Summary
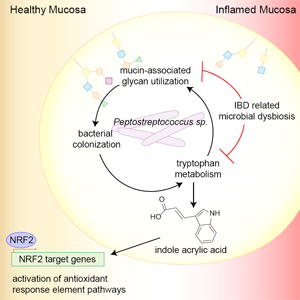
Host factors in the intestine help select for bacteria that promote health. Certain commensals can utilize mucins as an energy source, thus promoting their colonization. However, health conditions such as inflammatory bowel disease (IBD) are associated with a reduced mucus layer, potentially leading to dysbiosis associated with this disease. We characterize the capability of commensal species to cleave and transport mucin-associated monosaccharides and identify several Clostridiales members that utilize intestinal mucins. One such mucin utilizer, Peptostreptococcus russellii, reduces susceptibility to epithelial injury in mice. Several Peptostreptococcus species contain a gene cluster enabling production of the tryptophan metabolite indoleacrylic acid (IA) that promotes intestinal epithelial barrier function and mitigates inflammatory responses. Furthermore, metagenomic analysis of human stool samples reveals that the genetic capability of microbes to utilize mucins and metabolize tryptophan is diminished in IBD patients. Our data suggest that stimulating IA production could promote anti-inflammatory responses and have therapeutic benefits.
Introduction
The gastrointestinal tract microbiota are well appreciated for their importance to human health by aiding in immune development, metabolism, and pathogen clearance. Dysbiosis in this bacterial community is linked to the pathophysiology of a plethora of human illnesses, including inflammatory bowel disease (IBD) (Gevers et al., 2014), intestinal cancers (Kostic et al., 2013), obesity (Turnbaugh et al., 2009), diabetes (Kostic et al., 2015; Vatanen et al., 2016), Parkinson’s disease (Sampson et al., 2016), and rheumatoid arthritis (Zhang et al., 2015). Despite a growing understanding of this microbial community through deep sequencing (Human Microbiome Project, 2012), there is still little mechanistic insight into the relative contribution of each bacterial species towards promoting intestinal health. Concurrently, host factors are critical in establishing an ecosystem that promotes a beneficial relationship with the microbiota, including maintenance of anatomical and chemical barriers such as the mucus layer, production of bacterial nutrient sources, and bacterial recognition by immune cells (Hooper et al., 2012).
The intestinal mucus layer lies at the interface between bacteria and host epithelial and immune cells. Intestinal mucin is mainly composed of a single, highly O-glycosylated protein called mucin 2 (MUC2), which is produced by goblet cells and secreted into the intestinal lumen in a TLR- and NLRP6-dependent manner (Birchenough et al., 2016; Johansson et al., 2008; Wlodarska et al., 2014). In the large intestine, secreted mucins form a two-layered structure: the inner mucus layer, which is devoid of bacteria, and the outer mucus layer (Johansson et al., 2008). Within the outer mucus layer, commensals bind to and utilize mucin-associated sugars, which provide a rich source of nutrients for the bacteria.
The capacity of a commensal bacterium to utilize mucins as an energy source likely plays an important role in establishing a mucosal-associated niche. The initial isolation of the beneficial bacterium Akkermansia muciniphila based on its capacity to use mucin as a sole carbon source (Derrien et al., 2004) is an indication that further understanding of bacterial mucin utilization could be an effective strategy to identify potential health-associated commensals. Mucin is a glycosylated protein, with fucose or sialic acid terminating the glycan chains (Tailford et al., 2015a). The fucosidase and sialidase activity of certain symbiotic bacteria, such as Bacteroides thetaiotaomicron, can liberate mucosal glycans without compromising the integrity of the mucus layer to support colonization (Martens et al., 2008; Martens et al., 2009). Further, sensing of microbial ligands by type 3 innate lymphoid cells stimulates the addition of fucose to mucin through the action of IL-22, which further supports colonization of commensal bacteria (Goto et al., 2014; Pickard et al., 2014).
Once established in the intestine, the microbiota influence host metabolism and immunity through metabolic activities such as the production of short-chain fatty acids from carbohydrate metabolism and tryptophan metabolites produced from amino acid metabolism (Wikoff et al., 2009). In mice, dietary lack of tryptophan leads to impaired intestinal immunity and dysbiosis of the microbiota (Hashimoto et al., 2012). Bacterial tryptophan metabolism leads to the production of potent bioactive metabolites, such as indole-3-aldehyde, indole 3-propionate, and indole-3-acetic acid, that affect intestinal barrier integrity and immune cells in mice through the activation of the pregnane X receptor (PXR) and the aryl hydrocarbon receptor (AhR) (Lamas et al., 2016; Venkatesh et al., 2014; Zelante et al., 2013). In a human study, patients with IBD exhibited reduced tryptophan metabolism, presumably due to an altered bacterial gut community (Lamas et al., 2016). IBD patients also have a thinner inner mucus layer and reduced glycosylation of MUC2 (Fyderek et al., 2009; Larsson et al., 2011). These alterations in mucus production likely contribute to reduced commensal fitness, driving the microbial dysbiosis in IBD patients. Together, these observations suggest the relevance of intestinal mucins in establishing bacterial colonization and maintaining health-promoting functions of the microbiome, such as tryptophan metabolism (Fyderek et al., 2009; Larsson et al., 2011).
In this study, we hypothesized that a global understanding of bacterial mucin utilization in the intestine could yield insights into currently uncharacterized bacterial species with the potential to beneficially impact human health. We computationally identified commensal bacteria with the genetic capacity to utilize intestinal mucins as an energy source. We validated the computational prediction that members of the Clostridiales order can be prolific mucin degraders by functional in vitro growth assays. We identified a mucin-degrading bacterium, Peptostreptococcus russellii, that significantly reduces susceptibility to chemically induced colitis in mice. Mechanistically, we show that several Peptostreptococcus species metabolize tryptophan to produce the metabolite indoleacrylic acid (IA), which has a beneficial effect on intestinal epithelial barrier function and mitigates inflammatory responses by immune cells. Finally, we show that the genetic capability of microbes to utilize mucins as an energy source and to metabolize tryptophan is altered in IBD patients.
Results
Targeted Computational Approach Identifies Mucin-Utilizing Bacteria in the Healthy Intestinal Microbiome
We analyzed metagenomic sequencing data to determine the genetic capacity of healthy human fecal-associated microbes to utilize intestinal mucin. Using foundational literature describing mucus utilization by commensal bacteria (Corfield et al., 1992; Marcobal et al., 2011; Martens et al., 2009; Nakayama-Imaohji et al., 2012), we manually generated a list of genes likely to be involved in mucin-associated O-glycan utilization. Mucin-utilizing genes were grouped into two families: those that cleave mucin-associated sugars (cleavage genes) and those required for uptake of the cleaved sugars (transporter genes). Secreted mucin-associated O-glycans have long branched structures made up of N-acetylgalactosamine, N-acetylglucosamine, and galactose, which are terminated by fucose or sialic acid residues (Figure 1A). We focused on these five monosaccharides and their transporters to assemble the seed gene list used in downstream analysis (Table S1).
Figure 1. Computational approach to predict microbial mucin utilization.
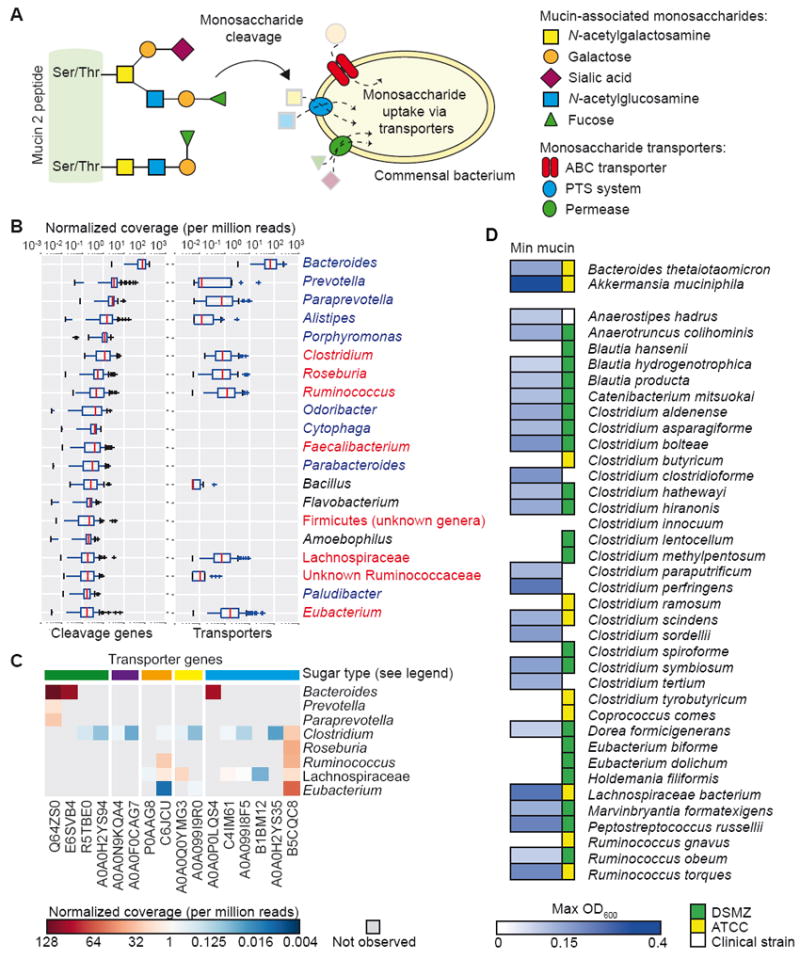
(A) Microbial scavenging of monosaccharides derived from mucin-associated oligosaccharides. Mucin 2 peptide is heavily glycosylated at Ser/Thr residues with oligosaccharides composed of N-acetylgalactosamine, galactose, sialic acid, N-acetylglucosamine, and fucose. Bacterial genes relevant for mucin oligosaccharide utilization by commensal bacteria are listed in Table S1.
(B) Genus-level contribution to normalized read coverage from 90 HMP subjects for cleavage genes (left) and transporters (right). Blue font, order Bacteroidales; red font, order Clostridiales.
(C) Distribution of transporter gene abundance in each genus. Colors at top indicate sugar transported by indicated gene: green, fucose; purple, sialic acid; orange, galactose; blue, N-acetylglucosamine.
(D) Screen for in vitro growth in minimal medium containing mucin as the sole carbon source. Max OD600 values are shown for each strain as averaged across 3 independent experiments.
See also Figure S1 and Table S1.
We randomly selected 90 healthy subjects from the publicly available Human Microbiome Project (HMP) data set for analysis of their stool metagenomic data (Figure S1A). First, mucin cleavage and transporter seed genes were searched against all available bacterial genomes on NCBI, complete or draft, using BLAT (Kent, 2002) protein sequence search. Of the 30 most abundant genes (relative abundance of each gene in each species for each subject in the HMP data set), 14 were fucosidases or fucose transporters, suggesting that fucose cleavage and uptake is essential in the healthy microbiome (Figure S1B and Table 1). Surprisingly, mucin-associated cleavage genes were abundant in most taxa, whereas transporter genes showed selectivity for members of the Bacteroidales and Clostridiales orders (Figure 1B). Members of the Bacteroidales order encoded a high gene abundance of fucose and N-acetylglucosamine transporters, whereas members of the Clostridiales order showed a greater distribution and diversity of transporter gene abundance, encoding transporters for fucose, sialic acid, galactose, N-acetylgalactosamine, and N-acetylglucosamine (Figure 1C and Table 2).
Table 1. Mucin cleavage and transporter genes found in HMP subjects.
30 most abundant mucin utilization genes found in HMP subjects.
| UniProt ID | Gene Name | Organism |
|---|---|---|
| D7J3U9 | Alpha-L-fucosidase 2 | Bacteroides sp. |
| Q64NW2 | ATP-dependent Clp protease proteolytic subunit/serine-type endopeptidase activity* | Bacteroides fragilis |
| R7KV56 | Alpha-L-fucosidase | Bacteroides |
| B7GNN8 | Alpha-1,3/4-fucosidase, putative | Bifidobacterium longum |
| D7KA95 | Alpha-L-fucosidase 1 | Bacteroides sp. |
| Q89ZI2 | O-GlcNAcase BT_4395 | Bacteroides |
| Q64ZS0 | L-fucose permease | Bacteroides fragilis |
| A0A0P0LQS4 | N-acetylglucosamine related transporter, NagX | Bacteroides vulgatus |
| Q9L5W0 | Mucin-desulfating sulfatase MdsA | Prevotella sp. |
| E6SVB4 | L-fucose transporter | Bacteroides helcogenes |
| D4LLN2 | Alpha-L-fucosidase | Ruminococcus sp. |
| Q8A3I4 | Alpha-L-fucosidase | Bacteroides |
| Q8A5P6 | Putative lipoprotein, alpha-L-fucosidase activity* | Bacteroides |
| D4LLN1 | Alpha-L-fucosidase | Ruminococcus sp. |
| R5N5G7 | Alpha-L-fucosidase | Ruminococcus sp. |
| B5CQC8 | PTS system, N-acetylglucosamine-specific IIBC | Ruminococcus lactaris |
| B7GNN7 | Uncharacterized protein, alpha-L-fucosidase activity* | Bifidobacterium longum |
| R7EDR1 | Alpha-L-fucosidase | Roseburia sp. |
| A0A0Q0YMG3 | Pts N-acetylgalactosamine transporter subunit IID | Clostridium butyricum |
| A7B8B7 | Alpha-L-fucosidase | Ruminococcus gnavus |
| C6JCU1 | Galactose/methyl galactoside import ATP-binding | Ruminococcus sp. |
| P0AAG8 | Galactose/methyl galactoside import ATP-binding | Escherichia coli |
| U2NJT6 | PTS system mannose-specific transporter subunit IID | Clostridium intestinale |
| B2UN42 | Exo-alpha-sialidase | Akkermansia muciniphila |
| B2UPI5 | Exo-alpha-sialidase | Akkermansia muciniphila |
| B2UQL7 | Glycosyl hydrolase family 109 protein 2 | Akkermansia muciniphila |
| A8ANB2 | Peptidase E | Citrobacter koseri |
| B1JI47 | Beta-hexosaminidase | Yersinia pseudotuberculosis |
| P27297 | Protein bax, amidase activity* | Escherichia coli |
| P42640 | Putative phosphoethanolamine transferase YhbX | Escherichia coli |
Table 2. Transporter genes found in the Bacteroidales and Clostridiales order.
| UniProt ID | Gene Name | Organism |
|---|---|---|
| Q64ZS0 | L-fucose permease | Bacteroides fragilis |
| E6SVB4 | L-fucose transporter | Bacteroides helcogenes |
| R5TBE0 | Transporter putative fucose permease | Clostridium hathewayi |
| A0A0H2YS94 | L-fucose:H+ symporter permease | Clostridium perfringens |
| A0A0N9KQA4 | Putative sialic acid transporter | Clostridium perfringens |
| A0A0F0CAG7 | Sialic acid TRAP transporter permease | Clostridium sp. |
| P0AAG8 | Galactose/methyl galactoside import ATP-binding | Escherichia coli |
| C6JCU1 | Galactose/methyl galactoside import ATP-binding | Ruminococcus sp. |
| A0A0Q0YMG3 | Pts N-acetylgalactosamine transporter subunit IID | Clostridium butyricum |
| A0A099I9R0 | Pts acetylgalactosamine transporter subunit IID | Clostridium innocuum |
| A0A0P0LQS4 | N-acetylglucosamine related transporter, NagX | Bacteroides vulgatus |
| C4IM61 | Pts system, N-acetylglucosamine-specific iibc | Clostridium butyricum |
| A0A099I8F5 | Pts N-acetylglucosamine transporter subunit IIBC | Clostridium innocuum |
| B1BM12 | Pts system, N-acetylglucosamine-specific IIBC | Clostridium perfringens |
| A0A0H2YS35 | Pts system, N-acetylglucosamine-specific IIBC | Clostridium perfringens |
| B5CQC8 | Pts system, N-acetylglucosamine-specific IIBC | Ruminococcus lactaris |
| A0A099I4W2 | Pts mannose transporter subunit IID | Clostridium innocuum |
| U2NJT6 | Pts system mannose-specific transporter subunit IID | Clostridium intestinale |
To test the computational prediction that members of the Clostridiales order are prolific utilizers of mucin-associated glycans, we screened 36 strains of bacteria belonging to the Clostridiales order for growth in minimal medium containing commercially available pig-derived gastric mucin as a sole carbon source. Using B. thetaiotaomicron and A. muciniphila as positive controls (Derrien et al., 2004; Martens et al., 2009), we found that 64% of the tested species were able to grow in the minimal medium, suggesting that the mucus-utilizing capacity of the intestinal microbiome has been underappreciated (Figure 1D).
Mucin Utilization Screen Leads to Identification of Beneficial Bacterial Species
To determine whether mucin was a preferential energy source for these species, the 15 strains that grew to a maximum OD600 greater than 0.15 within 48 h of growth in minimal mucin media were selected for growth comparisons in a minimal M9T medium or M9T medium containing 0.25% mucin (Figure 2A). A. muciniphila, Anaerotruncus colihominis, Marvinbryantia formatexigens, and Peptostreptococcus russellii showed enhanced growth in the presence of mucin (Figures 2A and 2B), suggesting that these strains robustly utilize mucin as an energy source. P. russellii exhibited a biphasic growth curve, suggesting exhaustion of a primary carbon source followed by a switch to mucin utilization (Figure 2B). To test the hypothesis that these mucin-utilizing commensals could serve protective functions in the intestine, we examined the effects of these four strains in the context of dextran sodium sulfate (DSS)-induced colitis by administering bacteria to mice by oral gavage before and during DSS exposure (Figure 2C). As A. muciniphila is known to promote intestinal wound repair and to have restitutive effects during DSS-induced colitis, we used this strain as a positive control (Alam et al., 2016). As a negative control, we used Clostridium butyricum, a Clostridiales species that did not grow in minimal mucin media (Figure 1D). As expected, administration of A. muciniphila resulted in reduced weight loss (p = 0.050) and reduced colon pathology (Figures 2D-2F). In addition, mice administered P. russellii showed significant protection from weight loss (Figure 2D) and reduced colon pathology, including reduced ulceration and decreased immune cell infiltration of the mucosa compared to PBS-treated mice (Figures 2E and 2F).
Figure 2. In vitro mucin utilization screen identifies Peptostreptococcus species that protects against DSS-induced colitis.
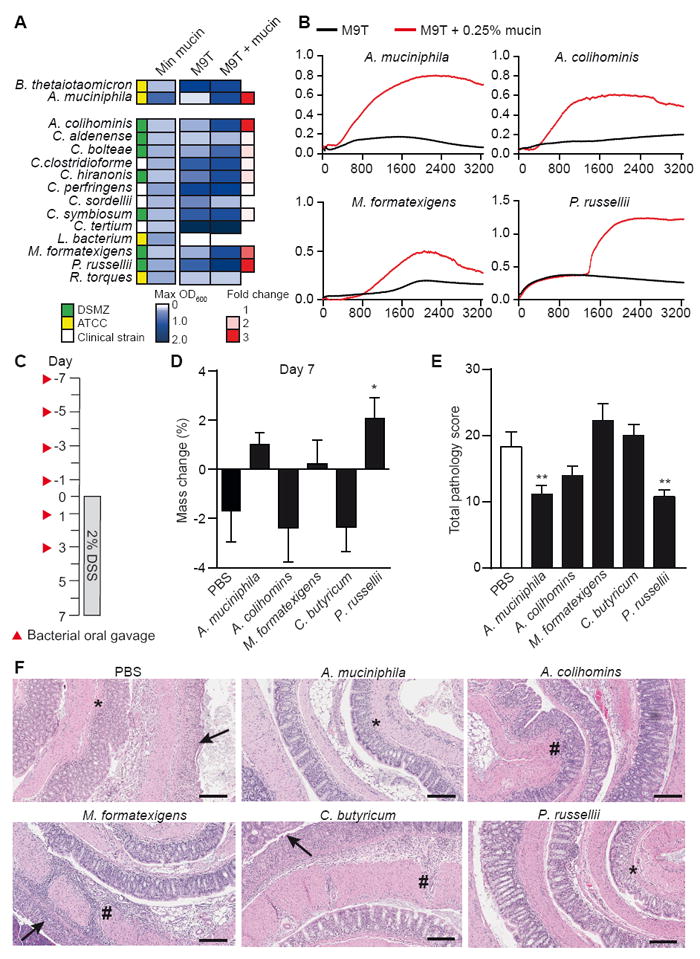
(A) Bacterial strains were grown in minimal media containing glucose and 0.5% tryptone (M9T medium) or M9T supplemented with 0.25% mucin. Max OD600 values were compared to determine the fold change in growth.
(B) Growth curves of bacterial strains with greater than 2-fold increase in growth from panel A in M9T (black) compared to M9T + mucin (red). Data are representative of 2 independent experiments.
(C) Schematic for oral gavage of mice and 2% DSS time course.
(D) Percent mass change at day 7 of DSS exposure relative to weight at day 0. *P = 0.0128.
(E) Total histopathology score of the colon on day 7 of DSS exposure. A. muciniphila (**P = 0.0060); P. russellii (**P = 0.0036).
(F) Representative images of H&E-stained colon sections from mice orally gavaged with PBS or indicated bacterial strains. Arrows, ulceration; #, transmural inflammation; *, mucosal immune infiltrate. Scale bar, 100 μm. Data represent one experiment, n = 10 per group. Significance determined using one-way ANOVA with Fisher’s LSD test and expressed as mean + SEM (D, E).
To more fully characterize the protective effect of P. russellii, we explored the role of this strain in regulating mucosal immunity prior to DSS exposure. Administration of P. russellii resulted in increased expression of the proliferation marker Ki67, goblet cell-specific secreted protein MUC2 (Muc2), and glycosyltransferase enzymes, Fut2, St6gal1 and B3gnt6, however, the expression of goblet cell lineage transcription factors were unaffected (Figure 3A and S2A-B). Goblet cell staining in colon tissue with an anti-MUC2 antibody and the lectin Ulexeuropaeus agglutinin I (UEA-I), which labels α-1,2 fucose residues, showed increased numbers of Muc2+ and Muc2+UEA-I+ goblet cells in the colonic mucosa of P. russellii and A. muciniphila treated mice (Figure 3B-D). Flow cytometry analysis of colonic epithelial cells showed that administration of P. russellii led to an increase in the percentage of α-1,2 fucose-positive (UEA-I reactive) goblet cells, suggesting an increase in mucin-associated fucose residues (Figure 3E). To confirm successful intestinal colonization of P. russellii after repeated oral gavage, we performed qPCR on DNA extracted from stool samples and observed a two-fold increase in the abundance of P. russellii in gavaged mice compared to control mice (Figure S2C). These data implicate P. russellii as a protective commensal that increases goblet cell number and mucin fucosylation, and mediates protection from DSS-induced colitis.
Figure 3. P. russellii promotes goblet cell differentiation and function in vivo.
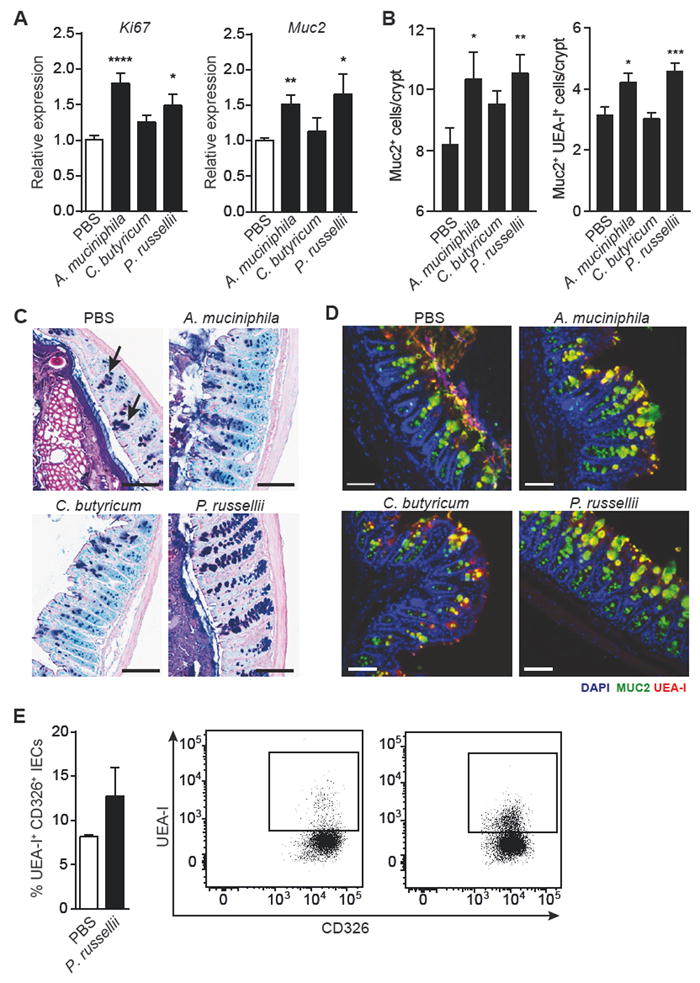
Mice were orally gavaged with PBS, A. muciniphila, C. butyricum, or P. russellii every other day for 2 weeks and the distal large intestine was harvested for analysis.
(A) Quantitative RT-PCR showing expression of proliferation marker Ki67 (****P < 0.0001; *P = 0.0105) and goblet cell-derived mucin 2 (Muc2; **P = 0.0020; *P = 0.0275), relative to Gapdh. n = 10 per group. Data pooled from 2 independent experiments.
(B) Quantification of Muc2+ (**P = 0.0088; *P = 0.0418) and Muc2+UEA-I+ (***P = 0.0009; *P = 0.0184) goblet cell number per crypt in the distal colon, n = 5 per group.
(C) Representative alcian blue and periodic acid-Schiff (AB/PAS)-stained distal colon sections showing goblet cell staining within the mucosa. Goblet cells are indicated by black arrows. Scale bar, 100 μm.
(D) Representative epifluorescence staining for goblet cells in the distal colon using an antibody against Muc2 (green) and the lectin UEA-I (red) with DAPI (blue) as the counter stain. Scale bar, 100 μm.
(E) Intracellular flow cytometry analysis of isolated colonic epithelial cells (CD326+) shows increased presence of UEA-1+CD326+ double-positive cells in P. russellii-treated animals. The lectin UEA-1 binds α-1,2 fucose. Significance determined using Student’s t-test and expressed as mean + SEM.
See also Figure S2.
Peptostreptococcus Species Containing the fldAIBC Gene Cluster Produce Tryptophan Metabolites that Promote Barrier Function and Immune Tolerance
We used a comparative genomics approach to identify potential mechanisms underlying the protective effect of P. russellii. We used PacBio sequencing to obtain complete genome sequences of A. muciniphila, A. colihominis, M. formatexigens, C. butyricum, and P. russellii. We noted the unique presence of the phenyllactate dehydratase gene cluster (fldAIBC) only in P. russellii. This gene cluster was homologous to the fldAIBC cluster in Clostridium sporogenes, which is required for metabolism of phenylalanine and is hypothesized to be involved in the metabolism of other aromatic amino acids, including tryptophan (Dickert et al., 2002) (Figure 4A). Intriguingly, C. sporogenes metabolizes tryptophan to indole-3-propionic acid (IPA), a metabolite that protects mice from DSS-induced colitis (Venkatesh et al., 2014).
Figure 4. IPA and IA are produced by some Peptostreptococcus species and IA exhibits anti-inflammatory effects.
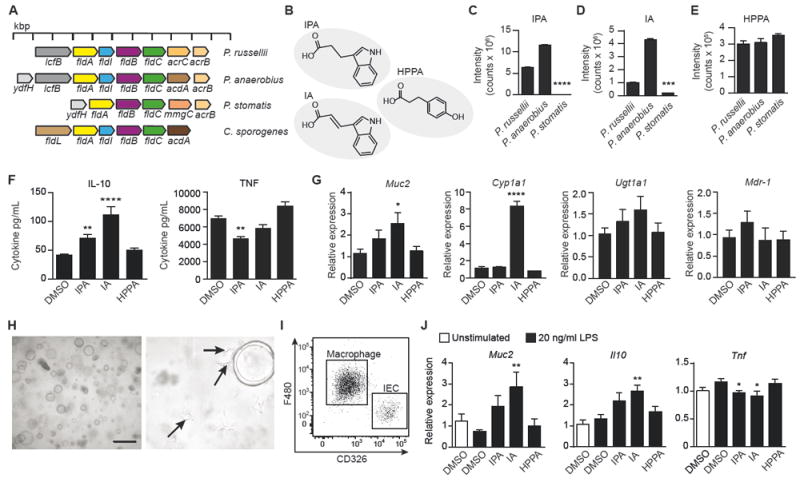
(A) Bacterial genomic comparison showing phenyllactate operon in Peptostreptococcus species compared to a previously identified operon in C. sporogenes.
(B) MS QTOF analysis of culture supernatant from Peptostreptococcus species identified tryptophan metabolites (IPA, indole-3-propionic acid; IA, indoleacrylic acid) and a phenylalanine metabolite (HPPA, 3-hydroxyphenyl propionic acid).
(C-E) Ion chromatograms extracted at m/z and retention times of IPA (C), IA (D), and HPPA (E) were used to determine the relative abundance of each metabolite produced by 3 indicated strains. See also Figure S3A-S3C. ****P < 0.0001, ***P = 0.0003. Data representative of 3 experiments and expressed as mean + SEM.
(F) Isolated and cultured BMDMs were treated with 100 μM of each metabolite or 0.1% DMSO control for 6 h, followed by 24 h of IL-4 conditioning, and then stimulated with 20 ng/mL LPS for 18 h. Supernatants were analyzed for IL-10 (IPA, **P = 0.0032; IA, ****P = 0.0001) and TNF (**P = 0.004) production. Data pooled from 4 independent experiments.
(G) Quantitative RT-PCR showing expression of Muc2 (*P = 0.0263), the AhR target gene Cyp1a1 (****P = 0.0001), and the PXR target genes, Ugt1a1 and MDR-1, relative to Actb and compared to 0.1% DMSO control. Data pooled from 2 independent experiments using 4-5 independent colonic spheroid lines.
(H) Representative light phase image of large intestinal spheroids co-cultured with macrophages in Matrigel at 5X (left) and 40X (right) magnification. Arrows indicate macrophages with extended pseudopods.
(I) Representative flow cytometry dot plot of F480+ macrophages and CD326+ epithelial cells in the co-culture system indicating the percentage of macrophages (80.0 ± 4.3%) and epithelial cells (16.0 ± 4.5%) after 3 days of co-culture, expressed as mean ± SD.
(J) Quantitative RT-PCR showing expression of Muc2 (**P = 0.0077), Il10 (**P = 0.0072), and Tnf (*P = 0.0477 for IPA; *P = 0.0148 for IA) in co-culture of large intestinal spheroids and BMDMs after 48 h of treatment with 100 μM IPA, IA, HPPA, or 0.1% DMSO control followed by 20 ng/mL LPS stimulation for 24 h. Data are pooled from 2 independent experiments using 4-5 independent colonic spheroid lines. Significance determined using one-way ANOVA with Fisher’s LSD test and expressed as mean + SEM.
See also Figures S3-S4.
We hypothesized that the presence of the complete fldAIBC gene cluster in P. russellii would enable it to metabolize aromatic amino acids, specifically tryptophan. To investigate this possibility, we used quadrupole time-of-flight (QTOF) mass spectrometry (MS) to analyze acetonitrile-extracted supernatant from P. russellii grown in rich medium. We detected the production of two tryptophan metabolites, indole 3-propionic acid (IPA, 190.087 m/z) and 3-indoleacrylic acid (IA, 188.071 m/z), and one phenylalanine metabolite, 3-hydroxyphenyl propionic acid (HPPA, 167.071 m/z) (Figures 4B and S3A-S3C), and confirmed the identification of these metabolites using collision induced-MS/MS analysis (Figures S3D-S3F).
To determine whether the fldAIBC gene cluster is conserved in the Peptostreptococcus genus, we analyzed the genomes of two other Peptostreptococcus species found in the human intestinal microbiota, P. anaerobius and P. stomatis. P. anaerobius encoded the complete fldAIBC gene cluster, whereas P. stomatis lacked the initiator fldI but encoded the phenyllactate dehydratase fldABC (Figure 4A). Using QTOF MS, we found that like P. russellii, P. anaerobius also produced the tryptophan metabolites IPA and IA, whereas P. stomatis produced significantly lower levels, consistent with our genomic analysis (Figure 4C and 4D). All three species produced equivalent amounts of the phenylalanine metabolite HPPA (Figure 4E).
Bacterial tryptophan metabolites such as IPA and indole acetic acid can affect intestinal barrier integrity and immune cells in mice through the activation of AhR or PXR transcription factors (Lamas et al., 2016; Venkatesh et al., 2014; Zelante et al., 2013). In the intestine, these metabolites are produced in the context of both the intestinal epithelium and immune cells; therefore, we tested the effect of IPA, IA, and HPPA on inflammatory responses in epithelial cells and macrophages. To determine whether these metabolites have an anti-inflammatory function, we treated bone marrow-derived macrophages (BMDMs) with IPA, IA, or HPPA followed by stimulation with lipopolysaccharide (LPS) and examined the production of IL-10, an anti-inflammatory cytokine, and TNF, a proinflammatory cytokine. Both IPA and IA significantly enhanced IL-10 production after LPS stimulation, and IPA significantly reduced TNF production (Figure 4F). Next we examined the effect of IPA, IA, and HPPA on epithelial cell differentiation and activation of AhR and PXR target genes. Murine-derived colon crypts were cultured in Matrigel to produce spheroids as previously described (Miyoshi and Stappenbeck, 2013). Colonic spheroids were allowed to differentiate in the presence of IPA, IA, or HPPA for 72 hours, after which epithelial cells were examined for gene expression changes. No overt changes in spheroid growth or size were evident (Figure S4A); however, genes involved in goblet cell function, such as Muc2, showed increased expression after treatment with IA (Figures 4G and S4B). Further, only IA induced the expression of the AhR target gene Cyp1a1 and neither IA or IPA induced expression of PXR target genes, Ugt1a1 and Mdr-1.
The ability of IA to enhance goblet cell function during epithelial differentiation and to promote IL-10 production in LPS-stimulated macrophages led us to investigate if the effect of IA on goblet cells would be maintained in the context of an inflammatory response. Given that a characteristic of intestinal inflammation is goblet cell exhaustion resulting in decreased mucus secretion, it is of interest to identify metabolites that can simultaneously promote anti-inflammatory cytokine production while promoting goblet cell differentiation and function. To study these potential effects in the context of both innate immune cell and epithelial inflammation, we established an in vitro co-culture system. Since inflammatory cytokines produced by innate immune cells in response to LPS (e.g., TNFα) reduce epithelial barrier integrity, this co-culture system allows the study of epithelial-immune cell crosstalk during the inflammatory response. We thus co-cultured BMDMs with colonic spheroids in Matrigel under conditions that yielded normal macrophage and spheroid growth. In this culture system, colonic spheroids grew normally and macrophages could be seen extending pseudopodia within the Matrigel (Figures 4H, 4I, and S4C) even after treatment with metabolites (IPA, IA, or HPPA) and stimulation by LPS. Following LPS stimulation, RT-qPCR revealed that IA-treated co-cultures had increased Muc2 expression compared to the LPS-stimulated DMSO control (Figure 4J). IA treatment also had a potent anti-inflammatory effect on cytokines in the co-culture system, as shown by enhanced expression of Il10 and reduced expression of Tnf after LPS stimulation (Figure 4J). Consistent with these results, cytokine bead array analysis revealed that IPA and IA suppressed secretion of IL-6 and TNF and stimulated secretion of IL-10 in the co-culture (Figure S4D). Interestingly, in this co-culture system BMDMs produced 4-fold higher levels of IL-10 in response to LPS compared to when these cells were cultured independently (Figure S4D). Further, we verified that in co-culture only IA induces expression of the AhR target gene, Cyp1a1, and not PXR target genes (Figure S4E).
Indoleacrylic Acid Inhibits pro-inflammatory cytokine production in Human PBMCs
Serum metabolomics analyses of germ-free versus conventional mice have identified multiple microbial metabolites, including IPA, that presumably originate in the gut, suggesting that microbially produced metabolites can have a profound effect on both mucosal and circulating leukocytes (Wikoff et al., 2009). To understand the effect of IPA and IA metabolites on human immune cells, we used peripheral blood mononuclear cells (PBMCs) isolated from healthy individuals. PBMCs were treated with a dose titration of IPA and IA and then stimulated with LPS, after which cytokine production was assessed. In contrast to our findings in murine BMDMs, IPA had no effect on cytokine production in human PBMCs. IA had a significant effect on secretion of IL-1β (43 ± 14% reduction; Figure 5A) and IL-6 (33 ± 13% reduction; Figure 5B), but no effect on secretion of TNF (Figure 5C). More global analysis by RNA sequencing showed that IA treatment resulted in downregulation of genes involved in inflammation and oxidative stress (e.g., CD14, CCL2, MT2A, CYBB, IL6, and PTAFR) as well as genes involved in innate immune cell activation and/or differentiation (e.g., FPR2, LRRC25, CPM, MS4A7, and SLC7A7; Figure 5D). Gene set enrichment analysis confirmed a potent anti-inflammatory response induced by IA, also revealing enrichment of antioxidant pathways (Figure 5E).
Figure 5. IA suppresses cytokine secretion by human PBMCs in response to LPS.
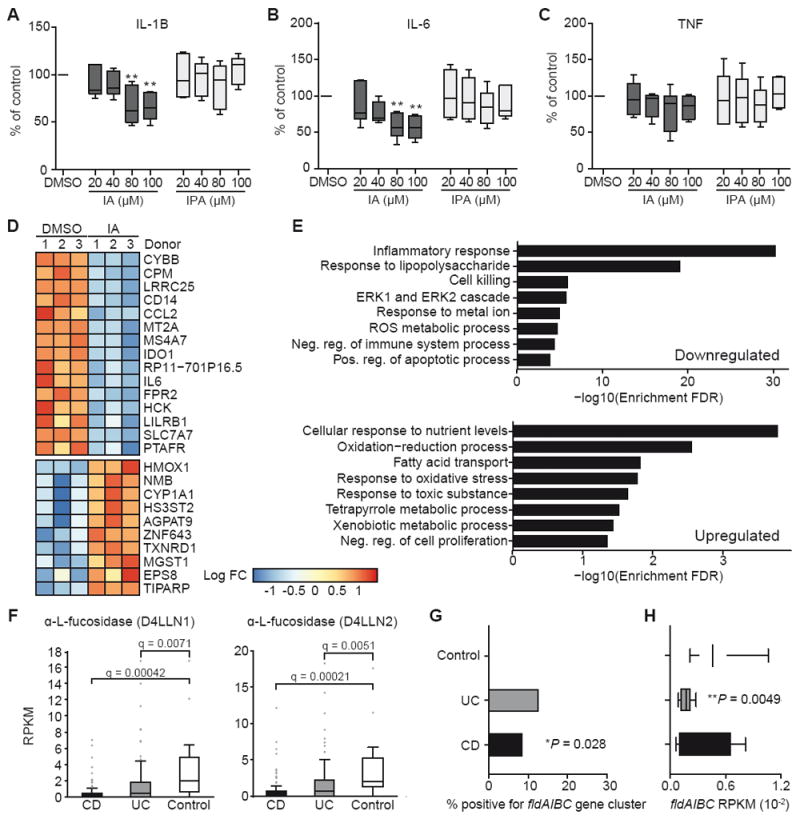
(A-C) Human PBMCs were pre-incubated with IA or IPA for 45 min followed by LPS stimulation for 20 h. Collected supernatants were assessed by CBA for secretion of IL-1β (A), IL-6 (B), and TNF (C). Data are normalized to 0.1% DMSO control samples and pooled from 6 human healthy donors. **P < 0.01 by one-way ANOVA with Fisher’s LSD test; data shown as box and whisker plots with mean and min max values.
(D) Heatmap showing differentially expressed genes as identified by RNA-Seq (FDR < 0.05) for human PBMCs treated with (IA) or without (DMSO). Donor effects have been removed and values from replicate measurements from the same treatment and donors are averaged.
(E) Gene set enrichment analysis from RNA-Seq results on ranked lists of upregulated and downregulated genes.
(F) Abundance of mucin utilization genes in metagenomic data from controls and individuals with CD or UC. Significance determined using student’s t-test and FDR adjusted using Hochberg-Benjamini procedure.
(G) Incidence of phenyllactate gene cluster in controls and individuals with CD or UC; *P = 0.028, CD vs. control comparison by Chi2 contingency table test.
(H) Abundance of phenyllactate gene cluster in controls and individuals with CD or UC; **P = 0.0049, UC vs. control comparison by student’s t-test.
See also Figure S5.
IA treatment led to increased expression of target genes in the NF-E2-related factor 2 (NRF2)-mediated antioxidant pathway (HMOX1, HS3ST2, TXNRD1, MGST1, ZNF643, EPS8, and TIPARP) and upregulation of the AhR-inducible gene CYP1A1 (Figure 5D). NRF2 is a transcription factor that activates cellular antioxidant response element (ARE) pathways, suppresses pro-inflammatory signaling pathways, and activates AhR signaling (Hayes et al., 2009; Li et al., 2008; Tsuji et al., 2012; Zhang et al., 2014). We validated the specific activation of NRF2 by IA (and not by IPA) in our in vitro murine colonic spheroids and BMDM co-culture system and using an NRF2-luciferase reporter in the RAW macrophage cell line (Figure S5A and S5B). In the murine co-culture system, LPS stimulation results in oxidative stress and IA treatment leads to significantly increased expression of NRF2 target genes including, heme oxygenase 1 (Hmox1) and NADPH:quinone dehydrogenase (Nqo1); as well as NRF2 target genes involved in tripeptide glutathione synthesis, glutamate-cysteine ligase, modifier subunit (Gclm), and glutamate-cysteine ligase, catalytic subunit (Gclc; Figure S5A). Thus, microbial production of the tryptophan metabolite IA likely promotes antioxidant and anti-inflammatory immune responses at least in part through the activation of the NRF2-ARE pathway.
The relevance of NRF2 in IBD (Arisawa et al., 2008), coupled with a reduced availability of mucin-associated sugars in IBD patients (Fyderek et al., 2009; Larsson et al., 2011), motivated us to examine stool metagenomic samples from an IBD cohort to determine whether there was a difference in prevalence of mucus-utilization genes. Using a cross-sectional cohort of IBD patients, we performed whole-genome sequencing (WGS) of fecal samples from 34 non-IBD controls, 56 individuals with ulcerative colitis (UC), and 72 individuals with Crohn’s disease (CD). Similar to our HMP analysis, we used the mucin cleavage and transporter seed gene list to determine the abundance of genes predicted to be involved in mucin utilization (Table S1). Remarkably, both UC and CD patients had a reduced microbial genetic capacity to cleave mucin-associated fucose, with decreased abundance of the α-L-fucosidases D4LLN1 and D4LLN2 (Figure 5F, Table S1). Overall, individuals with CD showed extensive changes in the ability of their intestinal bacteria to utilize mucins (Figures S5C and S5D). During active IBD, the mucus layer has reduced barrier function and decreased glycosylation of the mucin peptide (Fyderek et al., 2009; Johansson et al., 2014; Larsson et al., 2011), and it has been postulated that the microbial changes seen in IBD are in part due to bacterial adaptation to the changes in mucin glycosylation (Png et al., 2010). Furthermore, we found increased abundance of transporters for N-acetylgalactosamine, N-acetylglucosamine, and galactose as well as increased abundance of peptidase E, a serine peptidase (Figures S5C and S5D). Together these observations suggest an increased availability of smaller mucin oligosaccharides (Larsson et al., 2011) and increased depolymerization of MUC2 through the action of serine peptidases in this cohort of IBD patients (Abreu et al., 2016; McGuckin et al., 2011).
We speculated that this alteration in mucin utilization would correlate with reduced colonization of the subset of microbes that metabolize tryptophan (using the fldAIBC gene cluster) in the intestine. We therefore assessed the extent of bacterial tryptophan metabolism in these samples by searching for the phenyllactate gene (fldAIBC) cluster in these fecal WGS data sets. The phenyllactate gene cluster was detectable in 9 of 34 (26.5%) non-IBD control samples, 7 of 56 (12.5%) UC patient samples, and 6 of 72 (8.3%) CD patient samples (Figure 5G). To determine the abundance of the gene cluster in samples that were positive for fldAIBC, we calculated the RPKM of each fld gene in the cluster and used the median to represent each individual’s fldAIBC RPKM (Figure S5E). Individuals with UC showed a significantly reduced abundance of the phenyllactate gene cluster (Figure 5H). Thus, there is a reduced presence and abundance of the fldAIBC gene cluster in IBD patients.
Discussion
Previous studies on mucus utilization by B. thetaiotaomicron and A. muciniphila have suggested that mucus utilization is reserved for a subset of the intestinal microbiota (Derrien et al., 2004; Sonnenburg et al., 2005; Tailford et al., 2015a). However, we found that 64% of the tested strains belonging to the Clostridiales order were capable of growing on mucin as a sole carbon source. These findings suggest that the mucin-utilizing capacity of the intestinal microbiome has been underappreciated.
We assessed several mucin-utilizing bacteria for their ability to confer a protective phenotype to mice treated with DSS and found a difference in the protective capabilities of various bacteria in this model. Indeed, not all mucus-utilizing bacteria are beneficial. For example, mice on low fiber diets have an increase in microbial degradation of mucus, resulting in compromised intestinal barrier function (Desai et al., 2016). Furthermore, Ruminococcus gnavus can cleave sialic acid from mucin oligosaccharides in a manner that prevents other bacteria from utilizing the cleaved sugar, suggesting that this “selfish” glycan utilization strategy could contribute to the increased prevalence of R. gnavus in the mucus-limited environment of the inflamed intestine in IBD patients (Tailford et al., 2015b). Nevertheless, P. russellii exhibited a protective effect against DSS-mediated colitis in mice.
We found that P. russellii uniquely encodes the phenyllactate gene cluster, homologous to one found in C. sporogenes. Mass spectrometry analysis of spent medium from P. russellii revealed that it is capable of metabolizing tryptophan to produce IPA and IA. The protective effect of P. russellii treatment during DSS colitis may be in part due to the production of these tryptophan metabolites, potentially leading to the increased differentiation and expression of goblet cell-associated genes such as Muc2. Initial stages of DSS-induced colitis are characterized by a compromise in mucus barrier integrity (Johansson et al., 2010), to which P. russellii-treated mice may be more resilient. Previous studies have also shown that tryptophan metabolites signal through AhR to promote IL-22 secretion or through PXR to regulate mucosal integrity (Lamas et al., 2016; Monteleone et al., 2011; Zelante et al., 2013). Interestingly, these studies also showed that AhR and PXR regulated pathways are relevant for Muc2 gene expression in the intestine. PXR-deficient (Nr1i2 knockout) mice exhibit a leaky gut and reduced expression of Muc2 in the small intestine (Venkatesh et al., 2014), whereas Ido1 knockout mice, which exhibit increased production of bacterially derived tryptophan metabolites, have increased expression of Muc2 in the vagina (Zelante et al., 2013). Here, we show that treatment of murine-derived colonic spheroids with IA results in potent activation of AhR and upregulation of Muc2 gene expression, further suggesting a link between AhR activation and goblet cell function.
A co-culture system consisting of mini-gut spheroid cultures and BMDMs allowed us to demonstrate that the effect of IA on Muc2 gene expression was maintained in an assay designed to simulate intestinal inflammation. This co-culture system showed that both IA and IPA promote secretion of IL-10 and inhibit TNF production following LPS stimulation. When BMDMs were cultured independently, IA had a minimal effect on TNF secretion and four-fold less IL-10 was produced compared to levels measured in co-culture, suggesting that the co-culture system allows cross-talk between epithelial cells and BMDMs that affects cytokine secretion in response to LPS. Even in the context of the LPS-stimulated co-culture, IA maintained its effect on Muc2 gene expression and increased IL-10 production, suggesting that IA may have an important anti-inflammatory function in the intestine. IL-10 is critical to maintain goblet cell production of mucins (Hasnain et al., 2013) and mutations in IL-10 receptors are associated with IBD (Begue et al., 2011). We have found that bacterially-derived IA can increase both IL-10 production and mucin gene expression.
Importantly, we confirmed that the anti-inflammatory functions of IA and IPA are relevant in human PBMCs. Surprisingly, IPA showed an anti-inflammatory effect in murine BMDMs but not in human PBMCs, suggesting that signaling pathways are not conserved in murine and human cells. To date most studies assessing tryptophan metabolite signaling through AhR and PXR have used murine models, and consequently the relevance of these pathways in human cells has been largely overlooked. We found that treatment of human PBMCs with IA, but not IPA, led to reduced IL-6 and IL-1β secretion in response to LPS stimulation and activation of the NRF2-ARE pathway, in addition to AhR activation, suggesting that IA may have an anti-oxidative and anti-inflammatory function in humans. We validated the activation of the NRF2 pathway in our murine co-culture system and find that IA treatment results in upregulation of i) NRF2 target genes, including Hmox1, ii) the AhR target gene, Cyp1a1, and iii) genes involved in tripeptide glutathione synthesis, Gclm and Gclc. Glutathione is one of the most important intracellular antioxidants and the enzymes involved in glutathione synthesis are deficient in IBD patients likely contributing to increased oxidative stress in the intestinal mucosa (Sido et al., 1998). Bacteria that encode the phenyllactate gene cluster can produce IA which reduces oxidative stress in the intestine, increasing microbial fitness while benefiting the host by promoting antioxidant processes.
Polymorphisms in NRF2 are associated with development of UC (Arisawa et al., 2008), and HMOX1, an NRF2 target gene, is upregulated during treatment with 5-aminosalicylic acid, a standard therapy for IBD patients (Horvath et al., 2008; Paine et al., 2010). NRF2 activation requires displacement of the cytosolic NRF2-KEAP1 (kelch-like ECH-associated protein 1) complex through covalent modification of cysteine thiols of KEAP1 by electrophiles (Hur et al., 2010). Although IPA and IA are structurally very similar, the double bond present in IA provides an electrophilic carbon that would be essential for NRF2 activation, similar to the mechanism of other NRF2-activating electrophiles such as tBHQ (Hur et al., 2010; O’Connell et al., 2016). These findings suggest targeting the NRF2-ARE pathway may have therapeutic benefit in IBD patients.
A recent study reported that IBD patients have reduced levels of fecal indole-3-acetic acid, suggesting that reduced tryptophan metabolism may contribute to the pathoetiology of IBD (Lamas et al., 2016). We add to this observation by showing that UC and CD patients show significantly reduced abundance of bacteria that can cleave terminal fucose residues from intestinal mucins using α-L-fucosidases, a finding that correlates with reduced abundance of the phenyllactate gene cluster involved in the production of IA and IPA. We suggest that mucins provide the sugar substrates to direct the colonization of health-promoting bacteria, such as tryptophan metabolizers. Supporting this hypothesis, recent studies have shown that microbial tryptophan metabolites such as indole-3-aldehyde, indole 3-propionate, and indole-3-acetic acid signal through AhR to increase IL-22 secretion by innate-like lymphoid cells and T cells, leading to protection from chemically induced colitis (Lamas et al., 2016; Monteleone et al., 2011; Zelante et al., 2013). IL-22 signaling leads to increased fucosylation of intestinal mucin (Pickard et al., 2014), perhaps creating a feedback loop in which the host promotes the fucosidase activity of specific commensals that in turn metabolize tryptophan to produce beneficial metabolites such as IA. We propose that increasing production of IA by restoring tryptophan metabolism in the intestine could have a therapeutic effect in IBD patients by promoting anti-inflammatory responses.
STAR Methods
CONTACT FOR REAGENT AND RESOURCE SHARING
Further information and requests for reagents may be directed to, and will be fulfilled by the corresponding author Ramnik Xavier (xavier@molbio.mgh.harvard.edu).
EXPERIMENTAL MODELS AND SUBJECT DETAILS
Computational Analysis of HMP and PRISM Subjects
The mucin cleavage and transporter seed genes were first searched against all available bacterial genomes on NCBI, complete or draft, using BLAT (Kent, 2002) protein sequence search. Matches with more than 40% amino acid sequence identity, less than 1×10-5 e-value, and more than 50% aligned length were selected as a gene in the expanded gene pool. To quantify the relative abundance of each gene in each species for each subject in the cross-sectional HMP study (Human Microbiome Project, 2012), reads were removed for human DNA sequences and trimmed for quality as previously described (Gevers et al., 2014). Bowtie2 (Langmead and Salzberg, 2012) was then employed to map the reads onto the expanded gene pool using default settings. Paired reads with proper orientation and mapping quality more than 15 were counted towards coverage. GreenGene taxonomy was used to guide aggregation of coverage for higher levels such as genus and family. To quantify the relative abundance of genes, we first discarded the 100-bp regions near the 3′ and 5′ ends as they suffer bias introduced by hanging reads, we then used RPKM (reads per kilobase per million) as a proxy of gene relative abundance to account for different sequencing depths among samples and different gene length. PRISM samples were searched against the expanded gene pool in the same manner, and the number of reads were counted using the same thresholds and normalized to RPKM. The genes were hierarchically clustered using Pearson’s correlation coefficient over Euclidean distance on their relative abundance. Phenyllactate gene cluster presence and abundance was calculated by limiting that all 4 fld genes must be within a 6-gene radius to be considered part of the gene cluster. Each gene’s RPKM was determined and the median value was used as the sample’s fld gene cluster RPKM.
Human Subjects
Stools samples from control individuals or patients with CD or UC were drawn from the Prospective Registry in IBD Study at MGH (PRISM). Details of this patient registry have been described previously (Ananthakrishnan et al., 2014). All subjects were adult >18 years of age at enrollment. The specific cohort used for these analyses had samples from 56 UC, 72 CD, and 35 control individuals. When relevant, active CD was defined as a Harvey Bradshaw index > 4 or with active UC defined as a Simple Clinical Colitis Activity Index > 5. As the PRISM cohort is population based, and IBD is known to affect both genders and all ethnic and racial categories, no subgroup is excluded from study. The net incidence of IBD overall is about 1:1 for the genders and that is represented in the samples studies here. As the incidence of IBD is roughly equal between male and female patients, and there has been no previous association found with gender and IBD microbiome, we did not include this variable in our analysis. Controls are pre-screened for eligibility and recruited prior to appointment. The study was approved by the Institutional Review Board of Partners Healthcare and all patients provided informed consent.
Mice and Dextran Sulfate Sodium Experiments
All animal studies were conducted under protocols approved by the Institutional Animal Care and Use Committee (IACUC) at Novartis Institutes for Biomedical Research (NIBR). Mice were maintained in specific-pathogen-free (SPF) facilities at NIBR facilities in Cambridge, MA and housed in a single room under standard SPF conditions with 5 mice per cage. For all studies 8-12 week old female C57Bl/6J mice purchased from Jackson Laboratories were used. DSS was supplied in drinking water at 2% (w/v) for 7 days. For oral gavage, bacterial strains were grown in BHI plus medium (agar plates or 30 mL liquid culture) and after 48 h of growth, bacteria were scraped or centrifuged and re-suspended in reduced anaerobic PBS. Mice were orally gavaged with 100 μL of each indicated bacterial strain resuspended to an OD600 of 2.0, corresponding to 0.6-2 × 108 CFU. Unless otherwise stated, mice were subjected to oral gavage every other day with each bacterial strain or PBS for two weeks. Mice were allocated to experimental groups randomly. Investigators were blinded during weight loss and pathology assessment in DSS experiments. No statistical tests were used to predetermine sample size.
3D Spheroid and RAW Cell Cultures
Primary colonic epithelial stem cells were isolated from female C57Bl/6J mice, grown, and maintained as 3D spheroid cultures in Matrigel (BD Biosciences) as previously described(Miyoshi and Stappenbeck, 2013). Cells were kept in 50% L-WRN CM. Media were changed every 2 days and cells were passaged every 3 days (1:3 split). In experiments testing the effect of metabolites on spheroid gene expression, spheroids were split and 12,000 epithelial cell clusters were seeded in 30 μL matrigel in a 48-well plate. 100 μM of metabolite or 0.1% DMSO was added to 50% L-WRN CM and spheroids developed over 24 h. Media was changed to 5% L-WRN CM, also supplemented with 100 μM of metabolite or 0.1% DMSO, and spheroids were given 48 h to differentiate in this medium. 5% L-WRN CM led to differentiation of spheroids as previously described(Miyoshi and Stappenbeck, 2013). Media was removed, washed 1x with PBS, and RLT buffer (Qiagen) was added to lyse cells for RNA extraction per the manufacturer’s instructions. RAW264.7 cells were obtained from ATCC and cell lines were tested for mycoplasma contamination every six months.
BMDM culture
To obtain murine BMDMs, femurs and tibias were harvested from female C57Bl/6J mice and marrow cores were flushed using syringes filled with RPMI 1640/10% FBS. Cells were triturated and red blood cells (RBCs) were lysed with 1X RBC lysis solution (eBioscience). Cells were washed once in media, then plated and cultured in RPMI 1640 supplemented with 0.1 mg/mL penicillin/streptomycin, 10 mM HEPES, 0.001% β-mercaptoethanol, 1 mM sodium pyruvate, 2 mM L-glutamine, 0.1 mM non-essential amino acids, and 10% FBS and in the presence of 30 ng/ml rmM-CSF (PeproTech). Cells were incubated for 7 days prior to use. On day 7, cells were scraped and washed with PBS and placed in RPMI/10% FBS. 96-well plates were seeded with 105 BMDMs/well and incubated with 100 μM IA, 100 μM IPA, 100 μM HPPA, or 0.1% DMSO for 6 h, then 20 ng/mL rmIL-4 (PeproTech) was added for 24 h, followed by 20 ng/mL LPS stimulation for 18 h. Supernatants were collected and analyzed using a cytokine bead array mouse inflammation kit (BD Biosciences) according to the manufacturer’s instructions.
Co-Culture
Spheroids and BMDMs were prepared as described above. 6000 epithelial cell clusters and 105 BMDMs were seeded in 30 μL matrigel in a 48-well plate. 100 μM of metabolite or 0.1% DMSO was added to 50% L-WRN CM and maintained in this medium for 24 h. Media was changed to 5% L-WRN CM, also supplemented with 100 μM of metabolite or 0.1% DMSO, and spheroids were given 24 h to differentiate after which the co-culture was stimulated with 20 ng/mL LPS for an additional 24 h. Supernatant was collected for cytokine analysis as above. Remaining medium was removed, cells were washed 1x with PBS, and RLT buffer (Qiagen) was added to lyse cells for RNA extraction per the manufacturer’s instructions. For flow cytometry analysis, cells were dissociated in 0.25% (w/v) trypsin solution for 15 min with intermittent vigorous pipetting at 37°C, then washed in DMEM/F12 containing 10% FBS, 0.1 mg/mL penicillin/streptomycin, and 2 mM L-glutamine.
Human PBMC Assays
Blood buffy coats were obtained from healthy male and female volunteers after informed consent. The study protocol and any amendments were reviewed and approved by an independent review board (New England IRB, Newton, MA, USA) before the start of the study. The study was conducted according to the ethical principles of the Declaration of Helsinki. PBMCs were freshly isolated from blood by Ficoll-Hypaque gradient centrifugation as previously described(Nair et al., 2012). PBMCs (105) were incubated in the presence of IA or IPA for 45 min and then stimulated with 1 ng/mL LPS purified from E. coli ECOR2 for 20-22 h. Supernatants were collected and analyzed using the cytokine bead array human inflammation kit (BD Biosciences) according to the manufacturer’s instructions.
METHOD DETAILS
Bacterial strains
Bacterial strains used in this study are as follows: Clostridium butyricum ATCC 19398, Clostridium ramosum ATCC 25582, Clostridium scindens ATCC 35704, Clostridium tyrobutyricum ATCC 25755, Coprococcus comes ATCC 27758, Lachnospiraceae bacterium ATCC BAA2281, Ruminococcus gnavus ATCC 29148, Ruminococcus torques ATCC 27756, Bacteroides thetaiotaomicron ATCC 29148, Akkermansia muciniphila ATCC BAA835, Anaerotruncus colihominis DSM 17241, Blautia hansenii DSM 20583, Blautia hydrogenotrophica DSM 10507, Blautia producta DSM 2950, Catenibacterium mitsuokai DSM 15897, Clostridium aldenense DSM 19262, Clostridium asparagiforme DSM 15981, Clostridium bolteae DSM 15670, Clostridium hathewayi DSM 13479, Clostridium hiranonis DSM 13275, Clostridium lentocellum DSM 5427, Clostridium methylpentosum DSM 5476, Clostridium spiroforme DSM 1552, Clostridium symbiosum DSM 934, Dorea formicigenerans DSM 3992, Eubacterium biforme DSM 3989, Eubacterium dolichum DSM 3991, Holdemania filiformis DSM 12042, Marvinbryantia formatexigens DSM 14469, Peptostreptococcus anaerobius DSM 2949, Peptostreptococcus russellii DSM 23041, Peptostreptococcus stomatis DSM 17678, and Ruminococcus obeum DSM 25238.
Bacterial Growth Medium
Minimal mucin media was made with 0.5% porcine mucin (M2378, Sigma), 100 mM KH2PO4 (pH 7.2), 15 mM NaCl, 8.5 mM (NH4)2SO4, 4 mM L-cysteine, 1% vitamin K1-hemin solution (Becton Dickinson), 100 μM MgCl2, 1.4 μM FeSO4·7H2O, 50 μM CaCl2, 1% trace mineral supplement (ATCC), and 1% vitamin supplement (ATCC). Minimal M9 media was purchased (Teknova) and supplemented with 0.5% tryptone, plus or minus 0.25% porcine mucin (M2378, Sigma). Brain heart infusion medium (BHI) was purchased (Becton Dickinson) as agar or broth powder and made according to manufacturer’s instructions and supplemented with 1% vitamin K1-hemin solution (Becton Dickinson). BHI plus medium was supplemented with 5% heat inactivated fetal bovine serum, 1% vitamin K1-hemin solution (Becton Dickinson), 1% trace mineral supplement (ATCC), 1% vitamin supplement (ATCC), 3 mM D-(+)-cellubiose, 3 mM D-(+)-maltose, 6 mM D-(+)-fructose) and 4 mM L-cysteine. Media were filter sterilized using a Millipore Express filter unit (0.22-μm pore diameter) with the exception of porcine mucin, which was autoclaved in dH20 and added after sterile filtration. A flexible anaerobic chamber (Coy Laboratory Products) containing 20% CO2, 5% H2, and 75% N2 and maintained at 37°C was used for all anaerobic microbiology steps.
Chemicals
Identified bacterial tryptophan and phenylalanine metabolites trans-indole-3-acrylic acid (IA, Sigma), indole 3-propionic acid (IPA, Sigma), and 3-(4-hydroxyphenyl)propionic acid (HPPA, Alfa Aesar), were purchased and dissolved in DMSO to a 100 mM stock concentration.
Histology and Immunofluorescence
The terminal 5 mm of the colon were excised, immediately submerged in Ethanol-Carnoy’s fixative at 4°C for 2 h and then placed into 100% ethanol. Fixed colon tissues were embedded in paraffin and cut into 5 μm sections. Tissues were stained with Alcian blue/PAS to enumerate goblet cells per crypt. Paraffin embedded tissues Carnoys-fixed were deparaffinized and rehydrated. Antigen retrieval was performed in 10 mM citric acid pH 6.0 at 90-100°C. Immunostaining was carried out using an antibody against Muc2 (H-300, Santa Cruz) followed by incubation with an Alexa-conjugated secondary antibody (Invitrogen) or the FITC conjugated UEA-I lectin (EY laboratories). Tissues were mounted using ProLong Gold® Antifade (Molecular Probes/Invitrogen) that contains 4’,6’-diamidino-2-phenylindole (DAPI) for DNA staining.
Histopathological Scoring
The entire large intestine (colon) was removed, cut longitudinally and rolled into a Swiss Roll for histological analysis. Swiss rolls were fixed in 10% buffered formalin and placed into 70% ethanol followed by paraffin embedding, sectioned and stained with hematoxylin and eosin (H&E). Swiss rolls were assessed for pathology along the entire length of the colon and every focus of inflammation was scored. Regions of ulceration larger than 1 mm were considered large ulcers and given a score of 5; small ulcers (< 1 mm) were given a score of 4. Regions of immune infiltration but an intact surface epithelium (no ulceration present) were scored based on whether immune cell infiltrate was seen only in the mucosa (low score = 1), infiltrate in the mucosa and submucosa with evident mucosal damage (medium score = 2), or transmural immune infiltration with evident mucosal damage (high score = 3).
LC/MS Analysis of Peptostreptococcus Species Metabolites
Bacterial strains were grown in 5 mL BHI medium supplemented with 1% vitamin K1- hemin solution (Becton Dickinson). After 48 h, cultures were centrifuged at 3000 x rpm for 5 min to pellet bacteria. 4 mL of supernatant was mixed with acetonitrile in a 1:1 ratio. Magnesium sulfate (0.2 g/mL) and sodium acetate (0.05 g/mL) were added and mixed by vortexing and gentle shaking for 10 min followed by centrifugation at 3000 x rpm for 10 min. The organic phase was removed and placed in a pre-weighed vial and dried in a Genevac. Accurate mass LC/MS was performed on an AB Sciex TripleTOF 5600 operating in IDA mode. Delivery of mobile phases was performed using a Shimadzu Prominence series HPLC system with 0.1% formic acid in water (v:v) as mobile phase A and 0.1% formic acid in acetonitrile (v:v) as mobile phase B. Total organic extract of Peptostreptococcus spp. culture was solubilized at 1 mg/mL in starting mobile phase (95:5, A:B) and 25 μg of material was separated over a Waters Acquity BEH C18 column (2.1 × 100 mm, 1.7 μm) coupled to a Waters Acquity BEH C18 VanGuard pre-column (2.1 × 5 mm, 1.7 μm). The column temperature was maintained at 40°C and samples were eluted with a binary gradient: 0-2 min, 5% B; 2-22min, 40% B; 22-28 min, 100% B; 28-32 min, 100% B; 32-32.5 min, 5% B, followed by an additional 3 min at 5% B for post-run equilibration. Flow rate was maintained at 0.3 mL/min throughout the run. Data analysis was performed using PeakView 1.2 software. IPA, IA, and HPPA were absent from media blanks (data not shown).
Quantitative RT-PCR
The terminal 2-3 mm of the colon were excised, immediately submerged in RNAlater (Qiagen) and stored at 4°C overnight and then at -80°C for subsequent RNA extraction. RNA was extracted using RNeasy Mini kit (Qiagen) according to the manufacturer’s instructions. RNA concentration was determined using a NanoDrop ND-1000 (NanoDrop Technologies) and reverse transcription was performed with the SuperScript III reverse transcriptase kit (Invitrogen) according to the manufacturer’s instructions. qPCR was performed with TaqMan Fast Advanced Master Mix (Invitrogen) using the following TaqMan probes (Invitrogen): Gapdh Mm99999915, Actb Mm02619580, Ki67 Mm01278617, Muc2 Mm01276696, IL10 Mm01288386, IL22 Mm01226722, Tnf Mm00443258, Fut2 Mm00490152, B3gnt6 Mm04204642, C1galt1 Mm01167001, St6gal1 Mm00486119, Spedf Mm00600221, Cyp1a1 Mm00487218, Ugt1a1 Mm02603337, Abcb1b (Mdr-1) Mm00440736, Hmox1 Mm00516005, Gclm Mm00514996, Gclc Mm00802655, and Nqo1 Mm01253561. The reactions were run on a ViiA7 real-time PCR machine (Applied Biosystems) and changes in gene expression were calculated relative to Gapdh (tissue) or Actb (spheroid and co-culture) and the fold difference in expression was calculated as 2-∆∆Ct.
Flow cytometry
For staining of mouse cells (mouse or spheroid-derived), cells were stained with viability dye Zombie NIR (BioLegend) followed by blocking of Fc receptors with anti-mouse or anti-human CD16/32 (BioLegend). Cells were stained with surface markers CD326 (G8.8, BD Pharmingen) and F480 (T45-2342, BD Pharmingen), washed, and fixed with fixation buffer (eBioscience). Cells were permeabilized with 1x permeabilization buffer (eBioscience) and goblet cells were stained intracellularly with lectin from Ulex europaeus, UEA-I, TRITC conjugate (Sigma).
Quantifying P. russellii from mouse stool
Bacterial genomic DNA was extracted from fecal pellets using MoBio PowerSoil DNA Isolation Kit. Quantitative PCR was performed using Power SYBR Green PCR Master Mix (Applied Biosystems) and a CFX96 C1000 Touch Thermal Cycler (Bio-Rad). To identify a P. russellii-specific marker sequences for qPCR, we first called open reading frames (ORFs) in the assembled P. russellii genomic contigs using Prodigal (Hyatt et al., 2010) (default settings), identifying 1,806 ORFs. We then performed nucleotide-level search (Altschul et al., 1990) of ORFs against a databases of prokaryotic coding sequences (Huang et al., 2014). If an ORF had any hit in this process, it was flagged as non-unique. 725 P. russellii ORFs had such hits, leaving 1,081 ORFs as candidate P. russellii-specific markers. From among these candidates, we selected a putative HD superfamily hydrolase for qPCR. We further evaluated the uniqueness of this marker by checking primer specificity using Primer-BLAST (Ye et al., 2012) against all bacterial genomes and no other targets were found. Universal primers (5’-GGTGAATACGTTCCCGG-3’ and 5’-TACGGCTACCTTGTTACGACTT-3’) (Atarashi et al., 2011) were used to measure total 16S rRNA per sample. P. russellii strain-specific primer set 298 (5’-GGAGAGCGAATATGAAGCCT-3’ and 5’-CCCAGAATAAGCCATTTCCCT-3’) was designed by Geneious 10.0.9 for the amplification of a putative HD superfamily hydrolase. A standard curve was generated with six 10-fold serial dilutions of P. russellii genomic DNA starting at 10 ng per reaction. Primer set 298 was screened against closely related species, P. anaerobis and P. stomatis, to validate the specificity for P. russellii. Quantitative PCR was performed with 10 ng DNA per sample with the universal primer set and primer set 298. Abundance of P. russellii DNA and total bacterial DNA was calculated with the linear equation from the standard curve and were expressed as P. russellii putative HD superfamily hydrolase or universal 16S rRNA gene copies per 10 ng DNA.
RNA Sequencing
RNA was isolated from human PBMCs using an RNeasy Plus Mini Kit (Qiagen). The smart-seq2 protocol (Picelli et al., 2013) was modified for 20 ng total input RNA to generate cDNA libraries, which were read on a NextSeq500 (Illumina, San Diego, CA) per the manufacturer’s instructions. Paired end raw (fastq) reads from RNASeq were aligned against the Homo sapiens (USCS hg19) genome using TopHat + Bowtie (Langmead et al., 2009). HTSeq-count (Anders et al., 2015) was used to count the transcripts associated with each gene, and a counts matrix containing the number of counts for each gene across different samples and stimulations was obtained. The samples were normalized using the voom package (Law et al., 2014) and statistically analysed using linear models from the limma package (Ritchie et al., 2015). Differentially expressed genes between treatments were found using a linear model that included both treatment and donor effect and the significance threshold FDR < 0.05 was applied. In the heatmap representation, the donor effect was removed from the data and the values from replicate measurements from the same treatment and donor were averaged. Gene set enrichment analysis was performed using g:Profiler toolkit (Reimand et al., 2016) on the 1,000 most up- and down-regulated genes, with ordered query option and filtering out GO terms with more than 1,000 and fewer than 50 genes.
NRF2 reporter
RAW264.7 cells were transduced with a lentivirus encoding a NRF2-luciferase reporter construct (O’Connell et al., 2016). 100,000 cells per well were stimulated with 300 μM IA, 300 μM IPA, 12.5 μM tert-Butylhydroquinone (tBHQ), or DMSO for 45 min followed addition of 20 ng/μl LPS and overnight incubation in a 96-well plate. The assay was quantified with the Steadylite Reporter system (Perkin Elmer) and fluorescence was measured with a Synergy H4 Hybrid Reader (Biotek).
QUANTIFICATION AND STATISTICAL ANALYSIS
Unless otherwise indicated, statistical significance was calculated by two-tailed Student’s t-test using GraphPad Prism 7. For gene coverage in the HMP and PRISM cohort, each gene was tested for between-group significance using two-tailed Student’s t-test first, and p values were corrected for FDR using the Hochberg-Benjamini procedure. Unless otherwise indicated, ****p < 0.0001; ***p < 0.001; **p < 0.01; *p ≤ 0.05; ns (not significant) p > 0.05. Unless otherwise indicated, results are expressed as mean values with standard error of the mean. Reported “n” in animal studies represents the number of animals used and in experiments with murine spheroids “n” represents the number of unique lines used for the experiments multiplied by the experiment replication number.
DATA AND SOFTWARE AVAILABILITY
All raw and normalized gene expression data presented in this manuscript is available at GEO: GSE98884.
Supplementary Material
KEY RESOURCES TABLE.
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| anti-mouse CD16/32 | BioLegend | Cat#101301 |
| anti-mouse CD326, G8.8 | BioLegend | Cat#118207 |
| anti-mouse F480, clone T45-2342 | BD Biosciences | Cat#565410 |
| TRITC Ulex europaeus, UEA-I, lectin | Sigma | Cat#L4889 |
| FITC conjugated UEA-I lectin | EY laboratories | Cat#F-2201-2 |
| Muc 2 antibody, H-300 | Santa Cruz | Cat#sc-15334 |
| Secondary Antibody, Alexa Fluor 647 | ThermoFisher | Cat#A31573 |
| Bacterial and Virus Strains | ||
| Clostridium butyricum | ATCC | ATCC 19398 |
| Clostridium ramosum | ATCC | ATCC 25582 |
| Clostridium scindens | ATCC | ATCC 35704 |
| Clostridium tyrobutyricum | ATCC | ATCC 25755 |
| Coprococcus comes | ATCC | ATCC 27758 |
| Lachnospiraceae bacterium | ATCC | ATCC BAA2281 |
| Ruminococcus gnavus | ATCC | ATCC 29148 |
| Ruminococcus torques | ATCC | ATCC 27756 |
| Bacteroides thetaiotaomicron | ATCC | ATCC 29148 |
| Akkermansia muciniphila | ATCC | ATCC BAA835 |
| Anaerotruncus colihominis | DSMZ | DSM 17241 |
| Blautia hansenii | DSMZ | DSM 20583 |
| Blautia hydrogenotrophica | DSMZ | DSM 10507 |
| Blautia producta | DSMZ | DSM 2950 |
| Catenibacterium mitsuokai | DSMZ | DSM 15897 |
| Clostridium aldenense | DSMZ | DSM 19262 |
| Clostridium asparagiforme | DSMZ | DSM 15981 |
| Clostridium bolteae | DSMZ | DSM 15670 |
| Clostridium hathewayi | DSMZ | DSM 13479 |
| Clostridium hiranonis | DSMZ | DSM 13275 |
| Clostridium lentocellum | DSMZ | DSM 5427 |
| Clostridium methylpentosum | DSMZ | DSM 5476 |
| Clostridium spiroforme | DSMZ | DSM 1552 |
| Clostridium symbiosum | DSMZ | DSM 934 |
| Dorea formicigenerans | DSMZ | DSM 3992 |
| Eubacterium biforme | DSMZ | DSM 3989 |
| Eubacterium dolichum | DSMZ | DSM 3991 |
| Holdemania filiformis | DSMZ | DSM 12042 |
| Marvinbryantia formatexigens | DSMZ | DSM 14469 |
| Peptostreptococcus anaerobius | DSMZ | DSM 2949 |
| Peptostreptococcus russellii | DSMZ | DSM 23041 |
| Peptostreptococcus stomatis | DSMZ | DSM 17678 |
| Ruminococcus obeum | DSMZ | DSM 25238 |
| Chemicals, Peptides, and Recombinant Proteins | ||
| Dextran Sulfate Sodium salt, colitis grade | MP Biomedicals | Cat#02160110 |
| rmIL-4 | Peprotech | Cat#214-14 |
| rmM-CSF | Peprotech | Cat#315-02 |
| trans-indole-3-acrylic acid, IA | Sigma | Cat#I3807 |
| indole 3-propionic acid, IPA | Sigma | Cat#57400 |
| 3-(4-hydroxyphenyl)propionic acid, HPPA | Alfa Aesar | Cat#A14567 |
| porcine mucin | Sigma | Cat#M2378 |
| 1% vitamin K1-hemin solution | Becton Dickinson | Cat#212354 |
| 1% vitamin supplement | ATCC | Cat#MD-VS |
| 1% trace mineral supplement | ATCC | Cat#MD-TMS |
| Zombie NIR | BioLegend | Cat#423105 |
| Permeabilization buffer | eBioscience | Cat#00-8333-56 |
| Fixation/Permeabilization concentrate | eBioscience | Cat#00-5123-43 |
| tert-Butylhydroquinone, tBHQ | Sigma | Cat#112941 |
| Critical Commercial Assays | ||
| Mouse Inflammation Cytokine Bead Array | BD biosciences | Cat#552364 |
| SuperScript III reverse transcriptase kit | Invitrogen | Cat#18080051 |
| TaqMan Fast Advanced Master Mix | Invitrogen | Cat#4444557 |
| Human Inflammation Cytokine Bead Array | BD biosciences | Cat#551811 |
| Deposited Data | ||
| Human PBMCs RNA seq data set | GEO database | Accession#GSE98884 |
| Experimental Models: Cell Lines | ||
| RAW264.7 | ATCC | Cat#TIB-71 |
| Oligonucleotides | ||
| See Table S2 | N/A | N/A |
| Software and Algorithms | ||
| Bowtie 2 | Langmead and Salzberg, 2012 | http://bowtie-bio.sourceforge.net/bowtie2/index.shtml |
| BLAT | Kent, 2002 | http://genome.ucsc.edu/cgi-bin/hgBlat |
| Other | ||
| Qiagen RNeasy Plus Mini Kit | Qiagen | 74136 |
Highlights.
Computational analysis identifies Peptostreptococcus russellii as a mucin-utilizer
Peptostreptococcus species produce the tryptophan metabolite indoleacrylic acid (IA)
IA promotes intestinal epithelial barrier function and mitigates inflammatory responses
Microbes of IBD patients have reduced ability to cleave mucins and metabolize tryptophan
Acknowledgments
We thank Tiffany Poon for help in sequence production and sample management, Paola Capodieci (Novartis) for providing histology services, Christine Petersen and Junmei Yao for experimental assistance, Mukund Varma for help processing RNA Seq data, Dan Graham and Kara Lassen (Broad Institute) for helpful discussions, and Natalia Nedelsky (Massachusetts General Hospital) for invaluable editorial help.
Footnotes
Author Contributions
M.W. devised the concept and performed experiments involving bacterial growth analysis, BMDMs, spheroids, co-culture system, PBMCs, and mouse modeling. C.L. designed and performed computational analysis. M.W., C.L., E.A.F., and C.H. generated the list of mucus utilization genes. R.K. analyzed RNA-sequencing data. E.H. performed some experiments involving PBMCs. C.E.H. aided in BMDM generation and some experiments involving PBMCs and mice. J.W.A., P.K., and E.K.S. performed LC/MS analysis and identified metabolites. A.S.O. assisted with Muc2/UEA-I staining. E.A.C. assessed AhR signaling in colonic spheroid experiments. A.L.G. and D.J.O. performed assays with the NRF2 reporter. S.M. aided in qPCR analysis of murine colonic tissue. S.A. analyzed Pac-bio sequences for gene cluster identification. T.D.A. performed qPCR to quantify P. russellii in mouse stool. L.O.M. supervised the studies. M.W., H.V., and R.J.X. wrote the paper. M.W., H.V., H.J.H., J.A.P., and R.J.X. served as project leaders.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Abreu AG, Abe CM, Nunes KO, Moraes CT, Chavez-Duenas L, Navarro-Garcia F, Barbosa AS, Piazza RM, Elias WP. The serine protease Pic as a virulence factor of atypical enteropathogenic Escherichia coli. Gut Microbes. 2016;7:115–125. doi: 10.1080/19490976.2015.1136775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alam A, Leoni G, Quiros M, Wu H, Desai C, Nishio H, Jones RM, Nusrat A, Neish AS. The microenvironment of injured murine gut elicits a local pro-restitutive microbiota. Nat Microbiol. 2016;1:15021. doi: 10.1038/nmicrobiol.2015.21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- Ananthakrishnan AN, Huang H, Nguyen DD, Sauk J, Yajnik V, Xavier RJ. Differential effect of genetic burden on disease phenotypes in Crohn’s disease and ulcerative colitis: analysis of a North American cohort. Am J Gastroenterol. 2014;109:395–400. doi: 10.1038/ajg.2013.464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anders S, Pyl PT, Huber W. HTSeq--a Python framework to work with high-throughput sequencing data. Bioinformatics. 2015;31:166–169. doi: 10.1093/bioinformatics/btu638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arisawa T, Tahara T, Shibata T, Nagasaka M, Nakamura M, Kamiya Y, Fujita H, Yoshioka D, Okubo M, Sakata M, et al. Nrf2 gene promoter polymorphism is associated with ulcerative colitis in a Japanese population. Hepatogastroenterology. 2008;55:394–397. [PubMed] [Google Scholar]
- Atarashi K, Tanoue T, Shima T, Imaoka A, Kuwahara T, Momose Y, Cheng G, Yamasaki S, Saito T, Ohba Y, et al. Induction of colonic regulatory T cells by indigenous Clostridium species. Science. 2011;331:337–341. doi: 10.1126/science.1198469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Begue B, Verdier J, Rieux-Laucat F, Goulet O, Morali A, Canioni D, Hugot JP, Daussy C, Verkarre V, Pigneur B, et al. Defective IL10 signaling defining a subgroup of patients with inflammatory bowel disease. Am J Gastroenterol. 2011;106:1544–1555. doi: 10.1038/ajg.2011.112. [DOI] [PubMed] [Google Scholar]
- Birchenough GM, Nystrom EE, Johansson ME, Hansson GC. A sentinel goblet cell guards the colonic crypt by triggering Nlrp6-dependent Muc2 secretion. Science. 2016;352:1535–1542. doi: 10.1126/science.aaf7419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corfield AP, Wagner SA, Clamp JR, Kriaris MS, Hoskins LC. Mucin degradation in the human colon: production of sialidase, sialate O-acetylesterase, N-acetylneuraminate lyase, arylesterase, and glycosulfatase activities by strains of fecal bacteria. Infect Immun. 1992;60:3971–3978. doi: 10.1128/iai.60.10.3971-3978.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Derrien M, Vaughan EE, Plugge CM, de Vos WM. Akkermansia muciniphila gen. nov., sp. nov., a human intestinal mucin-degrading bacterium. Int J Syst Evol Microbiol. 2004;54:1469–1476. doi: 10.1099/ijs.0.02873-0. [DOI] [PubMed] [Google Scholar]
- Desai MS, Seekatz AM, Koropatkin NM, Kamada N, Hickey CA, Wolter M, Pudlo NA, Kitamoto S, Terrapon N, Muller A, et al. A Dietary Fiber-Deprived Gut Microbiota Degrades the Colonic Mucus Barrier and Enhances Pathogen Susceptibility. Cell. 2016;167:1339–1353 e1321. doi: 10.1016/j.cell.2016.10.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dickert S, Pierik AJ, Buckel W. Molecular characterization of phenyllactate dehydratase and its initiator from Clostridium sporogenes. Mol Microbiol. 2002;44:49–60. doi: 10.1046/j.1365-2958.2002.02867.x. [DOI] [PubMed] [Google Scholar]
- Fyderek K, Strus M, Kowalska-Duplaga K, Gosiewski T, Wedrychowicz A, Jedynak-Wasowicz U, Sladek M, Pieczarkowski S, Adamski P, Kochan P, et al. Mucosal bacterial microflora and mucus layer thickness in adolescents with inflammatory bowel disease. World J Gastroenterol. 2009;15:5287–5294. doi: 10.3748/wjg.15.5287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gevers D, Kugathasan S, Denson LA, Vazquez-Baeza Y, Van Treuren W, Ren B, Schwager E, Knights D, Song SJ, Yassour M, et al. The treatment-naive microbiome in new-onset Crohn’s disease. Cell Host Microbe. 2014;15:382–392. doi: 10.1016/j.chom.2014.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goto Y, Obata T, Kunisawa J, Sato S, Ivanov II, Lamichhane A, Takeyama N, Kamioka M, Sakamoto M, Matsuki T et al. Innate lymphoid cells regulate intestinal epithelial cell glycosylation. Science. 2014;345:1254009. doi: 10.1126/science.1254009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hashimoto T, Perlot T, Rehman A, Trichereau J, Ishiguro H, Paolino M, Sigl V, Hanada T, Hanada R, Lipinski S, et al. ACE2 links amino acid malnutrition to microbial ecology and intestinal inflammation. Nature. 2012;487:477–481. doi: 10.1038/nature11228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hasnain SZ, Tauro S, Das I, Tong H, Chen AC, Jeffery PL, McDonald V, Florin TH, McGuckin MA. IL-10 promotes production of intestinal mucus by suppressing protein misfolding and endoplasmic reticulum stress in goblet cells. Gastroenterology. 2013;144:357–368 e359. doi: 10.1053/j.gastro.2012.10.043. [DOI] [PubMed] [Google Scholar]
- Hayes JD, Dinkova-Kostova AT, McMahon M. Cross-talk between transcription factors AhR and Nrf2: lessons for cancer chemoprevention from dioxin. Toxicol Sci. 2009;111:199–201. doi: 10.1093/toxsci/kfp168. [DOI] [PubMed] [Google Scholar]
- Hooper LV, Littman DR, Macpherson AJ. Interactions between the microbiota and the immune system. Science. 2012;336:1268–1273. doi: 10.1126/science.1223490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horvath K, Varga C, Berko A, Posa A, Laszlo F, Whittle BJ. The involvement of heme oxygenase-1 activity in the therapeutic actions of 5-aminosalicylic acid in rat colitis. Eur J Pharmacol. 2008;581:315–323. doi: 10.1016/j.ejphar.2007.12.004. [DOI] [PubMed] [Google Scholar]
- Huang K, Brady A, Mahurkar A, White O, Gevers D, Huttenhower C, Segata N. MetaRef: a pan-genomic database for comparative and community microbial genomics. Nucleic Acids Res. 2014;42:D617–624. doi: 10.1093/nar/gkt1078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Human Microbiome Project C. Structure, function and diversity of the healthy human microbiome. Nature. 2012;486:207–214. doi: 10.1038/nature11234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hur W, Sun Z, Jiang T, Mason DE, Peters EC, Zhang DD, Luesch H, Schultz PG, Gray NS. A small-molecule inducer of the antioxidant response element. Chem Biol. 2010;17:537–547. doi: 10.1016/j.chembiol.2010.03.013. [DOI] [PubMed] [Google Scholar]
- Hyatt D, Chen GL, Locascio PF, Land ML, Larimer FW, Hauser LJ. Prodigal: prokaryotic gene recognition and translation initiation site identification. BMC Bioinformatics. 2010;11:119. doi: 10.1186/1471-2105-11-119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johansson ME, Gustafsson JK, Holmen-Larsson J, Jabbar KS, Xia L, Xu H, Ghishan FK, Carvalho FA, Gewirtz AT, Sjovall H, et al. Bacteria penetrate the normally impenetrable inner colon mucus layer in both murine colitis models and patients with ulcerative colitis. Gut. 2014;63:281–291. doi: 10.1136/gutjnl-2012-303207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johansson ME, Gustafsson JK, Sjoberg KE, Petersson J, Holm L, Sjovall H, Hansson GC. Bacteria penetrate the inner mucus layer before inflammation in the dextran sulfate colitis model. PLoS ONE. 2010;5:e12238. doi: 10.1371/journal.pone.0012238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johansson ME, Phillipson M, Petersson J, Velcich A, Holm L, Hansson GC. The inner of the two Muc2 mucin-dependent mucus layers in colon is devoid of bacteria. Proc Natl Acad Sci U S A. 2008;105:15064–15069. doi: 10.1073/pnas.0803124105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kent WJ. BLAT--the BLAST-like alignment tool. Genome Res. 2002;12:656–664. doi: 10.1101/gr.229202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kostic AD, Chun E, Robertson L, Glickman JN, Gallini CA, Michaud M, Clancy TE, Chung DC, Lochhead P, Hold GL, et al. Fusobacterium nucleatum potentiates intestinal tumorigenesis and modulates the tumor-immune microenvironment. Cell Host Microbe. 2013;14:207–215. doi: 10.1016/j.chom.2013.07.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kostic AD, Gevers D, Siljander H, Vatanen T, Hyotylainen T, Hamalainen AM, Peet A, Tillmann V, Poho P, Mattila I, et al. The dynamics of the human infant gut microbiome in development and in progression toward type 1 diabetes. Cell Host Microbe. 2015;17:260–273. doi: 10.1016/j.chom.2015.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamas B, Richard ML, Leducq V, Pham HP, Michel ML, Da Costa G, Bridonneau C, Jegou S, Hoffmann TW, Natividad JM, et al. CARD9 impacts colitis by altering gut microbiota metabolism of tryptophan into aryl hydrocarbon receptor ligands. Nat Med. 2016;22:598–605. doi: 10.1038/nm.4102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langmead B, Salzberg SL. Fast gapped-read alignment with Bowtie 2. Nat Methods. 2012;9:357–359. doi: 10.1038/nmeth.1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langmead B, Trapnell C, Pop M, Salzberg SL. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 2009;10:R25. doi: 10.1186/gb-2009-10-3-r25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larsson JM, Karlsson H, Crespo JG, Johansson ME, Eklund L, Sjovall H, Hansson GC. Altered O-glycosylation profile of MUC2 mucin occurs in active ulcerative colitis and is associated with increased inflammation. Inflamm Bowel Dis. 2011;17:2299–2307. doi: 10.1002/ibd.21625. [DOI] [PubMed] [Google Scholar]
- Law CW, Chen Y, Shi W, Smyth GK. voom: Precision weights unlock linear model analysis tools for RNA-seq read counts. Genome Biol. 2014;15:R29. doi: 10.1186/gb-2014-15-2-r29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li W, Khor TO, Xu C, Shen G, Jeong WS, Yu S, Kong AN. Activation of Nrf2-antioxidant signaling attenuates NFkappaB-inflammatory response and elicits apoptosis. Biochem Pharmacol. 2008;76:1485–1489. doi: 10.1016/j.bcp.2008.07.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marcobal A, Barboza M, Sonnenburg ED, Pudlo N, Martens EC, Desai P, Lebrilla CB, Weimer BC, Mills DA, German JB, et al. Bacteroides in the infant gut consume milk oligosaccharides via mucus-utilization pathways. Cell Host Microbe. 2011;10:507–514. doi: 10.1016/j.chom.2011.10.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martens EC, Chiang HC, Gordon JI. Mucosal glycan foraging enhances fitness and transmission of a saccharolytic human gut bacterial symbiont. Cell Host Microbe. 2008;4:447–457. doi: 10.1016/j.chom.2008.09.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martens EC, Roth R, Heuser JE, Gordon JI. Coordinate regulation of glycan degradation and polysaccharide capsule biosynthesis by a prominent human gut symbiont. J Biol Chem. 2009;284:18445–18457. doi: 10.1074/jbc.M109.008094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGuckin MA, Linden SK, Sutton P, Florin TH. Mucin dynamics and enteric pathogens. Nat Rev Microbiol. 2011;9:265–278. doi: 10.1038/nrmicro2538. [DOI] [PubMed] [Google Scholar]
- Miyoshi H, Stappenbeck TS. In vitro expansion and genetic modification of gastrointestinal stem cells in spheroid culture. Nat Protoc. 2013;8:2471–2482. doi: 10.1038/nprot.2013.153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monteleone I, Rizzo A, Sarra M, Sica G, Sileri P, Biancone L, MacDonald TT, Pallone F, Monteleone G. Aryl hydrocarbon receptor-induced signals up-regulate IL-22 production and inhibit inflammation in the gastrointestinal tract. Gastroenterology. 2011;141:237–248, 248 e231. doi: 10.1053/j.gastro.2011.04.007. [DOI] [PubMed] [Google Scholar]
- Nair S, Archer GE, Tedder TF. Isolation and generation of human dendritic cells. Curr Protoc Immunol. 2012;Chapter 7(Unit7):32. doi: 10.1002/0471142735.im0732s99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakayama-Imaohji H, Ichimura M, Iwasa T, Okada N, Ohnishi Y, Kuwahara T. Characterization of a gene cluster for sialoglycoconjugate utilization in Bacteroides fragilis. J Med Invest. 2012;59:79–94. doi: 10.2152/jmi.59.79. [DOI] [PubMed] [Google Scholar]
- O’Connell DJ, Kolde R, Sooknah M, Graham DB, Sundberg TB, Latorre IJ, Mikkelsen TS, Xavier RJ. Simultaneous Pathway Activity Inference and Gene Expression Analysis Using RNA Sequencing. Cell Syst. 2016;2:323–334. doi: 10.1016/j.cels.2016.04.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paine A, Eiz-Vesper B, Blasczyk R, Immenschuh S. Signaling to heme oxygenase-1 and its anti-inflammatory therapeutic potential. Biochem Pharmacol. 2010;80:1895–1903. doi: 10.1016/j.bcp.2010.07.014. [DOI] [PubMed] [Google Scholar]
- Picelli S, Bjorklund AK, Faridani OR, Sagasser S, Winberg G, Sandberg R. Smart-seq2 for sensitive full-length transcriptome profiling in single cells. Nat Methods. 2013;10:1096–1098. doi: 10.1038/nmeth.2639. [DOI] [PubMed] [Google Scholar]
- Pickard JM, Maurice CF, Kinnebrew MA, Abt MC, Schenten D, Golovkina TV, Bogatyrev SR, Ismagilov RF, Pamer EG, Turnbaugh PJ, et al. Rapid fucosylation of intestinal epithelium sustains host-commensal symbiosis in sickness. Nature. 2014;514:638–641. doi: 10.1038/nature13823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Png CW, Linden SK, Gilshenan KS, Zoetendal EG, McSweeney CS, Sly LI, McGuckin MA, Florin TH. Mucolytic bacteria with increased prevalence in IBD mucosa augment in vitro utilization of mucin by other bacteria. Am J Gastroenterol. 2010;105:2420–2428. doi: 10.1038/ajg.2010.281. [DOI] [PubMed] [Google Scholar]
- Reimand J, Arak T, Adler P, Kolberg L, Reisberg S, Peterson H, Vilo J. g:Profiler-a web server for functional interpretation of gene lists (2016 update) Nucleic Acids Res. 2016;44:W83–89. doi: 10.1093/nar/gkw199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ritchie ME, Phipson B, Wu D, Hu Y, Law CW, Shi W, Smyth GK. limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 2015;43:e47. doi: 10.1093/nar/gkv007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sampson TR, Debelius JW, Thron T, Janssen S, Shastri GG, Ilhan ZE, Challis C, Schretter CE, Rocha S, Gradinaru V, et al. Gut Microbiota Regulate Motor Deficits and Neuroinflammation in a Model of Parkinson’s Disease. Cell. 2016;167:1469–1480 e1412. doi: 10.1016/j.cell.2016.11.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sido B, Hack V, Hochlehnert A, Lipps H, Herfarth C, Droge W. Impairment of intestinal glutathione synthesis in patients with inflammatory bowel disease. Gut. 1998;42:485–492. doi: 10.1136/gut.42.4.485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sonnenburg JL, Xu J, Leip DD, Chen CH, Westover BP, Weatherford J, Buhler JD, Gordon JI. Glycan foraging in vivo by an intestine-adapted bacterial symbiont. Science. 2005;307:1955–1959. doi: 10.1126/science.1109051. [DOI] [PubMed] [Google Scholar]
- Tailford LE, Crost EH, Kavanaugh D, Juge N. Mucin glycan foraging in the human gut microbiome. Front Genet. 2015a;6:81. doi: 10.3389/fgene.2015.00081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tailford LE, Owen CD, Walshaw J, Crost EH, Hardy-Goddard J, Le Gall G, de Vos WM, Taylor GL, Juge N. Discovery of intramolecular trans-sialidases in human gut microbiota suggests novel mechanisms of mucosal adaptation. Nat Commun. 2015b;6:7624. doi: 10.1038/ncomms8624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsuji G, Takahara M, Uchi H, Matsuda T, Chiba T, Takeuchi S, Yasukawa F, Moroi Y, Furue M. Identification of ketoconazole as an AhR-Nrf2 activator in cultured human keratinocytes: the basis of its anti-inflammatory effect. J Invest Dermatol. 2012;132:59–68. doi: 10.1038/jid.2011.194. [DOI] [PubMed] [Google Scholar]
- Turnbaugh PJ, Hamady M, Yatsunenko T, Cantarel BL, Duncan A, Ley RE, Sogin ML, Jones WJ, Roe BA, Affourtit JP, et al. A core gut microbiome in obese and lean twins. Nature. 2009;457:480–484. doi: 10.1038/nature07540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vatanen T, Kostic AD, d’Hennezel E, Siljander H, Franzosa EA, Yassour M, Kolde R, Vlamakis H, Arthur TD, Hamalainen AM, et al. Variation in Microbiome LPS Immunogenicity Contributes to Autoimmunity in Humans. Cell. 2016;165:842–853. doi: 10.1016/j.cell.2016.04.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Venkatesh M, Mukherjee S, Wang H, Li H, Sun K, Benechet AP, Qiu Z, Maher L, Redinbo MR, Phillips RS, et al. Symbiotic bacterial metabolites regulate gastrointestinal barrier function via the xenobiotic sensor PXR and Toll-like receptor 4. Immunity. 2014;41:296–310. doi: 10.1016/j.immuni.2014.06.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wikoff WR, Anfora AT, Liu J, Schultz PG, Lesley SA, Peters EC, Siuzdak G. Metabolomics analysis reveals large effects of gut microflora on mammalian blood metabolites. Proc Natl Acad Sci U S A. 2009;106:3698–3703. doi: 10.1073/pnas.0812874106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wlodarska M, Thaiss CA, Nowarski R, Henao-Mejia J, Zhang JP, Brown EM, Frankel G, Levy M, Katz MN, Philbrick WM, et al. NLRP6 inflammasome orchestrates the colonic host-microbial interface by regulating goblet cell mucus secretion. Cell. 2014;156:1045–1059. doi: 10.1016/j.cell.2014.01.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye J, Coulouris G, Zaretskaya I, Cutcutache I, Rozen S, Madden TL. Primer-BLAST: a tool to design target-specific primers for polymerase chain reaction. BMC Bioinformatics. 2012;13:134. doi: 10.1186/1471-2105-13-134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zelante T, Iannitti RG, Cunha C, De Luca A, Giovannini G, Pieraccini G, Zecchi R, D’Angelo C, Massi-Benedetti C, Fallarino F, et al. Tryptophan catabolites from microbiota engage aryl hydrocarbon receptor and balance mucosal reactivity via interleukin-22. Immunity. 2013;39:372–385. doi: 10.1016/j.immuni.2013.08.003. [DOI] [PubMed] [Google Scholar]
- Zhang T, Kimura Y, Jiang S, Harada K, Yamashita Y, Ashida H. Luteolin modulates expression of drug-metabolizing enzymes through the AhR and Nrf2 pathways in hepatic cells. Arch Biochem Biophys. 2014;557:36–46. doi: 10.1016/j.abb.2014.05.023. [DOI] [PubMed] [Google Scholar]
- Zhang X, Zhang D, Jia H, Feng Q, Wang D, Liang D, Wu X, Li J, Tang L, Li Y, et al. The oral and gut microbiomes are perturbed in rheumatoid arthritis and partly normalized after treatment. Nat Med. 2015;21:895–905. doi: 10.1038/nm.3914. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


