Abstract
Non-structural protein 5B (NS5B) is the RNA-dependent RNA polymerase that catalyzes replication of the hepatitis C virus (HCV) RNA genome and therefore is central for its life cycle. NS5B interacts with the intrinsically disordered domain 2 of NS5A (NS5A-D2), another essential multifunctional HCV protein that is required for RNA replication. As a result, these two proteins represent important targets for anti-HCV chemotherapies. Despite this importance and the existence of NS5B crystal structures, our understanding of the conformational and dynamic behavior of NS5B in solution and its relationship with NS5A-D2 remains incomplete. To address these points, we report the first detailed NMR spectroscopic study of HCV NS5B lacking its membrane anchor (NS5BΔ21). Analysis of constructs with selective isotope labeling of the δ1 methyl groups of isoleucine side chains demonstrates that, in solution, NS5BΔ21 is highly dynamic but predominantly adopts a closed conformation. The addition of NS5A-D2 leads to spectral changes indicative of binding to both allosteric thumb sites I and II of NS5BΔ21 and induces long-range perturbations that affect the RNA-binding properties of the polymerase. We compared these modifications with the short- and long-range effects triggered in NS5BΔ21 upon binding of filibuvir, an allosteric inhibitor. We demonstrate that filibuvir-bound NS5BΔ21 is strongly impaired in the binding of both NS5A-D2 and RNA. NS5A-D2 induces conformational and functional perturbations in NS5B similar to those triggered by filibuvir. Thus, our work highlights NS5A-D2 as an allosteric regulator of the HCV polymerase and provides new insight into the dynamics of NS5B in solution.
Keywords: allosteric regulation, nuclear magnetic resonance (NMR), protein dynamic, protein-protein interaction, RNA-protein interaction
Introduction
Hepatitis C virus (HCV)8 is a small enveloped RNA virus belonging to the Hepacivirus genus in the Flaviridae family. It was identified as the non-A, non-B hepatitis causative agent in the late 1980s and is now estimated to have chronically infected ∼71 million people worldwide (1). HCV infection may be asymptomatic, but 75–85% of individuals with acute hepatitis will develop a chronic infection that can lead to severe liver damage, such as cirrhosis or hepatocellular carcinoma, and finally to death (2). Currently, it is estimated that about 400,000 people worldwide die from hepatitis C each year. Although the incidence of HCV infections is decreasing in developed countries, the number of deaths associated with this liver disease is expected to rise within the next 15 years. Depending on the nucleotide sequence of its RNA genome (∼9,600 nucleotides), HCV is classified into seven genotypes (3) that are unevenly distributed over the world. Genotypes 1 and 3 are responsible for ∼45 and ∼30% of HCV infections worldwide, respectively. Whereas genotype 1 is the most prevalent in developed countries, genotypes 4 and 5 are essentially present in developing countries (4).
The HCV-positive sense single-stranded RNA genome ((+)ssRNA) contains a unique open reading frame between 5′- and 3′-non-translated regions that are required for translation and replication. The open reading frame encodes a polyprotein precursor, which is subsequently cleaved by host and viral proteases into 10 mature proteins: the structural proteins (core, E1, and E2) that constitute the virus particle and the non-structural (NS) proteins (p7, NS2, NS3, NS4A, NS4B, NS5A, and NS5B) (5, 6). p7 (viroporin) and NS2 (cysteine protease) are required for the assembly of new virions, whereas NS3 to NS5B, all anchored to the endoplasmic reticulum (ER) host membrane, constitute a replicase complex catalyzing RNA replication (7). This complex has been localized in induced ER-derived membrane structures, named the double-membrane vesicles (8, 9). NS3 carries both serine protease (with its NS4A cofactor peptide) and helicase activity (10). NS4B is involved in cellular membrane rearrangements leading to double-membrane vesicle formation (9, 11). NS5A is required for RNA replication (12, 13) and is involved in viral particle production (14). Finally, NS5B carries the RNA-dependent RNA polymerase (RdRp) enzymatic activity (15, 16), which is central in the viral RNA replication process.
Tremendous progress has been made in the development of anti-HCV drugs since the introduction, in 2011, of the first two direct-acting antiviral agents (DAA) that target the NS3/4A serine protease (17). Nowadays, efficient DAAs that inhibit either NS3/4A, NS5A, or NS5B proteins have been approved for therapeutic use. These all-oral, interferon-free DAAs, mainly used in combination, have enabled the achievement of cure rates of >90% (18, 19). Bourlière et al. (20) recently reported that, following the administration for 12 weeks of a combination of an NS5B inhibitor (sofosbuvir) with an NS5A inhibitor (velpatasvir) and an NS3/4A inhibitor (voxilaprevir) to HCV-infected patients previously treated with a DAA-containing regimen, the sustained virologic response was 96–98%.
HCV NS5B constitutes the enzymatic core of the viral replicase complex, as it carries the RdRp activity required for RNA replication. In this process, NS5B catalyzes the polymerization of ribonucleoside tri-phosphates (rNTPs) to synthesize an intermediate (−)ssRNA molecule from its initial template, the (+)ssRNA genome. Then the newly synthesized molecule serves as a template to produce numerous (+)ssRNA molecules that will later be used either for polyprotein translation, production of infectious particles, or synthesis of new intermediates for RNA replication (21). NS5B (591 residues, 65 kDa) is anchored to the ER membrane via its 21 C-terminal residues (22, 23), which form a hydrophobic helix (24). Its polymerase domain is located in the cytosol. The crystallographic structure of NS5BΔ21 (i.e. without its C-terminal membrane anchor) revealed that the polymerase resembles a closed right hand with three subdomains termed the palm, fingers, and thumb. NS5B also contains three additional structural features: 1) two loops (Λ1 and Λ2) of the fingers subdomain, which make extensive contacts with the top of the thumb and thus encircle the active site; 2) a β-loop in the thumb subdomain, which protrudes into the active site; and 3) a C-terminal disordered linker that folds back into the active site (25–27) (see Fig. 6 in Ref. 28 and Fig. 3 in Ref. 29). As such, the closed polymerase structure cannot accommodate double-stranded RNA due to steric hindrance, and it has been proposed that NS5B alternates between at least two conformations, one “closed” and one “open” (30–34). Recently, Appleby et al. (35) solved several crystallographic structures of NS5B mutants in complex with RNA template, RNA primer, and ribonucleotides and showed that, concomitantly to the opening of the polymerase (via the rotation of the thumb subdomain), the β-loop and the C-terminal linker progressively move away from the active site during the RNA enzymatic replication process. This conformational rearrangement, a rate-limiting event, is believed to take place between the de novo initiation and the processive elongation steps of RNA replication.
Figure 6.
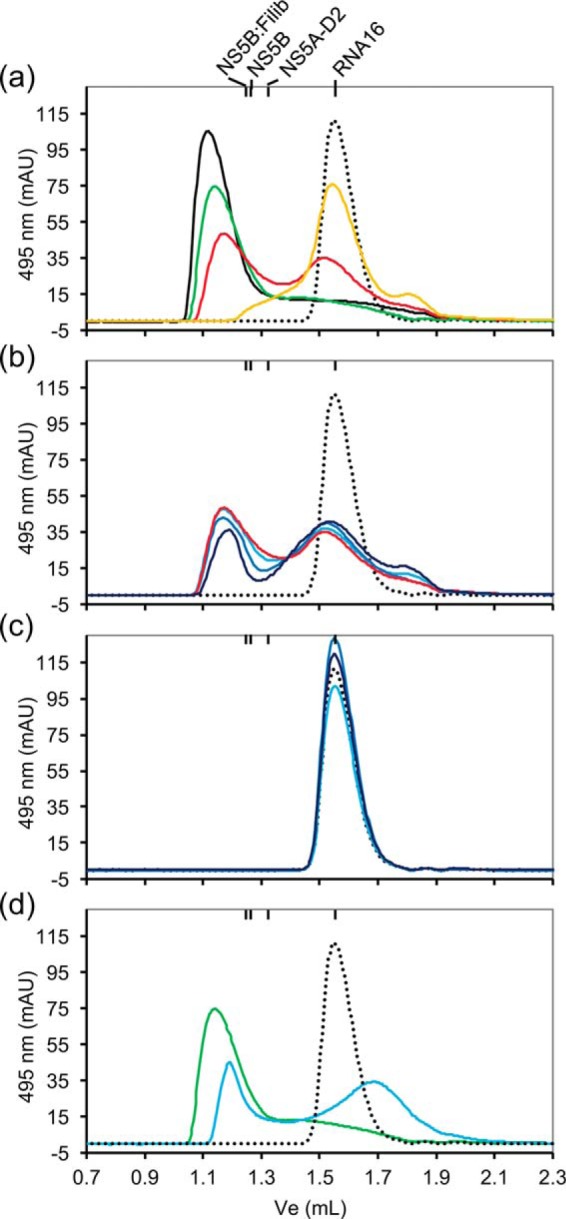
Inhibition of the NS5B–RNA16 interaction. A fluorescent RNA oligonucleotide (6FAM-RNA16, 10 μm) was analyzed by size-exclusion chromatography (Superdex 75 3.2/300) in the absence or the presence of NS5B and/or NS5A-D2. The elution of 6FAM-RNA16 was followed by absorption at 495 nm. a, interaction with NS5BΔ21. 10 μm 6FAM-RNA16 (dotted black line) was mixed with 3 μm (yellow line), 7 μm (red line), 10 μm (green line), and 15 μm (black line) concentrations of NS5BΔ21, respectively. b, inhibition of the interaction between NS5BΔ21 and RNA16 by NS5A-D2. 10 μm 6FAM-RNA16 (dotted black line) was mixed with either 7 μm NS5BΔ21 (red line, as in a), 7 μm NS5BΔ21 + 5 μm NS5A-D2 (light blue line), 7 μm NS5BΔ21 + 15 μm NS5A-D2 (medium blue line), or 7 μm NS5BΔ21 + 50 μm NS5A-D2 (dark blue line), respectively. c, interaction with NS5A-D2. 10 μm 6FAM-RNA16 (dotted black line) was mixed with 5 μm (light blue line), 15 μm (medium blue line), or 50 μm (dark blue line) of NS5A-D2, respectively. d, inhibition of the interaction between NS5BΔ21 and RNA16 by filibuvir. 10 μm 6FAM-RNA16 (dotted black line) was mixed with either 10 μm NS5BΔ21 (green line, as in a) or 10 μm NS5BΔ21–filibuvir (light blue line). The respective elution volumes of isolated RNA-16, NS5A-D2, NS5BΔ21, and NS5BΔ21–filibuvir were monitored by UV at 260/280 nm and are indicated above the chromatograms. mAU, milliabsorbance units.
Figure 3.
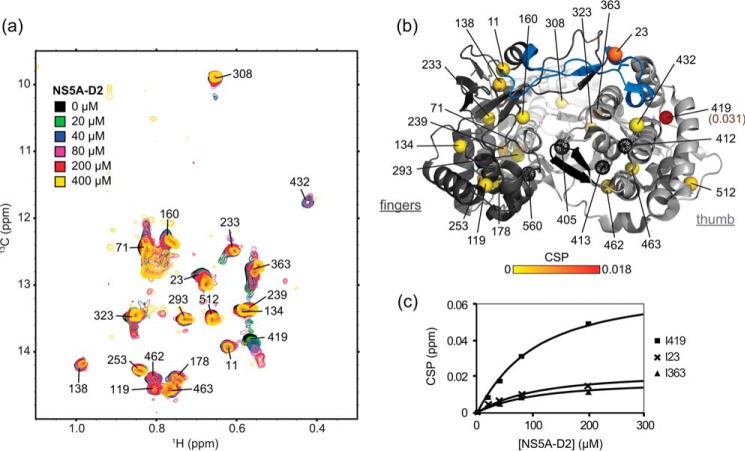
NS5A-D2 interacts with NS5BΔ21. a, superposition of methyl-TROSY spectra of U-15N,2H-,Ile δ1-13C1H3-labeled NS5BΔ21 (40 μm) recorded in the presence of increasing concentrations (0, 20, 40, 80, 200, and 400 μm) of unlabeled NS5A-D2. b, 1H,13C combined CSPs observed in the NS5B–NS5A-D2 (1:2 molar ratio in a) spectrum mapped, from yellow (0 ppm) to red (0.018 ppm), onto the Ile δ1 methyl groups of the NS5B structure (PDB code 2XXD). The CSP for Ile-419 is 0.031 ppm (above the defined upper limit) and is highlighted. Methyl groups that were not observed are shown as black dotted spheres. The figure was prepared using PyMOL. c, titration curves corresponding to the experiments in a. 1H,13C combined CSPs observed for Ile-419, Ile-23, and Ile-363 residues in NS5BΔ21 are plotted as a function of increasing concentrations of NS5A-D2. Corresponding dissociation constants were obtained by fitting the experimental data to the theoretical equation corresponding to a reversible complex.
NS5B is targeted by two types of DAAs (19): nucleoside/nucleotide inhibitors that act as substrate analogues at the active site of the polymerase during the enzymatic reaction and non-nucleoside inhibitors (NNIs, also termed allosteric inhibitors) that bind away from the active site (36). At least four NNI-binding sites have been identified on NS5B: thumb site I (NNI-1), thumb site II (NNI-2), palm site I (NNI-3), and palm site II (NNI-4) (37). Despite the numerous crystallographic structures of NS5B bound to NNIs that are available and the wealth of data they have generated, the precise mechanism(s) of action of these NNIs remains to be elucidated. Because it has been proposed that NNIs interfere with the conformation and/or dynamics of NS5B (30, 36, 38–41), experimental techniques allowing for the study of NS5B in solution (42–44) should be highly valuable to decipher the molecular mechanisms upon binding of allosteric inhibitors.
NS5A is a peculiar multifunctional phosphoprotein (45). This protein is required for viral RNA replication (12, 13), is involved in infectious particle production (14), and has been shown to modulate numerous viral and host processes (46, 47), but so far no enzymatic activity has been identified. NS5A has RNA-binding properties and establishes a large number (>100) of interactions with cellular and viral proteins (48). NS5A is anchored to the ER membrane via an N-terminal amphipathic α-helix and has three cytoplasmic domains (D1, D2, and D3) interconnected by two low-complexity sequences (LCS-1 and -2). NS5A-D1 is well structured and is mostly responsible for the RNA-binding properties of the protein (49, 50). Indeed, its first crystallographic structure revealed a homodimer with a basic groove that may be suitable for RNA binding (12). Later crystallographic studies highlighted different homodimer organizations for NS5A-D1 (51, 52). In contrast, NS5A-D2 and -D3 have been shown to be mainly intrinsically disordered (53–57) despite the presence of some (residual) structures as well as weak long-range tertiary contacts (58). Using different biophysical techniques, we recently proposed an overall structural model of HCV NS5A (59). NS5A-D1 and -D2 are mandatory for RNA replication (13), whereas NS5A-D3 is involved in the production of new virions (14, 60). One of the main functional roles of NS5A may be to guide the viral RNA all along the HCV replication process, from synthesis to encapsidation. However, NS5A also exerts biological functions through protein-protein interactions. Its intrinsically disordered domain 2 (NS5A-D2) is of particular interest here. We and others have shown that NS5A-D2 interacts with cyclophilin A (CypA) (61–63), a host factor involved in viral replication. Recently, we identified in NS5A-D2 a small conserved structural motif, termed PW-turn, which is essential for HCV RNA replication and is involved in CypA binding (64). Moreover, we have shown that the CypA-binding region in NS5A-D2 (residues 306–323) is also involved in the direct interaction with NS5B (65). This result was later confirmed by Ngure et al. (66). Using NMR spectroscopy, we have identified, at the per-residue level, three NS5A-D2 regions (residues 250–262, 274–287, and 306–333) that interact with the HCV polymerase. Concomitantly, exploiting the first 1H-15N NMR spectrum of NS5BΔ21, we have proposed that the NS5A-D2–binding site on NS5BΔ21 overlaps at least partially with that of a thiophene-2–carboxylic acid inhibitor (NNI-2) (65). The interaction of NS5B with NS5A has long been identified as crucial for viral replication (67–69), but its functional consequences remain to be deciphered. Indeed, NS5A has been shown to either stimulate (43, 69, 70) or inhibit (66, 69, 71, 72) NS5B polymerase activity. Because molecular data describing the NS5B–NS5A-D2 complex are presently scarce, mainly due to the disordered nature of NS5A-D2 and the rather low affinity of the complex, the functional consequences for NS5B of NS5A-D2 binding could not be explained by structural data until now.
In this work, we addressed the functional and structural consequences of NS5A-D2 binding to NS5BΔ21. To achieve this, we performed the first detailed NMR spectroscopic study of NS5BΔ21, using selective labeling of δ1 methyl groups of isoleucine residues. We first characterized the behavior of the HCV polymerase in solution at the molecular level. Second, we studied the interaction of NS5BΔ21 with NS5A-D2 and mapped the region of the polymerase involved in this interaction. Then we found a perturbation of the RNA-binding properties of NS5BΔ21 upon NS5A-D2 binding. Finally, to understand the molecular basis of this observation, we performed an NMR study of NS5BΔ21 bound to filibuvir, a known allosteric inhibitor (NNI-2), and monitored its effects on the interaction with both RNA and NS5A-D2. The similarities between the effects of NS5A-D2 and filibuvir upon NS5BΔ21 binding show that the former acts as an allosteric regulator of the HCV polymerase.
Results
Ile δ1 methyl groups as probes for the NMR analysis of the HCV polymerase NS5B
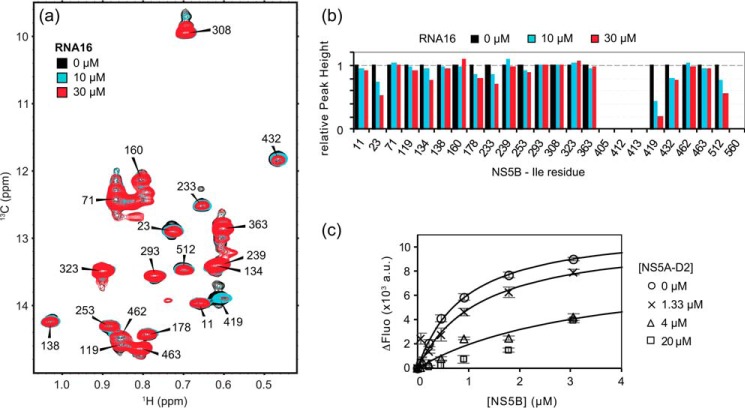
Initially, we used the classical approach to study HCV NS5BΔ21 (strain JFH-1, genotype 2a) by NMR spectroscopy (i.e. recording a 1H,15N HSQC spectrum of the protein and assigning it via sequential connectivity in a set of 3D experiments). Using a triply 2H,15N,13C-labeled NS5BΔ21 sample combined with a TROSY pulse sequence (73) and a high-field 900-MHz spectrometer equipped with a cryoprobe, we obtained a heteronuclear spectrum of good quality (65). However, due to the high molecular mass of the polymerase (65 kDa), its limited solubility (∼90 μm), and finally its requirement of highly saline buffers, we did not obtain 3D 1H,15N,13C NMR spectra of sufficient quality (low signal/noise ratio) for sequential resonance assignment. We then resorted to selective labeling of methyl groups and methyl-TROSY NMR spectroscopy (74, 75), an approach that has successfully been used to study a growing number of large protein assemblies. Because the 24 isoleucine residues of NS5B are well distributed all over the polymerase structure, we selectively labeled the δ1 methyl groups (13C1H3) (76) of the isoleucine side chains in NS5BΔ21. The 1H,13C TROSY HMQC (methyl-TROSY) spectrum acquired at 900-MHz 1H Larmor frequency on a 90 μm U-15N,2H-, Ile δ1-13C1H3-labeled NS5BΔ21 sample exhibits 20 mostly well-resolved resonances out of the 24 expected ones (Fig. 1a). To obtain site-specific information, we assigned the NMR resonances via the combined use of point mutations, distance measurements, and chemical shift predictions based on crystal structures. First, we produced 14 single-amino acid (Ile to Val) mutants of NS5BΔ21 and compared their respective methyl-TROSY spectra with that of NS5BΔ21 WT (see supplemental Fig. S1). This allowed for the assignment of 10 Ile δ1 resonances of NS5BΔ21. Strikingly, the NMR spectra of the I405V, I412V, I413V, and I560V NS5B mutants contain the same number of resonances as that of the WT, showing that the resonances of residues Ile-405, Ile-412, Ile-413, and Ile-560 are broadened beyond detection in the NS5BΔ21 spectrum. To assign the remaining resonances, we recorded 3D NOESY-HMQC and 3D HMQC-NOESY-HMQC spectra and compared the derived intermethyl distances with those measured in the crystallographic structure of NS5BΔ21 (PDB code 2XXD, in a closed conformation) (77) (see supplemental Fig. S2 and Table S1). Chemical shift predictions performed with ShiftX2 (78) on this crystallographic structure were used to resolve cases of ambiguous assignments. The assignment of the 20 Ile δ1 methyl resonances of HCV NS5BΔ21 (strain JFH-1) has been deposited in the Biological Magnetic Resonance Data Bank (BMRB accession number 27073; see supplemental Table S2).
Figure 1.
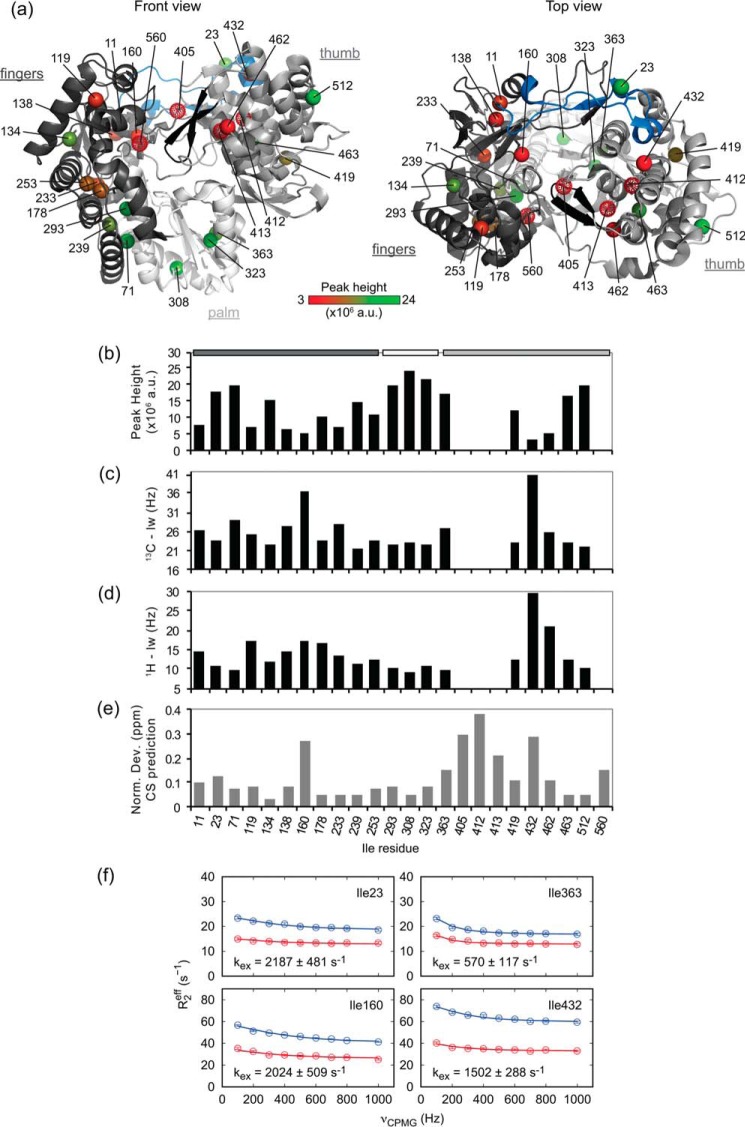
NMR analysis of the HCV RNA-dependent RNA-polymerase. a, 1H,13C 2D methyl TROSY spectrum of U-15N,2H-, Ile δ1-13C1H3-labeled NS5BΔ21 from the HCV JFH1 strain recorded at 900 MHz. b, structure of NS5B (PDB code 2XXD) shown in a schematic representation with its three subdomains: fingers (in black, residues 1–188 and 225–284), palm (in white, residues 189–224 and 285–358), and thumb (in gray, residues 359–530). The δ1 methyl group of each Ile residue is shown as a sphere and is colored green if the corresponding resonance is observed in the NMR spectrum in a and in red if not. Figures were prepared using PyMOL (PyMOL Molecular Graphics System, version 1.8, Schrödinger, LLC, New York).
The analysis of the spectra of single Ile-to-Val NS5BΔ21 mutants (see supplemental Fig. S1) used for resonance assignment highlights both short- and long-range perturbations induced by the mutations in the HCV polymerase (see supplemental Table S3). Amino acid point substitutions are expected to influence their direct chemical environment, roughly in a sphere of 5 Å. However, Ile to Val amino acid substitution at position 23, close to the apex of the fingers loop Λ1, induces spectral perturbations on the resonances of Ile-432 and Ile-160, which are at distances of 16.6 and 30.5 Å from the substitution, respectively, in the crystallographic structure (PDB code 2XXD, closed conformation) (Fig. 1b). Ile-432 is located on the edge of the hydrophobic pocket at the top of the thumb subdomain, where the apex of the fingers loop Λ1 makes extensive contacts. This interaction is important for the stabilization of the closed conformation of NS5B and is abolished in the presence of thumb site I inhibitors (79). Ile-160 is located in the fingers, and its side chain is oriented toward the RNA-binding site. It is thus facing the thumb subdomain. When Ile-413, for which no peak was detected in the spectrum, is substituted by valine, NMR signal perturbations are induced in residues Ile-462, -432, -419, and -160, which are 4.7, 18.1, 20.3, and 22.2 Å away, respectively. Altogether, the data suggest that the different subdomains of NS5B are connected via a conformational and/or dynamical network that can be probed using the assigned methyl-TROSY spectrum of the polymerase.
Ile δ1 methyl resonances as probes for the dynamics of NS5B
When analyzing the intensities of the resonances in the methyl-TROSY spectrum of NS5BΔ21 with respect to the position of the corresponding residues in the structure of NS5B, we clearly observed some correlations. The four isoleucine residues for which no NMR resonances could be observed are all located at the interface between the thumb and the fingers subdomains of the polymerase (Fig. 1b) and in direct proximity of the β-loop (residues 443–455) that protrudes from the thumb into the active site. Residue 405 has previously been identified as a molecular switch involved both in the stabilization of a closed NS5B conformation for de novo initiation and in the opening of the polymerase for subsequent elongation (80). Residues 412 and 413 are located in the same secondary structure element (Helix P (25)) in the thumb, whereas Ile-560 belongs to the C-terminal linker that folds back into the active site of NS5B. Furthermore, the NMR resonances with the weakest intensities are those surrounding the RNA-binding cleft, at the interface between the fingers and the thumb (Fig. 2, a and b), whereas the most intense NMR resonances correspond to residues in the palm and on the outside face of the fingers and thumb subdomains. As expected, the peak intensity variations correlate with line broadening in both 1H and 13C dimensions (Fig. 2, c and d).
Figure 2.
NMR properties of Ile methyl groups in NS5B. a, structure of NS5B (PDB code 2XXD) shown in a schematic representation (as in Fig. 1). Ile δ1 methyl groups, shown as spheres, are colored from red to green according to peak heights of the corresponding resonances in the methyl-TROSY spectrum (shown in b). Methyl groups that are not observed are shown as red dotted spheres. Two important structural features of NS5B are highlighted: the Λ1 loop (fingertips; residues 12–44) in blue and the β-flap (residues 443–454) in black. The figures were prepared using PyMOL. b–d, peak height (b) and line width at half-height in the 13C dimension (13C lw) (c) and in the 1H dimension (1H lw) (d) of Ile methyl (δ1) groups in the methyl-TROSY spectrum of NS5BΔ21. NS5B subdomains are indicated at the top with the same color code as in a. e, normalized deviations (Equation 3) of 1H,13C combined chemical shift predictions of Ile methyl (δ1) groups calculated with SHIFTX2 on available crystallographic structures of NS5B from the HCV JFH1 strain (see supplemental Table S4 and Fig. S3). f, methyl-TROSY MQ CPMG relaxation dispersion data for NS5BΔ21 Ile residues 23 (fingertips), 160 (fingers), 363 (palm), and 432 (thumb). Red, 600 MHz; blue, 900 MHz. Exchange rates kex fitted for each individual residue are indicated in the panels. Residues 23 and 432 were fitted with 1H and 13C Δω allowing to vary; residues 160 and 363 were fitted allowing only 13C Δω to vary. a.u., arbitrary units.
NMR resonances that are broadened beyond detection typically indicate peculiar conformational dynamics on the micro- to millisecond time scale (intermediate exchange) for the corresponding residues. To explore this possibility, we performed methyl chemical shift predictions (see supplemental Fig. S3), using ShiftX2 (78), on eight crystallographic structures of apo-NS5BΔ21 (JFH-1 strain) (see supplemental Table S4), of which five are in closed and three in open conformations. Interestingly, the residues with the highest normalized deviations of combined 1H and 13C chemical shift predictions correspond to those that are broadened beyond detection (Ile-405, -412, -413, and -560) or simply broadened (Ile-160 and -432) in the NMR spectrum of NS5BΔ21 (Fig. 2e). The more variable chemical shift predictions of these residues are due to significant differences in their predicted methyl chemical shifts for open and closed states of the polymerase. Whereas proton chemical shifts are highly sensitive to their chemical environment (e.g. aromatic ring currents), a major contribution to the isoleucine 13Cδ1 chemical shift is the conformation of its side chain and especially its χ2 dihedral angle (81). The two main conformations of Ile χ2 (i.e. trans (180°) and gauche (−60°)) can be distinguished based on their extreme 13Cδ1 chemical shift values of 14.8 and 9.3 ppm, respectively. We extracted the χ2 dihedral angles of isoleucine residues in the eight crystallographic structures that were used for chemical shift predictions (see above). Most isoleucine residues adopt the trans conformation or a mix of trans and other conformations, whereas Ile-308 and -432 are always in gauche conformation (see supplemental Fig. S4). This is consistent with their low 13Cδ1 chemical shift values (9.55 and 11.80 ppm, respectively) in the methyl-TROSY spectrum (Fig. 1a). Isoleucine residues 160, 405, and, to a lesser extent, 23 and 432, have different χ2 conformations, depending on the closed or open state of NS5B. The side chain of Ile-405 is in the gauche conformation in the closed polymerase, whereas it is in trans in the open structures. Thus, the difference between its 13Cδ1 chemical shift values (Δω) in closed and open states of the polymerase is predicted to be large. Dynamic exchange between these states could then explain why its resonance is broadened beyond detection in the methyl-TROSY spectrum. Isoleucine residues 412 and 413 have similar χ2 conformations regardless of the conformational state of NS5B, in agreement with the similar 13Cδ1 chemical shift predictions for these states. On the other hand, the 1H chemical shift predictions for these two residues are significantly different, depending on the conformation of NS5B. Indeed, Ile-412 and -413 are surrounded by aromatic residues (Trp-408, Tyr-415, Trp-420, Phe-429, Phe-430, Phe-445, Tyr-452, Trp-550, and Phe-551), and their relative orientations and/or distances differ between open and closed conformations of NS5B. This observation suggests that their resonances are not visible in the methyl-TROSY spectrum due to chemical exchange on the micro- to millisecond time scale between two states with a large 1H frequency difference. Because Ile-560 is not visible in the electronic density of the open NS5B structures, no chemical shift predictions or χ2 dihedral angle values could be obtained for this residue in the open polymerase. However, already in the closed structures, the Ile-560 side chain can adopt either trans or gauche conformations with their respective extreme 13C chemical shift values, which could explain the broadening of its resonance. In addition, the fact that this residue is invisible in crystallographic data of open NS5B suggests that it undergoes structural changes between open and closed states of the polymerase.
Altogether, these results indicate that residues Ile-23, -160, -405, -412, -413, -432, and -560 should be sensitive to the open-closed dynamics of NS5B in NMR experiments. Comparisons of experimental NS5BΔ21 13Cδ1 chemical shifts with those predicted for either all (8), open (3), or closed (5) NS5B structures show that for all residues in the thumb subdomain, and also for Ile-363, which is in the hinge with the palm, the experimental chemical shift values are closer to predictions corresponding to NS5B in a closed state (see supplemental Fig. S5). In contrast, the data for Ile-160 would rather argue in favor of an open conformation. Nevertheless, these observations suggest that, in solution, the polymerase mainly populates a closed conformation but is highly dynamic and also samples open states. Moreover, it appears that it is mostly the thumb subdomain that is affected along this dynamic transition (residues 405, 412, 413, 432, and 560), as well as residues in the fingers subdomain at the interface with the thumb (residues 23 and 160; see supplemental Figs. S3–S5).
Micro- to millisecond time scale dynamics in NS5B
Differences in 1H and 13C line widths were observed for several resonances of NS5BΔ21 in methyl-TROSY experiments acquired at 600- and 900-MHz 1H Larmor frequency (see supplemental Fig. S6). This strongly suggests the presence of a dynamic process on the micro- to millisecond time scale. To further characterize the dynamics of NS5BΔ21 in solution, we performed methyl-TROSY 13C-1H multiple-quantum (MQ) Carr-Purcell-Meiboom-Gill (CPMG) relaxation dispersion experiments. This type of NMR experiment is sensitive to dynamics on the micro- to millisecond time scale and allows for extraction of the exchange rate involved as well as the populations of and the chemical shift differences between the exchanging states (82). Statistically significant non-flat dispersion curves indicative of micro- to millisecond time scale exchange were observed for 7 of the 20 visible Ile δ1 resonances (Fig. 2f; see supplemental Fig. S7). These resonances correspond to residues in many different sites of the polymerase. Ile-71, -160, and -233 belong to the fingers subdomain; Ile-23 is in the fingertips; Ile-363 is in the palm; and, finally, Ile-432 and -462 are located in the thumb of the polymerase. Exchange rates (kex) for individual residues, fitted assuming a two-state exchange model, fall into one of three ranges, ∼600, ∼1,450, and >2,000 s−1, respectively, with no apparent correlation between kex and location on the structure (see supplemental Table S5). Populations (pB) of the minor exchanging states, as well as their chemical shift differences from the major state (Δω), are often ill-defined from the individual-residue fits. A fit of all dispersion data to a global two-state exchange model is possible, yielding an exchange rate of 1,309 ± 135 s−1. The minor state population is then found to be about 6% (6.1 ± 6.6%). 13C chemical shift changes between exchanging states are between 0.3 and 0.7 ppm; additional sizable 1H chemical shift changes are found for residues 23, 160, and 462. These results suggest that the apo-HCV polymerase may undergo global concerted motion on the millisecond time scale in solution. However, the rather large increase in the reduced χ2 measure of goodness of fit for the global compared with the individual-residue fit (see supplemental Table S5), as well as the large variation in individual-residue kex values, instead indicate that different dynamic processes are at play in different residues. Comparison of kex and Δω values places these processes in the intermediate-to-fast time scale (kex > Δω) for most residues. Consequently, Ile residues 405, 412, 413, and 560, whose resonances are invisible in spectra, are probably involved in a slower dynamic process and/or exhibit larger chemical shift differences between the different states of the polymerase. To our knowledge, these NMR relaxation dispersion data represent the first experimental measurement of the dynamics of NS5B in solution.
Altogether, our NMR data show that, in solution, the apo-HCV polymerase is highly dynamic and probably alternates between closed and open conformations, with the closed conformation predominantly occupied. The finding that 4 of the 24 Ile resonances are broadened beyond detection, as well as the NMR relaxation dispersion data (Fig. 2f; see supplemental Fig. S7), confirm the presence of dynamic processes on the micro- to millisecond time scale.
Interaction of NS5B with NS5A-D2
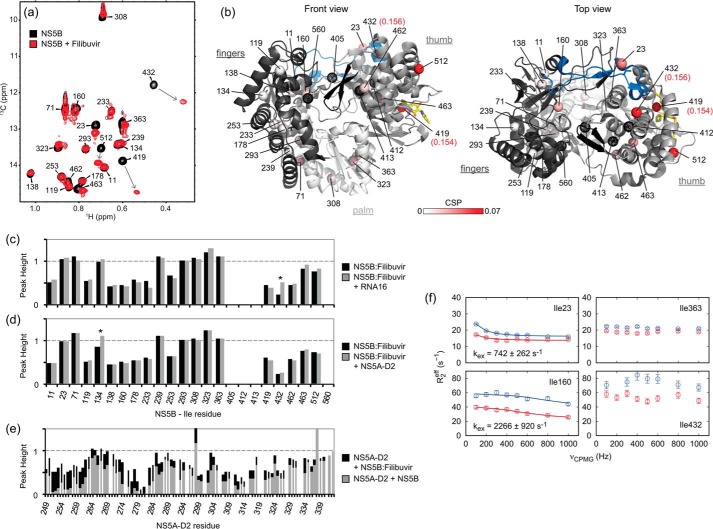
We next used the assigned methyl-TROSY NMR spectrum of NS5BΔ21 to investigate the interaction of the viral polymerase with the intrinsically disordered domain 2 of the HCV NS5A protein (NS5A-D2). HCV NS5B and NS5A are two central proteins of the viral RNA replication complex that have been shown to interact directly. The precise identification of the interaction site(s) on each protein is rather challenging due to the intrinsically disordered nature of NS5A-D2, which prevents crystallization of the complex. Previously, using the 1H,15N HSQC NMR spectrum of NS5A-D2, we identified, at the per-residue level, the three regions of NS5A-D2 (residues 250–262, 274–287, and 306–333; see Fig. 1C in Ref. 65) that are involved in the interaction with NS5B (65). In this work, we investigated the interaction between NS5B and NS5A-D2 by analyzing the methyl-TROSY NMR spectrum of NS5BΔ21 in the presence of NS5A-D2. Increasing concentrations (0–400 μm) of unlabeled NS5A-D2 were added to a 40 μm U-15N,2H-, Ile δ1-13C1H3-labeled NS5BΔ21 sample, and a methyl-TROSY spectrum was acquired for each point of the titration (Fig. 3a). Upon the addition of NS5A-D2, we observed changes in both resonance intensities and frequencies. The resonances corresponding to residues Ile-419, Ile-432, and Ile-23 were broadened. Significant 1H and 13C combined chemical shift perturbations (methyl-CSPs) were measured for residues Ile-419, Ile-23, and Ile-363 (Fig. 3, a and b). From the gradual methyl-CSPs along the titration, a dissociation constant (KD) of 80 μm was estimated for the interaction between NS5BΔ21 and NS5A-D2 in solution (Fig. 3c), a value in line with our previous study, in which we found a KD of 21 μm by surface plasmon resonance (65).
The resonance of Ile-419, which is located in the NNI-2–binding pocket of NS5B, is most affected by NS5A-D2 binding (Fig. 3) and should thus indicate the main binding site of NS5A-D2. This finding is coherent with our previous study (65), in which we proposed that the NS5A-D2–binding site at least partially overlaps with that of TCA, an NNI-2 inhibitor. The residues whose resonances are perturbed upon NS5A-D2 binding are rather widely distributed in the polymerase structure. Residues Ile-419 and Ile-432 are in the thumb, Ile-23 is in the fingers loop Λ1, and Ile-363 is in the palm (in the hinge with the thumb domain) (Fig. 3b).
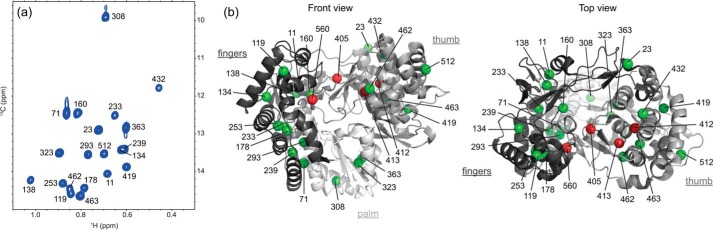
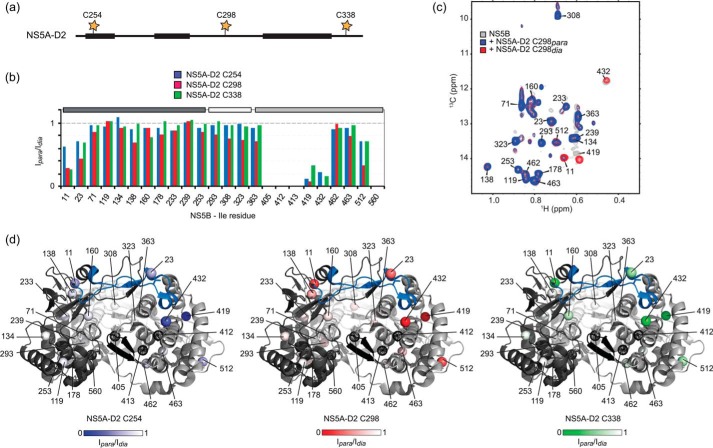
Taking into account the possibility that NS5BΔ21 spectral perturbations upon NS5A-D2 binding may be triggered by direct short-range contacts and/or by long-range induced conformational changes, we additionally performed intermolecular NMR paramagnetic relaxation enhancement (PRE) experiments (Fig. 4) (83). Paramagnetic broadening of NMR signals, induced by an unpaired electron, is proportional to the 〈r−6〉 distance between the paramagnetic label and the nucleus under study. The paramagnetic spin label (nitroxide) was incorporated in the cysteine side chain of three different NS5A-D2 single cysteine mutants (Fig. 4a). Each of these spin-labeled NS5A-D2 constructs (C254, C298, and C338) was added to Ile δ1-13C1H3-labeled NS5BΔ21 in a 1:1 ratio. Then methyl-TROSY spectra were acquired in the absence (paramagnetic sample) and in the presence of vitamin C (diamagnetic sample). A ratio of peak intensities in paramagnetic and diamagnetic conditions (Ipara/Idia) < 1 indicates NS5B residues that are close (up to ∼25 Å (84)) to the paramagnetic center (Fig. 4, b and c). Regardless of the spin label position on NS5A-D2, the maximal PREs were measured on Ile-419 and Ile-432 resonances as well as, to a lesser extent, on those of Ile-11, Ile-23, and Ile-512. The PRE effects following binding of each NS5A-D2 mutant were mapped onto the NS5B structure. The residues affected upon binding of NS5A-D2 are essentially located in the thumb and in the fingers loop Λ1 of the HCV polymerase (Fig. 4, b and d). No significant PREs were observed for residues surrounding the RNA–binding site or for those in the palm. Ile-419 and Ile-432, for which a drastic PRE was observed, are located in the binding sites of thumb site II and thumb site I allosteric inhibitors (NNI-2 and NNI-1), respectively, and are ∼20 Å away from each other. Comparison of PRE data acquired with the spin label attached to either the N-terminal, the central, or the C-terminal region of NS5A-D2 did not allow us to define a unique mode of binding of this disordered domain to NS5B. However, given the dynamics of the disordered NS5A-D2 chain as well as the fact that three different regions of NS5A-D2 (residues 250–262, 274–287, and 306–333), spanning about half its length, are implicated in NS5B binding (65), the observation of binding-related PRE effects in different parts of NS5B should not come as a surprise. There may well be more than one binding mode, possibly with exchange between different modes (i.e. NS5A-D2 and the polymerase may form a fuzzy complex) (85). Nevertheless, the Ile-11 and Ile-23 resonances are less broadened when the spin label is at position 254 in NS5A-D2 (Fig. 4, b and d), suggesting a preferred orientation of NS5A-D2 in which its N-terminal region is, on average, further away from the fingers loop Λ1 than from the other affected regions. The observation of a relatively strong PRE on Ile-11 and a much weaker one on the nearby Ile-138 may be explained if NS5A-D2 binding to the thumb I site, similar to inhibitor binding at this site, displaces the fingers loop Λ1 containing Ile-11. Taken together, our solution NMR data show that the thumb II site as well as the thumb I site of HCV NS5B constitute the main binding sites for NS5A-D2.
Figure 4.
Localization of the NS5A-D2 binding site on NS5BΔ21. a, schematic representation of NS5A-D2. Cysteine residues (one in each of the mutants C254, C298, and C338) labeled with a paramagnetic relaxation agent (3-(2-iodoacetamido)-PROXYL) are indicated by yellow stars. The three NS5A-D2 regions found to interact with NS5B are indicated by black bars. b, ratios of methyl-TROSY peak intensities (Ipara/Idia) of Ile methyl (δ1) groups of NS5BΔ21 measured in the presence of paramagnetic and diamagnetic versions of each of the NS5A-D2 mutants C254 (blue), C298 (red), and C338 (green), respectively. NS5B subdomains are indicated at the top with the same color code as in Fig. 1b. c, superposition of methyl-TROSY spectra of 13Cδ11H3-Ile, U-2H-labeled NS5BΔ21 (72 μm) alone (gray) and with equimolar amounts of paramagnetic (blue) and diamagnetic (red) NS5A-D2 C298. d, PRE results (Ipara/Idia relative intensities) obtained with NS5A-D2 C254, NS5A-D2 C298, and NS5A-D2 C338 mapped onto the NS5B structure (PDB code 2XXD). Methyl groups that were not observed are shown as black dotted spheres. Figures were prepared using PyMOL.
NS5A-D2 binding decreases the thermal stability of NS5B
When increasing concentrations of NS5A-D2 were added in the titration experiment (Fig. 3), we observed small quantities of white precipitate in the NMR tube. We thus tested the thermal stability of NS5BΔ21 in the absence and the presence of NS5A-D2 using differential scanning fluorimetry (DSF) (86). In the absence of NS5A-D2, the melting temperature (Tm) was 41.4 °C. The addition of NS5A-D2 shifted the Tm toward lower temperatures (see supplemental Fig. S8). The Tm of NS5BΔ21 (30 μm) was 38.5, 36.7, and 36.1 °C in the presence of 30, 60, and 180 μm NS5A-D2, respectively. NS5A-D2 thus seems to decrease the overall stability of NS5BΔ21. Alternatively, it may stabilize a less stable open conformation of the polymerase.
NS5A-D2 interferes with the RNA-binding properties of NS5B
Using the methyl-TROSY spectrum of Ile δ1-13C1H3-labeled NS5BΔ21, we monitored the interaction of the HCV polymerase with a 16-mer RNA oligonucleotide (RNA16) (Fig. 5, a and b). To do so, we had to find a buffer composition saline enough to keep NS5BΔ21 soluble at working NMR concentrations (i.e. ∼50 μm) but still compatible with a nucleic acid interaction. This was achieved using R/E buffer (100 mm arginine, 100 mm glutamic acid, pH 7.0, 100 mm NaCl, 2 mm MgCl2, 1 mm EDTA, 4 mm THP). Unfortunately, the residues for which the largest effect upon RNA binding would be expected (i.e. Ile-405, -412, -413, and -560 near the RNA-binding cleft) are not visible in the methyl-TROSY spectrum of NS5BΔ21, and this did not change upon the addition of RNA16. Nevertheless, the addition of increasing concentrations of RNA16 resulted in a progressive reduction in peak intensity of several NS5B resonances. Residues whose peak intensity is reduced in the presence of RNA16 are Ile-419, Ile-432, and Ile-512 in the thumb and Ile-23, Ile-134, Ile-178, and Ile-233 in the fingers (Fig. 5, a and b).
Figure 5.
Interaction of NS5BΔ21 with an RNA oligonucleotide and effect of NS5A-D2. a, superposition of methyl-TROSY spectra of U-15N,2H-,Ile δ1-13C1H3-labeled NS5BΔ21 (50 μm) recorded in the presence of increasing concentrations (0, 10, and 30 μm) of RNA16. b, ratios of peak heights of Ile methyl (δ1) resonances in methyl-TROSY spectra of NS5BΔ21 obtained in the presence and absence of different concentrations of RNA16. Peak heights in individual spectra were first normalized to that of the invariant Ile-293 resonance to correct for the observed sample precipitation over the course of the titration. c, titration of a fluorescent RNA oligonucleotide (6FAM-RNA16, 50 nm) with increasing concentrations of NS5BΔ21. The increase in fluorescence intensity (515–525 nm) of 6FAM-RNA16 is plotted as a function of NS5BΔ21 concentration. The titration experiment was performed in the absence and in the presence of NS5A-D2 (0, 1.33, 4, and 20 μm). Error bars, one S.D. value of three replicates. Corresponding dissociation constants were obtained by fitting the experimental data to the theoretical equation corresponding to a reversible complex.
Some of the resonances that are perturbed in the presence of RNA-16 are outside of the RNA-binding cleft of NS5B and were also shifted upon interaction with NS5A-D2 (Fig. 3). Thus, we hypothesized that NS5A-D2 might regulate the RNA-binding properties of NS5B. We performed titration experiments with NS5BΔ21 on a RNA16 oligonucleotide with a fluorescent 6-carboxyfluorescein (6-FAM) label at its 5′ end (6FAM-RNA16) (87) and monitored the change in fluorescence intensity in the absence and in the presence of NS5A-D2. These assays were performed in the same R/E buffer as used in the NMR experiments (see above). The data show that NS5A-D2 lowers the affinity of the interaction between NS5BΔ21 and RNA (Fig. 5c). By fitting the experimental fluorescence data with a quadratic equation, KD values of 0.8, 1.2, and 3.3 μm were estimated in the absence and in the presence of 1.33 and 4 μm of NS5A-D2, respectively, representing a >3-fold decrease in the affinity of NS5BΔ21 for RNA over this range of NS5A-D2 concentrations. No KD value was extracted from the experiment performed in the presence of 20 μm NS5A-D2 because these fluorescence data did not follow a regular quadratic KD curve (Fig. 5c). Control experiments, using both fluorescence and NMR spectroscopy, show that NS5A-D2 does not directly interact with RNA16 in our experimental conditions (see supplemental Fig. S9). Thus, the possibility that NS5A-D2 reduces the apparent binding affinity of NS5BΔ21 for RNA16 via a direct competition for RNA16 was excluded.
Taking advantage of the absorbance of 6FAM-RNA16 at 495 nm, we also analyzed its interaction with NS5BΔ21 in the presence and absence of NS5A-D2 using analytical size exclusion chromatography (SEC). NS5BΔ21 efficiently binds 6FAM-RNA16 to form the expected complex (Fig. 6a). The elution volume (Ve) of this complex (Ve = 1.13 ml) is lower than that of NS5BΔ21 alone (Ve = 1.26 ml), showing an increase in hydrodynamic radius. NS5A-D2 triggers a dose-dependent reduction of NS5BΔ21–6FAM-RNA16 complex formation (Fig. 6b) and does not directly interact with 6FAM-RNA16 (Fig. 6c). The data obtained by SEC are thus in full agreement with the results of the fluorescence assays and indicate that the affinity of the polymerase for RNA is reduced in the presence of NS5A-D2 (Figs. 5c and 6b).
Interaction of NS5B with filibuvir, an allosteric inhibitor
Our data so far have shown that the thumb site II of NS5B is a main site for interaction with NS5A-D2 and that this interaction can interfere with the RNA-binding properties of the polymerase, possibly via an allosteric network. To consolidate this finding, we next investigated the binding of filibuvir, a thumb site II inhibitor, to NS5B and examined its effect on the interaction between NS5B and either RNA or NS5A-D2.
Filibuvir (88) has potent in vitro activity against HCV replicons and decreased the viral RNA load in a phase 2 clinical trial (89, 90). Both its profile of NS5B resistance mutations (91) and its crystallographic structure bound to the polymerase (PDB code 3FRZ (89)) have shown that filibuvir is an allosteric inhibitor (NNI-2) that binds to NS5B in its thumb site II. In the context of the crystal, the inhibitor does not cause significant changes in the conformation of NS5B. Here, we used NMR spectroscopy to characterize, in solution, the binding of filibuvir to NS5BΔ21 and its allosteric effects. Filibuvir, as a powder, was added in large excess to a δ1-13C1H3-labeled NS5BΔ21 sample (∼80 μm). Then, a methyl-TROSY spectrum was acquired and compared with that obtained in the absence of the inhibitor (Fig. 7). Upon binding, filibuvir induces both peak intensity changes and chemical shift perturbations. Resonances corresponding to Ile-419, Ile-432, and Ile-512 are broadened (intensity ratios of 0.29, 0.73, and 0.74, respectively), whereas those of Ile-23, Ile-71, and Ile-363 are more intense (ratio of 1.14, 1.35, and 1.26, respectively) in the presence of the inhibitor. The two largest methyl-CSPs were measured for the resonances of Ile-419 and Ile-432 (Fig. 7, a and b). A second set of resonances exhibits smaller, but still significant, CSPs (residues 23, 71, 160, 308, 323, 363, 462, 463, and 512). The main perturbations were detected in the thumb subdomain, but some are also present in both fingers (Ile-23, -71, and -160) and palm (Ile-308, -323, and -363).
Figure 7.
NS5B and its inhibition by filibuvir. a, superposition of methyl-TROSY spectra of NS5BΔ21 (in black) and the NS5BΔ21–filibuvir complex (in red). b, 1H,13C combined CSPs following filibuvir binding are mapped, from white (0 ppm) to red (0.07 ppm), onto the Ile methyl groups of the NS5B structure (PDB code 2XXD). CSPs for Ile-419 and Ile-432 are 0.154 and 0.156 ppm (above the defined upper limit) and are highlighted. Methyl groups that were not observed are depicted as black dotted spheres. Figures were prepared using PyMOL. c, effect of filibuvir on the NS5B interaction with RNA16. Relative peak heights of Ile methyl (δ1) resonances in 1H,13C TROSY spectra of the NS5BΔ21–filibuvir complex (50 μm) obtained in the absence and in the presence of RNA16 (30 μm) are shown. For each spectrum, peak intensities are normalized to the Ile-293 signal. d and e, effect of filibuvir on the NS5B interaction with NS5A-D2. d, relative peak heights of Ile methyl (δ1) resonances in 1H,13C TROSY spectra of the NS5BΔ21–filibuvir complex (50 μm) obtained in the absence and in the presence of NS5A-D2 (50 μm) are shown. For each spectrum, peak intensities are normalized to the Ile-293 signal. e, relative peak heights of backbone amide resonances in 1H,15N HSQC spectra of 15N-labeled NS5A-D2 (20 μm) in the presence of unlabeled NS5BΔ21 (100 μm; gray bars) or unlabeled NS5BΔ21–filibuvir complex (100 μm; black bars), normalized to peak heights in the absence of NS5B. f, methyl-TROSY MQ CPMG relaxation dispersion data for NS5BΔ21 Ile residues 23 (fingertips), 160 (fingers), 363 (palm), and 432 (thumb) in the presence of filibuvir. Red, 600 MHz; blue, 900 MHz. For residues 23 and 160 exhibiting non-flat dispersion curves, exchange rates (kex) fitted individually are indicated in the panels. Fits were performed allowing only 13C Δω to vary.
According to crystallographic data, Ile-419, which is located in the thumb site II, is the only isoleucine residue directly involved in the interaction with filibuvir (NNI-2), via hydrophobic contacts (Fig. 7b). The second most affected resonance corresponds to the δ1-methyl of Ile-432, which is at a distance of about 19 Å from the nearest atom of the inhibitor. Ile-432 is part of the thumb site I of NS5B, known to bind NNI-1. Thus, our NMR data show that the binding of an inhibitor to the NNI-2–binding site induces conformational and/or dynamical perturbations in the NNI-1–binding site. This communication could propagate via the α-helix Q (according to the nomenclature in Ref. 25), which contains both Ile-419 and Ile-432. Helix T may also play a role in this communication network, as a significant NMR perturbation was detected on the resonance of Ile-512, although it is at a distance of >16 Å from filibuvir in the crystallographic structure. In the NS5B structure, helix T is situated between the NNI-2– and NNI-1–binding sites. Upon binding of filibuvir to the NNI-2 site, the allosteric communication with the NNI-1 site triggers modifications in the behavior of the fingers loop Λ1, as illustrated by the NMR perturbations of the Ile-23 resonance (Fig. 7, a and b). As the fingers loop Λ1 is directly involved in the open/closed dynamics of the polymerase (25, 35), the perturbation of this structural element might explain the effects that were observed upon filibuvir binding on residues in the fingers (Ile-71 and Ile-160) and palm subdomains (Ile-308, Ile-323, and Ile-363), all of which are >26 Å away from the NNI-2 site.
To quantify the exchange dynamics of NS5B in the presence of the inhibitor, we performed methyl-TROSY MQ CPMG relaxation dispersion experiments on a filibuvir-bound NS5BΔ21 sample (Fig. 7f; see supplemental Fig. S7) and compared the data with those obtained on the apo-polymerase. In apo-NS5BΔ21, seven resonances exhibit micro- to millisecond time scale exchange detectable by CPMG relaxation dispersion (see Fig. 2f). In contrast, in the presence of the inhibitor, statistically significant dispersion was only observed for three of these residues, namely Ile-23, -71, and -160, all of which are in the fingers subdomain, with Ile-23 in its Λ1 loop (Fig. 7f; see supplemental Fig. S7). The exchange rates kex observed for Ile-71 and Ile-160 are not significantly different in both apo- and filibuvir-bound NS5BΔ21 (see supplemental Table S5). In contrast, Ile-23 close to the apex of the fingertips loop, for which a kex of about 2,200 s−1 was measured in free NS5BΔ21, exhibits markedly slowed exchange dynamics in the presence of filibuvir (742 ± 262 s−1). In thumb and palm subdomains, including in the hinge in between (Ile-363), micro- to millisecond time scale dynamics appear to be quenched when filibuvir is bound. Together with the slowing of Ile-23 exchange dynamics, this suggests a rigidification of the polymerase on the micro- to millisecond time scale by filibuvir binding. On the other hand, an overall increase in R2,eff values at high CPMG frequencies as well as lower signal intensities, especially for residues 233, 419, 432, and 512, suggest an increase in faster (microsecond) time scale motion in these residues, probably of restricted amplitude.
Overall, the picture that emerges is that binding of the NNI-2 filibuvir reduces the extent of conformational dynamics of the thumb subdomain in NS5B. Our solution NMR data highlight an allosteric network in NS5B that links the NNI-2 and NNI-1 sites and, via perturbation of the fingers loop Λ1, induces a more global effect throughout the polymerase.
Filibuvir impairs NS5B binding to NS5A-D2 and RNA
We next investigated whether the binding of filibuvir to NS5B, in its NNI-2 site, might have an effect on the interaction between the polymerase and either RNA16 or NS5A-D2. First, a δ1-13C1H3-labeled NS5BΔ21 sample (50 μm) was incubated with filibuvir for a few hours; then a methyl-TROSY spectrum was acquired in the absence and in the presence of RNA16 (30 μm). The only notable difference between the two spectra is a higher signal intensity for the resonance corresponding to Ile-432 (Fig. 7c). This result suggests that NS5BΔ21 does not interact with RNA16 in the presence of filibuvir. To confirm this observation, we resorted to SEC analyses. Fluorescent 6FAM-RNA16 (10 μm) was mixed either with NS5BΔ21 (10 μm) or with the preformed NS5BΔ21–filibuvir complex (10 μm), and then the sample was injected onto the column (Fig. 6d). When filibuvir is bound to the polymerase, the amount of 6FAM-RNA16 that interacts with the polymerase is strongly reduced. However, a residual interaction between NS5BΔ21 and the oligonucleotide was still observed. Nevertheless, the data show that filibuvir severely impairs the RNA-binding properties of the HCV polymerase via an allosteric mechanism.
We also compared the methyl-TROSY spectra of the NS5BΔ21–filibuvir complex (50 μm) in the absence and in the presence of unlabeled NS5A-D2 (50 μm). The spectra are nearly identical with small variations in peak intensities, notably the resonance corresponding to residue 134, which is more intense in the presence of NS5A-D2 (Fig. 7d). To confirm the apparent absence of an NS5B–NS5A-D2 interaction in the presence of filibuvir, we performed the inverse experiment. In other words, we compared the 1H,15N HSQC spectrum of 15N-labeled NS5A-D2 (20 μm) alone with that recorded in the presence of either unlabeled NS5BΔ21 (100 μm) or unlabeled NS5BΔ21–filibuvir complex (100 μm). Upon the addition of NS5BΔ21, we observed broadening of resonances corresponding to residues located in the three NS5A-D2–interacting regions (Fig. 7e) that we have previously identified (65). Upon the addition of the NS5BΔ21–filibuvir complex, broadening of these resonances was still visible, but to a significantly smaller extent (Fig. 7e). Thus, filibuvir interferes with the interaction between NS5BΔ21 and NS5A-D2. This finding is consistent with our observation that the filibuvir and NS5A-D2–binding sites of the HCV polymerase overlap. Interestingly, when filibuvir is bound to NS5BΔ21, the interaction with NS5A-D2 is primarily perturbed in the N-terminal region of this disordered domain, in agreement with the preferential orientation of NS5A-D2 on NS5B that we deduced from PRE experiments. Taken together, our data show that filibuvir severely reduces, but does not abolish, NS5B binding of the oligonucleotide RNA16 and the intrinsically disordered NS5A-D2 domain.
Discussion
NS5B is the catalytic core of the HCV RNA replication complex, as it carries the RNA-dependent RNA polymerase activity, allowing for the synthesis of new RNA molecules from a template RNA and incoming nucleotides. As such, NS5B is one of the main targets for antiviral therapy (19). Despite its importance, some of the molecular mechanisms involved in enzymatic RNA replication, from initiation to processive elongation, as well as in its inhibition, notably by allosteric inhibitors, remain elusive. This is mainly due to the fact that these processes rely on subtle conformational and dynamical changes that could not be fully resolved by X-ray crystallography, the technique that so far has yielded the most insight in structural studies of the HCV polymerase. We here performed the first detailed NMR spectroscopic study of HCV NS5BΔ21 (JFH-1, g2a) in solution, using a methyl-TROSY strategy with selective isotope labeling of δ1 methyl groups of isoleucine side chains (Fig. 1). We showed that NS5BΔ21 is in conformational exchange on the millisecond time scale in solution (Fig. 2; see supplemental Fig. S7) and mainly adopts a closed conformation (see supplemental Figs. S3–S5). We provided experimental evidence that the disordered domain 2 of the HCV NS5A protein binds to both thumb site II and thumb site I of NS5BΔ21 and induces long-range perturbations (Figs. 3 and 4), leading to reduced RNA binding by the polymerase (Figs. 5 and 6). We also characterized, by NMR spectroscopy, the short- and long-range effects triggered in NS5BΔ21 by binding of filibuvir, an allosteric inhibitor. Filibuvir binds to the thumb site II and induces chemical shift perturbations throughout NS5BΔ21 via an allosteric pathway that comprises the thumb site I and the fingertips loop Λ1 (Fig. 7). It also reduces micro- to millisecond time scale dynamics present in the polymerase, most notably in the thumb subdomain. Finally, we showed that filibuvir-bound NS5BΔ21 is strongly impaired in the binding of both NS5A-D2 and RNA (Figs. 6 and 7). Numerous similarities were observed between the binding of NS5A-D2 and that of filibuvir to NS5BΔ21: occupation of thumb site II, perturbations of thumb site I and fingertips loop Λ1, and reduction of the affinity of the polymerase for RNA. This leads us to conclude that NS5A-D2 acts as an allosteric regulator of the HCV polymerase NS5B.
Numerous studies have suggested that apo-NS5B adopts different conformations in solution. These conformations are mainly characterized by the extent of opening of the polymerase, following rotation of the thumb subdomain with respect to palm and fingers. For example, 1) a crystal structure of NS5B has revealed the presence of two conformations (30), one being more open than the other; 2) NS5B has been shown to be able to bind a circular RNA template (92), which implies opening of its encircled closed active site; and 3) molecular simulations have suggested that NS5B may sample a conformational space in which the intersubdomain angle and the RNA channel width can vary significantly (36). Recently, Deredge et al. have shown using hydrogen/deuterium exchange followed by MS that fingers, palm, and, to a lesser extent, the core of the thumb are highly exchange-protected regions of apo-NS5B (42). These regions match those for which we observe the most intense resonances in the methyl-TROSY spectrum of NS5BΔ21 (Fig. 2, a and b). In this spectrum, 4 of the 24 isoleucine δ1 methyl group resonances are missing. Such NMR signal broadening is evidence of dynamics on the micro- to millisecond time scale. The corresponding residues are Ile-405, -412, -413, and -560, all located around the β-loop at the interface between the thumb and fingers subdomains. Broadening of NMR signals of these residues is thus in line with a dynamic opening and closing mechanism of NS5BΔ21 on the micro- to millisecond time scale. Relaxation dispersion NMR experiments indeed showed the presence of dynamics on this time scale in the polymerase (Fig. 2f; see supplemental Fig. S7). At present, it is not possible to establish a formal link between these dynamics data and a well-identified conformational dynamics event in the polymerase, such as an opening/closing process. Nevertheless, the hypothesis of the presence of a dynamic open/closed equilibrium in NS5BΔ21 is also supported by the fact that experimental δ1-methyl chemical shifts do not fully match those predicted for the fully open or closed polymerase, but rather indicate a population-weighted average (Figs. 1 and 2; see supplemental Figs. S3–S5), as well as by the quenching of micro- to millisecond time scale dynamics in thumb and palm subdomains upon binding of filibuvir (Fig. 7f; see supplemental Fig. S7), which has been shown to reduce functional dynamics in NS5B (36, 41, 42).
Following our observation that in solution the equilibrium of NS5BΔ21 structures is shifted toward closed conformations, we investigated NS5B mutants that have been described to adopt a more open conformation. First, we acquired a methyl-TROSY spectrum of an Ile δ1-13C1H3-labeled NS5BΔ21 L30S mutant (supplemental Fig. S10). In the NS5B L30S mutant, which is largely inactive (93), the fingertips loop Λ1 is displaced from the thumb (94) and becomes more flexible and sensitive to proteolysis (44). This polymerase mutant has been suggested to be more open (31, 79) or alternatively to sample both open and closed conformations more rapidly (36). Second, we studied the spectrum of a truncated polymerase (NS5BΔ60) (supplemental Fig. S11) that lacks both its C-terminal membrane anchor and the preceding linker that folds back into its active site (38). Third, we analyzed the methyl-TROSY spectrum of an Ile δ1-13C1H3-labeled NS5BΔ21 sample comprising five amino acid substitutions (S15G, E86Q, E87Q, C223H, and V321I), designated NS5BΔ21 5mut (supplemental Fig. S12). The structure of this mutant (PDB code 4OBC) (34) showed a polymerase in a more open conformation, as in the crystal structure of NS5B in complex with RNA template, RNA primer, and incoming nucleotide (PDB code 4WTL) (35). From our experiments, we did not obtain an unambiguous NMR signature of an open conformation for NS5B. However, we clearly observed that the resonances corresponding to residues Ile-23, -160, -419, and -432 are always perturbed when dynamics and/or conformation of the thumb subdomain are altered.
Among the residues for which no NMR resonances were observed, Ile-405, -412, and -413 have been associated with the same functional region in a molecular dynamics study of NS5B (36) and are all located in direct proximity of the β-loop, whereas Ile-560 belongs to the C-terminal linker of the polymerase. Both of these structural motifs have been shown to be regulatory elements and are required for the inhibitory activity of thumb site II inhibitors (95).
We have shown that filibuvir, upon binding to the thumb site II of NS5B, elicits both short- and long-range perturbations in the NMR spectrum of the HCV polymerase. Indeed, we identified an allosteric network by which perturbations at the NNI-2 site can propagate throughout the polymerase via both the NNI-1 site and the fingertips loop Λ1 (Fig. 7). The allosteric communication that we observed between NNI-2– and NNI-1–binding sites could correspond to the perturbations in helix T identified in crystallographic structures of NS5B when bound to different NNI-2s (thiophene-based) (40). The identification of NNI-2 resistance-associated mutations in the NNI-2–binding site (position 419), but also in the NNI-1–binding site (positions 494 and 432), in the fingertips (19), in the palm (position 338), and in the β-loop (positions 441, 442, and 445), further supports the global NS5B perturbations that we monitored by NMR spectroscopy upon filibuvir binding (see Ref. 38 and references therein). Our NMR data are also in line with those of Deredge et al. (42), which showed that, upon filibuvir binding, several regions of NS5B, including the NNI-2 site and the fingers loops Λ1 and Λ2, were more protected from a hydrogen/deuterium exchange process. Protection at the NNI-2 site can be explained by direct binding of the inhibitor, whereas for other regions, it is linked to restriction of conformational fluctuations, depending on allosteric mechanism(s). The authors showed that NNI-2 binding reduces conformational flexibility of the fingers, the fingertips, and part of the thumb, notably the NNI-2–binding site, and that there is a correlation between conformational sampling restriction and inhibitor potency. In parallel, they also showed that neither initiation nor elongation of RNA synthesis was significantly inhibited by NNI-2s. They thus proposed that NNI-2s instead block the conformational transition required between these two steps (41).
Similarly, molecular dynamics studies have proposed that NNI-2s may restrict the conformational sampling of NS5B, leading to more closed or more open conformations (36). Our NMR relaxation dispersion data show that, in the presence of filibuvir, the thumb of the polymerase is less dynamic on the micro- to millisecond time scale, because only flat dispersion profiles were seen in this subdomain after filibuvir binding (Fig. 7f and supplemental Fig. S7). However, despite reduced NS5B dynamics in the presence of filibuvir, we did not detect any new resonances in the methyl-TROSY spectrum that might correspond to those missing in the apo-NS5BΔ21 spectrum. In the filibuvir-bound NS5BΔ21 sample, the 13C chemical shift of Ile-23 (in the fingers loops Λ1) is shifted further away from the random coil value than in apo-NS5BΔ21 (Fig. 7). Ile-23 also exhibits reduced dynamics on the micro- to millisecond time scale when filibuvir is bound (Fig. 7f and supplemental Fig. S7). This is in agreement with the idea that filibuvir stabilizes a closed conformation of NS5B (36, 41, 95) and that the fingers loop Λ1 is insensitive to proteolysis when the polymerase is bound to a thumb site II inhibitor (44).
Conflicting results have been reported about the inhibition of the NS5B polymerase activity mediated by filibuvir. This NNI-2 has been shown to either inhibit primer-dependent RNA synthesis, with no effect or only a modest effect on de novo initiation (90, 95, 96), or inhibit neither initiation nor elongation significantly, but rather block the transition between the two (41). In this study, we show that the RNA-binding capacity of NS5BΔ21 bound to filibuvir is strongly altered but not completely abolished (Fig. 6d). A similar finding has been obtained by Winquist et al. (97) using surface plasmon resonance experiments. Faster release of RNA from the elongation complex in the presence of NNI-2s has also been described (41). Overall, our experimental data are compatible with a filibuvir-mediated NS5B inhibition mechanism in which binding to the thumb site II, via an allosteric network, leads to stiffening of the thumb as well as stabilization of a closed conformation, via the fingers loop Λ1, and results in an overall less dynamic polymerase with reduced RNA-binding capacity. Based on our NMR data, involvement of the β-loop and the C-terminal linker in this process remains elusive, because no resonances corresponding to residues in these structural elements could be observed. Moreover, as we have shown here that filibuvir perturbs the interaction between NS5B and the disordered domain 2 of NS5A (Fig. 7, d and e), it is possible that, in vivo, filibuvir exerts its inhibitory effect, at least partially, through perturbation of this protein–protein interaction.
Both HCV NS5B and NS5A proteins are required for viral RNA replication and, as such, are interesting targets for antivirals. We and others have shown that these two proteins interact (65, 67–69). However, the structural and functional relationships between them still remain to be uncovered. Depending on the experimental assay used, NS5A has been reported to inhibit (66, 69, 71, 72) and/or activate (43, 69, 70) polymerase activity of NS5B. A major drawback of these in vitro studies is that the protein constructs used were devoid of their membrane anchors to make them soluble. Recently, Cho et al. (43) reported an elegant functional assay of full-length NS5B anchored in a lipid bilayer. In this context, NS5A, devoid of its N-terminal amphipathic helix, increased polymerase activity after an NS5A-dependent lag time. Moreover, NS5A-induced NS5B activation was more efficient if NS5A was added after the RNA. To understand the functional interaction of NS5B and NS5A, detailed molecular and structural data characterizing this interaction are required. Achieving this goal has been so far hampered by the highly dynamic nature of the interaction with NS5A, which contains large intrinsically disordered domains. Using NMR spectroscopy, we previously identified three regions in disordered NS5A-D2 that are involved in direct interaction with NS5B (65). In this work, we used the methyl-TROSY NMR spectrum of the NS5BΔ21 isoleucine δ1 methyl groups to identify polymerase regions involved in the interaction with NS5A-D2 (Fig. 3a), and we investigated both its conformational and functional consequences. From methyl-CSPs, a KD of 80 μm was calculated for the interaction between NS5BΔ21 and NS5A-D2 in solution (Fig. 3c). This KD value agrees well with that of ∼60 μm, which we estimated by comparison with the CypA–NS5A-D2 interaction in solution, and is close to the value of 21 μm that we obtained by surface plasmon resonance (65). In contrast, this value is significantly different from the KD of 14.5 nm reported by Ngure et al. (66) using FRET data. However, these authors also reported a KD of 67.5 nm for the CypA–NS5A-D2 interaction, which we measured as 64 μm by NMR (61). The source of this discrepancy is unknown; however, the data agree in the sense that NS5A-D2 interacts with CypA and NS5B with similar affinities.
NMR spectral perturbations and intermolecular PRE analyses show that the disordered domain 2 of HCV NS5A binds to both the thumb site II and thumb site I of NS5BΔ21 and induces long-range perturbations (Figs. 3 and 4). This is in agreement with our initial observation that the NS5A-D2–binding site on NS5BΔ21 overlaps partially with that of an NNI-2 inhibitor (65). The experimental data described here are compatible with NS5A-D2 anchored to both NNI-2– and NNI-1–binding sites and wrapped around the thumb subdomain of NS5B. Its N- and C-terminal extremities remain flexible and may transiently interact with the surrounding NS5B surface, notably with the fingers loop Λ1. Upon binding of NS5A-D2, most NMR perturbations identified are in the thumb of NS5BΔ21, whereas, using an MS footprinting approach, Ngure et al. (66) have found protected peptides mainly in the fingers subdomain. Both NMR and MS footprinting methods can lead to the identification of regions directly involved in the molecular interaction, but also of regions indirectly affected by induced conformational/dynamic changes. Our intermolecular PRE experiments (Fig. 4) allowed us to differentiate between these cases and confirmed the direct implication of both thumb site II and thumb site I of NS5B in the interaction with NS5A-D2. Moreover, we showed that filibuvir (NNI-2) interferes with the binding of NS5B to NS5A-D2 (Fig. 7, d and e). The direct involvement of the NS5B thumb subdomain is consistent with the strongly reduced interaction between NS5A-D2 and an NS5B R380E mutant that has been reported (66). Residue Arg-380 is located at the base of the thumb, ∼10 Å below thumb site II.
For the interaction between NS5A-D2 and NS5B, we identified three regions in the former and at least three distinct regions (NNI-2– and NNI-1–binding sites and the fingers loop Λ1) in the polymerase. Our NMR data do not define a unique binding mode of the two partners, which probably is not present anyway, given the dynamic nature of disordered NS5A-D2. These two viral partners may thus form a fuzzy complex (98). However, a set of concordant observations argue in favor of the N-terminal extremity of NS5A-D2 being located in proximity of the NS5B thumb site II: a reduced PRE effect on Ile-11 in the fingers loop Λ1 when the paramagnetic label is near the N terminus of NS5A-D2 (Fig. 4) and reduced broadening of signals from N-terminal NS5A-D2 residues when the thumb site II is occupied by filibuvir (Fig. 7e). Concerning the effect of NS5A-D2 on the NS5B fingers loop Λ1, we cannot be certain whether the interaction strengthens its contacts with the thumb or whether it rather displaces this loop. However, a line of evidence argues in favor of an opening of NS5B upon NS5A-D2 binding: 1) the interaction induces NS5BΔ21 spectral perturbations (Figs. 3 and 4) in the same set of residues that exhibit changes in NS5B mutants (L30S, Δ60, 5mut; see supplemental Figs. S5–S7) known to be more open; 2) in the presence of NS5A-D2, the thermal stability of NS5BΔ21 is lowered (see supplemental Fig. S8), as observed for NS5B C-terminal deletions and for NS5B bound to NNI-1s (95); 3) when increasing concentrations of NS5A-D2 were added to NS5BΔ21, a white precipitate appeared, similar to what was described by Jin et al. (32) during the formation of an active elongation complex. Nevertheless, in the presence of NS5A-D2, we did not detect any new resonance in the methyl-TROSY spectrum of NS5BΔ21 that could correspond to Ile-560. This might have been expected, given that this residue is situated in the C-terminal linker, which should be released upon opening of the polymerase, as proposed previously (35). However, it is possible that a released C-terminal linker still interacts with the remainder of NS5B in the NMR intermediate exchange regime, which may induce excessive broadening of the Ile-560 resonance.
The NNI-2–binding site, which is involved in the interaction with NS5A-D2, is rather well conserved across HCV genotypes (99, 100), supporting its important functional role. Using both fluorescence and size exclusion chromatography analyses, we showed here that NS5A-D2 binding reduces the RNA-binding properties of the HCV polymerase (Figs. 5 and 6). Moreover, this effect of NS5A-D2 on NS5B occurs although, as we have shown, NS5A-D2 neither binds RNA (Fig. 6c; see supplemental Fig. S9) nor interacts with the RNA-binding site in NS5B (Fig. 4). A direct interaction between NS5A-D2 (from the HCV Con1 strain, genotype 1b) and RNA has recently been reported (66); however, the basic residues identified as essential (304RRSRKFPR311) are not conserved among all HCV genotypes (see Fig. 1 in Ref. 61) and in particular are absent in the JFH-1 strain (g2a) used in this study (300LPRSGFPR307). Nevertheless, our results are consistent with those of Ngure et al. (66), who have shown an altered affinity of NS5B for single-stranded RNA in the presence of NS5A-D2, which decreased RNA polymerase activity.
In conclusion, we have shown that, upon binding, NS5A-D2 induces conformational and functional perturbations in NS5B similar to those triggered by NNIs. Thus, our experimental data strongly suggest that NS5A-D2 acts as an allosteric regulator of the HCV polymerase. This intrinsically disordered domain, through its binding to the NS5B NNI-2– and NNI-1–binding sites and displacement of the fingers loop Λ1, may regulate the open/closed conformational dynamics of the polymerase, altering its RNA-binding properties and thus its enzymatic activity. Our NMR study of the HCV polymerase paves the way for further detailed molecular descriptions of the mechanisms that affect the conformational sampling of NS5B, which governs its functions.
Experimental procedures
Expression and purification of unlabeled NS5BΔ21 (JFH-1)
Expression and purification of unlabeled recombinant NS5BΔ21 from the HCV JFH-1 strain (GenBankTM number AB047639) were performed as described previously (65) with a pET21-NS5BΔ21 plasmid kindly provided by Dr. S. Bressanelli (28), which corresponds to residues 1–570 of NS5B from JFH-1, containing extra MA- and -HHHHHH extensions at N and C termini, respectively. Following purification, the protein was flash-frozen in liquid nitrogen and stored at −80 °C.
Expression and purification of Ile δ1-13C1H3-labeled NS5BΔ21 (JFH-1)
U-15N,2H-, Ile δ1-13C1H3-labeled NS5BΔ21 was expressed in Escherichia coli BL21(DE3) cells carrying the pET21- NS5BΔ21 plasmid. Cells were grown in deuterated M9-based semi-rich medium (M9 medium in 99.98% D2O with d-glucose-d7 (3 g/liter), 15NH4Cl (1 g/liter), and 15N,2H-rich medium (Isogro 15N,D, Isotec) (0.5 g/liter)) at 37 °C until the culture reached A600 of ∼0.8. Then 2-ketobutryric acid-4-13C,4,4-d2 sodium salt (60 mg/liter) (Sigma-Aldrich) was added to the medium to label the δ1 methyl group of isoleucine residues (Ile δ1-13C1H3). One hour later, the temperature was lowered to 22 °C, and protein expression was induced by 0.4 mm isopropyl 1-β-d-thiogalactopyranoside for 22 h. Cells were harvested by centrifugation and resuspended in 50 ml of lysis buffer (for 1 liter of culture) (20 mm Tris buffer, pH 7.0, 500 mm NaCl, 10% (v/v) glycerol, 1 cOmplete, EDTA-free protease inhibitor tablet (Roche Applied Science)). Purification by nickel affinity chromatography and storage were as for unlabeled NS5BΔ21.
Expression and purification of unlabeled and 15N-labeled NS5A-D2 (JFH-1)
Expression, purification, and storage of unlabeled and 15N-labeled recombinant NS5A-D2 from the HCV JFH-1 strain were performed as described previously (61). NS5A-D2 used in this study corresponds to residues 248–341 from the HCV JFH-1 strain containing additional M- and -LQHHHHHH extensions at the N and C termini, respectively.
Site-directed mutagenesis
Site-directed mutagenesis for both NS5BΔ21 and NS5A-D2 single-site mutations was done either by GeneCust (Luxembourg) or in-house using a QuikChange II site-directed mutagenesis kit (Agilent Technologies, Santa Clara, CA).
Nitroxide labeling of NS5A-D2
Wild-type NS5A-D2 from the JFH-1 strain contains two natural cysteines at positions 298 and 338, according to full-length NS5A numbering. Three single-cysteine constructs of NS5A-D2 were obtained by site-directed mutagenesis: NS5A-D2 C254, NS5A-D2 C298, NS5A-D2 C338. NS5A-D2 C298 and NS5A-D2 C338 were obtained by substituting cysteines 338 and 298 for serine, respectively. NS5A-D2 C254 was obtained by substituting both natural cysteine residues for serine and by the substitution of methionine 254 for cysteine. These mutants were expressed and purified as described for unlabeled wild-type NS5A-D2. Before labeling, the buffer was exchanged to ammonium carbonate buffer (100 mm NH4HCO3, 50 mm NaCl, 1 mm THP) using a Zeba Spin desalting column (7,000 molecular weight cut-off, 2 ml; Thermo Scientific). Protein samples were incubated for 45 min at room temperature to allow for the reduction of the cysteines. Then cysteine residues were labeled with a nitroxide group by the addition of a 25-fold molar excess of 3-(2-iodoacetamido)-PROXYL (Sigma-Aldrich) and an incubation period of 3 h at room temperature (protected from light). Unreacted reagent was quenched with 5 mm 1,4-dithiothreitol. Finally, the reaction buffer was exchanged with the NMR buffer of the partner protein using a Zeba Spin desalting column (7,000 molecular weight cut-off, 2 ml; Thermo Scientific).
Preparation of the NS5B–filibuvir complex
Filibuvir (a thumb II site inhibitor) was purchased from Sigma-Aldrich. To prepare the NS5BΔ21–filibuvir complex, 1 mg of the inhibitor was directly added in powder form to 400 μl of protein solution. The sample mixture was gently shaken (300 rpm) for several hours (at least 5 h and up to overnight) at 15 °C. Excess solid ligand was removed by filtration at 0.2 μm.
Nuclear magnetic resonance experiments
NMR experiments performed on NS5B samples were acquired at 305 K on a 900-MHz Avance III HD spectrometer equipped with a CPTCI 5-mm cryoprobe (Bruker Biospin) unless stated otherwise. 13C,1H HMQC spectra (1H acquisition time of 81 ms, 13C acquisition time of 28 ms) were acquired on 300-μl samples in 5-mm Shigemi tubes. Ile δ1-13C1H3-labeled NS5BΔ21 samples (40–80 μm) were exchanged to phosphate buffer (300 mm Na2HPO4/NaH2PO4, pH 6.8, 50 mm NaCl, 2 mm MgCl2, 1 mm EDTA, and 4 mm THP in 99.9% D2O) or R/E buffer (100 mm arginine, 100 mm glutamic acid, pH 7.0, 100 mm NaCl, 2 mm MgCl2, 1 mm EDTA, 4 mm THP in 99.9% D2O) using a desalting column (Zeba Spin, Thermo Scientific) just before NMR experiments.
For resonance assignment, 13C,1H methyl-TROSY HMQC spectra were acquired on ∼30 μm Ile-to-Val NS5B mutants (2,048 × 128 real data points, 1H acquisition time of 81 ms, 13C acquisition time of 28 ms, 32 scans). In addition, 3D 1H,1H,13C NOESY-HMQC (2,048 × 166 × 48 data points, 16 scans, 900-ms mixing time) and 1H,13C,13C HMQC-NOESY-HMQC spectra (2,048 × 66 × 66 data points, 16 scans, 1-s mixing time) were acquired on an Ile δ1-13C1H3-labeled NS5BΔ21 sample.
Intermolecular PRE NMR experiments (methyl-TROSY spectra set up as above, but using 112 scans) were performed with samples containing 72 μm Ile δ1-13C1H3-labeled NS5BΔ21 and an equimolar amount of nitroxide-labeled NS5A-D2 in phosphate buffer. 4 mm ascorbic acid was added to an independent and identical sample to reduce the paramagnetic label and obtain a diamagnetic control spectrum acquired under the same conditions. This was done for all three single-cysteine constructs of NS5A-D2.
Spectra containing 15N-labeled NS5A-D2 were acquired at 298 K on a 600-MHz Avance I spectrometer equipped with a CPQCI cryoprobe (Bruker Biospin). NS5A-D2 samples were prepared in phosphate buffer (300 mm Na2HPO4/NaH2PO4, pH 6.8, 50 mm NaCl, 2 mm MgCl2, 1 mm EDTA, and 4 mm THP, 10% D2O) for experiments with NS5BΔ21 and in Tris buffer (50 mm Tris-d7, pH 6.4, 30 mm NaCl, 2 mm MgCl2, 0.5 mm EDTA, and 4 mm THP) for experiments with RNA16.
Proton chemical shifts were referenced using the methyl signal of sodium 3-trimethylsilyl-[2,2,3,3-d4]-propionate at 0 ppm. Spectra were acquired and processed using TopSpin version 3.5 (Bruker Biospin). Visualization and analysis of spectra was performed using the NMRFAM-Sparky software package (101). CSPs in methyl-HMQC spectra were calculated using Equation 1,
| (1) |
where Δδ(1H) and Δδ(13C) correspond to chemical shift perturbations in 1H and 13C dimensions, respectively, and α is a scaling factor corresponding to the relative chemical shift dispersion in both dimensions. In this work, a value of 0.16 was used, corresponding to the relative ranges of Ile-δ1 1H and 13C chemical shifts.
Methyl-TROSY 13C-1H MQ CPMG relaxation dispersion (RD) experiments (82) were recorded on Ile δ1-13C1H3-labeled NS5BΔ21 NMR samples with and without filibuvir on both 600- and 900-MHz spectrometers. 10 or 12 points, with one or two repeat experiments for error analysis, were recorded for each dispersion curve, corresponding to CPMG frequencies (νCPMG) between 100 and 1,000 Hz, using a constant-time relaxation delay T = 20 ms. Spectra were acquired using acquisition times of 82 and 32 ms as well as 2,048 and 128 real points in 1H and 13C dimensions, respectively (at 900 MHz), and 256–360 scans. Spectra were processed using NMRPipe (102) and analyzed in Sparky (101). RD profiles, R2,eff(νCPMG), were calculated from peak heights using Equation 2,
| (2) |
with I(νCPMG) the peak height in the spectrum plane recorded with CPMG frequency νCPMG and I0 the peak height in a reference spectrum recorded without the CPMG pulse train. Errors in R2,eff values were calculated as described (103) based on repeat measurements. A minimum error of 0.5 s−1 was assumed. RD data were fit to an analytical model of MQ CPMG relaxation dispersion under two-site exchange (82) using the software cpmg_fitd9 (104) kindly provided by Dmitry Korzhnev as well as to a model assuming no exchange (i.e. a flat horizontal line). Data were retained for further analysis if the model assuming exchange improved the fit at the 98% confidence level according to F test statistics and if the difference between R2,eff values at minimum and maximum νCPMG frequencies was at least 2 s−1. Fits to individual residues were performed, allowing for 13C and 1H chemical shift differences between states (Δω) to vary, as well as with 1H Δω fixed to zero. The additional 1H Δω fit parameter did not improve fits in a statistically significant way, but in several cases, errors of fit parameters decreased substantially. In these cases, the results from fits allowing both 1H and 13C Δω to vary are reported; otherwise, results from fits in which only 13C Δω was adjustable are given. In grouped fits of several residues to a single exchange process, both 1H and 13C Δω were always allowed to vary for each residue.
Chemical shift predictions
NMR chemical shift predictions (13C and 1H) for δ1-methyl groups of isoleucine residues in NS5B were obtained from crystallographic data using ShiftX2 (78) with the following parameters: per-deuterated protein, pH 7, and 305 K. Predictions were obtained for both closed (PDB codes 2XXD (77), 2XYM (77), 3I5K (28), 4AEP (80), and 4AEX (80)) and open (PDB codes 4E76 (33), 4OBC (34), and 4WT9 (35)) apo structures of NS5B from the JFH-1 strain (see supplemental Table S4; only chain A was kept for predictions when PDB files contained multiple chains). A normalized deviation (σ) across the different predictions for combined 13C and 1H shifts of each Ile δ1-methyl group was calculated using Equation 3,
| (3) |
where σH and σC correspond to the S.D. value of 1H and 13C predictions, respectively, and α is the carbon scaling factor used in the calculation of chemical shift perturbations (0.16; see above).
Fluorescence spectroscopy
Binding of RNA16 by NS5BΔ21 was assayed by fluorescence spectroscopy at 294 K on a fluorescence spectrometer (PTI, Monmouth Junction, NJ). The RNA16 molecule (5′-CUAAGAUGCUCGCUGC-3′) with a 6-FAM label at the 5′ end (87) was obtained from GeneCust (Luxembourg). Excitation of the 6-FAM label was performed at 492 nm, and the emission spectrum was scanned from 500 to 530 nm (1 nm stepwise, 2 s/scan). Excitation and emission slit widths were set to 1 nm. Fluorescence intensities were integrated from 515 to 525 nm. The assay was carried out in R/E buffer (20 mm Tris·Cl, pH 7.5, 100 mm NaCl, 80 mm arginine, 80 mm glutamate, 5 mm MgCl2, 1 mm DTT) in 1-cm path-length cells. The fluorescence of 50 nm 6-FAM-labeled RNA16 was measured in the presence of increasing NS5BΔ21 concentrations. This binding assay was performed in the absence and in the presence of NS5A-D2 (0–20 μm). The dissociation constant (KD) was determined by using a least-squares fitting procedure between the experimental data and the quadratic Equation 4,
| (4) |
where F is the measured fluorescence intensity, Fm is the maximum fluorescence, [R] the concentration of RNA16 and [5B] the concentration of NS5BΔ21.
Analytical size-exclusion chromatography
Size-exclusion chromatography analyses were performed on an ÄKTA Purifier FPLC system (GE Healthcare) equipped with a multiwavelength detector using a Superdex 75 3.2/300 column (GE Healthcare) equilibrated in 20 mm Tris·Cl, pH 7.0, 100 mm arginine, 100 mm glutamate, 100 mm NaCl, 5 mm MgCl2, 2 mm THP, with a flow rate of 50 μl/min and at room temperature. A loop of 40 μl was used to inject the samples, all containing 10 μm 6FAM-RNA16 (same molecule as used for fluorescence spectroscopy; see above). Elution of RNA16 was followed by monitoring absorbance at 495 nm. UV absorbance at 260 and 280 nm were also collected at the same time. Elution of RNA16 was analyzed in the presence of increasing concentrations of 1) NS5BΔ21 or 2) NS5A-D2; 3) in the presence of a fixed NS5BΔ21 concentration and increasing amounts of NS5A-D2; and 4) in the presence of the NS5BΔ21–filibuvir complex. The proteins were prepared in the running buffer and were mixed with RNA16 just before injection onto the column.
Differential scanning fluorimetry
DSF was carried out on a Stratagene Mx3005P quantitative PCR thermocycler (Agilent Technologies) with a 40-μl reaction volume in R/E buffer (20 mm Tris, 100 mm NaCl, 100 mm arginine, 100 mm glutamate, 5 mm MgCl2, and 2 mm THP) containing Sypro orange (1:1,000, v/v) fluorophore (Thermo Fisher Scientific). Thermal denaturation was monitored by measuring fluorescence intensity at 516 nm (excitation at 492 nm) during an increase in temperature from 298 to 368 K at a rate of 1 K/min. The inflection point of the melting curves, which represents the melting temperature (Tm), was determined by fitting the obtained intensities, up to the plateau, to a Boltzmann model (Equation 5) (86),
| (5) |
where Fmax and Fbase are the maximum and minimum recorded fluorescence intensities, respectively, and a is the slope of the curve. DSF was performed on a 30 μm NS5BΔ21 sample alone or in the presence of increasing concentrations of NS5A-D2 (0–180 μm) and finally on a 30 μm NS5BΔ21–filibuvir complex sample. A control experiment performed with 180 μm of NS5A-D2 and without NS5BΔ21 showed that NS5A-D2 does not contribute to the measured fluorescence.
Accession numbers
The Ile-δ1 methyl resonance assignments of HCV NS5BΔ21 (strain JFH-1) have been deposited in the Biological Magnetic Resonance Data Bank (BMRB) under accession number 27073.
Author contributions
X. H. designed the research. L. M. B., H. L., M. D., and X. H. performed protein expression and purification, mutagenesis, fluorescence, SEC, and DSF experiments. L. M. B., H. L., F.-X. C., G. L., I. L., R. S., and X. H. performed and analyzed NMR experiments. X. H. and R. S. wrote the paper. All authors reviewed the results and approved the final version of the manuscript.
Supplementary Material
Acknowledgments
The NMR facility was supported by the European Community, the CNRS (IR RMN THC, FR-3050), the University of Lille, the Région Hauts-de-France (France), and the Pasteur Institute of Lille (IPL).
This work was supported by French National Agency for Research (ANR) Grant ANR-11-JSV8-005 and French National Agency for Research on AIDS and Viral Hepatitis (ANRS) Grant A02014-2. This work was also supported by FRABIO (CNRS, FR-3688). The authors declare that they have no conflicts of interest with the contents of this article.
This article contains supplemental Tables S1–S5 and Figs. S1–S12.
- HCV
- hepatitis C virus
- NS
- non-structural protein
- ER
- endoplasmic reticulum
- RdRp
- RNA-dependent RNA polymerase
- DAA
- direct acting antivirals
- NNI
- non-nucleoside inhibitor
- NS5BΔ21
- recombinant protein representing amino acids 1–570 of NS5B with a C-terminal SHHHHHH extension
- NS5A-D2
- domain 2 of the NS5A protein
- g2a
- genotype 2a
- CypA
- cyclophilin A
- HSQC
- heteronuclear single quantum coherence
- HMQC
- heteronuclear multiple-quantum coherence
- TROSY
- transverse relaxation optimized spectroscopy
- methyl-TROSY
- 1H,13C TROSY HMQC spectrum centered on the resonances of the δ1 methyl group (13C1H3) of the isoleucine side chains
- CPMG
- Carr-Purcell-Meiboom-Gill
- CSP
- chemical shift perturbation
- methyl-CSP
- 1H and 13C combined chemical shift perturbations of the Ile δ1 methyl group
- DSF
- differential scanning fluorimetry
- RNA16
- a 16-mer RNA oligonucleotide
- 6-FAM
- fluorescent 6-carboxyfluorescein
- 6FAM-RNA16
- a fluorescent RNA16
- SEC
- size exclusion chromatography
- MQ
- multiple-quantum
- PRE
- paramagnetic relaxation enhancement
- RD
- relaxation dispersion
- PDB
- Protein Data Bank
- BMRB
- Biological Magnetic Resonance Data Bank
- THP
- Tris(hydroxypropyl)phosphine.
References
- 1. World Health Organization (2017) Hepatitis C: fact sheet, World Health Organization, Geneva [Google Scholar]
- 2. Stanaway J. D., Flaxman A. D., Naghavi M., Fitzmaurice C., Vos T., Abubakar I., Abu-Raddad L. J., Assadi R., Bhala N., Cowie B., Forouzanfour M. H., Groeger J., Hanafiah K. M., Jacobsen K. H., James S. L., et al. (2016) The global burden of viral hepatitis from 1990 to 2013: findings from the Global Burden of Disease Study 2013. Lancet 388, 1081–1088 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Smith D. B., Bukh J., Kuiken C., Muerhoff A. S., Rice C. M., Stapleton J. T., and Simmonds P. (2014) Expanded classification of hepatitis C virus into 7 genotypes and 67 subtypes: updated criteria and genotype assignment web resource. Hepatology 59, 318–327 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Messina J. P., Humphreys I., Flaxman A., Brown A., Cooke G. S., Pybus O. G., and Barnes E. (2015) Global distribution and prevalence of hepatitis C virus genotypes. Hepatology 61, 77–87 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Moradpour D., Penin F., and Rice C. M. (2007) Replication of hepatitis C virus. Nat. Rev. Microbiol. 5, 453–463 [DOI] [PubMed] [Google Scholar]
- 6. Moradpour D., and Penin F. (2013) Hepatitis C virus proteins: from structure to function. Curr. Top. Microbiol. Immunol. 369, 113–142 [DOI] [PubMed] [Google Scholar]
- 7. Lohmann V. (2013) Hepatitis C virus RNA replication. Curr. Top. Microbiol. Immunol. 369, 167–198 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Romero-Brey I., Merz A., Chiramel A., Lee J.-Y., Chlanda P., Haselman U., Santarella-Mellwig R., Habermann A., Hoppe S., Kallis S., Walther P., Antony C., Krijnse-Locker J., and Bartenschlager R. (2012) Three-dimensional architecture and biogenesis of membrane structures associated with hepatitis C virus replication. PLoS Pathog. 8, e1003056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Paul D., Hoppe S., Saher G., Krijnse-Locker J., and Bartenschlager R. (2013) Morphological and biochemical characterization of the membranous hepatitis C virus replication compartment. J. Virol. 87, 10612–10627 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Raney K. D., Sharma S. D., Moustafa I. M., and Cameron C. E. (2010) Hepatitis C virus non-structural protein 3 (HCV NS3): a multifunctional antiviral target. J. Biol. Chem. 285, 22725–22731 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Egger D., Wölk B., Gosert R., Bianchi L., Blum H. E., Moradpour D., and Bienz K. (2002) Expression of hepatitis C virus proteins induces distinct membrane alterations including a candidate viral replication complex. J. Virol. 76, 5974–5984 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Tellinghuisen T. L., Marcotrigiano J., and Rice C. M. (2005) Structure of the zinc-binding domain of an essential component of the hepatitis C virus replicase. Nature 435, 374–379 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Tellinghuisen T. L., Foss K. L., Treadaway J. C., and Rice C. M. (2008) Identification of residues required for RNA replication in domains II and III of the hepatitis C virus NS5A protein. J. Virol. 82, 1073–1083 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Appel N., Zayas M., Miller S., Krijnse-Locker J., Schaller T., Friebe P., Kallis S., Engel U., and Bartenschlager R. (2008) Essential role of domain III of nonstructural protein 5A for hepatitis C virus infectious particle assembly. PLoS Pathog. 4, e1000035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Lohmann V., Körner F., Herian U., and Bartenschlager R. (1997) Biochemical properties of hepatitis C virus NS5B RNA-dependent RNA polymerase and identification of amino acid sequence motifs essential for enzymatic activity. J. Virol. 71, 8416–8428 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Yamashita T., Kaneko S., Shirota Y., Qin W., Nomura T., Kobayashi K., and Murakami S. (1998) RNA-dependent RNA polymerase activity of the soluble recombinant hepatitis C virus NS5B protein truncated at the C-terminal region. J. Biol. Chem. 273, 15479–15486 [DOI] [PubMed] [Google Scholar]
- 17. Schlütter J. (2011) Therapeutics: new drugs hit the target. Nature 474, S5–S7 [DOI] [PubMed] [Google Scholar]
- 18. Rice C. M., and Saeed M. (2014) Hepatitis C: treatment triumphs. Nature 510, 43–44 [DOI] [PubMed] [Google Scholar]
- 19. Götte M., and Feld J. J. (2016) Direct-acting antiviral agents for hepatitis C: structural and mechanistic insights. Nat. Rev. Gastroenterol. Hepatol. 13, 338–351 [DOI] [PubMed] [Google Scholar]
- 20. Bourlière M., Gordon S. C., Flamm S. L., Cooper C. L., Ramji A., Tong M., Ravendhran N., Vierling J. M., Tran T. T., Pianko S., Bansal M. B., de Lédinghen V., Hyland R. H., Stamm L. M., Dvory-Sobol H., et al. (2017) Sofosbuvir, Velpatasvir, and Voxilaprevir for previously treated HCV infection. N. Engl. J. Med. 376, 2134–2146 [DOI] [PubMed] [Google Scholar]
- 21. Lindenbach B. D., and Rice C. M. (2005) Unravelling hepatitis C virus replication from genome to function. Nature 436, 933–938 [DOI] [PubMed] [Google Scholar]
- 22. Schmidt-Mende J., Bieck E., Hugle T., Penin F., Rice C. M., Blum H. E., and Moradpour D. (2001) Determinants for membrane association of the hepatitis C virus RNA-dependent RNA polymerase. J. Biol. Chem. 276, 44052–44063 [DOI] [PubMed] [Google Scholar]
- 23. Ivashkina N., Wölk B., Lohmann V., Bartenschlager R., Blum H. E., Penin F., and Moradpour D. (2002) The hepatitis C virus RNA-dependent RNA polymerase membrane insertion sequence is a transmembrane segment. J. Virol. 76, 13088–13093 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Brass V., Gouttenoire J., Wahl A., Pal Z., Blum H. E., Penin F., and Moradpour D. (2010) Hepatitis C virus RNA replication requires a conserved structural motif within the transmembrane domain of the NS5B RNA-dependent RNA polymerase. J. Virol. 84, 11580–11584 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Bressanelli S., Tomei L., Roussel A., Incitti I., Vitale R. L., Mathieu M., De Francesco R., and Rey F. A. (1999) Crystal structure of the RNA-dependent RNA polymerase of hepatitis C virus. Proc. Natl. Acad. Sci. U.S.A. 96, 13034–13039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Ago H., Adachi T., Yoshida A., Yamamoto M., Habuka N., Yatsunami K., and Miyano M. (1999) Crystal structure of the RNA-dependent RNA polymerase of hepatitis C virus. Structure 7, 1417–1426 [DOI] [PubMed] [Google Scholar]
- 27. Lesburg C. A., Cable M. B., Ferrari E., Hong Z., Mannarino A. F., and Weber P. C. (1999) Crystal structure of the RNA-dependent RNA polymerase from hepatitis C virus reveals a fully encircled active site. Nat. Struct. Biol. 6, 937–943 [DOI] [PubMed] [Google Scholar]
- 28. Simister P., Schmitt M., Geitmann M., Wicht O., Danielson U. H., Klein R., Bressanelli S., and Lohmann V. (2009) Structural and functional analysis of hepatitis C virus strain JFH1 polymerase. J. Virol. 83, 11926–11939 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Sesmero E., and Thorpe I. F. (2015) Using the hepatitis C virus RNA-dependent RNA polymerase as a model to understand viral polymerase structure, function and dynamics. Viruses 7, 3974–3994 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Biswal B. K., Cherney M. M., Wang M., Chan L., Yannopoulos C. G., Bilimoria D., Nicolas O., Bedard J., and James M. N. (2005) Crystal structures of the RNA-dependent RNA polymerase genotype 2a of hepatitis C virus reveal two conformations and suggest mechanisms of inhibition by non-nucleoside inhibitors. J. Biol. Chem. 280, 18202–18210 [DOI] [PubMed] [Google Scholar]
- 31. Chinnaswamy S., Yarbrough I., Palaninathan S., Kumar C. T., Vijayaraghavan V., Demeler B., Lemon S. M., Sacchettini J. C., and Kao C. C. (2008) A locking mechanism regulates RNA synthesis and host protein interaction by the hepatitis C virus polymerase. J. Biol. Chem. 283, 20535–20546 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Jin Z., Leveque V., Ma H., Johnson K. A., and Klumpp K. (2012) Assembly, purification, and pre-steady-state kinetic analysis of active RNA-dependent RNA polymerase elongation complex. J. Biol. Chem. 287, 10674–10683 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Mosley R. T., Edwards T. E., Murakami E., Lam A. M., Grice R. L., Du J., Sofia M. J., Furman P. A., and Otto M. J. (2012) Structure of hepatitis C virus polymerase in complex with primer-template RNA. J. Virol. 86, 6503–6511 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Lam A. M., Edwards T. E., Mosley R. T., Murakami E., Bansal S., Lugo C., Bao H., Otto M. J., Sofia M. J., and Furman P. A. (2014) Molecular and structural basis for the roles of hepatitis C virus polymerase NS5B amino acids 15, 223, and 321 in viral replication and drug resistance. Antimicrob. Agents Chemother. 58, 6861–6869 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Appleby T. C., Perry J. K., Murakami E., Barauskas O., Feng J., Cho A., Fox D. 3rd, Wetmore D. R., McGrath M. E., Ray A. S., Sofia M. J., Swaminathan S., and Edwards T. E. (2015) Structural basis for RNA replication by the hepatitis C virus polymerase. Science 347, 771–775 [DOI] [PubMed] [Google Scholar]
- 36. Davis B. C., Brown J. A., and Thorpe I. F. (2015) Allosteric inhibitors have distinct effects, but also common modes of action, in the HCV polymerase. Biophys. J. 108, 1785–1795 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Eltahla A. A., Luciani F., White P. A., Lloyd A. R., and Bull R. A. (2015) Inhibitors of the hepatitis C virus polymerase: mode of action and resistance. Viruses 7, 5206–5224 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Caillet-Saguy C., Simister P. C., and Bressanelli S. (2011) An objective assessment of conformational variability in complexes of hepatitis C virus polymerase with non-nucleoside inhibitors. J. Mol. Biol. 414, 370–384 [DOI] [PubMed] [Google Scholar]
- 39. Davis B. C., and Thorpe I. F. (2013) Thumb inhibitor binding eliminates functionally important dynamics in the hepatitis C virus RNA polymerase. Proteins 81, 40–52 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Biswal B. K., Wang M., Cherney M. M., Chan L., Yannopoulos C. G., Bilimoria D., Bedard J., and James M. N. (2006) Non-nucleoside inhibitors binding to hepatitis C virus NS5B polymerase reveal a novel mechanism of inhibition. J. Mol. Biol. 361, 33–45 [DOI] [PubMed] [Google Scholar]
- 41. Li J., and Johnson K. A. (2016) Thumb site 2 inhibitors of hepatitis C viral RNA-dependent RNA polymerase allosterically block the transition from initiation to elongation. J. Biol. Chem. 291, 10067–10077 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Deredge D., Li J., Johnson K. A., and Wintrode P. L. (2016) Hydrogen/deuterium exchange kinetics demonstrate long range allosteric effects of thumb site 2 inhibitors of hepatitis C viral RNA-dependent RNA polymerase. J. Biol. Chem. 291, 10078–10088 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Cho N.-J., Pham E. A., Hagey R. J., Lévêque V. J., Ma H., Klumpp K., and Glenn J. S. (2016) Reconstitution and functional analysis of a full-length hepatitis C virus NS5B polymerase on a supported lipid bilayer. ACS Cent. Sci. 2, 456–466 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Rigat K., Wang Y., Hudyma T. W., Ding M., Zheng X., Gentles R. G., Beno B. R., Gao M., and Roberts S. B. (2010) Ligand-induced changes in hepatitis C virus NS5B polymerase structure. Antiviral Res. 88, 197–206 [DOI] [PubMed] [Google Scholar]
- 45. Ross-Thriepland D., and Harris M. (2015) Hepatitis C virus NS5A: enigmatic but still promiscuous 10 years on! J. Gen. Virol. 96, 727–738 [DOI] [PubMed] [Google Scholar]
- 46. Reiss S., Rebhan I., Backes P., Romero-Brey I., Erfle H., Matula P., Kaderali L., Poenisch M., Blankenburg H., Hiet M.-S., Longerich T., Diehl S., Ramirez F., Balla T., Rohr K., et al. (2011) Recruitment and activation of a lipid kinase by hepatitis C virus NS5A is essential for integrity of the membranous replication compartment. Cell Host Microbe 9, 32–45 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Ramière C., Rodriguez J., Enache L. S., Lotteau V., André P., and Diaz O. (2014) Hexokinase activity is increased by its interaction with Hepatitis C virus protein NS5A. J. Virol. 10.1128/JVI.02862-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. de Chassey B., Navratil V., Tafforeau L., Hiet M. S., Aublin-Gex A., Agaugué S., Meiffren G., Pradezynski F., Faria B. F., Chantier T., Le Breton M., Pellet J., Davoust N., Mangeot P. E., Chaboud A., et al. (2008) Hepatitis C virus infection protein network. Mol. Syst. Biol. 4, 230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Huang L., Hwang J., Sharma S. D., Hargittai M. R., Chen Y., Arnold J. J., Raney K. D., and Cameron C. E. (2005) Hepatitis C virus nonstructural protein 5A (NS5A) is an RNA-binding protein. J. Biol. Chem. 280, 36417–36428 [DOI] [PubMed] [Google Scholar]
- 50. Foster T. L., Belyaeva T., Stonehouse N. J., Pearson A. R., and Harris M. (2010) All three domains of the hepatitis C virus nonstructural NS5A protein contribute to RNA binding. J. Virol. 84, 9267–9277 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Love R. A., Brodsky O., Hickey M. J., Wells P. A., and Cronin C. N. (2009) Crystal structure of a novel dimeric form of NS5A domain I protein from hepatitis C virus. J. Virol. 83, 4395–4403 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Lambert S. M., Langley D. R., Garnett J. A., Angell R., Hedgethorne K., Meanwell N. A., and Matthews S. J. (2014) The crystal structure of NS5A domain 1 from genotype 1a reveals new clues to the mechanism of action for dimeric HCV inhibitors. Protein Sci. 23, 723–734 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Liang Y., Ye H., Kang C. B., and Yoon H. S. (2007) Domain 2 of nonstructural protein 5A (NS5A) of hepatitis C virus is natively unfolded. Biochemistry 46, 11550–11558 [DOI] [PubMed] [Google Scholar]
- 54. Hanoulle X., Badillo A., Verdegem D., Penin F., and Lippens G. (2010) The domain 2 of the HCV NS5A protein is intrinsically unstructured. Protein Pept. Lett. 17, 1012–1018 [DOI] [PubMed] [Google Scholar]
- 55. Hanoulle X., Verdegem D., Badillo A., Wieruszeski J.-M., Penin F., and Lippens G. (2009) Domain 3 of non-structural protein 5A from hepatitis C virus is natively unfolded. Biochem. Biophys. Res. Commun. 381, 634–638 [DOI] [PubMed] [Google Scholar]
- 56. Verdegem D., Badillo A., Wieruszeski J.-M., Landrieu I., Leroy A., Bartenschlager R., Penin F., Lippens G., and Hanoulle X. (2011) Domain 3 of NS5A protein from the hepatitis C virus has intrinsic α-helical propensity and is a substrate of cyclophilin A. J. Biol. Chem. 286, 20441–20454 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Feuerstein S., Solyom Z., Aladag A., Favier A., Schwarten M., Hoffmann S., Willbold D., and Brutscher B. (2012) Transient structure and SH3 interaction sites in an intrinsically disordered fragment of the hepatitis C virus protein NS5A. J. Mol. Biol. 420, 310–323 [DOI] [PubMed] [Google Scholar]
- 58. Sólyom Z., Ma P., Schwarten M., Bosco M., Polidori A., Durand G., Willbold D., and Brutscher B. (2015) The disordered region of the HCV protein NS5A: conformational dynamics, SH3 binding, and phosphorylation. Biophys. J. 109, 1483–1496 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Badillo A., Receveur-Brechot V., Sarrazin S., Cantrelle F.-X., Delolme F., Fogeron M.-L., Molle J., Montserret R., Bockmann A., Bartenschlager R., Lohmann V., Lippens G., Ricard-Blum S., Hanoulle X., and Penin F. (2017) Overall structural model of NS5A protein from hepatitis C virus and modulation by mutations confering resistance of virus replication to cyclosporin A. Biochemistry 56, 3029–3048 [DOI] [PubMed] [Google Scholar]
- 60. Tellinghuisen T. L., Foss K. L., and Treadaway J. (2008) Regulation of hepatitis C virion production via phosphorylation of the NS5A protein. PLoS Pathog. 4, e1000032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Hanoulle X., Badillo A., Wieruszeski J. M., Verdegem D., Landrieu I., Bartenschlager R., Penin F., and Lippens G. (2009) Hepatitis C virus NS5A protein is a substrate for the peptidyl-prolyl cis/trans isomerase activity of cyclophilins A and B. J. Biol. Chem. 284, 13589–13601 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Grisé H., Frausto S., Logan T., and Tang H. (2012) A conserved tandem cyclophilin-binding site in hepatitis C virus nonstructural protein 5A regulates Alisporivir susceptibility. J. Virol. 86, 4811–4822 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Foster T. L., Gallay P., Stonehouse N. J., and Harris M. (2011) Cyclophilin A interacts with domain II of hepatitis C virus NS5A and stimulates RNA binding in an isomerase-dependent manner. J. Virol. 85, 7460–7464 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Dujardin M., Madan V., Montserret R., Ahuja P., Huvent I., Launay H., Leroy A., Bartenschlager R., Penin F., Lippens G., and Hanoulle X. (2015) A proline-tryptophan turn in the intrinsically disordered domain 2 of NS5A protein is essential for hepatitis C virus RNA replication. J. Biol. Chem. 290, 19104–19120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Rosnoblet C., Fritzinger B., Legrand D., Launay H., Wieruszeski J.-M., Lippens G., and Hanoulle X. (2012) Hepatitis C virus NS5B and host cyclophilin A share a common binding site on NS5A. J. Biol. Chem. 287, 44249–44260 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Ngure M., Issur M., Shkriabai N., Liu H.-W., Cosa G., Kvaratskhelia M., and Götte M. (2016) Interactions of the disordered domain II of hepatitis C virus NS5A with cyclophilin A, NS5B, and viral RNA show extensive overlap. ACS Infect. Dis. 2, 839–851 [DOI] [PubMed] [Google Scholar]
- 67. Shimakami T., Hijikata M., Luo H., Ma Y. Y., Kaneko S., Shimotohno K., and Murakami S. (2004) Effect of interaction between hepatitis C virus NS5A and NS5B on hepatitis C virus RNA replication with the hepatitis C virus replicon. J. Virol. 78, 2738–2748 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Qin W., Yamashita T., Shirota Y., Lin Y., Wei W., and Murakami S. (2001) Mutational analysis of the structure and functions of hepatitis C virus RNA-dependent RNA polymerase. Hepatology 33, 728–737 [DOI] [PubMed] [Google Scholar]
- 69. Shirota Y., Luo H., Qin W., Kaneko S., Yamashita T., Kobayashi K., and Murakami S. (2002) Hepatitis C virus (HCV) NS5A binds RNA-dependent RNA polymerase (RdRP) NS5B and modulates RNA-dependent RNA polymerase activity. J. Biol. Chem. 277, 11149–11155 [DOI] [PubMed] [Google Scholar]
- 70. Quezada E. M., and Kane C. M. (2009) The hepatitis C virus NS5A stimulates NS5B during in vitro RNA synthesis in a template-specific manner. Open Biochem. J. 3, 39–48 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Chinnaswamy S., Murali A., Li P., Fujisaki K., and Kao C. C. (2010) Regulation of de novo-initiated RNA synthesis in hepatitis C virus RNA-dependent RNA polymerase by intermolecular interactions. J. Virol. 84, 5923–5935 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Ranjith-Kumar C. T., Wen Y., Baxter N., Bhardwaj K., and Cheng Kao C. (2011) A cell-based assay for RNA synthesis by the HCV polymerase reveals new insights on mechanism of polymerase inhibitors and modulation by NS5A. PLoS One 6, e22575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Nietlispach D. (2005) Suppression of anti-TROSY lines in a sensitivity enhanced gradient selection TROSY scheme. J. Biomol. NMR 31, 161–166 [DOI] [PubMed] [Google Scholar]
- 74. Tugarinov V., Hwang P. M., Ollerenshaw J. E., and Kay L. E. (2003) Cross-correlated relaxation enhanced 1H-13C NMR spectroscopy of methyl groups in very high molecular weight proteins and protein complexes. J. Am. Chem. Soc. 125, 10420–10428 [DOI] [PubMed] [Google Scholar]
- 75. Ollerenshaw J. E., Tugarinov V., and Kay L. E. (2003) Methyl TROSY: explanation and experimental verification. Magn. Reson. Chem. 41, 843–852 [Google Scholar]
- 76. Tugarinov V., Kanelis V., and Kay L. E. (2006) Isotope labeling strategies for the study of high-molecular-weight proteins by solution NMR spectroscopy. Nat. Protoc. 1, 749–754 [DOI] [PubMed] [Google Scholar]
- 77. Schmitt M., Scrima N., Radujkovic D., Caillet-Saguy C., Simister P. C., Friebe P., Wicht O., Klein R., Bartenschlager R., Lohmann V., and Bressanelli S. (2011) A comprehensive structure-function comparison of hepatitis C virus strain JFH1 and J6 polymerases reveals a key residue stimulating replication in cell culture across genotypes. J. Virol. 85, 2565–2581 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Han B., Liu Y., Ginzinger S. W., and Wishart D. S. (2011) SHIFTX2: significantly improved protein chemical shift prediction. J. Biomol. NMR 50, 43–57 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Di Marco S., Volpari C., Tomei L., Altamura S., Harper S., Narjes F., Koch U., Rowley M., De Francesco R., Migliaccio G., and Carfí A. (2005) Interdomain communication in hepatitis C virus polymerase abolished by small molecule inhibitors bound to a novel allosteric site. J. Biol. Chem. 280, 29765–29770 [DOI] [PubMed] [Google Scholar]
- 80. Scrima N., Caillet-Saguy C., Ventura M., Harrus D., Astier-Gin T., and Bressanelli S. (2012) Two crucial early steps in RNA synthesis by the hepatitis C virus polymerase involve a dual role of residue 405. J. Virol. 86, 7107–7117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Hansen D. F., Neudecker P., and Kay L. E. (2010) Determination of isoleucine side-chain conformations in ground and excited states of proteins from chemical shifts. J. Am. Chem. Soc. 132, 7589–7591 [DOI] [PubMed] [Google Scholar]
- 82. Korzhnev D. M., Kloiber K., Kanelis V., Tugarinov V., and Kay L. E. (2004) Probing slow dynamics in high molecular weight proteins by methyl-TROSY NMR spectroscopy: application to a 723-residue enzyme. J. Am. Chem. Soc. 126, 3964–3973 [DOI] [PubMed] [Google Scholar]
- 83. Iwahara J., and Clore G. M. (2006) Detecting transient intermediates in macromolecular binding by paramagnetic NMR. Nature 440, 1227–1230 [DOI] [PubMed] [Google Scholar]
- 84. Battiste J. L., and Wagner G. (2000) Utilization of site-directed spin labeling and high-resolution heteronuclear nuclear magnetic resonance for global fold determination of large proteins with limited nuclear Overhauser effect data. Biochemistry 39, 5355–5365 [DOI] [PubMed] [Google Scholar]
- 85. Tompa P., and Fuxreiter M. (2008) Fuzzy complexes: polymorphism and structural disorder in protein–protein interactions. Trends Biochem. Sci. 33, 2–8 [DOI] [PubMed] [Google Scholar]
- 86. Niesen F. H., Berglund H., and Vedadi M. (2007) The use of differential scanning fluorimetry to detect ligand interactions that promote protein stability. Nat. Protoc. 2, 2212–2221 [DOI] [PubMed] [Google Scholar]
- 87. Cramer J., Jaeger J., and Restle T. (2006) Biochemical and pre-steady-state kinetic characterization of the hepatitis C virus RNA polymerase (NS5BΔ21, HC-J4). Biochemistry 45, 3610–3619 [DOI] [PubMed] [Google Scholar]
- 88. Shi S. T., Herlihy K. J., Graham J. P., Nonomiya J., Rahavendran S. V., Skor H., Irvine R., Binford S., Tatlock J., Li H., Gonzalez J., Linton A., Patick A. K., and Lewis C. (2009) Preclinical characterization of PF-00868554, a potent nonnucleoside inhibitor of the hepatitis C virus RNA-dependent RNA polymerase. Antimicrob. Agents Chemother. 53, 2544–2552 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89. Li H., Tatlock J., Linton A., Gonzalez J., Jewell T., Patel L., Ludlum S., Drowns M., Rahavendran S. V., Skor H., Hunter R., Shi S. T., Herlihy K. J., Parge H., Hickey M., et al. (2009) Discovery of (R)-6-cyclopentyl-6-(2-(2,6-diethylpyridin-4-yl)ethyl)-3-((5,7-dimethyl-[1,2,4]triazolo[1,5-a]pyrimidin-2-yl)methyl)-4-hydroxy-5,6-dihydropyran-2-one (PF-00868554) as a potent and orally available hepatitis C virus polymerase inhibitor. J. Med. Chem. 52, 1255–1258 [DOI] [PubMed] [Google Scholar]
- 90. Yi G., Deval J., Fan B., Cai H., Soulard C., Ranjith-Kumar C. T., Smith D. B., Blatt L., Beigelman L., and Kao C. C. (2012) Biochemical study of the comparative inhibition of hepatitis C Virus RNA polymerase by VX-222 and Filibuvir. Antimicrob. Agents Chemother. 56, 830–837 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91. Troke P. J. F., Lewis M., Simpson P., Gore K., Hammond J., Craig C., and Westby M. (2012) Characterization of resistance to the nonnucleoside NS5B inhibitor Filibuvir in hepatitis C virus-infected patients. Antimicrob. Agents Chemother. 56, 1331–1341 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92. Ranjith-Kumar C. T., and Kao C. C. (2006) Recombinant viral RdRps can initiate RNA synthesis from circular templates. RNA 12, 303–312 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93. Labonté P., Axelrod V., Agarwal A., Aulabaugh A., Amin A., and Mak P. (2002) Modulation of hepatitis C virus RNA-dependent RNA polymerase activity by structure-based site-directed mutagenesis. J. Biol. Chem. 277, 38838–38846 [DOI] [PubMed] [Google Scholar]
- 94. Eastman K. J., Parcella K., Yeung K.-S., Grant-Young K. A., Zhu J., Wang T., Zhang Z., Yin Z., Beno B. R., Sheriff S., Kish K., Tredup J., Jardel A. G., Halan V., Ghosh K., et al. (2017) The discovery of a pan-genotypic, primer grip inhibitor of HCV NS5B polymerase. Med. Chem. Commun. 8, 796–806 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95. Boyce S. E., Tirunagari N., Niedziela-Majka A., Perry J., Wong M., Kan E., Lagpacan L., Barauskas O., Hung M., Fenaux M., Appleby T., Watkins W. J., Schmitz U., and Sakowicz R. (2014) Structural and regulatory elements of HCV NS5B polymerase, β-loop and C-terminal tail, are required for activity of allosteric thumb site II inhibitors. PLoS One 9, e84808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96. Eltahla A. A., Tay E., Douglas M. W., and White P. A. (2014) Cross-genotypic examination of hepatitis C virus polymerase inhibitors reveals a novel mechanism of action for thumb binders. Antimicrob. Agents Chemother. 58, 7215–7224 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97. Winquist J., Abdurakhmanov E., Baraznenok V., Henderson I., Vrang L., and Danielson U. H. (2013) Resolution of the interaction mechanisms and characteristics of non-nucleoside inhibitors of hepatitis C virus polymerase. Antiviral Res. 97, 356–368 [DOI] [PubMed] [Google Scholar]
- 98. Fuxreiter M., and Tompa P. (2012) Fuzzy complexes: a more stochastic view of protein function. Adv. Exp. Med. Biol. 725, 1–14 [DOI] [PubMed] [Google Scholar]
- 99. Di Maio V. C., Cento V., Mirabelli C., Artese A., Costa G., Alcaro S., Perno C. F., and Ceccherini-Silberstein F. (2014) Hepatitis C virus genetic variability and the presence of NS5B resistance-associated mutations as natural polymorphisms in selected genotypes could affect the response to NS5B inhibitors. Antimicrob. Agents Chemother. 58, 2781–2797 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100. Wang M., Ng K. K.-S., Cherney M. M., Chan L., Yannopoulos C. G., Bedard J., Morin N., Nguyen-Ba N., Alaoui-Ismaili M. H., Bethell R. C., and James M. N. G. (2003) Non-nucleoside analogue inhibitors bind to an allosteric site on HCV NS5B polymerase. J. Biol. Chem. 278, 9489–9495 [DOI] [PubMed] [Google Scholar]
- 101. Lee W., Tonelli M., and Markley J. L. (2015) NMRFAM-SPARKY: enhanced software for biomolecular NMR spectroscopy. Bioinformatics 31, 1325–1327 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102. Delaglio F., Grzesiek S., Vuister G. W., Zhu G., Pfeifer J., and Bax A. (1995) NMRPipe: a multidimensional spectral processing system based on UNIX pipes. J. Biomol. NMR 6, 277–293 [DOI] [PubMed] [Google Scholar]
- 103. Ishima R., and Torchia D. A. (2005) Error estimation and global fitting in transverse-relaxation dispersion experiments to determine chemical-exchange parameters. J. Biomol. NMR 32, 41–54 [DOI] [PubMed] [Google Scholar]
- 104. Korzhnev D. M., Neudecker P., Mittermaier A., Orekhov V. Y., and Kay L. E. (2005) Multiple-site exchange in proteins studied with a suite of six NMR relaxation dispersion experiments: an application to the folding of a Fyn SH3 domain mutant. J. Am. Chem. Soc. 127, 15602–15611 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.