Abstract
The outer membrane (OM) of Gram-negative is a unique lipid bilayer containing LPS in its outer leaflet. Because of the presence of amphipathic LPS molecules, the OM behaves as an effective permeability barrier that makes Gram-negative bacteria inherently resistant to many antibiotics. This review focuses on LPS biogenesis and discusses recent advances that have contributed to our understanding of how this complex molecule is transported across the cellular envelope and is assembled at the OM outer leaflet. Clearly, this knowledge represents an important platform for the development of novel therapeutic options to manage Gram-negative infections.
Keywords: ABC transporter, gram-negative bacteria, lipopolysaccharide (LPS), outer membrane, protein-protein interaction, LPS transport
Introduction
Gram-negative diderm bacteria possess a double-membrane system as part of their envelope structure. Although the cytoplasmic or inner membrane (IM)2 is a symmetrical lipid bilayer made of phospholipids, the outer membrane (OM) is asymmetrical, containing phospholipids in the inner leaflet and a complex glycolipid, LPS, in the outer leaflet (1). These two membranes are separated by an aqueous compartment, the periplasm, which contains a thin layer of peptidoglycan, a polymer that protects the cell from bursting by its internal turgor and maintains cell shape (2) (Fig. 1).
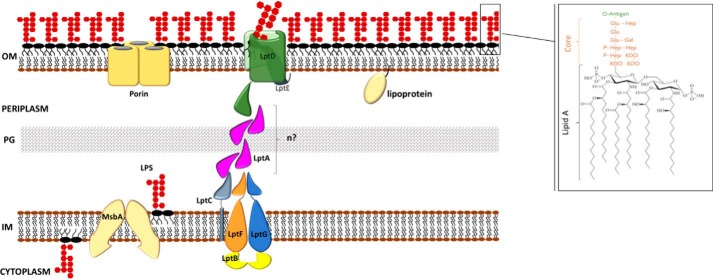
Figure 1.
The LPS export pathway in Gram-negative bacteria. Following flipping across the IM by the ABC transporter MsbA, LPS is extracted from the IM and transported across the periplasm to the OM at the expense of ATP hydrolysis by the transenvelope Lpt protein machine composed, in E. coli, by seven essential proteins (LptA–G). See text for details. Only the lipid A–core moiety is represented, whereas the O-antigen repeat moiety is omitted. PG, peptidoglycan. Inset. the structure of E. coli LPS with a K-12 core region is shown. The lipid A moiety and the core oligosaccharide are indicated in black and orange, respectively. The chemical composition of the O-antigen is not shown. Gal, d-galactose; Glu, d-glucose; Hep, l-glycero-d-manno-heptose; Kdo, 3-deoxy-d-manno-octulosonic acid. n, number of LptA monomers.
The OM is a remarkable lipid bilayer differing in many aspects from the IM. Like all biological membranes, IM and OM contain a wide variety of proteins; however, in the IM, integral proteins cross the membrane as α-helices almost entirely composed of hydrophobic residues, whereas the vast majority of integral proteins in the OM (outer membrane proteins) consist of amphipathic β-strands that adopt a β-barrel structure (3, 4). Porins represent an abundant class of outer membrane proteins and play an important role in OM function: their β-barrel structures form both specific and nonspecific channels that orchestrate the flux of small hydrophilic molecules across the OM (4). Contrary to the IM, the OM is not an energized membrane, and therefore the transport of molecules/nutrients across this lipid bilayer either is governed by concentration gradient (5) or occurs via energy-coupled transporters (6). Lipoproteins are a very diverse group of proteins anchored to bacterial membranes via an N-terminal lipid moiety (7). In the OM, lipoproteins can either extend into the periplasm or be exposed at the surface of the cell, whereas in the IM, they are exclusively anchored at the periplasmic site of the lipid bilayer. Due to their diversity and subcellular localization, lipoproteins serve several functions including formation and maintenance of cell shape, biogenesis of the OM, transport of a variety of molecules, signal transduction, and cell motility (8).
Structure and functions of LPS
The peculiar lipid asymmetry of the OM is the consequence of the presence of LPS exclusively in the outer leaflet (9). LPS is an unusual glucosamine-based saccharolipid that has a tripartite structure: lipid A, the hydrophobic moiety that anchors LPS to the OM; a core oligosaccharide; and an O-antigen made of repeating oligosaccharide units (Fig. 1).
The lipid A is the most conserved part of the molecule; in Escherichia coli and in many Enterobacteriaceae, it typically consists of a β-1′-6-linked glucosamine disaccharide that is phosphorylated at the 1 and 4′ positions to which six fatty acyl chains are attached (10). The oligosaccharide core is attached at the 6′ position of the disaccharide of lipid A and can be divided into a conserved inner core that contains at least a residue of 3-deoxy-d-manno-oct-2-ulosonic acid (Kdo) and of l-glycero-d-manno-heptose (heptose) and a more variable outer core (11). The O-antigen polysaccharide chain of variable length is the most distal portion of the molecule (12); notably, it is not produced by many E. coli laboratory strains (K-12 derivatives) due to a mutation in the rfb locus where the genes responsible for the O-antigen biosynthesis are clustered (13, 14). LPS is essential in many Gram-negative bacteria with several notable exceptions, namely Neisseria meningitidis (15), Moraxella catarrhalis (16), and Acinetobacter baumannii (17, 18), which can survive without LPS.
The OM is positioned at the frontline of the cell's interaction with its environment/host, and the LPS molecule plays a crucial role in such an interaction. Indeed, the most conserved moiety, lipid A, also known as endotoxin, is a potent stimulator of the innate immune response and serves as an early warning signal of bacterial infection (19, 20). Instead, the outermost and variable portion of the molecule, the O-antigen, helps bacteria to avoid phagocytosis and to resist the lytic action of the complement system (21).
The unique structure and composition of OM make it a formidable permeability barrier able to exclude many toxic compounds such as bile salts, detergents, and antibiotics and thus enabling Gram-negative bacteria to survive in many hostile environments (22). LPS molecules mainly contribute to the permeability barrier properties of the OM. Indeed, the LPS outer layer is strongly stabilized by the presence of divalent cations such as Mg2+ and Ca2+ that interact with the negative charges of lipid A and oligosaccharide inner core. The resulting tightly packed LPS layer makes the OM an impermeable surface to hydrophobic molecules and large hydrophilic compounds that cannot permeate through the narrow porin channels (22, 23).
Although bacteria that survive without LPS compensate for the loss of LPS through alterations of the cell envelope (18, 24, 25), in species in which LPS is essential, impairment of LPS biogenesis leads to OM permeability defects. In mutants defective in LPS biosynthesis or transport at the cell surface, phospholipids migrate in the OM outer leaflet, resulting in the formation of patches that are more susceptible to the influx of hydrophobic, toxic molecules (23, 26). However, some of these mutants also become hyper-susceptible to large hydrophilic antibiotics such as vancomycin and bacitracin that cannot permeate through OM hydrophobic patches (27–29). It has been proposed that these large molecules diffuse through transient imperfections or “cracks” of the defective OM (23). This view has been recently supported by the observation that E. coli cells become susceptible to vancomycin at low temperatures. Surprisingly, mutations in LPS biosynthesis affecting the composition of the oligosaccharide core restore vancomycin resistance. This apparent paradox is explained by assuming that during cold stress the decreased membrane fluidity causes “cracks” in the OM and that inhibition of LPS biosynthesis results in an OM that is resistant to these perturbations (30). Overall, these data help explain the intrinsic resistance of Gram-negative bacteria to many otherwise clinically useful antibiotics (22, 31, 32).
LPS biogenesis
The biosynthesis of LPS is a well-known process. The lipid A–core moiety is synthesized at the interface between the IM and the cytoplasm by a conserved pathway (10, 33). O-antigen repeat units are synthesized in the cytoplasm and then flipped to the periplasmic face of the IM attached to the lipid carrier undecaprenyl diphosphate (12). Formation of the mature LPS molecule occurs at the periplasmic site of the IM where O-antigen repeat units are polymerized and then ligated to the lipid A-core by the WaaL ligase (12).
The assembly of the LPS layer on the surface of the Gram-negative bacteria is instead a very challenging process for the cell, and the details of this process have only recently emerged. Indeed, three fundamental aspects need to be taken into consideration: (i) the heterogeneous chemical nature of the LPS molecule and the different physico-chemical characteristics of the envelope compartments to be crossed (two lipid membranes and the aqueous periplasmic space); (ii) the unidirectionality of the transport, occurring against a concentration gradient from the site of synthesis at the IM up to the OM, whose outer layer is constituted by tightly packed LPS molecules; and (iii) the fact that LPS transport must not perturb the integrity of the OM. To meet these requirements, the bacterial cell has evolved a two-step process. The lipid A–core moiety synthesized at the inner leaflet of the IM is initially translocated across the IM by the MsbA transporter. The mature LPS molecule synthesized at the periplasmic side of the IM is then ferried at the cell surface by the Lpt (lipopolysaccharide transport) system, a multiprotein complex that spans the entire envelope (Fig. 1). This strategy not only provides the energy to overcome thermodynamic issues such as LPS detachment from the IM and transport to the cell surface, in an environment normally devoid of energy sources, but also ensures that the integrity of the OM is preserved during transport.
LPS translocation across the IM: The MsbA flippase
MsbA, the first component of the LPS export pathway that has been discovered (34, 35), catalyzes the flipping of the lipid A-core moiety across the IM. MsbA is an integral IM protein member of the large ATP-binding cassette (ABC) superfamily of proteins (36). Similarly to other ABC exporters, MsbA, often termed as “half-transporter,” functions as a homodimer as assessed by biochemical (37–39) and crystallographic (40) studies. In vitro, MsbA ATPase activity is highly stimulated by hexa-acylated lipid A species, strongly suggesting that they might be the substrates (41). When reconstituted in proteoliposomes, MsbA behaves as a lipid flippase catalyzing the flipping of several fluorescently labeled phospholipids derivatives (42), showing the potential of this transporter to handle, at least in vitro, a wide variety of substrates.
Following MsbA-dependent translocation, the lipid A–core is anchored at the outer leaflet of the IM where O-antigen repeats may be added. The journey of the mature LPS molecule to the cell surface continues with the assistance of the Lpt molecular machine as detailed below.
LPS transport across the periplasm to the cell surface: The Lpt molecular machine
In E. coli, the Lpt machinery is composed by seven essential proteins (LptABCDEFG) that are organized in two subassemblies: LptB2CFG and LptDE, located at the IM and at the OM, respectively, which are connected by the periplasmic protein LptA (43–49). These proteins form a transenvelope bridge that spans the entire cell from the cytoplasm to the OM (Fig. 1). Importantly, as detailed in the following paragraphs, the assembly of the Lpt molecular machine is mediated by the oligomerization of a structural motif shared by all the Lpt proteins that have a periplasmic domain.
The bulk of knowledge of the LPS transport has been obtained mainly using E. coli as a model system, and the process seems to be structurally conserved among Gram-negative bacteria (50–54). However, emerging evidence indicates that there are LPS export systems that do not adhere to the canonical E. coli model, and a few examples will be discussed in the following sections.
The Lpt proteins operate in concert as a unique device in LPS transport. The first evidence supporting this notion came from membrane fractionation experiments upon depletion of the different Lpt proteins. These experiments revealed that breaking transport at any level within the cell leads to LPS accumulation at the periplasmic leaflet of the IM in a step downstream of the MsbA-mediated flipping (44, 47) (see above). This evidence was later confirmed by the observation that all seven Lpt proteins physically interact and co-fractionate in a cellular fraction distinct from IM and OM (the so-called light OM, OML) in sucrose density gradient fractionations (55). The relevance of the transenvelope architecture for LPS transport was further corroborated by the observation that depletion of LptC and LptDE components leads to LptA degradation (56), pointing to a quality control role for LptA–LptC and LptA–LptD interactions in the Lpt complex assembly.
The LPS transport from the IM to the OM can be conceptually divided into three steps involving different Lpt players: (i) LPS detachment from the IM; (ii) LPS transport across the periplasm; and (iii) LPS insertion and assembly in the OM at the cell surface.
LPS detachment from the IM
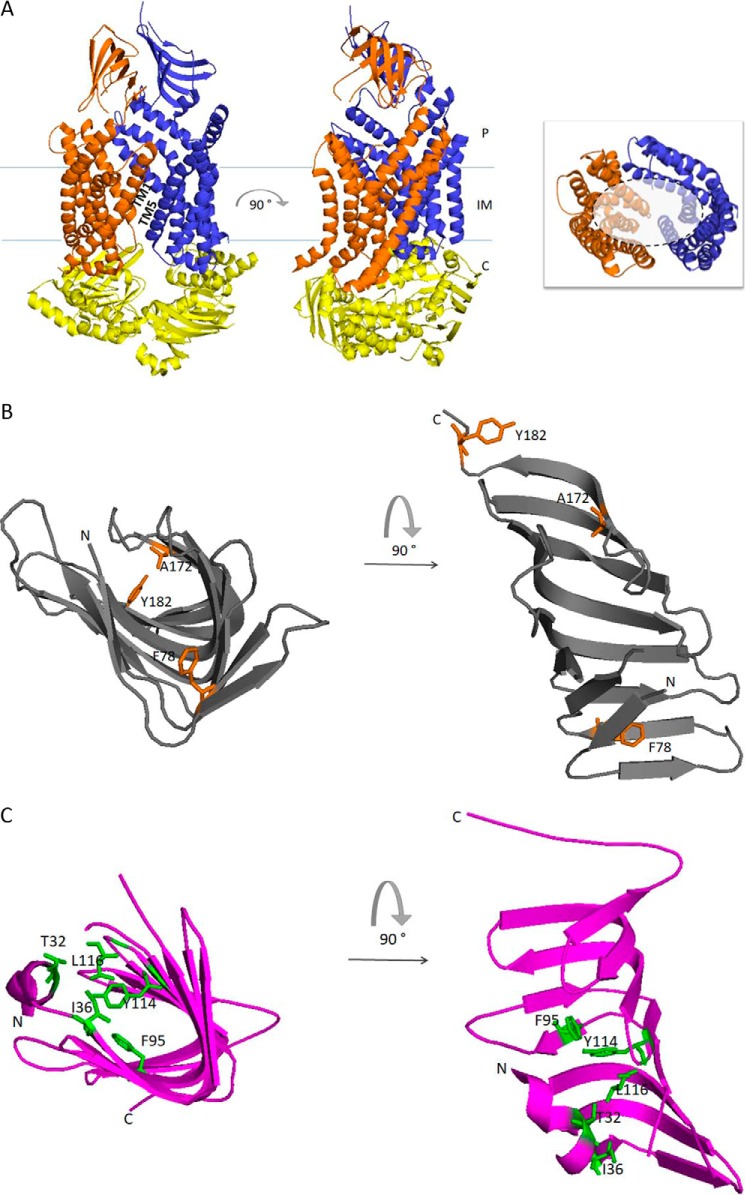
The first step of LPS extraction from the IM is carried out by the LptB2FG protein complex. LptB2FG is an IM ABC transporter composed of the heterodimeric transmembrane domain (TMD) subunit LptFG (44) and the cytoplasmic homodimeric nucleotide-binding domain LptB2 (57). LptF and LptG each contain six transmembrane helices, a large periplasmic domain that adopts a β-jellyroll architecture, and a coupling helix that interacts with one monomer of LptB on the cytoplasmic side (Fig. 2A) (58). LptB2FG is an unusual transporter in that it does not translocate its LPS substrate across the IM, but rather, it extracts LPS from the IM outer leaflet and delivers it to LptC (59), an IM-anchored protein that stably associates to the LptB2FG complex (43).
Figure 2.
Graphic representation of the crystal structures of the seven Lpt proteins and structural comparisons. A, LptB2FG complex from P. aeruginosa (Protein Data Bank (PDB) 5X5Y). LptB, LptF and LptG are represented in yellow, orange, and blue, respectively. The 12 TM helices of LptF–G bent outwards, creating a V-shaped central cavity at the membrane–periplasm interface, which would function as an LPS-binding pocket. TM1 of LptF with TM5 of LptG and TM1 of LptG with TM5 of LptF constitute the lateral gates that would allow the entry of LPS into the central cavity of the transporter (58). Inset, top view of the V-shaped central cavity highlighted by a dotted circle; for clarity, periplasmic β-jellyroll-like domains of LptF–G and LptB dimer have been removed. B, LptC from E. coli (gray, PDB 3M2Y). Residues in LptC involved in LPS binding as assessed by photo-crosslinking experiments are depicted in orange (59). Thr-47 residue is not shown because the electron density of this region is absent in the structure. C, LptA from E. coli (magenta, PDB 2R19). Residues in LptA involved in LPS binding as assessed by photo-crosslinking experiments are depicted in green (59). D, LptDE complex from K. pneumoniae (PDB 5IV9). LptD is colored green, and LptE, inserted in the LptD lumen, is colored blue. During transport, lipid A moiety of LPS is bound by the N-terminal domain of LptD and inserted directly into the membrane through an intramembrane hole (highlighted by gray dotted circle). The saccharide portion of LPS passes through the lumenal gate formed by β-strands 1 and 26 of the C-terminal domain of LptD. Inset, residues in the LptD N-terminal domain involved in LPS binding have been experimentally detected in S. enterica serovar Typhimurium (79), and the corresponding residues identified by structural alignment with the LptD homologue from K. pneumoniae are depicted in magenta. The model structures were generated with PyMOL using the corresponding PDB files. E–G, the β-jellyroll domain of LptF (orange, PDB 5X5Y) superimposed on LptC (gray, PDB 3M2Y) (E); LptC (gray, PDB 3M2Y) superimposed on LptA (magenta, PDB 2R19) (F); and LptA (magenta, PDB 2R19) superimposed on the N-terminal β-jellyroll domain of LptD (green, PDB 5IV9) (G). Superimpositions were obtained manually using the Maestro software. E, extra-cytoplasmic milieu; P, periplasm; C, cytoplasm.
An important breakthrough in understanding how LPS leaves the IM came from an elegant work from Kahne's group using in vivo photo-crosslinking on right-side-out membrane vesicles devised to trap LPS into LptC and LptA, and to characterize the dependence of the transport on ATP, LptB2FG, and LptC (59). This work showed that LPS is extracted from the IM and loaded on LptC by LptB2FG at the expense of ATP hydrolysis, and that LPS transfer from LptC to LptA requires a second round of ATP hydrolysis (see below) (59).
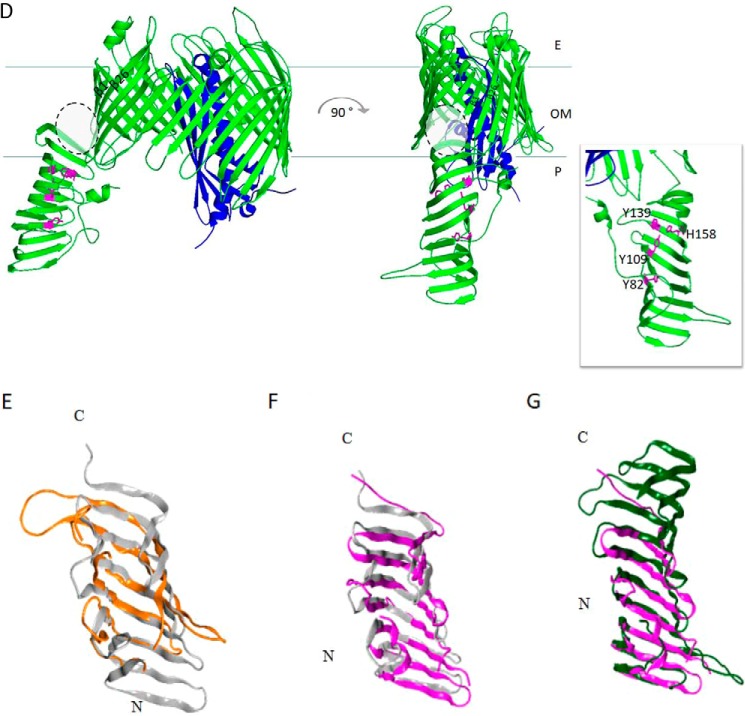
How does LptB2 ATPase transmit energy? The resolution of the three-dimensional structure of the LptB2 dimer coupled to mutagenesis and photo-crosslinking experiments (57, 60) revealed that the structure of LptB dimer experiences a global movement upon ATP binding and hydrolysis, which is transmitted to LptF and LptG subunits through their coupling helices. This movement seems to be essential for coordinating the energy of ATP hydrolysis with LPS extraction (60). Interestingly, the characterization of mutants in the coupling helices revealed that LptF and LptG might have distinct roles in coupling ATP hydrolysis in the cytoplasm with LPS extraction at the periplasmic leaflet of the IM. This observation is reminiscent of the distinct roles played by the TMDs of the Lol system for lipoprotein trafficking at the OM in E. coli (61). Such biochemical and genetic studies were nicely complemented by the resolution of the crystal structure of the LptB2FG complex from Pseudomonas aeruginosa in the nucleotide-free state (58). Notably, the arrangement of the TMDs of LptF and LptG defines a cavity whose surface is mainly hydrophobic, with the exception of the IM–periplasm interface, which is positively charged. This cavity appears to be large enough to accommodate LPS, which, anchored at the IM outer leaflet, could enter via the lateral gates formed by the TMDs of LptF and LptG (Fig. 2A) (58). The P. aeruginosa LptB2FG structure is thought to represent the resting state of the transporter, with the lateral gates that could further open upon ATP hydrolysis to allow LPS loading into the LPS-binding cavity. Overall, the model proposed for LPS extraction by the LptB2FG transporter postulates that ATP binding induces LptB dimerization and that the associated conformational changes are transmitted to LptFG via the coupling helices, triggering the lateral entry of LPS into the internal cavity of LptFG. ATP hydrolysis is hypothesized to induce the conformational switch back to the resting state, which may result in LPS delivery into the periplasmic β-jellyroll domains of LptFG (58) (Fig. 3). The hydrophobicity of the internal cavity formed by LptF and LptG TMDs and the presence of positively charged residues at the IM–periplasm interface suggest that only the lipid A portion of LPS may be loaded into the LptFG cavity from the periplasmic side of the IM, whereas the sugar portion of the molecule would remain accessible from the periplasm (58) (Fig. 3). Once LPS is extracted from the IM, the energy of ATP hydrolysis empowers LPS loading into the periplasmic protein bridge and its flow to the cell surface.
Figure 3.
Models for LPS extraction from the IM and assembly at the OM by the LptB2FG and LptDE complexes, respectively. Lower panel, the LptB2FG transporter in the nucleotide-free form is in the resting state. Binding of ATP triggers a conformational change of TMDs of LptF and LptG, allowing entry of LPS from the IM outer leaflet into the central hydrophobic cavity of the transporter through the lateral gates formed by the TMDs of LptF and LptG. Upon ATP hydrolysis, LPS is extracted from the IM and delivered to the β-jellyroll domains of LptF or LptG. ADP release may promote the return of the LptB2FG transporter to nucleotide-free form resting state (adapted from Ref. 53). Upper panel, the LPS molecule is thought to be delivered from the LptA to LptDE complex for insertion into the OM. The lipid A moiety is delivered to the β-jellyroll domain of LptD. Upon arrival of the LPS at the inner face of the OM, the LptD β-barrel is thought to open laterally, allowing lipid A to be inserted directly into the membrane, whereas the sugar chain is supposed to transit vertically through the β-barrel lumen of LptD.
The periplasmic Lpt protein bridge
LptA is the periplasmic protein that connects IM and OM (46, 62). The N-terminal domain of LptA connects to LptC, a bitopic IM protein with a single transmembrane helix and a large periplasmic domain (47, 56), whereas the LptA C-terminal region makes contact with the N-terminal periplasmic domain of LptD (62).
The crystal structure of both LptA and LptC has been solved (Fig. 2, B and C). Both proteins share the same β-jellyroll fold present in the periplasmic domains of LptF and LptG (58, 63, 64). Importantly, the same fold is also present in the periplasmic N-terminal domain of LptD (65, 66) (Fig. 2D), indicating that the assembly of the periplasmic protein bridge occurs via small structurally homologous domains adopting the same basic module, the so-called “Lpt fold” (Fig. 2, E–G). LptA and LptC both bind LPS (59, 64, 67, 68); therefore the Lpt fold also plays a crucial role in the transport of LPS. Inspection of the β-jellyroll structure of LptC reveals a cavity containing hydrophobic residues oriented toward the interior that could form a possible pocket for LPS binding (64). A hydrophobic pocket large enough to accommodate LPS molecule is not immediately evident in the monomeric structure of LptA. However, LptA tends to form head-to-tail oligomers in the presence of LPS (63) and in solution in a concentration-dependent manner (69, 70). In this type of LptA oligomeric organization, cavities with a larger size are formed (63, 71).
The flow of LPS inside the periplasmic bridge has been dissected by in vivo photo-crosslinking experiments suggesting that lipid A moiety crosses the periplasm inside the open hydrophobic groove formed by juxtaposition of the hydrophobic cavities of the β-jellyroll domains of LptC and LptA, whereas the hydrophilic portion of the molecule would be exposed in the periplasm (59) (Fig. 2, B–D). LPS interaction with Lpt proteins has also been explored by NMR, highlighting that LPS binds at LptA–LptC and at LptA–LptA intermolecular cavities that form only when the proteins composing the periplasmic bridge are assembled (71). Similarly, studies on LPS–LptA interaction using site-specific EPR spectroscopy suggest that LptA oligomerization is required to efficiently move LPS through the protein (68). Although the physiological significance of LptA oligomerization and the number of LptA molecules present in the Lpt bridge are not known yet, these data seem to indicate that conformational movements are needed for LPS transport to occur across the periplasm.
The insertion of LPS at the OM
The β-barrel protein LptD and the lipoprotein LptE constitute the OM translocon that assembles LPS at the OM outer leaflet (72). LptD and LptE form a complex with a peculiar two-protein plug-and-barrel architecture (73). To date, the structures of LptDE complexes from five pathogenic species (Salmonella enterica, Shigella flexneri, Yersinia pestis, P. aeruginosa, and Klebsiella pneumoniae) are available (65, 66, 74). The five structures are remarkably similar despite the divergent primary sequences: the LptD C-terminal domain forms a 26-stranded β-barrel, the largest reported so far, whereas LptE is almost entirely inserted in the β-barrel, thus confirming the prediction made based on photo-crosslinking experiments (73), whereas the N-terminal periplasmic region assumes the canonical Lpt fold (Fig. 2D) (65, 66).
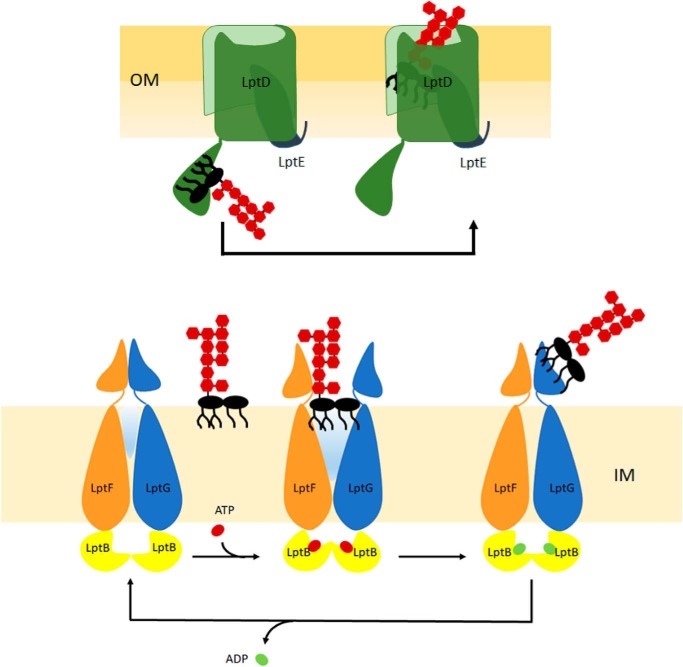
Following transport across the periplasmic bridge, LPS is thought to be delivered from LptA to LptDE for insertion into the OM. It is not known yet whether release of LPS from LptA to LptD requires ATP hydrolysis, but it has been demonstrated that the cell tightly controls whether a functional translocon has correctly assembled into the OM before making contact with LptA. This control is ensured by the presence of a couple of disulfide bonds within LptD (75), connecting the periplasmic and the β-barrel domains, whose correct formation allows the N-terminal domain of LptD to orient toward the periplasm to interact with LptA (62, 66, 76). The lipoprotein LptE plays a crucial role in controlling the correct maturation of the LptD disulfide bonds (76). The coordinated maturation of LptD and LptE, whose assembly into the OM requires different biogenesis pathways (namely, the β-barrel assembly machinery for LptD (77) and the Lol system for LptE (8)), is a system adopted by the cell to ensure, on the one hand, that LptE is inserted into the lumen of LptD and, on the other hand, that only a mature LptDE translocon makes contact with LptA, thus enabling the establishment of a functional transenvelope bridge. Based on evidences obtained from E. coli, LptE insertion into LptD has a dual role within the Lpt machinery: LptE plugs the otherwise too large lumen of LptD (72, 78) but also has a direct involvement in the assembly of LPS into the outer leaflet of the OM (51), (see below).
The availability of the crystal structure of the LptDE complex has greatly contributed to our understanding of how LPS is assembled at the outer leaflet of the OM. In the β-barrel structure of LptD, two highly conserved proline residues in the β1 and β2 strands appear to perturb the secondary structure of the strands, thus preventing the formation of the typical β-sheet hydrogen bonds between the β1 and β26 strands, and generating a local gap that provides gateways for the lateral migration of the LPS molecules between the LptDE complex and the membrane (66) (Fig. 3). According to structural data, the hydrophilic lumen of LptD is large enough to accommodate the sugar moiety of LPS. Thus, once LPS is delivered to the N-terminal domain of LptD, a conformational change in LptDE would occur, enabling the saccharide portion of the LPS molecules to enter the β-barrel and travel to the cell surface, passing through the lateral gate of LptD. On the contrary, it has been proposed that the lipid A domain of LPS would pass first inside the β-jellyroll structure of the LptD N-terminal domain and then through the hydrophobic intramembrane open between the N-terminal and the β-barrel domains of LptD (66, 79, 80) (Fig. 2D). It is worth mentioning that the orientation of the N-terminal domain of LptD relative to its barrel domain, ensured by the correct formation of the nonconsecutive disulfide bonds, would secure the correct positioning of the LPS substrate to allow insertion of the lipid A moiety of LPS into the membrane while maintaining the sugar moiety within the LptD lumen (66).
During the transfer of LPS across the LptDE translocon, conformational changes must be triggered in the translocon, allowing LptE to directly interact with LPS (72). In fact, in E. coli, LptE–LPS interaction could be functional in disrupting LPS aggregates during LPS assembly at the outer leaflet of the OM and in preventing LPS mislocalization in the inner leaflet of the OM (51). Notably, LptE does not seem to be directly involved in LPS transport in N. meningitidis but instead is only implicated in assisting LptD biogenesis (81). Structural differences have also been shown in LptDE complex from P. aeruginosa (82), underscoring the relevance of studying conserved mechanisms in different model systems.
The enigmatic role of LptC in the Lpt machinery
The overall picture of the LPS transport from the IM to the cell surface has now been clarified together with the molecular role of each component of the Lpt machinery with the exception of LptC. The LptC protein stably associates with the purified LptB2FG transporter, but fails to modulate the ATPase activity of the transporter in vitro (43). Several lines of evidence demonstrate that LptC interaction with LptA is essential for LPS transport (56, 59, 62, 83). Nevertheless, the apparent structural and functional redundancy between LptA and LptC makes it hard to differentiate their roles in LPS transport. Recent reports have described different mechanisms to overcome defects in LptC or even lptC deletion. The C-terminal region of LptC is involved in binding to LptA (56, 62). However, truncation of this region of LptC can be suppressed by overexpression of LptB, resulting in structural stabilization of the truncated LptC (54). How LptB overexpression rescues LPS transport across the periplasm is not clear. It is possible that the truncated LptC mutant retains the ability to interact with LptA, although with low efficiency, and that the overexpression of LptB stabilizes the LptB2FG transporters containing the LptC mutant, allowing the formation of the periplasmic protein bridge. These observations fit well with the hypothesis that the assembly of LptC into the IM LptB2FG complex is a quality control step for transenvelope bridge formation. This hypothesis is further supported by several lines of evidence: (i) a mutant in the N-terminal region of the LptC periplasmic domain that is impaired in its association with LptB2FG does not recruit LptA in vivo, although it maintains an intact interaction interface (83); (ii) LptC from P. aeruginosa cannot fully substitute for E. coli LptC due to defective interaction with the LptB2FG complex (54), whereas hybrid Lpt machineries containing P. aeruginosa LptA are functional in E. coli (52), suggesting that interspecies LptA–LptC interaction occurs; and (iii) the LptB2FG complex from P. aeruginosa cannot functionally substitute for E. coli LptB2FG (58), possibly due to inability to interact with E. coli LptC. Overall, LptC appears to be a species-specific component of the Lpt machinery. We do not know how LptB2FG interacts with LptC, but the presence of the Lpt fold in the periplasmic domains of the LptFG and LptC (Fig. 2) strongly implicates these domains in forming the stable LptB2FGC IM complex. Accordingly, the TM domain of LptC does not seem to be implicated in LptB2FG binding as LptC missing the TM domain is functional and assembles to LptB2FG (83). Additional evidence pointing to a regulative role for LptC is the isolation of a class of mutants lacking LptC and expressing LptF mutants carrying amino acid substitutions at a unique residue of the periplasmic domain. This class of mutants behaves similarly to the wild type, suggesting that a six-component machinery ensures the core functions for LPS transport (84). Further characterization of this mutant will offer new insights into the molecular role of LptC in LPS transport.
Conclusions
The overall picture of the LPS transport system proposed by Kahne and co-workers (85) depicts the LPS transport apparatus as a PEZ “candy dispenser” in which the energy of ATP hydrolysis is used to push a continuous stream of LPS molecules in the periplasmic hydrophobic groove up to the OM. The recently solved structure of the LptB2FG complex completes the puzzle of the LPS export pathway, proposing how LPS is extracted from IM and propelled into the periplasmic hydrophobic channel (58). However, there are still important molecular details on the functioning of the Lpt machine that remain unanswered. We lack the structural description of the periplasmic protein bridge as well as the molecular details on how these proteins bind LPS. Also we do not yet know how the large and bulky O-antigen is transported across the periplasmic space, which contains the peptidoglycan layer; note that most of the studies on LPS transport have been conducted either on strains missing the O-antigen (E. coli) or on strains that do not produce it (N. meningitidis).
Elucidating the molecular mechanism of LPS biogenesis has the potential to advance the development of novel therapies against Gram-negative pathogens. Notably, peptidomimetics specifically targeting P. aeruginosa LptD (86, 87) and molecules that inhibit the ATPase activity of LptB (88) are the first inhibitors of the LPS export pathway. As the OM is a crucial permeability barrier, compounds inhibiting the LPS export pathway not only can be used as antibiotics but can also be used as a means to manipulate OM permeability.
Acknowledgment
We thank Dr. Luca Palazzolo for help in the preparation of structural alignments.
This work was supported by the Train2Target project (Grant 721484) granted from the European Union's Horizon 2020. The authors declare that they have no conflicts of interest with the contents of this article.
- IM
- inner membrane
- OM
- outer membrane
- ABC
- ATP-binding cassette
- TM
- transmembrane
- TMD
- transmembrane domain.
References
- 1. Silhavy T. J., Kahne D., and Walker S. (2010) The bacterial cell envelope. Cold Spring Harb. Perspect. Biol. 2, a000414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Vollmer W., Blanot D., and de Pedro M. A. (2008) Peptidoglycan structure and architecture. FEMS Microbiol. Rev. 32, 149–167 [DOI] [PubMed] [Google Scholar]
- 3. Fairman J. W., Noinaj N., and Buchanan S. K. (2011) The structural biology of β-barrel membrane proteins: a summary of recent reports. Curr. Opin. Struct. Biol. 21, 523–531 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Schulz G. E. (2002) The structure of bacterial outer membrane proteins. Biochim. Biophys. Acta 1565, 308–317 [DOI] [PubMed] [Google Scholar]
- 5. Delcour A. H. (2003) Solute uptake through general porins. Front. Biosci. 8, d1055–1071 [DOI] [PubMed] [Google Scholar]
- 6. Braun V. (2006) Energy transfer between biological membranes. ACS Chem. Biol. 1, 352–354 [DOI] [PubMed] [Google Scholar]
- 7. Sankaran K., and Wu H. C. (1994) Lipid modification of bacterial prolipoprotein: transfer of diacylglyceryl moiety from phosphatidylglycerol. J. Biol. Chem. 269, 19701–19706 [PubMed] [Google Scholar]
- 8. Narita S. I., and Tokuda H. (2017) Bacterial lipoproteins; biogenesis, sorting and quality control. Biochim. Biophys. Acta 1862, 1414–1423 [DOI] [PubMed] [Google Scholar]
- 9. Kamio Y., and Nikaido H. (1976) Outer membrane of Salmonella typhimurium: accessibility of phospholipid head groups to phospholipase c and cyanogen bromide activated dextran in the external medium. Biochemistry 15, 2561–2570 [DOI] [PubMed] [Google Scholar]
- 10. Raetz C. R., and Whitfield C. (2002) Lipopolysaccharide endotoxins. Annu. Rev. Biochem. 71, 635–700 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Holst O. (2007) The structures of core regions from enterobacterial lipopolysaccharides: an update. FEMS Microbiol. Lett. 271, 3–11 [DOI] [PubMed] [Google Scholar]
- 12. Greenfield L. K., and Whitfield C. (2012) Synthesis of lipopolysaccharide O-antigens by ABC transporter-dependent pathways. Carbohydr. Res. 356, 12–24 [DOI] [PubMed] [Google Scholar]
- 13. Liu D., and Reeves P. R. (1994) Presence of different O antigen forms in three isolates of one clone of Escherichia coli. Genetics 138, 6–10 [PMC free article] [PubMed] [Google Scholar]
- 14. Stevenson G., Neal B., Liu D., Hobbs M., Packer N. H., Batley M., Redmond J. W., Lindquist L., and Reeves P. (1994) Structure of the O antigen of Escherichia coli K-12 and the sequence of its rfb gene cluster. J. Bacteriol. 176, 4144–4156 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Steeghs L., den Hartog R., den Boer A., Zomer B., Roholl P., and van der Ley P. (1998) Meningitis bacterium is viable without endotoxin. Nature 392, 449–450 [DOI] [PubMed] [Google Scholar]
- 16. Peng D., Hong W., Choudhury B. P., Carlson R. W., and Gu X. X. (2005) Moraxella catarrhalis bacterium without endotoxin, a potential vaccine candidate. Infect. Immun. 73, 7569–7577 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Moffatt J. H., Harper M., Harrison P., Hale J. D., Vinogradov E., Seemann T., Henry R., Crane B., St Michael F., Cox A. D., Adler B., Nation R. L., Li J., and Boyce J. D. (2010) Colistin resistance in Acinetobacter baumannii is mediated by complete loss of lipopolysaccharide production. Antimicrob. Agents Chemother. 54, 4971–4977 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Boll J. M., Crofts A. A., Peters K., Cattoir V., Vollmer W., Davies B. W., and Trent M. S. (2016) A penicillin-binding protein inhibits selection of colistin-resistant, lipooligosaccharide-deficient Acinetobacter baumannii. Proc. Natl. Acad. Sci. U.S.A. 113, E6228–E6237 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Miyake K. (2004) Innate recognition of lipopolysaccharide by Toll-like receptor 4-MD-2. Trends Microbiol. 12, 186–192 [DOI] [PubMed] [Google Scholar]
- 20. Park B. S., and Lee J. O. (2013) Recognition of lipopolysaccharide pattern by TLR4 complexes. Exp. Mol. Med. 45, e66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Lerouge I., and Vanderleyden J. (2002) O-antigen structural variation: mechanisms and possible roles in animal/plant-microbe interactions. FEMS Microbiol. Rev. 26, 17–47 [DOI] [PubMed] [Google Scholar]
- 22. Nikaido H. (2003) Molecular basis of bacterial outer membrane permeability revisited. Microbiol. Mol. Biol. Rev. 67, 593–656 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Nikaido H. (2005) Restoring permeability barrier function to outer membrane. Chem. Biol. 12, 507–509 [DOI] [PubMed] [Google Scholar]
- 24. Henry R., Vithanage N., Harrison P., Seemann T., Coutts S., Moffatt J. H., Nation R. L., Li J., Harper M., Adler B., and Boyce J. D. (2012) Colistin-resistant, lipopolysaccharide-deficient Acinetobacter baumannii responds to lipopolysaccharide loss through increased expression of genes involved in the synthesis and transport of lipoproteins, phospholipids, and poly-β-1,6-N-acetylglucosamine. Antimicrob. Agents Chemother. 56, 59–69 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Steeghs L., de Cock H., Evers E., Zomer B., Tommassen J., and van der Ley P. (2001) Outer membrane composition of a lipopolysaccharide-deficient Neisseria meningitidis mutant. EMBO J. 20, 6937–6945 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Ruiz N., Wu T., Kahne D., and Silhavy T. J. (2006) Probing the barrier function of the outer membrane with chemical conditionality. ACS Chem. Biol. 1, 385–395 [DOI] [PubMed] [Google Scholar]
- 27. Ruiz N., Falcone B., Kahne D., and Silhavy T. J. (2005) Chemical conditionality: a genetic strategy to probe organelle assembly. Cell 121, 307–317 [DOI] [PubMed] [Google Scholar]
- 28. Sutterlin H. A., Zhang S., and Silhavy T. J. (2014) Accumulation of phosphatidic acid increases vancomycin resistance in Escherichia coli. J. Bacteriol. 196, 3214–3220 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Sutterlin H. A., Shi H., May K. L., Miguel A., Khare S., Huang K. C., and Silhavy T. J. (2016) Disruption of lipid homeostasis in the Gram-negative cell envelope activates a novel cell death pathway. Proc. Natl. Acad. Sci. U.S.A. 113, E1565–E1574 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Stokes J. M., French S., Ovchinnikova O. G., Bouwman C., Whitfield C., and Brown E. D. (2016) Cold stress makes Escherichia coli susceptible to glycopeptide antibiotics by altering outer membrane integrity. Cell Chem. Biol. 23, 267–277 [DOI] [PubMed] [Google Scholar]
- 31. Pagès J. M., James C. E., and Winterhalter M. (2008) The porin and the permeating antibiotic: a selective diffusion barrier in Gram-negative bacteria. Nat. Rev. Microbiol. 6, 893–903 [DOI] [PubMed] [Google Scholar]
- 32. Zabawa T. P., Pucci M. J., Parr T. R. Jr, and Lister T. (2016) Treatment of Gram-negative bacterial infections by potentiation of antibiotics. Curr. Opin. Microbiol. 33, 7–12 [DOI] [PubMed] [Google Scholar]
- 33. Raetz C. R. H., Reynolds C. M., Trent M. S., and Bishop R. E. (2007) Lipid A modification systems in Gram-negative bacteria. Annu. Rev. Biochem. 76, 295–329 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Polissi A., and Georgopoulos C. (1996) Mutational analysis and properties of the msbA gene of Escherichia coli, coding for an essential ABC family transporter. Mol. Microbiol. 20, 1221–1233 [DOI] [PubMed] [Google Scholar]
- 35. Zhou Z., White K. A., Polissi A., Georgopoulos C., and Raetz C. R. (1998) Function of Escherichia coli MsbA, an essential ABC family transporter, in lipid A and phospholipid biosynthesis. J. Biol. Chem. 273, 12466–12475 [DOI] [PubMed] [Google Scholar]
- 36. Locher K. P. (2016) Mechanistic diversity in ATP-binding cassette (ABC) transporters. Nat. Struct. Mol. Biol. 23, 487–493 [DOI] [PubMed] [Google Scholar]
- 37. Eckford P. D., and Sharom F. J. (2008) Functional characterization of Escherichia coli MsbA: interaction with nucleotides and substrates. J. Biol. Chem. 283, 12840–12850 [DOI] [PubMed] [Google Scholar]
- 38. Borbat P. P., Surendhran K., Bortolus M., Zou P., Freed J. H., and Mchaourab H. S. (2007) Conformational motion of the ABC transporter MsbA induced by ATP hydrolysis. PLoS Biol. 5, e271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Dong J., Yang G., and McHaourab H. S. (2005) Structural basis of energy transduction in the transport cycle of MsbA. Science 308, 1023–1028 [DOI] [PubMed] [Google Scholar]
- 40. Ward A., Reyes C. L., Yu J., Roth C. B., and Chang G. (2007) Flexibility in the ABC transporter MsbA: alternating access with a twist. Proc. Natl. Acad. Sci. U.S.A. 104, 19005–19010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Doerrler W. T., and Raetz C. R. (2002) ATPase activity of the MsbA lipid flippase of Escherichia coli. J. Biol. Chem. 277, 36697–36705 [DOI] [PubMed] [Google Scholar]
- 42. Eckford P. D., and Sharom F. J. (2010) The reconstituted Escherichia coli MsbA protein displays lipid flippase activity. Biochem. J. 429, 195–203 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Narita S., and Tokuda H. (2009) Biochemical characterization of an ABC transporter LptBFGC complex required for the outer membrane sorting of lipopolysaccharides. FEBS Letters 583, 2160–2164 [DOI] [PubMed] [Google Scholar]
- 44. Ruiz N., Gronenberg L. S., Kahne D., and Silhavy T. J. (2008) Identification of two inner-membrane proteins required for the transport of lipopolysaccharide to the outer membrane of Escherichia coli. Proc. Natl. Acad. Sci. U.S.A. 105, 5537–5542 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Sperandeo P., Pozzi C., Dehò G., and Polissi A. (2006) Non-essential KDO biosynthesis and new essential cell envelope biogenesis genes in the Escherichia coli yrbG-yhbG locus. Res. Microbiol. 157, 547–558 [DOI] [PubMed] [Google Scholar]
- 46. Sperandeo P., Cescutti R., Villa R., Di Benedetto C., Candia D., Dehò G., and Polissi A. (2007) Characterization of lptA and lptB, two essential genes implicated in lipopolysaccharide transport to the outer membrane of Escherichia coli. J. Bacteriol. 189, 244–253 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Sperandeo P., Lau F. K., Carpentieri A., De Castro C., Molinaro A., Dehò G., Silhavy T. J., and Polissi A. (2008) Functional analysis of the protein machinery required for transport of lipopolysaccharide to the outer membrane of Escherichia coli. J. Bacteriol. 190, 4460–4469 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Bos M. P., Tefsen B., Geurtsen J., and Tommassen J. (2004) Identification of an outer membrane protein required for the transport of lipopolysaccharide to the bacterial cell surface. Proc. Natl. Acad. Sci. U.S.A. 101, 9417–9422 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Wu T., McCandlish A. C., Gronenberg L. S., Chng S. S., Silhavy T. J., and Kahne D. (2006) Identification of a protein complex that assembles lipopolysaccharide in the outer membrane of Escherichia coli. Proc. Natl. Acad. Sci. U.S.A. 103, 11754–11759 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Bos M. P., Robert V., and Tommassen J. (2007) Biogenesis of the Gram-negative bacterial outer membrane. Annu. Rev. Microbiol. 61, 191–214 [DOI] [PubMed] [Google Scholar]
- 51. Malojčić G., Andres D., Grabowicz M., George A. H., Ruiz N., Silhavy T. J., and Kahne D. (2014) LptE binds to and alters the physical state of LPS to catalyze its assembly at the cell surface. Proc. Natl. Acad. Sci. U.S.A. 111, 9467–9472 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Bollati M., Villa R., Gourlay L. J., Benedet M., Dehò G., Polissi A., Barbiroli A., Martorana A. M., Sperandeo P., Bolognesi M., and Nardini M. (2015) Crystal structure of LptH, the periplasmic component of the lipopolysaccharide transport machinery from Pseudomonas aeruginosa. FEBS J. 282, 1980–1997 [DOI] [PubMed] [Google Scholar]
- 53. Putker F., Bos M. P., and Tommassen J. (2015) Transport of lipopolysaccharide to the Gram-negative bacterial cell surface. FEMS Microbiol. Rev. 39, 985–1002 [DOI] [PubMed] [Google Scholar]
- 54. Martorana A. M., Benedet M., Maccagni E. A., Sperandeo P., Villa R., Dehò G., and Polissi A. (2016) Functional interaction between the cytoplasmic ABC protein LptB and the inner membrane LptC protein, components of the lipopolysaccharide transport machinery in Escherichia coli. J. Bacteriol. 198, 2192–2203 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Chng S.-S., Gronenberg L. S., and Kahne D. (2010) Proteins required for lipopolysaccharide assembly in Escherichia coli form a transenvelope complex. Biochemistry 49, 4565–4567 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Sperandeo P., Villa R., Martorana A. M., Samalikova M., Grandori R., Dehò G., and Polissi A. (2011) New insights into the Lpt machinery for lipopolysaccharide transport to the cell surface: LptA-LptC interaction and LptA stability as sensors of a properly assembled transenvelope complex. J. Bacteriol. 193, 1042–1053 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Sherman D. J., Lazarus M. B., Murphy L., Liu C., Walker S., Ruiz N., and Kahne D. (2014) Decoupling catalytic activity from biological function of the ATPase that powers lipopolysaccharide transport. Proc. Natl. Acad. Sci. U.S.A. 111, 4982–4987 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Luo Q., Yang X., Yu S., Shi H., Wang K., Xiao L., Zhu G., Sun C., Li T., Li D., Zhang X., Zhou M., and Huang Y. (2017) Structural basis for lipopolysaccharide extraction by ABC transporter LptB2FG. Nat. Struct. Mol. Biol. 24, 469–474 [DOI] [PubMed] [Google Scholar]
- 59. Okuda S., Freinkman E., and Kahne D. (2012) Cytoplasmic ATP hydrolysis powers transport of lipopolysaccharide across the periplasm in E. coli. Science 338, 1214–1217 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Simpson B. W., Owens T. W., Orabella M. J., Davis R. M., May J. M., Trauger S. A., Kahne D., and Ruiz N. (2016) Identification of residues in the lipopolysaccharide ABC transporter that coordinate ATPase activity with extractor function. mBio 7, e01729–16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Okuda S., and Tokuda H. (2009) Model of mouth-to-mouth transfer of bacterial lipoproteins through inner membrane LolC, periplasmic LolA, and outer membrane LolB. Proc. Natl. Acad. Sci. U.S.A. 106, 5877–5882 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Freinkman E., Okuda S., Ruiz N., and Kahne D. (2012) Regulated assembly of the transenvelope protein complex required for lipopolysaccharide export. Biochemistry 51, 4800–4806 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Suits M. D. L., Sperandeo P., Dehò G., Polissi A., and Jia Z. (2008) Novel structure of the conserved Gram-negative lipopolysaccharide transport protein A and mutagenesis analysis. J. Mol. Biol. 380, 476–488 [DOI] [PubMed] [Google Scholar]
- 64. Tran A. X., Dong C., and Whitfield C. (2010) Structure and functional analysis of LptC, a conserved membrane protein involved in the lipopolysaccharide export pathway in Escherichia coli. J. Biol. Chem. 285, 33529–33539 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Qiao S., Luo Q., Zhao Y., Zhang X. C., and Huang Y. (2014) Structural basis for lipopolysaccharide insertion in the bacterial outer membrane. Nature 511, 108–111 [DOI] [PubMed] [Google Scholar]
- 66. Botos I., Majdalani N., Mayclin S. J., McCarthy J. G., Lundquist K., Wojtowicz D., Barnard T. J., Gumbart J. C., and Buchanan S. K. (2016) Structural and functional characterization of the LPS transporter LptDE from Gram-negative pathogens. Structure 24, 965–976 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Sestito S. E., Sperandeo P., Santambrogio C., Ciaramelli C., Calabrese V., Rovati G. E., Zambelloni L., Grandori R., Polissi A., and Peri F. (2014) Functional characterization of E. coli LptC: interaction with LPS and a synthetic ligand. Chembiochem 15, 734–742 [DOI] [PubMed] [Google Scholar]
- 68. Schultz K. M., Lundquist T. J., and Klug C. S. (2017) Lipopolysaccharide binding to the periplasmic protein LptA. Protein Sci. 26, 1517–1523 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Merten J. A., Schultz K. M., and Klug C. S. (2012) Concentration-dependent oligomerization and oligomeric arrangement of LptA. Protein Sci. 21, 211–218 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Santambrogio C., Sperandeo P., Villa R., Sobott F., Polissi A., and Grandori R. (2013) LptA assembles into rod-like oligomers involving disorder-to-order transitions. J. Am. Soc. Mass Spectrom. 24, 1593–1602 [DOI] [PubMed] [Google Scholar]
- 71. Laguri C., Sperandeo P., Pounot K., Ayala I., Silipo A., Bougault C. M., Molinaro A., Polissi A., and Simorre J. P. (2017) Interaction of lipopolysaccharides at intermolecular sites of the periplasmic Lpt transport assembly. Sci. Rep. 7, 9715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Chng S. S., Ruiz N., Chimalakonda G., Silhavy T. J., and Kahne D. (2010) Characterization of the two-protein complex in Escherichia coli responsible for lipopolysaccharide assembly at the outer membrane. Proc. Natl. Acad. Sci. U.S.A. 107, 5363–5368 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Freinkman E., Chng S. S., and Kahne D. (2011) The complex that inserts lipopolysaccharide into the bacterial outer membrane forms a two-protein plug-and-barrel. Proc. Natl. Acad. Sci. U.S.A. 108, 2486–2491 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Dong H., Xiang Q., Gu Y., Wang Z., Paterson N. G., Stansfeld P. J., He C., Zhang Y., Wang W., and Dong C. (2014) Structural basis for outer membrane lipopolysaccharide insertion. Nature 511, 52–56 [DOI] [PubMed] [Google Scholar]
- 75. Ruiz N., Chng S. S., Hiniker A., Kahne D., and Silhavy T. J. (2010) Nonconsecutive disulfide bond formation in an essential integral outer membrane protein. Proc. Natl. Acad. Sci. U.S.A. 107, 12245–12250 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Chng S. S., Xue M., Garner R. A., Kadokura H., Boyd D., Beckwith J., and Kahne D. (2012) Disulfide rearrangement triggered by translocon assembly controls lipopolysaccharide export. Science 337, 1665–1668 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Noinaj N., Gumbart J. C., and Buchanan S. K. (2017) The β-barrel assembly machinery in motion. Nat. Rev. Microbiol. 15, 197–204 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Chimalakonda G., Ruiz N., Chng S. S., Garner R. A., Kahne D., and Silhavy T. J. (2011) Lipoprotein LptE is required for the assembly of LptD by the β-barrel assembly machine in the outer membrane of Escherichia coli. Proc. Natl. Acad. Sci. U.S.A. 108, 2492–2497 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Gu Y., Stansfeld P. J., Zeng Y., Dong H., Wang W., and Dong C. (2015) Lipopolysaccharide is inserted into the outer membrane through an intramembrane hole, a lumen gate, and the lateral opening of LptD. Structure 23, 496–504 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Li X., Gu Y., Dong H., Wang W., and Dong C. (2015) Trapped lipopolysaccharide and LptD intermediates reveal lipopolysaccharide translocation steps across the Escherichia coli outer membrane. Sci. Rep. 5, 11883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Bos M. P., and Tommassen J. (2011) The LptD chaperone LptE is not directly involved in lipopolysaccharide transport in Neisseria meningitidis. J. Biol. Chem. 286, 28688–28696 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Moehle K., Kocherla H., Bacsa B., Jurt S., Zerbe K., Robinson J. A., and Zerbe O. (2016) Solution structure and dynamics of LptE from Pseudomonas aeruginosa. Biochemistry 55, 2936–2943 [DOI] [PubMed] [Google Scholar]
- 83. Villa R., Martorana A. M., Okuda S., Gourlay L. J., Nardini M., Sperandeo P., Dehò G., Bolognesi M., Kahne D., and Polissi A. (2013) The Escherichia coli Lpt transenvelope protein complex for lipopolysaccharide export is assembled via conserved structurally homologous domains. J. Bacteriol. 195, 1100–1108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Benedet M., Falchi F. A., Puccio S., Di Benedetto C., Peano C., Polissi A., and Dehò G. (2016) The lack of the essential LptC protein in the trans-envelope lipopolysaccharide transport machine is circumvented by suppressor mutations in LptF, an inner membrane component of the Escherichia coli transporter. PLoS One 11, e0161354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Okuda S., Sherman D. J., Silhavy T. J., Ruiz N., and Kahne D. (2016) Lipopolysaccharide transport and assembly at the outer membrane: the PEZ model. Nat. Rev. Microbiol. 14, 337–345 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86. Srinivas N., Jetter P., Ueberbacher B. J., Werneburg M., Zerbe K., Steinmann J., Van der Meijden B., Bernardini F., Lederer A., Dias R. L., Misson P. E., Henze H., Zumbrunn J., Gombert F. O., et al. (2010) Peptidomimetic antibiotics target outer-membrane biogenesis in Pseudomonas aeruginosa. Science 327, 1010–1013 [DOI] [PubMed] [Google Scholar]
- 87. Werneburg M., Zerbe K., Juhas M., Bigler L., Stalder U., Kaech A., Ziegler U., Obrecht D., Eberl L., and Robinson J. A. (2012) Inhibition of lipopolysaccharide transport to the outer membrane in Pseudomonas aeruginosa by peptidomimetic antibiotics. Chembiochem 13, 1767–1775 [DOI] [PubMed] [Google Scholar]
- 88. Sherman D. J., Okuda S., Denny W. A., and Kahne D. (2013) Validation of inhibitors of an ABC transporter required to transport lipopolysaccharide to the cell surface in Escherichia coli. Bioorg. Med. Chem. 21, 4846–4851 [DOI] [PMC free article] [PubMed] [Google Scholar]