Abstract
Mitochondria are organelles indispensable for maintenance of cellular energy homeostasis. Most mitochondrial proteins are nuclearly encoded and are imported into the matrix compartment where they are properly folded. This process is facilitated by the mitochondrial heat shock protein 70 (mtHsp70), a chaperone contributing to mitochondrial protein quality control. The affinity of mtHsp70 for its protein clients and its chaperone function are regulated by binding of ATP/ADP to mtHsp70's nucleotide-binding domain. Nucleotide exchange factors (NEFs) play a crucial role in exchanging ADP for ATP at mtHsp70's nucleotide-binding domain, thereby modulating mtHsp70's chaperone activity. A single NEF, Mge1, regulates mtHsp70's chaperone activity in lower eukaryotes, but the mammalian orthologs are unknown. Here, we report that two putative NEF orthologs, GrpE-like 1 (GrpEL1) and GrpEL2, modulate mtHsp70's function in human cells. We found that both GrpEL1 and GrpEL2 associate with mtHsp70 as a hetero-oligomeric subcomplex and regulate mtHsp70 function. The formation of this subcomplex was critical for conferring stability to the NEFs, helped fine-tune mitochondrial protein quality control, and regulated crucial mtHsp70 functions, such as import of preproteins and biogenesis of Fe–S clusters. Our results also suggested that GrpEL2 has evolved as a possible stress resistance protein in higher vertebrates to maintain chaperone activity under stress conditions. In conclusion, our findings support the idea that GrpEL1 has a role as a stress modulator in mammalian cells and highlight that multiple NEFs are involved in controlling protein quality in mammalian mitochondria.
Keywords: 70 kilodalton heat shock protein (Hsp70), cell culture, mitochondria, mitochondrial transport, protein import, protein translocation, yeast, nucleotide exchange factor, mitochondrial Hsp70
Introduction
Mitochondria are essential organelles involved in multiple aspects of physiological functions, including energy metabolism, redox homeostasis, apoptosis, and metabolic switch. The constitutive functions of mitochondria are regulated by a plethora of proteins, acting together to maintain cellular and organellar homeostasis. Although mitochondria contain their own genome, most of the mitochondrial proteins are nuclearly encoded and translocated inside the organelle through dedicated translocases present on the outer and inner membranes (1–6). The inner membrane translocase, also known as presequence translocase, comprises TIM23-core complex and matrix-directed import motor subunit. The mtHsp70 forms the core of the import motor, which translocates matrix-targeted proteins with the assistance of J-proteins and nucleotide exchange factors (NEFs)4 followed by subsequent folding into their native conformation, and thus maintains proteostasis and organellar health (4, 5, 7, 8).
The import motor functions through a regulated cycling of mtHsp70 at the import channel, and its chaperoning activity is modulated by the presence of J-proteins and NEFs (7, 9, 10). During the import process, binding and release of polypeptides occurs at the mtHsp70 substrate-binding domain in a cyclic manner, which is governed by the sequestration of ADP or ATP at the N-terminal nucleotide-binding pocket (7, 8, 11–13). In the ATP-bound state, mtHsp70 has low affinity for the substrate with higher association rate (Kon). Subsequent hydrolysis of bound ATP to ADP triggered by peptide binding and J-proteins transforms the substrate-binding domain into its high-affinity state with lower dissociation rate (Koff) (13–18). The NEF functions by exchanging ADP with ATP at the nucleotide-binding pocket. This promotes release of substrate and makes the chaperone ready for the next phase of the substrate binding-release cycle (13, 19–22). Through the transformation of high/low-affinity states, which depends upon cyclic hydrolysis of ATP, mtHsp70 vectorially pulls the polypeptide into the matrix. This highlights the important synergistic action of J-proteins and NEFs in the import process and overall mitochondrial function (12, 18, 21, 23–25).
Recent discoveries highlight moonlighting functions for various components of the mitochondrial protein transport machinery. Differential expression of DnaJC15, a cochaperone of mtHsp70 at the import motor, is responsible for altering the sensitivity of patients to chemotherapeutic drugs. A recent report has linked it as a modulator of cell death pathways through regulation of mitochondrial permeability transition (26, 27). DnaJC15 also regulates mitochondrial respiration as an inhibitor of respiratory complex I (28). Its paralog, DnaJC19, which has housekeeping functions in protein import, independently forms a complex with prohibitins to maintain cardiolipin metabolism in mitochondrial membranes (29–31). Magmas, which regulates mtHsp70's activity at the import channel, additionally functions as a reactive oxygen species (ROS) regulator and assists in maintenance of redox homeostasis (30, 32). Interestingly, preliminary reports in yeast have shown that activity of the NEF Mge1 is sensitive to oxidative stress (33). This further supports the view of high involvement of human NEFs in multiple pathways with potential moonlighting functions in mitochondria.
Previous in vitro reconstitution studies have shown that GrpEL1 functions as a nucleotide exchange factor for mtHsp70 in humans that could replace ADP with ATP and promote mtHsp70 chaperone activity (34). Interestingly, in silico and experimental evidence from the mammalian system reveals the existence of a second mitochondrial NEF, GrpEL2 (35). This is an intriguing observation because other species except mammals encode for a single NEF for the mitochondrial function. Despite having two NEFs as cochaperones for mtHsp70 machinery, their specific role in fine-tuning import and folding pathways in the maintenance of organellar homeostasis in humans is still elusive.
The current studies highlight the nucleotide exchange abilities of two NEFs, EL1 and EL2, in regulating human mtHsp70's function. The human NEFs showed significant difference in their complex organization as compared with the yeast NEF Mge1. Human NEFs EL1 and EL2 function as a heterosubcomplex at the import channel by stabilizing the aggregation-prone individual homo-oligomers. Strikingly, our findings reveal that EL2 has evolved as a stress resistance protein in higher vertebrates to regulate threshold mtHsp70 activity and thus modulates overall organellar function under stress conditions. Together, we propose that the interplay between the two human NEFs is utilized by the cell to regulate mitochondrial functions.
Results
EL1 and EL2 regulate human mtHsp70's functions
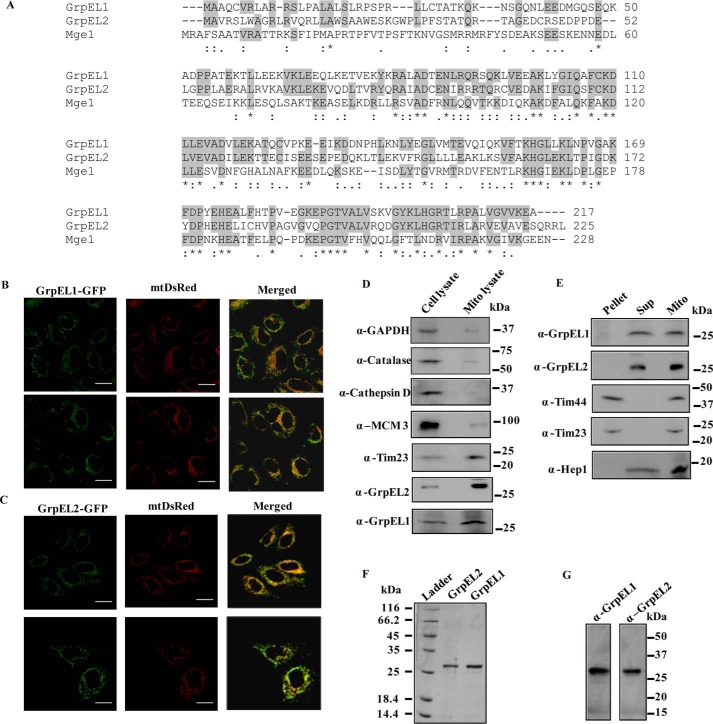
The nucleotide exchange ability of EL1 for human mtHsp70 has been documented previously using purified proteins (34, 35). A sequence alignment utilizing the UniProt database also proposed the existence of two NEF paralogs, EL1 and EL2, in human mitochondria in contrast to a single yeast ortholog, Mge1 (Fig. 1A). Sequence alignment of NEF homologs revealed that yeast Mge1 shares 39% identity with EL1 and 33% identity with EL2, whereas EL1 and EL2 share 45% sequence identity with each other (Fig. 1A). Foremost, to establish the physiological relevance of two NEF paralogs, the precise subcellular location of EL1 and EL2 in human cells was determined by microscopic analysis. Both EL1 and EL2 fused with C-terminal GFP tags were expressed separately in HeLa cells. The cells were cotransfected with mitochondrially targeted mtDsRed to counterstain the organelle. An overlap of GFP-EL1 fluorescence with mtDsRed marker establishes its mitochondrial localization (Fig. 1B). Similarly, the GFP-EL2 overlapped with red fluorescence of mtDsRed, indicating mitochondrial distribution of the protein (Fig. 1C). The cellular localization of EL1 and EL2 was further confirmed by an alternative approach using subcellular fractionation analysis. The mitochondrially enriched fraction was isolated, and the relative distribution of EL1 and EL2 was determined through Western blotting using protein-specific antibodies. The blot was probed with antibodies specific for different mitochondrial, nuclear, and cytosolic markers. Both EL1 and EL2 were enriched in the mitochondrial fraction along with another organelle-specific marker (Tim23), thereby confirming the mitochondrial localization of EL1 and EL2 (Fig. 1D). As negative controls, the blot was probed for marker proteins such as anti-GAPDH (cytosol), anti-cathepsin D (lysosome), anti-catalase (peroxisome), and anti-MCM3 antibody (nucleus) (Fig. 1D). To further establish the submitochondrial location of EL1 and EL2, the membrane and matrix fractions of mitochondria were separated using ultracentrifugation and immunoblotted using specific antibodies. Both EL1 and EL2 were separated in the matrix fraction similarly to Hep1, which served as control for matrix-targeted proteins, and membrane proteins Tim23 and Tim44 were probed as controls for inner membrane-associated proteins (Fig. 1E).
Figure 1.
A, sequence alignment. Predicted amino acid sequences of NEF orthologs from yeast (Mge1) and human (GrpEL1 and GrpEL2) are aligned using the ClustalW2 program. The identical and similar amino acid residues are highlighted. An asterisk (*) represents positions having fully conserved residue. A colon (:) represents conservation between group of residues with strongly similar properties. A period (.) represents conservation between group of residues with weakly similar properties. B–D, cellular localization of EL1/EL2. Confocal microscopic images of HeLa cells expressing EL1-GFP (B) and EL2-GFP (C) green fluorescence (left panels) and mitochondria labeled with red fluorescence cotransfected with mtDsRed marker (middle panels) are shown. Colocalization of green and red fluorescence is indicated in the merged panels. Scale bar, 10 μm. Similarly, 50 μg of protein from total cell lysate and lysates prepared from purified mitochondria were separated by SDS-PAGE and subjected to Western blot analysis using organelle-specific markers. Anti-catalase (60 kDa) for peroxisomes, -GAPDH (37 kDa) and -cathepsin D (33 kDa) for the cytosolic fraction, and -MCM3 (100 kDa) for the nuclear fraction were used as negative controls. Tim23 (23 kDa) was probed as a positive mitochondrial control. The enrichment of EL1/EL2 in mitochondria was probed using anti-GrpEL1 and -GrpEL2 antibodies (D). E, submitochondrial fractionation. Mitochondria isolated from HEK293T cells were subjected to fractionation analysis. The pellet, supernatant (Sup), and intact mitochondria (Mito) were resolved by SDS-PAGE followed by immunodecoration with specific antibodies. Mitochondrial protein Hep1 (18 kDa) was probed as a matrix control, and anti-Tim23 (23 kDa) and -Tim44 (44 kDa) antibodies were used as controls for mitochondrial inner membrane proteins. The separation of EL1/EL2 in the supernatant matrix fraction was probed using anti-GrpEL1 and -GrpEL2 antibodies. F and G, protein (EL1 and EL2) purity analysis by SDS-PAGE and Western blotting. Ni-NTA affinity-purified EL1 and EL2 were separated by SDS-PAGE, which shows the presence of single bands corresponding to their respective molecular weights (F). Mitochondria were isolated from HEK293T cells, and the lysate was used to analyze specificity of anti-GrpEL1 and anti-GrpEL2 antibodies. The Western blot analysis followed by immunodecoration with protein-specific antibodies indicates specificity of anti-GrpEL1 and -GrpEL2 antibodies (G). For all Western blots and SDS-PAGE, Precision Plus dual-color protein standard from Bio-Rad was used as standard molecular mass markers.
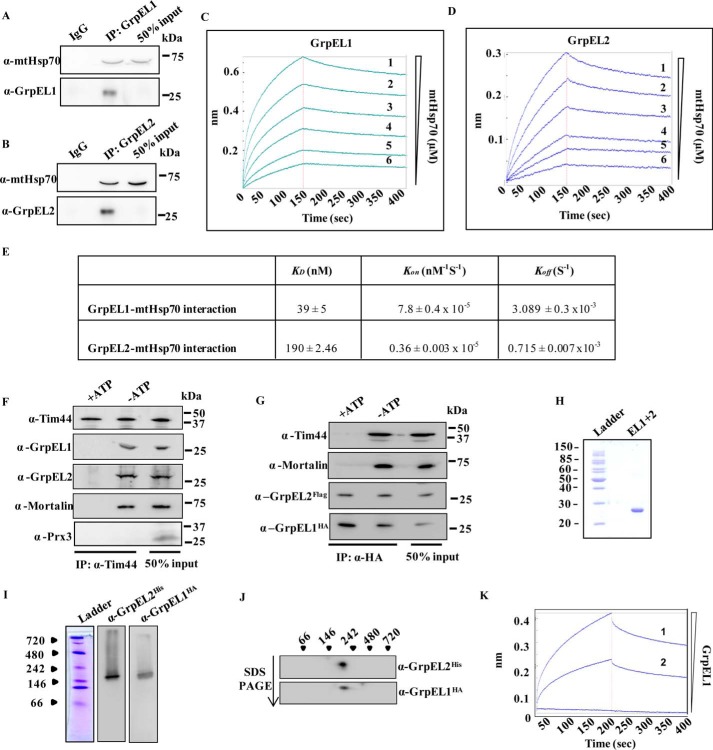
The nucleotide exchange activity of NEF is dependent on a physical interaction with Hsp70 (22). To establish this, His6-GrpEL1 and His6-GrpEL2 proteins were affinity-purified to homogeneity using Ni-NTA chromatography (Fig. 1F). The purified proteins were also used to generate protein-specific antibodies. The specificity of the respective antibodies was further confirmed in the mitochondrial lysate, which showed a single band corresponding to protein EL1 or EL2 (Fig. 1G). To determine physical interaction between mtHsp70 and EL1 or EL2, in vitro coimmunoprecipitation (coIP) analysis was performed using purified EL1/EL2 and mtHsp70. Indeed, equimolar mixtures of EL1/EL2 and mtHsp70 resulted in coimmunoprecipitation of mtHsp70 with antibody specific for EL1 or EL2, thus indicating that both NEFs physically associate with mtHsp70 (Fig. 2, A and B). The relative affinity of interaction of EL1 and EL2 with mtHsp70 was estimated through biolayer interferometry. Both EL1 and EL2 interacted robustly with mtHsp70. The affinity of EL1 for mtHsp70 (KD = 39 nm) was comparable with the affinity of bacterial GrpE for DnaK (KD = 30 nm) (36). However, EL2 showed a 5-fold lower affinity for mtHsp70 (KD = 190 nm) (Fig. 2, C–E). The reduced affinity of EL2 is mainly attributed to an ∼4-fold slower dissociation rate (Koff) and ∼21-fold slower association rate (Kon) as compared with EL1 (Fig. 2, C–E).
Figure 2.
A–E, in vitro interaction analysis. A and B, in vitro coimmunoprecipitation of purified EL1/EL2 with purified mtHsp70 using anti-EL1 (A) and anti-EL2 (B) antibodies. 50% input served as a loading control. Affinity of mtHsp70 for EL1 (C) and EL2 (D) was determined through biolayer interferometry using increasing concentrations of mtHsp70 analyte (1, 5 μm; 2, 2.5 μm; 3, 1.25 μm; 4, 0.625 μm; 5, 0.3125 μm; 6, 0.156 μm) as a function of time. E, the affinity constant (KD), association constant (Kon), and dissociation constant (Koff) for the EL1/EL2 interaction with mtHsp70 was calculated by fitting the data from the biolayer interferometry experiment using Forte Bio BioOctate software. F and G, coimmunoprecipitation. The recruitment of EL1 and EL2 to the mitochondrial import motor was analyzed by immunoprecipitation using anti-Tim44 antibody in the isolated mitochondrial lysates in the presence of 2 mm ATP (+ATP) or in the presence of 5 mm EDTA (−ATP). The blots were immunodetected using the indicated import motor subunit-specific antibodies. Anti-peroxiredoxin 3 (Prx3) antibody was used as a negative control to probe non-import motor-related protein (F). Co-recruitment of EL1 (HA tag) and EL2 (FLAG tag) at the import motor was analyzed through immunoprecipitation in isolated mitochondria using an anti-HA antibody in the presence or absence of ATP. The blot was probed with anti-HA, anti-FLAG, anti-Tim44, and anti-mtHsp70 antibodies (G). H–J, copurification of EL1-EL2 complex and in vitro interaction analysis. Coexpressed EL1-EL2 complex was purified from E. coli by Ni-NTA chromatography, and the eluate was analyzed by SDS-PAGE to check the purity (H). The EL1-EL2 complex was further resolved by native PAGE (I) and 2D SDS-PAGE (J) followed by immunoblotting with anti-His (tagging GrpEL2) and anti-HA (tagging GrpEL1) antibodies. The positions of standard molecular mass markers are indicated. K, direct interaction between EL1 and EL2 was determined by biolayer interferometry using increasing concentrations (1, 5 μm; 2, 2.5 μm) of GrpEL2 analyte as a function of time.
Regulation of human mitochondrial import by EL1-EL2 hetero-oligomeric complex
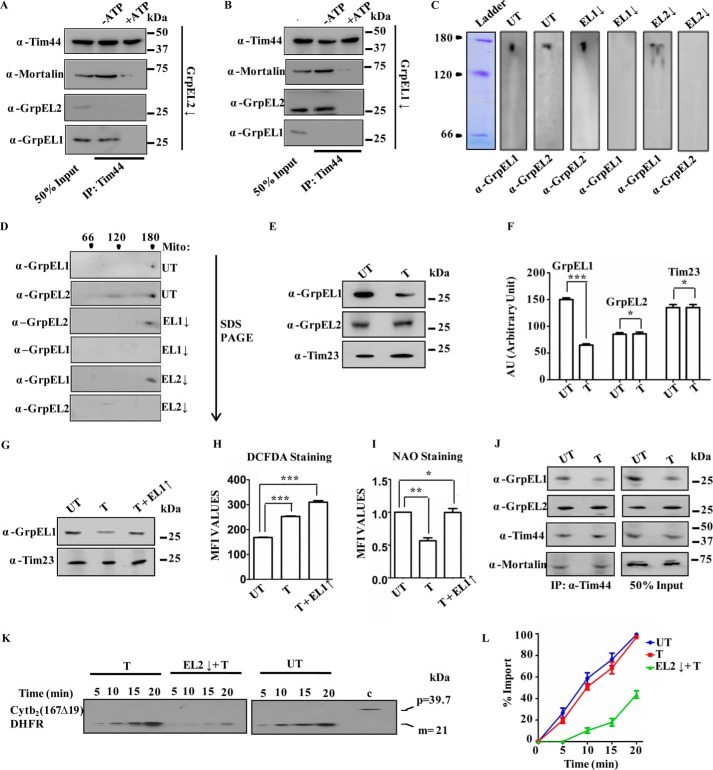
The mitochondrial import motor consists of chaperone mtHsp70 as the core component together with a heterodimer of cochaperones J-proteins/J-like proteins, NEF, and scaffold protein Tim44. Optimum activity of the mitochondrial import motor for the efficient translocation of precursor proteins requires synchronized recruitment of ATP-mtHsp70 and NEF on the Tim44 scaffold (23, 37, 38). The presence of two NEFs in human mitochondria raises an important question, whether both NEF paralogs are recruited to the import site through interaction with mtHsp70, which is tethered to Tim23 channel via Tim44 scaffold. The mode of association of NEFs with the import channel was first assessed by testing whether the NEFs are recruited to the channel individually or as a heteromeric complex. To assess their recruitment to the import channel, a coIP analysis was performed using an anti-Tim44 antibody in the purified mitochondrial lysates. CoIP analysis of EL1/EL2 together with mtHsp70 and Tim44 suggested the recruitment of both NEFs to the mitochondrial import motor. Because interaction of NEF-mtHsp70-Tim44 complex is known to be ATP-sensitive in nature, the coIP was performed in the presence of 2 mm ATP. None of the import motor components were immunoprecipitated in the presence of hydrolyzable nucleotides (ATP), suggesting that co-recruitment of EL1-EL2 is very specific at the import channel. Peroxiredoxin 3, a mitochondrial matrix protein that is not a component of the human import motor, was probed as a negative control (Fig. 2F).
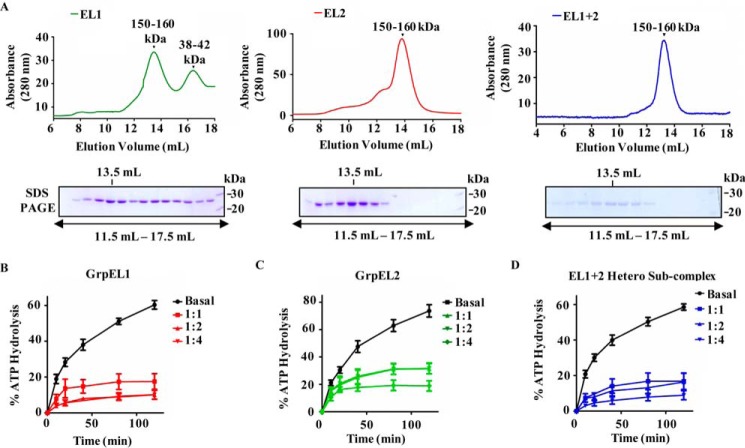
The association of both EL1 and EL2 with the import channel suggests that both NEF paralogs are co-recruited to the import motor as homo- or heterosubcomplexes. To analyze the nature of their association, coIP was performed in the mitochondrial lysates by coexpressing hemagglutinin (HA)-tagged EL1 (EL1-HA) and FLAG-tagged EL2 (EL2-FLAG) at the C terminus. The coIP analysis was performed using an anti-HA antibody that showed the presence of both EL1-HA and EL2-FLAG together with mtHsp70 and Tim44. Addition of ATP in the immunoprecipitates resulted in the dissociation of mtHsp70 and Tim44, but the EL1-EL2 subcomplex remained stable, indicating that in humans the NEF paralogs associate with the import motor as a heterosubcomplex (Fig. 2G). To further validate the direct association between these two NEFs, EL1-HA and N-terminal His6-tagged EL2 (His6-EL2) were cloned in a bicistronic vector, pRSFduet, and expressed in Escherichia coli. The complex was purified from bacterial lysate using Ni-NTA chromatography followed by an ATP wash to remove any bacterial DnaK contamination. The EL1-EL2 complex was copurified to homogeneity as determined by SDS-PAGE, which showed the presence of a single band corresponding to ∼25 kDa (Fig. 2H). The complex was further resolved by native PAGE followed by Western blot analysis. Probing with anti-HA and anti-His antibodies resulted in detection of both EL1 and EL2 at a molecular size higher than that of their heterodimer or individual homodimer (Fig. 2I). Conversely, separation of the observed complex on a second dimension denaturing gel revealed the existence of both EL1 and EL2 as a part of a single subcomplex, which potentially existed in a higher-order oligomeric complex (Fig. 2J). In His tag-dependent affinity chromatography, the detection of HA tag is possible if the two proteins are physically interacting with each other. A direct physical interaction between EL1 and EL2, resulting from the formation of heterocomplex, was further established by biolayer interferometry using biotinylated EL1 as a ligand on a sensor chip. A robust interaction was observed as a function of time upon using two concentrations of EL2 as analyte (Fig. 2K). The oligomeric state of human NEFs was determined by size exclusion chromatography of purified proteins. The size exclusion experiment was performed in the presence of ATP to rule out any possible interference of residual bacterial DnaK. The purified individual proteins EL1 and EL2 and EL1-EL2 heterosubcomplex eluted as a higher molecular mass oligomeric complex (150–160 kDa), which is consistent with native and 2D SDS-PAGE results (Fig. 3A).
Figure 3.
A, analyses of oligomeric nature of NEFs by gel filtration chromatography. Equimolar concentrations of purified GrpEL1 alone (left panel), GrpEL2 alone (middle panel), and EL1-EL2 heterosubcomplex (right panel) were resolved by gel filtration chromatography. Fractions corresponding to elution volumes 11.5–17.5 ml were subjected to SDS-PAGE, and the maximum amount of protein was detected at 13.5-ml elution volume in all the samples as indicated. B–D, measurement of nucleotide exchange activity of NEFs. Preformed radiolabeled mtHsp70-ATP (2 μm) complex was incubated with the indicated different molar ratios of either EL1 alone (B), EL2 alone (C), or EL1-EL2 complex (D), and the rate of ATP hydrolysis was monitored under single-turnover conditions. The percentage of ATP to ADP conversion is plotted against different time intervals. Error bars are derived from three independent sets of experiments and are represented as mean ± S.E. p (two-tailed) < 0.0001.
The recruitment of EL1 and EL2 as a hetero-oligomer raises an important question, whether the heterosubcomplex retains the ability to function as an NEF in the chaperone cycle. The major function of NEFs is to replenish ATP by exchanging it with ADP at the nucleotide-binding site of mtHsp70 in a chaperone cycle. Therefore, the nucleotide exchange ability of EL1, EL2, and EL1-EL2 complex was assessed by monitoring the rate of ATP hydrolysis using radiolabeled [γ-32P]ATP-mtHsp70 complex under single turnover conditions. The inhibition of the [γ-32P]ATP hydrolysis rate due to exchange with unlabeled cold ATP was measured as the exchange activity of NEFs (34). In the presence of cold ATP, both EL1 and EL2 showed similar exchange activity and inhibited ATP hydrolysis at saturating concentrations (Fig. 3, B and C). However, at equivalent concentrations, EL1 retained better exchange activity as compared with EL2 due to its higher affinity for mtHsp70. Stoichiometric association of both NEFs as a heterosubcomplex retained robust exchange activity across the range of concentrations tested (Fig. 3D). This indicates the ability of the EL1-EL2 heterosubcomplex to efficiently perform the cochaperoning function during the import as well as the folding of proteins across the mitochondrial matrix.
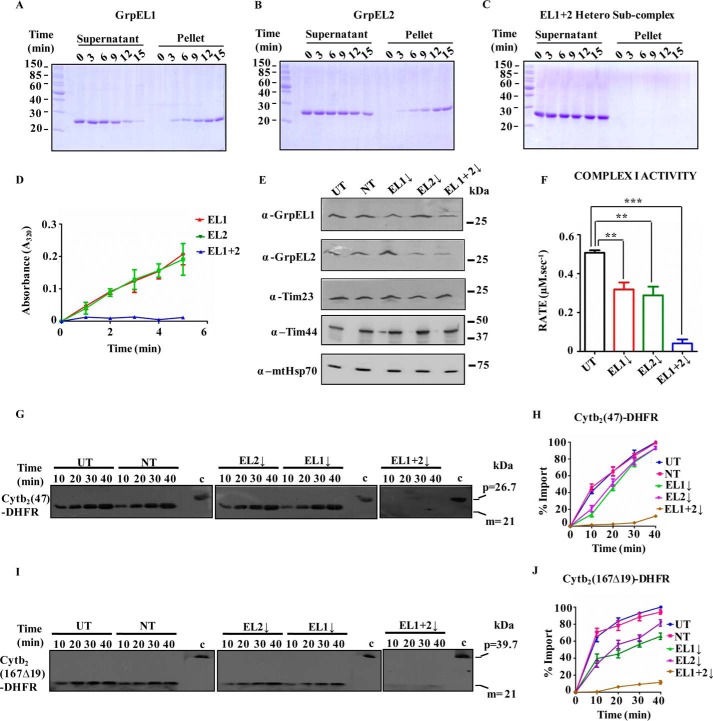
To address the cellular relevance of functional EL1-EL2 heterosubcomplex at the import channel, the biochemical property of the subcomplex was assessed by measuring the stability using protein aggregation through in-gel analysis and spectrophotometry. The individual proteins (EL1 or EL2) or heterosubcomplex (EL1-EL2) was incubated at physiological temperature (37 °C). The reaction mixture was then subjected to centrifugation to separate soluble and pellet fractions and analyzed by SDS-PAGE. In-gel analysis showed accumulation of aggregates over a period of time for both EL1 and EL2 alone in pellet fractions. However, the aggregation propensity of EL2 was found to be 1.5-fold lower than that of EL1 (Fig. 4, A and B, compare pellet fractions). In contrast, the EL1-EL2 heterosubcomplex remained in the soluble fraction throughout the experimental condition, indicating that the hetero-oligomeric state enhances the stability of both proteins (Fig. 4C and compare pellet fractions of B and C). The importance of the heterosubcomplex in maintaining the stability of individual NEFs was further validated by measuring the formation of protein aggregates spectrophotometrically at 320 nm. Similar to our in-gel analysis, an increase in the aggregation (as a measure of absorbance at 320 nm) was observed upon incubating the individual protein, EL1 or EL2, alone at 37 °C. At the same time, the association of both NEFs enhanced the stability of the heterosubcomplex with no apparent detection of protein aggregates (Fig. 4D).
Figure 4.
A–D, stability analysis of NEFs. 2.5 μm EL1 alone (A), EL2 alone (B), or EL1-EL2 complex (C) was incubated at 37 °C at the indicated time intervals and subjected to centrifugation to separate supernatant and pellet fractions followed by SDS-PAGE analysis. The reduced aggregation propensity of the above proteins was analyzed spectrophotometrically under similar experimental conditions by measuring the absorbance at 320 nm as a function of time (D). Data are represented as mean ± S.E. (n = 3). Error bars represents S.E. p (two-tailed) < 0.001. E, silencing of NEFs by siRNA. Western blot of HEK293T cells in which EL1, EL2, or EL1-EL2 was down-regulated by siRNA. Untransfected (UT) cells and non-targeting siRNA (NT) were used as negative controls for siRNA-mediated down-regulation. Anti-Tim23 antibody was used as a loading control. The blots were probed for import motor components (Tim44 and mtHsp70) as positive controls. F, activity of electron transport chain complex I in mitochondria isolated after down-regulation of NEFs. Student's t test was used to compare the rate in cells with down-regulated EL1 (EL1↓), EL2 (EL2↓), or EL1-EL2 together (EL1+2↓) with that in untransfected cells. Data are represented as mean ± S.E. (n = 3). Error bars represents S.E. ***, p (two-tailed) < 0.0001; **, p (two-tailed) < 0.05. G–J, in vitro protein import kinetics. Import activity in the isolated intact mitochondria from untransfected cells (UT), cells transfected with non-targeting siRNA (NT) (left panels), and cells showing down-regulation (↓) for either EL1 alone, EL2 alone (middle panels), or EL1-EL2 together (right panels) was measured at the indicated time intervals using mitochondrion-specific substrates Cytb2(47)-DHFR (G) and Cytb2(167Δ19)-DHFR (I). Precursor protein of 39.7 kDa and mature protein of 21 kDa are represented as “p” and “m.” “c” represents control. Blots were developed with identical exposure, quantitated by densitometry, and represented graphically as percentage of import by setting the highest import point as 100% in each case (H and J). Data represented as mean ± S.E. (n = 3). Error bars represents S.E. p (two-tailed) < 0.0001.
Complementary function of EL1 and EL2 in mitochondrial homeostasis
To determine the specific role of EL1 and EL2 in the maintenance of mitochondrial function, EL1 and EL2 were down-regulated individually or together in HEK293T cells using siRNA (Fig. 4E). Interestingly, an enhancement in the expression level of EL1 was observed when EL2 was down-regulated and vice versa (Fig. 4E), suggesting the possible complementation of import functions. To confirm that compensatory up-regulation of EL1 upon down-regulation of EL2 and vice versa is not due to changes in overall import motor composition, the blots were probed with anti-Tim44 and anti-mtHsp70 antibodies. No apparent change in the expression level of these other import motor components was observed, thus further supporting the idea of complementarity in the functions of both NEFs.
As NEFs assist mtHsp70 in its different cellular functions, the requirement of EL1 and EL2 in two physiological roles of mtHsp70 was assessed using mitochondria with reduced expression of EL1, EL2, or both. The two functions of mtHsp70 utilized for analysis are 1) Fe–S cluster biogenesis and 2) mitochondrial import. To evaluate the effect of EL1-EL2 down-regulation on Fe–S cluster biogenesis, the activity of respiratory complex I in mitochondria was measured. As the mtHsp70 chaperone together with J-protein and NEFs aids in transfer of Fe–S cluster for the formation of active holoenzyme, any defect in Fe–S cluster biogenesis will lead to loss of complex I activity. As shown in Fig. 4F, an ∼30% reduction in the activity of complex I was observed upon down-regulation of either EL1 or EL2, whereas a complete loss of activity was observed when both NEFs were depleted, thereby highlighting their importance in Fe–S cluster biogenesis.
The role of EL1 and EL2 in mitochondrial import was assessed by measuring in vitro protein import kinetics. The relative import rates in isolated mitochondria were measured using model precursor substrates dihydrofolate reductase (DHFR) conjugated with the 167-amino acid-long signal sequence of cytochrome b2 (Cytb2(167Δ19)-DHFR) and DHFR conjugated with the 47-amino acid-long signal sequence of cytochrome b2 (Cytb2(47)-DHFR). As indicated in Fig. 4, G–J, reductions up to ∼20 or ∼40% in overall import efficiency were observed when the expression level of either EL2 or EL1, respectively, was depleted. However, import of the precursors was significantly impaired (up to >90%) in cells in which EL1 and EL2 were down-regulated together. These results highlight that the presence of either of the NEFs at threshold levels is necessary for the optimum import of mitochondrial precursor proteins into the organelle, and both NEFs complement each other's import function in the mtHsp70-mediated chaperone cycle.
To further assess whether the perceived import activity is due to the ability of both NEFs to be individually recruited to the import motor, coIP was performed using the anti-Tim44 antibody in mitochondria isolated from cells depleted either for EL1 alone, EL2 alone, or EL1 and EL2 together. It was observed that, in the case of EL1 down-regulation, only EL2 was immunoprecipitated with Tim44. In contrast, down-regulation of EL2 did not have any effect on the association of EL1 with the import channel (Fig. 5, A and B). This result supports the previous observation that down-regulation of either of the NEFs does not translate into a significant import defect because a depleted amount of one protein is compensated by the recruitment of the other NEF to the import motor.
Figure 5.
A and B, coimmunoprecipitation. Immunoprecipitation was performed in isolated mitochondria from HEK293T cells silenced for EL2 (A) and EL1 (B) in the presence of 2 mm ATP (+ATP) or in the presence of 5 mm EDTA (−ATP) using anti-Tim44 antibody, separated by SDS-PAGE, and immunodecorated with the indicated specific antibodies. 50% of the sample served as input control. C and D, analyses of the oligomeric nature of NEFs in mitochondria. Mitochondrial lysates prepared from untransfected cells (UT) or cells treated with siRNA to down-regulate (↓) EL1 and EL2 were separated by blue native PAGE (C) and 2D SDS-PAGE (D) and immunodetected by anti-GrpEL1 and -GrpEL2 antibodies. The second dimension direction for SDS-PAGE is marked by an arrow, and the positions of molecular mass markers are indicated. E–G, expression of NEFs under oxidative stress. Mitochondrial lysates prepared from untreated (UT) cells or cells treated (T) with 100 μm NaAsO2 were analyzed by SDS-PAGE followed by immunodetection using anti-GrpEL1 and -GrpEL2 antibodies (E). The blots were quantitated by densitometry and are presented as a bar chart. Student's t test was used to compare the expression of EL1, EL2, or Tim23 between untreated and treated samples. Data are represented as mean ± S.E. (n = 3). ***, p (two-tailed) < 0.0001; **, p (two-tailed) < 0.05 (F). Similarly, mitochondrial lysates prepared from untransfected cells (UT) or cells treated with 100 μm NaAsO2 in the absence (T) or presence of exogenously overexpressed (↑) GrpEL1 were separated by SDS-PAGE and subjected to immunoblotting with the indicated antibodies (G). H and I, ROS and mitochondrial mass measurements. Generation of total ROS and mitochondrial mass were measured in untransfected cells (UT) or cells treated with 100 μm NaAsO2 (T) or in the presence of exogenously overexpressed (↑) GrpEL1 by DCFDA dye (H) and NAO staining (I) using flow cytometry. The increment in total cellular ROS levels is presented in terms of mean fluorescence intensity (MFI) over untransfected HEK293T cells (UT). Two sets of data (UT with T and UT with T + EL1↑) were compared using Student's t test. Data are represented as mean ± S.E. (n = 3). Error bars represents S.E. ***, p (two-tailed) < 0.0001 (panel H); **, p (two-tailed) < 0.001; *, p (two-tailed) < 0.01 (panel I). J–L, coimmunoprecipitation and protein import under oxidative stress. Immunoprecipitation using anti-Tim44 antibody was performed in the mitochondrial lysates prepared from untreated HEK293T cells (UT) or HEK293T cells treated with 100 μm NaAsO2 followed by SDS-PAGE and immunoblotting using the indicated specific antibodies (J). 50% of the sample served as input control. The protein import kinetics was measured in the isolated intact mitochondria from untransfected cells (UT) or cells treated with 100 μm NaAsO2 (T) alone or together with EL2 down-regulation (T + EL2↓) using Cytb2(167Δ19)-DHFR as precursor protein (K). Blots were developed with identical exposure and quantitated by densitometry and are represented graphically as percentage of import by setting the highest import point as 100% in each case (L). Data are represented as mean ± S.E. (n = 3). Error bars represents S.E. p (two-tailed) < 0.0001.
Because both proteins were recruited to the import motor as a heterosubcomplex, the oligomeric status of the individual NEFs was further assessed under silenced condition. Mitochondrial lysate isolated from HEK293T cells after down-regulation of either of the NEFs was resolved by blue native PAGE. The Western blot analysis indicated the ability of both EL1 and EL2 to exist individually in oligomeric states similar to that of the EL1-EL2 heterosubcomplex. The oligomeric homosubcomplex was further separated by second dimension SDS-PAGE, resulting in the detection of a single NEF paralog under knockdown condition of the other (Fig. 5, C and D). These findings indicate that although both NEFs possess an ability to form a homosubcomplex the EL1-EL2 heterosubcomplex is preferred physiologically at the import channel.
Together, these experiments provide compelling evidence that the presence of either EL1 or EL2 is sufficient to maintain the threshold mtHsp70 function due to up-regulation and that one NEF is recruited as a compensatory response to depletion of the other NEF. However, import of preproteins and Fe–S cluster biogenesis are compromised when levels of both proteins are reduced.
Evolution of stress-resistant EL2 for maintenance of protein import
In yeast, a single NEF, Mge1, assists in the function of Ssc1 (mtHsp70) for mitochondrial import of precursor proteins (39). Although our results suggest the existence of two NEFs in the mammalian system for the constitutive functions, the additional importance of two NEFs in mitochondrial homeostasis was further explored. Apart from many vital functions, mitochondria are a major hub of reactive oxygen species production and cellular redox maintenance (40). Initial evidence suggested that yeast Mge1 was sensitive to increased cellular peroxides (33). Considering the functional similarity of EL1 and EL2 with yeast Mge1 in import of mitochondrial protein, we analyzed the possible role of EL1 and EL2 in cellular redox homeostasis. HEK293T cells were treated with 100 μm NaAsO2 to induce oxidative stress and analyzed for the effect of oxidative damage on NEF levels. Interestingly, a ∼50% reduction in EL1 expression was observed in cells exposed to oxidative stress with no apparent change in the expression levels of EL2 (Fig. 5, E and F). To understand the physiological relevance of oxidative stress-mediated reduction in EL1 expression, the EL1 protein levels in stressed cells were restored by exogenously expressing EL1 under a constitutive promoter (Fig. 5G). Importantly, upon oxidative stress, an enhanced accumulation of cellular ROS levels was observed in the cells where the EL1 expression level was exogenously maintained as compared with cells showing intrinsically lower levels of EL1 (Fig. 5H). Because mitochondria are the major source of cellular ROS production, the oxidative environment of the cell and mitochondrial biogenesis are intricately balanced. As an adaptive mechanism, cells exposed to oxidative stress are usually associated with alterations in the mitochondrial volume. Therefore, the total mitochondrial volume was measured through nonyl acridine orange (NAO) staining using flow cytometry analysis (41, 42). A reduction in the mitochondrial volume was observed after the treatment with NaAsO2 that was concomitant with a decline in EL1 expression levels. However, restoration of EL1 expression prevented the stress-dependent reduction in the mitochondrial content, which was observed to be comparable with that of untreated cells even in the presence of oxidative stress (Fig. 5I).
To establish a correlation between mitochondrial biogenesis and import, the effect of oxidative stress on recruitment of EL1 or EL2 at the import channel was analyzed. Through coIP analysis using the anti-Tim44 antibody, a reduced association of EL1 was observed with the import channel in cells exposed to oxidative stress as compared with untreated controls (Fig. 5J). However, the reduced recruitment of EL1 did not translate into a decreased import rate as revealed by in vitro import kinetics using Cytb2(167Δ19)-DHFR as a substrate. As indicated in Fig. 5K, there is no impairment in the import of precursor proteins upon oxidative stress as compared with untreated control, possibly due to complementation of EL1 function by EL2 in the maintenance of protein import. To validate this hypothesis, EL2 was down-regulated using siRNA in cells treated with NaAsO2. A marked defect in import of Cytb2(167Δ19)-DHFR was observed under the condition of reduced EL2 (caused by siRNA mediated down-regulation) and EL1 (caused by oxidative stress) as compared with control (Fig. 5L). In conclusion, these results suggest that EL2 maintains the basal import activity in cells during the stress-induced reduction in EL1 expression.
In summary, it can be inferred that, in the mammalian system, the mitochondrial Hsp70 machinery is more evolved to utilize an oxidative stress-resistant EL2 to maintain the import of mitochondrially targeted proteins under stress condition. At the same time, EL1 is required for the maintenance of redox homeostasis by altering the mass of mitochondria, which are major ROS-producing organelles.
EL1 elicits evolutionarily conserved function in yeast
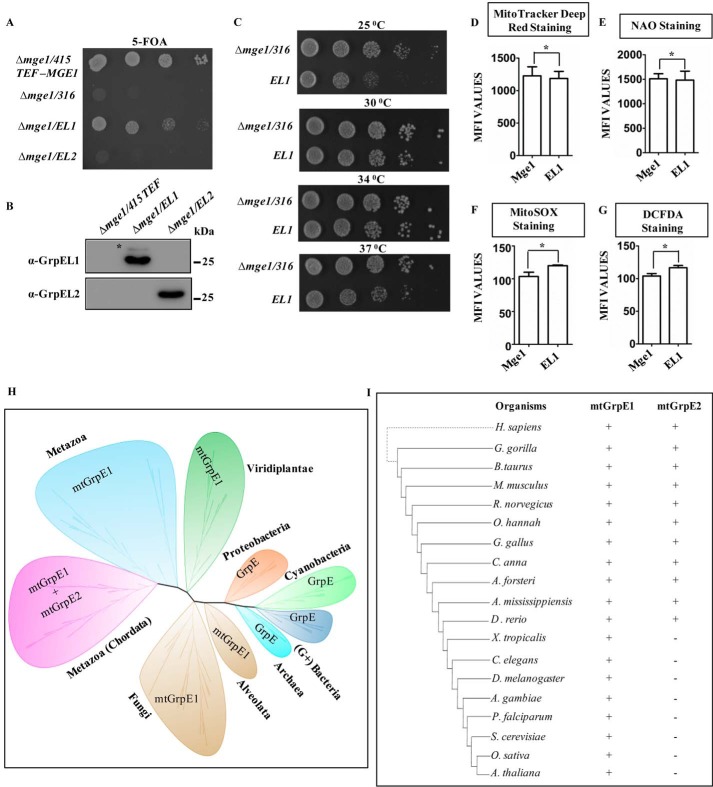
It is evident from our results that during evolution a single NEF from lower eukaryotes diversified in humans into two NEFs, EL1 and EL2, that share a common function in protein transport with a supplemental role of EL1 in ROS sensing. Given the comparable sequence similarity between human and yeast NEFs, an attempt was made to delineate whether EL1 and EL2 are able to retain their evolutionarily conserved function in the yeast system. Deletion of Mge1 gene is lethal in Saccharomyces cerevisiae (39). The functional complementation of Mge1 was tested by transforming pRS414 plasmid containing ORFs for EL1 and EL2 in Δmge1 yeast cells. The transformants were selected on a minimal medium lacking tryptophan and counterselected on 5-fluoroorotic acid (5-FOA) plates against wild-type MGE1 under a URA marker. Interestingly, yeast cells transformed with EL1 could complement the inviability of Δmge1 on 5-FOA medium, but at the same time cells harboring EL2 were inviable (Fig. 6A), suggesting that EL1 could rescue the evolutionarily conserved function of Mge1 in yeast. To investigate whether the observed cellular lethality in EL2-transformed cells is due to insufficient protein levels, cellular expression of EL1 and EL2 was measured after transformation. A comparable protein level was observed upon Western blot analysis, thereby further revealing that the inability of functional complementation of Δmge1 cells by EL2 is not the consequence of its expression levels (Fig. 6B).
Figure 6.
A–C, growth complementation and expression analysis. MGE1-deleted yeast cells harboring a copy of MGE1 in pRS316 plasmid were transformed with pRS415 TEF-MGE1, -EL1, or -EL2 and subjected to drop dilution analysis on 5-FOA medium followed by incubation for 72 h at 30 °C (A). The expression of EL1 (Δmge1/EL1) and EL2 (Δmge1/EL2) in transformed yeast cells was analyzed by immunoblotting and probed with anti-GrpEL1 or -GrpEL2 antibodies. The cells transformed with empty vector were used as a negative control (Δmge1/415 TEF). *, band corresponding to yeast Mge1 due to cross-reactivity with EL1 antibody (B). Temperature-dependent growth complementation by EL1 was analyzed on YPD medium by incubating the plates at 25, 30, 34, and 37 °C for 72 h (C). D–G, mitochondrial functional analysis. The following mitochondrial functional parameters were measured in yeast cells expressing EL1 or Mge1 using flow cytometry: membrane potential by MitoTracker Deep Red (D), total mass by NAO (E), mitochondrial ROS levels by MitoSOX (F), and total cellular ROS levels by DCFDA (G). The mean fluorescence intensity (MFI) for Mge1 in comparison with EL1 is plotted in each case and analyzed using Student's t test. No significant differences in ROS levels, mitochondrial function, and mass were observed between Mge1- and EL1-expressing cells. Data are represented as mean ± S.E. (n = 3). Error bars represents S.E. *, p (two-tailed) < 0.1. H and I, phylogenetic analysis. An unrooted phylogenetic tree for GrpE orthologs across phylogeny was constructed using MEGA software with the Jones-Tailor-Thornton amino acid substitution model and ML heuristic method. Each clade wrapped in a bubble represents a phylum and kingdom level representation of the phylogeny (H). The inference obtained from the unrooted phylogenetic tree (H) was compiled descriptively with selected well studied species to represent the presence (+) and absence (−) of mtGrpE1 and mtGrpE2 across taxonomy (I).
Drop dilution analysis of cells expressing EL1 showed an identical growth pattern on YPD medium compared with Mge1 at all temperatures except 25 °C at which a slightly slower growth was observed (Fig. 6C). To validate the functional complementation of Mge1 by EL1, mitochondrion-specific functional parameters such as mitochondrial mass (by NAO staining), membrane potential (by MitoTracker® Deep Red staining), and relative leakage of free oxygen radicals (by MitoSOXTM and DCFDA staining) were measured (Fig. 6, D–G). No significant differences were observed for mitochondrial functional parameters between cells expressing Mge1 and EL1. Together, these results highlight a functional overlap between EL1 and its yeast ortholog Mge1, thereby suggesting EL1 to be evolutionarily more closely related to Mge1 and that the specialized function of EL2 is limited to the mammalian system. This idea was further supported by in silico phylogenetic reconstruction of NEF diversification across primitive prokaryotes to advanced multicellular organisms. Although proteins similar to Mge1 largely constitute the unicellular organisms (such as yeast) and recent land plants, EL1 is conserved across all metazoans and phylogenetically closer to Mge1, thus supporting its ability to rescue Δmge1 yeast. However, during chordate evolution, the EL1 lineage branched into a separate clade, EL2, which evolved in parallel across higher vertebrates. Hence, the vertebrate-specific evolution of GrpEL2 differentiates the NEF-dependent regulation in mammals from that in lower organisms (Fig. 6, H and I).
Discussion
Mitochondria play an essential role in regulation of cellular metabolic and stress conditions. To adapt in a complex cellular environment in a mammalian system, mitochondria have diversified their protein import machinery (4, 5, 43). It is quite well known that relative distribution of certain molecules in the organelle reprograms mitochondrial functions as a part of homeostatic mechanisms within the cell (4, 43–45). Because the majority of these proteins localize in the mitochondrial matrix, regulated functioning of mtHsp70 as a core component of the import motor is critical. Evidently, the functional diversity of mtHsp70 in humans is enhanced by the presence of two J-proteins, which suitably modulate its activity for its housekeeping role or response to xenobiotic stress (26, 27, 31). mtHsp70 mainly functions through a cycle of substrate binding and release at its substrate-binding domain, which in turn depends on ATP binding and hydrolysis in the nucleotide-binding domain. Exchange of fresh ATP by NEF in mtHsp70's nucleotide-binding domain makes it available for the next series of the cycle, highlighting the importance of NEFs in maintenance of mitochondrial homeostasis (18, 21, 24).
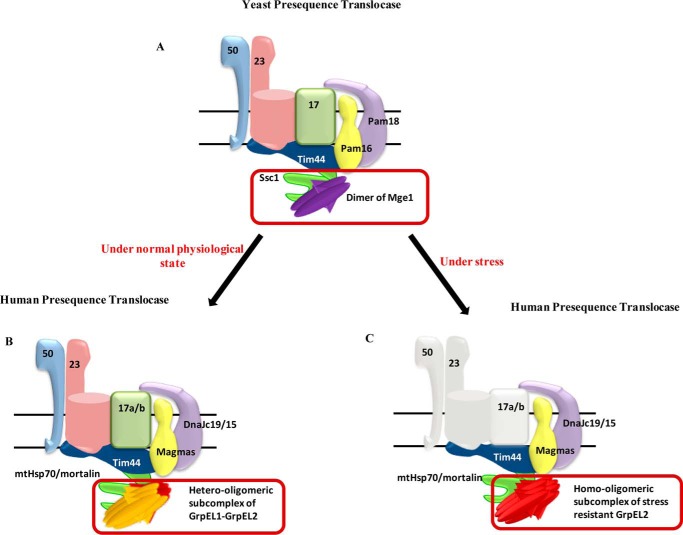
In the present study, we propose diversification of human NEF into two paralogs, EL1 and EL2, for better maintenance of mitochondrial functions. It is evident from our results that EL1 has a higher affinity for mtHsp70 as compared with EL2. Under normal cellular conditions, EL1 and EL2 function as a higher-order oligomer in contrast to NEF dimers traditionally observed in lower organisms (Fig. 7). The individual paralogs, along with the EL1-EL2 heterosubcomplex, exhibit similar nucleotide exchange ability at saturating concentrations with the nucleotide-bound state of mtHsp70. However, at stoichiometric concentrations, EL1 and EL1-EL2 heterosubcomplex showed a better exchange of nucleotides than EL2. The physiological significance of formation of this heterosubcomplex is to maintain the aggregation-prone human NEFs in the soluble state. At the same time, the aggregation propensity of EL1 and EL2 did not reflect on their respective ability to maintain NEF function.
Figure 7.
Organization of yeast and human mitochondrial presequence translocases. A and B, yeast, a dimer of single NEF Mge1 maintains import motor function (A), whereas in humans a hetero-oligomeric subcomplex of two NEFs, GrpEL1 and GrpEL2, regulates the mtHsp70 cycle under normal physiological conditions (B). C, the evolution of another stress-resistant NEF GrpEL2 in human mitochondria maintains the import process and organellar homeostasis under stress conditions.
Both NEFs have the ability to be recruited at the import channel independently or as heterosubcomplex to modulate mtHsp70 chaperone activity. Moreover, co-recruitment of EL1 and EL2 provides the first evidence for the mutual complementation of transport functions to maintain threshold levels of protein import by both NEFs. The maintenance of one NEF's function in the absence of the other is probably contributed by the high-turnover rate and feedback overexpression observed in these proteins under stress conditions (Fig. 7). Besides, both NEFs can perform the cochaperone function in assisting mtHsp70 chaperone-mediated folding of precursor proteins in the matrix compartment, thereby maintaining quality control and organellar homeostasis.
Although the constitutive functions are performed by a single NEF in lower eukaryotes, the presence of two orthologs in higher eukaryotes raises a beguiling question, whether it provides additional physiological significance in the mammalian system. Being a complex organism, human cells are vulnerable to many external stresses, such as oxidative stress, which is a major by-product of mitochondrial metabolism (46, 47). Although ROS play an important role in various physiological functions within the cell, exposure to excessive stress is harmful for cell survival (40, 48). The defense response toward accumulation of ROS might trigger the cell to fine-tune the biogenesis of mitochondria, the major ROS-producing organelles. Indeed, it has been previously discussed in several studies on the role of different transcription factors, such as Nrf1, Nrf2, and PGC-1α, in regulating biogenesis of mitochondria in response to oxidative stress (49). Under stress, the less aggregation-prone stress-resistant EL2 performs the threshold level of activity at comparatively lower efficiency due to stress-induced loss of EL1 expression and subsequent dissociation from the heterosubcomplex (Fig. 7). This in turn is reflected in the reduced biogenesis of mitochondria under stress conditions, thereby maintaining redox homeostasis. The idea is supported by unaltered mitochondrial biogenesis even under oxidative stress when the levels of EL1 and EL1-EL2 heterosubcomplex formation are maintained by their exogenous expression.
In the mammalian system, moonlighting functions have been reported for the components of the inner mitochondrial membrane import machinery. For example, two translocation machineries are involved in protein import, namely translocase A, which contains Tim17a and DnaJC15, and translocase B with Tim17b and DnaJC19 paralogs. The subunits of translocase B are essential for constitutive organellar functions. In contrast, the components of translocase A, Tim17a and DnaJC15 paralogs, are involved in cellular oncogenesis and development of chemoresistance, respectively (26, 31, 50). Such moonlighting secondary functions in proteins are usually incorporated as a result of gene duplication or due to random genetic drift, providing the organism a selective advantage for better adaptation in adverse environmental conditions (51). Therefore, we envision that the evolutionary incorporation of two NEFs in human mitochondria occurred to regulate the organellar stress response without altering the mitochondrial import process under stress.
In summary, we report here the existence of two NEFs in humans that are potentially utilized by the import machinery in regulating mitochondrial proteostasis. Together, we propose that dynamic association of EL1 and EL2, in cooperation with their differential nucleotide exchange rates, is utilized by the cell for reprogramming the mitochondrial life cycle and cellular energy demand. However, it will be imperative to understand the underlying molecular interactions between the human NEFs and other stress-sensitive factors that govern the overall redox regulatory processes. In addition, our observations will also expose new avenues to understand the novel regulators of the mitochondrial stress response.
Materials and methods
Cell culture and transfection
HEK293T cells and HeLa cells were cultured in DMEM (Invitrogen) supplemented with 10% fetal bovine serum (Invitrogen), 5% penicillin/streptomycin (Invitrogen), and 5% GlutaMAX (Invitrogen). Cells were grown at 37 °C in the presence of 5% CO2. Transfection of both plasmid and siRNA was carried out using Lipofectamine 2000 (Invitrogen) according to the manufacturer's protocol. Cells were harvested after 48 h of plasmid transfection for various experiments. For down-regulation of GrpEL1 and GrpEL2, siRNA pools were obtained from Integrated DNA Technologies. After 48 h of transfection with siRNA, a second round of transfection was performed to get maximum down-regulation. Cells were harvested for various experiments after 36 h of second transfection.
Yeast strains and complementation
GrpEL1 or GrpEL2 constructs in pRS414 vector were transformed in Δmge1 strains carrying a copy of WT MGE1 in pRS316 plasmid. The transformants were selected on minimal medium lacking tryptophan (Trp− medium) followed by drop dilution analysis on medium containing 5-FOA to observe the complementation. The viable cells from 5-FOA medium were then revived using YPD medium.
Plasmids and cloning
GrpEL1 and GrpEL2 ORFs were amplified from a HeLa cell cDNA library (Stratagene). For testing the subcellular localization of the proteins, GrpEL1 and GrpEL2 were inserted at the N terminus of GFP in pEGFP-N1 vector. The proteins were individually purified by cloning C-terminal His6-tagged EL1 or EL2 in MCS2 of bicistronic vector pRSFduet. For copurification, HA-tagged EL1 and His6-tagged EL2 were cloned in MCS1 and MCS2, respectively. Yeast complementation analysis was performed by generating pRS414 constructs carrying ORFs of human GrpEL1 and GrpEL2 under TEF promoter. To replenish the level of GrpEL1 in mammalian cells under NaAsO2 treatment, GrpEL1 ORF with C-terminal FLAG tag was ligated to mammalian expression vector pCINeo.
Localization experiments
To confirm the localization of GrpEL1 and GrpEL2, plasmids carrying C-terminal GFP-tagged EL1 or EL2 were transfected in HeLa cells. Cells were cotransfected with mtDsRed to counterstain mitochondria for the purpose of colocalization. After 72 h of transfection, cells transfected with GrpEL1-GFP or GrpEL2-GFP and mtDsRed were mounted on ProLong Gold antifade reagent (Invitrogen) and observed using a Leica SP5 confocal microscope under 63× objective lens.
Isolation of mitochondria and mitochondrial fractionation
Mitochondria were isolated from HEK293T cells. Briefly, cells were harvested using 0.25% trypsin, EDTA and washed with 1× PBS. Cells were resuspended in hypotonic solution (10 mm Tris, pH 7.5, 10 mm NaCl, 3 mm MgCl2, 1 mm EDTA) containing protease inhibitor mixture (Sigma) and incubated on ice for 30 min. Cells were then lysed by homogenization using a Dounce homogenizer. The cellular debris was pelleted at 600 × g for 15 min. The mitochondrial fraction was obtained by centrifuging the supernatant at 13,000 rpm for 15 min. The mitochondrion-enriched fraction was resuspended in SEM buffer (250 mm sucrose, 400 mm EDTA, 10 mm MOPS-KOH, pH 7.2). Submitochondrial fractionation was performed according to the previously published protocol (30).
Protein purification and generation of antibodies
Both GrpEL1 and GrpEL2 with N-terminal His6 tag were expressed in E. coli Rosetta strain. Briefly, cells were grown at 30 °C until midlog phase followed by induction with 0.5 mm isopropyl 1-thio-d-galactopyranoside for 12 h. Cells were lysed in lysis buffer (150 mm Tris, pH 8, 200 mm NaCl, 5% glycerol, 50 mm imidazole) containing 0.5 mg/ml lysozyme for 45 min at 4 °C followed by treatment with 0.2% deoxycholate for 15 min. Cell lysate was centrifuged at 18,000 rpm for 45 min to separate soluble and pellet fractions. The supernatant containing the protein of interest was incubated with Ni-NTA-Sepharose beads at 4 °C for 2 h. The unbound protein was washed off with lysis buffer. To remove bacterial DnaK contamination, beads were washed with lysis buffer containing 1 mm ATP and 1 mm MgCl2 three times. Protein was eluted using 250 mm imidazole and stored at −80 °C. The purified proteins (EL1 and EL2) were used separately to raise antibodies against the respective proteins using rabbit at Imgenex Corp. The antisera obtained were further affinity-purified against the respective purified proteins.
Immunoprecipitation
Mitochondria (450 μg) were lysed in lysis buffer (250 mm sucrose, 80 mm KCl, 20 mm MOPS-KOH, pH 7.2, 0.2% Triton X-100, 5 mm EDTA) for 45 min at 4 °C. Antibody against the required protein (Tim44 or HA) (BD Biosciences) was added to mitochondrial lysate and incubated for 1 h at 4 °C. The antibody-lysate mixture was then added to protein G-Sepharose beads, equilibrated with lysis buffer, and incubated at 4 °C for 2 h. After binding, beads were washed with lysis buffer, resuspended in 2× SDS sample buffer, and resolved by SDS-PAGE followed by Western blotting.
Mitochondrial import
Mitochondrial protein import was measured using isolated mitochondria. The two substrates used in the assay were Cytb2(47)-DHFR and Cytb2(167Δ19)-DHFR, which lacks the 19-amino acid transmembrane region, as matrix-localized substrates. These substrates were made through fusion of the nucleus-localized protein DHFR and the N-terminal presequence of Cytb2. The experiment was performed as described previously (30). The import of mature protein was quantified by ImageJ and plotted in GraphPad Prism5.
Gel filtration
For gel filtration analysis of proteins, 500 μl of protein samples (1 mg/ml) were filtered through a 0.22-μm filter and subjected to analysis using a Superdex-200 10/300 GL column (GE Healthcare) in buffer A (50 mm Tris-Cl, pH 8, 100 mm NaCl, 100 mm imidazole, 1 mm DTT, 5% glycerol) at 4 °C. The column was calibrated using standard molecular mass markers from GE Healthcare (aprotinin (6.5 kDa), ribonuclease A (13.7 kDa), carbonic anhydrase (29 kDa), ovalbumin (43 kDa), conalbumin (75 kDa), aldolase (158 kDa), ferritin (440 kDa), and thyroglobulin (669 kDa)). The void volume (V0) was calculated by running blue dextran 2000 (2000 kDa). The elution volume (Ve) of standard markers and proteins (EL1, EL2, and EL1-EL2 subcomplex) was determined under identical temperature, buffer, and pH conditions as mentioned before. The molecular weight was calculated from the standard curve of Ve/V0 versus the log of the molecular weight of standard molecular weight markers.
ATPase assay
A complex of human mtHsp70 with ATP was prepared using γ-32P-labeled ATP (250 μCi) and 2 μm mtHsp70. A Sephadex G-50 Nick column (GE Healthcare) pre-equilibrated with buffer A (25 mm HEPES-KOH, pH 7.5, 100 mm KCl, 10 mm Mg(OAc)2) was used for isolation of the complex. In a single-turnover assay, GrpEL1, GrpEL2, or EL1-EL2 heterosubcomplex was mixed with ATP-mtHsp70 complex in a 1:1 ratio. The reaction was performed at 25 °C at 550 rpm. At each time point, the reaction was stopped using stop solution (5 m LiCl, 17 m HCOOH, 0.1 m ATP in a ratio of 10:6:9), and 1.5 μl of reaction mixture were spotted on a PEI-cellulose plate followed by separation by thin-layer chromatography for 40 min using a 1:1 (v/v) mixture of 1 m LiCl and 2 m formic acid as solvent. The plate was dried and exposed to phosphorimaging cassettes. Images were acquired in Typhoon FLA 9000 (GE Healthcare), and postacquisition quantification was done using Multi Gauge V3.0 software.
In vitro aggregation assay
The aggregation propensity of NEFs was analyzed by two different methods.
In-gel analysis
2.5 μm protein (EL1, EL2, or copurified EL1-EL2) was incubated at 37 °C for 15 min in analysis buffer (150 mm Tris, pH 8, 200 mm NaCl, 5% glycerol, 50 mm imidazole). Aliquots (10-μl volume) corresponding to different time points were centrifuged at 13,000 rpm for 2 min to separate the aggregates in the pellet fraction from the soluble supernatant fraction. Both pellet and supernatant were mixed with 2× SDS sample buffer, heated at 90 °C, and loaded on the gel.
Spectroscopic analysis
A 2.5 μm concentration of the respective protein was incubated at 37 °C for 15 min followed by spectroscopic measurement of absorbance at 320 nm at different time points.
Flow cytometry
For FACS analysis of mammalian cells, 50,000 cells were harvested, washed with 1× PBS, and stained with respective dyes (DCFDA, 15 μm; MitoSOX, 5 μm; NAO, 5 nm; MitoTracker Deep Red, 5 nm). Following staining, the cells were again washed with 1× PBS. The acquisition and analysis were done in a BD FACSCanto II. The flow cytometry experiments to analyze cellular ROS, mitochondrial superoxide level, mitochondrial content, and functional mass in yeast were performed using 0.03 OD cells. The cells were incubated with respective dyes (DCFDA, 100 μm; MitoSOX,15 μm; NAO, 20 nm; MitoTracker Deep Red, 20 nm) followed by acquisition and analysis in the BD FACSCanto II.
Biolayer interferometry
For analyzing interaction of EL1 and EL2 with mtHsp70, both NEFs were biotinylated for 3 h at 4 °C and immobilized as ligand on streptavidin sensors in two separate experiments. Increasing concentrations of MtHsp70 protein were used as analyte. Similarly, for the analysis of EL1 and EL2 interaction, biotinylated GrpEL1 was immobilized as ligand, and increasing concentrations of GrpEL2 were used as analyte. Sensorgrams were recorded as a function of time. The affinity constant (KD), dissociation rate (Koff), and association rate (Kon) were calculated by fitting the data using Forte Bio BioOctate software.
Electron transport chain complex I activity
The activity of complex I was measured according to the protocol described by Spinazzi et al. (52).
Proteome-wide GrpE search and phylogenetic analysis
An extensive search was done to identify GrpE from whole proteomes of 5862 species using the UniProt reference proteome data set (release 2016_09; October 5, 2016). Evolutionarily important species across phyla were selected to perform phylogenetic analysis.
Hidden Markov model (HMM) profiles
HMM profiles were developed for GrpE orthologs (bacterial GrpE, eukaryote GrpE, and archaeal GrpE) and paralogs (mtGrpE1 and mtGrpE2) among all three kingdoms using the build profile module of Hmmer-3.1b2-linux. Well annotated and literature-reviewed sequences were used as seed sequences for multiple sequence alignment using the MUSCLE alignment program and to generate the HMM profiles for all orthologs and paralogs of GrpE.
HMM search
GrpE was searched using the HMM profiles created from the above method to identify GrpE-like proteins for each species from the whole UniProt reference proteome data set followed by paralog- and ortholog-specific HMM searches against respective HMM profiles. A few in-house Perl scripts were developed to compare the HMM outputs, and the short-listed proteins predicted as GrpE were ranked according to respective ortholog and paralog using taxonomic lineage, sequence length, domain organization, gene identity, score thresholds, and E-value from the HMM search output. The resulting output was a list of non-redundant GrpE homologs for each species.
Data set
Orthologs of GrpE indentified from the whole reference proteome of 5862 species through the above method were compared taxonomically and further scrutinized to remove similar genera. Only well established model organisms were selected for further analyses. The final short-listed data contain GrpEs from 41 bacterial, eight archaeal, and 145 eukaryotic species.
Phylogenetic analysis
Multiple sequence alignment was done using the MUSCLE sequence alignment program for a total of 194 species using Mega-CC software (version 7.0.14). Phylogenetic reconstruction analysis was done using the Mega-CC program with statistical method “maximum likelihood,” bootstrap test of phylogeny, substitution type “amino acid,” substitution model “Jones-Tailor-Thornton (JTT),” maximum likelihood (ML) heuristic method “nearest-neighbor interchange,” and initial tree for ML “neighbor joining” used to build the phylogeny. FigTree was used for analysis.
Statistical analysis
Experiments were performed in multiple sets, and results were analyzed for statistical significance using two-tailed Student's t test utilizing GraphPad Prism5 software. p values are given in the figure legends.
Author contributions
S. S. and P. D. conceived the study and designed the experiments. S. S., M. A. S., and D. S. performed the experiments. A. B. performed the ATPase assay. V. R. performed all in silico analysis. P. P. S. performed GFC. S. S. and P. D. analyzed the data. S. S., D. S., and P. D. wrote the manuscript.
Acknowledgments
We thank Prof. Elizabeth A. Craig for yeast strains and yeast-specific antibodies. We thank Prof. H. S. Savithri for generous help in conducting the gel filtration chromatography experiment. We acknowledge the FACS facility of Indian Institute of Science-Bangalore for performing flow cytometry experiments.
This work was supported by Department of Science and Technology (DST) Swarnajayanti Fellowship DST/SJF/LS-01/2011–2012, a Lady Tata Memorial Trust Young Researcher Award, Department of Biochemistry-Indian Institute of Science (DBT-IISc) Partnership Program Grant DBT/BF/PR/INS/2011–12/IISc, DST-Fund for Improvement of Science and Technology Infrastructure (FIST) Program Grant SR/FST/LSII-023/2009, and University Grants Commission (UGC)-Centre of Advanced Study (CAS) Special Assistance Programme (SAP)-II program Grant UGC LT. F. 5-2/2012.SAP-II) (to P. D.); a DST-Innovation in Science Pursuit for Inspired Research (INSPIRE) fellowship (to S. S.); a Council of Scientific and Industrial Research fellowship (to P. P. S. and A. B.); and a UGC-D. S. Kothari postdoctoral fellowship (to A. M. S.). The authors declare that they have no conflicts of interest with the contents of this article.
- NEF
- nucleotide exchange factor
- mtHsp70
- mitochondrial heat shock protein 70
- GrpEL
- GrpE-like
- ROS
- reactive oxygen species
- Ni-NTA
- nickel-nitrilotriacetic acid
- coIP
- coimmunoprecipitation
- Cytb2
- cytochrome b2
- DHFR
- dihydrofolate reductase
- NAO
- nonyl acridine orange
- 5-FOA
- 5-fluoroorotic acid
- YPD
- yeast extract-peptone-dextrose
- DCFDA
- 2′,7′-dichlorofluorescin diacetate
- HMM
- hidden Markov model
- ML
- maximum likelihood
- EL1
- GrpEL1
- EL2
- GrpEL2.
References
- 1. Bolender N., Sickmann A., Wagner R., Meisinger C., and Pfanner N. (2008) Multiple pathways for sorting mitochondrial precursor proteins. EMBO Rep. 9, 42–49 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Hoogenraad N. J., and Ryan M. T. (2001) Translocation of proteins into mitochondria. IUBMB Life 51, 345–350 [DOI] [PubMed] [Google Scholar]
- 3. Pfanner N., and Geissler A. (2001) Versatility of the mitochondrial protein import machinery. Nat. Rev. Mol. Cell Biol. 2, 339–349 [DOI] [PubMed] [Google Scholar]
- 4. Baker M. J., Frazier A. E., Gulbis J. M., and Ryan M. T. (2007) Mitochondrial protein-import machinery: correlating structure with function. Trends Cell Biol. 17, 456–464 [DOI] [PubMed] [Google Scholar]
- 5. Chacinska A., Koehler C. M., Milenkovic D., Lithgow T., and Pfanner N. (2009) Importing mitochondrial proteins: machineries and mechanisms. Cell 138, 628–644 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Neupert W., and Herrmann J. M. (2007) Translocation of proteins into mitochondria. Annu. Rev. Biochem. 76, 723–749 [DOI] [PubMed] [Google Scholar]
- 7. Liu Q., D'Silva P., Walter W., Marszalek J., and Craig E. A. (2003) Regulated cycling of mitochondrial Hsp70 at the protein import channel. Science 300, 139–141 [DOI] [PubMed] [Google Scholar]
- 8. Neupert W., and Brunner M. (2002) The protein import motor of mitochondria. Nat. Rev. Mol. Cell Biol. 3, 555–565 [DOI] [PubMed] [Google Scholar]
- 9. D'Silva P. D., Schilke B., Walter W., Andrew A., and Craig E. A. (2003) J protein cochaperone of the mitochondrial inner membrane required for protein import into the mitochondrial matrix. Proc. Natl. Acad. Sci. U.S.A. 100, 13839–13844 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. D'Silva P. R., Schilke B., Hayashi M., and Craig E. A. (2008) Interaction of the J-protein heterodimer Pam18/Pam16 of the mitochondrial import motor with the translocon of the inner membrane. Mol. Biol. Cell 19, 424–432 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Amick J., Schlanger S. E., Wachnowsky C., Moseng M. A., Emerson C. C., Dare M., Luo W. I., Ithychanda S. S., Nix J. C., Cowan J. A., Page R. C., and Misra S. (2014) Crystal structure of the nucleotide-binding domain of mortalin, the mitochondrial Hsp70 chaperone. Protein Sci. 23, 833–842 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Matouschek A., Pfanner N., and Voos W. (2000) Protein unfolding by mitochondria. The Hsp70 import motor. EMBO Rep. 1, 404–410 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Kim Y. E., Hipp M. S., Bracher A., Hayer-Hartl M., and Hartl F. U. (2013) Molecular chaperone functions in protein folding and proteostasis. Annu. Rev. Biochem. 82, 323–355 [DOI] [PubMed] [Google Scholar]
- 14. Mayer M. P., Brehmer D., Gässler C. S., and Bukau B. (2001) Hsp70 chaperone machines. Adv. Protein Chem. 59, 1–44 [DOI] [PubMed] [Google Scholar]
- 15. Laufen T., Mayer M. P., Beisel C., Klostermeier D., Mogk A., Reinstein J., and Bukau B. (1999) Mechanism of regulation of hsp70 chaperones by DnaJ cochaperones. Proc. Natl. Acad. Sci. U.S.A. 96, 5452–5457 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Goloubinoff P., and De Los Rios P. (2007) The mechanism of Hsp70 chaperones: (entropic) pulling the models together. Trends Biochem. Sci. 32, 372–380 [DOI] [PubMed] [Google Scholar]
- 17. Rüdiger S., Buchberger A., and Bukau B. (1997) Interaction of Hsp70 chaperones with substrates. Nat. Struct. Biol. 4, 342–349 [DOI] [PubMed] [Google Scholar]
- 18. McCarty J. S., Buchberger A., Reinstein J., and Bukau B. (1995) The role of ATP in the functional cycle of the DnaK chaperone system. J. Mol. Biol. 249, 126–137 [DOI] [PubMed] [Google Scholar]
- 19. Brehmer D., Rüdiger S., Gässler C. S., Klostermeier D., Packschies L., Reinstein J., Mayer M. P., and Bukau B. (2001) Tuning of chaperone activity of Hsp70 proteins by modulation of nucleotide exchange. Nat. Struct. Biol. 8, 427–432 [DOI] [PubMed] [Google Scholar]
- 20. Harrison C. (2003) GrpE, a nucleotide exchange factor for DnaK. Cell Stress Chaperones 8, 218–224 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Westermann B., Prip-Buus C., Neupert W., and Schwarz E. (1995) The role of the GrpE homologue, Mge1p, in mediating protein import and protein folding in mitochondria. EMBO J. 14, 3452–3460 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Voos W., Gambill B. D., Laloraya S., Ang D., Craig E. A., and Pfanner N. (1994) Mitochondrial GrpE is present in a complex with hsp70 and preproteins in transit across membranes. Mol. Cell. Biol. 14, 6627–6634 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. D'Silva P., Liu Q., Walter W., and Craig E. A. (2004) Regulated interactions of mtHsp70 with Tim44 at the translocon in the mitochondrial inner membrane. Nat. Struct. Mol. Biol. 11, 1084–1091 [DOI] [PubMed] [Google Scholar]
- 24. Laloraya S., Dekker P. J., Voos W., Craig E. A., and Pfanner N. (1995) Mitochondrial GrpE modulates the function of matrix Hsp70 in translocation and maturation of preproteins. Mol. Cell. Biol. 15, 7098–7105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Bolliger L., Deloche O., Glick B. S., Georgopoulos C., Jenö P., Kronidou N., Horst M., Morishima N., and Schatz G. (1994) A mitochondrial homolog of bacterial GrpE interacts with mitochondrial hsp70 and is essential for viability. EMBO J. 13, 1998–2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Sinha D., and D'Silva P. (2014) Chaperoning mitochondrial permeability transition: regulation of transition pore complex by a J-protein, DnaJC15. Cell Death Dis. 5, e1101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Schusdziarra C., Blamowska M., Azem A., and Hell K. (2013) Methylation-controlled J-protein MCJ acts in the import of proteins into human mitochondria. Hum. Mol. Genet. 22, 1348–1357 [DOI] [PubMed] [Google Scholar]
- 28. Hatle K. M., Gummadidala P., Navasa N., Bernardo E., Dodge J., Silverstrim B., Fortner K., Burg E., Suratt B. T., Hammer J., Radermacher M., Taatjes D. J., Thornton T., Anguita J., and Rincon M. (2013) MCJ/DnaJC15, an endogenous mitochondrial repressor of the respiratory chain that controls metabolic alterations. Mol. Cell. Biol. 33, 2302–2314 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Richter-Dennerlein R., Korwitz A., Haag M., Tatsuta T., Dargazanli S., Baker M., Decker T., Lamkemeyer T., Rugarli E. I., and Langer T. (2014) DNAJC19, a mitochondrial cochaperone associated with cardiomyopathy, forms a complex with prohibitins to regulate cardiolipin remodeling. Cell Metab. 20, 158–171 [DOI] [PubMed] [Google Scholar]
- 30. Sinha D., Joshi N., Chittoor B., Samji P., and D'Silva P. (2010) Role of Magmas in protein transport and human mitochondria biogenesis. Hum. Mol. Genet. 19, 1248–1262 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Sinha D., Srivastava S., Krishna L., and D'Silva P. (2014) Unraveling the intricate organization of mammalian mitochondrial presequence translocases: existence of multiple translocases for maintenance of mitochondrial function. Mol. Cell. Biol. 34, 1757–1775 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Srivastava S., Sinha D., Saha P. P., Marthala H., and D'Silva P. (2014) Magmas functions as a ROS regulator and provides cytoprotection against oxidative stress-mediated damages. Cell Death Dis. 5, e1394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Marada A., Allu P. K., Murari A., PullaReddy B., Tammineni P., Thiriveedi V. R., Danduprolu J., and Sepuri N. B. (2013) Mge1, a nucleotide exchange factor of Hsp70, acts as an oxidative sensor to regulate mitochondrial Hsp70 function. Mol. Biol. Cell 24, 692–703 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Goswami A. V., Chittoor B., and D'Silva P. (2010) Understanding the functional interplay between mammalian mitochondrial Hsp70 chaperone machine components. J. Biol. Chem. 285, 19472–19482 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Naylor D. J., Stines A. P., Hoogenraad N. J., and Høj P. B. (1998) Evidence for the existence of distinct mammalian cytosolic, microsomal, and two mitochondrial GrpE-like proteins, the co-chaperones of specific Hsp70 members. J. Biol. Chem. 273, 21169–21177 [DOI] [PubMed] [Google Scholar]
- 36. Harrison C. J., Hayer-Hartl M., Di Liberto M., Hartl F., and Kuriyan J. (1997) Crystal structure of the nucleotide exchange factor GrpE bound to the ATPase domain of the molecular chaperone DnaK. Science 276, 431–435 [DOI] [PubMed] [Google Scholar]
- 37. Moro F., Okamoto K., Donzeau M., Neupert W., and Brunner M. (2002) Mitochondrial protein import: molecular basis of the ATP-dependent interaction of MtHsp70 with Tim44. J. Biol. Chem. 277, 6874–6880 [DOI] [PubMed] [Google Scholar]
- 38. Rassow J., Maarse A. C., Krainer E., Kübrich M., Müller H., Meijer M., Craig E. A., and Pfanner N. (1994) Mitochondrial protein import: biochemical and genetic evidence for interaction of matrix hsp70 and the inner membrane protein MIM44. J. Cell Biol. 127, 1547–1556 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Laloraya S., Gambill B. D., and Craig E. A. (1994) A role for a eukaryotic GrpE-related protein, Mge1p, in protein translocation. Proc. Natl. Acad. Sci. U.S.A. 91, 6481–6485 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Sena L. A., and Chandel N. S. (2012) Physiological roles of mitochondrial reactive oxygen species. Mol. Cell 48, 158–167 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Petit J. M., Maftah A., Ratinaud M. H., and Julien R. (1992) 10N-Nonyl acridine orange interacts with cardiolipin and allows the quantification of this phospholipid in isolated mitochondria. Eur. J. Biochem. 209, 267–273 [DOI] [PubMed] [Google Scholar]
- 42. Septinus M., Seiffert W., and Zimmermann H. W. (1983) Hydrophobic acridine dyes for fluorescence staining of mitochondria in living cells. 1. Thermodynamic and spectroscopic properties of 10-n-alkylacridine orange chlorides. Histochemistry 79, 443–456 [DOI] [PubMed] [Google Scholar]
- 43. Chan D. C. (2006) Mitochondria: dynamic organelles in disease, aging, and development. Cell 125, 1241–1252 [DOI] [PubMed] [Google Scholar]
- 44. Gogvadze V., Orrenius S., and Zhivotovsky B. (2008) Mitochondria in cancer cells: what is so special about them? Trends Cell Biol. 18, 165–173 [DOI] [PubMed] [Google Scholar]
- 45. Gough D. J., Corlett A., Schlessinger K., Wegrzyn J., Larner A. C., and Levy D. E. (2009) Mitochondrial STAT3 supports Ras-dependent oncogenic transformation. Science 324, 1713–1716 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. D'Autréaux B., and Toledano M. B. (2007) ROS as signalling molecules: mechanisms that generate specificity in ROS homeostasis. Nat. Rev. Mol. Cell Biol. 8, 813–824 [DOI] [PubMed] [Google Scholar]
- 47. Brand M. D. (2010) The sites and topology of mitochondrial superoxide production. Exp. Gerontol. 45, 466–472 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Shigenaga M. K., Gimeno C. J., and Ames B. N. (1989) Urinary 8-hydroxy-2′-deoxyguanosine as a biological marker of in vivo oxidative DNA damage. Proc. Natl. Acad. Sci. U.S.A. 86, 9697–9701 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Dinkova-Kostova A. T., and Abramov A. Y. (2015) The emerging role of Nrf2 in mitochondrial function. Free Radic. Biol. Med. 88, 179–188 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Fernández-Cabezudo M. J., Faour I., Jones K., Champagne D. P., Jaloudi M. A., Mohamed Y. A., Bashir G., Almarzooqi S., Albawardi A., Hashim M. J., Roberts T. S., El-Salhat H., El-Taji H., Kassis A., O'Sullivan D. E., et al. (2016) Deficiency of mitochondrial modulator MCJ promotes chemoresistance in breast cancer. JCI Insight 1, e86873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Copley S. D. (2014) An evolutionary perspective on protein moonlighting. Biochem. Soc. Trans. 42, 1684–1691 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Spinazzi M., Casarin A., Pertegato V., Salviati L., and Angelini C. (2012) Assessment of mitochondrial respiratory chain enzymatic activities on tissues and cultured cells. Nat. Protoc. 7, 1235–1246 [DOI] [PubMed] [Google Scholar]