Abstract
Photochemical and other reactions on DNA cause damage and corrupt genetic information. To counteract this damage, organisms have evolved intricate repair mechanisms that often crosstalk with other DNA-based processes, such as transcription. Intriguing observations in the late 1980s and early 1990s led to the discovery of transcription-coupled repair (TCR), a subpathway of nucleotide excision repair (NER). TCR, found in all domains of life prioritizes for repair lesions located in the transcribed DNA strand, directly read by RNA polymerase. Here we give a historical overview of developments in the field of bacterial TCR, starting from the pioneering work of Evelyn Witkin and Aziz Sancar, which led to the identification of the first transcription-repair coupling factor (the Mfd protein), to recent studies that have uncovered alternative TCR pathways and regulators.
INTRODUCTION
Protecting genomic integrity and the faithful transmission of genetic information, from DNA to RNA to protein, is essential for all life. The double-helical nature of DNA, the length of genes (particularly in higher eukaryotes), and the ubiquitous presence of endogeneous and exogeneous DNA-damaging agents can pose significant topological and information processing challenges to the cell. This is not only due to the chemical modification of the DNA itself that can lead to mutations and can be fixed by replication, but also due to cytotoxic effects, which can impact DNA-based processes such as transcription, chromatin remodeling, and more generally, cellular signaling (1). One of the most prevalent sources of DNA damage, UV radiation, is notable for several reasons. It leads to a variety of distinct lesions, including cyclobutane pyrimidine dimers (CPDs) and to a lesser extent single- and double-stranded breaks, crosslinks, and especially pyrimidine (6-4) pyrimidone photoproducts (6-4PPs) and related Dewar valence isomers. Relatively minor oxidatively-generated damage consisting mostly of oxidized bases and single strand breaks are also formed (2). These are largely repaired via a universally conserved and remarkably versatile “cut and patch” mechanism, in which a large repertoire of chemically distinct lesions are excised as part of an oligonucleotide. This is followed by gap filling, and ligation to restore the integrity of the strand. This nucleotide excision repair (NER) mechanism, also known as dark (light-independent) repair – to distinguish it from photoreactivation, an alternative mechanism for repairing CPDs and 6-4PP via the action of photolyases – was the first of the three main DNA repair mechanisms to be described. In 1964, landmark papers on the disappearance of thymine dimers in bacteria were published. These combined biophysical studies on UV radiation with the characterization of UV-sensitive repair-defective strains of Escherichia coli K12 (3–5). These papers suggested a repair model that was rather surprising at the time – that DNA had to be first cut to be repaired! CPDs could be released from DNA and this error-correcting mechanism involved non-semiconservative DNA synthesis in patches. Around the same time, Howard-Flanders and Boyce identified three genetic loci in E. coli that were important for damage excision (6). It was essential to develop cell free assays to further characterize this process. The biochemical purification of the proteins responsible for the bacterial NER activity did not prove an easy task, and was not accomplished until more than a decade later through the combined work of Erling Seeberg, Peter Strike, and Aziz Sancar, then a postdoctoral in the laboratory of Dean Rupp (7–12). It was Sancar who purified and reconstituted the Uvr(A)BC machinery and established it as an excinuclease, which could doubly incise the DNA, excise the damage and undergo complicated dynamics (12). While the eukaryotic NER system proved considerably more baroque, with factors that differ at the sequence level from their bacterial counterparts, in essence, the process with its (1) damage detection and (2) verification steps, (3) dual incision, (4) excision, (5) repair synthesis, and (6) ligation is universally conserved. A long list of laboratories have contributed to our understanding of NER in all three domains of life, including at the structural and single-molecule level, but here we will focus on the bacterial pathway.
FROM NER TO TCR – EXCISION REPAIR CAN BE BIASED TOWARDS THE TEMPLATE STRAND
The assembly of the core NER machinery, composed of products of the uvr genes (uvrA, uvrB, uvrC) occurs stepwise. Multiple events of ATP hydrolysis by several of these factors (UvrA, UvrB, UvrD) play crucial roles in the assembly of the various intermediates, in damage sensing and damage verification (13). While these are still subject of active investigation, we now know that the initial search for DNA lesions is accomplished by a heterotetrameric UvrA2B2 complex (14, 15), which was seen, using quantum-dot labeled UvrA/UvrB, to “slide” during its search for DNA damage (16). Once a lesion is located, UvrA dissociates, the cryptic helicase function of UvrB is activated, a tight “pre-incision” complex forms. This locally melts the DNA around the lesion and this opening of the DNA helix by UvrB is required for damage verification (13). The DNA-UvrB complex then recruits UvrC, a multi-domain nuclease, which first incises the DNA 3′ to the lesion using its GIY-YIG-like endonuclease domain, and then 5′ to the lesion using its C-terminal region. This encompasses an unusual RNAse H-like domain (13). The UvrD helicase then displaces the damaged strand, which is followed by repair synthesis by Pol I, and ligation of the repaired patch by DNA ligase.
While mechanistic details of NER across domains of life were beginning to unravel in the mid-1980s, two key observations regarding the repair of UV damage were made in the context of fibroblasts isolated from individuals with a rare progeroid disorder called Cockayne syndrome. First, UV irradiation of these cells, led to a defect in transcription, which s inhibited immediately after irradiation in a dose and time-dependent manner. This failure to easily recover RNA synthesis in Cockayne syndrome cells eventually activate pro-apoptotic signals. Second, the two strands of the chromosome were repaired asymmetrically. Intriguing reports from the Hanawalt lab demonstrated that in Chinese hamster ovary cells photoproducts were removed more rapidly from transcribed sequences than non-transcribed DNA (17). Repair was detected by treating restriction enzyme digested DNA with T4 endonuclease V, which incises DNA at pyrimidine dimers, followed by Southern hybridization. Soon afterwards, work from the same laboratory proposed that a bias towards repair of the transcribed strand underlay the preferential repair of active genes (18). Hence a direct connection between repair and transcription was established – and this preferential repair of the transcribed strand became known as transcription-coupled repair (TCR). In addition to the previously noted compromise in the recovery of RNA synthesis after UV irradiation, this biased form of repair was also defective in Cockayne syndrome cells (19), and was hypothesized to explain the accelerated aging process noted in patients with Cockayne syndrome.
A few years later, through studies of the lac operon, TCR was extended to bacteria, and was demonstrated to be dependent on the induction of this operon (20). TCR was subsequently studied in a variety of other model organisms, including Saccharomyces cerevisiae (21–25), mice (26) and Caenorhabditis elegans (27, 28), and very recently, archaea (29), pointing to the universality of the process. Quantitatively, the bias towards repair of the template strand has been found to be widely organism and gene-specific, and also dependent on the transcriptional status (high versus low) of the gene. For example, Bohr and colleagues measured a five-fold bias in the DHFR gene of Chinese hamster ovary cells (17), while Mellon and Hanawalt estimated that TCR mediated a tenfold faster repair of the transcribed strand of the lactose operon (20). Assays in halophilic archaebacteria detected TCR in several genes (29), while previous work in Sulfolobus solfataricus had failed to indicate any preferential repair (30, 31). TCR was also reported to be cell-type dependent. Embryonic fibroblasts isolated from a TCR deficient mouse were hypersensitive to UV and displayed impaired recovery of RNA synthesis post UV irradiation (26). In contrast, embryonic stem cells appeared relatively resistant to UV in the absence of a functional TCR pathway (32). This argues for complex regulation that remains inadequately understood at the organismal level.
The defining feature of TCR is its dependence on active transcription. TCR was not observed in the presence of the transcription elongation inhibitor α-amanitin (33) or the bacterial RNAP inhibitor, rifampin (34), nor with a temperature-sensitive allele of yeast Rbp1 (35), the large catalytic subunit of RNAPII. Taken together, these data indicated that the trigger of TCR is not any RNAP, such as promoter-proximal molecule, but rather RNAPs that have become stalled by DNA damage in the transcribed strand. In agreement with this, CPDs, the most prominent lesion repaired by TCR, constitute an almost complete block to transcription elongation in vitro when present on the template strand (36–38). TCR was also not seen close to transcriptional start sites, only past the region where RNAP undergoes the transition from abortive initiation to processive elongation (39). These emerging data were consistent with the hypothesis, first put forth by Mellon and Hanawalt, that the trigger eliciting TCR was an arrested RNA polymerase (20). Other concepts explaining this form of biased repair were also considered. First, an open conformation of chromatin could directly increase the accessibility of DNA repair enzymes. Secondly, transcription is associated with special topological changes – overwinding of DNA in front of RNAP and underwinding in its wake. These were thought to create a high affinity site for the excision proteins.
In humans, the connection between TCR and Cockayne syndrome, a rare autosomal recessive disease associated with mutations in either of two TCR genes (CSA and CSB) (19) gave an important impetus to work. Although a rare disease, Cockayne syndrome has received significant attention from the scientific community as it constituted, along with xeroderma pigmentosum (40, 41), a prototypical DNA repair disease (Figure 1). Cockayne syndrome manifestations are complex and multi-systemic, and include dwarfism, sensorineural hearing loss, contractures, gait ataxia and, in general, pronounced neurodegeneration due to an increased inflammatory response in the area surrounding oligodendrocytes within myelinated axons (42). This eventually leads to nerve demyelination. While Cockayne syndrome had initially been exclusively viewed as a nucleotide excision repair syndrome, more recent studies implicating the TCR proteins, CSA and CSB, in transcriptional regulation (43), repair of oxidative damage in the mitochondria (44) and chromatin remodeling (45, 46), suggest that the precise molecular mechanisms behind Cockayne syndrome may be more complex than initially anticipated. Nonetheless, the DNA repair asymmetry provided by TCR processes has now been established to have far-reaching implications, including in the evolution of lagging strand genes (vide infra), but also in the development of tumors (47–49). It is outside the scope of this review to provide a comprehensive account of the massive body of work that has been done on eukaryotic TRCFs. These studies have been the subject of several excellent reviews (50, 51, 43) to which the reader is referred. Instead, in this perspective, we will focus on the discovery and mechanistic studies of so-called “transcription-repair coupling factors” or TRCFs from bacteria, starting with the pioneering studies of recent Lasker Award recipient, Evelyn Witkin, Phil Hanawalt and recent Nobel Prize laureate, Aziz Sancar to the more recent identification of alternative TRCFs.
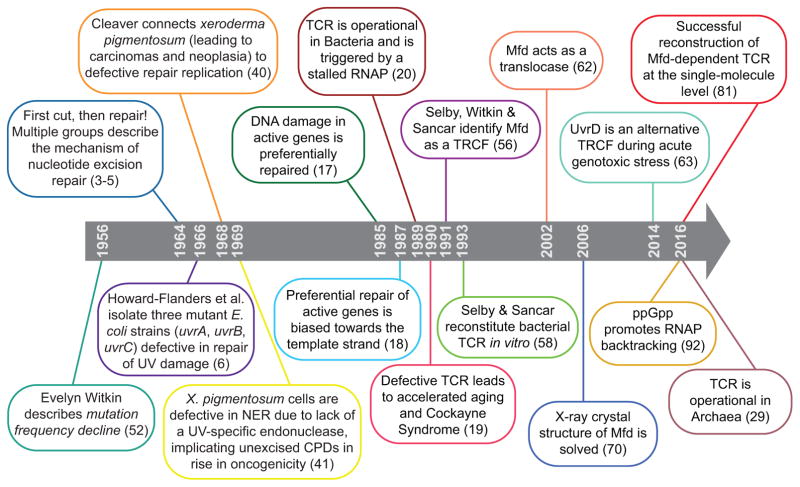
Figure 1.
A timeline of key developments in the TCR field.
WHY TCR IS AN ELEGANT SOLUTION TO THE DNA DAMAGE RECOGNITION PROBLEM
With a direct connection made between repair and transcription, the obvious question was whether RNA polymerase alone could direct the NER machinery to the template strand. Given the development of in vitro transcription and repair systems, this was readily tested by the Sancar lab, and the conclusion was that RNA polymerase inhibited, rather than stimulated DNA repair due to the burying of the DNA lesion under the RNAP footprint (34). Thus, the topological effects on superhelical DNA exerted by transcription, first considered in explaining preferential repair, fell out of favor, and instead the search for a transcription repair “coupling” factor (TRCF), missing from the well-defined in vitro system (consisting of DNA, RNA polymerase, UvrAB(C), UvrD, DNA polymerase I and DNA ligase as well as nucleotide factors) began. A strong first candidate was photolyase. This could stimulate excision on its own, but was ultimately found not to have a stimulatory effect on NER in the presence of RNA polymerase (34). Interestingly, decades before TCR was discovered, Evelyn Witkin had described a phenomenon called mutation frequency decline, which she defined as the decrease of damage-induced suppressor mutations that occurs under specific conditions – when protein synthesis was transiently inhibited after UV irradiation (52). Mutation frequency decline was abolished in uvr strains, suggesting it was due to a form of dark repair, but also by mutations in a different gene, which became known as mfd (53, 54). Bockrath and coworkers then demonstrated that mutation frequency decline was strand-specific and that it involved excision repair of premutational lesions located in the transcribed strand only (55). Thus, given this strand specificity, the mfd gene appeared a likely candidate for a TRCF. Indeed, cell extracts from mfd strains proved deficient for TCR. Partial purification of the relevant activity led to strand-specific incorporation of radioactivity in repair patches and allowed for in vitro complementation with Mfd protein, jointly reported by Sancar and Witkin in 1991 (56). Rapid progress followed. Mfd – now renamed TRCF – was a large SF2-type ATPase that could bind to RNA polymerase as well as the UvrA subunit of the NER machinery, and dsDNA (57, 58). Importantly, the action of TRCF appeared to be dependent on ATP hydrolysis. DNA binding was observed in the presence of poorly-hydrolyzable ATP analogs, while hydrolysis appeared to lead to dissociation of TRCF off the DNA with concomitant release of RNA polymerase (59).
How coupling occurred remained unknown – did it follow a sequential model, with TRCF remaining bound to DNA after removal of RNA polymerase to recruit UvrA, or a concerted reaction, with TRCF binding to the elongation complex leading to a high molecular weight assembly that included UvrA and possibly UvrB? Nonetheless, the solution nature had found was elegant – it took advantage of the remarkable ability of RNAP to translate template heterogeneity, chemical and structural, into discontinuity of DNA tracking (transcriptional arrest and pausing). Pervasive transcription accounts for 80% coverage of the genome, as demonstrated by the ENCODE project (60), and hence by their intrinsic scanning function, RNAPs are unusually efficient genome-wide sensors of DNA damage. This modified view of RNAP as a DNA damage recognition protein was consistent with in vitro behavior. RNAPs have uniquely long half-lives at particular lesions, which in the case of CDPs are on the order of hours (61, 37), allowing factors ample timeframe to achieve binding and regulation.
Once RNAP is stably bound to the lesion, three distinct mechanistic strategies could potentially resolve the repair inhibition posed by the stalled RNAP. First, the polymerase could be released off the DNA, freeing the lesion for repair and terminating transcript synthesis; this is accomplished via the action of the Mfd ATPase (Figures 2, 3a, and 3b), as described below (62, 58). Secondly, the polymerase could undergo backtracking or slide backwards, making the lesion temporarily accessible to NER enzymes. This mechanism has only recently been identified, and involves a TRCF distinct from Mfd, the UvrD helicase. This “pulls” RNAP backwards (63); backwards translocation is then followed by factor-mediated forward translocation to restore the catalytic alignment of the RNA with the enzyme’s active site, possibly via Mfd (Figure 4). Unlike the first mechanism, UvrD-dependent TCR ultimately rescues transcript elongation (64). Finally, a third scenario would be that the polymerase could be forced to translocate past the lesion, possibly resulting in misincorporation during RNA synthesis. This process has been described as transcriptional mutagenesis and has been addressed elsewhere (65, 66).
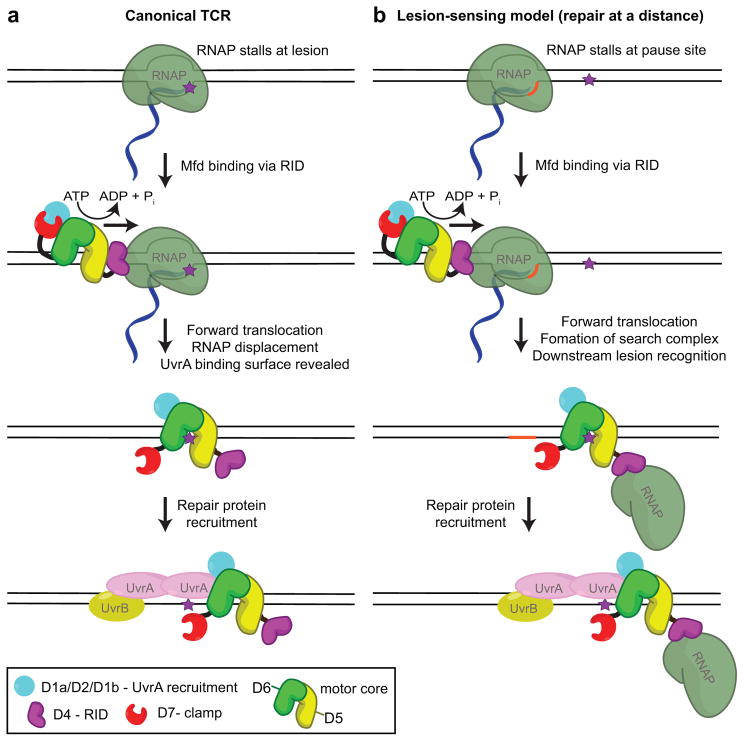
Figure 2. Mfd-dependent TCR and repair at a distance.
(a) In the canonical model, Mfd binds upstream of the RNAP stalled by a lesion (purple star) and translocates in the 3′ to 5′ direction with respect to the transcribed strand. This translocation activity serves to ‘push’ RNAP forward resulting in reannealing of the upstream edge of the transcription bubble and unwinding the RNA/DNA hybrid to dissociate stalled transcription elongation complexes. Mfd likely remains associated with the lesion to recruit the NER machinery through direct interaction with UvrA. TCR joins general the NER pathway at the damage verification step. (b) In the lesion-sensing (repair at a distance) model, Mfd (in complex with a RNAP that has been displaced from the nucleic acid chains) scans downstream from an RNAP pause site (orange sequence) until it reaches a lesion. It then recruits the NER machinery and joins general NER at the damage verification step.
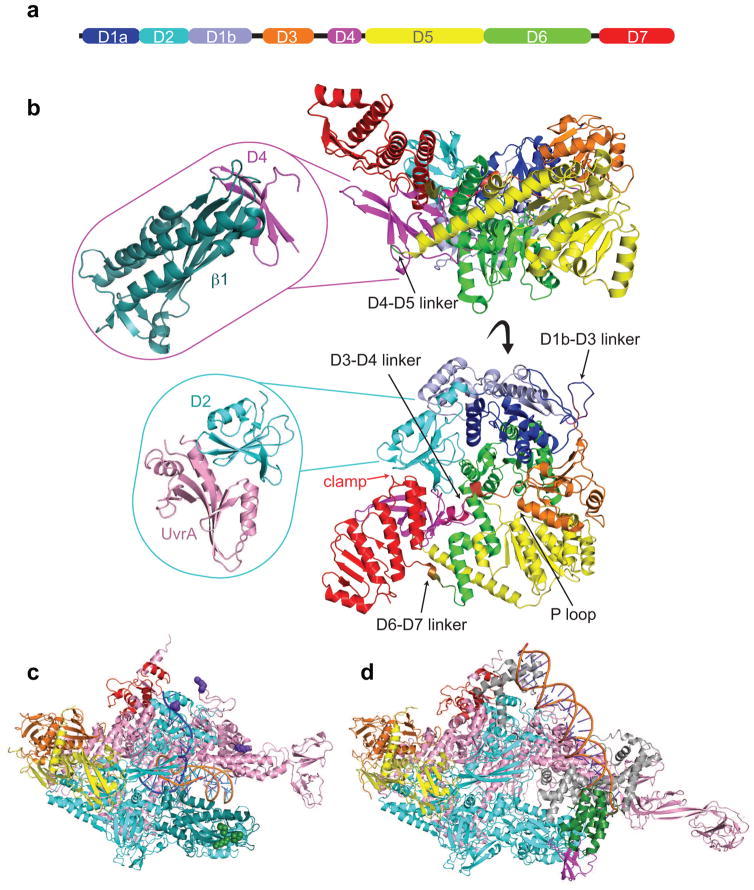
Figure 3. Structural features of key TCR players.
a) Schematic representation of Mfd domain architecture colored as follows: D1a in blue, D2 in cyan, D1b in slate, D3 in orange, RID in magenta, D5 in yellow, D6 in green, and D7 in red. The horizontal bar represents the 1148-residue primary sequence with colored blocks indicating domains and black lines highlighting flexible connecting linkers.
b) Top and side views showing an α carbon backbone ribbon representation of the E. coli apo Mfd structure (PDB ID 2EYQ) with domains colored as in (a). Top inset shows an α carbon ribbon representation of Thermus sp. core Mfd-RID/RNAP-β complex (PDB ID 3MLQ). Bottom inset shows an α carbon representation of E. coli core Mfd/UvrA complex (PDB ID 4DFC).
c) Overall architecture of Thermus thermophilus RNAP elongation complex (PDB ID 2O5I) with RNAP subunits colored as follows: α1 and α2 in orange and yellow, β in cyan, β′ in pink, ω in red. The N-terminal region of the β subunit interacting with Mfd-RID is colored in deep teal. Residues involved in direct contacts with Mfd-RID (the “IKE” motif) are shown as green spheres (β subunit I108, K109, and E110). Residues involved in direct contacts with UvrD are shown as purple spheres (β subunit K781, β′ subunit K28 and R67).
d) Overall architecture of Thermus aquaticus transcription initiation complex bound to CarD (PDB ID 4XLS). RNAP subunits are colored as in (c) and the σ factor in gray. The CarD-RID is magenta and CarD-CTD is green.
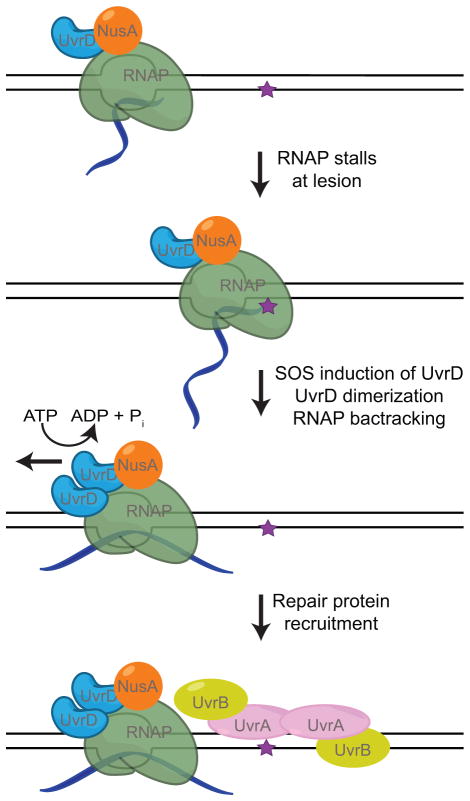
Figure 4. UvrD-dependent TCR.
UvrD and NusA associate with RNAP throughout transcription elongation. Upon induction of the DNA damage response the concentration of UvrD increases (113, 114) resulting in UvrD dimerization and activation of its helicase activity. With the help of NusA, UvrD pushes RNAP backwards without collapsing the transcription bubble possibly using its helicase activity to unwind the upstream edge of the bubble. As with the Mfd-dependent TCR pathway, UvrD-dependent TCR joins general NER at the damage verification step. However, transcription elongation can resume after UvrD-dependent TCR with the assistance of anti-backtracking factors.
Mfd CAUSES RNA POLYMERASE TO FORWARD TRANSLOCATE, ALLOWING REPAIR TO OCCUR
The Mfd protein, isolated by the pioneering work of the Sancar lab, was able to terminate transcription in an ATP-dependent manner and had clear homology to other characterized DNA helicases, such as RecG (58). An intriguing distinction at that time was that Mfd appeared to lack DNA or DNA/RNA unwinding activity (59), unlike RecG or Rho, the only other enzymatic transcription terminator known at the time. Instead, beautiful biochemical work from the Roberts laboratory demonstrated, a decade later, that Mfd was functioning as a dsDNA translocase, which docked at the upstream edge of the transcription bubble, and occupied an ~26 bp footprint (62). By substituting dATP for ATP as a substrate for Mfd, Park et al. completely decoupled ATP utilization by Mfd from ATP utilization by RNAP, and in combination with an immobilized assay for assessing release of RNA from magnetic bead-bound transcription elongation complexes proved that Mfd was using ATP hydrolysis to forward translocate nucleotide-starved RNAP in the downstream direction (62). Importantly, in the presence of substrate NTPs, elongation competed with Mfd-mediated release, suggesting two outcomes. In the absence of physical blockage (e.g. lesion on the template strand, a protein roadblock), but in the presence of NTPs, Mfd would rescue arrested, backtracked complexes, while in the presence of a lesion (or lack of NTPs), Mfd would release the transcript. Notably, while a large dsDNA segment upstream of the bubble was required for RNA release (due to Mfd binding), only little downstream DNA (3–4 bp according to exonuclease III accessibility assays) was necessary for this process. Altogether these data had a dual implication. First, Mfd likely translocated on dsDNA when bound to RNAP, at least over short distances. Second, Mfd did not dissociate arrested complexes from the backtracked position, but rather from the forward position where the RNA 3′ end is poised for elongation. Topologically, the consequence of Mfd translocation in the presence of tethering to RNAP via protein-protein interactions is that Mfd action could potentially impose torque on the upstream DNA, rewinding the upstream edge of the transcription bubble, with eventual collapse and complete dissociation of the elongation complex. This model was directly tested by employing heteroduplex elongation complexes that were assembled on templates carrying substitutions in the upstream edge of the bubble. As anticipated, Mfd released RNA more slowly from heteroduplex DNA complexes than from homoduplex DNA complexes due to its inability to rewind the heteroduplex portion of the transcription bubble, and pointed to mechanistic similarities with both intrinsic and Rho-mediated termination (67).
While dsDNA translocation is the one activity that is key for all known Mfd functions, the structural mechanism utilized by Mfd for translocation has remained elusive. It has been well established that duplex DNA binds across the central ATP-binding region of Mfd (Figure 3b), but no residues directly involved in DNA binding have been reported. In addition, homology modeling with structures of DNA-bound helicases/translocases are problematic due to the large conformational changes within this region that likely occur upon nucleotide binding/hydrolysis. These are diversely modulated by family-specific domains. Recent studies of Mfd binding to DNA in the presence of nucleotides using single-molecule magnetic trapping and permanganate footprinting have suggested that Mfd alone distorts DNA by wrapping and not unwinding (68, 69), consistent with Selby and Sancar’s DNaseI footprinting assays reported more than twenty years before (57). Since only the structure of nucleotide-free Mfd (Figure 3b), incompetent for DNA binding, is available (70, 71), crystal structures of nucleotide-bound and DNA-bound Mfd will be required for understanding the conformational changes associated with DNA binding and translocation and are, therefore, eagerly awaited.
While this body of work indicated that bubble rewinding likely played a key role in destabilization of the elongation complex, a purely mechanical model to explain Mfd action appeared too simple-minded. Several emerging lines of evidence gradually pointed to the process as involving multiple, distinct structural intermediates involving conformational changes in both Mfd and RNAP. First, the crystal structure of full-length Mfd (70) as well as of its N-terminal region (72) revealed a complex enzyme containing an UvrB-homology module (domains D1a-D2-D1b), a RNAP interaction domain (D4 or RID, interacting with the β subunit of RNAP, top inset in Figure 3b and Figure 3c), an ATP-dependent motor core (D5 and D6) and a regulatory C-terminal domain D7 (Figure 3b). The RID does not constitute a RNAP interaction structural module unique to TRCFs, but is also found in an emerging family of essential mycobacterial transcription factors called CarD (Figure 3d). Like Mfd, CarD utilizes its RID domain to bind to the β1 region of RNAP (73, 74), and together with its DNA-binding C-terminal domain, stabilizes open promoter complexes by inserting a conserved Trp wedge at the dsDNA/ssDNA junction within the bubble, preventing its collapse (74). There is no evidence that Mfd might interact with fork junctions, and CarD-RID does not make contacts with the DNA in the reported crystal structure (74).
The multi-modular architecture of Mfd with flexible loops (highlighted in Figure 3a) was suggestive of large conformational changes. What was striking was that the region of highest sequence homology to UvrB, where UvrA was believed to dock, was completely occluded due to a “clamp” interaction with domain D7. Savery and colleagues showed that this “clamp” was inhibitory – interfering with this interaction by domain deletion resulted in Mfd hyperactivity increased ATPase rates as well as the ability of Mfd to translocate on naked DNA (as opposed to in the presence of RNAP, as assayed using triplex displacement assays) (75). At the same time, deletion of D7 led to hypersensitivity to UV, presumably due to unscheduled sequestration of UvrA and its diversion from global NER (75, 59, 57). A subsequent crystal structure of a core Mfd-UvrA complex confirmed this model (Figure 3b, bottom inset). UvrA bound Mfd via a surface located in D2 that was largely occluded in the context of full-length Mfd by domain D7 (76), and UvrA inhibited in a dose-dependent manner the formation of an engineered D2–D7 interdomain crosslink (76). It follows then that for UvrA to dock, D7 would have to undergo displacement.
What triggers UvrA recruitment? The engineered D2–D7 crosslink did not substantially inhibit RNA release (76), implying that repositioning of D7 was not required for activating the translocase function of Mfd when bound to RNAP. This argued for a sequential model of UvrA recruitment in which opening of the clamp was not required for RNA release and short-range forward translocation of RNAP (and Mfd), but likely only occurred later, possibly upon Mfd engaging the DNA lesion (Figure 2). Precise spatiotemporal regulation could be achieved this way to prevent unscheduled displacement of RNAP at pause sites rather than DNA damage sites. At the same time, this mechanistic scheme implied that Mfd participated in lesion recognition. Indeed, TCR and global genome NER differ in their requirements for damage recognition by UvrA. NER and TCR assays conducted with UvrA variants defective in damage recognition [UvrA K37A affecting the Walker A motif of the proximal ATPase site; UvrAG502D (77, 78); and UvrA Δ290–400, lacking the insertion domain, important for DNA damage specificity (77)] indicated that these mutations had a more detrimental effect on global NER than on TCR (79). The corollary of these findings was that Mfd acted at an early stage of DNA repair, likely during loading of UvrA rather than later, such as at the stage of destabilizing the UvrA2:B2:DNA complex as had been suggested by Sancar (57, 58). Given that during NER, the UvrA2B2 complex sequentially probes both strands of the DNA via wrapping by each of the two UvrB subunits, as shown using atomic force microscopy (80), it is attractive to speculate that during TCR, an asymmetric UvrA2:UvrB:Mfd:DNA complex might form. This might explain the asymmetry of repair characteristic of TCR. In fact, coupling intermediates, either RNAP:DNA:RNA:Mfd:UvrA(B) or Mfd:UvrA:DNA that would help discriminate between the concerted and sequential model of recruitment have been difficult to trap via traditional hydrodynamic methods. Single-molecule nanomanipulation combined with single-molecule fluorescence have only very recently suggested that binding of both UvrA and UvrB to Mfd-RNAP causes eviction of both Mfd and RNAP from the macromolecular intermediate formed on the DNA (81).
The RNA:DNA hybrid within the RNAP elongation complex plays a crucial role in conferring its stability (82). Therefore, it has been generally surmised that once the transcript was released by Mfd, the elongation complex would soon dissociate, and RNAP would diffuse away. In fact, single-molecule magnetic trap experiments coupled with single-molecule fluorescence experiments demonstrated that this might not be so. First, after initial Mfd binding to RNAP, a long intermediate is formed in which two thirds of the transcription bubble is rewound and DNA is bent by 90° (68). For its resolution (e.g. RNA release), pulse chase experiments showed that the binding on a new ATP molecule is necessary. In a subsequent study, Strick and coworkers were able to demonstrate by fluorescently labeling Mfd and RNAP, that once the transcript was released, and RNAP dissociated off the template, Mfd formed a “search complex” with RNAP (Figure 2b). This search complex likely forms via protein-protein interactions of the RID domain in Mfd with the “IKE” motif in the N-terminal region of the β subunit of RNAP (top inset in Figure 3a and Figure 3b) (62, 70, 69), conferring Mfd processivity on DNA (69, 83).
If Mfd-RNAP interactions via the RID are sufficiently strong to stably tether the two molecules, one could imagine that Mfd might be able to dock even onto actively elongating or paused transcription complexes. In a series of transcription assays performed on templates carrying a pause site, followed by a template strand DNA lesion hundreds of basepairs downstream, Mfd appeared to bind to paused (rather than lesion-stalled) RNAP and mediate UvrA-dependent, strand-specific repair of the downstream lesion (83). This repair mechanism “at a distance” (illustrated in Figure 2b) relies on active translocation by Mfd as part of the search complex, and implies that Mfd does not recognize, at least initially, a specific structural state of a lesion stalled RNAP, but, in fact, may bind to and possibly travel with RNAP throughout the elongation phase. In fact, crystallographic studies of RNAPII stalled by a CPD showed only subtle conformational changes compared to a RNAPII stalled by nucleotide deprivation (84). Transcription initiation complexes are resistant to Mfd (62), likely because the sigma promoter specificity subunit high-affinity binding site on the upstream DNA coincides with the region recognized by Mfd. Then why are only stalled complexes susceptible to Mfd-mediated release? The answer might lie in the kinetics of the pathway. While initial docking of Mfd to RNAP is fast, the subsequent mechanistic step (formation of the stable intermediate containing a partially rewound bubble) is remarkably slow, and may involve slow Mfd conformational changes. This kinetic selection mechanism would ensure that Mfd does not act on the majority of actively elongating or paused transcription complexes, which would be deleterious to the cell.
UvrD INDUCES RNA POLYMERASE BACKTRACKING, ALLOWING REPAIR TO OCCUR
Besides forward translocation with repair exemplified by Mfd-dependent TCR, the second possible fate for a stalled TEC is for it to be temporarily backtracked. This would allow for assembly of repair factors at the exposed DNA lesion, and possibly, subsequent resumption of elongation via anti-backtracking mechanisms (Figure 4). This may seem like a rather complicated mechanism, but, at the same time, it is particularly attractive in the case of highly transcribed genes. Here, arrays of multiple, densely packed RNAPs would prevent binding and action of Mfd at the upstream edge of the transcription bubble. Indeed, Kunala and Brash noticed quite early on that Mfd-dependent repair occurs primarily under conditions of basal transcription, and not upon full induction of the lac operon (85). The UV sensitivity of the mfd strain is quite modest, raising the possibility that additional, Mfd-independent pathways operate in cells, particularly at UV fluences high enough to induce the SOS response. Recent work from the Walker and Nudler laboratories implicate a well-known transcriptional regulator, NusA, and a component of the core NER system, UvrD, in this alternative pathway. The sensitivity of uvrD and nusA strains to UV and DNA damaging agents is significantly greater than that of the mfd strain. In addition, mutations in the β-subunit of RNA polymerase that confer sensitivity or resistance to the DNA-damaging agent nitrofurazone can be suppressed by mutations in uvrA and nusA, but not in the mfd gene (86, 87).
Given its more prominent role in resistance to UV, why was UvrD only recently implicated in TCR? UvrD, which belongs to the superfamily 1 (SF1) of helicases/translocases, displaces the damaged, excised oligonucleotide during global genome NER and Mfd-dependent TCR. UvrD unwinds dsDNA by translocating on ssDNA in the 3′ to 5′ direction, thereby pealing away the damage-containing oligonucleotide. Its essential role in global genome NER and Mfd-dependent TCR made it difficult to link it to an alternative TCR pathway, as its deletion would impair all of these processes. It was interactome studies suggesting that UvrD binds at abundant levels to RNAP (63) that led investigators to take a closer look at UvrD and investigate the possibility that it may interact directly with RNAP and function as a TRCF. Nudler and coworkers showed that UvrD does indeed form a stable interaction with the β and β′ subunits of RNAP, and that again, like Mfd, it binds at the upstream edge of the transcription bubble. However, unlike Mfd, UvrD preferentially binds to single-stranded/duplex DNA junctions and may promote unwinding of the upstream edge of the bubble (88). This promotes backtracking of RNA polymerase in vivo and in vitro, even in the absence of pause sites or lesions (63). Another difference between Mfd and UvrD is that UvrD likely acts as a dimer in this context, while Mfd as a monomer. Indeed, titration experiments have shown that UvrD-mediated backtracking dependence on UvrD concentrations is sigmoidal (63).
Notably, UvrD is an abundant protein, with a copy number of ~3000, comparable to RNAP copy numbers, which range between 1600–8000 depending on the growth stage (89). These data, together with the stability and abundance of UvrD-RNAP complexes observed by the Nudler lab (63), suggest that RNAP might be frequently pre-loaded with UvrD in non-stressed cells, even before DNA damage is encountered. This would promote backtracking and TCR even under conditions of high transcription, characterized by the presence of highly-packed RNAP arrays, which would be inhibitory to Mfd loading onto individual elongation complexes within the array. Upon DNA damage, UvrD levels rise even higher due to Lex-dependent upregulation (90). This increase in intracellular UvrD concentrations could promote UvrD dimerization onto RNAP, thus activating the backtracking function and triggering TCR. Consequently, UvrD-dependent TCR might be prevalent during acute genotoxic stress, while Mfd-dependent TCR might prevail during normal growth.
While UvrD translocase function fulfills one of the functions essential for a TRCF, it does not bind UvrA. So how are the rest of the repair enzymes recruited in this UvrD-dependent pathway? The answer is unclear at the moment, but might involve direct physical interaction between the well-known transcriptional regulator NusA and UvrA (86) or direct interaction between UvrD and UvrB (91).
THE ALARMONE ppGpp: THE SMALL MOLECULE “COUPLING” AND PRO-BACKTRACKING FACTOR
Two key events are required for UvrD-dependent TCR. First, UvrD must bind to RNAP. This appears an easy enough task as the RNAP-UvrD interaction is high-affinity (Kd=35 nM) (92), and UvrD is relatively abundant, found at approximately a 1:1 ratio with RNAP in the absence of stress (93). Second, for UvrD to become competent for DNA unwinding, it must dimerize (63, 94). UvrD self-association equilibria are such that these dimers are not stable (95), and likely form transiently, even upon induction of uvrD during stress (93). One solution to this problem could be to modify RNAP in such a way as to lower the energy barrier to backtracking. Indeed, we now know of both small molecule as well as protein pro-backtracking factors. NusA, already implicated in recruiting UvrA to nitrofurazone-stalled RNAPs, is a well-studied example (96), but equally interesting is the small-molecule alarmone guanosine-3′,5′-(bis)pyrophosphate (ppGpp). ppGpp plays a crucial role in the stringent response. Upon starvation, intracellular levels of ppGpp rapidly rise, which results in a global reprogramming of gene expression, primarily due to ppGpp directly targeting RNAP and destabilizing open promoter complexes (97, 98). Structural analyses have suggested that ppGpp modifies RNAP structure in such a way as to make it more prone to backtracking – it may widen its claw pincers (98). Interestingly, cells deficient in the enzymes producing ppGpp, relA and spoT, are sensitive to certain DNA damaging agents, such as UV, 4-nitroquinoline-1-oxide and nitrofurazone (99, 92, 100). Recently, work from the laboratory of Evgeny Nudler found that in ppGpp-deficient cells, the repair of the transcribed strand of the lac operon was slowed down and occurred at levels comparable to that seen for the nontemplate strand, supporting a key role for ppGpp in TCR. In vitro, addition of ppGpp stimulated UvrD-mediated backtracking, and in vivo, ppGpp induction also enhanced RNAP backtracking (92). Together with a rapid decline of ppGpp after the genotoxic stress, this ensures a rapid activation of repair after backtracking, but also a rapid recovery, in which anti-backtracking factors (DksA, ribosomes, and Mfd) could act upon the RNAP to promote continued elongation.
CONCLUSIONS AND PERSPECTIVES
For roughly two decades since discovery of TCR in bacteria and the identification of Mfd as a TRCF, it has been surmised that damage detection takes place by proxy, with RNAP fulfilling, at least partially, the role of the DNA damage recognition subunit of the Uvr(A)BC excinuclease. It is becoming clear that DNA damage detection and verification during TCR is a more complex process than anticipated, and that TRCFs such as Mfd might have an active role in this process. We are still far away from fully understanding the stoichiometry and order of assembly of TCR intermediates, but single-molecule approaches that can track specific components throughout the multi-step TCR pathway as well structure-informed biochemistry have started to shed light onto these questions (69, 68, 81).
Factors such as Mfd have roles beyond TCR that stem from their motor protein activity disrupting elongation complexes stalled by various roadblocks, including replication forks (101). This leads to relief of conflicts (head-on collisions) between the transcription and replication machineries, and prevents the appearance of deleterious dsDNA breaks (102). This process has even been reconstituted in vitro (101). However, when replication-transcription collisions are promoted by inversion of the highly-transcribed ribosomal rrn operon, mfd inactivation shows no significant effect on resolving head-on collisions (103). Instead, alternative pathways involving replication fork reversal and RecBC(D) as well as the subsequent action of accessory replicative helicases such as Rep, UvrB and DinG appear to have a more pronounced effect (103). These data are seemingly contradictory, but the failure to notice mfd effects in the latter case could be due to the high transcription of the rrn operon and failure of Mfd to load onto densely packed RNAP arrays (vide supra).
The interplay between replication and transcription via Mfd can also result in accelerated evolution, specifically of lagging-strand genes (104). In Bacillus subtilis, mfd is epistatic to both uvrA and the error-prone DNA polymerase polY1 (105). In the B. subtilis sporangium, the interplay between mfd and TLS polymerases also plays a key role in protection from DNA damage as inactivation of the yqjH and yqjW genes, both encoding TLS polymerases, suppresses UV-induced mutations in the mfd knockout strain (106). At least under certain conditions, gap filling during TCR appears carried out by TLS polymerases rather than family A DNA polymerases. This is not without precedent. The use of TLS polymerases during NER has been well documented in eukaryotes (107).
While many of the details of the TCR pathway have been fleshed out during the last decade, structurally, the Mfd system remains relatively poorly characterized. How does Mfd translocate on DNA? What are the mutually induced conformational changes in Mfd and RNAP? Without doubt, X-ray crystallography and the use of new technological developments in electron microscopy, will prove instrumental in providing structural snapshots of the molecular players and in reconstructing the underlying mechanochemistry of the TCR system. mfd is not an essential gene and its deletion only modestly affects UV sensitivity, but the Mfd factor has roles beyond TCR, and is gradually emerging as playing an important role in mediating adaptive mutagenesis (108), in virulence and the development of antibiotic resistance in several human pathogens, including Helicobacter pylori (109), Campylobacter jejuni (110) and Clostridium difficile (111). Most recently, Mfd has also been implicated in protection from reactive nitrogen species, which are produced by the host immune response following infection (112). These new data suggest that the study of Mfd roles in bacterial physiology might still yield surprises, and that Mfd could be targeted for development of a broad-spectrum antibiotic.
The long-held idea of Mfd as the only bacterial TRCF has been overturned. UvrD, a protein known for decades to be involved in genome-wide NER, can also promote backtracking of stalled RNAP and strand-specific DNA repair. This newly identified role for UvrD raises the question of whether additional TRCFs are yet to be identified, and also signals the necessity of renaming TRCF back to Mfd, as this no longer is the sole bacterial coupling factor.
Acknowledgments
A.M.D. is a recipient of the Medical Research Grant No. 20133965 from The Rhode Island Foundation and of the RI-INBRE Pilot Project and Career Development Awards from the National Institute of General Medical Sciences of the National Institutes of Health under grant No. P20GM103430. We thank Dr. Art Landy for generous support in starting the Deaconescu laboratory, and our anonymous reviewers for constructive feedback.
Footnotes
This article is part of the Special Issue highlighting Dr. Aziz Sancar’s outstanding contributions to various aspects of the repair of DNA photodamage in honor of his recent Nobel Prize in Chemistry.
References
- 1.Ljungman M, Lane DP. Transcription - Guarding the Genome by Sensing DNA Damage. Nat Rev Cancer. 2004;4:727–737. doi: 10.1038/nrc1435. [DOI] [PubMed] [Google Scholar]
- 2.Cadet J, Wagner JR. DNA Base Damage by Reactive Oxygen Species, Oxidizing Agents, and UV Radiation. Cold Spring Harb Perspect Biol. 2013:5. doi: 10.1101/cshperspect.a012559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Setlow RB, Carrier WL. The Disappearance of Thymine Dimers from DNA: An Error-Correcting Mechanism. Proc Natl Acad Sci U S A. 1964;51:226–231. doi: 10.1073/pnas.51.2.226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Boyce RP, Howard-Flanders P. Release of Ultraviolet Light-Induced Thymine Dimers from DNA in E. Coli K-12. Proc Natl Acad Sci U S A. 1964;51:293–300. doi: 10.1073/pnas.51.2.293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Pettijohn D, Hanawalt P. Evidence for Repair-Replication of Ultraviolet Damaged DNA in Bacteria. J Mol Biol. 1964;9:395–410. doi: 10.1016/s0022-2836(64)80216-3. [DOI] [PubMed] [Google Scholar]
- 6.Howard-Flanders P, Boyce RP, Theriot L. Three Loci in Escherichia Coli K-12 That Control the Excision of Pyrimidine Dimers and Certain Other Mutagen Products from DNA. Genetics. 1966;53:1119–1136. doi: 10.1093/genetics/53.6.1119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Seeberg E, Nissen-Meyer J, Strike P. Incision of Ultraviolet-Irradiated DNA by Extracts of E. Coli Requires Three Different Gene Products. Nature. 1976;263:524–526. doi: 10.1038/263524a0. [DOI] [PubMed] [Google Scholar]
- 8.Seeberg E, Strike P. Excision Repair of Ultraviolet-Irradiated Deoxyribonucleic Acid in Plasmolyzed Cells of Escherichia Coli. J Bacteriol. 1976;125:787–795. doi: 10.1128/jb.125.3.787-795.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sancar A, Wharton RP, Seltzer S, Kacinski BM, Clarke ND, Rupp WD. Identification of the Uvra Gene Product. J Mol Biol. 1981;148:45–62. doi: 10.1016/0022-2836(81)90234-5. [DOI] [PubMed] [Google Scholar]
- 10.Sancar A, Kacinski BM, Mott DL, Rupp WD. Identification of the UvrC Gene Product. Proc Natl Acad Sci U S A. 1981;78:5450–5454. doi: 10.1073/pnas.78.9.5450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sancar A, Clarke ND, Griswold J, Kennedy WJ, Rupp WD. Identification of the UvrB Gene Product. J Mol Biol. 1981;148:63–76. doi: 10.1016/0022-2836(81)90235-7. [DOI] [PubMed] [Google Scholar]
- 12.Sancar A, Rupp WD. A Novel Repair Enzyme: UvraABC Excision Nuclease of Escherichia Coli Cuts a DNA Strand on Both Sides of the Damaged Region. Cell. 1983;33:249–260. doi: 10.1016/0092-8674(83)90354-9. [DOI] [PubMed] [Google Scholar]
- 13.Kisker C, Kuper J, Van Houten B. Prokaryotic Nucleotide Excision Repair. Cold Spring Harb Perspect Biol. 2013;5:a012591. doi: 10.1101/cshperspect.a012591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Pakotiprapha D, Jeruzalmi D. Small-Angle X-Ray Scattering Reveals Architecture and A(2)B(2) Stoichiometry of the UvrA-UvrB DNA Damage Sensor. Proteins. 2013;81:132–139. doi: 10.1002/prot.24170. [DOI] [PubMed] [Google Scholar]
- 15.Pakotiprapha D, Liu Y, Verdine GL, Jeruzalmi D. A Structural Model for the Damage-Sensing Complex in Bacterial Nucleotide Excision Repair. J Biol Chem. 2009;284:12837–12844. doi: 10.1074/jbc.M900571200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kad NM, Wang H, Kennedy GG, Warshaw DM, Van Houten B. Collaborative Dynamic DNA Scanning by Nucleotide Excision Repair Proteins Investigated by Single- Molecule Imaging of Quantum-Dot-Labeled Proteins. Mol Cell. 2010;37:702–713. doi: 10.1016/j.molcel.2010.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bohr VA, Smith CA, Okumoto DS, Hanawalt PC. DNA Repair in an Active Gene: Removal of Pyrimidine Dimers from the Dhfr Gene of Cho Cells Is Much More Efficient Than in the Genome Overall. Cell. 1985;40:359–369. doi: 10.1016/0092-8674(85)90150-3. [DOI] [PubMed] [Google Scholar]
- 18.Mellon I, Spivak G, Hanawalt PC. Selective Removal of Transcription-Blocking DNA Damage from the Transcribed Strand of the Mammalian Dhfr Gene. Cell. 1987;51:241–249. doi: 10.1016/0092-8674(87)90151-6. [DOI] [PubMed] [Google Scholar]
- 19.Venema J, Mullenders LH, Natarajan AT, van Zeeland AA, Mayne LV. The Genetic Defect in Cockayne Syndrome Is Associated with a Defect in Repair of Uv-Induced DNA Damage in Transcriptionally Active DNA. Proc Natl Acad Sci U S A. 1990;87:4707–4711. doi: 10.1073/pnas.87.12.4707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mellon I, Hanawalt PC. Induction of the Escherichia Coli Lactose Operon Selectively Increases Repair of Its Transcribed DNA Strand. Nature. 1989;342:95–98. doi: 10.1038/342095a0. [DOI] [PubMed] [Google Scholar]
- 21.Li S. Transcription Coupled Nucleotide Excision Repair in the Yeast Saccharomyces Cerevisiae: The Ambiguous Role of Rad26. DNA Repair (Amst) 2015;36:43–48. doi: 10.1016/j.dnarep.2015.09.006. [DOI] [PubMed] [Google Scholar]
- 22.van Gool AJ, Verhage R, Swagemakers SM, van de Putte P, Brouwer J, Troelstra C, Bootsma D, Hoeijmakers JH. Rad26, the Functional S. Cerevisiae Homolog of the Cockayne Syndrome B Gene Ercc6. EMBO J. 1994;13:5361–5369. doi: 10.1002/j.1460-2075.1994.tb06871.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Li S, Smerdon MJ. Rpb4 and Rpb9 Mediate Subpathways of Transcription-Coupled DNA Repair in Saccharomyces Cerevisiae. EMBO J. 2002;21:5921–5929. doi: 10.1093/emboj/cdf589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Li S, Smerdon MJ. Dissecting Transcription-Coupled and Global Genomic Repair in the Chromatin of Yeast Gal1-10 Genes. J Biol Chem. 2004;279:14418–14426. doi: 10.1074/jbc.M312004200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Terleth C, van Sluis CA, van de Putte P. Differential Repair of UV Damage in Saccharomyces Cerevisiae. Nucleic Acids Res. 1989;17:4433–4439. doi: 10.1093/nar/17.12.4433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.van der Horst GT, van Steeg H, Berg RJ, van Gool AJ, de Wit J, Weeda G, Morreau H, Beems RB, van Kreijl CF, de Gruijl FR, Bootsma D, Hoeijmakers JH. Defective Transcription-Coupled Repair in Cockayne Syndrome B Mice Is Associated with Skin Cancer Predisposition. Cell. 1997;89:425–435. doi: 10.1016/s0092-8674(00)80223-8. [DOI] [PubMed] [Google Scholar]
- 27.Lans H, Marteijn JA, Schumacher B, Hoeijmakers JH, Jansen G, Vermeulen W. Involvement of Global Genome Repair, Transcription Coupled Repair, and Chromatin Remodeling in UV DNA Damage Response Changes During Development. PLoS Genet. 2010;6:e1000941. doi: 10.1371/journal.pgen.1000941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lans H, Vermeulen W. Tissue Specific Response to DNA Damage: C. Elegans as Role Model. DNA Repair (Amst) 2015;32:141–148. doi: 10.1016/j.dnarep.2015.04.025. [DOI] [PubMed] [Google Scholar]
- 29.Stantial N, Dumpe J, Pietrosimone K, Baltazar F, Crowley DJ. Transcription-Coupled Repair of UV Damage in the Halophilic Archaea. DNA Repair (Amst) 2016;41:63–68. doi: 10.1016/j.dnarep.2016.03.007. [DOI] [PubMed] [Google Scholar]
- 30.Romano V, Napoli A, Salerno V, Valenti A, Rossi M, Ciaramella M. Lack of Strand-Specific Repair of Uv-Induced DNA Lesions in Three Genes of the Archaeon Sulfolobus Solfataricus. J Mol Biol. 2007;365:921–929. doi: 10.1016/j.jmb.2006.10.045. [DOI] [PubMed] [Google Scholar]
- 31.Dorazi R, Gotz D, Munro S, Bernander R, White MF. Equal Rates of Repair of DNA Photoproducts in Transcribed and Non-Transcribed Strands in Sulfolobus Solfataricus. Mol Microbiol. 2007;63:521–529. doi: 10.1111/j.1365-2958.2006.05516.x. [DOI] [PubMed] [Google Scholar]
- 32.de Waard H, Sonneveld E, de Wit J, Esveldt-van Lange R, Hoeijmakers JH, Vrieling H, van der Horst GT. Cell-Type-Specific Consequences of Nucleotide Excision Repair Deficiencies: Embryonic Stem Cells Versus Fibroblasts. DNA Repair (Amst) 2008;7:1659–1669. doi: 10.1016/j.dnarep.2008.06.009. [DOI] [PubMed] [Google Scholar]
- 33.Christians FC, Hanawalt PC. Inhibition of Transcription and Strand-Specific DNA Repair by Alpha-Amanitin in Chinese Hamster Ovary Cells. Mutat Res. 1992;274:93–101. doi: 10.1016/0921-8777(92)90056-9. [DOI] [PubMed] [Google Scholar]
- 34.Selby CP, Sancar A. Transcription Preferentially Inhibits Nucleotide Excision Repair of the Template DNA Strand in Vitro. J Biol Chem. 1990;265:21330–21336. [PubMed] [Google Scholar]
- 35.Sweder KS, Hanawalt PC. Preferential Repair of Cyclobutane Pyrimidine Dimers in the Transcribed Strand of a Gene in Yeast Chromosomes and Plasmids Is Dependent on Transcription. Proc Natl Acad Sci U S A. 1992;89:10696–10700. doi: 10.1073/pnas.89.22.10696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Tornaletti S, Donahue BA, Reines D, Hanawalt PC. Nucleotide Sequence Context Effect of a Cyclobutane Pyrimidine Dimer Upon RNA Polymerase Ii Transcription. J Biol Chem. 1997;272:31719–31724. doi: 10.1074/jbc.272.50.31719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Tornaletti S, Reines D, Hanawalt PC. Structural Characterization of RNA Polymerase Ii Complexes Arrested by a Cyclobutane Pyrimidine Dimer in the Transcribed Strand of Template DNA. J Biol Chem. 1999;274:24124–24130. doi: 10.1074/jbc.274.34.24124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Tornaletti S, Hanawalt PC. Effect of DNA Lesions on Transcription Elongation. Biochimie. 1999;81:139–146. doi: 10.1016/s0300-9084(99)80046-7. [DOI] [PubMed] [Google Scholar]
- 39.Selby CP, Sancar A. Mechanisms of Transcription-Repair Coupling and Mutation Frequency Decline. Microbiol Rev. 1994;58:317–329. doi: 10.1128/mr.58.3.317-329.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Cleaver JE. Defective Repair Replication of DNA in Xeroderma Pigmentosum. Nature. 1968;218:652–656. doi: 10.1038/218652a0. [DOI] [PubMed] [Google Scholar]
- 41.Setlow RB, Regan JD, German J, Carrier WL. Evidence That Xeroderma Pigmentosum Cells Do Not Perform the First Step in the Repair of Ultraviolet Damage to Their DNA. Proc Natl Acad Sci U S A. 1969;64:1035–1041. doi: 10.1073/pnas.64.3.1035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Jaarsma D, van der Pluijm I, de Waard MC, Haasdijk ED, Brandt R, Vermeij M, Rijksen Y, Maas A, van Steeg H, Hoeijmakers JH, van der Horst GT. Age-Related Neuronal Degeneration: Complementary Roles of Nucleotide Excision Repair and Transcription-Coupled Repair in Preventing Neuropathology. PLoS Genet. 2011;7:e1002405. doi: 10.1371/journal.pgen.1002405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Velez-Cruz R, Egly JM. Cockayne Syndrome Group B (CSB) Protein: At the Crossroads of Transcriptional Networks. Mech Ageing Dev. 2013;134:234–242. doi: 10.1016/j.mad.2013.03.004. [DOI] [PubMed] [Google Scholar]
- 44.Kamenisch Y, Berneburg M. Mitochondrial CSA and CSB: Protein Interactions and Protection from Ageing Associated DNA Mutations. Mech Ageing Dev. 2013;134:270–274. doi: 10.1016/j.mad.2013.03.005. [DOI] [PubMed] [Google Scholar]
- 45.Lake RJ, Geyko A, Hemashettar G, Zhao Y, Fan HY. UV-Induced Association of the Csb Remodeling Protein with Chromatin Requires Atp-Dependent Relief of N-Terminal Autorepression. Mol Cell. 2010;37:235–246. doi: 10.1016/j.molcel.2009.10.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Citterio E, Van Den Boom V, Schnitzler G, Kanaar R, Bonte E, Kingston RE, Hoeijmakers JH, Vermeulen W. ATP-Dependent Chromatin Remodeling by the Cockayne Syndrome B DNA Repair-Transcription-Coupling Factor. Molecular & Cellular Biology. 2000;20:7643–7653. doi: 10.1128/mcb.20.20.7643-7653.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Stephens PJ, Tarpey PS, Davies H, Van Loo P, Greenman C, Wedge DC, Nik-Zainal S, Martin S, Varela I, Bignell GR, Yates LR, Papaemmanuil E, Beare D, Butler A, Cheverton A, Gamble J, Hinton J, Jia M, Jayakumar A, Jones D, Latimer C, Lau KW, McLaren S, McBride DJ, Menzies A, Mudie L, Raine K, Rad R, Chapman MS, Teague J, Easton D, Langerod A, Lee MT, Shen CY, Tee BT, Huimin BW, Broeks A, Vargas AC, Turashvili G, Martens J, Fatima A, Miron P, Chin SF, Thomas G, Boyault S, Mariani O, Lakhani SR, van de Vijver M, van ‘t Veer L, Foekens J, Desmedt C, Sotiriou C, Tutt A, Caldas C, Reis-Filho JS, Aparicio SA, Salomon AV, Borresen-Dale AL, Richardson AL, Campbell PJ, Futreal PA, Stratton MR. The Landscape of Cancer Genes and Mutational Processes in Breast Cancer. Nature. 2012;486:400–404. doi: 10.1038/nature11017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Nik-Zainal S, Alexandrov LB, Wedge DC, Van Loo P, Greenman CD, Raine K, Jones D, Hinton J, Marshall J, Stebbings LA, Menzies A, Martin S, Leung K, Chen L, Leroy C, Ramakrishna M, Rance R, Lau KW, Mudie LJ, Varela I, McBride DJ, Bignell GR, Cooke SL, Shlien A, Gamble J, Whitmore I, Maddison M, Tarpey PS, Davies HR, Papaemmanuil E, Stephens PJ, McLaren S, Butler AP, Teague JW, Jonsson G, Garber JE, Silver D, Miron P, Fatima A, Boyault S, Langerod A, Tutt A, Martens JW, Aparicio SA, Borg A, Salomon AV, Thomas G, Borresen-Dale AL, Richardson AL, Neuberger MS, Futreal PA, Campbell PJ, Stratton MR. Mutational Processes Molding the Genomes of 21 Breast Cancers. Cell. 2012;149:979–993. doi: 10.1016/j.cell.2012.04.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Nik-Zainal S, Van Loo P, Wedge DC, Alexandrov LB, Greenman CD, Lau KW, Raine K, Jones D, Marshall J, Ramakrishna M, Shlien A, Cooke SL, Hinton J, Menzies A, Stebbings LA, Leroy C, Jia M, Rance R, Mudie LJ, Gamble SJ, Stephens PJ, McLaren S, Tarpey PS, Papaemmanuil E, Davies HR, Varela I, McBride DJ, Bignell GR, Leung K, Butler AP, Teague JW, Martin S, Jonsson G, Mariani O, Boyault S, Miron P, Fatima A, Langerod A, Aparicio SA, Tutt A, Sieuwerts AM, Borg A, Thomas G, Salomon AV, Richardson AL, Borresen-Dale AL, Futreal PA, Stratton MR, Campbell PJ. The Life History of 21 Breast Cancers. Cell. 2012;149:994–1007. doi: 10.1016/j.cell.2012.04.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Marteijn JA, Lans H, Vermeulen W, Hoeijmakers JH. Understanding Nucleotide Excision Repair and Its Roles in Cancer and Ageing. Nat Rev Mol Cell Biol. 2014;15:465–481. doi: 10.1038/nrm3822. [DOI] [PubMed] [Google Scholar]
- 51.Vermeulen W, Fousteri M. Mammalian Transcription-Coupled Excision Repair. Cold Spring Harb Perspect Biol. 2013;5:a012625. doi: 10.1101/cshperspect.a012625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Witkin EM. Time, Temperature, and Protein Synthesis: A Study of Ultraviolet-Induced Mutation in Bacteria. Cold Spring Harbor Symposia on Quantitative Biology. 1956;21:123–140. doi: 10.1101/sqb.1956.021.01.011. [DOI] [PubMed] [Google Scholar]
- 53.Witkin EM. Radiation-Induced Mutations and Their Repair. Science. 1966;152:1345–1353. doi: 10.1126/science.152.3727.1345. [DOI] [PubMed] [Google Scholar]
- 54.George DL, Witkin EM. Ultraviolet Light-Induced Responses of an Mfd Mutant of Escherichia Coli B/R Having a Slow Rate of Dimer Excision. Mutat Res. 1975;28:347–354. doi: 10.1016/0027-5107(75)90229-8. [DOI] [PubMed] [Google Scholar]
- 55.Bockrath RC, Palmer JE. Differential Repair of Premutational Uv-Lesions at Trna Genes in E. Coli. Mol Gen Genet. 1977;156:133–140. doi: 10.1007/BF00283485. [DOI] [PubMed] [Google Scholar]
- 56.Selby CP, Witkin EM, Sancar A. Escherichia Coli Mfd Mutant Deficient in “Mutation Frequency Decline” Lacks Strand-Specific Repair: In Vitro Complementation with Purified Coupling Factor. Proc Natl Acad Sci U S A. 1991;88:11574–11578. doi: 10.1073/pnas.88.24.11574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Selby CP, Sancar A. Structure and Function of Transcription-Repair Coupling Factor. I. Structural Domains and Binding Properties. J Biol Chem. 1995;270:4882–4889. doi: 10.1074/jbc.270.9.4882. [DOI] [PubMed] [Google Scholar]
- 58.Selby CP, Sancar A. Molecular Mechanism of Transcription-Repair Coupling. Science. 1993;260:53–58. doi: 10.1126/science.8465200. [DOI] [PubMed] [Google Scholar]
- 59.Selby CP, Sancar A. Structure and Function of Transcription-Repair Coupling Factor. Ii. Catalytic Properties. J Biol Chem. 1995;270:4890–4895. doi: 10.1074/jbc.270.9.4890. [DOI] [PubMed] [Google Scholar]
- 60.Kellis M, Wold B, Snyder MP, Bernstein BE, Kundaje A, Marinov GK, Ward LD, Birney E, Crawford GE, Dekker J, Dunham I, Elnitski LL, Farnham PJ, Feingold EA, Gerstein M, Giddings MC, Gilbert DM, Gingeras TR, Green ED, Guigo R, Hubbard T, Kent J, Lieb JD, Myers RM, Pazin MJ, Ren B, Stamatoyannopoulos JA, Weng Z, White KP, Hardison RC. Defining Functional DNA Elements in the Human Genome. Proc Natl Acad Sci U S A. 2014;111:6131–6138. doi: 10.1073/pnas.1318948111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Lindsey-Boltz LA, Sancar A. Rna Polymerase: The Most Specific Damage Recognition Protein in Cellular Responses to DNA Damage? Proc Natl Acad Sci U S A. 2007;104:13213–13214. doi: 10.1073/pnas.0706316104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Park JS, Marr MT, Roberts JW. E. Coli Transcription Repair Coupling Factor (Mfd Protein) Rescues Arrested Complexes by Promoting Forward Translocation. Cell. 2002;109:757–767. doi: 10.1016/s0092-8674(02)00769-9. [DOI] [PubMed] [Google Scholar]
- 63.Epshtein V, Kamarthapu V, McGary K, Svetlov V, Ueberheide B, Proshkin S, Mironov A, Nudler E. UvrD Facilitates DNA Repair by Pulling Rna Polymerase Backwards. Nature. 2014;505:372–377. doi: 10.1038/nature12928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Kamarthapu V, Nudler E. Rethinking Transcription Coupled DNA Repair. Curr Opin Microbiol. 2015;24:15–20. doi: 10.1016/j.mib.2014.12.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Morreall JF, Petrova L, Doetsch PW. Transcriptional Mutagenesis and Its Potential Roles in the Etiology of Cancer and Bacterial Antibiotic Resistance. J Cell Physiol. 2013;228:2257–2261. doi: 10.1002/jcp.24400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Deaconescu AM. RNA Polymerase between Lesion Bypass and DNA Repair. Cell Mol Life Sci. 2013 doi: 10.1007/s00018-013-1384-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Park JS, Roberts JW. Role of DNA Bubble Rewinding in Enzymatic Transcription Termination. Proc Natl Acad Sci U S A. 2006;103:4870–4875. doi: 10.1073/pnas.0600145103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Howan K, Smith AJ, Westblade LF, Joly N, Grange W, Zorman S, Darst SA, Savery NJ, Strick TR. Initiation of Transcription-Coupled Repair Characterized at Single-Molecule Resolution. Nature. 2012;490:431–434. doi: 10.1038/nature11430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Graves ET, Duboc C, Fan J, Stransky F, Leroux-Coyau M, Strick TR. A Dynamic DNA-Repair Complex Observed by Correlative Single-Molecule Nanomanipulation and Fluorescence. Nat Struct Mol Biol. 2015;22:452–457. doi: 10.1038/nsmb.3019. [DOI] [PubMed] [Google Scholar]
- 70.Deaconescu AM, Chambers AL, Smith AJ, Nickels BE, Hochschild A, Savery NJ, Darst SA. Structural Basis for Bacterial Transcription-Coupled DNA Repair. Cell. 2006;124:507–520. doi: 10.1016/j.cell.2005.11.045. [DOI] [PubMed] [Google Scholar]
- 71.Deaconescu AM, Savery N, Darst SA. The Bacterial Transcription Repair Coupling Factor. Curr Opin Struct Biol. 2007;17:96–102. doi: 10.1016/j.sbi.2007.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Assenmacher N, Wenig K, Lammens A, Hopfner KP. Structural Basis for Transcription-Coupled Repair: The N Terminus of Mfd Resembles UvrB with Degenerate ATPase Motifs. J Mol Biol. 2006;355:675–683. doi: 10.1016/j.jmb.2005.10.033. [DOI] [PubMed] [Google Scholar]
- 73.Weiss LA, Harrison PG, Nickels BE, Glickman MS, Campbell EA, Darst SA, Stallings CL. Interaction of CarD with RNA Polymerase Mediates Mycobacterium Tuberculosis Viability, Rifampin Resistance, and Pathogenesis. J Bacteriol. 2012;194:5621–5631. doi: 10.1128/JB.00879-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Bae B, Chen J, Davis E, Leon K, Darst SA, Campbell EA. CarD Uses a Minor Groove Wedge Mechanism to Stabilize the Rna Polymerase Open Promoter Complex. Elife. 2015:4. doi: 10.7554/eLife.08505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Smith AJ, Szczelkun MD, Savery NJ. Controlling the Motor Activity of a Transcription-Repair Coupling Factor: Autoinhibition and the Role of RNA Polymerase. Nucleic Acids Res. 2007;35:1802–1811. doi: 10.1093/nar/gkm019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Deaconescu AM, Sevostyanova A, Artsimovitch I, Grigorieff N. Nucleotide Excision Repair (Ner) Machinery Recruitment by the Transcription-Repair Coupling Factor Involves Unmasking of a Conserved Intramolecular Interface. Proc Natl Acad Sci U S A. 2012;109:3353–3358. doi: 10.1073/pnas.1115105109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Timmins J, Gordon E, Caria S, Leonard G, Acajjaoui S, Kuo MS, Monchois V, McSweeney S. Structural and Mutational Analyses of Deinococcus Radiodurans UvrA2 Provide Insight into DNA Binding and Damage Recognition by Uvras. Structure. 2009;17:547–558. doi: 10.1016/j.str.2009.02.008. [DOI] [PubMed] [Google Scholar]
- 78.Thiagalingam S, Grossman L. Both Atpase Sites of Escherichia Coli UvrA Have Functional Roles in Nucleotide Excision Repair. J Biol Chem. 1991;266:11395–11403. [PubMed] [Google Scholar]
- 79.Manelyte L, Kim YI, Smith AJ, Smith RM, Savery NJ. Regulation and Rate Enhancement During Transcription-Coupled DNA Repair. Mol Cell. 2010;40:714–724. doi: 10.1016/j.molcel.2010.11.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Verhoeven EE, Wyman C, Moolenaar GF, Goosen N. The Presence of Two UvrB Subunits in the UvrAB Complex Ensures Damage Detection in Both DNA Strands. EMBO J. 2002;21:4196–4205. doi: 10.1093/emboj/cdf396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Fan J, Leroux-Coyau M, Savery NJ, Strick TR. Reconstruction of Bacterial Transcription-Coupled Repair at Single-Molecule Resolution. Nature. 2016;536:234–237. doi: 10.1038/nature19080. [DOI] [PubMed] [Google Scholar]
- 82.Sidorenkov I, Komissarova N, Kashlev M. Crucial Role of the Rna:DNA Hybrid in the Processivity of Transcription. Mol Cell. 1998;2:55–64. doi: 10.1016/s1097-2765(00)80113-6. [DOI] [PubMed] [Google Scholar]
- 83.Haines NM, Kim YI, Smith AJ, Savery NJ. Stalled Transcription Complexes Promote DNA Repair at a Distance. Proc Natl Acad Sci U S A. 2014;111:4037–4042. doi: 10.1073/pnas.1322350111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Brueckner F, Hennecke U, Carell T, Cramer P. CPD Damage Recognition by Transcribing RNA Polymerase Ii. Science. 2007;315:859–862. doi: 10.1126/science.1135400. [DOI] [PubMed] [Google Scholar]
- 85.Kunala S, Brash DE. Intragenic Domains of Strand-Specific Repair in Escherichia Coli. J Mol Biol. 1995;246:264–272. doi: 10.1006/jmbi.1994.0082. [DOI] [PubMed] [Google Scholar]
- 86.Cohen SE, Lewis CA, Mooney RA, Kohanski MA, Collins JJ, Landick R, Walker GC. Roles for the Transcription Elongation Factor Nusa in Both DNA Repair and Damage Tolerance Pathways in Escherichia Coli. Proc Natl Acad Sci U S A. 2010;107:15517–15522. doi: 10.1073/pnas.1005203107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Cohen SE, Walker GC. New Discoveries Linking Transcription to DNA Repair and Damage Tolerance Pathways. Transcription. 2011;2:37–40. doi: 10.4161/trns.2.1.14228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Tomko EJ, Jia H, Park J, Maluf NK, Ha T, Lohman TM. 5′-Single-Stranded/Duplex DNA Junctions Are Loading Sites for E. Coli UvrD Translocase. Embo j. 2010;29:3826–3839. doi: 10.1038/emboj.2010.242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Shepherd N, Churchward G, Bremer H. Synthesis and Function of Ribonucleic Acid Polymerase and Ribosomes in Escherichia Coli B/R after a Nutritional Shift-Up. J Bacteriol. 1980;143:1332–1344. doi: 10.1128/jb.143.3.1332-1344.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Easton AM, Kushner SR. Transcription of the UvrD Gene of Escherichia Coli Is Controlled by the LexA Repressor and by Attenuation. Nucleic Acids Res. 1983;11:8625–8640. doi: 10.1093/nar/11.24.8625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Ahn B. A Physical Interaction of UvrD with Nucleotide Excision Repair Protein Uvrb. Molecules and Cells. 2000;10:592–597. doi: 10.1007/s10059-000-0592-5. [DOI] [PubMed] [Google Scholar]
- 92.Kamarthapu V, Epshtein V, Benjamin B, Proshkin S, Mironov A, Cashel M, Nudler E. ppGpp Couples Transcription to DNA Repair in E. Coli. Science. 2016;352:993–996. doi: 10.1126/science.aad6945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Arthur HM, Eastlake PB. Transcriptional Control of the UvrD Gene of Escherichia Coli. Gene. 1983;25:309–316. doi: 10.1016/0378-1119(83)90235-4. [DOI] [PubMed] [Google Scholar]
- 94.Maluf NK, Fischer CJ, Lohman TM. A Dimer of Escherichia Coli UvrD Is the Active Form of the Helicase in Vitro. J Mol Biol. 2003;325:913–935. doi: 10.1016/s0022-2836(02)01277-9. [DOI] [PubMed] [Google Scholar]
- 95.Maluf NK, Lohman TM. Self-Association Equilibria of Escherichia Coli UvrD Helicase Studied by Analytical Ultracentrifugation. J Mol Biol. 2003;325:889–912. doi: 10.1016/s0022-2836(02)01276-7. [DOI] [PubMed] [Google Scholar]
- 96.Bar-Nahum G, Epshtein V, Ruckenstein AE, Rafikov R, Mustaev A, Nudler E. A Ratchet Mechanism of Transcription Elongation and Its Control. Cell. 2005;120:183–193. doi: 10.1016/j.cell.2004.11.045. [DOI] [PubMed] [Google Scholar]
- 97.Barker MM, Gaal T, Josaitis CA, Gourse RL. Mechanism of Regulation of Transcription Initiation by ppGpp. I. Effects of ppGpp on Transcription Initiation in Vivo and in Vitro. J Mol Biol. 2001;305:673–688. doi: 10.1006/jmbi.2000.4327. [DOI] [PubMed] [Google Scholar]
- 98.Zuo Y, Wang Y, Steitz TA. The Mechanism of E. Coli RNA Polymerase Regulation by Ppgpp Is Suggested by the Structure of Their Complex. Mol Cell. 2013;50:430–436. doi: 10.1016/j.molcel.2013.03.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Trautinger BW, Jaktaji RP, Rusakova E, Lloyd RG. RNA Polymerase Modulators and DNA Repair Activities Resolve Conflicts between DNA Replication and Transcription. Molecular Cell. 2005;19:247–258. doi: 10.1016/j.molcel.2005.06.004. [DOI] [PubMed] [Google Scholar]
- 100.Madison KE, Jones-Foster EN, Vogt A, Kirtland Turner S, North SH, Nakai H. Stringent Response Processes Suppress DNA Damage Sensitivity Caused by Deficiency in Full-Length Translation Initiation Factor 2 or Pria Helicase. Mol Microbiol. 2014;92:28–46. doi: 10.1111/mmi.12538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Pomerantz RT, O’Donnell M. Direct Restart of a Replication Fork Stalled by a Head-on Rna Polymerase. Science. 2010;327:590–592. doi: 10.1126/science.1179595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Dutta D, Shatalin K, Epshtein V, Gottesman ME, Nudler E. Linking RNA Polymerase Backtracking to Genome Instability in E. Coli. Cell. 2011;146:533–543. doi: 10.1016/j.cell.2011.07.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.De Septenville AL, Duigou S, Boubakri H, Michel B. Replication Fork Reversal after Replication-Transcription Collision. PLoS Genet. 2012;8:e1002622. doi: 10.1371/journal.pgen.1002622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Paul S, Million-Weaver S, Chattopadhyay S, Sokurenko E, Merrikh H. Accelerated Gene Evolution through Replication-Transcription Conflicts. Nature. 2013;495:512–515. doi: 10.1038/nature11989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Million-Weaver S, Samadpour AN, Moreno-Habel DA, Nugent P, Brittnacher MJ, Weiss E, Hayden HS, Miller SI, Liachko I, Merrikh H. An Underlying Mechanism for the Increased Mutagenesis of Lagging-Strand Genes in Bacillus Subtilis. Proc Natl Acad Sci U S A. 2015;112:E1096–1105. doi: 10.1073/pnas.1416651112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Ramirez-Guadiana FH, Del Carmen Barajas-Ornelas R, Ayala-Garcia VM, Yasbin RE, Robleto E, Pedraza-Reyes M. Transcriptional Coupling of DNA Repair in Sporulating Bacillus Subtilis Cells. Mol Microbiol. 2013;90:1088–1099. doi: 10.1111/mmi.12417. [DOI] [PubMed] [Google Scholar]
- 107.Kunkel TA, Van Houten B. Survival Choices. Nat Cell Biol. 2006;8:547–549. doi: 10.1038/ncb0606-547. [DOI] [PubMed] [Google Scholar]
- 108.Ross C, Pybus C, Pedraza-Reyes M, Sung HM, Yasbin RE, Robleto E. Novel Role of Mfd: Effects on Stationary-Phase Mutagenesis in Bacillus Subtilis. J Bacteriol. 2006;188:7512–7520. doi: 10.1128/JB.00980-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Lee GH, Jeong JY, Chung JW, Nam WH, Lee SM, Pak JH, Choi KD, Song HJ, Jung HY, Kim JH. The Helicobacter Pylori Mfd Protein Is Important for Antibiotic Resistance and DNA Repair. Diagn Microbiol Infect Dis. 2009;65:454–456. doi: 10.1016/j.diagmicrobio.2009.08.007. [DOI] [PubMed] [Google Scholar]
- 110.Han J, Sahin O, Barton YW, Zhang Q. Key Role of Mfd in the Development of Fluoroquinolone Resistance in Campylobacter Jejuni. PLoS Pathog. 2008;4:e1000083. doi: 10.1371/journal.ppat.1000083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Willing SE, Richards EJ, Sempere L, Dale AG, Cutting SM, Fairweather NF. Increased Toxin Expression in a Clostridium Difficile Mfd Mutant. BMC Microbiol. 2015;15:280. doi: 10.1186/s12866-015-0611-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Guillemet E, Lereec A, Tran SL, Royer C, Barbosa I, Sansonetti P, Lereclus D, Ramarao N. The Bacterial DNA Repair Protein Mfd Confers Resistance to the Host Nitrogen Immune Response. Sci Rep. 2016;6:29349. doi: 10.1038/srep29349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Kumura K, Sekiguchi M. Identification of the UvrD Gene Product of Escherichia Coli as DNA Helicase Ii and Its Induction by DNA-Damaging Agents. J Biol Chem. 1984;259:1560–1565. [PubMed] [Google Scholar]
- 114.Siegel EC. The Escherichia Coli UvrD Gene Is Inducible by DNA Damage. Mol Gen Genet. 1983;191:397–400. doi: 10.1007/BF00425753. [DOI] [PubMed] [Google Scholar]