Abstract
Dynamic nuclear polarization provides sensitivity improvements that make NMR a viable method for following metabolic conversions in real time. There are now many in vivo applications to animal systems and even to diagnosis of human disease. However, application to microbial systems is rare. Here we demonstrate its application to the pathogenic protozoan, Trypanosoma brucei, using hyperpolarized 13C1 pyruvate as a substrate and compare the parasite metabolism with that of commonly cultured mammalian cell lines, HEK-293 and Hep-G2. Metabolic differences between insect and bloodstream forms of T. brucei were also investigated. Significant differences are noted with respect to lactate, alanine, and CO2 production. Conversion of pyruvate to CO2 in the T. brucei bloodstream form provides new support for the presence of an active pyruvate dehydrogenase in this stage.
Keywords: lactic acid, metabolism, nuclear magnetic resonance (NMR), pyruvate, Trypanosoma brucei, dynamic nuclear polarization
Introduction
Dynamic nuclear polarization (DNP),2 particularly as implemented in dissolution DNP (1), offers tremendous potential as a means of following metabolic conversions of substrates, in living systems, in real time. There are now many examples of its application to perfused organs, small animals, and even humans (2, 3). There are fewer examples of its application to cultured cells (4–8), but results in this case have a special appeal in that the metabolic pathways being utilized can be better defined, and metabolite supply and environmental conditions can be more easily controlled. A cell type that has received little attention is that of single cell eukaryotic organisms (protozoa) such as Trypanosoma brucei, which belong to the group of parasites that cause African trypanosomiasis or sleeping sickness in humans and Nagana in animals (9, 10). T. brucei has two life-cycle stages that are easily grown in culture: the procyclic form (PCF) found in the insect vector (tsetse fly) and the bloodstream form (BSF) found in its animal host. The metabolic pathways utilized by these parasites are quite distinct from those of their hosts and even differ between the PCF and the BSF. Investigation of metabolic processes in these forms can potentially uncover steps that are distinct from those in humans and susceptible to targeting by drugs that could combat disease (11, 12).
NMR is a very versatile analytical technique, allowing the identification of many metabolites, often from just chemical shift and scalar coupling patterns of spin-[1/2] magnetic nuclei. It also can be highly quantitative, with signal intensity being directly proportional to metabolite concentrations (13–15). For metabolic studies, it is advantageous to use 13C-enriched (∼99%) metabolites so that products from substrate introduced at a particular point in time can easily be distinguished from pre-existing background.
NMR, as practiced in most metabolic applications, is not a very sensitive technique and can require hours of signal averaging to achieve an adequate signal to noise ratio. This is incompatible with real-time metabolic conversions in cells, which can be very fast (seconds to minutes) and give low levels of signal when cells occupy less than 10% of the sample volume. DNP solves this sensitivity problem by using low-temperature polarization (<2 K) and transfer of polarization from a more highly polarized free radical to prepare a hyperpolarized metabolic substrate (2, 16). In dissolution DNP, the hyperpolarized sample is quickly warmed, dissolved, and transferred to the system under study. The length of time that hyperpolarization is available depends on spin-lattice relaxation times (T1). T1 values reach many tens of seconds for non-protonated 13C sites, and observation can be continued for several T1 values. Hence, use of substrates such as pyruvic acid (pyruvate as a salt) has become very common. It has two non-protonated carbon sites, a C1 carboxyl and a C2 carbonyl, and compounds enriched in 13C at these sites are readily available. Transfer of these sites to other compounds by enzymatic action can be easily detected, and kinetic constants can be extracted by analysis of the time course of transfer.
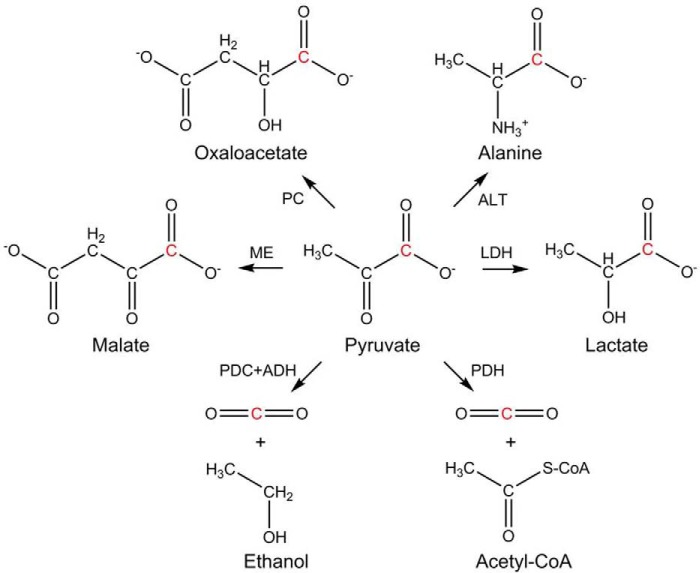
Pyruvate is the end product of glycolysis, a process common to almost all organisms. It can undergo a variety of conversions as illustrated in Scheme 1. For aerobic organisms its principal use is conversion to acetyl-CoA by pyruvate dehydrogenase (PDH) or oxaloacetate by pyruvate carboxylase. Oxaloacetate is a key element in the citric acid cycle; acetyl-Co-A also enters the citric acid cycle after producing NADH and releasing CO2 during its synthesis. Under anaerobic or oxygen-poor conditions, the need for NADH is partially satisfied during conversion of pyruvate to lactate by lactate dehydrogenase (LDH). There are different isoforms of LDH and different cells express different levels of these isoforms (17, 18). T. brucei lacks all forms of LDH (19), and the fate of pyruvate differs in PCF and BSF. Although PCF can convert pyruvate to alanine using l-alanine aminotransferase (ALT) and to acetyl-CoA using PDH (19), most pyruvate is excreted in BSF to maintain intracellular pH (20). Recent work has, however, shown the presence of a low PDH activity in BSF lysates (21, 22), although no evidence for its in vivo activity was reported except for the detection of acetate as a product of glucose or threonine metabolism (22).
Scheme 1.
Metabolic conversions of pyruvate. The C1 position of pyruvate is highlighted in red. PC, pyruvate carboxylase (EC 6.4.1.1); ALT, alanine aminotransferase (EC 2.6.1.2); ME, malate dehydrogenase (EC 1.1.1.40); LDH, lactate dehydrogenase (EC 1.1.1.27); PDC, pyruvate decarboxylase (EC 4.1.1.1); ADH, alcohol dehydrogenase (EC 1.1.1.1); PDH, pyruvate dehydrogenase complex (EC 1.2.4.1).
In our demonstration of the applicability of dissolution DNP to the analysis of metabolic processes, we expected to see a dramatic difference in product profile for cultures of T. brucei and some representative mammalian cells. We chose HEK-293 and Hep-G2 because culture conditions differ significantly (HEK-293 cells are in suspension and Hep-G2 cells are adherent on microbeads), and we felt it important to demonstrate an ability to monitor metabolism under both conditions. We will show a distinct conversion of hyperpolarized pyruvate to alanine in T. brucei as opposed to lactate in mammalian cells and produce kinetic constants for these conversions. We also show CO2 formation from pyruvate in both PCF and BSF, further supporting the presence of an active PDH in BSF.
Results
Signal enhancement of metabolic substrates is critical in these experiments. Because the formulation of the mixture being polarized can affect both the rate of polarization and the level of polarization (23–25), we explored several options for polarizing pyruvic acid. Total signal from a 13C-labeled sample placed in the polarization capsule can be monitored directly, and this is illustrated in Fig. 1. The signal is dominated by the 13C-enriched pyruvic acid in each case, and because the amount of pyruvate is similar in each case, the signal is proportional to the percentage of polarization. Because pyruvic acid is a self-glassing reagent, a control with only pyruvic acid was tested first. The rise is relatively fast (the time constant is 7 min), and a clear plateau is achieved. A glycerol water mixture is a common polarization medium in many applications (26–28). Analysis of data for this mixture yielded a time constant of 27 min and a plateau at approximately twice the value observed with pyruvic acid alone. Eutectic mixtures, including that of pyruvic acid and choline, have been found to be particularly useful polarization media because they solidify as an intimate mixture of the components, maintaining a random dispersion of pyruvate that is conducive to polarization transfer (9). A time constant of 44 min and a plateau of approximately four times the value found with pyruvic acid alone was found for a choline–pyruvic acid mixture. An enhancement factor can be extracted for any of these systems by comparing NMR signals after dissolution to a signal averaged spectrum taken after polarization has returned to equilibrium conditions at 25 °C. Correcting for differences in pulse widths, number of acquisitions, and extrapolation back to the time of dissolution, an enhancement factor of ∼19,000 was obtained for the choline–pyruvic acid mixture.
Figure 1.
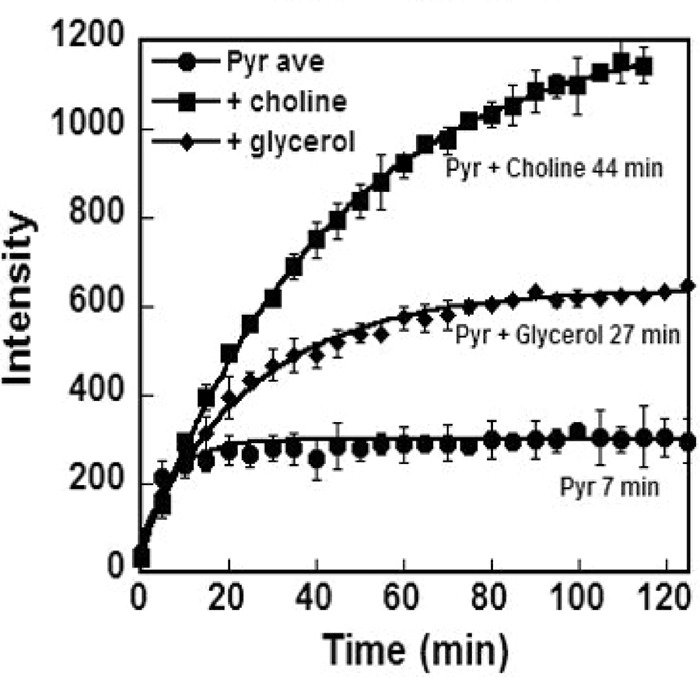
Polarization profile of 13C1 pyruvic acid (Pyr) comparing pure pyruvic acid (circles) to 1:1 mixtures with glycerol (diamonds) and choline (squares). The data points are averages of three or four runs, and error bars are standard deviations. Polarization time constants were obtained by fitting the data to an exponential function (lines). Intensity is in arbitrary units but is consistent across samples.
In choosing a polarization medium, a trade-off is often made between a high limiting polarization that is reached slowly and a lesser limiting polarization that can be reached quickly (23). In our case, signal enhancement is more crucial than the time required for full polarization; hence, we chose to use the 1:1 mixture with choline in all experiments. Choline also produces a single by-product, as opposed to the multiple products found when using glycerol. The single peak at 179.4 ppm is consistent with that expected for the ester carbonyl of pyruvyl choline, and we have identified peaks in the mass spectrum of the polarized mixture consistent with the molecular weight of this compound. We do not believe that the ∼5 mm choline in the dissolution buffer or choline containing side products at much lower levels has a significant effect on the metabolism of the cells studied here.
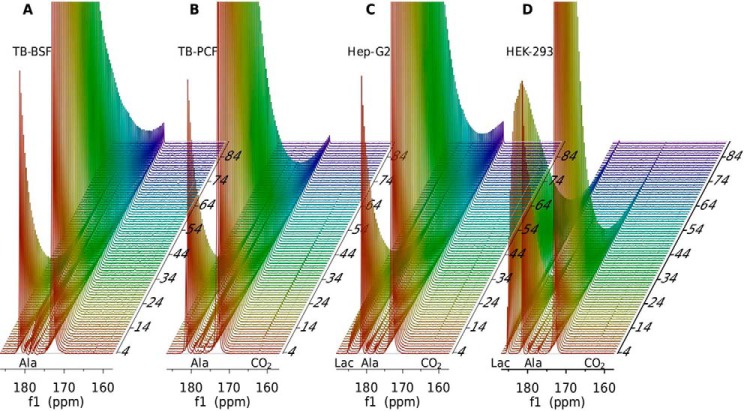
Fig. 2 shows a stacked plot of one-dimensional 13C spectra of hyperpolarized pyruvate taken at 2.0-s intervals after injection into cultures of BSF (Fig. 2A), PCF (Fig. 2B), Hep-G2 (Fig. 2C), and HEK-293 cells (Fig. 2D). Note that the cell number for T. brucei BSF, PCF, Hep-G2, and HEK-293 is 25, 100, 50, and 100 million, respectively. The major peaks at 172.9 and 181.4 correspond to the keto and hydrate forms of pyruvate. The small peak at 179.4 from pyruvyl choline and another at 176.3 from a contaminant present in all pyruvate-containing samples appear to build quickly and then decay, much like the pyruvate peaks. The build-up is a result of injection over a finite period of time (<4 s) and redistribution of material in the NMR tube; a similar build-up can be seen in the pyruvate hydrate peaks, as well. The peak at 178.6 ppm rises more slowly and persists for a much longer time. This peak is assigned to the C1 carbon of alanine, a product of the alanine transaminase reaction. There is a peak at 185.2 ppm that rises and decays much like the alanine peak for both Hep-G2 and HEK-293 cells, but the chemical shift is indicative of the C1 carbon of lactate. This peak is clearly smaller in the Hep-G2 sample. The minor peak at 163.3 ppm is assigned to CO2, which is most apparent in the T. brucei PCF forms, Hep-G2 and HEK-293 cells. Clearly the metabolic processes in these cell types are quite different.
Figure 2.
Stacked spectra of 13C1 pyruvic acid and its metabolic products in T. brucei. A, BSF (25 million cells). B, PCF forms (100 million cells). C, Hep-G2 (50 million cells. D, HEK-293 cells (100 million cells). Spectra are collected every 2 s (vertical axis is time in s). Spectral intensities were normalized to the maximum observed for pyruvic acid 13C1 in each case. Labels for alanine (Ala), lactate (Lac), and CO2 are placed at corresponding chemical shift positions for their 13C1 resonances along the horizontal axis (f1 (ppm)).
It is potentially useful to treat the conversion of pyruvate to alanine or lactate in these samples more quantitatively. Here, we only take the major product (>90%) into account, alanine for T. brucei and lactate for mammalian cells. The time courses for both substrate (pyruvate) and product (alanine or lactate) can be fit to a model that is composed of two parts: a part that relates directly to time-dependent concentrations of metabolites caused by effective transport and chemical conversion and a part that converts concentrations to NMR signals, correcting for the repetitive effects of radio frequency pulses. Treating these parts separately is advantageous because the first is described by continuous functions, and the second is described by discrete sets of observation points. The differential Equations 1–3 below describe the transport and chemistry part.
| (1) |
| (2) |
| (3) |
| (4) |
Although injection of polarized substrate actually occurs over a few seconds, the equations implicitly assume instantaneous injection. Therefore, we take zero time to be the point at the midpoint of injection and allow Psubout(0) to be fit using all data points following the point of maximum amplitude for the pyruvate signal (typically 6–8 s from the beginning of injection). kT in Equations 1 and 2 is introduced primarily to account for transport of substrate across cell membranes but may also absorb some variation associated with injection and mixing of substrate with cells. The T1 values are spin relaxation times for substrate (sub) and product (pro), and kenz is an effective first order rate constant for conversion of substrate to product by enzymes within a cell. The model assumes that the total time over which polarization is observable (∼60 s) is short enough to neglect subsequent reactions involving the initial product and reversal of substrate conversion. The three differential equations were solved simultaneously to get a time course for the polarization of substrate outside and inside the cells (Psubout(t) and Psubin(t), respectively), as well as the product (Ppro(t)) in terms of the several unknowns. Because we do not expect to resolve signals from substrate inside and outside the cells, the total substrate to be observed is given by Equation 4.
The signals observed in NMR observation at time points (n) are given in Equations 5 and 6 for substrate and product (Ssub(n) and Spro(n), respectively). The fraction of polarization remaining after sampling by a radio frequency pulse of angle θ is given by cos(θ). These equations were fit to experimental data using a non-linear regression routine to evaluate variables. Because the time between pulses was short in our application and the pulse angle was relatively large (nominally 15 or 18°), the decay of substrate signal is primarily due to a loss of magnetization on pulsing and not due to T1 decay. Hence, T1 values could not be determined precisely and were constrained in ranges of 45–60 s for pyruvic acid and 20–45 s for products during fitting.
| (5) |
| (6) |
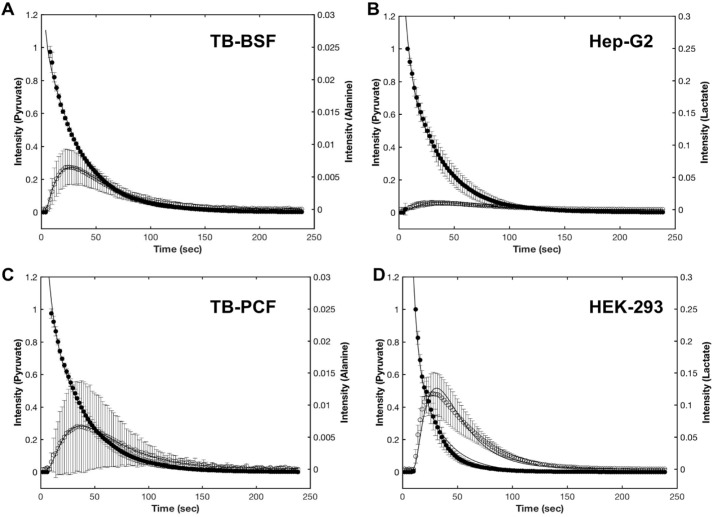
The averaged data (normalized to the first fit point) and best-fit curves for experiments on BSF and PCF samples, as well as HEK-293 and Hep-G2 samples, are shown in Fig. 3. The parameters corrected for variation in the number of cells are summarized in Table 1.
Figure 3.
The time courses of pyruvate consumption (solid circles) and metabolite production (open circles) in T. brucei BSF (A), Hep-G2 (B), T. brucei PCF (C), and HEK-293 cells (D). The average data from duplicate measurements on each of two batches of BSF and PCF cells or measurements on three batches of HEK-293 and Hep-G2 cells are shown. The error bars represent the standard deviation of these repeats. For each cell line, the data points were normalized to the maximum peak height of pyruvic acid in the first data point. The solid lines are from the best fit theoretical equations as described under “Experimental Procedures.” Note that the cell numbers are 25, 50, 100, and 100 million for A–D, respectively.
Table 1.
Parameters extracted by fitting equations 1–6 to experimental data
The errors are standard deviations over three biological replicates for Hep-G2 and HEK-293 cells and two technical replicates on each of two biological replicates for PCF and BSF cells.
| Parameters |
T. brucei |
Human |
||
|---|---|---|---|---|
| PCF | BSF | Hep-G2 | HEK-293 | |
| θ (°) | 11 ± 1 | 14 ± 1 | 12 ± 1 | 15 ± 2 |
| kT (s−1)a | 0.95 ± 0.91 | 5.1 ± 1.7 | 3.3 ± 0.8 | 1.8 ± 0.3 |
| kenz (ms−1)a | 0.7 ± 0.7 | 3.5 ± 0.6 | 5.1 ± 1.8 | 21 ± 6 |
| T1 (sub) (s) | 46 ± 2 | 59 ± 2 | 50 ± 8 | 49 ± 7 |
| T1 (pro) (s) | 35 ± 1 | 21 ± 2 | 30 ± 9 | 33 ± 3 |
| Psubout(0) | 58 ± 35 | 93 ± 18 | 82 ± 38 | 87 ± 45 |
a Kinetic constants have been adjusted to 100 million for all runs. The kenz values correspond to rates of alanine production for PCF and BSF and lactate production for Hep-G2 and HEK-293.
Although kT was introduced to account for transport across cell membranes, we chose not to interpret variations because of the possible inclusion of injection and mixing effects in this parameter. The pulse angles (θ) vary slightly from set values (15 or 18°) because probe tuning is done prior to substrate injection and tube positioning, and the amounts of sample in the coil affect tuning. Variations in Psubout(0) reflect differences in polarization, not concentrations of pyruvate. As stated previously, the T1 values are not well determined because of attenuation of signal caused by repetitive pulsing.
Of most interest are the rate constants (kenz) for conversion of product to substrate. These are in most cases well determined and show significant variation from cell type to cell type. The constants for T. brucei correspond to the production of alanine. Higher values are reached in BSF than in PCF. Although the errors for PCF cultures are large, differences between BSF and PCF are well outside the margin of error.
The rates constants for Hep-G2 and HEK-293 cells are for conversion of pyruvate to lactate. On average they show higher rates of lactate production in HEK-293 cells (approximately four times higher). The total lactate produced (obtained by integrating the area under the lactate curve) is also ∼4-fold greater in HEK-293 than Hep-G2. The viability of both cell lines was at approximately the same level (>90%), both before and ∼10 min after experiments. Hence, there appears to be a fundamental difference in lactate production for the two cell types.
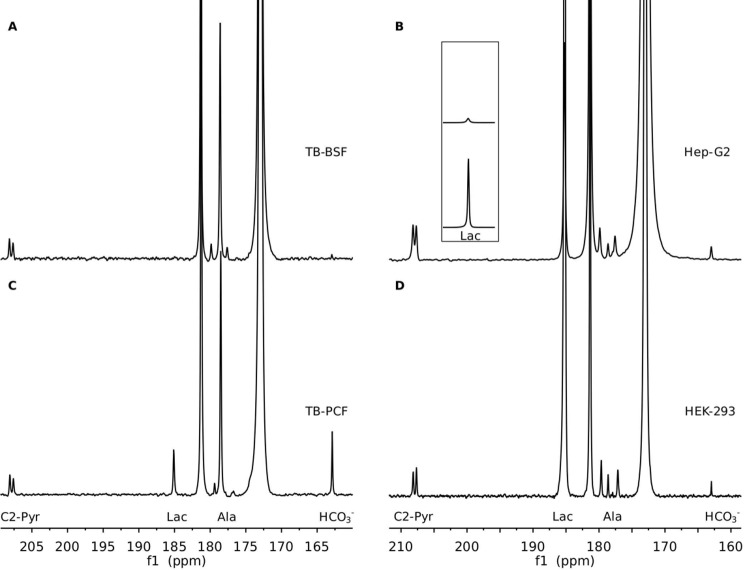
Products of metabolic conversion that occur at lower concentrations can be detected by summing spectra over the time course of our experiment. A comparison of total production of products in the two trypanosome forms is presented in the summed spectra (60 fids) presented in Fig. 4. In addition to the peak at 208 ppm assigned to the natural abundance C2 carbon of pyruvate (it shows the expected 58 Hz 13C-13C scalar coupling to the enriched C1 site), there is a peak at 163.3 ppm in both BSF (Fig. 4A) and PCF (Fig. 4C) spectra. This is where a peak from bicarbonate (HCO3−) would be expected. Bicarbonate is a by-product of conversion of pyruvate to acetyl-CoA by PDH. Although the peak from BSF cells is small, the observation is significant because of previous questions about the activity of PDH in T. brucei BSF. The CO2 released from the polarized pyruvic acid and converted to bicarbonate in the PCF sample is, nevertheless, substantially greater than that in the BSF sample. It is also noteworthy that a lactate signal is observable in the summed spectra of PCF.
Figure 4.
Comparison of summed 13C spectra for metabolic products from 13C1 pyruvate in T. brucei (A, BSF; C, PCF) and mammalian cells (B, Hep-G2; D, HEK-293). The spectra were summed from the time course spectra in Fig. 2. The inset shows a scaled region for the lactate resonances of B and D, allowing comparison of intensities. Peak intensity was in all cases normalized to the natural abundance pyruvic acid 13C2 resonances.
Summing the time course data for Hep-G2 (Fig. 4B) and HEK-293 (Fig. 4D) cells produced no peaks in addition to those from lactate, alanine, and HCO3−. In general, alanine is at a lower level than lactate for both Hep-G2 cells and HEK 293 cells. The HCO3− production in either HEK-293 or Hep-G2 cells is not as efficient as lactate, probably because the non-perfused samples in our setting may have been oxygen-poor, resulting in dominance by the anaerobic pathways. Despite the lower number of cells, the average amount of both alanine and CO2 produced in Hep-G2 is approximately twice that in HEK-293.
Discussion
The data presented above demonstrate that hyperpolarization of metabolic substrates using DNP is a useful means of investigating key aspects of pathways in parasitic organisms such as T. brucei and uncovering ways in which these pathways differ from those of their hosts. Differences in types of products produced, as well as variations in kinetic parameters, have been observed for pyruvate metabolism in T. brucei and two types of mammalian cells. Pyruvate was chosen for these investigations primarily because of its demonstrated utility in the investigation of human metabolism (2, 3, 29, 30) and because its fate was likely to depend on the life-cycle stage of T. brucei.
Some of the differences in pyruvate metabolism seen between mammalian cells and T. brucei might have been expected. In human cells pyruvate can be converted to lactate, alanine, CO2, acetyl-CoA, oxaloacetate, and malate, in each case employing just a single enzymatic step. In T. brucei lactate and oxaloacetate production from pyruvate is not expected because of the lack of LDH and pyruvate carboxylase, although detection of a small amount of lactate has been reported in PCF (19). Our results support this observation and suggest that there may be other pathways capable of producing lactate. For example, Trypanosoma cruzi possess an α-hydroxyacid deshydrogenase that can produce small amounts of phenyllactic acid and p-hydroxyphenyllactic acid arising from the catabolism of the aromatic amino acids phenylalanine and tyrosine (31), although this activity was not found in T. brucei (32).
Few kinetic parameters for enzymes involved in pyruvate metabolism are available, especially for human and T. brucei cells. According to the records in the BRENDA database (http://www.brenda-enzymes.org/), turnover numbers range broadly among all the current documented organisms, from 21 to 884/s for LDH (EC 1.1.1.27) 0.67–136/s for ALT (EC 2.6.1.2), 0.077–486/s for PDH (EC 1.2.4.1), and 8.3–284.1/s for malate dehydrogenase (EC 1.1.1.40). Unfortunately, there are no data collected in human cells for malate dehydrogenase, and for trypanosomes the only kinetic data are in a single report with a turnover number of 113/s for one isoform (ME1) and 34/s for another (ME2) in T. cruzi (33). Thus, with the addition of pyruvate isotopically labeled at C1, observation of signals from alanine, CO2, and malate in both human cells and T. brucei might be expected, and lactate is expected only in the cases of HEK-293 and Hep-G2. Our observation of alanine and CO2 in T. brucei and lactate in both HEK-293 and Hep-G2 is reasonable. However, we were unable to detect malate or oxaloacetate in any of the cells studied.
We also observed some differences in lactate production on comparing our two mammalian cell lines, HEK-293 versus Hep-G2, with our HEK cells producing more. Although these levels may depend strongly on the levels of oxygen available, we made every effort to treat cell preparations in an identical manner. Hence, we look to differences in typical expression levels for the relevant enzymes in these two cell types. LDH has five isoforms, and the type V encoded by the LDHA gene is dominant in liver and skeletal muscle. Our Hep-G2 cells are from a liver cell line. On the other hand, LDHB gene has higher expression in HEK-293 cells according to the Human Protein Atlas database (www.proteinatlas.org) (17). Our observation that lactate production is higher in HEK cells could correlate with differences in the activity of these isoforms or their expression levels. Also, we observed some enhanced level of production of alanine in Hep-G2 cells. This is consistent with the Human Protein Atlas database, indicating an enhanced RNA level of ALT in the Hep-G2 cell line.
The different forms of T. brucei, PCF and BSF, also are expected to differ dramatically in their metabolism. Although PCFs have well developed mitochondria with a respiratory chain, oxidative phosphorylation, and a partial Krebs cycle, BSFs lack a respiratory chain and Krebs cycle enzymes; they rely exclusively on glycolysis, generating ATP in the cytosol (22). Pyruvate is used in PCF to produce acetyl-CoA. This could be converted into acetate by the acetate:succinyl-CoA transferase enzyme, generating succinyl-CoA, which in turn could generate ATP by substrate level phosphorylation (19). Acetyl-CoA could also be used directly in the cytosol for fatty acid synthesis (19). In contrast, in BSF pyruvate is mostly excreted to the environment as a mechanism to regulate its intracellular pH (20). However, recent proteomic studies have detected the presence of all subunits of PDH in BSF (34, 35), and a low PDH activity was detected in lysates from this stage (21, 22). Mitochondrial PDH was proposed to be important for mitochondrial fatty acid synthesis (FAS II pathway) (21), which is a source of myristic acid needed for the formation of the glycosylphosphatidylinositol anchor of the variant surface glycoprotein and for the synthesis of lipoic acid, a cofactor of PDH (36). Although we observe a higher level of PDH activity in T. brucei PCF, we do observe activity in T. brucei BSF. Our results are the first to provide direct evidence that PDH is active in live T. brucei BSF.
The studies using pyruvate can clearly be improved to provide more quantitative data. Although fitting time-course data produce reliable kinetic constants, extracted values vary from sample to sample more than we wish, and we continue to work on improvements in protocol, including automation of injection procedures. It is also possible to adapt the perfusion apparatus to allow better control and monitoring of sample conditions (37). However, this may be difficult with very small cells such as those of T. brucei. The information returned can also be increased by considering substrates other than pyruvate. Trypanosomes make use of other substrates for their energy and synthesis needs, in particular amino acids that appear to be readily available in the insect gut. Many different substrates can be hyperpolarized, and sites for isotopic labeling can be chosen to prolong polarization storage. Ultimately, one would hope to identify bottlenecks in critical pathways that are unique to pathogenic organisms. These may be especially sensitive targets for disruption by drugs designed to control or eliminate infection. Typically substrates for enzymes at these bottlenecks build up, and an analysis of kinetic profiles can identify these sites. Trypanosomes are convenient organisms for the development and testing of this strategy. However, the strategy is clearly applicable to many other pathogenic organisms, and successful development could have a significant impact on our ability to combat disease.
Experimental procedures
Sample preparation and hyperpolarization
Pyruvic acid (13C1, 99% enriched) was purchased from Cambridge Isotope Laboratories (Tewksbury, MA). 1.25 μl of a 14.1 m solution was mixed with glycerol at a volume ratio of 1:1 or with choline chloride at a molar ratio of 1:1. The mixture was doped with OXO63 (a stable trityl radical derivative; Oxford Instruments America Inc.) to reach a final concentration of 15 mm. The samples were cooled to 1.4 K and polarized using a HyperSense DNP polarizer (Oxford Instruments, Abingdon, UK). Transfer of polarization to 13C-enriched sites was accomplished by irradiation at 93.997 GHz with 100-milliwatt power in a field of 3.35 T for ∼150 min.
Cell culture
HEK-293 and Hep-G2 cells were grown under suspension conditions or on plates in Freestyle and DMEM/F-12 medium, respectively, at 37 °C with 5% CO2 in a humidified incubation chamber. The density of HEK-293 cells was maintained in the range of 1–3 × 106 cells/ml. The Hep-G2 cells were maintained following ATCC instructions, after which microbeads (catalog no. 3779; Corning Enhanced Attachment Microcarriers) were used to scale up the cell number in a limited culturing volume. The cell number and viability were evaluated using a CountessTM automated cell counter (Invitrogen) immediately before and ∼10 min after DNP experiments. Approximately 100 million HEK-293 and 50 million Hep-G2 cells with viability greater than 90% were used for all experiments. PBS was applied to wash cells, and ∼0.5 ml of the concentrated cell (or microbead) suspensions were transferred to an 8-mm NMR tube.
Wild-type T. brucei from Lister strain 427 in both PCF and BSF were used for this study. PCFs were cultivated in SDM-79 medium and supplemented with hemin (7.5 μg/ml) and heat-inactivated fetal bovine serum (10%) at 27 °C (21). BSFs were cultivated in HMI-9 medium supplemented with heat-inactivated fetal bovine serum (10%) at 37°C with 5% CO2 (21). Cell concentrations were determined using a Neubauer Chamber. Approximately 109 PCF cells or 5 × 108 BSF cells were used per experiment. The cells were washed in PBS for PCF and buffer A with glucose (116 mm NaCl, 5.4 mm KCl, 0.8 mm MgSO4, 50 mm Hepes, pH 7.3, 5.5 mm glucose) for BSF, before loading in the NMR probe. The volume of the cell suspension in residual buffer was ∼0.5 ml.
NMR measurements
All experiments were performed using an Varian INOVA 500 MHz spectrometer equipped with an 8-mm HX probe at 37°C. Dissolution of the polarized sample was manually initiated after reaching desired polarization levels in the HyperSense polarizer. A neutralizing buffer containing 40 mm HEPES, 140 mm NaCl, pH 7.5, in 100% H2O was injected in sufficient amounts to reach a final concentration of pyruvic acid of 5 mm, and this solution was captured in a receiving vessel, which allows outgassing and homogenizing of the substrate solution. 1.5 ml of the polarized and dissolved substrate solution was then transferred to an 8-mm NMR tube by manual injection. The total time from the beginning of dissolution to completion of injection was typically 10 s. Data collection started automatically on an electronic trigger from the polarizer once the dissolution process was complete (∼6 s from initiation) and continued for ∼4 min. One-dimensional carbon spectra were acquired repetitively using a standard one-pulse sequence (a 1-s delay followed with a 15 or 18° pulse and 1 s of acquisition without proton decoupling). All experiments were repeated three to four times. Error estimates correspond to standard deviations from measurements on multiple samples.
Data analysis
NMR experimental data were processed using MesReNova (version 8.1). Exponential multiplication with a time constant approximating transverse decay of magnetization was applied to increase the signal to noise ratio. The peak heights of pyruvic acid and its product (lactate or alanine) were extracted from each spectrum in the acquisition array for data fitting using MATLAB routines (version R2016b). First, differential equations describing kinetic processes were solved using the dsolve routine. Simultaneous curve fitting, for both the substrate and product (described as Equation 5 and 6) by its non-linear least squares solver (lsqnonlin) was then used to extract optimum values for kinetic constants.
Author contributions
Y. Z. and J. H. P. designed the experiments and analyzed the data. Y. Z. and C. D. C. conducted the experiments. Y. Z. and J. H. P. wrote the majority of the manuscript, with specific sections contributed by C. D. C., and R. D., J. H. P., and R. D. supervised the work and contributed to the analysis of experiments.
Acknowledgment
The DNP instrumentation used in this study was supported by the NIGMS, National Institutes of Health Grant GM103390.
This work was supported, in whole or in part, by National Institutes of Health Grants AI108222 (to R. D.) and EB017862 (to J. H. P.). The authors declare that they have no conflicts of interest with the contents of this article. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
- DNP
- dynamic nuclear polarization
- PCF
- procyclic form
- BSF
- bloodstream form
- PDH
- pyruvate dehydrogenase
- LDH
- lactate dehydrogenase
- ALT
- l-alanine aminotransferase.
References
- 1. Ardenkjaer-Larsen J. H. (2016) On the present and future of dissolution-DNP. J. Magn Reson. 264, 3–12 [DOI] [PubMed] [Google Scholar]
- 2. Lumata L., Yang C., Ragavan M., Carpenter N., DeBerardinis R. J., and Merritt M. E. (2015) Hyperpolarized 13C magnetic resonance and its use in metabolic assessment of cultured cells and perfused organs. Methods Enzymol. 561, 73–106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Chaumeil M. M., Najac C., and Ronen S. M. (2015) Studies of metabolism using 13C MRS of hyperpolarized probes. Methods Enzymol. 561, 1–71 [DOI] [PubMed] [Google Scholar]
- 4. Meier S., Solodovnikova N., Jensen P. R., and Wendland J. (2012) Sulfite action in glycolytic inhibition: in vivo real-time observation by hyperpolarized 13C NMR spectroscopy. Chembiochem 13, 2265–2269 [DOI] [PubMed] [Google Scholar]
- 5. Christensen C. E., Karlsson M., Winther J. R., Jensen P. R., and Lerche M. H. (2014) Non-invasive in-cell determination of free cytosolic [NAD+]/[NADH] ratios using hyperpolarized glucose show large variations in metabolic phenotypes. J. Biol. Chem. 289, 2344–2352 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Canapè C., Catanzaro G., Terreno E., Karlsson M., Lerche M. H., and Jensen P. R. (2015) Probing treatment response of glutaminolytic prostate cancer cells to natural drugs with hyperpolarized [5-13C]glutamine. Magn. Reson. Med. 73, 2296–2305 [DOI] [PubMed] [Google Scholar]
- 7. Yang C., Harrison C., Jin E. S., Chuang D. T., Sherry A. D., Malloy C. R., Merritt M. E., and DeBerardinis R. J. (2014) Simultaneous steady-state and dynamic 13C NMR can differentiate alternative routes of pyruvate metabolism in living cancer cells. J. Biol. Chem. 289, 6212–6224 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Lerche M. H., Jensen P. R., Karlsson M., and Meier S. (2015) NMR insights into the inner workings of living cells. Anal. Chem. 87, 119–132 [DOI] [PubMed] [Google Scholar]
- 9. Bern C., Kjos S., Yabsley M. J., and Montgomery S. P. (2011) Trypanosoma cruzi and Chagas' disease in the United States. Clin. Microbiol. Rev. 24, 655–681 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Malvy D., and Chappuis F. (2011) Sleeping sickness. Clin. Microbiol. Infect. 17, 986–995 [DOI] [PubMed] [Google Scholar]
- 11. Urbina J. A., and Docampo R. (2003) Specific chemotherapy of Chagas disease: controversies and advances. Trends Parasitol. 19, 495–501 [DOI] [PubMed] [Google Scholar]
- 12. Docampo R., and Moreno S. N. (2003) Current chemotherapy of human African trypanosomiasis. Parasitol. Res. 90, S10–S13 [DOI] [PubMed] [Google Scholar]
- 13. Smolinska A., Blanchet L., Buydens L. M., and Wijmenga S. S. (2012) NMR and pattern recognition methods in metabolomics: from data acquisition to biomarker discovery: a review. Anal. Chim. Acta 750, 82–97 [DOI] [PubMed] [Google Scholar]
- 14. Larive C. K., Barding G. A. Jr, and Dinges M. M. (2015) NMR spectroscopy for metabolomics and metabolic profiling. Anal. Chem. 87, 133–146 [DOI] [PubMed] [Google Scholar]
- 15. Barb A. W., Hekmatyar S. K., Glushka J. N., and Prestegard J. H. (2013) Probing alanine transaminase catalysis with hyperpolarized 13CD3-pyruvate. J. Magn. Reson. 228, 59–65 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Ardenkjaer-Larsen J. H., Fridlund B., Gram A., Hansson G., Hansson L., Lerche M. H., Servin R., Thaning M., and Golman K. (2003) Increase in signal-to-noise ratio of >10,000 times in liquid-state NMR. Proc. Natl. Acad. Sci. U.S.A. 100, 10158–10163 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Uhlén M., Fagerberg L., Hallström B. M., Lindskog C., Oksvold P., Mardinoglu A., Sivertsson Å., Kampf C., Sjöstedt E., Asplund A., Olsson I., Edlund K., Lundberg E., Navani S., Szigyarto C. A., et al. (2015) Proteomics: tissue-based map of the human proteome. Science 347, 1260419. [DOI] [PubMed] [Google Scholar]
- 18. Uhlen M., Oksvold P., Fagerberg L., Lundberg E., Jonasson K., Forsberg M., Zwahlen M., Kampf C., Wester K., Hober S., Wernerus H., Björling L., and Ponten F. (2010) Towards a knowledge-based Human Protein Atlas. Nat. Biotechnol. 28, 1248–1250 [DOI] [PubMed] [Google Scholar]
- 19. Ebikeme C., Hubert J., Biran M., Gouspillou G., Morand P., Plazolles N., Guegan F., Diolez P., Franconi J. M., Portais J. C., and Bringaud F. (2010) Ablation of succinate production from glucose metabolism in the procyclic trypanosomes induces metabolic switches to the glycerol 3-phosphate/dihydroxyacetone phosphate shuttle and to proline metabolism. J. Biol. Chem. 285, 32312–32324 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Vanderheyden N., Wong J., and Docampo R. (2000) A pyruvate-proton symport and an H+-ATPase regulate the intracellular pH of Trypanosoma brucei at different stages of its life cycle. Biochem. J. 346, 53–62 [PMC free article] [PubMed] [Google Scholar]
- 21. Huang G., Vercesi A. E., and Docampo R. (2013) Essential regulation of cell bioenergetics in Trypanosoma brucei by the mitochondrial calcium uniporter. Nat. Commun 4, 2865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Mazet M., Morand P., Biran M., Bouyssou G., Courtois P., Daulouède S., Millerioux Y., Franconi J. M., Vincendeau P., Moreau P., and Bringaud F. (2013) Revisiting the central metabolism of the bloodstream forms of Trypanosoma brucei: production of acetate in the mitochondrion is essential for parasite viability. PLoS Negl Trop. Dis. 7, e2587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Lama B., Collins J. H., Downes D., Smith A. N., and Long J. R. (2016) Expeditious dissolution dynamic nuclear polarization without glassing agents. NMR Biomed. 29, 226–231 [DOI] [PubMed] [Google Scholar]
- 24. Lumata L., Merritt M. E., Malloy C. R., Sherry A. D., and Kovacs Z. (2012) Impact of Gd3+ on DNP of [1–13C]pyruvate doped with trityl OX063, BDPA, or 4-oxo-TEMPO. J. Phys. Chem. A 116, 5129–5138 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Bowen S., and Ardenkjaer-Larsen J. H. (2013) Formulation and utilization of choline based samples for dissolution dynamic nuclear polarization. J. Magn. Reson. 236, 26–30 [DOI] [PubMed] [Google Scholar]
- 26. Lelli M., Rossini A. J., Casano G., Ouari O., Tordo P., Lesage A., and Emsley L. (2014) Hydrophobic radicals embedded in neutral surfactants for dynamic nuclear polarization of aqueous environments at 9.4 Tesla. Chem. Commun. (Camb.) 50, 10198–10201 [DOI] [PubMed] [Google Scholar]
- 27. Akbey Ü., Franks W. T., Linden A., Lange S., Griffin R. G., van Rossum B. J., and Oschkinat H. (2010) Dynamic nuclear polarization of deuterated proteins. Angew. Chem. Int. Ed. Engl. 49, 7803–7806 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Day S. E., Kettunen M. I., Gallagher F. A., Hu D. E., Lerche M., Wolber J., Golman K., Ardenkjaer-Larsen J. H., and Brindle K. M. (2007) Detecting tumor response to treatment using hyperpolarized 13C magnetic resonance imaging and spectroscopy. Nat. Med. 13, 1382–1387 [DOI] [PubMed] [Google Scholar]
- 29. Sriram R., Van Criekinge M., DeLos Santos J., Keshari K. R., Wilson D. M., Peehl D., Kurhanewicz J., and Wang Z. J. (2016) Non-invasive differentiation of benign renal tumors from clear cell renal cell carcinomas using clinically translatable hyperpolarized 13C pyruvate magnetic resonance. Tomography 2, 35–42 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Lau J. Y., Chen A. P., Gu Y. P., and Cunningham C. H. (2016) Voxel-by-voxel correlations of perfusion, substrate, and metabolite signals in dynamic hyperpolarized 13 C imaging. NMR Biomed. 29, 1038–1047 [DOI] [PubMed] [Google Scholar]
- 31. Montemartini M., Santomé J. A., Cazzulo J. J., and Nowicki C. (1994) Production of aromatic α-hydroxy acids by epimastigotes of Trypanosma cruzi, and its possible role in NADH reoxidation. FEMS Microbiol. Lett. 118, 89–92 [DOI] [PubMed] [Google Scholar]
- 32. Taylor M. B., and Gutteridge W. E. (1986) The occurrence and distribution of α-hydroxy-acid dehydrogenase in some members of the order kinetoplastida. FEBS Lett. 199, 237–241 [DOI] [PubMed] [Google Scholar]
- 33. Leroux A. E., Maugeri D. A., Opperdoes F. R., Cazzulo J. J., and Nowicki C. (2011) Comparative studies on the biochemical properties of the malic enzymes from Trypanosoma cruzi and Trypanosoma brucei. FEMS Microbiol. Lett. 314, 25–33 [DOI] [PubMed] [Google Scholar]
- 34. Roldán A., Comini M. A., Crispo M., and Krauth-Siegel R. L. (2011) Lipoamide dehydrogenase is essential for both bloodstream and procyclic Trypanosoma brucei. Mol. Microbiol. 81, 623–639 [DOI] [PubMed] [Google Scholar]
- 35. Gunasekera K., Wüthrich D., Braga-Lagache S., Heller M., and Ochsenreiter T. (2012) Proteome remodelling during development from blood to insect-form Trypanosoma brucei quantified by SILAC and mass spectrometry. BMC Genomics 13, 556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Stephens J. L., Lee S. H., Paul K. S., and Englund P. T. (2007) Mitochondrial fatty acid synthesis in Trypanosoma brucei. J. Biol. Chem. 282, 4427–4436 [DOI] [PubMed] [Google Scholar]
- 37. Sriram R., Van Criekinge M., Hansen A., Wang Z. J., Vigneron D. B., Wilson D. M., Keshari K. R., and Kurhanewicz J. (2015) Real-time measurement of hyperpolarized lactate production and efflux as a biomarker of tumor aggressiveness in an MR compatible three-dimensional cell culture bioreactor. NMR Biomed. 28, 1141–1149 [DOI] [PMC free article] [PubMed] [Google Scholar]