Abstract
Small noncoding RNAs play a pivotal role in the regulation of gene expression, and are key regulators of animal development. Freshwater planarian exhibits an extraordinary ability to regenerate any missing body parts, representing an emerging model for studying mechanism underlying stem cell regulation and tissue regeneration.
Here, we utilized next-generation sequencing (NGS) to identify small RNAs that are expressed in planarian adult stem cells, and are implicated in tissue regeneration. We profiled microRNAs (miRNAs), piwi-interacting RNA (piRNAs), small rDNA-derived RNAs (srRNAs) and endogenous interfering RNAs (endo-siRNAs) population from size 18–30 nt, measured the expression of 244 conserved miRNAs, and identified 41 novel miRNAs and 64 novel endo-siRNAs. Expression profiling analyses revealed that most piRNAs and srRNAs are up-regulated during regeneration, and that the most abundantly expressed srRNAs are from 5.8s and 28s rRNA. Furthermore, a target prediction method was adopted to investigate the anti-correlation of small RNAs and mRNA expression. We built up a gene regulatory network based on the genes that are targeted by dynamically changed small RNAs.
These results expand the known small RNA repertoire in planarian, and provide valuable insights and a rich resource for understanding the small RNAs landscape in stem cell-mediated regeneration.
1. Introduction
Freshwater planarians are bilaterally symmetrical flatworms, and are capable of regenerate any missing body parts, from even tiny fragment within a few days. The extraordinary regenerative capability was driven by a population of adult stem cells called neoblasts, which are pluripotent in nature and are sensitive to irradiation treatment [1].
The model planarian Schmidtea mediterranea (S. mediterranea) is diploid, exists in sexual and asexual strains. It is widely chosen for modern molecular biology research and high-throughput sequencing [2]. In addition, hundreds of genes identified in planarian are linked to regeneration and stem cell biology. Many of these genes are conserved in human [3].
Small RNAs have emerged as important regulators of development in both plants and animals. Using combinations of next-generation sequencing (NGS) technologies, small RNA (sRNA) population was profiled in varieties of cell types [4]. In particular, microRNAs (miRNAs) and Piwi-interacting RNA (piRNA) have been implicated in various aspects of biological context. miRNAs are endogenous noncoding RNAs with approximately 18–25 nucleotides (nts) in length, which regulate gene expression through inhibiting translation or promoting degradation of target mRNAs by base pairing with specific mRNA targets [5]. piRNA is another class of small non-coding RNA, which is thought to have germ-line restricted expression in most animals. Recent results show that miRNAs and piRNAs are enriched in planarian neoblasts, and are dynamically expressed during regeneration [6], [7], [8], [9], [10]. Knockdown of key regulators of miRNAs and piRNA pathway revealed pivotal roles of small RNA pathway in regulating regeneration and stem cell function [11], [12], [13].
In addition to the miRNA and piRNA, new classes of small RNAs continue to be discovered due to the technical advancement. For instance, in previous high-throughput sequencing studies, short RNA sequences mapping to ribosomal RNAs (rRNAs) were considered to be degradation products, and therefore been discarded. However, recent study indicated that some small rDNA-derived RNAs (srRNAs) are associated with human disease, such as diabetes, and are involved in regulation of metabolism and other biological processes [14].
Another class of small RNA, endogenous interfering RNAs (endo-siRNAs) were identified in flies and mammals. They are derived from exogenous double-stranded RNA (dsRNA). In addition to suppressing viral infection, they silence selfish genetic elements in the fly somatic cells [15]. They have function in post-transcriptional regulation of transcripts and transposons, and also can transcriptionally silence gene expression [16].
To understand the biological function of small non-coding RNAs in stem cell regulation and tissue regeneration, we devised a strategy to systematically profile critical small RNAs involved in regeneration and stem cell regulation in both sexual and asexual planarian species. We profiled small RNAome before and after amputation from both asexual and sexual strains. Furthermore, to characterize the small RNAs specifically expressed in stem cell population and in differentiated cells, we compared untreated samples to the samples that were treated with two conditions known to drastically eliminate the stem cell population. The first condition is RNAi knockdown of Histone deacetylase 1 (HDAC-1 KD), which is specifically expressed in stem cell compartment, and is required for maintaining stem cell population in previous studies [9], [10]. The second one is one-day after lethal irradiation, which has been shown to specifically eliminate stem cell population [1].
We analyzed small RNA high-throughput datasets and calculated the small RNA expression in these different conditions. Moreover, the network of genes targeted by dynamically-changed small RNAs was also investigated. Taken together, our results comprehensively profiled small RNA species in stem cell-mediated regeneration, representing an important resource for understanding the roles of small RNAs in stem cell regulation and tissue regeneration.
2. Materials and methods
2.1. Planarian cultures and sample preparation
S. mediterranea (smed) CIW4 asexual strain and S2F2 sexual strain were maintained in 1x Montjuic salts as described [17]. Animals were starved for 10 days prior to any experiments. Sample 1 and 2 are from sexual strain of intact and amputated animals. Samples A to D are from asexual strain of intact, amputated, HDAC1 KD and irradiated animals, respectively. Sample 2 and B were transversely cut at the pre- and post-pharyngeal regions. Sample 2 was harvested 72 h after amputation, while B was harvested 36 h after amputation. Sample C was Smed-HDAC1 knockdown (KD), and was harvested after 10 days' RNAi. Sample D was exposed to 10 gray (Gy) of X-ray, and was harvested one day after irradiation.
2.2. Small RNA cloning and sequencing
S. mediterranea total RNA was extracted using Trizol (Invitrogen). The quality and integrity of the total RNA was evaluated by electrophoresis on 1.2% agarose gel followed by Agilent 2100 BioAnalyzer (Agilent). Small RNAs ranging from 18 to 30 nt were gel-purified and ligated to the 3′ adaptor and 5′ adaptor oligonucleotides. Samples were allowed for deep sequencing on the Illumina GAII platform.
2.3. Fluorescence-activated cell sorting
The procedures of fluorescence-activated cell sorting were mainly performed as described [18]. Planarians were diced with a razor blade on ice-cold dishes, and the tissue mash were collected in CMFB (400 mg/L NaH2PO4, 800 mg/L NaCl, 1200 mg/L KCl, 800 mg/L NaHCO3, 240 mg/L glucose, 1% BSA, 15 mM HEPES pH 7.3) supplemented with 1 mg/mL collagenase (Sigma). After digestion for 45 min under agitation at room temperature, cell suspensions were passed through a 35 μm cell-strainer cap (BD Biosciences), and pelleted. Then the cells were stained with Hoechst 33342 (Invitrogen) and propidium iodide and filtered again. Cells were sorted on a MoFlo (Beckman-Coulter), and Hoechst blue versus red plots were used to identify the X1 fraction that is high in DNA content.
2.4. qRT-PCR
qRT-PCR was performed as previously described. Briefly, total RNAs of the regenerating pieces were isolated using TRIZOL (Invitrogen). M-MLV Reverse Transcriptase (Promega) was used to synthesize cDNA from 1 μg of total RNAi. Gene specific primers were designed using Primer3 (http://frodo.wi.mit.edu/primer3/). qPCRs were performed with SYBR Green quantitative PCR master mix (Toyobo Co.) on a quantitative PCR system (7900HT Fast Real-Time PCR System, Applied Biosystems). When detecting miRNA, stem-loop RT primer were used in reverse transcription system first, and each sample was normalized on the basis of its endogenous ura4. Three biological replicates were performed for each group. The relative mRNA expression was plotted with GraphPad Prism.
2.5. Bioinformatics analysis of miRNA data
Reads were aligned to the genome of S. mediterranea using megablast. Draft assembly of the S. mediterranea S2F2 genome was used for sexual S2F2 strain, while SmedAsxl_v1.1 was used for asexual CIW4 strain (https://www.ncbi.nlm.nih.gov/Traces/wgs/?val=AAWT01&display=contigs&page=1,
https://www.ncbi.nlm.nih.gov/Traces/wgs/?val=AUVC01#contigs). Mapped reads were aligned to miRBase Release 20 (http://www.mirbase.org/).
2.6. Comparing sets of piRNAs
Reads ≥ 24 nt were selected as piRNAs candidates. We then mapped our piRNA candidates against the dataset of S. mediterranea piRNA identified by Palakodeti et al. [19] (http://rnajournal.cshlp.org/content/suppl/2008/05/02/rna.1085008.DC1/Supplemental_Table_1.pdf).
2.7. Annotating rRNA and mRNA gene
First, we obtained all available rRNA sequences from ten flatworms in GenBank: Dugesia gonocephala, Dugesia sicula, Dugesia tigrina, Dugesia japonica, Neppia sp., Spathula alba, Dugesia ryukyuensis, Dolichoplana sp., Geoplana ladislavii and Novibipalium venosum (https://www.ncbi.nlm.nih.gov/nuccore/DQ666002.1, https://www.ncbi.nlm.nih.gov/nuccore/DQ665965.1, https://www.ncbi.nlm.nih.gov/nuccore/KF308693.1, https://www.ncbi.nlm.nih.gov/nuccore/DQ665969.1, https://www.ncbi.nlm.nih.gov/nuccore/AF013157.2,
https://www.ncbi.nlm.nih.gov/nuccore/AY216702.1, https://www.ncbi.nlm.nih.gov/nuccore/D83382.1, https://www.ncbi.nlm.nih.gov/nuccore/DQ665966.1, https://www.ncbi.nlm.nih.gov/nuccore/DQ665999.1, https://www.ncbi.nlm.nih.gov/nuccore/DQ665982.1, https://www.ncbi.nlm.nih.gov/nuccore/DQ665991.1, https://www.ncbi.nlm.nih.gov/nuccore/DQ666006.1, https://www.ncbi.nlm.nih.gov/nuccore/DQ665968.1, https://www.ncbi.nlm.nih.gov/nuccore/AF050433.1, https://www.ncbi.nlm.nih.gov/nuccore/DQ666003.1, https://www.ncbi.nlm.nih.gov/nuccore/DQ665971.1, https://www.ncbi.nlm.nih.gov/nuccore/DQ666005.1, https://www.ncbi.nlm.nih.gov/nuccore/DQ665975.1, https://www.ncbi.nlm.nih.gov/nuccore/DQ665981.1, https://www.ncbi.nlm.nih.gov/nuccore/DQ666019.1). Then, we blasted these sequences against the sexual and asexual S. mediterranea genome [2]. The matched sequences were annotated and further compiled into a S. mediterranea rRNA database. Planarian expressed sequence tag (EST) sequences were used for annotating mRNA. In case of some reads mapped to more than one type of annotation. The following priority rule was used: miRNA > mRNA > srRNA > piRNA > endo-siRNA.
2.8. Normalizing RNA-Seq data
Normalizatiaon (TPM): normalized expression = actual miRNA count/total count of clean reads × 1,000,000. Here, unique reads refer to different types of reads, and redundant reads refer to total reads.
2.9. Novel miRNA prediction
Small RNA sequences were mapped to the genome and then each exact sequence that match along 100 bases in either flanking side was fetched. mfold was further utilized (http://mfold.rna.albany.edu/?q=mfold/RNA-Folding-Form) to predict the secondary structure. If: 1) the read is 18–25 nt long, 2) its precursor sequence could form a stem-loop structure, and 3) it has not been registered in miRBase Release 20, we considered it as a novel miRNA.
2.10. Novel endo-siRNA prediction
Reads were aligned themselves by using megablast. We only fetched the plus/minus paired reads. Candidates were mapped to the transcriptome SRR955099 and SRR955511. By using mfold, the secondary structure of transcriptome was predicted. The paired reads so as to leave 3′, two-nucleotide overhangs, and on the long double-stranded transcriptomes was identified as novel endo-siRNAs.
2.11. Network construction
The differential expression of miRNAs was calculated by DeSeq software (http://bioconductor.org/packages/release/bioc/html/DESeq.html). By using miRanda (http://cbio.mskcc.org/miRNA2003/miranda.html) software, we predicted the targets of small RNAs. The regulatory networks were constructed by cytoscape (http://www.cytoscape.org/).
3. Results
3.1. High-throughput sequencing and annotation of small RNA in different samples
3.1.1. Small RNA sequencing and mapping
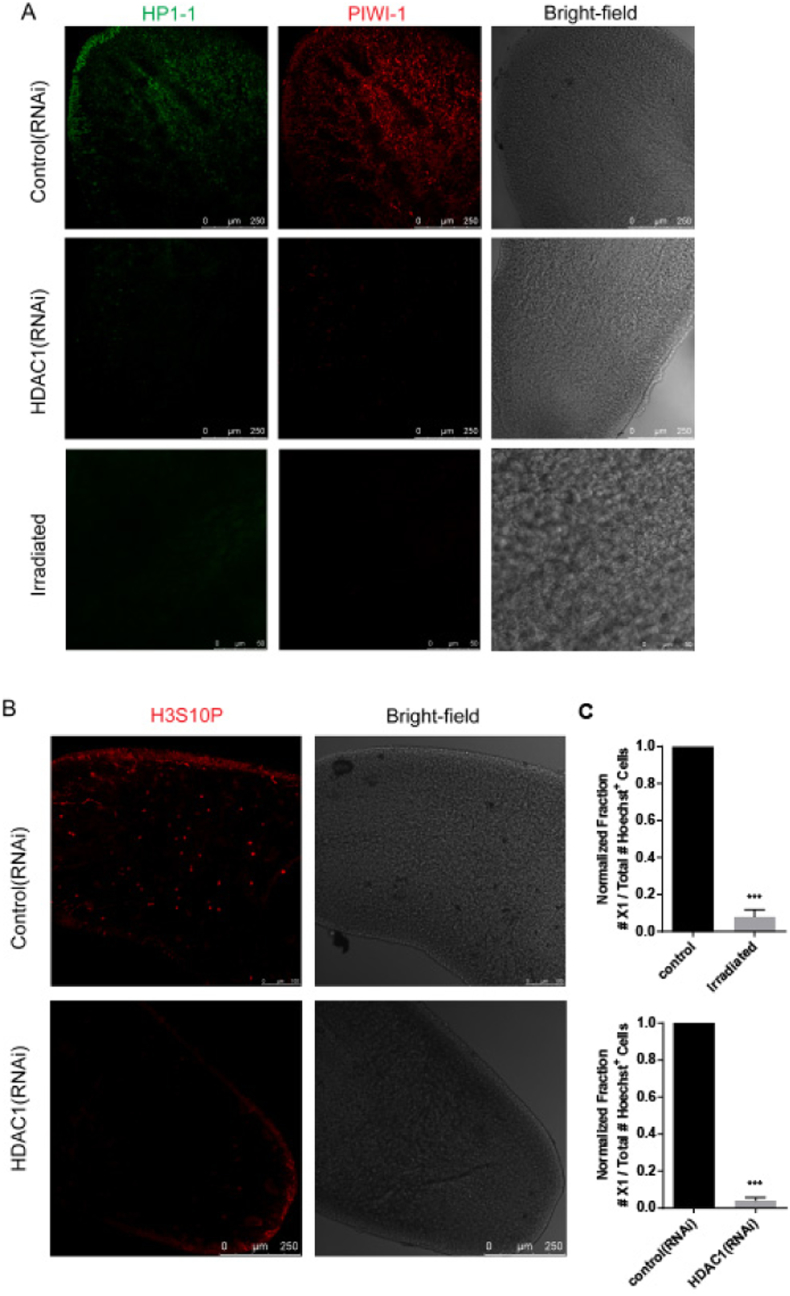
To comprehensively profile expression differences of small RNAs, we prepared samples from intact animals of both sexual and asexual biotypes. In order to identify small RNAs that are enriched in stem cells, and are dynamically expressed in regeneration, we also prepared the animals devoid of neoblasts with either RNAi knockdown of HDAC1 or irradiation treatment, and animals in which stem cells are maximally activated with injury. Immunostaining of stem cell markers, including HP1-1, PIWI-1 and H3S10P, showed that stem cell population were drastically removed after HDAC1 knockdown or irradiation (Fig. 1A and B). It was also confirmed by flow cytometry analyses of the neoblast number following irradiation or HDAC1 RNAi (Fig. 1C), which is consistent with previous report [20]. Given that regenerative response in S. mediterranea involves two mitotic waves, and the maximal stem cell response happens after 48 h of amputation, we harvested samples at 72 h of amputation [21]. Therefore, totally six small RNA libraries were generated and subjected to sequencing (Table S1A).
Fig. 1.
Stem cell depletion by HDAC1 RNAi and irradiation. (A) Immunostaining of stem cell marker expression with PIWI-1 and HP-1 antibodies after gene known-down or irradiation. (B) Immunostaining of mitotic marker H3S10P after gene known-down. (C) The number of neoblasts decreased following irradiation or HDAC1 RNAi. Error bars represent SD; ⁎⁎⁎equals p < 0.0001; significance determined with Student's t-test.
Numbers of high quality reads are 7,801,678, 7,946,054, 14,045,499, 12,921,705, 13,267,244, 12,279,559, which correspondingly represent 6,876,211, 6,954,731, 10,856,210, 9,610,090, 10,115,706, 9,388,917 small RNA reads. These data show that an unprecedented depth of small RNA profile was obtained. After blasting against the planarian S. mediterranea genome, we obtained 4,139,073, 3,801,369, 6,949,694, 6,464,729, 6,256,702, 6,294,189 reads that can be found in the genome locus. Among these reads, 17.1%, 26.3%, 21.3%, 18.8%, 42.1% and 29.5% were mapped to the rfam (ribosomal fragments), whereas 71.9%, 56.4%, 53.6%, 59.9%, 31.1% and 45.6% can be mapped to the miRNAs. These data indicate that miRNAs and srRNAs are two major small RNA populations in S. mediterranea.
Table S1B shows the annotation of unique reads. Surprisingly, those corresponding to unannotated reads are relatively high (87.9%, 87.6%, 71.2%, 71.0%, 70.4%, 71.1%), suggesting that there are still a large number of unannotated small RNAs species.
3.1.2. Length distribution of small RNAs and miRNAs
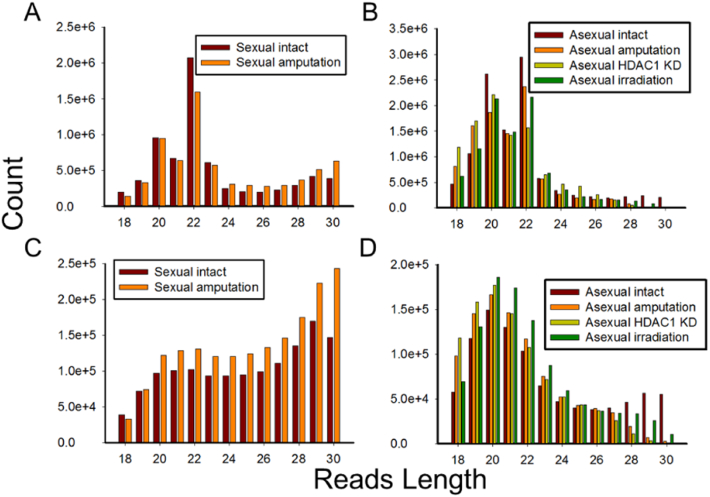
Length distribution of total redundant small RNAs of sexual and asexual S. mediterranea are shown in Fig. 2A and B, which exhibit two distinct peaks at nucleotide 20 and 22. Majority of small RNAs are between 19 and 22 nt in length, corresponding to the size of miRNAs. Length distribution of unique small RNAs is shown in Fig. 2C and D. In sexual samples, reads have highest expression at 29 and 30 nt, whereas in asexual samples, they are 19 to 21 nt, suggesting there are more varieties of small RNAs around 29 nt in sexual samples, typically the size of piRNAs.
Fig. 2.
Length distribution of small RNAs. (A) Redundant small RNAs in sexual intact and amputation samples. (B) Redundant small RNAs in asexual intact, amputation, HDAC1 KD and irradiation samples. (C) Unique small RNAs in sexual intact and amputation samples. (D) Unique small RNAs in asexual intact, amputation, HDAC1 KD and irradiation samples.
3.2. Expression profile of miRNAs during regeneration and after stem cell depletion
3.2.1. Normalized miRNAs reads counts
All annotated miRNAs measured by reads counts are displayed in Table S2. Notably, of 257 known S. mediterranea miRNAs deposited in miRBase, 244 mature were sequenced at least once, as well as 216 were detected in at least five of the six small RNA libraries. These samples employ almost the entire repertoire of the known miRNAs. Top 20 of the most abundantly expressed miRNAs were listed in Table S3.
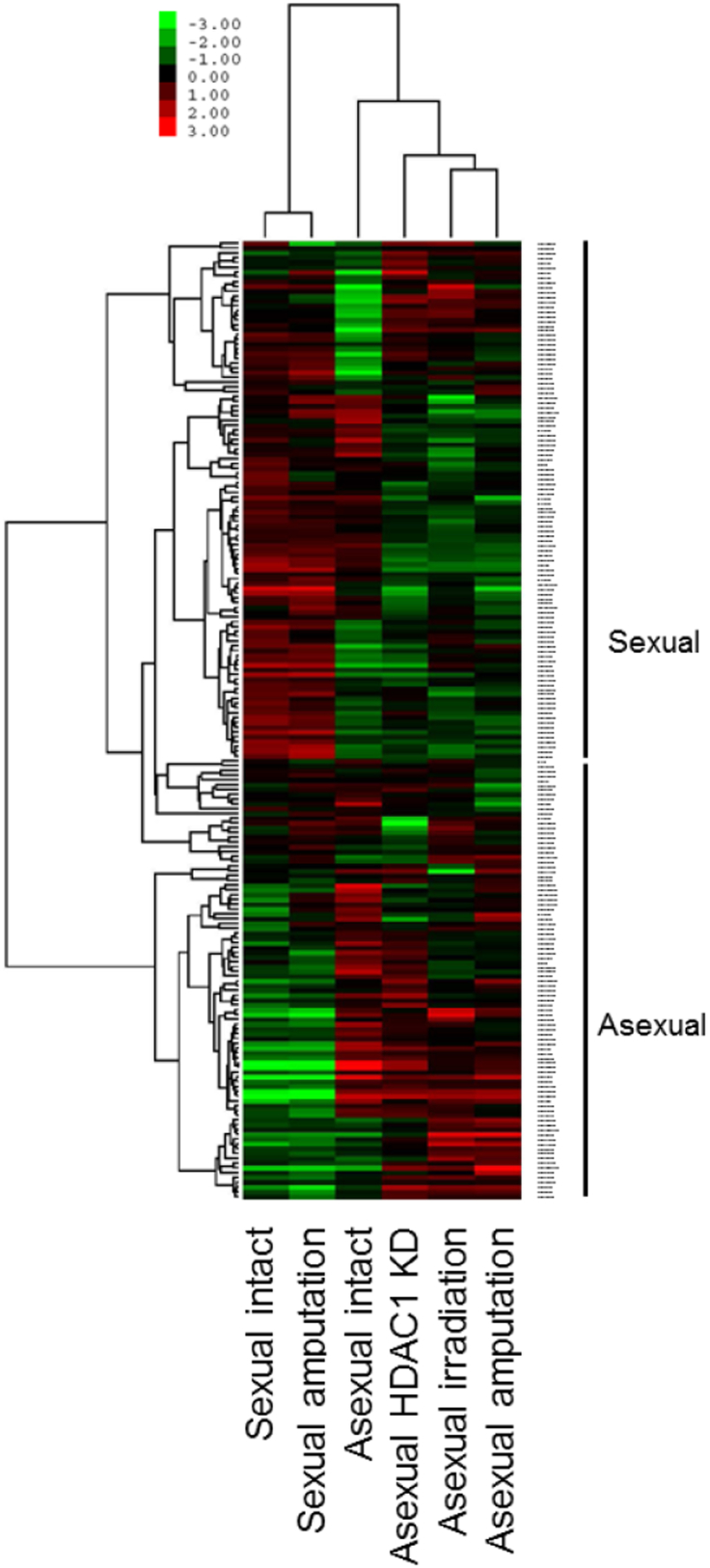
Heatmap made by the total normalized miRNA was presented in Fig. 3. Two prominent classes of genes were revealed by the most highly expressed miRNAs: sexual and asexual specific. Among asexual samples, amputated was closer to irradiated sample.
Fig. 3.
Heatmap of total miRNAs in sexual intact, amputation; asexual intact, HDAC1 KD, irradiation and amputation samples.
3.2.2. Differentially expressed miRNAs in different groups
We further used the program DeSeq [22] to normalize counts and test for differential expression of small RNAs. Normalized miRNAs in different groups were listed in Table S4. Here, | log2 fold change | ≥ 0.73 was selected as the criteria for up or down-regulated, and small RNAs with minimum reads ≥ 5000 were used for further analysis. Heatmaps of differentially expressed small RNAs were shown in Fig. S1.
In sexual amputated samples, miR-277d-5p was up-regulated; however, miR-36a, c-3p and miR-125a-5p were down-regulated (Fig. S1A).
As neoblasts are the only proliferative cell type in asexual planarian, here we focused on asexual samples [23]. miR-13-3p, miR-2157-5p and miR-31a-5p were up-regulated, whereas miR-125a-5p, miR-190a-3p and bantam-a were down-regulated upon amputation (Fig. S1B). miR-125a-5p were down-regulated upon amputation in both sexual or asexual samples, suggesting that it may target genes that are functional essential for regeneration.
After HDAC1 knockdown, the number of stem cell in planarian decreased significantly, and planarian failed to maintain tissue homeostasis and to regenerate missing body parts (Fig. 1). miR-2157-5p, miR-190a-5p and miR-13-3p were up-regulated, on the other hand, miR-125a-5p, miR-71c-5p, bantam-a and let-7d were down-regulated in HDAC1 KD sample (Fig. S1C). We further subjected animals to irradiation, another treatment that could specifically eliminate stem cells. One day after irradiation, miR-2157-5p, miR-31a-5p, miR-13-3p were up-regulated; while miR-125a-5p, miR-190a-3p, bantam-a and let-7d were down-regulated (Fig. S1D).
As shown in heatmap (Fig. S1), in both treatments where stem cell been depleted, miR-2157-5p and miR-13-3p were up-regulated, whereas miR-125a-5p, bantam-a and let-7d were down- regulated, suggesting that these miRNAs are highly enriched in stem cells, and may target functional important genes in stem cells. However, the expression profiles of small RNAs from asexual HDAC1 KD and the asexual irradiation are different, possibly reflecting the differences of those two treatments on stem cell regulation. Irradiation may directly kill neoblasts as observed by Pellettieri [24], while HDAC1 RNAi initially inhibits differentiation process, and finally indirectly leads to stem cell depletion, as suggested in our previous studies [17].
3.3. Scatter plots of miRNAs in different groups
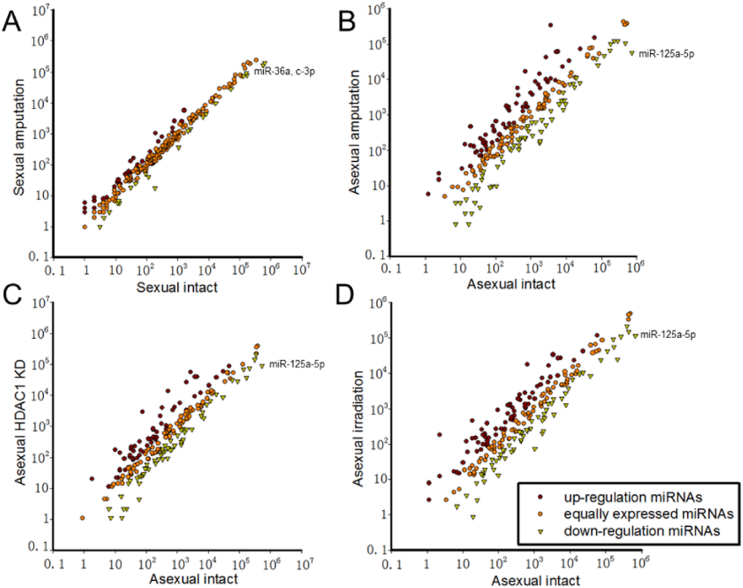
By using DeSeq software, we analyze the changes of miRNAs in different groups. We reason that the highly expressed miRNA should have more important function [25]. Here, | log2 fold change | ≥ 0.73 was selected as the criteria for selection [25]. Scatter plots for the four groups was generated (Fig. 4).
Fig. 4.
Scatter plots of miRNAs in different samples. (A) Sexual amputation compared with intact sample. (B) Asexual amputation compared with intact sample. (C) Asexual HDAC1 knockdown compared with intact sample. (D) Asexual irradiation compared with intact sample.
In sexual sample, among 226 miRNAs detected, 37 (16.4%) were up-regulated and 24 (10.6%) were down-regulated upon amputation. Most miRNAs did not show significant changes between two samples. The most abundant miRNAs, miR-36a, c-3p, were down-regulated; suggesting that it may have important function in sexual regeneration (Fig. 4A).
In asexual samples, about 29.8% of miRNAs were up-regulated and 30.7% were down-regulated upon amputation. The highest expressed miR-125a-5p was down-regulated, suggesting they may play a vital role in asexual regeneration (Fig. 4B). Interestingly, the extent of miRNA change in asexual is greater than in sexual strain, highlighting the unappreciated differences between different strains of S. mediterranea.
HDAC1 knockdown leads to 27.9% of miRNAs up-regulated, while 30.3% down-regulated. However, there were 31.6% miRNAs up-regulated and 30.2% down-regulated after irradiation. The highest expressed miR-125a-5p was down-regulated, hinting that they may have important role in stem cells (Fig. 4C and D). The list of all up-regulated and down-regulated miRNAs is shown in Table S4.
3.4. Target prediction and network of down and up-regulated miRNA
3.4.1. Target prediction of down and up-regulated miRNA
miRNAs silence mRNA through base-pairing with complementary sequences within mRNA molecules; therefore, there is a negative correlation between miRNA and target mRNA expression. Open source software miRanda was used to predict the targets of those differentially expressed miRNAs [25]. The changes of mRNAs were based on a recent microarray gene expression profile generated in our lab. Here, only the highly expressed miRNAs were selected for analysis. We only focused on up-regulated miRNA vs. down-regulated ESTs, and vice versa. The correlation between miRNA and ESTs is shown in Table S5.
By using Blastx, we searched database of Non-redundant protein sequences (nr) and annotated the matched ESTs (Table S6). In sexual and asexual amputated samples, histone H2B and H3 were up-regulated, whereas lectin was down-regulated, suggesting these mRNAs may have function in regeneration.
In asexual strain with HDAC1 knockdown or irradiation treatment: DjGluR1 and GLI pathogenesis related-1were up-regulated; whereas serine protease inhibitor-1, Rap55, TPA, etc. were down-regulated, indicating these mRNAs may play important roles in stem cell function.
Together, these results suggest that many mRNAs are altered in response to treatment, and the expression of miRNAs show strong anti-correlation with mRNA.
3.4.2. Networks of down and up-regulated miRNA targets
We further built up gene regulatory networks based on the dynamically expressed miRNAs and their predicted targets (Fig. S2). In sexual amputated sample, Pax6A and Runt-like 1 were targeted by down-regulated miR-2157-5p, let-7d (Fig. S2A), suggesting up-regulation of these genes are essential for regeneration, which is consistent with the published results [26]. In asexual regenerating sample, zinc finger protein was targeted by down-regulated miR-190a-3p, while lectin-like protein was targeted by up-regulated miR-13-3p; implying these genes many have biological function in regeneration (Fig. S2B).
MFS transporter DHA1 family solute carrier family 18 was targeted by down-regulated miR-71c-5p in asexual HDAC1 knockdown sample, whereas collagen alpha-1(IV) chain was targeted by down-regulated miR-190a-3p in asexual irradiated sample (Fig. S2C and D), suggesting that they may play a vital role during stem cell depletion. Moreover, we verified the dynamic change of some small RNA targets by qRT-PCR. Following HDAC RNAi, miRNA let-7a-5p expression decreased dramatically, meanwhile, the expression of its two targets gi | 116035649 and gi | 84606196 increased significantly (Fig. S2E), suggesting that our sequencing data could be used for identifying novel networks controlling regeneration.
3.5. Prediction of novel miRNAs by mfold
Deep sequencing of small RNAs is especially effective in the discovery of novel transcripts. To identify novel miRNAs, we further analyzed unannotated small RNAs with mfold. Small RNAs with at least 100 reads were counted. Finally, totally 41 novel miRNAs were identified. These novel miRNAs show a typical hairpin loop secondary structure as shown in Fig. S3. The discovery of novel miRNAs suggests that there are still some lowly-expressed miRNAs remained to be discovered. Sequences of these novel miRNAs were shown in Table S7.
3.6. Expression profile of piRNAs during regeneration and after stem cell depletion
Of the 4500 piRNAs previously reported in the S. mediterranea piRNA datasets, we found that there are 1324 (30%) were recovered in our dataset, possibly because we have restricted our small RNA library to the size of 18 to 30 nts, which could have removed most of piRNAs longer than 30 nts. Expression profiling of piRNAs in six datasets were listed in Table S8. Top 20 of the most abundant piRNAs were presented in Table S9.
In both sexual and asexual strains, smed_1612_0_1, smed_6643_0_1 and smed_6807_0_12 were up-regulated after amputation. The genome locations and fold change of the abundantly expressed piRNAs are listed in Table S10. As expected, most of piRNAs could map to more than one chromosomal region, indicating that they are from repeat sequences. Our expression profiling demonstrated that piRNAs are also abundantly expressed in S. mediterranea.
3.7. Scatter plots of piRNAs in different groups
Scatter plots of piRNAs were shown in Fig. 5. Interestingly, in sexual amputated sample, 159 (67.7%) out of 235 piRNAs were up-regulated, and none of them was down-regulated (Fig. 5A). The highest expressed piRNA smed_1612_1_0 was up-regulated after amputation. These data suggest that most of piRNAs were up-regulated in sexual strain of regenerating sample.
Fig. 5.
Scatter plots of piRNAs in different samples. (A) Sexual amputation compared with intact sample. (B) Asexual amputation compared with intact sample. (C) Asexual HDAC1 knockdown compared with intact sample. (D) Asexual irradiation compared with intact sample.
In asexual regenerating sample, about 38.7% piRNAs were up-regulated; while 37.4% were down-regulated. (Fig. 5B). In asexual HDAC1 knockdown and irradiated sample, 50.2% and 54.5% piRNAs were up-regulated, while 35.3% and 27.2% were down-regulated. The most abundant piRNAs, smed_1612_0_1 in HDAC1 knockdown sample and smed_8238_1_0 in irradiated sample, were both up-regulated (Fig. 5C, D).
The scatter plots are more discrete in asexual samples compared to sexual sample, suggesting that, after amputation, the change of piRNAs in asexual samples is greater than in sexual sample. Most of piRNAs were up-regulated after injury, suggesting that they might play important regulatory roles. The list of the up-regulated and down-regulated piRNAs is shown in Table S11.
3.8. Networks of piRNAs targets
Targets of up-regulated piRNAs were predicted by miRanda. Regulatory networks of up-regulated piRNAs were constructed in Fig. S4. TPA: Y box protein 4-like protein was targeted by smed_6643_0_1 and smed_6807_0_12 in both sexual and asexual amputated samples, indicating these genes were down-regulated during the process of regeneration (Fig. S4A and B).
EST 84589886, etc. were targeted by up-regulated smed_6643_0_1 and smed_6807_0_12 in asexual HDAC1 knockdown sample (Fig. S4C). Tubulin alpha chain was targeted by smed_20261_0_1 in asexual irradiated sample (Fig. S4D). These data imply that these genes were down-regulated after stem cell depletion. The correlation between piRNA and their target ESTs is shown in Table S12.
3.9. Expression profile of srRNAs in different samples
Recent study shows that the expression of some srRNAs is associated with disease, and srRNAs have potential functions in biological and pathological processes [14]. Here, we investigated the expression profiling and fold change of srRNAs in different groups (Table S13). The srRNAs were named according to their position in ribosomal RNA and length.
Table S14 shows top 20 of the most abundantly expressed srRNAs. Expression profiling of these srRNAs were shown in Fig. S5. Fig. S5A displays the srRNAs in sexual sample, while Fig. S5B shows asexual samples. sme5.8s-1610-20 and sme5.8s-1610-19 are the most abundant srRNAs in all samples. Most of srRNAs are from 5.8s and 28s rRNAs, and are up-regulated after amputation. To our knowledge, these data demonstrated, for the first time, that srRNA is a novel small RNA class highly enriched in S. mediterranea.
3.10. Analysis of srRNAs in different samples
3.10.1. Positions of srRNAs in different type of ribosomal RNA
Positions of srRNAs in different type of ribosomal RNAs were shown in Fig. 6. In sme5.8s, the highest expressed srRNAs in all samples are sme5.8s-1610-20. Moreover, they have similar expression pattern in the genome loci (Fig. 6A). In sme28s, gti28s and nve18s, the highest expressed srRNAs in asexual samples after wounding are the same. They are sme28s-1554-20, gti28s-531-18 and nve18s-1185-20, respectively (Fig. 6B–D). All these srRNAs displayed similar distribution pattern.
Fig. 6.
Positions of srRNAs in different type of ribosomal RNAs. (A) sme5.8s. (B) sme28s. (C) gti28s. (D)nve18s.
These data suggest that srRNAs are not mainly from random degradation, rather come from specific genomic loci and changed dynamically during the process of regeneration and stem cell depletion.
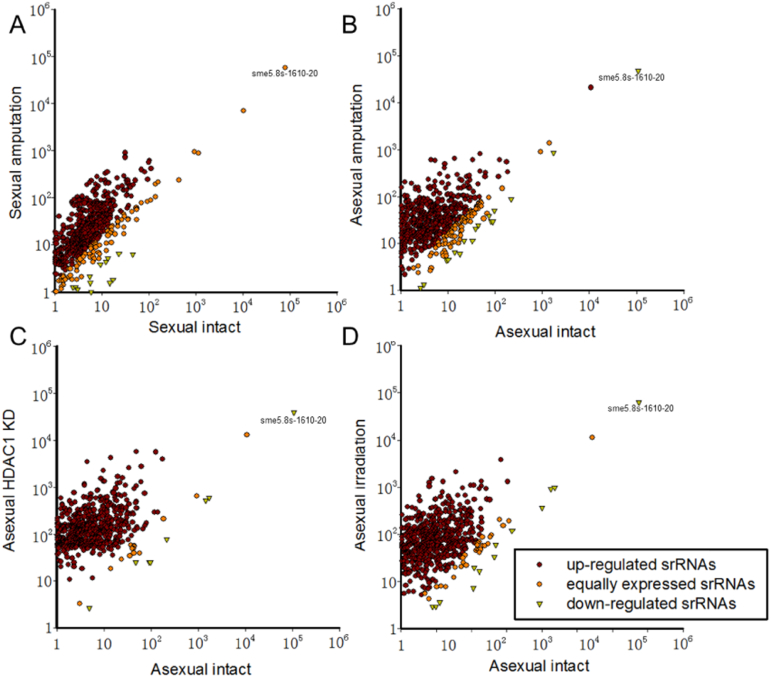
3.10.2. Scatter plots of srRNAs in different samples
Scatter plots of srRNAs were shown in Fig. 7. In sexual and asexual amputated samples: 590 (82.2%) and 610 (84.7%) out of 720 srRNAs are up-regulated, while 4.6% and 2.7% were down-regulated after amputation. The highest expressed srRNA sme5.8s-1610-20 remains unchanged in sexual strain, but is down-regulated in asexual strain (Fig. 7A and B).
Fig. 7.
Scatter plots of srRNAs in different samples. (A) Sexual amputation compared with intact sample. (B) Asexual amputation compared with intact sample. (C) Asexual HDAC1 knockdown compared with intact sample. (D) Asexual irradiation compared with intact sample.
In asexual HDAC1 knockdown and irradiated samples, about 96.5% and 93.6% were up-regulated, while 1.1% and 1.9% were down-regulated after amputation. The most abundant srRNA sme5.8s-1610-20 was down-regulated in both these conditions (Fig. 7C and D).
> 80% of srRNAs were up-regulated, while < 5% were down-regulated, indicating that most srRNAs were up-regulated after wounding. The obvious abundantly expressed srRNAs implied that they may have biological function in regeneration. The list of up-regulated and down-regulated srRNAs was shown in Table S15.
3.11. Networks of srRNAs targets
Targets of the altered srRNAs were predicted by miRanda. Regulatory networks of differentially expressed srRNAs targets were further built (Fig. S6). In sexual and asexual amputated samples, only sme5.8s-1612-18 was down-regulated. Most of other srRNAs were up-regulated, suggesting that their target genes were down-regulated during regeneration (Fig. S6A and B).
S1P and beta-tubulin were targeted by up-regulated gti28s-1554-22 in sexual regenerating sample (Fig. S6A). Bmp-like protein was targeted by up-regulated nve18s-1185-20 in asexual regenerating sample (Fig. S6B). All of these genes may play a pivotal role in regeneration. Piwi-like protein 1 was targeted by up-regulated sme28s-935-19 in asexual HDAC1 knockdown and irradiated samples (Fig. S6C, D). All of these genes may have important function to support stem cell function, as demonstrated in previous studies [12]. The correlation between srRNA and their target ESTs were shown in Table S16.
3.12. Expression profile of siRNAs in different samples
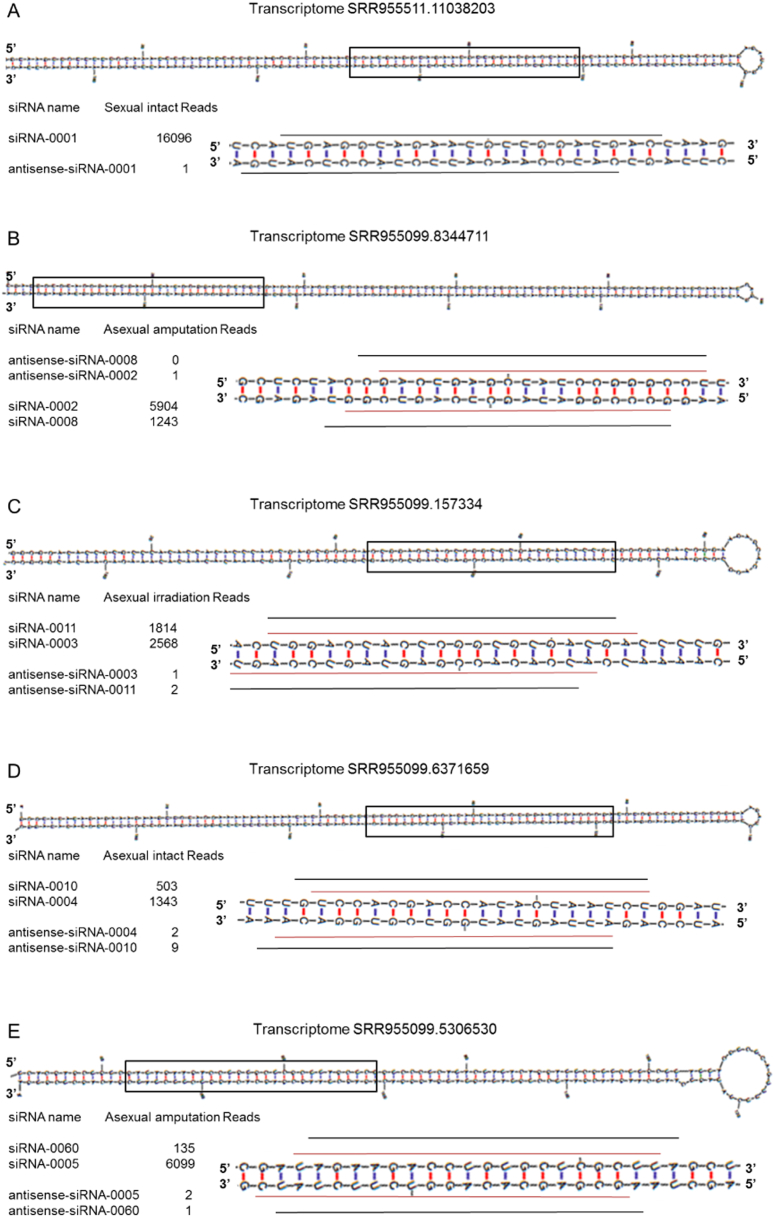
Somatic cells produce siRNAs from exogenous double-stranded RNA (dsRNA) as a defense mechanism against viral infection. The secondary structures of detected endogenous siRNAs (reads number > 200) were shown in Fig. 8 and Fig. S7. All of these siRNAs duplexes derive from long dsRNA, comprise two ~ 21 nt strands, paired so as to leave 3′, two-nucleotides overhangs. The most abundantly expressed siRNA-0001 is derived from transcript SRR955511.11038203.
Fig. 8.
The secondary structure of endogenous siRNA. (A) siRNA-0001 on transcriptome SRR955511.11038203. (B) siRNA-0002, 0008 on transcriptome SRR955099.8344711. (C) siRNAs-0003, 0011 on transcriptome SRR955099.157334. (D) siRNA-0004, 0010 on transcriptome SRR955099.6371659. (E) siRNA-0004, 0060 on transcriptome SRR955099.5306530.
siRNA sequences, antisense sequences and normalized reads counts were shown in Table S17. siRNA-0001 was down-regulated in both asexual HDAC1 knockdown and irradiated samples, suggesting that it's enriched in stem cells. siRNA-0002 and 0003 were down-regulated, and siRNA-0004 were up-regulated after wounded in all the samples. siRNA-0006, 0009 and 0012 were only expressed in asexual HDAC1 knockdown sample. Therefore, the data suggests that endo-siRNAs are not exclusively existed in C. elegans and flies, but also are expressed in S. mediterranea. The expression level of different endo-siRNAs also varies in different conditions, implying their biological significance.
3.13. Networks of endogenous siRNAs targets
Endo-siRNAs complementary to messenger RNAs (mRNAs). These siRNAs mapped to the complementary regions of overlapping mRNAs predicted to form double-stranded RNA in vivo [15]. By using miRanda, we predict the targets of differentially expressed siRNAs.
In sexual amputated sample, EST 21308675 was targeted by down-regulated siRNA-0004 (Fig. S8A). In asexual amputated sample, Src substrate cortactin was targeted by the down-regulated siRNA-0007. HDAC1 knockdown and irradiated samples, suggesting these proteins may have critical functions in regeneration and stem cell depletion (Fig. S8B, C and D).
4. Discussion
Planarian S. mediterranea exhibits an extraordinary ability to regenerate lost body parts. Here, we comprehensively profiled small RNAome before and after tissue amputation, aiming at identifying small RNA molecules that are implicated in regeneration [27]. By comparing small RNAome in different conditions, we measured the expression of known and novel small RNA species, including miRNA, piRNA, srRNA and endo-siRNA in both sexual and asexual strains.
4.1. Identification and characterization of conserved small RNA and candidate novel small RNAs in planarian S. mediterranea
The small RNAs reads in our study were trimmed for 30 nt in length. In Fig. 2, the prominent ~ 22 nt peak represents miRNAs, and the size distribution of the srRNAs was mainly peaked in 20–22 nt [14]. piRNAs are ranged from 24 to 32 nt in length [27]. Although 31 and 32 nt piRNAs are not retrieved, most of the other small RNAs were included in this study. Endo-siRNAs are around 21 nt in length, and our data indicated that novel endo-siRNAs are ranged from 18 to 26 nt.
piRNA, srRNA and siRNA are recently found to have biological function in gene regulation. Our data suggested that these three classes of small RNAs species might be functionally involved in stem cell-mediated regeneration, which requires complex regulation of gene expression. Functions of these small RNAs species can be further studied by knockdown.
4.2. Dynamic regulation of small RNA expression during stem cell depletion and stem cell activation
miR-36a and miR36c-3p have the highest reads counts in our dataset, and they are both down-regulated after amputation. Recent work shows that it was down-regulated after irradiation, and miR-36 family exists in helminthes including B. malayi [5], [28], suggesting that they regulate the parasitic life cycle. Knocking down of miR-36, by anti-miRNAs for example, in regeneration will therefore provide important insights into the role of this family of miRNAs.
miR-13-3p, miR-124 families and miR-31-5p were up-regulated in asexual strain after amputation. Interestingly, the miR-13 family has been found to be protostome-specific, and inversely correlated with target gene expression in Drosophila [29], [30]. miR-124 is highly conserved from worm to human; it also controls gene expression in the sensory nervous system of Caenorhabditis elegans [31], [32]. miR-31 is a key regulator for promoting keratinocyte proliferation and migration during wound healing, and it functions as a tumor suppressor [33], [34].
miR-125-5p and let-7 families were all down-regulated in all four groups after wounding. miR-125-5p regulates the activation of macrophages and inflammation; it is a prognostic biomarker in breast and lung cancer [35], [36], [37]. The expression of let-7 is down-regulated in many cancer types and it was characterized as a tumor suppressor [38].
miR-bantam, miR-190 and miR-10b-5p were all down-regulated in asexual strain after amputation and irradiation. Bantam facilitates cellular proliferation in the hematopoietic system of Drosophila [39]. miR-190 enhances cell survival by preventing apoptosis and relieving G0/G1 cell cycle, and significantly suppresses tumor metastasis [40], [41]. miRNA-10b promotes migration and invasion in human cancer [42]. The clusters of miR-71 were conserved from planarian to parasitic flatworms and were expanded in free-living S. mediterranea [43]. miR-71c-5p was down-regulated when HDAC1 Knockdown. Therefore, it will be worth investigating these up-regulated or down-regulated miRNAs by RNAi knockdown them or their target genes.
4.3. Small RNA/targets comprise a possible regulatory network in regulating stem cell function and regeneration
Pax6A and HMG protein TCF/LEF were predicted to be the target of miR-2157-5p. Pax6A has been shown to be required for neuronal progenitor cell proliferation [44]. A TCF/LEF-mediated Wnt signal axis is critical for animal development, and its mis-regulation leads to disease such as cancer; where over-activated Wnt signaling drives LEF/TCFs to transform cells [45]. Lectin-like protein is the target of miR-13-3p, and it is transcriptionally activated during sexual development [46]. Given that these genes have broad function in development and diseases, it would be interesting to further pursue their function during regeneration.
SMEDWI-2 is a PIWI-like protein that regulates planarian stem cells [47]. It's also predicted to be a piRNA targets, suggesting that there may be a PIWI-piRNA feedback loop involved in regeneration.
Taken together, we have comprehensively surveyed small RNA species with deep sequencing in six biological contexts, and built up a regulatory network based on genes targeted by differentially expressed small RNAs. The dynamic expression of four species of small RNAs and their target genes was further characterized, indicating the importance of small RNAs in fine-tuning gene expression in planarian stem cells and regeneration process. This study provides an important resource for understanding the roles of small RNAs in stem cell regulation and tissue regeneration.
Conflict of interest statement
We declare that we have no financial and personal relationships with other people or organizations that can inappropriately influence our work; there is no professional or other personal interest of any nature or kind in any product, service and/or company that could be construed as influencing the position presented in, or the review of this manuscript.
Acknowledgments
Acknowledgement
This work was supported by the National Key Research and Development Program of China (2017YFA0103700) and the National Natural Science Foundation of China (91339205, 31401238).
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.gdata.2017.10.004.
Contributor Information
Yue Li, Email: yueli7@126.com.
Bairong Shen, Email: bairong.shen@suda.edu.cn.
Qing Jing, Email: qjing@sibs.ac.cn.
Appendix A. Supplementary data
Supplementary material
Scatter plots of miRNAs by DeSeq.
Targets of miRNAs.
Scatter plots of normalized piRNAs.
Targets of piRNAs.
Scatter plots of normalized srRNAs.
Targets of srRNAs.
Profiling of endogenous siRNAs.
Targets of siRNAs.
References
- 1.Reddien P.W., Alvarado A.S. Fundamentals of planarian regeneration. Annu. Rev. Cell Dev. Biol. 2004;20:725–757. doi: 10.1146/annurev.cellbio.20.010403.095114. [DOI] [PubMed] [Google Scholar]
- 2.Friedlander M.R., Adamidi C., Han T., Lebedeva S., Isenbarger T.A., Hirst M., Marra M., Nusbaum C., Lee W.L., Jenkin J.C., Sanchez Alvarado A., Kim J.K., Rajewsky N. High-resolution profiling and discovery of planarian small RNAs. Proc. Natl. Acad. Sci. U. S. A. 2009;106:11546–11551. doi: 10.1073/pnas.0905222106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sánchez Alvarado A. Stem cells in animal models of regeneration. StemBook. 2008:1–24. [PubMed] [Google Scholar]
- 4.Li Y., Zhang Z., Liu F., Vongsangnak W., Jing Q., Shen B. Performance comparison and evaluation of software tools for microRNA deep-sequencing data analysis. Nucleic Acids Res. 2012;40:4298–4305. doi: 10.1093/nar/gks043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Liu G., Ding M., Wang H., Huang J., Jing Q., Shen B. Pathway analysis of microRNAs in mouse heart development. Int. J. Bioinforma. Res. Appl. 2010;6:12–20. doi: 10.1504/IJBRA.2010.031289. http://www.ncbi.nlm.nih.gov/pubmed/20110206 [DOI] [PubMed] [Google Scholar]
- 6.Sasidharan V., Lu Y.C., Bansal D., Dasari P., Poduval D., Seshasayee A., Resch A.M., Graveley B.R., Palakodeti D. Identification of neoblast- and regeneration-specific miRNAs in the planarian Schmidtea mediterranea. RNA. 2013;19:1394–1404. doi: 10.1261/rna.038653.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lu Y.-C., Smielewska M., Palakodeti D., Lovci M.T., Aigner S., Yeo G.W., Graveley B.R. Deep sequencing identifies new and regulated microRNAs in Schmidtea mediterranea. RNA. 2009;15:1483–1491. doi: 10.1261/rna.1702009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Palakodeti D., Smielewska M., Graveley B.R. 2006. MicroRNAs From the Planarian Schmidtea mediterranea: A Model System for Stem Cell Biology; pp. 1640–1649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Friedländer M.R., Adamidi C., Han T., Lebedeva S., Isenbarger T.A., Hirst M., Marra M., Nusbaum C., Lee W.L., Jenkin J.C., Alvarado A.S., Kim J.K., Rajewsky N. High-resolution profiling and discovery of planarian small RNAs. Proc. Natl. Acad. Sci. 2009;106:11546–11551. doi: 10.1073/pnas.0905222106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Shibata N., Kashima M., Ishiko T., Nishimura O., Rouhana L., Misaki K., Yonemura S., Saito K., Siomi H., Siomi M.C., Agata K. Inheritance of a nuclear PIWI from pluripotent stem cells by somatic descendants ensures differentiation by silencing transposons in planarian. Dev. Cell. 2016;37:226–237. doi: 10.1016/j.devcel.2016.04.009. [DOI] [PubMed] [Google Scholar]
- 11.Palakodeti D., Smielewska M., Lu Y.-C., Yeo G.W., Graveley B.R. The PIWI proteins SMEDWI-2 and SMEDWI-3 are required for stem cell function and piRNA expression in planarians. RNA (New York, NY). 2008;14:1174–1186. doi: 10.1261/rna.1085008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Reddien P.W. SMEDWI-2 is a PIWI-like protein that regulates planarian stem cells. Science. 2005;80 −( 310):1327–1330. doi: 10.1126/science.1116110. [DOI] [PubMed] [Google Scholar]
- 13.Li Y.-Q., Zeng A., Han X.-S., Wang C., Li G., Zhang Z.-C., Wang J.-Y., Qin Y.-W., Jing Q. Argonaute-2 regulates the proliferation of adult stem cells in planarian. Cell Res. 2011;21:1750–1754. doi: 10.1038/cr.2011.151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wei H., Zhou B., Zhang F., Tu Y., Hu Y., Zhang B., Zhai Q. Profiling and identification of small rDNA-derived RNAs and their potential biological functions. PLoS One. 2013;8 doi: 10.1371/journal.pone.0056842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ghildiyal M., Seitz H., Horwich M.D., Li C., Du T., Lee S., Xu J., Kittler E.L.W., Zapp M.L., Weng Z., Zamore P.D. Endogenous siRNAs derived from transposons and mRNAs in drosophila somatic cells. Science. 2008;320:1077–1081. doi: 10.1126/science.1157396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ghildiyal M., Zamore P.D. Small silencing RNAs: an expanding universe. Nat. Rev. Genet. 2009;10:94–108. doi: 10.1038/nrg2504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zeng A., Li Y.Q., Wang C., Han X.S., Li G., Wang J.Y., Li D.S., Qin Y.W., Shi Y., Brewer G., Jing Q. Heterochromatin protein 1 promotes self-renewal and triggers regenerative proliferation in adult stem cells. J. Cell Biol. 2013;201:409–425. doi: 10.1083/jcb.201207172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.van Wolfswinkel J.C., Wagner D.E., Reddien P.W. Single-cell analysis reveals functionally distinct classes within the planarian stem cell compartment. Cell Stem Cell. 2014;15:326–339. doi: 10.1016/j.stem.2014.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Palakodeti D., Smielewska M., Lu Y.C., Yeo G.W., Graveley B.R. The PIWI proteins SMEDWI-2 and SMEDWI-3 are required for stem cell function and piRNA expression in planarians. RNA. 2008;14:1174–1186. doi: 10.1261/rna.1085008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Eisenhoffer G.T., Kang H., Alvarado A.S. Molecular analysis of stem cells and their descendants during cell turnover and regeneration in the planarian Schmidtea mediterranea. Cell Stem Cell. 2008;3:327–339. doi: 10.1016/j.stem.2008.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wenemoser D., Reddien P.W. Planarian regeneration involves distinct stem cell responses to wounds and tissue absence. Dev. Biol. 2010;344:979–991. doi: 10.1016/j.ydbio.2010.06.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Anders S., Huber W. Differential expression analysis for sequence count data. Genome Biol. 2010;11:R106. doi: 10.1186/gb-2010-11-10-r106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Solana J., Kao D., Mihaylova Y., Jaber-Hijazi F., Malla S., Wilson R., Aboobaker A. Defining the molecular profile of planarian pluripotent stem cells using a combinatorial RNAseq, RNA interference and irradiation approach. Genome Biol. 2012;13:R19. doi: 10.1186/gb-2012-13-3-r19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Pellettieri J., Fitzgerald P., Watanabe S., Mancuso J., Green D.R., Sanchez Alvarado A. Cell death and tissue remodeling in planarian regeneration. Dev. Biol. 2010;338:76–85. doi: 10.1016/j.ydbio.2009.09.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Qin Y.F., Zhao J.M., Bao Z.X., Zhu Z.Y., Mai J., Huang Y.B., Li J.B., Chen G., Lu P., Chen S.J., Su L.L., Fang H.M., Lu J.K., Zhang Y.Z., Zhang S.T. Identification of small non-coding RNAs in the planarian Dugesia japonica via deep sequencing. Genomics. 2012;99:315–321. doi: 10.1016/j.ygeno.2012.03.001. [DOI] [PubMed] [Google Scholar]
- 26.Wenemoser D., Lapan S.W., Wilkinson A.W., Bell G.W., Reddien P.W. A molecular wound response program associated with regeneration initiation in planarians. Genes Dev. 2012;26:988–1002. doi: 10.1101/gad.187377.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Resch A.M., Palakodeti D. Small RNA pathways in Schmidtea mediterranea. Int J Dev Biol. 2012;56:67–74. doi: 10.1387/ijdb.113436ar. [DOI] [PubMed] [Google Scholar]
- 28.Poole C.B., Davis P.J., Jin J., McReynolds L.A. Cloning and bioinformatic identification of small RNAs in the filarial nematode, Brugia malayi. Mol. Biochem. Parasitol. 2010;169:87–94. doi: 10.1016/j.molbiopara.2009.10.004. [DOI] [PubMed] [Google Scholar]
- 29.Boutla A., Delidakis C., Tabler M. Developmental defects by antisense-mediated inactivation of micro-RNAs 2 and 13 in drosophila and the identification of putative target genes. Nucleic Acids Res. 2003;31:4973–4980. doi: 10.1093/nar/gkg707. http://www.ncbi.nlm.nih.gov/pubmed/12930946 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.de Souza Gomes M., Donoghue M.T., Muniyappa M., Pereira R.V., Guerra-Sa R., Spillane C. Computational identification and evolutionary relationships of the microRNA gene cluster miR-71/2 in protostomes. J. Mol. Evol. 2013;76:353–358. doi: 10.1007/s00239-013-9563-2. [DOI] [PubMed] [Google Scholar]
- 31.Clark A.M., Goldstein L.D., Tevlin M., Tavare S., Shaham S., Miska E.A. The microRNA miR-124 controls gene expression in the sensory nervous system of Caenorhabditis elegans. Nucleic Acids Res. 2010;38:3780–3793. doi: 10.1093/nar/gkq083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Peng X.H., Huang H.R., Lu J., Liu X., Zhao F.P., Zhang B., Lin S.X., Wang L., Chen H.H., Xu X., Wang F., Li X.P. MiR-124 suppresses tumor growth and metastasis by targeting Foxq1 in nasopharyngeal carcinoma. Mol. Cancer. 2014;13:186. doi: 10.1186/1476-4598-13-186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Li D., Li X., Wang A., Meisgen F., Pivarcsi A., Sonkoly E., Stahle M., Landen N.X. MicroRNA-31 promotes skin wound healing by enhancing keratinocyte proliferation and migration. J Invest Dermatol. 2015 doi: 10.1038/jid.2015.48. [DOI] [PubMed] [Google Scholar]
- 34.Kim H.S., Lee K.S., Bae H.J., Eun J.W., Shen Q., Park S.J., Shin W.C., Yang H.D., Park M., Park W.S., Kang Y.K., Nam S.W. MicroRNA-31 functions as a tumor suppressor by regulating cell cycle and epithelial-mesenchymal transition regulatory proteins in liver cancer. Oncotarget. 2015 doi: 10.18632/oncotarget.3512. http://www.ncbi.nlm.nih.gov/pubmed/25797269 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hsieh T.H., Hsu C.Y., Tsai C.F., Long C.Y., Chai C.Y., Hou M.F., Lee J.N., Wu D.C., Wang S.C., Tsai E.M. miR-125a-5p is a prognostic biomarker that targets HDAC4 to suppress breast tumorigenesis. Oncotarget. 2015;6:494–509. doi: 10.18632/oncotarget.2674. http://www.ncbi.nlm.nih.gov/pubmed/25504437 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wang R.J., Zheng Y.H., Wang P., Zhang J.Z. Serum miR-125a-5p, miR-145 and miR-146a as diagnostic biomarkers in non-small cell lung cancer. Int. J. Clin. Exp. Pathol. 2015;8:765–771. http://www.ncbi.nlm.nih.gov/pubmed/25755772 [PMC free article] [PubMed] [Google Scholar]
- 37.Banerjee S., Cui H., Xie N., Tan Z., Yang S., Icyuz M., Thannickal V.J., Abraham E., Liu G. miR-125a-5p regulates differential activation of macrophages and inflammation. J. Biol. Chem. 2013;288:35428–35436. doi: 10.1074/jbc.M112.426866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Huang T., Yang J., Cai Y.D. Novel candidate key drivers in the integrative network of genes, MicroRNAs, methylations, and copy number variations in squamous cell lung carcinoma. Biomed. Res. Int. 2015;2015:358125. doi: 10.1155/2015/358125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lam V., Tokusumi T., Tokusumi Y., Schulz R.A. Bantam miRNA is important for drosophila blood cell homeostasis and a regulator of proliferation in the hematopoietic progenitor niche. Biochem. Biophys. Res. Commun. 2014;453:467–472. doi: 10.1016/j.bbrc.2014.09.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Cramer E.M., Shao Y., Wang Y., Yuan Y. miR-190 is upregulated in Epstein-Barr virus type I latency and modulates cellular mRNAs involved in cell survival and viral reactivation. Virology. 2014;464–465:184–195. doi: 10.1016/j.virol.2014.06.029. [DOI] [PubMed] [Google Scholar]
- 41.Hao Y., Yang J., Yin S., Zhang H., Fan Y., Sun C., Gu J., Xi J.J. The synergistic regulation of VEGF-mediated angiogenesis through miR-190 and target genes. RNA. 2014;20:1328–1336. doi: 10.1261/rna.044651.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Xiao H., Li H., Yu G., Xiao W., Hu J., Tang K., Zeng J., He W., Zeng G., Ye Z., Xu H. MicroRNA-10b promotes migration and invasion through KLF4 and HOXD10 in human bladder cancer. Oncol. Rep. 2014;31:1832–1838. doi: 10.3892/or.2014.3048. [DOI] [PubMed] [Google Scholar]
- 43.Jin X., Lu L., Su H., Lou Z., Wang F., Zheng Y., Xu G.T. Comparative analysis of known miRNAs across platyhelminths. FEBS J. 2013;280:3944–3951. doi: 10.1111/febs.12395. [DOI] [PubMed] [Google Scholar]
- 44.Thummel R., Enright J.M., Kassen S.C., Montgomery J.E., Bailey T.J., Hyde D.R. Pax6a and Pax6b are required at different points in neuronal progenitor cell proliferation during zebrafish photoreceptor regeneration. Exp. Eye Res. 2010;90:572–582. doi: 10.1016/j.exer.2010.02.001. S0014-4835(10)00046-1 [pii]\r10.1016/j.exer.2010.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Arce L., Yokoyama N.N., Waterman M.L. Diversity of LEF/TCF action in development and disease. Oncogene. 2006;25:7492–7504. doi: 10.1038/sj.onc.1210056. [DOI] [PubMed] [Google Scholar]
- 46.Nowrousian M., Cebula P. The gene for a lectin-like protein is transcriptionally activated during sexual development, but is not essential for fruiting body formation in the filamentous fungus Sordaria macrospora. BMC Microbiol. 2005;5:64. doi: 10.1186/1471-2180-5-64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Reddien P.W., Oviedo N.J., Jennings J.R., Jenkin J.C., Sánchez Alvarado A. SMEDWI-2 is a PIWI-like protein that regulates planarian stem cells. Science. 2005;310:1327–1330. doi: 10.1126/science.1116110. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material
Scatter plots of miRNAs by DeSeq.
Targets of miRNAs.
Scatter plots of normalized piRNAs.
Targets of piRNAs.
Scatter plots of normalized srRNAs.
Targets of srRNAs.
Profiling of endogenous siRNAs.
Targets of siRNAs.