Abstract
Single-cell techniques have a long history of unveiling fundamental paradigms in biology. Recent improvements in the throughput, resolution, and availability of microfluidics, computational power, and genetically encoded fluorescence have led to a modern renaissance in microbial physiology. This resurgence in research activity has offered new perspectives on physiological processes such as growth, cell cycle, and cell size of model organisms such as Escherichia coli. We expect these single-cell techniques, coupled with the molecular revolution of biology’s recent half-century, to continue illuminating unforeseen processes and patterns in microorganisms, the bedrock of biological science. In this article we review major open questions in single-cell physiology, provide a brief introduction to the techniques for scientists of diverse backgrounds, and highlight some pervasive issues and their solutions.
Keywords: cell cycle, growth, microbiology, microfluidics, systems biology, quantitative biology
INTRODUCTION
The late François Jacob (89) is said to have remarked, “The dream of every cell is to become two cells.” There are several basic steps for cellular reproduction: growth of the cell, replication and segregation of the genetic materials, and cell division. The physiological study of microorganisms aims to understand the fundamental controls underlying these processes. In particular, a single-cell approach can reveal causal relationships that are inaccessible to population-level experiments, and provide key insights into the underlying control mechanisms. Researchers were interested in single-cell measurements of microorganisms nearly a century ago (15). However, it is only in the last decade that the field has come of age with the rapid development of video microscopy and lab-on-a-chip technologies, along with computers that are powerful enough to analyze large amounts of data.
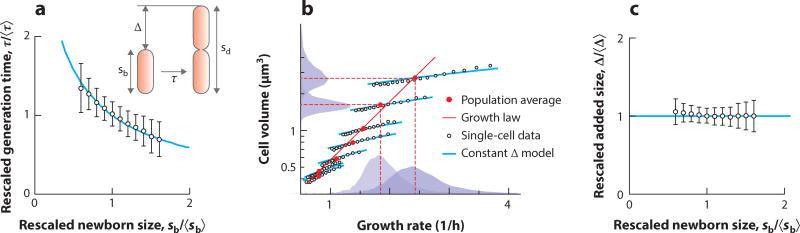
A telling example is the study of cell size. In the classic literature, cell size was measured by optical density of a growing culture and colony counts, assuming that light absorbance per cell was proportional to the cell size (79). Data from batch culture, however, do not disclose cell-to-cell variations and may not reveal important correlations. See, for instance, the relationship between the size of newborn Escherichia coli cells and their respective generation time between birth and division (Figure 1a). They are negatively correlated; i.e., cells born smaller than the population average take longer to divide than cells born larger than the population average. This finding is a strong indication for a mechanism of cell size control, which would not have been possible to measure in batch culture.
Figure 1. Single-cell experiments of Escherichia coli reveal the size control mechanism by parameters that are not accessible in a population average experiment.
(a) Generation time, τ, is negatively correlated with newborn size, sb, ruling out the timer model. (b) Single-cell data show a systematic deviation from the growth law. The population average data (red lines and red symbols) confirm the classic work of Schaechter et al. (79), but single-cell data (black symbols) systematically deviate from the growth law. This deviation is related to how cells control their size in a steady-state growth condition. (c) Cells add a constant size, Δ, from birth to division, independent of their size at birth, sb. Figure based on data from Wang et al. (98).
Control of gene expression is another important example for which single-cell approaches have been immensely valuable. The study of gene expression mechanisms dates back to the 1950s, such as the analysis of the lac operon induction levels by Novick & Weiner (75). This experiment revealed that the smooth, correlated increase in protein expression observed in batch culture experiments was not due to the equal and gradual induction of all cells. Instead, discrete subpopulations of cells transition from fully repressed to fully induced states. Novick & Weiner’s observation inspired modern systems biology studies on bistability of gene expression at the single-cell level (77).Noise in gene expression has also benefited from single-cell methods (34, 76). The revelation of new behaviors and mechanisms through single-cell experiments is integral to biology.
We anticipate two audiences will read this review article. The first audience consists of researchers trained in quantitative fields of science and engineering who want to enter biology and tackle long-standing problems in cell physiology. We introduce key scientific questions in single-cell physiology that can benefit from the tools and ways of thinking from the physical sciences and engineering. The second audience consists of biologists who want to use single-cell technologies for their research. Similarly, we introduce key technical approaches that can complement existing molecular assays. Like any technology, single-cell methods produce good data only when used appropriately. Unfortunately, virtually all current techniques suffer from varying degrees of artifacts and physiological compromises. It can take years of painstaking control experiments to discover and (hopefully) correct these problems. For both audiences, we discuss some of the most common problems and how to troubleshoot them.
Although this is a review article, space constraints mean that we must focus and impose our own view of this important branch of basic science through the selection of material. As such, we use examples from bacterial cell physiology that share the same natural core with eukaryotes. We also apologize to our colleagues whose work should have been cited otherwise. We provide a partial solution by introducing excellent review articles for the important materials we are unable to discuss.
FUNDAMENTAL QUESTIONS IN SINGLE-CELL PHYSIOLOGY
Growing cells have been the subject of endless fascination since their first observation by Antony van Leeuwenhoek (30). Many individuals have recorded with great care and accuracy the basic properties of cells. In the 1940s, Jacques Monod (71) studied cells utilizing multiple carbon sources in a growth medium. His study on cells transitioning from metabolizing glucose to lactose, known as the diauxic shift, was one of the major stepping stones for the later study with Jacob and Lwoff on the mechanism of gene expression—the lac operon (49).
For those trained at the interface between biological and physical sciences, reading the literature from the early-twentieth century until the beginning of the 1970s is a refreshing experience. See, for example, the work by Schaechter et al. (79) on bacterial growth in the late 1950s. They established the basic principle in bacterial physiology known as the growth law on the basis of meticulous measurements of cell mass, RNA, and DNA content with respect to nutrient-imposed growth rate. The growth law states that the average cell size increases exponentially with respect to nutrient-imposed growth rate. Remarkably, this exponential relationship is independent of the chemical details of the growth media. Similar and equally important quantitative approaches to bacterial physiology and the bacterial cell cycle continued in the 1960s. Koch & Schaechter (59), among many others, based their study of cell size distributions on the elongation model of individual cells. Cooper & Helmstetter (28) established the phenomenological kinematic rule of the bacterial cell cycle, which explained the timing of chromosome replication and cell division under a wide range of nutrient-imposed growth rates.
Although most classic studies relied on population-level measurements, their interests were deeply rooted in single-cell behavior. However, the lack of single-cell data has hindered our ability to answer many fundamental questions because population averages can conceal important causal insights and because throughput suffers when resolution is increased (see Figure 1). Some of these questions include, How do cells coordinate growth and cell cycle? How do cells control their size and shape? How do cells know when to replicate their chromosome and when to divide? How do cells die? We follow with how single-cell methods are needed to address these questions.
Growth and Cell Size
Cell size control is a long-standing, multifaceted problem in biology. First, many single-celled microorganisms, including yeast (52) and bacteria (79), change their average size with respect to growth conditions. The growth law shows that the average cell size increases exponentially as the nutrient-imposed growth rate increases. Second, even in a steady-state growth condition, cell size, among other cell cycle parameters, exhibits significant variability. Using single-cell measurements, we now know that the size of E. coli varies about 15% at division (92, 98). Although this finding implies cell size is subject to stochastic fluctuation, the size distribution of a population remains unchanged over generations, indicating that cells respond to fluctuations to maintain size homeostasis.
Two classes of models, sizer and timer, account for size control and homeostasis in microorganisms. Sizer presumes cells trigger division at a specific size, whereas timer assumes cell division occurs after a set elapsed time. Thus, sizer predicts that the division size is independent of the birth size, whereas timer indicates that generation time is constant irrespective of the birth size. Testing these predictions requires single-cell data, in which the growth of a large population of individual cells can be tracked to find correlations between growth parameters. Data collected from 105 individual cells (presented in Figure 1a) preclude the timer model since the generation time is negatively correlated with newborn size. Similar analysis (92) of single-cell data showed that size at division is correlated with size at birth, precluding sizer.
Our analysis of the growth law at a single-cell level led to a surprising discovery: The growth law breaks down at a single-cell level (92) (Figure 1b). That is, the population average data confirm the original growth law, but single-cell data systematically deviate from it. These deviations are related to how individual cells control their size in the presence of fluctuations in a steady-state growth condition: Cells add a constant size from birth to division, independent of their size at birth. This fundamental and simple size homeostasis mechanism operates in both E. coli and Bacillus subtilis (see Figure 1c for E. coli). Following this “adder” principle, cell size deviations are restored after several generations by geometric regression to the mean (23, 53, 92, 97).
New developments in molecular physiology have created opportunities to potentially perturb cell size control mechanisms. Levin and colleagues (46, 101) have reported a metabolic sensor in B. subtilis that links nutrient availability with division machinery, specifically the FtsZ constriction ring. This metabolic sensor is thought to regulate an inhibitor of constriction ring assembly to control cell size in a nutrient-dependent manner. How the metabolic sensor integrates with other molecular mechanisms and ultimately leads to a quantitative relationship between the average cell size and the nutrient-imposed growth rate is still unknown. Kinetic information is needed if we are to understand these processes. By revisiting classic nutrient shift-up experiments (26, 79) to measure the response in size and the cell cycle of individual cells during the transition from one steady-state growth condition to another, researchers may discriminate among these hypotheses.
Cell Cycle
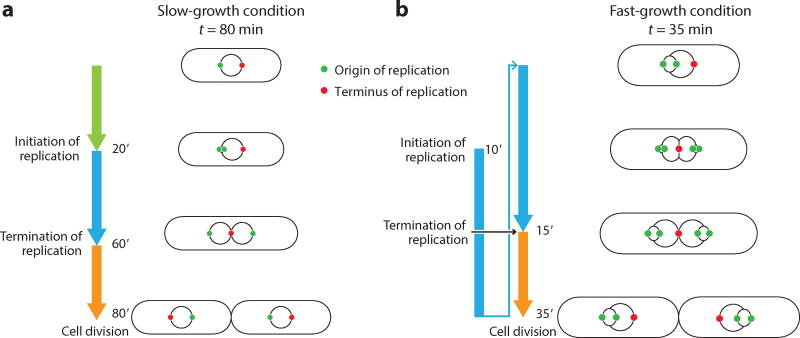
The cell cycle is composed of a set of processes that must be faithfully performed in order to create two viable daughter cells. Undergraduate biology textbooks focus on the cell cycle of eukaryotes, in which the chromosome is duplicated once and partitioned among the two daughter cells and checkpoints arrest the cycle if the process goes awry (3, 69). Unlike eukaryotes, most prokaryotes are capable of multifork replication, i.e., of maintaining multiple overlapping rounds of DNA synthesis (28), as shown in Figure 2. Significant questions remain pertaining to how these cells can coordinate growth, size, DNA replication, and division to create two functioning daughter cells without incident.
Figure 2. Cell cycle of Escherichia coli cells.
(a) In slow-growth conditions (generation time larger than C and D periods combined), the cell cycle has three distinct steps: the time between birth and initiation of chromosome replication (B period, green arrow; length depends on generation time), the time of chromosome replication (C period, blue arrow; 40 min), and the gap between termination of chromosome replication and division (D period, orange arrow; 20 min). (b) In fast-growth conditions, multiple cell cycles must overlap because the period C + D is constant and longer than the generation time. The illustration shows chromosome replication spanning two generations. Under all growth conditions, one round of the replication cycle must be coupled to one round of the division cycle.
In a series of studies in the 1960s,Helmstetter, Cooper, and colleagues (26, 28, 44, 45) developed experimental methods to measure the duration of chromosome replication, known as C period (analogous to S phase in eukaryotes), in the model organism E. coli. For a wide range of growth conditions at 37°C, C period is constant, approximately 40 min for their strain of E. coli B/r. In addition, a constant amount of time known as D period (20 min; analogous to G2 phase in eukaryotes) passes between replication termination and cell division. Thus, C and D periods combined last for approximately 60 min, although E. coli can divide as frequently as every 20 min. This seeming conflict, namely that DNA replication can take longer than the generation time, led Cooper & Helmstetter to conclude that cell cycles in E. coli must overlap. Their influential model predicted that E. coli initiates DNA replication C + D minutes prior to each cell division.
An obvious question is, What is the relationship between cell cycle timing and cell size? In an important theoretical study, Donachie (32) showed that the Helmstetter-Cooper model together with the growth law by Schaechter et al. implies that replication initiates when cell mass per replication origin reaches a critical value, independent of the growth condition. The notion of critical mass, however, remains controversial (14, 27).
Single-cell data can provide fundamental insights into cell cycle control mechanisms not available from population-level data. For example, the negative correlations between the generation time and the newborn size (Figure 1) clearly show the presence of a size control mechanism. Similarly, the following questions can be answered only by single-cell experiments, which were not possible when the Helmstetter-Cooper model was developed in the 1960s: How variable are C and D periods under constant growth conditions? Will replication always initiate when the cell reaches a critical mass as Donachie predicted? More generally, how is the cell cycle maintained when cells deviate from the average growth rate or size of the population or when cells experience stochastic variation at the initiation of replication? Subsequent analyses of various E. coli strains revealed deviations in the duration and arrangement of different growth periods (73). In consideration of these differences even within one species, how general are the aforementioned models to diverse strains and organisms that perform multifork replication? Rigorous single-cell measurements of replication timing will shed light on the above questions and may reveal whether fundamental processes underlie cell cycle control across diverse microbes.
Metabolism
Intimately connected with growth, metabolism represents the entire complex chemical network underlying living organisms. Despite ongoing work to characterize and model these networks, metabolism in vivo remains woefully unquantified. This is partially due to the difficulty of simultaneously measuring the kinetics, abundances, and fluctuations of the many players in these complex systems. Can we quantitatively understand metabolism without measuring seemingly endless parameters? This dilemma captures one of the central problems of systems biology: how to bridge bottom-up and top-down methods in order to uncover guiding principles.
Single-cell data can provide fundamental insights into the general properties of metabolism. Recently, Kiviet et al. (57) continuously and simultaneously monitored instantaneous growth rate and metabolic production (e.g., lac expression) in single cells. This exposed fluctuations at timescales far below that of the generation time and allowed them to measure how growth and gene expression influence each other. By computing cross correlations between measured growth rate and the level of gene expression, they showed that noise propagates bidirectionally. Because correlations in fluctuations were also observed in the Krebs cycle at metabolic nodes distant from the lac operon, these observations are likely a general feature of noise propagation in metabolism. It also gives us hope that single-cell studies can provide general and critical insights into even the most complex systems.
Cell Shape
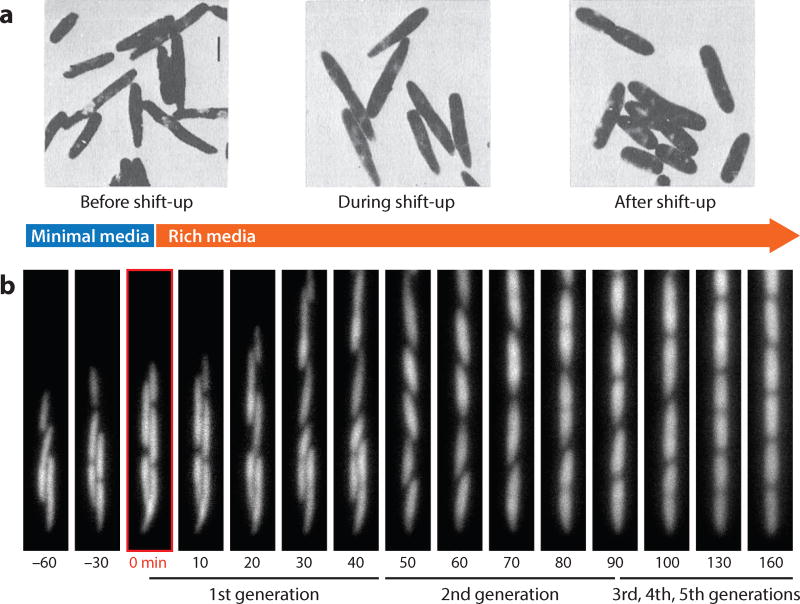
Cell shape may be considered a topic in cell biology or mathematical biology rather than physiology. Early attempts to quantify shape focused on the physical constraints on the forms unicellular organisms can take (58).More recently, the role of bacterial cell walls and cytoskeletal components has become an important avenue of investigation to understand cell shape control. We believe the subject can greatly benefit from a physiological approach that focuses on responses of cell shape to changing growth conditions. Woldringh et al. (104) addressed how E. coli cell shape changes during the transition from one steady-state growth condition to another, i.e., from slow growth in minimal glycerol to fast growth in synthetic rich media. Their critical observation was that cell morphology changes anisotropically; although both cell length and cell width increased immediately after nutrient shift-up, many cells continued to possess a tapered old pole (Figure 3a). This observation suggests that the old pole is metabolically inert relative to the sides of the cell wall in terms of the turnover rate of constituent materials. We performed a nutrient shift-up experiment and continuously monitored cell shape changes at the single-cell level (Figure 3b; S. Jun & P. Wang, unpublished results). The tapered pole was indeed the result of the slow change of the old pole dimensions compared with the faster increase in size of the side walls, consistent with the previous anisotropic observations. However, the tapered old pole eventually achieved normal shape and dimensions, indicating that the cell pole is not completely inert—an observation not possible without modern microfluidic techniques.
Figure 3. Shape changes of Escherichia coli cells during the transition from slow-growth to fast-growth conditions.
(a) Anisotropic change of cell morphology observed by Woldringh et al. (104) leading to the conclusion that the old pole is metabolically inert in terms of the turnover rate of constituent materials relative to the sides of the cell wall. (b) A nutrient shift-up experiment in a microfluidic device consistent with and extending the anisotropic interpretation. The tapered pole was indeed the result of the old pole dimensions changing slower than the size of the adjacent side walls. The tapered old pole eventually regained normal shape and dimensions, indicating the cell pole is not completely inert. Panel a is reprinted from Reference 104, with permission from Elsevier.
This type of single-cell growth experiment can be combined with modern imaging techniques that can directly visualize cell wall synthesis (31, 37, 96). Furthermore, we can now easily manipulate the shape of individual cells using controllable forces (4, 99) or geometrical constraints of microfabricated devices (67, 93). How does a cell maintain the correct shape in the face of a fluctuating physical environment? A lesson from these recent single-cell studies is that growth and physiology are important for understanding cell shape maintenance.
Senescence and Death
Whether the two daughter cells resulting from cell division are physiologically and genetically identical and whether they are indistinguishable from the mother cell remain important open questions. These questions have often been addressed in the context of senescence, which has required single-cell approaches beginning with the first experiments. The classic study by Mortimer & Johnston (72) discovered that morphologically asymmetric dividing budding yeast accumulate bud scars on the mother cell and that she dies after several dozen divisions. Experimental reports indicate that even for bacteria the two daughter cells are different in both morphologically asymmetric and symmetric organisms (1, 90). Specifically, these authors found that the growth rate of the daughter cells continuously decreases with respect to the replicative age of the cells. Follow-up studies by several groups (65, 103) reported correlations between protein aggregation (formation of inclusion bodies) and decrease of growth rate, although whether protein aggregation actually reduces growth rate has not been shown.
A new class of experiments using microfluidic devices has painted a different picture (70). Our group (98) used a simple microfluidic habitat (the mother machine) to observe the long-term growth patterns of many single E. coli cells. Cells grew and replicated while the growth medium was continuously replenished in the device. E. coli mother cells trapped in narrow, dead-end growth channels grew without any decrease in growth rates for several hundred generations in steady-state conditions. Instead, the frequency of filamentation (and by inference, SOS induction) had a dramatic effect on the rate of death. To determine the heritable material driving an increased rate of cell death or filamentation, a combination of single-cell cultivation and monitoring by single-cell DNA sequencing will ultimately resolve questions about the nature of bacterial senescence. We feel that this is a field where microfluidics and single-cell technology are already mature and ready for more serious genetics and physiological approaches.
Evolution
Single-cell approaches can unexpectedly impact the study of evolution and the genetic origin of physiology (54). An early implementation of single-cell sequencing that targeted discrete loci observed the genetic progression of mouse tumor cell lineages and linked specific mutations to the appearance of tumor phenotypes (36). Because mutations and selection happen at the individual level, efforts to quantify the rate of evolution are somewhat futile without single-cell resolution. This is particularly intriguing considered in the context of directed evolution in microfluidic devices (106). Single-cell evolution experiments could decouple selection scheme design from growth rate, which is impossible in traditional adaptive laboratory evolution. A method to rapidly sequester individual cells could possibly be combined with a single-cell-level phenotypic assay (e.g., quantitative mass spectrometry), eliminating the need to couple the analyte of interest with continued rapid growth and division. A chosen cell could subsequently seed a new round of growth and selection, and sequencing could trace the effects of evolution on physiology.
SINGLE-CELL EXPERIMENTS WITH MICROFLUIDICS: LESSONS LEARNED AND SHARED
We have outlined fundamental questions in cell physiology that can benefit from quantitative single-cell approaches. Unfortunately, implementation of these approaches is not an easy feat. Observation of growth in individual cells has been recorded since at least 1932 (15) but micrographs alone portray limited information. Two elegant technologies developed in the mid-twentieth century, the baby machine and the Coulter counter, were an enormous leap forward (22, 28, 42, 44). Today we are fortunate to be armed with even more precise and robust technology for direct tracking and manipulation of individual cells.
Microfluidics paired with microscopy has become chief among these tools, enabling many studies that were not possible with bulk culture. Its adoption has been accelerated by advances in the miniaturization of electronics and parallel liquid sample handling methods for chemical analysis (80, 102). Low-cost and accessible soft-lithography techniques pioneered and matured by the Whitesides laboratory (105) were major contributions to the field. Rapid development of a complex microfluidic manipulation system by the Quake laboratory (95) has thrust microfluidics to the forefront of biology. A salient example of a previously inaccessible study is Balaban et al. (10), which characterized the bacterial persistence phenotype as a nongenetic switch between growth and nongrowth. Monitoring growth of single cells, the authors were able to directly observe individual persistent cells that survive antibiotic treatment without developing genetic resistance to the antibiotic. Other efforts to characterize cellular growth took advantage of the mother machine (98). Studies of both rod-shaped bacteria such as B. subtilis (74) and single-cell eukaryotes such as fission yeast (74, 87) utilized the high-throughput and long-term experimental capabilities of this device. In addition to answering existing questions, the use of microfluidic devices has enabled researchers to observe dynamic systems that were otherwise relegated to theory. For instance, it is now possible to directly observe competition and rapid evolution within a population of single cells (55).
With awareness of the limitations imposed by microfluidic techniques, it is possible to conceive, design, and successfully execute exciting experiments. Our philosophical compass for making decisions herein is guided by three central tenets: (a) Results must be reproducible; (b) quantitative distributions, as opposed to means or cherry-picked images, must be presented; and (c) averages calculated from the distributions must agree with population-level control experiments.
In the following sections we review useful technologies, pitfalls we faced in the past, and lessons we have learned along the way. In our experience, the application of these practices with great care dramatically improves the reliability and reproducibility of experimental data.
Design Strategy
The growth environment of microfluidic-based single-cell experiments is different from that of batch culture or continuous culture devices, e.g., chemostats and turbidostats. One consideration is that the volume of the liquid environment surrounding the cells is reduced such that a single cell can be in constant contact with several sides of the vessel’s walls. Thus, cells growing in microchannels can be influenced by the physical presence of channel walls, a phenomenon absent in liquid culture experiments. These forces along with fluid dynamics may drive unexpected behavior that may be anticipated at the device design stage (88).
Most of the current single-cell microfluidic devices are based on polydimethylsiloxane (PDMS) annealed to a coverslip, allowing microscopy from one side of the glass. PDMS-based devices are robust, reproducible, and economic. However, PDMS is relatively stiff, which can introduce mechanical stress to cells under specific circumstances. Cluzel and colleagues (70) designed a microfluidic device based on printing micron-scale channels on a block of agarose gel. The soft, transparent, and porous nature of agarose gel makes it favorable for cell trapping; the physical pressure on cells is minimal while the gel allows delivery of nutrients. Another advantage of agarose gel over PDMS is its optical properties. With an index of refraction almost similar to that of water, agarose gel improves the quality of transmission light microscopy. Unfortunately, agarose itself is less stable than PDMS over time; agarose-based microfluidic devices are thus less suitable for long-term experiments.
The isolation of individual cells is often a prerequisite for single-cell studies. Although some laboratory strains have lost the ability to form biofilms, their occurrence can clog a microfluidic device and render the experiment useless. Clever and elaborate device designs have been critical in resolving these challenges. Quake and colleagues (11) created a lab-on-a-chip bioreactor that uses microfluidic plumbing networks to allow long-term growth of small populations of E. coli while actively preventing biofilm formation. Even organisms that do not form biofilms can spontaneously adhere to surfaces. We have empirically found that media flow rates on the order of 10 mm/s inside the mother machine will successfully evacuate cells (98).
Microfluidic habitats currently have long development cycles. Complex designs that alleviate biological problems require longer development cycles before the question of interest is actually addressed. Direct 3D printing and two-photon fabrication are two technologies with the potential to dramatically reduce the time from design to implementation to a single day. In addition, they remove the need for clean room photolithography. Current 3D printing technology allows feature scales as small as tens of microns, whereas the feature size attainable by two-photon systems is limited by optical resolution to 200–300 nm. These two technologies can in principle be combined to design 3D devices that cannot be realized by traditional photolithography techniques. For interested readers, we recommend recent reviews (48, 56, 87).
Growth Media and Culturability
Not all organisms and cells grow in a microfluidic device. Microorganisms grown under seemingly identical nutrient conditions sometimes show different growth rates depending on the cultivation methods, e.g., liquid culture, surfaces, or microfluidic devices. For example, in rich media such as lysogeny broth (LB), prototrophic B. subtilis strains grow well in both a microfluidic device and batch culture and show the same average growth rate. However, in defined minimal media (e.g., S750), the same strains grow much more slowly, if at all, in a microfluidic device compared with the batch culture of the same medium.
The barrier to growth might be traced to the one of the advantages of microfluidics itself. The precise control over media composition—including oxygenation (2)—possible in microfluidics can also be the source of artifacts and challenges. Molecules secreted by cells to improve their environment are immediately washed away, whereas in batch culture they persist and are available to the population. To troubleshoot this type of growth problem in a microfluidic device, we suggest trying conditioned, filtered medium prepared from batch culture in exponential or stationary phase. If the cells grow in a conditioned medium in a microfluidic device, it is likely that small molecules secreted by cells in batch culture are important for growth. Such molecules are constantly removed in a typical microfluidic device and cells are likely to experience nutrient conditions akin to infinite dilution.
Once the cause of the problem is identified, the next step is to add the essential molecules that are present in batch culture but not in the defined media. This step can be painstaking and requires a priori knowledge of the physiological demands of the specific strain or organism. In the case of B. subtilis grown in S750, the addition of sodium citrate and ferric chloride to S750 medium immediately revived growth. Indeed, the secretion of siderophores to increase the availability of metal salts, particularly iron, is essential for growth (62, 78). Other secreted agents such as autoinducer-2 (29), indole (63), and outer membrane vesicles (68) can build to levels in batch and turbidostat cultures that can alter transcription or physiology. The effect of these environment-conditioning agents on data obtained from various culture conditions is largely unexplored, and microfluidic growth devices are a powerful tool for investigators in this field.
Physical Manipulation of Single Cells
The physical capture of single cells is a prerequisite for some advanced analytics but it poses significant challenges. For example, aside from the unbiased amplification of DNA, the physical isolation of cells is a major hurdle in single-cell sequencing. Unfortunately, best practices in single-cell capture may not be intuitively obvious and are not refined enough to be immediately successful for new investigators. Microfluidic solutions are preferable to alternative methods such as flow sorting, micropipetting, or laser dissection. Although FACS (fluorescence-activated cell sorter) is high throughput and unbiased, large amounts of cells are needed, precluding studies of individual microorganisms that have ceased growth. Micropipetting and laser microdissection are low-throughput manual methods subject to operator bias. On the other hand, microfluidic techniques can be high throughput, provide visual confirmation of capture, and can be automated to diminish operator bias (66). The small reagent volumes required also reduce costs and suppress the frequency of contamination.
Actuated microfluidics and optical traps (also known as laser tweezers) offer rapidly maturing solutions that are often employed hand-in-hand. Actuated microfluidics is a highly controllable fluid flow system that utilizes micron-scale valves and pressure gradients that can sequester cells without undue stress. Researchers are developing increasingly sophisticated devices that can capture single cells in nanoliter volumes and deliver picoliter volume reagents to reaction chambers in a defined manner (64). Droplet microfluidics is another emerging technique that allows for the manipulation of individually isolated nanoliter volumes of liquid that can contain cells or reagents (41). Optical traps were applied to biological studies the year after their unveiling (7, 8). They can isolate specific single cells, in contrast to actuated microfluidics, in which liquid and molecules surrounding cells are cotransported. Complete optical trap systems can be purchased, but advanced do-it-yourself systems can be constructed for significantly less (J. Xu, personal communication). Intrigued readers might start with the book chapter on setting up an optical trap for cell isolation by Block (20) and a recent review by Landry et al. (61).
Imaging and Phototoxicity
Fluorescence imaging is powerful and often essential in single-cell studies, but it can also severely compromise the physiological state of individual cells. Phototoxicity is arguably the most serious and yet poorly understood problem associated with fluorescence microscopy for single-cell experiments. Upon excitation with a fluorescent light source, live cells will grow more slowly or stop growing altogether. The primary mechanism of phototoxicity seems to be the production of radical oxygen species. Radical oxygen species are highly reactive and can bind to and damage any number of important cellular constituents. In many cases this can be attributed directly to the fluorescent protein itself (39) but also to naturally occurring metabolites (e.g., NAD+, flavins) that might otherwise confound the use of the green spectrum for detailed quantitative studies (16, 25).
Intense illumination is associated with growth defects in a number of organisms and is no less important in studies of bacteria, although the few quantitative analyses of the effect that have been performed are often relegated to supplementary information (91). To better understand the adverse effect of fluorescence illumination on bacterial physiology and, in particular, bacterial growth, we examined the effect of fluorescent illumination on the doubling rate of E. coli (D. McIntosh, unpublished results). Using a strain of E. coli that constitutively expresses a GFP fusion of a single-stranded binding protein, we took phase-contrast and fluorescence images 20 times per cell division in slow-growth media using a GFP filter set (see Figure 4). The growth rate of the cells is attenuated with increasing illumination intensity. Clearly, fluorescence imaging has a dramatic effect on the physiology of the cells.
Figure 4. Strong effect of fluorescence imaging on growth.
(a) A phase contrast/GFP overlay of a single field of view of Escherichia coli cells expressing Ssb-GFP (a GFP fusion of a single-stranded binding protein) grown in steady-state conditions. Cells imaged approximately 20 times per cell cycle for approximately 3 days with a fluorescence excitation illumination gradient across the field of view (cells on the left-hand side experience minimal illumination, whereas the exposure is maximum on the right-hand side). (b) A slice from an image taken with the same optical alignment of an FITC (fluorescein isothiocyanate) solution showing the intensity gradient. (c) Co-plot of FITC fluorescence intensity (a.u.) and growth rate (1/h) versus position in the field of view. As fluorescence intensity increases, the growth rate decreases. The insets show the intensity profile of the fluorescence image along a line passing through cells from different channels, as they experience different excitation light intensity.
Phototoxicity is correlated primarily with the illumination power delivered to the cell. Assuming that the illumination, exposure, camera triggering, and shutter opening have been optimized to the lowest possible levels that still permit accurate quantification, it will be necessary to upgrade to the most sensitive detectors available. Other methods to consider include (a) illuminating with longer wavelength light for two-photon excitation to reduce the unwanted excitation of metabolites, reducing the burden of associated reactive oxygen species (84, 107); (b) limiting the excitation area to only a small region of interest within the specimen, sparing the remainder of the specimen from unnecessary damage (47); and (c) avoiding chromophore overexcitation and thus phototoxicity (51) by using a pulsed laser with an interpulse time exceeding the lifetime of the triplet excited state (33).
Although most single-cell techniques rely on light microscopy, alternatives to visual observation that provide high-resolution insights do exist. Manalis and colleagues (85) designed a novel microchannel resonator to measure the ultralow volume of particles passing through a suspended resonator. Together with the Kirschner laboratory (38), they studied growth of single cells by measuring the buoyant mass of cells at femtogram resolution. Their experimental data based on B. subtilis, E. coli, S. cerevisiae, and mouse lymphoblasts were consistent with exponential growth of cells during the cell cycle.
Choice of Fluorescent Proteins
Fluorescence microscopy can provide valuable information about protein interactions and localizations and metabolic activities of cells. With the advent of modern genetics, fluorescent proteins became more and more popular for tracking gene expression and protein interactions (81). The efficiency of the method and the possibility of live-cell visualization outpaced immunofluorescence techniques, the previous gold standard. A review article by Tsien and colleagues (82) still provides a nice guideline for choosing fluorescent proteins.
The drawback of fusion fluorescent proteins is that fusion can affect the function of the tagged protein. Sometimes the artifact leads to an obvious phenotypic problem, such as slow growth rate or unusual morphology (82). But the dangerous artifacts are those that are not obvious. In a remarkable report, Landgraf et al. (60) performed extensive and extremely careful studies on a previously reported ClpX protein localization pattern. They used two methods to validate the localization: (a) tracking the downstream processes in the daughter cells in both wild-type and tagged cells and (b) using alternative fluorescent proteins. They confirmed the observed foci were an artifact of fluorescent protein dimerization. An important lesson from this study is that scientists must construct and test different reporter proteins for any serious experiments employing fluorescent fusion proteins. In addition, results based on fluorescent fusion proteins should be validated by independent means.
Temperature Control
Temperature affects the growth rate of the cells as well as the brightness of most fluorescent proteins. Most laboratories use a heated microscope stage or a thermal incubator made of plexiglass coupled to a feedback-controlled heated air blower. These methods work to an extent, but we recommend careful measurements of the temperature for important physiological experiments. Sometimes the timescale for cyclic heating and cooling of the incubator can be similar to the timescale of the physiological parameters of interest. This makes it difficult, when interpreting the data, to decouple otherwise orthogonal external (e.g., environmental fluctuations) and internal (e.g., physiological variability) factors that can influence the physiological state of the cells. A carefully designed and regulated environmental chamber in which a microscope, a microscope table, and other small equipment can be stationed at a constant temperature is an excellent solution to solve all problems related to temperature fluctuations. In contrast to the quest for uniformity, Groisman et al. (40) designed a microfluidic chemostat that provided unique temperature and pressure control for growing microorganisms in micron-sized traps.
Passivity of the Cellular Environment
The problems of passivation of microfluidic devices, motility of cells, and the infusion rate of growth media are often related in a typical single-cell experiment. If the internal surfaces of the microfluidic device are not properly passivated, cells can stick to the walls and affect the growth of other cells. For PDMS-based microfluidic devices, we find preloading with 50 mg/ml BSA (bovine serum albumin) for several minutes and 0.5 mg/ml BSA in the supplied growth medium is sufficient for passivation. Another method is to add mild neutral detergents such as Tween, which prevents cells from sticking to the glass surface (94). If the strain is motile, the cells may swim against incoming nutrient flow and can eventually clog the inlet and the tubing of the microfluidic device. We have found that keeping flow rates high (>10 mm/s), as well as simply knocking out motility genes, can abrogate this problem (see Strain Selection, below).
Communication Among the Electronics
Consider typical time-lapse imaging of multiple fields of view, or fast fluorescent particle tracking in vivo using a microscope equipped with a motorized stage, a mechanical shutter, fluorescence filter cubes or filter wheels, and a white fluorescence illumination source such as a mercury or xenon lamp. The microscope software sends an instruction from the computer to the microscope to (a) move the stage to the preregistered position, (b) find the focal plane, (c) open the mechanical shutter, (d) acquire an image, (e) send the image back to the computer, and (f) close the mechanical shutter. For multicolor imaging, additional steps cause further delay while the filter cube or filter wheel changes the illumination color and takes additional images. Each of these steps involves communication between equipment and waiting time, which all add up and can cause a significant discrepancy between the actual exposure time and the nominal exposure time (from tens to hundreds of milliseconds). Such a discrepancy can affect both the reliability of the acquired data and the physiology of the cells because of phototoxicity. To prevent these problems, we recommend fast communication methods such as TTL (transistor-transistor logic) and replacing mechanical components with electronic components where possible. For example, use LEDs or lasers with multiband filter cubes instead of mechanical shutters and use mercury lamps with single-band filter wheels.
Primary Image Analysis
Although bioinformatics is popularly considered the domain of next-generation sequencing, we propose that it includes the analysis of data produced by any high-throughput experimental technique and in particular the processing of time-lapse video data. In our view, this is one of the most difficult challenges of high-throughput single-cell experiments. Despite the ubiquity of publicly available image analysis software such as microbeTracker (83), Cell Profiler (24), and the mother machine–specific MMJ (6), we have not found a one-size-fits-all solution for high-throughput image analysis of the images generated from microfluidics-based single-cell experiments. The wide range of experimental conditions, the requirements dictated by the types of hypotheses under consideration, and the vagaries of superficially simple issues such as the perceived sharpness of focus all contribute to complex and evolving requirements for data analysis. Developing dedicated image analysis software can take several years of effort in addition to constant customization and maintenance for different experimental configurations. The science as a whole will benefit from community-level efforts to improve the available algorithms. We are optimistic that, like sequencing, image analysis will increasingly become the realm of engineering rather than art.
Strain Selection
Since its landmark sequencing, E. coli MG1655 (19) has become one of the most widely used E. coli strains. The related strains W3110 (43) and BW25113 have also been used extensively; BW25113 is the basis for the enormously useful Keio knockout collection (9). Unfortunately, there are increasingly well-known problems with the anabolic pathways of these and other strains derived from W1485, particularly pyrimidine biosynthesis (50), amino acid biosynthesis (5), and carbon metabolism (21). This picture is further complicated by MG1655’s lack of lambda prophage and the emerging importance of phage elements in mediating the host’s resistance to external stresses (13, 100). If MG1655 is nonetheless required, it is imperative to ensure that the correct substrain of MG1655 is selected (12, 35).
Our prototrophic strain of choice is the K12 derivative NCM3722 by the late Sydney Kustu. NCM3722 has been corrected for several defects found in MG1655 (86). To further improve this strain’s broad suitability for physiology studies, we recently derived an F– NCM3722 strain (SJ358), which is available from our laboratory and from the E. coli Genetic Stock Center.
When physical constraints do not fix the position of cells, natural motility systems can interfere with long-term monitoring. Genetically engineered defects in the flagellar system of E. coli can render cells nonmotile but will have unequal consequences on the physiology of the cell (17, 18). This is in addition to potential differences in the natural biological factors (e.g., insertion sequences) that contribute tomotility (12). We have employed ΔmotA and ΔfliC strains in the past but either of these may not be desirable depending on the nature of the assay under consideration.
CONCLUSIONS
Addressing fundamental questions in cellular physiology requires single-cell techniques that can capture the growth and cell cycle events of individual unicellular organisms. Given the extent of the intrinsic fluctuations in biological systems, population-level measurements mask important causal relationships underlying the biological phenomena. In this article we reviewed some of the questions from different aspects of bacterial physiology, including cell cycle, cell size, shape, metabolism, and cell death. Although recent advances in single-cell technologies have allowed us to answer some of the long-standing problems, many fundamental problems remain to be understood.
Among various single-cell techniques, microfluidics combined with high-resolution time-lapse imaging is popular and powerful. They not only allow for the investigation of individual cells, but also enable high-throughput observation such that researchers can draw quantitative conclusions about a cell’s physiology. However, as all technology is blind to its implementation, it is up to the prudent investigator to responsibly apply experimental techniques in order to minimize confounding artifacts. The hurdle here is not of desire but of painstaking trial and error. We hope the lessons we have shared in this article save the reader aggravation and encourage discovery in this exciting field.
SUMMARY POINTS.
Despite decades of study, many basic aspects of microbial physiology remain poorly understood. Single-cell data can greatly improve our understanding of these aspects.
Single-cell approaches help us unravel cell-to-cell variations and correlations that are otherwise averaged out in population-level measurements.
Microfluidic devices coupled with high-resolution microscopy imaging are a powerful means available to physiologists.
When collecting data and interpreting results from single-cell experiments, researchers must follow the three tenets: (a) Results must be reproducible; (b) quantitative distributions, as opposed to means or cherry-picked images, must be presented; and (c) averages calculated from the distributions must agree with population-level control experiments.
FUTURE ISSUES.
Progression of the cell cycle involves a set of processes that lead to cell division. Precise description of how these processes are coordinated is an open question that relies on future single-cell data.
The growth law, one of the first quantitative rules in bacterial physiology, states that cell size is exponentially related to growth rate. How individual cells recognize growth rate and their size remains to be understood.
Recent single-cell studies revealed that bacteria, including E. coli and B. subtilis, maintain size homeostasis by the “adder” principle. That is, cells add a constant size to each cell cycle. Yet, we need to find out how this principle results from growth and cell cycle coordination, especially in the context of evolution.
Current imaging techniques can disrupt the physiology of living cells. Phototoxicity is a major challenge that needs to be solved.
Acknowledgments
This work was supported by the Paul G. Allen Foundation, the Pew Charitable Trusts, and the National Science Foundation CAREER Award (to S.J.).
Glossary
- Diauxic shift
a state of growth characterized by the exclusive metabolism of one energy source to exhaustion followed by a switch to exclusive metabolism of another energy source
- Growth law
an exponential dependence of the average cell size on the nutrient-imposed growth rate
- Sizer
a model for cell size control based on a critical size for triggering division
- Timer
a model for cell size control based on a constant time period between divisions
- “Adder” principle
a model for cell size homeostasis based on added size from birth to division
- Mother machine
a microfluidic device with many narrow parallel growth channels that has one closed end and one open end exposed to fresh media
- SOS
a group of bacterial functions that act to repair and recover from DNA damage
- Baby machine
a device based on membrane elution to produce synchronized populations of cells
- Turbidostat
a continuous bulk culture device that maintains a constant concentration of cells by exchanging old media for new media at a variable rate
- PDMS
polydimethylsiloxane
- LB
lysogeny broth
- Prototroph
an organism capable of synthesizing all its metabolites from inorganic material, requiring no organic nutrients
- FACS
fluorescence-activated cell sorting
- BSA
bovine serum albumin
- TTL
transistor-transistor logic
Footnotes
DISCLOSURE STATEMENT
The authors are not aware of any affiliations, memberships, funding, or financial holdings that might be perceived as affecting the objectivity of this review.
LITERATURE CITED
- 1.Ackermann M, Stearns SC, Jenal U. Senescence in a bacterium with asymmetric division. Science. 2003;300:1920. doi: 10.1126/science.1083532. [DOI] [PubMed] [Google Scholar]
- 2.Adler M, Erickstad M, Gutierrez E, Groisman A. Studies of bacterial aerotaxis in a microfluidic device. Lab Chip. 2012;12:4835–47. doi: 10.1039/c2lc21006a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Alberts B, Johnson A, Lewis J, Raff M, Roberts K, Walter P. Molecular Biology of the Cell. New York: Garland Science; 2003. [Google Scholar]
- 4.Amir A, Babaeipour F, McIntosh DB, Nelson DR, Jun S. Bending forces plastically deform growing bacterial cell walls. PNAS. 2014;111:5778–83. doi: 10.1073/pnas.1317497111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Andersen DC, Swartz J, Ryll T, Lin N, Snedecor B. Metabolic oscillations in an E. coli fermentation. Biotechnol. Bioeng. 2001;75:212–18. doi: 10.1002/bit.10018. [DOI] [PubMed] [Google Scholar]
- 6.Arnoldini M, Vizcarra IA, Peña-Miller R, Stocker N, Diard M, et al. Bistable expression of virulence genes in Salmonella leads to the formation of an antibiotic-tolerant subpopulation. PLOS Biol. 2014;12:e1001928. doi: 10.1371/journal.pbio.1001928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ashkin A, Dziedzic J, Bjorkholm JE, Chu S. Observation of a single-beam gradient-force optical trap for dielectric particles in air. Opt. Lett. 1986;22:816–18. doi: 10.1364/ol.11.000288. [DOI] [PubMed] [Google Scholar]
- 8.Ashkin A, Dziedzic J, Yamane T. Optical trapping and manipulation of single cells using infrared laser beams. Nature. 1987;330:769–71. doi: 10.1038/330769a0. [DOI] [PubMed] [Google Scholar]
- 9.Baba T, Ara T, Hasegawa M, Takai Y, Okumura Y, et al. Construction of Escherichia coli K-12 in-frame, single-gene knockout mutants: the Keio collection. Mol. Syst. Biol. 2006;2:2006.0008. doi: 10.1038/msb4100050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Balaban NQ, Merrin J, Chait R, Kowalik L, Leibler S. Bacterial persistence as a phenotypic switch. Science. 2004;305:1622–25. doi: 10.1126/science.1099390. [DOI] [PubMed] [Google Scholar]
- 11.Balagadde F, You L, Hansen C, Arnold F, Quake S. Long-term monitoring of bacteria undergoing programmed population control in a microchemostat. Science. 2005;309:137–40. doi: 10.1126/science.1109173. [DOI] [PubMed] [Google Scholar]
- 12.Barker CS, Prüss BM, Matsumura P. Increased motility of Escherichia coli by insertion sequence element integration into the regulatory region of the flhD operon (MG1655) J. Bacteriol. 2004;186:7529–37. doi: 10.1128/JB.186.22.7529-7537.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Barondess J, Beckwith J. bor gene of phage λ involved in serum resistance, encodes a widely conserved outer membrane lipoprotein. J. Bacteriol. 1995;177:1247–53. doi: 10.1128/jb.177.5.1247-1253.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bates D, Kleckner N. Chromosome and replisome dynamics in E. coli: Loss of sister cohesion triggers global chromosome movement and mediates chromosome segregation. Cell. 2005;121:899–911. doi: 10.1016/j.cell.2005.04.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bayne-Jones S, Adolph EF. Growth in size of micro-organisms measured from motion pictures: II. Bacillus megatherium. J. Cell. Comp. Physiol. 1932;1:388–407. [Google Scholar]
- 16.Benson RC, Meyer RA, Zaruba ME, McKhann GM. Cellular autofluorescence—Is it due to flavins? J. Histochem. Cytochem. 1979;27:44–48. doi: 10.1177/27.1.438504. [DOI] [PubMed] [Google Scholar]
- 17.Berg HC. The rotary motor of bacterial flagella. Annu. Rev. Biochem. 2003;72:9–54. doi: 10.1146/annurev.biochem.72.121801.161737. [DOI] [PubMed] [Google Scholar]
- 18.Blair DF, Berg HC. The MotA protein of E. coli is a proton-conducting component of the flagellar motor. Cell. 1990;60:439–49. doi: 10.1016/0092-8674(90)90595-6. [DOI] [PubMed] [Google Scholar]
- 19.Blattner FR, Plunkett GI, Bloch CA, Perna NT, Burland V, et al. The complete genome sequence of Escherichia coli K-12. Science. 1997;277:1453–62. doi: 10.1126/science.277.5331.1453. [DOI] [PubMed] [Google Scholar]
- 20.Block SM. Construction of optical tweezers. In: Spector D, Goldman R, Leinwand L, editors. Cells: A Laboratory Manual. Vol. 3. Cold Spring Harbor, NY: Cold Spring Harbor Press; 1998. pp. 81.1–81.14. [Google Scholar]
- 21.Brinkkötter A, Klöss H, Alpert CA, Lengeler JW. Pathways for the utilization of N-acetylgalactosamine and galactosamine in Escherichia coli. Mol. Microbiol. 2000;37:125–35. doi: 10.1046/j.1365-2958.2000.01969.x. [DOI] [PubMed] [Google Scholar]
- 22.Campbell A. Synchronization of cell division. Bacteriol. Rev. 1957;21:263–72. doi: 10.1128/br.21.4.263-272.1957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Campos M, Surovtsev IV, Kato S, Paintdakhi A, Beltran B, et al. A constant size extension drives bacterial cell size homeostasis. Cell. 2014;159:1433–466. doi: 10.1016/j.cell.2014.11.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Carpenter AE, Jones TR, Lamprecht MR, Clarke C, Kang IH, et al. Cell Profiler: image analysis software for identifying and quantifying cell phenotypes. Genome Biol. 2006;7:R100. doi: 10.1186/gb-2006-7-10-r100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chance B, Cohen P, Jobsis F, Schoener B. Intracellular oxidation-reduction states in vivo. Science. 1962;137:499–508. doi: 10.1126/science.137.3529.499. [DOI] [PubMed] [Google Scholar]
- 26.Cooper S. Cell division and DNA replication following a shift to a richer medium. J. Mol. Biol. 1969;43:1–11. doi: 10.1016/0022-2836(69)90074-6. Shows that chromosome replication initiates once accumulation of hypothetical initiator(s) reaches a threshold; rate of accumulation depends on growth rate. [DOI] [PubMed] [Google Scholar]
- 27.Cooper S. Regulation of DNA synthesis in bacteria: analysis of the Bates/Kleckner licensing/initiation-mass model for cell cycle control. Mol. Microbiol. 2006;62:303–7. doi: 10.1111/j.1365-2958.2006.05342.x. [DOI] [PubMed] [Google Scholar]
- 28.Cooper S, Helmstetter CE. Chromosome replication and the division cycle of Escherichia coli B/r. J. Mol. Biol. 1968;31(3):519–40. doi: 10.1016/0022-2836(68)90425-7. Demonstrates that periods of chromosome replication and the gap between termination of replication and cell division are constant, independent of growth rate. [DOI] [PubMed] [Google Scholar]
- 29.DeLisa M, Valdes J, Bentley W. Mapping stress-induced changes in autoinducer AI-2 production in chemostat-cultivated Escherichia coli K-12. J. Bacteriol. 2001;183:2918–28. doi: 10.1128/JB.183.9.2918-2928.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Dobell C. Antony Van Leeuwenhoek and His “Little Animals”. New York: Harcourt, Brace; 1923. [Google Scholar]
- 31.Domínguez-Escobar J, Chastanet A, Crevenna AH, Fromion V, Wedlich-Söldner R, Carballido-Lóez R. Processive movement of MreB-associated cell wall biosynthetic complexes in bacteria. Science. 2011;333:225–28. doi: 10.1126/science.1203466. [DOI] [PubMed] [Google Scholar]
- 32.Donachie W. Relationship between cell size and time of initiation of DNA replication. Nature. 1968;219:1077–79. doi: 10.1038/2191077a0. Introduces notion of critical mass at initiation of chromosome replication: Cells initiate replication at a constant cell mass per origin of replication. [DOI] [PubMed] [Google Scholar]
- 33.Donnert G, Eggeling C, Hell SW. Triplet-relaxation microscopy with bunched pulsed excitation. Photochem. Photobiol. Sci. 2009;8:481–85. doi: 10.1039/b903357m. [DOI] [PubMed] [Google Scholar]
- 34.Elowitz M, Levine A, Siggia E, Swain P. Stochastic gene expression in a single cell. Science. 2002;297:1183–86. doi: 10.1126/science.1070919. Categorizes sources of noise in genetic networks as intrinsic and extrinsic noise. [DOI] [PubMed] [Google Scholar]
- 35.Freddolino PL, Amini S, Tavazoie S. Newly identified genetic variations in common Escherichia coli MG1655 stock cultures. J. Bacteriol. 2012;194:303–6. doi: 10.1128/jb.06087-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Frumkin D, Wasserstrom A, Itzkovitz S, Stern T, Harmelin A, et al. Cell lineage analysis of a mouse tumor. Cancer Res. 2008;68:5924–31. doi: 10.1158/0008-5472.CAN-07-6216. [DOI] [PubMed] [Google Scholar]
- 37.Garner EC, Bernard R, Wang W, Zhuang X, Rudner DZ, Mitchison T. Coupled, circumferential motions of the cell wall synthesis machinery and MreB filaments in B. subtilis. Science. 2011;333:222–25. doi: 10.1126/science.1203285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Godin M, Delgado FF, Son S, Grover WH, Bryan AK, et al. Using buoyant mass to measure the growth of single cells. Nat. Methods. 2010;7:387–90. doi: 10.1038/nmeth.1452. Uses buoyant mass to measure the relationship between cell mass and mass increase rate. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Greenbaum L, Rothmann C, Lavie R, Malik Z. Green fluorescent protein photobleaching: a model for protein damage by endogenous and exogenous singlet oxygen. Biol. Chem. 2000;381:1251–58. doi: 10.1515/BC.2000.153. [DOI] [PubMed] [Google Scholar]
- 40.Groisman A, Lobo C, Cho H. A microfluidic chemostat for experiments with bacterial and yeast cells. Nat. Methods. 2005;2:685–89. doi: 10.1038/nmeth784. [DOI] [PubMed] [Google Scholar]
- 41.Guo MT, Rotem A, Heyman JA, Weitz DA. Droplet microfluidics for high-throughput biological assays. Lab Chip. 2012;12:2146–55. doi: 10.1039/c2lc21147e. [DOI] [PubMed] [Google Scholar]
- 42.Harvey R, Marr A, Painter P. Kinetics of growth of individual cells of Escherichia coli and Azotobacter agilis. J. Bacteriol. 1967;93:605–17. doi: 10.1128/jb.93.2.605-617.1967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Hayashi K, Morooka N, Yamamoto Y, Fujita K, Isono K, et al. Highly accurate genome sequences of Escherichia coli K-12 strains MG1655 and W3110. Mol. Syst. Biol. 2006;2:2006.0007. doi: 10.1038/msb4100049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Helmstetter C. Rates of DNA synthesis during the division cycle of Escherichia coli B/r. J. Mol. Biol. 1967;24:417–27. [Google Scholar]
- 45.Helmstetter CE, Cummings DJ. Bacterial synchronization by selection of cells at division. PNAS. 1963;50:767–74. doi: 10.1073/pnas.50.4.767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Hill NS, Buske PJ, Shi Y, Levin PA. A moonlighting enzyme links Escherichia coli cell size with central metabolism. PLOS Genet. 2013;9:e1003663. doi: 10.1371/journal.pgen.1003663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Hoebe RA, Van Oven CH, Gadella TWJ, Dhonukshe PB, Van Noorden CJF, Manders EMM. Controlled light-exposure microscopy reduces photobleaching and phototoxicity in fluorescence live-cell imaging. Nat. Biotechnol. 2007;25:249–53. doi: 10.1038/nbt1278. [DOI] [PubMed] [Google Scholar]
- 48.Hribar KC, Soman P, Warner J, Chung P, Chen S. Light-assisted direct-write of 3D functional biomaterials. Lab Chip. 2014;14:268–75. doi: 10.1039/c3lc50634g. [DOI] [PubMed] [Google Scholar]
- 49.Jacob F, Monod J. Genetic regulatory mechanisms in the synthesis of proteins. J. Mol. Biol. 1961;3(3):318–56. doi: 10.1016/s0022-2836(61)80072-7. [DOI] [PubMed] [Google Scholar]
- 50.Jensen KF. The Escherichia coli K-12 “wild types” W3110 and MG1655 have an rph frameshift mutation that leads to pyrimidine starvation due to low pyrE expression levels. J. Bacteriol. 1993;175:3401–7. doi: 10.1128/jb.175.11.3401-3407.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Johnson ID. Practical considerations in the selection and application of fluorescent probes. In: Pawley JB, editor. Handbook of Biological Confocal Microscopy. New York: Springer; 2006. pp. 353–67. [Google Scholar]
- 52.Johnston GC, Ehrhardt CW, Lorincz A, Carter BLA. Regulation of cell size in the yeast Saccharomyces cerevisiae. J. Bacteriol. 1979;137:1–5. doi: 10.1128/jb.137.1.1-5.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Jun S, Taheri-Araghi S. Cell-size maintenance: universal strategy revealed. Trends Microbiol. 2014;23:4–6. doi: 10.1016/j.tim.2014.12.001. [DOI] [PubMed] [Google Scholar]
- 54.Kalisky T, Quake SR. Single-cell genomics. Nat. Methods. 2011;8:311–14. doi: 10.1038/nmeth0411-311. [DOI] [PubMed] [Google Scholar]
- 55.Keymer JE, Galajda P, Muldoon C, Park S, Austin RH. Bacterial metapopulations in nanofabricated landscapes. PNAS. 2006;103:17290–95. doi: 10.1073/pnas.0607971103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Kitson PJ, Rosnes MH, Sans V, Dragone V, Cronin L. Configurable 3D-printed millifluidic and microfluidic ‘lab on a chip’ reactionware devices. Lab Chip. 2012;12:3267–71. doi: 10.1039/c2lc40761b. [DOI] [PubMed] [Google Scholar]
- 57.Kiviet DJ, Nghe P, Walker N, Boulineau S, Sunderlikova V, Tans SJ. Stochasticity of metabolism and growth at the single-cell level. Nature. 2014;514:376–79. doi: 10.1038/nature13582. [DOI] [PubMed] [Google Scholar]
- 58.Koch AL. Bacterial Growth and Form. New York: Springer; 2001. [Google Scholar]
- 59.Koch AL, Schaechter M. A model for statistics of the cell division process. J. Gen. Microbiol. 1962;29:435–54. doi: 10.1099/00221287-29-3-435. [DOI] [PubMed] [Google Scholar]
- 60.Landgraf D, Okumus B, Chien P, Baker TA, Paulsson J. Segregation of molecules at cell division reveals native protein localization. Nat. Methods. 2012;9:480–82. doi: 10.1038/nmeth.1955. Explicitly demonstrates potential confounding effects of fluorescent fusion proteins in physiologic investigations. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Landry ZC, Giovanonni SJ, Quake SR, Blainey PC. Optofluidic cell selection from complex microbial communities for single-genome analysis. Methods Enzymol. 2013;531:61–69. doi: 10.1016/B978-0-12-407863-5.00004-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Lankford C, Byers B. Bacterial assimilation of iron. Crit. Rev. Microbiol. 1973;2:273–331. [Google Scholar]
- 63.Lee JH, Lee J. Indole as an intercellular signal in microbial communities. FEMS Microbiol. Rev. 2010;34:426–44. doi: 10.1111/j.1574-6976.2009.00204.x. [DOI] [PubMed] [Google Scholar]
- 64.Leung K, Zahn H, Leaver T, Konwar KM, Hanson NW, et al. A programmable droplet-based microfluidic device applied to multiparameter analysis of single microbes and microbial communities. PNAS. 2012;109:7665–70. doi: 10.1073/pnas.1106752109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Lindner AB, Madden R, Demarez A, Stewart EJ, Taddei F. Asymmetric segregation of protein aggregates is associated with cellular aging and rejuvenation. PNAS. 2008;105:3076–81. doi: 10.1073/pnas.0708931105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Macaulay IC, Voet T. Single cell genomics: advances and future perspectives. PLOS Genet. 2014;10:e1004126. doi: 10.1371/journal.pgen.1004126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Männik J, Driessen R, Galajda P, Keymer JE, Dekker C. Bacterial growth and motility in submicron constrictions. PNAS. 2009;106:14861–66. doi: 10.1073/pnas.0907542106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Mashburn-Warren LM, Whiteley M. Special delivery: vesicle trafficking in prokaryotes. Mol. Microbiol. 2006;61:839–46. doi: 10.1111/j.1365-2958.2006.05272.x. [DOI] [PubMed] [Google Scholar]
- 69.Mitchison JM. The Biology of the Cell Cycle. Cambridge, UK: Cambridge Univ. Press; 1971. [Google Scholar]
- 70.Moffitt JR, Lee JB, Cluzel P. The single-cell chemostat: an agarose-based, microfluidic device for high-throughput, single-cell studies of bacteria and bacterial communities. Lab Chip. 2012;12:1487–94. doi: 10.1039/c2lc00009a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Monod J. The growth of bacterial cultures. Annu. Rev. Microbiol. 1949;3:371–94. [Google Scholar]
- 72.Mortimer R, Johnston J. Life span of individual yeast cells. Nature. 1959;183:1751–52. doi: 10.1038/1831751a0. [DOI] [PubMed] [Google Scholar]
- 73.Nanninga N, Woldringh C. Molecular Cytology of Escherichia coli: Cell Growth, Genome Duplication, and Cell Division. London: Academic; 1985. [Google Scholar]
- 74.Norman TM, Lord ND, Paulsson J, Losick R. Memory and modularity in cell-fate decision making. Nature. 2013;503:481–86. doi: 10.1038/nature12804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Novick A, Weiner M. Enzyme induction as an all-or-none phenomenon. PNAS. 1957;43:553–66. doi: 10.1073/pnas.43.7.553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Ozbudak EM, Thattai M, Kurtser I, Grossman AD, van Oudenaarden A. Regulation of noise in the expression of a single gene. Nat. Genet. 2002;31:69–73. doi: 10.1038/ng869. Shows that protein copy number noise is a function of translation (stochastic ribosomal activity) as opposed to transcription (gene expression). [DOI] [PubMed] [Google Scholar]
- 77.Ozbudak EM, Thattai M, Lim HN, Shraiman BI, Van Oudenaarden A. Multistability in the lactose utilization network of Escherichia coli. Nature. 2004;427:737–40. doi: 10.1038/nature02298. [DOI] [PubMed] [Google Scholar]
- 78.Rolfe MD, Rice CJ, Lucchini S, Pin C, Thompson A, et al. Lag phase is a distinct growth phase. J. Bacteriol. 2012;194:686–701. doi: 10.1128/JB.06112-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Schaechter M, Maaløe O, Kjeldgaard NO. Dependency on medium and temperature of cell size and chemical composition during balanced growth of Salmonella typhimurium. J. Gen. Microbiol. 1958;19:592–606. doi: 10.1099/00221287-19-3-592. Introduces growth law stating that cell size is exponentially related to growth rate. [DOI] [PubMed] [Google Scholar]
- 80.Schaller RR. Moore’s law: past, present, and future. IEEE Spectrosc. 1997;6:52–59. [Google Scholar]
- 81.Shaner NC, Patterson GH, Davidson MW. Advances in fluorescent protein technology. J. Cell Sci. 2007;120:4247–60. doi: 10.1242/jcs.005801. [DOI] [PubMed] [Google Scholar]
- 82.Shaner NC, Steinbach PA, Tsien RY. A guide to choosing fluorescent proteins. Nat. Methods. 2005;2:905–9. doi: 10.1038/nmeth819. [DOI] [PubMed] [Google Scholar]
- 83.Sliusarenko O, Heinritz J, Emonet T, Jacobs-Wagner C. High-throughput, subpixel precision analysis of bacterial morphogenesis and intracellular spatio-temporal dynamics. Mol. Microbiol. 2011;80:612–27. doi: 10.1111/j.1365-2958.2011.07579.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.So P, Dong C. Two-photon excitation fluorescence microscopy. Annu. Rev. Biomed. Eng. 2000;2:399–429. doi: 10.1146/annurev.bioeng.2.1.399. [DOI] [PubMed] [Google Scholar]
- 85.Son S, Grover W, Burg T, Manalis S. Suspended microchannel resonators for ultralow volume universal detection. Anal. Chem. 2008;80:4757–60. doi: 10.1021/ac800307a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Soupene E, van Heeswijk WC, Plumbridge J, Stewart V, Bertenthal D, et al. Physiological studies of Escherichia coli strain MG1655: growth defects and apparent cross-regulation of gene expression. Am. Soc. Microbiol. 2003;185:5611–26. doi: 10.1128/JB.185.18.5611-5626.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Spivey EC, Xhemalce B, Shear JB, Finkelstein IJ. 3D-printed microfluidic microdissector for high-throughput studies of cellular aging. Anal. Chem. 2014;86:7406–12. doi: 10.1021/ac500893a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Squires T, Quake SR. Microfluidics: fluid physics at the nanoliter scale. Rev. Modern Phys. 2005;77:977–1026. [Google Scholar]
- 89.Starka J. Avant-propos (foreword) Ann. Microbiol. 1974;125B:133–34. [Google Scholar]
- 90.Stewart EJ, Madden R, Paul G, Taddei F. Aging and death in an organism that reproduces by morphologically symmetric division. PLOS Biol. 2005;3:e45. doi: 10.1371/journal.pbio.0030045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Strack RL, Hein B, Bhattacharyya D, Hell SW, Keenan RJ, Glick BS. A rapidly maturing far-red derivative of DsRed-Express2 for whole-cell labeling. Biochemistry. 2009;48:8279–81. doi: 10.1021/bi900870u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Taheri-Araghi S, Bradde S, Sauls JT, Hill NS, Levin PA, et al. Cell size control in bacteria. Curr. Biol. 2015 doi: 10.1016/j.cub.2014.12.009. http://dx.doi.org/10.1016/j.cub.2014.12.009 Demonstrates that bacteria maintain size homeostasis by adding a constant size in each cell cycle. [DOI] [PMC free article] [PubMed]
- 93.Takeuchi S, DiLuzio WR, Weibel DB, Whitesides GM. Controlling the shape of filamentous cells of Escherichia coli. Nano Lett. 2005;5:1819–23. doi: 10.1021/nl0507360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Turner L, Zhang R, Darnton NC, Berg HC. Visualization of flagella during bacterial swarming. J. Bacteriol. 2010;192:3259–67. doi: 10.1128/JB.00083-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Unger MA, Chou H-P, Thorsen T, Scherer A, Quake SR. Monolithic microfabricated valves and pumps by multilayer soft lithography. Science. 2000;288:113–16. doi: 10.1126/science.288.5463.113. [DOI] [PubMed] [Google Scholar]
- 96.Van Teeffelen S, Wang S, Furchtgott L, Huang KC, Wingreen NS, et al. The bacterial actin MreB rotates, and rotation depends on cell-wall assembly. PNAS. 2011;108:15822–27. doi: 10.1073/pnas.1108999108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Voorn WJ, Koppes LJ, Grover N. Mathematics of cell division in Escherichia coli. Curr. Top. Mol. Genet. 1993:187–94. [Google Scholar]
- 98.Wang P, Robert L, Pelletier J, Dang WL, Taddei F, et al. Robust growth of Escherichia coli. Curr. Biol. 2010;20:1099–103. doi: 10.1016/j.cub.2010.04.045. Overturned widespread belief that cell age and growth rate were inversely correlated by using microfluidic growth chambers instead of agarose pads. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Wang S, Arellano-Santoyo H, Combs PA, Shaevitz JW. Actin-like cytoskeleton filaments contribute to cell mechanics in bacteria. PNAS. 2010;107:9182–85. doi: 10.1073/pnas.0911517107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Wang X, Kim Y, Ma Q, Hong SH, Pokusaeva K, et al. Cryptic prophages help bacteria cope with adverse environments. Nat. Commun. 2010;1:147. doi: 10.1038/ncomms1146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Weart RB, Lee AH, Chien AC, Haeusser DP, Hill NS, Levin PA. A metabolic sensor governing cell size in bacteria. Cell. 2007;130:335–47. doi: 10.1016/j.cell.2007.05.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Whitesides GM. The origins and the future of microfluidics. Nature. 2006;442:368–73. doi: 10.1038/nature05058. [DOI] [PubMed] [Google Scholar]
- 103.Winkler J, Seybert A, König L, Pruggnaller S, Haselmann U, et al. Quantitative and spatiotemporal features of protein aggregation in Escherichia coli and consequences on protein quality control and cellular ageing. EMBO J. 2010;29:910–23. doi: 10.1038/emboj.2009.412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Woldringh C, Grover N, Rosenberger R, Zaritsky A. Dimensional rearrangement of rod-shaped bacteria following nutritional shift-up. II. Experiments with Escherichia coli B/r. J. Theor. Biol. 1980;86:441–54. doi: 10.1016/0022-5193(80)90344-6. [DOI] [PubMed] [Google Scholar]
- 105.Xia Y, Whitesides GM. Soft lithography. Annu. Rev. Mater. Sci. 1998;28:153–84. [Google Scholar]
- 106.Zhang Q, Lambert G, Liao D, Kim H, Robin K, et al. Acceleration of emergence of bacterial antibiotic resistance in connected microenvironments. Science. 2011;333:1764–67. doi: 10.1126/science.1208747. [DOI] [PubMed] [Google Scholar]
- 107.Zipfel WR, Williams RM, Webb WW. Nonlinear magic: multiphoton microscopy in the biosciences. Nat. Biotechnol. 2003;21:1369–77. doi: 10.1038/nbt899. [DOI] [PubMed] [Google Scholar]