Abstract
Ribonucleotides are the natural analogs of deoxyribonucleotides, which can be misinserted by DNA polymerases, leading to the most abundant DNA lesions in genomes. During replication, DNA polymerases tolerate patches of ribonucleotides on the parental strands to different extents. The majority of human DNA polymerases have been reported to misinsert ribonucleotides into genomes. However, only PrimPol, DNA polymerase α, telomerase, and the mitochondrial human DNA polymerase (hpol) γ have been shown to tolerate an entire RNA strand. Y-family hpol η is known for translesion synthesis opposite the UV-induced DNA lesion cyclobutane pyrimidine dimer and was recently found to incorporate ribonucleotides into DNA. Here, we report that hpol η is able to bind DNA/DNA, RNA/DNA, and DNA/RNA duplexes with similar affinities. In addition, hpol η, as well as another Y-family DNA polymerase, hpol κ, accommodates RNA as one of the two strands during primer extension, mainly by inserting dNMPs opposite unmodified templates or DNA lesions, such as 8-oxo-2′-deoxyguanosine or cyclobutane pyrimidine dimer, even in the presence of an equal amount of the DNA/DNA substrate. The discovery of this RNA-accommodating ability of hpol η redefines the traditional concept of human DNA polymerases and indicates potential new functions of hpol η in vivo.
Keywords: DNA damage, DNA enzyme, DNA polymerase, reverse transcription, RNA, DNA enzyme, DNA enzymes, replication initiation, reverse transcriptase
Introduction
DNA replication has been targeted for treatment of cancer because cancer cells proliferate rapidly with a high level of replication. Multiple chemotherapeutic drugs are designed to block DNA replication to stop the growth of tumor cells. The alkylating agents cisplatin and carboplatin directly introduce DNA damage in genomes, resulting in arrested replication forks (1). However, some DNA polymerases are tolerant of lesions on the templates. Y-family DNA polymerase hpol2 η replicates past platinum-induced DNA lesions, allowing cells to escape from arrested cell cycles (2, 3). High expression levels of hpol η in tumors are correlated with drug resistance (4). Some other chemotherapeutic drugs (e.g. cytarabine and gemcitabine) are designed to mimic nucleosides. After phosphorylation, these analogs can be inserted into genomes by DNA polymerases, leading to termination of replication (1). Interestingly, a class of deoxynucleotide analogs, ribonucleotides, is naturally present in cells, and the cellular concentrations of rNTPs are much higher than those of dNTPs (5).
DNA polymerases sometimes misinsert rNMPs into genomes (5). At least 12 of the 17 known human DNA polymerases (6) have been reported to tolerate ribonucleotides to some extent. PrimPol, pol α, and telomerase accommodate RNA by their very natures, and the other nine polymerases have been shown to incorporate ribonucleotides into DNA, including the replicative polymerases hpol δ and ϵ (nuclear genome replication) (7, 8) and pol γ (mitochondrial DNA replication) (9). In addition to base discrimination for forming Watson–Crick base pairs, sugar selectivity varies among polymerases. Most of the polymerases prefer dNTPs, with orders of magnitude higher efficiency compared with the ribo-counterparts, except hpol μ and terminal deoxynucleotidyl transferase, which have little sugar discrimination ability (10, 11). These polymerases are also able to extend a primer with a ribonucleotide at the 3′-end in vitro. Although RNase H2-dependent and topoisomerase-dependent pathways both contribute to remove embedded ribonucleotides from genomes, it is still possible that replication forks encounter ribonucleotides on the parental strands (5). hpol δ and ϵ can replicate past patches of several ribonucleotides in DNA templates, albeit with decreased catalytic efficiencies (12). Interestingly, hpol γ, the major polymerase in mitochondria, is capable of conducting reverse transcription by using an RNA strand as a template to synthesize DNA in vitro (13).
Y-family DNA polymerases have open and large active sites that can tolerate DNA lesions on template strands during replication (14). In general, polymerases in this family have relatively poor base selectivity and low fidelity compared with the replicative DNA polymerases. hpol η belongs to this family and has a particular role in bypass of the UV-induced DNA lesion cyclobutane pyrimidine dimer (CPD). Defects in hpol η cause patients to be highly sensitive to UV light and have a significantly increased incidence of skin cancer (15–17). Recently, hpol η has been shown to incorporate ribonucleotides opposite either undamaged bases or DNA lesions (e.g. CPD and 8-oxo-2′-deoxyguanosine (8-oxodG)), albeit with 103 times lower catalytic efficiencies compared with those for 2′-deoxyribonucleotides (18).
Here we report that hpol η, as well as another Y-family DNA polymerase, hpol κ, surprisingly accommodates an entire RNA strand as the primer during strand synthesis opposite a DNA template without sacrificing base selectivity, even with CPD or 8-oxodG in the template. In addition, hpol η is able to reverse-transcribe an RNA strand. These discoveries indicate potential new functions of hpol η beyond its established role as a translesion synthesis DNA polymerase.
Results
hpol η extends RNA primers
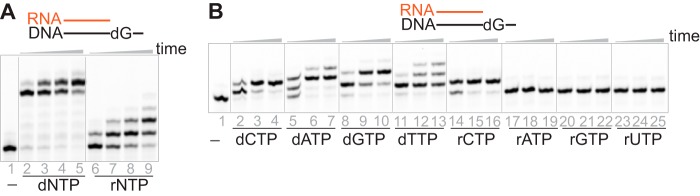
rNTPs and dNTPs are both present in cells and can be incorporated by hpol η (18). hpol η is able to extend a DNA primer even after several ribonucleotides are incorporated (18), leading to the question: How many ribonucleotides can hpol η tolerate in a primer? To push this case to an extreme, an 18-mer RNA oligonucleotide was used as the primer with a 23-mer DNA template. Surprisingly, hpol η was able to extend the RNA primer in the presence of either dNTPs or rNTPs (Fig. 1A). The RNA primer was elongated to full length within 5 min with a mixture of dNTPs (1 mm each dNTP) (Fig. 1A, lanes 2–5), whereas typically only 1–2 ribonucleotides were inserted after 4 h of incubation with the mixture of rNTPs (Fig. 1A, lanes 6–9). This result indicates that hpol η preferred dNTPs instead of rNTPs as the incoming nucleotides for polymerization of the RNA/DNA substrate. Single nucleotide insertion and steady-state kinetic assays both showed that the order of nucleotide incorporation by hpol η at the RNA primer end is as follows: paired dNTP > non-paired dNTP ≥ paired rNTP > non-paired rNTP (Fig. 1B and Tables 1 and 2), similar to previous observations for a DNA/DNA duplex (18). hpol η incorporated rCTP 130-fold less efficiently compared with dCTP opposite dG with an RNA strand as the primer, whereas for the extension of a DNA primer, the incorporation difference was 770-fold (Table 1). In addition, with dT on the template, 5′ next to the double-stranded portion, the dATP insertion efficiency was 710 times higher than that for rATP with an RNA primer, whereas the difference was 3,400-fold with a DNA primer (Table 2). These results indicated that both sugar selectivity and base pairing effect contribute to the extension by hpol η regardless of the backbone of the primer. In summary, hpol η was able to tolerate an RNA strand as the primer, and the sugar moiety of the incoming nucleotide was an important factor for the primer extension by hpol η.
Figure 1.
hpol η extends an RNA primer opposite a DNA template. The fluorescence-labeled oligonucleotide 5′-FAM-rCrGrG rGrCrU rCrGrU rArArG rCrGrU rCrArU-3′ was annealed with the template 5′-TCA TGA TGA CGC TTA CGA GCC CG-3′. A, hpol η (1.2 μm) extended an RNA primer opposite a DNA template (dG) (5 μm) in the presence of a mixture of dNTPs or rNTPs (1 mm concentration of each nucleotide). The reaction times were 5, 30, 55, and 240 min. B, hpol η (500 nm) extended an RNA primer opposite a DNA template (dG) (5 μm) in the presence of a single dNTP or rNTP (1 mm). The reaction times were 5, 30, and 55 min.
Table 1.
Steady-state kinetics of hpol η insertion of nucleotides opposite dG, 8-oxodG, and rG
| Primer/template | dNTP/rNTP | Km | kcat | kcat/Km | fa | 1/f |
|---|---|---|---|---|---|---|
| μm | min−1 | μm−1 min−1 | ||||
| RNA/DNA (dG) | dCTP | 117 ± 13 | 180 ± 6 | 1.5 ± 0.2 | 1 | 1 |
| dATP | 89 ± 11 | 2.0 ± 0.1 | 0.022 ± 0.003 | 0.015 | 67 | |
| dGTP | 80 ± 10 | 3.1 ± 0.1 | 0.039 ± 0.005 | 0.026 | 38 | |
| dTTP | 430 ± 36 | 4.8 ± 0.1 | 0.011 ± 0.001 | 0.0073 | 140 | |
| rCTP | 333 ± 29 | 4.0 ± 0.1 | 0.012 ± 0.001 | 0.0080 | 130 | |
| DNA/DNA (dG)b | dCTP | 2.4 ± 0.3 | 119 ± 3 | 50 ± 6 | 1 | 1 |
| dATP | 84 ± 9 | 11 ± 1 | 0.13 ± 0.02 | 0.0026 | 380 | |
| dGTP | 58 ± 7 | 12 ± 1 | 0.21 ± 0.03 | 0.0042 | 240 | |
| dTTP | 170 ± 16 | 58 ± 2 | 0.34 ± 0.03 | 0.0068 | 150 | |
| rCTP | 188 ± 14 | 12 ± 1 | 0.064 ± 0.007 | 0.0013 | 770 | |
| RNA/DNA (8-oxodG) | dCTP | 98 ± 17 | 168 ± 9 | 1.7 ± 0.3 | 1 | 1 |
| dATP | 56 ± 4 | 59 ± 1 | 1.1 ± 0.1 | 0.65 | 1.5 | |
| dGTP | 53 ± 4 | 19 ± 1 | 0.36 ± 0.03 | 0.21 | 5 | |
| dTTP | 185 ± 17 | 7.9 ± 0.2 | 0.043 ± 0.004 | 0.025 | 40 | |
| rCTP | 633 ± 68 | 2.3 ± 0.1 | 0.0036 ± 0.0004 | 0.0021 | 480 | |
| DNA/RNA (rG) | dCTP | 3.5 ± 0.5 | 80 ± 4 | 23 ± 3 | 1 | 1 |
| dATP | 146 ± 16 | 8.6 ± 0.3 | 0.059 ± 0.007 | 0.0026 | 380 | |
| dGTP | 159 ± 20 | 5.0 ± 0.2 | 0.031 ± 0.004 | 0.0013 | 770 | |
| dTTP | 404 ± 50 | 15 ± 1 | 0.037 ± 0.005 | 0.0016 | 630 | |
| rCTP | 34 ± 3 | 0.78 ± 0.02 | 0.023 ± 0.002 | 0.0010 | 1,000 |
a Misinsertion frequency: f = (kcat/Km)incorrect/(kcat/Km)correct.
b This part of the results has been published (18).
Table 2.
Steady-state kinetics of hpol η insertion of nucleotides opposite dT or CPD lesion
| Primer/template | dNTP/rNTP | Km | Kcat | kcat/Km | fa | 1/f |
|---|---|---|---|---|---|---|
| μm | min−1 | μm−1 min−1 | ||||
| RNA/DNA (dT) | dCTP | 387 ± 59 | 3.0 ± 0.2 | 0.0078 ± 0.0013 | 0.0087 | 110 |
| dATP | 141 ± 16 | 127 ± 5 | 0.90 ± 0.11 | 1 | 1 | |
| dGTP | 109 ± 5 | 13 ± 1 | 0.12 ± 0.01 | 0.13 | 8 | |
| dTTP | 393 ± 77 | 5.7 ± 0.4 | 0.015 ± 0.003 | 0.017 | 59 | |
| rATP | 290 ± 30 | 0.39 ± 0.01 | 0.0013 ± 0.0001 | 0.0014 | 710 | |
| DNA/DNA (dT)b | dCTP | 127 ± 8 | 19 ± 1 | 0.15 ± 0.01 | 0.0048 | 210 |
| dATP | 2.5 ± 0.2 | 77 ± 2 | 31 ± 3 | 1 | 1 | |
| dGTP | 9.1 ± 0.6 | 27 ± 1 | 3.0 ± 0.2 | 0.097 | 10 | |
| dTTP | 132 ± 15 | 27 ± 1 | 0.20 ± 0.02 | 0.0065 | 150 | |
| rATP | 278 ± 37 | 2.5 ± 0.1 | 0.0090 ± 0.0012 | 0.00029 | 3,400 | |
| RNA/DNA (CPD) | dCTP | 235 ± 23 | 15 ± 1 | 0.064 ± 0.008 | 0.019 | 53 |
| dATP | 35 ± 3 | 118 ± 2 | 3.4 ± 0.3 | 1 | 1 | |
| dGTP | 145 ± 14 | 32 ± 1 | 0.22 ± 0.02 | 0.065 | 15 | |
| dTTP | 200 ± 17 | 25 ± 1 | 0.13 ± 0.01 | 0.038 | 26 | |
| rATP | 282 ± 27 | 0.56 ± 0.02 | 0.0020 ± 0.0002 | 0.00059 | 1,700 | |
| DNA/DNA (CPD)b | dCTP | 31 ± 3 | 27 ± 1 | 0.87 ± 0.09 | 0.026 | 38 |
| dATP | 1.7 ± 0.2 | 57 ± 1 | 34 ± 4 | 1 | 1 | |
| dGTP | 34 ± 3 | 40 ± 1 | 1.2 ± 0.1 | 0.035 | 29 | |
| dTTP | 23 ± 2 | 32 ± 1 | 1.4 ± 0.1 | 0.041 | 24 | |
| rATP | 295 ± 35 | 7.0 ± 0.3 | 0.024 ± 0.003 | 0.00071 | 1,400 |
a Misinsertion frequency: f = (kcat/Km)incorrect/(kcat/Km)correct.
b This part of the results has been published (18).
hpol η extends an RNA primer opposite a DNA template containing CPD or 8-oxodG
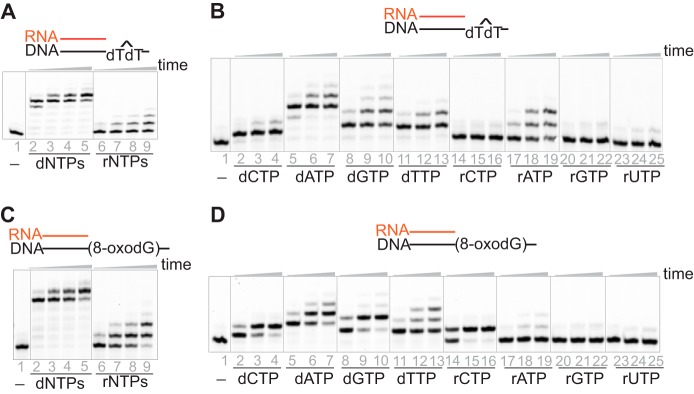
hpol η is a translesion DNA polymerase, and bypass of the UV-induced DNA lesion CPD is one of its most important roles in cells (14). We included CPD and another common DNA lesion, 8-oxodG, in our study. hpol η was able to extend RNA primers opposite both lesions with a mixture of either dNTPs or rNTPs (1 mm each nucleotide), and the extension patterns were similar to that for the RNA/DNA (dG) substrate (Figs. 1A and 2 (A and C)). In addition, the preferred insertion order showed little difference among substrates with different templates: undamaged bases, CPD, or 8-oxodG (Tables 1 and 2 and Figs. 1B and 2 (B and D)). In summary, hpol η had the ability to extend an RNA primer opposite lesions on the DNA template.
Figure 2.
hpol η elongates an RNA primer opposite CPD or 8-oxodG. A and C, hpol η (1.2 μm) elongated an RNA primer opposite CPD (A) or 8-oxodG (C) (5 μm) in the presence of a mixture of dNTPs or rNTPs (1 mm concentration of each nucleotide) for 5, 30, 55, and 240 min. B and D, hpol η (500 nm) was incubated with RNA/DNA (CPD) (B) and RNA/DNA (8-oxodG) (D) (5 μm) in the presence of 1 mm concentrations of each single dNTP or rNTP for 5, 30, and 55 min.
hpol η elongates RNA primers in the presence of physiological concentrations of dNTPs or rNTPs
Cellular concentrations of rNTPs and dNTPs vary and change depending on cell types and cell cycles. In general, concentrations of rNTPs are much higher than those of dNTPs (5). In considering physiological relevance, we tested primer extension by hpol η in the presence of typical cellular concentrations of dNTPs and rNTPs (19, 20). The RNA primers were extended to full length after 5 min of incubation with physiological concentrations of dNTPs against either a native DNA template or one containing a CPD lesion. However, insertion of ribonucleotides was slower: only one or two were inserted after 60 min (Fig. 3, A and B). These results indicated that hpol η probably extends an RNA primer by mainly inserting dNTPs instead of rNTPs in a cellular environment, thus resembling the activity of pol α at the initiation of replication or the origin of each Okazaki fragment. Cellular RNA extension by hpol η remains to be investigated.
Figure 3.
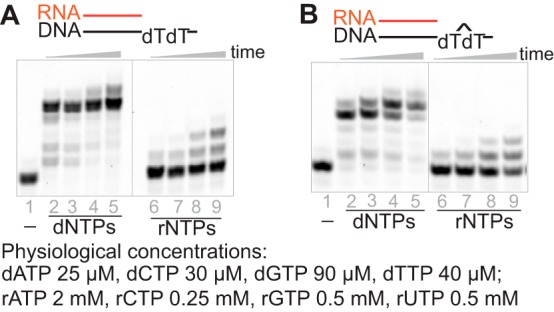
hpol η extends an RNA primer opposite DNA in the presence of physiological concentrations of nucleotides. hpol η (500 nm) and RNA/DNA (dT) (A) or RNA/DNA (CPD) (B) (5 μm) were incubated with physiological concentrations of dNTPs or rNTPs (19, 20) for 5, 10, 30, and 60 min.
hpol η is a reverse transcriptase
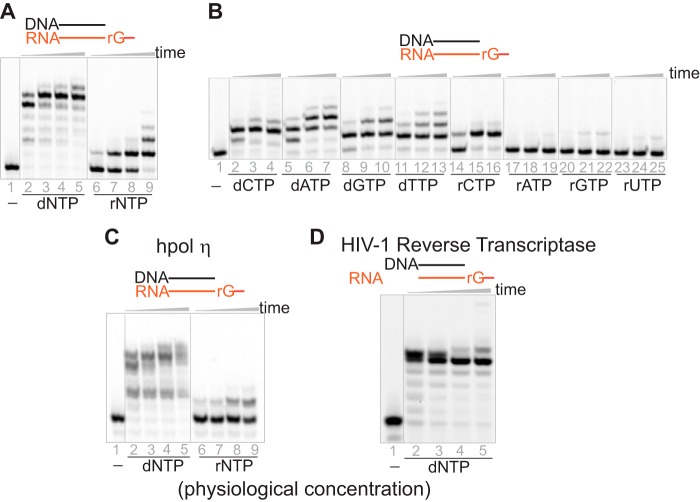
DNA polymerases not only misinsert ribonucleotides into genomes but also conduct translesion synthesis past inserted rNMPs during replication (12). Yeast pol α, pol ϵ, and pol δ have been shown to have weak activities for bypass of ribonucleotides on the template strand (≤4 nucleotides) in vitro (5, 21). To assess the capability of hpol η to bypass a series of ribonucleotides, we utilized an extreme condition by testing a DNA/RNA substrate, a DNA primer paired opposite an RNA template. The elongation pattern for DNA/RNA was very similar to that for the RNA/DNA complex. The DNA primer was elongated to the end by hpol η within 5 min against an RNA template with a mixture of dNTPs (1 mm concentration of each dNTP), whereas only 1–3 nucleotides were incorporated with a mixture of rNTPs (Fig. 4A). The catalytic efficiency for dCTP incorporation opposite rG was 23 μm−1 min−1, 1,000-fold higher than that for rCTP and several hundred times higher than those for non-paired dNTPs. The kinetic data for non-paired rNTPs were too low to be quantitated. This preferred order of insertion for DNA/RNA was consistent with RNA/DNA and DNA/DNA complexes (Table 1 and Fig. 4B). Together, these results indicated that the change of template or primer from DNA to RNA did not significantly affect the base and sugar selectivity of hpol η (Table 1).
Figure 4.
hpol η has reverse transcription activity. A, hpol η (1.2 μm) extended a DNA primer paired with an RNA template (rG) (5 μm) with a mixture of dNTPs or rNTPs (1 mm each nucleotide). The reactions were conducted for 5, 30, 55, and 240 min. B, single nucleotide incorporation assays were conducted by incubation of hpol η (500 nm) and DNA/RNA (rG) (5 μm) as well as a single nucleotide (1 mm) for 5, 30, and 55 min. C, hpol η (500 nm) and DNA/RNA (rG) (5 μm) were incubated with physiological concentrations of dNTPs or rNTPs (19, 20) for 5, 10, 30, and 60 min. D, HIV-1 RT (500 nm) was incubated with DNA/RNA (rG) (5 μm) in the presence of physiological concentrations of dNTPs (19, 20) for 5, 10, 30, and 60 min.
Noticeably, the catalytic efficiencies were similar for hpol η to extend the DNA primers by inserting dCTP opposite rG on an RNA template and dG on a DNA template: 23 and 50 μm−1 min−1, respectively. The 2.2-fold difference between DNA/RNA and DNA/DNA complexes was much smaller than the 33-fold difference between RNA/DNA and DNA/DNA, indicating that the backbone of the primer might play a more important role in determining the catalytic efficiency of hpol η compared with that of the template (Table 1). In summary, hpol η had reverse transcription activity, and its catalytic efficiency for the DNA/RNA complex was similar to that for DNA/DNA.
In addition, to estimate the reverse transcription activity of hpol η, HIV-1 reverse transcriptase (HIV-1 RT) was included for comparison. With the mixture of physiological concentrations of dNTPs, equal concentrations (500 nm) of hpol η and HIV-1 RT were both able to extend ≥90% of the primers within 5 min, although the elongation patterns were slightly different, demonstrating that hpol η is an effective reverse transcriptase in vitro (Fig. 4, C and D).
hpol η elongates the primers of RNA/DNA and DNA/RNA hybrids regardless of the presence of an equal amount of DNA/DNA
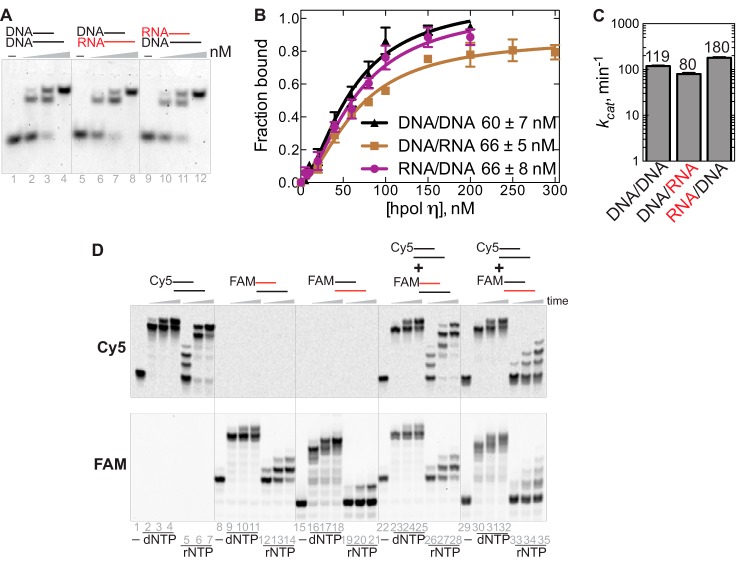
hpol η has similar binding affinities to DNA/DNA, DNA/RNA, and RNA/DNA complexes (Fig. 5, A and B). By comparing the steady-state kinetic results, an interesting phenomenon was observed; the kcat values of the paired dNTP (dCTP) incorporation were very similar among all of the substrates: 119 ± 3 min−1 for DNA/DNA (dG), 80 ± 4 min−1 for DNA/RNA (rG), and 180 ± 6 min−1 for RNA/DNA (dG) (Fig. 5C and Table 1). The differences in the catalytic efficiencies (kcat/Km) were mainly due to the Km values (Table 1), indicating that concentrations of incoming nucleotides played an important role in determining the rate of strand extension for each substrate by hpol η. To test this hypothesis, two fluorescent dyes with non-overlapping spectral properties were used, FAM and Cy5. A Cy5-labeled DNA/DNA substrate (Cy5-DNA/DNA) was mixed with FAM-RNA/DNA or FAM-DNA/RNA at a molar ratio of 1:1, and these annealed duplex mixtures were tested in extension assays with hpol η before scanning in the two corresponding spectral channels. Comparing lanes 22–28 and lanes 1–7 in the top panel of Fig. 5D, the primers of Cy5-DNA/DNA were extended to the same extent, regardless of whether the duplex was mixed with FAM-RNA/DNA or present by itself. Also, the primer elongation patterns of FAM-RNA/DNA were nearly identical regardless of the presence of Cy5-DNA/DNA (Fig. 5D, bottom, lanes 22–28 and 8–14). In addition, hpol η inserted dNMPs into the primers of FAM-DNA/RNA at similar levels with or without the presence of Cy5-DNA/DNA (Fig. 5D, lanes 16–18 and 30–32). However, in the presence of rNTPs, the elongation patterns were slightly changed when incubating with the mixture of FAM-DNA/RNA and Cy5-DNA/DNA compared with that of each substrate alone (Fig. 5D, lanes 5–7, 19–21, and 33–35). These results showed that hpol η was able to extend the primer of RNA/DNA or DNA/RNA regardless of the presence of an equal amount of the DNA/DNA substrate.
Figure 5.
Substrate binding and competition assay for hpol η. A, annealed substrates DNA/DNA, DNA/RNA, and RNA/DNA (20 nm) were incubated with 0, 50, 100, and 200 nm hpol η, respectively, at 30 °C for 30 min. B, titration of hpol η (0–300 nm) with the different substrates (20 nm). The quantified data were best fit with the equation, Y = Bmax × Xh/(Kdh + Xh) (in GraphPad Prism). The maximum fraction bound values, Bmax, were close to unity for all three substrates, and the Hill coefficients were 1.6 ± 0.2, 1.7 ± 0.2, and 1.7 ± 0.2 for DNA/DNA, DNA/RNA, and RNA/DNA complexes, respectively, indicative of multiple proteins binding with a molecule of DNA. The Kd value for each substrate is indicated in the figure. C, comparison of kcat values from steady-state kinetics for each substrate by hpol η. D, Cy5-DNA/DNA was mixed with FAM-RNA/DNA or FAM-DNA/RNA at a molar ratio of 1:1. hpol η (500 nm) and annealed substrates (5 μm) were reacted with the mixture of dNTPs or rNTPs (1 mm concentration of each nucleotide) for 5, 30, and 60 min. The top panel was scanned in the Cy5 channel, and the bottom panel was scanned in the FAM channel (GE Typhoon system).
hpol κ extends the primers of RNA/DNA and DNA/RNA
hpol η can accommodate RNA for strand elongation, possibly due to its open active site on the major groove side (14). Another Y-family human DNA polymerase, hpol κ, with an open active site in the minor groove, also drew our attention, and we therefore undertook similar experiments with hpol κ. hpol κ barely inserted any ribonucleotides, but the enzyme was able to incorporate 2′-deoxyribonucleotides at the primer ends of RNA/DNA, DNA/RNA, and DNA/DNA substrates, even with an 8-oxodG in the template strand (Fig. 6). However, the catalytic efficiencies were reduced dramatically for RNA/DNA (dG) and DNA/RNA (rG), by 260- and 2,400-fold, respectively, compared with the DNA/DNA (dG) complex (Table 3). In summary, hpol κ is another DNA polymerase that can accommodate RNA strands for primer elongation.
Figure 6.
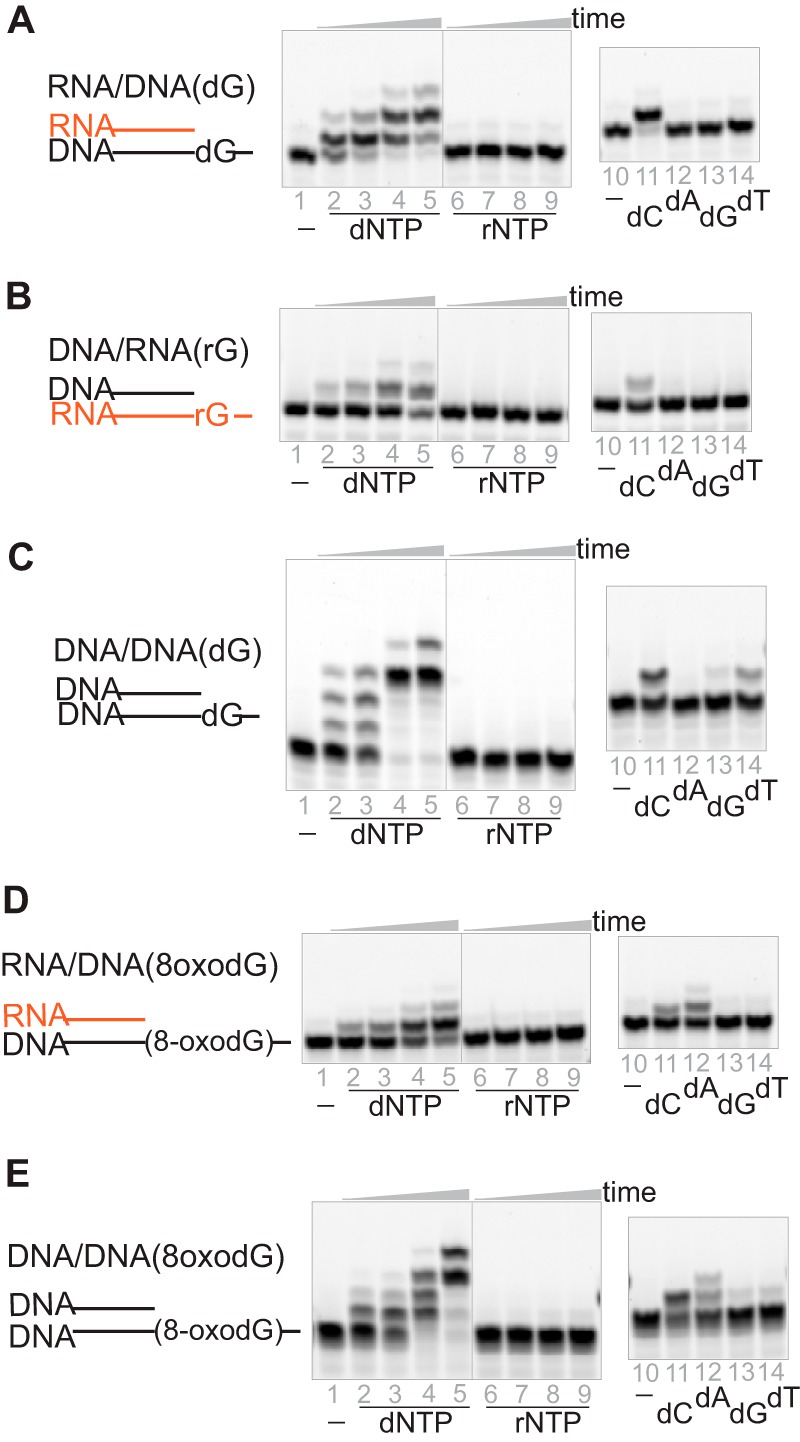
hpol κ can also accommodate an RNA strand during primer extension. hpol κ (500 nm) was incubated with 5 μm RNA/DNA (dG) (A), DNA/RNA (rG) (B), DNA/DNA (dG) (C), RNA/DNA (8-oxodG) (D), and DNA/DNA (8-oxodG) (E) in the presence of either a mixture of dNTPs or rNTPs (1 mm each nucleotide) or a single dNTP (1 mm). The reaction times were 5, 10, 30, and 60 min with the mixtures or 5 min with a single dNTP.
Table 3.
Steady-state kinetics of hpol κ incorporation of nucleotides
| Primer/template | dNTP | Km | kcat | kcat/Km | -Fold changea | fb |
|---|---|---|---|---|---|---|
| μm | min−1 | μm−1 min−1 | -fold | |||
| RNA/DNA (dG) | dCTP | 330 ± 42 | 4.5 ± 0.2 | 0.014 ± 0.002 | 260 | |
| DNA/RNA (rG) | dCTP | 726 ± 145 | 1.1 ± 0.1 | 0.0015 ± 0.0003 | 2,400 | |
| DNA/DNA (dG) | dCTP | 0.47 ± 0.04 | 1.7 ± 0.1 | 3.6 ± 0.4 | ||
| RNA/DNA (8-oxodG) | dCTP | 618 ± 86 | 1.3 ± 0.1 | 0.0021 ± 0.0003 | 1 | |
| RNA/ DNA (8-oxodG) | dATP | 497 ± 81 | 1.1 ± 0.1 | 0.0022 ± 0.0004 | 1.05 | |
| DNA/DNA (8-oxodG) | dCTP | 8.9 ± 0.9 | 2.1 ± 0.1 | 0.24 ± 0.03 | 1 | |
| DNA/DNA (8-oxodG) | dATP | 0.75 ± 0.08 | 1.7 ± 0.1 | 2.3 ± 0.3 | 9.6 |
a -Fold change = (kcat/Km)hybrid/(kcat/Km)DNA/DNA.
b Misinsertion frequency: f = (kcat/Km)incorrect/(kcat/Km)correct.
In addition, steady-state kinetic data showed that hpol κ extended the primers of RNA/DNA (dG), DNA/RNA (rG), and DNA/DNA (dG) with similar kcat values of 4.5, 1.1, and 1.7 min−1, respectively (Table 3). This phenomenon of similar kcat values was also observed for hpol η. The substrate competition assays with Cy5-DNA/DNA and FAM-RNA/DNA or FAM-DNA/RNA were also tested with hpol κ. The results showed that the ability of hpol κ to extend the primer of RNA/DNA or DNA/RNA was not significantly disrupted in the presence of an equal amount of DNA/DNA, as observed for hpol η (supplemental Fig. S1).
Discussion
The human DNA polymerase Y family includes hpol η, hpol κ, hpol ι, and REV1, all of which have been widely studied during the past 2 decades. These polymerases, with large spacious active sites and relatively poor fidelity, are often responsible for generating mutations, which may lead to cancer and other diseases. In addition, they have the ability to bypass DNA lesions on templates during replication, thereby rescuing the cell cycle by restarting replication forks (14). Also, previous studies reported that hpol η and hpol ι were able to insert ribonucleotides into DNA but with low catalytic efficiencies compared with those for 2′-deoxyribonucleotides (18, 22). This finding with hpol η can be explained by inspecting crystal structures of hpol η–DNA-rNTP complexes; the incoming rNTP adopts a conformation that avoids a clash with the steric gate residue, Phe-18, leading to a longer distance between the primer 3′-end and the α-phosphate of the incoming nucleotide and therefore a higher energy barrier for the reaction compared with dNTP (18).
Our results demonstrate that hpol η, as well as hpol κ, is able to manifest a variety of unexpected functions by accommodating RNA for strand extension. Extending an RNA primer by incorporation of 2′-deoxyribonucleotides opposite a DNA template is a typical process conducted by hpol α, commonly seen at the origin of replication and each Okazaki fragment. The ability of hpol η to utilize RNA/DNA as a substrate indicates that it may replace hpol α in extending an RNA primer when encountering a CPD lesion during the replication initiation process. Another human replication initiation polymerase, PrimPol, has been shown to play a role in cell survival upon UV irradiation and to conduct translesion synthesis opposite DNA lesions (23, 24). However, PrimPol is more efficient in the bypass of another type of UV-induced DNA damage, 6-4 photoproducts, instead of CPD (23, 24). In fact, whether PrimPol can replicate past CPD is still controversial. One study showed that PrimPol only extended the DNA primer after a CPD lesion (23) instead of inserting 2′-deoxyribonucleotides opposite CPD. However, another study reported that it mainly stopped at the CPD, and only a small percentage of the DNA primer escaped for further extension by PrimPol (24). Of note, the primers used in both studies were DNA strands. On the basis of all of these results, we believe that hpol η is a strong candidate for bypass of CPD lesions by replacing hpol α during replication initiation (Fig. 7A).
Figure 7.
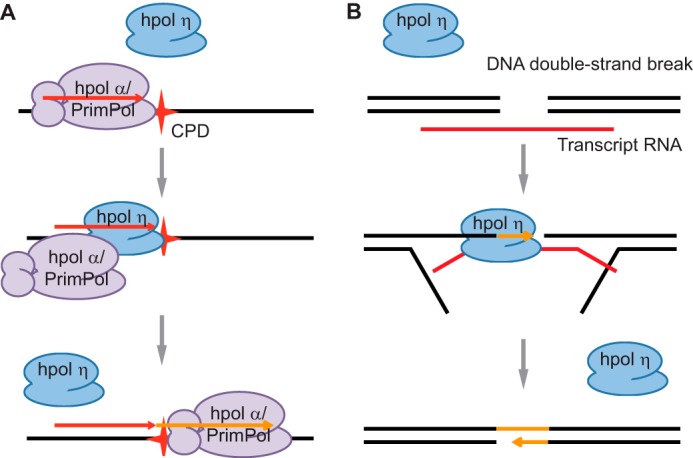
hpol η is possibly involved in two additional biological processes due to its RNA-accommodating ability. A, model in which hpol η replaces hpol α or PrimPol to conduct translesion bypass CPD by extending an RNA primer at the origin of replication or an Okazaki fragment. B, a model depicting how hpol η might use the transcript RNA strand as a template to synthesize DNA at a DNA double-strand break. RNA is in red, DNA is in black, and the newly synthesized strand is shown in orange. The red star in A represents a CPD lesion.
In addition to extending an RNA primer opposite a DNA template, hpol η and hpol κ can perform strand elongation with DNA/RNA substrates by mainly inserting 2′-deoxyribonucleotides, thus demonstrating their reverse transcription activities. In a previous report, the catalytic efficiency of HIV-1 RT for a DNA/RNA substrate was 46 μm−1 min−1 (13), only slightly higher than that of hpol η, 23 μm−1 min−1 in our study here (Table 1). To make a more careful comparison, HIV-1 RT was also included. The side-by-side extension experiments by hpol η and HIV-1 RT showed that the two polymerases extended the DNA/RNA substrate with similar efficiencies (Fig. 4, C and D). In summary, hpol η is a relatively efficient reverse transcriptase.
It has been shown that unknown enzymes can utilize RNA as a template to synthesize DNA during double-strand break repair (12, 25, 26). Y-family DNA polymerase hpol η is possibly the one to fill this gap due to its reverse transcription activity, especially in the presence of physiological concentrations of nucleotides, according to our results (Figs. 4 and 7B).
hpol η, as well as hpol κ, prefers to insert 2′-deoxyribonucleotides instead of ribonucleotides regardless of the sugar-phosphate backbone of substrates. The preferred order of insertion of hpol η (paired dNTP > non-paired dNTP ≥ paired rNTP > non-paired rNTP) is consistent among all of the substrates we studied: RNA/DNA, DNA/RNA, and DNA/DNA. An interesting observation is that the kcat values for paired dNTPs were similar among substrates for both hpol η and κ: less than a 2.3-fold difference for hpol η and at most 4-fold for hpol κ (Fig. 5C and Tables 1 and 3). The difference in the catalytic efficiencies (kcat/Km) is mainly derived from the Km values for both polymerases, suggesting that local concentrations of dNTPs may play a role in determining the rates of hpol η or hpol κ for extension of the substrates. At physiological concentrations of nucleotides, hpol η is able to effectively conduct strand extension for both hybrid substrates (Figs. 3 and 4C), indicating the possibility that hpol η can extend the RNA strand opposite a DNA template or the DNA strand opposite an RNA template in vivo.
In summary, our studies demonstrate unexpected activities of hpol η and hpol κ in accommodating RNA for strand extension, without sacrificing base and sugar discrimination ability and regardless of the presence of an equal amount of the DNA/DNA substrate. Our results provide evidence for other functions of these two polymerases in addition to their well-known roles in translesion synthesis.
Materials and methods
Oligonucleotide substrates
The oligonucleotide containing the CPD lesion was purchased from TriLink BioTechnologies (San Diego, CA), and the others were from Integrated DNA Technologies (Coralville, IA). All of these oligonucleotides were purified by HPLC (reversed phase) by the manufacturers. The fluorescence-labeled primers 5′-FAM-CGG GCT CGT AAG CGT CAT-3′ and 5′-FAM-rCrGrG rGrCrU rCrGrU rArArG rCrGrU rCrArU-3′ were annealed with template oligonucleotides in a 1:1 molar ratio of (i) 5′-TCA (CPD)A TGA CGC TTA CGA GCC CG-3′ (where CPD indicates cyclobutane pyrimidine dimer), (ii) 5′-TCA TTA TGA CGC TTA CGA GCC CG-3′, (iii) 5′-TCA TGA TGA CGC TTA CGA GCC CG-3′, and (iv) 5′-TCA T(8-oxodG)A TGA CGC TTA CGA GCC CG-3′. In addition, the template oligonucleotides 5′-rUrCrA rUrGrA rUrGrA rCrGrC rUrUrA rCrGrA rGrCrC rCrG-3′ were annealed with 5′-FAM-CGG GCT CGT AAG CGT CAT-3′. 5′-Cy5-CGG GCT CGT AAG CGT CAT-3′ and 5′-TCA TGA TGA CGC TTA CGA GCC CG-3′ were annealed for substrate competition assays.
Protein expression and purification
The catalytic cores of wild-type hpol η (amino acids 1–432) and hpol κ (amino acids 19–526) were expressed in Escherichia coli and purified as reported previously (27–29). HIV-1 RT (full-length p66 subunit and p51 subunit with C-terminal deletion of 13 amino acids) was a gift from Dr. Kenneth A. Johnson (University of Texas, Austin, TX) (30).
Extension, single nucleotide incorporation, and steady-state kinetics assays
hpol η, as well as hpol κ or HIV-1 reverse transcriptase, was incubated with annealed 5′-FAM–labeled primer/template substrates in the reaction buffer (40 mm Tris-HCl (pH 7.5), 10 mm DTT, 0.1 mg/ml bovine serum albumin, 5% glycerol (v/v), 5 mm MgCl2, and 100 mm KCl) at 37 °C for 5 min before adding the mixtures of nucleotides or a single nucleotide. Reactions were conducted for the indicated times and stopped by the addition of a quench buffer (95% formamide (v/v) and 10 mm EDTA). The reaction products were loaded onto 18% (w/v) denaturing polyacrylamide gels, separated, and visualized using a Typhoon system (GE Healthcare).
For steady-state kinetic assays, the formation of product was always kept <20% of the initial substrate concentration. The results were quantified using ImageJ software, with (hyperbolic, nonlinear regression) fitting to the Michaelis–Menten equation using the program Prism (GraphPad, La Jolla, CA) (28). The results are presented as values ± S.D. using the program Prism.
Electrophoretic mobility shift assay
FAM-labeled annealed DNA/DNA, RNA/DNA, or DNA/RNA duplexes (20 nm) were incubated with 0–300 nm hpol η in the presence of the buffer containing 20 mm HEPES (pH 7.5), 100 mm KCl, 5 mm MgCl2, 5% glycerol (v/v), 0.25 mg/ml bovine serum albumin, and 10 mm DTT at 30 °C for 30 min before adding 15% FicollTM (w/v). The products were loaded on 5% native polyacrylamide gels, separated using a current of 10 mA for 30 min, and visualized with a Typhoon system.
The results were quantified with ImageJ software and fit with a Hill equation, Y = Bmax × Xh/(Kdh + Xh) where X is the protein concentration, Y is the fraction bound, Bmax is the maximum bound fraction, h is the Hill coefficient, and Kd is the protein concentration needed to reach half-maximal fractional binding.
Substrate competition assay
Annealed Cy5-labeled DNA/DNA substrate was mixed with FAM-labeled RNA/DNA or DNA/RNA at a molar ratio of 1:1. The substrate (5 μm) was preincubated with 500 nm hpol η in the reaction buffer (same as for the extension assays) at 37 °C for 5 min, and a mixture of dNTPs or rNTPs (1 mm each nucleotide) was added to initiate the reaction. Reaction times were 5, 30, and 60 min. Products were separated on denaturing polyacrylamide gels and scanned for FAM and Cy5 signals separately with a Typhoon system. All experiments were done at least twice.
Author contributions
Y. S. designed and conducted the experiments and analyzed the results. Y. S., F. P. G., and M. E. conceived the studies and wrote the paper.
Supplementary Material
Acknowledgments
We thank Dr. Kenneth A. Johnson for providing HIV-1 RT. We also thank Dr. David Cortez (Vanderbilt University) for discussion of some of the results and K. Trisler for assistance in preparation of the manuscript.
This work was supported, in whole or in part, by National Institutes of Health Grants R01 ES026955 (to F. P. G. and M. E.), R01 ES010546 (to F. P. G.), and P01 CA160032 (to M. E.). The authors declare that they have no conflicts of interest with the contents of this article. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
This article contains supplemental Fig. S1.
- hpol
- human (DNA) polymerase
- pol
- (DNA) polymerase
- CPD
- cyclobutane pyrimidine dimer
- FAM
- 6-carboxyfluorescein
- 8-oxodG
- 8-oxo-2′-deoxyguanosine
- rNMPs
- ribonucleoside monophosphates
- rNTPs
- ribonucleoside triphosphates.
References
- 1. Cheung-Ong K., Giaever G., and Nislow C. (2013) DNA-damaging agents in cancer chemotherapy: serendipity and chemical biology. Chem. Biol. 20, 648–659 [DOI] [PubMed] [Google Scholar]
- 2. Vaisman A., Masutani C., Hanaoka F., and Chaney S. G. (2000) Efficient translesion replication past oxaliplatin and cisplatin GpG adducts by human DNA polymerase η. Biochemistry 39, 4575–4580 [DOI] [PubMed] [Google Scholar]
- 3. Gregory M. T., Park G. Y., Johnstone T. C., Lee Y. S., Yang W., and Lippard S. J. (2014) Structural and mechanistic studies of polymerase eta bypass of phenanthriplatin DNA damage. Proc. Natl. Acad. Sci. U.S.A. 111, 9133–9138 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Srivastava A. K., Han C., Zhao R., Cui T., Dai Y., Mao C., Zhao W., Zhang X., Yu J., and Wang Q. E. (2015) Enhanced expression of DNA polymerase η contributes to cisplatin resistance of ovarian cancer stem cells. Proc. Natl. Acad. Sci. U.S.A. 112, 4411–4416 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Williams J. S., Lujan S. A., and Kunkel T. A. (2016) Processing ribonucleotides incorporated during eukaryotic DNA replication. Nat. Rev. Mol. Cell Biol. 17, 350–363 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. van Loon B., Woodgate R., and Hübscher U. (2015) DNA polymerases: biology, diseases and biomedical applications. DNA Repair 29, 1–3 [DOI] [PubMed] [Google Scholar]
- 7. Göksenin A. Y., Zahurancik W., LeCompte K. G., Taggart D. J., Suo Z., and Pursell Z. F. (2012) Human DNA polymerase ϵ is able to efficiently extend from multiple consecutive ribonucleotides. J. Biol. Chem. 287, 42675–42684 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Clausen A. R., Zhang S., Burgers P. M., Lee M. Y., and Kunkel T. A. (2013) Ribonucleotide incorporation, proofreading and bypass by human DNA polymerase δ. DNA Repair 12, 121–127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Kasiviswanathan R., and Copeland W. C. (2011) Ribonucleotide discrimination and reverse transcription by the human mitochondrial DNA polymerase. J. Biol. Chem. 286, 31490–31500 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Ruiz J. F., Juárez R., García-Díaz M., Terrados G., Picher A. J., González-Barrera S., Fernández de Henestrosa A. R., and Blanco L. (2003) Lack of sugar discrimination by human pol μ requires a single glycine residue. Nucleic Acids Res. 31, 4441–4449 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Boulé J.-B., Rougeon F., and Papanicolaou C. (2001) Terminal deoxynucleotidyl transferase indiscriminately incorporates ribonucleotides and deoxyribonucleotides. J. Biol. Chem. 276, 31388–31393 [DOI] [PubMed] [Google Scholar]
- 12. Storici F., Bebenek K., Kunkel T. A., Gordenin D. A., and Resnick M. A. (2007) RNA-templated DNA repair. Nature 447, 338–341 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Murakami E., Feng J. Y., Lee H., Hanes J., Johnson K. A., and Anderson K. S. (2003) Characterization of novel reverse transcriptase and other RNA-associated catalytic activities by human DNA polymerase γ: importance in mitochondrial DNA replication. J. Biol. Chem. 278, 36403–36409 [DOI] [PubMed] [Google Scholar]
- 14. Yang W. (2014) An overview of Y-family DNA polymerases and a case study of human DNA polymerase η. Biochemistry 53, 2793–2803 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Masutani C., Kusumoto R., Yamada A., Dohmae N., Yokoi M., Yuasa M., Araki M., Iwai S., Takio K., and Hanaoka F. (1999) The XPV (xeroderma pigmentosum variant) gene encodes human DNA polymerase η. Nature 399, 700–704 [DOI] [PubMed] [Google Scholar]
- 16. Inui H., Oh K. S., Nadem C., Ueda T., Khan S. G., Metin A., Gozukara E., Emmert S., Slor H., Busch D. B., Baker C. C., DiGiovanna J. J., Tamura D., Seitz C. S., Gratchev A., et al. (2008) Xeroderma pigmentosum-variant patients from America, Europe, and Asia. J. Invest. Dermatol. 128, 2055–2068 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Johnson R. E., Kondratick C. M., Prakash S., and Prakash L. (1999) hRAD30 mutations in the variant form of xeroderma pigmentosum. Science 285, 263–265 [DOI] [PubMed] [Google Scholar]
- 18. Su Y., Egli M., and Guengerich F. P. (2016) Mechanism of ribonucleotide incorporation by human DNA polymerase η. J. Biol. Chem. 291, 3747–3756 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Crespan E., Furrer A., Rösinger M., Bertoletti F., Mentegari E., Chiapparini G., Imhof R., Ziegler N., Sturla S. J., Hübscher U., van Loon B., and Maga G. (2016) Impact of ribonucleotide incorporation by DNA polymerases β and λ on oxidative base excision repair. Nat. Commun. 7, 10805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Mentegari E., Crespan E., Bavagnoli L., Kissova M., Bertoletti F., Sabbioneda S., Imhof R., Sturla S. J., Nilforoushan A., Hübscher U., van Loon B., and Maga G. (2017) Ribonucleotide incorporation by human DNA polymerase η impacts translesion synthesis and RNase H2 activity. Nucleic Acids Res. 45, 2600–2614 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Clausen A. R., Murray M. S., Passer A. R., Pedersen L. C., and Kunkel T. A. (2013) Structure-function analysis of ribonucleotide bypass by B family DNA replicases. Proc. Natl. Acad. Sci. U.S.A. 110, 16802–16807 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Donigan K. A., McLenigan M. P., Yang W., Goodman M. F., and Woodgate R. (2014) The steric gate of DNA polymerase ι regulates ribonucleotide incorporation and deoxyribonucleotide fidelity. J. Biol. Chem. 289, 9136–9145 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Bianchi J., Rudd S. G., Jozwiakowski S. K., Bailey L. J., Soura V., Taylor E., Stevanovic I., Green A. J., Stracker T. H., Lindsay H. D., and Doherty A. J. (2013) PrimPol bypasses UV photoproducts during eukaryotic chromosomal DNA replication. Mol. Cell 52, 566–573 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Mourón S., Rodriguez-Acebes S., Martínez-Jiménez M. I., García-Gómez S., Chocrón S., Blanco L., and Méndez J. (2013) Repriming of DNA synthesis at stalled replication forks by human PrimPol. Nat. Struct. Mol. Biol. 20, 1383–1389 [DOI] [PubMed] [Google Scholar]
- 25. Keskin H., Shen Y., Huang F., Patel M., Yang T., Ashley K., Mazin A. V., and Storici F. (2014) Transcript-RNA-templated DNA recombination and repair. Nature 515, 436–439 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Mazina O. M., Keskin H., Hanamshet K., Storici F., and Mazin A. V. (2017) Rad52 inverse strand exchange drives RNA-templated DNA double-strand break repair. Mol. Cell 67, 19–29.e3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Biertümpfel C Zhao Y., Kondo Y., Ramón-Maiques S., Gregory M., Lee J. Y., Masutani C., Lehmann A. R., Hanaoka F., and Yang W. (2010) Structure and mechanism of human DNA polymerase η. Nature 465, 1044–1048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Su Y., Patra A., Harp J. M., Egli M., and Guengerich F. P. (2015) Roles of residues Arg-61 and Gln-38 of human DNA polymerase η in bypass of deoxyguanosine and 7,8-dihydro-8-oxo-2′-deoxyguanosine. J. Biol. Chem. 290, 15921–15933 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Irimia A., Eoff R. L., Guengerich F. P., and Egli M. (2009) Structural and functional elucidation of the mechanism promoting error-prone synthesis by human DNA polymerase κ opposite the 7,8-dihydro-8-oxo-2′-deoxyguanosine adduct. J. Biol. Chem. 284, 22467–22480 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Kati W. M., Johnson K. A., Jerva L. F., and Anderson K. S. (1992) Mechanism and fidelity of HIV reverse transcriptase. J. Biol. Chem. 267, 25988–25997 [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.