Molecular biology has transformed the treatment of solid tumours over the last decade. By improving through better biomarkers for traditional standard of care (Mulligan et al, 2014), delivering the promise of targeted therapeutics (Tan and Lynch, 2012) or opening the avenue of immune checkpoint therapeutics (Sharma and Allison, 2015), the landscape of oncology treatment has changed substantially, and numerous new targeted therapies and predictive diagnostics have been added to the known ER/Tamoxifen and Her2/Trastuzumab examples. Genomic medicine, imperfect as it may be (Tannock and Hickman, 2016), is providing some patients with longer, better lives. And yet, ∼1 in 4 patients with cancer will have these new therapeutics denied in the near future because of the way national grant agencies and research-focused charities are prioritising the administration of scientific resources. The work by Alvi et al (2017), published in this issue of the BJC, supported by a generous donation from a cancer patient, is an example of the difficulty of aligning competitive funds to a neglected area of cancer research.
Although some of the issues regarding rare cancers are shared by many other diseases, there are specific questions in relation to funding that are probably specific to rare cancers in adults. Indeed, rare cancers comprise a staggering 198 varieties (a complete list can be obtained at http://www.rarecarenet.eu/rarecarenet/index.php/cancerlist) and are primarily grouped according to the site of origin. (Rare Cancers Europe, 2017; RARECARE, 2017). Some rare cancers are clear-cut independent disease entities of low frequency (e.g., small bowel adenocarcinoma with only ∼3000 cases diagnosed yearly in the US) whereas others are unusual subtypes of mainstream cancers (e.g., colorectal signet ring cell carcinoma comprises ∼1% of all colorectal carcinomas; Nitsche et al, 2013). The latter is important because of the way we typically undertake research – such unusual subtypes are subsumed in large cohorts of common cancers, with no statistical power to discern their potential genetic uniqueness and very often they simply remain undetected, hidden amongst the common subtypes. Although there is no agreement on a precise definition, rare cancers are defined as those with an incidence rate of <6 per 100 000 persons per year, whereas in the US it is <15 per 100 000 (Greenlee et al, 2010; Cancer Research UK, 2016; RARECARE, 2017). Collectively, rare cancers represent ∼22% of all cancer cases diagnosed in the EU and 27% of these in the USA (National Cancer Institute Epidemiology and Genetics Research, 2017; Rare Cancers Europe, 2017). The development of clinical trials in rare cancers in both early and advanced disease settings has been a major challenge, but the International Rare Cancers Initiative has recently catalysed clinical trials across the UK, Europe, N. America, Japan and Australasia in a variety of rare tumours (Bogaerts et al, 2015 and see http://www.irci.info/).
Grant funding agencies and grant reviewers (most of whom may not have had any experience in rare cancer research) face a major problem in judging the quality of rare cancer research proposals. Indeed, it is expected that such applications should contain all the components that make a successful application in the common cancer paradigm: adequate sample calculations for biomarker discovery and validation, preliminary in vitro evidence of the research question, cell lines and animal models to test the research hypothesis, adequately designed and powered clinical trials and so on. While some of these shortcomings can be tackled with international consortia, others are very difficult if not possible to address in the space of rare cancer research. As a result, when looking at the main expenditure in cancer research by leading research funding agencies, the amount of funding is generally related to the single most prevalent forms, and does not include significant funding for rare cancer research (https://www.everydayhealth.com/cancer/cancer-research-where-funding-goes.aspx).
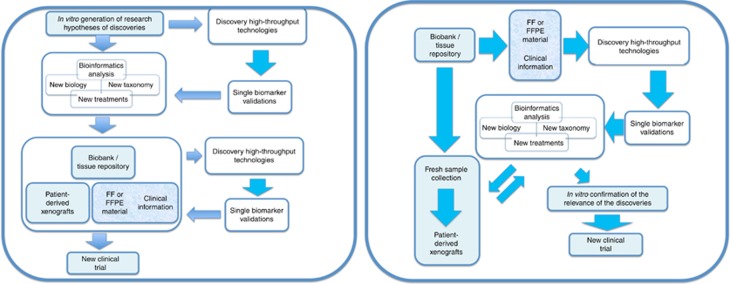
Figure 1 depicts the 2 main models of research design when interrogating cancer samples. On the right the ‘classic’ model is shown, where the in vitro analysis of cell lines and mouse models generates interesting hypotheses on disease mechanisms, disease characterisation and novel targets. This is then confirmed with analysis of clinical samples, either directly or via xenograft models, and the confirmation of the clinical relevance of those findings informs a possible clinical trial. The figure on the left provides a schematic of the ‘alternative’ model, where direct interrogation of the clinical materials generates the hypotheses that are then confirmed in vitro, ahead of clinical trial design.
Figure 1.
Schematic paths for research design in the context of translational research in cancer. In either model, we have highlighted in blue the activities that may need to be revisited (i.e. how we power a study in rare diseases) or avoided altogether (for instance, the use of animal models or cell lines that are usually not available for individual rare cancers) when designing discovery work in this field.
In either model, we have highlighted in blue the portions that become significantly challenged when undertaking research in rare cancer.
Despite the relative inability of cell culture models to reflect ‘clinical genomics’ (Gillet et al, 2013), or the lack of predictability of clinical outcomes associated with the use of mouse models (Vandamme 2014), both seem to appear as conditio sine qua non to fund a research programme in any cancer type. Similarly, patient derived xenografts seem to be essential for the monitoring of cancer dynamics and complexity (Aparicio et al, 2015). However, we know that, in rare cancers, generating these resources in sufficient numbers or variety to make them meaningful will be an extremely difficult, if not impossible, task. Indeed, although the utility of these models needs to be recognised, the generation of comprehensive ‘omics’ from well-curated clinical sample collections is a reality today and may make some of the uses of these models redundant, particularly when the main goal of the programme is to find actionability/druggability or to describe dynamic determinants of therapeutics such as the cancer immune contexture.
The characterisation of large cohorts of clinical samples to power biomarker discovery studies in rare cancers is another difficult task, which can be in part mitigated by large, multi-national consortia. The problem here is that samples coming from multiple centres, from a disease that often does not have a nationally or internationally agreed standard of care, typically leads to samples with diverse therapeutic interventions and pathology curation, thus lacking the relative homogeneity of large cohorts of common cancers. This, together with the lack of existing clinical trials in rare cancers that could be informed by the results of genetic studies, makes the overall landscape of research programme argumentation difficult and frustrating.
In theory, the scientific community should not water down the definition of good research because of aspects that are operational rather than conceptual. And, yet, there is a strong need in the area of rare cancers to ‘start testing’, start collecting meaningful genomics information from whatever cohorts we may have, as imperfect as they may be, to inform better science in the future and start understanding alternative therapeutic interventions. Our current era has seen the acceptance of adaptive trial designs (Kaplan et al, 2013 and see http://www.focus4trial.org/), drug approvals based solely on early phase trial results (Kwak et al, 2010), and the ability to obtain whole genomic/transcriptomic/epigenetic information on small cancer cohorts that inform the overall nature of cancer to a point never achieved before with a candidate-based approach (Alvi et al, 2015). At the same time, we should not judge scientific proposals and scientific merit in rare cancer studies in the same way as we do for mainstream malignancies, as this may not only be scientifically flawed, but may also represent a strategic mistake that will deny therapeutic advancement to almost a quarter of all cancer patients.
Footnotes
The authors declare no conflict of interest.
References
- Alvi MA, Loughrey MB, Dunne P, McQuaid S, Turkington R, Fuchs MA, McGready C, Bingham V, Pang B, Moore W, Maxwell P, Lawler M, James JA, Murray GI, Wilson RH, Salto-Tellez M (2017) Molecular profiling of signet ring cell colorectal cancer provides a strong rationale for genomic targeted and immune checkpoint inhibitor therapies. BJC 117: 203–209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alvi MA, McArt DG, Kelly P, Fuchs MA, Alderdice M, McCabe CM, Bingham V, McGready C, Tripathi S, Emmert-Streib F, Loughrey MB, McQuaid S, Maxwell P, Hamilton PW, Turkington R, James JA, Wilson RH, Salto-Tellez M (2015) Comprehensive molecular pathology analysis of small bowel adenocarcinoma reveals novel targets with potential for clinical utility. Oncotarget 6: 20863–20874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aparicio S, Hidalgo M, Kung AL (2015) Examining the utility of patient-derived xenograft mouse models. Nat Rev Cancer 15: 311–316. [DOI] [PubMed] [Google Scholar]
- Bogaerts J, Sydes MR, Keat N, McConnell A, Benson A, Ho A, Roth A, Fortpied C, Eng C, Peckitt C, Coens C, Pettaway C, Arnold D, Hall E, Marshall E, Sclafani F, Hatcher H, Earl H, Ray-Coquard I, Paul J, Blay JY, Whelan J, Panageas K, Wheatley K, Harrington K, Licitra L, Billingham L, Hensley M, McCabe M, Patel PM, Carvajal R, Wilson R, Glynne-Jones R, McWilliams R, Leyvraz S, Rao S, Nicholson S, Filiaci V, Negrouk A, Lacombe D, Dupont E, Pauporté I, Welch JJ, Law K, Trimble T, Seymour M (2015) Clinical trial designs for rare diseases: Studies developed and discussed by the International Rare Cancers Initiative. Eur J Cancer 51(3): 271–281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cancer Research UK (2016) Available at http://www.cancerresearchuk.org.
- Gillet JP, Varma S, Gottesman MM (2013) The clinical relevance of cancer cell lines. J Natl Cancer Inst 105(7): 452–458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greenlee RT, Goodman MT, Lynch CF, Platz CE, Havener LA, Howe HL (2010) The occurrence of rare cancers in United States adults, 1995-2004. Public Health Rep 125: 28–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaplan R, Maughan T, Crook A, Fisher D, Wilson RH, Brown L, Parmar M (2013) Evaluating many treatments and biomarkers in oncology: a new design. J Clin Oncol 31(36): 4562–4568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwak EL, Bang YJ, Camidge DR, Shaw AT, Solomon B, Maki RG, Ou SH, Dezube BJ, Jänne PA, Costa DB, Varella-Garcia M, Kim WH, Lynch TJ, Fidias P, Stubbs H, Engelman JA, Sequist LV, Tan W, Gandhi L, Mino-Kenudson M, Wei GC, Shreeve SM, Ratain MJ, Settleman J, Christensen JG, Haber DA, Wilner K, Salgia R, Shapiro GI, Clark JW, Iafrate AJ (2010) Anaplastic lymphoma kinase inhibition in non-small-cell lung cancer. N Engl J Med 363: 1693–1703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mulligan JM, Hill LA, Deharo S, Irwin G, Boyle D, Keating KE, Raji OY, McDyer FA, O'Brien E, Bylesjo M, Quinn JE, Lindor NM, Mullan PB, James CR, Walker SM, Kerr P, James J, Davison TS, Proutski V, Salto-Tellez M, Johnston PG, Couch FJ, Paul Harkin D, Kennedy RD (2014) Identification and validation of an anthracycline/cyclophosphamide-based chemotherapy response assay in breast cancer. J Natl Cancer Inst. 106: djt335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- National Cancer Institute Epidemiology and Genetics Research (2017) Synergizing epidemiologic research on rare cancers. Available at http://epi.grants.cancer.gov/events/rare-cancers/.
- Nitsche U, Zimmermann A, Spath C, Muller T, Maak M, Schuster T, Slotta-Huspenina J, Käser SA, Michalski CW, Janssen KP, Friess H, Rosenberg R, Bader FG (2013) Mucinous and signet-ring cell colorectal cancers differ from classical adenocarcinomas in tumor biology and prognosis. Ann Surg 258: 775–782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rare Cancers Europe (2017) Available at http://www.rarecancerseurope.org/.
- RARECARE (2017) Available at http://www.rarecarenet.eu/rarecarenet/.
- Sharma P, Allison JP (2015) The future of immune checkpoint therapy. Science 348: 56–61. [DOI] [PubMed] [Google Scholar]
- Salto-Tellez M (2013) Overview of Molecular Tests and Personalized Cancer Medicine. In: Principles of Molecular Diagnostics and Personalized Cancer Medicine. Tan D, Lynch HT (eds). Wolters Kluwer Health: Philadelphia, PA, USA, pp 196–2015.
- Tannock IF, Hickman JA (2016) Limits to personalized cancer medicine. N Engl J Med 375: 1289–1294. [DOI] [PubMed] [Google Scholar]
- Vandamme TF (2014) Use of rodents as models of human diseases. J Pharm Bioallied Sci 6(1): 2–9. [DOI] [PMC free article] [PubMed] [Google Scholar]