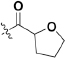
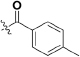
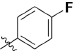
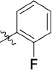
Abstract
Poly (ADP-ribose) polymerase 1 (PARP1) is overexpressed in a variety of cancers, especially in breast and ovarian cancers; tumor cells that are deficient in breast cancer gene 1/2 (BRCA1/2) are highly sensitive to PARP1 inhibition. In this study, we identified a series of 2,4-difluorophenyl-linker analogs (15–55) derived from olaparib as novel PARP1 inhibitors. Four potent analogs 17, 43, 47, and 50 (IC50=2.2–4.4 nmol/L) effectively inhibited the proliferation of Chinese hamster lung fibroblast V-C8 cells (IC50=3.2–37.6 nmol/L) in vitro, and showed specificity toward BRCA-deficient cells (SI=40–510). The corresponding hydrochloride salts 56 and 57 (based on 43 and 47) were highly water soluble in pH=1.0 buffered salt solutions (1628.2 μg/mL, 2652.5 μg/mL). In a BRCA1-mutated xenograft model, oral administration of compound 56 (30 mg·kg-1·d-1, for 21 d) exhibited more prominent tumor growth inhibition (96.6%) compared with the same dose of olaparib (56.3%); in a BRCA2-mutated xenograft model, oral administration of analog 43 (10 mg·kg-1·d-1, for 28 d) significantly inhibited tumor growth (69.0%) and had no negative effects on the body weights. Additionally, compound 56 exhibited good oral bioavailability (F=32.2%), similar to that of olaparib (F=45.4%). Furthermore, the free base 43 of the hydrochloride salt 56 exhibited minimal hERG inhibition activity (IC50=6.64 μmol/L). Collectively, these data demonstrate that compound 56 may be an excellent drug candidate for the treatment of cancer, particularly BRCA-deficient tumors.
Supplementary information
The online version of this article (doi:10.1038/aps.2017.104) contains supplementary material, which is available to authorized users.
Keywords: PARP-1 inhibitor, olaparib, breast cancer, BRCA-deficient tumors
Introduction
Poly (ADP-ribose) polymerases (PARPs) are ADP-ribosyltransferases that cleave nicotinamide adenine dinucleotide (NAD+) into ADP-ribose and nicotinamide and then transfer the ADP-ribose units to a variety of target proteins (such as histones, topoisomerases, DNA polymerases, and DNA ligases) or to themselves1,2,3,4. PARPs are involved in a number of important biological processes, such as genome integrity surveillance, cell cycle progression, DNA damage response initiation, apoptosis, and transcription regulation5. PARP1 is the most abundant among the PARP family members and plays an essential role in the repair of DNA single-strand breaks (SSB)6,7. PARP1 is responsible for more than 90% of ADP-ribosylation occurring in cells and is evolutionarily conserved in all advanced eukaryotes8,9. PARP1 is a 113 kDa enzyme that contains three major structural domains, including a 42 kDa DNA-binding domain in the N-terminal region with two zinc fingers, a 55 kDa catalytic domain in the C-terminal region and an auto-modification domain in the central region10,11,12. The DNA-binding domain can recognize, and bind damaged DNA single-strand breaks and consequently stimulate the polymerization of ADP-ribose, thus leading to unwinding of DNA from histones and exposing the damaged DNA for repair. The catalytic domain utilizes NAD+ as a substrate to construct linear and branched polymers of ADP-ribose on its targets, and thus performs a pivotal role in DNA damage repair.
PARP1 is overexpressed in a variety of cancers, and the expression of PARP1 has been associated with overall prognosis in cancers, especially breast and ovarian cancers11,12; therefore, PARP1 has emerged as an attractive anticancer drug target for anchoring DNA damage. Cell lines deficient in the breast cancer genes 1/2 (BRCA1/2, two known tumor suppressor genes whose mutations are associated with breast, ovarian, prostate, and pancreatic cancers) were 1000-fold more sensitive to PARP inhibition than either wild-type or heterozygous mutant cells. Therefore, the inhibition of PARP1 induces highly selective killing of BRCA1/2-deficient tumor cells. In this regard, many small molecules targeting PARP1 have been developed as chemosensitizers used in combination with ionizing radiation, as DNA-damaging chemotherapeutic agents or as stand-alone therapies (synthetic lethal) to kill cancer cells defective in DNA repair mechanisms (especially BRCA1/2 mutant breast or ovarian tumors)13,14,15,16,17,18,19,20,21.
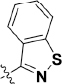
Many PARP inhibitors have previously been described, most of which are analogs of nicotinamide7,22,23. Several PARP inhibitors (Figure 1) have been used in late-phase (phases 2 and 3) clinical trials and approved for some ovarian cancers2; these inhibitors include olaparib (1, AZD-2281; AstraZeneca/Kudos)24, rucaparib (2, AG-014699; Clovis/Pfizer)25,26, niraparib (3, MK-4827; Tesaro/Merck)27,28, veliparib (4, ABT-888; AbbVie)29,30,31, and talazoparib (5, BMN 673; BioMarin/LEAD/Medivation)32.
Figure 1.
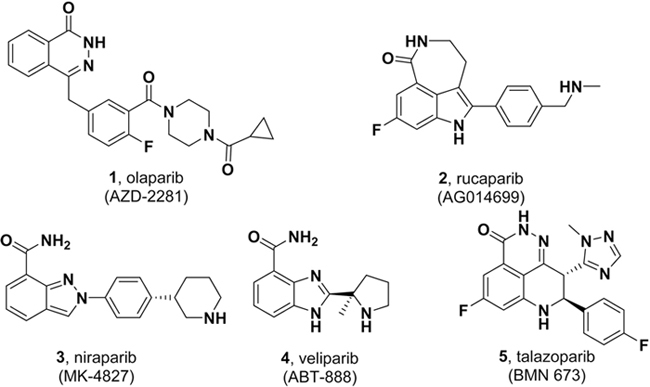
PARP inhibitors currently in late-phase (phases 2 and 3) clinical trials or already approved for clinical use.
The fluorine atom plays an important and unique role in influencing molecular conformation. From the perspective of steric effects, the influence of fluorine is anticipated to be marginal, and fluorine is a small atom with a van der Waals radius of 1.47 Å, which is close to the 1.20 Å radius of hydrogen33,34,35. Moreover, the high electronegativity of fluorine results in highly polarized C–F bonds and in new potential hydrogen bond interactions between the fluorine atom and drug targets. In addition, fluorine is a powerful tool for modulating the pKa of proximal functional groups and the electron densities of (hetero)aromatic rings36,37,38. The pKa of an ionizable center in a drug molecule determines the lipophilic profile, which in turn influences solubility, permeability, and protein binding. Changes in pKa can manifest as changes in potency, selectivity, toxicity, and pharmacokinetic (PK) properties, including absorption, distribution, metabolism, and excretion (ADME)39,40,41,42,43. Evidently, the incorporation of fluorine into drug molecules is a commonly used strategy to increase pharmacological activity and optimize the ADME profile44.
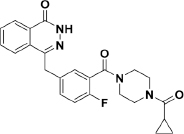
The discovery and investigation of olaparib have indicated that the phthalazine fragment is essential and that the piperazine segment is flexible to support PARP1 inhibition potency24. Given the therapeutic potential of olaparib as a PARP1 inhibitor, we designed a novel series of 2,4-difluoro-linkers for analog 15 derived from olaparib (Figure 2) to obtain novel olaparib analogs. Under the premise of minimizing structural changes, we introduced the 4-F substituent on the phenyl linker. The molecular docking studies (described below) showed that not only the fluorine atom at position 4 but also the fluorine atom at position 2 of analog 15 formed a hydrogen bond with Glu763 and Ser864, respectively, thus indicating that the 2,4-difluorophenyl-linker analogs of 1 might improve biological activity. To explore the SAR (structure-activity relationship) of the 2,4-difluorophenyl-linker analogs of 1, we designed a novel series of 2,4-difluorophenyl-linker analogs (15–55) derived from 1 and comprehensively evaluated whether a specific type of substituent (R, Figure 2) on the terminal nitrogen of piperazine could improve pharmacological activity and modulate water solubility. Finally, a highly potent PARP1 enzymatic inhibitor, 56, which has excellent in vivo anti-tumor efficacy, good oral bioavailability, enhanced water solubility and a good safety profile was identified as a unique candidate compound for the development of antitumor drugs.
Figure 2.
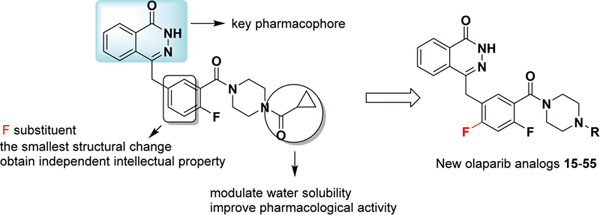
Chemical modification strategies (15–55) for lead compound olaparib.
Materials and methods
Reagents
Reagents and solvents were purchased from Adamas-Beta, J&K, Energy Chemical, Target Molecule, TCI, Alfa Aesar, and Acros, and were used without further purification. Analytical thin-layer chromatography (TLC) was performed with HSGF 254 (150–200 μm thickness; Yantai Huiyou Co, China), and the spots were visualized with UV light. The yields were not optimized. Melting points were measured in a capillary tube with an SGW X-4 melting point apparatus without correction. Nuclear magnetic resonance (NMR) spectroscopy was performed on a Bruker AMX-400 NMR (IS as TMS). Chemical shifts were determined in parts per million (ppm, δ) downfield from tetramethylsilane. Proton coupling patterns were described as follows: singlet (s), doublet (d), triplet (t), quartet (q), multiplet (m), or doublet of doublets (dd). High-resolution mass spectra (HRMS) were determined with electric ionization (EI) and electrospray ionization (ESI) produced by Waters GCT Premier and Waters LCT mass spectrometers. HPLC analysis of analogs 15–55 was performed on an Agilent 1100 with a quaternary pump and a diode array detector (DAD). The peak purity was verified on the basis of UV spectra. All analogs were confirmed to be ≥95% pure.
Figure 3.
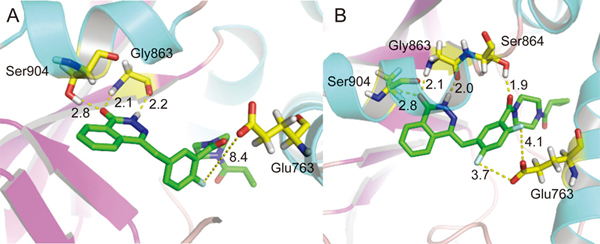
Molecular docking studies on 1 and 15. Docked poses of 1 (green sticks) (A) and 15 (green sticks) (B) in the NAD+ binding site (cyan and pink cartoon). Hydrophilic interacting residues are represented with yellow carbons. Hydrogen bonding distances (Å) are represented with black numbers. Hydrogen bonds are shown with yellow dash lines.
Chemistry
As outlined in Scheme 1, 1,5-difluoro-2-iodo-4-methylbenzene (7) was prepared from commercially available 2,4-difluoro-1-methylbenzene (6) in the presence of N-iodosuccinimide in trifluoroacetic acid at 0 °C for 24 h, and a 73% yield was achieved. Further treatment of compound 7 with cuprous cyanide from N,N-dimethylformamide under reflux for 20 h gave rise to 2,4-difluoro-5-methylbenzonitrile (8), with a 62% yield. The oxidation of compound 8 with chromic anhydride from the mixture of acetic anhydride and glacial acetic acid at 0 °C for 4 h yielded compound 9, with a 30% yield. The hydrolysis of compound 9 provided 2,4-difluoro-5-formylbenzonitrile (10). Dimethyl (3-oxo-1,3-dihydroisobenzofuran-1-yl)phosphonate (12) was prepared from the reaction of 2-formyl-benzoic acid with dimethyl phosphonate and methanesulfonic acid in sodium methoxide at 0 °C for 6 h, with a 52% yield. The coupling of compound 10 and compound 12 afforded compound 13, through use of triethylamine and tetrahydrofuran at room temperature for 5 h, with an 86% yield. The 2,4-difluoro-5-((4-oxo-3,4-dihydroph-thalazin-1-yl)methyl)benzoic acid (14) was prepared from compound 13 with hydrazine hydrate and sodium hydroxide at 70–90 °C for 21 h, with a 75% yield. The subsequent condensation of acid 14 with an appropriate piperazine substrate in the presence of HOBt, EDCI and dichloro-methane produced the target analogs 15–55, with yields of 10%–60%25. All analogs were confirmed to be ≥95% pure (Supplementary Table S1).
Figure 5.

Synthesis of analogues 15–55. aReagents and conditions: (a) NIS, TFA, 0 °C, 24 h; (b) DMF, CuCN, reflux, 20 h; (c) CrO3, (CH3CO)2O, CH3COOH, 0 °C, 4 h; (d) EtOH, H2O, reflux, 2 h; (e) MeONa, (CH3O)2PHO, CH3SO3H, 0 °C, 2 h; (f) THF, Et3N, 15 °C, 1 h; (g) (i) NaOH, 90 °C, 1 h; (ii) NH2-NH2·H2O, 70 °C, 18 h; (h) HOBt, EDCI, DIPEA, piperazine, DMA, overnight.
PARP1 enzyme inhibition assays
The inhibition of the tested compounds on PARP1 enzymatic activity in a cell-free system was measured with enzyme-linked immunosorbent assays (ELISA)45,46,47. Briefly, NAD+ (6 μmol/L) and the activator deoxyoligonucleotide (100 μg/mL) were diluted in 70 μL of reaction buffer and added to each well of a histone-pre-coated 96-well plate, and this was followed by the addition of 10 μL of the compound or a solvent control. The reaction was started by addition of 20 μL of recombinant human PARP1 (10 ng/well) for 1.5 h. After cells were washed with PBST, 100 μL of anti-PARP polyclonal antibody (Trevigen) was added, and the mixture was incubated for another 1.5 h. After washing the cells, the second antibody, goat anti-rabbit IgG horseradish peroxidase, was added, and the mixture incubated for an additional 30 min. Finally, a solution of 0.03% H2O2 and 2 mg/mL OPD in 0.1 mol/L citrate buffer (pH 5.4) was added, and the mixture was incubated for 10 min. The reaction was stopped by the addition of 2 mol/L H2SO4. A490 was measured with a multi-well spectrophotometer (SPECTRA MAX190). The inhibition rate of PARP1 enzymatic activity was calculated as (A490control-A490treated/A490control)×100%. The concentration required to inhibit 50% of PARP1 enzymatic activity (IC50) was determined with the Logit method.
Cytotoxicity assays
The V-C8 and V79 cells obtained from Leiden University were seeded on 96-well plates, cultured overnight and treated with gradient concentrations of the tested compounds for 72 h. The IC50 values were measured with a Cell Counting Kit-8 (CCK-8) assay (Dojindo Laboratories, Japan)47,48.
The UWB1.289 and UWB1.289+BRCA1 cells obtained from ATCC were seeded on 96-well plates, cultured overnight and treated with gradient concentrations of the tested compounds for 168 h. The IC50 values were measured with a sulforhodamine B colorimetric assay.
Determination of water solubility
The water solubility of 1, 17, 43, 47, 50, and 56–58 was determined through an HPLC method. Stock solutions (800 μg/mL) of the samples were prepared in methanol. Then, 10 μL of the diluted solutions (50, 100, 200, 400, and 800 μg/mL) were injected into the HPLC system to assess linearity. Calibration curves were plotted as peak area versus the concentration of the sample. Next, 10 mg of the sample was added to a 5-mL centrifuge tube, and 1 mL of pure water or a buffered salt solution (pH=7.4 or 1.0) was pipetted into the tube. If the solution was unsaturated and remained clear, more testing was needed. After the solution was stirred for 24 h, it was filtered with a syringe, and the HPLC system was injected with the same 10 μL. Water solubility was calculated by comparison of the peak area of the tested compound to the calibration curves.
Pharmacokinetic analysis
Compounds 1 and 56 were tested in pharmacokinetic studies using Sprague-Dawley (male) rats obtained from Shanghai Sippr-BK Laboratory Animal Co Ltd. The animals were housed in a room with controlled temperature and humidity and were allowed free access to food and water. For studying compounds 1 and 56, the rats were divided into 2 groups (3 rats/group), and each group received the compounds intravenously at a dose of 10 mg/kg or orally at a dose of 20 mg/kg, respectively. Compounds 1 and 56 were dissolved in DMSO (5%) and 15% HP-β-CD water (95%) to make a 5 mg/mL stock concentration. Blood samples from the orally treated rats and the intravenously injected rats were collected at 0.083, 0.25, 0.5, 1, 2, 4, 6, 8, and 24 h post-dose. All blood samples were centrifuged at 8000 revolutions per minute at 4 °C for 6 min. The plasma was collected and stored at -80 °C. All plasma samples were analyzed within one week after collection. Sample analysis was performed via LC-MS/MS using an Agilent 1100 chromatography system with a Phenomenex Gemini-NX C18 column (5 μm, 50 mm×2.1 mm). The mobile phase consisted of mixtures of 0.1% formic acid in acetonitrile and 0.1% formic acid in water and was run in gradient-mode at a flow rate of 0.6 mL/min. Mass spectra were detected with an API4000 triple quadruple equipped with an ESI source. Internal standards were used to track the responses of analytes in plasma samples. Plasma concentration-time data were analyzed, and the pharmacokinetic parameters were calculated.
Xenograft experiments
Human breast cancer MDA-MB-436 xenografts in nude mice were used to evaluate the anticancer activities of 1, 56 and 57, and human pancreatic cancer Capan-1 xenografts were used to evaluate the anticancer activities of 5 and 43. The model was established by the transplantation of 5×106 MDA-MB-436 cells or Capan-1 cells that were subcutaneously injected into the right armpit of each of the 4-5-week-old female BALB/cA nude mice. When the average tumor volume reached approximately 150 mm3, the mice were selected on the basis of tumor volume and randomly assigned to vehicle, 1, 5, 56, 57 and 43 groups. For 21 consecutive days, the animals in the 56 and 57 groups were given daily oral doses of 56 and 57 (1 mg·kg-1·d-1, 10 mg·kg-1·d-1 and 30 mg·kg-1·d-1, respectively). The animals in the positive control (1) group were given an oral dose of olaparib (30 mg/kg). For 28 consecutive days, the animals in the 43 group were given an oral dose of 43 (10 mg·kg-1·d-1) daily. For 10 consecutive days, the animals in the positive control (5) group were given an oral dose of 5 (0.5 mg/kg) daily. The animals in the vehicle group received a daily oral treatment of 0.5% carboxymethylcellulose sodium. During the treatment period, the implanted tumors were measured with a caliper twice weekly. The maximum width (X) and length (Y) of the tumors were measured, and the volume (V) was calculated using the formula: V=(X2Y)/2. Then, relative tumor volume (RTV) was calculated as follows: RTV=Vt/V0, where V0 represents the tumor volume at the beginning of the treatments, and Vt represents the tumor volume after treatment. The animal body weights were also measured at the same time. The experiments followed the Institutional Ethical Guidelines of the Animal Care and Use Committee (Shanghai Institute of Materia Medica, Chinese Academy of Sciences, China).
Results and discussion
Lead optimization and SAR
All synthesized analogs were first evaluated for PARP1 enzymatic activity inhibition by using olaparib as the reference standard in the assay. Given the importance of the fluorine atom, analog 15 was designed by replacing the middle 2-fluorophenyl linker with a 2,4-difluorophenyl linker. As a result, analog 15 not only displayed good enzymatic potency compared with that of olaparib (IC50=16.1 nmol/L vs 7.3 nmol/L) but also showed better cell inhibition activity (IC50=167.1 nmol/L vs 263.6 nmol/L) in the V-C8 cytotoxicity assay (Table 1). Encouraged by this result, we turned our attention to explore SAR by incorporating various substituents (R) on the N-atom of the piperazine ring of analog 15.
Table 1.
In vitro activity of analogues 15–30.
 | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| No | R | PARP1 enzyme IC50 (nmol/L) | Cell activity IC50 (nmol/L)a | No | R | PARP1 enzyme IC50 (nmol/L) | Cell activity IC50 (nmol/L)a | ||
| V-C8 (BRCA2−/−) | V79 (BRCA2+/+) | V-C8 (BRCA2−/−) | V79 (BRCA2+/+) | ||||||
| 1 | 7.3±0.5 | 263.6 | >10000 | ||||||
| 15 |
|
16.1±2.4 | 167.1 | >10000 | 23 |
|
24.1±15.1 | ||
| 16 |
|
30.0±5.1 | 24 |
|
15.3±1.4 | ||||
| 17 |
|
3.5±0.3 | 13.9 | 5106.9 | 25 |
|
10.4±0.9 | ||
| 18 |
|
12.5±3.3 | 26 |
|
21.1±2.7 | ||||
| 19 |
|
14.0±3.2 | 27 |
|
34.6±4.8 | ||||
| 20 |
|
4.6±1.4 | 99.2 | 2693.5 | 28 |
|
35.2* | ||
| 21 |
|
157.4±29.4 | 29 |
|
49.4* | ||||
| 22 |
|
61.3±20.9 | 30 |
|
8.9±5.7 | 1135.8 | 5734.0 | ||
*Single test results.
First, a small set of analogs with various acyl substituents (16–20, Table 1) on the terminal nitrogen of the piperazine ring were examined. The enzymatic inhibitory activity of the acetyl analog 16 (IC50=30.0 nmol/L) was 2-fold less than that of analog 15. Replacing the cyclopropane with tetrahydrofuran or morpholine resulted in analogs 18 and 19, respectively, which had similar enzymatic inhibitory activities (IC50=12.5 nmol/L and 14.0 nmol/L, respectively). Interestingly, the benzoyl (17) and 4-methylbenzoyl (20) analogs showed greater enzymatic potencies than that of olaparib (IC50=3.5 nmol/L and 4.6 nmol/L vs 7.3 nmol/L).
To further develop this 2,4-difluorophenyl linker, we next explored analogs 21–25, in which the acyl fragment was replaced by alkyl groups (Table 1). Analogs 24 and 25 showed similar enzymatic activities to that of 15 (IC50=15.3 nmol/L and 10.4 nmol/L vs 16.1 nmol/L). However, analogs 21–23 replaced the acyl fragment with smaller methyl, ethyl, and propyl groups through enzymatic potency reductions that were approximately 2–10-fold. These data indicated a favorable role for bulky substituents on the terminal nitrogen of the piperazine ring. Thus, we next introduced either arylmethyl (26–29) or aryl (30) groups on the terminal nitrogen of the piperazine ring (Table 1). Analog 26, which had a benzyl substitution, demonstrated an enzymatic inhibitory activity (IC50=21.1 nmol/L) similar to that of 15. Analogs 27–29, which had heteroarylmethyl substitutions, showed approximately 2–3-fold less enzymatic inhibitory activity than that of 15. Significantly, analog 30, which had a phenyl ring on the terminal nitrogen of the piperazine ring, displayed an enzymatic potency (IC50=8.9 nmol/L) similar to that of olaparib.
Considering the phenyl substituent advantage at the terminal nitrogen of the piperazine ring, we next tested the efficacy of different substituted phenyl analogs (31–42, Table 2). Unexpectedly, electron-withdrawing or electron-donating substituents on the phenyl ring did not differentially affect activity (36 vs 38 vs 41). However, substituted positions within the phenyl ring substantially improved the activity, and substitution at the para-position of the phenyl ring showed the best enzymatic inhibitory activity, as compared with the substitutions at the ortho-position (31 vs 33, 39 vs 40 and 41 vs 42) and meta-position (35 vs 37).
Table 2.
In vitro activity of analogues 31–46.
 | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| No | R | PARP1 enzyme IC50 (nmol/L) | Cell activity IC50 (nmol/L)a | No | R | PARP1 enzyme IC50 (nmol/L) | Cell activity IC50 (nmol/L)a | ||
| V-C8 (BRCA2−/−) | V79 (BRCA2+/+) | V-C8 (BRCA2−/−) | V79 (s) | ||||||
| 1 | 7.3±0.5 | 263.6 | >10000 | ||||||
| 31 |
|
12.0±4.6 | 37 |
|
7.0±1.1 | 8579.9 | 2745.8 | ||
| 32 |
|
27.9±11.8 | 38 |
|
2.1±1.7 | 1585.1 | 1593.4 | ||
| 33 |
|
64.1±25.5 | 39 |
|
5.8±2.2 | 1108.4 | 852.5 | ||
| 34 |
|
33.9±4.1 | 40 |
|
24.3±7.7 | ||||
| 35 |
|
6.8±4.8 | 2916.4 | 1549.4 | 41 |
|
3.1±1.6 | 3254.2 | >10000 |
| 36 |
|
3.1±2.3 | 72.9 | 568.4 | 42 |
|
10.6±2.7 | ||
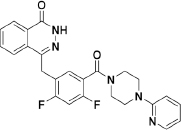
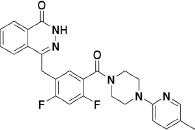
To extend the substitution at the terminal nitrogen of the piperazine, we replaced the phenyl ring with other heteroaryl rings (43–55, Table 3). Adding a pyrid-2-yl group (43) significantly enhanced the enzymatic inhibitory activity (43 vs 1 and 30). Thus, we explored the inhibitory effects of different substitutions on the pyridine ring (44–49, Table 3). The incorporation of a substituent, either electron-donating (47–49) or electron-withdrawing (44–46), at diverse positions in the pyridine ring was not beneficial and resulted in various losses of enzymatic inhibitory activity. Intriguingly, we observed that the double heteroatom-containing heteroaryl rings (50, pyrimidinyl and 54, thiazolyl) exhibited more promising potencies than that of analog 43, and analog 54 had the strongest effect. Not surprisingly, the size of the heteroaryl ring was very important for the enzymatic inhibitory activity, and substitutions (52–53) as well as fused aromatic rings (55) were not tolerable.
Table 3.
In vitro activity of analogues 47–55.
 | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| No | R | PARP1 enzyme IC50 (nmol/L) | Cell activity IC50 (nmol/L)a | No | R | PARP1 enzyme IC50 (nmol/L) | Cell activity IC50 (nmol/L)a | ||
| V-C8 (BRCA2−/−) | V79 (BRCA2+/+) | V-C8 (BRCA2−/−) | V79 (BRCA2+/+) | ||||||
| 1 | 7.3±0.5 | 263.6 | >10000 | ||||||
| 43 |
|
2.7±0.6 | 37.6 | 2737.8 | 50 |
|
2.2±1.3 | 3.2 | 1632.9 |
| 44 |
|
4.6±2.3 | 41.9 | 978.6 | 51 |
|
14.8±4.8 | ||
| 45 |
|
27.9±7.8 | 52 |
|
23.1±7.1 | ||||
| 46 |
|
9.1±4.5 | 774.8 | >10000 | 53 |
|
33.7±14.2 | ||
| 47 |
|
4.4±2.1 | 9.6 | 385.1 | 54 |
|
1.6±0.7 | 615.6 | >10000 |
| 48 |
|
3.9±2.1 | 115.1 | 3481.6 | 55 |
|
4.3±1.8 | 267.0 | 2662.1 |
| 49 |
|
33.9±15.7 | |||||||
Analysis of the data shown in Tables 1, 2, 3 revealed some noteworthy observations of the SAR for analogs 1 and 15–55: (1) replacement of the middle 2-fluorophenyl linker with the 2,4-difluorophenyl linker was tolerated (1 vs 15); (2) an R substituent at the terminal nitrogen of the piperazine ring was critical for high potency, and (aryl)alkyl substituents (21–29) were not beneficial, but acyl (especially aroyl for 17 and 20), aryl (especially para-substituted phenyl for 36, 38, and 41) and heteroaryl (especially unsubstituted heteroaryl for 43, 50, and 54) substituents improved the inhibitory activity; (3) among the studied sets of the R substituents (Tables 1, 2, 3), the potency increased in the order of heteroaryl>aryl>acyl>(aryl)alkyl.
Cell proliferation inhibition of potent analogs
From the results described above, seventeen analogs (17, 20, 30, 35–39, 41, 43–44, 46–48, 50, and 54–55) were identified as potent PARP1 inhibitors with IC50 values lower than 10 nmol/L. Therefore, these compounds were further evaluated for their inhibition of cell proliferation. As shown in Tables 1, 2, 3, the selected analogs together with the reference compound (1, olaparib) were tested in a matched pair of well-characterized Chinese hamster lung fibroblast V79 (wild type) and V-C8 (BRCA2 deficient) cell lines.
Interestingly, the benzoyl and 4-methylbenzoyl analogs (17 and 20) showed similar enzymatic potency, but analog 17 had a better cell inhibitory activity (IC50=13.9 nmol/L vs 99.2 nmol/L) and a better selectivity against BRCA2-deficient cells (SI=367 vs 27). Although analogs 30, 35, 37, and 46 demonstrated enzymatic inhibitory activity (IC50=6.8–9.1 nmol/L) similar to that of 1, they had much less potent cellular activity in the V-C8 assays (IC50>700 nmol/L). Moreover, a similar trend was also observed for analogs 38–39, 41, and 54, which displayed good enzymatic potency (IC50=1.6–5.8 nmol/L), although the cellular activity determined in the V-C8 assays was decreased 3–12-fold compared with that of olaparib. Additionally, analogs 36, 44, 48 and 55 presented both good enzymatic and cellular inhibitory activities. However, they also showed modest proliferation inhibition in wild-type V79 cells (IC50<3500 nmol/L), thus leading to worse selectivity against BRCA2-deficient cells compared with that of olaparib (SI=∼38 vs 8–30). Four of the most potent inhibitors (17, 43, 47, and 50) not only displayed better enzymatic (IC50=2.2–4.4 nmol/L) and cellular (IC50=3.2–37.6 nmol/L) inhibitory activities but also were more selective (SI=40–510), thus confirming that the tested PARP1 inhibitors selectively killed BRCA2-deficient cells. Therefore, we focused on these four analogs for further development.
Selectivity of analogs on BRCA1−/− and BRCA1+/+ cells
To further investigate the selectivity of six analogs (1, 30, 35, 43–44, 47, and 50), we tested their inhibitory activity against both the BRCA1-deficient human ovarian carcinoma cells UWB1.289 and BRCA1-complemented human ovarian carcinoma UWB1.289+BRCA1 cells. As shown in Table 4, these analogs selectively killed the BRCA1-deficient human ovarian carcinoma cells, especially analog 47, which had excellent selectivity compared with that of 1 [IC50(BRCA1+/+)/IC50(BRCA1−/−)=5.6 vs 10.2].
Table 4.
Selectivity of analogues on BRCA1−/− and BRCA1+/+ cells.
| No | Cell activity IC50 (μmol/L) (mean SD) | IC50 (BRCA1+/+)/IC50 (BRCA1−/−) | |
|---|---|---|---|
| UWB1.289 (BRCA1−/−) | UWB1.289+BRCA1 (BRCA2+/+) | ||
| 1 | 4.3±0.05 | 44.2±5.8 | 10.2 |
| 30 | 23.1±11.6 | 25.3±1.8 | 1.1 |
| 35 | 2.5±0.3 | 14.0±4.3 | 5.6 |
| 43 | 2.7±0.04 | 11.3±1.1 | 4.1 |
| 44 | 0.4±0.1 | 0.5±0.1 | 1.3 |
| 47 | 0.008±0.002 | 0.5±0.2 | 65.6 |
| 50 | 0.4±0.05 | 2.3±0.07 | 6.1 |
Water solubility evaluation
Because compound water solubility is an important property of drug potency, we next tested the water solubility of the four potent PARP1 inhibitors in pure water (17, 43, 47, and 50). As shown in Table 5, all four analogs showed poor solubility compared with that of olaparib (1.9–2.3 μg/mL vs 103.9 μg/mL), and the lower solubility may have been due to the higher number of lipophilic terminal (hetero)aryl groups. Considering the basic groups (tertiary amine) in the structures of analogs 43, 47, and 50, we explored the solubility of the hydrochloride salts to improve their water solubility. As expected, the corresponding hydrochloride salts of analogs 43 and 47 (56 and 57) presented significantly increased solubility values, which were 2193.0 μg/mL and 3601.0 μg/mL, respectively. The solubility was increased almost 21- and 35-fold compared with that of olaparib, yet the water solubility of the hydrochloride salt 58 was still less than 10 μg/mL. Next, we further tested the solubility of 43, 47, 50, 56, 57 and 58 in pH=7.4 and pH=1.0 buffered salt solutions (BSS). As shown in Table 6, the solubility of each pair of compounds in pH=7.4 was identical; however, the corresponding salts (56 and 57) displayed better solubility than their bases (43 and 47) in pH=1.0 buffer (414.4 μg/mL vs 1628.2 μg/mL, 1526.2 μg/mL vs 2652.5 μg/mL, respectively). Hence, we presumed that 56 and 57 would have accelerated absorption in the stomach (pH=1–3). Considering their excellent in vitro biological and water solubility profiles, 56 and 57 were further evaluated in an in vivo xenograft tumor model.
Table 5.
The water solubility of selected analogues.
| No | Analog | S (μg/mL) | No | Analog | S (μg/mL) |
|---|---|---|---|---|---|
| 1 |
|
103.9 | 50 |
|
0.4 |
| 17 |
|
1.9 | 56 |
|
2193.0 |
| 43 |
|
2.3 | 57 |
|
3601.0 |
| 47 |
|
2.0 | 58 |
|
3.0 |
Table 6.
The water solubility of selected analogues in pH=7.4 and pH=1.0 buffer.
| No | S (μg/mL) | No | S (μg/mL) | |
|---|---|---|---|---|
| pH=7.4 | ||||
| 43 | 1.9 | 56 | 2.4 | |
| 47 | 4.7 | 57 | 4.7 | |
| 50 | <1.0 | 58 | <1.0 | |
| pH=1.0 | ||||
| 43 | 414.4 | 56 | 1628.2 | |
| 47 | 1526.2 | 57 | 2652.5 | |
| 50 | 132.3 | 58 | 145.3 | |
In vivo antitumor studies
To assess the in vivo antitumor effects of 56 and 57 as single agents, nude mice with subcutaneous BRCA1-mutated human breast cancer MDA-MB-436 xenografts were treated with 56 or 57 once daily for 21 consecutive days (Figure 4). Oral administration of a 30 mg/kg dose of 56 significantly inhibited the growth of MDA-MB-436 xenografts, with an excellent growth inhibition rate of 96.6% at the end of treatment. Remarkably, a 10 mg/kg dose of 56 still showed a growth inhibition rate of 79.5% at the end of treatment, and this treatment was more effective than olaparib (1) at the 30 mg/kg dose (which resulted in a growth inhibition rate of 56.3%). When the dose of 56 was decreased to 1 mg/kg, the treatment resulted in a modest growth inhibition rate (43.2%), which was only slightly lower than that with the 30 mg/kg dose of olaparib. Those data were consistent with the in vitro cytotoxicity results of compounds 1 and 56 (IC50: 90.5 nmol/L for 1 and 15.7 nmol/L for 56) in MDA-MB-436 cells.
Figure 4.
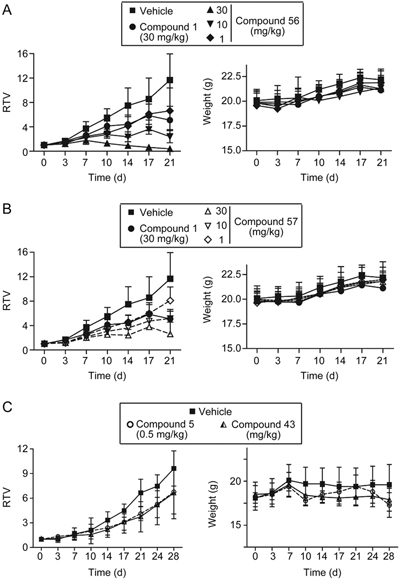
Analogues 56 and 57 exhibited antitumor activity against BRCA1-mutated MDA-MB-436 xenografts in nude mice. And 43 also showed anti-tumor effect against BRCA2-mutated Capan-1 xenografts. Nude mice with MDA-MB-436 and Capan-1 xenografts received and oral dose of the vehicle (0.2 mL/20 g body weight), the positive control (1), 56 or 57 once each day for 21 consecutive days and positive control (5), 43 once each day for 28 consecutive days. Tumor volume and the body weight of the nude mice were measured at the indicated time points. A (56), B (57), and C (43) represent the changes in relative tumor volume (RTV; calculated from the corresponding tumor volume) over the treatment time and the changes in body weight over the treatment time. Note: The same controls (vehicle and olaparib) were used for 56 and 57, but for clarity and readability, we plotted the data from the 56 groups and the 57 groups separately.
The mice treated with oral 57 at a 30 mg/kg dose showed a potent effect against tumor growth, with a growth inhibition rate of 77.6% at the 21-day time-point, an effect relatively weaker than that of 56 overall. Furthermore, 56 and 57 at the 1 mg/kg, 10 mg/kg and 30 mg/kg doses were well tolerated, and no lethality or significant weight loss was observed during the 21 consecutive treatment days (Figure 4).
Because 56 displayed excellent antitumor efficacy in subcutaneous BRCA1-mutated human breast cancer MDA-MB-436 xenografts, we next evaluated the inhibitory activities of 43 (the corresponding base of 56) and 56 against Capan-1 (BRCA2−/−) cells. The results showed that 43 presented had inhibitory activity than 56 (IC50=5.6 vs 34.4 nmol/L); therefore, we further assessed the in vivo antitumor effects of 43 in the subcutaneous BRCA2-mutated human pancreatic cancer Capan-1 xenografts. Nude mice with subcutaneous BRCA2-mutated human pancreatic cancer Capan-1 xenografts were treated with 43 once daily for 28 consecutive days (Figure 4C). Oral administration of a 10 mg/kg dose of 43 significantly inhibited the growth of Capan-1 xenografts, with a partial growth inhibition rate of 69.0% at the end of treatment, similarly to the results for Talazoparib (5) at the 0.5 mg/kg dose (which resulted in a growth inhibition rate of 70.4%). However, the mice in the 0.5 mg/kg dose group that were treated with Talazoparib (5) were in poor condition and experienced weight loss at the 10-day time-point; therefore, the administration was discontinued. The mice were allowed to recover until day 21, at which point treatment was administered until the end of the experiment.
Pharmacokinetic and in vitro safety properties
The excellent in vitro and in vivo biological properties of the hydrochloride salt 56 demonstrated that 56 was a good drug candidate that targeted PARP1. Therefore, we next assessed the pharmacokinetic properties of 56 in an in vivo rat model. As shown in Table 7, 56 and olaparib (1) demonstrated similar oral bioavailability (F) after oral exposure in Sprague-Dawley rats, with oral bioavailability values of 32.2% and 45.4%, respectively. Compared with olaparib, 56 had a slightly poorer oral half-life (0.6 h vs 1.0 h), displayed faster plasma clearance (3.0 L·h-1·kg-1 vs 1.8 L·h-1·kg-1) and had a larger volume distribution (3.5 L/kg vs 3.0 L/kg). Moreover, the safety evaluation experiment (see the detailed assay in the Supplementary Information) showed that the corresponding base 43 possessed low hERG inhibition activity (IC50=6.64 μmol/L). Altogether, these data demonstrate that 56 may be a viable candidate for discovering potential therapeutic drugs with specificity toward BRCA-deficient tumors.
Conclusions
In summary, we discovered a novel series of 2,4-difluorophenyl-linker analogs (15–55) derived from olaparib for use as PARP1 inhibitors. On the basis of the structure of the lead compound, olaparib, 41 completely novel 2,4-difluoro-based analogs were synthesized and tested in a PARP1 enzymatic inhibitory assay. Seventeen analogs, 17, 20, 30, 35–39, 41, 43, 44, 46–48, 50, 54 and 55, showed potent PARP1 inhibitory activities (IC50<10 nmol/L). The most potent analog (54) demonstrated a PARP1 inhibitory capability approximately five times higher than that of olaparib. Preliminary SARs showed that the 2,4-difluorophenyl linker of the new analogs inherited the PARP1 inhibitory activity, which was contributed by a 2-fluorophenyl linker. Additionally, terminal small steric aroyl, para-substituted phenyl, and unsubstituted heteroaryl substituents substantially improved the potency.
Cellular inhibition assays with a matched pair of V79 (wild-type) and V-C8 (BRCA2-deficient) cells further confirmed that four analogs (17, 43, 47, and 50) were potent PARP1 inhibitors (IC50=2.2–4.4 nmol/L) and effectively inhibited the proliferation of V79 cells (IC50=3.2–37.6 nmol/L) and had specificity toward BRCA-deficient cells (SI=40–510). Moreover, six selected analogs, 30, 35, 43, 44, 47, and 50, selectively kill BRCA1-deficient human ovarian carcinoma cells, as determined by evaluation of the inhibitory activities against both UWB1.289+BRCA1−/− cells and UWB1.289+BRCA1 cells. Analog 47 was particularly potent [IC50 (BRCA1+/+)/ IC50 (BRCA1−/−)=65.6]. Among the four potent analogs (17, 43, 47, and 50), the hydrochloride salts of analogs 43 and 47 (56 and 57) were found to have high water solubility in pH=1.0 buffered salt solutions (1628.3 μg/mL and 2652.5 μg/mL, respectively). The in vivo pharmacological results showed that 56 (30 mg/kg, 96.6%; 10 mg/kg, 79.5%) had a more potent tumor growth inhibition than did 1 (30 mg/kg, 56.3%). Analog 43 presented better inhibitory activity against Capan-1 (BRCA2−/−) than did the corresponding salt 56 (IC50=5.6 vs 34.4 nmol/L), and 43 (10 mg/kg, 69.0%) significantly inhibited tumor growth in the BRCA2-mutated xenograft model without negatively influencing the mice body weights. Promisingly, 56 had good oral bioavailability (F=32.2%), similar to that of olaparib (F=45.4%). Furthermore, the corresponding free base 43 exhibited minimal hERG inhibition activity (IC50=6.64 μmol/L). Overall, 56 has the potential to be developed as an antitumor drug, especially for treatment of BRCA-deficient tumors, by inhibiting PARP1. Further structural optimization of 56 is currently still being conducted in our laboratory.
Supplementary information
The synthetic procedures and related spectroscopic data for compounds 7–55, HPLC reports for the purity analysis of analogs 15–55, the hERG inhibition assay methods and the molecular docking details are available on the Acta Pharmacologica Sinica website.
Supplementary information
Supplementary Information (DOC 180 kb)
Acknowledgements
Financial support of this research was provided by the National Natural Science Foundation of China (Grants 21672064, 8152200403 and 21372001), the “Shu Guang” Project supported by the Shanghai Municipal Education Commission and the Shanghai Education Development Foundation (Grant 14SG28), the Science and Technology Commission of Shanghai Municipality (No 15431901200), and the Fundamental Research Funds for the Central Universities.
PowerPoint slides
Footnotes
Supplementary information is available on the website of Acta Pharmacologica Sinica.
Wen-hua Chen, Shan-shan Song and Ming-hui Qi: These authors contributed equally to this work.
Contributor Information
Guo-bin Ren, Email: rgb@ecust.edu.cn.
Ze-hong Miao, Email: zhmiao@simm.ac.cn.
Jian Li, Email: jianli@ecust.edu.cn.
References
- 1.He JX, Yang CH, Miao ZH. Poly(ADP-ribose) polymerase inhibitors as promising cancer therapeutics. Acta Pharmacol Sin. 2010;31:1172–80. doi: 10.1038/aps.2010.103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Wang YQ, Wang PY, Wang YT, Yang GF, Zhang A, Miao ZH. An update on poly(adp-ribose)polymerase-1 (PARP-1) inhibitors: opportunities and challenges in cancer therapy. J Med Chem. 2016;59:9575–98. doi: 10.1021/acs.jmedchem.6b00055. [DOI] [PubMed] [Google Scholar]
- 3.Burkle A. Poly (ADP-ribose). The most elaborate metabolite of NAD+ FEBS J. 2005;272:4576–89. doi: 10.1111/j.1742-4658.2005.04864.x. [DOI] [PubMed] [Google Scholar]
- 4.Krishnakumar R, Kraus WL. The PARP side of the nucleus: molecular actions, physiological outcomes, and clinical targets. Mol Cell. 2010;39:8–24. doi: 10.1016/j.molcel.2010.06.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hassa PO, Hottiger MO. The diverse biological roles of mammalian PARPS, a small but powerful family of poly-ADP-ribose polymerases. Front Biosci. 2008;13:3046–82. doi: 10.2741/2909. [DOI] [PubMed] [Google Scholar]
- 6.Rouleau M, Patel A, Hendzel MJ, Kaufmann SH, Poirier GG. PARP inhibition: PARP1 and beyond. Nat Rev Cancer. 2010;10:293–301. doi: 10.1038/nrc2812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Curtin NJ, Szabo C. Therapeutic applications of PARP inhibitors: anticancer therapy and beyond. Mol Aspects Med. 2013;34:1217–56. doi: 10.1016/j.mam.2013.01.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Yélamos J, Schreiber V, Dantzer F. Toward specific functions of poly(ADP-ribose) polymerase-2. Trends Mol Med. 2008;14:169–78. doi: 10.1016/j.molmed.2008.02.003. [DOI] [PubMed] [Google Scholar]
- 9.Yelamos J, Farres J, Llacuna L, Ampurdanes C, Martin-Caballero J. PARP-1 and PARP-2: New players in tumour development. Am J Cancer Res. 2011;1:328–46. [PMC free article] [PubMed] [Google Scholar]
- 10.Sandhu SK, Yap TA, de Bono JS. Poly(ADP-ribose) polymerase inhibitors in cancer treatment: a clinical perspective. Eur J Cancer. 2010;46:9–20. doi: 10.1016/j.ejca.2009.10.021. [DOI] [PubMed] [Google Scholar]
- 11.Langelier MF, Servent KM, Rogers EE, Pascal JM. A third zinc-binding domain of human poly(ADP-ribose) polymerase-1 coordinates DNA-dependent enzyme activation. J Biol Chem. 2008;283:4105–14. doi: 10.1074/jbc.M708558200. [DOI] [PubMed] [Google Scholar]
- 12.Langelier MF, Ruhl DD, Planck JL, Kraus WL, Pascal JM. The Zn3 domain of human poly(ADP-ribose) polymerase-1 (PARP-1) functions in both DNA-dependent poly(ADP-ribose) synthesis activity and chromatin compaction. J Biol Chem. 2010;285:18877–87. doi: 10.1074/jbc.M110.105668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wang Q. Cancer predisposition genes: molecular mechanisms and clinical impact on personalized cancer care: examples of Lynch and HBOC syndromes. Acta Pharmacol Sin. 2016;37:143–9. doi: 10.1038/aps.2015.89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Schreiber V, Dantzer F, Ame JC, de Murcia G. Poly(ADP-ribose): novel functions for an old molecule. Nat Rev Mol Cell Biol. 2006;7:517–28. doi: 10.1038/nrm1963. [DOI] [PubMed] [Google Scholar]
- 15.Peralta-Leal A, Rodríguez MI, Oliver FJ. Poly(ADP-ribose)polymerase-1 (PARP-1) in carcinogenesis: potential role of PARP inhibitors in cancer treatment. Clin Transl Oncol. 2008;10:318–23. doi: 10.1007/s12094-008-0207-8. [DOI] [PubMed] [Google Scholar]
- 16.Ferraris DV. Evolution of poly (ADP-ribose) polymerase-1 (PARP-1) inhibitors. From concept to clinic. J Med Chem. 2010;53:4561–84. doi: 10.1021/jm100012m. [DOI] [PubMed] [Google Scholar]
- 17.Amir E, Seruga B, Serrano R, Ocana A. Targeting DNA repair in breast cancer: a clinical and translational update. Cancer Treat Rev. 2010;36:557–65. doi: 10.1016/j.ctrv.2010.03.006. [DOI] [PubMed] [Google Scholar]
- 18.Heitz F, Harter P, Ewald-Riegler N, Papsdorf M, Kommoss S, du Bois A. Poly(ADP-ribosyl)ation polymerases: mechanism and new target of anticancer therapy. Expert Rev Anticancer Ther. 2010;10:1125–36. doi: 10.1586/era.10.53. [DOI] [PubMed] [Google Scholar]
- 19.Glendenning J, Tutt A. PARP inhibitors–current status and the walk towards early breast cancer. Breast. 2011;20:S12–9. doi: 10.1016/S0960-9776(11)70288-0. [DOI] [PubMed] [Google Scholar]
- 20.Wang Z, Wang F, Tang T, Guo C. The role of PARP1 in the DNA damage response and its application in tumor therapy. Front Med. 2012;6:156–64. doi: 10.1007/s11684-012-0197-3. [DOI] [PubMed] [Google Scholar]
- 21.Basu B, Yap TA, Molife LR, de Bono JS. Targeting the DNA damage response in oncology: past, present and future perspectives. Curr Opin Oncol. 2012;24:316–24. doi: 10.1097/CCO.0b013e32835280c6. [DOI] [PubMed] [Google Scholar]
- 22.De Vos M, Schreiber V, Dantzer F. The diverse roles and clinical relevance of PARPs in DNA damage repair: current state of the art. Biochem Pharmacol. 2012;84:137–46. doi: 10.1016/j.bcp.2012.03.018. [DOI] [PubMed] [Google Scholar]
- 23.Kummar S, Chen A, Parchment RE, Kinders RJ, Ji J, Tomaszewski JE. Advances in using PARP inhibitors to treat cancer. BMC Med. 2012;10:25. doi: 10.1186/1741-7015-10-25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ekblad T, Camaioni E, Schüler H, Macchiarulo A. PARP inhibitors: polypharmacology versus selective inhibition. FEBS J. 2013;280:3563–75. doi: 10.1111/febs.12298. [DOI] [PubMed] [Google Scholar]
- 25.Lupo B, Trusolino L. Inhibition of poly(ADP-ribosyl)ation in cancer: old and new paradigms revisited. Biochim Biophys Acta Rev Cancer. 2014;1846:201–15. doi: 10.1016/j.bbcan.2014.07.004. [DOI] [PubMed] [Google Scholar]
- 26.Menear KA, Adcock C, Boulter R, Cockcroft X, Copsey L, Cranston A. 4-[3-(4-cyclopropanecarbonylpiperazine-1-carbonyl)-4-fluorobenzyl]-2H- phthalazin-1-one: A novel bioavailable inhibitor of poly (ADP-ribose) polymerase-1. J Med Chem. 2008;51:6581–91. doi: 10.1021/jm8001263. [DOI] [PubMed] [Google Scholar]
- 27.Thomas HD, Calabrese CR, Batey MA, Canan S, Hostomsky Z, Kyle S. Preclinical selection of a novel poly (ADP-ribose) polymerase inhibitor for clinical trial. Mol Cancer Ther. 2007;6:945–56. doi: 10.1158/1535-7163.MCT-06-0552. [DOI] [PubMed] [Google Scholar]
- 28.Canan-Koch SS, Thoresen LH, Tikhe JG, Maegley KA, Yu XH, Zook SE. Novel tricyclic poly(ADP-ribose) polymerase-1 inhibitors with potentanticancer chemopotentiating activity: Design, synthesis, and X-ray cocrystal structure. J Med Chem. 2002;45:4961–74. doi: 10.1021/jm020259n. [DOI] [PubMed] [Google Scholar]
- 29.Jones P, Altamura S, Boueres J, Ferrigno F, Fonsi M, Giomini C. Discovery of 2-{4-[(3S)-piperidin-3-yl]phenyl}-2H-indazole- 7-carboxamide (MK-4827): A novel oral poly(ADP-ribose) polymerase (PARP) inhibitor efficacious in BRCA-1 and -2 mutant tumors. J Med Chem. 2009;52:7170–85. doi: 10.1021/jm901188v. [DOI] [PubMed] [Google Scholar]
- 30.Jones P, Wilcoxen K, Rowley M, Toniatti C. Niraparib: a poly (ADP-ribose) polymerase (PARP) inhibitor for the treatment of tumors with defective homologous recombination. J Med Chem. 2015;58:3302–14. doi: 10.1021/jm5018237. [DOI] [PubMed] [Google Scholar]
- 31.Donawho CK, Luo Y, Luo Y, Penning TD, Bauch JL, Bouska JJ. ABT-888, an orally active poly(ADP-ribose) polymerase inhibitor that potentiates DNA-damaging agents in preclinical tumor models. Clin Cancer Res. 2007;13:2728–37. doi: 10.1158/1078-0432.CCR-06-3039. [DOI] [PubMed] [Google Scholar]
- 32.Penning TD, Zhu GD, Gandhi VB, Gong J, Thomas S, Lubisch W. Discovery and SAR of 2-(1-propylpiperidin-4-yl)-1H-benzimidazole- 4-carbox-amide: A potent inhibitor of poly (ADP-ribose) polymerase (PARP) for the treatment of cancer. Bioorg Med Chem. 2008;16:6965–75. doi: 10.1016/j.bmc.2008.05.044. [DOI] [PubMed] [Google Scholar]
- 33.Shen Y, Rehman FL, Feng Y, Boshuizen J, Bajrami I, Elliott R. BMN 673, a novel and highly potent PARP1/2 inhibitor for the treatment of human cancers with DNA repair deficiency. Clin Cancer Res. 2013;19:5003–15. doi: 10.1158/1078-0432.CCR-13-1391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Böhm HJ, Banner D, Bendels S, Kansy M, Kuhn B, Müller K. Fluorine in medicinal chemistry. ChemBioChem. 2004;5:637–43. doi: 10.1002/cbic.200301023. [DOI] [PubMed] [Google Scholar]
- 35.O'Hagan D. Understanding organofluorine chemistry. An introduction to the C-F bond. Chem Soc Rev. 2008;37:308–19. doi: 10.1039/b711844a. [DOI] [PubMed] [Google Scholar]
- 36.Hunter L. The C–F bond as a conformational tool in organic and biological chemistry. Beilstein J Org Chem. 2010;6:38. doi: 10.3762/bjoc.6.38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Shah P, Westwell AD. The role of fluorine in medicinal chemistry. J Enzyme Inhib Med Chem. 2007;22:527–40. doi: 10.1080/14756360701425014. [DOI] [PubMed] [Google Scholar]
- 38.Eastman KJ, Gillis EP, Meanwell NA . Tactical applications of fluorine in drug design and development. Fluorine in Heterocyclic Chemistry, Volume 1, 5-Membered Heterocycles and Macrocycles; Nenajdenko V. Ed.; Springer International: Cham, Switzerland, 2014; 1–54.
- 39.Meanwell NA. Improving drug candidates by design: a focus on physicochemical properties as a means of improving compound disposition and safety. Chem Res Toxicol. 2011;24:1420–56. doi: 10.1021/tx200211v. [DOI] [PubMed] [Google Scholar]
- 40.Hopkins AL, Keserü GM, Leeson PD, Rees DC, Reynolds CH. The role of ligand efficiency metrics in drug discovery. Nat Rev Drug Discov. 2014;13:105–21. doi: 10.1038/nrd4163. [DOI] [PubMed] [Google Scholar]
- 41.Ritchie TJ, Macdonald SJF. How drug-like are “ugly” drugs: do drug-likeness metrics predict ADME behaviour in humans? Drug Discov Today. 2014;19:489–95. doi: 10.1016/j.drudis.2014.01.007. [DOI] [PubMed] [Google Scholar]
- 42.Wager TT, Kormos BL, Brady JT, Will Y, Aleo MD, Stedman DB. Improving the odds of success in drug discovery: choosing the best compounds for in vivo toxicology studies. J Med Chem. 2013;56:9771–9. doi: 10.1021/jm401485p. [DOI] [PubMed] [Google Scholar]
- 43.Tarcsay A, Keserú GM. Contributions of molecular properties to drug promiscuity. J Med Chem. 2013;56:1789–95. doi: 10.1021/jm301514n. [DOI] [PubMed] [Google Scholar]
- 44.Yusof I, Segall MD. Considering the impact drug-like properties have on the chance of success. Drug Discov Today. 2013;18:659–66. doi: 10.1016/j.drudis.2013.02.008. [DOI] [PubMed] [Google Scholar]
- 45.Ye N, Chen CH, Chen T, Song Z, He JX. Huan XJ et al. Design, synthesis, and biological evaluation of a series of benzo[de][1,7] naphthyridin-7(8H)-ones bearing a functionalized longer chain appendage as novel PARP1 inhibitors. J Med Chem. 2013;56:2885–903. doi: 10.1021/jm301825t. [DOI] [PubMed] [Google Scholar]
- 46.Yuan B, Ye N, Song SS, Wang YT, Song Z, Chen HD. Poly(ADPribose)polymerase (PARP) inhibition and anticancer activity of simmiparib, a new inhibitor undergoing clinical trials. Cancer Lett. 2017;386:47–56. doi: 10.1016/j.canlet.2016.11.010. [DOI] [PubMed] [Google Scholar]
- 47.He JX, Wang M, Huan XJ, Chen CH, Song SS, Wang YQ. Novel PARP1/2 inhibitor mefuparib hydrochloride elicits potent in vitro and in vivo anticancer activity characteristic of high tissue distribution. Oncotarget. 2017;8:4156–68. doi: 10.18632/oncotarget.13749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Yang Zhong-min, Liao Xue-mei, Chen Yi, Shen Yan-yan, Yang Xin-ying, Su Yi, Sun Yi-ming, Gao Ying-lei, Ding Jian, Zhang Ao, He Jin-xue, Miao Ze-hong. Combining 53BP1 with BRCA1 as a biomarker to predict the sensitivity of poly(ADP-ribose) polymerase (PARP) inhibitors. Acta Pharmacologica Sinica. 2017;38(7):1038–1047. doi: 10.1038/aps.2017.8. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Information (DOC 180 kb)