Abstract
Glycogen synthase kinase-3 (GSK-3) is a constitutively active, ubiquitously expressed protein kinase that regulates multiple signaling pathways. In vitro kinase assays and genetic and pharmacological manipulations of GSK-3 have identified more than 100 putative GSK-3 substrates in diverse cell types. Many more have been predicted on the basis of a recurrent GSK-3 consensus motif ((pS/pT)XXX(S/T)), but this prediction has not been tested by analyzing the GSK-3 phosphoproteome. Using stable isotope labeling of amino acids in culture (SILAC) and MS techniques to analyze the repertoire of GSK-3–dependent phosphorylation in mouse embryonic stem cells (ESCs), we found that ∼2.4% of (pS/pT)XXX(S/T) sites are phosphorylated in a GSK-3–dependent manner. A comparison of WT and Gsk3a;Gsk3b knock-out (Gsk3 DKO) ESCs revealed prominent GSK-3–dependent phosphorylation of multiple splicing factors and regulators of RNA biosynthesis as well as proteins that regulate transcription, translation, and cell division. Gsk3 DKO reduced phosphorylation of the splicing factors RBM8A, SRSF9, and PSF as well as the nucleolar proteins NPM1 and PHF6, and recombinant GSK-3β phosphorylated these proteins in vitro. RNA-Seq of WT and Gsk3 DKO ESCs identified ∼190 genes that are alternatively spliced in a GSK-3–dependent manner, supporting a broad role for GSK-3 in regulating alternative splicing. The MS data also identified posttranscriptional regulation of protein abundance by GSK-3, with ∼47 proteins (1.4%) whose levels increased and ∼78 (2.4%) whose levels decreased in the absence of GSK-3. This study provides the first unbiased analysis of the GSK-3 phosphoproteome and strong evidence that GSK-3 broadly regulates alternative splicing.
Keywords: alternative splicing, embryonic stem cell, glycogen synthase kinase 3 (GSK-3), leukemia, phosphoproteomics, spliceosome, Wnt signaling, SILAC, lithium
Introduction
Glycogen synthase kinase-3 (GSK-3)2 is a ubiquitously expressed, highly conserved serine/threonine kinase that plays a central role in multiple signaling pathways, most prominently as an antagonist of insulin/AKT and Wnt/β-catenin signaling pathways (1). Unlike most protein kinases, GSK-3 is constitutively active and in general is inhibited by upstream signaling. GSK-3 was first identified as a protein kinase that phosphorylates and inhibits glycogen synthase, the rate-limiting enzyme in glycogen synthesis (2) and was later shown to antagonize canonical Wnt signaling (1). However, GSK-3 is now known to regulate a broad range of cellular processes, including cytoskeletal organization, circadian rhythm, cell growth and survival, immune responses, and developmental processes (1).
In metazoans, GSK-3 is encoded by two similar genes, Gsk3a and Gsk3b, with 98% amino acid sequence identity in the catalytic domains of mammalian GSK-3α and GSK-3β. The two genes are partially redundant, and mice with complete loss of Gsk3a are viable due to compensation by Gsk3b. However, Gsk3b loss-of-function mutations in mice are embryonic or neonatal lethal (3, 4), and the Gsk3a;Gsk3b DKO is lethal in early embryogenesis (5, 6). Furthermore, DKO mouse ESCs maintain expression of pluripotency markers and are unable to differentiate into most embryonic lineages either in vitro (as embryoid bodies) or in vivo (as teratomas) (6). Here, we will refer to the two genes together as Gsk3 (or GSK-3 for the protein) unless referring to isoform-specific features.
Hormones and growth factors, such as insulin, activate receptor tyrosine kinases that in turn initiate PI3K/AKT signaling. AKT phosphorylates serine 21 of GSK-3α and serine 9 of GSK-3β, thereby inhibiting activity. GSK-3 tends to phosphorylate substrates that are prephosphorylated (“primed”) at the +4-position; the phosphorylated N terminus of GSK-3 mimics these primed substrates, creating a pseudosubstrate inhibitor of catalytic activity (7). The (S/T)XXX(S/T) consensus was first demonstrated in glycogen synthase; phosphorylation by casein kinase II at serine 657 leads to processive phosphorylation by GSK-3 at serines 653, 649, 645, and 641 (8). The requirement of the consensus sequence has since been demonstrated for multiple GSK-3 substrates and is often used to predict GSK-3 phosphorylation sites (7, 9–12). However, several established GSK-3 substrates do not have a serine or threonine at the +4-position, including cyclin D1, c-Myc, and protein phosphatase-1 inhibitor-2.
GSK-3 is also inhibited by mechanisms that are independent of its N-terminal phosphorylation. Most importantly, Wnt-mediated inhibition of GSK-3 catalytic activity does not require N-terminal phosphorylation, as mice with serine-to-alanine knock-in mutations in Gsk3a and Gsk3b display normal Wnt signaling (6, 13, 14). Inhibition of GSK-3 by Wnts may instead involve rapid dissociation of APC (15, 16), interaction with the phosphorylated C terminus of LRP5/6 (17), and, with slower kinetics, sequestration of GSK-3 into multivesicular bodies (18).
GSK-3 phosphorylation regulates multiple functions, including protein stability, enzymatic activity, microtubule polymerization, and protein–protein interactions (11, 12, 19). To date, over 100 putative substrates of GSK-3 have been identified in diverse cell types, including multiple transcription factors, regulators of translation, cytoskeletal proteins, and other protein kinases (1, 11, 12, 19).
Although GSK-3 has been proposed to phosphorylate many substrates, these experiments were conducted using a variety of approaches and experimental settings, including in vitro kinase assays and pharmacological inhibition in diverse cell types. In vitro protein kinase assays may not recapitulate in vivo regulation, especially for substrates that require priming phosphorylation by unknown protein kinases, and off-target effects of small molecule inhibitors (20) can confound the analysis. Furthermore, the range of substrates and cellular processes regulated by GSK-3 in a single cell type has not previously been examined using specific inhibition of GSK-3 (e.g. by gene knockouts). We therefore used a large-scale proteomic approach to characterize the GSK-3 phosphoproteome and determine the range of GSK-3 substrates within a single cell type, comparing wild type with cells with a complete loss of Gsk3 (6). Specifically, stable isotope labeling by amino acids in culture (SILAC) (21) was used to metabolically label the proteome of wild-type and Gsk3a/b double-knock-out (DKO) mouse embryonic stem cells (ESCs). To our knowledge, this is the first study to conduct a global analysis of GSK-3–dependent phosphorylation.
Results
Global phosphoproteomic analysis of GSK-3–dependent phosphorylation
To characterize the GSK-3 phosphoproteome, we performed SILAC and quantitative MS using wild-type and Gsk3 DKO mouse ESCs (Fig. 1A). DKO ESCs were cultured in heavy isotope-labeled amino acids for at least 4 passages to achieve complete labeling of the proteome (supplemental Fig. 1). Wild-type ESCs were cultured in parallel in light amino acid medium. Before protein isolation, expression of the stem cell markers Oct4 and Nanog was assessed by flow cytometry to establish homogeneity of the two-cell populations; in the three replicates, 70–92% of wild-type cells and 75–90% of DKO cells expressed high levels of both markers (Fig. 1B). Deletion of GSK-3α and GSK-3β and activation of Wnt signaling by β-catenin accumulation were confirmed by Western blotting (Fig. 1C). Whole-cell lysates in biological triplicate were digested with trypsin, and phosphorylated peptides were enriched using titanium dioxide (TiO2) chromatography and analyzed by LC-MS/MS on an LTQ-orbitrap (Fig. 1A). The unbound peptides from the TiO2 flow-through were used for non-modified, proteomic identification. The level of each phosphopeptide was normalized to total protein abundance. (All MS raw files are available at https://chorusproject.org,3 project number 1290.)
Figure 1.
Global phosphoproteome analysis using SILAC. A, experimental work flow. Cells were cultured for four passages in medium supplemented with either heavy (Arg 10/Lys 8) or light (Arg 0/Lys 0) amino acids to ensure complete incorporation. Whole-cell lysates were digested with trypsin, and phosphorylated peptides were enriched using titanium oxide columns. Flow-through proteins were used to identify the proteome. B, flow cytometry for the ESC pluripotency markers Oct4 and Nanog indicates homogeneity of the wild-type and DKO cell populations. C, Western blotting to assess Gsk3 knock-out (middle) and Wnt signaling activation by β-catenin protein accumulation (top). GAPDH is a loading control. D, overview of phosphorylation sites identified in the screen.
Overall, we identified 5858 phosphosites belonging to 1939 phosphoproteins. Most phosphosites were identified from peptides carrying a single phosphorylation (Fig. 1D, right). The relative frequency of phosphorylation of serine/threonine/tyrosine was 88:11:0.4, comparable with the 80:20:1 distribution known from most mammalian reports (Fig. 1D, left) (22). The overwhelming majority (80%) of identified phosphorylation sites could be localized with a probability of ≥85% to specific positions on the amino acid sequence (supplemental Fig. 2).
Known GSK-3 substrates were identified in this analysis, with reduced phosphorylation in DKO cells observed for the microtubule associated proteins MAP1b, MAP2, and tau (although tau was expressed at low abundance in ESCs, and the phosphopeptide was detected in only one biological replicate) as well as the GSK-3 substrates c-Jun and cryptochrome-2 (phosphorylated Cry2 was present in wild-type cells but was undetectable in cells lacking Gsk3). Furthermore, the level of β-catenin protein increased in DKO cells, as predicted and consistent with the Western blotting (Fig. 1C). However, β-catenin phosphopeptides were not detected in our analysis, most likely because the phosphorylated form of β-catenin is rapidly degraded by the proteasome (23).
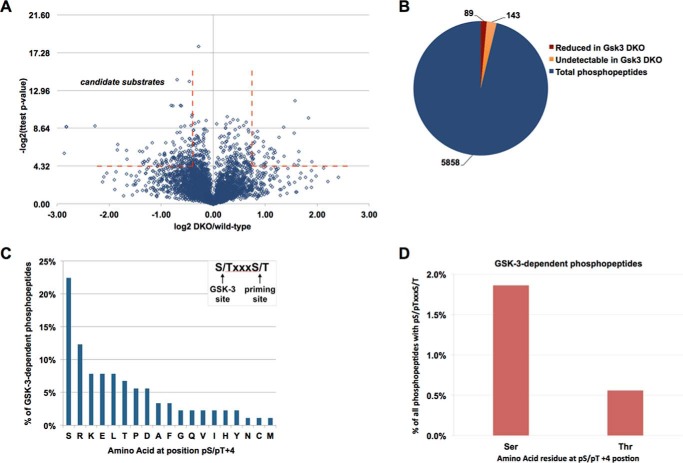
Using a p value < 0.05, we found 404 phosphopeptides representing 269 unique proteins that were differentially phosphorylated in DKO compared with wild-type ESCs. Filtering the data set to include only phosphopeptides with an absolute -fold change of ≥1.5-fold, we found 89 phosphopeptides from 65 unique proteins (3.4% of all phosphoproteins identified) that had significantly reduced phosphorylation in DKO ESCs compared with wild type (Fig. 2 (A and B) and supplemental Table 1). Of these phosphorylation events, 88% were on a serine residue and 12% were on threonine. It should be noted that SILAC does not distinguish whether these phosphorylation events are mediated directly or indirectly by GSK-3; for example, we also detected an increase in phosphorylation of 72 phosphopeptides representing 59 unique proteins in the DKO ESCs (Fig. 2A and supplemental Table 2), consistent with indirect regulation of these proteins by GSK-3.
Figure 2.
Gsk3 null ESC phosphoproteome identifies candidate substrates of GSK-3. A, volcano plot showing log2(-fold change) of each phosphopeptide (DKO/wild type) versus −log2(p value). Broken red lines indicate peptides with an absolute change of 1.5-fold or greater, p value ≤ 0.05. B, distribution of phosphopeptides that were significantly reduced in Gsk3 DKO (red), phosphopeptides present in wild type but undetectable in DKO (yellow), and phosphopeptides that did not change significantly (blue). C, frequency of each amino acid at the +4-position relative to the GSK-3–dependent phosphorylation site (inset shows reported GSK-3 consensus sequence with serine or threonine at +4-position (potential priming sites)). D, frequency of GSK-3–dependent phosphopeptides with serine (1.9%) or threonine (0.6%) at the +4-position as a percentage of all 1075 identified phosphopeptides with serine or threonine at the +4-position.
Notably, 143 phosphopeptides from 120 proteins were identified in wild-type cells in two or more biological replicates but were undetectable in Gsk3 DKO ESCs (Fig. 2B). The absence of these phosphopeptides in DKO cells may indicate an absolute requirement for GSK-3–dependent phosphorylation and may further suggest that as many as 185 proteins are phosphorylated in a GSK-3–dependent manner in ESCs. In addition, 65 phosphopeptides from 53 proteins were identified in Gsk3 DKO ESCs but were undetectable in wild-type cells, which may represent indirect regulation by GSK-3.
Phosphoproteome analysis identifies novel candidate GSK-3 substrates
To analyze GSK-3–dependent phosphorylation events further, we selected the 89 phosphosites that were reduced in DKO compared with wild type as high-confidence GSK-3–dependent phosphorylation sites. GSK-3 commonly engages with “primed” substrates that have been prephosphorylated at a serine or threonine 4 residues C-terminal to the GSK-3 site, providing the loose consensus (S/T)XXX(S/T) in which the first serine or threonine (S/T) is a GSK-3 site and (S/T) at the +4-position has been previously phosphorylated by a “priming” protein kinase (Fig. 2C). However, the prevalence of this consensus has not previously been addressed in a systematic manner in a single cell type. We therefore asked how many of the high-confidence sites identified in ESCs contain the putative GSK-3 consensus sequence. Among GSK-3–regulated phosphopeptides, serine is the most common residue at the +4-position, occurring in 20 phosphopeptides, and threonine is present at the +4-position in an additional six phosphopeptides (Fig. 2C). These data indicate that 29% of the GSK-3–dependent phosphosites have the potential to be primed by phosphorylation at the +4-position. (The data in Fig. 2C refer to phosphopeptides that were reduced but still detectable in DKO cells). Similarly, 30% of phosphopeptides had a serine or threonine at the +4-position when we included the 143 phosphopeptides that were detected only in Gsk3 wild-type cells (data not shown). In contrast, serine and threonine together accounted for 18% of residues at the +4-position of GSK-3–independent phosphopeptides (supplemental Fig. 3A). These data suggest a modest enrichment for serine and threonine at the +4-position relative to GSK-3–dependent phosphorylation sites.
We confirmed this analysis using the pLogo generator tool, which employs iterative comparisons of selected peptide sequences to background peptide sequences to extract significantly enriched motifs (24, 25). Although we did not observe statistically significant enrichment of any residues, the most common residues at the +4-position in the set of GSK-3–dependent phosphopeptides were serine and threonine (supplemental Fig. 3B). Taken together, these analyses suggest a bias for serine or threonine at the +4-position but also indicate that the majority of GSK-3–dependent phosphorylation sites do not contain the (S/T)XXX(S/T) consensus.
As an alternative approach, we performed in vitro GSK-3 phosphorylation using denatured cell lysates from DKO ESCs as a source of substrates. With this approach, substrates that require prephosphorylation at the +4-position should be phosphorylated by endogenous kinases, and GSK-3 sites should be unphosphorylated. To distinguish sites phosphorylated by GSK-3, we used [γ-18O]ATP for in vitro labeling. Filtering for a ≥2-fold increase in 18O-phosphoryl groups (p < 0.05), we identified 370 peptides that could be phosphorylated by GSK-3 in vitro, and 23% of these had serine or threonine at the +4-position. (These data are available at https://chorusproject.org,3 project 1290.) Thus, consistent with the SILAC data and published data, GSK-3 readily phosphorylates substrates lacking the (S/T)XXX(S/T) motif.
As 20% of the human proteome contains a (S/T)XXX(S/T) motif, a large proportion of the proteome has been predicted to include potential GSK-3 substrates (10, 18). We therefore asked whether the (S/T)XXX(S/T) motif predicts GSK-3–dependent phosphorylation. Similar to published predictions, 1075 (18%) of all phosphopeptides detected here have a serine or threonine at the +4-position, but only 26 (2.4%) of these phosphopeptides showed reduced phosphorylation in DKO ESCs (Fig. 2D).
We next conducted a gene ontology (GO) analysis using the Database for Annotation, Visualization, and Integrated Discovery (DAVID, version 6.8) to categorize the 65 candidate GSK-3 substrates based on previously reported functional terms (Fig. 3A and supplemental Table 9) (26). Significantly enriched (p < 0.05) GO terms include the following: mRNA processing (10 genes), RNA splicing (8 genes), regulation of transcription (14 genes), cell cycle (7 genes), and DNA repair (6 genes). Similar categories were enriched when GO and DAVID analyses were repeated to include the 120 phosphoproteins detected in wild-type but not in Gsk3 DKO cells (supplemental Table 9), including mRNA processing (14 genes), RNA splicing (12 genes), negative regulation of transcription (19 genes), transcription regulation (35 genes), and cell cycle (13 genes).
Figure 3.
Proteins with a broad range of functions are phosphorylated in a GSK-3–dependent manner in mouse ESCs. A, gene ontology analysis of proteins with significantly reduced phosphorylation in Gsk3 DKO cells using DAVID. B, STRING analysis of proteins enriched in the GO analysis generates three networks of proteins with related functions. Darker hue indicates greater reduction in phosphorylation in DKO cells. The red asterisk indicates phosphorylation sites with GSK-3 consensus motif. Phosphorylation of proteins in this analysis was reduced >1.5-fold with p value ≤ 0.05.
To identify potential relationships between the identified substrates, we analyzed direct and indirect interactions among the proteins enriched in the GO analysis using the Search Tool for the Retrieval of Interacting Genes/Proteins (STRING) database (27). This tool provides a confidence score for each interaction based on known experimental data, pathway knowledge from curated databases, text mining, and co-expression. The analysis enriched for three networks of proteins with related function: alternative splicing, transcriptional regulators, and cell division (Fig. 3B).
Identification of GSK-3–dependent phosphoproteins associated with transcriptional regulation is consistent with published observations that multiple transcription factors are phosphorylated by GSK-3, leading to changes in protein stability, DNA binding, and subcellular localization (12, 28). For example, the transcription factor c-Jun, a known GSK-3 substrate, was included in this cluster; phosphorylation by GSK-3 inhibits c-Jun DNA binding activity and also targets c-Jun for proteasomal degradation (29–32). Although GSK-3 phosphorylation of c-Jun at threonine 239 has been shown previously to target the protein for proteasomal degradation, we observed GSK-dependent phosphorylation of serine 73, a commonly phosphorylated site in c-Jun that regulates transcription activity (31–33). The transcription regulation substrate cluster also included the plant homeodomain finger protein 6 (PHF6), which suppresses ribosomal RNA synthesis and interacts with splicing factors (34, 35) (phosphorylation at serine 155 reduced 4.8-fold); the SWI/SNF chromatin remodeling factor ATRX (phosphorylation of serine 1088 reduced 1.8-fold); and Baz2a, a bromodomain-containing epigenetic silencer of ribosomal RNA genes (phosphorylation of serine 695 reduced 1.6-fold).
We also identified a network of candidate substrates with functions related to cell division (Fig. 3B, Cell Division). Ajuba (Jub), a regulator of Aurora A kinase (36, 37), was one of the strongest candidates in this cluster, with a 3-fold reduction in GSK-3–dependent phosphorylation at serine 129 (Fig. 3B), a site previously shown to be phosphorylated by other protein kinases to regulate entry into mitosis (37, 38). Septin-9 (SEPT9), a member of the septin family of cytoskeletal proteins that regulates multiple functions, including cytokinesis and chromosome segregation (39, 40), is phosphorylated at serine 85, also a GSK-3 consensus site, and this phosphorylation was reduced almost 2-fold in DKO ESCs. A weaker signal suggestive of GSK-3–dependent phosphorylation was also observed at serine 82 in SEPT9, and, although not detected in all replicates, reduced phosphorylation was also observed in SEPT1 serine 247 and SEPT5 serine 225. Other candidates included cyclin-dependent kinase 1 (CDK-1) and CDC20, an activator of the anaphase-promoting complex and component of the spindle assembly checkpoint.
Phosphoproteome analysis identifies a role for GSK-3 in alternative splicing
Multiple proteins involved in alternative mRNA splicing were identified in the STRING analysis (Fig. 3B). RNA splicing and mRNA processing were also the most significantly enriched terms in GO analysis (Fig. 3A) and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway analysis. Moreover, previous work has shown that GSK-3 phosphorylates PSF, a proline- and glutamine-rich splicing factor, to regulate alternative splicing of CD45 in human T-cells (41), but broader roles for GSK-3 in the phosphorylation of splicing factors and regulation of alternative splicing have not been addressed.
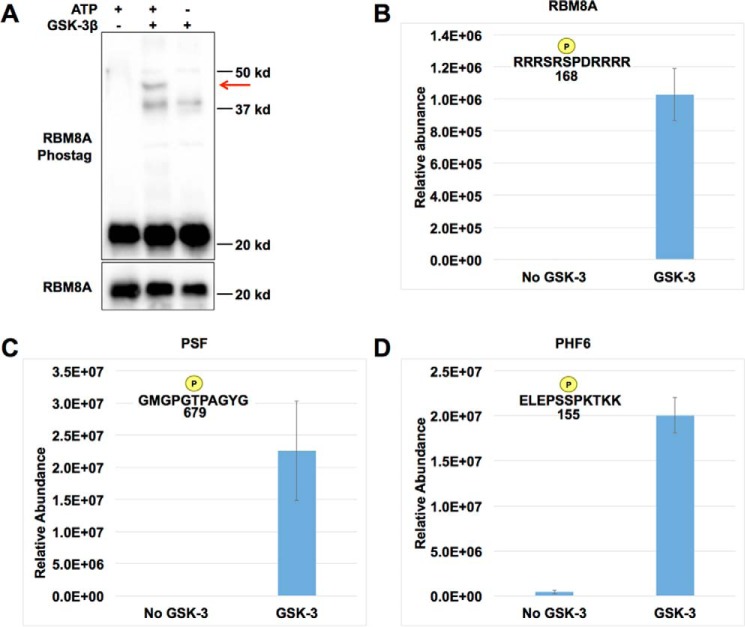
Many of the identified splicing factors contain arginine-serine (RS) dipeptide motifs, including multiple SR family members, SR family-related factors, and other RS-containing splicing factors. These include, for example, RNA-binding protein 8A (RBM8A, also known as Y14), SRSF9, TRA2B (SRSF10), CELF1, and RBM39 (Fig. 3B and Table 1). We selected RBM8A, a component of the exon-junction complex, for further characterization. The SILAC/MS data identified GSK-3–dependent phosphorylation of RBM8A (without a change in overall protein level, supplemental Fig. 4) at serine 166 and/or serine 168, which lie within adjacent RS motifs. Prior work has shown that phosphorylation of these residues inhibits RBM8A interaction with other exon-junction complex components (42, 43). As these sites do not have a +4 priming site, GSK-3 should be able to phosphorylate recombinant RBM8A in vitro without requiring prephosphorylation. Recombinant RBM8A was phosphorylated by GSK-3, as detected by reduced mobility (Fig. 4A, red arrow) in a Phos-tag/polyacrylamide gel, in which a phosphate-binding moiety slows mobility of phosphorylated species relative to non-phosphorylated forms (44).
Table 1.
GSK-3–dependent phosphorylation associated with RS motifs in splicing factors detected by SILAC
Phosphorylation sites are underlined, and overlapping or adjacent RS dipeptide motifs are in boldface type. (MS could not distinguish phosphorylation of RBM8A at serine 166 from serine 168, but both sites have been reported previously to be phosphorylated (42, 43). Similarly, MS could not distinguish phosphorylation of TRA2B at serine 83 from serine 85 and/or serine 87. For a complete list of phosphopeptides, see supplementary Table 1A).
| Splicing factor | Site | Sequence |
|---|---|---|
| RBM8A | Ser-166, -168 | RRGGRRRSRSPDRRRR |
| RBM39 | Ser-125 | IKLSRRRSRSKSPFRK |
| SRRM2 | Ser-1864 | RSVNRRRSRSRASPVS |
| TRA2B | Ser-83, -85, -87 | RSHRRSRSRSYSRDYR |
| TRA2B | Ser-39 | ARHTPARSRSKEDSRR |
| SRSF9 | Ser-190 | IRVYPERSTSYGYSRS |
Figure 4.
GSK-3 directly phosphorylates RBM8A, PSF, and PHF6. A, in vitro GSK-3–dependent phosphorylation of recombinant RBM8A protein (red arrow) resolved by Phos-tag polyacrylamide electrophoresis and Western blotting. B–D, mass spectrometry demonstrates GSK-3 phosphorylation of RBM8A at serine 168 (B), PSF at threonine 679 (C), and PHF6 at serine 155 (D). Error bars, S.D.
To confirm phosphorylation within the RS motif, we performed MS of in vitro phosphorylated RBM8A and identified serine 168 as a site of direct phosphorylation by GSK-3 (Fig. 4B and supplemental Table 3), consistent with the observations from SILAC. GSK-3 also phosphorylated SRSF9 in vitro, as detected by reduced mobility in standard SDS-PAGE (not shown) and by MS (supplemental Table 3). In parallel, we also assessed GSK-3 phosphorylation of PSF (41). Prior work had shown that PSF function is sensitive to a small molecule GSK-3 inhibitor, and mutagenesis studies suggested threonine 679 (equivalent to threonine 687 in the standard human nomenclature) as a likely site of phosphorylation. MS after in vitro phosphorylation of recombinant PSF confirmed that threonine 679 is phosphorylated directly by GSK-3 (Fig. 4C and supplemental Table 3). We also detected in vitro phosphorylation of PSF at serine 33 and threonine 468 (supplemental Table 3). Overall, we demonstrate that GSK-3 directly phosphorylates three splicing factors, RBM8A, SRSF9, and PSF.
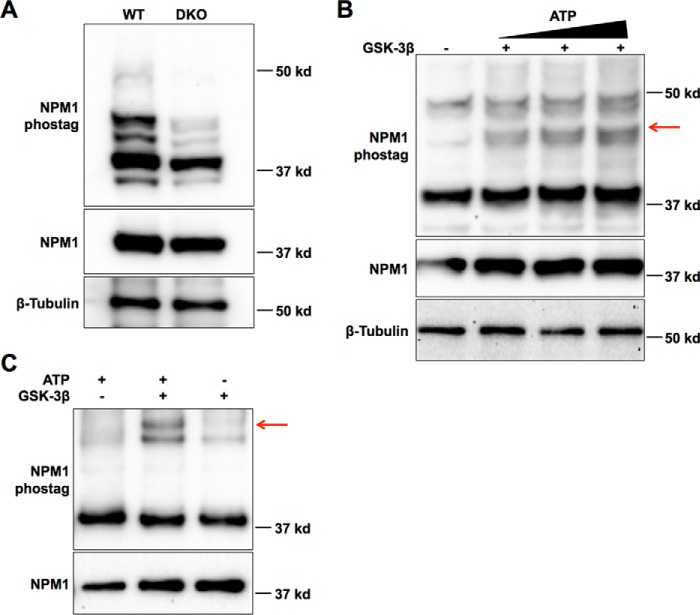
We also selected nucleophosmin 1 (NPM1) and PHF6 for further validation, as they appeared in multiple enriched functional categories related to RNA biosynthesis, and both are commonly mutated in acute myeloid leukemia (AML). NPM1 is a nucleolar protein with diverse functions in the cell, including ribosome biogenesis, chromatin remodeling, and mRNA processing, in which it has been reported to function as a negative regulator of alternative splicing (45, 46). We observed 11 NPM1 phosphopeptides that have also been identified in previous phosphoproteomic screens (47–49). Furthermore, phosphorylation at one of these sites, serine 225, was significantly reduced in Gsk3 DKO cells. We confirmed a reduction in NPM1 phosphorylation in DKO compared with wild-type cell lysates using Phos-tag gels and immunoblotting for NPM1 (Fig. 5A). The appearance of multiple bands with reduced mobility compared with the non-phosphorylated band (37 kDa) is consistent with the known multiple phosphorylated species of NPM1 in ESCs (47–49). We next performed in vitro kinase assays using DKO cell lysates and recombinant GSK-3 and detected a GSK-3–dependent increase in a phosphorylated form of NPM1 (Fig. 5B, indicated by a red arrow, compare lanes 1–4). We also observed GSK-3–dependent phosphorylation of recombinant NPM1 in an in vitro kinase assay, confirming that NPM1 can be directly phosphorylated by GSK-3 (Fig. 5C), although MS detected in vitro phosphorylation at serine 220 rather than serine 225. MS analysis also confirmed direct, GSK-3–mediated phosphorylation of PHF6 at serine 155 after in vitro phosphorylation (Fig. 4D and supplemental Table 3).
Figure 5.
GSK-3 directly phosphorylates NPM1. A, multiple phosphorylated forms of NPM1 were detected by Western blotting of wild-type and Gsk3 DKO cell lysates resolved by Phos-tag gel (top). NPM1 migrates as a single species in standard electrophoresis (middle). Bottom, β-tubulin loading control. B, Gsk3 DKO cell lysates were added to an in vitro protein kinase reaction with recombinant GSK-3 with or without ATP for 1 h. The reaction was then resolved by Phos-tag gel, and NPM1 was visualized by Western blotting. A slower-migrating species detected only in the presence of ATP, representing a GSK-3–phosphorylated form of NPM1, is indicated by the red arrow. C, GSK-3 phosphorylation of recombinant NPM1 in vitro resolved by Phos-tag (top) and standard electrophoresis (bottom).
GSK-3 regulates alternative splicing of multiple genes
To address whether GSK-3 modulates alternative splicing in ESCs, as observed previously using the GSK-3 inhibitor SB216763 in T-cells (41, 50), we examined global changes in splicing after complete loss of both Gsk3 isoforms by performing deep sequencing of poly(A) selected RNA from wild-type and Gsk3 DKO ESCs. We then used MAJIQ software to identify and quantify local splice variants (LSVs) and to measure the relative LSV abundance (change in percentage spliced in (dPSI)) between wild-type and DKO ESCs (Fig. 6A). This analysis identified 194 significant splicing differences (dPSI >20) in 188 genes (supplemental Table 4). When we reduced the stringency of detection to dPSI > 10, we observed 620 splicing events altered between wild-type and DKO ESCs (supplemental Table 5). Comparison of overall differential gene expression between wild type and DKO identified 321 genes with a log2 -fold change > 1.5 (adjusted p < 0.05) in mRNA levels (supplemental Table 6). Expression of multiple known Wnt target genes was increased in DKO cells, as expected, including brachyury (T), Sp5, Axin2, Cdx1, Snail1, Wif1, Ido1 and -2, and Dkk1. Notably, only six genes exhibited GSK-3–dependent regulation of both splicing and mRNA abundance (Fig. 6B), indicating that the splicing changes we detected are not a secondary effect of altered transcription or mRNA stability. Cassette exons, intron retention, and alternative first exon use were among the most common splicing changes in wild-type versus DKO ESCs (Fig. 6C). Whereas the latter could be driven by alternative promoter choice due to changes in transcription factors, regulation of cassette exons and intron retention most likely result from altered activity of RNA-binding proteins. Taken together, these data confirm that GSK-3 regulates alternative splicing in mouse ESCs, as observed previously with pharmacological inhibition of GSK-3 in human T cells (41, 50).
Figure 6.
Analysis of gene expression changes in wild-type and Gsk3 DKO mESCs. A, work flow of alternative splicing (AS) and differential expression (DE) analysis from RNA-Seq. B, total number of genes differing in alternative splicing or differential expression between wild-type and DKO cells. The six genes observed in both populations are indicated below the Venn diagram. C, categories of alternative splicing for the 194 changing splicing events detected by MAJIQ. Categories are defined as indicated by the schematics, followed by number of events in that category. Events not falling in any single category are defined as “Complex.”
Protein abundance in Gsk3 WT and DKO ESCs
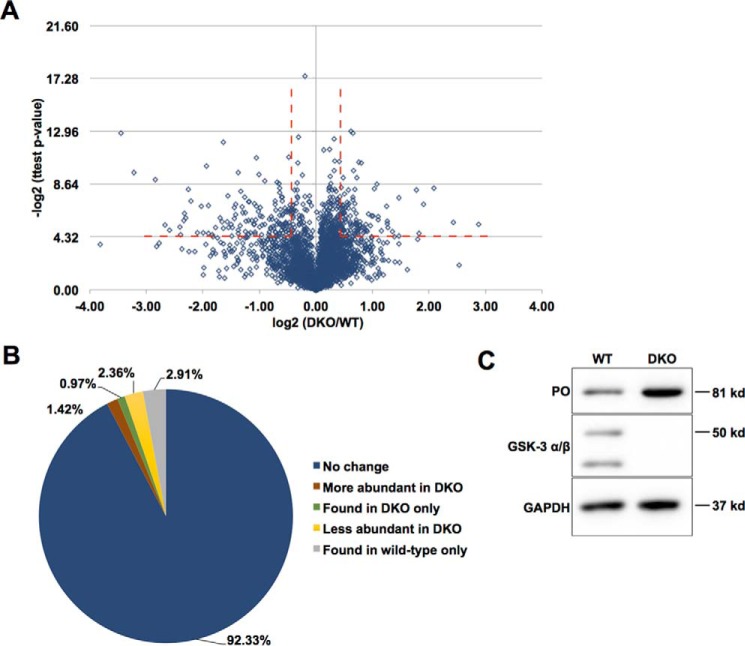
As GSK-3 regulates the stability of many of its target proteins (23, 51), we also analyzed changes in total protein abundance between the wild-type and DKO ESCs. Overall, our analysis identified 3299 proteins, and 459 of these had significantly different abundance in wild type compared with DKO. To select high-confidence candidates, we filtered the data for an absolute -fold change of ≥1.5 and then removed candidates that showed a significant (p < 0.05) corresponding change in mRNA abundance in the RNA-Seq data. This analysis identified 47 proteins (1.4%) with increased abundance in the DKO and 78 (2.4%) with reduced abundance in DKO, compared with wild type (Fig. 7 (A and B) and supplemental Table 7). In addition, 32 proteins were detected only in DKO ESCs, and 96 proteins were detected only in Gsk3 wild-type ESCs, without a significant difference in RNA abundance (Fig. 7B and supplemental Table 8). The fraction of proteins with increased abundance in DKO cells agrees closely with reports showing GSK-3–dependent degradation or polyubiquitination of ∼0.4–1.3% of queried proteins (23, 52). The decrease in abundance of proteins in DKO cells is also consistent with previous reports that GSK-3 phosphorylation stabilizes some targets, including the nuclear hormone Reverbα and the NF-κB1 subunit p105 (10, 53, 54).
Figure 7.
Protein abundance changes in DKO compared with wild type. A, volcano plot of log2(-fold change in protein abundance) in DKO/wild-type versus −log2(p value) demonstrates multiple proteins that are both increased and decreased in abundance in a Gsk3 dependent manner. B, percentage of proteins that are increased (red) or decreased (yellow) 1.5-fold (p < 0.05) in abundance in Gsk3 DKO cells without a statistically significant change in RNA abundance. The percentage of proteins detected only in DKO cells (green) or only in Gsk3 wild-type ESCs (gray) without a statistically significant change in RNA abundance is also shown. C, PREP/PO total protein levels are increased in DKO compared with wild type without a significant change in RNA abundance.
Of the proteins that increase upon Gsk3 deletion, we detected β-catenin, a well-known substrate of GSK-3 (Fig. 1C), which increased ∼4-fold in DKO cells without a change in mRNA abundance. We also observed an increase in prolyl oligopeptidase (PREP or PO), which in other cell types modulates sensitivity to lithium, a well-characterized inhibitor of GSK-3 as well as inositol monophosphatase (55). Prep RNA abundance did not change significantly by RNA-Seq, consistent with posttranscriptional regulation of PREP protein abundance, although a small increase in RNA abundance that did not achieve statistical significance could also account for this increase in PREP. MS did not detect a change in PREP phosphorylation in DKO cells compared with WT, which could indicate that, similar to β-catenin, the phosphorylated form of PREP is highly unstable and falls below our limit of detection, or it may indicate that the regulation of PREP abundance by GSK-3 is indirect.
Discussion
GSK-3 is a central regulator of multiple cellular processes and signaling pathways, and the full range of GSK-3 regulated functions is not known. The identification of GSK-3 substrates has so far been based on a diverse range of assays performed in different experimental models, leaving an incomplete picture of the overall GSK-3 phosphoproteome. We have used SILAC to characterize the GSK-3 phosphoproteome in ESCs. To identify global changes in phosphorylation, we compared the phosphorylation state of proteins in ESCs lacking Gsk3a and Gsk3b with wild-type ESCs. This approach provides a quantitative, unbiased picture of the spectrum of GSK-3–dependent phosphoproteins. Phosphorylation of 65 proteins was significantly reduced in DKO ESCs compared with wild-type cells, and phosphorylation of an additional 120 proteins was detected only in wild-type ESCs, consistent with an absolute requirement for GSK-3–mediated phosphorylation. The identified GSK-3–dependent phosphoproteins span a wide array of functions, including splicing factors, translational initiation factors and other RNA-regulatory proteins, microtubule-associated proteins, transcription factors, chromatin modifiers, and regulators of cell division.
One of the most striking findings in the phosphoproteomic data set was the identification of multiple proteins involved in RNA splicing, including RBM8A, RBM39 (also known as CAPER), SRRM1, SRRM2, SRSF9, TRA2B (SRSF10), CELF1, WBP11/SIPP1 (splicing factor that interacts with PQBP-1 and PP1), PPP4R2, and CDK13. We used in vitro phosphorylation assays to confirm that GSK-3β can directly phosphorylate the splicing factors SRSF9 and RBM8a, a component of the exon junction complex that is involved in nonsense-mediated decay in addition to its role in alternative splicing. We also confirmed direct GSK-3β phosphorylation of the splicing factor PSF at threonine 679 (equivalent to threonine 687 in humans), shown previously to regulate the alternative splicing of CD45 (41). In that work, GSK-3 was proposed to phosphorylate threonine 687 of PSF in unstimulated T-cells, promoting its interaction with TRAP150. Upon activation of T-cell receptor signaling, GSK-3 is inhibited, reducing PSF phosphorylation and allowing PSF to dissociate from TRAP150, bind to CD45 preRNA, and suppress variable exon inclusion (41). Here we have demonstrated direct phosphorylation of PSF by GSK-3β and confirmed phosphorylation at the predicted threonine residue. An important question for future study is whether GSK-3α is also able to phosphorylate these substrates directly; although the amino acid sequence of GSK-3α is 98% identical to GSK-3β in the catalytic domain, the two genes are not completely redundant, as Gsk3a KO mice are viable, whereas the Gsk3b KO is embryonic or perinatal lethal (3–5).
The finding that multiple splicing factors are phosphorylated in a GSK-3–dependent manner led us to ask whether GSK-3 might play a more general role in alternative splicing. Deep sequencing of wild-type and Gsk3a/b DKO ESCs revealed 188 genes that exhibit ≥20% dPSI in splice form expression (>95% probability) between wild-type and Gsk3 KO cells. These data demonstrate that GSK-3 regulates alternative splicing of multiple genes in mouse ES cells and confirm previous observations using pharmacological inhibition of GSK-3 in human T cells (41, 50). Formally, one possibility is that GSK-3 affects alternative splicing through changing transcription rates, given that GSK-3 phosphorylates a number of transcription factors (Fig. 3B), and transcription efficiency can affect splicing patterns (56, 57). However, only 6 of the 188 genes that are alternatively spliced in the DKO ESCs exhibit changes in mRNA abundance between DKO and wild-type ESCs. This separation of differential expression and differential splicing is typical of what has been observed in cellular conditions known to directly regulate splicing (58, 59) and is consistent with a direct effect of GSK-3 on RNA-binding proteins that we observe here.
These findings are also consistent with a previous report that GSK-3 can phosphorylate SRSF2 (SC35) in vitro and that inhibition of GSK-3 in cultured neurons increases the inclusion of exon 10 in the microtubule associated protein tau (60). Taken together, the data presented here identifying GSK-3–dependent phosphorylation of multiple splicing factors, published work demonstrating GSK-3 phosphorylation of the specific factors PSF and SRSF2, and our finding that genetic loss of Gsk3 significantly disrupts alternative splicing of >190 genes argue strongly for a broad role for GSK-3 in the regulation of alternative splicing. Interestingly, both insulin and Wnt signaling, which function by inhibiting GSK-3, alter splicing of 149 (insulin) and 85 (Wnt/wingless) genes in Drosophila S2 cells (61), raising the possibility that these pathways could regulate gene expression through modulation of GSK-3 activity and alternative splicing in other organisms.
Somatic mutations in several of the splicing factors identified in our screen occur frequently in AML and myelodysplastic syndrome (MDS) (62–67). Knock-out of Gsk3 (DKO) in hematopoietic cells in mice causes a dramatic myeloproliferative and myelodysplastic disorder with features of AML (68). Whereas the signaling networks downstream of GSK-3 that mediate these neoplastic phenotypes are probably complex, disruption of splicing factor function through altered GSK-3–dependent phosphorylation, with subsequent disruption of splicing of mRNAs that encode hematopoietic regulators, could contribute to the progression of these neoplasms. Consistent with this, mRNA splicing is disrupted in MDS and AML progenitor cell populations compared with normal hematopoietic stem and progenitor cells, and drugs that modulate alternative splicing have been proposed as therapies for MDS and AML (67, 69, 70). Furthermore, inhibition of GSK-3 in primary human T cells alters the splicing of RUNX1 and ASXL1, which are both commonly mutated in AML. GSK-3 inhibition increases the frequency of splice forms that encode nonfunctional proteins, with 80% of alternatively spliced ASXL1 transcripts encoding an early termination codon that probably generates either a nonfunctional protein or an mRNA subject to nonsense-mediated decay (50). Predominance of these nonfunctional mRNAs could mimic somatic mutations and similarly contribute to AML pathogenesis. Thus, the hematopoietic neoplasms that arise from Gsk3 DKO mouse bone marrow could be due, in part, to disruption of splicing factor function.
We also identified GSK-3–dependent phosphorylation of NPM1, one of the most commonly mutated genes in AML, and PHF6, which is mutated in ∼3% of AML (62). Although the functions of NPM1 and PHF6 are not well-characterized, both are nucleolar proteins that regulate rRNA biosynthesis and may also affect splicing (34, 35, 45).
GSK-3 is a core negative regulator of canonical Wnt signaling with a primary role to phosphorylate β-catenin and target it for proteasomal degradation. However, emerging data suggest that “canonical” Wnt signaling (Wnt signaling that depends on the Axin–APC–GSK-3 complex) regulates multiple processes that are independent of β-catenin and the transcriptional response to β-catenin/Tcf activation, including mitotic spindle orientation (71), mTORC1 activity (15, 72, 73), and protein stability (18, 23, 51, 52, 74). For example, Wnt-dependent stabilization of proteins (Wnt/STOP), which occurs primarily at the onset of mitosis, leads to stabilization of multiple proteins in addition to β-catenin, consistent with a role for GSK-3 in the canonical pathway that bypasses β-catenin/TCF transcription (51) (23, 51, 52, 75, 76). However, the overlap between the set of GSK-3–dependent phosphopeptides and the group of proteins that either increased (12 proteins) or decreased (4 proteins) in abundance was limited. In cases where GSK-3 phosphorylation targets proteins for degradation, this may indicate that the phosphorylated form is below the limit of detection by SILAC, as observed with β-catenin, but it could also represent indirect regulation of target stability by GSK-3.
The observation that 20% of proteins in the human proteome contain the (S/T)XXX(S/T) motif led to the suggestion that the stability of a large number of proteins could depend on GSK-3 phosphorylation (10, 18). In support of Wnt stabilization of proteins, a microarray analysis of ubiquitinated proteins identified 119 of >9000 interrogated proteins (1.3%) that showed GSK-3–dependent polyubiquitination, indicative of proteasomal targeting (52). Similarly, an in vitro screen for GSK-3–dependent degradation identified 42 proteins (∼0.6% of those screened) that were stabilized by GSK-3 inhibitors (23). In close agreement with these studies, we identified 47 proteins whose abundance increased >1.5-fold in Gsk3 DKO ESCs without a significant increase in the mRNAs encoding those proteins (based on RNA-Seq data) as well as 32 proteins that were detected only in DKO cells, consistent with GSK-3–mediated destabilization of 1.4–2.4% of the proteome. However, GSK-3 phosphorylation also stabilizes multiple proteins, including the circadian regulators Reverbα (53), Timeless (77), and cryptochrome (78) as well as the apoptosis signal-regulating kinase-1 (ASK1) (79) and the p105 precursor of the NF-κB p50 subunit (54). Consistent with a positive role for GSK-3 in protein stabilization, we found 78 proteins that were reduced in abundance by Gsk3 deletion (without a significant change in the corresponding mRNAs) and another 96 proteins that were only detectable in cells expressing Gsk3. Therefore, up to 5% of the proteome may be stabilized by GSK-3. Small changes in RNA that do not reach statistical significance in the RNA-Seq data could, in principle, account for statistically significant changes in protein abundance. Nevertheless, overall, the analysis presented here together with published work from others indicates that the abundance of a substantial fraction of the proteome is posttranscriptionally regulated by GSK-3.
The priming mechanism for GSK-3 substrates has been elegantly established for glycogen synthase (80), β-catenin (81), and a handful of other substrates, and the structural basis for GSK-3 recognition of primed substrates has also been worked out (7, 82–84). The identification of a loose consensus has led to identification of additional proteins that contain this consensus sequence but may also have biased the search for GSK-3 substrates to proteins that contain the consensus. Our unbiased phosphoproteome analysis revealed that 26 of 68 of the high-confidence candidate substrates contain the consensus sequence, with either serine or threonine at the +4-position, supporting the importance of the (S/T)XXX(S/T) consensus sequence but also indicating that this consensus is neither essential nor sufficient to direct GSK-3–dependent phosphorylation. Moreover, of all of the phosphopeptides identified by SILAC, 1075 peptides have a serine or threonine at the +4-position, and only 4.6% of these demonstrated GSK-3–dependent phosphorylation. Published evidence also supports GSK-3–dependent phosphorylation of sites that lack serine or threonine at the +4-position, including c-Myc, the protein phosphatase-1 inhibitor-2 (I-2), cyclin D1, and tau. In a published tabulation of over 100 putative GSK-3 substrates (12), only 27% were known to be primed, consistent with our finding of ∼70% of GSK-3–dependent phosphorylation occurring at sites lacking the consensus sequence. These observations suggest additional complexity to the regulation of GSK-3–dependent phosphorylation.
Our analysis of in vivo phosphorylation does not distinguish direct from indirect phosphorylation. We therefore showed direct phosphorylation of RBM8A, SRSF9, PHF6, PSF, and NPM1 by GSK-3. We were able to test these by in vitro phosphorylation because the tested sites do not require a priming step. An in vitro analysis of substrates that absolutely require priming will be challenging without first identifying the priming kinase. In addition, in vitro kinase reactions are inherently limited by the artificial conditions of the assay. Therefore, these assays can only establish that a putative substrate can be phosphorylated by GSK-3, and establishing direct phosphorylation in vivo is a more challenging task that, to our knowledge, has not yet been feasible for GSK-3 substrates. Our phosphoproteomic screen nevertheless identifies multiple targets that demonstrate GSK-3–dependent phosphorylation. These will be interesting to pursue in more depth in the setting of Wnt signaling, stem cell homeostasis, leukemia pathogenesis, and other GSK-3–regulated biological processes.
Experimental procedures
Cell culture
Gsk-3a/b double knock-out and wild-type (floxed) E14 ESCs (6) were kindly provided by Drs. Bradley Doble (McMaster Stem Cell and Cancer Institute, Hamilton, Canada) and James Woodgett (Lunenfeld-Tanenbaum Research Institute, Toronto, Canada). All media and supplements were obtained from Gibco (Invitrogen) unless otherwise noted. E14 mESCs were cultured at 37 °C in 5% CO2 in DMEM supplemented with 15% FBS (HyClone Defined), 0.1 mm Eagle's minimum essential medium nonessential amino acids, 2 mm Glutamax, 0.055 mm β-mercaptoethanol, and 1000 units/ml LIF (Chemicon) on 0.1% gelatin-coated plates with irradiated mouse embryonic fibroblasts (Global Stem). Cells were routinely tested for mycoplasma using the MycoAlert detection kit (Lonza). For SILAC labeling experiments, cells were cultured with DMEM without arginine or lysine supplemented with 0.798 mm isotope heavy l-lysine (13C6, 15N2) and 0.398 mm heavy l-arginine (13C6, 15N4) (Cambridge Isotope Laboratories) and 15% KnockOut Serum Replacement with the same medium components as standard culture. In case of light media, standard l-lysine and l-arginine were used. During isolation, ES cells were dissociated into a single cell suspension with TrypLE, and mouse embryo fibroblasts were removed by absorption to plastic dishes for two consecutive 30-min incubations. Proteomic experiments were conducted in biological triplicate. For protein stabilization experiments, cells were treated with MG-132 or cycloheximide at the indicated concentrations and times. MG-132 and cycloheximide were added to mouse embryo fibroblast medium during feeder separation.
Proteomics and phosphoproteomics analysis using nLC-MS/MS
All chemicals used for preparation of nLC-MS/MS samples were of at least sequencing grade and were purchased from Sigma-Aldrich, unless otherwise stated. Light and heavy labeled cells were separately lysed using 6 m urea, 2 m thiourea, 50 mm ammonium bicarbonate, pH 8.2, and phosphatase and protease inhibitor mix (Thermo Fisher Scientific) by vortexing. After protein quantification using Bradford, equal amounts of light and heavy protein lysates were mixed. Proteins were first digested using endopeptidase Lys-C (Wako, MS grade) for 3 h, after which the solution was diluted 10 times with 20 mm ammonium bicarbonate. Subsequently, samples were reduced using 10 mm DTT for 1 h at room temperature and alkylated with 20 mm iodoacetamide in the dark for 30 min at room temperature. Further digestion was performed using trypsin (Promega) at an enzyme/substrate ratio of ∼1:50 overnight at room temperature. After digestion, the samples were concentrated to ∼100 μl by lyophilization. Phosphopeptide enrichment using TiO2 chromatographic resin was performed as described previously (85). The lyophilized phosphorylated peptide samples were reconstituted in 0.1% TFA and desalted using Poros Oligo R3 RP (PerSeptive Biosystems) P200 columns. Unbound peptides from the TiO2 flow-through and subsequent TiO2 washes were combined and lyophilized to produce the non-modified peptide fraction. The non-modified peptide fraction was resuspended in 0.1% TFA and desalted using a Sep-Pak tC18 Plus Light Cartridge (Waters). Afterward, samples were dried to completely remove traces of acetonitrile from stage tip elution. Dried samples were resuspended in buffer A (0.1% formic acid) and loaded onto an Easy-nLC system (Thermo Fisher Scientific), coupled online with a Q-Exactive or an Orbitrap Fusion Tribrid mass spectrometer (both from Thermo Scientific). Peptides were loaded into a picofrit fused silica capillary column (75-μm inner diameter) packed in-house with reversed-phase Repro-Sil Pur C18-AQ 3-μm resin of about 18 cm. A gradient of 165 min was set for peptide elution from 2 to 28% buffer B (100% acetonitrile, 0.1% formic acid) at a flow rate of 300 nl/min. Both instruments were programmed in a data-dependent acquisition mode. For the Q-Exactive, the full MS scan range was 360–1200 m/z in the orbitrap with a resolution of 70,000 (200 m/z) and an AGC target of 5 × 10e5. MS/MS was performed in the orbitrap selecting the top 12 ions, a resolution of 17,500, an AGC target of 5 × 10e4, and a collision energy of 22. For the Orbitrap Fusion Tribrid, the full MS scan was 350–1200 m/z in the orbitrap with a resolution of 120,000 (200 m/z) and an AGC target of 5 × 10e5. MS/MS was performed in the ion trap using the top speed mode (3 s), an AGC target of 10e4, and a higher-energy collisional dissociation collision energy of 32. MS raw files were analyzed by the MaxQuant software version 1.5.2.8. MS/MS spectra were searched by the Andromeda search engine against the mouse UniProt FASTA database (version November 2015) (86, 87). The search for total proteome included variable modifications of methionine oxidation and N-terminal acetylation and fixed modification of carbamidomethyl cysteine. Analysis of the phosphoproteome included carbamidomethylation on cysteine residues as a fixed modification, whereas phosphorylation on serine, threonine, and tyrosine residues was set as variable modification. Trypsin was specified as the digestive enzyme. SILAC labeling was used as quantification. Match between runs was enabled and set to a 1-min window. All other values were kept as default. Protein tables were filtered to eliminate the identifications from the reverse database, only identified by site and common contaminants. All MS raw files are available at https://chorusproject.org3 at project number 1290.
Statistical analysis and downstream bioinformatics
Each analysis was performed with three biological replicates. Heteroscedastic t test was used to assess the significant differences in peptide/protein abundance (p value < 0.05). Data distribution was assumed to be normal, but this was not formally tested. The protein interaction network was extracted from the STRING database (27) and visualized using Cytoscape (88). GO and KEGG pathway analysis were performed using DAVID version 6.8. The list of proteins with GSK-3–dependent phosphorylation (supplemental Table 1) was submitted as the gene list, and automatic Mus musculus background was used. Categories with a p value of <0.05 and DAVID enrichment score >1.0 were selected.
RNA-Seq
Total RNA was isolated by TRIzol from wild-type and DKO mESC cells after a similar number of passages. RNAs were then further purified by DNase treatment and the RNeasy kit (Qiagen). Purity was confirmed by bioanalyzer (RNA integrity number (RIN) >8). RNAs were sent to the Genomics Division of the Iowa Institute of Human Genetics for poly(A)-selection, generation of stranded Illumina RNA-Seq library, and sequencing on three lanes of a HiSeq4000. 269 million unique, mappable reads (402 million total) were obtained for wild-type mESCs, and 255 million unique, mappable reads (383 million total) were obtained for DKO mESCs.
Quantification of alternative spliced and differential expressed genes
Alternative splicing and differential expression analyses relied on RNA-Seq reads mapped to the reference mouse genome (mm10) using STAR with the option alignSJoverhangMin 8 (89) (see above for numbers). Alternative splicing events were analyzed using MAJIQ and VOILA with the default parameters (90). Briefly, uniquely mapped, junction-spanning reads (217 million for wild-type and 203 million for DKO mESCs) were used by MAJIQ to construct splice graphs for transcripts by using the Ensembl transcriptome annotation (release 82) supplemented with de novo-detected junctions. Here, de novo refers to junctions that were not in the Ensembl transcriptome database but had sufficient evidence in the RNA-Seq data (default: at least three reads mapping to at least two different start positions). The resulting gene splice graphs were analyzed for all identified LSVs, defined as splits in a splice graph to or from a given exon. For every junction in each LSV, MAJIQ then quantified the expected PSI value in control and GSK3DKO samples and expected dPSI between WT and GSK3DKO samples. Results from VOILA were then filtered for high-confidence changing LSVs (whereby one or more junctions had at least a 95% probability of expected dPSI of at least an absolute value of 20 PSI units (noted as “20% dPSI”) between WT and GSK3DKO) and candidate changing LSVs (95% probability, 10% dPSI).
Finally, to correct for overlapping cases where two LSVs capture the same alternativity in isoform usage at one locus, LSVs with a changing junction were grouped together into events based on shared changing junctions. Each group was assigned an event ID. For the high confidence results (dPSI ≥ 20%), the events were further categorized as single-exon cassette, multiexon cassette, alternative 5′- and/or 3′-splice site, de novo intron removal, intron retention, or alternate transcript start or end events. No mutually exclusive exon usage events were observed. All remaining events that did not fit into these categories were classified as complex.
Counts of STAR-aligned reads that overlapped transcript exons were quantified with the GenomicRanges package in R (count mode: Union) (91). Counts were then normalized, and differentially expressed genes between WT and GSK3 DKO with a log2 -fold change of ≥1.5 and a Benjamini and Hochberg corrected p value of <0.05 were identified using default DESeq parameters in R (92).
Western blot analysis
Cells were lysed in buffer containing 20 mm Tris, pH 7.5, 140 mm NaCl, 1 mm EDTA, 10% glycerol, 1% Triton X-100, 1 mm DTT, 50 mm NaF with protease inhibitor mixture (Sigma) and phosphatase inhibitor mixtures 2 and 3 (Sigma) used at 1:100. Supernatants were collected after centrifugation at 14,000 rpm for 10 min at 4 °C. Protein was quantified by a Bradford assay. Standard SDS-PAGE and Western blot protocols were followed. The following antibodies were used at 1:1000, unless otherwise listed: GAPDH (Cell Signaling), β-catenin (Cell Signaling), proyl oligopeptidase (Abcam), phosphorylated ribosomal protein S6 (Cell Signaling), ribosomal protein S6 (Cell Signaling), and GSK-3α/β (Cell Signaling).
Phos-tag electrophoresis and immunoblotting
Cells were lysed in 6 m urea, 2 m thiourea, 50 mm ammonium bicarbonate, pH 8.2, with protease inhibitor mixture (Sigma) and phosphatase inhibitor mixtures 2 and 3 (Sigma), used at 1:100. Supernatants were collected after centrifugation at 14,000 rpm for 10 min at 4 °C. Protein was quantified by a Bradford assay. Samples were diluted in standard 2× Laemmli sample buffer and heated at 95 °C for 5 min before PhosTag analysis. PhosTag gels were prepared using PhosTagTM acrylamide reagent (Wako Pure Chemical Industries, Ltd., catalogue no. AAL-107) with ZnCl2 based on the manufacturer's protocol (44). The composition for optimal separation of NPM1 was 30 μm PhosTagTM acrylamide reagent and 10% acrylamide using a acrylamide/bisacrylamide solution (29:1). The composition for optimal separation of RBM8A was 30 μm PhosTagTM acrylamide reagent and 12% acrylamide. Gels were run at constant 70 V for 3–4 h, until dye front was off the gel. PhosTag gels were soaked in transfer buffer with 1 mm EDTA for 15 min and then washed for 15 min in transfer buffer without EDTA. Gels were transferred using PVDF membrane, at 150 mA for 16 h. Gels were blocked with 5% nonfat milk. The following antibodies were used at 1:1000: NPM1 (Bethyl Laboratories) and RBM8A (Bethyl Laboratories).
In vitro protein kinase assays
Recombinant proteins were incubated in kinase reaction buffer (100 mm Tris, pH 7.5, 5 mm DTT, 10 mm MgCl2) with 800 μm ATP for 60 min at 30 °C. Reactions were stopped by adding standard 2× Laemmli sample buffer and incubating at 95 °C for 5 min. Samples were run on PhosTag gels following the reaction. 0.4 μg of recombinant protein was used; RBM8A (ProSpecBio), NPM1 (ProSpecBio), PHF6 (Origene), and PSF (41) were expressed in and purified from bacteria, and SRSF9 (Abnova) was translated in vitro in wheat germ extract and then purified. Recombinant GSK-3 was used at 50 units/20-μl reaction (New England Biolabs). For MS analysis, MS proteolysis methods were followed starting at the 10 mm DTT step. Peptides were analyzed following similar methods as indicated under “Proteomics and phosphoproteomics analysis using nLC-MS/MS.”
For in vitro phosphorylation of proteins in ESC cell lysates, wild-type and DKO ESCs (in triplicate) were lysed in 8 m urea (6 m urea, 2 m thiourea, 50 mm ammonium bicarbonate, pH 8.2, phosphatase and protease inhibitors) as described above for SILAC. Samples were diluted in protein kinase buffer (50 mm Tris, pH 7.5, 10 mm MgCl2, 0.1 mm EDTA, and 2 mm DTT with 0.8 mm [γ-18O]ATP (Cambridge Isotopes catalog no. OLM-7858-20)) to a final protein concentration of ∼0.5 mg/ml in a 200-μl total reaction volume (urea <0.4 m). Recombinant GSK-3β (25 units) or water (control) was added, and samples were incubated for 60 min at 30 °C and then reduced, alkylated, trypsinized, and processed for MS as described above for SILAC.
Intracellular flow cytometry
Up to 5 million cells were fixed in 1.6% paraformaldehyde for 20 min at 37 °C and washed in FACS buffer. (Cells were permeabilized with 1× saponin buffer (Biolegend 421002).) To stain for Oct4 and Nanog, cells for were incubated with primary antibody at 1:200 for 1 h at room temperature. Antibodies were as follows: Oct4 (Santa Cruz Biotechnology, Inc.) and Nanog (Abcam). Cells were incubated with secondary antibody at 1:400 for 30 min at room temperature. Antibodies were as follows: goat anti-mouse IgG-PE and IgG-CFL488 (both from Santa Cruz Biotechnology). Samples were resuspended in FACS buffer (1× PBS + 0.5% BSA, 0.05% azide). Analysis was performed using a FACSCanto flow cytometer (BD Biosciences), and data were analyzed using FlowJo software (Treestar).
Author contributions
M. Y. S. participated in experimental design, carried out experiments, and analyzed data for all figures and co-wrote the manuscript. S. S. participated in experimental design, carried out and analyzed all MS experiments, and wrote a portion of the manuscript. K. K. contributed to design, execution, and analysis of MS experiments. M. J. M. contributed to experiments on splicing proteins and analysis of RNAs. C. M. R. contributed to experimental design and analysis of RNA-Seq data. A. R. and R. L. M. provided technical assistance and analyzed experiments. Y. B. supervised computational analysis of RNA-Seq/splicing data and contributed to experimental design. K. W. L. supervised work on splicing, contributed to experimental design and interpretation of data, and contributed to writing and editing of the manuscript. B. A. G. supervised MS experiments and contributed to experimental design and interpretation of data and editing of the manuscript. P. S. K. conceived and coordinated the study, contributed to experimental design and interpretation of data, and wrote the manuscript. All authors reviewed the results and approved the final version of the manuscript.
Supplementary Material
Acknowledgments
We thank Drs. Bradley Doble and James Woodgett for generously providing the wild-type and DKO mESCs. We also thank Drs. Judy Meinkoth, Paul Gadue, and Ed Morrisey for helpful discussions. Data presented herein were obtained at the Genomics Division of the Iowa Institute of Human Genetics, which is supported, in part, by the University of Iowa Carver College of Medicine. RNA-Seq data were deposited in the Gene Expression Omnibus repository (GEO accession number GSE90921). MS raw files are available at https://chorusproject.org3 at project number 1290.
This work was supported by National Institutes of Health Grants 1R01MH100923, 1R01HL110806, and 1R01GM115517; a pilot grant from the Hematological Malignancies Translational Center of Excellence of the Abramson Cancer Center; and a pilot grant from the Institute for Translational Medicine and Applied Therapeutics at the University of Pennsylvania (to P. S. K.). This work was also supported by National Institutes of Health Grants R01GM110174, R01AI118891, and P01CA196539 (to B. A. G.), R35GM118048 (to K. W. L.), and R01AG046544 (to Y. B.). The authors declare that they have no conflicts of interest with the contents of this article. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
This article was selected as one of our Editors' Picks.
This article contains supplemental Tables 1–9 and Figs. 1–4.
Please note that the JBC is not responsible for the long-term archiving and maintenance of this site or any other third party hosted site.
- GSK-3
- glycogen synthase kinase-3
- PSI
- percentage spliced in
- dPSI
- change in PSI
- SILAC
- stable isotope labeling by amino acids in culture
- DKO
- double knock-out
- ESC
- embryonic stem cell
- mESC
- mouse ESC
- DAVID
- Database for Annotation, Visualization, and Integrated Discovery
- STRING
- Search Tool for the Retrieval of Interacting Genes/Proteins
- KEGG
- Kyoto Encyclopedia of Genes and Genomes
- AML
- acute myeloid leukemia
- LSV
- local splice variant
- PREP
- prolyl oligopeptidase
- MDS
- myelodysplastic syndrome
- nLC
- nano-LC.
References
- 1. Doble B. W., and Woodgett J. R. (2003) GSK-3: tricks of the trade for a multi-tasking kinase. J. Cell Sci. 116, 1175–1186 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Picton C., Woodgett J., Hemmings B., and Cohen P. (1982) Multisite phosphorylation of glycogen synthase from rabbit skeletal muscle: phosphorylation of site 5 by glycogen synthase kinase-5 (casein kinase-II) is a prerequisite for phosphorylation of sites 3 by glycogen synthase kinase-3. FEBS Lett. 150, 191–196 [DOI] [PubMed] [Google Scholar]
- 3. Hoeflich K. P., Luo J., Rubie E. A., Tsao M. S., Jin O., and Woodgett J. R. (2000) Requirement for glycogen synthase kinase-3β in cell survival and NF- κB activation. Nature 406, 86–90 [DOI] [PubMed] [Google Scholar]
- 4. Liu K. J., Arron J. R., Stankunas K., Crabtree G. R., and Longaker M. T. (2007) Chemical rescue of cleft palate and midline defects in conditional GSK-3β mice. Nature 446, 79–82 [DOI] [PubMed] [Google Scholar]
- 5. Barrell W. B., Szabo-Rogers H. L., and Liu K. J. (2012) Novel reporter alleles of GSK-3α and GSK-3β. PLoS One 7, e50422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Doble B. W., Patel S., Wood G. A., Kockeritz L. K., and Woodgett J. R. (2007) Functional redundancy of GSK-3α and GSK-3β in Wnt/β-catenin signaling shown by using an allelic series of embryonic stem cell lines. Dev. Cell 12, 957–971 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Frame S., Cohen P., and Biondi R. M. (2001) A common phosphate binding site explains the unique substrate specificity of GSK3 and its inactivation by phosphorylation. Mol. Cell 7, 1321–1327 [DOI] [PubMed] [Google Scholar]
- 8. Fiol C. J., Mahrenholz A. M., Wang Y., Roeske R. W., and Roach P. J. (1987) Formation of protein kinase recognition sites by covalent modification of the substrate: molecular mechanism for the synergistic action of casein kinase II and glycogen synthase kinase 3. J. Biol. Chem. 262, 14042–14048 [PubMed] [Google Scholar]
- 9. Zhang W., DePaoli-Roach A. A., and Roach P. J. (1993) Mechanisms of multisite phosphorylation and inactivation of rabbit muscle glycogen synthase. Arch Biochem. Biophys. 304, 219–225 [DOI] [PubMed] [Google Scholar]
- 10. Xu C., Kim N. G., and Gumbiner B. M. (2009) Regulation of protein stability by GSK3 mediated phosphorylation. Cell Cycle 8, 4032–4039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Kaidanovich-Beilin O., and Woodgett J. R. (2011) GSK-3: functional insights from cell biology and animal models. Front. Mol. Neurosci. 4, 40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Sutherland C. (2011) What are the bona fide GSK3 substrates? Int. J. Alzheimers Dis. 2011, 505607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. McManus E. J., Sakamoto K., Armit L. J., Ronaldson L., Shpiro N., Marquez R., and Alessi D. R. (2005) Role that phosphorylation of GSK3 plays in insulin and Wnt signalling defined by knockin analysis. EMBO J. 24, 1571–1583 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Ding V. W., Chen R. H., and McCormick F. (2000) Differential regulation of glycogen synthase kinase 3beta by insulin and Wnt signaling. J. Biol. Chem. 275, 32475–32481 [DOI] [PubMed] [Google Scholar]
- 15. Valvezan A. J., Zhang F., Diehl J. A., and Klein P. S. (2012) Adenomatous polyposis coli (APC) regulates multiple signaling pathways by enhancing glycogen synthase kinase-3 (GSK-3) activity. J. Biol. Chem. 287, 3823–3832 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Tran H., and Polakis P. (2012) Reversible modification of adenomatous polyposis coli (APC) with K63-linked polyubiquitin regulates the assembly and activity of the β-catenin destruction complex. J. Biol. Chem. 287, 28552–28563 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Piao S., Lee S. H., Kim H., Yum S., Stamos J. L., Xu Y., Lee S. J., Lee J., Oh S., Han J. K., Park B. J., Weis W. I., and Ha N. C. (2008) Direct inhibition of GSK3β by the phosphorylated cytoplasmic domain of LRP6 in Wnt/β-catenin signaling. PLoS One 3, e4046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Taelman V. F., Dobrowolski R., Plouhinec J. L., Fuentealba L. C., Vorwald P. P., Gumper I., Sabatini D. D., and De Robertis E. M. (2010) Wnt signaling requires sequestration of glycogen synthase kinase 3 inside multivesicular endosomes. Cell 143, 1136–1148 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Valvezan A. J., and Klein P. S. (2012) GSK-3 and Wnt signaling in neurogenesis and bipolar disorder. Front. Mol. Neurosci. 5, 1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Bain J., Plater L., Elliott M., Shpiro N., Hastie C. J., McLauchlan H., Klevernic I., Arthur J. S., Alessi D. R., and Cohen P. (2007) The selectivity of protein kinase inhibitors: a further update. Biochem. J. 408, 297–315 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Ong S. E., Blagoev B., Kratchmarova I., Kristensen D. B., Steen H., Pandey A., and Mann M. (2002) Stable isotope labeling by amino acids in cell culture, SILAC, as a simple and accurate approach to expression proteomics. Mol. Cell Proteomics 1, 376–386 [DOI] [PubMed] [Google Scholar]
- 22. Olsen J. V., Blagoev B., Gnad F., Macek B., Kumar C., Mortensen P., and Mann M. (2006) Global, in vivo, and site-specific phosphorylation dynamics in signaling networks. Cell 127, 635–648 [DOI] [PubMed] [Google Scholar]
- 23. Kim N. G., Xu C., and Gumbiner B. M. (2009) Identification of targets of the Wnt pathway destruction complex in addition to β-catenin. Proc. Natl. Acad. Sci. U.S.A. 106, 5165–5170 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. O'Shea J. P., Chou M. F., Quader S. A., Ryan J. K., Church G. M., and Schwartz D. (2013) pLogo: a probabilistic approach to visualizing sequence motifs. Nat. Methods 10, 1211–1212 [DOI] [PubMed] [Google Scholar]
- 25. Schwartz D., and Gygi S. P. (2005) An iterative statistical approach to the identification of protein phosphorylation motifs from large-scale data sets. Nat. Biotechnol. 23, 1391–1398 [DOI] [PubMed] [Google Scholar]
- 26. Dennis G. Jr., Sherman B. T., Hosack D. A., Yang J., Gao W., Lane H. C., and Lempicki R. A. (2003) DAVID: Database for Annotation, Visualization, and Integrated Discovery. Genome Biol. 4, P3. [PubMed] [Google Scholar]
- 27. Szklarczyk D., Franceschini A., Wyder S., Forslund K., Heller D., Huerta-Cepas J., Simonovic M., Roth A., Santos A., Tsafou K. P., Kuhn M., Bork P., Jensen L. J., and von Mering C. (2015) STRING v10: protein-protein interaction networks, integrated over the tree of life. Nucleic Acids Res. 43, D447–D452 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Beurel E., Grieco S. F., and Jope R. S. (2015) Glycogen synthase kinase-3 (GSK3): regulation, actions, and diseases. Pharmacol. Ther. 148, 114–131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Boyle W. J., Smeal T., Defize L. H. K., Angel P., Woodgett J. R., Karin M., and Hunter T. (1991) Activation of protein kinase C decreases phosphorylation of c-Jun at sites that negatively regulate its DNA-binding activity. Cell 64, 573–584 [DOI] [PubMed] [Google Scholar]
- 30. Troussard A. A., Tan C., Yoganathan T. N., and Dedhar S. (1999) Cell-extracellular matrix interactions stimulate the AP-1 transcription factor in an integrin-linked kinase- and glycogen synthase kinase 3-dependent manner. Mol. Cell. Biol. 19, 7420–7427 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Wei W., Jin J., Schlisio S., Harper J. W., and Kaelin W. G. Jr. (2005) The v-Jun point mutation allows c-Jun to escape GSK3-dependent recognition and destruction by the Fbw7 ubiquitin ligase. Cancer Cell 8, 25–33 [DOI] [PubMed] [Google Scholar]
- 32. Morton S., Davis R. J., McLaren A., and Cohen P. (2003) A reinvestigation of the multisite phosphorylation of the transcription factor c-Jun. EMBO J. 22, 3876–3886 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Lopez-Bergami P., Huang C., Goydos J. S., Yip D., Bar-Eli M., Herlyn M., Smalley K. S., Mahale A., Eroshkin A., Aaronson S., and Ronai Z. (2007) Rewired ERK-JNK signaling pathways in melanoma. Cancer Cell 11, 447–460 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Todd M. A., and Picketts D. J. (2012) PHF6 interacts with the nucleosome remodeling and deacetylation (NuRD) complex. J. Proteome Res. 11, 4326–4337 [DOI] [PubMed] [Google Scholar]
- 35. Wang J., Leung J. W., Gong Z., Feng L., Shi X., and Chen J. (2013) PHF6 regulates cell cycle progression by suppressing ribosomal RNA synthesis. J. Biol. Chem. 288, 3174–3183 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Schimizzi G. V., and Longmore G. D. (2015) Ajuba proteins. Curr. Biol. 25, R445–R446 [DOI] [PubMed] [Google Scholar]
- 37. Hirota T., Lipp J. J., Toh B. H., and Peters J. M. (2005) Histone H3 serine 10 phosphorylation by Aurora B causes HP1 dissociation from heterochromatin. Nature 438, 1176–1180 [DOI] [PubMed] [Google Scholar]
- 38. Chen X., Stauffer S., Chen Y., and Dong J. (2016) Ajuba phosphorylation by CDK1 promotes cell proliferation and tumorigenesis. J. Biol. Chem. 291, 14761–14772 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Fung K. Y., Dai L., and Trimble W. S. (2014) Cell and molecular biology of septins. Int. Rev. Cell Mol. Biol. 310, 289–339 [DOI] [PubMed] [Google Scholar]
- 40. Oh Y., and Bi E. (2011) Septin structure and function in yeast and beyond. Trends Cell Biol. 21, 141–148 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Heyd F., and Lynch K. W. (2010) Phosphorylation-dependent regulation of PSF by GSK3 controls CD45 alternative splicing. Mol. Cell 40, 126–137 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Ishigaki Y., Nakamura Y., Tatsuno T., Ma S., and Tomosugi N. (2015) Phosphorylation status of human RNA-binding protein 8A in cells and its inhibitory regulation by Magoh. Exp. Biol. Med. 240, 438–445 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Hsu I. W., Hsu M., Li C., Chuang T. W., Lin R. I., and Tarn W. Y. (2005) Phosphorylation of Y14 modulates its interaction with proteins involved in mRNA metabolism and influences its methylation. J. Biol. Chem. 280, 34507–34512 [DOI] [PubMed] [Google Scholar]
- 44. Kinoshita E., Yamada A., Takeda H., Kinoshita-Kikuta E., and Koike T. (2005) Novel immobilized zinc(II) affinity chromatography for phosphopeptides and phosphorylated proteins. J. Sep. Sci. 28, 155–162 [DOI] [PubMed] [Google Scholar]
- 45. Tarapore P., Shinmura K., Suzuki H., Tokuyama Y., Kim S. H., Mayeda A., and Fukasawa K. (2006) Thr199 phosphorylation targets nucleophosmin to nuclear speckles and represses pre-mRNA processing. FEBS Lett. 580, 399–409 [DOI] [PubMed] [Google Scholar]
- 46. Box J. K., Paquet N., Adams M. N., Boucher D., Bolderson E., O'Byrne K. J., and Richard D. J. (2016) Nucleophosmin: from structure and function to disease development. BMC Mol. Biol. 17, 19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Santamaria A., Wang B., Elowe S., Malik R., Zhang F., Bauer M., Schmidt A., Sillje H. H., Korner R., and Nigg E. A. (2011) The Plk1-dependent phosphoproteome of the early mitotic spindle. Mol. Cell Proteomics 10.1074/mcp.M110.004457 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Grosstessner-Hain K., Hegemann B., Novatchkova M., Rameseder J., Joughin B. A., Hudecz O., Roitinger E., Pichler P., Kraut N., Yaffe M. B., Peters J. M., and Mechtler K. (2011) Quantitative phospho-proteomics to investigate the polo-like kinase 1-dependent phospho-proteome. Mol. Cell Proteomics 10.1074/mcp.M111.008540 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Humphrey S. J., Yang G., Yang P., Fazakerley D. J., Stöckli J., Yang J. Y., and James D. E. (2013) Dynamic adipocyte phosphoproteome reveals that Akt directly regulates mTORC2. Cell Metab. 17, 1009–1020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Martinez N. M., Agosto L., Qiu J., Mallory M. J., Gazzara M. R., Barash Y., Fu X. D., and Lynch K. W. (2015) Widespread JNK-dependent alternative splicing induces a positive feedback loop through CELF2-mediated regulation of MKK7 during T-cell activation. Genes Dev. 29, 2054–2066 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Acebron S. P., and Niehrs C. (2016) β-Catenin-independent roles of Wnt/LRP6 signaling. Trends Cell Biol. 26, 956–967 [DOI] [PubMed] [Google Scholar]
- 52. Acebron S. P., Karaulanov E., Berger B. S., Huang Y. L., and Niehrs C. (2014) Mitotic wnt signaling promotes protein stabilization and regulates cell size. Mol. Cell 54, 663–674 [DOI] [PubMed] [Google Scholar]
- 53. Yin L., Wang J., Klein P. S., and Lazar M. A. (2006) Nuclear receptor Rev-erbalpha is a critical lithium-sensitive component of the circadian clock. Science 311, 1002–1005 [DOI] [PubMed] [Google Scholar]
- 54. Demarchi F., Bertoli C., Sandy P., and Schneider C. (2003) Glycogen synthase kinase-3β regulates NF-κ B1/p105 stability. J. Biol. Chem. 278, 39583–39590 [DOI] [PubMed] [Google Scholar]
- 55. Williams R. S. B., Eames M., Ryves W. J., Viggars J., and Harwood A. J. (1999) Loss of a prolyl oligopeptidase confers resistance to lithium by elevation of inositol (1,4,5) trisphosphate. EMBO J. 18, 2734–2745 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Saldi T., Cortazar M. A., Sheridan R. M., and Bentley D. L. (2016) Coupling of RNA polymerase II transcription elongation with pre-mRNA splicing. J. Mol. Biol. 428, 2623–2635 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Naftelberg S., Schor I. E., Ast G., and Kornblihtt A. R. (2015) Regulation of alternative splicing through coupling with transcription and chromatin structure. Annu. Rev. Biochem. 84, 165–198 [DOI] [PubMed] [Google Scholar]
- 58. Cole B. S., Tapescu I., Allon S. J., Mallory M. J., Qiu J., Lake R. J., Fan H. Y., Fu X. D., and Lynch K. W. (2015) Global analysis of physical and functional RNA targets of hnRNP L reveals distinct sequence and epigenetic features of repressed and enhanced exons. RNA 21, 2053–2066 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Ip J. Y., Tong A., Pan Q., Topp J. D., Blencowe B. J., and Lynch K. W. (2007) Global analysis of alternative splicing during T-cell activation. RNA 13, 563–572 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Hernández F., Pérez M., Lucas J. J., Mata A. M., Bhat R., and Avila J. (2004) Glycogen synthase kinase-3 plays a crucial role in tau exon 10 splicing and intranuclear distribution of SC35: implications for Alzheimer's disease. J. Biol. Chem. 279, 3801–3806 [DOI] [PubMed] [Google Scholar]
- 61. Hartmann B., Castelo R., Blanchette M., Boue S., Rio D. C., and Valcárcel J. (2009) Global analysis of alternative splicing regulation by insulin and wingless signaling in Drosophila cells. Genome Biol. 10, R11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Cancer Genome Atlas Research Network, Ley T. J., Miller C., Ding L., Raphael B. J., Mungall A. J., Robertson A., Hoadley K., Triche T. J. Jr., Laird P. W., Baty J. D., Fulton L. L., Fulton R., Heath S. E., Kalicki-Veizer J., et al. (2013) Genomic and epigenomic landscapes of adult de novo acute myeloid leukemia. N. Engl. J. Med. 368, 2059–2074 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Inoue D., Bradley R. K., and Abdel-Wahab O. (2016) Spliceosomal gene mutations in myelodysplasia: molecular links to clonal abnormalities of hematopoiesis. Genes Dev. 30, 989–1001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Yoshida K., Sanada M., Shiraishi Y., Nowak D., Nagata Y., Yamamoto R., Sato Y., Sato-Otsubo A., Kon A., Nagasaki M., Chalkidis G., Suzuki Y., Shiosaka M., Kawahata R., Yamaguchi T., et al. (2011) Frequent pathway mutations of splicing machinery in myelodysplasia. Nature 478, 64–69 [DOI] [PubMed] [Google Scholar]
- 65. Papaemmanuil E., Cazzola M., Boultwood J., Malcovati L., Vyas P., Bowen D., Pellagatti A., Wainscoat J. S., Hellstrom-Lindberg E., Gambacorti-Passerini C., Godfrey A. L., Rapado I., Cvejic A., Rance R., McGee C., et al. (2011) Somatic SF3B1 mutation in myelodysplasia with ring sideroblasts. N. Engl. J. Med. 365, 1384–1395 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Gelsi-Boyer V., Trouplin V., Adélaïde J., Bonansea J., Cervera N., Carbuccia N., Lagarde A., Prebet T., Nezri M., Sainty D., Olschwang S., Xerri L., Chaffanet M., Mozziconacci M. J., Vey N., and Birnbaum D. (2009) Mutations of polycomb-associated gene ASXL1 in myelodysplastic syndromes and chronic myelomonocytic leukaemia. Br. J. Haematol. 145, 788–800 [DOI] [PubMed] [Google Scholar]
- 67. Lee S. C., Dvinge H., Kim E., Cho H., Micol J. B., Chung Y. R., Durham B. H., Yoshimi A., Kim Y. J., Thomas M., Lobry C., Chen C. W., Pastore A., Taylor J., Wang X., Krivtsov A., Armstrong S. A., Palacino J., Buonamici S., Smith P. G., Bradley R. K., and Abdel-Wahab O. (2016) Modulation of splicing catalysis for therapeutic targeting of leukemia with mutations in genes encoding spliceosomal proteins. Nat. Med. 22, 672–678 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Guezguez B., Almakadi M., Benoit Y. D., Shapovalova Z., Rahmig S., Fiebig-Comyn A., Casado F. L., Tanasijevic B., Bresolin S., Masetti R., Doble B. W., and Bhatia M. (2016) GSK3 deficiencies in hematopoietic stem cells initiate pre-neoplastic state that is predictive of clinical outcomes of human acute leukemia. Cancer Cell 29, 61–74 [DOI] [PubMed] [Google Scholar]
- 69. Crews L. A., Balaian L., Delos Santos N. P., Leu H. S., Court A. C., Lazzari E., Sadarangani A., Zipeto M. A., La Clair J. J., Villa R., Kulidjian A., Storb R., Morris S. R., Ball E. D., Burkart M. D., and Jamieson C. H. (2016) RNA splicing modulation selectively impairs leukemia stem cell maintenance in secondary human AML. Cell Stem Cell 19, 599–612 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Adamia S., Haibe-Kains B., Pilarski P. M., Bar-Natan M., Pevzner S., Avet-Loiseau H., Lode L., Verselis S., Fox E. A., Burke J., Galinsky I., Dagogo-Jack I., Wadleigh M., Steensma D. P., Motyckova G., et al. (2014) A genome-wide aberrant RNA splicing in patients with acute myeloid leukemia identifies novel potential disease markers and therapeutic targets. Clin. Cancer Res. 20, 1135–1145 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Schlesinger A., Shelton C. A., Maloof J. N., Meneghini M., and Bowerman B. (1999) Wnt pathway components orient a mitotic spindle in the early Caenorhabditis elegans embryo without requiring gene transcription in the responding cell. Genes Dev. 13, 2028–2038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Inoki K., Ouyang H., Zhu T., Lindvall C., Wang Y., Zhang X., Yang Q., Bennett C., Harada Y., Stankunas K., Wang C. Y., He X., MacDougald O. A., You M., Williams B. O., and Guan K. L. (2006) TSC2 integrates Wnt and energy signals via a coordinated phosphorylation by AMPK and GSK3 to regulate cell growth. Cell 126, 955–968 [DOI] [PubMed] [Google Scholar]
- 73. Valvezan A. J., Huang J., Lengner C. J., Pack M., and Klein P. S. (2014) Oncogenic mutations in adenomatous polyposis coli (Apc) activate mechanistic target of rapamycin complex 1 (mTORC1) in mice and zebrafish. Dis. Model. Mech. 7, 63–71 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Fuentealba L. C., Eivers E., Ikeda A., Hurtado C., Kuroda H., Pera E. M., and De Robertis E. M. (2007) Integrating patterning signals: Wnt/GSK3 regulates the duration of the BMP/Smad1 signal. Cell 131, 980–993 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Huang Y. L., Anvarian Z., Döderlein G., Acebron S. P., and Niehrs C. (2015) Maternal Wnt/STOP signaling promotes cell division during early Xenopus embryogenesis. Proc. Natl. Acad. Sci. U.S.A. 112, 5732–5737 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Koch S., Acebron S. P., Herbst J., Hatiboglu G., and Niehrs C. (2015) Post-transcriptional Wnt signaling governs epididymal sperm maturation. Cell 163, 1225–1236 [DOI] [PubMed] [Google Scholar]
- 77. Top D., Harms E., Syed S., Adams E. L., and Saez L. (2016) GSK-3 and CK2 kinases converge on timeless to regulate the master clock. Cell Rep. 16, 357–367 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Stoleru D., Nawathean P., Fernández M. P., Menet J. S., Ceriani M. F., and Rosbash M. (2007) The Drosophila circadian network is a seasonal timer. Cell 129, 207–219 [DOI] [PubMed] [Google Scholar]
- 79. Noh K. T., Park Y. M., Cho S. G., and Choi E. J. (2011) GSK-3β-induced ASK1 stabilization is crucial in LPS-induced endotoxin shock. Exp. Cell Res. 317, 1663–1668 [DOI] [PubMed] [Google Scholar]
- 80. Fiol C. J., Wang A., Roeske R. W., and Roach P. J. (1990) Ordered multisite protein phosphorylation: analysis of glycogen synthase kinase 3 action using model peptide substrates. J. Biol. Chem. 265, 6061–6065 [PubMed] [Google Scholar]
- 81. Liu C., Li Y., Semenov M., Han C., Baeg G. H., Tan Y., Zhang Z., Lin X., and He X. (2002) Control of β-catenin phosphorylation/degradation by a dual-kinase mechanism. Cell 108, 837–847 [DOI] [PubMed] [Google Scholar]
- 82. Dajani R., Fraser E., Roe S. M., Yeo M., Good V. M., Thompson V., Dale T. C., and Pearl L. H. (2003) Structural basis for recruitment of glycogen synthase kinase 3β to the axin-APC scaffold complex. EMBO J. 22, 494–501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Dajani R., Fraser E., Roe S. M., Young N., Good V., Dale T. C., and Pearl L. H. (2001) Crystal structure of glycogen synthase kinase 3β: structural basis for phosphate-primed substrate specificity and autoinhibition. Cell 105, 721–732 [DOI] [PubMed] [Google Scholar]
- 84. Frame S., and Cohen P. (2001) GSK3 takes centre stage more than 20 years after its discovery. Biochem. J. 359, 1–16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Thingholm T. E., and Larsen M. R. (2016) The use of titanium dioxide for selective enrichment of phosphorylated peptides. Methods Mol. Biol. 1355, 135–146 [DOI] [PubMed] [Google Scholar]
- 86. Cox J., and Mann M. (2008) MaxQuant enables high peptide identification rates, individualized p.p.b.-range mass accuracies and proteome-wide protein quantification. Nat. Biotechnol. 26, 1367–1372 [DOI] [PubMed] [Google Scholar]
- 87. Cox J., Neuhauser N., Michalski A., Scheltema R. A., Olsen J. V., and Mann M. (2011) Andromeda: a peptide search engine integrated into the MaxQuant environment. J. Proteome Res. 10, 1794–1805 [DOI] [PubMed] [Google Scholar]
- 88. Shannon P., Markiel A., Ozier O., Baliga N. S., Wang J. T., Ramage D., Amin N., Schwikowski B., and Ideker T. (2003) Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res. 13, 2498–2504 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89. Dobin A., Davis C. A., Schlesinger F., Drenkow J., Zaleski C., Jha S., Batut P., Chaisson M., and Gingeras T. R. (2013) STAR: ultrafast universal RNA-seq aligner. Bioinformatics 29, 15–21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90. Vaquero-Garcia J., Barrera A., Gazzara M. R., González-Vallinas J., Lahens N. F., Hogenesch J. B., Lynch K. W., and Barash Y. (2016) A new view of transcriptome complexity and regulation through the lens of local splicing variations. eLife 5, e11752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91. Lawrence M., Huber W., Pagès H., Aboyoun P., Carlson M., Gentleman R., Morgan M. T., and Carey V. J. (2013) Software for computing and annotating genomic ranges. PLoS Comput. Biol. 9, e1003118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92. Anders S., and Huber W. (2010) Differential expression analysis for sequence count data. Genome Biol. 11, R106. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.