Supplemental Digital Content is available in the text
Keywords: African American, black populations, blood pressure, discrimination, gene-environment interaction
Abstract
Both genomics and environmental stressors play a significant role in increases in blood pressure (BP). In an attempt to further explain the hypertension (HTN) disparity among African Americans (AA), both genetic underpinnings (selected candidate genes) and stress due to perceived racial discrimination (as reported in the literature) have independently been linked to increased BP among AAs. Although Gene x Environment interactions on BP have been examined, the environmental component of these investigations has focused more on lifestyle behaviors such as smoking, diet, and physical activity, and less on psychosocial stressors such as perceived discrimination.
The present study uses candidate gene analyses to identify the relationship between Everyday Discrimination (ED) and Major Life Discrimination (MLD) with increases in systolic BP (SBP) and diastolic BP (DBP) among AA in the Jackson Heart Study. Multiple linear regression models reveal no association between discrimination and BP after adjusting for age, sex, body mass index (BMI), antihypertensive medication use, and current smoking status.
Subsequent candidate gene analysis identified 5 SNPs (rs7602215, rs3771724, rs1006502, rs1791926, and rs2258119) that interacted with perceived discrimination and SBP, and 3 SNPs (rs2034454, rs7602215, and rs3771724) that interacted with perceived discrimination and DBP. Most notably, there was a significant SNP × discrimination interaction for 2 SNPs on the SLC4A5 gene: rs3771724 (MLD: SBP P = .034, DBP P = .031; ED: DBP: P = .016) and rs1006502 (MLD: SBP P = .034, DBP P = .030; ED: DBP P = .015).
This study supports the idea that SNP × discrimination interactions combine to influence clinically relevant traits such as BP. Replication with similar epidemiological samples is required to ascertain the role of genes and psychosocial stressors in the development and expression of high BP in this understudied population.
1. Introduction
Hypertension (HTN) is a common disease, affecting over 78 million adults in the US every year.[1] African Americans (AA) are disproportionately affected by this disease, with an earlier age of onset as well as higher rates of complications than other racial/ethnic groups.[2] The etiology of HTN is complex and multifactorial. Studies have identified social, biological, environmental, and genetic risk factors for HTN,[3] but few studies have considered the ways in which these risk factors interact or work together in elevating risk among AA. Social factors such as perceived discrimination related to skin color have been associated with high blood pressure (HBP) in diverse populations, especially in low-income groups.[4,5] Further, although self-reported race has been associated with poor mental and physical health outcomes in AA,[3] little is known about the degree to which genetic risk for elevated BP and HTN is shaped by environmental factors, including psychosocial stressors such as the experience of unfair treatment-environment interaction that may exacerbate poor outcomes.
Discrimination is a complex and multidimensional psychosocial stressor. Previous studies have defined the experience of discrimination as the perception or view that one has been treated unfairly. Self-report is the most common way of measuring perceptions of discrimination in survey research.[6] Two of the more widely used measures of perceived discrimination are Everyday Discrimination (ED) and Major Life Discrimination (MLD).[7,8] MLD refers to the experience of unfair treatment (during one's lifetime) that might block or forestall social mobility,[8,9] such as unfair treatment by the police or in the housing market. ED refers to “daily hassles” that are experienced in “everyday situations,” such as the frequency of being made to feel uncomfortable for unfair or unjust reasons, like being treated with less courtesy in public places than others.
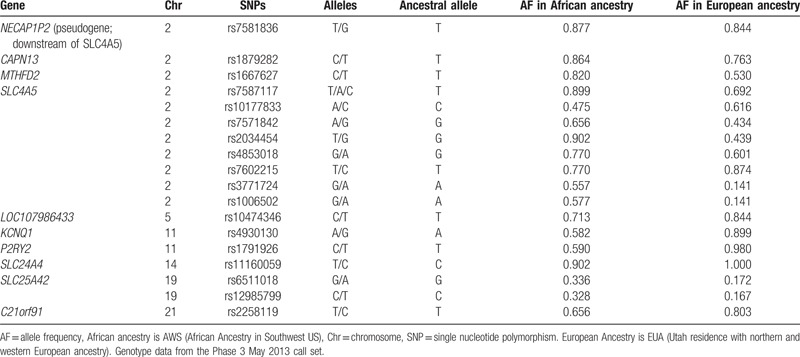
According to the National Survey of American Life (NSAL), 51% of AA have experienced at least some type of ED at least a few times per month and 59% have experienced a least 1 type of MLD at some point in their lifetime.[10] Studies also show a link between various measures of discrimination and HTN and related cardiovascular diseases (CVD).[11–14] These studies suggest that chronic experiences with discrimination are more consistently linked to BP and HTN than are subtle or acute experiences.[15] Although the relationship between genetic polymorphisms and biological factors such as drug metabolism, body mass index (BMI), or cardiac structure and function have been established, how the interaction between genetics and stressors, such as perceived discrimination, must continue to be examined. There is evidence of interactions between perceived discrimination and genetic risk on developing HBP.[13] In a sample of AA women in the Midwest (N = 137), 1 SNP (rs10177833) on the SLC4A5 gene was found to have a significant association with skin color on systolic BP (SBP).[16] This finding was replicated in a sample of 3 generations of West African women (N = 199), where the rs8179526 SNP on the SLC4A5 gene was associated with SBP.[17] In this study, we use a targeted candidate gene approach to examine the interaction between genetic polymorphisms, and perceived racial discrimination on BP among AA. Specific candidate genes were selected based on those that have been significantly associated with HBP previously in the literature as noted previously. The genes and SNPs proposed for investigation in the present study are those positional candidate genes that have been found most consistently to be statistically significantly associated with HBP in previous studies among AAs. A complete list of the genes and SNPs can be found in Table 1, and complete full description of functionality and differences in allele frequency by ancestry can be found in Taylor et al, 2016.[18] The purpose of the study was to examine the main effects of gene (SNP) and discrimination on SBP and diastolic BP (DBP), as well as gene (G) and discrimination (D) interaction effects (G × D) on SBP and DBP. This study extends the science from examining only individual main effects of perceived discrimination or genetic underpinnings on BP. Because it has been well established that genomics and perceived discrimination alone may contribute significantly to increases in BP among AAs, this study integrates both genomic and perceived discrimination interaction effects to advance the science to better understand the combinatorial effects of these factors on BP. If we can better understand the multiple factors and interactive effects that contribute to AAs having the highest incidence and prevalence of HTN in the US, we will be better equipped to develop more precise individualized interventions to reduce this health disparity.
Table 1.
Allele frequencies of relevant SNPs from The 1000 Genomes Project[19].
2. Methods
2.1. Study group
The Jackson Heart Study (JHS) enrolled 5301 AA living in the metropolitan Jackson, MI area between 2000 and 2004. For this study, we included only those who had both genotypic and phenotypic data available for analyses (N = 2937), of which 5 participants were missing SBP and DBP readings, so N = 2932 was the final sample size in this report. Adult men and women were recruited in 4 main ways: random community sampling, volunteer, family member participants, and through the Atherosclerosis Risk in Communities Study (ARIC). Participants ranged in age from 21 to 85 years and self-identified as AA. The overall purpose of the study was to examine genetic, environmental, and social factors contributing to CVD development in AA. Detailed study design and methods are discussed elsewhere.[20–23] The University of Mississippi Medical Center Institutional Review Board provided ethical approval for the JHS. The Jackson Heart Study Publications and Presentations Committee provided approval for this secondary analysis study as presented here.
3. Measures
3.1. Height, weight, and body mass index
Height was measured by stadiometer and weight by electronic balance. BMI was calculated using weight in kilograms divided by height in meters squared. BMI over 25 is defined as overweight, and BMI greater than 30 is defined as obese. BMI has been shown to be a valid predictor of adiposity calculated for weight and height.[24]
3.2. Blood pressure readings
BP measurements were taken with random-zero sphygmomanometers and cuffs appropriate for arm size. Three readings were measured in the right arm after the participant rested in the sitting position for at least 5 minutes according to The Seventh Report of the Joint National Committee on Prevention, Detection, Evaluation, and Treatment of High Blood Pressure (JNC-7) guidelines.[25] SBP and DBP were determined by the first and fifth phase Korotkoff sounds, respectively, with the last 2 BP readings averaged for the analyses. The diagnosis of HTN was established based on average BP levels measured at the study visit (>140/90 mm Hg) or a prior diagnosis of HTN and current treatment with antihypertensive medications. Briefly, during each visit, JHS participants presented the medications they had taken over the 2 weeks prior to the examination. These included both prescribed and over-the-counter medications as well as herbal preparations. For those who did not bring medications to the examination, medication lists for the participant were obtained by a telephone call to the participants following the visit or by phone call to the consenting participant's pharmacy.[26]
3.3. Perceived discrimination
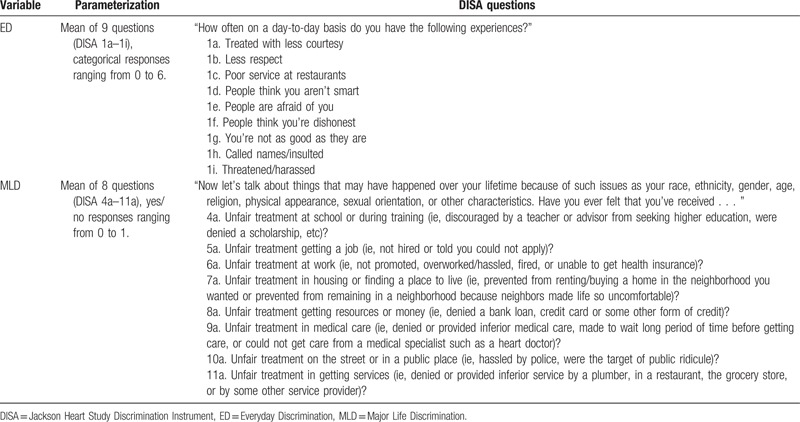
We measured personal experiences with discrimination using the Discrimination Instrument (DISA), a multidimensional instrument that has also been used in the NSAL.[27] The instrument measures the occurrence, frequency, attribution, and responses to discrimination, and has 2 subscales for everyday and MLD. We followed previous research in operationalizing ED and MLD.[27] Questions and responses for Everyday (ED) and MLD are presented in Table 2. ED was assessed via 9 questions about day-to-day exposures to discrimination, with responses ranging from 0 to 6, with higher values corresponding with a greater frequency of exposure (ie, 0 = never, 1 = less than a few times a year, 2 = a few times a year, 3 = a few times a month, 4 = at least once a week, 5 = almost every day, 6 = several times a day). The mean of these responses was used to create a summary scale for ED. The second scale, MLD, was created using the mean of 8 questions regarding unfair treatment due to a variety of physical or cultural characteristics, without attribution. These questions had yes (1) or no (0) coded responses. The overall instrument, as well as ED and MLD subscales demonstrate good reliability (Cronbach alpha = .78, .84, and .77, respectively).
Table 2.
Creation of summary variables for discrimination, Jackson Heart Study Discrimination Instrument (DISA).
In addition to ED and MLD, we further examined participants’ experiences of discrimination related specifically to race: EDAR (Experiences of Discrimination Attributed to Race) and CMLDAR (Chronic Major Life Discrimination Attributed to Race). As respondents were asked to attribute their ED to several factors, we examined the ED summary scale, and then, created EDAR to represent those experiences that were attributed to race, versus those that were attributed to age, gender, weight, or some other reason. The EDAR is the mean of 9 questions (1a–1i) on the ED instrument, for those who indicated “race” as the primary reason for discrimination. Similarly, CMLDAR was created to represent chronic experiences of MLD that were related to race. Participants who indicated a higher frequency of discriminatory experiences (several times a year to a few times a month) were categorized as “chronic- CMLD,” compared with those who reported a frequency of a few times a year or less. Among participants who had chronically experienced discrimination, we further categorized those who indicated that their experiences were related to race (and not age, gender, or another factor) as CMLDAR. CMLDAR is the mean of 4b to 11b for those who indicated “race” on 13a. Further details on scoring assessment of EDAR and CMLDAR can be found in Sims et al.[27]
3.4. Genotyping
Quality control of genotyped data (SNPs) was performed using the BROAD genetic analysis platform (GAP) that consists of PLINK[28] and Birdseed v1.33[29] software. Samples with genotyping success rate <95%, monomorphic SNPs, 1176 SNPs that mapped to several loci in the human genome, and SNPs with minor allele frequency (MAF) <1% were removed for quality control. We also removed samples with very low heterozygosity suggesting poor DNA quality and samples with very high heterozygosity suggesting sample contamination. In addition, we removed all pairs that shared ≥5% of their genome, as well as samples that did not cluster well when subjected to multidimensional scaling (MDS) or genome-wide “neighbor” analysis in PLINK. For the family-based subcohort of the JHS, early analytical assessment by CARE investigators found little effect on inflation factor due to familial correlation. Further, we removed SNPs for which genotype missingness could not be predicted by surrounding haplotypes. Mendelian inconsistence was checked for family data using PLINK and the corresponding SNPs were removed. No SNPs were removed due to significant deviation from Hardy-Weinberg equilibrium (HWE) because the AA population is an admixed population, which may result in departure from HWE.
Genotype imputation performed in CARe has been detailed elsewhere. Briefly, in CARe, imputation was performed using the MACH[30] program with HapMap phase 2 (build 36 release 22) as input. As the AA population is admixed with the proportion of European ancestry (EA) estimated to be ∼17% to 19%,[31,32] an artificial reference panel consisting of equal proportions of the YRI and CEU HapMap phased haplotypes (using only SNPs found in both YRI and CEU panels, ie, ∼2.2 M SNPs) was constructed. Hao et al suggested that the accuracy of using the mixed panel for AAs is comparable to the accuracy reported when imputing a population of Nigerians using YRI as a reference panel.[33]
3.5. Statistical methods
Data management, descriptive statistics for the covariates and outcome variables, and the regression analyses were conducted using the statistics software program R, version 3.2.3 (http://www.r-project.org/). Known predictors of BP, such as age, sex, BMI, antihypertensive use, and smoking status, were assessed in a multiple linear regression model including all predictors. We used similar models to assess associations between each perceived discrimination (including ED and MLD), top 10 principal components (PCs), and outcome variables (ie, SBP and DBP) adjusted for age, sex, BMI, antihypertensive use, and current smoking status. We used the same models to test for the association between each SNP and the BP phenotypes (ie, SBP and DBP). We tested each SNP for additive main effects on BP in a test with 1 degree of freedom. We also tested the SNP × perceived discrimination (ie, ED and MLD) interaction effects of SBP and DBP using linear mixed models with age, sex, BMI, antihypertensive medication use, and top 10 PCs of the Genome-Wide Association Study (GWAS) data as covariates. Data management, descriptive statistics for the covariates and outcome variables, and the regression analyses were conducted using the statistics software package R in version 3.2.3.[34]
To study Gene × Discrimination (G × D) interactions on BP where D represents the perceived discrimination, we applied the following linear regression model.
where Yi is the BP outcome (ie, SBP and DBP) for person i; Zi is a vector of adjustment variables including age, sex, BMI, antihypertensive medication use, current smoker, and the first 10 PCs of the common genome-wide SNPs; Di is the measure of perceived discrimination of person i; and SNPi is the additive genetic effect of any given SNP. β4 represents the effect of SNP × D interaction. In this study, we examined 18 SNPs from 10 candidate genes (Tables 1 and 3) on SBP and DBP in 2932 JHS AA participants. We also investigated the SNP × D interaction effects on SBP and DBP for women and men separately using the same statistical model without sex as a covariate.
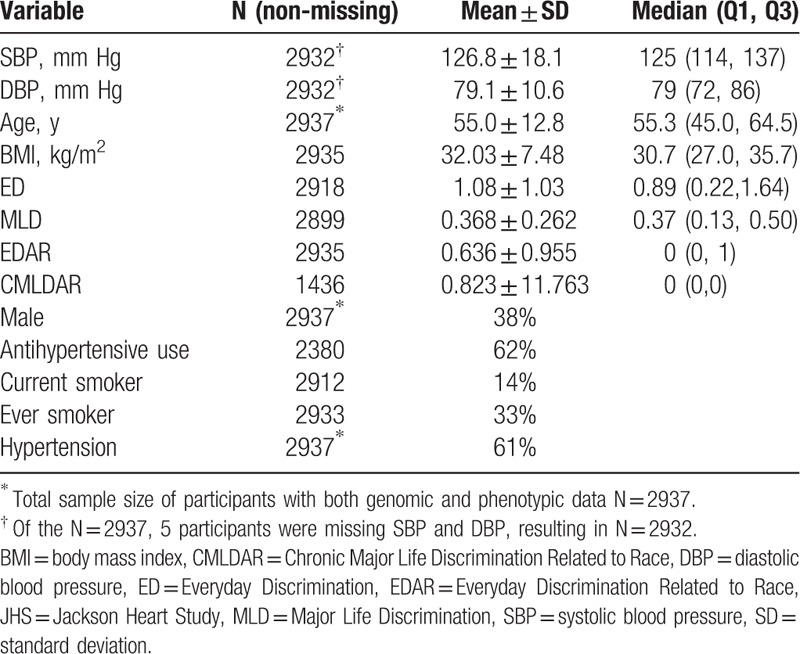
Table 3.
Descriptive statistics of the JHS sample with both phenotypic and genotypic data.
4. Results
In Table 3, we present descriptive statistics for the 2937 JHS participants with both phenotypic and genetic data. Five participants were missing SBP and DBP data, resulting in 2932 participants for examination. The sample had a mean age of 55 years, and the majority of participants was female, obese, nonsmokers, had HTN, and was on antihypertension medication. Mean SBP and DBP were 126.8 and 79.1 mm Hg, respectively. Overall, reports of discrimination reported among participants were low. The mean ED was 1.08 (range 0–6), mean EDAR was 0.63 (range 0–6), mean MLD was 0.36 (range 0–1).
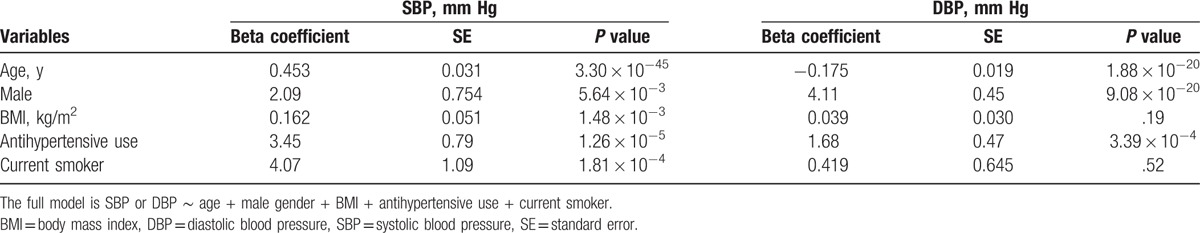
To assess the associations between SBP/DBP and known predictors, we conducted further multiple linear regression analyses. We confirmed that older age, male sex, and antihypertensive medication use were significantly associated with both SBP and DBP (Table 4). BMI and smoking status were significantly associated with SBP, but not DBP. These 5 variables were consistently used as covariates in the association analyses of discrimination measures and genetic factors described later.
Table 4.
The summary of multiple regression analysis of systolic (SBP) and diastolic (DBP) blood pressure.
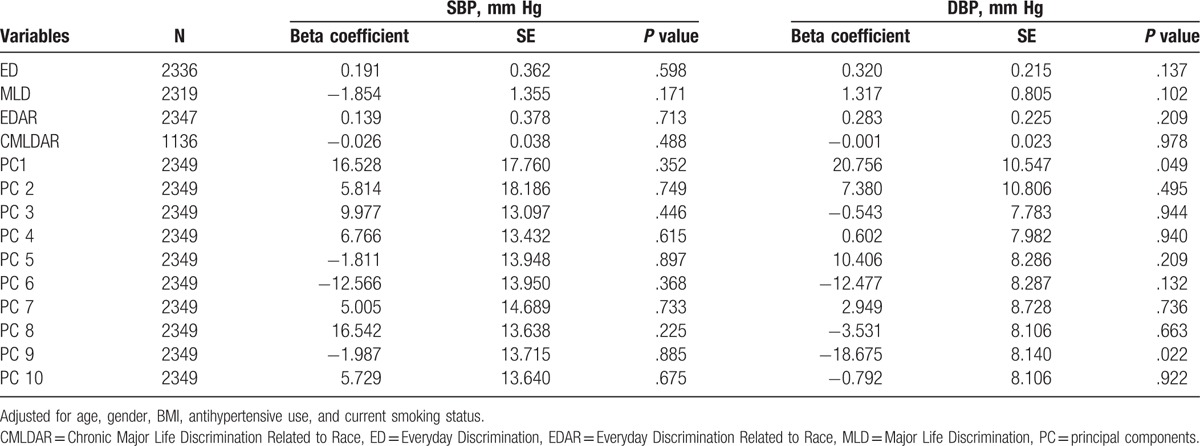
The association analyses of the 4 discrimination measures with SBP and DBP adjusted for age, sex, BMI, antihypertensive use, and current smoking status are presented in Table 5. ED, MLD, EDAR, and CMLDAR were examined in univariate analyses, and only ED and MLD were suggestively associated with SBP and DBP; thus they were the only 2 variables included in interaction models. Neither ED nor MLD was independently associated with SBP or DBP after adjustment for covariates. Additionally, the top PCs of GWAS data were tested for association with SBP and DBP (Table 5). At alpha level of 0.05, only PC1 and PC9 were associated with DBP after adjustment of covariates.
Table 5.
The associations of covariates and systolic and diastolic blood pressure.
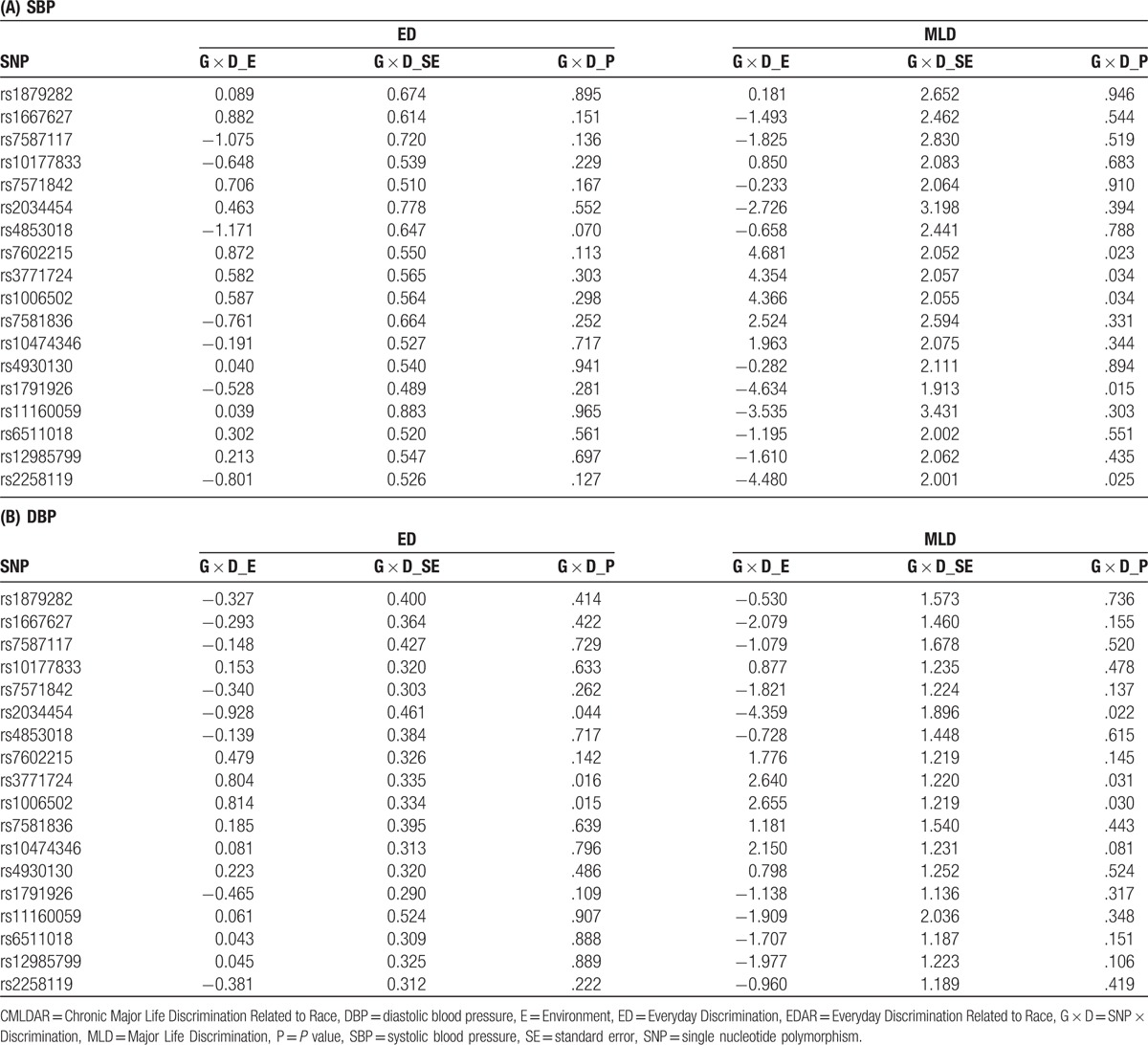
We obtained imputed dosage data of 20 SNPs based on previously reported candidate genes and SNPs (supplementary tables, sex stratified). All SNPs were coded as additive genetic effects. After filtering SNPs with poor imputed quality (ie, r2 <0.8), we tested G × D interaction of 18 SNPs, adjusted for age, sex, BMI, antihypertensive medication use, current smoking status, top 10 PCs of GWAS data, and main effects of each SNP and discrimination measure. The test statistics including beta coefficient (E), standard error (SE), and P values (P) of each pair of G × D are summarized in Table 6. For SBP (Table 6A), 5 G × MLD interaction terms were associated with SBP (rs7602215, rs3771724, rs1006502, rs1791926, and rs2258119) at alpha level of 0.05. However, none of G × ED interaction terms were associated with SBP. For DBP, 3 SNPs (rs2034454, rs7602215 and rs3771724) had consistently significant interaction effects of both ED and MLD at alpha level of 0.05. Notably, rs3771724 and rs1006502 had significant G × MLD interaction associated with both SBP and DBP (Table 6A and B). Supplementary tables are provided for sex-stratified G × D results. Interestingly, the significant G × D identified in the pooled analyses were only observed in women, not men. In a few cases, the effects were even more significant in women with a smaller sample size (see supplementary tables). After adjusting with Bonferroni methods at a significance threshold of 0.05, (0.05/18 SNPs = 0.0028), we did not identify any significant interaction effects. The marginally significant interactions account for 0.17% to 0.26% of total variation in BPs. A supplementary table has been added that includes partial R2 (percent of variance) of all tested SNPs with BP phenotypes.
Table 6.
Association of SNP × Discrimination (G × D) interaction with systolic (SBP) (A) and diastolic (DBP) (B) blood pressure.
5. Discussion
This study addressed the paucity of research examining the Gene × Discrimination interaction on BP in an AA population using data from the JHS. We identified a number of SNPs prime for further exploration in SNP-discrimination interactions on BP among AA. Our most robust finding included 2 SNPs (rs3771724 and rs1006502) that showed interaction with MLD on both SBP and DBP, and ED on DBP in this sample of AAs.
Interestingly, the rs3771724 and rs1006502 loci are linked (r2 = 1)[35] and located in the intronic region of the SLC4A5 (NBCe2) gene, previously shown to be associated with BP.[16,35–39] SLC4A5 is an electrogenic sodium-bicarbonate cotransporter in the kidney and liver. A mouse model lacking homolog Slc4a5 displays a hypertensive phenotype,[40] which may be a result of sodium-induced hyperaldosteronism via increased activity of the epithelial sodium channel in renal principal cells, though this has yet to be confirmed in human subjects.[41] High salt concentrations in cultured human renal cortical tissue showed increased SLC4A5 expression compared with low salt conditions in vitro.[42]
Several studies have similarly reported associations between the SLC4A5 gene and HBP among AA and European American (EA) adults. In one study, adults from the University of Virginia and HyperPath cohorts, polymorphisms in the SLC4A5 gene were associated with induction of salt-sensitive HTN among adults of EA ancestry.[43] AA typically have higher rates of salt sensitivity than EA, and rates of salt-sensitivity among hypertensive AA men and women have been reported at 73%, with normotensive AA men and women ranging 36% to 79%, and normotensive AA women ranging from 43% to 64%.[44] Genetic polymorphisms, such as SNPs, in the SLC4A5 gene have also been implicated in BP-related traits, such as resting pulse pressure, submaximal exercise pulse pressure, submaximal SBP, and submaximal rate pressure in EA.[35]
One other study reported an association between the rs1006502 (hcv8941031) locus of the SLC4A5 gene and HTN in AA[39] and with BP in EA.[37] The TT allele, which was the minor allele among their Utah-based population of EA, had a slight increase in plasma CO2 levels compared with participants with a CT or CC genotype.[37] Among AAs, the CC genotype is the minor allele, suggesting this population may have increased risk for elevated plasma CO2 levels (Table 6). The rs3771724 locus has been associated with submaximal exercise oxygen consumption (VO250) and submaximal exercise carbon dioxide expiration (VCO250) in subjects of EA.[35] To our knowledge, no mechanism of action for the genotype at the rs1006502 or the rs3771724 locus on the SLC4A5 gene product has been elucidated.
5.1. Strengths
Our data add to the evidence supporting an association between SNPs in the SLC4A5 gene, and increase in BP is on which a number of sample populations of AA are reporting similar results, thus showing the replicability of the SLC4A5 gene across various samples.[16,36,39] Additional studies point toward chromosome 2, where SLC4A5 is located, for regions associated with modulating BP.[45,46] Populations of EA with the rs1006502 SNP have been shown to be associated with increase in BP[37]: VO250 and VCO250.[35] As Hunt and colleagues were only using Utah residents of EA,[37] and research results published by Stutz and colleagues reported too few AA participants to be included in their analyses,[35] further research into how SLC4A5 polymorphisms affect AAs, a group with disproportionately high rates of HTN, is needed. Our data show a G × E relationship between SLC4A5 polymorphisms and racial discrimination in populations of AAs. Thus, further research into the potential mechanism of action by which the SLC4A5 gene, particularly the 2 SNPs rs1006502 and rs3771724, might have a SNP-discrimination interaction resulting in increases in BP and further explaining the health disparity between EA and AA population groups is needed.
This study supports the idea that SNP-discrimination interactions combine to influence clinically relevant traits such as BP. As AAs in the US experience high levels of both ED and MLD,[7,8] and (acute and) chronic experiences with discrimination are consistently linked to HBP and HTN,[15] it is important for clinicians to be mindful of this SNP-discrimination relationship when working with AA patients. Furthermore, these analyses suggest that more work on SNP-discrimination interactions is required for development and implementation of clinically relevant protocols for the identification and treatment of HBP to address a major health disparity affecting AAs.
5.2. Limitations
The use of self-report perceived racism and discrimination may be a limitation; however, this is the best noninvasive gold standard measure of examining racism and discrimination. Additionally, the DISA instrument used here has been validated in other samples of AAs in the NSAL and JHS. Replication with similar epidemiological samples is required to ascertain the role of genes and psychosocial stressors in the development and expression of HBP in this understudied population. The addition of other -omic methodologies such as epigenetic[47] and whole genome sequencing[48] among under-represented minority populations may also help to elucidate a more holistic picture of this health disparity.
Acknowledgment
The authors also wish to thank the staffs and participants of the JHS.
Supplementary Material
Footnotes
Abbreviations: AA = African Americans, ARIC = Atherosclerosis Risk in Communities Study, BMI = body mass index, BP = blood pressure, CMLDAR = Chronic Major Life Discrimination Attributed to Race, CVD = cardiovascular disease, DBP = diastolic blood pressure, DISA = Discrimination Instrument, EA = European ancestry, ED = Everyday Discrimination, EDAR = Experiences of Discrimination Attributed to Race, G × D = Gene × Discrimination interaction, GWAS = Genome-Wide Association Study, HBP = high blood pressure, HTN = hypertension, HWE = Hardy-Weinberg equilibrium, JHS = Jackson Heart Study, MAF = minor allele frequency, MDS = multidimensional scaling, MLD = Major Life Discrimination, NSAL = National Survey of American Life, PC = principal components, SBP = systolic blood pressure, SNP = single nucleotide polymorphism.
The Jackson Heart Study (JHS) is supported and conducted in collaboration with Jackson State University (HHSN268201300049C and HHSN268201300050C), Tougaloo College (HHSN268201300048C), and the University of Mississippi Medical Center (HHSN268201300046C and HHSN268201300047C) contracts from the National Heart, Lung, and Blood Institute (NHLBI) and the National Institute for Minority Health and Health Disparities (NIMHD) and NR013520 from the National Institute of Nursing Research and P30-AG-015281 from the National Institute on Aging.
The authors report no conflicts of interest.
Supplemental Digital Content is available for this article.
References
- [1].Go AS, Mozaffarian D, Roger VL, et al. Executive summary: heart disease and stroke statistics—2014 update: a report from the American Heart Association. Circulation 2014;129:399–410. [DOI] [PubMed] [Google Scholar]
- [2].Nwankwo T, Yoon SS, Burt V, Gu Q. Hypertension among adults in the United States: National Health and Nutrition Examination Survey, 2011-2012. NCHS Data Brief 2013;1–8. [PubMed] [Google Scholar]
- [3].Lewis TT, Cogburn CD, Williams DR. Self-reported experiences of discrimination and health: scientific advances, ongoing controversies, and emerging issues. Annu Rev Clin Psychol 2015;11:407–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Gravlee CC, Dressler WW. Skin pigmentation, self-perceived color, and arterial blood pressure in Puerto Rico. Am J Hum Biol 2005;17:195–206. [DOI] [PubMed] [Google Scholar]
- [5].Sweet E, McDade TW, Kiefe CI, et al. Relationships between skin color, income, and blood pressure among African Americans in the CARDIA Study. Am J Public Health 2007;97:2253–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Brown TN. Measuring self-perceived racial and ethnic discrimination in social surveys. Sociological spectrum 2001;21:377–92. [Google Scholar]
- [7].Williams DR, Yan Y, Jackson JS, et al. Racial differences in physical and mental health: socio-economic status, stress and discrimination. J Health Psychol 1997;2:335–51. [DOI] [PubMed] [Google Scholar]
- [8].Kessler RC, Mickelson KD, Williams DR. The prevalence, distribution, and mental health correlates of perceived discrimination in the United States. J Health Soc Behav 1999;40:208–30. [PubMed] [Google Scholar]
- [9].Ifatunji MA, Harnois CE. An explanation for the gender gap in perceptions of discrimination among African Americans: considering the role of gender bias in measurement. Sociology of race and ethnicity 2015;2:263–88. [Google Scholar]
- [10].Jackson JS, Neighbors HW, Nesse RM, et al. Methodological innovations in the National Survey of American Life. Int J Methods Psychiatr Res 2004;13:289–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Sims M, Diez-Roux AV, Gebreab SY, et al. Perceived discrimination is associated with health behaviours among African-Americans in the Jackson Heart Study. J Epidemiol Community Health 2016;70:187–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Sims M, Diez-Roux AV, Dudley A, et al. Perceived discrimination and hypertension among African Americans in the Jackson Heart Study. Am J Public Health 2012;102suppl 2:S258–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Dolezsar CM, McGrath JJ, Herzig AJ, et al. Perceived racial discrimination and hypertension: a comprehensive systematic review. Health Psychol 2014;33:20–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Krieger N, Sidney S. Racial discrimination and blood pressure: the CARDIA Study of young black and white adults. Am J Public Health 1996;86:1370–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Brondolo E, Love EE, Pencille M, et al. Racism and hypertension: a review of the empirical evidence and implications for clinical practice. Am J Hypertens 2011;24:518–29. [DOI] [PubMed] [Google Scholar]
- [16].Taylor JY, Wu CY, Darling D, et al. Gene-environment effects of SLC4A5 and skin color on blood pressure among African American women. Ethn Dis 2012;22:155–61. [PMC free article] [PubMed] [Google Scholar]
- [17].Clark AE, Adamian M, Taylor JY. An overview of epigenetics in nursing. Nurs Clin North Am 2013;48:649–59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Taylor JY, Wright ML, Crusto CA, et al. The Intergenerational Impact of Genetic and Psychological Factors on Blood Pressure (InterGEN) Study: Design and Methods for Complex DNA Analysis. Biol Res Nurs 2016;18:521–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Auton A, Brooks LD, et al. 1000 Genomes Project Consortium. A global reference for human genetic variation. Nature 2015;526:68–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Taylor HA, Jr, Wilson JG, Jones DW, et al. Toward resolution of cardiovascular health disparities in African Americans: design and methods of the Jackson Heart Study. Ethn Dis 2005;15suppl 6:S6-4-17. [PubMed] [Google Scholar]
- [21].Taylor HA., Jr The Jackson Heart Study: an overview. Ethn Dis 2005;15suppl 6:S6-1-3. [PubMed] [Google Scholar]
- [22].Wilson JG, Rotimi CN, Ekunwe L, et al. Study design for genetic analysis in the Jackson Heart Study. Ethn Dis 2005;15suppl 6:S6-30-37. [PubMed] [Google Scholar]
- [23].Payne TJ, Wyatt SB, Mosley TH, et al. Sociocultural methods in the Jackson Heart Study: conceptual and descriptive overview. Ethn Dis 2005;15suppl 6:S6-38-48. [PubMed] [Google Scholar]
- [24].Centers for Disease Control and Prevention (CDC), Body mass index. 2015; 2016. [Google Scholar]
- [25].Chobanian AV, Bakris GL, Black HR, et al. The Seventh Report of the Joint National Committee on Prevention, Detection, Evaluation, and Treatment of High Blood Pressure: the JNC 7 report. JAMA 2003;289:2560–72. [DOI] [PubMed] [Google Scholar]
- [26].Harman J, Walker ER, Charbonneau V, et al. Treatment of hypertension among African Americans: the Jackson Heart Study. J Clin Hypertens (Greenwich) 2013;15:367–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Sims M, Wyatt SB, Gutierrez ML, et al. Development and psychometric testing of a multidimensional instrument of perceived discrimination among African Americans in the Jackson Heart Study. Ethn Dis 2009;19:56–64. [PMC free article] [PubMed] [Google Scholar]
- [28].Purcell S, Neale B, Todd-Brown K, et al. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet 2007;81:559–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Korn JM, Kuruvilla FG, McCarroll SA, et al. Integrated genotype calling and association analysis of SNPs, common copy number polymorphisms and rare CNVs. Nat Genet 2008;40:1253–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Abecasis G, Li Y. MaCH: University of Michigan Center for Statistical Genetics. 2010; 2016. [Google Scholar]
- [31].Parra EJ, Marcini A, Akey J, et al. Estimating African American admixture proportions by use of population-specific alleles. Am J Hum Genet 1998;63:1839–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Zhu X, Luke A, Cooper RS, et al. Admixture mapping for hypertension loci with genome-scan markers. Nat Genet 2005;37:177–81. [DOI] [PubMed] [Google Scholar]
- [33].Hao K, Chudin E, McElwee J, et al. Accuracy of genome-wide imputation of untyped markers and impacts on statistical power for association studies. BMC Genet 2009;10:27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].R Development Core Team. R: a language and environment for statistical computing. R Foundation for Statistical Computing, Vienna, Austria. 2006; 2015. [Google Scholar]
- [35].Stutz AM, Teran-Garcia M, Rao DC, et al. Functional identification of the promoter of SLC4A5, a gene associated with cardiovascular and metabolic phenotypes in the HERITAGE Family Study. Eur J Hum Genet 2009;17:1481–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Taylor JY, Sampson D, Taylor AD, et al. Genetic and BMI risks for predicting blood pressure in three generations of West African Dogon women. Biol Res Nurs 2013;15:105–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Hunt SC, Xin Y, Wu LL, et al. Sodium bicarbonate cotransporter polymorphisms are associated with baseline and 10-year follow-up blood pressures. Hypertension 2006;47:532–6. [DOI] [PubMed] [Google Scholar]
- [38].Guo L, Liu F, Chen S, et al. Common variants in the Na(+)-coupled bicarbonate transporter genes and salt sensitivity of blood pressure: the GenSalt study. J Hum Hypertens 2016;30:543–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Barkley RA, Chakravarti A, Cooper RS, et al. Positional identification of hypertension susceptibility genes on chromosome 2. Hypertension 2004;43:477–82. [DOI] [PubMed] [Google Scholar]
- [40].Gröger N, Vitzthum H, Fröhlich H, et al. Targeted mutation of SLC4A5 induces arterial hypertension and renal metabolic acidosis. Hum Mol Genet 2012;21:1025–36. [DOI] [PubMed] [Google Scholar]
- [41].Wen D, Sansom SC. Physiological role of NBCe2 in the regulation of electrolyte transport in the distal nephron. Am J Physiol Renal Physiol 2015;309:F489–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].Gildea JJ, Xu P, Carlson JM, et al. The sodium-bicarbonate cotransporter NBCe2 (slc4a5) expressed in human renal proximal tubules shows increased apical expression under high-salt conditions. Am J Physiol Regul Integr Comp Physiol 2015;309:R1447–59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [43].Carey RM, Schoeffel CD, Gildea JJ, et al. Salt sensitivity of blood pressure is associated with polymorphisms in the sodium-bicarbonate cotransporter. Hypertension 2012;60:1359–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [44].Richardson SI, Freedman BI, Ellison DH, et al. Salt sensitivity: a review with a focus on non-Hispanic blacks and Hispanics. J Am Soc Hypertens 2013;7:170–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [45].Morrison AC, Cooper R, Hunt S, et al. Genome scan for hypertension in nonobese African Americans: The National Heart, Lung, and Blood Institute Family Blood Pressure Program. Am J Hypertens 2004;17:834–8. [DOI] [PubMed] [Google Scholar]
- [46].Cooper RS, Luke A, Zhu X, et al. Genome scan among Nigerians linking blood pressure to chromosomes 2, 3, and 19. Hypertension 2002;40:629–33. [DOI] [PubMed] [Google Scholar]
- [47].Barcelona de Mendoza V, Huang Y, Crusto C, et al. Effects of perceived racial discrimination and DNA methylation among African American women in the InterGEN study. Biological Research for Nursing, March 2018. In press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [48].Taylor JY, Barcelona de Mendoza V. Omics based research and precision health in minority populations: Recommendations for Nurse Scientists. Journal of Nursing Scholarship, 50:1, Jan 2018. In press. DOI: 10.1111/jnu.12358 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


