Abstract
Background
Exposure to drugs of abuse alters the epigenetic landscape of the brain’s reward regions such as the nucleus accumbens (NAc). We investigated how combinations of chromatin modifications affect genes that regulate responses to cocaine. We focused on autism-candidate 2 (Auts2), a gene linked to human evolution and cognitive disorders, which displays strong clustering of cocaine-induced chromatin modifications in this brain region.
Methods
We combined chromosome conformation capture (4C and 3C) and related approaches with behavioral paradigms relevant to cocaine phenotypes. Cell type-specific functions were assessed by FACS-sorting and viral-mediated overexpression in Cre-dependent mouse lines.
Results
We observed that Auts2 gene expression is increased by repeated cocaine administration specifically in D2-type medium spiny neurons (MSNs) in NAc, an effect seen in male but not female mice. Auts2 mRNA expression was also upregulated postmortem in NAc of male human cocaine addicts. We obtained evidence that chromosomal looping, bypassing 1,524 kb of linear genome, connects Auts2 to the calneuron 1 (Caln1) gene locus under baseline conditions. This looping was disrupted after repeated cocaine exposure, resulting in increased expression of both genes in D2 MSNs. Cocaine exposure reduces binding of CTCF, a chromosomal scaffolding protein, and increases histone and DNA methylation, at the Auts-Caln1 loop base in NAc. Cell type-specific overexpression of Auts2 or Calnl in D2 MSNs demonstrated that both genes promote cocaine reward.
Conclusions
These findings suggest that cocaine-induced alterations of neuronal 3D genome organization destabilize higher order chromatin at specific loci that regulate responses to the drug.
Keywords: Addiction, CTCF, histone methylation, DNA methylation, chromatin looping, nucleus accumbens
Introduction
Changes in chromatin architecture have been associated with gene expression since the early 20th century (1). The recent development of chromatin conformation capture techniques, in combination with state-of-the-art approaches in molecular biology, has allowed a conceptual link between genomic function and architecture (2; 3). These mechanisms are beginning to be examined in psychiatric conditions (4). However, the relationship between genomic architecture and drug addiction has not yet been explored.
Autism-candidate 2 (Auts2) is a rapidly evolving gene, which has been linked to evolution (5) as well as to autism (6), alcoholism (7), and heroin dependence (8). Recent work has shown that the Auts2 protein is part of the PRC2 polycomb complex (9) and alters Rac signaling. Consisting mostly of untranscribed regions, the large Auts2 gene locus may have structural functions, since most disease-related SNPs occur in non-coding segments of the gene.
We identified Auts2 as a key target for cocaine-induced chromatin modifications: analysis of published chromatin immunoprecipitation (ChIP)-sequencing data in NAc (10) revealed that this locus contains among the largest number of cocaine-induced chromatin changes genome-wide, yet its mRNA expression levels are not affected in this region based on RNA-sequencing data of whole NAc extracts. As a first step in investigating this paradox, we examined chromatin looping interactions formed by Auts2 in NAc, based on the notion that the high degree of chromatin modifications might influence the 3D structure of this locus. Using chromatin conformation capture approaches, we show that Auts2 indeed interacts with the calneuron 1 (Caln1) gene at baseline and that this interaction is disrupted by repeated cocaine administration. While cocaine-induced changes in Auts2 and Caln1 expression were not observed in whole NAc extracts (10), we demonstrate that increased Auts2 and Caln1 transcription occurs selectively in D2-type medium spiny neurons (MSNs) of NAc, with no effect seen in D1-type MSNs. We next show that the cocaine-induced change in interaction between the Auts2 and Caln1 genes is accompanied by changes in H3K4me3 (trimethylation of Lys4 of histone H3), in binding of CTCF (CCCTC binding factor), and in DNA methylation at these loci. By employing a novel CRISPR-epigenome editing approach, we show that targeting DNA methyltransferase (DNMT) 3a/3L to the Auts2 gene controls Caln1 expression in cell culture. Finally, viral-mediated overexpression of Auts2 or Caln1 selectively in D2 MSNs of NAc increases rewarding responses to cocaine. Together, these findings shed light on a novel mechanism by which cocaine-induced chromatin modifications underlie the complex regulation of an ensemble of genes in NAc to influence behavioral sensitivity to the drug.
Methods and Materials
Animals
Sprague-Dawley rats and C57BL/6J mice were purchased from Jackson, Bar Harbor, Maine. D1-tomato, D2-GFP, D1-Cre, and D2-Cre mice were obtained from N. Heintz (Rockefeller) and C. Gerfen (NIMH). Rats and mice were used for their unique strengths; see Results and supplementary material. All experiments were conducted on male animals, except where indicated.
Chromosome conformation capture (3C) and circularized chromosome-conformation capture (4C)
Cocaine self-administration
Rats containing chronic indwelling jugular catheters were trained for v. self-administration as described (14; 15).
Human brain tissue
NAc from cocaine-addicted or depressed male patients and matched male controls were acquired from McGill University. Psychological autopsies were performed as described (16).
FACS from D1-Tomato and D2-GFP mice
D1 and D2 MSNs were isolated from NAc punches by use of a BD FACS Aria II. RNA was extracted using the Direct-zol RNA miniprep kit (Zymo, Irvine, USA, #R2050).
Quantitative ChIP
qChIP was performed as described (17).
Bisulfite sequencing
Bisulfite conversation was conducted on purified DNA with the EZ DNA methylation kit (Zymo, Irvine, USA, #D5001). Libraries were prepared with a TruSeq Nano DNA Library Prep Kit (Illumina, San Diego, USA, #FC-121-4003) according to manufacturers’ instructions. Libraries were pooled and sequenced on an Illumina MiSeq sequencer, using a 600 cycle, V3-chemistry sequencing kit (Illumina, San Diego, USA, #MS-102-3003). Sequencing data were then analyzed using Bismark software (Reference: PMID: 21493656).
CRISPR epigenome-editing
A vector coding the dCas9-DNMT3ACD-DNMT3LCD-3xFLAG fusion gene was constructed by Gibson assembly. Guide RNAs were designed via a published protocol (https://www.addgene.org/crispr/church/).
miRNA vectors
were designed with the Thermo Fisher Scientific BLOCK-iT™ RNAi Designer tool. Constructs, purchased from IDT, were cloned into a pcDNA 6.2 GW/miR vector with the BLOCK- iT™ Pol II miR RNAi Expression Vector Kit (Thermo Fisher Scientific, Waltham, USA, #K493500).
Herpes simplex virus (HSV) vectors
Plasmids for Auts2 and Caln1 were purchased from Origene (Rockville, USA, # MR225787 and MR202484) and cloned into a p1005-LS1L-vector, then packaged.
Conditioned place preference
was carried out as described previously (18).
A complete description of methods is provided in supplementary material online.
Results
Auts2 and Caln1 genes interact at the chromatin level
Repeated exposure to cocaine induces numerous epigenetic modifications in NAc as assessed from genome-wide ChIP-sequencing analyses (10). To identify genes that are highly regulated by cocaine, we analyzed changes in several chromatin marks that are altered in this brain region 24 h after 7 days of cocaine and sorted for genes with the highest number of modifications (Table S1), revealing Auts2 as a top hit. In cultured cells, Auts2 has been shown to form chromatin loops with other genes (19). Recently developed databases for the prediction of chromatin interactions suggest genomic interactions of the Auts2 gene in mouse cortex as well (Yue Lab 3D Genome browser, http://promoter.bx.psu.edu). Auts2 is a large gene (1,099,500 bp in rat), which consists mostly of intronic regions, and it is in those regions that most cocaine-induced chromatin modifications occur. We therefore hypothesized that Auts2 might interact with other genes in NAc and that cocaine may alter this interaction profile.
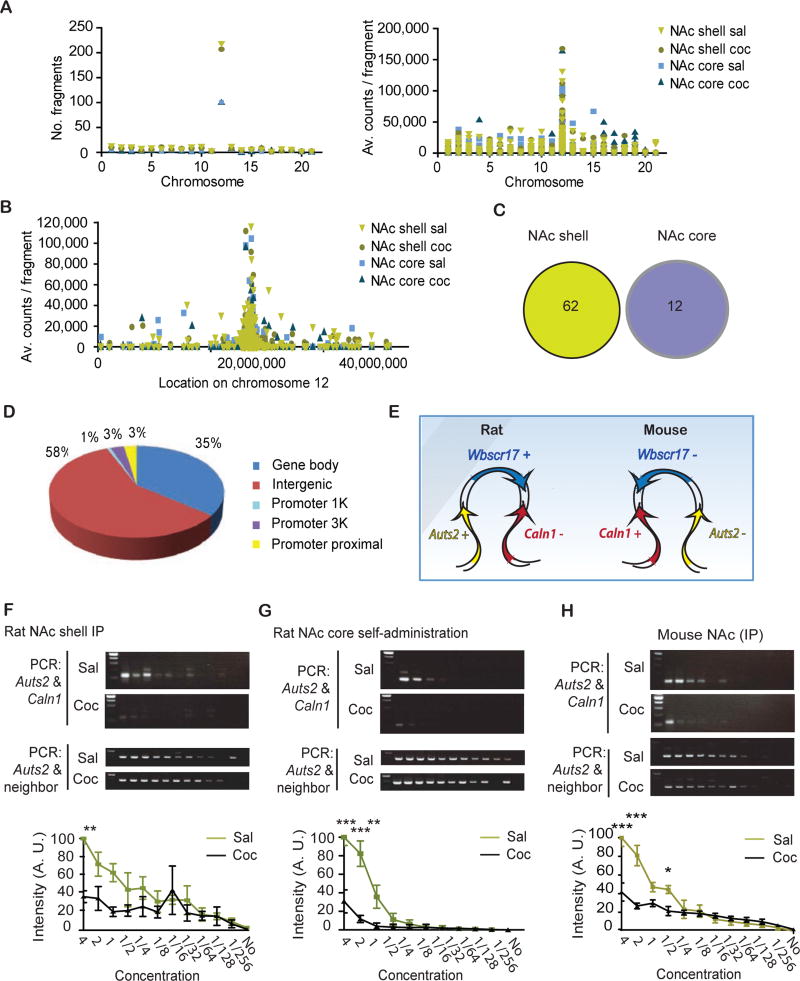
To test this hypothesis, we collected punches of NAc shell and core from rats that had received 7 daily IP injections of saline or 20 mg/kg cocaine and performed 4C with an Auts2 bait. 4C allows an unbiased approach to detect all chromatin interacting regions with a given locus of interest (see Supplementary methods for details). We targeted an un-transcribed region near the Auts2 promoter that fulfilled the criteria necessary for 4C baits (Fig. S1). Because high cell numbers are required to study chromatin interactions, we conducted the 4C experiment in rats. As NAc shell and core have different roles in cocaine and other drug responses (20–22), separate 4C libraries were made for the two subregions. Paired end sequencing of the libraries yielded fragments that are in good agreement with the 4C literature (23). Most 4C-interacting fragments were located on the same chromosome as Auts2 (chr12) and the number of fragments correlated with vicinity to the 4C bait (Fig. 1a-c; (Table S2). On average, 8,735,380 reads/sample were obtained for NAc core and 9,198,209 for NAc shell from 1,486 and 1,615 fragments, respectively. 4C fragments that were differentially regulated by cocaine showed no overlap between NAc shell and core (Fig. 1c, Fig. S2).This is in agreement with the specific functions of these NAc subregions in cocaine- related behaviors, as noted above. The majority (58%) of interacting fragments were intergenic and 35% were intragenic (Fig. 1d, Fig. S3). In NAc core, only 0.8% of all detected fragments were significantly affected by cocaine, while 3.8% were altered in NAc shell (no cutoff applied). The majority of these fragments comprised the Auts2 gene itself or proximal intergenic regions (data not shown). The next gene that interacted most with the Auts2 bait in NAc shell was Calnl, which notably was also one of the 10 genes with most cocaine-induced chromatin modifications, even after adjusting for gene length (Table S1, Fig. S4). Calnl is a brain-specific calcium-binding protein that regulates Golgi-to-plasma membrane trafficking of vesicles (24; 25) and under certain conditions is reported to alter D2-A2AR heteromerization (26). The predicted Calnl -fragment is located 1,524 kb away from the Auts2 bait. Between Auts2 and Caln17 lies the Wbscr17 gene, which has been associated with chromatin-breakage in Williams syndrome (27). Both in mice and rats, Caln1, Wbscr17 and Auts2 are neighboring genes, however, the direction of the genes is inverted between the two species (Fig. 1e). 3C confirmed a cocaine-induced reduction in interactions between Auts2 and Caln1 in NAc shell of IP-injected rats (F(1,48)=16.20, P<0.001, Fig. 1f). 3C on NAc core showed no signal. This is consistent with the fact that no interaction between Auts2 and Calnl was detected by 4C in NAc core even in saline conditions.
Figure 1.
Auts2 and Caln1 gene interactions are reduced by repeated cocaine administration. a, Number of detected fragments and number of counts per fragment plotted for chromosome location over the entire rat genome. Auts2 is located on chr12. Sal, saline; Coc, cocaine. b, Average counts per fragment on chr12. The 4C bait of Auts2 is located at 27,155 kbp. c, Pie chart of genomic locations of detected 4C fragments (shell and core combined). d, Venn diagram of significantly changed fragments in NAc shell and core. e, Overview over gene locations in mouse (chr5) and rat (chr12). f, 3C libraries were prepared from NAc shell punches of rats that had been I.P.-injected for 7 days with saline or 20 mg/kg cocaine and were analyzed 24 h later. N=3,3. Two-way ANOVA: interaction: F(11,48)=2.10, P<0.05; drug factor: F(1,48)=16.20, P<0.001; dilution factor: F(11,48)=6.56, P<0.0001. Bonferroni: sal vs. coc: **P<0.01; all following: P>0.05. g, 3C libraries were prepared from NAc core punches of rats that have acquired cocaine during self-administration for at least 7 days with 20 mg/kg cocaine. They were analyzed 24 h later. N=3,3. Two-way ANOVA: interaction: F(9,40)=11.40, P<0.0001; drug factor: F(1,40)=45.92, P<0.0001; dilution factor: F(9,40)=28.43, P<0.0001. Bonferroni: sal vs. coc: ***P<0.001; ***P<0.001; **P<0.01; all following: P>0.05. h, 3C libraries were prepared from NAc shell punches of rats that had been I.P.-injected for 7 days with saline or 20 mg/kg of cocaine and were analyzed 24 h later. N=3,3. Two-way ANOVA: interaction: F(11,48)=9.834, P<0.0001; drug factor: F(1,48)=31.34, P<0.0001; dilution factor: F(11,48)=35.06, P<0.0001. Bonferroni: sal vs. coc: ***P<0.001; ***P<0.001; all following: P>0.05. f-h, Average +/− SD shown.
Next, we assessed whether 3C changes occur after cocaine self-administration. Rats selfadministered cocaine for 10 days and were analyzed 24 h after the last session. We observed a reduction in 3C interactions in NAc core after self-administering cocaine vs. saline controls (F(1,40)=45.92, P<0.0001, Fig. 1g). This agrees with previous studies showing that the NAc core is more active in cocaine seeking paradigms (28). We also confirmed the cocaine-induced reduction in Auts2-Caln1 looping in mice (effect of drug: F(1,48)=31.34, P<0.0001) (Fig. 1h). Because of the high amounts of DNA required, this experiment was conducted on whole NAc. Because there is a high homology between Wbscr17 and the 4C-predicted Caln1 -fragment in rats, but not in mice, this experiment was particularly important to confirm the specificity of Caln1-Auts2 binding. In summary, we show that cocaine reduces chromatin interactions between Auts2 and Caln1, although in different NAc subregions with investigator- vs. self-administered cocaine.
Increased cocaine-induced Auts2 and Caln1 gene expression in D2 MSNs
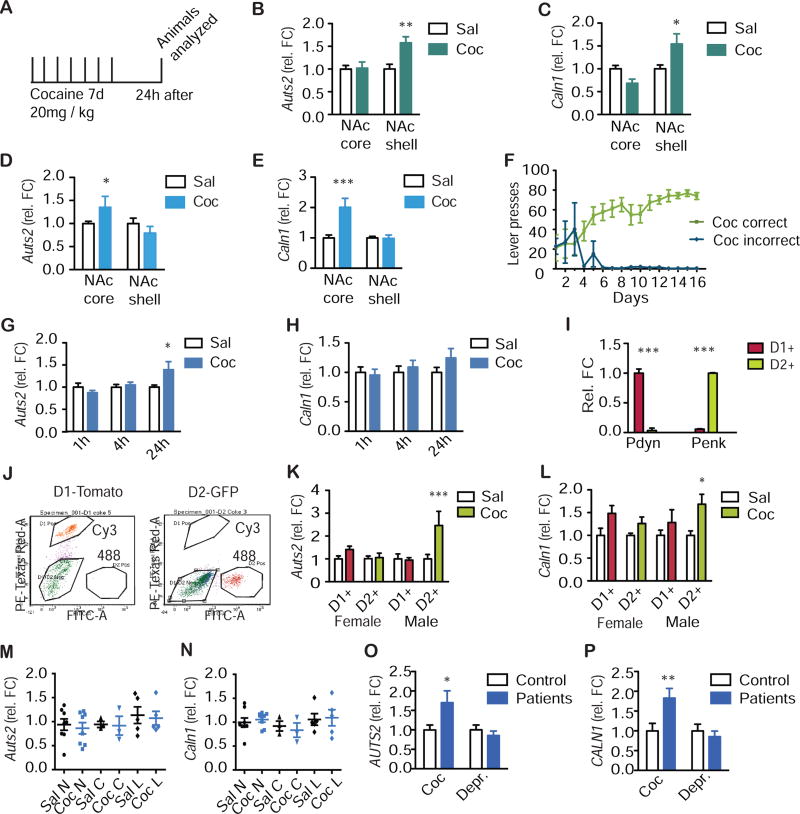
We next asked whether cocaine-induced changes in Auts2-Caln1 chromatin looping are associated with altered gene expression. Both Auts2 and Caln1 mRNA were increased in rat NAc shell after cocaine injections (Auts2: F(1,28)=6.71, P<0.05; Caln1: F(1,44)=10.68, P<0.01) and in NAc core after self-administration (Auts2: F(1,30)=9.76, P<0.01; Caln1: F(1,32)=9.06, P<0.01, Fig. 2a-f, Fig. S5). Auts2 mRNA was also increased in whole NAc of mice that had been injected for 7 days with cocaine and analyzed 24 h after the last dose (F(2,59)=3.80, P<0.05, Fig. 2g,h). No effect of cocaine was seen at earlier withdrawal time points. Caln1 expression showed a trend towards an increase at 24 h but this effect did not reach significance (P>0.05, Fig. 2h). The fact that increased Auts2 mRNA levels were detected in whole NAc of mouse, despite no effect seen earlier by RNA-sequencing under identical experimental conditions, illustrates false negative errors when using high stringency statistical analyses of RNA-sequencing data (see Discussion). It may also relate to isoform-specific alterations (Table S3).
Figure 2.
Expression of Auts2 and Caln1 is increased by repeated cocaine administration. a, Cocaine paradigm: Mice or rats were I.P.-injected for 7 days with saline (sal) or 20 mg/kg cocaine (coc) and analyzed 24 h later. b,c, Separate punches were taken from rat NAc shell and core. b, qPCR was conducted for Auts2. N=8,8,8,8. Two-way ANOVA: interaction: F(1,28)=5.83, P<0.05; drug factor: F(1,28)=6.71, P<0.05; brain region: F(1,28)=5.83, P<0.05. Bonferroni: sal vs. coc: NAcc: P>0.05; NAcs: **P<0.01. c, qPCR was conducted for Caln1. N=12,12,12,12. Two-way ANOVA: interaction: F(1,44)=10.68, P<0.01; drug factor: F(1,44)=0.16, P>0.05; brain region: F(1,44)=2.22, P<0.01. Bonferroni: sal vs. coc: NAcc: P>0.05; NAcs: *P<0.05. d, e, Rats self-administered cocaine for 10 days and were analyzed 24 h after the last session. Separate punches were taken from NAc shell and core. d, qPCR was conducted for Auts2. N=9,8,9,8. Two-way ANOVA: interaction: F(1,30)=9.76, P<0.01; drug factor: F(1,30)=2.22, P>0.05; brain region: F(1,30)=9.76, P<0.01. Bonferroni: sal vs. coc: P<0.05*; P>0.05. e, qPCR was conducted for Caln1. N=9,9,9,9. Two-way ANOVA: interaction: F(132)=9.57, P<0.01; drug factor: F(1,32)=9.06, P<0.01; brain region: F(1,32)=9.57, P<0.01**. Bonferroni: sal vs. coc: NAcc: ***P<0.001; NAcs: P>0.05. f, Acquisition curve for cocaine self-administration. Correct and incorrect lever presses for the cocaine group are shown. N=8 per group. Two-way ANOVA: interaction: F(121,182)=5.51, P<0.0001; drug factor: F(1,182)=328.9, P<0.0001; time factor: F(12,182)=4.36, P<0.0001. Bonferroni: sal vs. coc: Days −3: P>0.05; all other days: P<0.001. g,h, Punches were taken of whole NAc of male mice. g, qPCR was conducted for Auts2. N=10,11 ;11,11;11,11. Two-way ANOVA: interaction: F(2,59)=3.80, P<0.05; drug factor: F(1,59)=1.84, P>0.05; time factor: F(2,59)=3.80, P<0.05. Bonferroni: sal vs. coc: P>0.05; P>0.05; *P<0.05. h, qPCR was conducted for Caln1. N=10,11,11,11,11,11. 2 way ANOVA with Bonferroni: interaction: F(2,59)=0.87, P>0.05; drug factor: F(1,59)= 1.15, P>0.05; time factor: F(2,59)=0.84, P>0.05. Bonferroni: sal vs. coc: all P>0.05. i-l, FACS sorting on NAc punches of D1-GFP and D2-Tomato mice. i, Specificity of D1/D2-MSN markers was assessed by qPCR for Pdyn and Penk. N=36,36;30,30. Two-way ANOVA: interaction: F(1,128)=485.1, P<0.0001; gene factor: F(1,128)=0.07, P>0.05; cell factor: F(1,128)=0.07, P>0.05. Bonferroni: D1 vs. D2: ***P<0.001; ***P<0.001. j, FACS on Cy3 and Alexa488 lasers. k, qPCR was conducted for Auts2. N=10,10:7,7;8,6;9,7. Two-way ANOVA: interaction: F(3,56)=3.84, P<0.05; drug factor: F(3,56)=6.97, P<0.05; factor of sex and cell type: F(3,56)=3.79, P<0.05. Bonferroni: sal vs. coc: P>0.05; P>0.05; P>0.05; ***P<0.001. l, qPCR was conducted for Caln1. N=10,10;7,7;9,7;9,7. Two-way ANOVA: interaction: F(3,84)=0.72, P>0.05; drug factor: F(1,84)=13.38, P<0.001; factor of sex and cell type: F(3,84)=0.72, P>0.05. Bonferroni: sal vs. coc: P>0.05; P>0.05; P>0.05; *P<0.05. m, n, Punches were taken of whole NAc of female mice at nuclear (N), corneal (C), and lucidic (L) stages of their estrous cycle. m, qPCR was conducted for Auts2. N=8,8;3,3;5,5. Two-way ANOVA: interaction: F(2,26)=0.01, P>0.05; drug factor: F(1,26)=1.01, P>0.05; estrous factor: F(2,26)=0.19, P>0.05. Bonferroni: sal vs. coc: all P>0.05. n, qPCR was conducted for Caln1. N=8,8;3,3;5,5. Two-way ANOVA: interaction: F(2,26)=0.20, P>0.05; drug factor: F(1,26)=1.47, P>0.05; estrous factor: F(2,26)=0.01, P>0.05. Bonferroni: sal vs. coke: all P>0.05. o, p, RNA was extracted from NAc postmortem samples from human cocaine addicts or patients that suffered from depression and their matched controls (ctrl). o, qPCR was conducted for Auts2. N=15,14,14,14. Two-way ANOVA: interaction: F(1,53)=5.36, P<0.05; factor of disease: F(1,53)=2.74, P>0.05; factor of patients: F(1,53)=5.36, *P<0.05. Bonferroni: ctrl vs. patients: *P<0.05; P>0.05. p, qPCR was conducted for Caln1. N=14,13,14,14. Two-way ANOVA: interaction: F(1,51)=5.36, P<0.05; factor of disease: F(1,51)=5.47, P<0.05; factor of patients: F(1,51)=5.36, P<0.05,. Bonferroni: ctrl vs. patients: **P<0.01 ; P>0.05. b-i, k, l, o, p, Average +/− SD shown.
Roughly 95% of NAc neurons are MSNs, approximately half of which are D1-type vs. D2-type. Cells from NAc punches of D1-Tomato and D2-GFP mice with repeated cocaine or saline treatment were isolated by FACS. Both Auts2 and Caln1 mRNA levels were significantly increased in D2 MSNs of male mice (Auts2: F(3,56)=6.97, P<0.05; Caln1: F(1,84)=13.38, P<0.001, Fig. 2i-l), with no effect seen in D1 MSNs. The accuracy of FACS was confirmed by showing enrichment of Pdyn in D1 MSNs and Penk in D2 MSNs (Fig. 2i). This is in good agreement with a previous study, which observed a cocaine-induced D2-specific increase in Auts2 expression (29). In contrast, female mice displayed no effect of cocaine for either gene in either cell type. To rule out possible confounds of the estrous cycle, we tested whether mRNA levels of Auts2 or Caln1 fluctuated with the cycle (Fig. 2m,n); this was not the case (P>0.05).
We next tested whether Auts2 or Caln1 mRNAs are altered in NAc of humans with cocaine addiction examined postmortem. Auts2 and Caln1 expression was augmented in NAc of male cocaine addicts, while there were no changes in NAc of male patients with depression examined for comparison (Auts2: F(1,53)=5.36, P<0.05; Caln1 : F(1,51)=5.36, P<0.05, Fig. 2o, p, Table S4).
In summary, we observed an inverse correlation between chromatin 3D interactions of Auts2 and Caln1 and expression of these genes in NAc, and the induction of both in D2 MSNs by cocaine. Expression of Wbscr17, the gene located between Auts2 and Caln1, was not altered in all three species examined (rats, mice, and humans: P>0.05, data not shown).
Cocaine induces chromatin alterations on Auts2 and Calnl genes
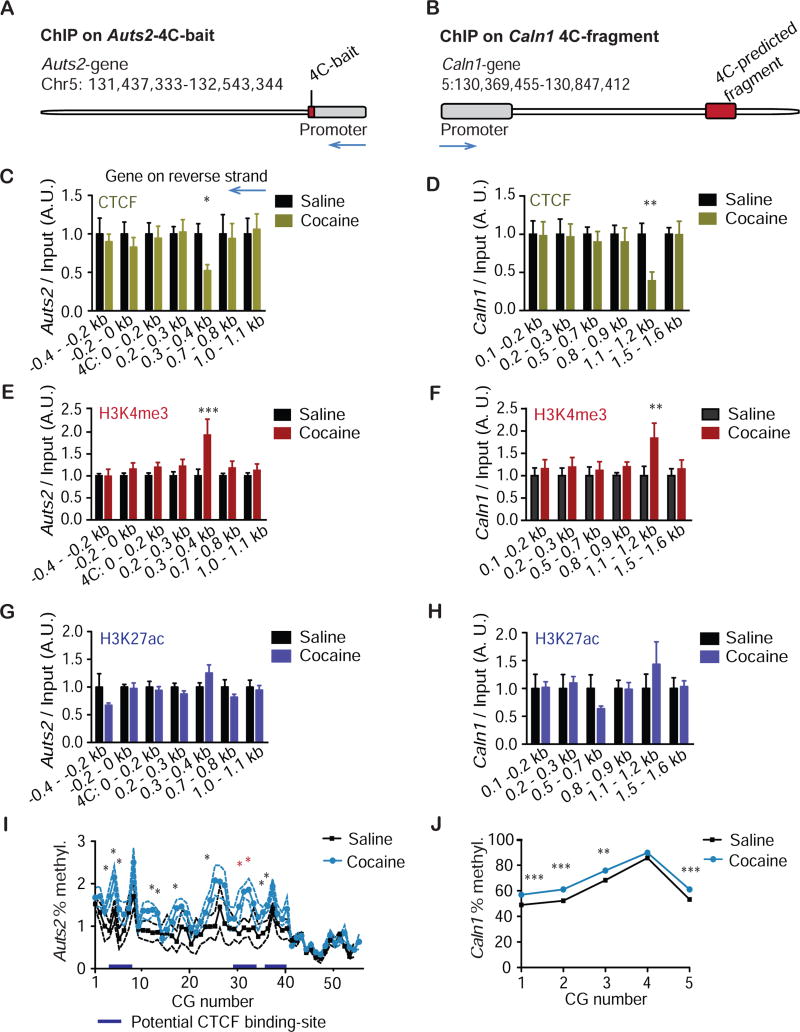
CTCF is a key mediator of chromatin interactions (30). To test the hypothesis that CTCF binding might be altered by cocaine in the vicinity of 4C interaction sites, qChIP was performed on NAc punches from IP-injected mice for CTCF. Levels of immunoprecipitated Auts2- and Caln1- fragments in the vicinity of the 4C sites were quantified by qPCR (Fig. 3a-d). CTCF binding was reduced at specific regions on both genes (Bonferroni posthoc after Two-way ANOVA: Auts2+2: P<0.05; Caln1 4C.5: P<0.01) in agreement with the reduced chromatin looping observed between Auts2 and Caln1 under these conditions. Next, we tested whether altered CTCF-binding is associated with altered histone marks. For instance, CTCF occupancy negatively correlates with the permissive histone mark H3K4me3 (31), and chromatin looping is favored at enhancers, which are enriched in H3K27 acetylation (H3K27ac). qChIP for H3K4me3 revealed a cocaine- induced increase in this mark at the same DNA regions that had displayed reduced CTCF-binding on both genes (Bonferroni posthoc after Two-way ANOVA: Auts2+2: P<0.001; Caln1 4C.5: P<0.01, Fig. 3e,f). qChIP against H3K27ac did not show cocaine-induced alterations at these regions (P>0.05 for all locations, Fig. 3g,h).
Figure 3.
Cocaine induces multiple epigenetic modifications on Auts2 and Caln1 genes. a, Overview of location of the 4C bait relative to the Auts2 gene (reverse strand) in mice. The mouse genomic area homologous to the 4C-bait from rat was identified using the UCSC genome browser BLAT tool. b, Overview of the location of the Caln1 4C-predicted fragment in mice. The 4C fragment from rat was determined for the mouse genome via the UCSC genome browser BLAT tool. c, d, Chromatin was purified from NAc of mice that had been injected for 7 days with saline (sal) or 20 mg/kg cocaine (coc) and analyzed 24 h later. Chromatin was precipitated with a CTCF antibody. c, qPCR was conducted for Auts2. N=4,4;4,4;10,9;8,7;15,16;4,4;4,4. Two-way ANOVA: interaction: F(6,83)=0.52, P>0.05; drug factor: F(1,83)=1.44, P>0.05; location factor: F(6,83)=0.52, P>0.05. Bonferroni: sal vs. coc: P>0.05; P>0.05; P>0.05; P>0.05; *P<0.05; P>0.05; P>0.05. X-axis refers to primer locations up- and downstream of the 4C-bait. d, qPCR was conducted for Caln1. N=4,4;4,4;8,7;8,8;9,8;8,7. Two-way ANOVA: interaction: F(5,67)=1.13, P>0.05; drug factor: F(1,67)=2.70, P>0.05; location factor: F(5, 67)= 1.13, P>0.05. Bonferroni: sal vs. coc: P>0.05; P>0.05; P>0.05; P>0.05; **P<0.01; P>0.05. X-axis refers to primer locations within the 4C-predicted fragment. e,f, like c,d, but chromatin was precipitated with a H3K4me3 antibody. e, qPCR was conducted for Auts2. N=8,8;14,10;26,26;8,8;14,10;8,8;8,8. Two-way ANOVA: interaction: F(6,142)=1.81, P>0.05; drug factor: F(1,142)=8.85, P<0.01; location factor: F(6,142)=1 80, P>0.05. Bonferroni: sal vs. coc: P>0.05; P>0.05; P>0.05; P>0.05; ***P<0.001; P>0.05; P>0.05. X-axis refers to primer locations up- and downstream of the 4C bait. f, qPCR was conducted for Caln1. N=4,4;10,9;8,8;17,17;4,4;4,4. Two-way ANOVA with Bonferroni: interaction: F(5,130)=0.99, P>0.05; drug factor: F(1,130)=5.80, P<0.05; dilution factor: F(5,130)=0.99, P>0.05. Bonferroni: sal vs. coc: P>0.05; P>0.05; P>0.05; P>0.05; **P<0.01; P>0.05. X-axis refers to primer locations within the 4C predicted fragment. g,h, like c,d, but chromatin was precipitated with a H3K27ac antibody. g, qPCR was conducted for Auts2. N=4,6;8,8;12,13;8,8;8,8;4,5;4,5. Two-way ANOVA: interaction: F(6,87)=1.39, P>0.05; drug factor: F(1,87)=1.72, P>0.05; location factor: F(6,87)=1.39, P>0.05. Bonferroni: sal vs. coc: all P>0.05. X- axis refers to primer locations up- and downstream of the 4C bait. h, qPCR was conducted for Auts2. N=4,6;4,6;4,6;8,8;8,8;4,6. Two-way ANOVA: interaction: F(5,60)=0.58, P>0.05; drug factor: F(1,60)=0.06, P<0.01; location factor: F(5,60)=0.58, P>0.05. Bonferroni: sal vs. coc: all P>0.05. X- axis refers to primer locations within the 4C-predicted fragment. i, j: Bisulfite sequencing was conducted for regions of Auts2 and Caln1 that were altered by 4C / ChIP in a cocaine-dependent manner. One-way ANOVA was conducted for each CG. i, Bisulfite sequencing with Auts2 primers: positions 4–6, 13–14, 18,26: *P<0.05; positions 32, 33: *P=0.01 (red, these CGs were targeted in later experiments by CRISPR-dCas9); all other positions: P>0.05. . j, Bisulfite sequencing with Caln1 primers CG 1–3, 6: ***P<0.0001; CG 4: **P<0.01; CG 5: P>0.05. c-h, Average +/− SD shown.
Another genomic mark that is linked to CTCF binding is DNA methylation, which generally inhibits CTCF binding (32). To assess whether cocaine alters DNA methylation at our genes of interest, we performed bisulfite sequencing spanning the vicinity of Auts2-Caln1 interaction regions (Fig. S1). The Caln1 region had much higher baseline CG methylation than Auts2 (Fig. 3a,b). In both genes, we detected a small but significant increase in DNA methylation after cocaine-exposure (Auts2: P<0.05 for 11 out of 55 CGs; Caln1: P<0.05 or less for 5 out of 6 CGs) (Fig. 3i,j). In Auts2, the largest increase occurred at a putative CTCF site (CCGCGCCGGGGATC, Fig. S6) (33). In summary we observed reduced CTCF binding, increased H3K4me3 enrichment, and increased DNA methylation in the vicinity of 4C interaction sites between Auts2 and Caln1.
Causality of DNA methylation and Caln1 expression regulation by Auts2
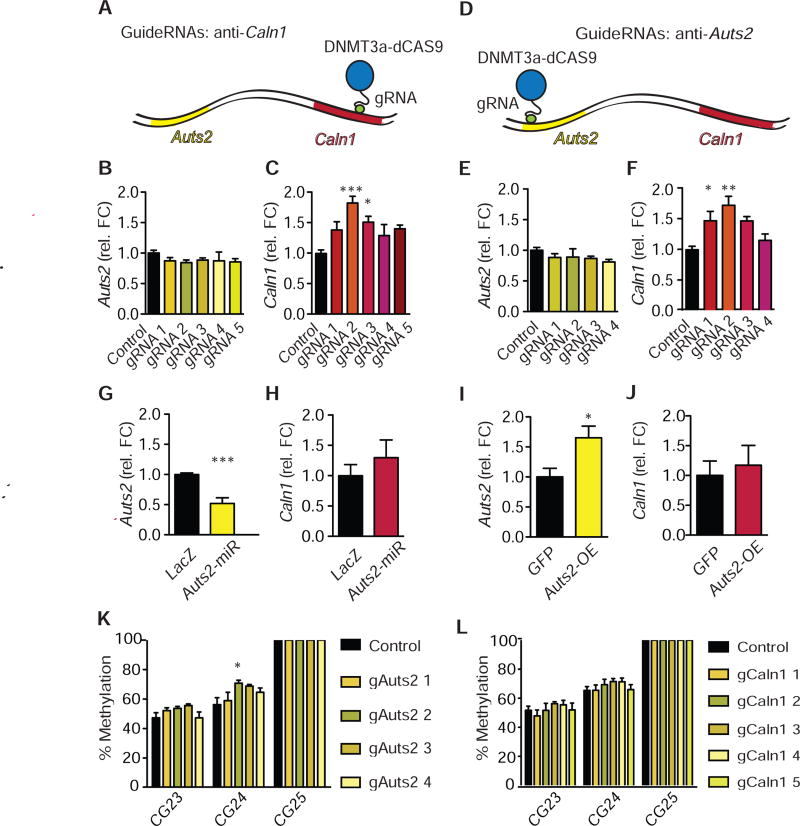
While data presented in the previous section demonstrated a correlation between DNA methylation and altered Auts2-Caln1 interactions and expression, we wanted to test whether these alterations can be causally linked. A dCas9-DNMT3a-DNMT3L-3xFlag CRISPR was designed, which targets deposition of DNA methylation at a particular DNA region of interest. Targeting of DNA methylation has been shown previously (34). Guide RNAs were created to target DNMT activity to the regions of Auts2 and Caln1 that had shown increased DNA methylation after cocaine administration (Fig. S1), which were located in the proximal promoter and intronic gene body of the two genes, respectively. Non-targeting p1005-guide RNA was designed to bind nowhere in the mouse genome and served as a control. This experiment was performed in cell culture because it is not yet feasible to do so in brain. First, Neuro2A cells were transfected with the dCas9-DNMT3A/3L and guide RNAs targeting Caln1 (gCaln1 1–5). We then extracted RNA and performed qPCR against both Auts2 and Caln1 (Fig. 4a-c). This targeted methylation increased gene expression of Caln1 (F(5,35)=5.50, P=0.001), without affecting Auts2 mRNA levels, indicating that DNA methylation changes at the Caln1 intronic region do not alter Auts2 expression (F(5,35)=0.59, P>0.05). Next, we co-transfected the dCas9-DNMT3A/3L with guide RNAs against Auts2 (Fig. 4d-f). Again, the dCas9-DNMT3A/3L did not affect Auts2 expression, suggesting that DNA methylation changes at the Auts2 promoter region are not causal for regulation of Auts2 expression (F(4,29)=0.86, P>0.05). However, qPCR for Caln1 revealed that CRISPR-dCas9 mediated epigenome-editing of Auts2 increased the expression of Caln1 (F(4,29)=6.41, P=0.001). This is direct evidence that manipulation of one gene (Auts2) can alter the expression of a distant chromatin-interaction partner. Additionally, co-transfection of Auts2 guide RNA with a dCas9-FLAG did not affect Caln1 expression, indicating that the effect is not caused by steric artifacts of the dCAS9 bound to the gene (interaction of gene and gRNA: F(1,8)=1 82, P>0.05; Fig S7). While guide RNAs + dCas9-DNMT3A/3L increased DNA methylation on the targeted genes (Fig. 4K,L, Fig. S8-10), selected guide RNAs also altered methylation on the looping partner (Fig. S8-10). The absence of DNA methylation changes in an unrelated control gene suggests that these effects were not due to lack of specificity of guide RNA binding (Fig. S11). Instead this molecular footprint provides further evidence that looping between Caln1 and Auts2 can result in shared chromatin regulation.
Figure 4.
Increased DNA methylation on Auts2 and Caln1 genes increase Caln1 expression. a, Overview over CRISPR-binding on Caln1. b,c, Neuro2A cells were co-transfected with dCas9-DNMT3ACD-DNMT3LCD-3xFLAG CRISPR and either sham (“p1005”) or Caln1 guide RNA. b, qPCR was conducted for Auts2. N=6 per group. One-way ANOVA: F(5,35)=0.59, P>0.05; Tukey’s test vs. p1005-ctrl: all P>0.05. c, qPCR was conducted for Caln1. N=6 per group. One-way ANOVA: F(5,35)=5.50, P=0.001; Tukey’s test vs. p1005-ctrl: P>0.05; ***P<0.001; *P<0.05; P>0.05; P>0.05. d, Overview over CRISPR-binding on Auts2 e, f, Neuro2A cells were co-transfected with dCas9-DNMT3ACD-DNMT3LCD-3xFLAG CRISPR and either sham (“p1005”) or Auts2 guide RNA. e, qPCR was conducted for Auts2. N=6 per group. One-way ANOVA: F(4,29)=0.86, P>0.05; Tukey’s test vs. p1005-control: all P>0.05. f, qPCR was conducted for Caln1. N=6 per group. One-way ANOVA: F(4,29)=6.41, P=0.001; Tukey’s test vs. p1005-control: *P<0.05; **P<0.01; P>0.05; P>0.05. g, h, Neuro2A cells were transfected with either Auts2-miR or LacZ control. g, qPCR was conducted for Auts2. N=9,6. Student’s t-test: t13=5.82, P<0.0001***. h, qPCR was conducted for Caln1. N=9,6. Student’s t-test: t13=0.90, P>0.05. i, j, NAc punches were obtained from D2-Cre mice that had been injected 3 days prior with an Auts2-p1005-LSIL-HSV or p1005- LSIL-HSV, which express Auts2 or GFP, respectively, in a Cre-dependent manner. i, qPCR was conducted for Auts2. N=5.3. Student’s t-test: t6=2.736, *P<0.05. j, qPCR was conducted for Caln1. N=5,3. Student’s t-test: t6=0.4314, P>0.05. k, l, DNA methylation on a putative CTCF-site of Auts2 is increased by dCas9-DNMT3A/3L combined with Auts2 guide-RNAs (k) but not by guide-RNAs against Caln1 (l). Statistics for k and l are given in Fig. S8 and S9. b, c, e-l, Average +/− SD are shown.
As repeated cocaine administration increases Auts2 expression (Fig. 2), we wanted to rule out that the effects on Caln1 are not mediated indirectly by increased Auts2 mRNA or protein. For instance, Auts2 regulates the PRC2-complex (9), which could conceivably control Caln1 expression. To that end, we designed a miR-Auts2 construct and expressed it in Neuro2A cells (Fig. 4g,h). While Auts2 expression was suppressed by Auts2-miR (t13=5.82, P<0.0001***), Caln1 expression was unaffected (t13=0.90, P>0.05). Similarly, overexpression of Auts2 with a Cre- dependent HSV vector in NAc of sterotaxically-injected D2-Cre mice increased Auts2 expression levels (t6=2.736, *P<0.05), but did not affect Caln1 levels (t6=0.4314, P>0.05, Fig. 4i,j).
Cell type-specific effects of Auts2 and Calnl overexpression on cocaine place preference
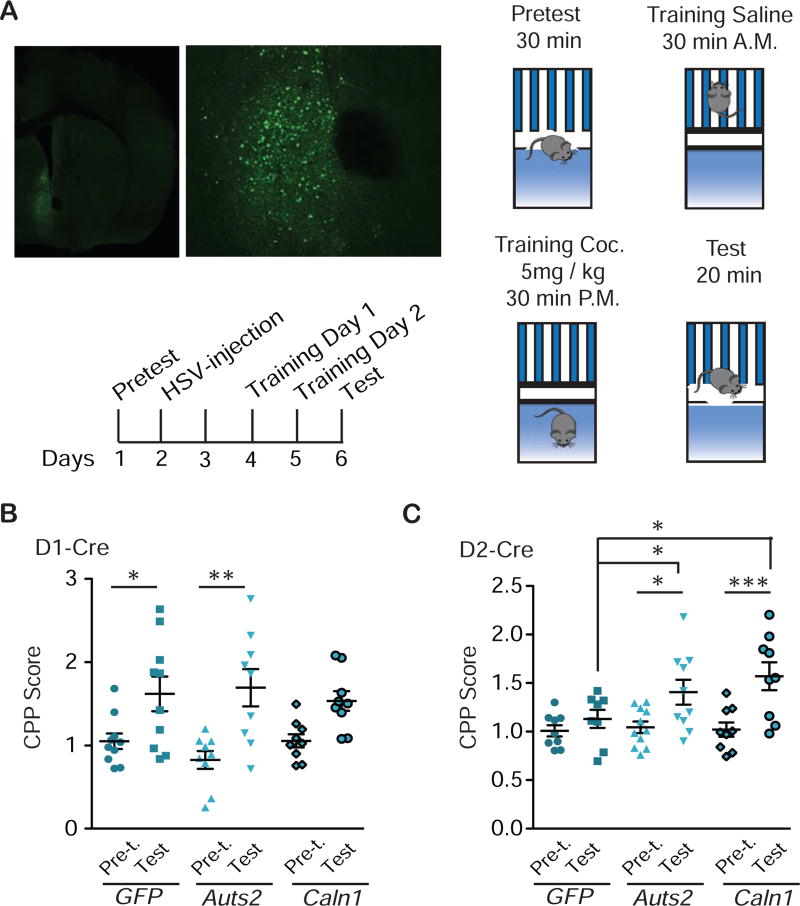
After establishing a pathway by which chromatin looping affects Caln1 expression via Auts2 in NAc, we tested whether altered expression of either gene affects behavioral responses to cocaine. We used Cre-dependent HSV vectors to overexpress Auts2 or Caln1 in a cell type- specific manner (Fig. 4i,j; Fig. S12). Overexpression of either gene in D1-Cre mice did not affect cocaine place preference (CPP), which provides an indirect measure of cocaine reward, relative to the expression of a control vector (F(5,71)=7.02, P<0.0001. Tukey’s test: pretest vs. test: GFP *P<0.05; Auts2: *P<0.05; Caln1: **P<0.01; gene factor within test: P>0.05) (Fig. 5a,b; Fig. S13). However, overexpression of Caln1 or Auts2 in D2-Cre mice increased cocaine CPP (F(5,61)=9.36, P<0.0001. Tukey’s test: pretest vs. test: GFP ***P<0.001; Auts2: ***P<0.0001; Caln1: **P<0.01; within test: GFP vs. Auts2: *P<0.05; GFP vs. Caln1: *P<0.05) (Fig. 5c). To avoid ceiling effects, CPP had been optimized on wild-type mice at 5 and 7.5 mg/kg doses. As CPP occurred at both doses, the lower dose was chosen for the HSV experiment (drug factor: F(1,36)=36.18, P<0.001; Bonferroni: saline vs. cocaine: 5 mg/kg: *P<0.05; 7.5 mg/kg: ***P<0.001, data not shown). CPP in D1-Cre mice after viral overexpression was repeated at a 3 mg/kg dose—due to small differences in baseline preference seen in the two transgenic mouse lines despite both being on the same background (see supplementary methods)—and again neither Auts2 nor Caln1 overexpression affected CPP (F(5,45)=0.54, P>0.05, data not shown). Together, this experiment suggests a permissive, cell type-specific role of Auts2 and Caln1 in cocaine-elicited behavior.
Figure 5.
D2-specific overexpression of Auts2 or Caln1 in NAc increases cocaine place conditioning. a, Procedure for cocaine conditioned place preference (CPP): On day 1, D1- or D2-Cre mice were subjected to 30 min pretest in the CPP chambers to balance for bias in preference. (On average, we use an unbiased procedure, where there is no overall preference for one side or the other.) On day 2, mice were stereotaxically injected into NAc with Auts2-p1005-LSIL, Caln1-p1005-LSIL or p1005-LSIL control HSV vectors (see photos - viral expression: green; DAPI: blue; magnifications clockwise: 10x, 20x, and 100x with 2x zoom). On days 4 and 5, mice were conditioned and on day 6, place preference was tested. b, CPP of D1-Cre mice. N=12,9,15. One-way ANOVA: F(5,71)=7.02, P<0.0001. Tukey’s test: pretest vs. test: GFP *P<0.05; Auts2: *P<0.05; Caln1: **P<0.01; gene factor within test: P>0.05. c, CPP of D2-Cre mice. N=8,14,9. One-way ANOVA: F(5,61)=9.36, P<0.0001. Tukey’s test: pretest vs. test: GFP ***P<0.001; Auts2: ***P<0.0001 ; Caln1: **P<0.01 ; within test: GFP vs. Auts2: *P<0.05; GFP vs. Caln1: *P<0.05. B,C, Average +/− SD shown.
Discussion
Cocaine and other drugs of abuse have been shown by genome-wide approaches to induce numerous chromatin modifications in NAc and other brain reward regions (35). While some of these alterations correlate with drug-induced changes in expression of the specific target genes affected, most of them are not correlated with altered gene transcription (10). These observations have raised questions about the functional role played by such changes in chromatin regulation. Results of the present study provide new and important insight into this question.
We provide evidence that one function of cocaine-induced chromatin alterations is to regulate the expression of a gene—not only directly by altering that gene’s promoter—but also by altering the 3D structure of chromatin around that gene which then alters its accessibility for transcription. Specifically, we demonstrate that repeated cocaine exposure induces several types of chromatin alterations at the Auts2 and Caln1 loci, including decreased CTCF binding and increased H3K4me3 deposition and DNA methylation, which are associated with reduced chromatin looping between these nearby genes and with their increased expression. By utilizing CRISPR-dCas9 mediated epigenome-editing, we demonstrate a causal link between altered DNA methylation on the Auts2 gene and altered expression of the Caln1 gene. This latter experiment was performed in cell culture due to current technical limitations; it will be important in future research to replicate these causal data in NAc as methodological improvements are made.
Our study, which focused on the Auts2 and Caln1 genes, provides a proof of principle for the ability of cocaine to control gene expression and behavior through the regulation of the 3D architecture of chromatin within a target brain area such as NAc. We propose that such regulation is likely far more widespread and involves numerous other genomic loci based on the relatively large number of genes that show strong chromatin modifications without an associated change in RNA expression (10). It would thus be interesting to study this question systematically in the drug addiction field to obtain a complete overview of drug-induced regulation of the 3D structure of the entire genome. HiC techniques provide such a genome-wide interrogation and are now being applied increasingly to numerous systems including brain (36). It is not yet feasible to apply them to small brain regions but recent advances suggest that the technique should become suitable for smaller amounts of tissue (37).
DNA methylation often correlates inversely with CTCF binding, which is implicated in controlling chromatin structure (32). Our data are in good agreement with this literature. DNA methylation has historically been associated with gene silencing, although more recent evidence has demonstrated a far more complex role based on the sequence and genomic region undergoing methylation and the other types of chromatin alterations that occur in concert (38). Our findings are in line with this latter view, as we found increased DNA methylation associated with reduced CTCF binding, increased H3K4me3 enrichment, and increased expression of Auts2 and Caln1. In cell culture we found that CRISPR-dCas9-mediated targeting of DNMT3A/3L to the Auts2 gene had no effect on that gene’s expression levels, but nevertheless induced Caln1, consistent with an indirect mechanism such as reduced chromatin looping. In contrast, targeting DNMT3A/3L to Caln1 induced that gene without affecting Auts2, suggesting a direct cis-effect of methylation at Caln1. The mechanism of looping between Auts2 and Caln1 is likely very complex, as DNA methylation is also known to prevent H3K4me3, and therefore other mechanisms in addition to weakening of the loop could play a role. Ultimately, further work is needed to define how increased methylation at these two loci induces gene expression albeit via different mechanisms. Further work is also needed to confirm the mechanism of action of dCas9-DNMT3A/3L, which could be achieved through other means such as recruiting DNMT3-binding proteins to the vicinity of the targeted sequences.
It is interesting that the ability of cocaine to control Auts2 and Caln1 expression in NAc occurs exclusively in male mice, with no effects seen in females. We do not know what mechanisms underlie these sex differences. Sex-specific effects in psychiatric models are becoming increasingly well known (39; 40). Hence, our observation deserves further investigation.
We identified Caln1 as a key interacting gene of Auts2 in NAc based on 3C and 4C data. However, in NAc shell, the Auts2 bait displayed interactions with several intergenic regions as well, and many of these interactions were highly regulated by repeated cocaine exposure. These regions could represent distal enhancers of additional genes in the genome and should be studied further, not only in the context of cocaine exposure, but also with respect to other Auts2- related conditions, including autism, alcoholism, and heroin dependence.
The Wbscr17 gene is located between Auts2 and Caln1, although with different orientations in mouse vs. rat. WBSCR17 is also sandwiched by AUTS2 and CALN1 in humans with an orientation more similar to rat. WBSCR17 is associated with Williams syndrome, a neurodevelopmental condition caused by chromosome breakage at this locus. Our model suggests that WBSCR17 may be exposed on a loop formed between AUTS2 and CALN1 under control conditions. Whether this contributes to an increased susceptibility for breakage remains to be studied.
To conclude, our data reveal a novel mechanism by which repeated cocaine administration affects the genome of NAc neurons, namely, through regulation of the 3D structure of chromatin at specific sites. Cocaine regulation of this chromatin looping occurs in concert with cocaine-induced alterations in several types of chromatin alterations and ultimately leads to expression changes of key genes, Auts2 and Caln1, which we demonstrate control behavioral responses to cocaine. These findings establish a new line of research into the molecular basis of cocaine’s effects on the genome.
Supplementary Material
Figure 6.
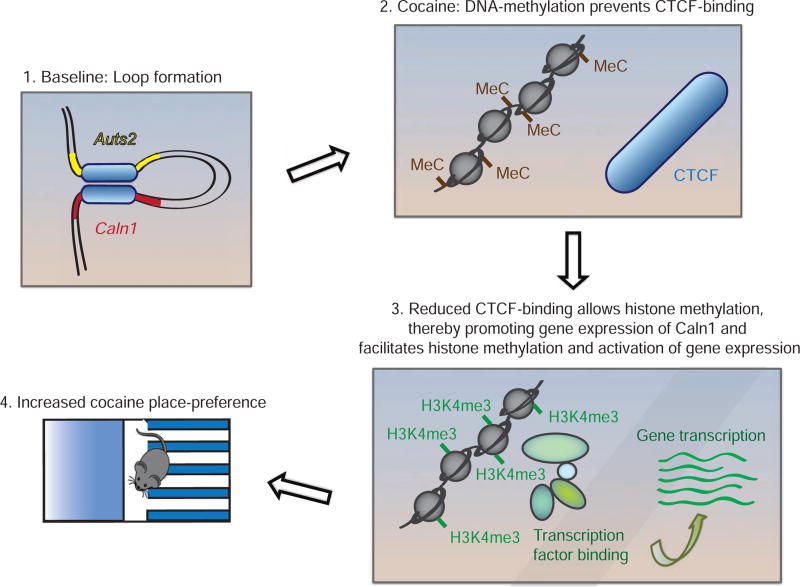
Model for cocaine regulation of the 3D genome in NAc. 3C/4C demonstrate that Auts2 and Caln1 form a loop, which is disrupted by repeated cocaine. CTCF, a mediator of looping, binds less to Auts2 and Caln1 after cocaine administration, with coincident increases in the permissive chromatin mark, H3K4me3. We propose that these changes are mediated by increased DNA methylation at a CTCF site and lead to opening of the Auts2-Caln1 loop, which mediates the cocaine-induced increase in their expression. Such induction of Auts2 and Caln1 then augments rewarding responses to cocaine. These mechanisms appear specific for NAc D2 MSNs.
Acknowledgments
This project was funded by grant P01 DA008227 (EJN); K99DA042111 and a NARSAD Young Investigator Award (ESC).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Financial disclosure:
The authors report no biomedical financial interests or potential conflicts of interest.
References
- 1.Cajal E. Un sencillo metodo de coloracion selectiva del reticulo protoplasmico y sus efectos en los diversos organos nerviosos de vertebrados e invertebrados. Trab Lab Investig Biol Univ Madr. 1903;2:129–221. [Google Scholar]
- 2.Dekker J, Rippe K, Dekker M, Kleckner N. Capturing chromosome conformation. Science. 2002;295:1306–11. doi: 10.1126/science.1067799. [DOI] [PubMed] [Google Scholar]
- 3.Tai PWL, Zaidi SK, Wu H, Grandy RA, Montecino M, Van Wijnen AJ, et al. The dynamic architectural and epigenetic nuclear landscape: Developing the genomic almanac of biology and disease. J Cell Physiol. 2014;229:711–727. doi: 10.1002/jcp.24508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Mitchell AC, Jiang Y, Peter C, Akbarian S. Transcriptional regulation of GAD1 GABA synthesis gene in the prefrontal cortex of subjects with schizophrenia. Schizophr Res. 2015;167:28–34. doi: 10.1016/j.schres.2014.10.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Green RE, Krause J, Briggs AW, Maricic T, Stenzel U, Kircher M, et al. A draft sequence of the Neandertal genome. Science. 2010;328:710–22. doi: 10.1126/science.1188021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sultana R, Yu C-E, Yu J, Munson J, Chen D, Hua W, et al. Identification of a Novel Gene on Chromosome 7q11.2 Interrupted by a Translocation Breakpoint in a Pair of Autistic Twins. Genomics. 2002;80:129–134. doi: 10.1006/geno.2002.6810. [DOI] [PubMed] [Google Scholar]
- 7.Schumann G, Coin LJ, Lourdusamy A, Charoen P, Berger KH, Stacey D, et al. Genome-wide association and genetic functional studies identify autism susceptibility candidate 2 gene (AUTS2) in the regulation of alcohol consumption. Proc Natl Acad Sci U S A. 2011;108:7119–24. doi: 10.1073/pnas.1017288108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chen YH, Liao DL, Lai CH, Chen CH. Genetic analysis of AUTS2 as a susceptibility gene of heroin dependence. Drug Alcohol Depend. 2013;128:238–242. doi: 10.1016/j.drugalcdep.2012.08.029. [DOI] [PubMed] [Google Scholar]
- 9.Gao Z, Lee P, Stafford JM, von Schimmelmann M, Schaefer A, Reinberg D. An AUTS2-Polycomb complex activates gene expression in the CNS. Nature. 2014;516:349–54. doi: 10.1038/nature13921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Feng J, Wilkinson M, Liu X, Purushothaman I, Ferguson D, Vialou V, et al. Chronic cocaine-regulated epigenomic changes in mouse nucleus accumbens. Genome Biol. 2014;15:R65. doi: 10.1186/gb-2014-15-4-r65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bharadwaj R, Peter CJ, Jiang Y, Roussos P, Vogel-Ciernia A, Shen EY, et al. Conserved higher-order chromatin regulates NMDA receptor gene expression and cognition. Neuron. 2014;84:997–1008. doi: 10.1016/j.neuron.2014.10.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mitchell AC, Jiang Y, Peter C, Akbarian S. Transcriptional regulation of GAD1 GABA synthesis gene in the prefrontal cortex of subjects with schizophrenia. Schizophr Res. 2015:167. doi: 10.1016/j.schres.2014.10.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Roussos P, Mitchell AC, Voloudakis G, Fullard JF, Pothula VM, Tsang J, et al. A Role for Noncoding Variation in Schizophrenia. Cell Rep. 2014;9:1417–1429. doi: 10.1016/j.celrep.2014.10.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Calipari ES, Ferris MJ, Siciliano CA, Zimmer BA, Jones SR. Intermittent cocaine self administration produces sensitization of stimulant effects at the dopamine transporter. J Pharmacol Exp Ther. 2014;349:192–8. doi: 10.1124/jpet.114.212993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Calipari ES, Ferris MJ, Siciliano CA, Jones SR. Differential influence of dopamine transport rate on the potencies of cocaine, amphetamine, and methylphenidate. ACS. Chem Neurosci. 2015;6:155–162. doi: 10.1021/cn500262x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hodes GE, Pfau ML, Purushothaman I, Ahn HF, Golden SA, Christoffel DJ, et al. Sex Differences in Nucleus Accumbens Transcriptome Profiles Associated with Susceptibility versus Resilience to Subchronic Variable Stress. J Neurosci. 2015;35:16362–76. doi: 10.1523/JNEUROSCI.1392-15.2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wook Koo J, Labonté B, Engmann O, Calipari ES, Juarez B, Lorsch Z, et al. Essential Role of Mesolimbic Brain-Derived Neurotrophic Factor in Chronic Social Stress-Induced Depressive Behaviors. Biol Psychiatry. 2015 doi: 10.1016/j.biopsych.2015.12.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Feng J, Shao N, Szulwach KE, Vialou V, Huynh J, Zhong C, et al. Role of Tet1 and 5-hydroxymethylcytosine in cocaine action. Nat Neurosci. 2015;18:536–544. doi: 10.1038/nn.3976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gheldof N, Witwicki RM, Migliavacca E, Leleu M, Didelot G, Harewood L, et al. Structural variation-associated expression changes are paralleled by chromatin architecture modifications. PLoS One. 2013:8. doi: 10.1371/journal.pone.0079973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chaudhri N, Sahuque LL, Schairer WW, Janak PH. Separable Roles of the Nucleus Accumbens Core and Shell in Context- and Cue-Induced Alcohol-Seeking. Neuropsychopharmacology. 2010;35:783–791. doi: 10.1038/npp.2009.187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Dumitriu D, LaPlant Q, Grossman YS, Dias C, Janssen WG, Russo SJ, et al. Subregional, Dendritic Compartment, and Spine Subtype Specificity in Cocaine Regulation of Dendritic Spines in the Nucleus Accumbens. J Neurosci. 2012;32:6957–6966. doi: 10.1523/JNEUROSCI.5718-11.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.West EA, Carelli RM. Nucleus Accumbens Core and Shell Differentially Encode Reward-Associated Cues after Reinforcer Devaluation. J Neurosci. 2016;36:1128–1139. doi: 10.1523/JNEUROSCI.2976-15.2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Dekker J, Marti-Renom MA, Mirny LA. Exploring the three-dimensional organization of genomes: interpreting chromatin interaction data. Nat Rev Genet. 2013;14:390–403. doi: 10.1038/nrg3454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wu YQ, Lin X, Liu CM, Jamrich M, Shaffer LG. Identification of a human brain-specific gene, calneuron 1, a new member of the calmodulin superfamily. Mol Genet Metab. 2001;72:343–350. doi: 10.1006/mgme.2001.3160. [DOI] [PubMed] [Google Scholar]
- 25.Hradsky J, Raghuram V, Reddy PP, Navarro G, Hupe M, Casado V, et al. Post-translational membrane insertion of tail-anchored transmembrane EF-hand Ca 2+ sensor calneurons requires the TRC40/Asna1 protein chaperone. J Biol Chem. 2011;286:36762–36776. doi: 10.1074/jbc.M111.280339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Navarro G, Aguinaga D, Moreno E, Hradsky J, Reddy PP, Cort??s A, et al. Intracellular calcium levels determine differential modulation of allosteric interactions within G protein-coupled receptor heteromers. Chem Biol. 2014;21:1546–1566. doi: 10.1016/j.chembiol.2014.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Merla G, Ucla C, Guipponi M, Reymond A. Identification of additional transcripts in the Williams-Beuren syndrome critical region. Hum Genet. 2002;110:429–438. doi: 10.1007/s00439-002-0710-x. [DOI] [PubMed] [Google Scholar]
- 28.Garcia-Keller C, Kupchik YM, Gipson CD, Brown RM, Spencer S, Bollati F, et al. Glutamatergic mechanisms of comorbidity between acute stress and cocaine selfadministration. Mol Psychiatry. 2016;21:1063–1069. doi: 10.1038/mp.2015.151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Heiman M, Schaefer A, Gong S, Peterson JD, Day M, Ramsey KE, et al. A Translational Profiling Approach for the Molecular Characterization of CNS Cell Types. Cell. 2008;135:738–748. doi: 10.1016/j.cell.2008.10.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ong C-T, Corces VG. CTCF: an architectural protein bridging genome topology and function. Nat Rev Genet. 2014;15:234–46. doi: 10.1038/nrg3663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gomes NP, Espinosa JM. Gene-specific repression of the p53 target gene PUMA via intragenic CTCF-Cohesin binding. Genes Dev. 2010;24:1022–1034. doi: 10.1101/gad.1881010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wang H, Maurano MT, Qu H, Varley KE, Gertz J, Pauli F, et al. Widespread plasticity in CTCF occupancy linked to DNA methylation. Genome Res. 2012;22:1680–1688. doi: 10.1101/gr.136101.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bell AC, Felsenfeld G. Methylation of a CTCF-dependent boundary controls imprinted expression of the Igf2 gene. Nature. 2000;405:482–485. doi: 10.1038/35013100. [DOI] [PubMed] [Google Scholar]
- 34.Kungulovski G, Jeltsch A. Epigenome Editing: State of the Art, Concepts, and Perspectives. Trends Genet. 2016:32. doi: 10.1016/j.tig.2015.12.001. [DOI] [PubMed] [Google Scholar]
- 35.Nestler EJ. Epigenetic mechanisms of drug addiction. Neuropharmacology. 2014:76. doi: 10.1016/j.neuropharm.2013.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Mitchell A, Roussos P, Peter C, Tsankova N, Akbarian S. Prog Mol Biol Transl Sci. 1. Vol. 128. Elsevier Inc; 2014. The future of neuroepigenetics in the human brain. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Nagano T, Lubling Y, Yaffe E, Wingett SW, Dean W, Tanay A, Fraser P. Single-cell HiC for genome-wide detection of chromatin interactions that occur simultaneously in a single cell. Nat Protoc. 2015;10:1986–2003. doi: 10.1038/nprot.2015.127. [DOI] [PubMed] [Google Scholar]
- 38.Jones PA. Functions of DNA methylation: islands, start sites, gene bodies and beyond. Nat Rev Genet. 2012;13:484–92. doi: 10.1038/nrg3230. [DOI] [PubMed] [Google Scholar]
- 39.Seney ML, Sibille E. Sex differences in mood disorders: perspectives from humans and rodent models. Biol Sex Differ. 2014;5:17. doi: 10.1186/s13293-014-0017-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Franceschelli A, Herchick S, Thelen C, Papadopoulou-Daifoti Z, Pitychoutis PM. Sex differences in the chronic mild stress model of depression. Behav Pharmacol. 2014;25:372–83. doi: 10.1097/FBP.0000000000000062. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.