Abstract
Cytochrome P450 2B6 (CYP2B6), mainly expressed in the liver and brain, is important for processing a number of widely used drugs. Variations in CYP2B6 expression are associated with decreased drug efficacy or adverse effects in some patients. Although CYP2B6 genetic variants are associated with its differential expression, epigenetic mechanisms affecting CYP2B6 gene regulation have not been established. Sequence analysis identified 29 domains in the CYP2B6 mRNA transcript that could be subject to regulation by microRNAs. Inverse correlations were found in human hepatocytes for the levels of the microRNAs hsa-miR-504-5p and hsa-miR-25-3p compared with CYP2B6 mRNA. Reporter gene assays showed that hsa-miR-25-3p suppresses CYP2B6 expression by targeting a specific sequence in the 3′-untranslated region of the mRNA transcript. Electrophoretic mobility shift assays confirmed that hsa-miR-25-3p forms stable complexes with its cognate mRNA sequence and that it recruits cellular factors, including Ago-4. Transfection of HepaRG cells with hsa-miR-25-3p mimics inhibited expression of the endogenous CYP2B6 gene and it also decreased rifampicin-dependent induction of CYP2B6 at the mRNA and protein levels.
In summary, in silico and in vitro analyses show that hsa-miR-25-3p suppresses CYP2B6 expression in human liver cells via an epigenetic mechanism.
Keywords: hsa-miR-25-3p, CYP2B6, Drug metabolizing enzymes, Pharmacogenomics, Inter-individual variability
1. Introduction
Human cytochrome P450 2B6 (CYP2B6), mainly expressed in human liver and brain, is a member of the cytochrome P450 family and it constitutes about 2–10% of the total hepatic CYP450 content [1]. As an important drug metabolizing enzyme (DME), CYP2B6 is involved in the metabolism of nearly 25% of drugs in the market today, including the anticancer drugs cyclophosphamide, ifosfamide, and tamoxifen and the central nervous system active agents mephobarbital, bupropion, and selegiline [2]. It is worthwhile to mention that the enzyme is highly inducible by several drugs, such as vitamin D, cyclophosphamide, and rifampicin [1,3]. In addition, the inter-individual variability in the expression of CYP2B6 in humans is relatively high [4]. We speculate that individual differences in the extent of CYP2B6 expression due to variations in genetic, epigenetic regulation, and drug induction may result in decreased efficacy or adverse effects for certain therapeutics. A more thorough knowledge of CYP2B6 regulation will be necessary to test this hypothesis.
Although our understanding of the mechanisms regulating CYP2B6 expression is somewhat incomplete, some genetic variants in CYP2B6 are known to influence gene regulation at the level of transcription [1,5,6]. One of the most important CYP2B6 haplotypes, CYP2B6*6, is comprised by two non-synonymous variants CYP2B6 785 A>G and CYP2B6 516 G>T. These sequence variants occur together, with or without additional variants in the CYP2B6 promoter region, notably −1456 T>C and −750 T>C. The CYP2B6*6 haplotype is associated with decreased CYP2B6 enzyme activity for metabolizing bupropion and efavirenz [7]. In addition, the CYP2B6*6 haplotype is associated with increased cyclophosphamide-induced liver toxicity [8]. However, regulatory pathways for CYP2B6 transcription or translation that may be affected by other CYP2B6 variants have not been elucidated extensively. The nuclear receptors pregnane X receptor (PXR) and constitutive androstane receptor (CAR) enhance CYP2B6 promoter activity through binding to their respective cognate response elements and these receptors play pivotal roles in CYP2B6 transcriptional regulation in response to exogenous stimuli [2,9,10]. Many CYP2B6 substrates, including cyclophosphamide, rifampicin, carbamazepine, phenobarbital, nevirapine, artemisinin, efavirenz, and phenytoin, have also been identified as inducers of CYP2B6 via interactions with PXR and CAR receptors [10,11]. Regulatory mechanisms for CYP2B6 involving CAR and PXR can be complex. For example, clotrimazole, a CAR deactivator but a PXR inducer, increases CYP2B6 expression and activity in human hepatocyte cultures [2,12]. Although DNA methylation affects the expression of multiple CYP genes (CYP1A1, 1A2, 1B1, 2A6, 2C19, 2D6, 2E1, 2J2, 2R1, 2S1 and 2W1) by epigenetic pathways [13], no methylation-sensitive CpG islands have been identified in the promoter or coding regions of the CYP2B6 gene that influence its expression.
MicroRNAs (miRNAs) are estimated to modulate the expression of 30% protein-coding genes in a post-transcriptional manner [14] and thereby influence important biologic processes, such as proliferation, differentiation and apoptosis [15]. Results of recent studies have emphasized the impact of miRNAs on gene expression involving drug absorption, distribution, metabolism, and excretion [16–19]. Several important human DME genes, such as CYP1B1, CYP2E1, CYP3A4, CYP24A1 and SUL1A1, are now known to be regulated by miRNAs [16,18,20]. In the simplest case, CYP gene expression could be regulated by miRNAs that interact directly with mRNA transcripts. Alternatively, miRNAs could act indirectly by influencing the expression of another gene that encodes a regulatory factor involved in transcription of the CYP gene. Examples of direct inhibition of CYP expression by miRNAs include: (a) CYP1B1 and CYP3A4, which are suppressed by miR-27b [21]; (b) CYP2E1, which is targeted by miR-378 [22]; (c) and CYP2C9, which is regulated by miR-128-3p [18]. An example of the alternative miRNA regulatory pathway is provided by nuclear receptor PXR, which is suppressed by miR-148a and leads to a decreased production of CYP3A4 and CYP2B6 [23]. If it is found that certain drugs modulate the levels of miRNAs that regulate DME gene expression, a novel miRNA-dependent mechanism for drug/drug interaction might be revealed.
Although miRNAs have been shown to regulate the expression of other important DME genes, a functional role for miRNA in the regulation of CYP2B6 has not been established. Swart and Dandra [24] used bioinformatics tools to predict potential miRNA regulatory interactions for a set of 11 DME genes that included CYP2B6. Interestingly, CYP2B6 was found to contain the largest number of potential regulatory sites among these 11 DME genes that might be sensitive to miRNAs. Although these discoveries are intriguing, experimental verification is still lacking to demonstrate a significant role for miRNAs in the regulation of CYP2B6 gene expression.
In the present study, in silico and in vitro methods were used in conjunction to investigate potential miRNA-dependent mechanisms for regulating the expression of CYP2B6. We selected 29 potential miRNA targeting sites within 3′-UTR of CYP2B6 using in silico methods, correlated the levels of these miRNAs with that of CYP2B6 mRNA in human liver samples, and then employed a series of biochemical assays to investigate the interaction between miRNA and CYP2B6 mRNA transcripts and their influence on gene expression.
2. Materials and methods
2.1. Cell Culture, transfection and chemical treatments
Human embryonic kidney (HEK) 293T cells were obtained from the American Type Culture Collection (ATCC, Manassas, VA). HEK293T cells were cultured in Dulbecco minimum essential medium (DMEM) supplemented with 10% fetal bovine serum (FBS), 100 mg/mL streptomycin, and 100 U/mL penicillin. Terminally differentiated HepaRG cells were purchased from Life Technologies (Carlsbad, CA). HepaRG cells were seeded at 5 × 105 cells per well in 24-multiwell plates, and maintained in William’s E medium supplemented with the Thaw, Plate, & General Purpose Medium Supplement (Life Technologies; Carlsbad, CA) for one day. The cells were then incubated for 7 additional days in William’s E medium supplemented with Maintenance/Metabolism Medium Supplement (Life Technologies) until the hepatocyte-like cells formed well-delineated trabeculae. Both cell lines were cultured at 37 °C in a humidified 5% CO2 atmosphere.
The hsa-miR-25-3p and hsa-miR-504-5p synthetic miRNA mimics and inhibitors used in this study were obtained from Thermo Scientific (Waltham, MA) and transiently transfected into HepaRG cells with the final concentration of 25 nmol/L using Lipo-fectamine 2000 reagent (Life Technologies). Rifampicin was obtained from Sigma–Aldrich (St. Louis, MO) and suspended in dimethyl sulfoxide (DMSO). HepaRG cells were treated for 24 h with 0 or 20 μmol/L rifampicin. Cells were harvested 48 h after transfection. Each assay was performed at least three times.
2.2. In silico analyses
Potential miRNA binding sites located in the 3′-UTR of CYP2B6 were predicted using the public database microRNA.org (http://www.microrna.org). The RNAhybrid program (http://bibiserv2.cebitec.uni-bielefeld.de/rnahybrid) was used to calculate the minimum free energy of hybridization for candidate miRNAs interacting with putative binding sites detected within the CYP2B6 mRNA sequence (NM_000767.4).
2.3. Analysis of RNA levels in human liver
The GSE22058 dataset (http://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE22058) includes mRNA profiles determined using DNA microarrays for 96 cases of human hepatocellular carcinoma and for adjacent non-tumor liver tissue samples. In addition, the GSE22058 dataset provides the miRNA levels detected in these samples using quantitative PCR. The RNA levels reported for CYP2B6 and 29 candidate miRNAs, including hsa-miR-25-3p and hsa-miR-504-5p, in 96 liver (non-tumor) tissues were extracted from the dataset and subjected to Pearson’s correlation analysis (vide infra).
2.4. Luciferase reporter gene assay
A wildtype DNA fragment derived from the core 3′-UTR region of the CYP2B6 gene between 1 and 1156, relative to the termination codon, was chemically synthesized by Genechem Company (Shanghai, China) and then subcloned in the pGL3-control vector (Promega, Madison, WI) according to the manufacturer’s recommendations. Similarly, mutant DNA fragments designed to be altered specifically at the hsa-miR-25-3p or/and hsa-miR-504-5p targeting sites were synthesized chemically and each was subcloned in pGL3-control vector plasmids. The resultant vectors containing the wildtype CYP2B6 3′-UTR, a defective hsa-miR-25-3p site, a defective hsa-miR-504-5p site, or both altered sites were designated as CYP2B6-W, CYP2B6-mut1, CYP2B6-mut2, and CYP2B6-D-mut, respectively. The nucleotide sequences of the four constructs were determined to confirm their authenticity. For luciferase reporter gene assays, HEK 293T cells were seeded into 96-multiwell plates and co-transfected with one of the constructed plasmids (100 ng/well) together with the hsa-miR-25-3p mimic, hsa-miR-504-5p mimic, or miRNA negative (all final concentration: 50 nmol/L), respectively. Renilla luciferase was used to standardize transfection efficiency. Three independent co-transfection experiments were carried out in triplicate.
2.5. RNA electrophoretic mobility shift assays (EMSA)
All oligonucleotides and primers used in this study were obtained from Integrated DNA Technologies (Coraville, IA). The oligonucleotides hsa-miR-25-3p: 5′-CAU UGC ACU UGU CUC GGU CUG A-3′ and hsa-miR-504-5p: 5′-AGA CCC UGG UCU GCA CUC UAU C-3′ were synthesized and 5′-labeled with cy5.5TM dye. The 2′-O-methyl-modified RNA oligonucleotides CYP2B6-miR-25: 5′-CCA GGC UGG AGU GCU AUG GUG CAA UU-3′ and CYP2B6-miR-504: 5′-GGG GGU CAA AGG AUU CCA GGG UCA-3′, corresponding to the hsa-miR-25-3p and hsa-miR-504-5p targeting sequences resident in 3′-UTR of CYP2B6, were 5′-labeled with IRDye®800 dye (IDT). The unlabeled oligonucleotides, including the miRNA negative control (5′-UCA CAA CCU CCU AGA AAG AGU AGA-3′), hsa-miR-25-3p and hsa-miR-504-5p, were used in competition assays.
Cytoplasmic extracts were prepared from HepaRG cells using NE-PER Nuclear and Cytoplasmic extraction reagents (Thermo Scientific). We performed the RNA EMSAs according to the manufacturer’s instructions (Thermo Scientific). Briefly, 1× RNA EMSA binding buffer, 5% glycerol, 200 mM KCl, 100 mM MgCl2, and 200 nmol synthetic miRNA or/and cognate mRNA oligonucleotides were mixed in 20 μL reactions. Cytoplasmic extract (2 μg) and non-specific tRNA (1 μg, included in the RNA EMSA kit) were incubated 20 min at 4 °C to allow RNA:protein interactions to develop. Subsequently, antibodies against Ago1, Ago2, Ago3, and Ago4 (Abcam, Cambridge, MA) were used in the super-shift assays. In addition, unlabeled probes at 50-fold molar excesses were added to the reaction before the addition of dye-labeled probes in competition assays. The reaction mixtures were separated on a 12% PAGE by electrophoresis at 4 °C and detected with an Odyssey CLx Infrared Imaging System (LI-COR Biosciences, Lincoln, NE).
2.6. RNA extraction and quantitative real-time PCR (qRT-PCR)
Total RNA was extracted from HEK 293T or HepaRG cells using the miRNeasy Mini Kits (Qiagen, Valencia, CA). Reverse-transcription reactions of miRNA or mRNA were performed using NCodeTM microRNA First-Strand cDNA Synthesis Kits (Life Technologies) or QuantiTect Reverse Transcription Kits (Qiagen), respectively. Quantitative real-time PCR was performed using an ABI Prism7900 Sequence Detection System (Applied Biosystems, Foster City, CA) based on the SYBR Green method, according to the QuantiFast SYBR® Green RT-PCR Kit (Qiagen) manufacturer’s instructions, with the primers CYP2B6-RT-F: 5′-GGC ACA CAG GCA AGT TTA C-3′ and CYP2B6-RT-R: 5′-TCA GTG CCA GCA AAG AAG-3′ for the CYP2B6 gene; and the primers GAPDH-RT-F: 5′-GAA ATC CCA TCA CCA TCT TCC AGG-3′ and GAPDH-RT-R: 5′-GAG CCC CAG CCT TCT CCA TG-3′ for the GAPDH gene. Quantitative real-time PCR for hsa-miR-25-3p, hsa-miR-504-5p or U6 was performed according to NCodeTM microRNA First-Strand cDNA Synthesis Kits, using the primers miR25-RT-F: 5′-CAT TGC ACT TGT CTC GGT CTG A-3′, miR504-RT-F: 5′-AGA CCC TGG TCT GCA CTC TAT C-3′, or U6-RT-F: 5′-CTC GCT TCG GCA GCA CA-3′ and U6-RT-R: 5′-AAC GCT TCA CGA ATT TGC GT-3′, respectively. The RNA expression levels for CYP2B6 mRNA or the miRNAs hsa-miR-25-3p and hsa-miR-504-5p were calculated relative to the expression of GAPDH or U6 small nuclear RNA, respectively.
2.7. Western blotting assay
Protein extracts from HEK 293T or HepaRG cells were prepared using RIPA lysis buffer (Thermo Scientific). Antibodies against CYP2B6 and GAPDH were purchased from Abcam (Cambridge. MA). Western blotting was performed according to the OdysseyTM Western Blotting Kit (LI-COR Biosciences) instructions and quantitative analyses were performed using the Odyssey CLx Infrared Imaging System.
2.8. Statistical analyses
All statistical analyses were performed using SigmaPlot, version 13.0. Pearson’s correlation analysis was used to analyze the original data of human liver tissues to evaluate the correlations between CYP2B6 mRNA levels and the levels of the miRNAs hsa-miR-25-3p and hsa-miR-504-5p. Student’s t-tests were used to compare the differences in CYP2B6, hsa-miR-25-3p and hsa-miR-504-5p protein or RNA levels between subgroups, and P < 0.05 was considered significant.
3. Results
3.1. Identification of potential miRNAs modulating CYP2B6
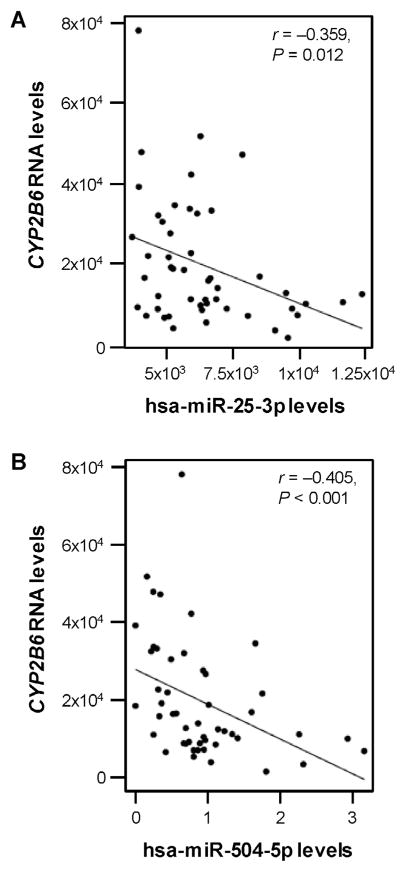
Using the microRNA.org database, we identified 29 miRNAs with potential binding sites within the 3′-UTR of CYP2B6 (Table 1). Subsequently, the free energy of association for each putative miRNA/mRNA hybrid duplex was calculated. Next, Pearson correlation analysis was used to evaluate the associations between CYP2B6 mRNA levels in normal liver tissues and levels of the 29 candidate miRNAs. In this way the miRNAs hsa-miR-25-3p and hsa-miR-504-5p were selected for further analyses because both exhibited (1) free energy of hybridization less than −20 kcal/mol (−26.7 kcal/mol for hsa-miR-25-3p, and −28.4 kcal/mol for hsa-miR-504-5p) which indicated a high binding affinity (Table 1) and (2) significantly negative correlations (r = −0.359 for hsa-miR-25-3p, and r = −0.405 for hsa-miR-504-5p) with CYP2B6 mRNA expression in liver tissues (Fig. 1A and B).
Table 1.
In silico prediction of microRNAs targeting 3′-UTR of CYP2B6.
| # | miRNAs | Position | Binding free energies (kcal/mol) |
|---|---|---|---|
| 1 | hsa-miR-92a | 1068 | −30.90 |
| 2 | hsa-miR-128a-3p | 397 | −29.00 |
| 3 | hsa-miR-504-5pa | 17 | −28.40 |
| 4 | hsa-miR-92b-3p | 1068 | −26.90 |
| 5 | hsa-miR-25-3pa | 1068 | −26.70 |
| 6 | hsa-miR-485-5p | 745 | −26.10 |
| 7 | hsa-miR-367-3p | 1072 | −26.00 |
| 8 | hsa-miR-363-3p | 1072 | −24.90 |
| 9 | hsa-miR-7-5p | 1374 | −24.40 |
| 10 | hsa-miR-134-5p | 1356 | −24.30 |
| 11 | hsa-miR-148b-3p | 1466 | −22.90 |
| 12 | hsa-miR-148a-3p | 1466 | −22.90 |
| 13 | hsa-miR-152-3p | 1467 | −22.10 |
| 14 | hsa-miR-421 | 1314 | −21.60 |
| 15 | hsa-miR-542-3p | 664/898 | −20.80/−21.90 |
| 16 | hsa-miR-216a-5p | 161 | −20.40 |
| 17 | hsa-miR-216b-5p | 1113 | −20.40 |
| 18 | hsa-miR-185 | 865 | −19.20 |
| 19 | hsa-miR-129b | 1472 | −19.10 |
| 20 | hsa-miR-32 | 1072 | −18.90 |
| 21 | hsa-miR-141 | 1254 | −17.90 |
| 22 | hsa-miR-200a | 1254 | −17.50 |
| 23 | hsa-miR-143 | 928 | −16.90 |
| 24 | hsa-miR-29a | 1473 | −16.40 |
| 25 | hsa-miR-29c | 1473 | −16.20 |
| 26 | hsa-miR-590-3p | 521/549/569 | −13.60/−14.50/−13.70 |
| 27 | hsa-miR-590-5p | 524 | −12.60 |
| 28 | hsa-miR-374b | 174 | −7.80 |
| 29 | hsa-miR-21 | 525 | −5.90 |
The correlations (r) between CYP2B6 RNA levels and hsa-miR-504-5p, or and hsa-miR-25-3p expression in normal liver tissues were −0.405 or −0.359, respectively.
Fig. 1.
Correlation between hsa-miR-25-3p, has-miR-504-5p and CYP2B6 transcripts in human liver samples. The levels of (A) hsa-miR-25-3p (r = −0.359, P = 0.012), and (B) hsa-miR-504-5p (r = −0.405, P < 0.001), were negatively correlated with CYP2B6 mRNA levels in non-tumor human liver samples (n = 96) included in the GSE22058 public dataset.
3.2. Hsa-miR-25-3p suppressed luciferase reporter gene activity produced by CYP2B6 3′-UTR
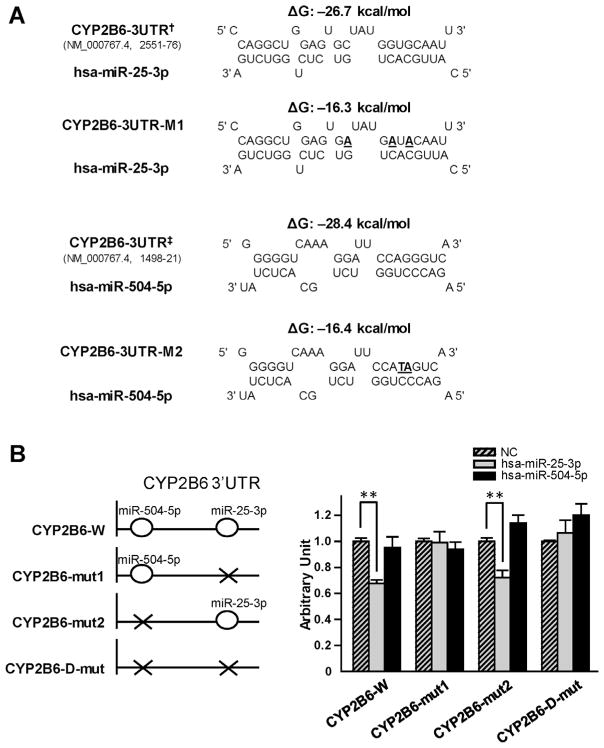
We created a reporter gene plasmid that contained the wild-type 3′-UTR of CYP2B6 and three additional plasmid constructs that contained mutations introduced at the putative target sites for hsa-miR-25-3p and/or hsa-miR-504-5p (Fig. 2A and B). HEK 293T cells were co-transfected with one each of the reporter gene constructs together with the miRNA mimics or the miRNA negative control. As shown in Fig. 2B, hsa-miR-25-3p mimics could significantly decrease the luciferase activity produced by the CYP2B6-W and CYP2B6-mut2 plasmids (33% and 28%, respectively, all P < 0.001), both containing the wildtype target sequences of hsa-miR-25-3p in the CYP2B6 3′-UTR. However, the hsa-miR-25-3p mimic did not inhibit luciferase activities for CYP2B6-mut1 or CYP2B6-D-mut constructs that contained altered targeting sequences for hsa-miR-25-3p, indicating that intact targeting sequences are necessary for hsa-miR-25-3p-dependent suppression of CYP2B6. No similar suppressive effect of the hsa-miR-504-5p mimics on luciferase activities was observed.
Fig. 2.
Hsa-miR-25-3p down-regulated luciferase reporter activities by targeting CYP2B6 3′-UTR. (A) Predicted hybrid complexes for hsa-miR-25-3p and hsa-miR-504-5p with sequences found in the wild-type CYP2B6 3′-UTR and with altered sequences (bold, underlined). † and ‡ indicated the nucleotide positions 2551–76 and 1498–21 at the CYP2B6 3′-UTR in NM_000767.4, counting from the transcription start site as nucleotide position 1. CYP2B6-3UTR-M1 and CYP2B6-3UTR-M2 indicated the mutated hsa-miR-25-3p and hsa-miR-504-5p targeting sequences based on the wild-type CYP2B6 3′-UTR, respectively. (B) The diagrams on the left depict the structures of the reporter gene constructs used in this study. CYP2B6-W construct contains the wild-type CYP2B6 3′-UTR. CYP2B6-mut1 and CYP2B6-mut2 constructs contain the 3′-UTR fragment with the CYP2B6-3UTR-M1 and CYP2B6-3UTR-M2 mutant sequences, respectively. The CYP2B6-D-mut contains the 3′-UTR with both the CYP2B6-3UTR-M1 and the CYP2B6-3UTR-M2 mutant sequences. The graph on the right shows that luciferase reporter gene activity in HEK 293T cells was suppressed by transfection with hsa-miR-25-3p mimics. The luciferase reporter plasmids (100 ng) were co-transfected into HEK 293T cells together with 50 nmol/L of miRNA negative controls (NC; striped bars), hsa-miR-25-3p (gray bars) or has-miR-504-5p mimics (black bars), respectively. Data are presented as mean ± S.D. (n = 9). **P < 0.001.
3.3. Hsa-miR-25-3p regulates CYP2B6 directly by an Ago4 dependent manner
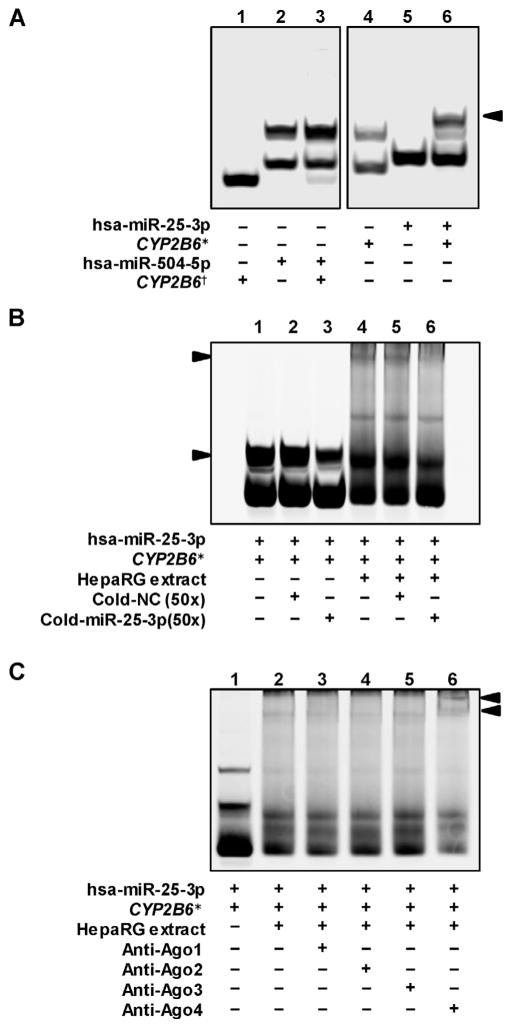
RNA EMSAs were performed to explore whether hsa-miR-25-3p or hsa-miR-504-5p is able to interact with its cognate CYP2B6 target sequence directly. As shown in Fig. 3A, hsa-miR-25-3p could bind its cognate CYP2B6 mRNA oligonucleotide to form a complex (lane 6). However, hsa-miR-504-5p could not create an electrophoretically stable miRNA/mRNA complex with its cognate CYP2B6 mRNA oligonucleotide (lane 3). Further, the hsa-miR-25-3p/CYP2B6 complex was able to interact with HepaRG cytoplasmic extracts to form new complexes (Fig. 3B, lane 4). Both the miR-25-3p/CYP2B6 mRNA complex and the miR-25-3p/CYP2B6 mRNA/protein complex can be eliminated by excess unlabeled hsa-miR-25-3p probes (Fig. 3B, lanes 3 and 6), also indicating the interaction between hsa-miR-25-3p and its cognate mRNA was sequence specific. The argonaute proteins associate with the miRNAs to interact with their target mRNAs. To investigate which argonaute protein can mediate the hsa-miR-25-3p/CYP2B6 RNA/protein complex, antibodies against Ago1, Ago2, Ago3 or Ago4 were used to capture the RNA–protein complex, respectively. As shown in Fig. 3C, antibody against Ago4 formed a new super-shift complex (lane 6, upper arrow) with the hsa-miR-25-3p/CYP2B6 RNA /protein complex.
Fig. 3.
hsa-miR-25-3p oligonucleotides interact directly with CYP2B6 mRNA oligonucleotides in an Ago4-dependent manner. (A) RNA EMSA shows that hsa-miR-25-3p interacts with CYP2B6 mRNA oligonucleotides to form an electrophoretically stable complex (Arrow, lane 6); no such an interaction was observed between hsa-miR-504-5p and CYP2B6 mRNA oligonucleotides (lane 5). (B) The interaction between hsa-miR-25-3p oligonucleotides and CYP2B6 mRNA oligonucleotides is sequence specific in the presence or absence of HepaRG protein extracts, since cold probe can reduce the density of the hsa-miR-25-3p/CYP2B6 complexes and the density of the hsa-miR-25-3p/CYP2B6/protein complex (Arrows). (C). Antibody against Ago4 was able to capture hsa-miR-25-3p/CYP2B6/protein complex (Upper arrow, supershift complex). *hsa-miR-25-3p targeting mRNA sequence in CYP2B6 3′-UTR. *hsa-miR-504-5p targeting mRNA sequence in CYP2B6 3′-UTR.
3.4. Hsa-miR-25-3p suppressed endogenous CYP2B6 expression
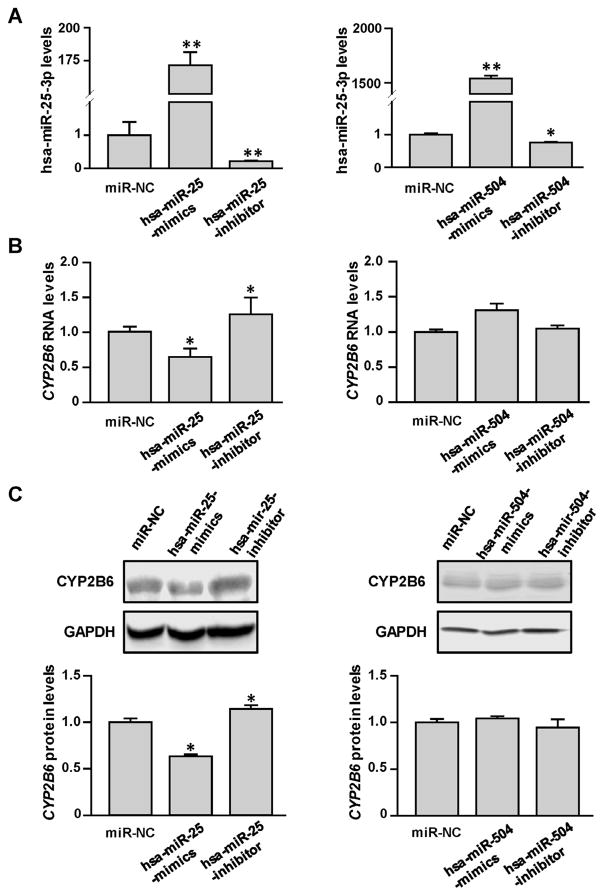
The HepaRG cell line, which can express DMEs at levels similar to primary hepatocytes, was selected to test the regulatory effects of hsa-miR-25-3p or hsa-miR-504-5p on expression of the endogenous CYP2B6 gene. As shown in Fig. 4A, both hsa-miR-25-3p and hsa-miR-504-5p levels in HepaRG cells were significantly elevated after transfection with the corresponding mimics (172 fold and 1542 fold, respectively, all P < 0.001), and decreased by their inhibitors (78% and 25%, respectively, all P < 0.05). However, only exogenous hsa-miR-25-3p mimics/inhibitors, but not hsa-miR-504-5p mimics/inhibitors, were observed to influence expression of the endogenous CYP2B6 gene significantly (35% and 37% decreased for RNA and protein levels after hsa-miR-25-3p mimics transfection, respectively, all P < 0.05; 1.26 and 1.24-fold increased for RNA and protein levels after hsa-miR-25-3p inhibitors transfection, respectively; all P < 0.05) (Fig. 4B and C).
Fig. 4.
Hsa-miR-25-3p inhibited endogenous CYP2B6 expression in HepaRG cells. Differentiated HepaRG cells were transiently transfected using 25 nmol/L miRNA negative controls or transfected with mimics or inhibitors for hsa-miR-25-3p and hsa-miR-504-5p. Together with the increased miRNA levels (A), decreased CYP2B6 mRNA levels (B) were observed in cells transfected with hsa-miR-25-3p, but not hsa-miR-504-5p. (C) Western blots show that CYP2B6 protein levels were decreased in HepaRG cells transfected with the hsa-miR-25-3p mimic but were increased in cells transfected with the hsa-miR-25-3p inhibitor. Each assay was carried out in triplicate. *P < 0.05; **P < 0.001; NC, miRNA negative control.
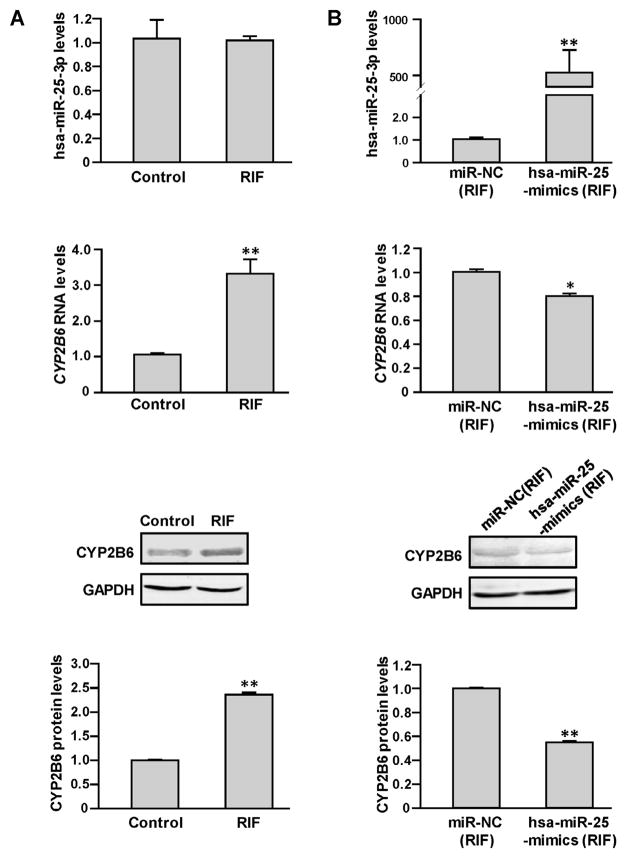
Further, we used rifampicin to induce CYP2B6 expression in HepaRG cells and then tested whether the elevated CYP2B6 mRNA and protein levels could be suppressed by transfection with hsa-miR-25-3p mimics. As shown in Fig. 5A, rifampicin significantly elevated CYP2B6 expression (3.32 and 2.42-fold changed for RNA and the proteins levels after 20 μM rifampicin treatment, respectively, all P < 0.05). Consequently, we found that transfection by hsa-miR-25-3p mimics was able to suppress the increased CYP2B6 expression induced rifampicin in HepaRG cells (21% and 45% decreased for RNA and the protein levels after 20 μM rifampicin treatment, respectively, all P < 0.05).
Fig. 5.
Hsa-miR-25-3p inhibited drug elevated CYP2B6 expression in HepaRG cells. Differentiated HepaRG cells were treated with 0 or 20 μmol/L rifampicin with or without transfection with miRNA negative control or hsa-miR-25-3p mimic. Treatment with rifampicin (A) led to increased levels of CYP2B6 mRNA in HepaRG cells but did not affect hsa-miR-25-3p levels. Transfection HepaRG cells treated with rifampicin with hsa-miR-25-3p mimics or miRNA negative control (B) showed that the induction of CYP2B6 rifampicin could be inhibited efficiently by hsa-miR-25-3p mimic transfection. Each assay was carried out in triplicate. *P < 0.05; **P < 0.001; NC, miRNA negative control; RIF, rifampicin.
4. Discussion
DMEs, drug transporters, and nuclear receptors play crucial roles in drug responses and clinical outcomes [24]. Many pharmacogenetic studies have concentrated on functional effects of genetic variations found within the exons, introns, and promoters of relevant genes [25–27]. Recent studies have shown that certain miRNAs serve as epigenetic regulatory agents to modulate DME expression [16]. MicroRNAs affect the expression of approximately one third of mammalian genes [14], including many phase I and II DMEs and drug transporters [24]. To illustrate, we reported that up-regulation of hsa-miR-128-3p suppresses the expression of CYP2C9 in liver cells and hepatocellular carcinoma [25] and we also reported that hsa-miR-29a-3p suppresses CYP2C19 expression in liver cells and tissue [18]. In the present study, we investigated the regulatory roles of miRNAs in the expression of CYP2B6. Here, using systematic in silico analyses, including miRNA target prediction, free energy calculation and correlation analyses between miRNAs and CYP2B6 mRNA levels in liver tissues, we selected hsa-miR-25-3p and hsa-miR-504-5p as potential candidate miRNAs that might regulate CYP2B6. Luciferase reporter assay indicated that hsa-miR-25-3p suppresses CYP2B6 expression by binding to its 3′-UTR region. RNA-EMSA confirmed that hsa-miR-25-3p can bind directly with specific sequences found in the CYP2B6 3′-UTR. In vitro experiments using overexpression or inhibition of miR-25-3p confirmed the suppressive effect of hsa-miR-25-3p on CYP2B6 expression. Moreover, we found that the Ago4 protein, but not the Ago1, Ago2, or Ago3 proteins, was detected in macromolecular complexes formed using hsa-miR-25-3p and CYP2B6 mRNA synthetic oligonucleotides incubated with cellular extracts. In addition, transfection with hsa-miR-25-3p mimics inhibited the increased expression of CYP2B6 induced by rifampicin. Our comprehensive results revealed that hsa-miR-25-3p negatively regulates the expression of CYP2B6 in liver cells.
It was reported that some novel small RNAs exhibit a selective bias for associating with human Ago1-RISC (the multi-protein RNA-induced silencing complex) or Ago2-RISC compared with other argonaute species [16,26]. Our previous study observed that Ago1, but not other argonaute proteins, could be detected in complexes formed with hsa-miR-29a-3p, supporting the notion that at least some miRNAs exhibit sorting ability to distinguish specific Ago-RISCs in mammals [16]. In the present study, Ago4, but not other argonaute proteins, was detected in complexes formed using hsa-miR-25-3p and CYP2B6 mRNA oligonucleotides and cellular extracts. Our results provide a new clue that, like hsa-miR-25-3p, some microRNAs may associate selectively with human Ago4-RISC.
Rifampicin is an antibiotic that is used widely in treating tuberculosis (first-line agent for treatment of all forms of active tuberculosis), leprosy, and Legionnaire’s disease [28]. Rifampicin is metabolized primarily by CYP2B6 and it acts via CAR or PXR mediation to induce CYP2B6 expression [2,29]. We exposed HepRG cells to rifampicin to induce CYP2B6 expression in our study, and then transfected HepaRG cells with hsa-miR-25-3p mimics and noted that hsa-miR-25-3p blocks the induction of CYP2B6 stimulated by the drug. This observation suggests that the expression status of hsa-miR-25-3p should be considered as a potential mediator of drug-drug interactions through CYP2B6 inhibition or induction.
Multiple drug regimens are often used in patient treatments. However, drug–drug interactions may compromise therapeutic efficacy or contribute to adverse drug reactions in some cases. The abnormal induction or inhibition of CYPs has been implicated in the mechanisms for many drug-drug interactions and adverse drug reactions [30]. The concomitant use of efavirenz and carbamazepine (CBZ) can create a drug–drug interaction which illustrates this point. Carbamazepine, widely used in the treatment of epilepsy, psychiatric disorders and trigeminal neuralgia [31], is almost completely (about 95%) metabolized in the liver [32]. CYP2B6 participates in 3-OH-CBZ formation together with CYP3A4 [31]. An interaction between efavirenz and CBZ is predictable due to involvements of CYP2B6 and CYP3A [28]. CBZ may increase the clearance of efavirenz by inducing CYP2B6/3A4, resulting in a time-dependent decrease in efavirenz exposure. Furthermore, efavirenz is also an inducer of CYP2B6/3A4; thus it could stimulate its own metabolism as well as the metabolism of co-medications that are substrates of CYP2B6/3A4 [33]. In view of our results, it is reasonable to speculate that an hsa-miR-25-3p inducer may inhibit CYP2B6 transcription and translation, resulting in decreased metabolism or clearance of CYP2B6 substrates such as CBZ and efavirenz, thus reducing efficacy, and perhaps provoking drug resistance. For example, Withaferin A (WFA), an effective drug for arthritis, is an inducer of hsa-miR-25-3p [34]. We speculate that management of arthritis using WFA could suppress CYP2B6 expression via WFA-dependent induction of hsa-miR-25-3p and that this scenario introduces the potential for drug–drug interactions for cancer or tuberculosis patients being treated with rifampicin, respectively. Thus, the drug efficacy and safety for multi-drug regimens should be considered carefully if drugs that increase or decrease the expression of hsa-miR-25-3p are prescribed together with drugs that are substrates of CYP2B6, such as rifampicin.
In summary, we demonstrated that hsa-miR-25-3p suppresses CYP2B6 expression in an AGO4-dependent manner by targeting a specific domain located within the 3′-UTR of CYP2B6. In this way we identified a novel epigenetic mechanism for regulating CYP2B6 gene expression. We also validated the interplay between hsa-miR-25-3p and CYP2B6 in response to rifampicin treatment.
Acknowledgments
This study was supported and funded by the National Center for Toxicological Research (Project E0752601) and U.S. Food and Drug Administration.
Abbreviations
- CYP
cytochrome P450
- miRNA
microRNA
- DMETs
drug metabolizing enzymes and transporters
- CAR
constitutive androstane receptor
- PXR
pregnane X receptor
- RE
response element
- 3′-UTR
3′-untranslated region
- SULT
sulfotransferase
- Ago
argonaute RISC catalytic component
- RISC
RNA-induced silencing complex
- FBS
fetal bovine serum
- EMSA
electrophoretic mobility shift assay
Footnotes
Author contributions
Participated in Study design: Ning, Yu, Tolleson, and Guo. Conducted experiments: Jin, Yu, Knox, Chen, and Ren. Performed data analysis: Jin, Yu, Guo, Deng, Wang and Ning. Wrote or contributed to the writing of the manuscript: Jin, Tolleson, Yu, Guo, Deng, and Ning.
Disclaimer
The information in these materials is not a formal dissemination of the U.S. Food and Drug Administration.
Conflict of interest
The authors have no conflict of interest.
References
- 1.Lamba V, Lamba J, Yasuda K, Strom S, Davila J, Hancock ML, et al. Hepatic CYP2B6 expression: gender and ethnic differences and relationship to CYP2B6 genotype and CAR (constitutive androstane receptor) expression. J Pharmacol Exp Therapeut. 2003;307:906–922. doi: 10.1124/jpet.103.054866. [DOI] [PubMed] [Google Scholar]
- 2.Wang H, Faucette S, Sueyoshi T, Moore R, Ferguson S, Negishi M, et al. A novel distal enhancer module regulated by pregnane X receptor/constitutive androstane receptor is essential for the maximal induction of CYP2B6 gene expression. J Biol Chem. 2003;278:14146–14152. doi: 10.1074/jbc.M212482200. [DOI] [PubMed] [Google Scholar]
- 3.Ning B, Dial S, Sun Y, Wang J, Yang J, Guo L. Systematic and simultaneous gene profiling of 84 drug-metabolizing genes in primary human hepatocytes. J Biomol Screen. 2008;13:194–201. doi: 10.1177/1087057108315513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Yang L, Price ET, Chang CW, Li Y, Huang Y, Guo LW, et al. Gene expression variability in human hepatic drug metabolizing enzymes and transporters. PLoS One. 2013;8:e60368. doi: 10.1371/journal.pone.0060368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Code EL, Crespi CL, Penman BW, Gonzalez FJ, Chang TK, Waxman DJ. Human cytochrome P4502B6: interindividual hepatic expression, substrate specificity, and role in procarcinogen activation. Drug Metab Dispos. 1997;25:985–993. [PubMed] [Google Scholar]
- 6.Lang T, Klein K, Fischer J, Nussler AK, Neuhaus P, Hofmann U, et al. Extensive genetic polymorphism in the human CYP2B6 gene with impact on expression and function in human liver. Pharmacogenetics. 2001;11:399–415. doi: 10.1097/00008571-200107000-00004. [DOI] [PubMed] [Google Scholar]
- 7.Desta Z, Saussele T, Ward B, Blievernicht J, Li L, Klein K, et al. Impact of CYP2B6 polymorphism on hepatic efavirenz metabolism in vitro. Pharmacogenomics. 2007;8:547–558. doi: 10.2217/14622416.8.6.547. [DOI] [PubMed] [Google Scholar]
- 8.Rocha V, Porcher R, Fernandes JF, Filion A, Bittencourt H, Silva W, Jr, et al. Association of drug metabolism gene polymorphisms with toxicities, graft-versus-host disease and survival after HLA-identical sibling hematopoietic stem cell transplantation for patients with leukemia. Leukemia. 2009;23:545–556. doi: 10.1038/leu.2008.323. [DOI] [PubMed] [Google Scholar]
- 9.Goodwin B, Moore LB, Stoltz CM, McKee DD, Kliewer SA. Regulation of the human CYP2B6 gene by the nuclear pregnane X receptor. Mol Pharmacol. 2001;60:427–431. [PubMed] [Google Scholar]
- 10.Thorn CF, Lamba JK, Lamba V, Klein TE, Altman RB. PharmGKB summary: very important pharmacogene information for CYP2B6. Pharmacogenet Genom. 2010;20:520–523. doi: 10.1097/fpc.0b013e32833947c2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zanger UM, Klein K, Saussele T, Blievernicht J, Hofmann MH, Schwab M. Polymorphic CYP2B6: molecular mechanisms and emerging clinical significance. Pharmacogenomics. 2007;8:743–759. doi: 10.2217/14622416.8.7.743. [DOI] [PubMed] [Google Scholar]
- 12.Willson TM, Kliewer SA. PXR, CAR and drug metabolism. Nat Rev Drug Discovery. 2002;1:259–266. doi: 10.1038/nrd753. [DOI] [PubMed] [Google Scholar]
- 13.Ingelman-Sundberg M, Sim SC, Gomez A, Rodriguez-Antona C. Influence of cytochrome P450 polymorphisms on drug therapies: pharmacogenetic, pharmacoepigenetic and clinical aspects. Pharmacol Ther. 2007;116:496–526. doi: 10.1016/j.pharmthera.2007.09.004. [DOI] [PubMed] [Google Scholar]
- 14.Lewis BP, Burge CB, Bartel DP. Conserved seed pairing, often flanked by adenosines, indicates that thousands of human genes are microRNA targets. Cell. 2005;120:15–20. doi: 10.1016/j.cell.2004.12.035. [DOI] [PubMed] [Google Scholar]
- 15.Cao JX, Lu Y, Qi JJ, An GS, Mao ZB, Jia HT, et al. MiR-630 inhibits proliferation by targeting CDC7 kinase, but maintains the apoptotic balance by targeting multiple modulators in human lung cancer A549 cells. Cell Death Dis. 2014;5:e1426. doi: 10.1038/cddis.2014.386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Koturbash I, Tolleson WH, Guo L, Yu D, Chen S, Hong H, et al. MicroRNAs as pharmacogenomic biomarkers for drug efficacy and drug safety assessment. Biomarkers Med. 2015;9:1153–1176. doi: 10.2217/bmm.15.89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yu D, Tolleson WH, Knox B, Jin Y, Guo L, Guo Y, et al. Modulation of ALDH5A1 and SLC22A7 by microRNA hsa-miR-29a-3p in human liver cells. Biochem Pharmacol. 2015;98:671–680. doi: 10.1016/j.bcp.2015.09.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yu D, Green B, Tolleson WH, Jin Y, Mei N, Guo Y, et al. MicroRNA hsa-miR-29a-3p modulates CYP2C19 in human liver cells. Biochem Pharmacol. 2015;98:215–223. doi: 10.1016/j.bcp.2015.08.094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yu D, Green B, Marrone A, Guo Y, Kadlubar S, Lin D, et al. Suppression of CYP2C9 by microRNA hsa-miR-128-3p in human liver cells and association with hepatocellular carcinoma. Sci Rep. 2015;5:8534. doi: 10.1038/srep08534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yokoi T, Nakajima M. MicroRNAs as mediators of drug toxicity. Annu Rev Pharmacol Toxicol. 2013;53:377–400. doi: 10.1146/annurev-pharmtox-011112-140250. [DOI] [PubMed] [Google Scholar]
- 21.Pan YZ, Gao W, Yu AM. MicroRNAs regulate CYP3A4 expression via direct and indirect targeting. Drug Metab Dispos. 2009;37:2112–2117. doi: 10.1124/dmd.109.027680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mohri T, Nakajima M, Fukami T, Takamiya M, Aoki Y, Yokoi T. Human CYP2E1 is regulated by miR-378. Biochem Pharmacol. 2010;79:1045–1052. doi: 10.1016/j.bcp.2009.11.015. [DOI] [PubMed] [Google Scholar]
- 23.Zanger UM, Schwab M. Cytochrome P450 enzymes in drug metabolism: regulation of gene expression, enzyme activities, and impact of genetic variation. Pharmacol Ther. 2013;138:103–141. doi: 10.1016/j.pharmthera.2012.12.007. [DOI] [PubMed] [Google Scholar]
- 24.Swart M, Dandara C. Genetic variation in the 3′-UTR of CYP1A2, CYP2B6, CYP2D6, CYP3A4, NR1I2, and UGT2B7: potential effects on regulation by microRNA and pharmacogenomics relevance. Front Genet. 2014;5:167. doi: 10.3389/fgene.2014.00167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kanke M, Jambor H, Reich J, Marches B, Gstir R, Ryu YH, et al. Oskar RNA plays multiple noncoding roles to support oogenesis and maintain integrity of the germline/soma distinction. RNA. 2015;21:1096–1109. doi: 10.1261/rna.048298.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Relling MV, Evans WE. Pharmacogenomics in the clinic. Nature. 2015;526:343–350. doi: 10.1038/nature15817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ning B, Su Z, Mei N, Hong H, Deng H, Shi L, et al. Toxicogenomics and cancer susceptibility: advances with next-generation sequencing. J Environ Sci Health C Environ Carcinog Ecotoxicol Rev. 2014;32:121–158. doi: 10.1080/10590501.2014.907460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Thorn CF, Leckband SG, Kelsoe J, Leeder JS, Muller DJ, Klein TE, et al. PharmGKB summary: carbamazepine pathway. Pharmacogenet Genom. 2011;21:906–910. doi: 10.1097/FPC.0b013e328348c6f2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.LeCluyse EL. Human hepatocyte culture systems for the in vitro evaluation of cytochrome P450 expression and regulation. Eur J Pharmaceut Sci. 2001;13:343–368. doi: 10.1016/s0928-0987(01)00135-x. [DOI] [PubMed] [Google Scholar]
- 30.de Jonge ME, Huitema AD, Rodenhuis S, Beijnen JH. Clinical pharmacokinetics of cyclophosphamide. Clin Pharmacokinet. 2005;44:1135–1164. doi: 10.2165/00003088-200544110-00003. [DOI] [PubMed] [Google Scholar]
- 31.Pearce RE, Lu W, Wang Y, Uetrecht JP, Correia MA, Leeder JS. Pathways of carbamazepine bioactivation in vitro. III. The role of human cytochrome P450 enzymes in the formation of 2,3-dihydroxycarbamazepine. Drug Metab Dispos. 2008;36:1637–1649. doi: 10.1124/dmd.107.019562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kim KA, Oh SO, Park PW, Park JY. Effect of probenecid on the pharmacokinetics of carbamazepine in healthy subjects. Eur J Clin Pharmacol. 2005;61:275–280. doi: 10.1007/s00228-005-0940-7. [DOI] [PubMed] [Google Scholar]
- 33.Zhu M, Kaul S, Nandy P, Grasela DM, Pfister M. Model-based approach to characterize efavirenz autoinduction and concurrent enzyme induction with carbamazepine. Antimicrob Agents Chemother. 2009;53:2346–2353. doi: 10.1128/AAC.01120-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kim JH, Kim SJ. Overexpression of microRNA-25 by withaferin A induces cyclooxygenase-2 expression in rabbit articular chondrocytes. J Pharmacol Sci. 2014;125:83–90. doi: 10.1254/jphs.13232fp. [DOI] [PubMed] [Google Scholar]