Abstract
In this study, we demonstrated that antisense transcripts of human cytomegalovirus (HCMV) UL123, UL21.5 and cellular GAPDH genes were present in highly purified virions. These virion RNAs were delivered into the host cells upon infection, and de novo synthesized ones appeared in the infected cell at the immediate early stage. Although the sequence of UL123 antisense transcripts in virions is uncertain, we found that these transcripts in Towne-infected human fibroblasts had novel transcriptional start sites (TSSs) with various 5′-terminal deletions of open reading frame (ORF) 59. These findings not only provide new insight into the composition of HCMV virions but also reveal a possible viral strategy for initiating latent infection and switching between latent and productive infections.
Human cytomegalovirus (HCMV) belongs to the subfamily Betaherpesvirinae of the family Herpesviridae. It contains a linear, double-stranded DNA genome of over 230 kb packed in a 100-nm icosahedral capsid. The tegument layer of the protein matrix resides between the capsid and the membrane envelope. Although classified as a DNA virus, RNA transcripts have been found in highly purified virions of HCMV laboratory strains AD169, Towne, and Davis, as well as in a clinical isolate, CL203 [1–4]. The presence of RNA has also been observed in the virions of other members of the Herpesviridae, such as herpes simplex virus type 1 (HSV-1) [5], Kaposi’s sarcoma-associated herpesvirus (KSHV, human herpesvirus type 8) [6, 7], murine gammaherpesvirus 68 [8], and Epstein-Barr virus (EBV) [9]. Little is known about the role of virion-associated RNAs during infection, but it is certain that these RNAs transported into the newly infected cells have the potential to be translated in the absence of viral gene transcription/expression [9].
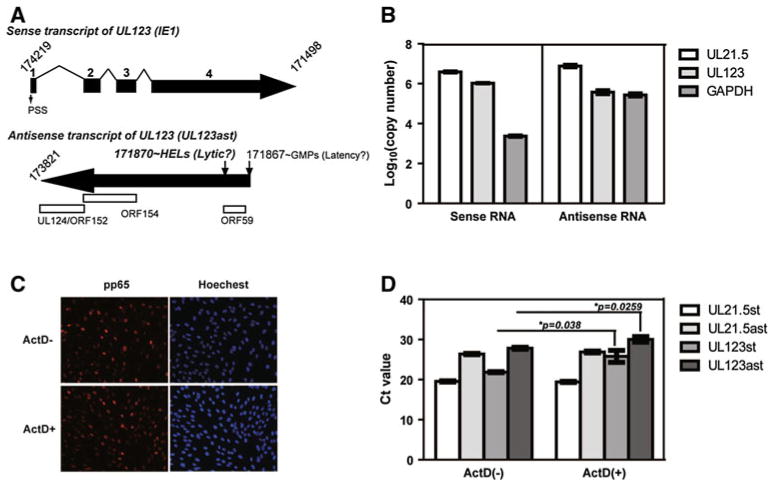
Natural antisense transcripts (NATs) are ubiquitous in prokaryotes and eukaryotes, contributing to regulation of gene expression by influencing RNA splicing, editing, stabilization, localization, and translation [10]. Viral NATs have been shown to be present in cells infected with a variety of viruses, such as human T-cell leukemia virus type 1 (HTLV-1) [11, 12], human immunodeficiency virus (HIV) [13], papillomavirus [14], KSHV [15], and HCMV. [16–18] To date, most of the work on HCMV NATs has focused on their roles in viral latent infection. The first reported NAT was UL123 antisense transcript (UL123ast) [16, 17]. UL123ast consists of the three open reading frames corresponding to ORF59, ORF152/UL124 and ORF154 and is unspliced and complementary to ie1/UL123 exons 2, 3 and 4 (Fig. 1A). This transcript has been found not only in experimentally infected monocytes and granulocyte macrophage progenitor (GM-P) cells, but also in bone marrow cells from healthy seropositive individuals, indicating that it may play a significant role in establishing/maintaining HCMV latency [16, 17]. Recent work by Zhang et al. [18] revealed that at least 55 % of cDNA clones were completely or partially antisense to known or predicted viral genes by genome-wide cDNA annotation in HCMV. Since it has been reported that both viral and cellular RNA transcripts are nonspecifically packed into HCMV in proportion to their intracellular concentration at the time of assembly [3], we thus investigated whether NATs, such as UL123ast, are incorporated into HCMV virions. A confluent monolayer of human embryonic lung fibroblast cells (HELs) was infected with HCMV strain Towne (ATCC VR-977) at an MOI of 2. At 96 hours postinfection (96 hpi), the supernatants were harvested and centrifuged at 3,600 g for 15 min to remove cell debris. The clarified medium was then laid onto a 20 % sorbitol cushion and centrifuged at 55,000g for 1.5 h to pellet the virus particles, including virions. The virus particle pellet was further purified by glycerol-tartrate gradient centrifugation to obtain the virions. The purified virion pellet was resuspended in phosphate-buffered saline (PBS) and treated sequentially with RNaseOne (Promega) and proteinase K [3, 19, 20]. The virion RNA was isolated using an RNeasy Mini Kit (QIAGEN), followed by RNase-free DNase treatment (RNase-Free DNase Set, QIAGEN) to remove contaminating DNA. Prior to RNase treatment, 400 ng of yeast DNA-free RNA was added prior to RNase treatment. A primer pair (forward primer, 5′-GAAGG-TAGTCAAAGAAGCCAAGATAGAAC-3′; reverse primer, 5′-TCCCAGGATT TGCCGAAAGAATGC-3′) specific for the yeast actin gene was used to determine the presence/absence of contaminating RNA bound to the outside of the virus. The quantitative real-time reverse transcriptase PCR (qRT-PCR) assay was applied to detect sense-antisense transcript pairs in virions using an iScript™ One-Step RT-PCR Kit with SYBR®Green (Bio-Rad). Twenty-five ng of the purified DNA-free virion RNA was added to 20 μl of reaction buffer containing 10 μl of 2x SYBR®Green RT-PCR reaction mix, 0.4 μl of Script reverse transcriptase for one-step RT-PCR and either 300 nM forward primer (for detection of antisense transcript) or reverse primer (for detection of sense transcript), followed by 50 °C for 10 min and 95 °C for 5 min to amplify the first-strand cDNA. After adding the corresponding primer (300 nM), the mix was subjected to the following PCR amplification cycles: 35 cycles of 95 °C for 10 s, followed by 60 °C for 30 s. Melt curve analysis was carried out with a protocol of 95 °C for 1 min, 55 °C for 1 min, and 10 s at 55 °C-95 °C. The primer pairs against both viral and cellular transcripts are listed in Table 1. No-template and no-reverse-transcriptase controls were included for each primer pair to monitor reagents and genomic DNA contamination, respectively. The inability to amplify yeast-actin RNA indicated the complete removal of any RNA contamination from virion RNA samples. Both sense and antisense RNAs of UL123, UL21.5 (a viral late gene), and GAPDH (a cellular gene) were found in purified virions (Fig. 1B). The copy numbers were calculated by comparison with a standard curve of the respective gene, and they were 1.05×106 (sense)/3.74×105 (antisense), 3.89×106 (sense)/7.69×106 (antisense), and 2.33×103 (sense)/2.71×105 (antisense), respectively.
Fig. 1.
HCMV antisense RNAs are packaged into virions. (A) Schematic diagram of the structure of sense and antisense transcripts of the UL123 region in laboratory strain Towne. The upper diagram shows the UL123 sense transcript, starting from the PSS in productive infection, encoding the initial and most abundant viral protein, referred to as IE1 (also named IE72). The lower diagram shows a summary of UL123ast in previously reported GM-Ps latently infected with AD169 (originating from nt 171,867) and novel UL123ast observed in HELs productively infected with Towne (originating from nt 171,870). (B) Sense-antisense RNA levels of the viral genes UL21.5, UL123 and the cellular gene GAPDH in virions. The y-axis represents the log10-transformed copy number of each RNA. (C) The virion protein pp65 was used as an indicator of HCMV entry by IFA assay. Cells on cover slips were stained with monoclonal antibodies to pp65 (left) and the nuclear dye Hoechst (right). (D) Sense-antisense RNAs levels of UL21.5 and UL123, either in the presence or absence of ActD by real time RT-PCR assay. The average Ct values, which are inversely proportional to the amount of the target nucleic acid in the samples, were compared for each RNA between these two groups. Data were analyzed using Student’s unpaired t-test (one-tailed) and are presented as mean ± standard deviation (SD) (n = 2). Values of *p<0.05 were considered statistically significant
Table 1.
Target genes and primers used in qRT-PCR and RACE
| Gene | Forward primer (5′ location, nt) | Reverse primer (5′ location, nt) |
|---|---|---|
| a UL123 | 5′CACGACGTTCCTGCAGACTA3′ (173048) | 5′TTTTCAGCATGTGCTCCTTG3′ (172655) |
| a UL21.5 | 5′GCTTTGGCGGCACCTTCTCA3′ | 5′TTCGCTGCCATCTCCGTCTGTA3′ |
| b GAPDH | 5′GAAGGTGAAGGTCGGAGTC3′ | 5′GAAGATGGTGATGGGATTTC3′ |
| a F-GSP primer | 5′CAGCGGGTCTCCCAGACTCAGCTGACT3′ (172982) | – |
| a F-nest-GSP primer | 5′GGTGACTGCAGAAAAGACCCATGGA3′ (173268) | – |
| a R-GSP primer | – | 5′GCGGGAGATGTGGAT GGCTTGTAT3′ (172462) |
| a R-nest-GSP primer | – | 5′GAGAGACAAGGTGCTCACGCACATTGA3′ (172168) |
Accession no. FJ616285.1
Accession no. is J04038.1
In order to determine whether the virion RNAs could be delivered into target cells, actinomycin D (ActD), an inhibitor of transcription, was applied at a concentration of 5 μg/ml to inhibit de novo RNA synthesis. Briefly, HEL cells were incubated with a high MOI of Towne (MOI = 10) at 4 °C for 1 h in the presence (ActD+) or absence of ActD (ActD−), and the cultures were then shifted to 37 °C for 1.5 h in the presence or absence of ActD. The cells were then collected for immunofluorescence assay (IFA) and qRT-PCR. In addition, a blank group without either Towne or ActD treatment was used as a control. pp65, the most abundant tegument protein, was stained at 1.5 hpi for tracking viral entry by IFA [21]. Virus entry was identical in cells either treated or untreated with ActD, as distinct nuclear localization of pp65 at the immediate early time (1.5 hpi) of HCMV infection (entry stage) was observed in both cases (Fig. 1C). Two hundred fifty ng of total RNA from each sample was used to determine the sense-anti-sense RNAs levels of UL21.5 and UL123 in the presence or absence of ActD using the qRT-PCR assay described above. As shown in Fig. 1D, the UL21.5 and UL123 sense-antisense pairs were still readily detectable when transcription was inhibited by treatment with ActD, with a Ct value of 19.42 (sense)/26.89 (antisense) and 25.77 (sense)/30.02 (antisense), respectively. This indicated that the sense-antisense RNA pairs are present in considerable amounts and that a variety of other RNAs exist in HCMV virions that can also be transported into infected cells. Furthermore, the levels of UL123 sense-antisense pairs were higher in the infected cells that were not treated with ActD, indicating that the UL123 sense-antisense RNA pairs were synthesized during HCMV infection, even at the immediate early infection stage (1.5 hpi). This strongly suggests that this transcript may be related to regulatory functions upon and during infection. A recent study has demonstrated that EBV virion RNAs can deploy immediate functions, enhancing the viruses’ capacity to transform primary B cells [9]. Whether the imported UL123ast also plays a similar role in viral infection needs to be further investigated in future work.
Having established that UL123ast exists in the HCMV virion and enters infected cells, the first step for the functional study was to investigate the structure(s) of the UL123asts. Since it is extremely difficult to obtain sufficient virion RNAs for identifying the UL123ast structure in HCMV virions, in order to determine the transcription start and end sites, HEL cells were infected with Towne (MOI = 5) in the presence of a protein synthesis inhibitor, cyclo-heximide (CHX, 100 μg/ml), for 1h, and total RNA was then extracted and subjected to RACE assays. To identify their 3′-ends, PCR products were TA cloned and four clones were selected for further analysis. All of them exhibited an identical polyadenylylation site, which was consistent with the results of previous studies using AD169-infected GM-Ps [17]. The site corresponded to nt 173,821 of the Towne sequence and nt 173,331 of the AD169 sequence, respectively (Fig. 1A). The identified sequence was 5′-AAATAATAAATGGGACCCCATCCTGTA-3′. To identify their 5′-ends, PCR products were TA cloned and 16 colonies were sequenced. Sequences of these colonies revealed that they had different 5′ ends, which were mapped to positions corresponding to nt 171,847 (one cDNA clone), nt 171,870 (five cDNA clones), nt 171,892 (four cDNA clones), nt 171,921 (three cDNA clones), nt 171 985 (two cDNA clones) and nt 172,003 (one cDNA clone), respectively, of the Towne sequence (Table 2). In contrast to our results, UL123ast has not yet been identified in lyticly infected human fibroblasts [17, 18] infected with a different laboratory strain, AD169. In this study, we also failed to identify the transcriptional start site of this transcript in AD169-infected HELs, although the results from qRT-PCR and 3′-RACE assays indicated that the antisense transcript of UL123 is present during productive infection, and its 3′ polyadenylation site is the same as that in latent infection [17] (data not shown). It is possible that these UL123asts exist in AD169-infected HELs, but the level is probably lower than in Towne-infected HELs. All clones of the 5′-RACE products that we identified initiate at a site downstream from the transcriptional start site found in latently infected cells (GM-Ps). It is uncertain whether such differential utilization of transcriptional start sites between latent and productive infection can be attributed to differences among the HCMV strains, but the sense MIE genes have been reported to be transcribed in a similar manner [17, 22]. It was found that the sense transcripts of MIE genes could be transcribed from either the productive infection transcription start site (PSS) or the latent infection transcription start sites (LSSs: LSS-1 and LSS-2) positioned about 300 nucleotides upstream of PSS (Fig. 1A). During latent infection, LSSs transcripts were dominant. During productive infection, transcripts initiated from PSS were detectable throughout the infection and were translated into UL122 (IE2) and UL123 (IE1) viral proteins. The levels of LSSs transcripts were very low and could not be detected at the very early times of the IE stage at 2 hpi, and it was only at very late times, such as 5 dpi and 7 dpi that they reached detectable levels [22]. These accumulated LSSs transcripts may regulate virus replication at the late stage during productive infection. In this study the infection was performed in the presence of a protein synthesis inhibitor, CHX, which was used to selectively accumulate IE transcripts [23, 24]. The creation of IE-like conditions may be the major reason for inducing the transcription of UL123ast downstream from the previously reported sites (designated as LSSs-ast) (Fig. 1A). At the immediate early stage of infection (IE stage), it is possible that the transcripts from the site downstream of LSSs-ast are the dominant products and, in contrast, no or only a small amount of UL123ast is transcribed from LSSs-ast. Compared to the transcripts starting from LSSs-ast, most of the transcripts identified in our study lacked the complete sequence of ORF59. A study of whether these UL123ast (originating from different TSSs) and their corresponding ORFs play different roles in regulation of HCMV infection will be carried out in the near future. It has been reported that both viral and cellular RNA transcripts are nonspecifically packed into HCMV virions in proportion to their intracellular concentration at the time of assembly [3]. We thus speculated that the UL123ast in virions is most likely a mixture of transcripts.
Table 2.
Determination of TSSs by 5′RACE analysis of UL123ast in Towne-infected HEL cells
| DNA sequence (5′-3′) | Start site | Number of clones |
|---|---|---|
| Tcggcgatggc--tcatcc--ctctca--tgggca--tggtta--tggccagca | 171847 | 1 |
|
|
171870 | 5 |
|
|
171892 | 4 |
|
|
171921 | 3 |
|
|
171985 | 2 |
|
|
172003 | 1 |
↓ Arrows indicate TSSs as determined in individual clones of the 5′RACE analysis
atg represents the start codon of ORF59
In conclusion, we report the coexistence of sense and antisense transcripts in HCMV virions. Our findings provide additional information about the composition of the virion and suggest a possible regulatory mode prior to initiation of transcription. In addition, we mapped alternative TSSs of UL123ast in productively infected cells. Although we have yet to characterize UL123ast during the late phase of infection when virion assembly occurs, it appears certain that UL123ast in virions is a complex mixture of RNAs transcribed from heterogenic TSSs.
Acknowledgments
This project was supported by the National Natural Science Foundation of China (31000090 and 81071350), National Program on Key Basic Research Project (973 Program, 2011CB504800), “Hundred Talents Program” of the Chinese Academy of Sciences (2010-88), and RO1 AI20211 (to ESM).
Contributor Information
Cui-Qing Yang, State Key Laboratory of Virology, Wuhan Institute of Virology, Chinese Academy of Sciences, Wuhan 430071, China.
Ling-Feng Miao, State Key Laboratory of Virology, Wuhan Institute of Virology, Chinese Academy of Sciences, Wuhan 430071, China.
Xing Pan, State Key Laboratory of Virology, Wuhan Institute of Virology, Chinese Academy of Sciences, Wuhan 430071, China.
Cong-Cong Wu, State Key Laboratory of Virology, Wuhan Institute of Virology, Chinese Academy of Sciences, Wuhan 430071, China.
Simon Rayner, State Key Laboratory of Virology, Wuhan Institute of Virology, Chinese Academy of Sciences, Wuhan 430071, China.
Edward S. Mocarski, Department of Microbiology and Immunology, Emory Vaccine Center, Emory University School of Medicine, Atlanta, GA 30322, USA
Han-Qing Ye, State Key Laboratory of Virology, Wuhan Institute of Virology, Chinese Academy of Sciences, Wuhan 430071, China.
Min-Hua Luo, State Key Laboratory of Virology, Wuhan Institute of Virology, Chinese Academy of Sciences, Wuhan 430071, China.
References
- 1.Bresnahan WA, Shenk T. A subset of viral transcripts packaged within human cytomegalovirus particles. Science. 2000;288(5475):2373–2376. doi: 10.1126/science.288.5475.2373. [DOI] [PubMed] [Google Scholar]
- 2.Greijer AE, Dekkers CA, Middeldorp JM. Human cytomegalovirus virions differentially incorporate viral and host cell RNA during the assembly process. J Virol. 2000;74(19):9078–9082. doi: 10.1128/jvi.74.19.9078-9082.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Terhune SS, Schroer J, Shenk T. RNAs are packaged into human cytomegalovirus virions in proportion to their intracellular concentration. J Virol. 2004;78(19):10390–10398. doi: 10.1128/JVI.78.19.10390-10398.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sarcinella E, Brown M, Tellier R, Petric M, Mazzulli T. Detection of RNA in purified cytomegalovirus virions. Virus Res. 2004;104(2):129–137. doi: 10.1016/j.virusres.2004.03.008. [DOI] [PubMed] [Google Scholar]
- 5.Sciortino MT, Suzuki M, Taddeo B, Roizman B. RNAs extracted from herpes simplex virus 1 virions: apparent selectivity of viral but not cellular RNAs packaged in virions. J Virol. 2001;75(17):8105–8116. doi: 10.1128/JVI.75.17.8105-8116.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bechtel J, Grundhoff A, Ganem D. RNAs in the virion of Kaposi’s sarcoma-associated herpesvirus. J Virol. 2005;79(16):10138–10146. doi: 10.1128/JVI.79.16.10138-10146.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lin X, Li X, Liang D, Lan K. MicroRNAs and unusual small RNAs discovered in Kaposi’s sarcoma-associated herpesvirus virions. J Virol. 2012;86(23):12717–12730. doi: 10.1128/JVI.01473-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cliffe AR, Nash AA, Dutia BM. Selective uptake of small RNA molecules in the virion of murine gammaherpesvirus 68. J Virol. 2009;83(5):2321–2326. doi: 10.1128/JVI.02303-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Jochum S, Ruiss R, Moosmann A, Hammerschmidt W, Zeidler R. RNAs in Epstein-Barr virions control early steps of infection. Proc Nat Acad Sci USA. 2012;109(21):E1396–E1404. doi: 10.1073/pnas.1115906109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zhang Y, Liu XS, Liu QR, Wei L. Genome-wide in silico identification and analysis of cis natural antisense transcripts (cis-NATs) in ten species. Nucleic Acids Res. 2006;34(12):3465–3475. doi: 10.1093/nar/gkl473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Arnold J, Yamamoto B, Li M, Phipps AJ, Younis I, Lairmore MD, Green PL. Enhancement of infectivity and persistence in vivo by HBZ, a natural antisense coded protein of HTLV-1. Blood. 2006;107(10):3976–3982. doi: 10.1182/blood-2005-11-4551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gaudray G, Gachon F, Basbous J, Biard-Piechaczyk M, Devaux C, Mesnard JM. The complementary strand of the human T-cell leukemia virus type 1 RNA genome encodes a bZIP transcription factor that down-regulates viral transcription. J Virol. 2002;76(24):12813–12822. doi: 10.1128/JVI.76.24.12813-12822.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Vanhee-Brossollet C, Thoreau H, Serpente N, D’Auriol L, Levy JP, Vaquero C. A natural antisense RNA derived from the HIV-1 env gene encodes a protein which is recognized by circulating antibodies of HIV+ individuals. Virology. 1995;206(1):196–202. doi: 10.1016/s0042-6822(95)80034-4. [DOI] [PubMed] [Google Scholar]
- 14.Higgins GD, Uzelin DM, Phillips GE, Burrell CJ. Presence and distribution of human papillomavirus sense and antisense RNA transcripts in genital cancers. J Gen Virol. 1991;72(Pt 4):885–895. doi: 10.1099/0022-1317-72-4-885. [DOI] [PubMed] [Google Scholar]
- 15.Xu Y, Ganem D. Making sense of antisense: seemingly noncoding RNAs antisense to the master regulator of Kaposi’s sarcoma-associated herpesvirus lytic replication do not regulate that transcript but serve as mRNAs encoding small peptides. J Virol. 2010;84(11):5465–5475. doi: 10.1128/JVI.02705-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kondo K, Kaneshima H, Mocarski ES. Human cytomegalovirus latent infection of granulocyte-macrophage progenitors. Proc Nat Acad Sci USA. 1994;91(25):11879–11883. doi: 10.1073/pnas.91.25.11879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kondo K, Xu J, Mocarski ES. Human cytomegalovirus latent gene expression in granulocyte-macrophage progenitors in culture and in seropositive individuals. Proc Nat Acad Sci USA. 1996;93(20):11137–11142. doi: 10.1073/pnas.93.20.11137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zhang G, Raghavan B, Kotur M, Cheatham J, Sedmak D, Cook C, Waldman J, Trgovcich J. Antisense transcription in the human cytomegalovirus transcriptome. J Virol. 2007;81(20):11267–11281. doi: 10.1128/JVI.00007-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Baldick CJ, Jr, Shenk T. Proteins associated with purified human cytomegalovirus particles. J Virol. 1996;70(9):6097–6105. doi: 10.1128/jvi.70.9.6097-6105.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Patrone M, Secchi M, Fiorina L, Ierardi M, Milanesi G, Gallina A. Human cytomegalovirus UL130 protein promotes endothelial cell infection through a producer cell modification of the virion. J Virol. 2005;79(13):8361–8373. doi: 10.1128/JVI.79.13.8361-8373.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Luo MH, Schwartz PH, Fortunato EA. Neonatal neural progenitor cells and their neuronal and glial cell derivatives are fully permissive for human cytomegalovirus infection. J Virol. 2008;82(20):9994–10007. doi: 10.1128/JVI.00943-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lunetta JM, Wiedeman JA. Latency-associated sense transcripts are expressed during in vitro human cytomegalovirus productive infection. Virology. 2000;278(2):467–476. doi: 10.1006/viro.2000.0666. [DOI] [PubMed] [Google Scholar]
- 23.Ma Y, Wang N, Li M, Gao S, Wang L, Ji Y, Qi Y, He R, Sun Z, Ruan Q. An antisense transcript in the human cytomegalovirus UL87 gene region. Virol J. 2011;8:515. doi: 10.1186/1743-422X-8-515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Snaar SP, Verdijk P, Tanke HJ, Dirks RW. Kinetics of HCMV immediate early mRNA expression in stably transfected fibroblasts. J Cell Sci. 2002;115(Pt 2):321–328. doi: 10.1242/jcs.115.2.321. [DOI] [PubMed] [Google Scholar]