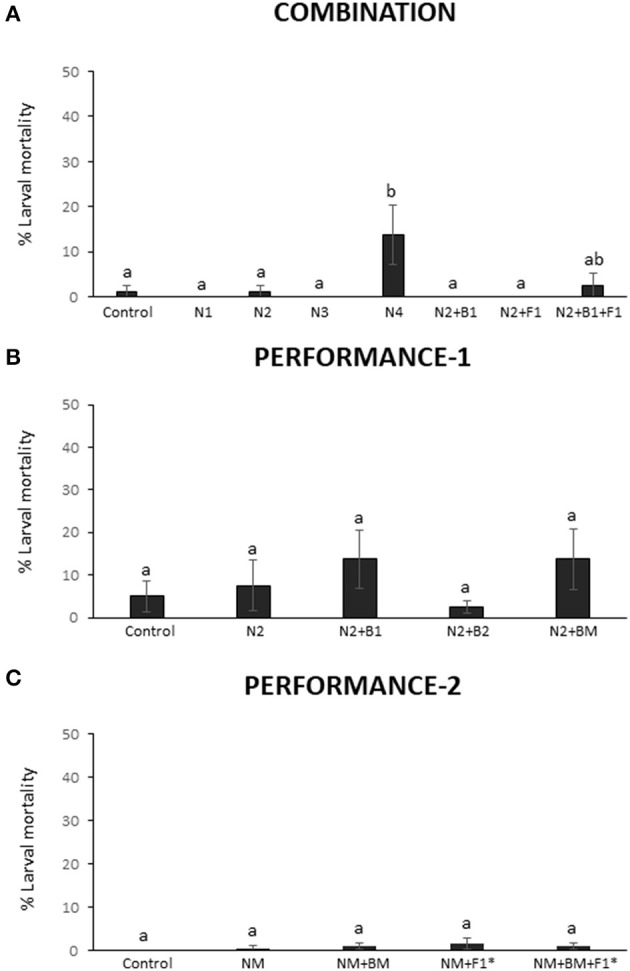
Activity of entomopathogenic nematodes (EPN) post application in three field trials. EPN activity was quantified by a Galleria mellonella larvae infection assay in soil samples from the (A) COMBINATION, (B) PERFORMANCE-1 and (C) PERFORMANCE-2 trials. Inoculants were Heterorhabditis megidis (N1), Heterorhabditis bacteriophora (N2), Steinernema carpocapsae (N3), and Steinernema feltiae (N4), individually or in combination with Pseudomonas protegens (B1), Pseudomonas chlororaphis (B2), and Rhizoglomus irregularis at two dosages (F1 and F1*). Mixtures of EPN or bacteria are indicated with NM and BM, respectively (for details see Figure S1). Bar graphs report means (± SEM). Significant differences between treatments were calculated with one-way ANOVA (significance level P < 0.05) followed by the Tukey post-hoc test. Different letters indicate statistical significance at P < 0.05.
In the PERFORMANCE-1 trial, we only augmented certain plots with H. bacteriophora. This EPN was detected in about 50% of the plots when applied alone or in combination with the bacterial inoculant P. chlororaphis and in about 75% of the plots when applied with the P. protegens in different combinations. No significant difference in EPN populations (qPCR measurements) and their activity (% larval mortality) was observed between plots where the EPN were applied alone or in combination with bacterial inoculants. Nor were they different from control plots (Figures 3B, 4B; Table 4). In both trials, COMBINATION and PERFORMANCE-1, there was a slight trend to detect more H. bacteriophora in the combined treatment with AMF and/or bacterial inoculants (N2+B1+F1 and N2+BM, respectively) (Figures 3A,B). The same trend was also observed for nematode activity (Figures 4A,B). In the PERFORMANCE-1 field trials, Steinernema affine (Table S1) was the dominant native EPN species in the soil of the experimental plots as determined by qPCR (Figure 3B).
In the PERFORMANCE-2 trial, the augmented EPN species (a mix of S. feltiae, H. megidis, and H. bacteriophora) could be detected in 100% of the plots inoculated with the three EPN, alone or in combination with the Pseudomonas inoculants, in 91% of the plots where they were combined with AMF and in only 44.4% of the plots when combined with both bacterial and AMF inoculants. Again, the species S. affine was dominant among the native taxa as displayed in the proportional chart, although, contrary to the PERFORMANCE-1 trial, native species were largely displaced in all the treatments where EPN were applied (Figure 3C). All plots with EPN application showed significantly higher total numbers of EPN than the control plots (Figure 3C; Table 4). All the augmented EPN species were detected in each of the plots, but S. feltiae and H. bacteriophora dominated. The nematode activity was low and did not significantly vary among treatments (Figure 4C; Table 4). The progeny from the activity tests belonged mainly to H. bacteriophora (62.5%), followed by S. feltiae (34.5%), in all the cases we found mixed EPN-free-living nematodes emergence as observed in previous studies in Swiss soils (Jaffuel et al., 2016, 2017).
In general, inoculated EPN persisted during the crop season and remained active until the time for wheat harvest, but with limited pest suppressive potential as measured with a Galleria larvae infection assay. We observed that EPN application increased the total numbers of EPN only in specific treatments, displacing at least partially the native populations (Figure 3). No long-term effect was observed with respect to soil organisms that can be expected to be modulated by EPN augmentation, such as nematophagous fungi and free-living nematodes. The combined application of EPN with other BeSO indicated compatibility with respect to their persistence, prevalence, and activity, when compared with the single EPN application, but some differences depending on EPN species and co-inoculant identity were observed. As for the bacterial and AMF inoculants, the success of EPN inoculants appeared to be context dependent.
Agronomic impact of the applied beneficial soil organisms
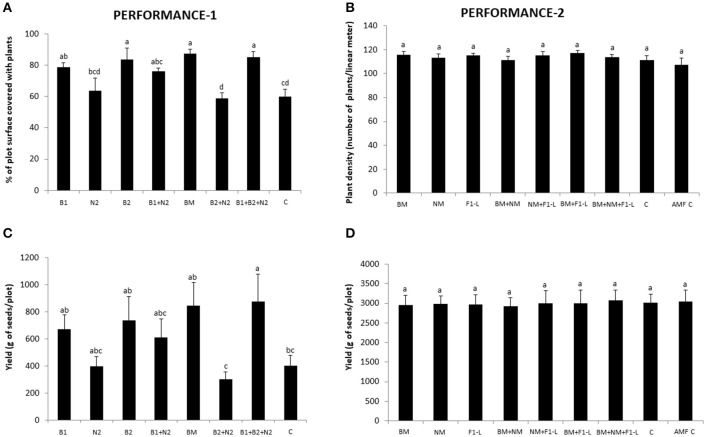
The 2014 trials (COMBINATION and PERFORMANCE-1) were intentionally not subjected to standard pesticide treatments and suffered from heavy attack by frit flies (Oscinella frit). For the small scale COMBINATION trial, insect damage was very patchy and therefore not agronomically representative and not included in the plant performance analyses. The larger plot sizes in the PERFORMANCE-1 trial permitted analysis of agronomically relevant plant density and seed yield data (Table 4). The % of plot surface covered with plants was significantly higher in augmentation plots than in the control treatment when the two bacterial inoculants, P. protegens and P. chlororaphis, were applied individually (treatments B1 and B2, respectively) or as a mixture with and without the EPN (treatments BM and B1+B2+N2, respectively) (Figure 5A). Seed yield per plot followed a similar pattern, but only the combined treatment with both bacterial strains and the EPN showed significantly higher values than the control (Figure 5C). AMF effects could not be examined in the PERFORMANCE-1 trial due to limited inoculum production. Nevertheless, the neighboring COMBINATION experiment indicated that seedling survival after frit fly attack tended to be higher in plots inoculated with R. irregulare (Figure S4). In the 2015 PERFORMANCE-2 trial, plots were subjected to pesticide treatment, no pest damage was observed and all plant traits were considered in the analysis. However, none of these measures, including plant density and seed yield per plot (Figures 5B,D) nor the other plant performance traits (Figure S5; Tables 3, 4) differed significantly from the control treatment.
Figure 5.
Impact of field inoculations with beneficial organisms on plant performance in the PERFORMANCE-1 (A,C) and PERFORMANCE-2 (B,D) trials. Plant performance was evaluated in terms of plant density (A,B) and yield (weight of wheat seeds) (C,D) for each plot. The PERFORMANCE-1 experiment was exposed to heavy natural infestation with the firt fly (Oscinella frit) causing significant plant damage. Plant density in the PERFORMANCE-1 trial was therefore determined by visual scoring the percentage of plot area covered by wheat plants in this experiment while it was determined by counting the number of plants per linear meter in the PERFORMANCE-2 experiment, which had no measurable frit fly damage. Inoculants were Pseudomonas protegens (B1), Pseudomonas chlororaphis (B2), individually or in combination with Heterorhabditis bacteriophora (N2) and Rhizoglomus irregularis (F1*). Mixtures of the two bacteria or of the entomopathogenic nematodes (Heterorhabditis megidis, Heterorhabditis bacteriophora, and Steinernema feltiae) are indicated with NM and BM, respectively (for details see Figure S1). C, non-inoculated control; AMF-C, substrate control for AMF inoculation. Bar graphs report means (± SEM). Significant differences between treatments were calculated with one-way ANOVA (significance level P < 0.05) followed by the Tukey post-hoc test. Different letters indicate statistical significance at P < 0.05.
In summary, when wheat was exposed to biotic stress (i.e., a heavy insect pest attack in 2014) a significant positive effect of the application of BeSO, notably Pseudomonas bacteria, on performance of the crop was observed. The presence of the EPN was only beneficial when combined with both bacteria together. In absence of a biotic stress conditions, as in the PERFORMANCE-2 trial in 2015, there was no measurable plant-beneficial effect of the presence of BeSO, highlighting the context dependence of their protective effect on the crops.
Discussion
Overall, the three field experiments showed consistent results: (1) the inoculated BeSO persisted until the end of the crop season, although their prevalence gradually declined with time; (2) in most of cases, the introduced BeSO in augmented plots were consistently present at higher levels than the native populations, without cross-contamination between plots; (3) the augmented BeSO integrated with or displaced the natural community to varying degrees depending on the strain/population and dosage; and (4) the combined application of Pseudomonas, EPN, and AMF showed only beneficial effects under conditions with an insect outbreak. In particular and contrary to our expectations, our current tripartite BeSO inoculant system (bacteria + EPN + AMF) did not provide clear additive or synergistic positive effects to allow a better performance of wheat than the application of the individual BeSO. Overall, our results are in agreement with the previous observation that the combination of various BeSO can lead to a beneficial effect under certain conditions (Frey-Klett et al., 2007; Ansari et al., 2010; Walker et al., 2011; Couillerot et al., 2012), but mainly have similar effects as single applications (Tarasco et al., 2011; Glare, Hurst, and Narciso, personal communication). We can conclude that there is still a large gap between the promising results from BeSO applications under controlled experiments (laboratory and greenhouse settings) and their performance under field conditions.
Many factors can explain this difference between applications in laboratory/greenhouse and field settings. The characteristics of a particular agroecosystem (i.e., soil type, soil geochemistry, humidity, plant genotype, climate, etc.) play a decisive role in determining the success of augmented BeSO. From a biogeographic point of view, the selection of the BeSO should take in consideration the biology and ecology of the BeSO. The soil and environmental conditions in the target soils should match the conditions within the range of the natural occurrence of the BeSO, in order to obtain the desired activity. The soil is a complex medium, with physicochemical and biological interactions that vary over time and space (Ritz and van der Putten, 2012). In the three trial, the general characteristics of the soil were largely similar (Table 2), although unnoticed microhabitat differences might patchily occur and produce internal stochasticity, a factor that is better controlled in any greenhouse experiment where often soils are homogenize first and treatments are confined to smaller experimental units such as pots. In a field experiment, fundamental differences in soil chemistry (acid soils vs. basic soils, presence of micronutrients, etc.) and soil physical properties (texture, pore size, compaction, available water, etc.), should be considered to select the most appropriate BeSO (Schlaeppi and Bulgarelli, 2015). For example, AMF mostly perform better in low nutrient soils (Pellegrino et al., 2012, 2015). Also the effects of AMF on crop productivity are highly dependent on the plant species or genotypes investigated (Lekberg and Koide, 2005): plants and crops with fine roots such as wheat (as in this study) are usually less responsive to AMF compared to species with thicker roots such as red clover (Köhl et al., 2016). Similarly, EPN species have ecological and habitat preferences that are largely determined by texture and moisture of soils (Campos-Herrera et al., 2013, 2016; El-Borai et al., 2016). Soil physico-chemical characteristics can also impact persistence and activity of Pseudomonas species (Natsch et al., 1996; Troxler et al., 2012; Mascher et al., 2014; Imperiali et al., 2017). Hence, locally adapted species might have an advantage in persistence over exotic organisms that are not present in the target soil (Schlaeppi et al., 2016).
In addition to the abiotic soil conditions, BeSO inoculants are also subjected to interactions with the resident soil organism community. The diversity of soil organisms can contribute to buffering, masking and silencing beneficial effects of inoculations. Again, this is a major difference with controlled experiments in the growth chamber or greenhouse where conditions usually limit or simplify the interactions of inoculant BeSO with the naturally present soil organisms and the target crop. Often laboratory or greenhouse experiments are conducted with sterilized soils, with entirely or greatly reduced abundance of native soil organisms. Under field conditions, there are also spatial and temporal differences in these effects on the augmented BeSO. This is particular relevant when considering naturally occurring populations of the BeSO. In our experiments, we observed that the native populations of AMF and EPN were displaced to varying degrees, depending on the BeSO species/population inoculated in the field plots. In agreement with Schlaeppi et al. (2016) and Jaffuel et al. (2017), we also observed that augmented BeSO species that also occurred naturally in the area performed better than those that were not represented or only at low numbers. The fact that virtually no cross-contaminations with inoculants occurred between plots and in many cases the displacement of native populations was corrected by the time of harvest, i.e., returning to the original numbers/presence of native populations, underscore that these introductions have only low and transient impacts on the native populations. Yet, more studies are needed to evaluate the potential long-term impacts of implementing inoculation strategies of single or combined BeSO, especially if inoculants are not native or no present in the area of application (Abate et al., 2017; Hardt et al., 2017).
Here we introduce a comprehensive toolbox to trace Pseudomonads, AMF, and EPN after application. Some of the BeSO did not reach the numbers known to be required to reach beneficial plant effects (Haas and Défago, 2005), did not persist well-after application (i.e., the EPN species H. megidis and S. carpocapsae), or did not establish following field inoculation (i.e., the AMF species F. mosseae and C. claroideum). Nevertheless, results for some isolates and combinations were highly promising. Under the experimental field settings, the bacterium P. protegens CHA0, the AMF R. irregularis and the EPN S. feltiae established very well. Under conditions with high biotic stress (frit fly infestation in the PERFORMANCE-1 trial), the combination of bacterial and EPN inoculants produced the highest yields. Because such ecological conditions will change from one season to another, the development of a pre-application diagnosis tool may help the choice of an optimized BeSO (Schlaeppi and Bulgarelli, 2015; Schlaeppi et al., 2016). For example, areas strongly impacted by plant diseases and pests might benefit from the integration of various Pseudomonas bacteria. Whereas, the presence of insect pests will better support the development and persistence of native and augmented EPN, thereby enhancing their protective effects. Finally, selecting BeOS, in particular AMF, that are compatible with local soil conditions (e.g. low or high nutrient content) is highly advisable (Pellegrino et al., 2015; Schlaeppi et al., 2016).
Advancing our understanding of the soil-plant interface in its broadest sense is critical to achieve sustainable agriculture (Adl, 2016). We evaluated the simultaneous application of three types of BeSO (bacteria, EPN, and AMF) and its impact on wheat productivity under realistic field conditions. While we confirmed the prevalence and persistence of the three organisms throughout the season, their beneficial effects were variable and differed between inoculant strains. Clear beneficial effects on wheat growth were observed only when the plants were exposed to high insect infestation. We learned that there is still a major gap in our understanding of the capacities of BeSO to enhance plant performance under well-controlled conditions and their performance and impacts when applied to the field. We believe that to close this gap and for the successful use of BeSO in agroecosystems there is an urgent need to unravel the context dependency of effective BeSO augmentations. Optimizations should go toward adapting and fine-tuning the selection of inoculant strains that are well-adapted to local abiotic and biotic soil conditions. Advancing such an integrative and context-dependent approach is vital before next-generation, sustainable agriculture, in which field crops are protected by applying beneficial soil organisms instead by agrochemicals becomes imaginable.
Author contributions
KS, MvdH, MM, FM, TT, CK, and RCH planned the experiments and supervised the study. NI, XC, KS, GJ, SB, FD, MF, RBP, DV, MvdH, MM, FM, CK, and RCH contributed in the field experiments and collected the data. NI, XC, KS, MF, CK, and RCH analyzed the data, discussed the main structure and wrote the manuscript. All authors contributed to revisions and commented on previous versions of the manuscript.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
The authors thank Angès Armunday, Dylan Baer, Michele Gusberti, Alain Held, Karent Paola Kupferschmied, Titouan Laessle, Larissa Mysko, Maria Péchy-Tarr, Jana Schneider, and Neil Villard for technical assistance in the field and laboratory. Also, we thank to Steve Breitenmoser, entomologist at Agroscope in Changins, for the identification of the frit fly in our trials in 2014. The project was funded by grants of the Swiss National Research Program 68 on the “Sustainable Use of Soil as a Resource” from the Swiss National Science Foundation (406840_143065 awarded to TT and FM, 406840_143141 awarded to CK and MM, 406840_143097 to MvdH, OF, and WC). GJ was supported by an assistantship from the University of Neuchâtel, and a Swiss Government Excellence Scholarships supported XC for Foreign Scholars. The Government of Portugal supports RCH with an Investigator Program award (IF/00552/2014).
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2017.01809/full#supplementary-material
References
- Abate E., Hussein S., Laing M., Mengistu F. (2017). Soil acidity under multiple land-uses: assessment of perceived causes and indicators, and nutrient dynamics in small-holders' mixed-farming system of northwest Ethiopia. Acta Agric. Scand. Sect. B 67, 134–147. 10.1080/09064710.2016.1230227 [DOI] [Google Scholar]
- Adams B. J., Fodor A., Koppenhöfer H. S., Stackebrandt E., Stock P. S., Klein M. G. (2006). Biodiversity and systematics of nematode-bacterium entomopathogens. Biol. Control 37, 32–49. 10.1016/j.biocontrol.2005.11.008 [DOI] [Google Scholar]
- Adl S. (2016). Rhizosphere food security, and climate change: a critical role for plant-soil research. Rhizosphere 1, 1–3. 10.1016/j.rhisph.2016.08.005 [DOI] [Google Scholar]
- Alkan N., Gadkar V., Yarden Q., Kapulnik Y. (2006). Analysis of quantitative interactions between two species of arbuscular mycorrhizal fungi, Glomus mosseae and G. intraradices, by real-time PCR. Appl. Environ. Microbiol. 72, 4192–4199. 10.1128/AEM.02889-05 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ansari M. A., Shah F. A., Butt T. M. (2010). The entomopathogenic nematode Steinernema kraussei and Metarhizium anisopliae work synergistically in controlling overwintering larvae of the black vine weevil, Otiorhynchus sulcatus, in strawberry growbags. Biocontrol Sci. Technol. 20, 99–105. 10.1080/09583150903420031 [DOI] [Google Scholar]
- Atkins S. D., Clark I. M., Pande S., Hirsch P. R., Kerry B. R. (2005). The use of real-time PCR and species-specific primers for the identification and monitoring of Paecilomyces lilacinus. FEMS Microbiol. Ecol. 51, 257–264. 10.1016/j.femsec.2004.09.002 [DOI] [PubMed] [Google Scholar]
- Bale J. S., van Lenteren J. C., Bigler F. (2008). Biological control and sustainable food production. Philos. Trans. R. Soc. Lond. B Biol. Sci. 363, 761–776. 10.1098/rstb.2007.2182 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bedding R. A., Akhurst R. J. (1975). A simple technique for the detection of insect parasitic rhabditid nematodes in soil. Nematologica 21, 109–110. [Google Scholar]
- Bender S. F., Wagg C., van der Heijden M. G. A. (2016). An underground revolution: biodiversity and soil ecological engineering for agricultural sustainability. Trends Ecol. Evol. 31, 440–444. 10.1016/j.tree.2016.02.016 [DOI] [PubMed] [Google Scholar]
- Berg G. (2009). Plant-microbe interactions promoting plant growth and health: perspectives for controlled use of microorganisms in agriculture. Appl. Microbiol. Biotechnol. 84, 11–18. 10.1007/s00253-009-2092-7 [DOI] [PubMed] [Google Scholar]
- Bertani G. (1951). Studies on lysogenesis. I. The mode of phage liberation by lysogenic Escherichia coli. J. Bacteriol. 62, 293–300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bommarco R., Kleijn D., Potts S. G. (2013). Ecological intensification: harnessing ecosystem services for food security. Trends Ecol. Evol. 28, 230–238. 10.1016/j.tree.2012.10.012 [DOI] [PubMed] [Google Scholar]
- Bustin S. A., Benes V., Garson J. A., Hellemans J., Huggett J., Kubista M., et al. (2009). The MIQE guidelines: minimum information for publication of quantitative real-time PCR experiments. Clin. Chem. 55, 611–622. 10.1373/clinchem.2008.112797 [DOI] [PubMed] [Google Scholar]
- Campos-Herrera R. (2015). Nematode Pathogenesis of Insects and Other Pests - Ecology and Applied Technologies for Sustainable Plant and Crop Protection. Series: Sustainability in Plant and Crop Protection Vol. 1, ed Ciancio A. (Springer; ), 530. [Google Scholar]
- Campos-Herrera R., Ali J. G., Díaz B. M., Duncan L. W. (2013). Analyzing spatial patterns linked to the ecology of herbivores and their natural enemies in the soil. Front. Plant Sci. 4:378. 10.3389/fpls.2013.00378 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campos-Herrera R., El–Borai F. E., Duncan L. W. (2012). Wide interguild relationships among entomopathogenic and free–living nematodes in soil as measured by real time qPCR. J. Invertebr. Pathol. 111, 126–135. 10.1016/j.jip.2012.07.006 [DOI] [PubMed] [Google Scholar]
- Campos-Herrera R., El-Borai F. E., Rodriguez Martin J. A., Duncan L. W. (2016). Entomopathogenic nematode food web assemblages in Florida natural areas. Soil Biol. Biochem. 93, 105–114. 10.1016/j.soilbio.2015.10.022 [DOI] [Google Scholar]
- Campos-Herrera R., El–Borai F. E., Stuart R. J., Graham J. H., Duncan L. W. (2011a). Entomopathogenic nematodes, phoretic Paenibacillus spp, and the use of real time quantitative PCR to explore soil food webs in Florida citrus groves. J. Invertebr. Pathol. 108, 30–39. 10.1016/j.jip.2011.06.005 [DOI] [PubMed] [Google Scholar]
- Campos-Herrera R., Jaffuel G., Chiriboga X., Blanco-Pérez R., Fesselet M., PuŽa V., et al. (2015a). Traditional and molecular detection methods reveal intense interguild competition and other multitrophic interactions associated with native entomopathogenic nematodes in Swiss tillage soils. Plant Soil 389, 237–255. 10.1007/s11104-014-2358-4 [DOI] [Google Scholar]
- Campos-Herrera R., Johnson E. G., El–Borai F. E., Stuart R. J., Graham J. H., Duncan L. W. (2011b). Long–term stability of entomopathogenic nematode spatial patterns measured by sentinel insects and real–time PCR assays. Ann. Appl. Biol. 158, 55–68. 10.1111/j.1744-7348.2010.00433.x [DOI] [Google Scholar]
- Campos-Herrera R., Půža V., Jaffuel G., Blanco-Pérez R., Cepulyte Rakauskiene R., Turlings T. C. J. (2015b). Unraveling the intraguild competition between Oscheius spp. nematodes and entomopathogenic nematodes: implications for their natural distribution in Swiss agricultural soils. J. Invertebr. Pathol. 132, 216–227. 10.1016/j.jip.2015.10.007 [DOI] [PubMed] [Google Scholar]
- Chin-A-Woeng T. F., Bloemberg G. V., van der Bij A. J., van der Drift K. M., Schripsema J., Kroon B., et al. (1998). Biocontrol by phenazine-1- carboxamide-producing Pseudomonas chlororaphis PCL1391 of tomato root rot caused by Fusarium oxysporum f. sp. radicis-lycopersici. Mol. Plant Microbe Interact. 11, 1069–1077. 10.1094/MPMI.1998.11.11.1069 [DOI] [Google Scholar]
- Couillerot O., Ramírez-Trujillo A., Walker V., von Felten A., Jansa J., Maurhofer M., et al. (2012). Comparison of prominent Azospirillum strains in Azospirillum-Pseudomonas-Glomus consortia for promotion of maize growth. Appl. Microbiol. Biotechnol. 97, 4639–4649. 10.1007/s00253-012-4249-z [DOI] [PubMed] [Google Scholar]
- de Rooij-van der Goes P. C. E. M., Putten W. H., Dijk C. (1995). Analysis of nematodes and soil-borne fungi from Ammophila arenaria (Marram grass) in Dutch coastal foredunes by multivariate techniques. Eur. J. Plant Pathol. 101, 149–162. 10.1007/BF01874761 [DOI] [Google Scholar]
- Dillman A. R., Chaston J. M., Adams B. J., Ciche T. A., Goodrich-Blair H., Stock S. P., et al. (2012). An entomopathogenic nematode by any other name. PLoS Pathog. 8:e1002527. 10.1371/journal.ppat.1002527 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duncan L. W., Stuart R. J., El–Borai F. E., Campos-Herrera R., Pathak E., Giurcanu M., et al. (2013). Modifying orchard planting sites conserves entomopathogenic nematodes, reduces weevil herbivory and increases citrus tree growth, survival and fruit yield. Biol. Control 64, 26–36. 10.1016/j.biocontrol.2012.09.006 [DOI] [Google Scholar]
- El-Borai F. E., Killiny N., Duncan L. W. (2016). Concilience in entomopathogenic nematode responses to water potential and their geospatial patterns in Florida. Front. Microbiol. 7:356. 10.3389/fmicb.2016.00356 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Enright M. R., Griffin C. T. (2005). Effects of Paenibacillus nematophilus on the entomopathogenic nematode Heterorhabditis megidis. J. Invertebr. Pathol. 88, 40–48. 10.1016/j.jip.2004.10.002 [DOI] [PubMed] [Google Scholar]
- Flury P., Aellen N., Ruffner B., Péchy-Tarr M., Fataar S., Metla Z., et al. (2016). Insect pathogenicity in plant-beneficial pseudomonads: phylogenetic distribution and comparative genomics. ISME J. 10, 2527–2542. 10.1038/ismej.2016.5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fogliano V., Ballio A., Gallo M., Woo S., Scala F., Lorito M. (2002). Pseudomonas lipodepsipeptides and fungal cell wall-degrading enzymes act synergistically in biological control. Mol. Plant Microbe Interact. 15, 323–333. 10.1094/MPMI.2002.15.4.323 [DOI] [PubMed] [Google Scholar]
- Frey-Klett P., Garbaye J., Tarkka M. (2007). The mycorrhiza helper bacteria revisited. New Phytol. 176, 22–36. 10.1111/j.1469-8137.2007.02191.x [DOI] [PubMed] [Google Scholar]
- Georgis R., Koppenhöfer A. M., Lacey L. A., Bélair G., Duncan L. W., Grewal P. S., et al. (2006). Successes and failures in the use of parasitic nematodes for pest control. Biol. Control 38, 103–123. 10.1016/j.biocontrol.2005.11.005 [DOI] [Google Scholar]
- Giménez M. J., Pistón F., Atienza S. G. (2011). Identification of suitable reference genes for normalization of qPCR data in comparative transcriptomics analyses in the Triticeae. Planta 233, 163–173. 10.1007/s00425-010-1290-y [DOI] [PubMed] [Google Scholar]
- Grewal P. S., Peters A. (2005). Formulation and quality control of entomopathogenic nematodes, in Nematodes as Biocontrol Agents, eds Grewal P. S., Ehlers R. U., Shapiro-Ilan D. (Wallingford: CABI Publishing; ), 79–90. [Google Scholar]
- Haas D., Défago G. (2005). Biological control of soil-borne pathogens by fluorescent pseudomonads. Nat. Rev. Microbiol. 3, 307–319. 10.1038/nrmicro1129 [DOI] [PubMed] [Google Scholar]
- Hardt M. M., Antunes P. M., Abbott L. (2017). Unknown risks to soil biodiversity from commercial fungal inoculants. Nat. Ecol. Evol. 1:0115 10.1038/s41559-017-0115 [DOI] [PubMed] [Google Scholar]
- Hijri M. (2016). Analysis of a large dataset of mycorrhiza inoculation field trials on potato shows highly significant increases in yield. Mycorrhiza 26, 209–214. 10.1007/s00572-015-0661-4 [DOI] [PubMed] [Google Scholar]
- Hol W. H., Bezemer T. M., Biere A. (2013). Getting the ecology into interactions between plants and the plant growth-promoting bacterium Pseudomonas fluorescens. Front. Plant Sci. 10:81 10.3389/fpls.2013.00081 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ijdo M., Cranenbrouck S., Declerck S. (2011). Methods for large-scale production of AM fungi: past, present, and future. Mycorrhiza 21, 1–16. 10.1007/s00572-010-0337-z [DOI] [PubMed] [Google Scholar]
- Imperiali N., Dennert F., Schneider J., Laessle T., Velatta C., Fesselet M., et al. (2017). Relationships between root pathogen resistance, abundance and expression of Pseudomonas antimicrobial genes, and soil properties in representative Swiss agricultural soils. Front. Plant Sci. 8:427. 10.3389/fpls.2017.00427 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaffuel G., Blanco-Pérez R., Büchi L., Mäder P., Fliessbach A., Charles R., et al. (2017). Effects of cover crops on the overwintering success of entomopathogenic nematodes and their antagonists. Appl. Soil Ecol. 114, 62–73. 10.1016/j.apsoil.2017.02.006 [DOI] [Google Scholar]
- Jaffuel G., Mader P., Blanco-Perez R., Chiriboga X., Fliessbach A., Turlings T. C. J., et al. (2016). Prevalence and activity of entomopathogenic nematodes and their antagonists in soils that are subject to different agricultural practices. Agric. Ecosys. Environ. 230, 329–340. 10.1016/j.agee.2016.06.009 [DOI] [Google Scholar]
- Jakobsen I., Abbott L. K., Robson A. D. (1992). External hyphae of vesicular–arbuscular mycorrhizal fungi associated with Trifolium subterraneum L. I: spread of hyphae and phosphorus inflow into roots. New Phytol. 120, 371–380. [Google Scholar]
- Jeffries P., Gianinazzi S., Perotto S., Turnau K., Barea J. M. (2003). The contribution of arbuscular mycorrhizal fungi in sustainable maintenance of plant health and soil fertility. Biol. Fertil. Soils 37, 1–16. 10.1007/s00374-002-0546-5 [DOI] [Google Scholar]
- Jenkins W. R. (1964). A rapid centrifugal-flotation technique for separating nematodes from soil. Plant Dis. Rep. 48, 692. [Google Scholar]
- Johnson S. N., Benefer C. M., Frew A., Griffiths B. S., Hartley S. E., Karley A. J., et al. (2016). New frontiers in belowground ecology for plant protection from root-feeding insects. Appl. Soil Ecol. 108, 96–107. 10.1016/j.apsoil.2016.07.017 [DOI] [Google Scholar]
- Kaya H. K., Aguillera M. M., Alumai A., Choo H. Y., de la Torre M., Foder A., et al. (2006). Status of entomopathogenic nematodes and their symbiotic bacteria from selected countries or regions of the world. Biol. Control 38, 134–155. 10.1016/j.biocontrol.2005.11.004 [DOI] [Google Scholar]
- King E. O., Ward M. K., Raney D. E. (1954). Two simple media for the demonstration of pyocyanin and fluorescein. J. Lab. Clin. Med. 44, 301–307. [PubMed] [Google Scholar]
- Köhl L., Lukasiewicz C. E., van der Heijden M. G. A. (2016). Establishment and effectiveness of mycorrhizal inoculation in agricultural soils. Plant Cell Environ. 39, 136–146. 10.1111/pce.12600 [DOI] [PubMed] [Google Scholar]
- Kupferschmied P., Maurhofer M., Keel C. (2013). Promise for plant pest control: root-associated pseudomonads with insecticidal activities. Front. Plant Sci. 4:287. 10.3389/fpls.2013.00287 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lacey L. A., Grzywacz D., Shapiro-Ilan D. I., Frutos R., Brownbridge M., Goettel M. S. (2015). Insect pathogens as biological control agents: back to the future. J. Invertebr. Pathol. 132, 1–41. 10.1016/j.jip.2015.07.009 [DOI] [PubMed] [Google Scholar]
- Lekberg Y., Koide R. T. (2005). Is plant performance limited by abundance of arbuscular mycorrhizal fungi? A meta-analysis of studies published between 1988 and 2003. New Phytol. 168, 189–204. 10.1111/j.1469-8137.2005.01490.x [DOI] [PubMed] [Google Scholar]
- Lichtfouse E., Navarrete M., Debaeke P., Souchère V., Alberola C., Ménassieu J. (2009). Agronomy for sustainable agriculture. A review. Agron. Sustain. Dev. 29, 1–6. 10.1051/agro:2008054 [DOI] [Google Scholar]
- Mäder P. A., Vierheilig H., Streitwolf-Engel R., Boller T., Frey B., Christie P., et al. (2000). Transport of 15N from a soil compartment separated by a polytetrafluoroethylene membrane to plant roots via the hyphae of arbuscular mycorrhizal fungi. New Phytol. 146, 155–161. 10.1046/j.1469-8137.2000.00615.x [DOI] [Google Scholar]
- Mascher F., Hase C., Bouffaud M. L., Défago G., Moënne-Loccoz Y. (2014). Cell culturability of Pseudomonas protegens CHA0 depends on soil pH. FEMS Microbiol. Ecol. 87, 441–450. 10.1111/1574-6941.12234. [DOI] [PubMed] [Google Scholar]
- Mercado-Blanco J., Bakker P. A. (2007). Interactions between plants and beneficial Pseudomonas spp.: exploiting bacterial traits for crop protection. Antonie van Leeuwenhoek 92, 367–389. 10.1007/s10482-007-9167-1 [DOI] [PubMed] [Google Scholar]
- Natsch A., Keel C., Pfirter H. A., Haas D., Défago G. (1994). Contribution of the global regulator gene gacA to persistence and dissemination of Pseudomonas fluorescens biocontrol strain CHA0 introduced into soil microcosms. Appl. Environ. Microbiol. 60, 2553–2560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Natsch A., Keel C., Troxler J., Zala M., Von Albertini N., Defago G. (1996). Importance of preferential flow and soil management in vertical transport of a biocontrol strain of Pseudomonas fluorescens in structured field soil. Appl. Environ. Microbiol. 62, 33–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oerke E. C. (2006). Crop losses to pests. J. Agric. Sci. 144, 31–43. 10.1017/S0021859605005708 [DOI] [Google Scholar]
- Pathak E., El-Borai F. E., Campos-Herrera R., Johnson E. G., Stuart R. J., Graham J. H., et al. (2012). Use of real-time PCR to discriminate parasitic and saprophagous behaviour by nematophagous fungi. Fungal Biol. 116, 563–573. 10.1016/j.funbio.2012.02.005 [DOI] [PubMed] [Google Scholar]
- Pellegrino E., Opik M., Bonari E., Ercoli L. (2015). Responses of wheat to arbuscular mycorrhizal fungi: a meta-analysis of field studies from 1975 to 2013. Soil Biol. Biochem. 84, 210–217. 10.1016/j.soilbio.2015.02.020 [DOI] [Google Scholar]
- Pellegrino E., Turrini A., Gamper H. A., Cafa G., Bonari E., Young J. P. W., et al. (2012). Establishment, persistence and effectiveness of arbuscular mycorrhizal fungal inoculants in the field revealed using molecular genetic tracing and measurement of yield components. New Phytol. 194, 810–822. 10.1111/j.1469-8137.2012.04090.x [DOI] [PubMed] [Google Scholar]
- Philippot L., Raaijmakers J. M., Lemanceau P., van der Putten W. (2013). Going back to the roots: the microbial ecology of the rhizophere. Nat. Rev. Microbiol. 11, 789–799. 10.1038/nrmicro3109 [DOI] [PubMed] [Google Scholar]
- Pierson E. A., Weller D. M. (1994). Use of mixtures of fluorescent pseudomonads to suppress take-all and improve the growth of wheat. Phytopathology 84, 940–947. [Google Scholar]
- Rasmann S., Turlings T. C. J. (2016). Root signals that mediate mutualistic interactions in the rhizosphere. Curr. Opin. Plant Biol. 32, 62–68. 10.1016/j.pbi.2016.06.017 [DOI] [PubMed] [Google Scholar]
- Ritz K., van der Putten W. H. (2012). The living soil and ecosystem services, in Soil Ecology and Ecosystem Services, eds Wall D. H., Bardgett R. D., Behan-Pelletier V., Herrick J. E., Jones T. H., Ritz K., Six J., Strong D. R., van der Putten W. H. (Oxford: Oxford University Press; ), 5–60. [Google Scholar]
- Ruffner B., Péchy-Tarr M., Ryffel F., Hoegger P., Obrist C., Rindlisbacher A., et al. (2013). Oral insecticidal activity of plant-associated pseudomonads. Enviro. Microbiol. 15, 751–763. 10.1111/j.1462-2920.2012.02884.x [DOI] [PubMed] [Google Scholar]
- Ruijter J. M., Ramakers C., Hoogaars W. M. H., Karlen Y., Bakker O., van den hoff M. J. B., et al. (2009). Amplification efficiency: linking baseline and bias in the analysis of quantitative PCR data. Nucleic Acids Res. 37:e45. 10.1093/nar/gkp045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scanferlato V. S., Orvos D. R., Lacy G. H., Cairns J. (1990). Enumerating low densities of genetically engineered Erwinia carotovora in soil. Lett. Appl. Microbiol. 10, 55–59. [DOI] [PubMed] [Google Scholar]
- Schlaeppi K., Bulgarelli D. (2015). The plant microbiome at work. Mol. Plant Microbe Interact. 28, 212–217. 10.1094/MPMI-10-14-0334-FI [DOI] [PubMed] [Google Scholar]
- Schlaeppi K., Bender S. F., Mascher F., Russo G., Patrignani A., Camenzind T., et al. (2016). High-resolution community profiling of arbuscular mycorrhizal fungi. New Phytol. 12, 780–791. 10.1111/nph.14070 [DOI] [PubMed] [Google Scholar]
- Schueßler A., Schwarzott D., Walker C. (2001). A new fungal phylum, the Glomero- mycota: phylogeny and evolution. Mycol. Res. 105, 1413–1421. 10.1017/S0953756201005196 [DOI] [Google Scholar]
- Sieverding E., Silva G. A., Berndt R., Oehl F. (2015). Rhizoglomus, a new genus of the Glomeraceae. Mycotaxon 129, 373–387. 10.5248/129.373 [DOI] [Google Scholar]
- Smith S. E., Smith F. A., Jakobsen I. (2004). Functional diversity in arbuscular mycorrhizal (AM) symbioses: the contribution of the mycorrhizal P uptake pathway is not correlated with mycorrhizal responses in growth or total P uptake. New Phytol. 162, 511–524. 10.1111/j.1469-8137.2004.01039.x [DOI] [Google Scholar]
- Stockwell V. O., Stack J. (2007). Using Pseudomonas spp. for integrated biological control. Phytopathology 97, 244–249. 10.1094/PHYTO-97-2-0244 [DOI] [PubMed] [Google Scholar]
- Stuart R. J., Barbercheck M. E., Grewal P. S. (2015). Entomopathogenic nematodes in the soil environment. Distributions, interactions and the influence of biotic and abiotic factors, in Nematode Pathogenesis of Insects and Other Pests: Ecology and Applied Technologies for Sustainable Plant and Crop Protection, Vol. 4, Series Sustainability in Plant and Crop Protection No. 1, ed Campos-Herrera (London: Springer Science+Business Media B.V.), 93–130. [Google Scholar]
- Stutz E. W., Défago G., Kern H. (1986). Naturally occurring fluorescent pseudomonads involved in suppression of black root rot of tobacco. Phytopathology 76, 181–185. 10.1094/Phyto-76-181 [DOI] [Google Scholar]
- Tarasco E., Santiago Alvarez C., Triggiani O., Moraga E. Q. (2011). Laboratory studies on the competition for insect haemocoel between Beauveria bassiana and Steinernema ichnusae recovered in the same ecological niche. Biocontrol Sci. Technol. 21, 693–704, 10.1080/09583157.2011.570428. [DOI] [Google Scholar]
- Thonar C., Erb A., Jansa J. (2012). Real-time PCR to quantify composition of arbuscular mycorrhizal fungal communities-marker design, verification, calibration and field validation. Mol. Ecol. Res. 12, 219–232. 10.1111/j.1755-0998.2011.03086.x [DOI] [PubMed] [Google Scholar]
- Troxler J., Svercela M., Natsch A., Zala M., Keel C., Moënne-Loccoz Y., et al. (2012). Persistence of a biocontrol Pseudomonas inoculant as high populations of culturable and non-culturable cells in 200-cm-deep soil profiles. Soil Biol. Biochem. 44, 122–129. 10.1016/j.soilbio.2011.09.020 [DOI] [Google Scholar]
- Vacheron J., Desbrosses G., Bouffaud M. L., Touraine B., Moënne-Loccoz Y., Muller D., et al. (2015). Plant growth-promoting rhizobacteria and root system functioning. Front. Plant Sci. 17:356 10.3389/fpls.2013.00356 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Heijden M. G. A., Martin F. M., Selosse M.-A. A., Sanders I. R. (2015). Mycorrhizal ecology and evolution: the past, the present, and the future. New Phytol. 205, 1406–1423. 10.1111/nph.13288 [DOI] [PubMed] [Google Scholar]
- van der Heijden M. G. A., Streitwolf-Engel R., Riedl R., Siegrist S., Neudecker A., Ineichen K., et al. (2006). The mycorrhizal contribution to plant productivity, plant nutrition and soil structure in experimental grassland. New Phytol. 172, 739–752. 10.1111/j.1469-8137.2006.01862.x [DOI] [PubMed] [Google Scholar]
- Venturi V., Keel C. (2016). Signaling in the rhizosphere. Trends Plant Sci. 21, 187–198. 10.1016/j.tplants.2016.01.005 [DOI] [PubMed] [Google Scholar]
- Walker V., Bertrand C., Bellvert F., Moënne-Loccoz Y., Bally R., Comte G. (2011). Host plant secondary metabolite profiling shows a complex, strain-dependent response of maize to plant growth-promoting rhizobacteria of the genus Azospirillum. New Phytol. 189, 494–506. 10.1111/j.1469-8137.2010.03484.x [DOI] [PubMed] [Google Scholar]
- Walker V., Couillerot O., Von Felten A., Bellvert F., Jansa J., Maurhofer M., et al. (2012). Variation of secondary metabolite levels in maize seedling roots induced by inoculation with Azospirillum, Pseudomonas and Glomus consortium under field conditions. Plant Soil 356, 151–163. 10.1007/s11104-011-0960-2 [DOI] [Google Scholar]
- White G. F. (1927). A method for obtaining infective nematode larvae from cultures. Science 66, 302–303. [DOI] [PubMed] [Google Scholar]
- Yigit F., Dikilitas M. (2007). Control of fusarium wilt of tomato by combination of fluorescent Pseudomonas, non-pathogen Fusarium and Trichoderma harzianum T-22 in greenhouse conditions. Plant Pathol. J. 6, 159–163. 10.3923/ppj.2007.159.163 [DOI] [Google Scholar]
- Zhang L. M., Liu X. Z., Zhu S. F., Chen S. Y. (2006). Detection of the nematophagous fungus Hirsutella rhossiliensis in soil by real-time PCR and parasitism bioassay. Biol. Control 36, 316–323. 10.1016/j.biocontrol.2005.08.002 [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.