Abstract
We have previously shown that the insulin-like growth factor 1 receptor (IGF-1R) translocates to the cell nucleus, where it binds to enhancer-like regions and increases gene transcription. Further studies have demonstrated that nuclear IGF-1R (nIGF-1R) physically and functionally interacts with some nuclear proteins, i.e. the lymphoid enhancer-binding factor 1 (Lef1), histone H3, and Brahma-related gene-1 proteins. In this study, we identified the proliferating cell nuclear antigen (PCNA) as a nIGF-1R-binding partner. PCNA is a pivotal component of the replication fork machinery and a main regulator of the DNA damage tolerance (DDT) pathway. We found that IGF-1R interacts with and phosphorylates PCNA in human embryonic stem cells and other cell lines. In vitro MS analysis of PCNA co-incubated with the IGF-1R kinase indicated tyrosine residues 60, 133, and 250 in PCNA as IGF-1R targets, and PCNA phosphorylation was followed by mono- and polyubiquitination. Co-immunoprecipitation experiments suggested that these ubiquitination events may be mediated by DDT-dependent E2/E3 ligases (e.g. RAD18 and SHPRH/HLTF). Absence of IGF-1R or mutation of Tyr-60, Tyr-133, or Tyr-250 in PCNA abrogated its ubiquitination. Unlike in cells expressing IGF-1R, externally induced DNA damage in IGF-1R-negative cells caused G1 cell cycle arrest and S phase fork stalling. Taken together, our results suggest a role of IGF-1R in DDT.
Keywords: insulin-like growth factor (IGF), phosphorylation, post-transcriptional regulation, proliferating cell nuclear antigen (PCNA), ubiquitin
Introduction
The insulin-like growth factor 1 receptor (IGF-1R)3 is a receptor tyrosine kinase. Upon ligand binding, membrane-bound receptors activate the phosphatidylinositol 3-kinase (PI3K)/Akt and mitogen-activated protein kinase (MAPK)/ERK signaling pathways (1).
We have previously shown that SUMOylation of IGF-1R (at Lys-1025, Lys-1100, and Lys-1120) leads to increased transcriptional activity due to nIGF-1R binding to enhancer-like sequences (2). Mutations of the three IGF1R SUMO-binding residues decreased nuclear transcriptional activity but did not affect the canonical signaling pathways (PI3K/Akt and MAPK/ERK) of the cell membrane IGF-1R. nIGF-1R has also been shown to associate with the LEF1 transcription factor and to phosphorylate histone H3 (3, 4). Although canonical IGF-1R signaling is well-characterized, the functional context of nIGF-1R is still poorly understood.
In this study, we sought to identify potential nIGF-1R-binding partners. For this purpose, we immunoprecipitated IGF-1R from human embryonic stem cells (hESCs) and analyzed receptor-associated proteins by mass spectrometry. One of the identified proteins was the proliferating cell nuclear antigen (PCNA), a nuclear protein that assembles in a homotrimeric ring structure encircling the DNA double helix and functions as a mobile sliding clamp to recruit other proteins (such as DNA polymerases and ligases) during DNA replication (5). If unresolved, replication fork stalling caused by replication stress or DNA damage agents could induce genomic instability. PCNA is a principal component in the cellular response to replication fork stalling, and its functionality is tightly regulated in this respect (6–8).
Ubiquitination of PCNA has been shown to regulate various DNA damage tolerance (DDT) mechanisms. PCNA monoubiquitination induces switching to low-fidelity DNA polymerases that bypass DNA lesions (translesion synthesis, TLS). Polyubiquitination is believed to initiate the more complex template switching operation, wherein the intact sister strand is utilized to extend past the lesion (9–11).
Mono- and polyubiquitination of PCNA are mediated by two distinct sets of E2 (ubiquitin-conjugating enzyme) and E3 (ubiquitin-protein ligase) enzymes that operate in a linear fashion (12). PCNA is first monoubiquitinated by RAD6 (E2) and RAD18 (E3) (13–15), followed by Lys-63 polyubiquitin linkage by UBC13-MMS2 (an E2 heterodimer) and HTLF or SHPRH (E3) (8, 9, 16–18).
In this study, we demonstrate that IGF-1R directly phosphorylates three PCNA tyrosine (Tyr-60, -133, and -250) residues. This phosphorylation leads to mono- and polyubiquitination. In addition, our results suggest that IGF-1R contribute to rescue of replication fork stalling in cells exposed to DNA damage.
Results
IGF-1R is expressed in cell nucleus of hESC
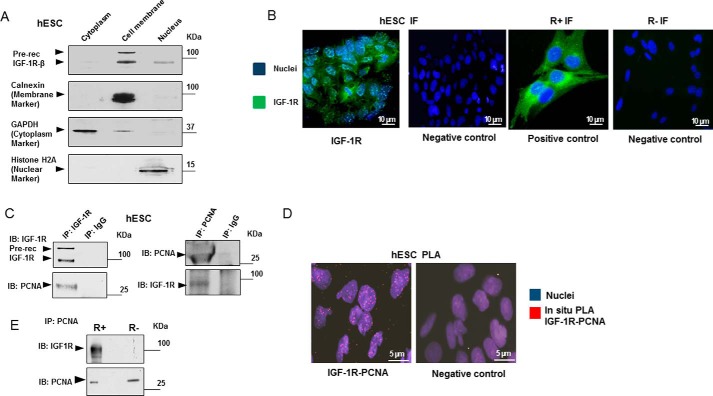
After subcellular fractionation of human embryonic stem cell line H1 (WA01) hESCs (designated hESC henceforth), IGF-1R was detected in both cell nuclear and membrane fractions (Fig. 1A) using immunoblotting. This was supported by immunofluorescence microscopy of IGF-1R, which showed clear fluorescence signals within the hESC nuclei (Fig. 1B). This result was further supported by analysis of R+ and R− cells. These cell lines are igf1r knock-out MEF cells, stably transfected with (R+) and not transfected with IGF1R (R−).
Figure 1.
IGF-1R translocates to cell nucleus and binds to PCNA. A, WA01 cells (hESC cell line), grown under feeder-free conditions, were harvested and subjected to cell fractionation. The obtained cytoplasm, cell membrane, and nuclear fractions were lysed and analyzed for IGF-1Rβ expression using immunoblotting (IB). Membranes were reblotted to determine the purity of the fractions (GAPDH for cytoplasm, calnexin for cell membrane, and histone H2A for nucleus). B, presence of nIGF-1R in hESC nuclei was confirmed using indirect immunofluorescence (two left panels). hESC cells were incubated with IGF-1R antibody (green) and counterstained with DAPI (blue) to visualize nuclei. The negative control was obtained by omitting the primary antibody. Two right panels show the same analysis on R+ (expressing human IGF-1R) and R− (IGF-1R negative) MEF cells. Scale bar, 10 μm. C, hESC cells were harvested, lysed, and immunoprecipitated for IGF-1R β-subunit (IGF-1R) and detected for PCNA (left panel) or immunoprecipitated for PCNA and detected for IGF-1Rβ (right panel). IgG was used as negative control. D, hESC cells were analyzed for subcellular localization of IGF-1R–PCNA complexes by in situ PLA (see under “Experimental procedures” for details). hESC Red dots indicate IGF-1R/PCNA interactions. Counterstaining with DAPI (blue) visualizes cell nuclei. The negative control was obtained by omitting one of the primary antibodies. Scale bar, 5 μm. E, R+ and R− cells were immunoprecipitated for PCNA and detected for IGF-1R and PCNA by immunoblotting. See also supplemental Fig. S2B showing this co-IP in comparison with the expression of IGF-1R and PCNA in the whole-cell lysates. Data are representative of three or more independent experiments.
IGF-1R associates with PCNA in the cell nucleus
To investigate the role of nIGF-1R, we sought to collect IGF-1R-binding partners by immunoprecipitation (IP) of IGF-1R from hESC and to identify them by liquid chromatography and mass spectrometry (LC-MS). IP of IgG served as a negative control. The immunoprecipitated proteins were eluted, separated by SDS-PAGE, and stained with Coomassie Blue. Ten prominent gel bands observed in the IGF-1R sample but not the IgG sample were excised (supplemental Fig. S1A). After LC-MS, multiple potential binding partners were identified, including e.g. 14-3-3 proteins (previously described to bind IGF-1R (19)), heat-shock-associated proteins, elongation factor 2 (proposed target of IGF-1R signaling (20)), histone H4, and PCNA (supplemental Fig. S1B). We decided to focus on PCNA because it exhibited several unique peptides and high coverage (supplemental Fig. S1B).
The association of IGF-1R with PCNA was confirmed using IP with anti-IGF-1R and reciprocal IP with anti-PCNA, followed by immunoblotting for PCNA and IGF-1R, respectively (Fig. 1C). Nuclear association was confirmed by co-IP after subcellular fractionation (supplemental Fig. S1, C and D) and in situ PLA of IGF-1R and PCNA (Fig. 1D). PLA showed clear signals abundantly located to the cell nuclei.
Next, we used R+ and R− cells to support the association between IGF-1R and PCNA (Fig. 1E). Although R+ exhibited this association, R−, as expected, did not. The percentage of cells in S phase and the expression level of PCNA (control blot) were similar in the two cell lines (supplemental Fig. S2, A and B). As the R+ cell line is a stable IGF1R transfectant produced from R− cells in the early 1990s (21, 22), we verified the observed IGF-1R/PCNA interaction in a newly transfected R− cell line (here denoted R-WT). R-EV represents empty vector control (supplemental Fig. S2C).
Because R+ and R− cells provide an excellent cell system for comparison of the effect in the presence and absence of IGF-1R, they were used as lead and control cell lines for investigation of IGF-1R/PCNA-dependent events. Selected findings were validated on other human and mouse non-engineered cell lines, including hESC.
IGF-1R directly phosphorylates PCNA on Tyr-60, Tyr-133, and Tyr-250
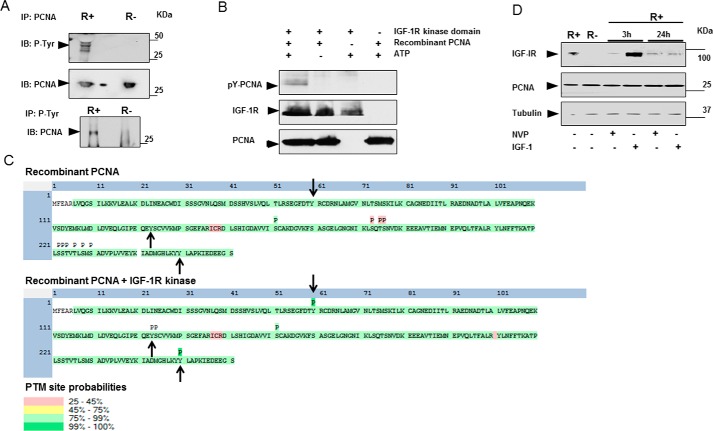
IGF-1R is an established receptor tyrosine kinase in the cell membrane, but it was recently shown to exert kinase activity also in the cell nucleus by directly phosphorylating histone H3Y41 (4). Consequently, we investigated whether nIGF-1R might phosphorylate PCNA. First, we compared PCNA tyrosine phosphorylation status in R+ with R− cells. IP of PCNA followed by detection of phosphotyrosine showed that PCNA is phosphorylated only in R+ and not in R− cells (Fig. 2A, top panel), as shown by several bands in R+ cells between 30 and 60 kDa. Reciprocal co-IP confirmed this finding (Fig. 2A, bottom panel). These results indicate that the expression of IGF-1R is necessary for tyrosine phosphorylation of PCNA, but they do not determine whether this is mediated through the canonical IGF-1R signaling or directly by the nuclear receptor.
Figure 2.
IGF-1R phosphorylates PCNA. A, R+ and R− cells, grown under basal conditions until subconfluency, were harvested, lysed, and immunoprecipitated for PCNA and detected for phosphotyrosine (P-Tyr) (top panel). Blot was stripped and incubated with anti-PCNA. Cells were also immunoprecipitated for p-Tyr and detected for PCNA (bottom panel). B, His tag recombinant PCNA was subjected to an in vitro phosphorylation reaction using recombinant IGF-1R kinase in the presence or absence of ATP. The reactions were terminated by boiling in SDS sample buffer. Separation of samples in 4–12% SDS-PAGE was followed by immunoblotting (IB) with a p-Tyr antibody. Reblotting was made for detection of IGF-1R kinase and His tag PCNA. As indicated, negative controls were obtained by omitting recombinant IGF-1R kinase, recombinant His-tagged PCNA, or ATP from the reactions. Data are representative of three or more independent experiments. C, diagrammatic representation of peptide coverage and post-translational modifications (PTM) in PCNA with (lower panel) or without (upper panel) IGF-1R kinase domain as observed by LC-MS/MS. Peptides and phosphorylation sites with high coverage are presented in green, and those with low coverage are presented in yellow or pink. Tyr-60, Tyr-133, and Tyr-240 are highlighted by arrows. D, R+ cells were treated with 1 μm NVP-AEW541 (NVP), 100 ng/ml IGF-1, or vehicle for 3 and 24 h, in comparison with non-treated R− cells. Cell lysates were separated on 4–12% BisTris SDS-PAGE and subjected to immunoblotting with a PCNA antibody. Tubulin was used as loading control.
To investigate whether IGF-1R phosphorylates PCNA directly, an in vitro kinase assay was used. As shown, recombinant IGF-1R kinase domain was able to tyrosine-phosphorylate recombinant PCNA (Fig. 2B). The presence or absence of ATP in the reactions was used as control. The specificity of IGF-1R phosphorylation was further confirmed by adding an IGF-1R inhibitor (NVP-AEW541) to the reaction (supplemental Fig. S3A). As a positive control for the IGF-1R kinase activity in vitro, we showed that it phosphorylated itself (a bona fide substrate) under such conditions (Fig. 3B).
Figure 3.
IGF-1R phosphorylation is essential for ubiquitination of PCNA. A, R+ and R− cells were harvested, lysed, and immunoprecipitated for PCNA and detected for ubiquitin (Ub). Blots were stripped and incubated with anti-PCNA to confirm equal loading. B, R+ cells were treated with either 1 μm NVP, 100 ng/ml IGF-1, or vehicle for 3 h, in comparison with non-treated R− cells. PCNA was immunoprecipitated from cells and blotted for Ub. Reblotting for p-Tyr and PCNA was made. C, R+ and R− cells were immunoprecipitated for PCNA and detected for SHPRH, HLTF, UBC13, RAD18, or pol η. Blots were stripped and incubated with anti-PCNA. R− was used as negative control. See supplemental Fig. S6A for expression of ligases in the whole-cell lysates. D, immunoprecipitation of green fluorescent protein (GFP) in R+ cells transfected with wild-type (WT) or mutated tyrosine residues (Y1–3) PCNA-GFP. Y1 = Y60F; Y2 = Y133F; Y3 = Y250F; and Y1–2-3 = triple mutant (Y60F/Y133F/Y250F). Mock-transfected R+ cells were used as a negative control. Membranes were immunoblotted (IB) for IGF-1R, Ub, p-Tyr, SHPRH, HLTF, RAD18, and PCNA. See supplemental Fig. S6B for expression of ligases in the whole-cell lysates. Data are representative of three or more independent experiments.
To identify the specific PCNA tyrosine residue(s) being phosphorylated by nIGF-1R, we utilized MS analysis of recombinant PCNA after in vitro phosphorylation by recombinant IGF-1R. Recombinant PCNA without IGF-1R was used as negative control. The result reveals that residues Tyr-60, Tyr-133, and Tyr-250 were phosphorylated by the IGF-1R kinase (Fig. 2C). These data indicate that IGF-1R is capable of directly phosphorylating PCNA, suggesting that PCNA is a substrate of the nuclear IGF-1R tyrosine kinase activity. To confirm these results, the potential function of the Tyr-60, Tyr-133, and Tyr-250 residues will be considered below.
IGF-1R activity does not affect PCNA stability
Nuclear epidermal growth factor receptor (EGFR) has previously been reported to control PCNA stability through Tyr-211 phosphorylation (23). To investigate whether IGF-1R-mediated PCNA phosphorylation affected PCNA stability, R+ cells were treated with either IGF-1 or NVP-AEW541 for 3 and 24 h. However, neither IGF-1R stimulation nor inhibition altered the PCNA levels (Fig. 2D). Additionally, R+ and R− cells showed similar PCNA levels. These data suggest that IGF-1R does not affect PCNA stability.
Nuclear IGF-1R-mediated phosphorylation marks PCNA for ubiquitination
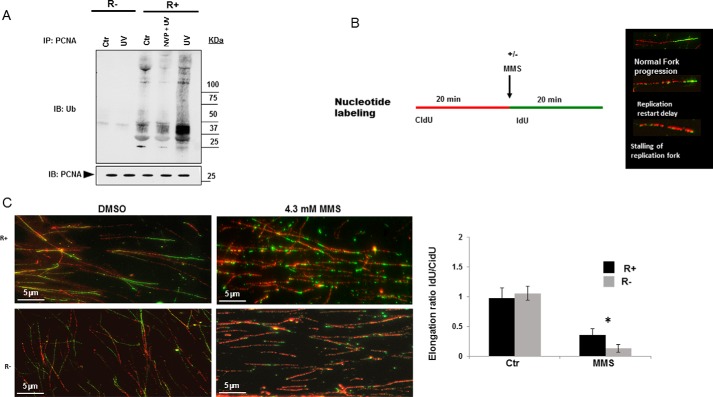
Post-translational modification of PCNA by mono- and polyubiquitination has been shown to control various PCNA functions, including the DDT pathway (see Introduction). Using co-IP experiments, we observed mono- and polyubiquitinated PCNA in R+ but not in R− cells (Fig. 3A).
To investigate the possible involvement of IGF-1R kinase activity in ubiquitination of PCNA, R+ cells were treated with either NVP-AEW541 or IGF-1 for 3 h, followed by immunoprecipitation of PCNA and detection of phosphotyrosine and ubiquitin. IGF-1 treatment clearly increased both PCNA phosphorylation and ubiquitination, whereas inhibition of IGF-1R reversed/decreased these effects (Fig. 3B).
To confirm the nuclear function of IGF-1R in inducing PCNA phosphorylation and subsequent ubiquitination, PCNA from the cytoplasmic and nuclear fractions of R+ cells was pulled down using IP. As shown in supplemental Fig. S5, A and B, PCNA only from the nuclear fraction was tyrosine-phosphorylated and ubiquitinated. Further confirmation was achieved by using R-TSM cells (R− transfected with triple SUMO-site-mutated IGF-1R cell line). Just recently we showed that R-TSM cells exhibited diminished nuclear gene regulatory effects of IGF-1R while retaining IGF-1R kinase-dependent signaling (2, 24).
IGF-1R from R+ and R-TSM was pulled down using IP, and equal amounts of it were subjected to in vitro kinase assay using recombinant PCNA as substrate (supplemental Fig. S4C). As shown, only IGF-1R from R-WT was able to phosphorylate PCNA (supplemental Fig. S4C). Incubation with TSM IGF-1R did not lead to any detectable phospho-PCNA signal. IP from R− was used as negative control. We then analyzed ubiquitination of PCNA in these cell lines. Only PCNA from R+ cells was phosphorylated and ubiquitinated (supplemental Fig. S4D). Taken together, the results suggest that nIGF-1R-induced phosphorylation of PCNA is involved in its ubiquitination. The inability of TSM IGF-1R to phosphorylate PCNA is in line with the observed decrease in phosphorylation of histone H3 in HeLa cells transfected with TSM IGF-1R (4).
Inhibition of the MAPK/ERK and the PI3K/Akt pathways using the inhibitors U0126 and LY2942002, respectively, did not decrease PCNA ubiquitination as compared with the untreated control (supplemental Fig. S5, A and B), which supports the notion that PCNA ubiquitination is mediated by nuclear IGF-1R and not by the canonical signaling of membrane IGF-1R.
PCNA Tyr-60, Tyr-133, and Tyr-250 are essential for recruitment of DDT-dependent ligases and ubiquitination
Mono- and polyubiquitinations of PCNA by RAD6 (E2)–RAD18 (E3) and UBC13 (E2)–SHPRH/HLTF (E3), respectively, have been shown to regulate PCNA function through the DDT pathway (9, 10). Using co-IP experiments, we showed the association of UBC13 (E2) and SHPRH/HLTF (E3) with PCNA in R+ but not R− cells (Fig. 3C). This suggests that nIGF-1R-dependent PCNA polyubiquitination may be mediated by these ligases. Similar results were also obtained for the monoubiquitin RAD18 (E3) ligase and the main TLS polymerase DNA polymerase η (Fig. 3C). As a control, the expression of ligases in whole-cell lysates was investigated by immunoblotting. Essentially similar levels are detected in R+ and R− cells (supplemental Fig. S6A). A separate experiment on the association of SHPRH, HTLF, and RAD18 with PCNA, and ligase expression in whole-cell lysates, is presented in supplemental Fig. S7A. This experiment also involved a random nuclear protein (i.e. p27), which did not co-IP with PCNA (supplemental Fig. S7A). IGF-1 stimulation increased the binding of SHPRH/HLTF, RAD18, and DNA polymerase η to PCNA, whereas inhibition with NVP-AWE541 decreased it (supplemental Fig. S7B).
The importance of PCNA Tyr-60, Tyr-113, and Tyr-250 phosphorylation sites for IGF-1R/PCNA interaction and subsequent modifications was then investigated by site-specific mutations. R+ cells were transfected with eGFP-PCNA constructs representing either wild-type PCNA or mutated PCNA, resulting in a tyrosine replaced by a phenylalanine (Y60F, Y133F, Y250F or triply mutated Y60F/Y133F/Y250F, denoted Y-Tpl). Exogenous PCNA was pulled down using co-IP with anti-GFP. In contrast to the exogenous wild-type PCNA, all four mutants abolished the IGF-1R/interaction, PCNA phosphorylation and ubiquitination, as well as binding of SHPRH/HLTF and RAD18 ligases (Fig. 3D). This suggests that phosphorylation of all three tyrosine residues is necessary for the nIGF-1R/PCNA interaction. Control blots of ligase expression in lysates of the transfected cells are shown in supplemental Fig. S6B.
Together with our findings that IGF-1R associates with PCNA in cell nuclei (Fig. 1), and has the capacity to directly phosphorylate PCNA (Fig. 2; supplemental Fig. S3), it appears that the nucleus-localized receptor serves as the kinase and thereby triggers the subsequent ubiquitination.
To verify the functional role of mutated PCNA constructs in DNA replication, we investigated GFP-gated Y-Tpl in R+ cells by FACS analysis after BrdU/DAPI labeling. Upon synchronization by serum starvation, the cells were released and analyzed at different time points (0–20 h). As shown in supplemental Fig. S8, the cells progressed from G1 to and through the S phase. After 20 h, many cells reentered the G1 phase. Similar results were seen for the other (single mutated) eGFP-PCNA transfected cell lines (data not shown).
IGF-1R regulates PCNA ubiquitination after DNA damage
Because PCNA ubiquitination has been coupled to the DDT pathway mainly after DNA damage that induced replication fork stalling, we next sought to explore the potential role of nIGF-1R-mediated PCNA ubiquitination in response to UV treatment. R+ and R− cells were exposed to UV irradiation, 30 mJ/cm2, for 30 s, followed by a 1-h exposure-free incubation. PCNA in R− cells did not exhibit any ubiquitination in response to UV irradiation (Fig. 4A). Conversely, the base-line ubiquitination of PCNA in R+ cells was increased substantially by UV light, but it was attenuated if NVP-AWE541 was added prior to the irradiation (Fig. 4A). Similarly, R+ cells showed a strong induction of PCNA ubiquitination after treatment with the alkylating agent methyl methanesulfonate (MMS) (supplemental Fig. S9). This effect was abolished by IGF-1R inhibition. Thus, DNA damage-induced ubiquitination of PCNA was dependent on IGF-1R activity.
Figure 4.
IGF-1R regulates ubiquitination of PCNA and replication fork progression after induced DNA damage. A, R+ and R− cells, grown under basal conditions, were treated with 1 μm NVP or vehicle for 1 h, followed by UV irradiation with 30 mJ/cm2 for 30 s and 1 h non-treatment incubation. Non-irradiated cells represent negative controls (Ctr). PCNA was immunoprecipitated from the cell lysates and subjected to immunoblotting (IB) for detection of Ub. Blots were stripped and incubated with anti-PCNA to confirm equal loading. B, left panel depicts a schematic diagram of protocol used for DNA fiber assay. Cells were pulse-labeled with halogenated nucleotides CidU (red) and IdU (green) for 20 min each. Using MMS, which interferes with DNA replication during the second pulse-labeling (IdU), fork elongation/stalling in response to replicative stress is detected. Right panel shows stained appearance of fork events as detected by DNA fiber assay. C, DNA fibers labeled with CidU (red) and IdU (green), as described in schematic diagram, were used to measure replication fork restart after treating R+ and R− cells with vehicle or 4.3 mm MMS for 20 min (left panel). For details see under “Experimental procedures.” Right panel shows graphical presentation of replication restart speed as the ratio of the lengths of nascent replication tracts after inducing DNA damage labeled with IdU (green) to the length of the replication tract before inducing damage labeled with CidU (red). At least 50 unidirectional forks labeled with both CidU and IdU were measured for each condition. Scale bar, 5 μm. Data represent mean ± S.D. of three independent experiments and considered significant (*) at p < 0.01, using one-way analysis of variance.
IGF-1R is involved in DDT pathway
Our next step was to investigate the role of IGF-1R in the DDT pathway. One method for assessing the efficiency of the DDT pathways is to measure the speed of replication restart after induction of DNA damage. This measures the cell's ability to tolerate DNA damage upon encountering polymerase-blocking lesions. To assess the functional role of IGF-1R in the DDT pathway, we applied the DNA fiber-labeling method to measure replication fork progression after MMS-induced DNA damage in R+ and R− cells. Briefly, cells were incubated with two different halogenated nucleotides, chlorodeoxyuridine (CidU) and iododeoxyuridine (IdU), which are incorporated into nascent DNA at replication forks before and after inducing DNA damage by MMS as schematically shown in Fig. 4B. The speed of DNA replication restart was estimated as the IdU/CidU ratios of the lengths of labeled nascent replication tracts before (CidU) and after (IdU) induction of DNA damage. MMS treatment delayed DNA replication by 60% in R+ cells and by as much as 90% in R− cells (Fig. 4C). Thus, R+ cells have a more efficient DDT pathway than R− cells.
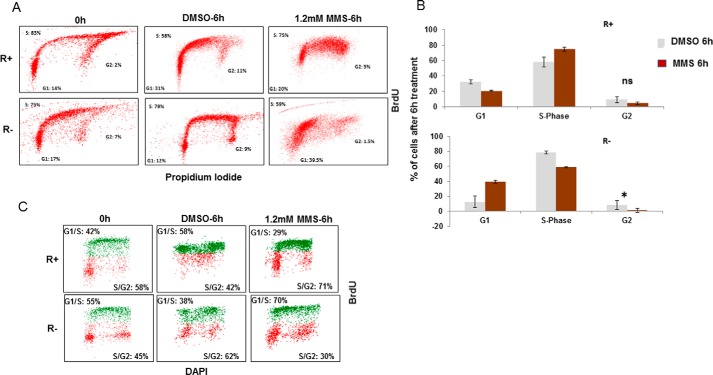
To further characterize the role of IGF-1R in the DDT pathway we used FACS analysis, after BrdU/PI labeling, to determine the effect of DNA damage upon cell cycle progression. R+ and R− cells were synchronized by serum depletion and then treated with MMS or DMSO (control) for 6 h. Upon serum repletion, the R+ and R− control cells progressed through the cell cycle (Fig. 5, A and B). Treatment with MMS slowed down the cell cycle progression only in R+ cells (Fig. 5, A and B). No increase in the percentage of cells in G1 was seen in R+ cells. In R− cells, MMS treatment substantially inhibited cell cycle progression by inducing G1-cell cycle arrest, decreasing the number of cells in S phase, and almost blocking the progression of cells into the G2 phase (Fig. 5, A and B). Further support for this response was obtained using unsynchronized cells. R+ and R− cells were pulse-labeled with BrdU for 1 h, followed by treatment with DMSO or MMS for 6 h (Fig. 5C). The BrdU-labeled cells were then gated (green dots), and the progression of the gated cells through S phase after treatment was investigated. In a pattern that was basically similar to that in synchronized cells, the R+ cells were able to progress from the G1/S phase to the S/G2 phase at a slower rate after MMS treatment, whereas R− cells collapsed in the G1/S phase, with minimal cell progression toward the S/G2 phase.
Figure 5.
IGF-1R is required for cell cycle progression after induced DNA damage. A, FACS analysis of R+ and R− cell lines, labeled with BrdU, and stained with PI after 6 h treatment with either 1.2 mm MMS or DMSO. Cells were synchronized at G1 by serum starvation for 24 h and then released in full medium for 12 h for maximal S phase synchronization before treatment. B, graphic presentation of cell cycle distributions of DMSO- and MMS-treated cells. Data represent mean ± S.D. of three independent experiments and considered significant (*) at p < 0.01, using two-sided t test. ns, not significant. C, unsynchronized R+ and R− cells were pulse-labeled with BrdU for 1 h, followed by washing the cells from BrdU labeling and the subsequent treatment with either MMS or DMSO for 6 h. Cells were fixed, stained for BrdU and DAPI, and processed to flow cytometry. The BrdU-labeled cells were gated (green dots) from non-BrdU-labeled cells (red dots).
To further investigate the role of IGF-1R in DDT, we analyzed the connection between IGF-1R activity and PCNA phosphorylation in cells entering the S phase. We used the same experimental condition as described in Fig. 5, A and B. Serum-starved R+ and R− cells were re-stimulated for 12 h for maximal S phase synchronization (supplemental Fig. S10A). We first investigated S phase cells under spontaneous fork stalling (i.e. without DNA damage exposures). After IP of PCNA, IGF-1R, p-Tyr, and PCNA were detected (supplemental Fig. S10B). In serum-starved R+ cells (indicated as G1), there was a slight IGF-1R/PCNA interaction and no PCNA phosphorylation. In contrast, R+ cells in S phase (indicated as S) showed a much stronger association between IGF-1R and PCNA as well as a clear PCNA phosphorylation. However, upon addition of NVP-AEW541, p-Tyr PCNA was almost deleted (supplemental Fig. S10B). No PCNA phosphorylation was seen in R− cells. Next, we investigated S phase cells under condition of fork stalling collapse, as induced by a 6-h MMS treatment (cf. Fig. 5, A and B). As shown in supplemental Fig. S10C, MMS treatment drastically increased phosphorylation of PCNA as compared with the negative controls. Simultaneous treatment with the IGF-1R inhibitor abrogated the MMS-induced response. No effects were observed in R− cells (supplemental Fig. S10C). These data support a role IGF-1R in DDT through phosphorylation of PCNA.
IGF-1R/PCNA interaction and PCNA ubiquitination in cells with endogenous IGF-1R
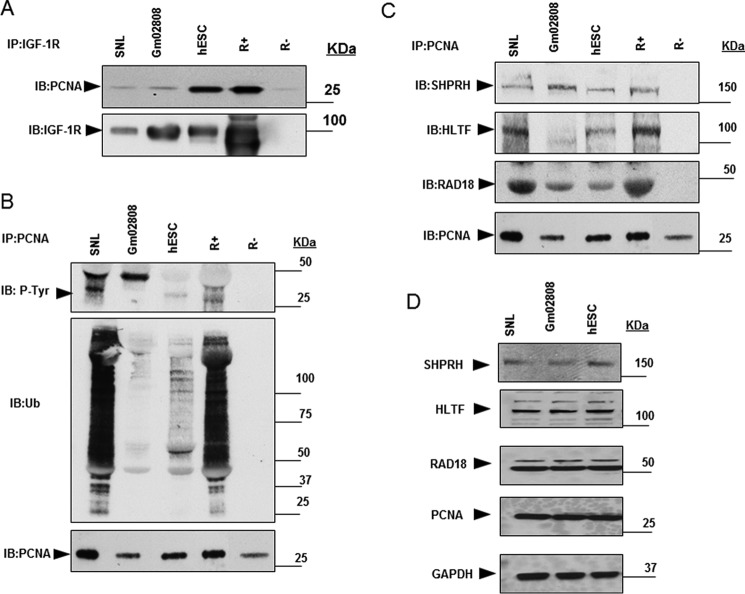
The cell lines SNL (an MEF cell line) and Gm02808 (human fibroblasts) were compared with hESC and R+/R− cells. Similar to hESC and R+, the IGF-1R–PCNA complex was detected in SNL and GM02808 (Fig. 6A). They all exhibited PCNA phosphorylation and ubiquitination in a pattern similar to R+ but at varying levels (Fig. 6B). Similar results were also shown regarding PCNA association with DDT-dependent E2/E3 ligases (Fig. 6C). The expression levels of the various E2/E3 ligases in whole-cell lysates of SNL, GM02808, and hESC cells are shown in Fig. 6D.
Figure 6.
IGF-1R/PCNA interaction and associated PCNA modifications in various cell lines. A, IGF-1R/PCNA interaction in indicated cell lines (SNL, Gm02808, and hESC) was determined by co-immunoprecipitation. Membranes were reblotted for IGF-1R for equal loading. R+ and R− were used as positive and negative controls, respectively. B and C, immunoprecipitates of PCNA from SNL, Gm02808, and hESC cells were immunoblotted (IB) for P-Tyr, Ub, and PCNA (B) and E2/E3 ligases (C). R+ and R− cells were used as controls. D, Western blotting analysis showing expression of the indicated ligases (cf. C) in whole-cell lysates. Data are representative of three or more independent experiments.
Discussion
The role of the nucleus-localized IGF-1R in normal physiology and disease is still poorly understood. Here, we identified and characterized a novel potential role of nIGF-1R. We found that IGF-1R interacts with and phosphorylates PCNA on three tyrosine residues (Tyr-60, Tyr-133, and Tyr-250), which was followed by ubiquitination. We also provided evidence that IGF-1R increases tolerance to DNA damage. The interaction between IGF-1R and PCNA may play a role in this respect, as we could show IGF-1R-dependent increase in phosphorylation of PCNA in cells entering S phase under conditions of spontaneous fork stalling and fork-stalling collapse as well. Together, our findings suggest a mechanistic link between the IGF-1R/PCNA interaction and DDT.
Our opinion that IGF-1R-dependent phosphorylation of PCNA is mediated by nIGF-1R and not through the canonical signaling pathways via the cell membrane receptor is supported by the followings findings: 1) IGF-1R and PCNA co-localize exclusively in cell nuclei; 2) IGF-1R phosphorylates PCNA in vitro in the absence of other kinases; 3) PCNA is phosphorylated in cells expressing WT-IGF-1R but not in those expressing TSM-IGF-1R; and 4) inhibition of PI3K/Akt and MAPK/ERK activity by specific inhibitors did not abrogate phosphorylation and ubiquitination of PCNA.
Other receptor tyrosine kinases have been shown to translocate to the nucleus and to phosphorylate PCNA on different sites (25). Nuclear EGFR was reported to phosphorylate PCNA on Tyr-211, regulating PCNA stability and inhibiting DNA mismatch repair (26). The tyrosine kinase c-ABL has also been shown to phosphorylate PCNA on Tyr-211, increasing DNA synthesis activity (27). These results are in line with the notion that PCNA phosphorylation may regulate different functions of PCNA in DNA repair and replication, and the discrepancies may be explained by the different tyrosine residues modified.
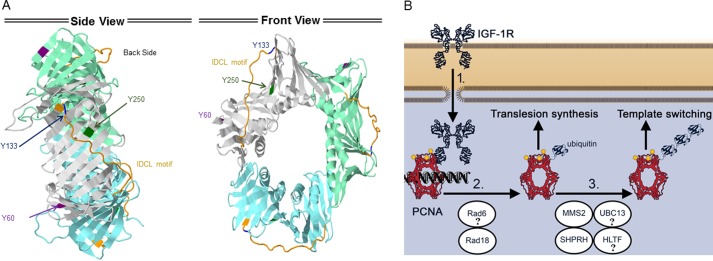
The identified tyrosine-phosphorylated sites (Tyr-60, Tyr-133, and Tyr-250) have been described in a number of previous studies mediating other functions. A study of global tyrosine phosphorylation identified a significant PCNA Tyr-250 phosphorylation after EGF stimulation in HeLa cells (28). PCNA Tyr-133 was shown to be important for PCNA interaction with MCM10, a key component of the pre-replication complex (29). It was shown that PCNA is phosphorylated prior to binding to DNA replication sites (30), suggesting a possible role in replication fork formation. Both Tyr-133 and Tyr-250 have previously been shown to participate in p21 interaction (31). Interestingly, our site-directed mutagenesis revealed that Tyr-60, Tyr-133, and Tyr-250 were all crucial for the IGF-1R/PCNA interaction and subsequent ubiquitination of PCNA. This points to a critical role for all three sites, as suggested by the location of Tyr-250 and Tyr-133 sites, which appear to lie within the hydrophobic groove on the front side adjacent to the interdomain-connecting loop where most of PCNA-interacting proteins bind (Fig. 7A) (32).
Figure 7.
Schematic model of suggested IGF-1R/PCNA regulation of DDT pathway. A, side and front views of the crystal structure of unmodified PCNA from Homo sapiens. The PCNA monomers are shaded in lime, gray, and cyan. Tyr-60 is shown in violet, the Tyr-133 in blue, and the Tyr-250 in green. The interdomain connecting loop (IDCL) is shown in orange. B, IGF-1R/PCNA regulation of DDT. See text.
Ubiquitination of PCNA has repeatedly been shown to regulate DDT. In response to replication fork stalling, PCNA polyubiquitination is thought to trigger the DDT pathway to avoid replication fork collapse (9, 11). Replication fork stalling could result from a variety of reasons, including DNA damage or defects in the replication machinery (33). In addition, recent studies have shown that nucleotide deprivation can induce replication fork stalling (33, 34). Thus increased proliferation caused by the high expression of IGF-1R may induce replication fork stalling through nucleotide deprivation. This might explain the presence of PCNA ubiquitination and association with E2/E3 ligases of the DDT pathway under basal growth conditions. The cell lines investigated by us (hESC, MEFs, and human fibroblast cell lines) all show a substantial expression IGF-1R, even in the absence of induced exogenous DNA damage. This is in line with the findings of detected PCNA ubiquitination in Xenopus extracts during unperturbed DNA replication (35) and in human cell lines not exposed to exogenous DNA damage (36). Thus, even in the absence of exogenous DNA-damaging agents, forks stalling occurs in a significant fraction of the cell division cycles (33).
Our finding that IGF-1R was required for PCNA ubiquitination in response to DNA damage by UV treatment is supported by previous studies showing that IGF-1R activation is associated with increased radioresistance both in vitro and in vivo (37, 38). However, it cannot be excluded that other protein kinases can phosphorylate and regulate DDT through ubiquitination of PCNA in other cell systems. In yeasts, which lack IGF-1R, PCNA ubiquitination by DDT E2/E3 ligases has been demonstrated (9–11).
The importance of PCNA in maintaining genomic stability has been demonstrated in a number of studies. An affected family that carried a homozygous PCNA missense mutation, resulting in PCNA S228I (39), showed a decreased ability to sustain UV radiation through a decreased PCNA affinity for DNA metabolic enzymes, with no effects on DNA replication. Clinically, this was reflected by genomic hypermutability. This suggests that the PCNA residue Ser-228 is important for DNA repair rather than proliferation. This is compatible with our finding that the Tyr-60, Tyr-133, and Tyr-250 mutants affect PCNA ubiquitination only, not DNA replication/proliferation.
Deficiencies in genes regulating the DDT pathway have been shown to be important for maintaining genomic stability. HLTF-deficient mouse embryonic fibroblasts showed elevated chromosome breaks and fusions after MMS treatment (8). Also, reduction of either SHPRH or HLTF expression enhanced spontaneous mutagenesis in human fibroblasts. Furthermore, mutation of the XPV gene, encoding DNA pol η in humans, results in a variant type of xeroderma pigmentosum (40). Patients with this disease variant are hypersensitive to UV damage, with reduced ability to elongate nascent DNA strands upon UV irradiation, and are predisposed to cancer (9, 41–43).
Genetic syndromes with deficiencies in genes regulating genomic stability are being increasingly recognized for their relevance in the aging process. The connection between IGF-1R signaling and longevity was made in genetically modified mice (44), and there is an ongoing discussion about the potential link between somatotropic axis signaling (GH/IGF), DNA repair, and DDT (45, 46). Although multiple factors may influence aging in genetically altered models, IGF-1R might be of some importance for normal aging by affecting DDT. A closer investigation on this matter is warranted.
In response to replication stress or external DNA damage, IGF-1R mediates PCNA ubiquitination, as proposed in the model shown in Fig. 7B. nIGF-1R phosphorylates PCNA on three tyrosine residues (Tyr-60, -133, and -250), which in turn targets PCNA for mono- and polyubiquitination, possibly by the DDT E2/E3 ligases. The phosphorylation of all three tyrosine residues (Tyr-60, -133, and -250) appears to be critical for subsequent nIGF-1R/PCNA interaction. IGF-1R aids in protecting the cells from replication fork stalling, and therefore it may contribute to maintaining genetic stability. The effect on DDT might be influenced by IGF-1R/PCNA interaction and associated PCNA modifications. Further studies are needed to more closely explore this interaction.
Experimental procedures
Reagents
NVP-AEW541 (IGF-1R tyrosine kinase inhibitor) was purchased from Cayman Chemical (Ann Arbor, MI). Insulin-like growth factor-1 (IGF-1), U0126 (MEK1/2 inhibitor), dimethyl sulfoxide (DMSO), LY2942002 (PI3K inhibitor), Polybrene, mitomycin C, and β-mercaptoethanol were all purchased from Sigma. Basic fibroblast growth factor (bFGF) was bought from R&D Systems (Minneapolis, MN), and MMS was from Santa Cruz Biotechnology (Santa Cruz, CA). NVP-AEW541, U0126, LY2942002, and MG132 were dissolved in DMSO and used at a concentration of 1, 5, 20, and 5 μm, respectively. IGF-1 and bFGF were dissolved in 0.1% bovine serum albumin (BSA) in PBS (Hyclone, Logan, UT) and used at a final concentration of 100 and 4 ng/ml, respectively. RPMI 1640 medium, F-12, Dulbecco's modified Eagle's medium (DMEM), collagenase, and cell culture supplements, including fetal bovine serum (FBS), non-essential amino acids (NEAA), knock-out serum replacement, l-glutamine, puromycin, and blasticidin were purchased from Invitrogen (Paisley, UK). mTeSRTM1 stem cell medium was from Stem Cell Technologies (Grenoble, France).
Cell cultures
Mitotically inactive feeder cells were established by treating mouse embryonic fibroblast feeder cells (SNL, kindly provided by Dr. K. Wiman, Karolinska Institutet, Stockholm, Sweden) with 10 μg/ml mitomycin C and cultured in DMEM with 10% FBS and 1% NEAA. H1 (WA01) hESC cells (WiCell Research Institute Inc., Madison, WI) were maintained as described in Ref. 47. Briefly, cells were seeded on inactivated SNL feeder cells in DMEM/F-12 with 20% knock-out serum replacement in 1 mm l-glutamine, 10 mm NEAA, 50 mm β-mercaptoethanol, and 4 ng/ml bFGF. All hESCs experiments were conducted between passage numbers 27 and 60. For feeder-free conditions, cells were passaged every 4–6 days using collagenase and plated on GeltrexTM matrix in mTeSRTM1 medium in feeder-free conditions as described in Ref. 47.
The platinum-A retroviral packaging cell line was purchased from Cell Biolabs Inc. (San Diego). MEF igf1r−/− (R−-) and R− cells overexpressing IGF-1R (R+) were from Dr. R. Baserga (Thomas Jefferson University, PA). The R-WT (R− with newly transfected WT IGF-1R), R-TSM (R− transfected with SUMO-mutated IGF-1R), and R−Puro (empty vector control) were obtained as described elsewhere (24). All cell lines were cultured in DMEM with 10% FBS, with the exception of Gm02808 being cultured in DMEM/F-12 with 20% FBS. Platinum-A cells were additionally supplied with 1 μg/ml puromycin and 10 μg/ml blasticidin.
All cell lines were maintained at 37 °C in a humidified atmosphere containing 5% CO2 and checked for mycoplasma using MycoalertTM kit (Lonza, Basel, Switzerland). All human cell lines were short tandem repeat-authenticated using AmpFLSTR® Identifier® plus kit (Applied Biosystems, Foster City, CA), except for Gm02808 due to lack of short tandem repeat reference.
Recombinant DNA techniques
PCNA cDNA sequence was PCR-amplified from pEGFP-PCNA-IRES-puro2b (Addgene plasmids ID:26461) and inserted into linearized pBabe-puro empty vector (Cell Biolabs, Inc.) in-between BamHI and EcoRI restriction sites using In-Fusion® Dry-Down PCR cloning kit (Clontech). The constructed pBabe-PCNA plasmid was expanded in Escherichia coli and extracted using PureLink® HiPure Plasmid Filter Maxiprep kit (Invitrogen). Site-specific mutated PCNA expression plasmids were generated using the QuikChange® site-directed mutagenesis kit (Agilent Technologies, Santa Clara, CA) using the primers listed in supplemental Table S1. The successful introductions of specific point mutations were confirmed by sequencing at GATC Biotech (Konstanz, Germany). Expression plasmids with multiple point mutations were created through several rounds of single point mutagenesis.
Stable transfections
Platinum-A virus packaging cells were plated in antibiotic-free medium 1 day before transfection, and a medium change was performed 6 h before transfection. Thirty μg of plasmid DNA was mixed with 500 μl of 250 mm CaCl2 and added to 500 μl of 2× BES-buffered saline (BBS) (Sigma). After incubation at room temperature for 30 min, the mixture with generated DNA precipitate was added dropwise to the cells, which were incubated in a CO2 incubator at 37 °C for 16 h, followed by medium change and an additional 24-h incubation.
R+ cells were seeded at low density in 6-well plates. Medium from the transfected platinum-A cells containing the packaged retrovirus was collected, supplemented with 8 μg/ml Polybrene, filtered through 0.45-μm polysulfonic filters (Pall Corp., Ann Arbor, MI), and added to the R+ cells for infection. A total of three infections were carried out with 24-h incubation intervals. Transfected R+ cells were obtained by antibiotic selection using 2 μg/ml puromycin. The stable clones were generated at limited dilution in 96-well plates.
Immunoblotting
Cells were lysed in modified RIPA buffer (50 mm Tris, pH 7.4, 150 mm NaCl, 1% Nonidet P-40, 1 mm EDTA, 0.25% sodium deoxycholate) containing protease (Roche Applied Science, Mannheim, Germany) and phosphatase (Sigma) inhibitors and processed as described previously (48). Primary antibodies used in this research included the following: rabbit anti-IGF-1R (catalog no. 3024), rabbit anti-IGF-1Rβ (catalog no. 3027), and mouse anti-PCNA (catalog no. 2586) from Cell Signaling Technology; mouse anti-phosphotyrosine (sc-7020), mouse anti-ubiquitin (sc-8017), and goat anti-GFP (sc-5385) from Santa Cruz Biotechnology; mouse anti-UBC13 (catalog no. 37-1100) from Zymed Laboratories Inc.; rabbit anti-PCNA (ab18197), rabbit anti-SHPRH (ab80129), rabbit anti-HLTF (ab155031), and rabbit anti-pol η (ab154330) from Abcam, Cambridge, UK; and mouse anti-Rad18 (WH0056852M1) from Sigma. Membranes were incubated with secondary anti-rabbit/mouse/goat IgG horseradish peroxidase-conjugated antibodies (NA934 and NA931 from GE Healthcare and 31402 from Pierce) followed by signal detection using enhanced luminescence Hyperfilm-ECL (GE Healthcare).
Cell fractionation
Subcellular fractionation for immunoprecipitation was prepared as described in Ref. 49 with some modifications. Briefly, the cytoplasmic fraction was achieved by lysing cells in 10 mm HEPES, pH 7.4, 10 mm KCl, 0.05% Nonidet P-40 buffer. The nuclear fraction was achieved by washing the pellet with same buffer and then lysing in modified RIPA buffer.
Fractionation of membranous, cytoplasmic, and nuclear components for Western blotting was done using the Qiagen Qproteome cell compartment kit (Qiagen, Hilden, Germany). Subcellular fractionations were confirmed by immunoblotting for membranous calnexin (rabbit anti-calnexin, catalog no. 2433 from Cell Signaling Technology), cytoplasmic GAPDH or Hsp-90 (rabbit anti-GAPDH, sc-25778 from Santa Cruz Biotechnology), mouse anti-Hsp90 (ab13492 from Abcam, Cambridge, UK), or nucleic histone H2A (rabbit anti-H2A) (catalog no. 2578 from Cell Signaling Technology) as described above.
Immunoprecipitation
The following antibodies were cross-linked to magnetic protein G Dynabeads using Dynabeads® antibody coupling kit 14311D (Invitrogen): mouse anti-IGF-1R (catalog no. 556000); mouse anti-IgG (catalog no. 555746, both from BD Biosciences); rabbit anti-PCNA (ab18197 from Abcam); and rabbit anti-IgG (sc-66931, Santa Cruz Biotechnology). Non-cross-linked antibodies included the following: rabbit anti-IGF-1Rβ (catalog no. 3027) and mouse anti-PCNA (catalog no. 2586, both from Cell Signaling Technology); mouse anti-IgG (catalog no. 554126 from BD Biosciences); and rabbit anti-IgG (sc-6693), goat anti-GFP (sc-5385), and mouse anti-p-Tyr (sc-7020, all from Santa Cruz Biotechnology).
After blocking with 0.1% BSA in PBS (Hyclone), 2–4 mg of cell lysates were incubated with cross-linked antibody at concentrations of 4–7 μg of antibody/mg beads for 1 h at 4 °C. For non-cross-linked antibodies, lysates were incubated overnight at 4 °C. The immune complexes were washed three times with lysis buffer and eluted by boiling in SDS sample buffer (Invitrogen). Subsequent immunoblottings were performed as described above.
In vitro kinase assay
1 μg of N-terminal His-tagged recombinant IGF-1R kinase domain fusion protein (Millipore, Bedford, MA) or endogenous IGF-1R pulled down using IP, was incubated with 2 μg of N-terminal His-tagged PCNA fusion protein (Invitrogen) at 30 °C for 30 min in 30 μl of kinase buffer (60 mm HEPES, pH 7.9, 5 mm MgCl2, 5 mm MnCl2, 30 μm Na3VO4, and 1.25 mm DTT) in the presence of 20 μm ATP (all from Sigma). Omission of the His-tagged PCNA fusion protein, the IGF-1R kinase domain, or ATP served as negative controls. The reactions were terminated by boiling in SDS sample buffer (Invitrogen), followed by separations in 10% SDS-PAGE and immunoblottings.
For analysis of PCNA post-translational modifications, in vitro kinase assays of His-tagged recombinant PCNA were carried out with or without the recombinant IGF-1R kinase domain. After SDS-PAGE separation and Coomassie Blue staining, the bands corresponding to PCNA were cut out and compared by liquid chromatography-tandem mass spectrometry (LC-MS/MS) analyses as described below.
Immunofluorescence
hESCs were seeded on coverslips pre-coated with Geltrex, fixed in 4% buffered paraformaldehyde, permeabilized using 0.1% Triton X-100, and incubated overnight at room temperature with rabbit anti-IGF-1Rβ (catalog no. 3027 from Cell Signaling Technology) or mouse anti-IGF-1Rβ (catalog no. 05-656 from Millipore). Cells were washed with PBS and incubated with goat anti-rabbit Alexa Fluor 594-conjugated antibody (catalog no. A11012) or goat anti-mouse Alexa Fluor 488-conjugated antibody (catalog no. A11029, both from Invitrogen) for 1 h at room temperature. The coverslips were mounted using Vectashield mounting medium containing DAPI (Vector Laboratories, Burlingame, CA), for counterstaining purposes, and examined using an Axioplan 2 fluorescence microscope (Zeiss, Jena, Germany).
In situ PLA
hESCs were cultured and fixed on coverslips as described above. Permeabilization was done with 0.2% Triton X-100 for 30 min at room temperature, followed by incubation with blocking buffer for 30 min (5% BSA, 5% donkey serum, 0.3% Triton X-100 in phosphate-buffered saline) and mouse anti-IGF-1Rβ (catalog no. 05-656 from Millipore) or rabbit anti-PCNA (catalog no. ab18197 from Abcam) at 4 °C overnight. Duolink in situ PLA was conducted as described previously (52), using PLA probe anti-mouse minus and PLA probe anti-rabbit plus (OLINK Bioscience, Uppsala, Sweden).
Mass spectrometry
For hESCs cells, total cell lysates were used for immunoprecipitation of IGF-1R or IgG (negative control). Precipitates were separated with SDS-PAGE and co-precipitated protein bands were visualized using Coomassie Blue. Ten prominent gel bands were exclusively seen in the IGF-1R lane (supplemental Fig. S1A) and excised for further processing. Gel pieces were cut into small pieces and shrunk in acetonitrile. The samples were reduced at 56 °C using 100 mm ammonium bicarbonate containing 10 mm DTT for 30 min. The supernatants were collected. Next, the samples were incubated in 100 mm ammonium bicarbonate containing 55 mm iodoacetamide at room temperature for 20 min in the dark. Then the gel pieces were shrunk once more, and the supernatant was collected. Gel pieces were destained in a 1:1 mixture of 100 mm ammonium bicarbonate and acetonitrile for 30 min, followed by shrinking and aspiration of the supernatant. The gel pieces were then incubated on ice for 2 h with 13 ng/μl trypsin (sequencing grade-modified, Pierce) in 10 mm ammonium bicarbonate and 10% acetonitrile. Digestion was carried out at 37 °C overnight. Peptides were extracted using 2 volumes of 1.67% formic acid in 67% acetonitrile at 37 °C for 15 min. The extracts were dried in a SpeedVac and resuspended in 3% acetonitrile, 0.1% formic acid.
LC-MS was performed using a hybrid LTQ-Orbitrap Velos mass spectrometer (Thermo Fisher Scientific). For each LC-MS/MS run, the autosampler (HPLC 1200 system, Agilent Technologies) dispensed 8 μl of solvent A, mixed for 10 min, and proceeded to inject 3 μl. Samples were trapped on a C18 guard desalting column (Agilent Technologies) and separated on a 15-cm-long C18 picoFrit column (100-μm internal diameter, 5-μm bead size, Nikkyo Technos, Tokyo, Japan) installed on the nano-electrospray ionization source. Solvent A was 97% water, 3% acetonitrile, and 0.1% formic acid; solvent B was 5% water, 95% acetonitrile, and 0.1% formic acid. At a constant flow of 0.4 μl/min, the curved gradient went from 2% B to 40% B in 45 min, followed by a steep increase to 100% B in 5 min.
The survey scan, performed in the Orbitrap with 30,000 resolution (and mass range 300–2000 m/z), was followed by data-dependent MS/MS (centroid mode) in two stages: first, the top five ions from the master scan were selected for collision-induced dissociation (CID), using 35% normalized collision energy with ion trap mass spectrometry detection; second, the same five ions underwent higher energy collision dissociation (HCD), using 37.5% normalized collision energy for iTRAQ or TMT-labeled samples, 32.5% for label-free samples (7,500 resolution) with detection in the Orbitrap (Fourier transform-MS). In addition, an ETD procedure was used where precursors underwent ETD fragmentation (instead of CID or HCD) provided they possessed sufficient charge density. Accordingly, only triple-charged precursor ions with m/z <650, quadruple-charged ions with m/z <900, quintuple-charged ions with m/z <950, or precursors with higher charge states were selected for ETD. Precursors were isolated with a 2 m/z window. Automatic gain control targets were 1 × 106 ions for MS and for MS/MS 3 × 104 (CID) and 5 × 104 (HCD). The software Proteome Discoverer version 1.4, including Sequest, was used to search the human Uniprot database for protein identification, limited to a false discovery rate of 1%.
Cell synchronization and cell cycle analysis
Cells were plated at low density and allowed to grow for 24 h. Then they were washed twice with PBS and incubated in serum-free medium for 24 h to achieve G0/G1 synchronization, followed by release for 12 h in medium containing 10% serum to synchronize cells at G1/S phase. Cells were then treated for 6 h with 1.2 mm MMS or vehicle.
The proportion of synchronized cells in S phase was measured by bromodeoxyuridine (BrdU, Sigma) incorporation, and G1 and G2 phases were measured by staining with PI (Sigma) as described previously (50). Briefly, cells were pulse-labeled with 100 μm BrdU for 1 h before harvest. For flow cytometry, cells were washed with PBS, fixed with 70% ethanol overnight, and stained with anti-BrdU (BD Biosciences), followed by FITC anti-mouse (DAKO, Glostrup, Denmark). Cells were then stained with a propidium iodide solution (50 μg/ml) containing RNase A (20 μg/ml) for 30 min at room temperature. The samples were processed using FACSCalibur (BD Biosciences), and the data were analyzed with the Cell Quest software (BD Biosciences).
DNA fiber assay
DNA replication fork dynamics were investigated on single DNA molecules as described previously (53). In brief, cells were pulse-labeled with 25 μm halogenated nucleotides CidU, washed, and treated with 4.3 mm MMS to induce replication fork stalling or DMSO as control and then incubated with 250 μm IdU (Sigma) for 20 min. Harvested cells were diluted to 106 cells/ml. Spreading and staining were performed as described previously (51). Fluorescence images were captured using Axioplan 2 fluorescence microscope (Zeiss) and analyzed using the ImageJ software. At least 50 unidirectional forks labeled with both CidU and IdU were measured for every condition. The elongation ratio was calculated by length of IdU/length of CidU, quantitatively reflecting the relative changes in replication fork efficiency.
Author contributions
A. W., Y. L., and D. W. performed the experiments. A. W., Y. L., E. A., and O. L. designed the experiments. A. W., Y. L., F. H., E. A., and O. L. wrote and edited the manuscript. O. L. was the principal investigator.
Supplementary Material
Acknowledgments
We thank Z. Elbeck for assisting with technical work and Dr. Arne Lindqvist for valuable comments on the manuscript.
This work was supported by the Swedish Cancer Foundation, the Swedish Research Council, the Cancer Society in Stockholm, the Swedish Children Cancer Society, Stockholm County Council, and the Karolinska Institutet. The authors declare that they have no conflicts of interest with the contents of this article.
This article contains supplemental Figs. S1–S10 and Table S1.
- IGF-1R
- insulin-like growth factor 1 receptor
- nIGF-1R
- nuclear IGF-1R
- PCNA
- proliferating cell nuclear antigen
- DDT
- DNA damage tolerance
- hESC
- human embryonic stem cell
- TLS
- translesion synthesis
- IP
- immunoprecipitation
- BisTris
- 2-[bis(2-hydroxyethyl) amino]-2-(hydroxymethyl)propane-1,3-diol
- CidU
- chlorodeoxyuridine
- MMS
- methyl methanesulfonate
- IdU
- iododeoxyuridine
- EGFR
- epidermal growth factor receptor
- BES
- 2-[bis(2-hydroxyethyl)amino]ethanesulfonic acid
- SUMO
- SUMOylation
- NEAA
- non-essential amino acid
- PI
- propidium iodide
- Ub
- ubiquitin
- CID
- collision-induced dissociation
- eGFP
- enhanced GFP
- MEF
- mouse embryonic fibroblast
- PLA
- proximity ligation assay
- HCD
- higher energy collision dissociation
- bFGF
- basic fibroblast growth factor
- ETD
- electron-transfer dissociation.
References
- 1. Pollak M. (2012) The insulin and insulin-like growth factor receptor family in neoplasia: an update. Nat. Rev. Cancer 12, 159–169 [DOI] [PubMed] [Google Scholar]
- 2. Sehat B., Tofigh A., Lin Y., Trocmé E., Liljedahl U., Lagergren J., and Larsson O. (2010) SUMOylation mediates the nuclear translocation and signaling of the IGF-1 receptor. Sci. Signal. 3, ra10. [DOI] [PubMed] [Google Scholar]
- 3. Warsito D., Sjöström S., Andersson S., Larsson O., and Sehat B. (2012) Nuclear IGF1R is a transcriptional co-activator of LEF1/TCF. EMBO Rep. 13, 244–250 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Warsito D., Lin Y., Gnirck A. C., Sehat B., and Larsson O. (2016) Nuclearly translocated insulin-like growth factor 1 receptor phosphorylates histone H3 at tyrosine 41 and induces SNAI2 expression via Brg1 chromatin remodeling protein. Oncotarget 7, 42288–42302 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Kelman Z., and O'Donnell M. (1995) Structural and functional similarities of prokaryotic and eukaryotic DNA polymerase sliding clamps. Nucleic Acids Res. 23, 3613–3620 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Carr A. M., Paek A. L., and Weinert T. (2011) DNA replication: failures and inverted fusions. Semin. Cell Dev. Biol. 22, 866–874 [DOI] [PubMed] [Google Scholar]
- 7. Liefshitz B., Steinlauf R., Friedl A., Eckardt-Schupp F., and Kupiec M. (1998) Genetic interactions between mutants of the “error-prone” repair group of Saccharomyces cerevisiae and their effect on recombination and mutagenesis. Mutat. Res. 407, 135–145 [DOI] [PubMed] [Google Scholar]
- 8. Motegi A., Liaw H. J., Lee K. Y., Roest H. P., Maas A., Wu X., Moinova H., Markowitz S. D., Ding H., Hoeijmakers J. H., and Myung K. (2008) Polyubiquitination of proliferating cell nuclear antigen by HLTF and SHPRH prevents genomic instability from stalled replication forks. Proc. Natl. Acad. Sci. U.S.A. 105, 12411–12416 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Chang D. J., and Cimprich K. A. (2009) DNA damage tolerance: when it's OK to make mistakes. Nat. Chem. Biol. 5, 82–90 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Friedberg E. C. (2005) Suffering in silence: the tolerance of DNA damage. Nat. Rev. Mol. Cell Biol. 6, 943–953 [DOI] [PubMed] [Google Scholar]
- 11. Unk I., Hajdú I., Blastyák A., and Haracska L. (2010) Role of yeast Rad5 and its human orthologs, HLTF and SHPRH in DNA damage tolerance. DNA Repair 9, 257–267 [DOI] [PubMed] [Google Scholar]
- 12. Andersen P. L., Xu F., and Xiao W. (2008) Eukaryotic DNA damage tolerance and translesion synthesis through covalent modifications of PCNA. Cell Res. 18, 162–173 [DOI] [PubMed] [Google Scholar]
- 13. Hoege C., Pfander B., Moldovan G. L., Pyrowolakis G., and Jentsch S. (2002) RAD6-dependent DNA repair is linked to modification of PCNA by ubiquitin and SUMO. Nature 419, 135–141 [DOI] [PubMed] [Google Scholar]
- 14. Stelter P., and Ulrich H. D. (2003) Control of spontaneous and damage-induced mutagenesis by SUMO and ubiquitin conjugation. Nature 425, 188–191 [DOI] [PubMed] [Google Scholar]
- 15. Watanabe K., Tateishi S., Kawasuji M., Tsurimoto T., Inoue H., and Yamaizumi M. (2004) Rad18 guides poleta to replication stalling sites through physical interaction and PCNA monoubiquitination. EMBO J. 23, 3886–3896 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Unk I., Hajdú I., Fátyol K., Hurwitz J., Yoon J. H., Prakash L., Prakash S., and Haracska L. (2008) Human HLTF functions as a ubiquitin ligase for proliferating cell nuclear antigen polyubiquitination. Proc. Natl. Acad. Sci. U.S.A. 105, 3768–3773 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Motegi A., Sood R., Moinova H., Markowitz S. D., Liu P. P., and Myung K. (2006) Human SHPRH suppresses genomic instability through proliferating cell nuclear antigen polyubiquitination. J. Cell Biol. 175, 703–708 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Unk I., Hajdú I., Fátyol K., Szakál B., Blastyák A., Bermudez V., Hurwitz J., Prakash L., Prakash S., and Haracska L. (2006) Human SHPRH is a ubiquitin ligase for Mms2-Ubc13-dependent polyubiquitylation of proliferating cell nuclear antigen. Proc. Natl. Acad. Sci. U.S.A. 103, 18107–18112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Parvaresch S., Yesilkaya T., Baer K., Al-Hasani H., and Klein H. W. (2002) 14-3-3 binding to the IGF-1 receptor is mediated by serine autophosphorylation. FEBS Lett. 532, 357–362 [DOI] [PubMed] [Google Scholar]
- 20. Niu M., Klingler-Hoffmann M., Brazzatti J. A., Forbes B., Akekawatchai C., Hoffmann P., and McColl S. R. (2013) Comparative proteomic analysis implicates eEF2 as a novel target of PI3Kγ in the MDA-MB-231 metastatic breast cancer cell line. Proteome Sci. 11, 4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Pietrzkowski Z., Sell C., Lammers R., Ullrich A., and Baserga R. (1992) Roles of insulin-like growth factor 1 (IGF-1) and the IGF-1 receptor in epidermal growth factor-stimulated growth of 3T3 cells. Mol. Cell. Biol. 12, 3883–3889 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Pietrzkowski Z., Lammers R., Carpenter G., Soderquist A. M., Limardo M., Phillips P. D., Ullrich A., and Baserga R. (1992) Constitutive expression of insulin-like growth factor 1 and insulin-like growth factor 1 receptor abrogates all requirements for exogenous growth factors. Cell Growth Differ. 3, 199–205 [PubMed] [Google Scholar]
- 23. Wang S. C., Nakajima Y., Yu Y. L., Xia W., Chen C. T., Yang C. C., McIntush E. W., Li L. Y., Hawke D. H., Kobayashi R., and Hung M. C. (2006) Tyrosine phosphorylation controls PCNA function through protein stability. Nat. Cell Biol. 8, 1359–1368 [DOI] [PubMed] [Google Scholar]
- 24. Lin Y., Liu H., Waraky A., Haglund F., Agarwal P., Jernberg-Wiklund H., Warsito D., and Larsson O. (2017) SUMO-modified insulin-like growth factor 1 receptor (IGF-1R) increases cell cycle progression and cell proliferation. J. Cell. Physiol. 232, 2722–2730 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Wang Y. N., and Hung M. C. (2012) Nuclear functions and subcellular trafficking mechanisms of the epidermal growth factor receptor family. Cell Biosci. 2, 13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Ortega J., Li J. Y., Lee S., Tong D., Gu L., and Li G. M. (2015) Phosphorylation of PCNA by EGFR inhibits mismatch repair and promotes misincorporation during DNA synthesis. Proc. Natl. Acad. Sci. U.S.A. 112, 5667–5672 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Zhao H., Chen M. S., Lo Y. H., Waltz S. E., Wang J., Ho P. C., Vasiliauskas J., Plattner R., Wang Y. L., and Wang S. C. (2014) The Ron receptor tyrosine kinase activates c-Abl to promote cell proliferation through tyrosine phosphorylation of PCNA in breast cancer. Oncogene 33, 1429–1437 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Olsen H., and Haldosén L. A. (2006) Peroxisome proliferator-activated receptor γ regulates expression of signal transducer and activator of transcription 5A. Exp. Cell Res. 312, 1371–1380 [DOI] [PubMed] [Google Scholar]
- 29. Das-Bradoo S., Ricke R. M., and Bielinsky A. K. (2006) Interaction between PCNA and diubiquitinated Mcm10 is essential for cell growth in budding yeast. Mol. Cell. Biol. 26, 4806–4817 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Prosperi E., Stivala L. A., Sala E., Scovassi A. I., and Bianchi L. (1993) Proliferating cell nuclear antigen complex formation induced by ultraviolet irradiation in human quiescent fibroblasts as detected by immunostaining and flow cytometry. Exp. Cell Res. 205, 320–325 [DOI] [PubMed] [Google Scholar]
- 31. Zhang P., Sun Y., Hsu H., Zhang L., Zhang Y., and Lee M. Y. (1998) The interdomain connector loop of human PCNA is involved in a direct interaction with human polymerase δ. J. Biol. Chem. 273, 713–719 [DOI] [PubMed] [Google Scholar]
- 32. Maga G., and Hubscher U. (2003) Proliferating cell nuclear antigen (PCNA): a dancer with many partners. J. Cell Sci. 116, 3051–3060 [DOI] [PubMed] [Google Scholar]
- 33. Pohlhaus J. R., and Kreuzer K. N. (2006) Formation and processing of stalled replication forks–utility of two-dimensional agarose gels. Methods Enzymol. 409, 477–493 [DOI] [PubMed] [Google Scholar]
- 34. de Feraudy S., Limoli C. L., Giedzinski E., Karentz D., Marti T. M., Feeney L., and Cleaver J. E. (2007) Pol η is required for DNA replication during nucleotide deprivation by hydroxyurea. Oncogene 26, 5713–5721 [DOI] [PubMed] [Google Scholar]
- 35. Leach C. A., and Michael W. M. (2005) Ubiquitin/SUMO modification of PCNA promotes replication fork progression in Xenopus laevis egg extracts. J. Cell Biol. 171, 947–954 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Yang X. H., and Zou L. (2009) Dual functions of DNA replication forks in checkpoint signaling and PCNA ubiquitination. Cell Cycle 8, 191–194 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Valenciano A., Henríquez-Hernández L. A., Moreno M., Lloret M., and Lara P. C. (2012) Role of IGF-1 receptor in radiation response. Transl. Oncol. 5, 1–9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Chitnis M. M., Lodhia K. A., Aleksic T., Gao S., Protheroe A. S., and Macaulay V. M. (2014) IGF-1R inhibition enhances radiosensitivity and delays double-strand break repair by both non-homologous end-joining and homologous recombination. Oncogene 33, 5262–5273 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Baple E. L., Chambers H., Cross H. E., Fawcett H., Nakazawa Y., Chioza B. A., Harlalka G. V., Mansour S., Sreekantan-Nair A., Patton M. A., Muggenthaler M., Rich P., Wagner K., Coblentz R., Stein C. K., et al. (2014) Hypomorphic PCNA mutation underlies a human DNA repair disorder. J. Clin. Invest. 124, 3137–3146 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Masutani C., Kusumoto R., Yamada A., Dohmae N., Yokoi M., Yuasa M., Araki M., Iwai S., Takio K., and Hanaoka F. (1999) The XPV (xeroderma pigmentosum variant) gene encodes human DNA polymerase η. Nature 399, 700–704 [DOI] [PubMed] [Google Scholar]
- 41. Stary A., and Sarasin A. (2002) The genetics of the hereditary xeroderma pigmentosum syndrome. Biochimie 84, 49–60 [DOI] [PubMed] [Google Scholar]
- 42. Stary A., and Sarasin A. (2002) Molecular mechanisms of UV-induced mutations as revealed by the study of DNA polymerase η in human cells. Res. Microbiol. 153, 441–445 [DOI] [PubMed] [Google Scholar]
- 43. Broughton B. C., Cordonnier A., Kleijer W. J., Jaspers N. G., Fawcett H., Raams A., Garritsen V. H., Stary A., Avril M. F., Boudsocq F., Masutani C., Hanaoka F., Fuchs R. P., Sarasin A., and Lehmann A. R. (2002) Molecular analysis of mutations in DNA polymerase η in xeroderma pigmentosum-variant patients. Proc. Natl. Acad. Sci. U.S.A. 99, 815–820 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Holzenberger M., Dupont J., Ducos B., Leneuve P., Géloën A., Even P. C., Cervera P., and Le Bouc Y. (2003) IGF-1 receptor regulates lifespan and resistance to oxidative stress in mice. Nature 421, 182–187 [DOI] [PubMed] [Google Scholar]
- 45. Castells-Roca L., Mueller M. M., and Schumacher B. (2015) Longevity through DNA damage tolerance. Cell Cycle 14, 467–468 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Ermolaeva M. A., Dakhovnik A., and Schumacher B. (2015) Quality control mechanisms in cellular and systemic DNA damage responses. Ageing Res. Rev. 23, 3–11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Waraky A., Aleem E., and Larsson O. (2016) Downregulation of IGF-1 receptor occurs after hepatic linage commitment during hepatocyte differentiation from human embryonic stem cells. Biochem. Biophys. Res. Commun. 478, 1575–1581 [DOI] [PubMed] [Google Scholar]
- 48. Waraky A., Akopyan K., Parrow V., Strömberg T., Axelson M., Abrahmsén L., Lindqvist A., Larsson O., and Aleem E. (2014) Picropodophyllin causes mitotic arrest and catastrophe by depolymerizing microtubules via insulin-like growth factor-1 receptor-independent mechanism. Oncotarget 5, 8379–8392 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Huang J., Huen M. S., Kim H., Leung C. C., Glover J. N., Yu X., and Chen J. (2009) RAD18 transmits DNA damage signalling to elicit homologous recombination repair. Nat. Cell Biol. 11, 592–603 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Aleem E., Kiyokawa H., and Kaldis P. (2005) Cdc2-cyclin E complexes regulate the G1/S phase transition. Nat. Cell Biol. 7, 831–836 [DOI] [PubMed] [Google Scholar]
- 51. Elvers I., Johansson F., Groth P., Erixon K., and Helleday T. (2011) UV stalled replication forks restart by re-priming in human fibroblasts. Nucleic Acids Res. 39, 7049–7057 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Söderberg O., Gullberg M., Jarvius M., Ridderstråle K., Leuchowius K. J., Jarvius J., Wester K., Hydbring P., Bahram F., Larsson L. G., and Landegren U. (2006) Direct observation of individual endogenous protein complexes in situ by proximity ligation. Nat. Methods 3, 995–1000 [DOI] [PubMed] [Google Scholar]
- 53. Schwab R. A., and Niedzwiedz W. (2011) Visualization of DNA replication in the vertebrate model system DT40 using the DNA fiber technique. J. Vis. Exp. 27, e3255. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.