Abstract
Chronic ethanol consumption is not only associated with the alteration of metabolic profiles in biofluids, but also the composition of the gut microbiome. Our understanding of the importance of the intestinal microbiota as well as the disturbances elicited by ethanol intervention is limited by the fact that previous analyses have primarily focused on biofluids and liver tissue metabolome; the metabolic profiles of the gastrointestinal (GI) contents are rarely investigated. In this study, we applied a metabonomics approach using a high performance liquid chromatography time of flight mass spectrometry (HPLC-TOFMS) and gas chromatography mass spectrometry (GC-MS) to characterize the metabolic alterations of the contents within the GI tract (stomach, duodenum, jejunum, ileum, cecum, colon, and rectum) in male Sprague Dawley rats following 8 weeks ethanol exposure. We obtained a snapshot of the distinct changes of the intestinal content metabolite composition in rats with ethanol exposure, which indicated a profound impact of ethanol consumption on the intestinal metabolome. Many metabolic pathways that are critical for host physiology were affected, including markedly altered bile acids, increased fatty acids and steroids, decreased carnitines and metabolites involved in lipid metabolism, a significant decrease of all amino acids and branched chain amino acids, and significantly decreased short chain fatty acids except for acetic acid which rapidly elevated as a product of ethanol metabolism. These results provide an improved understanding of the systemic alteration of intestinal metabolites in mammals and the interplay between the host and its complex resident microbiota, and may aid in the design of new therapeutic strategies that target these interactions.
Keywords: gastrointestinal tract, chronic ethanol consumption, short chain fatty acid, branched chain amino acid, metabonomics, gut microbiota, high performance liquid chromatography mass spectrometry, gas chromatography mass spectrometry
Table of Contents Synopsis
Ethanol consumption is associated with the alteration of metabolic profiles in the composition of the gut microbiome. Mass spectrometry based metabonomics study revealed distinct alterations of the intestinal content metabolites in the whole gastrointestinal tract (stomach, duodenum, jejunum, ileum, cecum, colon, and rectum) in male Sprague Dawley rats following 8 weeks ethanol exposure, which were characterized by increased fatty acids and steroids, a significant decrease of all amino acids and branched chain amino acids, and significantly decreased short chain fatty acids except for acetic acid.
Introduction
Chronic alcohol consumption is often associated with alcoholic liver diseases such as fatty liver, steatohepatitis, and cirrhosis,1, 2 which is a significant cause of morbidity and mortality globally.3, 4 An epidemiologic study has estimated that for every 1-liter increase in per capita alcohol consumption (independent of type of beverage), there was a 14% increase in cirrhosis in men and 8% increase in women,5 and in 2003, 44% of all deaths from liver disease were attributed to alcohol.6 According to a 2007 National Survey on Drug Use and Health (NSDUH), adult alcohol use is the most prevalent among Whites (59.8%), least among Asian Americans (38.0%), and of similar rates among Native Americans (i.e., American Indians and Alaska Natives; 47.8%), Hispanics (46.3%), and Blacks (43.8%). Native Americans have the highest prevalence (12.1%) of heavy drinking (i.e., five or more drinks on the same occasion for 5 or more of the past 30 days); followed by Whites (8.3%) and Hispanics (6.1%).7 In the last several decades, much research has been undertaken in rodent models to gain mechanistic insights into the biological effects of ethanol exposure.8–10 Recent metabonomics studies have been extensively conducted to profile global changes in endogenous urine, serum and liver metabolites in rodents in response to ethanol exposure11–16 and profound metabolic changes were observed, particularly in fatty acids13 and branched chain amino acids (BCAAs).17 It has been reported that chronic ethanol consumption is not only associated with the alteration of metabolic profiles in biofluids and liver, but also the content and composition of the gut microbiota.2, 18–21 It is also well documented that human gut microbiota can exert important health effects through the production of bacterial metabolites, including vitamins that are essential for human survival such as vitamins K and B,22 polyunsaturated fatty acids (PUFA) such as conjugated α-linolenic acid and conjugated linolenic acid, short chain fatty acids (SCFAs), neuroactive compounds such as 4-aminobutyric acid (GABA) and histamine,23 polysaccharide A and a variety of other proteins, peptides and nucleotides with immunomodulatory and anti-inflammatory properties.22 These changes of human enteric metabolites resulting from ethanol-induced alteration of the gut microbiota are important for understanding the disrupted whole-body metabolic homeostasis and the implication in the development of a variety of metabolic diseases such as liver conditions and cancer. Several works have been delineated the topographical metabolic variations in the intestinal contents and also demonstrated the potential of metabolic profiling as a useful approach for understanding host−microbiome interactions.24, 25 However, there are very few reports that systemically delineated the luminal content metabolic profiles in the whole gastrointestinal (GI) tract includes stomach, duodenum, jejunum, ileum, cecum, colon, and rectum, nor is there any information available on the dynamic changes of the metabolic profiles during chronic ethanol consumption that would allow us to understand such ethanol-mediated gut-liver metabolic interactions.
In this study, we applied a comprehensive metabolite profiling method using a high performance liquid chromatography time of flight mass spectrometry (HPLC-TOFMS) to characterize the metabolic alterations of the whole GI tract contents in stomach, duodenum, jejunum, ileum, cecum, colon, and rectum in male Sprague Dawley rats following 8 weeks ethanol consumption. BCAAs, closely related to ethanol induced liver injury,17 and SCFAs, the abundant intestinal metabolites, play important roles in the alcohol-induced pathophysiological process. However, current HPLC-TOFMS-based profiling approach may not be able to detect or generate accurate data due to the volatile properties of SCFAs and the very similar chemical and physical properties of leucine and isoleucine. Therefore, a targeted metabonomics approach using gas chromatography-mass spectrometry (GC-MS) was used to quantitatively measure specific metabolic panels of SCFAs and BCAAs, complementary to the HPLC-TOFMS-based profiling approach.
Materials and Methods
Chemicals and Reagents
All compound reference standards including SCFAs and BCAAs and propyl derivatives (propyl acetate, propyl propanoate, propyl isobutyrate, propyl butyrate, propyl isopentanoate, and propyl hexanoate) were purchased from Sigma-Aldrich (St. Louis, MO). Stable isotopes (acetic acid-d4, propanoic acid-d2, 2-methyl-butyric acid-d3, butyric acid-d2, pentanoic acid-d9, hexanoic acid-d3, heptanoic acid-d7, valine-d8, and leucine-d10) used for quantification were obtained from CDN Isotopes (Pointe-Claire, Quebec, Canada). HPLC grade propanol (PrOH), pyridine (Py), PCF, hexane, methanol, acetonitrile, water, ammonium acetate, and acetic acid were obtained from Sigma-aldrich (St. Louis, MO).
Animals and Ethanol Feeding Experiments
Animal experiments were carried out according to experimental procedures approved by the Institutional Animal Care and Use Committee. Three-month old male Sprague Dawley rats (Charles River, Wilmington, MA) were randomly divided into two groups: control group (n = 5) and ethanol consumption group (n = 9), and pair-fed with the modified Lieber-DeCarli control and ethanol liquid diet, respectively, for 8 weeks. The calories of the control liquid diet were derived 16% from protein, 34% from fat, and 50% from carbohydrate. The dietary compositions of protein and fat in the ethanol liquid diet were the same as in the control liquid diet, and the only difference was part of carbohydrate calories in the control liquid diet was replaced by ethanol in the ethanol liquid diet. To promote generation of alcohol toxicity, a step-wise ethanol feeding procedure was introduced for increasing alcohol intake and eliminating alcohol tolerance. The ethanol content (%, w/v) was 5%, 5.14%, 5.29% and 5.43% for 1–2, 3–4, 5–6 and 7–8 weeks, respectively. Accordingly, the ethanol calories consists of 35%, 36%, 37% and 38% total dietary calories for 1–2, 3–4, 5–6 and 7–8 weeks, respectively. The diet compositions of ingredients and calories are listed in Supplementary Table S1. To achieve equal daily calories intake, the ethanol group was fed ad libitum and the control group was pair-fed the amount consumed by the ethanol-fed mice in the prior day. All ingredients for the liquid diets were obtained from Dyets (Bethlehem, PA) with the exception of ethanol, which was purchased from Sigma-Aldrich (St. Louis, MO). Rats were anesthetized with isofluorane at the end of week 8 and GI contents were harvested for analysis. Duodenum was defined as pylorus to the ligament of Treitz. Because there are no anatomical boundaries apart jejunum and ileum, we followed a rough definition in a length ratio of 3:2 (jejunum : ileum) as described previously.26, 27
Assessment of Alcoholic Liver Injury
Blood samples were drawn from the dorsal vena cava. Serum was obtained by centrifuging the blood at 8,000 × g for 15 minutes at 4°C. Serum alanine aminotransferase (ALT) activity was colorimetrically measured using Infinity ALT Reagent provided by Thermo Scientific (Waltham, MA). Liver tissues were fixed in 10% formalin, and processed for paraffin embedding. Then paraffin sections were cut at 5 µm and processed for hematoxylin and eosin (H&E, Dako, Carpinteria, CA) staining to assess the histological features of steatosis and inflammation.
Metabolic Profiling
The metabolic profiling was performed according to our previous published method.28 GI contents (100 mg) were mixed with 500 µL of ice-cold water. The mixture was vortexed for 4 min and then centrifuged at 13,200 rpm at 4 °C for 10 min. A 300 µL aliquot of supernatant was transferred to a 2 mL tube and the pellets were further extracted with ice-cold methanol using the same protocol. Another 300 µL aliquot of supernatant was added to the same tube as the initial aliquot, and 10 µL of internal standard (5 µg/mL p-chlorophenylalanine in water) was added. The extraction was vortexed for 30 s and centrifuged at 13,000 rpm at 4 °C for 20 min. The resulting supernatant was used for LC-MS analysis.
An Agilent HPLC 1200 system equipped with a binary solvent delivery manager and a sample manager (Agilent Corporation, Santa Clara, CA) was used with chromatographic separations performed on a 4.6 × 150 mm 5 µm Agilent ZORBAX Eclipse XDB-C18 chromatography column. The LC elution conditions were optimized as follows: isocratic at 1% B (0–0.5 min), linear gradient from 1% to 20% B (0.5–9.0 min), 20–75% B (9.0–15.0 min), 75–100% B (15.0–18.0 min), isocratic at 100% B (18–19.5 min); linear gradient from 100% to 1% B (19.5–20.0 min) and isocratic at 1% B (20.0–25.0 min) with a flow rate of 0.4 mL/min. The column was maintained at 30 °C. A 5 µL aliquot sample was injected into the column. Mass spectral data was acquired using an Agilent model 6220 MSD TOF mass spectrometer equipped with a dual sprayer electrospray ionization source (Agilent Corporation, Santa Clara, CA). The system was tuned for optimum sensitivity and resolution before analysis. Agilent API-TOF reference mass solution kit was used to obtain accurate mass time-of-flight data in both positive and negative mode operation. The TOF mass spectrometry was operated with the following optimized conditions: (1) ES+ mode, capillary voltage 3.5 kV, nebulizer 45 psig, drying gas temperature 325 °C, drying gas flow 11 L/min, and (2) ES- mode, similar conditions as ES+ mode except the capillary voltage was adjusted to 3.0 kV. During metabolite profiling, both plot and centroid data were acquired for each sample from 50 to 1,000 Da over a 25 min analysis time.
The acquired data files from LC-TOF-MS were processed using Agilent MassHunter Qualitative Analysis Program (vB.05.00, Agilent) and XCMS package,29 respectively. The resulting data from the LC-MS platform was analyzed using multivariate statistical tools to establish characteristic metabolic profiles associated with different response phenotypes. Multivariate statistical analyses, including principal component analysis (PCA) and orthogonal partial least squares projection to latent structures-discriminant analysis (OPLS-DA) were performed by SIMCA-P 12.0 software (Umetrics, Umeå, Sweden). Variable importance in the projection (VIP) ranks the overall contribution of each variable to the OPLS-DA model, and those variables with VIP > 1.0 are considered relevant for group discrimination. In SIMCA-P package, a typical cross validation procedure was conducted by leaving 1/7th samples out in each round so as to validate the OPLS-DA model against overfitting. All of the differentially expressed compounds in GI content were selected by comparing the compounds in the ethanol intervention group with the control group using a univariate statistical analysis, Student’s t test. We regarded p values of < 0.05 as significant.
Metabolites annotation was performed by comparing the accurate mass (m/z) and retention time (Rt) of reference standards in our in-house library and the accurate mass of compounds obtained from the web-based resources such as the Human Metabolome Database (http://www.hmdb.ca/).
Quantitative Analysis of SCFAs and BCAAs
The quantitative analysis of SCFAs and BCAAs was performed according to our previous reported method.30
a) Sample preparation
Briefly, each accurate weighted GI content samples (50–150 mg) was mixed with a total of 1000 µL of 0.005 M aqueous NaOH containing IS (5 µg/mL hexanoic acid-d3), which then subjected to homogenization for 10 min and centrifuged at 13,200 g at 4 °C for 20 min. A 500 µL aliquot of supernatant was transferred into a 10 mL Corning disposable glass centrifuge tube, and 300 µL of water was added to this aliquot. An aliquot of 500 µL PrOH/Py mixture solvent (3:2, v/v) and 100 µL of PCF were subsequently added to the glass tube. After briefly vortexed (10 s), the derivatization reaction proceeded under ultrasonication for 1 min. After derivatization, the derivatives were extracted by a two-step extraction with hexane and an aliquot of 300 µL derivative extraction (upper hexane layer) was transferred to a sampling vial. An aliquot of 10 µL n-alkane series was added, serving as the retention index and quality control. The resultant mixture was briefly vortexed prior to GC-MS analysis.
b) GC-MS analysis
Each 1 µL aliquot of derivatives was injected in splitless mode into an Agilent 7890A gas chromatography system coupled to an Agilent 5975C inert XL EI/CI mass spectrometric detector (MSD, Agilent, Santa Clara, CA). Chromatographic separation was performed on an HP-5ms capillary column coated with 5% phenyl-95% methylpolysiloxane (30 m × 250 µm i.d., 0.25 µm film thickness, Agilent J & W Scientific, Folsom, CA) with helium as carrier gas at a constant flow rate of 1 mL/min. The initial oven temperature was held at 50 °C for 2 min, ramped to 70 °C at a rate of 10 °C min−1, to 85 °C at a rate of 3 °C min−1, to 110 °C at a rate of 5 °C min−1, to 290 °C at a rate of 30 °C min−1, and finally held at 290 °C for 8 min. The temperatures of the front inlet, transfer line, and electron impact (EI) ion source were set at 260, 290, and 230 °C, respectively. The electron energy was −70 eV, and the mass spectral data was collected in a full scan mode (m/z 30–600).
c) Data analysis
Raw GC-MS data files were converted to NetCDF files using the Agilent’s MSD ChemStation Data Analysis Application and subsequently the NetCDF data files were then imported to ChromaTOF (v4.32, Leco Corp., St. Joseph, MO) for extracting data information. Compound identification was performed by comparing both MS spectra and retention times with those of standard compounds. The peak area of each derivatized SCFA or BCAA was calculated using the unique mass selected by ChromaTOF and exported as a .csv file that included sample names, compounds, RT, quantification mass, and peak area for further statistical analysis. A student’s t test was used to investigate differences between the groups in metabolite measurements. We regarded p values of < 0.05 as significant.
Results
General Information about the Animal Experiment
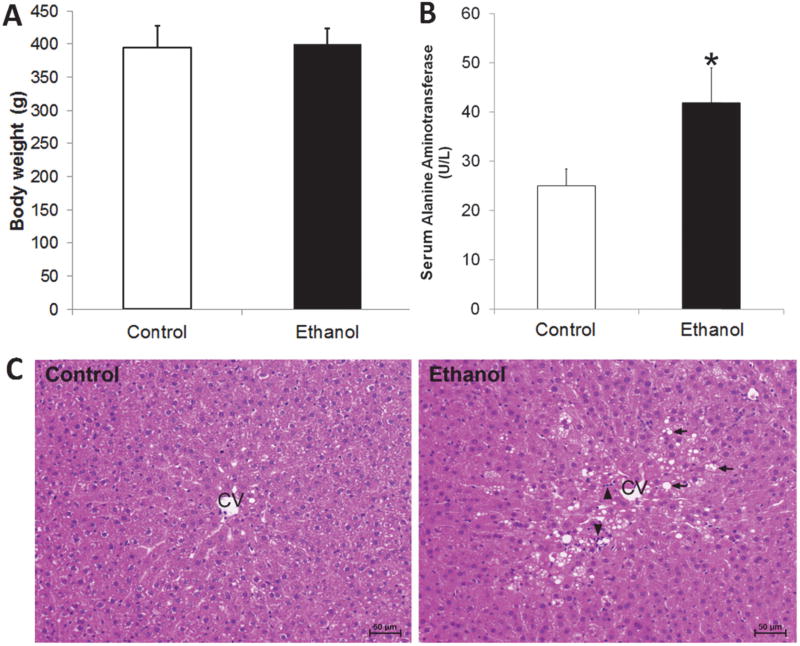
As indicated in Figure 1A, there was no statistically significant difference on the body weight between the control group and the chronic ethanol consumption group. The activity of serum alanine aminotransferase (U/L) was markedly increased due to ethanol consumption (p < 0.05, Figure 1B). Ethanol exposure caused lipid accumulation and inflammatory cell infiltration in the liver as indicated by hematoxylin and eosin (HE) staining (Figure 1C, arrowhead).
Figure 1.
Liver injury in rats chronically fed ethanol for 8 weeks. (A) The body weight of rats in control group and chronic ethanol consumption group; (B) The serum alanine aminotransferase activity of rats in chronic ethanol consumption group was significantly increased compared to controls (*, p < 0.05); (C) Liver histopathology (hematoxylin and eosin (HE) staining). Arrows indicate lipid droplets in hepatocytes and arrowheads indicate inflammatory cell infiltration. CV, central vein; Scale bar = 50 µm.
Ethanol Content in Serum and GI Contents
We measured the ethanol levels in serum and in the content of ileum, cecum and colon in control rats and rats with chronic ethanol consumption (Table 1). The ethanol content in ethanol consumption group was significantly increased compared to the control group. While within each group, there’s no significant difference in ethanol content among different GI regions.
Table 1.
Ethanol content in serum and gastrointestinal contents in control rats and rats under ethanol consumption for 8 weeks.
| Ethanol content (mean ± SD) | ||||
|---|---|---|---|---|
|
| ||||
| Group | Serum (mmol/L) | Ileum (mmol/g) | Cecum (mmol/g) |
Colon (mmol/g) |
| Control | 0.791 ± 0.596 | 1.128 ± 0.241 | 1.962 ± 1.024 | 1.467 ± 0.796 |
| Ethanol | 40.361 ± 4.658* | 37.574 ± 10.739* | 23.896 ± 6.454* | 31.153 ± 7.305* |
P<0.05, significantly different from control group.
Metabolic Profiles of GI Contents in Control Rats
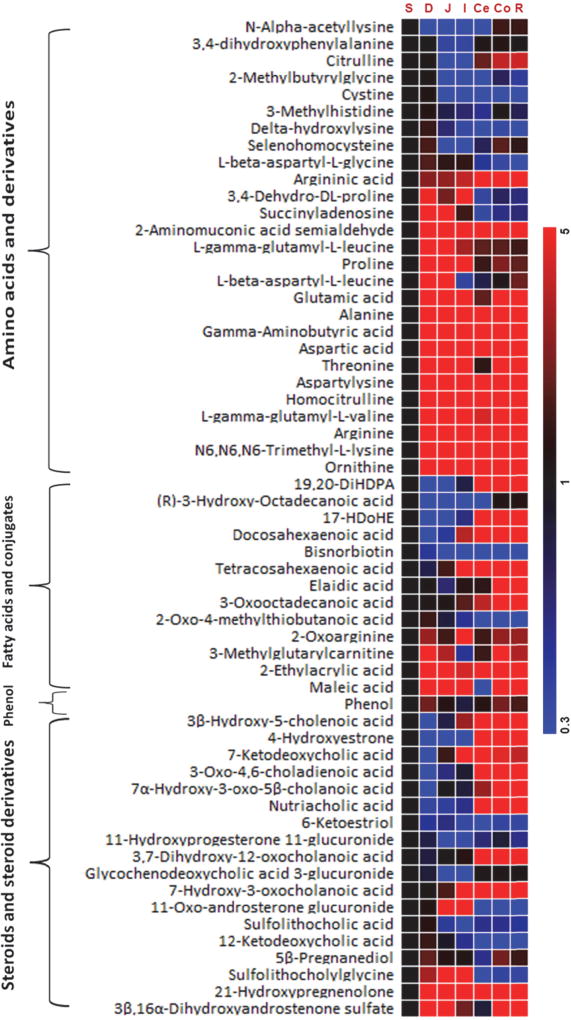
The metabolic profile of the GI content of control rats was systemically characterized by LC-TOFMS, which was delineated by the heatmap of the GI metabolites showing metabolic variations from stomach to small intestine and then large intestine in Figure 2. A PCA model was initially constructed for the content metabolic profiles of the whole GI tract (stomach, duodenum, jejunum, ileum, cecum, colon and rectum). The first two principal components accounted for 40% and 26% of the total variance in the combined multi-compartment data, respectively, and contributed to the separation of the small bowel from the large intestine, and among the ileum, the duodenum and the jejunum (Supplementary Figure S1A). The concentrations of amino acids, steroids, fatty acids, lipids, carnitines and phenols altered significantly from contents of the stomach to the duodenum, the jejunum, the ileum, the cecum, the colon, and then the rectum (Supplementary Figure S2–S4 and Supplementary Table S2). Significantly altered amino acids include alanine, arginine, argininic acid, aspartic acid, citrulline, γ-aminobutyric acid, glutamic acid, ornithine, and threonine. Altered steroids include 3,7-dihydroxy-12-oxocholanoic acid, 3β-hydroxy-5-cholenoic acid, 3-oxo-4,6-choladienoic acid, 4-hydroxyestrone, 7α-hydroxy-3-oxo-5β-cholanoic acid, 7-ketodeoxycholic acid, nutriacholic acid. Altered fatty acids include 3-methylglutarylcarnitine, (R)-3-hydroxy-octadecanoic acid, 2-ethylacrylic acid, 3-oxooctadecanoic acid, docosahexaenoic acid, elaidic acid, maleic acid, and tetracosahexaenoic acid. Altered metabolites involved in lipid metabolism are sphinganine, sphingosine, phytosphingosine, acetylcholine, and choline (Supplementary Figure S3). The levels of acetylcholine were significantly increased in jejunum, ileum, cecum, colon and rectum compared to it in stomach (p<0.05). The levels of sphinganine and phytosphingosine were higher in the contents of duodenum, jejunum, ileum, cecum, colon, and rectum compared to those in stomach. In addition, the levels of sphinganine were significantly increased in ileum and cecum contents (p<0.05), while for phytosphingosine, it was significantly increased in cecum, colon and rectum contents (p<0.05) compared to those in stomach. Altered carnitines include carnitine, 3-dehydroxycarnitine, and 3-methylglutarylcarnitine (Supplementary Figure S3). The levels of carnitine were significantly increased in the contents of jejunum and ileum, the levels of 3-dehydroxycarnitine were significantly increased in the contents of cecum, colon and rectum, and the level of 3-methylglutarylcarnitine was significantly increased in the contents of ileum, cecum and colon compared to those in stomach (p<0.05).
Figure 2.
Heatmap of the representative intestinal content metabolites in control rats. Heatmap shows changes in metabolites compared to stomach at stomach, duodenum, jejunum, ileum, cecum, colon and rectum. Shades of red and blue represent fold increase and fold decrease of a metabolite, respectively, in duodenum, jejunum, ileum, cecum, colon and rectum relative to stomach (see color scale). Each cell in the heatmap represents the fold change of a particular metabolite, which is the ratio of the concentration in the contents of duodenum, jejunum, ileum, cecum, colon or rectum to that in the stomach.
* S, stomach; D, Duodenum; J, Jejunum; I, Ileum; Ce, Cecum; Co, Colon; R, Rectum
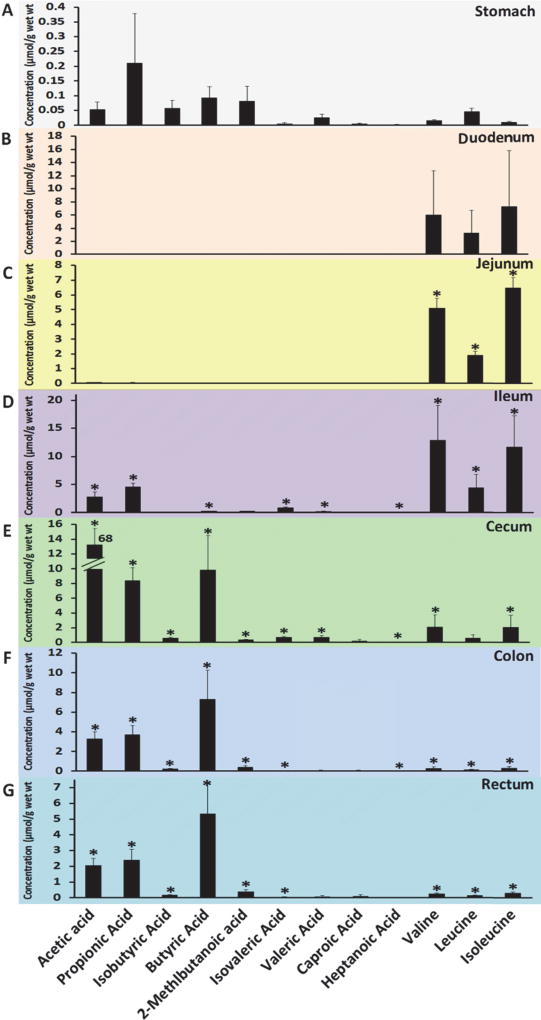
Specific metabolite panels, such as the SCFAs (acetic acid, propionic acid, isobutyric acid, butyric acid, 2-methlbutanoic acid, isovaleric acid, valeric acid, caproic acid, and heptanoic acid) and BCAAs (leucine, isoleucine and valine) were quantitatively measured by GC-MS in the GI contents in the stomach, duodenum, jejunum, ileum, cecum, colon, and rectum (Figure 3). The concentrations of all measured SCFAs in the contents of ileum, cecum, colon and rectum were significantly increased compared to those in stomach (p<0.05) and reach the highest in cecum, and then their levels were gradually decreased in colon and rectum contents (Figure 3 and Supplementary Table S3). The three SCFAs, namely acetic acid, propionic acid, and butyric acid, were most abundant in the entire intestinal content, particularly in ileum, cecum, colon, and rectum. For example, the concentration of acetic acid was the highest in the cecum content (68.03 µmol/g), but only a very low level of acetic acid was detected in the stomach (0.053 µmol/g) and duodenum and jejunum content (under detection limit). The concentrations of isobutyric acid, 2-methlbutanoic acid, isovaleric acid, valeric acid, caproic acid, and heptanoic acid were too low to be detected in duodenum and jejunum contents.
Figure 3.
Short chain fatty acids and branched chain amino acids profiles of the GI contents (A, stomach; B, Duodenum; C, Jejunum; D, Ileum; E, Cecum; F, Colon; G, Rectum) of control rats. Values are mean concentration (µmol/g intestinal contents) ± SEM measured using GC-MS. * P < 0.05 different from stomach concentration of the same SCFAs and BCAAs.
Contrary to SCFAs levels in GI contents, the levels of three BCAAs, valine, leucine and isoleucine, were predominant in the contents of duodenum, jejunum and ileum and of the highest concentration in ileum content. The BCAAs levels were relatively low in cecum, colon and rectum contents compared to those in the upper part of the intestine, but were all significantly higher compared to stomach (p<0.05, Figure 3 and Supplementary Table S4).
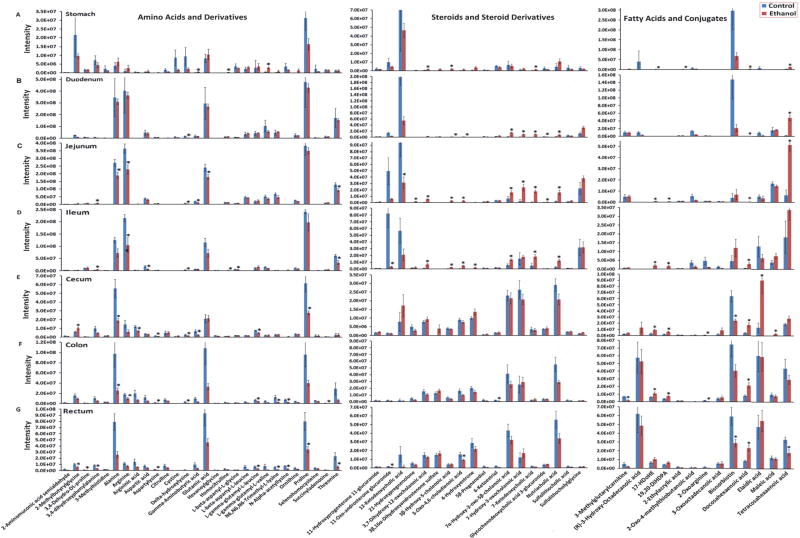
Chronic Ethanol Consumption Alters the Metabolic Profiles of GI Contents
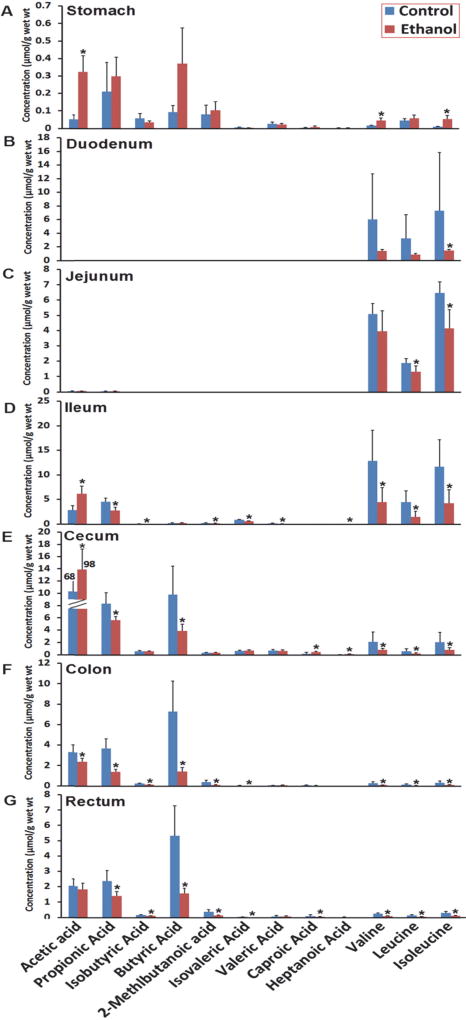
A PCA model was constructed for the metabolic profile of contents of the whole GI tract (stomach, duodenum, jejunum, ileum, cecum, colon and rectum) for control rats and rats under chronic ethanol consumption (Supplementary Figure S1B). Clear separation was also observed between the small bowel from the large intestine as well as the separation between the control group and the chronic ethanol consumption group. The bar plots in Figures 4 and 5 indicated distinct metabolite signatures for the contents of different intestinal regions in rats between chronic ethanol consumption group and control group. Interestingly, ethanol consumption resulted in a significant decrease in all amino acids in contents of the whole GI tract (Figure 5). The levels of steroids were significantly increased after ethanol consumption in stomach, duodenum, and jejunum contents to ileum. Most of the fatty acids detected were at higher levels after chronic ethanol consumption. There’s no significant alteration of phenol and derivatives after ethanol intervention (Supplementary Figure S4). Carnitines and metabolites involved in lipid metabolism were decreased after chronic ethanol consumption (Supplementary Figure S3).
Figure 4.
Alteration of short chain fatty acids and branched chain amino acids in the GI contents (A, stomach; B, Duodenum; C, Jejunum; D, Ileum; E, Cecum; F, Colon; G, Rectum) due to ethanol consumption. Values are mean concentration (µmol/g intestinal contents) ± SEM measured using GC-MS. *P < 0.05 vs. Control group.
Figure 5.
Alteration of metabolic profiles (A–G) in GI contents (A, stomach; B, Duodenum; C, Jejunum; D, Ileum; E, Cecum; F, Colon; G, Rectum) due to ethanol consumption. Values are mean intensities ± SEM measured using LC-MS. *P < 0.05 vs. Control group.
As described above, the concentration of SCFAs in control rats increased steadily from the stomach to cecum (0.05 to 68.30 µmol/g) and then decreased in colon and rectum (68.03 to 2.18 µmol/g). The same trend was observed in the ethanol group (0.32 to 98.50 µmol/g from stomach to cecum and 98.50 to 1.86 µmol/g) (Figure 4 and Supplementary Table S3). In the groups administrated with ethanol, acetic acid was significantly (p<0.05) elevated in stomach, ileum and cecum contents, whereas, propanoic acid, 2-methyl-propanoic acid, butyric acid, 2-methyl-butyric acid, 3-methyl-butyric acid, and heptanoic acid were significantly (p<0.05) decreased in ileum, colon, cecum and rectum contents. The SCFAs profiles for all GI tract contens in the ethanol-treated rats were abundant with acetic acid, propionic acid and butyric acid, with other SCFAs present in small amounts (15% in stomach, 0% in duodenum and jejunum, 10% in ileum, 3% in colon, 6% in cecum and 7% in rectum contents), so as in the control rats that the concentrations of acetic, propanoic, and butyric acid, were predominant (67% in stomach, 100% in duodenum and jejunum, 85% in ileum, 97% in colon, 94% in cecum and 93% in rectum) in all intestinal content samples (see pie plots in Supplementary Figure S5). Finally, a very low level of SCFAs was detected in the duodenums and jejunums contents of both control and ethanol-treated rats.
Similarly, the three BCAAs in luminal contents were steadily increased from stomach to ileum and then decreased from ileum to rectum (Figure 4 and Supplementary Table S4). Ethanol consumption led to significantly lower levels (p<0.05) of all three BCAAs in luminal contents of the ileum, cecum, colon and rectum relative to control rats.
Discussion
In this study, we examined the metabolic profiles of the GI contents in normal rats and determined how those metabolic profiles were impacted by ethanol consumption. Serum alanine aminotransferase, one of the widely used markers in evaluating the degree of liver injury,31 was markedly increased due to ethanol consumption. Consistent with our previous study,1 lipid accumulation and inflammatory cell infiltration in the liver caused by ethanol exposure was found by the liver histopathology result. Chronic ethanol consumption resulted in a global metabolite alteration including significantly altered amino acids, fatty acids, steroids, lipids, carnitine, SCFAs and BCAAs in GI tract. Due to the volatile properties of SCFAs and the very similar chemical and physical properties of leucine and isoleucine, it’s difficult to measure them accurately with the current LC-MS-based metabolic profiling approach. In view of the important role of SCFAs and BCAAs play in physiological and pathological processes, we applied the GC-MS to measure them quantitatively. The variation in metabolic profiles of both control rats and rats exposed to ethanol is indicative of the altered gut microbial composition leading to specific metabolic changes in each portion of the intestine, especially between small and large intestine.25,32, 33 The metabolites detected in the intestinal contents were mainly the endogenous metabolites of gut microbiota, metabolites excreted by epithelial cells of mammalian intestine, and food metabolites. Therefore, the dynamic changes of biochemical composition of the intestinal contents carry important information of ethanol-mediated gut microbial-host co-metabolism in the GI tract.
The levels of amino acids in the intestinal contents were decreased markedly from duodenum to jejunum and then ileum with a further decline when reached cecum and then remained unchanged from cecum to rectum (Supplementary Figure S2). This trend is consistent with the findings of previous report,24 which indicates that the absorption of amino acids in rats mainly occurred in jejunum and to a less extent in ileum.34 It was also reported that the gastric juice contains a coarse matrix of fat droplets together with major fragments of proteins and polysaccharides that are subsequently digested to both peptides and amino acids and monosaccharides respectively, which are absorbed in the duodenum and jejunum.35 Therefore, the observation of higher concentrations of amino acids in these tissues25 supports the essential role of the small intestine in catabolizing up to 50% of the dietary amino acids for energy production, intestinal de novo synthesis and maintenance of mucosa.34 These metabolic differences may have important implications for the utilization efficiency of dietary proteins and amino acids and their subsequent availability to extra-intestinal tissues. Notably, we observed high abundances of alanine, arginine, glutamic acid, proline and threonine in all the intestinal regions and a dramatic decrease after ethanol intervention (p<0.05). Since ethanol consumption can profoundly impact the gut microbial composition,20, 36 it is possible that the reduced abundance of amino acids in ethanol-treated rats is resulting from a disruption of the gut microbiota leading to a disrupted gut microbial-host co-metabolism.
BCAAs are essential nutrients obtained from food, as they cannot be synthesized de novo by mammals. Previous studies demonstrated that about 30% of the total ingested dietary leucine was extracted by dog small intestine37 and 20–30% of enterally available leucine was utilized in the first pass in human,38 suggesting substantial catabolism of dietary BCAA by the small intestinal mucosa in mammals. This also explains our observation that the high levels of BCAAs in duodenum, jejunum and ileum of both control and ethanol treated rats. Although not able to directly catabolize BCAAs, liver has a very active system for the degradation of the branched-chain α-keto acids derived from the corresponding BCAAs. The decreased BCAA levels in ethanol treated rats may be a result of the decreased BCAA catabolism due to decreased activity of the branched-chain α-keto acid dehydrogenase (BCKDH) complex in rat liver upon ethanol treatment.
SCFAs, such as acetic, propionic and butyric acids, are mainly produced by microbial fermentation of indigestible dietary fibers in the gut,39 or amino acids.40 SCFAs influence the gut microbiota by stimulating bifidobacteria growth while inhibiting gram-negative facultative and anaerobic bacteria.41 SCFAs are important energy sources for epithelial cells in the animal intestines, regulating the colonic and intracellular environment,42 and modulating cell proliferation and gene expression.43, 44 They also serve as fuel for active ion transportation in the large intestines.45 Consistent with previous reports,46 significantly elevated acetic acid levels after ethanol consumption is presumably due to the oxidation of ethanol to acetaldehyde and subsequently oxidized by the colonic mucosal or bacterial aldehyde dehydrogenase to acetic acid, which shown as protective effect against ethanol-induced damage. We also observed remarkable increases for SCFAs in the cecum compared with ileum and the levels of SCFAs appeared to be relatively high throughout the large intestine (Figure 3). This suggests that gut microbiota with capability of fermenting dietary fibers is mainly located in large intestine and may also be due to the increased degradation of amino acids in large intestine (especially in cecum). Since SCFAs in human colon are the products of anaerobic fermentation of nonabsorbed carbohydrate and, to a lesser extent, protein by colonic microbiota47 and ethanol consumption will alter the content and composition of gut microbiota,2, 18, 48 the alteration of SCFAs in rats of chronic ethanol consumption and in different GI regions may be due to the impact of ethanol on gut microbiota. Branched chain SCFAs, 2-methylpropanoic acid, 2-methylbutyric acid, and 3-methylbutyric acid are derived from the catabolism of BCAAs.49 The lower levels of branched chain SCFAs in small intestine may be a result of the decreased BCAA catabolism due to the fact of the predominant BCAAs (~90%) in small intestine. Since ethanol consumption further reduce the enteric BCAAs level, which may be a reason for the further decreased levels of branched chain SCFAs after ethanol consumption.
Since the large intestine (the cecum and colon) has the highest abundance of microbes, the increase in levels of steroids (unconjugated bile acids) from the ileum to the cecum in control rats is primarily due to the de-conjugation of taurine- and glycine-conjugated bile acids by the gut microbiota.50, 51 Moreover, since ethanol consumption can have a profound impact on the structure of gut microbiota,20, 36 a reduction of steroids levels is probably resulted from an ethanol-induced disruption of the gut microbiota.
Choline is an essential nutrient with a wide range of biological functions,52 which can be absorbed and converted into phosphatidylcholine53 by mammals or into trimethylamine by gut microbiota.54 The levels of choline in the intestinal contents were increased markedly from stomach to duodenum and then jejunum but decreased from jejunum to ileum with a further decline when reached cecum and then remained unchanged from cecum to rectum (Supplementary Figure S3). The altered level of choline with amino acids (Supplementary Figure S2) was probably associated with the absorption of choline in jejunum and ileum, being agreeable with the kinetic results for choline uptake in the small intestine of neonatal and adult rats.55 The altered choline level with chronic ethanol consumption is probably resulted from an ethanol-induced disruption of the gut microbiota. Sphingolipids are basic constituents of cellular membranes and are essential for numerous functions such as intracellular signalling. Dysregulated sphingolipid metabolism has been implicated in alcoholic liver disease and the reduced levels of sphingolipid metabolites, phytosphingosine and sphinganine in serum samples of liver injury mice15, 56 were observed.
Carnitine, an essential factor in fatty acid metabolism, plays a major role in transport of activated long-chain fatty acids to sites of β-oxidation in mitochondria. Carnitine can be a useful and safe drug in the liver pathology induced by chronic ethanol exposure.57 The reduced levels of carnitine in all GI regions in our study as well as its lower levels in heart, liver, brain and blood of rats58 observed by others may be an indication of the liver injury induced by chronic ethanol consumption. Carnitine can be degraded by intestinal bacteria and the relatively low carnitine level in ileum to rectum, particularly in colon in control rats (Supplementary Figure S3) may be due to the relatively higher abundance of microbes in the large intestine.
Human studies have shown that the oro-cecal transit time (OCTT) of patients with alcoholic cirrhosis and heavy drinkers was increased,59–61 indicating a gut motility disorder. However, the OCTT of moderate drinkers was not changed compared to the normal controls.60 Study with rat models of acute (one dose) and chronic (30 days) alcohol exposure demonstrated that gastric empting and small bowl transit were inhibited by acute alcohol intoxication but accelerated by chronic alcohol exposure.62 Although alcohol impact on the intestine may vary in different models (such as species, dosage and duration), it is noteworthy that ethanol consumption impacts on gut transit time and mobility,60 which in turn may account for the altered intestinal content metabolic profiles. Furthermore, a limitation of the Lieber-DeCarli liquid diet model used in this study is that part of carbohydrate (maltose dextrin) calories in the control diet must be replaced by ethanol in the ethanol diet to make up equal calories concentration. The less carbohydrate content in the ethanol diet may have impact on gut flora, and thereby the metabolites in the GI contents.
The aim of this study was to characterize the metabolic alterations of the GI tract (stomach, duodenum, jejunum, ileum, cecum, colon, and rectum) in male Sprague Dawley rats following 8 weeks ethanol exposure. The results of our study have indicated that the metabolite compositions of intestinal contents have unique signatures for different compartments under normal physiological conditions. A disruption in gut microbiota by chronic ethanol consumption will lead to remarkable changes in metabolite composition in intestinal contents of the concerned regions. It is thus conceivable that metabonomic analysis of the GI tract contents including fecal metabolome may be of significant importance for disease diagnosis as well as prognosis at a specific region of the GI tract. However, the ethanol can be originated from both exogenous and endogenous origin from bacteria and yeasts. We were not able to differentiate ethanol content of different origins, which is a limitation of the current study.
Conclusions
In conclusion, metabonomics analysis revealed a distinct profile of metabolites in the contents of rat stomach, duodenum, jejunum, ileum, cecum, colon and rectum, which were dependent on the topographical locations of the GI tract. Ethanol consumption has a profound impact on the metabolic profiles in GI contents. These differences in GI contents among intestinal regions and between controls and chronic ethanol-fed rats are also associated with the intestinal functions such as nutrient absorptions and activities of gut microbiota in different compartments of GI tract. This work provided a global metabolic profile of the GI contents in normal rats and how this profile was impacted by chronic ethanol consumption, demonstrating that the metabonomic analysis of intestinal contents may serve as a powerful tool for investigating the interactions between mammals and their gut microbiota.
Supplementary Material
Acknowledgments
This work was financially supported by National Institutes of Health/National Institute on Alcohol Abuse and Alcoholism Grant R01 AA020212.
Footnotes
Supporting Information Available
Compositions of modified Lieber-DeCarli control and ethanol liquid diets for step-wise feeding procedure; metabolites identified in gastrointestinal contents derived from LC-TOFMS analysis; short chain fatty acids in gastrointestinal contents of control rats and rats with ethanol consumption for 8 weeks (µmol/g wet wt); branched chain amino acids in gastrointestinal contents of control rats and rats with ethanol consumption for 8 weeks (µmol/g wet wt); PCA scores plot obtained by analysis of contents from 7 gastrointestinal segments including stomach, Duodenum, Jejunum, Ileum, Cecum, Colon, and Rectum of control rats (A) and rats under chronic ethanol consumption (B); Metabolic profiles (A–G) in GI contents (A, stomach; B, Duodenum; C, Jejunum; D, Ileum; E, Cecum; F, Colon; G, Rectum) of control rats. Values are mean intensities ± SEM measured using LC-MS. * P < 0.05 different from stomach concentration of the same metabolite; Metabolic profiles (A–G) in GI contents (A, stomach; B, Duodenum; C, Jejunum; D, Ileum; E, Cecum; F, Colon; G, Rectum) of control rats and rats with chronic ethanol consumption; Alteration of phenol and derivatives profiles (A–G) in GI contents (A, stomach; B, Duodenum; C, Jejunum; D, Ileum; E, Cecum; F, Colon; G, Rectum) due to ethanol consumption; The composition of short chain fatty acids in gastrointestinal contents of control rats (upper panel) and ethanol treated rats (under panel). This material is available free of charge via the Internet at http://pubs.acs.org.
References
- 1.Zhong W, Zhao YT, Tang YN, Wei XL, Shi X. Chronic alcohol exposure stimulates adipose tissue lipolysis in mice: role of reverse triglyceride transport in the pathogenesis of alcoholic steatosis. American Journal of Pathology. 2012;180(3):998–1007. doi: 10.1016/j.ajpath.2011.11.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Jesus Delgado-Villa M, Luisa Ojeda M, Maria Rubio J, Luisa Murillo M, Carreras Sanchez O. Beneficial Role of Dietary Folic Acid on Cholesterol and Bile Acid Metabolism in Ethanol-Fed Rats. Journal of Studies on Alcohol and Drugs. 2009;70(4):615–622. doi: 10.15288/jsad.2009.70.615. [DOI] [PubMed] [Google Scholar]
- 3.Beier JI, Arteel GE, McClain CJ. Advances in alcoholic liver disease. Curr Gastroenterol Rep. 2011;13(1):56–64. doi: 10.1007/s11894-010-0157-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gao B, Bataller R. Alcoholic liver disease: pathogenesis and new therapeutic targets. Gastroenterology. 2011;141(5):1572–85. doi: 10.1053/j.gastro.2011.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Corrao G, Ferrari P, Zambon A, Torchio P. Are the recent trends in liver cirrhosis mortality affected by the changes in alcohol consumption? Analysis of latency period in European countries. J Stud Alcohol. 1997;58(5):486–94. doi: 10.15288/jsa.1997.58.486. [DOI] [PubMed] [Google Scholar]
- 6.Yoon YH, Yi HY. Surveillance report #75: Liver Cirrhosis Mortality in the United States, 1970–2003. Bethesda, MD: 2006. [Google Scholar]
- 7.SAMHSA. 2007 National Survey on Drug Use and Health, d. t., tobacco product and alcohol use, table 2.46B [article online] [Accessed January 14, 2010];2008c Available at: http://oas.samhsa.gov/NSDUH/2k7NSDUH/tabs/Sect2peTabs43to84.htm#Tab2.46B.
- 8.Tsukamoto H, Mkrtchyan H, Dynnyk A. Intragastric ethanol infusion model in rodents. Methods Mol Biol. 2008;447:33–48. doi: 10.1007/978-1-59745-242-7_3. [DOI] [PubMed] [Google Scholar]
- 9.Shinohara M, Ji C, Kaplowitz N. Differences in betaine-homocysteine methyltransferase expression, endoplasmic reticulum stress response, and liver injury between alcohol-fed mice and rats. Hepatology. 2010;51(3):796–805. doi: 10.1002/hep.23391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.DeCarli LM, Lieber CS. Fatty liver in the rat after prolonged intake of ethanol with a nutritionally adequate new liquid diet. J Nutr. 1967;91(3):331–6. doi: 10.1093/jn/91.3_Suppl.331. [DOI] [PubMed] [Google Scholar]
- 11.Manna SK, Patterson AD, Yang Q, Krausz KW, Li H, Idle JR, Fornace AJ, Jr, Gonzalez FJ. Identification of noninvasive biomarkers for alcohol-induced liver disease using urinary metabolomics and the Ppara-null mouse. J Proteome Res. 2010;9(8):4176–88. doi: 10.1021/pr100452b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Manna SK, Patterson AD, Yang Q, Krausz KW, Idle JR, Fornace AJ, Gonzalez FJ. UPLC-MS-based Urine Metabolomics Reveals Indole-3-lactic Acid and Phenyllactic Acid as Conserved Biomarkers for Alcohol-induced Liver Disease in the Ppara-null Mouse Model. Journal of Proteome Research. 2011;10(9):4120–4133. doi: 10.1021/pr200310s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Loftus N, Barnes A, Ashton S, Michopoulos F, Theodoridis G, Wilson I, Ji C, Kaplowitz N. Metabonomic investigation of liver profiles of nonpolar metabolites obtained from alcohol-dosed rats and mice using high mass accuracy MSn analysis. J Proteome Res. 2011;10(2):705–13. doi: 10.1021/pr100885w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fernando H, Kondraganti S, Bhopale KK, Volk DE, Neerathilingam M, Kaphalia BS, Luxon BA, Boor PJ, Shakeel Ansari GA. (1)H and (3)(1)P NMR lipidome of ethanol-induced fatty liver. Alcohol Clin Exp Res. 2010;34(11):1937–47. doi: 10.1111/j.1530-0277.2010.01283.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Li SF, Liu HX, Jin YB, Lin SH, Cai ZW, Jiang YY. Metabolomics study of alcohol-induced liver injury and hepatocellular carcinoma xenografts in mice. Journal of Chromatography B-Analytical Technologies in the Biomedical and Life Sciences. 2011;879(24):2369–2375. doi: 10.1016/j.jchromb.2011.06.018. [DOI] [PubMed] [Google Scholar]
- 16.Gao XF, Zhao AH, Zhou MM, Lin JC, Qiu YP, Su MM, Jia W. GC/MS-based urinary metabolomics reveals systematic differences in metabolism and ethanol response between Sprague-Dawley and Wistar rats. Metabolomics. 2011;7(3):363–374. [Google Scholar]
- 17.Vazquez-Fresno R, Llorach R, Alcaro F, Rodriguez MA, Vinaixa M, Chiva-Blanch G, Estruch R, Correig X, Andres-Lacueva C. 1H-NMR-based metabolomic analysis of the effect of moderate wine consumption on subjects with cardiovascular risk factors. Electrophoresis. 2012;33(15):2345–2354. doi: 10.1002/elps.201100646. [DOI] [PubMed] [Google Scholar]
- 18.Purohit V, Bode JC, Bode C, Brenner DA, Choudhry MA, Hamilton F, Kang YJ, Keshavarzian A, Rao R, Sartor RB, Swanson C, Turner JR. Alcohol, intestinal bacterial growth, intestinal permeability to endotoxin, and medical consequences: summary of a symposium. Alcohol. 2008;42(5):349–61. doi: 10.1016/j.alcohol.2008.03.131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bull-Otterson L, Feng W, Kirpich I, Wang Y, Qin X, Liu Y, Gobejishvili L, Joshi-Barve S, Ayvaz T, Petrosino J, Kong M, Barker D, McClain C, Barve S. Metagenomic analyses of alcohol induced pathogenic alterations in the intestinal microbiome and the effect of Lactobacillus rhamnosus GG treatment. PLoS One. 2013;8(1):e53028. doi: 10.1371/journal.pone.0053028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mutlu EA, Gillevet PM, Rangwala H, Sikaroodi M, Naqvi A, Engen PA, Kwasny M, Lau CK, Keshavarzian A. Colonic microbiome is altered in alcoholism. American Journal of Physiology-Gastrointestinal and Liver Physiology. 2012;302(9):G966–G978. doi: 10.1152/ajpgi.00380.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yan AW, Fouts DE, Brandl J, Starkel P, Torralba M, Schott E, Tsukamoto H, Nelson KE, Brenner DA, Schnabl B. Enteric dysbiosis associated with a mouse model of alcoholic liver disease. Hepatology. 2011;53(1):96–105. doi: 10.1002/hep.24018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Collado MC, Isolauri E, Salminen S, Sanz Y. The impact of probiotic on gut health. Curr Drug Metab. 2009;10(1):68–78. doi: 10.2174/138920009787048437. [DOI] [PubMed] [Google Scholar]
- 23.Ross RP, Mills S, Hill C, Fitzgerald GF, Stanton C. Specific metabolite production by gut microbiota as a basis for probiotic function. International Dairy Journal. 2010;20(4):269– 276. [Google Scholar]
- 24.Tian Y, Zhang L, Wang Y, Tang H. Age-related topographical metabolic signatures for the rat gastrointestinal contents. J Proteome Res. 2012;11(2):1397–411. doi: 10.1021/pr2011507. [DOI] [PubMed] [Google Scholar]
- 25.Martin FP, Wang Y, Yap IK, Sprenger N, Lindon JC, Rezzi S, Kochhar S, Holmes E, Nicholson JK. Topographical variation in murine intestinal metabolic profiles in relation to microbiome speciation and functional ecological activity. J Proteome Res. 2009;8(7):3464–74. doi: 10.1021/pr900099x. [DOI] [PubMed] [Google Scholar]
- 26.Duan LP, Wang HH, Wang DQH. Cholesterol absorption is mainly regulated by the jejunal and ileal ATP-binding cassette sterol efflux transporters Abcg5 and Abcg8 in mice. Journal of Lipid Research. 2004;45(7):1312–1323. doi: 10.1194/jlr.M400030-JLR200. [DOI] [PubMed] [Google Scholar]
- 27.Hu M, Cheng ZQ, Zheng LX. Functional and molecular characterization of rat intestinal prolidase. Pediatric Research. 2003;53(6):905–914. doi: 10.1203/01.PDR.0000064903.33501.FB. [DOI] [PubMed] [Google Scholar]
- 28.Fordahl S, Cooney P, Qiu Y, Xie G, Jia W, Erikson KM. Waterborne manganese exposure alters plasma, brain, and liver metabolites accompanied by changes in stereotypic behaviors. Neurotoxicol Teratol. 2012;34(1):27–36. doi: 10.1016/j.ntt.2011.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Smith CA, Want EJ, O'Maille G, Abagyan R, Siuzdak G. XCMS: processing mass spectrometry data for metabolite profiling using nonlinear peak alignment, matching, and identification. Anal Chem. 2006;78(3):779–87. doi: 10.1021/ac051437y. [DOI] [PubMed] [Google Scholar]
- 30.Zheng XJ, Qiu YP, Zhong W, Baxter S, Su MM, Li Q, Xie GX, Ore BM, Qiao SL, Spencer MD, Zeisel SH, Zhou ZX, Zhao AH, Jia W. A targeted metabolomic protocol for short-chain fatty acids and branched-chain amino acids. Metabolomics. 2013 doi: 10.1007/s11306-013-0500-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Niemela O. Biomarkers in alcoholism. Clin Chim Acta. 2007;377(1–2):39–49. doi: 10.1016/j.cca.2006.08.035. [DOI] [PubMed] [Google Scholar]
- 32.Wang Y, Tang H, Holmes E, Lindon JC, Turini ME, Sprenger N, Bergonzelli G, Fay LB, Kochhar S, Nicholson JK. Biochemical characterization of rat intestine development using high-resolution magic-angle-spinning 1H NMR spectroscopy and multivariate data analysis. J Proteome Res. 2005;4(4):1324–9. doi: 10.1021/pr050032r. [DOI] [PubMed] [Google Scholar]
- 33.Wang Y, Cloarec O, Tang H, Lindon JC, Holmes E, Kochhar S, Nicholson JK. Magic angle spinning NMR and 1H-31P heteronuclear statistical total correlation spectroscopy of intact human gut biopsies. Anal Chem. 2008;80(4):1058–66. doi: 10.1021/ac701988a. [DOI] [PubMed] [Google Scholar]
- 34.Wu G. Intestinal mucosal amino acid catabolism. J Nutr. 1998;128(8):1249–52. doi: 10.1093/jn/128.8.1249. [DOI] [PubMed] [Google Scholar]
- 35.Hooper LV, Midtvedt T, Gordon JI. How host-microbial interactions shape the nutrient environment of the mammalian intestine. Annual Review of Nutrition. 2002;22:283– 307. doi: 10.1146/annurev.nutr.22.011602.092259. [DOI] [PubMed] [Google Scholar]
- 36.Isabel Queipo-Ortuno M, Boto-Ordonez M, Murri M, Miguel Gomez-Zumaquero J, Clemente-Postigo M, Estruch R, Cardona Diaz F, Andres-Lacueva C, Tinahones FJ. Influence of red wine polyphenols and ethanol on the gut microbiota ecology and biochemical biomarkers. American Journal of Clinical Nutrition. 2012;95(6):1323–1334. doi: 10.3945/ajcn.111.027847. [DOI] [PubMed] [Google Scholar]
- 37.Yu YM, Wagner DA, Tredget EE, Walaszewski JA, Burke JF, Young VR. Quantitative role of splanchnic region in leucine metabolism: L-[1-13C,15N]leucine and substrate balance studies. Am J Physiol. 1990;259(1 Pt 1):E36–51. doi: 10.1152/ajpendo.1990.259.1.E36. [DOI] [PubMed] [Google Scholar]
- 38.Hoerr RA, Matthews DE, Bier DM, Young VR. Effects of protein restriction and acute refeeding on leucine and lysine kinetics in young men. Am J Physiol. 1993;264(4 Pt 1):E567–75. doi: 10.1152/ajpendo.1993.264.4.E567. [DOI] [PubMed] [Google Scholar]
- 39.Cummings JH. Short chain fatty acids in the human colon. Gut. 1981;22(9):763–79. doi: 10.1136/gut.22.9.763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Macfarlane GT, Cummings JH, Allison C. PROTEIN-DEGRADATION BY HUMAN INTESTINAL BACTERIA. Journal of General Microbiology. 1986;132:1647–1656. doi: 10.1099/00221287-132-6-1647. [DOI] [PubMed] [Google Scholar]
- 41.Andoh A, Tsujikawa T, Fujiyama Y. Role of dietary fiber and short-chain fatty acids in the colon. Current Pharmaceutical Design. 2003;9(4):347–358. doi: 10.2174/1381612033391973. [DOI] [PubMed] [Google Scholar]
- 42.Wong JMW, de Souza R, Kendall CWC, Emam A, Jenkins DJA. Colonic health: Fermentation and short chain fatty acids. Journal of Clinical Gastroenterology. 2006;40(3):235–243. doi: 10.1097/00004836-200603000-00015. [DOI] [PubMed] [Google Scholar]
- 43.Blottiere HM, Buecher B, Galmiche JP, Cherbut C. Molecular analysis of the effect of short-chain fatty acids on intestinal cell proliferation. Proceedings of the Nutrition Society. 2003;62(1):101–106. doi: 10.1079/PNS2002215. [DOI] [PubMed] [Google Scholar]
- 44.Sanderson IR. Short chain fatty acid regulation of signaling genes expressed by the intestinal epithelium. Journal of Nutrition. 2004;134(9):2450S–2454S. doi: 10.1093/jn/134.9.2450S. [DOI] [PubMed] [Google Scholar]
- 45.Grubb BR. Avian cecum: role of glucose and volatile fatty acids in transepithelial ion transport. Am J Physiol. 1991;260(5 Pt 1):G703–10. doi: 10.1152/ajpgi.1991.260.5.G703. [DOI] [PubMed] [Google Scholar]
- 46.Israel Y, Orrego H, Carmichael FJ. Acetate-mediated effects of ethanol. Alcoholism-Clinical and Experimental Research. 1994;18(1):144–148. doi: 10.1111/j.1530-0277.1994.tb00894.x. [DOI] [PubMed] [Google Scholar]
- 47.Bugaut M. Occurrence, absorption and metabolism of short chain fatty acids in the digestive tract of mammals. Comp Biochem Physiol B. 1987;86(3):439–72. doi: 10.1016/0305-0491(87)90433-0. [DOI] [PubMed] [Google Scholar]
- 48.Dumas ME, Barton RH, Toye A, Cloarec O, Blancher C, Rothwell A, Fearnside J, Tatoud R, Blanc V, Lindon JC, Mitchell SC, Holmes E, McCarthy MI, Scott J, Gauguier D, Nicholson JK. Metabolic profiling reveals a contribution of gut microbiota to fatty liver phenotype in insulin-resistant mice. Proc Natl Acad Sci U S A. 2006;103(33):12511–6. doi: 10.1073/pnas.0601056103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Macfarlane S, Macfarlane GT. Regulation of short-chain fatty acid production. Proceedings of the Nutrition Society. 2003;62(1):67–72. doi: 10.1079/PNS2002207. [DOI] [PubMed] [Google Scholar]
- 50.Martin FPJ, Dumas ME, Wang YL, Legido-Quigley C, Yap IKS, Tang HR, Zirah S, Murphy GM, Cloarec O, Lindon JC, Sprenger N, Fay LB, Kochhar S, van Bladeren P, Holmes E, Nicholson JK. A top-down systems biology view of microbiome-mammalian metabolic interactions in a mouse model. Mol Syst Biol. 2007;3 doi: 10.1038/msb4100153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Ridlon JM, Kang DJ, Hylemon PB. Bile salt biotransformations by human intestinal bacteria. Journal of Lipid Research. 2006;47(2):241–259. doi: 10.1194/jlr.R500013-JLR200. [DOI] [PubMed] [Google Scholar]
- 52.Zeisel SH, Blusztajn JK. Choline and human nutrition. Annu Rev Nutr. 1994;14:269– 96. doi: 10.1146/annurev.nu.14.070194.001413. [DOI] [PubMed] [Google Scholar]
- 53.Pelech SL, Vance DE. Regulation of phosphatidylcholine biosynthesis. Biochim Biophys Acta. 1984;779(2):217–51. doi: 10.1016/0304-4157(84)90010-8. [DOI] [PubMed] [Google Scholar]
- 54.Zeisel SH, daCosta KA, Youssef M, Hensey S. Conversion of dietary choline to trimethylamine and dimethylamine in rats: dose-response relationship. J Nutr. 1989;119(5):800–4. doi: 10.1093/jn/119.5.800. [DOI] [PubMed] [Google Scholar]
- 55.Sheard NF, Zeisel SH. An in vitro study of choline uptake by intestine from neonatal and adult rats. Pediatr Res. 1986;20(8):768–72. doi: 10.1203/00006450-198608000-00014. [DOI] [PubMed] [Google Scholar]
- 56.Clugston RD, Jiang H, Lee MX, Piantedosi R, Yuen JJ, Ramakrishnan R, Lewis MJ, Gottesman ME, Huang LS, Goldberg IJ, Berk PD, Blaner WS. Altered hepatic lipid metabolism in C57BL/6 mice fed alcohol: a targeted lipidomic and gene expression study. J Lipid Res. 2011;52(11):2021–31. doi: 10.1194/jlr.M017368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Kepka A, Szajda SD, Waszkiewicz N, Pludowski P, Chojnowska S, Rudy M, Szulc A, Ladny JR, Zwierz K. Carnitine: function, metabolism and value in hepatic failure during chronic alcohol intoxication. Postepy Higieny I Medycyny Doswiadczalnej. 2011;65:645–653. doi: 10.5604/17322693.962226. [DOI] [PubMed] [Google Scholar]
- 58.Sakvarelidze EPE. Change of concentration of L-carnitine in blood and other tissues in rats on a background of the alcohol intake and influence of mildronate on its level. Georgian medical news. (137):94–6. [PubMed] [Google Scholar]
- 59.Huppe D, Tonissen R, Hofius M, Kuntz HD, May B. Effect of chronic alcohol drinking and liver cirrhosis on oro-cecal transit time (H2 breath test) Z Gastroenterol. 1989;27(10):624–8. [PubMed] [Google Scholar]
- 60.Papa A, Tursi A, Cammarota G, Certo M, Cuoco L, Montalto M, Cianci R, Papa V, Fedeli P, Fedeli G, Gasbarrini G. Effect of moderate and heavy alcohol consumption on intestinal transit time. Panminerva Medica. 1998;40(3):183–5. [PubMed] [Google Scholar]
- 61.Bode C, Bode JC. Effect of alcohol consumption on the gut. Best practice & research. Clinical gastroenterology. 2003;17(4):575–92. doi: 10.1016/s1521-6918(03)00034-9. [DOI] [PubMed] [Google Scholar]
- 62.Izbeki F, Wittmann T, Csati S, Jeszenszky E, Lonovics J. Opposite effects of acute and chronic administration of alcohol on gastric emptying and small bowel transit in rat. Alcohol Alcohol. 2001;36(4):304–8. doi: 10.1093/alcalc/36.4.304. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.