Abstract
Myosin-5B is a ubiquitous molecular motor that transports cargo vesicles of the endomembrane system in intracellular recycling pathways. Myosin-5B malfunction causes the congenital enteropathy microvillus inclusion disease, underlining its importance in cellular homeostasis. Here we describe the interaction of myosin-5B with F-actin, nucleotides, and the pyrazolopyrimidine compound myoVin-1. We show that single-headed myosin-5B is an intermediate duty ratio motor with a kinetic ATPase cycle that is rate-limited by the release of phosphate. The presence of a second head generates strain and gating in the myosin-5B dimer that alters the kinetic signature by reducing the actin-activated ADP release rate to become rate-limiting. This kinetic transition into a high-duty ratio motor is a prerequisite for the proposed transport function of myosin-5B in cellular recycling pathways. Moreover, we show that the small molecule compound myoVin-1 inhibits the enzymatic and functional activity of myosin-5B in vitro. Partial inhibition of the actin-activated steady-state ATPase activity and sliding velocity suggests that caution should be used when probing the effect of myoVin-1 on myosin-5–dependent transport processes in cells.
Keywords: actin, ATPase, inhibitor, kinetics, myosin, myoVin-1
Introduction
Double-headed class 5 myosins are evolutionary ancient, prototypic actin-based molecular motors implicated in the spatiotemporal segregation and the transport of organelles across phylogeny (1–10). A prerequisite for efficient transport is that a single motor can either take multiple discrete steps on an actin filament without detaching, a property also referred to as processivity, or that the motor works in an ensemble with other myosins-5 to efficiently move a cargo. These differences in physiological capacities and limitations are directly linked to the mechanoenzymatic signatures of class 5 myosins (11, 12).
Mammals express three myosin-5 genes (MYO5A, MYO5B, and MYO5C) in almost all cells (6, 13). Compared with the well studied paralog myosin-5A that exhibits single molecule processivity and the nonprocessive myosin-5C, little is known about the kinetic features of myosin-5B (14–18). Myosin-5B is best known for the transport of Rab GTPase positive vesicles of the endomembrane system along the F-actin cytoskeleton to the plasma membrane (4, 19–21). The long-range vesicle transport system is a prerequisite for diverse physiological processes including axon development, postsynaptic plasticity, cell polarization, and the prevention of polyspermy (19, 22–25). The mutation-induced loss of function of MYO5B is manifested in debilitating pathologies including microvillus inclusion disease that results in death in neonates and children (20, 26, 27). As reviewed recently, the majority of the >40 disease-associated mutations in MYO5B cluster in the coding region for the motor and the tail domain of the myosin heavy chain (26). The motor domain harbors a prototypic nucleotide-binding pocket and a binding region for F-actin (Fig. 1A). The tail domain contains a coiled-coil region that promotes the dimerization of two myosin heavy chains and terminates in a distal globular tail domain that constitutes the binding site for Rab GTPase family members and other adaptor proteins (4, 13, 25, 28). Both domains are separated by the neck that contains six IQ motifs, predicted binding regions for calmodulin (CaM)3 or calmodulin-like light chains (Fig. 1A) (29, 30). The distribution of mutations in the myosin heavy chain suggests that either (i) the nucleotide-dependent interaction between the motor domain and the actin filament may be altered or (ii) the interaction between the molecular motor and its cargo vesicle may be altered. Both scenarios are expected to result in the aberrant vesicle trafficking patterns in patients with microvillus inclusion disease (31, 32).
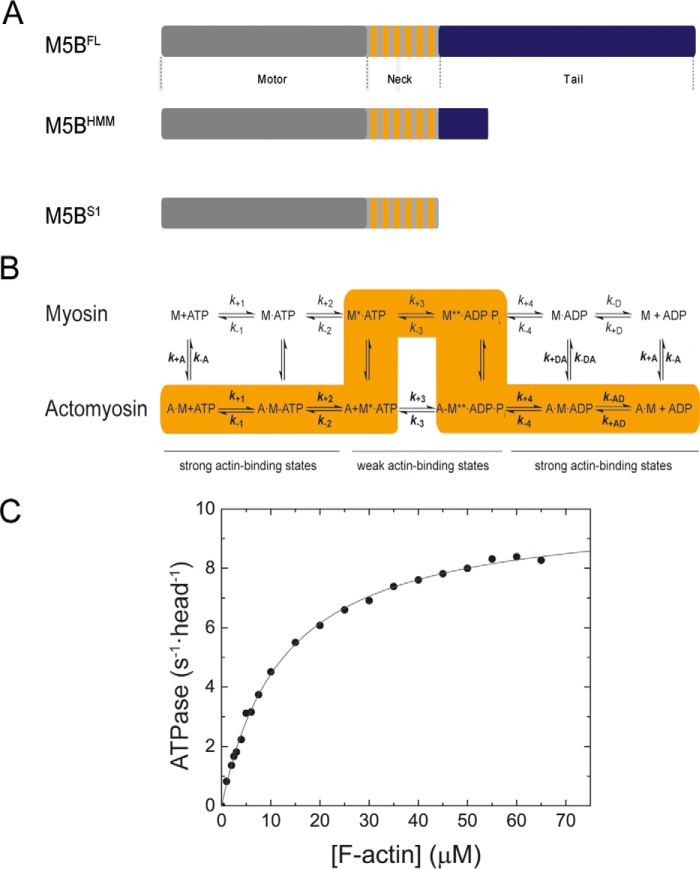
Figure 1.
Domain organization, kinetic scheme, and steady-state ATPase activity of M5BS1. A, domain organization of myosin-5B and expression constructs used in this work. The myosin motor domain in the myosin heavy chain is shown in gray. The six IQ motifs in the myosin neck domain are shown in orange, and the tail domain is in blue. The tail domain mediates the dimerization of M5BFL and M5BHMM by means of a coiled-coil and interacts with binding partners. B, simplified kinetic scheme of the myosin and actomyosin ATPase cycle. The events of ATP binding, ATP hydrolysis, and phosphate release are shown for myosin in the actin detached (top row) and attached (bottom row) states. The main flux through the pathway is highlighted in orange, and the weak and strong actin-binding states are indicated. The notation distinguishes between the kinetic constants in the presence and absence of F-actin by using regular versus bold type; subscripts A and D refer to F-actin (KA) and ADP (KD), respectively. Dissociation equilibrium constants were calculated as Kx = k−x/k+x. M, myosin; A, actin. C, steady-state ATPase activity of M5BS1. Increasing concentrations of F-actin activate the steady-state ATPase activity to a kcat of 10.0 ± 0.1 s−1 with a Kapp of 12.6 ± 0.4 μm.
To understand the mechanoenzymatic basis of the actin-based transport powered by myosin-5B in normalcy, we studied the interaction between the motor, nucleotides, and F-actin (Fig. 1B) with ensemble solution kinetics and functional assays in vitro. Our experiments show that single-headed myosin-5B has an intermediate duty ratio and that a second head is required to establish a high-duty ratio, a kinetic prerequisite for single molecule processivity. The data also allows for the comparative analysis of kinetic signatures in the class 5 myosome. Moreover, we show that the small molecule myoVin-1 inhibits the enzymatic and functional activity of myosin-5B in vitro. The observed effects resemble the previously described effect on myosin-5A and suggest that myoVin-1 may inhibit all class 5 myosins.
Results
Design of expression constructs for kinetic experiments
We recombinantly overproduced and purified the single-headed M5BS1 that contains all six IQ motifs and the double-headed M5BHMM (Fig. 1A) in complex with the light chain CaM in milligram quantities in the baculovirus/Sf9 insect cell system. The proteins allowed us to directly compare the mechanoenzymatic properties of single-headed and double-headed myosin-5B constructs and their interaction with nucleotides, F-actin, and the small molecule myoVin-1.
Enzymatic activity of M5BS1 under steady-state conditions
We first determined the enzymatic activity of M5BS1 with an NADH-coupled assay under steady-state conditions. As shown in Fig. 1C, F-actin potently activates the basal ATPase activity of M5BS1 from a kbasal of 0.07 ± 0.01 s−1 more than 2 orders of magnitude. Fitting a Michaelis–Menten equation
| (1) |
to the data results in a kcat of 10.0 ± 0.1 s−1. The Kapp is 12.6 ± 0.4 μm, the catalytic efficiency kcat/Kapp ∼0.8 μm−1 s−1. The steady-state kinetic parameters of M5BS1 are compiled in Table 1 and compared with previously studied class 5 myosins.
Table 1.
Steady-state kinetic parameters of mouse M5BS1 in comparison with previously characterized class-5 myosins
ND, not determined.
| Steady-state ATPase parameter | Signal or calculation | Mm M5BS1a | Gg M5AS1 (35) | Hs M5CS1 (16) | Dm M5S1 (47) |
|---|---|---|---|---|---|
| kbasal (s−1) | NADH assay | 0.07 ± 0.01 | 0.03 | 0.05 ± 0.01 | ND |
| kcat (s−1) | NADH assay | 10.0 ± 0.1 | 15 | 1.8 ± 0.3 | 12.5 ± 0.8 |
| Kapp (μm) | NADH assay | 12.6 ± 0.4 | 1.4 | 42.5 ± 10.6 | 9.9 ± 0.6 |
| kcat/Kapp (μm−1 s−1) | NADH assay | ∼0.8 | ∼11 | ∼0.04 | ∼1.3 |
a In this study.
ATP binding to myosin
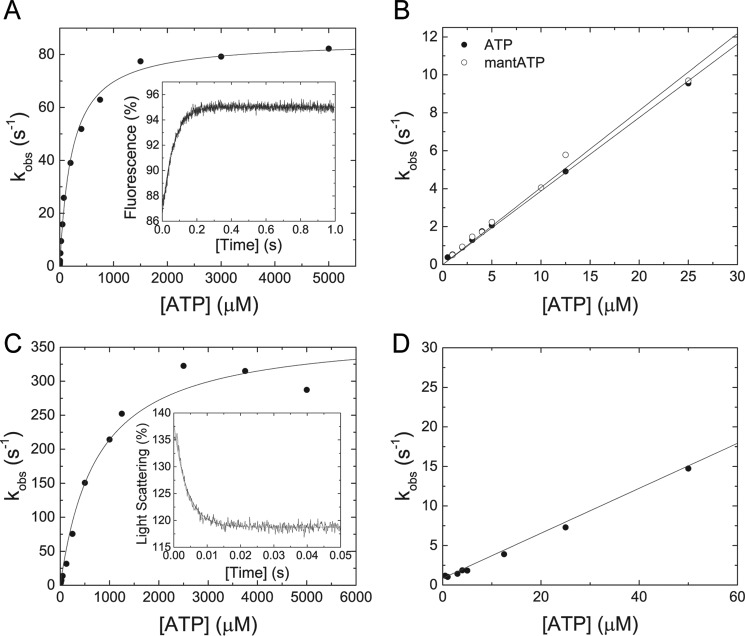
To determine the rate-limiting step of the M5BS1 kinetic cycle, we measured the individual parameters that describe the events of ATP binding, ATP hydrolysis and product release in the F-actin attached and detached states (Fig. 1B) in a stopped-flow spectrophotometer. As shown in the inset of Fig. 2A, ATP binding to M5BS1 causes a transient increase in the intrinsic tryptophan fluorescence that is best described by a single exponential fit. The observed rate constants kobs, obtained by varying the ATP concentration, follow a hyperbolic dependence according to
| (2) |
and approach a maximum kmax value of 85.4 ± 1.5 s−1, representing the ATP hydrolysis rate k+3 + k−3. Half-saturation (1/K0.5) is observed at an ATP concentration of 225.1 ± 14.7 μm (Fig. 2A). At low ATP concentrations, the dependence of the observed rate constant on ATP is linear. The slope of the straight line yields the apparent second-order rate constant for ATP binding, K1k+2 = 0.39 ± 0.01 μm−1·s−1 (Fig. 2B). An identical value of 0.4 ± 0.01 μm−1 s−1 is obtained after binding of the nucleotide analog mantATP to M5BS1 under pseudo-first order conditions. Fig. 2B shows the binding data of both nucleotides up to a concentration of 25 μm. All transient-state kinetic parameters of M5BS1 are compiled in Table 2 and compared with previously studied single-headed myosins-5.
Figure 2.
Transient kinetic interaction between MB5S1 and actoM5S1 with ATP and ATP analogs. A, the observed rate constant kobs depends hyperbolically on the ATP concentration and converges a value k+3 + k−3 of 85.4 ± 1.5 s−1. Half-saturation (1/K0.5) is reached at 225.1 + 14.7 μm ATP. Inset, the transient fluorescence increase represents the interaction between 50 μm ATP and 0.5 μm M5BS1 and has an observed rate constant kobs of 15.84 s−1. B, linear dependence of the observed rate constant kobs on [ATP] at low nucleotide concentrations. Binding of ATP or the nucleotide analog mantATP results in identical second-order rate constants of 0.39 ± 0.01 and 0.4 ± 0.01 μm−1 s−1, respectively. C, in the presence of F-actin, the observed rate constants kobs of the ATP-induced dissociation of the actomyosin complex show a hyperbolic dependence on the ATP concentration and converge to a value k+2 of 373.5 ± 18.7 s−1 with half-saturation (1/K0.5 = 1/K1) at 745.6 ± 120.7 μm ATP. Inset, time-dependent decrease in light scattering signal upon mixing 0.3 μm M5BS1 and 0.5 μm F-actin with 1250 μm ATP. The transient is best described with a single exponential fit with a kobs of 252.22 s−1. D, the observed rate constant kobs of the ATP-induced dissociation of the actomyosin complex linearly depends on the ATP concentration up to 50 μm substrate, defining K1k+2 to 0.28 ± 0.01 μm−1 s−1.
Table 2.
Transient state kinetic parameters of mouse M5BS1 in comparison with previously characterized class-5 myosins
Numbering of the kinetic constants refers to Fig. 1B. ND, not determined. NA, not experimentally accessible.
| Parameter | Signal or calculationa | Mm M5BS1 | Gg M5AS1 (35) | Hs M5CS1 (16) | Dm M5S1 (47) |
|---|---|---|---|---|---|
| ATP interaction | |||||
| K1k+2 (μm−1 s−1) | Tryptophan | 0.39 ± 0.01 | 1.6 ± 0.2 | ND | 1.31 ± 0.52 |
| K1k+2 (μm−1 s−1) | mantATP | 0.40 ± 0.01 | 1.5 ± 0.2 | ND | 1.31 ± 0.52 |
| 1/K0.5 (μm) | Tryptophan | 225.1 ± 14.7 | ND | ND | ND |
| k3 + k−3 (s−1) | Tryptophan | 85.4 ± 1.4 | 750 ± 45 | 90 | 68 ± 4.7 |
| K1k+2 (μm−1 s−1) | Light scattering | 0.28 ± 0.01 | 0.9 | 0.82 ± 0.03 | 0.36 ± 0.006 |
| 1/K0.5 (μm) | Light scattering | 745.6 ± 120.7 | ND | ND | >500 |
| k+2 (s−1) | Light scattering | 373.5 ± 18.7 | 870 | 287 | >180 |
| ADP interaction | |||||
| k+D (μm−1 s−1) | mantADP | 3.05 ± 0.16 | 4.6 | ND | 2.2 ± 0.3 |
| k−D (s−1) | mantADPb | 14.3 ± 1.61 | ND | ND | 37.9 ± 2.4 |
| k−D (s−1) | mantADPc | 18.0 ± 0.29 | 1.2 | ND | 38.3 ± 5.7 |
| k−D (s−1) | Tryptophanc | 18.9 ± 0.09 | ND | ND | ND |
| KD (μm) | k−D/k+D | 4.7–6.2 | 0.27 | ND | 17 |
| k+D (μm−1 s−1) | mantADP | 2.71 ± 0.11 | 12.6 | 56–72 | 4.7 |
| k−AD (s−1) | mantADPb | 21.9 ± 0.9 | ND | ND | ND |
| k−AD (s−1) | Light scatteringc | 17.13 ± 0.14 | 16 | 15.8 ± 0.8 | ND |
| k−AD (s−1) | mantADPc | 16.69 ± 0.11 | 12 | 15.6 ± 0.6 | 150 |
| KAD (μm) | k−AD/k+AD | 6.1–8.1 | 0.93 | 0.25 ± 0.02 | 32.54 |
| Actin interaction | |||||
| k+A (μm−1 s−1) | Light scatteringd | 8.39 ± 0.16 | 73 | 0.66 ± 0.02 | 2.5 ± 0.29 |
| k−A (s−1) | Pyrene-actind | NA | 0.00036 | 0.019 | 0.04 ± 0.001 |
| KA (nm) | k−A/k+A | NA | ND | 29 | 16 |
| k+AD (μm−1 s−1) | Light scattering | 5.98 ± 0.19 | 4.2 | 1.17 ± 0.03 | 2.3 ± 0.14 |
| k−AD (s−1) | Pyrene-actind | NA | 0.032 | 0.051 | 0.43 ± 0.01 |
| KAD (nm) | k−AD/k+AD | NA | ND | 0.044 | 190 |
a In this study.
b From y intercept.
c From chasing experiment.
d Samples were treated with apyrase prior to the assay.
ATP binding to actomyosin
The time-dependent change in light scattering signal was used to assay the interaction between actomyosin and ATP. Mixing of 0.3 μm actoM5BS1 with excess ATP results in a single exponential decrease in light scattering signal, as shown in the inset of Fig. 2C. The observed rate constant kobs values follow a hyperbolic dependence as a function of ATP that is best described by the following equation.4
| (3) |
The fit converges toward a maximum k+2 value of 373.5 ± 18.7 s−1. Half-saturation (1/K0.5 = 1/K1) is observed at 745.6 ± 120.7 μm ATP (Fig. 2C). At low [ATP] up to 50 μm, the data set is best fit with a linear function, representing the apparent second-order binding rate constant K1k+2 = 0.28 ± 0.01 μm−1 s−1 (Fig. 2D).
ADP binding and release from myosin
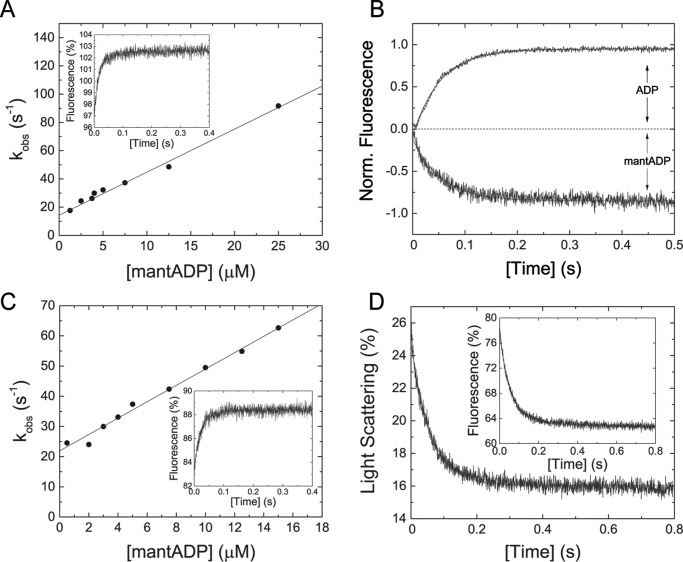
Binding kinetics of the fluorescent nucleotide analog mantADP to M5BS1 were determined in a stopped-flow spectrophotometer. Mixing of 0.5 μm M5BS1 under pseudo-first order conditions with mantADP results in a single exponential fluorescence increase (Fig. 3A, inset). A linear fit to the data set according to
| (4) |
results in a second-order binding rate constant k+D of 3.05 ± 0.16 μm−1 s−1 (Fig. 3A). Extrapolation of the fit to [mantADP] = 0 determines the ADP release rate constant k−D = 14.3 ± 1.61 s−1. This value is an order of magnitude higher than for previously characterized class 5A myosins (Table 2). As an independent measure, we directly determined k−D in chasing experiments in which mantADP is displaced from the myosin·mantADP complex with excess ATP. Independently, we measured the ADP release rate constant from the myosin·ADP complex when chased with excess ATP. The latter experimental design uses the intrinsic tryptophan fluorescence of myosin as readout. Both experimental setups result in almost identical values for k−D of 18.0 ± 0.29 s−1 (mantADP) and 18.9 ± 0.09 s−1 (ADP) that are in good agreement to the determined value from the binding data (k−D = 14.3 ± 1.61 s−1) (Fig. 3B and Table 2). From the ratio of the ADP release and binding rate constants (KD = k−D/k+D), the dissociation equilibrium constant of ADP was calculated to KD = 4.7–6.2 μm.
Figure 3.
Transient kinetic interaction between M5S1 and actoM5S1 with ADP and ADP analogs. A, the observed rate constants upon rapidly mixing M5BS1 and mantADP follow a linear dependence in the concentration range tested. The straight line describes the second-order rate constant for ADP-binding k+D to 3.05 ± 0.16 μm−1 s−1. From the y intercept, the ADP release rate k−D was determined to be 14.3 ± 1.61 s−1. Inset, representative fluorescence increase after mixing 0.5 μm M5BS1 and 12.5 μm mantADP in a stopped-flow spectrophotometer. Single exponential fit to the transient results in a kobs of 48.65 s−1. B, ADP release kinetics from M5BS1. Shown are the normalized data of the time-dependent increase in the intrinsic fluorescence signal after mixing 0.5 μm M5BS1 in the presence of a 25 μm ADP with 1.5 mm ATP. The transient was fit to a single exponential, yielding the ADP release rate constant k−D = 18.9 s−1. Similar, rapidly mixing 0.5 μm M5BS1 in the presence of a 50-fold molar excess of ADP (5 μm mantADP and 20 μm ADP) with 1.5 mm ATP resulted in a k−D of 18.0 s−1. C, mantADP dependence of the observed rate constant kobs after mixing with actoM5BS1 under pseudo-first order conditions. The solid line describes the second-order binding rate constant k+AD = 2.71 ± 0.11 μm−1 s−1 and the ADP release rate constant k−AD = 21.9 ± 0.9 s−1. Inset, time-dependent fluorescence increase after mixing 0.5 μm actoM5BS1 with 7.5 μm mantADP. Single exponential fit yields a kobs = 42.39 s−1. D, dissociation of 0.5 μm actomyosin in the presence of 40 μm ADP with 2 mm ATP. A single exponential fit to the time course of the light scattering signal yields a k−AD of 17.13 s−1. Inset, representative transient of the interaction between 0.5 μm actomyosin and 40 μm ADP (2.5 μm mantADP and 37.5 μm ADP) with 2 mm ATP. A k−AD of 16.69 s−1 was determined from a single exponential fit to the data.
ADP binding to actomyosin
ADP binding to the actomyosin complex was probed by mixing mantADP under pseudo-first order conditions with actoM5BS1. The associated time-dependent fluorescence increase is best described by a single exponential fit (Fig. 3C). The plot of the observed rate constants kobs versus [mantADP] follows a linear trend that is best described by the following equation.
| (5) |
The slope of the line defines the ADP-binding rate constant k+AD = 2.71 ± 0.11 μm−1 s−1 (Fig. 3C). The ADP release rate constant k−AD was extracted at [mantADP] = 0 to 21.9 ± 0.9 s−1 (Fig. 3C).
ADP release from actomyosin
The ADP release rate constant k−AD was directly determined by chasing the actomyosin·ADP complex with excess ATP. Two different experimental strategies were employed. First, mantADP was chased from actomyosin. Second, the change in light scattering upon the ATP-induced dissociation of the actomyosin·ADP complex was detected. In this indirect experimental setup, the ADP release limits the dissociation of the actomyosin complex. The former results in an ADP release rate k−AD of 16.69 ± 0.11 s−1, and the latter results in a k−AD of 17.13 ± 0.14 s−1 (Fig. 3D). These independent measurements suggest that the fast ADP release rate is a kinetic signature of actoM5BS1. The kinetic coupling ratio, the efficiency of F-actin to activate the ADP release, k−AD/k−D = ∼1–1.5, is low. Accordingly, the dissociation equilibrium constant KAD = k−AD/k+AD = 6.1–8.1 μm is similar to the respective constant in the absence of F-actin (KD = 4.7–6.2 μm; Table 2).
Interaction with F-actin
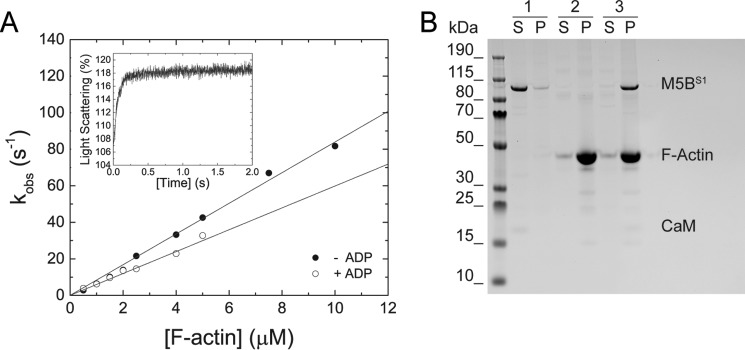
The binding kinetics of M5BS1 to F-actin in the presence and absence of excess ADP was probed by light scattering. The transients are best described with single exponential fits as exemplarily shown in the inset of Fig. 4A. The obtained kobs increase linearly within the F-actin concentration range tested. Linear fit of the data to
| (6) |
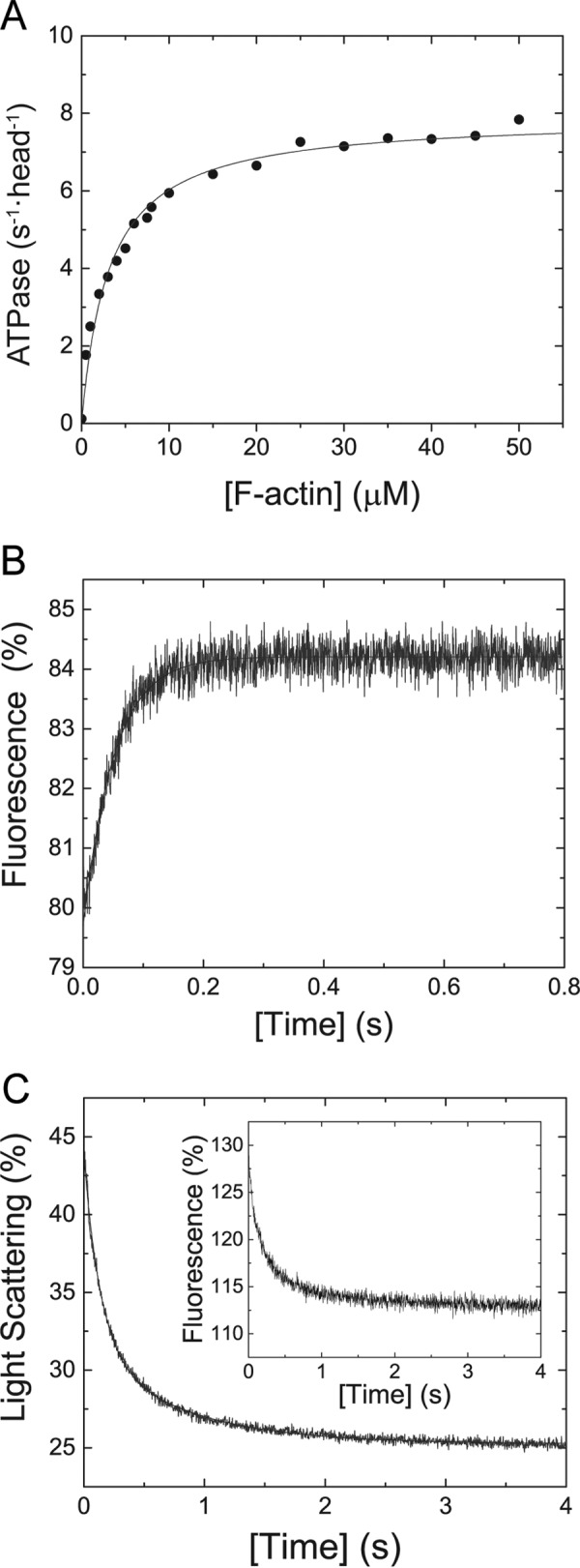
results in the second-order binding rate constants k+A of 8.39 ± 0.16 μm−1 s−1 and k+DA of 5.98 ± 0.19 μm−1 s−1 in the absence and presence of ADP, respectively (Fig. 4A). The dissociation rate constants k−A and k−DA are close to 0 and do not allow for a firm determination of the respective parameters (Fig. 4A). M5BS1 does not significantly quench pyrene fluorescence upon binding to pyrene-labeled F-actin. This observation precludes the use of this fluorescence label to directly determine the F-actin affinities in the presence and absence of ADP and the F-actin release rate constants, respectively. It is of note that the inability of M5BS1 to quench pyrene fluorescence is not an indication that the actomyosin complex is not formed, because a light scattering signal is observed under identical assay conditions (Fig. 4A, inset). Concomitantly, we find that the actoM5BS1 complex is formed under identical buffer conditions in high-speed cosedimentation assays (Fig. 5B).
Figure 4.
Transient kinetic interaction between M5BS1 and F-actin in the presence and absence of ADP. A, increasing concentrations of F-actin were mixed with M5BS1 in a stopped-flow spectrophotometer in the presence or absence of saturating ADP. Linear fits to the data sets describe the second-order binding rate constants k+A and k+DA to 8.39 ± 0.16 and 5.98 ± 0.19 μm−1 s−1, respectively. Inset, time-dependent change in light scattering after rapidly mixing 0.5 μm M5BS1 with 4 μm F-actin in the presence of 80 μm ADP. Single exponential fit to the data set results in a kobs of 13.61 s−1. B, a cosedimentation assay shows that an actomyosin complex is formed in SF buffer. Sample 1, 2 μm M5BS1 (control); sample 2, 10 μm F-actin (control); sample 3, 2 μm M5BS1 and 10 μm F-actin. Abbreviations S and P refer to supernatant and pellet, respectively.
Figure 5.
Steady-state ATPase activity and key kinetic parameters of M5BHMM. A, the Michaelis–Menten parameters kcat = 7.91 ± 0.15 s−1 and Kapp = 3.12 ± 0.26 μm for M5BHMM were determined under steady-state conditions for M5BHMM. B, the transient fluorescence increase shows the interaction of 0.25 μm M5BHMM in the presence of 80 μm ADP after rapid mixing with 2 mm ATP. Single exponential fit to the data set yields a k−D,HMM of 19.53 s−1. C, dissociation of the actoM5BHMM·ADP complex (0.25 μm M5BHMM, 0.5 μm F-actin, 40 μm ADP) with 2 mm ATP results in a double-exponential decrease in light scattering signal (k−AD,HMM,fast = 7.24 s−1, k−AD,HMM,slow = 1.27 s−1). The fast phase has an amplitude of −12.4%, the slow phase an amplitude of −6.19%. Inset, similar ADP release rates and amplitudes (k−AD,HMM,fast = 7.1 s−1, k−AD,HMM,slow = 1.1 s−1, Afast = −10.67%, Aslow = −4.4%) are obtained after rapidly mixing the actoM5BHMM·mADP complex (0.25 μm M5BHMM, 0.5 μm F-actin, 1.25 μm mADP, 18.75 μm ADP) with 1 mm ATP.
Duty ratio of M5BS1
The steady-state and transient kinetic characterization of single-headed M5BS1 allowed us to calculation the duty ratio, the time of its kinetic cycle M5BS1 spends strongly bound to actin (Fig. 1B). Values of ∼0.46–0.6 were obtained with the following equation
| (7) |
based on the determined kinetic constants k−AD = 16.69 ± 0.11–21.9 ± 0.9 s−1 and kcat = 10.0 ± 0.1 s−1 (Tables 1 and 2) under saturating conditions. It is of note that this equation neglects that the duty ratio depends on [F-actin] and the [ATP] concentration (29, 33, 34).
The duty ratio values obtained for M5BS1 are lower than for single-headed constructs of other myosin-5 paralogs and unconventional myosins associated with cellular transport function (35–37). A duty ratio >0.5/head is a theoretical cutoff value and a minimum requirement for processivity, but duty ratios >0.95 are required for efficient single molecule processivity of a double-headed myosin (11, 35, 38). Calculation of the duty ratio of a myosin-5B dimer (39, 40) based on
| (8) |
neglecting any steric factors, gating, and strain between the two motor domains results in values of 0.71–0.84.
Steady-state kinetic properties and key transient-state kinetic parameters of M5BHMM
The steady- and transient-state kinetic parameters of M5BS1 measured in the absence of load indicate that the motor is unlikely to function as an efficient cargo transporter inside cells (Tables 1 and 2). This finding conflicts with the described physiological function of myosin-5B as transporter of membrane vesicles along the F-actin cytoskeleton (4, 19–21). We therefore tested whether a second head, as it is found in the myosin-5B dimer inside the cell, would change the enzymatic signatures and render the molecule a high-duty ratio motor. First, we measured the kinetic properties of M5BHMM under steady-state conditions. F-actin activates the basal ATPase activity kbasal from 0.12 ± 0.02 s−1 ∼65-fold to a maximum kcat value of 7.91 ± 0.15 s−1 per head (Fig. 5A), a 20% reduction in kcat when compared with M5BS1 (Tables 2 and 3). The Kapp is with 3.12 ± 0.26 μm 60% lower than the respective parameter obtained with M5BS1, the catalytic efficiency kcat/Kapp with ∼2.5 elevated (Tables 1 and 3).
Table 3.
Steady-state and key transient-state kinetic parameters of mouse M5BHMM in comparison with previously characterized class-5 myosins
Numbering of the transient-state kinetic constants refers to Fig. 1B. ND, not determined.
| Parameter | Signal or calculationa | Mm M5BHMM | Mm/Gg M5AHMM (80, 81) | Hs M5CHMM (15) | Dm M5HMM (47) |
|---|---|---|---|---|---|
| Steady-state ATPase | |||||
| kbasal (s−1) | NADH assay | 0.12 ± 0.02 | ND | ND | ND |
| kcat (s−1) | NADH assay | 7.91 ± 0.15 | 3.8 ± 0.6 | 2.31 ± 0.55 | 12.6 ± 0.5 |
| Kapp (μm) | NADH assay | 3.12 ± 0.26 | 0.15 ± 0.06 | 5.1 ± 1.8 | 10.8 ± 1.2 |
| kcat/Kapp (μm−1 s−1) | NADH assay | ∼2.5 | ∼25 | ∼0.45 | ∼1.2 |
| ADP interaction | |||||
| k−D,HMM (s−1) | ADPb | 19.53 ± 0.25 | ND | ND | ND |
| k−AD,HMM,fast (s−1) | Light scatteringb | 7.24 ± 0.1 | 42 ± 9 | 5.5 ± 0.2 | ND |
| k−AD,HMM,slow (s−1) | Light scatteringb | 1.27 ± 0.02 | ND | 0.026 ± 0.9 | ND |
a In this study.
b From chasing experiment.
The difference in kcat must be reflected in a transient kinetic parameter that rate-limits the actomyosin ATPase cycle and led us to test whether the presence of a second head would influence the ADP release rate from M5BHMM. As shown in Fig. 5B, the ADP release rate from M5BHMM in the presence of a 320-fold molar excess ADP (k−D = 19.53 ± 0.25 s−1; Fig. 5B) is identical to the parameter measured for single-headed M5BS1 (k−D = 18.9 ± 0.09 s−1) in the absence of F-actin (Fig. 3D and Table 2). Moreover, the fluorescence transient is best described by a single exponential, indicating that the presence of a second head per se does not affect the kinetics in the absence of F-actin. When bound to F-actin, the ADP release rate from M5BHMM and the transient change in light scattering signal is double exponential with a fast rate k−AD,HMM,fast of 7.24 s−1 and a slow phase k−AD,HMM,slow of 1.27 s−1 (Fig. 5C). The fast rate is in good agreement with the titrated kcat of 7.91 ± 0.15 s−1 obtained in the steady-state ATPase assay (Fig. 5A and Table 3) and probably represents the ADP release rate from the positively strained trail head, and the slow rate probably represents the ADP release rate from the negatively strained lead head (18). Taken together, the steady-state and transient-state kinetic experiments indicate that the actin-activated ADP rate-limits the M5BHMM but not the M5BS1 kinetic cycle. Accordingly, the duty ratio of the lead head is high and approaches unity based on the calculation according to Equation 7 with the measured rates k−AD,HMM,fast = 7.24 ± 0.1 s−1 and kcat = 7.91 ± 0.15 s−1.
Chemical inhibition of M5BS1 with myoVin-1
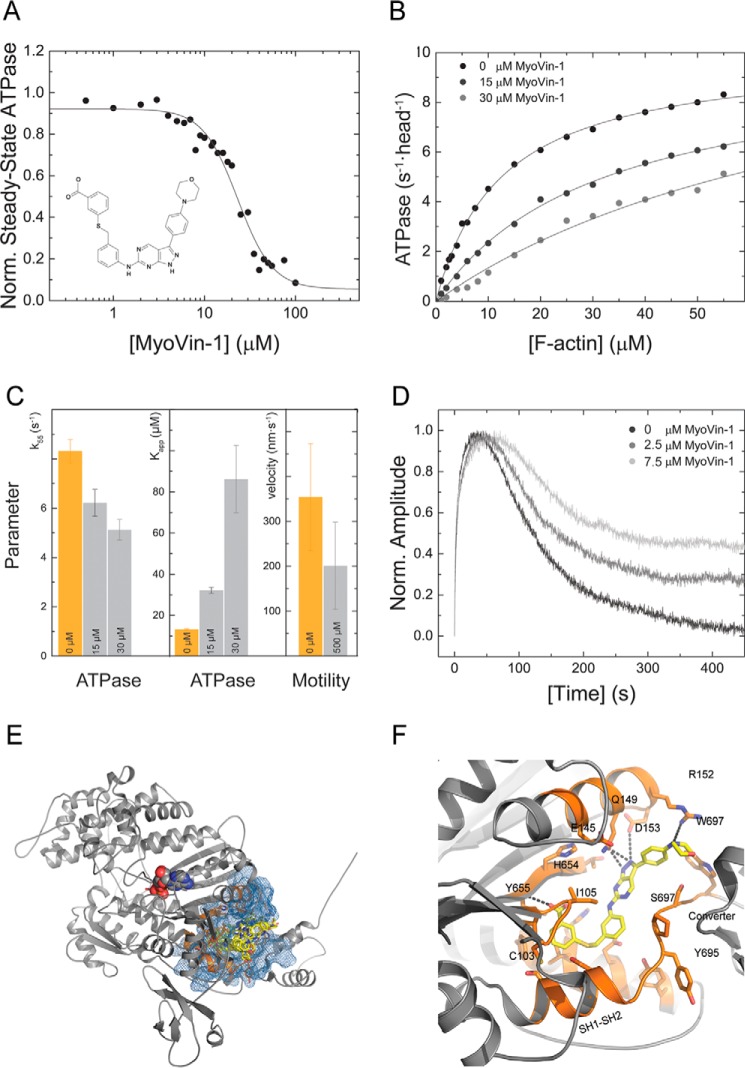
Our detailed kinetic characterization of M5BS1 and M5BHMM allowed us to probe the effect of the small molecule inhibitor myoVin-1 on its mechanoenzymatic properties. The pyrazolopyrimidine compound (Fig. 6A) was previously identified as an uncompetitive inhibitor for myosins-5A in vitro and has been used in cell biological studies to probe for myosin-5 motor function (41–45). To address whether the small molecule has selectivity for myosin-5A, we measured its effect in the actin-activated steady-state ATPase activity of M5BS1. The addition of myoVin-1 to M5BS1 leads to a partial inhibition of the steady-state ATPase activity at a fixed F-actin concentration of 30 μm. The KI, obtained after fitting the data, normalized to the ATPase activity in the absence of inhibitor, to a dose-response function is 23.12 ± 2.07 μm.
Figure 6.
Chemical inhibition of M5BS1 with the small molecule myoVin-1. A, myoVin-1 inhibits the steady-state ATPase activity of M5BS1 at a fixed actin concentration of 30 μm in a dose-dependent manner with a KI of 23.12 ± 2.07 μm. The data are normalized to the uninhibited control. The structure of the pyrazolopyrimidine compound myoVin-1 is shown in the inset. B, myoVin-1 changes the kcat and the Kapp of the M5BS1 actin-activated ATPase activity in a dose-dependent manner. C, effect of myoVin-1 on the steady-state ATPase activity of at 55 μm F-actin (k55) and Kapp of M5BS1 and gliding velocity. Orange bars represent the uninhibited control (0 μm myoVin-1). For k55 and Kapp, light gray bars represent the measured parameters in the presence of 15 and 30 μm myoVin-1 as indicated. For the in vitro motility data, the light gray bar represents the actin gliding velocity in the presence of 500 μm myoVin-1 as indicated. D, single-turnover measurements (0.25 μm M5BS1 and 0.15 μm mantATP) in the presence of 0, 2.5, and 7.5 myoVin-1. The data show that myoVin-1 predominantly decreases the release of the mantATP hydrolysis products in a concentration-dependent manner. E, favorable binding pocket of myoVin-1 in the homology model of the human myosin-5B motor domain. The site is located at a distance of 17 Å to the nucleotide-binding pocket. The myosin-5B motor domain is shown in gray cartoon representation, the nucleotide is in spheres, and the predicted myoVin-1 binding pocket is in orange. The pharmacophore is highlighted in blue, and myoVin-1 is in yellow stick representation. The best pose has a binding energy of −10.5 kcal/mol. Of the 10 best scored poses, 8 ligands bind to the highlighted binding pocket and vary in their binding energy by only 1 kcal/mol. F, close-up view of the best binding conformation of myoVin-1 in the predicted binding pocket. The small molecule is coordinated by residues of the seven-stranded β-sheets of the transducer and hydrophobic residues including Trp-697 from the converter and hydrophobic methylene group of Arg-152. The pyrazolopyrimidine group of myoVin-1 is further stabilized by Gln-149 of the α-helix that connects the seven-stranded β-sheet transducer. Color coding is according to E.
To address whether the partial reduction in the steady-state ATPase activity is caused by a decrease in kcat or Kapp, we measured the ATPase activity of M5BS1 as a function of [F-actin] at increasing myoVin-1 concentrations up to 30 μm. Higher inhibitor concentrations resulted in precipitation in the presence of high F-actin concentrations. MyoVin-1 inhibits the actin-activated ATPase activity of M5BS1 in a dose-dependent manner by reducing kcat (Fig. 6, B and C). The almost linear dependence of the ATPase activity on [F-actin] at high concentrations of myoVin-1 additionally suggests that the compound causes an increase in Kapp and kcat (Fig. 6, B and C).
Next, we tested whether myoVin-1 would inhibit the speed of actin filament gliding over a M5BHMM-decorated surface in the in vitro motility assay. M5BHMM smoothly moves actin filaments with a speed of 354 ± 119 nm s−1. The presence of 500 μm myoVin-1 reduces the gliding speed to 201 ± 97 nm s−1 without completely inhibiting the motion, and many actin filaments move less regularly with periods of pauses (Fig. 6C). This observation is in line with the increased Kapp observed in steady-state kinetic assays and suggests that the actin-activated ADP release is reduced in the presence of the compound (Fig. 6, B and C).
To investigate in more detail the transient kinetic transitions that are modulated by myoVin-1, we performed single-turnover measurements. By rapidly mixing M5BS1 with substoichiometric concentrations of mantATP, the molecular events of nucleotide binding, hydrolysis, and the release of the hydrolysis products can be monitored by the fluorescence increase, plateau, and decrease of the mant moiety. As shown in Fig. 6D, myoVin-1 predominantly reduces the release of mantADP in a concentration-dependent manner.
To explore how myoVin-1 modulates M5BS1 motor activity at the structural level, we performed in silico blind docking for unbiased mapping of the binding site and binding patterns in a homology model of the human myosin-5B motor domain with Mg·ADP·VO4 in the active site. The nucleotide analog was included to account for the noncompetitive mode of action of myoVin-1. Of the top 10 ranked binding conformations, 7 ligands clustered near and around the relay helix, the thiol-reactive region (SH1–SH2), three of the seven-stranded β-sheets of the transducer, and the converter region of the myosin motor domain (Fig. 6, E and F) at a distance of 17 Å from the active site. The close-up view of energetically most favorable binding pose (binding energy = −10.5 kcal/mol) of myoVin-1 shows that the ligand interacts closely with interfaces formed by the seven-stranded β-sheets of the transducer on one side and hydrophobic residues including Trp-697 from the converter and the hydrophobic methylene group of Arg-152. The pyrazolopyrimidine group of myoVin-1 is further stabilized by Gln-149 of the α-helix that connects the seven-stranded β-sheet transducer (Fig. 6F). The predicted binding site in great distance to the active site supports previous reports that identified myoVin-1 as an uncompetitive myosin inhibitor (41).
Discussion
We chose to determine the kinetic and regulatory features of mammalian myosin-5B with single-headed and double-headed constructs that contain the entire neck domain with its six IQ motifs (Fig. 1A). The M5BS1 construct is therefore considerably longer than the myosin-5 constructs commonly studied (16, 35, 46–48). We chose this approach because recent studies indicate that the light chains bound to the myosin neck region are (i) not passive components of the holoenzyme, (ii) can directly interact with the myosin motor domain and each other in some cases, and (iii) may therefore directly influence myosin motor function (29, 30). Moreover, the length of the neck domain, which defines the number of IQ motifs in the myosin heavy chain, determines the kinetic and mechanical activity of the protein when artificially altered (49–51). The M5BS1 construct with its six IQ motifs allows a direct comparison between with the single-headed and double-headed myosin constructs and the contribution of a second head on myosin motor function and intramolecular gating. Moreover, our detailed kinetic characterization allows for the comparative analysis of kinetic signatures in the class 5 myosome, the description of the interaction of myosin-5B with the small molecule inhibitor myoVin-1, and the prediction of the impact of disease-causing mutations on myosin-5B motor function.
Steady-state and transient kinetic signatures of M5BS1
With thorough transient and steady-state kinetic assays, we show that myosin-5B is a bona fide actin-based molecular motor with a high actin-activated ATPase activity under steady-state conditions. The transient kinetic signatures of M5BS1 include low thermodynamic (KAD/KD = ∼1–1.7) and kinetic coupling ratios (k−AD/k−D = ∼1–1.5), a high k+AD/K1k+2 = ∼10 ratio, and an intermediate duty ratio of ∼0.46–0.6 (Table 2). Other characteristics of the M5BS1 ATPase cycle are a low kcat/k+2 ratio of 0.027 that favors the dissociation of the actomyosin complex and a catalytic efficiency of kcat/Kapp ∼0.8 μm−1 s−1 that indicates a low number of ATP turnovers per F-actin encounter (48). The kinetic signatures of M5BS1 indicate that the molecule has an intermediate duty ratio, in between the parameters determined for the nonprocessive M5CS1 and the processive M5AS1 (Tables 1 and 2).
Our results disagree to some extent with an initial report on the kinetic properties of human myosin-5B (52). Although this study did not include a detailed description of the myosin kinetic cycle in the actin-detached states and the interaction of M5BS1 with F-actin, we find that the presented second-order binding rate constants for ATP and the Michaelis–Menten parameter of the steady-state ATPase activity agree with the parameters determined in this study (52). Discrepancies were found in the nucleotide release rates that are slightly reduced when compared with our work and result in a higher duty ratio for a single-headed M5BS1 construct (52). In summary, the kinetic characteristics of M5BS1 presented in this study suggest that myosin-5B has a limited processive ability and is likely to work in an ensemble of motors to efficiently transport vesicles along the cellular F-actin cytoskeleton.
The second head in M5BHMM establishes the kinetic prerequisite for single molecule processivity
Single molecule processivity is a feature of higher eukaryotic myosins-5A (17, 18). This behavior is in striking contrast to mammalian myosin-5C, Drosophila myosin-5, and the Saccharomyces myosins-5 Myo2p and Myo4p that have mechanoenzymatic properties that do not support single molecule processivity in biochemical in vitro assays (16, 47, 53). To either power or maximize the efficiency of myosin-5–based transport functions in the three-dimensional actin cytoskeleton of a cell, the motors cluster on the surface of cargo vesicles in densities up to >100 molecules to establish processivity (1, 54). This number may be crucial to maintain trafficking function, especially to compensate for either a reduced number of myosins-5 resulting in low surface densities or motors with compromised mechanoenzymatic activity as they are found in some disease states (32, 55). This makes the information regarding whether the myosin-5B dimer possesses the kinetic prerequisites for single molecule processivity essential to understand its function in normalcy and pathology.
To address the question of whether mammalian myosin-5B has the kinetic prerequisites to establish single molecule processivity, we explored the kinetic properties of double-headed M5BHMM. The presence of a second head induces gating in the dimer that reduces the actin-activated ADP release rate k−AD of ∼65% and the kcat of ∼20% when compared with M5BS1 under identical assay conditions (Tables 1–3). Our transient and steady-state kinetic ensemble measurements further indicate that the ADP release is most likely not the rate-limiting step in the M5BS1 kinetic cycle but the rate-limiting step in the M5BHMM kinetic cycle. This observation highlights that a second head in M5BHMM influences the kinetics of the first head, as has been shown for the gating between the two heads of other unconventional myosins associated with transport function (18, 56–59).
As reported previously, myosins that move processively, either as dimers or as filaments, typically have a calculated duty ratio >0.95 (35, 38). Based on our duty ratio calculations on M5BHMM that neglect every form of communication and steric constraints between the two heads, we speculate that the myosin-5B dimer is weakly processive and that a higher duty ratio and hence robust processivity may be achieved in the presence of actin-binding proteins or when stain is induced by a vesicle load. Both regulatory effects on the mechanical processivity have been described for some myosins-5, including nonprocessive myosins-5, and will be addressed for myosin-5B in future studies (60–63).
Potential impact of disease-associated mutations in the myosin-5B motor domain
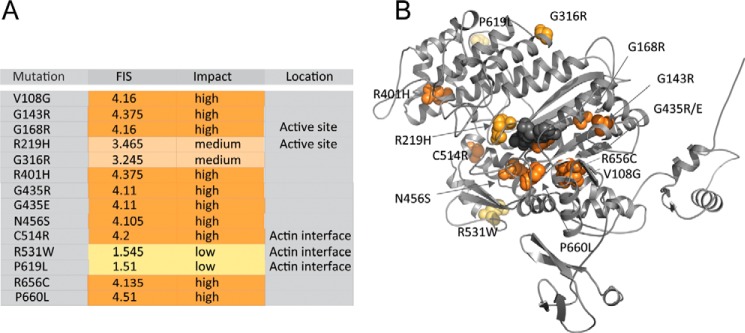
14 mutations in MYO5B result in amino acid changes in the motor domain at the protein level (26). To explore the possible effect of these mutations on myosin motor function, we predicted the functional impact of the mutations, mapped the mutations in a homology model of myosin-5B, and linked the structural analysis to our kinetic data (64).
The functional impact score of the mutations is shown in Fig. 7A, the respective location in the myosin motor domain in Fig. 7B. Of the 14 mutations, 2 have low, 2 have medium, and 10 have high functional impact scores (Fig. 7A), thereby ranking their predicted impact on myosin motor function (64). The mutations are distributed throughout the entire myosin motor domain including the active site (G168R/R219H) and the actin-binding region (C514R/R531W/P619L) (Fig. 7B). The other mutations are located outside the functional structural elements in allosteric regions of the motor domain: mutations V108G, G316R, R401H, N456S, and R656C were previously attributed to play crucial roles in allosteric communication pathways in the myosin motor domain, whereas mutations G143R, G435(R/E), and R656C are predicted to induce protein instability and misfolding (26).
Figure 7.
Location and impact of microvillus inclusion disease causing mutations in the myosin-5B motor domain. A, mutations in the myosin motor domain and their functional impact score and impact on myosin motor function. The localization to structural elements in the myosin motor domain to either the active site or the actin-binding region is indicated when applicable. B, the homology model of the human myosin-5B motor domain (gray cartoon representation) shows the localization of the respective disease-associated mutation. The mutations are colored according to severity from yellow to orange according to A. The nucleotide is highlighted in gray spheres. Mutations G168R and R219H are in the active site, and C514R/R531W/P619L is in the actin-binding region. Mutations V108G, G316R, R401H, N456S, and R656C are predicted to have allosteric effects on myosin motor function, whereas G143R, G435(R/E), and R656C may interfere with protein folding and induce protein instability (26).
Among the mutations with a medium or high functional impact score are G168R and R219H in the conserved P-loop and switch-1 loop in the myosin active site. Both structural elements play crucial roles in the coordination of the nucleotide throughout the myosin kinetic cycle (Fig. 1B). Switch-1 mutations at equivalent positions to the highly conserved Arg-219 have previously been described to reduce the actin-activated ATPase activity and motile properties of class 5 and 2 myosins by interfering with the hydrolysis of ATP (65–67). The resulting low-duty ratio motors are expected to have a reduced processivity when compared with the wild-type protein. This property may be of physiological significance in the myosin-5B R219H mutant that, based on our kinetic characterization of the wild-type protein, is not expected to have robust single molecule processivity. A reduction in duty ratio and processivity may be reflected in the aberrant vesicle trafficking and segregation patterns in patients with microvillus inclusion disease.
Similarly, the replacement of a negatively charged with a bulky residue in the myosin-5B mutant R531W is expected to weaken the interaction with the actin filament that prominently involves electrostatic interactions between positively charges residues in the myosin motor domain and the negatively charged surface of the actin filament. A reduced F-actin interaction is expected to weaken the affinity of the motor domain for its track in the presence of ATP, thereby limiting the degree of processivity (59).
Why does myosin-5B binding not quench the fluorescence of pyrene-labeled F-actin?
Apart from a kinetic description of single- and double-headed myosin-5B constructs, this study revealed that the motor does not significantly quench pyrene fluorescence in transient-kinetic in vitro assays. Site-specific labeling of actin on Cys-374 is a powerful method to measure binding of most myosins to F-actin (68–71). Cys-374 is located on the surface of the actin subunit of a filament in the vicinity of the myosin-binding site. The fluorescence of pyrene-actin is only quenched when myosin binds in the strong binding state (AM or AM·ADP; Fig. 1B), but not in the weak binding state (AM·ATP and AM·ADP·Pi; Fig. 1B) (34). The light scattering signal in contrast can be used as readout for the binding of myosin to actin in both the strong and the weak states (34). Our observation that the interaction between M5BS1 and pyrene-actin does not quench fluorescence but induces a change in light scattering signal further supports the results from the transient kinetic analysis that indicate that the motor binds F-actin weakly. However, actin sedimentation assays in the same buffer indicate that the actomyosin complex can form under the experimental conditions and that the lack of pyrene signal is caused by different interactions at the actomyosin interface when compared with other myosins-5. A similar result has been obtained for other myosins, including Drosophila indirect flight muscle myosin-2, rat myosin-9B, and also a truncated, single-headed myosin-5A construct (48, 72, 73).
Chemical inhibition with myoVin-1
The pyrazolopyrimidine compound myoVin-1 was identified as an uncompetitive inhibitor for myosin-5A based on privileged chemical scaffold design. The small molecule did not inhibit skeletal muscle myosin-2, nonmuscle myosin-2, and myosin-6 in vitro, suggesting selectivity for class 5 myosins (41). In this work, we show that myoVin-1 inhibits M5BS1 (KI = 23.12 ± 2.07 μm) with an almost identical KI as a single-headed myosin-5A construct (KI = 24 ± 4 μm) (41). Similar to the effects reported for myosin-5A, we find that myoVin-1 partially inhibits the actin-activated ATPase activity of M5BS1 (41). Partial inhibition is also observed in the in vitro motility assay, suggesting that a decrease in the actin-activated ADP release is the kinetic step in the actomyosin mechanoenzymatic cycle that is altered in the presence of the small molecule compound.
MyoVin-1 has been used to inhibit the myosin-5B–dependent trafficking of podocalyxin to apical membrane insertion sites and myosin-5–dependent intersynaptic vesicle exchange in live cells (43, 44). Vesicle velocity and directionality are slightly reduced in the presence of myoVin-1, whereas the frequency of vesicle pausing increases (43). These quantitative in vivo results agree with the observed partial inhibition of the myosin ATPase activity and functional activity and the repetitive motion of actin filaments in the in vitro motility assay in this study (Fig. 5, A–D). Intriguingly, myosin-5 moves vesicles with a speed of 320 ± 80 nm s−1 in vivo that drops to 280 ± 60 nm s−1 in the presence of 30 μm myoVin-1 (43). This effect is very similar to the speeds measured in the in vitro motility assay (354 ± 119 nm s−1 (no myoVin-1); 201 ± 97 nm s−1 (500 μm myoVin-1) (Fig. 6C). It is also of note that myoVin-1 inhibits the activity of skeletal muscle myosin-2 in prothrombin activation, a finding that contrasts the above mentioned selectivity for class 5 myosins (74).
Based on our blind docking results, we predict the inhibitor binding pocket to be located near the converter region in the myosin motor domain (Fig. 6, E and F). The predicted binding pocket is remote from the active site, underlining the uncompetitive mode of inhibition of myoVin-1. Interestingly, the same pocket was recently identified in structural studies as the binding site for the myosin-2 inhibitor CK-571, suggesting that the converter/SH1-SH2/relay interface is a promising site to target myosin motor function with small molecules (75).
In summary, the inhibition of the kinetic and functional activity of myosin-5B constructs in biochemical in vitro assays at high inhibitor concentrations, together with the results of previous in vitro and in vivo studies, suggest that myoVin-1 is not a potent inhibitor of class 5 myosins. The lack of selectivity for a myosin-5 paralogs, the almost identical KI values for myosins-5A and -5B in vitro, and the partial inhibition of the sliding velocity and steady-state ATPase activity of potentially all three mammalian myosins-5 in a cell suggest that caution should be used when interpreting the results from cell biological experiments in which myoVin-1 is used.
Experimental procedures
Construction of expression constructs, recombinant protein production, and preparation
cDNA constructs encoding for murine M5BS1 (amino acids 1–906, molecular mass of 107 kDa) and M5BHMM (amino acids 1–1095, molecular mass of 127 kDa) were inserted in a modified pFastBac1 encoding for a C-terminal Flag tag with standard cloning techniques. Rat CaM cDNA (molecular mass of 17 kDa) that is at the protein level 100% identical with murine CaM was expressed from pFastBac1. Recombinant baculovirus generation and gene expression were performed as recommended by the manufacturer (Thermo Fisher Scientific). For protein production, Sf9 insect cells were infected with recombinant baculoviruses encoding M5BS1/M5BHMM and CaM, and the protein complexes were purified as described via Flag capture (76). M5BHMM was further purified to electrophoretic homogeneity by size exclusion chromatography on a HiLoad Superdex 16/600 pg (GE Healthcare Life Sciences). Actin was prepared from rabbit skeletal muscle acetone powder (Pel-Freez Biologicals) and labeled with N-(1-prenyl)iodoacetamide (pyrene) as described (69, 77).
Steady-state and transient state kinetic assays
Steady-state kinetic assays were performed in a Cary 60 spectrophotometer (Agilent Technologies) at a temperature of 25 °C in buffer containing 10 mm MOPS, pH 7.0, 2 mm ATP, 50 mm NaCl, 2 mm MgCl2, 0.15 mm EGTA, 40 units/ml l-lactic dehydrogenase, 200 units/ml pyruvate kinase, 200 μm NADH, and 1 mm phosphoenolpyruvate at a protein concentration of <50 nm M5BS1 or M5BHMM as described (76). Transient kinetic assays were carried out on a Sf-61 DX2 stopped-flow (Hi-Tech Scientific) equipped with a 75-watt mercury–xenon arc lamp at a temperature of 20 °C in SF buffer containing 25 mm MOPS, pH 7.0, 100 mm KCl, 5 mm MgCl2, and 0.1 mm EGTA unless stated otherwise. Concentrations throughout the text represent post-mixing concentrations. Light scattering was excited at a wavelength of 320 nm, and emission was measured at a fixed angle of 90°. The intrinsic tryptophan fluoresce was excited at a wavelength of 297 nm, and the emission was recorded after passage through a WG320 filter. The fluorescence of mant-nucleotides was either directly excited at a wavelength of 365 nm or indirectly after energy transfer from tryptophan residues, and the emission was detected after passage through a LP390 filter.
The small molecule myoVin-1 (Calbiochem) was diluted in DMSO to a concentration of 50 mg/ml (92.83 mm) and stored at −20 °C. Kinetic measurements in the presence of myoVin-1 were carried out at a constant concentration of 2% DMSO. Reaction mixtures were incubated for 10 min at room temperature protected from light prior to the assay.
Cosedimentation assay
Proteins (2 μm M5BS1, 10 μm F-actin) were incubated in SF buffer for 10 min at room temperature. The samples were centrifuged (30 min, 100,000 × g, 4 °C), and supernatant and pellet fractions were separated on a 4–12% Bis-Tris gel (Thermo Scientific). The gel was stained with PageBlue (Thermo Scientific), destained with water, and documented on an Odyssey scanner (LI-COR Biosciences).
Functional in vitro motility assay
The in vitro motility assay was performed with M5BHMM as described previously in 20 mm MOPS, pH 7.4, 2 mm MgCl2, 0.1 mm EGTA, 50 mm KCl, 1 mm ATP, 2.5 μg/ml glucose oxidase, 45 μg/ml catalase, 2.5 mg/ml glucose, and 50 mm DTT at a temperature of 30 °C (49).
In silico homology modeling and docking
The human myosin-5B motor domain (amino acids 1–767) was modeled using the human myosin-5C motor domain (Protein Data Bank code 4ZG4) pre-powerstroke state structure as a template. Both proteins share 68% sequence identity and 81% homology. The model was built using Modeler 9.18 (78). For the myoVin-1–binding site analysis, the human myosin-5B motor domain model bound to Mg·ADP·VO4 was used for docking studies. AutodockTools was used to prepare the ligands and to assign Gasteiger charges (79). Blind docking was performed using autodock Vina by defining a cubic grid size of 79 Å. The exhaustiveness level was set at 50, and default options were used for the remaining parameters. The top 10 best docked ligand poses were selected based on the free energy predictions (−10.5 kcal/mol to −8.5 kcal/mol).
Author contributions
S. M. H. cloned expression plasmids, produced and purified proteins, and performed and analyzed the kinetic experiments. K. C. performed and analyzed the in silico homology modeling and docking studies. J. R. S. performed and analyzed the in vitro motility assay. All authors wrote and approved the final version of the manuscript.
This work was supported by the Intramural Research Program of the National Heart, Lung, and Blood Institute of the National Institutes of Health (HL004229). The authors declare that they have no conflicts of interest with the contents of this article. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
The notation used throughout this article distinguishes between the kinetic constants in the presence and absence of F-actin by using regular (K1) versus bold (K1) type; subscripts A and D refer to F-actin (KA) and ADP (KD), respectively.
- CaM
- calmodulin.
References
- 1. Gross S. P., Tuma M. C., Deacon S. W., Serpinskaya A. S., Reilein A. R., and Gelfand V. I. (2002) Interactions and regulation of molecular motors in Xenopus melanophores. J. Cell Biol. 156, 855–865 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Hammer J. A. 3rd, and Sellers J. R. (2011) Walking to work: roles for class V myosins as cargo transporters. Nat. Rev. Mol. Cell Biol. 13, 13–26 [DOI] [PubMed] [Google Scholar]
- 3. Jacobs D. T., Weigert R., Grode K. D., Donaldson J. G., and Cheney R. E. (2009) Myosin Vc is a molecular motor that functions in secretory granule trafficking. Mol. Biol. Cell 20, 4471–4488 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Lapierre L. A., Kumar R., Hales C. M., Navarre J., Bhartur S. G., Burnette J. O., Provance D. W. Jr., Mercer J. A., Bähler M., and Goldenring J. R. (2001) Myosin vb is associated with plasma membrane recycling systems. Mol. Biol. Cell 12, 1843–1857 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Nascimento A. A., Roland J. T., and Gelfand V. I. (2003) Pigment cells: a model for the study of organelle transport. Annu. Rev. Cell Dev. Biol. 19, 469–491 [DOI] [PubMed] [Google Scholar]
- 6. Rodriguez O. C., and Cheney R. E. (2002) Human myosin-Vc is a novel class V myosin expressed in epithelial cells. J. Cell Sci. 115, 991–1004 [DOI] [PubMed] [Google Scholar]
- 7. Wagner W., Brenowitz S. D., and Hammer J. A. 3rd (2011) Myosin-Va transports the endoplasmic reticulum into the dendritic spines of Purkinje neurons. Nat. Cell Biol. 13, 40–48 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Wu X., Rao K., Bowers M. B., Copeland N. G., Jenkins N. A., and Hammer J. A. 3rd (2001) Rab27a enables myosin Va-dependent melanosome capture by recruiting the myosin to the organelle. J. Cell Sci. 114, 1091–1100 [DOI] [PubMed] [Google Scholar]
- 9. Ji H. H., Zhang H. M., Shen M., Yao L. L., and Li X. D. (2015) The motor function of Drosophila melanogaster myosin-5 is activated by calcium and cargo-binding protein dRab11. Biochem. J. 469, 135–144 [DOI] [PubMed] [Google Scholar]
- 10. Johnston G. C., Prendergast J. A., and Singer R. A. (1991) The Saccharomyces cerevisiae MYO2 gene encodes an essential myosin for vectorial transport of vesicles. J. Cell Biol. 113, 539–551 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Heissler S. M., and Sellers J. R. (2016) Kinetic adaptations of myosins for their diverse cellular functions. Traffic 17, 839–859 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Heissler S. M., and Sellers J. R. (2016) Various themes of myosin regulation. J. Mol. Biol. 428, 1927–1946 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Roland J. T., Lapierre L. A., and Goldenring J. R. (2009) Alternative splicing in class V myosins determines association with Rab10. J. Biol. Chem. 284, 1213–1223 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Sladewski T. E., Krementsova E. B., and Trybus K. M. (2016) Myosin Vc is specialized for transport on a secretory superhighway. Curr. Biol. 26, 2202–2207 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Gunther L. K., Furuta K., Bao J., Urbanowski M. K., Kojima H., White H. D., and Sakamoto T. (2014) Coupling of two non-processive myosin 5c dimers enables processive stepping along actin filaments. Sci. Rep. 4, 4907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Takagi Y., Yang Y., Fujiwara I., Jacobs D., Cheney R. E., Sellers J. R., and Kovács M. (2008) Human myosin Vc is a low duty ratio, nonprocessive molecular motor. J. Biol. Chem. 283, 8527–8537 [DOI] [PubMed] [Google Scholar]
- 17. Mehta A. D., Rock R. S., Rief M., Spudich J. A., Mooseker M. S., and Cheney R. E. (1999) Myosin-V is a processive actin-based motor. Nature 400, 590–593 [DOI] [PubMed] [Google Scholar]
- 18. Sakamoto T., Webb M. R., Forgacs E., White H. D., and Sellers J. R. (2008) Direct observation of the mechanochemical coupling in myosin Va during processive movement. Nature 455, 128–132 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Cheeseman L. P., Boulanger J., Bond L. M., and Schuh M. (2016) Two pathways regulate cortical granule translocation to prevent polyspermy in mouse oocytes. Nat. Commun. 7, 13726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Knowles B. C., Roland J. T., Krishnan M., Tyska M. J., Lapierre L. A., Dickman P. S., Goldenring J. R., and Shub M. D. (2014) Myosin Vb uncoupling from RAB8A and RAB11A elicits microvillus inclusion disease. J. Clin. Invest. 124, 2947–2962 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Vogel G. F., Klee K. M., Janecke A. R., Müller T., Hess M. W., and Huber L. A. (2015) Cargo-selective apical exocytosis in epithelial cells is conducted by Myo5B, Slp4a, Vamp7, and Syntaxin 3. J. Cell Biol. 211, 587–604 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Liu Y., Xu X. H., Chen Q., Wang T., Deng C. Y., Song B. L., Du J. L., and Luo Z. G. (2013) Myosin Vb controls biogenesis of post-Golgi Rab10 carriers during axon development. Nat. Commun. 4, 2005. [DOI] [PubMed] [Google Scholar]
- 23. Schuh M. (2011) An actin-dependent mechanism for long-range vesicle transport. Nat. Cell Biol. 13, 1431–1436 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Wang Z., Edwards J. G., Riley N., Provance D. W. Jr., Karcher R., Li X. D., Davison I. G., Ikebe M., Mercer J. A., Kauer J. A., and Ehlers M. D. (2008) Myosin Vb mobilizes recycling endosomes and AMPA receptors for postsynaptic plasticity. Cell 135, 535–548 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Roland J. T., Bryant D. M., Datta A., Itzen A., Mostov K. E., and Goldenring J. R. (2011) Rab GTPase-Myo5B complexes control membrane recycling and epithelial polarization. Proc. Natl. Acad. Sci. U.S.A. 108, 2789–2794 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. van der Velde K. J., Dhekne H. S., Swertz M. A., Sirigu S., Ropars V., Vinke P. C., Rengaw T., van den Akker P. C., Rings E. H., Houdusse A., and van Ijzendoorn S. C. (2013) An overview and online registry of microvillus inclusion disease patients and their MYO5B mutations. Hum. Mutat. 34, 1597–1605 [DOI] [PubMed] [Google Scholar]
- 27. Müller T., Hess M. W., Schiefermeier N., Pfaller K., Ebner H. L., Heinz-Erian P., Ponstingl H., Partsch J., Röllinghoff B., Köhler H., Berger T., Lenhartz H., Schlenck B., Houwen R. J., Taylor C. J., et al. (2008) MYO5B mutations cause microvillus inclusion disease and disrupt epithelial cell polarity. Nat. Genet. 40, 1163–1165 [DOI] [PubMed] [Google Scholar]
- 28. Pylypenko O., Attanda W., Gauquelin C., Lahmani M., Coulibaly D., Baron B., Hoos S., Titus M. A., England P., and Houdusse A. M. (2013) Structural basis of myosin V Rab GTPase-dependent cargo recognition. Proc. Natl. Acad. Sci. U.S.A. 110, 20443–20448 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. De La Cruz E. M., Wells A. L., Sweeney H. L., and Ostap E. M. (2000) Actin and light chain isoform dependence of myosin V kinetics. Biochemistry 39, 14196–14202 [DOI] [PubMed] [Google Scholar]
- 30. Heissler S. M., and Sellers J. R. (2014) Myosin light chains: teaching old dogs new tricks. Bioarchitecture 4, 169–188 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Vogel G. F., Janecke A. R., Krainer I. M., Gutleben K., Witting B., Mitton S. G., Mansour S., Ballauff A., Roland J. T., Engevik A. C., Cutz E., Müller T., Goldenring J. R., Huber L. A., and Hess M. W. (2017) Abnormal Rab11-Rab8-vesicles cluster in enterocytes of patients with microvillus inclusion disease. Traffic 18, 453–464 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Szperl A. M., Golachowska M. R., Bruinenberg M., Prekeris R., Thunnissen A. M., Karrenbeld A., Dijkstra G., Hoekstra D., Mercer D., Ksiazyk J., Wijmenga C., Wapenaar M. C., Rings E. H., and van IJzendoorn S. C. (2011) Functional characterization of mutations in the myosin Vb gene associated with microvillus inclusion disease. J. Pediatr. Gastroenterol. Nutr. 52, 307–313 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Henn A., and De La Cruz E. M. (2005) Vertebrate myosin VIIb is a high duty ratio motor adapted for generating and maintaining tension. J. Biol. Chem. 280, 39665–39676 [DOI] [PubMed] [Google Scholar]
- 34. De La Cruz E. M., and Ostap E. M. (2009) Kinetic and equilibrium analysis of the myosin ATPase. Methods Enzymol. 455, 157–192 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. De La Cruz E. M., Wells A. L., Rosenfeld S. S., Ostap E. M., and Sweeney H. L. (1999) The kinetic mechanism of myosin V. Proc. Natl. Acad. Sci. U.S.A. 96, 13726–13731 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Heissler S. M., and Manstein D. J. (2012) Functional characterization of the human myosin-7a motor domain. Cell Mol. Life Sci. 69, 299–311 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Heissler S. M., Selvadurai J., Bond L. M., Fedorov R., Kendrick-Jones J., Buss F., and Manstein D. J. (2012) Kinetic properties and small-molecule inhibition of human myosin-6. FEBS Lett. 586, 3208–3214 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Nagy A., Takagi Y., Billington N., Sun S. A., Hong D. K., Homsher E., Wang A., and Sellers J. R. (2013) Kinetic characterization of nonmuscle myosin IIb at the single molecule level. J. Biol. Chem. 288, 709–722 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Veigel C., Wang F., Bartoo M. L., Sellers J. R., and Molloy J. E. (2002) The gated gait of the processive molecular motor, myosin V. Nat. Cell Biol. 4, 59–65 [DOI] [PubMed] [Google Scholar]
- 40. Katsimitsoulia Z., and Taylor W. R. (2012) Coarse-grained simulation of myosin-V movement. Comput. Math. Methods Med. 2012, 781456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Islam K., Chin H. F., Olivares A. O., Saunders L. P., De La Cruz E. M., and Kapoor T. M. (2010) A myosin V inhibitor based on privileged chemical scaffolds. Angew. Chem. Int. Ed Engl. 49, 8484–8488 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Avasthi P., Onishi M., Karpiak J., Yamamoto R., Mackinder L., Jonikas M. C., Sale W. S., Shoichet B., Pringle J. R., and Marshall W. F. (2014) Actin is required for IFT regulation in Chlamydomonas reinhardtii. Curr. Biol. 24, 2025–2032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Gramlich M. W., and Klyachko V. A. (2017) Actin/myosin-V- and activity-dependent inter-synaptic vesicle exchange in central neurons. Cell Rep. 18, 2096–2104 [DOI] [PubMed] [Google Scholar]
- 44. Richards M., Hetheridge C., and Mellor H. (2015) The formin FMNL3 controls early apical specification in endothelial cells by regulating the polarized trafficking of podocalyxin. Curr. Biol. 25, 2325–2331 [DOI] [PubMed] [Google Scholar]
- 45. Zorca C. E., Kim L. K., Kim Y. J., Krause M. R., Zenklusen D., Spilianakis C. G., and Flavell R. A. (2015) Myosin VI regulates gene pairing and transcriptional pause release in T cells. Proc. Natl. Acad. Sci. U.S.A. 112, E1587–E1593 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Rosenfeld S. S., Houdusse A., and Sweeney H. L. (2005) Magnesium regulates ADP dissociation from myosin V. J. Biol. Chem. 280, 6072–6079 [DOI] [PubMed] [Google Scholar]
- 47. Tóth J., Kovács M., Wang F., Nyitray L., and Sellers J. R. (2005) Myosin V from Drosophila reveals diversity of motor mechanisms within the myosin V family. J. Biol. Chem. 280, 30594–30603 [DOI] [PubMed] [Google Scholar]
- 48. Trybus K. M., Krementsova E., and Freyzon Y. (1999) Kinetic characterization of a monomeric unconventional myosin V construct. J. Biol. Chem. 274, 27448–27456 [DOI] [PubMed] [Google Scholar]
- 49. Sakamoto T., Wang F., Schmitz S., Xu Y., Xu Q., Molloy J. E., Veigel C., and Sellers J. R. (2003) Neck length and processivity of myosin V. J. Biol. Chem. 278, 29201–29207 [DOI] [PubMed] [Google Scholar]
- 50. Sakamoto T., Yildez A., Selvin P. R., and Sellers J. R. (2005) Step-size is determined by neck length in myosin V. Biochemistry 44, 16203–16210 [DOI] [PubMed] [Google Scholar]
- 51. Moore J. R., Krementsova E. B., Trybus K. M., and Warshaw D. M. (2004) Does the myosin V neck region act as a lever? J. Muscle Res. Cell Motil. 25, 29–35 [DOI] [PubMed] [Google Scholar]
- 52. Watanabe S., Mabuchi K., Ikebe R., and Ikebe M. (2006) Mechanoenzymatic characterization of human myosin Vb. Biochemistry 45, 2729–2738 [DOI] [PubMed] [Google Scholar]
- 53. Reck-Peterson S. L., Tyska M. J., Novick P. J., and Mooseker M. S. (2001) The yeast class V myosins, Myo2p and Myo4p, are nonprocessive actin-based motors. J. Cell Biol. 153, 1121–1126 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Miller K. E., and Sheetz M. P. (2000) Characterization of myosin V binding to brain vesicles. J. Biol. Chem. 275, 2598–2606 [DOI] [PubMed] [Google Scholar]
- 55. Dong W., Chen X., Chen P., Yue D., Zhu L., and Fan Q. (2012) Inactivation of MYO5B promotes invasion and motility in gastric cancer cells. Dig. Dis. Sci. 57, 1247–1252 [DOI] [PubMed] [Google Scholar]
- 56. De La Cruz E. M., Ostap E. M., and Sweeney H. L. (2001) Kinetic mechanism and regulation of myosin VI. J. Biol. Chem. 276, 32373–32381 [DOI] [PubMed] [Google Scholar]
- 57. Ropars V., Yang Z., Isabet T., Blanc F., Zhou K., Lin T., Liu X., Hissier P., Samazan F., Amigues B., Yang E. D., Park H., Pylypenko O., Cecchini M., Sindelar C. V., Sweeney H. L., and Houdusse A. (2016) The myosin X motor is optimized for movement on actin bundles. Nat. Commun. 7, 12456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Forgacs E., Cartwright S., Sakamoto T., Sellers J. R., Corrie J. E., Webb M. R., and White H. D. (2008) Kinetics of ADP dissociation from the trail and lead heads of actomyosin V following the power stroke. J. Biol. Chem. 283, 766–773 [DOI] [PubMed] [Google Scholar]
- 59. Rosenfeld S. S., and Sweeney H. L. (2004) A model of myosin V processivity. J. Biol. Chem. 279, 40100–40111 [DOI] [PubMed] [Google Scholar]
- 60. Bao J., Huck D., Gunther L. K., Sellers J. R., and Sakamoto T. (2013) Actin structure-dependent stepping of myosin 5a and 10 during processive movement. PLoS One 8, e74936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Clayton J. E., Pollard L. W., Sckolnick M., Bookwalter C. S., Hodges A. R., Trybus K. M., and Lord M. (2014) Fission yeast tropomyosin specifies directed transport of myosin-V along actin cables. Mol. Biol. Cell 25, 66–75 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Hodges A. R., Krementsova E. B., Bookwalter C. S., Fagnant P. M., Sladewski T. E., and Trybus K. M. (2012) Tropomyosin is essential for processive movement of a class V myosin from budding yeast. Curr. Biol. 22, 1410–1416 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Sckolnick M., Krementsova E. B., Warshaw D. M., and Trybus K. M. (2016) Tropomyosin isoforms bias actin track selection by vertebrate myosin Va. Mol. Biol. Cell 27, 2889–2897 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Reva B., Antipin Y., and Sander C. (2011) Predicting the functional impact of protein mutations: application to cancer genomics. Nucleic Acids Res. 39, e118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Forgacs E., Sakamoto T., Cartwright S., Belknap B., Kovács M., Tóth J., Webb M. R., Sellers J. R., and White H. D. (2009) Switch 1 mutation S217A converts myosin V into a low duty ratio motor. J. Biol. Chem. 284, 2138–2149 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Li X. D., Rhodes T. E., Ikebe R., Kambara T., White H. D., and Ikebe M. (1998) Effects of mutations in the γ-phosphate binding site of myosin on its motor function. J. Biol. Chem. 273, 27404–27411 [DOI] [PubMed] [Google Scholar]
- 67. Shimada T., Sasaki N., Ohkura R., and Sutoh K. (1997) Alanine scanning mutagenesis of the switch I region in the ATPase site of Dictyostelium discoideum myosin II. Biochemistry 36, 14037–14043 [DOI] [PubMed] [Google Scholar]
- 68. Adamek N., and Geeves M. A. (2014) Use of pyrene-labelled actin to probe actin-myosin interactions: kinetic and equilibrium studies. EXS 105, 87–104 [DOI] [PubMed] [Google Scholar]
- 69. Criddle A. H., Geeves M. A., and Jeffries T. (1985) The use of actin labelled with N-(1-pyrenyl)iodoacetamide to study the interaction of actin with myosin subfragments and troponin/tropomyosin. Biochem. J. 232, 343–349 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Geeves M. A. (1989) Dynamic interaction between actin and myosin subfragment 1 in the presence of ADP. Biochemistry 28, 5864–5871 [DOI] [PubMed] [Google Scholar]
- 71. Kouyama T., and Mihashi K. (1981) Fluorimetry study of N-(1-pyrenyl)iodoacetamide-labelled F-actin. Local structural change of actin protomer both on polymerization and on binding of heavy meromyosin. Eur. J. Biochem. 114, 33–38 [PubMed] [Google Scholar]
- 72. Nalavadi V., Nyitrai M., Bertolini C., Adamek N., Geeves M. A., and Bähler M. (2005) Kinetic mechanism of myosin IXB and the contributions of two class IX-specific regions. J. Biol. Chem. 280, 38957–38968 [DOI] [PubMed] [Google Scholar]
- 73. Silva R., Sparrow J. C., and Geeves M. A. (2003) Isolation and kinetic characterisation of myosin and myosin S1 from the Drosophila indirect flight muscles. J. Muscle Res. Cell Motil. 24, 489–498 [DOI] [PubMed] [Google Scholar]
- 74. Deguchi H., Sinha R. K., Marchese P., Ruggeri Z. M., Zilberman-Rudenko J., McCarty O. J. T., Cohen M. J., and Griffin J. H. (2016) Prothrombotic skeletal muscle myosin directly enhances prothrombin activation by binding factors Xa and Va. Blood 128, 1870–1878 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Sirigu S., Hartman J. J., Planelles-Herrero V. J., Ropars V., Clancy S., Wang X., Chuang G., Qian X., Lu P. P., Barrett E., Rudolph K., Royer C., Morgan B. P., Stura E. A., Malik F. I., et al. (2016) Highly selective inhibition of myosin motors provides the basis of potential therapeutic application. Proc. Natl. Acad. Sci. U.S.A. 113, E7448–E7455 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Heissler S. M., Chinthalapudi K., and Sellers J. R. (2015) Kinetic characterization of the sole nonmuscle myosin-2 from the model organism Drosophila melanogaster. FASEB J. 29, 1456–1466 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Lehrer S. S., and Kerwar G. (1972) Intrinsic fluorescence of actin. Biochemistry 11, 1211–1217 [DOI] [PubMed] [Google Scholar]
- 78. Sali A., and Blundell T. L. (1993) Comparative protein modelling by satisfaction of spatial restraints. J. Mol. Biol. 234, 779–815 [DOI] [PubMed] [Google Scholar]
- 79. Morris G. M., Huey R., Lindstrom W., Sanner M. F., Belew R. K., Goodsell D. S., and Olson A. J. (2009) AutoDock4 and AutoDockTools4: Automated docking with selective receptor flexibility. J. Comput. Chem. 30, 2785–2791 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Wang F., Chen L., Arcucci O., Harvey E. V., Bowers B., Xu Y., Hammer J. A. 3rd, Sellers J. R. (2000) Effect of ADP and ionic strength on the kinetic and motile properties of recombinant mouse myosin V. J. Biol. Chem. 275, 4329–4335 [DOI] [PubMed] [Google Scholar]
- 81. Olivares A. O., Chang W., Mooseker M. S., Hackney D. D., and De La Cruz E. M. (2006) The tail domain of myosin Va modulates actin binding to one head. J. Biol. Chem. 281, 31326–31336 [DOI] [PubMed] [Google Scholar]