Abstract
Affinity monolith chromatography, or AMC, is a liquid chromatographic method in which the support is a monolith and the stationary phase is a biological binding agent or related mimic. AMC has become popular for the isolation of biochemicals, for the measurement of various analytes, and for studying biological interactions. This review will examine the principles and applications of AMC. The materials that have been used to prepare AMC columns will be discussed, which have included various organic polymers, silica, agarose and cryogels. Immobilization schemes that have been used in AMC will also be considered. Various binding agents and applications that have been reported for AMC will then be described. These applications will include the use of AMC for bioaffinity chromatography, immunoaffinity chromatography, dye-ligand affinity chromatography, and immobilized metal-ion affinity chromatography. The use of AMC with chiral stationary phases and as a tool to characterize biological interactions will also be examined.
Keywords: affinity monolith chromatography, bioaffinity chromatography, chiral separations, immunoaffinity chromatography, dye-ligand affinity chromatography
1 Introduction
1.1 Overview of affinity monolith chromatography
Affinity chromatography is a liquid chromatographic method in which the applied or injected compounds are separated based on their binding to an immobilized biological agent or related mimic (i.e., the “affinity ligand”) [1–5]. This method makes use of the reversible and strong interactions that are present between many biological agents and chemicals. Some examples of these interactions are the binding by an antibody with an antigen and the binding of an enzyme with its substrate. The strength and selectivity of this binding is the result of several simultaneous forces or interactions that are often present between the immobilized agent and its target, such as coulombic forces, hydrogen bonds, dipole-dipole interactions, and steric effects [1,4].
Affinity chromatography can be used to isolate biochemicals such as enzymes, antibodies and other proteins. This method can also be used with other techniques for the separation and analysis of target compounds [1–6]. In addition, affinity chromatography can be employed to determine the number of binding sites, thermodynamics, and kinetics of an interaction between a target and its binding agent [7–12]. These features have made affinity chromatography a popular method for use in the isolation of biochemicals and chemicals, clinical or biochemical assays, biotechnology, and the study of biological processes [4–13].
Various supports can be used with the immobilized binding agent in affinity chromatography. In the past, particulate-based materials such as agarose beads, silica particles, glass beads, and particulate supports made from various organic polymers have been used for this purpose [1,2,6,14,15]. However, there has also been growing interest in using affinity chromatography with monolithic supports [15–20]. This combination is often referred to as affinity monolith chromatography, or AMC [18–20].
This review will discuss the principles of AMC, as well as common supports, binding agents and immobilization methods that have been used to make affinity monoliths. Recent applications of AMC will then be examined, including its use in bioaffinity chromatography, immunoaffinity chromatography, dye-ligand affinity chromatography, immobilized metal-ion affinity chromatography, chiral separations, and the study of biological interactions. The possible advantages and potential limitations of AMC will be considered as part of this discussion, as well as possible future directions in this field.
1.2 General principles of AMC
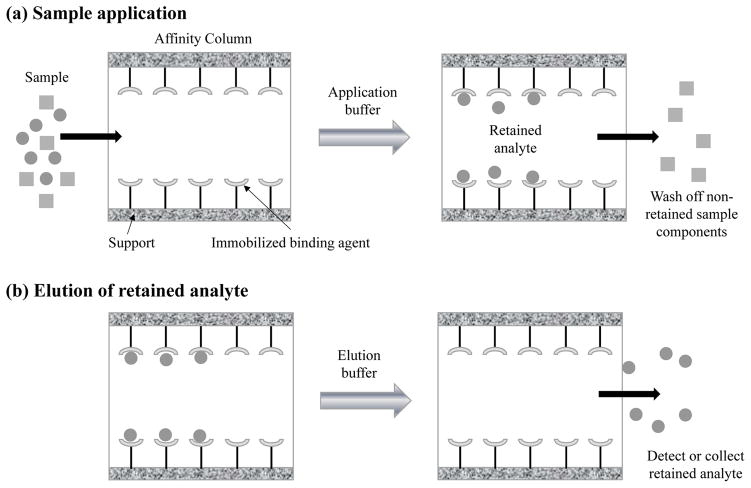
Figure 1 shows a typical scheme that is used in AMC or traditional affinity chromatography for compound isolation and measurement. This scheme is often called the “on/off” mode of affinity chromatography [4,21]. In this mode, a solution or sample that contains the desired analyte or target compound is passed through the affinity column under conditions that will allow the target to bind to the immobilized agent [1,4,21]. This first step is carried out in the presence of an application buffer that has a pH and composition that will allow strong binding by the target to the immobilized agent in the column. During this step, any non-retained or weakly-retained sample components are washed from the column. An elution buffer is next passed through the column to release the target. As the target is eluted, it can be collected for further use or passed through an on-line detector. In many cases, the column can be regenerated by again applying the application buffer and allowing the immobilized binding agent to return to its initial state before another sample is applied to the column [1,4,21].
Figure 1.
The on/off mode of affinity chromatography, including the steps for (a) sample application and (b) elution of the retained analyte or target compound.
The on/off format shown in Figure 1 is often employed in AMC and affinity chromatography when there are relatively strong interactions between the immobilized binding agent and the target [4,17–21]. This approach is often employed with targets and immobilized agents that have binding constants of at least 106 M−1 [21]. The elution buffer used in this mode may have a different pH, ionic strength, polarity, or additive content from the application buffer to cause dissociation of the target from the binding agent. This approach is known as “non-specific elution” and can be useful in situations where rapid release of the target is desired [1,21]. An alternative approach is to use an elution buffer that contains a competing agent to promote release of the target from the immobilized binding agent. This second method is known as “biospecific elution” and is often used when gentler elution conditions are desired [1,21]. If the target and immobilized agent have only weak or moderate strength binding (i.e., a binding constant of 105–106 M−1 or less), it may be possible to elute the target under isocratic conditions. This last approach is sometimes known as weak affinity chromatography (WAC) [21–24].
There are many factors that can contribute to the overall success of a separation that is based on affinity chromatography or AMC. One important factor is the immobilized agent that is placed in the column and used as the stationary phase [1,4]. This binding agent is the main factor that determines which targets will bind to the column and the retention of these targets. This binding agent is often of biological origin [4,25]. Examples are immunoaffinity chromatography (IAC), which uses an antibody or related agent as the stationary phase, and lectin affinity chromatography, in which the stationary phase is a lectin [25–27]. It is also possible to use binding agents that are synthetic or non-biological in nature. Examples of these latter binding agents include the use of chelated metal ions (e.g., in immobilized metal-ion affinity chromatography, or IMAC) and dyes [28–30]. The use of these binding agents and methods in AMC will be discussed later in this review.
Another factor that is important in affinity chromatography is the support that is used in the column [14,15]. Agarose and other carbohydrate-based supports have been popular in traditional affinity chromatography for target isolation and sample pretreatment because these materials have low non-specific binding towards many biological molecules, good pH stability, and relatively large pore diameters (i.e., allowing their use with large binding agents and targets) [1,2,15]. Materials that may be used in HPLC-based affinity chromatography (i.e., high-performance affinity chromatography, or HPAC) include silica particles, glass beads, and various organic polymers (e.g., modified polystyrene supports) [1,6,14,15]. These materials can be used to directly immobilize binding agents through covalent coupling methods (e.g., amine-based schemes) [18,31,32] or non-covalent methods (e.g., the use of entrapment or molecular imprinting) [32]. The following sections of this review will look at how these supports and immobilization strategies have been adapted for use in AMC.
2 Supports for AMC
2.1 General properties of supports for AMC
As its name implies, AMC is based on the use of a monolith as the support for an immobilized binding agent [15–20]. Monoliths are supports which consist of a single, continuous piece of a porous material [33,34]. An early example of a monolithic support was based on poly(ethylene glycol methacrylate) and used for size-exclusion chromatography in 1967 [35]. Other early monoliths, such as those based on polyurethane foams, often had low permeabilities and excessive swelling in some solvents, which made them difficult to use in HPLC [36–39]. Alternative types of monolithic columns were later developed based on macroporous disks, membranes or compressed polyacrylamide gels [40–44]. However, these materials still had low sample capacities and required specific solvents for their use [45].
Another type of monolith appeared in the early 1990s that was prepared by polymerizing monomers in the presence of porogenic solvents [45]. This approach helped to create materials with various pore sizes that depended on the types of porogenic solvents that were used and on the relative amount of each porogenic solvent that was added to the polymerization mixture [18,45]. It was found that this type of monolith could provide both large pores for solvent flow and small pores for efficient movement of the applied solutes to and from the stationary phase. The result was a set of supports that could provide a sufficient efficiency, sample capacity, and mechanical stability for use in HPLC [46–48].
There are several advantages in using modern monoliths as supports for affinity chromatography [15–20]. For instance, some of these monoliths can have higher permeability, better efficiencies and/or lower back pressures than particle-based supports. These properties can be useful for affinity-based separations that require work at high flow rates (e.g., rapid flow-based immunoassays) or in providing high efficiencies for applications such as chiral separations and high-throughput drug screening [17–20].
Various types of monoliths have been used in AMC (see Table 1). As will be shown in the following section, these monoliths have ranged from supports that are based on organic polymers or silica to supports that are based on agarose or cryogels [15–20]. These supports can be adapted for use with a number of immobilization methods [18–20] and can be made in several forms, including columns, disks, capillaries, and microchips [15–20]. These properties have made it possible to use affinity monoliths not only in traditional HPLC systems but also as part of microanalytical devices or in combination with techniques such as mass spectrometry and capillary electrophoresis [17–19].
Table 1.
Types of supports used in AMC
| General type of support | Examples of supportsa |
|---|---|
| Organic monolith | GMA/EDMA, GMM/EDMA, GMA/DVB, GMA/TRIM, DVB |
| Silica monoliths | Silica converted to an epoxy or diol form, aminopropyl silica |
| Carbohydrate-based monoliths | Agarose, agarose-chitosan composites, agarose/chitosan-PVA hybrids |
| Cryogel monoliths | Agarose, HEMA |
Abbreviations: DVB, divinylbenzene; EDMA, ethylene dimethacrylate; GMA, glycidyl methacrylate; GMM, glyceryl monomethacrylate; HEMA, 2-hydroxyethyl methacrylate; PVA, poly(vinyl alcohol); TRIM, trimethylolpropane trimethacrylate
2.2 Organic monoliths
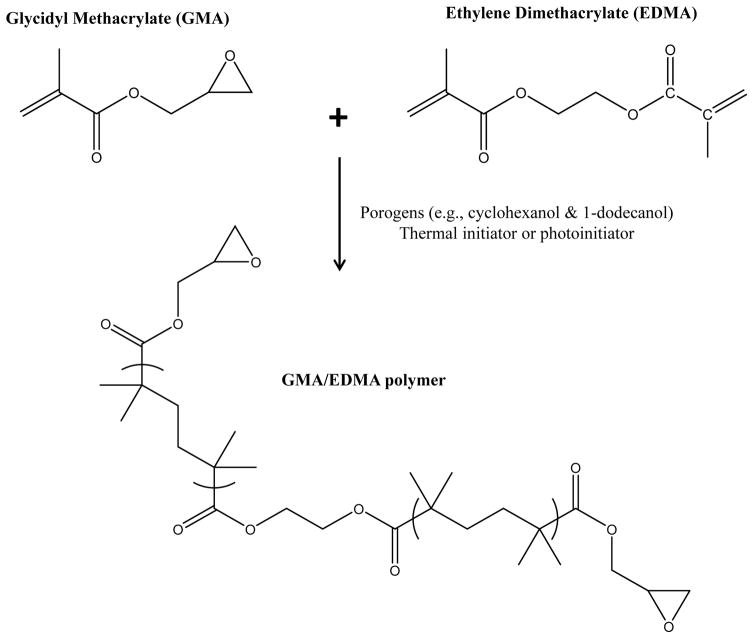
Various organic monoliths have been utilized in AMC [17–20,46]. One common type is based on a co-polymer of glycidyl methacrylate (GMA) and ethylene dimethacrylate (EDMA), as shown in Figure 2 [18–20]. GMA/EDMA monoliths are easy to synthesize and can be converted into a hydrophilic form that has low non-specific binding for many biological agents. GMA/EDMA monoliths can also be made with a variety of shapes, surface areas, and pore sizes, which is useful when adapting these supports for use with relatively large binding agents such as proteins [18,20]. In addition, these monoliths can be utilized with various immobilization methods (see Section 3). One disadvantage of GMA/EDMA monoliths is that they tend to have a smaller surface area than other types of monoliths or particulate supports, which can limit the amount of binding agent that can be immobilized to such a support and placed in an AMC column [17–20,49–51].
Figure 2.
General process for preparing a GMA/EDMA monolith.
In the general process for preparing a GMA/EDMA monolith, the monomers and porogens are combined, placed into the desired mold, and allowed to polymerize. The most common porogens for preparing these monoliths in AMC are 1-dodecanol and cyclohexanol [18–20,49–52], and either thermal initiators or photoinitiators can be employed [20,46,52]. One way the pore size and surface area of a GMA/EDMA monolith can be modified is by adjusting the ratio of the porogens that are used in its preparation [49,51,52]. The type of thermal initiator that is used and the polymerization temperature can also affect the structure of the final monolith [17–19,49,52]. Photoinitiators tend to result in faster polymerization than thermal initiators [52,53], which can be useful when placing in monoliths within specific locations (e.g., within a section of a microfluidic device) [52].
Other organic monoliths can also be created in which alternative monomers are used in place of GMA or EDMA. For instance, glyceryl monomethacrylate (GMM) has been used with EDMA to make monoliths that contain immobilized lectins [54]. GMA also has been combined with divinylbenzene (DVB) to create monoliths that contain immobilized chelating agents and Cu2+ or Fe3+ as a stationary phase [55,56]. Trimethylolpropane trimethacrylate (TRIM) has been used with GMA to create organic monoliths which contained HSA as a chiral stationary phase [57]. In addition, DVB has been employed to make monoliths that contain entrapped metal oxide nanoparticles [58].
2.3 Silica monoliths
Silica monoliths have also been employed in AMC [18–20]. This type of monolith can have several advantages when compared to silica particles, including their ability to provide lower backpressures and their potential to give better mass transfer and improved column efficiencies [47,59–61]. Another advantage of silica monoliths is that their surfaces can be modified by using many of the same reactions for immobilizing biological agents that have been developed for silica particles [18–20]. In addition, silica monoliths can have larger surface areas when compared to GMA/EDMA monoliths [20,59], which can allow for more of a binding agent to be placed into silica-based affinity monoliths. A disadvantage of silica monoliths is that they are only stable over a relatively limited pH range (pH 2–8) [15]. Another disadvantage is that silica monoliths must be transferred to their final housing after they have been prepared, due to shrinkage that can occur during their formation [18–20].
Silica monoliths tend to be more difficult to make than GMA/EDMA monoliths, but such supports are now available commercially [18,19,49,60,61]. AMC has been carried out using both bare silica monoliths and aminopropyl silica monoliths as the initial support [18–20,62–64]. When starting with a bare silica monolith, this support is often converted to an epoxy or diol form before it is used for the immobilization of a binding agent (see examples of such methods in Section 3) [18]. The most common way to prepare a silica monolith is the sol-gel method [19,59–61]. The starting mixture in this method typically consists of tetramethoxysilane (TMOS), poly(ethylene oxide) (PEO), and an acid catalyst in water. The PEO will hydrolyze under these conditions, and a phase separation will occur between the silica and solvent. Gelation of the silica also occurs during this process. The phase separation and gelation is what determines the pore sizes that will make up the final silica monolith. After aging has been allowed to take place, the silica monolith is dried to remove the excess solvent and placed into its final housing in a process known as “cladding” [19,59–61].
Silica monoliths have been used in AMC and related methods for various applications. Some studies have used HSA or AGP which were immobilized onto silica monoliths for work in chiral separations and drug-protein binding studies [62–65]. The same types of columns have been used to measure drug-protein dissociation rates [66,67]. Related applications for silica monoliths have included their use with immobilized enzymes for high-throughput drug screening or protein digestion [68,69].
2.4 Carbohydrate-based monoliths
Another group of monoliths that have been examined for use in AMC are those based on carbohydrates such as agarose [15,16,18–20]. Agarose has been used for many years as a support in traditional affinity chromatography due to its good stability over a broad pH range, its low non-specific binding for many biological compounds, its large pore size, and the ease with which it can be modified for the immobilization of binding agents [1,2,15]. A disadvantage of agarose is that it tends to have low mechanical stability, which has made it difficult to use in applications involving HPLC [1,15]. However, agarose can be crosslinked or combined with other polymers to strengthen it and allow its use in relatively high-performance separations [15].
Agarose is a polysaccharide made of chains of agarobiose, which in turn is composed of repeating units of D-galactose and 3,6-anhydro-L-galactose [15]. To develop an agarose monolith, agarose powder is dissolved in water and heated to 90–100 °C [15,16,18]. The agarose mixture is then mixed with a water-immiscible organic solution which contains an organic solvent and a surfactant to control the pore sizes of the final support. This mixture is shaken to obtain an emulsion and is placed inside a mold. The agarose is then allowed to cool slowly and washed to provide the final monolith [15,16,18].
The most common use of agarose monoliths in AMC has been in the purification of biological targets [15,18–20]. For example, agarose monoliths have been developed that contain an immobilized NAD+ derivative or dyes such as Cibacron Blue for use in enzyme and protein purification [70,71]. Monoliths that contain agarose-chitosan composites have been employed for the purification of immunoglobulin G (IgG) and glycoproteins [72,73]. Hybrid monoliths based on agarose or chitosan and poly(vinyl alcohol), or PVA, have been used with a synthetic mimic of protein A for the isolation of monoclonal antibodies from cell culture extracts [74].
2.5 Cryogel monoliths
A cryogel is another type of support that can be employed in AMC for the immobilization and purification of biological ligands [18–20,75,76]. These monoliths are spongy gel matrices that are formed when their monomeric or polymeric precursors are nearly frozen in an aqueous solution. The ice crystals that are present act as a porogen to allow formation of a porous bed within the polymer [18,19,75]. The final material can have good mechanical and chemical stability and results in a material with wide pores, as might be needed for large targets. A disadvantage of many cryogels is their relatively low surface area, which can limit the amount of a binding agent that can be immobilized onto this type of support [18–20].
Cryogel monoliths can be made with monomers that range from agarose to 2-hydroxyethyl methacrylate (HEMA) [18,19]. These monoliths have been employed in a variety of applications that have involved protein or enzyme purification and cell separations [18–20,75,76]. These supports have been combined with such binding agents as lectins, antibodies, and immobilized metal-ions [18–20,75–79]. A HEMA-based cryogel containing immobilized protein A has also been used for the removal of immunoglobulin M (IgM)-antibodies from human plasma [80].
3 Immobilization Methods for AMC
The immobilization method used to place a binding agent in a monolith is another factor to consider in AMC. The ideal immobilization method should not alter or denature the binding agent and should not adversely affect this agent’s activity [1,32]. Examples of undesirable effects that can occur during immobilization are improper orientation or multisite attachment of the binding agent, as well as steric hindrance of the target as it attempts to interact with the immobilized agent [1]. There are several approaches that have been used for the immobilization of binding agents in AMC. These approaches include covalent immobilization, coordination-based immobilization, entrapment, and molecular imprinting [18–20,32].
3.1 Covalent and coordination-based immobilization methods
Many binding agents are placed in affinity monoliths by using covalent immobilization [18–20]. This process is carried out after forming the initial monolith, which is then activated and placed in contact with the desired binding agent [18]. For example, this can be accomplished by circulating the activating reagents and binding agent through the monolith [49,50] or by dipping the monolith into solutions of the reagents and binding agent [18].
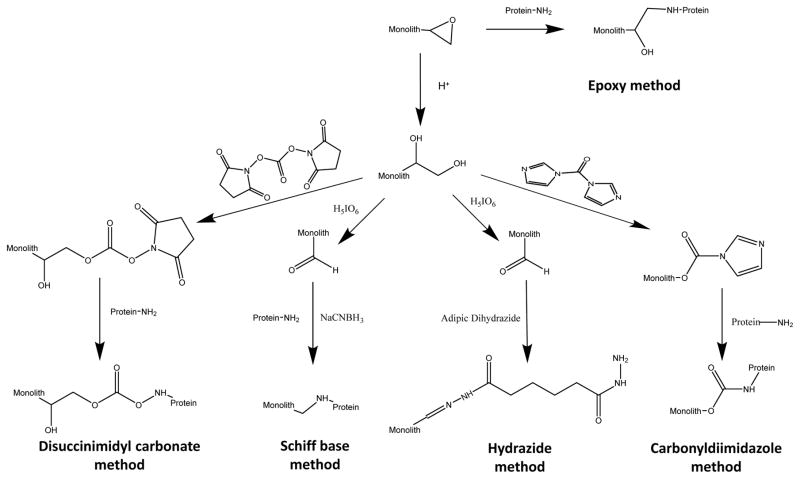
Figure 3 shows several common examples of covalent immobilization methods that have been employed in AMC [18–20]. One of these is the epoxy method, in which an epoxy group on the monolith couples with amine groups on a binding agent such as a protein or ligand to generate a secondary amine bond [18,32]. The epoxy method is easy to use with GMA/EDMA monoliths due to the epoxy groups that are present on GMA [18–20]. This method can also be used with other types of supports, such as silica monoliths, by first converting these supports into an epoxy-activated form [20]. Although this method can be completed in one step, it does tend to result in lower amounts of an immobilized binding agent than can be obtained with other covalent coupling techniques [50]. Examples of binding agents that have been coupled to monoliths by the epoxy method include amino acids (e.g., histidine) and proteins such as protein A or protein G, human serum albumin (HSA) and antibodies [18–20,49,50,81,82].
Figure 3.
Examples of common covalent immobilization techniques that have been used in AMC, including the epoxy method, Schiff base method, carbonyldiimidazole method, disuccinimidyl carbonate method, and hydrazide method.
Another common immobilization technique in AMC is the Schiff base method [18–20]. This method has been used with GMA/EDMA monoliths and silica monoliths and involves placing or using epoxy groups on these supports, converting these groups into diols, and then oxidizing the diols to form aldehyde groups [18–20,32]. These aldehyde groups are then reacted with a binding agent that contains one or more primary amines to form a reversible Schiff base. The Schiff base is then reduced upon its formation by cyanoborohydride to give a secondary amine. The remaining aldehyde groups can later be removed by reducing them or by reacting them with a small amine-containing capping agent [32,83]. This method tends to give a higher content for an active binding agent than the epoxy method but does involve the use of a reducing agent and multiple steps for the immobilization process [32,50]. Binding agents that have been immobilized within GMA/EDMA or silica monoliths by the Schiff base method include HSA, protein A, and antibodies [18–20,49,50,64,82].
The glutaraldehyde immobilization method is similar to the Schiff base method but instead starts with a support that contains amine groups [20,82,83]. These amine groups are reacted with glutaraldehyde to produce aldehyde groups on the support’s surface. This activated support is then used in the same manner as described earlier for the aldehyde-activated support in the Schiff base method. This glutaraldehyde-based scheme has been employed for the immobilization of protein A and trypsin on GMA/EDMA monoliths [82,84].
Another approach for covalent immobilization in AMC is the carbonyldiimidazole (CDI) method [18]. This method makes use of diol groups that are present on or that have been placed onto the monolith. These groups are then reacted with 1,1′-carbonyldiimidazole to produce imidazolyl carbamate groups. These activated sites can then react with primary amines on a binding agent such as a protein or peptide through a nucleophilic substitution reaction to generate a stable amide linkage [83]. This method is simpler to carry out than the Schiff base method but also tends to produce lower activities than this other approach [32,49,50]. The CDI method has been used with GMA/EDMA monoliths and for the immobilization of L-histidine, HSA and antibodies [49,50,82].
The disuccinimidyl carbonate (DSC) immobilization method can also be employed for binding agents that contain amine groups [18]. This technique often involves activation of a monolith in a diol-bonded form with DSC to place succinimidyl carbonate groups on the support’s surface; however, an amine-containing monolith can also be used for this reaction [18–20]. The activated form of the monolith is then combined with a binding agent that contains primary amine groups to form a carbamate linkage [32]. This technique has been used with GMA/EDMA monoliths to immobilize HSA and with a silica monolith to immobilize β-glucuronidase [50,85].
The hydrazide method is a covalent immobilization method that can be used with glycoproteins and carbohydrate-containing agents [20,32,83,86,87]. The first step in this method is the same as in the Schiff base method and results in an aldehyde-activated support. This material is then reacted with a dihydrazide, followed by the use of this support to immobilize a carbohydrate-containing binding agent that has been oxidized under mild conditions to form aldehyde groups. The result is a hydrazone bond between the binding agent and the support; the remaining aldehyde groups on the support can then be reduced or capped with a small hydrazide agent [32,86]. This method has been used with silica monoliths to immobilize α1-acid glycoprotein (AGP) [62].
The cyanogen bromide (CNBr) method has also been employed in AMC for immobilization [20]. This method is often used with polysaccharide-based supports such as agarose and involves the support’s activation with CNBr. The activated support can be coupled with an amine-containing binding agent [1,20,32,83]. However, this method has several potential disadvantages. For instance, care must be used in work with CNBr, and the immobilized binding agents are not as stable as those obtained with other common coupling techniques [32]. In addition, this method can generate ion-exchange sites on the support that may lead to non-specific binding [32,83]. This approach has been utilized to immobilize analogs of NAD+ and L-chlorosuccinamic acid on agarose monoliths and coated ceramic monoliths [70,88].
Immobilization in AMC is sometimes carried out by using coordination chemistry [20]. This occurs in the method of IMAC, as is discussed in Section 4.4. IMAC involves the interaction of targets such as proteins with metal-ions that have been placed onto the support in the form of a metal chelate [28–30]. The metal ion (e.g., Cu2+ or Ni2+) is held in place by allowing it to form a complex with a chelating agent like iminodiacetic acid or nitrilotriacetic acid that is covalently coupled to the support [30]. Supports for IMAC have been developed by using GMA/EDMA monoliths, GMA/DVB monoliths, and cryogels and have been used for the separation of various histidine-containing proteins [18,89,90].
3.2 Non-covalent immobilization methods
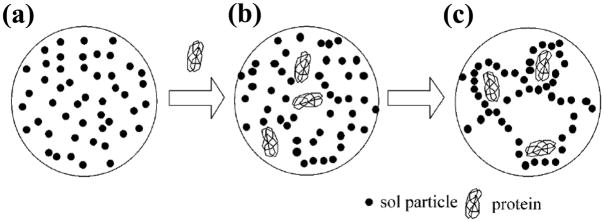
Entrapment is a non-covalent immobilization method that involves the physical containment of a binding agent in a support [32]. This typically involves the generation of a support that contains pores or openings that are smaller than the entrapped binding agent or that contains a highly cross-linked polymer network. This immobilization approach is usually achieved in AMC by incorporating the binding agent as part of a polymerization mixture, with the monolith entrapping the binding agent as the support is formed [32,65,68]. Such a procedure has often been used with sol-gel materials that can be made in aqueous solutions [65,68]. An advantage of this approach is it can be used to entrap an agent such as a protein in a soluble form without significant denaturation of this agent or loss of this agent’s activity [32,65,68]. Disadvantages of this method when using sol-gels include problems with controlling the pore size of support, the high degree of shrinkage of sol-gels, and the loss of protein activity that can occur if the correct silanes are not used to prepare the sol-gel [32,68].
Affinity monoliths that have been made through entrapment with sol-gels have been used in both chiral separations and in studies of biological interactions [65,68]. For instance, the proteins bovine serum albumin (BSA) and ovomucoid have been entrapped in sol-gels and tested for their stereoselectivity in separating the enantiomers for benzoin, chlorpheniramine, eperisone, and tryptophan [65]. A protein-compatible silica precursor, diglycerylsilane, has also been used with PEO to entrap dihydrofolate reductase in small monoliths; these columns were then used to screen the binding of this enzyme with various small molecules by using frontal affinity chromatography and mass spectrometry [68].
Molecular imprinting is another non-covalent method that has been used to create affinity monoliths [91]. This method makes use of a template molecule to create cavities in the support that are complementary to the shape and functional groups that are found in the template [91–95]. The resulting material is known as a molecularly imprinted polymer, or MIP. To make a MIP, the monomers are chosen to give interactions with the functional groups of the imprint molecule and are polymerized in the presence of this molecule [93,94]. This general strategy has been used to prepare a cryogel monolith that was imprinted with protein G, with the final support then being employed to capture a recombinant form of this protein from cell lysates of Escherichia coli [91].
4 Binding Agents and Applications of AMC
4.1 Bioaffinity chromatography
Bioaffinity chromatography is a subset of affinity chromatography which uses a biological binding agent as the stationary phase [25]. This is the most common type of both AMC and traditional affinity chromatography and can make use of numerous binding agents (see Table 2 for binding agents that are used in the various forms of AMC) [4,17–20,25]. Examples of binding agents that have been used in AMC-based applications of bioaffinity chromatography are immunoglobulin-binding proteins, enzymes, lectins and carbohydrates [17–20].
Table 2.
Types of binding agents and targets used in AMC
| General form of AMC | Binding agents (targets) employed in AMCa |
|---|---|
| Bioaffinity chromatography | Protein A or protein G (immunoglobulins/antibodies) Enzymes (inhibitors or substrates) Lectins (glycoproteins, glycopeptides, glycolipids)β-Cyclodextrin or complex carbohydrate (chiral or positional isomers, influenza virus) |
| Immunoaffinity chromatography | Antibodies or antibody-related agents (various targets, including blood group antigens, haptoglobulin, horseradish peroxidase, fluorescein, aflatoxin B1, insecticides) |
| Dye-ligand affinity chromatographyImmobilized metal-ion affinity chromatography | Cibacron Blue F3GA (various enzymes and proteins, such as lactate dehydrogenase and albumin)Immobilized chelate of a metal ion, such as Ni2+, Cu2+ or Fe3+ (Histidine-tagged proteins, cytochrome C, tumor necrosis factor-alpha, phosphopeptides) |
| Chiral separations | β-Cyclodextrin, BSA, HSA, AGP, ovomucoid (various chiral drugs and amino acids) |
Abbreviations: AGP, α1-acid glycoprotein; BSA, bovine serum albumin; HSA, human serum albumin
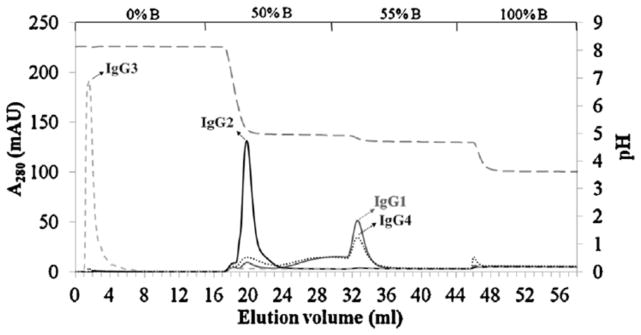
Two types of immunoglobulin-binding proteins that have been used in AMC are protein A and protein G [17,19,20,80,96–102]. These are bacterial cell wall proteins that can bind to the Fc regions of many types of immunoglobulins and antibodies, making them versatile agents for the purification, analysis, or biospecific adsorption of such targets [25,26]. For instance, commercial monoliths containing protein A have been used to separate IgG into its subclasses (see Figure 5) [96] and to measure IgG in purified samples or in supernatants obtained from Chinese hamster ovary cells [97]. A cryogel containing immobilized protein A has been used to remove IgM-antibodies (i.e., rheumatoid factor) from human plasma [80] and a GMA/EDMA/TRIM monolith containing protein A was used to measure IgG in human serum [102]. In addition, a GMA/EDMA monolith containing immobilized protein G has been employed with an anion-exchange monolith for measuring IgG, insulin and transferrin in cell cultures [98].
Figure 5.
Separation of subclasses 1 through 4 of human immunoglobulin G (IgG) by using step gradient elution on a protein A monolith column. Reproduced with permission from Ref. [96].
AMC has also been used with enzymes; however, most work combining monoliths with these agents has focused on the development of enzyme reactors [20,103]. In one study, dihydrofolate reductase was entrapped in a sol-gel monolith to screen for possible inhibitors for this enzyme [68]. Trypsin has been placed in silica monoliths, GMA/EDMA monoliths, and GMA/DVB monoliths for protein digestion [20,103,104], and such reactors have been used with liquid chromatography-mass spectrometry to analyze proteins [103,104]. Various enzymes have also been placed in agarose-based monoliths [20].
Lectins have been used in bioaffinity columns for AMC as well [20]. Lectins are non-immune system proteins that can bind to certain carbohydrate groups [25]. This property has made lectins valuable for the isolation or analysis of glycoproteins, glycopeptides, and glycolipids [20,25,105–107]. Two lectins that have been used for this purpose are concanavalin A (con A) and wheat germ agglutinin (WGA) [105–107]. One report used con A in a GMA/EDMA monolith to isolate high mannose glycans, which were then analyzed by tandem mass spectrometry [105]. In another report, WGA and Lens culinaris agglutinin were immobilized in GMA/EDMA monoliths and used for metabolomics and proteomics by combining these supports with nano-liquid chromatography or capillary electrophoresis [107].
Carbohydrates are another group of agents that have been used as stationary phases in bioaffinity chromatography and AMC [20,25,108–110]. Some studies have demonstrated and evaluated the use of immobilized β-cyclodextrin in silica monoliths for the separation of positional isomers (e.g., for naphthalenedisulfonic acids or cresols) and chiral solutes [107,108]. In another study, the influenza virus was isolated by using a complex carbohydrate that had been immobilized within a methacrylate-based monolith [110].
4.2 Immunoaffinity chromatography
Immunoaffinity chromatography (IAC) is a special type of bioaffinity chromatography in which the stationary phase is an antibody or antibody-related reagent [18–20,26,27,49,111–113]. This method can be employed for the purification or isolation of a given target compound (e.g., a method sometimes referred to as “immunoextraction”) or as a means for the direct or indirect analysis of this compound [26,27,111–113]. Antibodies have been useful as binding agents in traditional immunoaffinity chromatography and AMC-based methods because of the wide range of targets for which they can be used, along with the high affinity and selectivity that they often have for these targets [18–20,26,27]. In addition, antibodies can be used with various types of supports in AMC, including GMA/EDMA, agarose monoliths, and silica monoliths [18–20,49,114,115].
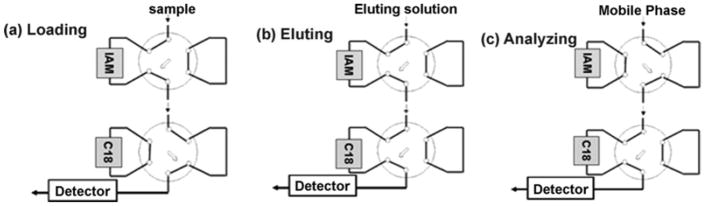
Immunoaffinity monoliths based on GMA/EDMA or other organic polymers have been utilized in a number of studies [49,114–120]. For instance, antibodies on GMA/EDMA disks have been used to purify various blood group antigens from a cell culture supernatant [114]. In another study, GMA/EDMA was used to make an immunoaffinity monolith for the extraction and on-line HPLC analysis of pyrethroid insecticides such as deltamethrin, flucythrinate, flumethrin, and permethrin (see Figure 6) [115]. It has been shown that antibodies placed within small GMA/EDMA disks can be used for the ultrafast immunoextraction of low mass targets (e.g., fluorescein) on the millisecond time scale [49]. Horseradish peroxidase and antibodies against this enzyme have further been employed as a model system to evaluate the final antibody content and to optimize the conditions used to make antibody-containing GMA/EDMA monoliths [116]. In addition, both antibodies and antibody-mimics such as aptamers have been placed in GMA/EDMA monoliths for use in immunoextraction or immunoaffinity-based AMC [17–20,117,118]. One study used antibodies against aflatoxin B1 in GMA/EDMA disks for the on-line extraction of this compound and its analysis by HPLC with fluorescence detection [119]. A monolith based on GMM and pentaerythritol triacrylate was prepared and tested for use with anti-haptoglobin antibodies to capture haptoglobin from serum [120].
Figure 6.
Scheme for coupling an immunoaffinity monoloith (IAM) for sample extraction and pretreatment with a C18 monolith for separation of the extracted sample components. Reproduced with permission from Ref. [115].
4.3 Dye-ligand affinity chromatography
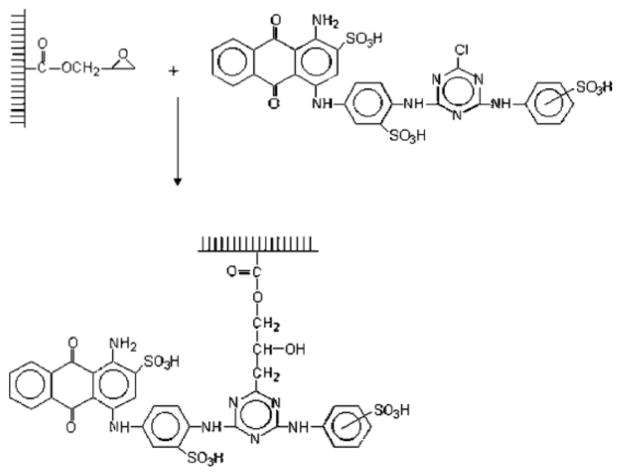
Another subset of affinity chromatography that has been used in AMC is dye-ligand affinity chromatography [20,30,121–123]. Dye-ligand affinity chromatography (or “dye-ligand chromatography”) uses a synthetic dye as the immobilized binding agent [20]. Triazine dyes make up the majority of the dye-ligands that are used for this method, and the most common of these is Cibacron Blue F3GA (see Figure 7) [30]. Advantages of using dyes as binding agents is that they are robust, inexpensive, easy to immobilize, and can result in a column with a high binding capacity [18–20,30]. Dye-ligand affinity chromatography is most often used to purify proteins [30,121], but it can also be utilized to remove proteins from samples (e.g., the use of Cibracron Blue F3GA for albumin depletion from serum or plasma) [30,123].
Figure 7.
Immobilization of Cibacron Blue F3GA within a GMA/EDMA monolith for use in dye-ligand affinity chromatography. Reproduced with permission from Ref. [122].
Immobilized dyes have been utilized as stationary phases in AMC with agarose, cryogels and organic-based monoliths [20,71,122,123]. One report used an agarose monolith with Cibacron Blue F3GA to purify the enzyme lactate dehydrogenase from a bovine heart extract [71]. Another study used GMA/EDMA as a monolithic support for dye-ligand affinity chromatography [122]. In addition, a cryogel based on poly(HEMA) was employed with Cibacron Blue F3GA to remove albumin from human serum samples [123].
4.4 Immobilized metal-ion affinity chromatography
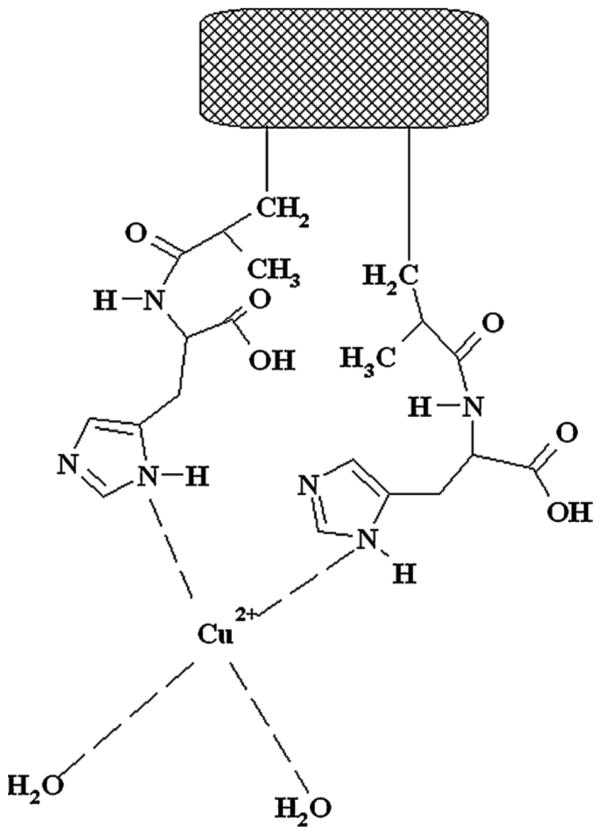
Another type of affinity method that has been combined with AMC is IMAC [20]. As was mentioned in Section 3.1, this method makes use of an immobilized metal ion (e.g., Cu2+ or Ni2+) as the stationary phase (see example in Figure 8) [20,28–30]. This type of stationary phase can be useful in binding to certain types of amino acids, peptides and proteins (e.g., those containing histidines, such as an added sequence of six histidine residues), as well as some modified forms of these biochemicals and DNA [28–30]. Strategies for chelating these metal ions with various monolithic supports have been reported, ranging from organic-based monoliths to cryogels and silica monoliths [18,89,90,124–132].
Figure 8.
A monolithic support for IMAC based on Cu2+ that is chelated with a cryogel consisting of poly(hydroxyethyl methacrylate-N-methacrylol-L-histidine methyl ester, or PHEMAH. Reproduced with permission from Ref. [129].
IMAC and AMC have been employed in several studies for the purification and analysis of biochemicals such as proteins and DNA [129–132]. As an example, a cryogel containing a chelate of Cu2+ was used to isolate cytochrome C from rat liver cells (see Figure 8) [129]. Recombinant histidine-tagged organophosphate hydrolase has been isolated from Escherichia coli by employing polyacrylamide-based cryogels that contained Co2+ [132]. Commercial methacrylate-based monoliths have also been tested for use in the purification of tumor necrosis factor-alpha and histidine-tagged green fluorescent protein [131]. Plasmid DNA has been purified by using GMA/EDMA disks that contained Cu2+ [130].
Another popular application for IMAC in AMC has been in the extraction or isolation of phosphopeptides in the field of phosphoproteomics [124–128]. This work has made use of columns that contain chelates of Fe3+ or Ti4+, as well as iron oxide nanoparticles [20,125–128]. These separations have been carried out using silica monoliths, organic monoliths, and organic-inorganic silica hybrid monoliths [124–128]. Specific examples of where IMAC and AMC have been employed include an analysis of the mitochondrial phosphoproteome [125] and an examination of phosphopeptides that were obtained from digests of α-casein or β-casein [126–128].
4.5 Chiral separations
Chiral separations are another set of applications in which AMC has been used [18–20,51,54,62,63,65,108,109,133,134]. An example that has already been mentioned is the use of β-cyclodextrin in silica monoliths as a chiral stationary phase [108,109]. This particular type of material has been evaluated for separating the enantiomers of baclofen, carprofen, and several types of dansylated amino acids by capillary electrochromatography [108]. In addition, β-cyclodextrin in monoliths has been tested for use in the chiral separation of chromakalim, methadone, norgestrel, prominal, metoprolol, and oxazepam, among others [109].
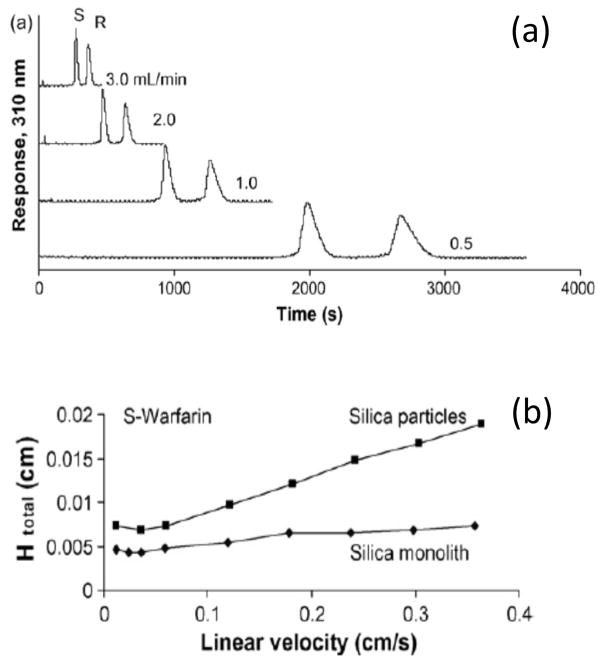
A number of proteins have also been employed in AMC for use in chiral separations. Examples include AGP, BSA, HSA, and ovomucoid [20,51,54,62,63,65,133,134]. One study used BSA in silica monoliths to separate the chiral forms of D/L-tryptophan, pantoprazole, and atenolol [133]. Organic-based monoliths based on GMA/EDMA or GMA/TRIM have been optimized and used with HSA for the rapid chiral separation of R/S-warfarin or D/L-tryptophan [50,54]. GMA/EDMA monoliths have also been made with HSA and used in the separation of other D/L-amino acids [134]. Silica monoliths containing AGP or HSA have been prepared and tested for the separation of chiral solutes such as R/S-warfarin, D/L-tryptophan and R/S-propranolol (see Figure 6) [62,63]. In addition, sol-gel monoliths have been prepared with entrapped BSA or ovomucoid and evaluated for use in the chiral separation of solutes such as R/S-benzoin, R/S-chlorpheniramine, R/S-eperisone, and D/L-tryptophan [65].
4.6 Biointeraction studies
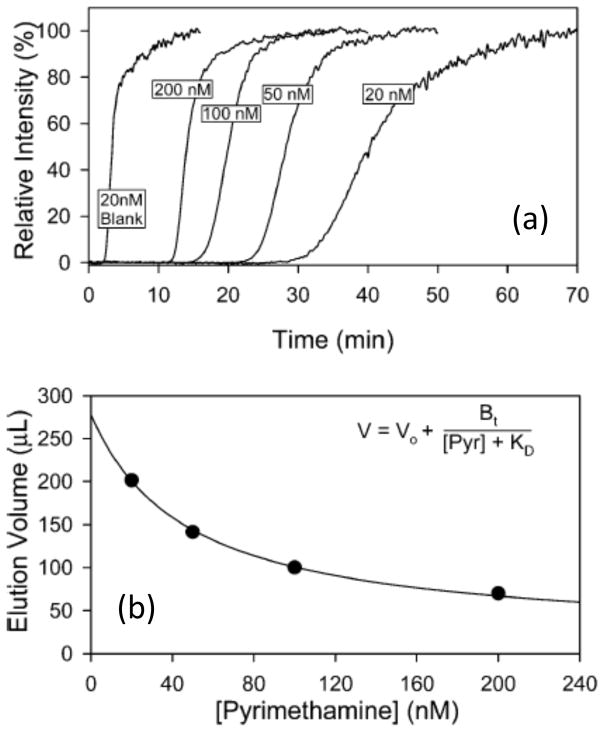
The study of biological interactions is another way in which AMC can be employed [18–20]. Frontal analysis (or “frontal affinity chromatography”) is one way this can be accomplished [8–11,62,63,68,135]. This technique is performed by continuously applying a known concentration of an analyte (or, in some cases, a mixture of analytes) to a column that contains the binding agent of interest. As the analyte begins to occupy binding sites in the column, the result is a breakthrough curve that can provide information on the amount of binding sites that are present and the equilibrium constant for the analyte as it binds to the immobilized ligand [8–11]. Frontal analysis has been employed in several studies to evaluate the binding capacities or binding strengths of new affinity monoliths [49,62,63,100]. This method has also been used to attain the binding constants for various systems, such as the interactions of carbamazepine with serum proteins such as HSA and AGP [62,63] and the binding of naproxen with BSA [135]. This approach has further been combined with mass spectrometry and a sol-gel column to screen for the binding of various small molecules to dihydrofolate reductase (see Figure 10) [68]. Frontal analysis and AMC have also been employed in characterizing the binding of IgG to immobilized forms of protein A, protein G or protein L [81,100].
Figure 10.
(a) Breakthrough curves obtained by frontal analysis chromatography-mass spectrometry for four applied solutes applied at various concentrations to silica monoliths containing dihydrofolate reductase and a blank monolith with no binding agent present, and (b) analysis of the breakthrough results for pyrimethamine on such columns at various concentrations of this solute, [Pyr], to determine the dissociation equilibrium constant (KD, in units of μM, in this case) for this agent with the immobilized dihydrofolate reductase. Other terms: Bt, picomoles of active binding agent in the column; V, retention or breakthrough volume seen at a given concentration of an applied solute (in μL); and Vo, void volume (in μL). Adapted with permission from Ref. [68].
Zonal elution is another approach that has been used in AMC for biointeraction studies [9–11,18–20,50,51,62,63]. In this method, a small plug of a target compound is injected onto the affinity column [9–11,18–20]. The retention factor of this target is then determined and used to obtain information on the strength of binding by this compound to the column or the amount of active binding sites that are present for the immobilized binding agent [9–11]. This approach has been used to optimize various types of monoliths containing HSA or AGP and to study the binding of these proteins to solutes such as R/S-warfarin, D/L-tryptophan, and R/S-propranolol [50,51,62,63].
Some reports have used AMC to provide information on the kinetics of a biological interaction. For instance, a monolith containing HSA as a chiral stationary phase has been used to study the enzyme kinetics for D-amino acid oxidase when it is combined with racemic mixtures of various amino acids [134]. In addition, a method know as peak decay analysis has been used with small monolith columns containing HSA or AGP to measure the dissociation rates of various drugs from these serum proteins [66,67].
5 Concluding Remarks
This review examined the development and applications of AMC. An overview of this method was first presented, followed by a discussion of its general principles. It was then shown how various types of supports have been used in this method, including organic-based monoliths, silica monoliths, carbohydrate-based monoliths and cryogels. Techniques for immobilizing binding agents within these supports were also discussed. These methods ranged from covalent and coordination-based techniques to non-covalent methods such as entrapment. Finally, the various types of binding agents that can be used in this method were described, along with some applications of AMC that make use of these agents. The categories of AMC that were discussed included bioaffinity chromatography, immunoaffinity chromatography, dye-ligand affinity chromatography, immobilized metal-ion affinity chromatography, chiral separations, and the use of AMC to study biological interactions. Applications that have been described for these methods span from protein and biochemical purification to sample pretreatment and separations for targets such as drugs, environmental agents, proteins, immunoglobulins, and phosphorylated peptides or proteins, among many others. As further developments occur in the future in the types of supports, immobilization methods and binding agents that can be used in AMC, an even greater set of applications is expected for this technique in the purification or analysis of chemicals and biochemicals.
Figure 4.
General scheme for protein entrapment in a sol-gel, including (a) initial formation of the sol particles, (b) addition of the protein or binding agent, and (c) creation of the final sol-gel that includes the entrapped binding agent. Reproduced with permission from Ref. [65].
Figure 9.
(a) Chiral separation of R- and S-warfarin at several flow rates on a silica monolith containing immobilized AGP and (b) comparison of the total plate heights (Htotal) measured at various linear velocities for S-warfarin on this monolith column versus an AGP column that was based on silica particles. Reproduced with permission from Ref. [62].
Acknowledgments
This work was supported, in part, by the National Institutes of Health under grants R01 GM044931 and R01 DK069629 and by the National Science Foundation under grant CHE 1309806. A. Pekarek was supported through a fellowship from the University of Nebraska Molecular Mechanisms of Disease program.
Footnotes
Conflict of interest statement
The authors have no financial or commercial conflicts of interest to declare regarding the material presented in this paper.
References
- 1.Walters RR. Anal Chem. 1985;57:1099A–1114A. doi: 10.1021/ac00288a001. [DOI] [PubMed] [Google Scholar]
- 2.Cuatrecasas P, Wilchek M, Anfinsen CB. Proc Natl Acad Sci USA. 1968;61:636–643. doi: 10.1073/pnas.61.2.636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ettre LS. Pure Appl Chem. 1992;65:819–872. [Google Scholar]
- 4.Hage DS, editor. Handbook of Affinity Chromatography. 2. Taylor & Francis; New York: 2006. [Google Scholar]
- 5.Hage DS, Carr JD. Analytical Chemistry and Quantitative Analysis. Chap. 22 Pearson; New Jersey: 2011. [Google Scholar]
- 6.Schiel JE, Mallik R, Soman S, Joseph KS, Hage DS. J Sep Sci. 2006;29:719–737. doi: 10.1002/jssc.200500501. [DOI] [PubMed] [Google Scholar]
- 7.Chaiken IM, editor. Analytical Affinity Chromatography. Boca Raton: CRC Press; 1987. [Google Scholar]
- 8.Schriemer DS. Anal Chem. 2004;76:440A–448A. doi: 10.1021/ac041684m. [DOI] [PubMed] [Google Scholar]
- 9.Hage DS, Anguizola JA, Bi C, Li R, Matsuda R, Papastavros E, Pfaunmiller E, Vargas J, Zheng X. J Pharm Biomed Anal. 2012;69:93–105. doi: 10.1016/j.jpba.2012.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hage DS, Anguizola JA, Jackson AJ, Matsuda R, Papastavros E, Pfaunmiller E, Tong Z, Vargas-Badillas J, Yoo MJ, Zheng X. Anal Methods. 2011;3:1449–1460. doi: 10.1039/C1AY05068K. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Patel S, Wainer IW, Lough WJ. In: Handbook of Affinity Chromatography. 2. Hage DS, editor. Chap. 24 Taylor & Francis; New York: 2006. [Google Scholar]
- 12.Schiel JE, Joseph KS, Hage DS. In: Advances in Chromatography. Grinsberg N, Grushka E, editors. Vol. 48. Chap. 4 Taylor & Francis; New York: 2009. [Google Scholar]
- 13.Hage DS. Clin Chem. 1999;45:593–615. [PubMed] [Google Scholar]
- 14.Larsson PO. Methods Enzymol. 1984;104:212–223. doi: 10.1016/s0076-6879(84)04091-x. [DOI] [PubMed] [Google Scholar]
- 15.Gustavsson PE, Larsson PO. In: Handbook of Affinity Chromatography, 2nd Edition. Hage DS, editor. Chap 2 Taylor & Francis; New York: 2006. [Google Scholar]
- 16.Gustavsson PE, Larsson PO. In: Monolithic Materials. Svec F, Tennikova TB, Deyl Z, editors. Elsevier; Amsterdam: 2003. pp. 121–141. [Google Scholar]
- 17.Tetala KK, van Beek TA. J Sep Sci. 2010;33:422–438. doi: 10.1002/jssc.200900635. [DOI] [PubMed] [Google Scholar]
- 18.Mallik R, Hage DS. J Sep Sci. 2006;29:1686–1704. doi: 10.1002/jssc.200600152. [DOI] [PubMed] [Google Scholar]
- 19.Yoo MJ, Hage DS. In: Monolithic Chromatography and Its Modern Applications. Wang P, editor. Chap. 1 ILM; St. Albans: 2010. [Google Scholar]
- 20.Pfaunmiller EL, Paulemond ML, Dupper CM, Hage DS. Anal Bioanal Chem. 2013;405:2133–2145. doi: 10.1007/s00216-012-6568-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hage DS, Xuan H, Nelson MA. In: Handbook of Affinity Chromatography, 2nd Edition. Hage DS, editor. Chap. 4 Taylor & Francis; New York: 2006. [Google Scholar]
- 22.Ohlson S, Lundblad A, Zopf D. Anal Biochem. 1988;15:204–208. doi: 10.1016/0003-2697(88)90275-8. [DOI] [PubMed] [Google Scholar]
- 23.Strandh M, Andersson HS, Ohlson S. Methods Mol Biol. 2000;147:7–23. doi: 10.1007/978-1-60327-261-2_2. [DOI] [PubMed] [Google Scholar]
- 24.Hage DS. In: Encyclopedia of Chromatography. 3. Cazes J, editor. Taylor & Francis; New York: 2010. pp. 33–36. [Google Scholar]
- 25.Hage DS, Bian M, Burks R, Karle E, Ohnmacht C, Wa C. In: Handbook of Affinity Chromatography, 2nd Edition. Hage DS, editor. Chap 5 Taylor & Francis; New York: 2006. [Google Scholar]
- 26.Hage DS, Phillips TM. In: Handbook of Affinity Chromatography. 2. Hage DS, editor. Chap. 6 Taylor & Francis; New York: 2006. [Google Scholar]
- 27.Hage DS. J Chromatogr B. 1998;715:3–28. doi: 10.1016/s0378-4347(97)00621-x. [DOI] [PubMed] [Google Scholar]
- 28.Todorova D, Vijayalakshmi MA. In: Handbook of Affinity Chromatography, 2nd Edition. Hage DS, editor. Chap. 10 Taylor & Francis; New York: 2006. [Google Scholar]
- 29.Block H, Maertens B, Spriestersbach A, Brinker N, Kubicek J, Fabis R, Labahn J, Schafer F. Methods Enzymol. 2009;463:439–473. doi: 10.1016/S0076-6879(09)63027-5. [DOI] [PubMed] [Google Scholar]
- 30.Labrou NE, Mazitsos K, Clonis YD. Dye-ligand and biomimetic affinity chromatography. In: Hage DS, editor. Handbook of Affinity Chromatography. 2. Chap. 9 Taylor & Francis; New York: 2006. [Google Scholar]
- 31.Hermanson GT, Mallia AK, Smith PK. Immobilized Affinity Ligand Techniques. Academic Press; Cambridge: 1992. [Google Scholar]
- 32.Kim HS, Hage DS. In: Handbook of Affinity Chromatography. 2. Hage DS, editor. Chap. 3 Taylor & Francis; New York: 2006. [Google Scholar]
- 33.Peters EC, Svec F, Frechet JM. Adv Mater. 1999;11:116–1181. [Google Scholar]
- 34.Svec F, Tennikova TB, Deyl Z, editors. Monolithic Materials: Preparation, Properties and Applications. Elsevier; Amsterdam: 2003. [Google Scholar]
- 35.Kubin M, Spacek P, Chromecek R. Coll Czechoslovak Chem Comm. 1967;32:3881–3887. [Google Scholar]
- 36.Lynn TR, Rushneck DR, Cooper AR. J Chromatogr Sci. 1974;12:76–79. [Google Scholar]
- 37.Hileman FD, Sievers RE, Hess GG, Ross WD. Anal Chem. 1973;45:1126–1130. [Google Scholar]
- 38.Ross WD, Jefferson RT. J Chromatogr Sci. 1970;8:386–389. [Google Scholar]
- 39.Hansen LC, Sievers RE. J Chromatogr. 1974;99:123–133. [Google Scholar]
- 40.Josic D, Reusch J, Loester K, Baum O, Reutter W. J Chromatogr. 1992;590:59–76. doi: 10.1016/0021-9673(92)87006-t. [DOI] [PubMed] [Google Scholar]
- 41.Merhar M, Podgornik A, Barut M, Zigon M, Strancer A. J Sep Sci. 2003;26:322–330. [Google Scholar]
- 42.Tennikova TB, Belenkii BG, Svec F. J Liq Chromatogr. 1990;13:63–70. [Google Scholar]
- 43.Tennikova TB, Svec F. J Chromatogr. 1993;646:279–288. [Google Scholar]
- 44.Hjerten S, Liao JL, Zhang R. J Chromatogr. 1989;473:273–275. [Google Scholar]
- 45.Svec F, Frechet JM. Anal Chem. 1992;64:820–822. doi: 10.1021/ac00035a008. [DOI] [PubMed] [Google Scholar]
- 46.Svec F. Chin J Chromatogr. 2005;23:585–594. [PubMed] [Google Scholar]
- 47.Guiochon G. J Chromatogr A. 2007;1168:101–168. doi: 10.1016/j.chroma.2007.05.090. [DOI] [PubMed] [Google Scholar]
- 48.Wang P, editor. Monolithic Chromatography and Its Modern Applications. ILM; St Albans: 2010. [Google Scholar]
- 49.Jiang T, Mallik R, Hage DS. Anal Chem. 2005;15:2362–2372. doi: 10.1021/ac0483668. [DOI] [PubMed] [Google Scholar]
- 50.Mallik R, Jiang T, Hage DS. Anal Chem. 2004;76:7013–7022. doi: 10.1021/ac049001q. [DOI] [PubMed] [Google Scholar]
- 51.Pfaunmiller EL, Hartmann M, Dupper CM, Soman S, Hage DS. J Chromatogr A. 2012;126:198–207. doi: 10.1016/j.chroma.2012.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Svec F. J Chromatogr A. 2010;1217:902–924. doi: 10.1016/j.chroma.2009.09.073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Kalashnikova I, Ivanova N, Tennikova T. Anal Chem. 2007;79:5173–5180. doi: 10.1021/ac0700629. [DOI] [PubMed] [Google Scholar]
- 54.Selvaraju S, El Rassi Z. J Sep Sci. 2012;35:1785–1795. doi: 10.1002/jssc.201200230. [DOI] [PubMed] [Google Scholar]
- 55.Rainer M, Najam-unl-Hag M, Bakry R, Huck CW, Bonn GK. J Proteome Res. 2007;6:382–386. doi: 10.1021/pr060426y. [DOI] [PubMed] [Google Scholar]
- 56.Aprilita NH, Huck CW, Bakry R, Feuerstein I, Stecher G, Morandell S, Huang HL, Stasyk T, Huber LA, Bonn GL. J Proteome Res. 2005;4:2312–2319. doi: 10.1021/pr050224m. [DOI] [PubMed] [Google Scholar]
- 57.Pfaunmiller EL. Development and Optimization of Organic Based Monoliths for use in Affinity Chromatography. University of Nebraska; Lincoln: 2011. [Google Scholar]
- 58.Rainer M, Sonderegger H, Bakry R, Huck CW, Morandell S, Huber LA, Gjerde DT, Bonn GK. Proteomics. 2008;8:4593–4602. doi: 10.1002/pmic.200800448. [DOI] [PubMed] [Google Scholar]
- 59.Nakanishi K. J Porous Mater. 1997;4:67–112. [Google Scholar]
- 60.Cabrera K. LC GC. 2012;4:30–35. [Google Scholar]
- 61.Lubda D, Cabrera K, Kraas W, Schaefer C, Cunningham D. LC GC. 2001;19:1186–1191. [Google Scholar]
- 62.Mallik R, Xuan H, Hage DS. J Chromatogr A. 2007;1149:294–304. doi: 10.1016/j.chroma.2007.03.063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Mallik R, Hage DS. J Pharm Biomed Anal. 2008;46:820–830. doi: 10.1016/j.jpba.2007.03.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Yoo MJ, Hage DS. J Sep Sci. 2009;32:2776–2785. doi: 10.1002/jssc.200900346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Kato M, Kato-Sakai K, Matsumoto N, Toyo’oka T. Anal Chem. 2002;74:1915–1921. doi: 10.1021/ac0112162. [DOI] [PubMed] [Google Scholar]
- 66.Yoo MJ, Hage DS. J Chromatogr A. 2011;1218:2072–2078. doi: 10.1016/j.chroma.2010.09.070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Yoo MJ, Hage DS. J Sep Sci. 2011;34:2255–2263. doi: 10.1002/jssc.201100280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Hodgson RJ, Chen Y, Zhang Z, Tleugabulova D, Long H, Zhao X, Organ M, Brook MA, Brennan JD. Anal Chem. 2004;76:2780–2790. doi: 10.1021/ac0352124. [DOI] [PubMed] [Google Scholar]
- 69.Besanger TR, Hodgson RJ, Green JR, Brennan JD. Anal Chim Acta. 2006;564:106–115. doi: 10.1016/j.aca.2005.12.066. [DOI] [PubMed] [Google Scholar]
- 70.Gustavsson PE, Larsson PO. J Chromatogr A. 1999;832:29–39. doi: 10.1016/s0021-9673(98)00979-0. [DOI] [PubMed] [Google Scholar]
- 71.Gustavsson PE, Larsson PO. J Chromatogr A. 2001;925:69–78. doi: 10.1016/s0021-9673(01)01027-5. [DOI] [PubMed] [Google Scholar]
- 72.Sun S, Tang Y, Fu Q, Liu X, Guo L, Zhao Y, Chang C. Int J Biol Macromol. 2012;50:1002–1007. doi: 10.1016/j.ijbiomac.2012.02.028. [DOI] [PubMed] [Google Scholar]
- 73.Sun S, Tang Y, Fu Q, Liu X, Wei D, Guo L, Zhao Y. J Sep Sci. 2012;35:893–900. doi: 10.1002/jssc.201100940. [DOI] [PubMed] [Google Scholar]
- 74.Barroso T, Casimiro T, Ferraria AM, Mattioli F, Aguiar-Ricardo A, Roque ACA. Adv Func Mater. 2014;24:4528–4541. [Google Scholar]
- 75.Dainiak MB, Galaev IY, Mattiasson B. J Chromatogr A. 2006;1123:145–150. doi: 10.1016/j.chroma.2006.05.089. [DOI] [PubMed] [Google Scholar]
- 76.Lozinsky VI, Plieva FM, Galaev IY, Mattiasson B. Biosep. 2002;10:163–188. doi: 10.1023/a:1016386902611. [DOI] [PubMed] [Google Scholar]
- 77.Arvidsson P, Plieva FM, Savina IN, Lozinsky VI, Fexby S, Bulow L, Galaev IY, Mattiasson B. J Chromatogr A. 2002;977:27–38. doi: 10.1016/s0021-9673(02)01114-7. [DOI] [PubMed] [Google Scholar]
- 78.Wang L, Shaochuan S, He X, Yun J, Yao K, Yao SJ. Biochem Eng J. 2008;42:237–242. [Google Scholar]
- 79.Derazshamshir A, Ergun B, Pesint G, Odabasi M. J Appl Polym Sci. 2008;109:2905–2913. [Google Scholar]
- 80.Alkan H, Bereli N, Baysal Z, Denizli A. Biochem Eng J. 2010;51:153–159. [Google Scholar]
- 81.Berruex LG, Freitag R, Tennikova TB. J Pharm Biomed Anal. 2000;24:95–104. doi: 10.1016/s0731-7085(00)00414-3. [DOI] [PubMed] [Google Scholar]
- 82.Luo Q, Zou H, Zhang Q, Xiao X, Ni J. Biotechnol Bioeng. 2002;80:481–489. doi: 10.1002/bit.10391. [DOI] [PubMed] [Google Scholar]
- 83.Hermanson GT, editor. Bioconjugate Techniques. Academic Press; San Diego: 1995. [Google Scholar]
- 84.Petro M, Svec F, Frechet JMJ. Biotechnol Bioeng. 1996;49:355–363. doi: 10.1002/(SICI)1097-0290(19960220)49:4<355::AID-BIT1>3.0.CO;2-O. [DOI] [PubMed] [Google Scholar]
- 85.Calleri E, Marrubini G, Massolini G, Lubada D, de Fazio SS, Fulanetto S, Wainer IW, Manzo L, Caccialanza G. J Pharm Biomed Anal. 2004;35:1179–1189. doi: 10.1016/j.jpba.2004.02.033. [DOI] [PubMed] [Google Scholar]
- 86.Ruhn PF, Garver S, Hage DS. J Chromatogr A. 1994;669:9–19. doi: 10.1016/0021-9673(94)80332-3. [DOI] [PubMed] [Google Scholar]
- 87.Xuan H, Hage DS. Anal Biochem. 2005;346:300–310. doi: 10.1016/j.ab.2005.08.025. [DOI] [PubMed] [Google Scholar]
- 88.Martin del Valle EM, Galan MA, Cerro RL. Biotechnol Prog. 2003;19:921–927. doi: 10.1021/bp0340234. [DOI] [PubMed] [Google Scholar]
- 89.Ren D, Penner NA, Slentz BE, Inerowicz HD, Rybalko M, Regnier FE. J Chromatogr A. 2004;1031:87–92. doi: 10.1016/j.chroma.2003.10.041. [DOI] [PubMed] [Google Scholar]
- 90.Vizioli NM, Rusell ML, Carbajal ML, Carducci CN, Grasselli M. Electrophoresis. 2005;26:2942–2948. doi: 10.1002/elps.200410416. [DOI] [PubMed] [Google Scholar]
- 91.Asliyuce S, Mattiason B, Mamo G. J Chromatogr B. 2016;1021:204–212. doi: 10.1016/j.jchromb.2015.12.060. [DOI] [PubMed] [Google Scholar]
- 92.Mayes AG, Mosbach K. Trends Anal Chem. 1997;16:321–332. [Google Scholar]
- 93.Alexander C, Andersson HS, Andersson LI, Ansell RJ, Kirsch N, Nicholls IA, O’Mahony J, Whitcombe MJ. J Mol Recognit. 2006;19:106–180. doi: 10.1002/jmr.760. [DOI] [PubMed] [Google Scholar]
- 94.Witcombe MJ, Kirsch N, Nicholls IA. J Mol Recognit. 2014;27:297–401. doi: 10.1002/jmr.2347. [DOI] [PubMed] [Google Scholar]
- 95.Haupt K. In: Handbook of Affinity Chromatography. 2. Hage DS, editor. Chap. 30 Taylor & Francis; New York: 2005. [Google Scholar]
- 96.Leblebici P, Leblebici ME, Ferreira-Silva F, Rodrigues AE, Pais LS. J Chromatogr B. 2014;962:89–93. doi: 10.1016/j.jchromb.2014.05.029. [DOI] [PubMed] [Google Scholar]
- 97.Tscheliessnig A, Jungbauer A. J Chromatogr A. 2009;1216:2676–2682. doi: 10.1016/j.chroma.2008.07.063. [DOI] [PubMed] [Google Scholar]
- 98.Ralla K, Anton F, Scheper T, Kasper C. J Chromatogr A. 2009;1216:2671–2675. doi: 10.1016/j.chroma.2008.09.102. [DOI] [PubMed] [Google Scholar]
- 99.Cernigoj U, Vidic U, Nemec B, Gaspersic J, Vidic J, Krajnc NL, Strancar A, Podgornik A. J Chromatogr A. 2016;1464:72–78. doi: 10.1016/j.chroma.2016.08.014. [DOI] [PubMed] [Google Scholar]
- 100.Herigstad MO, Dimartino S, Boi C, Sarti GC. J Chromatogr A. 2015;1407:130–138. doi: 10.1016/j.chroma.2015.06.045. [DOI] [PubMed] [Google Scholar]
- 101.Bakovic MP, Selman MHJ, Hoffmann M, Rudan I, Campbell H, Deelder AM, Lauc G, Wuhrer M. J Proteome Res. 2013;12:821–831. doi: 10.1021/pr300887z. [DOI] [PubMed] [Google Scholar]
- 102.Pan Z, Zou H, Mo W, Huang X, Wu R. Anal Chim Acta. 2002;466:141–150. [Google Scholar]
- 103.Temporini C, Perani E, Mancini F, Bartolini M, Calleri E, Lubada D, Felix G, Andrisano V, Massolini G. J Chromatogr A. 2006;1120:121–131. doi: 10.1016/j.chroma.2006.01.030. [DOI] [PubMed] [Google Scholar]
- 104.Calleri E, Temporini C, Perani E, Stella C, Rudaz S, Lubda D, Mellerio G, Veuthey JL, Caccialanza G, Massolini G. J Chromatogr A. 2004;1045:99–109. doi: 10.1016/j.chroma.2004.06.034. [DOI] [PubMed] [Google Scholar]
- 105.Bedair M, Oleschuk RD. Analyst. 2006;131:1316–1321. doi: 10.1039/b607359j. [DOI] [PubMed] [Google Scholar]
- 106.Feng S, Yang N, Pennathur S, Goodison S, Lubman DM. Anal Chem. 2009;81:3776–3783. doi: 10.1021/ac900085k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Okanda FM, El Rassi Z. Electrophoresis. 2006;27:1020–1030. doi: 10.1002/elps.200500766. [DOI] [PubMed] [Google Scholar]
- 108.Hsieh ML, Li GY, Chau LK, Hon YS. J Sep Sci. 2008;31:1819–1827. doi: 10.1002/jssc.200700631. [DOI] [PubMed] [Google Scholar]
- 109.Lubda D, Cabrera K, Nakanishi K, Lindner W. Anal Bioanal Chem. 2003;377:892–901. doi: 10.1007/s00216-003-2181-x. [DOI] [PubMed] [Google Scholar]
- 110.Kalashnikova I, Ivanova N, Tennikova T. Anal Chem. 2008;80:2188–2198. doi: 10.1021/ac702258t. [DOI] [PubMed] [Google Scholar]
- 111.Hage DS. J Chromatogr B. 1998;715:3–28. doi: 10.1016/s0378-4347(97)00621-x. [DOI] [PubMed] [Google Scholar]
- 112.Moser AC, Hage DS. Bioanalysis. 2010;2:769–790. doi: 10.4155/bio.10.31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Matsuda R, Rodriguez E, Suresh D, Hage DS. Bioanalysis. 2015;7:2947–2966. doi: 10.4155/bio.15.206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Monster A, Hiller O, Gruger D, Blasczyk R, Kasper C. J Chromatogr A. 2011;1218:706–710. doi: 10.1016/j.chroma.2010.12.009. [DOI] [PubMed] [Google Scholar]
- 115.Liang Y, Zhou S, Hu L, Li L, Zhao M, Liu H. J Chromatogr B. 2010;878:278–282. doi: 10.1016/j.jchromb.2009.06.007. [DOI] [PubMed] [Google Scholar]
- 116.Faye C, Chamieh J, Moreau T, Grainer F, Faure K, Dugas V, Demesmay C, Vandenabeele-Trambouze O. Anal Biochem. 2012;420:147–154. doi: 10.1016/j.ab.2011.09.016. [DOI] [PubMed] [Google Scholar]
- 117.Li L, Wang J, Zhao S, Zhao M. Anal Chim Acta. 2008;620:1–7. doi: 10.1016/j.aca.2008.05.036. [DOI] [PubMed] [Google Scholar]
- 118.Yang W, Sun X, Pan T, Woolley AT. Electrophoresis. 2008;29:3429–3435. doi: 10.1002/elps.200700704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Calleri E, Marrubini G, Brusotti G, Massolini G, Caccialanza G. J Pharm Biomed Anal. 2007;44:396–403. doi: 10.1016/j.jpba.2007.01.030. [DOI] [PubMed] [Google Scholar]
- 120.Gunasena DN, El Rassi Z. J Sep Sci. 2011;34:2097–2105. doi: 10.1002/jssc.201100353. [DOI] [PubMed] [Google Scholar]
- 121.Denizli A, Piskin E. J Biochem Biophys Methods. 2001;49:391–416. doi: 10.1016/s0165-022x(01)00209-3. [DOI] [PubMed] [Google Scholar]
- 122.Uzun L, Yavuz H, Say R, Ersoz A, Denizli A. Ind Eng Chem Res. 2004;43:6507–6513. [Google Scholar]
- 123.Andac M, Galaer I, Denizli A. J Sep Sci. 2012;35:1173–1182. doi: 10.1002/jssc.201101020. [DOI] [PubMed] [Google Scholar]
- 124.Josic D, Clifton JG. J Chromatogr A. 2007;1144:2–13. doi: 10.1016/j.chroma.2006.11.082. [DOI] [PubMed] [Google Scholar]
- 125.Hou C, Ma J, Tao D, Shan Y, Liang Z, Zhang L, Zhang Y. J Proteome Res. 2010;9:4093–4101. doi: 10.1021/pr100294z. [DOI] [PubMed] [Google Scholar]
- 126.Feng S, Pan C, Jiang X, Xu S, Zhou H, Ye M, Zou H. Proteomics. 2007;7:351–360. doi: 10.1002/pmic.200600045. [DOI] [PubMed] [Google Scholar]
- 127.Krenkova J, Foret F. J Sep Sci. 2011;34:2106–2112. doi: 10.1002/jssc.201100256. [DOI] [PubMed] [Google Scholar]
- 128.Saeed A, Maya F, Xiao DJ, Najam-ul-Haq M, Svec F, Britt DK. Adv Funct Mater. 2014;24:5790–5797. [Google Scholar]
- 129.Cimen D, Denizil A. Colloids Surf B. 2012;93:29–35. doi: 10.1016/j.colsurfb.2011.11.058. [DOI] [PubMed] [Google Scholar]
- 130.Shin MJ, Tan L, Jeong MH, Kim J, Choe W. J Chromtogr A. 2011;1218:5273–5278. doi: 10.1016/j.chroma.2011.06.040. [DOI] [PubMed] [Google Scholar]
- 131.Peterka M, Jarc M, Banjac M, Frankovic V, Bencina K, Merhar M, Gaberc-Porekar V, Menart V, Strancar A, Podgornik A. J Chromatogr A. 2006;1109:80–85. doi: 10.1016/j.chroma.2005.08.057. [DOI] [PubMed] [Google Scholar]
- 132.Efremanko E, Votchiseva Y, Plieva F, Galaev I, Mattiason B. Appl Microbiol Biotechnol. 2006;70:558–563. doi: 10.1007/s00253-005-0103-x. [DOI] [PubMed] [Google Scholar]
- 133.Hong T, Zheng Y, Hu W, Ji Y. Anal Biochem. 2014;464:43–50. doi: 10.1016/j.ab.2014.07.015. [DOI] [PubMed] [Google Scholar]
- 134.Yao C, Qi L, Qiao J, Zhang H, Wang F, Chen Y, Yang G. Talanta. 2010;82:1332–1337. doi: 10.1016/j.talanta.2010.06.041. [DOI] [PubMed] [Google Scholar]
- 135.Zacharis CK, Kalaitzantonakis EA, Podgornik A, Theodoridis G. J Chromatogr A. 2007;1144:126–134. doi: 10.1016/j.chroma.2006.12.081. [DOI] [PubMed] [Google Scholar]