Abstract
Application of CRISPR-Cas9 technology in diverse organisms has resulted in an explosion of genome modification efforts. To expand the toolbox of applications, we have created an E. coli Exonuclease I (sbcB) - Cas9 fusion that has altered enzymatic activity in zebrafish embryos. This Cas9 variant has increased mutation efficiency and favors longer deletions relative to wild type Cas9. We anticipate that this variant will allow for more efficient screening for F0 phenotypes and mutation of a larger spectrum of genomic targets including deletion of regulatory regions and creating loss of function mutations in transcription units with poor sequence conservation such as lncRNAs where larger deletions may be required for loss of function.
Keywords: Targeted Mutagenesis, S. pyogenes Cas9, Danio rerio
Introduction
Use of CRISPR-Cas9 mutagenesis has transformed reverse genetic approaches in the zebrafish and other organisms. Many labs adopted this method due to its efficiency and low entry cost. However, the efficiency of targeting is inconsistent between sgRNAs and it can be challenging to target some genes and non-coding regions (Carrington et al., 2015). Attempts at creating rules for sgRNA design have been challenging as well (Gagnon et al., 2014; Haeussler et al., 2016; Thyme et al., 2016; Varshney et al., 2015). The lesions created by repair of Cas9 induced double stranded breaks show a bias limiting the spectrum of mutations that can be recovered from wild type (WT) Cas9 induced breaks in some cases (Gagnon et al., 2014; Thomas et al., 2014). Cas9 induced NHEJ typically results in small deletions but creation of deletions on the order of tens or hundreds of base pairs would be very useful. These types of lesions can be used to create clear loss of function alleles, to delete gene regulatory regions and to delete large segments of non-coding RNAs. While these types of medium sized deletions can be accomplished by targeting two different regions of a DNA sequence (Xiao et al., 2013; Zhang et al., 2015), efficiency of this method is dependent on the presence of effective target sites and the efficiency of both guides.
Recent work on the latency of Cas9 binding to cleaved DNA described the 3’ end of the nontarget strand as being freed from Cas9 before dissociation of the rest of the DNA:sgRNA:Cas9 complex (Richardson et al., 2016). The cell cycle of the early zebrafish embryo is shorter than the half-life for release so Cas9 is likely to be displaced by the replication fork. However, we hypothesized that the long latency and asymmetry of dissociation of the Cas9 sgRNA complex from the target may make the genomic DNA accessible to enzymatic manipulation from Cas9 fusion proteins. Thus, we hypothesized that addition of an exonuclease to Cas9 would result in larger deletions in zebrafish embryos and might affect the mutagenesis rate of CRISPR-Cas9 by altering the structure of the DNA ends.
ExoI is encoded by sbcB (xonA) in E. coli and it was originally shown to be an inhibitor of homologous recombination (Kushner et al., 1971), specifically blocking protein binding in the RecF pathway (Phillips et al., 1988). Its exonuclease activity prefers a single stranded free 3’ hydroxyl end of a deoxy-ribo oligonucleotide at a rate of about 40,000 greater than intact, double-stranded DNA (Lehman and Nussbaum, 1964). There is no evidence in the literature that it must interact with a binding partner to initiate this exonuclease activity making it an excellent candidate to add an exonuclease activity to the endonuclease Cas9. We fused the 3’ to 5’ exonuclease E. coli sbcB (ExoI) to the N terminus of Cas9 and assayed the mutagenesis rate of specific loci of the fusion protein “ExoCas9” relative to wild type Cas9 and characterized the lesions that resulted from the repair of these breaks. We found that embryos injected with ExoCas9 mRNA and gene specific sgRNAs had a higher rate of mutant phenotype, a higher rate of indel formation and created larger deletions relative to unmodified Cas9.
Results and Discussion
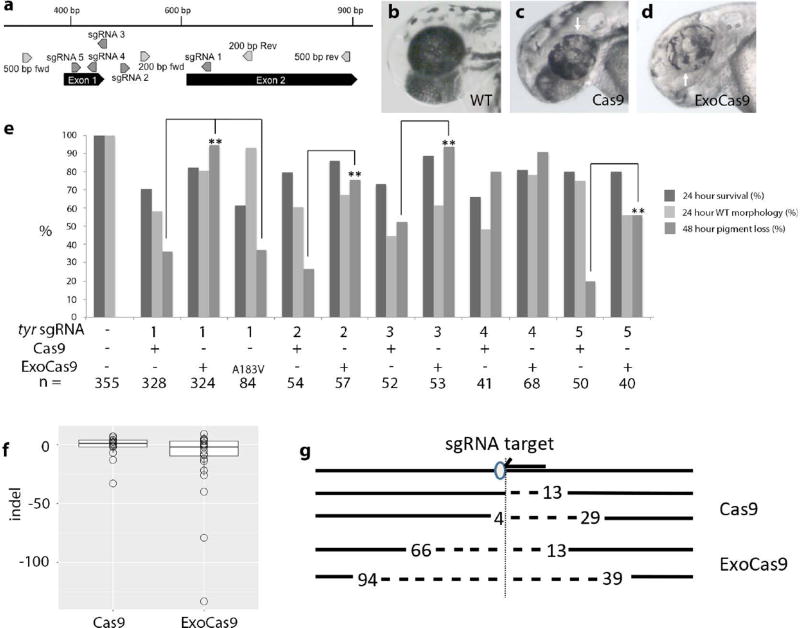
To test the activity of ExoCas9 (construction of ExoCas9 is described in the Methods section), we targeted tyrosinase (tyr) with a previously published sgRNA (Figure 1 a) (Jao et al., 2013). Tyrosinase is required for melanin production and bi-allelic mutation of this gene is readily visible 2 days post fertilization (dpf) as a loss of eye and body pigment (Figure 1 b–d). Microinjection of the same amount of ExoCas9 mRNA greatly increased the percentage of embryos displaying loss of pigment in the eye (Figure 1 e) relative to Cas9 (sgRNA 1 p < 1.0 E-5). This increased mutation rate was also confirmed with 4 additional sgRNAs for tyr (Figure 1 a&e) and only one sgRNA did not display a significant increase in efficiency (p = 7.2 E-5, 1.5 E-5, 0.26, and 1.6 E-3 for sgRNAs 2–5 respectively). Interestingly, introduction of the exonuclease activity did not result in widespread cell death and the survival was comparable if not better than the survival of Cas9 alone (Figure 1 e).
Figure 1. Visualization of CRISPR targeting, frequency of lesions produced by ExoCas9 and Cas9 in tyrosinase in the F0 generation, and quantification.
(a) Gene diagram for region of interest in tyrosinase. (b–d) tyrosinase targeting results in mosaic loss of pigment 2 dpf; wild type (b), Cas9 mutant (c), and ExoCas9 mutant (d). Areas of pigment loss in the eye are indicated by a white arrow in (c–d). (e) Survival and pigment loss induced with 5 different tyrosinase sgRNAs at 48 hpf for Cas9, ExoCas9 and A183V ExoCas9 (* = p < 0.05 and ** = p < 0.005). (f) Box and whisker plots of insertions and deletions produced by Cas9 (left) and ExoCas9 (right). The line in the middle of the box is the median value. The box itself represents surrounds the middle 50%, with the whiskers are first 25th (top) percentile and last 75th percentile (bottom). (g) Directionality of deletions shown relative sgRNA target (arrow) and protospacer adjacent motif (PAM, oval), followed by the two largest Cas9 deletions (dotted line and length relative to the PAM is indicated numerically), followed by the two largest ExoCas9 deletions
To confirm that the increased effectiveness is due to the Exo1 activity, we engineered and tested a point mutation in the exonuclease domain of ExoCas9, sbcB15 (A183V). This mutation has been shown to have 1.3% of wild type activity when tested in E. coli and is hypothesized to affect Mg2+ binding (Thoms et al., 2008). When tested with tyr sgRNA1, the 183 mutation did not show a significant increase in pigment loss when compared to Cas9 alone (p = 0.91), however, this mutation was not as efficient when compared to ExoCas9 (p < 1.0 E-5) (Figure 1 e), suggesting that the increase in activity is in fact due to the added ExoI activity.
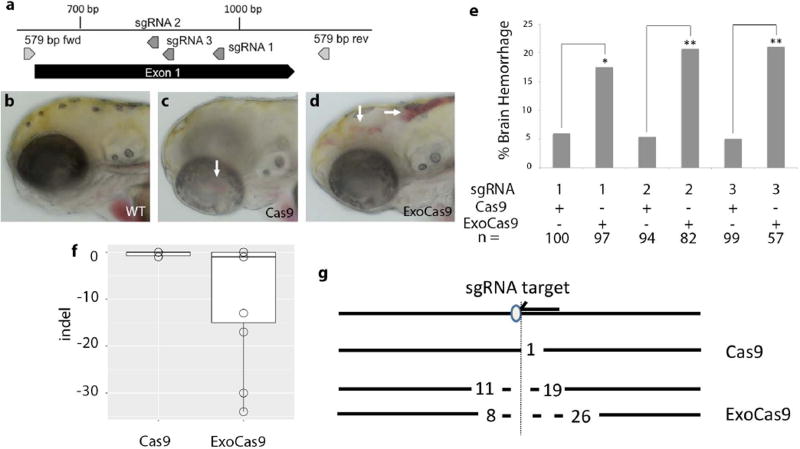
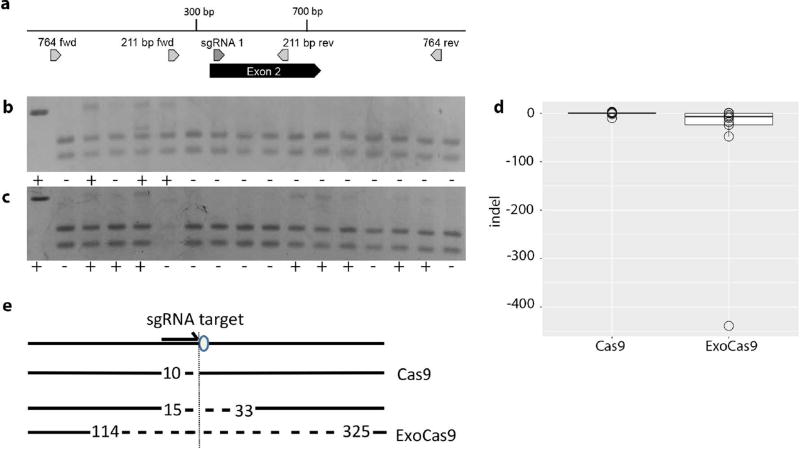
We hypothesized that local chromatin confirmation may be responsible for the increased effectiveness of ExoCas9 at the tyr locus. To test for locus dependence of ExoCas9, we confirmed an enhanced mutation rate by targeting the transforming growth factor, beta receptor II (tgfbr2b) (Figure 2 a). Tgfbr2 is required for brain angiogenesis in mouse and we hypothesized that mutation of tgfbr2b would result in brain hemorrhage in zebrafish embryos (Hirota et al., 2015; Nguyen et al., 2011). Microinjection of equal amounts of Cas9 and ExoCas9 mRNAs with an sgRNA targeting tgfbr2b resulted in a higher percentage of ExoCas9 embryos displaying brain hemorrhage (Figure 2 b–e) (p = 0.012). This increased activity was also confirmed with 2 additional sgRNAs for tgfbr2b (Figure 2 a&e) (p = 2.1 E-3 for both sgRNA 2 and 3), thus showing a significant increase in targeting for all the tgfbr2b sgRNAs tested. We also confirmed the increased effectiveness of ExoCas9 by targeting a third gene, ripk4 (Figure 3 a). This gene does not have an observable embryonic mosaic loss of function phenotype (TPC, DSW unpublished), so we performed a restriction fragment length polymorphism assay (RFLP assay) to confirm targeting in the F0 generation. A restriction enzyme site is adjacent to the protospacer adjacent motif (PAM) in the ripk4 sgRNA target site. Thus, creation of insertion or deletion (indel) mutations, this restriction is likely altered and therefore unable to be cut creating an RFLP. Embryos injected with ExoCas9 displayed a higher rate of targeting compared to that of Cas9 as confirmed through the RFLP assay (Figure 3 b–c) (p = 0.003).
Figure 2. Visualization of CRISPR targeting, frequency of lesions produced by ExoCas9 and Cas9 in tgfbr2b in the F0 generation, and quantification.
(a) Gene diagram for region of interest in tgfbr2b. (b–d) tgfbr2b targeting results in brain hemorrhage 3 dpf; wild type (b), Cas9 mutant (c), and ExoCas9 mutant (d). Areas of brain hemorrhage indicated by a white arrows in (c–d). (e) tgfbr2b CRISPR induced brain hemorrhage rates from 3–5 dpf. (f) Box and whisker plots of insertions and deletions produced by Cas9 (left) and ExoCas9 (right). (g) Directionality of deletions shown relative sgRNA target (arrow) and protospacer adjacent motif (PAM, oval), followed by the two largest Cas9 deletions (dotted line and length relative to the PAM is indicated numerically), followed by the two largest ExoCas9 deletions.
Figure 3. Frequency of lesions produced by ExoCas9 and Cas9 in ripk4 in the F0 generation and quantification.
(a) Gene diagram for region of interest in ripk4. (b) ripk4 Cas9 and (c) ripk4 ExoCas9 targeting is confirmed and quantified through RFLP analysis, positive (+) (WT uncut) and negative (−) in the first box to the left followed by the experimental runs to the right. (d). Box and whisker plots of insertions and deletions produced by Cas9 (left) and ExoCas9 (right). (e) Directionality of deletions shown relative sgRNA target (arrow) and protospacer adjacent motif (PAM, oval), followed by the largest Cas9 deletion (length relative to the PAM is indicated numerically), followed by the two largest ExoCas9 deletions.
We also analyzed the lesions made with both Cas9 variants to determine the nature of the lesions created by Cas9 and ExoCas9. We hypothesized that lesions produced by ExoCas9 would be large so we opted for sequencing of individual clones to limit bias toward smaller deletion that might occur from short read, high depth methods. Genomic DNA surrounding the sgRNA target sites from tyr, tgfbr2b, and ripk4 was amplified from injected embryos by PCR. This DNA was cloned and multiple clones were sequenced. We found that ExoCas9 induced longer deletions for tyr and ripk4 (Figures 1 f, 3d, and Table 1) (p = 0.018 and 0.011 respectively). Interestingly all larger deletions (> 40 bp) had a directional bias relative to the orientation of the guide RNA recognition sequence. We observed that the larger deletion was 3’ of the PAM site on the non-target strand indicating a bias in deletion direction for all of the loci that produced large deletions (< 40 bp) (Figures 1 g and 3 e). Although, the tgfbr2b locus did produce longer deletions using ExoCas9 none were longer than 40 bp and these results were not statistically significant (Figure 2f and Table 1) (p = 0.52).
Table 1. Summary and quantification of deletions produced using Cas9 and ExoCas9.
The number of clones analyzed, the largest deletion, and the range of deletions are quantified for both Cas9 and ExoCas9 for each gene target analyzed.
| Target | n= | Max deletion |
Deletions > 40 |
Deletions < 40 and > 10 |
Deletions < 10 and > 0 |
WT | Insertions |
|---|---|---|---|---|---|---|---|
| tyr Cas9 | 34 | 33 | 0 | 2 (5.9%) | 11 (32.4%) | 1 (2.9%) | 20 (58.9%) |
| tyr ExoCas9 | 32 | 133 | 3 (9.4%) | 5 (15.6%) | 11 (34.4%) | 1 (3.1%) | 12 (37.5%) |
| tgfbr2b Cas9 | 18 | 1 | 0 | 0 | 5 (27.8%) | 13 (72.2%) | 0 |
| tgfbr2b ExoCas9 | 15 | 34 | 0 | 2 (13.3%) | 5 (33.3%) | 4 (26.6%) | 4 (26.6%) |
| ripk4 Cas9 | 23 | 10 | 0 | 1 (4.3%) | 1 (4.3%) | 18 (78.3%) | 3 (13.0%) |
| ripk4 ExoCas9 | 32 | 439 | 2 (6.3%) | 4 (12.5%) | 2 (6.3%) | 23 (71.9%) | 1 (3.1%) |
Gene targeting with ExoCas9 resulted in a more efficient F0 mutant phenotype production, increased targeting rate, and longer deletions. We hypothesize that the 3’-5’ nuclease activity digests the 3’ ends before nonhomologous end joining (NHEJ) can repair the lesion. The lesion resulting from this digestion has a 5’ overhang, which may be resected, resulting in a large deletion.
We predict that ExoCas9 has a multitude of applications. The first being the capacity to target regions of DNA that are too difficult to target with Cas9 because sequence bias limits the ability to design multiple guide RNAs to promote efficient deletions. It may also be useful to target gene regulatory regions where redundancy and cooperative binding mean larger deletions are required to eliminate activity. Examples include transcription factor binding sites or UTR regulatory elements. Also, transcription units with poor sequence conservation such as long non-coding RNAs (lncRNAs) may require larger deletions to ensure essential domains are removed (Fatica and Bozzoni, 2014).
Methods
ExoCas9 was constructed by fusing the E. coli sdcB open reading frame in frame with Cas9 to replace the N-terminal nuclear localization signal in the pCS2+ nls Cas9 nls construct (Jao et al., 2013). This construct was assembled by Fastcloning (Li et al., 2011). Briefly, pCS2+ nlsCas9nls was amplified using the primers: TGCTTACCGTCATTCATCATGGTGGCGTCGACATTGGATC AGTACGCGGAAGAGATTGTCGGAAGCATGGATAAGAAGTA E. coli genomic DNA was amplified using the primers: GATCCAATGTCGACGCCACCATGATGAATGACGGTAAGCA TACTTCTTATCCATGCTTCCGACAATCTCTTCCGCGTACT PCR products were treated with DpnI and transformed into E .coli and ampicillin resistant colonies were screened by restriction digest. The sequence of the entire reading frame of the selected clone was confirmed by sequencing. This construct will be available for distribution following publication.
All zebrafish embryos were injected at the one-cell stage with in vitro transcribed sgRNA and Cas9 or ExoCas9 mRNA according to (Gagnon et al., 2014). sgRNAs were produced by synthesizing an oligo with bacteriophage promoter T7 (GCGCATGCTAATACGACTCACTATA) or Sp6 (GCGCATGCTATTTAGGTGACACTATA) promoter sequence at 5’ end followed by a sgRNA gene specific sequence (without PAM motif) and a 3’ backbone sequence (GTTTAAGAGCTATGCTGGAAAC). This oligonucleotide was annealed to a constant backbone sequence (AAAAGCACCGACTCGGTGCCACTTTTTCAAGTTGATAACGGACTAGCCTTATTTAAACTTGCTATGCTGTTTCCAGCATAGCTCTTAAAC) and extended according to (Gagnon et al., 2014). The sgRNAs backbone contains a base-pair switch (bold above) and extended hairpin (underlined above) according to (Chen et al., 2013). Concentrations of transcripts were confirmed via gel electrophesis and nanodrop measurements.
Specific sgRNA target sites are:
tyrosinase 1: GGACTGGAGGACTTCTGGGG
tyrosinase 2: GGGTGTGTGTGAAGCGTCTC
tyrosinase 3: GCAGAGTGTAAAGCCTCTCC
tyrosinase 4: GCAGTGTGTAAAGCATCTCC
tyrosinase 5: GAGAGAGTGTGTGCGTAAAG
tgfbr2b 1: TGTGGGAGACCCAAAGTGCC
tgfbr2b 2: GCAGGAGTACTTGATGCGCC
tgfbr2b 3: CTTGCTCAGCTGGGAAGACC
ripk4: GGGGCTGGTGATGGAGTACA
All work using live vertebrate animals was performed under protocols approved by the Rice University Institutional Animal Care and Use Committee. Wild type AB strain was used for all experiments. Embryos were maintained at 28 °C in E3 medium. Embryos were scored for pigment defects at 2 days post fertilization (dpf). To score for the brain hemorrhage phenotype, embryos were treated with 1× PTU (0.003%) in E3 at 1 dpf to prevent pigment formation and allow for unobscured visualization of the brain. Hemorrhages were scored at 3 and 4 dpf. Only embryos with robust circulation were scored and hemorrhages were counted once even if they persisted over multiple days as in (Hirota et al., 2015). Statistical analysis on the efficiency of phenotype production was performed using the n-1 proportion test as in (Hirota et al., 2015). Statistical analysis on the length of deletions produced was performed using the one-tailed Mann-Whitney U test.
Deletions were observed by lysing individual embryos at 3 dpf, pooling the embryos (n=8), amplifying the target region, and cloning the region into the TOPO Zero Blunt Vector (Invitrogen). Genotyping primers:
- tyrosinase
- 200 bp Fwd: GAAGCGTCTCACTCTCCTCG
- 200 bp Rev: ACAGAACCCTCGACCTGACT
- 500 bp Fwd: GTGTGTGTGTGCGAGAGAGA
- 500 bp Rev: CAGTTGGCACCGAAGAAGC
- tgfbr2b
- 579 bp Fwd: ACCCCTTTTAGGTGGGCAAG
- 579 bp Rev: AGCTGAAGTCTGCATGATTCTGA
- ripk4
- 211 bp Fwd: GGAGGCCAAGAAGATGGAGG
- 211 bp Rev: AGGTGGATTCATGCAGTGCA
- 764 bp Fwd: GAGGAAGGTCAATGGCAC
- 764 bp Rev: TCTCTGCTCTGTCGACTTTTGA
Acknowledgments
This work was supported by NIH R21 NS085688 to JHM and DSW.
Footnotes
The authors have no conflicts to declare.
References
- Carrington B, Varshney GK, Burgess SM, Sood R. CRISPR-STAT: an easy and reliable PCR-based method to evaluate target-specific sgRNA activity. Nucleic Acids Res. 2015 doi: 10.1093/nar/gkv802. gkv802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen B, Gilbert LA, Cimini BA, Schnitzbauer J, Zhang W, Li G-W, Park J, Blackburn EH, Weissman JS, Qi LS, et al. Dynamic Imaging of Genomic Loci in Living Human Cells by an Optimized CRISPR/Cas System. Cell. 2013;155:1479–1491. doi: 10.1016/j.cell.2013.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fatica A, Bozzoni I. Long non-coding RNAs: new players in cell differentiation and development. Nat. Rev. Genet. 2014;15:7–21. doi: 10.1038/nrg3606. [DOI] [PubMed] [Google Scholar]
- Gagnon JA, Valen E, Thyme SB, Huang P, Ahkmetova L, Pauli A, Montague TG, Zimmerman S, Richter C, Schier AF. Efficient Mutagenesis by Cas9 Protein-Mediated Oligonucleotide Insertion and Large-Scale Assessment of Single-Guide RNAs. PLoS ONE. 2014;9:e98186. doi: 10.1371/journal.pone.0098186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haeussler M, Schönig K, Eckert H, Eschstruth A, Mianné J, Renaud J-B, Schneider-Maunoury S, Shkumatava A, Teboul L, Kent J, et al. Evaluation of off-target and on-target scoring algorithms and integration into the guide RNA selection tool CRISPOR. Genome Biol. 2016;17:148. doi: 10.1186/s13059-016-1012-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirota S, Clements TP, Tang LK, Morales JE, Lee HS, Oh SP, Rivera GM, Wagner DS, McCarty JH. Neuropilin 1 balances β8 integrin-activated TGFβ signaling to control sprouting angiogenesis in the brain. Development. 2015;142:4363–4373. doi: 10.1242/dev.113746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jao L-E, Wente SR, Chen W. Efficient multiplex biallelic zebrafish genome editing using a CRISPR nuclease system. Proc. Natl. Acad. Sci. 2013 doi: 10.1073/pnas.1308335110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kushner SR, Nagaishi H, Templin A, Clark AJ. Genetic Recombination in Escherichia coli: The Role of Exonuclease I. Proc. Natl. Acad. Sci. 1971;68:824–827. doi: 10.1073/pnas.68.4.824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lehman IR, Nussbaum AL. The Deoxyribonucleases of Escherichia coli V. ON THE SPECIFICITY OF EXONUCLEASE I (PHOSPHODIESTERASE) J. Biol. Chem. 1964;239:2628–2636. [PubMed] [Google Scholar]
- Li C, Wen A, Shen B, Lu J, Huang Y, Chang Y. FastCloning: a highly simplified, purification-free, sequence- and ligation-independent PCR cloning method. BMC Biotechnol. 2011;11:92. doi: 10.1186/1472-6750-11-92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu D, Keck JL. Structural basis of Escherichia coli single-stranded DNA-binding protein stimulation of exonuclease I. Proc. Natl. Acad. Sci. U. S. A. 2008;105:9169–9174. doi: 10.1073/pnas.0800741105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen H-L, Lee YJ, Shin J, Lee E, Park SO, McCarty JH, Oh SP. TGF-β signaling in endothelial cells, but not neuroepithelial cells, is essential for cerebral vascular development. Lab. Investig. J. Tech. Methods Pathol. 2011;91:1554–1563. doi: 10.1038/labinvest.2011.124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phillips GJ, Prasher DC, Kushner SR. Physical and biochemical characterization of cloned sbcB and xonA mutations from Escherichia coli K-12. J. Bacteriol. 1988;170:2089–2094. doi: 10.1128/jb.170.5.2089-2094.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richardson CD, Ray GJ, DeWitt MA, Curie GL, Corn JE. Enhancing homology-directed genome editing by catalytically active and inactive CRISPR-Cas9 using asymmetric donor DNA. Nat. Biotechnol. 2016 doi: 10.1038/nbt.3481. advance online publication. [DOI] [PubMed] [Google Scholar]
- Thomas HR, Percival SM, Yoder BK, Parant JM. High-throughput genome editing and phenotyping facilitated by high resolution melting curve analysis. PloS One. 2014;9:e114632. doi: 10.1371/journal.pone.0114632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thoms B, Borchers I, Wackernagel W. Effects of Single-Strand DNases ExoI, RecJ, ExoVII, and SbcCD on Homologous Recombination of recBCD+ Strains of Escherichia coli and Roles of SbcB15 and XonA2 ExoI Mutant Enzymes. J. Bacteriol. 2008;190:179–192. doi: 10.1128/JB.01052-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thyme SB, Akhmetova L, Montague TG, Valen E, Schier AF. Internal guide RNA interactions interfere with Cas9-mediated cleavage. Nat. Commun. 2016;7:11750. doi: 10.1038/ncomms11750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varshney GK, Pei W, LaFave MC, Idol J, Xu L, Gallardo V, Carrington B, Bishop K, Jones M, Li M, et al. High-throughput gene targeting and phenotyping in zebrafish using CRISPR/Cas9. Genome Res. 2015 doi: 10.1101/gr.186379.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao A, Wang Z, Hu Y, Wu Y, Luo Z, Yang Z, Zu Y, Li W, Huang P, Tong X, et al. Chromosomal deletions and inversions mediated by TALENs and CRISPR/Cas in zebrafish. Nucleic Acids Res. 2013;41:e141–e141. doi: 10.1093/nar/gkt464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang L, Jia R, Palange NJ, Satheka AC, Togo J, An Y, Humphrey M, Ban L, Ji Y, Jin H, et al. Large Genomic Fragment Deletions and Insertions in Mouse Using CRISPR/Cas9. PLOS ONE. 2015;10:e0120396. doi: 10.1371/journal.pone.0120396. [DOI] [PMC free article] [PubMed] [Google Scholar]