Supplemental Digital Content is available in the text.
Keywords: epigenetics, gene expression and regulation, genomic imprinting, preeclampsia, trophoblasts
Abstract
Background:
Preeclampsia is a complex and common human-specific pregnancy syndrome associated with placental pathology. The human specificity provides both intellectual and methodological challenges, lacking a robust model system. Given the role of imprinted genes in human placentation and the vulnerability of imprinted genes to loss of imprinting changes, there has been extensive speculation, but no robust evidence, that imprinted genes are involved in preeclampsia. Our study aims to investigate whether disturbed imprinting contributes to preeclampsia.
Methods:
We first aimed to confirm that preeclampsia is a disease of the placenta by generating and analyzing genome-wide molecular data on well-characterized patient material. We performed high-throughput transcriptome analyses of multiple placenta samples from healthy controls and patients with preeclampsia. Next, we identified differentially expressed genes in preeclamptic placentas and intersected them with the list of human imprinted genes. We used bioinformatics/statistical analyses to confirm association between imprinting and preeclampsia and to predict biological processes affected in preeclampsia. Validation included epigenetic and cellular assays. In terms of human specificity, we established an in vitro invasion-differentiation trophoblast model. Our comparative phylogenetic analysis involved single-cell transcriptome data of human, macaque, and mouse preimplantation embryogenesis.
Results:
We found disturbed placental imprinting in preeclampsia and revealed potential candidates, including GATA3 and DLX5, with poorly explored imprinted status and no prior association with preeclampsia. As a result of loss of imprinting, DLX5 was upregulated in 69% of preeclamptic placentas. Levels of DLX5 correlated with classic preeclampsia markers. DLX5 is expressed in human but not in murine trophoblast. The DLX5high phenotype resulted in reduced proliferation, increased metabolism, and endoplasmic reticulum stress-response activation in trophoblasts in vitro. The transcriptional profile of such cells mimics the transcriptome of preeclamptic placentas. Pan-mammalian comparative analysis identified DLX5 as part of the human-specific regulatory network of trophoblast differentiation.
Conclusions:
Our analysis provides evidence of a true association among disturbed imprinting, gene expression, and preeclampsia. As a result of disturbed imprinting, the upregulated DLX5 affects trophoblast proliferation. Our in vitro model might fill a vital niche in preeclampsia research. Human-specific regulatory circuitry of DLX5 might help explain certain aspects of preeclampsia.
Clinical Perspective.
What Is New?
Unbiased analysis of genome-wide molecular and clinical data identifies DLX5 as an imprinted target gene with novel placental function in preeclampsia.
We observe that DLX5 is paternally imprinted in the human placenta and its expression is dysregulated in preeclampsia.
We provide evidence for a mechanistic coupling between preeclampsia and disturbed placental imprinting (loss of imprinting) as a causal role in preeclampsia.
Our data indicate that DLX5 has a role in trophoblast proliferation and differentiation (syncytialization).
DLX5-induced overexpression in trophoblasts can faithfully model preeclampsia in a cell culture system, signifying the contribution of a single transcription factor and providing a potential cellular model both for further research and for analysis of drugable targets.
DLX5 is expressed in human but not in mouse trophoblasts, underlining the human specificity of preeclampsia. Our study highlights the diverged cellular function of DLX5 during mammalian embryonic evolution.
What Are the Clinical Implications?
Our analysis supports the view that preeclampsia is not a single but a heterogeneous disease with disturbed imprinting commonplace.
Unsupervised clustering analysis identified 3 distinct transcriptomic classes of preeclampsia, not clustering with the intrauterine growth restriction. Our clusters are in conjunction with previously suggested classification of preeclampsia (early- versus late-onset) but subdivide early-onset preeclampsia further.
Our study identifies an early preeclampsia cluster that can be clearly characterized by elevated DLX5 levels, disturbed epigenetics, and similar clinical manifestation.
We find that levels of DLX5 correlate with a circulating biomarker, placental growth factor. Whether DLX5 will have utility as a biomarker is unclear because its loss of imprinting is not observed in all instances (69% of all preeclampsia cases).
Our cellular model has potential for further clinically related research, including analysis of drugable targets.
Preeclampsia is a complex, heterogeneous syndrome characterized by high blood pressure (≥140/90 mm Hg) after the 20th week of pregnancy in association with proteinuria (≥300 mg/L per 24 hours).1 Preeclampsia is the first sex-specific cardiovascular risk factor,2 affecting 2% to 8% of human pregnancies, and remains a leading cause of maternal and perinatal mortality.3
Despite considerable research efforts, the cause of preeclampsia is not fully understood. Preeclampsia is assumed to be associated with reduced fetal trophoblast invasion and impaired remodeling of maternal spiral arteries leading to poor uteroplacental perfusion. The improper placentation process triggers oxidative and endoplasmic reticulum (ER) stress and results in defective protein synthesis in the placenta. As a consequence, dysregulated expression of inflammatory, angiogenic, and antiangiogenic factors is observed.4 Currently, the only treatment is delivery, pinpointing the potentially central role of the placenta (or more generally the maternal-fetal interaction) in the disease.5
The placenta has unique epigenetic features, including low levels of genomic DNA methylation and a specific expression pattern of imprinted genes, which are the prime candidates to be associated with the evolution of intrauterine development.6 Because epigenetic disruption of imprinted gene was associated with certain diseases, the potential involvement of disturbed regulation of imprinted genes in preeclampsia has also been intensively discussed.7–11
Preeclampsia is not confirmed in other mammals and is considered to be human-specific (although a possible eclampsia event was observed in a gorilla).12 The human specificity of preeclampsia generates real challenges. First, despite intensive research, the genetic background of the human specificity and the pathogenesis of preeclampsia remain poorly understood. Second, although rodent models have been suggested, no animal models have proven suitable.
Because placentation is a diverse process even among primates,13 genes associated preferentially with placental expression (eg, imprinted genes) are potential candidates for understanding human specificity. This suggestion is centered largely on the understandings that the dosage of imprinted genes can have phenotypic impact, that the placenta is a hot spot for the activity of imprinted genes, and hence that loss of imprinting (LOI) will be likely to affect matters at the maternal-fetal interface.
Despite the abundant speculation that disruption of imprinting and preeclampsia might be mechanistically coupled, the evidence is at best limited. Indeed, a recent review of the genetics of preeclampsia14 reports no robust evidence for a mechanistic coupling with imprinting. What evidence there has been is circumstantial, often negative, or not replicable.15–19 A further barrier to effective research, until recently,20 has been the lack of a definitive list of genes imprinted in the human placenta.
To get a better understanding of this highly complex disease, we aim to confirm our hypotheses that preeclampsia is not a single but a heterogeneous disease of the placenta; that it is associated with improper trophoblast function; and that epigenetic turbulence can result in abnormal expression of imprinted genes, which in turn compromise proper maternal-fetal interaction and contribute to preeclampsia, specifically in humans. Our experimental strategy is based on a cross-disciplinary approach that uses the current catalog of human imprinted placental genes and combines the generation and analyses of genome-wide molecular data with well-characterized patient material followed by experimental validation. We use molecular and cellular technologies and bioinformatic analyses to predict biological processes affected in preeclampsia, followed by wet-bench validation. We also aimed at establishing an in vitro model to fill a vital niche in preeclampsia research, explicitly because there is no robust animal model system. Finally, we data-mine existing single-cell transcriptome data to possibly shed light on the human-specific nature of preeclampsia.
Methods
Detailed methods are provided in the online-only Data Supplement.
Patients
Microarray data of human placenta and decidua samples were described earlier.21 The study also consists of placental and decidual tissues from 56 preeclamptic women and 28 women with normotensive and uncomplicated pregnancies described earlier.22 The preeclamptic group was subdivided into preeclampsia with intrauterine growth restriction (n=14) and preeclampsia without intrauterine growth restriction (n=42). Furthermore, the preeclamptic group was divided into early-onset preeclampsia (delivery before gestational week 34) and late-onset preeclampsia (delivery during or after gestational week 34). The Regional Committee of Medical Research Ethics in South-Eastern Norway approved the study, and all the subjects gave informed written consent.
Healthy (n=5) and preeclamptic (n=5) primary trophoblast cells were isolated from human placenta samples obtained from HELIOS Klinikum in Berlin. Human placenta sampling was approved by the Regional Committee of the Medical Faculty of Charité Berlin.
Statistics
Data are presented as mean±SEM (for normally distributed data) or median with interquartile range (for nonnormally distributed data). Normality was assessed by Kolmogorov-Smirnov tests. Techniques for each analysis are specified in the figure legends. Values of P<0.05 were considered statistically significant.
Results
Cluster Analysis Identifies 3 Distinct Transcriptome Patterns of Preeclamptic Placenta
To determine which genes have aberrant expression in preeclampsia, we sought to first better understand whether preeclampsia is 1 disease or many and then to look for genes misregulated across all preeclamptic subtypes. We compared placenta and decidua samples from 24 patients with preeclampsia and 22 healthy women. Nineteen (of 24) preeclamptic placental samples generated 3 distinct molecular groups (PE_P1 through PE_P3; Figure 1A), whereas control samples (16 of 22) generated 2 groups (C1, C2). Five (of 24) preeclamptic and 4 (of 22) healthy placenta samples grouped with the opposite cluster. Whereas placental PE_P1 and PE_P3 contain samples from early-onset preeclampsia, PE_P2 included samples mainly from late-onset preeclampsia. We did not reveal any significant grouping for the intrauterine growth restriction phenotype. An alternative clustering method based on euclidean distances identified the same 3 preeclampsia clusters (Figure IA in the online-only Data Supplement). In contrast to placental samples, the preeclampsia and control decidua samples appeared to scatter randomly (Figure IB in the online-only Data Supplement), supporting that clinically established preeclampsia is a placental, not a decidual, disorder. The negative result for the decidua also provides a negative control for false/artifactual clustering.
Figure 1.
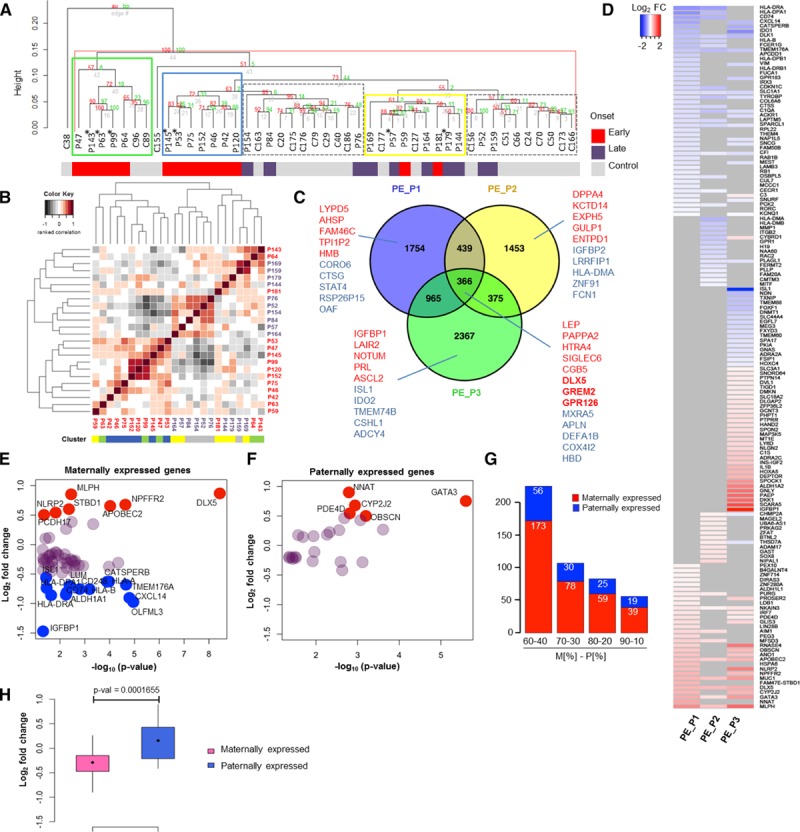
Gene expression analysis of preeclamptic and healthy placenta samples. A, Hierarchical clustering analysis of microarray data identified 3 preeclamptic groups in placenta, PE_P1 (blue), PE_P2 (yellow), and PE_P3 (green), and 2 control groups, C1 and C2 (gray, dashed) (control placenta, n=22; preeclamptic placenta, n=24). The onset of preeclampsia (by gestational age at delivery) for each sample is indicated. *Preeclampsia+intrauterine growth restriction samples. B, Clustering analysis of clinical data. Heat map representing pairwise correlation between patients based on the clinical data (Table I in the online-only Data Supplement). Hierarchical cluster dendrogram was calculated with ranked correlation and complete linkage method on relative values from the clinical variables (n=36). Height of dendrograms represents the euclidian distance. C, Venn diagram of differentially expressed genes (DEGs) in 3 preeclamptic clusters (DEG on P<0.05). Top upregulated (red) and downregulated (blue) genes for each cluster are shown. D, Imprinted genes in preeclampsia. Heat map representing differential expression of imprinted genes in the 3 preeclamptic clusters vs. controls (DEG P<0.05). Log2-fold change of differential expression of maternally (E) and paternally (F) expressed genes (MEGs and PEGs) in all preeclampsia cases (DEG P<0.05). Genes with log2-fold change ≥0.5 are shown in red and with ≤−0.5 change in blue. G, Distribution of differentially expressed MEGs and PEGs in preeclampsia according to different criteria for maternal to paternal allelic expression ratio (MEGs, M[%]-PEGs, P[%]): 60-40, 70-30, and 90-10. H, Wilcoxon test on the mean log2-fold change expression of differentially expressed MEGs and PEGs in preeclampsia.
A Subset of Early-Onset Preeclampsia Correlates With Clinical Symptoms
Is there an association between the preeclamptic gene clusters and clinical data? We clustered 36 patients’ clinical and biomarker parameters on their relative values (Table I in the online-only Data Supplement). Our analysis suggests that the gene expression profile can be related to clinical disease phenotyping, but only to a certain extent (Figure 1B and Figure IIA and IIB in the online-only Data Supplement). Besides a related transcriptome, PE_P1 shared a similar clinical profile. In contrast, most of the patients from PE_P2 generated a cluster together with preeclamptic samples that had diverse transcriptome, and patients from PE_P3 had a diverse clinical manifestation.
We also performed correlation analyses of clinical and gene expression data across all control and patient samples to identify genes associated with any of the clinical phenotypes. We found correlated gene expression with maternal placental growth factor (PlGF; eg, PAPPA2, SPAG4, ENG, and LEP) and sFlt1 (soluble fms-like tyrosine kinase-1) levels (eg, SPAG4, ENG, ANKRD37, and ERRFI1). Among the imprinted genes, the expression of both DLX5 and GATA3 correlated with diastolic blood pressure and anticorrelated with gestation age (Figure IIC in the online-only Data Supplement). DLX5 also correlated with sEndoglin (positive) and with baby weight (negative).
All Preeclamptic Clusters Have Disturbed Guidance Signaling
We then analyzed differentially expressed genes (DEGs) in each preeclampsia cluster compared with control samples. Each cluster contained >1000 unique DEGs with 366 dysregulated genes common to all 3 clusters (P<0.05; Figure 1C and Table II in the online-only Data Supplement). To characterize the clusters and to find possible interactions, data sets containing 3525, 2634, and 4073 DEGs corresponding to preeclamptic clusters PE_P1, PE_P2, and PE_P3, respectively, were analyzed with Ingenuity Pathway Analysis and GOrilla gene ontology tools. Axonal guidance was the top pathway in all 3 preeclamptic clusters, supporting the view that disturbed angiogenesis and disturbed cell–cell communication between endothelial cells and the trophoblasts via the mechanism of axonal guidance are common features of preeclampsia.23,24 Other than this universal property, the 3 clusters exhibited unique features (Figure III and Table II in the online-only Data Supplement).
Imprinted Genes Exhibit Differential Expression in Preeclamptic Placenta
Next, we focused on differentially expressed imprinted genes in preeclampsia. We used a merged list of genes of the Geneimprint database (184 validated and putative human imprinted genes) and the recent list of imprinted human placental genes (223; maternal:paternal allelic expression ratio, 80:20)20 (Table III in the online-only Data Supplement). Our analysis identified 150 imprinted/putatively imprinted genes that were dysregulated in at least 1 of the 3 preeclamptic clusters (P<0.05; Figure 1D and Table III in the online-only Data Supplement). Maternally expressed genes (MEGs) and paternally expressed genes (PEGs) were affected according to their relative commonality (23%, 59 of 257, versus 17%, 25 of 150; χ2=1.9, P=0.16; Figure 1F and Table III in the online-only Data Supplement). This was observed also when maternal/paternal allelic expression ratios were considered at different stringencies (Figure 1G). The mean expression of MEGs, however, was significantly downregulated compared with PEGs (Figure 1H). Nevertheless, among the most significantly deregulated genes, we identified both MEGs (eg, DLX5, APOBEC2, CD74) and PEGs (eg, GATA3, CYP2J2) (Figure 1E and 1F). In our study, the 2 most significantly dysregulated imprinted genes were GATA3 and DLX5 (P<2.51×10−6 and P<3.65×10−9, respectively). Furthermore, we focus on DLX5, which is a MEG in human lymphoblasts and brain,25 but its imprinted status in the placenta is not yet explored.
DLX5 Is Upregulated in Preeclamptic Placenta
We used quantitative reverse-transcription–polymerase chain reaction to confirm the upregulation of DLX5 in preeclampsia (Figure IVB in the online-only Data Supplement). We then confirmed these findings in a second independent patient cohort of 56 compared with 28 controls. Although DLX5 was significantly upregulated in both early-onset (P<0.0001) and late-onset (P<0.01) preeclampsia (Figure 2A) and in all 3 preeclamptic clusters, its expression level varied between placental samples. Nevertheless, altogether, 69% of preeclampsia samples (n=56) could be associated with DLX5 overexpression. We also confirmed increased DLX5 protein expression in preeclamptic placenta tissues by Western blotting (Figure IVC and IVD in the online-only Data Supplement). Correlation analysis of gestation age to DLX5 expression in control or preeclamptic placentas excluded gestation age–related changes in DLX5 differential expression (Figure IVE in the online-only Data Supplement). In tissues and cells derived from a pregnancy-related tissue panel, DLX5 expression was detected in placenta and in trophoblasts but was less pronounced in the decidua (Figure IVF and IVG in the online-only Data Supplement). Immunofluorescent staining detected coexpression of DLX5 with cytokeratin-7, a trophoblast-specific marker (Figure 2B). Immunohistochemistry confirmed elevated DLX5 protein expression in placental tissues of early-onset preeclampsia (Figure 2C).
Figure 2.
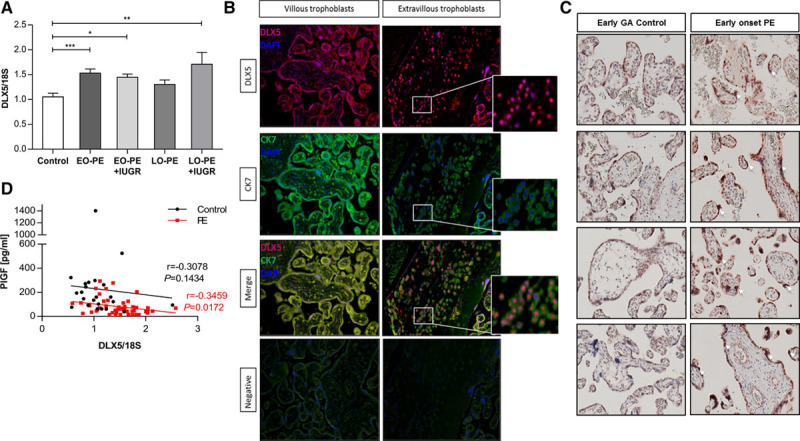
DLX5 is upregulated in preeclamptic placenta. A, Quantitative polymerase chain reaction confirmed increased DLX5 mRNA level in placentas of a second preeclamptic cohort (values are mean±SEM; control: 1.053±0.0745, n=28; early-onset (EO) preeclampsia+intrauterine growth restriction (IUGR): 1.447±0.066, n=9; late-onset (LO) preeclampsia+IUGR: 1.71±0.238, n=5; EO preeclampsia: 1.532±0.08, n=20; LO preeclampsia: 1.304±0.09, n=22). *P<0.05, **P<0.005, ***P<0.0005; ANOVA, Bonferroni multiple-comparisons test. B, Double immunofluorescence staining of term human placenta tissue indicated nuclear expression and colocalization of DLX5 to the cytokeratin 7 (CK7), a trophoblast-specific marker. DLX5 staining was positive in the nucleus of both villous and extravillous trophoblasts. C, Immunohistochemistry staining on placental villous tissue from healthy pregnancy (early control, gestation age [GA], 31 and 34 weeks) and preeclampsia (GA, 31 and 34 weeks) for human DLX5 confirmed increased DLX5 protein expression in early-onset preeclampsia (PE). D, Correlation of DLX5 expression and placental biomarker. Placental DLX5 expression significantly negatively correlated to the serum placental growth factor (PlGF) in the preeclampsia group but not in controls. Control, n=27, r=-0.3078, P=0.1434; preeclampsia, n=48, r=-0.3459, P=0.0172; Spearman rank correlation.
To associate our findings with clinical preeclampsia biomarkers, we compared the placental expression of DLX5 with the antiangiogenic sFlt1 and sFlt1/PlGF ratio and with proangiogenic PlGF concentration in maternal serum.26 Although there was no significant correlation between DLX5 expression and sFlt1 levels, we observed a negative correlation (r=−0.35, P=0.017) with PlGF concentrations comparing women with preeclampsia and healthy women (Figure 2D). This observation suggests that placental DLX5 expression is associated with decreased levels of a maternal circulating placental biomarker in preeclampsia.
Loss of Imprinting Results in Elevated Gene Expression of DLX5
To investigate whether alterations in the predicted imprinting status of DLX5 could be responsible for its overexpression in the preeclamptic placentas, we performed an LOI assay.27 We measured expression of the silenced allele in placental samples carrying the heterozygous DLX5 single nucleotide polymorphism (rs73708843). Primers are provided in Table IV in the online-only Data Supplement. Of 97 placental tissues, 42 were genotyped as heterozygous (43.3%), including 16 control (16.5%) and 26 preeclamptic (26.8%) placentas. The mean expression of the putatively inactive DLX5 allele was 58% in preeclampsia when the expression of the nonimprinted allele for each individual sample was set at 100%. Control samples also exhibited LOI but with significantly less frequent (19%) activation of the imprinted DLX5 allele (Figure 3A and 3B). Sequencing of cDNA through the single nucleotide polymorphism (rs73708843) on 3 placenta samples confirmed the single allelic expression from DLX5 (Figure 3C). We found a correlation (r=0.314, P=0.046) between LOI and DLX5 expression, suggesting that the overexpression phenotype of DLX5 was associated with its LOI (Figure 3D). We also inspected CpG methylation levels at the DLX5 locus in preeclamptic placental compared with healthy control samples (20 versus 20)28 and identified significant CpG hypomethylation in preeclampsia samples (Figure 3E and Table V in the online-only Data Supplement). Furthermore, the methylation level of several CpGs inversely correlated with DLX5 expression in these samples (8 versus 8; Figure 3F). Collectively, we interpreted our data as the altered methylation at the DLX5 locus in preeclampsia results in LOI and affects gene expression.
Figure 3.
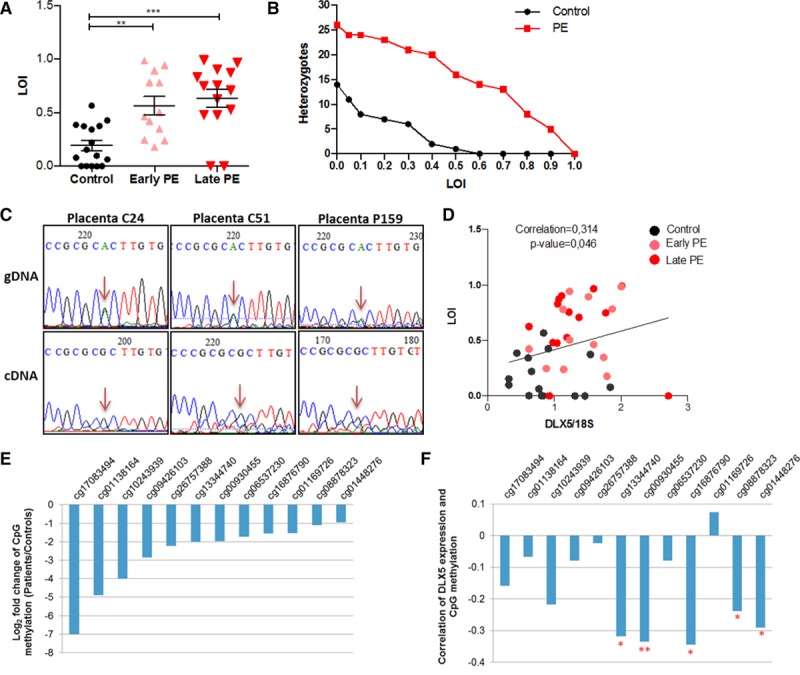
Loss of imprinting (LOI) of DLX5 in preeclamptic placenta. A, Analysis of mean LOI levels for DLX5 in healthy and preeclamptic placenta samples. Values are presented as a mean±SEM of LOI (control: 0.1943±0.04765, n=16; early-onset preeclampsia [PE]: 0.5661±0.08516, n=12; late-onset preeclampsia: 0.6349±0.08437, n=14). **P<0.001, ***P<0.0001, 1-way ANOVA, Bonferroni multiple-comparisons test. B, Distribution (number) of DLX5 heterozygocities exceeding a particular LOI. C, Allelic expression analysis of the imprinted DLX5 in placenta samples exhibiting the allele-specific expression but no LOI. cDNA of the heterozygous placenta samples for the single nucleotide polymorphism (rs73708843) were sequenced. D, LOI correlated with DLX5 expression in placenta. P=0.046, Spearman rank correlation. E, CpG methylation of DLX5 locus. Log2-fold change of CpG methylation level in preeclampsia (control, n=20; early-onset preeclampsia, n=20). Hypomethylated CpG sites are shown (differentially methylated region at adjusted P≤0.05). F, Pairwise Spearman rank correlation of CpG methylation and DLX5 expression in placenta (control, n=8; early-onset preeclampsia, n=8; false discovery rate: *P<0.05, **P<0.01). gDNA indicates genomic DNA.
Upregulated DLX5 Affects Genes Associated With Cell Growth, Proliferation, Survival, and Movement
To decipher the physiological effect of elevated DLX5 expression on trophoblasts, we stably overexpressed the human DLX5 protein in trophoblast cells in vitro. For the overexpression studies, we used the Sleeping Beauty transposon–derived expression system29 in SGHPL-4 cells, derived from first trimester extravillous trophoblasts (Figure VA–VC in the online-only Data Supplement). Immunohistochemistry revealed a predominant nuclear localization of DLX5 in the DLX5-overexpressing SGHPL-4 cells (DLX5High; Figure VD in the online-only Data Supplement). To observe the global effects of elevated DLX5 expression, we performed microarray transcriptome profiling of DLX5High and control cells. A total of 3650 DEGs (P<0.05; 771 genes at false discovery rate <0.05) were identified upon DLX5 overexpression (Figure VIA and Table VI in the online-only Data Supplement). Ingenuity pathway analysis revealed significant gene enrichment involved in cardiovascular system development and function. The most significant terms describing molecular and cellular functions include cellular growth and proliferation, cell death and survival, and cellular movement and development (Figure VIB in the online-only Data Supplement). The top pathways include interferon and death receptor signaling, and superpathway of cholesterol biosynthesis. Several affected pathways are common between preeclamptic placental samples and the in vitro model. These include deregulated axon guidance, interleukin-8, and neuregulin receptor signaling; thyroid hormone receptor/retinoid X receptor, retinoic acid receptor, and planar cell polarity pathway; antigen presentation pathway; unfolded protein response; and nuclear factor, erythroid 2 like 2-mediated oxidative stress responses (Table VI in the online-only Data Supplement).
Elevated Expression of DLX5 Models Certain Aspects of Preeclampsia
To test how well our DLX5high in vitro model mimics global transcriptional changes in preeclampsia, we compared transcriptome profiles of control and DLX5high cells with the 3 placental preeclamptic transcriptome clusters (PE_P1 through PE_P3). Hierarchical clustering of relative gene expression levels of control, DLX5high lines (6 versus 6), PE_P1 through PE_P3, and healthy placenta samples revealed that the transcriptome of DLX5high cells clusters with the 3 preeclamptic groups (PE_P1 through PE_P3), whereas control cells cluster with control placenta samples (Figure 4A). In addition, we asked if it was possible to correlate transcriptomes according to their DLX5 expression levels (Figure VIIA in the online-only Data Supplement). Placenta samples were ordered according to their DLX5 expression levels. The transcriptomes of DLX5high preeclampsia clustered with cultured DLX5high samples, whereas the low_DLX5 preeclamptic transcriptomes were clustered with control SGHPL-4 cells, suggesting that the overexpression of DLX5 in trophoblasts could model certain aspects of preeclampsia (Figure VIIB in the online-only Data Supplement).
Figure 4.
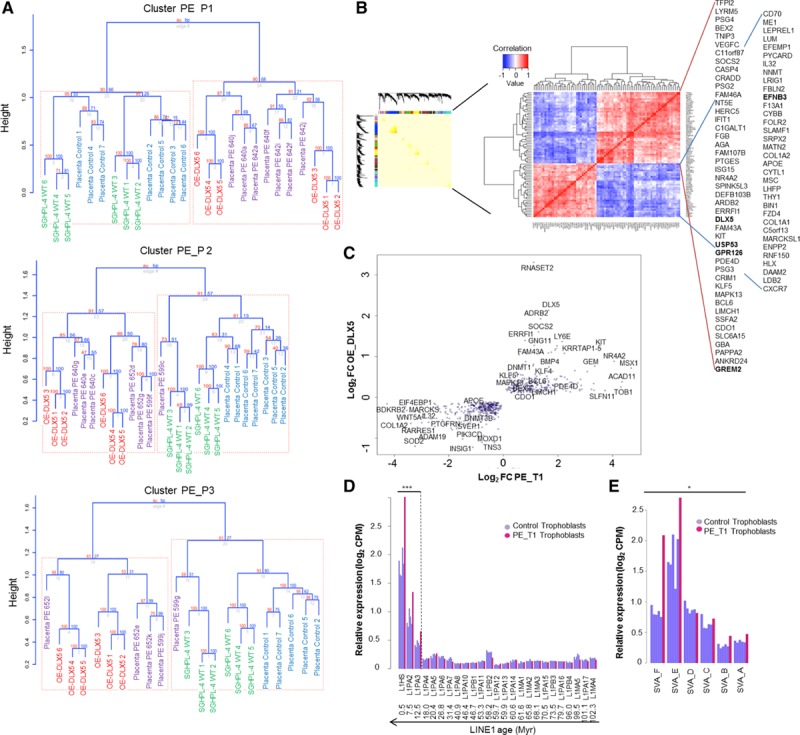
Intersection of preeclamptic transcriptomes with the DLX5high transcriptome. A, Hierarchical clustering (Spearman rank correlation, average linkage) and bootstrapping (1000 replicates) of the transcriptomes of SGHPL-4 cells with preeclamptic (PE) cluster PE_P1, PE_P2, and PE_P3 transcriptomes (Placenta PE) and control placenta samples (Placenta Control). The equal number of control placenta samples and samples in each preeclamptic cluster was chosen randomly. B, Weighted gene coexpression network analysis across 58 samples gave several modules containing a total of ≈3000 genes. Identification of DLX5 target genes in placenta and trophoblast cells. Clustered pairwise correlation matrix of identified 79 genes across 58 samples (Spearman rank correlation, threshold 0.6 and −0.55, P<0.05, euclidian distance). C, Comparison between the log2-fold change of the differentially expressed genes in trophoblast sample PE_T1 and genes differentially expressed on DLX5 overexpression in SGHPL-4 trophoblast cell line. Six hundred forty-one genes having the same differential expression pattern were common in both data sets. D and E, Relative expression of transposable LINE1 and SVA elements in control trophoblast samples and preeclamptic sample PE_T1. In PE_T1, the young members of the TE families L1PA3, L1PA2, L1HS, SVA-E, and SVA-F are upregulated. *P<0.05, ***P<0.005, Kolmogorov-Smirnov test, Benjamini and Hochberg false discovery rate. CMP indicates counts per million.
Identifying Genes With Correlated Expression Dynamic to DLX5
Because the target genes of the transcription factor DLX5 in placenta are not known, we thought to identify genes with an expression that is correlated with DLX5. Thus, we subjected a merged data set of our 2 microarrays (total 58 samples) to weighted gene correlation network analysis. We aimed to identify gene modules of correlated gene expression. This approach allowed us to detect several gene modules containing a total of 3000 genes. Using pairwise ranked correlation analysis of 79 genes associated with DLX5 across 58 samples (Figure 4B), we identified putative targets of DLX5. The list contains several genes previously associated with preeclampsia,30–35 including genes involved in cell growth, proliferation, and differentiation (eg, GREM2, KIT, ERRFI1), angiogenesis (eg, VEGFC, PAPPA2, GPR126), cytokine and growth hormone signaling (GBA, CXCR7), immune response (ISG15, HERC5, IFIT1), pregnancy-specific proteins (PSG2-4), and the paternally imprinted tissue factor pathway inhibitor-2 (TFPI2), involved in regulation of cell invasion and proliferation.
Upregulation of DLX5 in Trophoblasts Is Associated With Disturbed Epigenetics
To address the limitations of our initial approach using placental microarray data, we performed transcriptome analysis using RNA sequencing on freshly isolated, purified human trophoblasts from control and preeclamptic placentas (5 versus 5). In the PE_T1 trophoblast sample, DLX5 was highly upregulated, and we observed 1466 genes commonly dysregulated in both PE_T1 and DLX5high, with 641 genes exhibiting the same pattern of expression, including KIT, SOCS2, KLF5, BEX2, ERRFI1, and DNA methyltransferases (DNMT1 and DNMT3B; Figure 4C). In addition to common features, the PE_T1 sample exhibited further DEGs involved in the regulation of DNA methylation and histone modification such as the TET gene family (TET1-3), SETDB1, SIRT1, and histone deacetylases, indicating that a subset of preeclampsia might be associated with severe epigenetic disturbances. As a likely consequence, the PE_T1 sample is characterized by the deregulation of several imprinted genes (including DLX5) but also potentially mutagenic transposable elements such as LINE-1 and SVA36,37 (Figure 4D and 4E).
Upregulation of DLX5 Reduces Trophoblast Proliferation
The significant increase of DLX5 expression in preeclamptic placenta samples prompted us to explore the possible mechanism of DLX5 in the pathogenesis of preeclampsia. To characterize the DLX5high phenotype, we have performed several cellular assays inspired by the Pathway analyses. To determine whether the DLX5high phenotype affects trophoblast proliferation, we used time-lapse microscopy and observed reduced cell proliferation of DLX5high cells by 45% compared to control after 48 hours of incubation (Figure 5A). Significantly, reduced trophoblast proliferation of DLX5high was confirmed by a high-throughput sampler cell count and microtiter plate test colorimetric assay (Figure 5B and 5C). DLX5 overexpression had only a slight, not significant, effect on cell apoptosis as indicated by scoring apoptotic cells within 48 hours of incubation (Figure VIIIA and VIIIB in the online-only Data Supplement).
Figure 5.
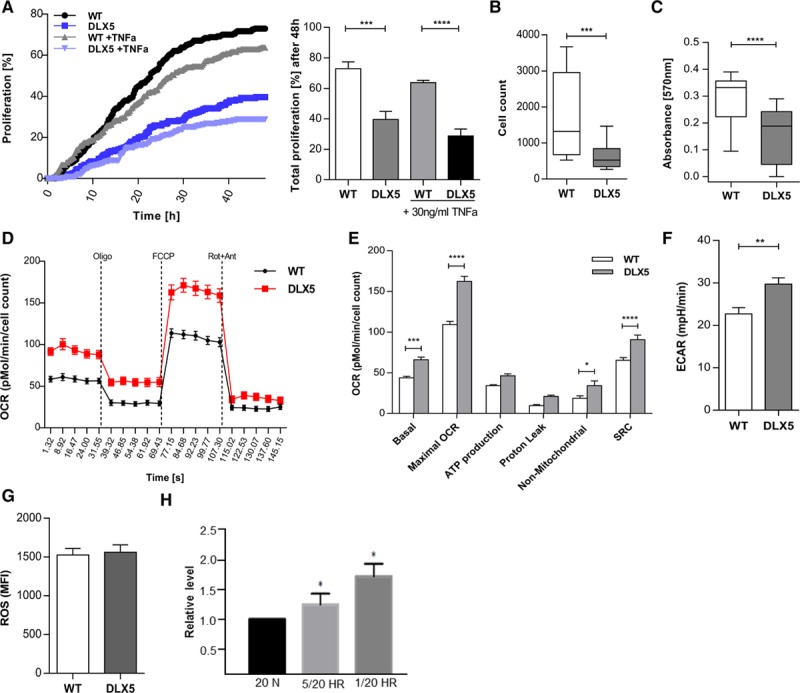
DLX5 decreases SGHPL-4 cell proliferation. A, DLX5high cells are less proliferative compared with wild-type (WT) cells as indicated by scoring dividing cells over 48 hours of incubation. After 48 hours of incubation, cell proliferation of DLX5high cells (39.58±5.34) is reduced by 45% compared with WT (72.92±4.49). ***P=0.001, ****P<0.0001, 2-way ANOVA, Bonferroni multiple-comparisons test. Tumor necrosis factor-α (TNFa) at a concentration of 30 ng/mL slightly decreased cell proliferation in both DLX5high and WT cells. P=NS. B, DLX5high cells exhibited decreased cell proliferation (n=6; median, 524.5; interquartile range [IQR], 343.8–843) compared with WT (n=6; median, 1320; IQR, 679.8–2955) confirmed by cell count high-throughput sampler assay. ***P<0.001, Mann-Whitney test. C, Microtiter plate test viability assay confirmed decreased cell proliferation in DLX5high cells (DLX5high: median, 0.1883; IQR, 0.0454–0.242 vs. WT: median, 0.3319; IQR, 0.2226–0.3565). ****P<0.0001, Mann-Whitney test. D, Effect of DLX5 on mitochondrial respiration in SGHPL-4 cells. Oxygen consumption rate (OCR) was measured under basal conditions followed by the sequential addition of oligomycin (Oligo; 0.75 µmol/L), FCCP (Carbonyl cyanide-4-[trifluoromethoxy]phenylhydrazone) (1 µmol/L), and antimycin A (Ant 1 µmol/L) plus rotenone (Rot; 0.1 µmol/L) in WT (n=6) and DLX5high cells (n=6). Data are normalized to the cell number. E, Individual parameters for basal respiration (WT vs. DLX5high, 43.901±1.705 vs. 66.082±3.213), maximal respiration (WT vs. DLX5high, 109.226±3.913 vs. 162.244±6.431), adenosine triphosphate (ATP) production (WT vs. DLX5high, 34.266±1.105 vs. 46.415±2.2), proton leak (WT vs. DLX5high, 9.635±1.307 vs. 20.85±1.687), nonmitochondrial OCR (WT vs. DLX5high, 18.72±2.989 vs. 34.247±5.773), and reserve capacity (WT vs. DLX5high, 65.325±3.381 vs. 90.866±5.455) were extracted from the assay. *P<0.05, ***P<0.001, ****P<0.0001, 2-way ANOVA, Bonferroni multiple-comparisons test. SRC indicates spare respiratory capacity. F, Mean basal extracellular acidification rate (ECAR) level in WT and DLX5high cells (DLX5high: median, 26.27; IQR, 24.27–36.35 vs. WT: median, 22.38; IQR, 17.05–26.99. **P=0.01, Mann-Whitney test. G, Reactive oxidative species (ROS) production in WT and DLX5high cells measured by fluorescent cell sorting. Data are presented as a mean fluorescent intensity (MFI) of the fluorescent signal from the dichlorodihydrofluorescein (DCF) oxidized by ROS. DCFH-DA diffuses into the cell and becomes deacetylated by cellular esterases to nonfluorescent 2’, 7’-Dichlorodihydrofluorescin (DCFH), which is next oxidized to fluorescent DCF by ROS. H, DLX5 expression level on induction of endoplasmic reticulum (ER) stress in BeWo cells. Quantification of DLX5 level on induction of ER stress indicates significant upregulation of its expression. 5/20 hypoxia-reoxygenation (HR) and 1/20 HR indicate cyclic condition of 6 hours of incubation in 5%/20% O2 and 1%/20% O2; and 20 N, normoxia. *P<0.01, Kruskal-Wallis test, Dunn multiple-comparisons test.
Elevated DLX5 Expression Affects the Metabolic Profile of the Trophoblast
Cell proliferation, growth, and metabolism are tightly linked processes. To determine whether reduced proliferation was associated with altered metabolism, we monitored metabolic parameters in DLX5high cells. We determined the extracellular acidification rate and oxygen consumption rate, indicators of mitochondrial respiration and glycolytic activity, respectively (Figure 5D–5F). Compared with control, metabolic profiling detected elevated level of extracellular acidification rate and maximal oxygen consumption rate values, suggesting an accelerated metabolism of DLX5high cells (Figure 5E and 5F). Furthermore, DLX5high cells displayed increased spare respiratory capacity values compared with control cells (Figure 5E). In principle, the increased energetic demand could reflect a response to increased stress or cell survival challenges.
DLX5 Expression Responds to ER Stress
Abnormal placentation in preeclampsia results in a series of biological stresses. To investigate the potential role of DLX5 in stress response, we monitored reactive oxidative species production and the effect of induced ER stress. Although we did not detect elevated reactive oxidative species production in DLX5high cells (Figure 5G), DLX5 expression was sensitive to induced ER stress in BeWo choriocarcinoma cells, expressing DLX5 at a readily detectable level. In a hypoxia-reoxygenation challenge assay, DLX5 expression increased significantly on ER stress in a severity-dependent manner (Figure 5H and Figure IXA in the online-only Data Supplement). Furthermore, in our DLX5high transcriptome, we observed upregulation of several genes involved in the unfolded protein response pathway that were associated with ER stress response (Figure IXB in the online-only Data Supplement). Eight of these genes (INSIG1, SREBF1, HSP90B1, ATF6, MBTPS1, PPP1R15A, XBP1, and HSPA2) were also dysregulated in our preeclamptic placenta samples (Figure IXC in the online-only Data Supplement). Although enhanced DLX5 level appears to trigger the cellular stress response, DLX5 expression increased with syncytium formation as evidenced by Forskolin treatment of BeWo cells, suggesting a potential role of DLX5 during the syncytialization process (Figure IXD in the online-only Data Supplement).
Dlx5 Expression Shifts From Postimplantation to Preimplantation Stage of Embryogenesis During Evolution
DLX5 is known primarily as a transcription factor regulating morphogenesis and tissue homeostasis,38–40 and it is mostly characterized during postimplantation embryogenesis. Because we observed DLX5 is expressed in trophoblasts, we sought to monitor its expression pattern in preimplantation embryos. We performed comparative single-cell RNA sequencing data analysis on mouse, macaque, and human embryos collected at early embryonic developmental stages.41–43 In human embryos, the expression of DLX5 appears at the transition from embryonic day 4 to 5 stage (Figure 6A), rendering DLX5 one of the earliest expressed genes in the human trophectoderm (Figure 6B). Dlx5 expression is shifted toward late trophectoderm in macaque embryos (Figure 6C) and is not detectable in murine preimplantation embryos (Figure 6A). To check whether Dlx5 is expressed at later stages of placenta development in mice, we performed placental immunostaining (embryonic day 14.5 and 15.5) in Dlx5-LacZ+/− animals. Although we observed a weak positive LacZ staining in the Dlx5-LacZ+/− animals on the external muscular layer of the placenta, the signal was not significantly different from the control (Figure X in the online-only Data Supplement), suggesting that Dlx5 is not involved in murine placentation. Surprisingly, only 33 common trophectoderm marker genes could be identified between mice and humans. In humans, among the early trophectoderm markers, DLX5 exhibits the highest activation of expression, followed by RGS13, NDRG2, ODAM, and SLC38A1, as well as ID3, HAND1, DLX3, and GCM1 (Figure 6D). Among the genes expressed differentially in human versus mouse preimplantation embryos, GREM2, GPR126, USP53, and EFNB3 are putative targets of DLX5 (Figure 5B). GREM2, GPR126, and USP53 are also upregulated in preeclampsia (Figure 1B), suggesting that the dysregulation of maternally expressed DLX5 and its putative targets might explain certain features of the human-specific nature of preeclampsia. The genes expressed in the same clusters might share transcriptional networks (Figure 6E and 6F and Table VII in the online-only Data Supplement). MEGs but not PEGs form characteristic clusters during human embryogenesis.
Figure 6.
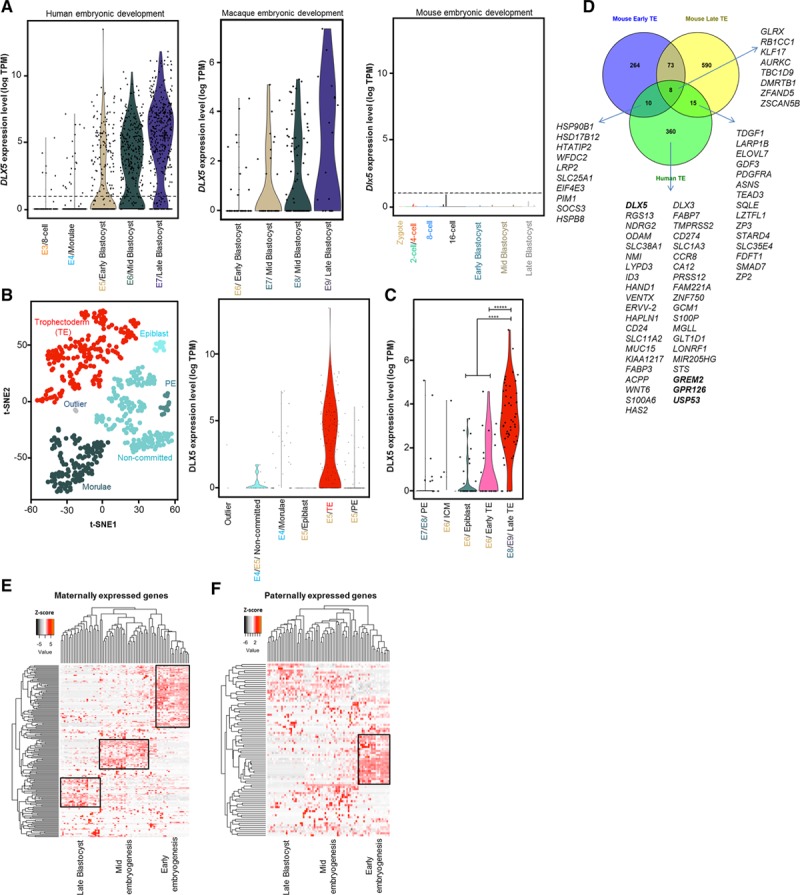
DLX5 expression in human, macaque, and mouse early embryonic development. A, Violin plots display the expression levels of DLX5 during different stages of human, macaque, and mouse early embryonic development. In humans, DLX5 starts to be expressed at embryonic day (E) E4/E5 stage of development. In macaques, DLX5 is expressed at E6 stage. Dlx5 is not expressed in mouse preimplantation embryo. B, Single-cell transcriptome analysis of human preimplantation embryo reveals trophectoderm (TE)-specific DLX5 expression. t-SNE analysis on human 353 single cells from E4 to E5 stages when inner cell mass (ICM) and trophectoderm split from Morulae. We defined clusters of cell populations to identify genes expressing exclusively in those clusters. We defined the cell type for each cluster according to the known markers. C, Single-cell transcriptome analysis of macaque preimplantation embryo demonstrates enriched DLX5 expression in the late trophectoderm. *****P=2.602E−12, ****P=1.387E-06, Wilcoxon test. D, Comparative analysis between human and mouse trophectoderm markers. For human trophectoderm markers, genes with log2-fold change >3 are shown. Single-cell transcriptome analysis of human embryogenesis for maternally (E) and paternally (F) expressed genes. Clustering analysis of imprinted genes expressed during early (oocyte, zygote, 2-cell, and 4-cell stage) and mid (8-stage, Morulae) embryogenesis and late blastocyst (179 of 257 maternally expressed genes, 86 of 150 paternally expressed genes). PE indicates primitive endoderm; TPM, transcripts per million; and t-SNE, t-distributed stochastic neighbor embedding.
Discussion
There certainly has been much speculation that disturbed regulation of imprinted genes might be involved in the development of preeclampsia; however, prior evidence could not establish a significant association between them.14–19 Here, we provide robust evidence for a mechanistic coupling between preeclampsia and disturbed placental imprinting. Our experimental strategy first aimed at identifying DEGs in preeclamptic placentas by analyzing genome-wide molecular data on well-characterized patient material. The list of DEGs was then intersected with the current catalog of human imprinted placental genes. Using the novel set of genes could clarify certain important issues concerning the long-term debated list of imprinted genes in the human placenta. Our strategy revealed several potential candidates, supporting the hypothesis that disturbed imprinting and preeclampsia could indeed be associated. Our candidate list included imprinted genes that were previously associated with preeclampsia, but their expression deregulation could not be convincingly connected to LOI.15 CYP2J2 and CD74 belonged to a category of genes whose deregulation was already implicated in preeclampsia, but their epigenetic disturbance was not considered as a contributing factor.21,22 Our strategy also identified genes that were not yet implicated in preeclampsia, and their imprinted status is poorly explored in placenta (APOBEC2, GATA3, DLX5). APOBEC2, an enzyme involved in controlling DNA-based parasites such as viruses and transposable elements, appeared on the list of MEGs deregulated in all 3 clusters of preeclampsia. The most significantly affected imprinted genes in preeclampsia were GATA3 and DLX5. GATA3 could be an excellent candidate for further research because it was previously reported to inhibit trophoblast invasion,44 thought to be a key process in preeclampsia. Here, we focus on DLX5, a transcription factor of the Distal-less homeobox protein family. DLX5 is involved in developmental processes of the limb, brain, and bone in both mice and humans.38,45 However, its placental function is not characterized.
We show that DLX5 is expressed in human villous and extravillous trophoblast and is controlled by imprinting. DLX5 is upregulated in ≈70% of patients with preeclampsia. The upregulation of DLX5 in preeclampsia is associated with its leaky expression from the imprinted allele (LOI). In contrast to previous studies,27,46 we find a correlation between expression of an imprinted gene and its LOI in preeclampsia. Our data mining28,47 also reveals differential CpG methylation of the DLX5 locus in preeclamptic placentas. Although our analysis does not rule out other mechanisms of DLX5 expression regulation such as transcriptional or microRNA regulation, we provide evidence of an association between disturbed imprinting gene expression and preeclampsia.
The spatial and temporal regulation of cell proliferation and differentiation is crucial for successful pregnancy. The first half of gestation is characterized by a series of trophoblast proliferation and differentiation processes, building mature villi and extravillous structures.48,49 Upregulation of DLX5 resulted in reduced (≈45%) proliferation of the trophoblast. The decreased trophoblast cell proliferation was accompanied by increased oxygen consumption. Why might poorly proliferating trophoblast cells require an accelerated metabolism? We hypothesized that DLX5 overexpression could sensitize trophoblasts to stress. As a result, the cells require increased metabolic activity to overcome this state of affairs. Indeed, the transcriptome analysis of DLX5high trophoblasts revealed several affected pathways acting as stress inducers, such as unfolded protein response pathways and increased interferon and death receptor signaling. Thus, the upregulation of DLX5 could be a factor contributing to an accelerated placenta aging process and elevated ER stress, resulting in stressed syncytiotrophoblast and consequently increased shedding of inflammatory factors into the maternal circulation.50,51 Although an enhanced DLX5 level triggers the cellular stress response, DLX5 expression increases with syncytium formation, suggesting a role of DLX5 in regulating the syncytialization. Our single-cell transcriptome data mining of human preimplantation embryos establishes DLX5 as a key marker of trophectoderm differentiation. We propose that DLX5 is involved in regulating a delicate balance between the proliferation and differentiation processes of the trophoblast. A disturbance of this key process has been previously associated with preeclampsia.52,53
In contrast with trophoblasts, DLX5 overexpression is associated with enhanced cell proliferation in various cancer cells.54–56 Thus, DLX5 might affect proliferation either negatively or positively during early development or in cancer, respectively. The response to overexpressed DLX5 possibly depends on cell type–specific target genes. Either way, DLX5 appears to be a key gene in determining the developmental decisions of trophoblast cells.
We modeled the effect of DLX5 upregulation in an in vitro system, overexpressing DLX5 in SGHPL-4 trophoblast cells (DLX5high). Because artificial, exogenous overexpression of a gene in cells could alter normal cellular function as a result of the accumulation of unprocessed proteins, we asked how faithfully DLX5high cells mimic preeclampsia. The transcriptome of DLX5high cells resembled that of the preeclamptic transcriptomes, and several dysregulated pathways were commonly seen both in vitro (DLX5high) and in vivo (preeclampsia samples). Thus, the DLX5high phenotype can model several features of preeclampsia in vitro in a cell culture system, signifying the impact of the deregulated DLX5 in the pathogenic phenotype of preeclampsia.
Given the barriers in analysis of preeclampsia resulting from its human specificity, our in vitro model system has considerable potential for downstream analyses. Nevertheless, our study do not suggest that there could be a single explanation for preeclampsia. We identified 3 distinct transcriptomic clusters of preeclamptic placenta (PE_P1 through PE_P3). That the clusters could be related to previously established categories of preeclampsia such as early and late onset of preeclampsia57 supports the view that preeclampsia is a heterogeneous placental disease and indeed comes in several discrete forms. Although PE_P2 matches late-onset preeclamptic placentas, PE_P1 and PE_P3 can be considered subdivisions of early-onset preeclampsia. The uncovered heterogenic nature of preeclampsia would call for validating a panel of subclass-specific biomarkers for future diagnostic procedures. The DLX5 overexpression phenotype is detectable in all the 3 clusters but is most pronounced in PE_P1 and PE_P2. Levels of DLX5 correlated with the placenta-derived PlGF circulating biomarker. Whether DLX5 will have utility as a biomarker is unclear because its LOI was not observed in all instances (69% of preeclampsia cases). Although the PE_P2 and PE_P3 clusters have no clear clinical pattern, patients in a PE_P1 cluster exhibit characteristic clinical phenotypes. Nevertheless, more samples need to be analyzed to securely relate the characteristics of the PE_P1 cluster to clinical disease phenotyping.
Our RNA sequencing data analysis revealed that a subset of preeclampsia is connected to disturbed epigenetic gene regulation. The global epigenetic turmoil is likely associated with the observed differential expression of genes regulating DNA methylation, resulting in the deregulation of transposable retroelements and imprinted genes. The mechanisms of regulating imprinting and repressing retroelements by DNA methylation share several common features.58 In fact, genomic imprinting is speculated to be a byproduct of the defense mechanisms of the genome against retroviruses and retroelements.59,60 A domesticated retroelement-derived gene (syncytin-1), implicated to have a key role in placental development,61–63 has been associated with preeclampsia.64,65 Here, in a subset of preeclampsia, we observed the reactivation of the human-specific L1_HS and SVA-F elements, capable of transposition in the human genome.37,66
Besides trophoblasts, dysregulation of DLX5 expression in other tissues has been reported to contribute to diseases. Downregulation of DLX5 in endometrial glands could also complicate early stages of pregnancy that could later manifest in intrauterine growth restriction or preeclampsia.67 LOI of the maternally expressed DLX5 in lymphoblastoid cells contributes to Rett syndrome, a disorder associated with GABAergic dysfunction.68,69 Dysregulation of DLX5 impairs the differentiation of GABAergic neurons.70 GABA can increase human chorionic gonadotrophin secretion in human placenta,71 indicating possible placenta-brain endocrine interactions regulated by imprinting. In contrast with Rett syndrome, we do not detect differential expression of DLX6 in preeclampsia, suggesting that the dysregulation of DLX5 in preeclampsia is not associated with expressional changes of DLX6 (not regulated by imprinting). We propose that DLX5 and DLX6 are regulated differently in brain and placenta.
DLX5 is expressed in human but not mouse trophoblast. Comparative single-cell transcriptome analysis revealed a differential expression of DLX5 between human, macaque, and mouse preimplantation embryogenesis, highlighting the diverged cellular function of DLX5 during mammalian embryonic evolution. Although the DLX5 gene is highly conserved across different mammalian species (>95% exon sequence similarity in human, macaque, and mouse), its upstream 2- to 10-kb (potential regulatory) region is much faster evolving (>80% and <10% sequence similarity for human versus macaque and for human versus mouse, respectively). Indeed, despite the conserved coding structure, DLX5 expression appears to be gradually shifted toward earlier developmental stages during mammalian evolution. Although DLX5 is not expressed in preimplantation embryos in mice, its expression peaks at the stage of trophectoderm and inner cell mass/epiblast separation, marks trophectoderm-committed cells, and is regulated by imprinting in humans. In addition to DLX5, we could identify further DEGs between human and mice trophectoderm. Among these genes, GREM2, GPR126, USP53, and EFNB3 are also putative targets of DLX5, suggesting that the function of a DLX5-regulated circuitry has been redefined during mammalian evolution. GREM2, GPR126, and USP53 are also upregulated in preeclampsia and thus might be associated with the human-specific nature of preeclampsia.
Acknowledgments
The authors thank Juliane Anders and Ute Gerhard for their excellent technical assistance. They also thank technician Lise Øhra Levy, postdoctoral researcher Meryam Sugulle, and Oslo University Hospital PhD students for valuable assistance in patient recruitment and biobank handling at Oslo University Hospital, Norway.
Sources of Funding
The Deutsche Forschungsgemeinschaft supported Drs Herse (HE 6249/1-2, HE 6249/4-1), Dechend (DE 631/9-1), and Müller. The German Center for Cardiovascular Research supported Dr Müller. This study was supported by the Wellcome Trust (grant 084804/2/08/Z). The Oslo Pregnancy Biobank work was supported by grants from the South-Eastern Norway Regional Health Authority, Norway, and from Oslo University Hospital. Dr Izsvák is funded by European Research Council Advanced (ERC-2011-AdG 294742). Dr LD Hurst is funded by ERC Advanced Grant ERC-2014-ADG 669207.
Disclosures
None.
Supplementary Material
Footnotes
Dr Zadora and M. Singh contributed equally.
Drs Hurst, Dechend, and Izsvák contributed equally.
The online-only Data Supplement is available with this article at http://circ.ahajournals.org/lookup/suppl/doi:10.1161/CIRCULATIONAHA.117.028110/-/DC1.
Circulation is available at http://circ.ahajournals.org.
References
- 1.ACOG. Hypertension in pregnancy: report of the American College of Obstetricians and Gynecologists’ Task Force on Hypertension in Pregnancy. Obstet Gynecol. 2013;122:1122–1131.. doi: 10.1097/01.AOG.0000437382.03963.88. [DOI] [PubMed] [Google Scholar]
- 2.Mosca L, Benjamin EJ, Berra K, Bezanson JL, Dolor RJ, Lloyd-Jones DM, Newby LK, Piña IL, Roger VL, Shaw LJ, Zhao D, Beckie TM, Bushnell C, D’Armiento J, Kris-Etherton PM, Fang J, Ganiats TG, Gomes AS, Gracia CR, Haan CK, Jackson EA, Judelson DR, Kelepouris E, Lavie CJ, Moore A, Nussmeier NA, Ofili E, Oparil S, Ouyang P, Pinn VW, Sherif K, Smith SC, Jr, Sopko G, Chandra-Strobos N, Urbina EM, Vaccarino V, Wenger NK. Effectiveness-based guidelines for the prevention of cardiovascular disease in women–2011 update: a guideline from the American Heart Association. Circulation. 2011;123:1243–1262. doi: 10.1161/CIR.0b013e31820faaf8. doi: 10.1161/CIR.0b013e31820faaf8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Steegers EA, von Dadelszen P, Duvekot JJ, Pijnenborg R. Pre-eclampsia. Lancet. 2010;376:631–644. doi: 10.1016/S0140-6736(10)60279-6. doi: 10.1016/S0140-6736(10)60279-6. [DOI] [PubMed] [Google Scholar]
- 4.Redman CW, Sargent IL. Immunology of pre-eclampsia. Am J Reprod Immunol. 2010;63:534–543. doi: 10.1111/j.1600-0897.2010.00831.x. doi: 10.1111/j.1600-0897.2010.00831.x. [DOI] [PubMed] [Google Scholar]
- 5.Roberts JM, Escudero C. The placenta in preeclampsia. Pregnancy Hypertens. 2012;2:72–83. doi: 10.1016/j.preghy.2012.01.001. doi: 10.1016/j.preghy.2012.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Reik W, Walter J. Genomic imprinting: parental influence on the genome. Nat Rev Genet. 2001;2:21–32. doi: 10.1038/35047554. doi: 10.1038/35047554. [DOI] [PubMed] [Google Scholar]
- 7.Graves JA. Genomic imprinting, development and disease:is pre-eclampsia caused by a maternally imprinted gene? Reprod Fertil Dev. 1998;10:23–29. doi: 10.1071/r98014. [DOI] [PubMed] [Google Scholar]
- 8.Haig D. Genetic conflicts in human pregnancy. Q Rev Biol. 1993;68:495–532. doi: 10.1086/418300. [DOI] [PubMed] [Google Scholar]
- 9.Haig D. Genomic imprinting and kinship: how good is the evidence? Annu Rev Genet. 2004;38:553–585. doi: 10.1146/annurev.genet.37.110801.142741. doi: 10.1146/annurev.genet.37.110801.142741. [DOI] [PubMed] [Google Scholar]
- 10.Hollegaard B, Byars SG, Lykke J, Boomsma JJ. Parent-offspring conflict and the persistence of pregnancy-induced hypertension in modern humans. PLoS One. 2013;8:e56821. doi: 10.1371/journal.pone.0056821. doi: 10.1371/journal.pone.0056821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Haig D. The kinship theory of genomic imprinting. Ann Rev Ecol Systematics. 2000;31:9–32.. [Google Scholar]
- 12.Robillard PY, Hulsey TC, Dekker GA, Chaouat G. Preeclampsia and human reproduction: an essay of a long term reflection. J Reprod Immunol. 2003;59:93–100. doi: 10.1016/s0165-0378(03)00040-8. [DOI] [PubMed] [Google Scholar]
- 13.Wildman DE, Chen C, Erez O, Grossman LI, Goodman M, Romero R. Evolution of the mammalian placenta revealed by phylogenetic analysis. Proc Natl Acad Sci U S A. 2006;103:3203–3208. doi: 10.1073/pnas.0511344103. doi: 10.1073/pnas.0511344103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Williams PJ, Broughton Pipkin F. The genetics of pre-eclampsia and other hypertensive disorders of pregnancy. Best Pract Res Clin Obstet Gynaecol. 2011;25:405–417. doi: 10.1016/j.bpobgyn.2011.02.007. doi: 10.1016/j.bpobgyn.2011.02.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yu L, Chen M, Zhao D, Yi P, Lu L, Han J, Zheng X, Zhou Y, Li L. The H19 gene imprinting in normal pregnancy and pre-eclampsia. Placenta. 2009;30:443–447. doi: 10.1016/j.placenta.2009.02.011. doi: 10.1016/j.placenta.2009.02.011. [DOI] [PubMed] [Google Scholar]
- 16.Huang GQ, Hu YY, Wang XD. Placental PHLDA2 gene imprinting in patients with pre-eclampsia [in Chinese]. Sichuan Da Xue Xue Bao Yi Xue Ban. 2015;46:104–107, 128.. [PubMed] [Google Scholar]
- 17.Berends AL, Bertoli-Avella AM, de Groot CJ, van Duijn CM, Oostra BA, Steegers EA. STOX1 gene in pre-eclampsia and intrauterine growth restriction. BJOG. 2007;114:1163–1167. doi: 10.1111/j.1471-0528.2007.01414.x. doi: 10.1111/j.1471-0528.2007.01414.x. [DOI] [PubMed] [Google Scholar]
- 18.Iglesias-Platas I, Monk D, Jebbink J, Buimer M, Boer K, van der Post J, Hills F, Apostolidou S, Ris-Stalpers C, Stanier P, Moore GE. STOX1 is not imprinted and is not likely to be involved in preeclampsia. Nat Genet. 2007;39:279–280. doi: 10.1038/ng0307-279. author reply 280. doi: 10.1038/ng0307-279. [DOI] [PubMed] [Google Scholar]
- 19.Kivinen K, Peterson H, Hiltunen L, Laivuori H, Heino S, Tiala I, Knuutila S, Rasi V, Kere J. Evaluation of STOX1 as a preeclampsia candidate gene in a population-wide sample. Eur J Hum Genet. 2007;15:494–497. doi: 10.1038/sj.ejhg.5201788. doi: 10.1038/sj.ejhg.5201788. [DOI] [PubMed] [Google Scholar]
- 20.Hamada H, Okae H, Toh H, Chiba H, Hiura H, Shirane K, Sato T, Suyama M, Yaegashi N, Sasaki H, Arima T. Allele-specific methylome and transcriptome analysis reveals widespread imprinting in the human placenta. Am J Hum Genet. 2016;99:1045–1058. doi: 10.1016/j.ajhg.2016.08.021. doi: 10.1016/j.ajhg.2016.08.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Herse F, Lamarca B, Hubel CA, Kaartokallio T, Lokki AI, Ekholm E, Laivuori H, Gauster M, Huppertz B, Sugulle M, Ryan MJ, Novotny S, Brewer J, Park JK, Kacik M, Hoyer J, Verlohren S, Wallukat G, Rothe M, Luft FC, Muller DN, Schunck WH, Staff AC, Dechend R. Cytochrome P450 subfamily 2J polypeptide 2 expression and circulating epoxyeicosatrienoic metabolites in preeclampsia. Circulation. 2012;126:2990–2999. doi: 10.1161/CIRCULATIONAHA.112.127340. doi: 10.1161/CIRCULATIONAHA.112.127340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Przybyl L, Haase N, Golic M, Rugor J, Solano ME, Arck PC, Gauster M, Huppertz B, Emontzpohl C, Stoppe C, Bernhagen J, Leng L, Bucala R, Schulz H, Heuser A, Weedon-Fekjær MS, Johnsen GM, Peetz D, Luft FC, Staff AC, Müller DN, Dechend R, Herse F. CD74-downregulation of placental macrophage-trophoblastic interactions in preeclampsia. Circ Res. 2016;119:55–68. doi: 10.1161/CIRCRESAHA.116.308304. doi: 10.1161/CIRCRESAHA.116.308304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Carmeliet P, Tessier-Lavigne M. Common mechanisms of nerve and blood vessel wiring. Nature. 2005;436:193–200. doi: 10.1038/nature03875. doi: 10.1038/nature03875. [DOI] [PubMed] [Google Scholar]
- 24.Liao WX, Laurent LC, Agent S, Hodges J, Chen DB. Human placental expression of SLIT/ROBO signaling cues: effects of preeclampsia and hypoxia. Biol Reprod. 2012;86:111. doi: 10.1095/biolreprod.110.088138. doi: 10.1095/biolreprod.110.088138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Okita C, Meguro M, Hoshiya H, Haruta M, Sakamoto YK, Oshimura M. A new imprinted cluster on the human chromosome 7q21-q31, identified by human-mouse monochromosomal hybrids. Genomics. 2003;81:556–559. doi: 10.1016/s0888-7543(03)00052-1. [DOI] [PubMed] [Google Scholar]
- 26.Stepan H, Herraiz I, Schlembach D, Verlohren S, Brennecke S, Chantraine F, Klein E, Lapaire O, Llurba E, Ramoni A, Vatish M, Wertaschnigg D, Galindo A. Implementation of the sFlt-1/PlGF ratio for prediction and diagnosis of pre-eclampsia in singleton pregnancy: implications for clinical practice. Ultrasound Obstet Gynecol. 2015;45:241–246. doi: 10.1002/uog.14799. doi: 10.1002/uog.14799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lambertini L, Diplas AI, Lee MJ, Sperling R, Chen J, Wetmur J. A sensitive functional assay reveals frequent loss of genomic imprinting in human placenta. Epigenetics. 2008;3:261–269. doi: 10.4161/epi.3.5.6755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Blair JD, Yuen RK, Lim BK, McFadden DE, von Dadelszen P, Robinson WP. Widespread DNA hypomethylation at gene enhancer regions in placentas associated with early-onset pre-eclampsia. Mol Hum Reprod. 2013;19:697–708. doi: 10.1093/molehr/gat044. doi: 10.1093/molehr/gat044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mátés L, Chuah MK, Belay E, Jerchow B, Manoj N, Acosta-Sanchez A, Grzela DP, Schmitt A, Becker K, Matrai J, Ma L, Samara-Kuko E, Gysemans C, Pryputniewicz D, Miskey C, Fletcher B, VandenDriessche T, Ivics Z, Izsvák Z. Molecular evolution of a novel hyperactive Sleeping Beauty transposase enables robust stable gene transfer in vertebrates. Nat Genet. 2009;41:753–761. doi: 10.1038/ng.343. doi: 10.1038/ng.343. [DOI] [PubMed] [Google Scholar]
- 30.Vaiman D, Calicchio R, Miralles F. Landscape of transcriptional deregulations in the preeclamptic placenta. PLoS One. 2013;8:e65498. doi: 10.1371/journal.pone.0065498. doi: 10.1371/journal.pone.0065498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Srinivas SK, Morrison AC, Andrela CM, Elovitz MA. Allelic variations in angiogenic pathway genes are associated with preeclampsia. Am J Obstet Gynecol. 2010;202:445.e1–445.11. doi: 10.1016/j.ajog.2010.01.040. doi: 10.1016/j.ajog.2010.01.040. [DOI] [PubMed] [Google Scholar]
- 32.Xiong Y, Zhou Q, Jiang F, Zhou S, Lou Y, Guo Q, Liang W, Kong D, Ma D, Li X. Changes of plasma and placental tissue factor pathway inhibitor-2 in women with preeclampsia and normal pregnancy. Thromb Res. 2010;125:e317–e322. doi: 10.1016/j.thromres.2010.02.017. doi: 10.1016/j.thromres.2010.02.017. [DOI] [PubMed] [Google Scholar]
- 33.Jebbink JM, Boot RG, Keijser R, Moerland PD, Aten J, Veenboer GJ, van Wely M, Buimer M, Ver Loren van Themaat E, Aerts JM, van der Post JA, Afink GB, Ris-Stalpers C. Increased glucocerebrosidase expression and activity in preeclamptic placenta. Placenta. 2015;36:160–169. doi: 10.1016/j.placenta.2014.12.001. doi: 10.1016/j.placenta.2014.12.001. [DOI] [PubMed] [Google Scholar]
- 34.Lu J, Zhou WH, Ren L, Zhang YZ. CXCR4, CXCR7, and CXCL12 are associated with trophoblastic cells apoptosis and linked to pathophysiology of severe preeclampsia. Exp Mol Pathol. 2016;100:184–191. doi: 10.1016/j.yexmp.2015.12.013. doi: 10.1016/j.yexmp.2015.12.013. [DOI] [PubMed] [Google Scholar]
- 35.Moore T, Dveksler GS. Pregnancy-specific glycoproteins: complex gene families regulating maternal-fetal interactions. Int J Dev Biol. 2014;58:273–280. doi: 10.1387/ijdb.130329gd. doi: 10.1387/ijdb.130329gd. [DOI] [PubMed] [Google Scholar]
- 36.Hancks DC, Mandal PK, Cheung LE, Kazazian HH., Jr. The minimal active human SVA retrotransposon requires only the 5’-hexamer and Alu-like domains. Mol Cell Biol. 2012;32:4718–4726. doi: 10.1128/MCB.00860-12. doi: 10.1128/MCB.00860-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hancks DC, Goodier JL, Mandal PK, Cheung LE, Kazazian HH., Jr. Retrotransposition of marked SVA elements by human L1s in cultured cells. Hum Mol Genet. 2011;20:3386–3400. doi: 10.1093/hmg/ddr245. doi: 10.1093/hmg/ddr245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Acampora D, Merlo GR, Paleari L, Zerega B, Postiglione MP, Mantero S, Bober E, Barbieri O, Simeone A, Levi G. Craniofacial, vestibular and bone defects in mice lacking the Distal-less-related gene Dlx5. Development. 1999;126:3795–3809. doi: 10.1242/dev.126.17.3795. [DOI] [PubMed] [Google Scholar]
- 39.Davideau JL, Demri P, Gu TT, Simmons D, Nessman C, Forest N, MacDougall M, Berdal A. Expression of DLX5 during human embryonic craniofacial development. Mech Dev. 1999;81:183–186. doi: 10.1016/s0925-4773(98)00227-5. [DOI] [PubMed] [Google Scholar]
- 40.Merlo GR, Paleari L, Mantero S, Zerega B, Adamska M, Rinkwitz S, Bober E, Levi G. The Dlx5 homeobox gene is essential for vestibular morphogenesis in the mouse embryo through a BMP4-mediated pathway. Dev Biol. 2002;248:157–169. doi: 10.1006/dbio.2002.0713. [DOI] [PubMed] [Google Scholar]
- 41.Deng Q, Ramsköld D, Reinius B, Sandberg R. Single-cell RNA-seq reveals dynamic, random monoallelic gene expression in mammalian cells. Science. 2014;343:193–196. doi: 10.1126/science.1245316. doi: 10.1126/science.1245316. [DOI] [PubMed] [Google Scholar]
- 42.Petropoulos S, Edsgärd D, Reinius B, Deng Q, Panula SP, Codeluppi S, Plaza Reyes A, Linnarsson S, Sandberg R, Lanner F. Single-cell RNA-seq reveals lineage and X chromosome dynamics in human preimplantation embryos. Cell. 2016;165:1012–1026. doi: 10.1016/j.cell.2016.03.023. doi: 10.1016/j.cell.2016.03.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Nakamura T, Okamoto I, Sasaki K, Yabuta Y, Iwatani C, Tsuchiya H, Seita Y, Nakamura S, Yamamoto T, Saitou M. A developmental coordinate of pluripotency among mice, monkeys and humans. Nature. 2016;537:57–62. doi: 10.1038/nature19096. doi: 10.1038/nature19096. [DOI] [PubMed] [Google Scholar]
- 44.Chiu YH, Chen H. GATA3 inhibits GCM1 activity and trophoblast cell invasion. Sci Rep. 2016;6:21630. doi: 10.1038/srep21630. doi: 10.1038/srep21630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Sowińska-Seidler A, Socha M, Jamsheer A. Split-hand/foot malformation: molecular cause and implications in genetic counseling. J Appl Genet. 2014;55:105–115. doi: 10.1007/s13353-013-0178-5. doi: 10.1007/s13353-013-0178-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Pozharny Y, Lambertini L, Ma Y, Ferrara L, Litton CG, Diplas A, Jacobs AR, Chen J, Stone JL, Wetmur J, Lee MJ. Genomic loss of imprinting in first-trimester human placenta. Am J Obstet Gynecol. 2010;202:391.e1–391.e8. doi: 10.1016/j.ajog.2010.01.039. doi: 10.1016/j.ajog.2010.01.039. [DOI] [PubMed] [Google Scholar]
- 47.Chu T, Bunce K, Shaw P, Shridhar V, Althouse A, Hubel C, Peters D. Comprehensive analysis of preeclampsia-associated DNA methylation in the placenta. PLoS One. 2014;9:e107318. doi: 10.1371/journal.pone.0107318. doi: 10.1371/journal.pone.0107318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Bulmer JN, Morrison L, Johnson PM. Expression of the proliferation markers Ki67 and transferrin receptor by human trophoblast populations. J Reprod Immunol. 1988;14:291–302. doi: 10.1016/0165-0378(88)90028-9. [DOI] [PubMed] [Google Scholar]
- 49.Reister F, Frank HG, Kingdom JC, Heyl W, Kaufmann P, Rath W, Huppertz B. Macrophage-induced apoptosis limits endovascular trophoblast invasion in the uterine wall of preeclamptic women. Lab Invest. 2001;81:1143–1152. doi: 10.1038/labinvest.3780326. [DOI] [PubMed] [Google Scholar]
- 50.Redman CW, Staff AC. Preeclampsia, biomarkers, syncytiotrophoblast stress, and placental capacity. Am J Obstet Gynecol. 2015;213(suppl):S9.e1, S9–S11. doi: 10.1016/j.ajog.2015.08.003. doi: 10.1016/j.ajog.2015.08.003. [DOI] [PubMed] [Google Scholar]
- 51.Redman CW, Sargent IL, Staff AC. IFPA Senior Award Lecture: making sense of pre-eclampsia: two placental causes of preeclampsia? Placenta. 2014;35(suppl):S20–S25. doi: 10.1016/j.placenta.2013.12.008. doi: 10.1016/j.placenta.2013.12.008. [DOI] [PubMed] [Google Scholar]
- 52.Redline RW, Patterson P. Pre-eclampsia is associated with an excess of proliferative immature intermediate trophoblast. Hum Pathol. 1995;26:594–600. doi: 10.1016/0046-8177(95)90162-0. [DOI] [PubMed] [Google Scholar]
- 53.Newhouse SM, Davidge ST, Winkler-Lowen B, Demianczuk N, Guilbert LJ. In vitro differentiation of villous trophoblasts from pregnancies complicated by intrauterine growth restriction with and without pre-eclampsia. Placenta. 2007;28:999–1003. doi: 10.1016/j.placenta.2007.04.008. doi: 10.1016/j.placenta.2007.04.008. [DOI] [PubMed] [Google Scholar]
- 54.Kato T, Sato N, Takano A, Miyamoto M, Nishimura H, Tsuchiya E, Kondo S, Nakamura Y, Daigo Y. Activation of placenta-specific transcription factor distal-less homeobox 5 predicts clinical outcome in primary lung cancer patients. Clin Cancer Res. 2008;14:2363–2370. doi: 10.1158/1078-0432.CCR-07-1523. doi: 10.1158/1078-0432.CCR-07-1523. [DOI] [PubMed] [Google Scholar]
- 55.Xu J, Testa JR. DLX5 (distal-less homeobox 5) promotes tumor cell proliferation by transcriptionally regulating MYC. J Biol Chem. 2009;284:20593–20601. doi: 10.1074/jbc.M109.021477. doi: 10.1074/jbc.M109.021477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Tan Y, Cheung M, Pei J, Menges CW, Godwin AK, Testa JR. Upregulation of DLX5 promotes ovarian cancer cell proliferation by enhancing IRS-2-AKT signaling. Cancer Res. 2010;70:9197–9206. doi: 10.1158/0008-5472.CAN-10-1568. doi: 10.1158/0008-5472.CAN-10-1568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Lisonkova S, Joseph KS. Incidence of preeclampsia: risk factors and outcomes associated with early- versus late-onset disease. Am J Obstet Gynecol. 2013;209:544.e1–544.e12. doi: 10.1016/j.ajog.2013.08.019. doi: 10.1016/j.ajog.2013.08.019. [DOI] [PubMed] [Google Scholar]
- 58.Suzuki S, Ono R, Narita T, Pask AJ, Shaw G, Wang C, Kohda T, Alsop AE, Marshall Graves JA, Kohara Y, Ishino F, Renfree MB, Kaneko-Ishino T. Retrotransposon silencing by DNA methylation can drive mammalian genomic imprinting. PLoS Genet. 2007;3:e55. doi: 10.1371/journal.pgen.0030055. doi: 10.1371/journal.pgen.0030055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Barlow DP. Methylation and imprinting: from host defense to gene regulation? Science. 1993;260:309–310. doi: 10.1126/science.8469984. [DOI] [PubMed] [Google Scholar]
- 60.McDonald JF, Matzke MA, Matzke AJ. Host defenses to transposable elements and the evolution of genomic imprinting. Cytogenet Genome Res. 2005;110:242–249. doi: 10.1159/000084958. doi: 10.1159/000084958. [DOI] [PubMed] [Google Scholar]
- 61.Varela M, Spencer TE, Palmarini M, Arnaud F. Friendly viruses: the special relationship between endogenous retroviruses and their host. Ann N Y Acad Sci. 2009;1178:157–172. doi: 10.1111/j.1749-6632.2009.05002.x. doi: 10.1111/j.1749-6632.2009.05002.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Rawn SM, Cross JC. The evolution, regulation, and function of placenta-specific genes. Annu Rev Cell Dev Biol. 2008;24:159–181. doi: 10.1146/annurev.cellbio.24.110707.175418. doi: 10.1146/annurev.cellbio.24.110707.175418. [DOI] [PubMed] [Google Scholar]
- 63.Sugimoto J, Schust DJ. Review: human endogenous retroviruses and the placenta. Reprod Sci. 2009;16:1023–1033. doi: 10.1177/1933719109336620. doi: 10.1177/1933719109336620. [DOI] [PubMed] [Google Scholar]
- 64.Ruebner M, Strissel PL, Ekici AB, Stiegler E, Dammer U, Goecke TW, Faschingbauer F, Fahlbusch FB, Beckmann MW, Strick R. Reduced syncytin-1 expression levels in placental syndromes correlates with epigenetic hypermethylation of the ERVW-1 promoter region. PLoS One. 2013;8:e56145. doi: 10.1371/journal.pone.0056145. doi: 10.1371/journal.pone.0056145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Vargas A, Toufaily C, LeBellego F, Rassart É, Lafond J, Barbeau B. Reduced expression of both syncytin 1 and syncytin 2 correlates with severity of preeclampsia. Reprod Sci. 2011;18:1085–1091. doi: 10.1177/1933719111404608. doi: 10.1177/1933719111404608. [DOI] [PubMed] [Google Scholar]
- 66.Hancks DC, Kazazian HH., Jr. SVA retrotransposons: evolution and genetic instability. Semin Cancer Biol. 2010;20:234–245. doi: 10.1016/j.semcancer.2010.04.001. doi: 10.1016/j.semcancer.2010.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Bellessort B, Le Cardinal M, Bachelot A, Narboux-Nême N, Garagnani P, Pirazzini C, Barbieri O, Mastracci L, Jonchere V, Duvernois-Berthet E, Fontaine A, Alfama G, Levi G. Dlx5 and Dlx6 control uterine adenogenesis during post-natal maturation: possible consequences for endometriosis. Hum Mol Genet. 2016;25:97–108. doi: 10.1093/hmg/ddv452. doi: 10.1093/hmg/ddv452. [DOI] [PubMed] [Google Scholar]
- 68.Horike S, Cai S, Miyano M, Cheng JF, Kohwi-Shigematsu T. Loss of silent-chromatin looping and impaired imprinting of DLX5 in Rett syndrome. Nat Genet. 2005;37:31–40. doi: 10.1038/ng1491. doi: 10.1038/ng1491. [DOI] [PubMed] [Google Scholar]
- 69.Chao HT, Chen H, Samaco RC, Xue M, Chahrour M, Yoo J, Neul JL, Gong S, Lu HC, Heintz N, Ekker M, Rubenstein JL, Noebels JL, Rosenmund C, Zoghbi HY. Dysfunction in GABA signalling mediates autism-like stereotypies and Rett syndrome phenotypes. Nature. 2010;468:263–269. doi: 10.1038/nature09582. doi: 10.1038/nature09582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Stühmer T, Anderson SA, Ekker M, Rubenstein JL. Ectopic expression of the Dlx genes induces glutamic acid decarboxylase and Dlx expression. Development. 2002;129:245–252. doi: 10.1242/dev.129.1.245. [DOI] [PubMed] [Google Scholar]
- 71.Licht P, Harbarth P, Merz WE. Gaba-mediated stimulation of hcg secretion suggests a parallelism in the control of central-nervous and placental gonadotropin release. Placenta. 1992;13:151–161. [Google Scholar]


