Abstract
Background:
Patients affected by gliomas have a poor prognosis. Astrocytoma is a subtype of glioma. Identification of biomarkers could be an effective way to an early diagnosis of tumor or to distinguish more aggressive tumors that need more intensive therapy. In this study, we investigated whether the expression of miR-362 was increased or decreased in patients with different grades of astrocytoma.
Materials and Methods:
miR-362 expression was compared in 25 patients with astrocytoma with that of 4 normal nonneoplastic brain tissues.
Results:
In all tumor tissues, the expression of miR-362 was significantly decreased relative to its expression in normal brain tissues. However, there was no significant difference between miR-362 expressions in high and low grades of astrocytoma.
Conclusions:
In conclusion, miR-362 showed a down-regulation pattern in astrocytoma tissues that was different from the pattern obtained from previously published microarray studies.
Keywords: Astrocytoma, brain tumors, expression, miR-362
Introduction
Glioma is the most prevalent type of tumors in the primary central nervous system and accounts for 2% of cancers. Despite advancements in treating these type of tumors, there is still a high rate of morbidity and mortality associated with gliomas.[1] The word “glioma” includes all tumors that are originated from glial cells; that is to say, astrocytomas, oligodendrogliomas, ependymomas, mixed gliomas, and malignant gliomas.[2] The most common type of glioma is astrocytoma (60–70%) which is mainly forms from inseparable astrocytes and is responsible for 75% of neuroepithelial cases.[3] Astrocytoma is classified using the most common grading system, World Health Organization classification system, based on the growth rate of the tumor. According to this system, astrocytomas are classified as astrocytoma grade I (pilocytic astrocytoma, low-grade, slow growth, biologically least aggressive, and benign), II (diffuse astrocytoma, low-grade, tendency to recurrence), III (anaplastic astrocytoma, mid-grade), and IV (glioblastoma [GBM], high-grade, rapidly growing, and highly malignant).[4,5]
Like other lesions that occupy space in the central nervous system, a patient affected by an astrocytic tumor may present symptoms because of intracranial pressure posed by a tumor or general dysfunction of the brain. These symptoms include a headache, vomiting, papilledema, fluctuations in the level of consciousness and focal or generalized convulsions. The most malignant part of the tumor is the basis of diagnosis.[6] Hence, sampling a tumor in adequate amount is determinant in identifying its type and malignancy. Considering the facts that grading of tumor based on sampling would be a minimum grading because the more malignant regions may be in areas which has not been sampled and that chemo- or radiation therapy makes it very difficult to type and grade the tumor, to reach a precise, definite, and easy-to-do diagnosis, it is more beneficial to use biomarkers such as cell proliferation, cell death and genetic factors.
Genetic factors play significant roles in the pathogenesis of tumors. In this regard, the biologists are trying to understand the genetic abnormality and cellular mechanisms which influence the features of tumors.[7] Nowadays, it is recognized that the classification of gliomas based on genetic changes and profiles serves as a diagnostic or predictive tool.[8] In this respect, there have been some attempts in identifying involved genes in astrocytomas. Many engaged genes are recognized in some grades. For example, in a study, it was observed that inactivation of Retinoblastoma susceptibility gene by mutations contributes to the occurrence of high grade astrocytomas.[9] In another study, it was shown that in patients affected by primary GBM, loss of heterozygosity of 10q, amplification of epidermal growth factor receptor, deletion of p16INK4a, and PTEN mutations has occurred.[10] Overexpression of platelet-derived growth factor ligand and receptor has also been reported in astrocytoma.[11] In addition, the involvement of another class of genes, that is, micro RNA (miRNA) coding genes in the pathogenesis of astrocytomas has been recently observed.[12,13]
miRNAs are small, noncoding, and single-stranded RNA molecules which are of 19–25 nucleotides length and regulate different genes’ expressions and protein production via matching to mRNA 3’UTR.[14] Broad research over last years showed that miRNAs are more common than previously thought, and they play many underlying roles in the regulation of biological processes.[15] According to available evidence, dysregulation of miRNAs causes diseases especially cancers by acting as oncogenes or tumor suppressors.[16,17] In different studies, the involvement of different miRNAs in astrocytoma pathogenesis is reported. For instance, in a study by Huse et al., they observed that the expression of 29 miRNAs including miR-362 increased by at least 2 folds.[18] In some other studies deregulated miRNAs were identified in malignant astrocytoma using genome-wide expression profiling.[12,13,19]
The aim of this study was identifying the miR-362 expression pattern in tumor samples obtained from patients affected by astrocytoma of different grades and determining whether there is any difference in its expression between different grades of astrocytoma.
Materials and Methods
Patients and sampling
Tumor samples were collected from patients affected by any types of brain tumors who were operated at Alzahra Hospital, Isfahan, Iran. Tissues were received freshly from neurosurgical operating rooms and stored at −70°C for subsequent RNA extraction. After being histopathologically confirmed the diagnosis of astrocytoma by using part of the tissue samples, 25 specimens were finally selected. Fresh nonneoplastic brain tissues were taken from 4 donors who died 2 h before surgery. Informed consents were taken from all donors before sampling. The study was approved by the Ethics Committee of Isfahan University of Medical Sciences. A total of 25 samples including 5 samples with astrocytoma grade II, 2 samples with astrocytoma Grade III, and 18 samples with astrocytoma grade IV were used in the current study.
RNA extraction from samples and complementary DNA synthesis
First, total RNA was extracted from tissue samples using solvent TRIzol (Invitrogen, USA) according to manufacturer's protocol and solved in RNase-free water. Then, RNA quality and quantity was examined by ultraviolet spectrophotometry and gel electrophoresis. Finally, 2 μl of extracted RNA was converted to complementary DNA (cDNA) using miRCURY LNA™ miRNA polymerase chain reaction (PCR), polyadenylation and cDNA synthesis kit II (Exiqon, Denmark).
Real-time-polymerase chain reaction
Following synthesis of cDNA, real-time PCR was accomplished with ExiLENT SYBR® Green master mix kit (Exiqon, Denmark) using StepOne Plus™ quantitative real-time PCR detection system (Applied Biosystems, Thermo Fisher Scientific corporation, USA). Ready-to-use primers were from Exiqon. U6 small nuclear RNA primers were used for normalizing the data. All reactions were performed in triplicates for each sample.
Statistical analysis
For expression analysis, the experiment applied the nontumor tissue as the control, so the relative quantification of miR-362 in tumor tissue was calculated using the equation:
Amount of target = 2−ΔΔCt, ΔΔCt = (CtmiR-362– CtU6) tumor − (CtmiR-362– CtU6) normal.
For the nontumor tissue control sample, ΔΔCt was considered zero and 2−ΔΔct was set as 1. After finishing the reaction, using SPSS software version 20.0 (SPSS, Chicago, IL, USA), the expression of miR-362 was quantitatively analyzed to compare the mean of relative miR-362 expression in two groups of astrocytoma tumor and normal tissues and also in different grades.
Results
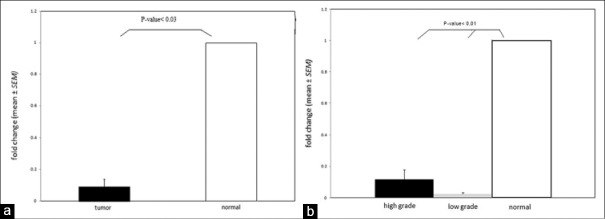
The expression of miR-362 was evaluated in 25 astrocytoma samples and 4 normal nonneoplastic tissues using real-time PCR method. miR-362 expression was significantly decreased in astrocytoma samples compared to normal samples (P = 0.043) [Figure 1a]. Moreover, the expression of miR-362 was assessed in high-grade and low-grade samples relative to the normal tumor (P = 0.01) [Figure 1b]. A brief description of patients, characteristics is given in Table 1.
Figure 1.
miR-362 expression in astrocytoma. (a) miR-362 expression also decreased in all of 25 astrocytoma samples compared with 4 normal tissues (P < 0.03). (b) miR-362 expression decreased between 7 low grades and 18 high grades of astrocytoma samples relative to 4 normal tissues (P < 0.01)
Table 1.
A brief description of patients’ characteristics

In addition, to determine the association of miR-362 with cancer progression, we compared miR-362 expression in low-grade astrocytoma samples compared with high-grade ones. Our results revealed that there was no significant difference between two groups (P > 0.05) [Figure 2].
Figure 2.
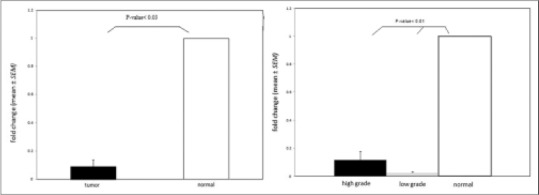
Comparison of miR-362 expression in low and high grades of astrocytoma. miR-362 expression decreased nonsignificantly in low grades relative to high grades (P > 0.05)
Discussion
Nowadays, evaluation of miRNA expression profiles in different types of cancers is performed for finding diagnostic and prognostic tools as well as for developing targeted avenues for intervening therapies.[20] Alternations of miRNAs expression have been studied by different methods including microarray and real-time reverse transcription-PCR.[21] For the first time, Chan et al. in 2005, showed that miR-21 was strongly overexpressed in GBM.[22] Then, several other studies were conducted on this issue.
In a study by Xia et al. it was observed that miR-362 overexpression in gastric cancer increases NF-κB activity and expression of NF-κB-regulated genes.[23] NF-κB signaling pathway is more active in GBM tissues than in non-GBM ones.[24]
Rao et al. profiled the expression of miRNAs in 26 GBM and 13 anaplastic astrocytoma and 7 normal brain samples using microarray technique and observed that 55 miRNAs including miR-362 were up-regulated, and 29 miRNAs were down-regulated in 39 astrocytoma samples in comparison with 7 normal tissues. Then, they assessed expression of miRNAs to see if they can identify the minimal number of miRNAs which can be used to distinguish anaplastic astrocytoma and GBM and they observed that there is a 23 miRNA signature discriminating these two kinds of astrocytoma.[12] Furthermore, in another study, a 29 miRNA signature was identified in 3 GBM samples using microarray, and it was observed that expression of 29 miRNAs including miR-362 is elevated relative to normal brain tissues by at least 2 folds.[18] Lang MF et al. profiled the expression of miRNAs using microarray technique in GBM multiforme (GBM) and identified a miRNA expression signatures including miR-362 that, in was up-regulated.[13]
In our data analysis, inconsistent with previous microarray data, miR-362 expression decreased significantly in 25 astrocytoma tumor tissues compared with normal tissues (P = 0.043). In other word, in contrast to the prior observation that reported its 1–3 fold overexpression in GBM,[12,13,18,19] our analysis showed that miR-362 was significantly under expressed both in high grades and in low grades of astrocytoma samples relative to normal samples.
In the same way, diverse expression patterns of miR-362 have been reported in other cancers. For example, Christensen et al. showed that in patients affected by colorectal cancer, miR-362 expression was higher in patients with no recurrence compared with patients with recurrence. In addition, they observed that there was a significant association between miR-362 and increased free disease survival.[25] While, Shen et al. observed that induced expression of miR-362-3p in human hepatocellular cells and tissues increased proliferation and anchorage-independent soft agar growth.[26] These observations may be an indicator of the possible different expression patterns and effects of miR-362 according to different tissue contexts.
In addition, to investigate whether miR-362 is differentially expressed in astrocytomas with different grades, we compared its expression pattern between low (II) and high (III and IV) grades astrocytomas and determined that miR-362 expression did not change significantly among different grades of astrocytoma [Figure 2].
In conclusion, previous microarray data showed that miR-362 expression was mostly overexpressed in astrocytoma samples specifically, GBM, but our data analysis could not confirm previous microarray data and showed that its expression has decreased significantly in astrocytoma tissues.
Conclusion
Previous microarray data showed that miR-362 expression was mostly overexpressed in astrocytoma samples specifically, GBM, but our data analysis could not confirm previous microarray data and showed that its expression has decreased significantly in astrocytoma tissues.
Financial support and sponsorship
This study was supported by Isfahan University of Medical Sciences, Isfahan, Iran.
Conflicts of interest
There are no conflicts of interest.
Acknowledgment
This study was supported by Isfahan University of Medical Sciences, Isfahan, Iran.
References
- 1.Burnet NG, Jefferies SJ, Benson RJ, Hunt DP, Treasure FP. Years of life lost (YLL) from cancer is an important measure of population burden – And should be considered when allocating research funds. Br J Cancer. 2005;92:241–5. doi: 10.1038/sj.bjc.6602321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ostrom QT, Gittleman H, Liao P, Rouse C, Chen Y, Dowling J, et al. CBTRUS statistical report: Primary brain and central nervous system tumors diagnosed in the United States in 2007-2011. Neuro Oncol. 2014;16(Suppl 4):iv1–63. doi: 10.1093/neuonc/nou223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Schwartzbaum JA, Fisher JL, Aldape KD, Wrensch M. Epidemiology and molecular pathology of glioma. Nat Clin Pract Neurol. 2006;2:494–503. doi: 10.1038/ncpneuro0289. [DOI] [PubMed] [Google Scholar]
- 4.Louis DN, Ohgaki H, Wiestler OD, Cavenee WK, Burger PC, Jouvet A, et al. The 2007 WHO classification of tumours of the central nervous system. Acta Neuropathol. 2007;114:97–109. doi: 10.1007/s00401-007-0243-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Daumas-Duport C, Scheithauer B, O’Fallon J, Kelly P. Grading of astrocytomas. A simple and reproducible method. Cancer. 1988;62:2152–65. doi: 10.1002/1097-0142(19881115)62:10<2152::aid-cncr2820621015>3.0.co;2-t. [DOI] [PubMed] [Google Scholar]
- 6.Ichimura K, Ohgaki H, Kleihues P, Collins VP. Molecular pathogenesis of astrocytic tumours. J Neurooncol. 2004;70:137–60. doi: 10.1007/s11060-004-2747-2. [DOI] [PubMed] [Google Scholar]
- 7.Reifenberger G, Louis DN. Oligodendroglioma: Toward molecular definitions in diagnostic neuro-oncology. J Neuropathol Exp Neurol. 2003;62:111–26. doi: 10.1093/jnen/62.2.111. [DOI] [PubMed] [Google Scholar]
- 8.von Deimling A, Eibl RH, Ohgaki H, Louis DN, von Ammon K, Petersen I, et al. p53 mutations are associated with 17p allelic loss in grade II and grade III astrocytoma. Cancer Res. 1992;52:2987–90. [PubMed] [Google Scholar]
- 9.Henson JW, Schnitker BL, Correa KM, von Deimling A, Fassbender F, Xu HJ, et al. The retinoblastoma gene is involved in malignant progression of astrocytomas. Ann Neurol. 1994;36:714–21. doi: 10.1002/ana.410360505. [DOI] [PubMed] [Google Scholar]
- 10.Ohgaki H, Kleihues P. Genetic pathways to primary and secondary glioblastoma. Am J Pathol. 2007;170:1445–53. doi: 10.2353/ajpath.2007.070011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ranza E, Facoetti A, Morbini P, Benericetti E, Nano R. Exogenous platelet-derived growth factor (PDGF) induces human astrocytoma cell line proliferation. Anticancer Res. 2007;27:2161–6. [PubMed] [Google Scholar]
- 12.Rao SA, Santosh V, Somasundaram K. Genome-wide expression profiling identifies deregulated miRNAs in malignant astrocytoma. Mod Pathol. 2010;23:1404–17. doi: 10.1038/modpathol.2010.135. [DOI] [PubMed] [Google Scholar]
- 13.Lang MF, Yang S, Zhao C, Sun G, Murai K, Wu X, et al. Genome-wide profiling identified a set of miRNAs that are differentially expressed in glioblastoma stem cells and normal neural stem cells. PLoS One. 2012;7:e36248. doi: 10.1371/journal.pone.0036248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bartel DP. MicroRNAs: Target recognition and regulatory functions. Cell. 2009;136:215–33. doi: 10.1016/j.cell.2009.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Macfarlane LA, Murphy PR. MicroRNA: Biogenesis, Function and Role in Cancer. Curr Genomics. 2010;11:537–61. doi: 10.2174/138920210793175895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Barbarotto E, Schmittgen TD, Calin GA. MicroRNAs and cancer: Profile, profile, profile. Int J Cancer. 2008;122:969–77. doi: 10.1002/ijc.23343. [DOI] [PubMed] [Google Scholar]
- 17.Zhang B, Pan X, Cobb GP, Anderson TA. MicroRNAs as oncogenes and tumor suppressors. Dev Biol. 2007;302:1–12. doi: 10.1016/j.ydbio.2006.08.028. [DOI] [PubMed] [Google Scholar]
- 18.Huse JT, Brennan C, Hambardzumyan D, Wee B, Pena J, Rouhanifard SH, et al. The PTEN-regulating microRNA miR-26a is amplified in high-grade glioma and facilitates gliomagenesis in vivo. Genes Dev. 2009;23:1327–37. doi: 10.1101/gad.1777409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Roth P, Wischhusen J, Happold C, Chandran PA, Hofer S, Eisele G, et al. Aspecific miRNA signature in the peripheral blood of glioblastoma patients. J Neurochem. 2011;118:449–57. doi: 10.1111/j.1471-4159.2011.07307.x. [DOI] [PubMed] [Google Scholar]
- 20.Subramanian S, Lui WO, Lee CH, Espinosa I, Nielsen TO, Heinrich MC, et al. MicroRNA expression signature of human sarcomas. Oncogene. 2008;27:2015–26. doi: 10.1038/sj.onc.1210836. [DOI] [PubMed] [Google Scholar]
- 21.Lu J, Getz G, Miska EA, Alvarez-Saavedra E, Lamb J, Peck D, et al. MicroRNA expression profiles classify human cancers. Nature. 2005;435:834–8. doi: 10.1038/nature03702. [DOI] [PubMed] [Google Scholar]
- 22.Chan JA, Krichevsky AM, Kosik KS. MicroRNA-21 is an antiapoptotic factor in human glioblastoma cells. Cancer Res. 2005;65:6029–33. doi: 10.1158/0008-5472.CAN-05-0137. [DOI] [PubMed] [Google Scholar]
- 23.Xia JT, Chen LZ, Jian WH, Wang KB, Yang YZ, He WL, et al. MicroRNA-362 induces cell proliferation and apoptosis resistance in gastric cancer by activation of NF-κB signaling. J Transl Med. 2014;12:33. doi: 10.1186/1479-5876-12-33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Atkinson GP, Nozell SE, Benveniste ET. NF-kappaB and STAT3 signaling in glioma: Targets for future therapies. Expert Rev Neurother. 2010;10:575–86. doi: 10.1586/ern.10.21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Christensen LL, Tobiasen H, Holm A, Schepeler T, Ostenfeld MS, Thorsen K, et al. MiRNA-362-3p induces cell cycle arrest through targeting of E2F1, USF2 and PTPN1 and is associated with recurrence of colorectal cancer. Int J Cancer. 2013;133:67–78. doi: 10.1002/ijc.28010. [DOI] [PubMed] [Google Scholar]
- 26.Shen H, Li W, Tian Y, Xu P, Wang H, Zhang J, et al. Upregulation of miR-362-3p modulates proliferation and anchorage-independent growth by directly targeting Tob2 in hepatocellular carcinoma. J Cell Biochem. 2015;116:1563–73. doi: 10.1002/jcb.25110. [DOI] [PubMed] [Google Scholar]