Abstract
In the present study, knockdown of E2F1 impaired the migration and invasion of osteosarcoma cells. Further analysis showed that E2F1 knockdown decreased the expression of discoidin domain receptor 1 (DDR1) which plays a crucial role in many fundamental processes such as cell differentiation, adhesion, migration and invasion. Luciferase and ChIP assays confirmed that E2F1 silencing attenuated the expression of DDR1 through disrupting E2F1-mediated transcription of DDR1 in osteosarcoma cells. Similarly with the effect of E2F1 silencing, DDR1 knockdown weakened the migratory and invasive capabilities of osteosarcoma cells; while overexpression of DDR1 resulted in a significant increase of cell motility and invasiveness, even after knocking down E2F1. Interestingly, inactivation of E2F1/DDR1 pathway by shRNA weakened STAT3 signaling and subsequently suppressed the epithelial-mesenchymal transition (EMT) of osteosarcoma cells, as shown with decreased vimentin, MMP2, MMP9, and increased E-cadherin. Consistently, high expressions of E2F1 and DDR1 observed in osteosarcoma tissues were related to TNM stage and metastasis. In addition, high level of E2F1 or DDR1 was associated with poor prognosis in osteosarcoma patients. These results suggest that E2F1/DDR1/STAT3 pathway is critical for malignancy of osteosarcoma, which may provide a novel prognostic indicator or approach for osteosarcoma therapy.
Keywords: osteosarcoma, E2F1, discoidin domain receptor 1, migration, invasion
Introduction
Osteosarcoma is the most common malignant bone tumor that occurs primarily in children and adolescents. Despite surgery combined with chemotherapy has been widely applied for osteosarcoma treatment, an overwhelming tendency for invasion and early metastasis lead to poor prognosis. The 5-year survival rate of patients with recur rent and metastasis remains at only 20% (1). The mechanisms for osteosarcoma development has drawn great attention (2,3). However, due to lack of effective drug targets, a breakthrough for osteosarcoma treatment has not yet been achieved.
As a well-documented driver of cell cycle, E2F1 regulates the expression of genes required in the G1→S phase transition (4). However, recent studies showed a role of E2F1 in metastasis and invasion of several cancer types. E2F1-induced metastasis was verified by clinical studies showing that high expression of E2F1 and its related target genes predicted the transformation from superficial to invasive bladder cancer (5). In addition, it was demonstrated that the depletion of endogenous E2F1 attenuated tumor invasion and pulmonary metastasis in therapy-resistant metastatic melanoma xenografts (6). Recently, Liang et al reported a novel function of E2F1 as enhancer of tumor invasion and migration in prostate cancer via regulating the expression of CD147 (7). However, the mechanisms underlying the metastasis promoted by E2F1 remain unclear.
DDR1 belongs to discoidin domain receptor (DDR) family which consists of two highly homologous members, DDR1 and DDR2. DDRs function as the unique receptor tyrosine kinases (RTKs) which are activated by collagen, a major extracellular matrix (ECM) component (8). Previous studies have reported abnormal expression of DDR1 in some high-invasive tumors, such as lung, breast, and prostate cancers (9–11). Growing evidence suggested that DDR1 was closely associated with lymph node metastasis and shorter survival, and overexpression of DDR1 promoted cell mobility and invasion (12,13). Although DDR1 has been verified to be involved in tumor development, dysregulation of DDR1 as well as the roles of DDR1 in tumor aggressiveness is poorly understood.
In the present study, E2F1 was found to be critical for the migration and invasion of osteosarcoma cells through transactivating DDR1. Furthermore, DDR1 enhances the phosphorylation of STAT3 which drives the epithelial-mesenchymal transition (EMT) of osteosarcoma cells. Finally, in vivo experiments and clinical analysis confirm that E2F1 and DDR1 are important for maintaining the malignant features of osteosarcoma. Together, these findings suggest a novel mechanism for E2F1-dependent migration and invasion in osteosarcoma, and provide a new understanding of E2F1-driven tumor progression.
Materials and methods
Cell culture and transfection
Two osteosarcoma cell lines, U2OS (TP53-WT; RB1-WT) and SaOs-2 (TP53-Mut; RB1-Mut), and an immortalized osteoblast cell line hFOB1.19 (TP53-WT; RB1-WT) were purchased from the Institute of Biochemistry and Cell Biology of the Chinese Academy of Sciences (Shanghai, China) and grown in Dulbeco's modified Eagle's medium supplemented with 10% fetal bovine serum, and the cells were incubated at 37°C and 5% CO2. Before experiments, cells were cultured for 3–5 passages. Cell transfection was performed using Lipofectamine 3000 reagent (Thermo Fisher Scientific, Waltham, MA, USA) according to the manufacturer's instructions. The U2OS and SaOs-2 cells with stable knockdown of E2F1 or DDR1 were established with indicated shRNA and maintained by G418 (Sigma, St. Louis, MO, USA).
Antibodies and reagents
Antibodies against E2F1 (sc-251), DDR1 (sc-532), E-cadherin (sc-8426), vimentin (sc-6260), MMP2 (sc-10736), MMP9 (sc-10737), and GAPDH (sc-32233) were from Santa Cruz Biotechnology (Santa Cruz, CA, USA). The p-AKT (S473) (#4051), AKT (#2966), p-STAT3 (Y705) (#4113), and STAT3 (#9139) antibodies were from Cell Signaling Technology (Beverly, MA, USA). The siRNAs targeting STAT3 were from Santa Cruz.
Western blotting
Cells were lysed in RIPA buffer supplemented with PMSF, and the protein lysates were separated by SDS-PAGE and transferred to nitrocellulose membranes (Whatman, Maidstone, UK). Then the membranes were incubated overnight at 4°C by using primary antibodies. After incubation with goat anti-rabbit (926-32211; 926-68071) or goat anti-mouse (926-32210; 926-68070) IgG secondary antibodies (1:10,000; LI-COR, Lincoln, NE, USA) at room temperature for 1 h, the fluorescence intensity was visualized by the Odyssey Infrared Imaging system (LI-COR).
Cell viability assay
Cell viability was detected using the Cell Counting Kit-8 assay (Dojindo, Kumamoto, Japan). Transfected cells were dispensed in triplicate into 96-well plates and incubated overnight at 37°C. After indicated time, 10 µl of CCK-8 kit solution was added to the cells, which were then incubated for 2.5 h at 37°C. Absorbance was then measured by a microplate reader at 450 nm. Data were obtained from at least three separate experiments done in triplicate.
Quantitative real-time PCR analysis
Total RNA was prepared using the RNAiso Plus (Takara, Kyoto, Japan). cDNA was synthesized from total RNA using PrimeScript™ RT-PCR kit (RR014B; Takara). Quantitative real-time PCR (qRT-PCR) was performed by the 7500 Real-Time PCR system (Applied Biosystems, Foster City, CA, USA) with GAPDH being used as an internal control. The results are representative of at least three independent experiments.
RNA-seq analysis
RNA from U2OS cells with or without E2F1 knockdown was extracted and purified for quantification, RNA-seq library preparation, and sequencing. The libraries were sequenced on the illumina HiSeq 2500 platform. The reads containing adapter or poly-N and reads of low quality were removed from raw data to generate clean reads for further analyses. Based on the clean reads, the Q20 (>90%), Q30 (>85%), and error rate (<0.1%) of the clean data were required. Then mapped reads were obtained by Tophat2 through aligning clean reads to the human genome reference (hg19). The number of mapped clean reads for each unigene was counted and normalized into a reads per kb per million reads (RPKM) to calculate the expression level of the unigene.
Luciferase reporter assay
Various lengths of DDR1 promoter were amplified by PCR and inserted into the pGL3 vector. The reporter construct with a mutated E2F1 binding site was generated by subsequent PCR-based cloning. These reporter plasmids were transfected into osteosarcoma cells, and pRL-TK was co-transfected as an internal control for transfection efficiency. At 48 h post-transfection, cells were lysed and the luciferase activities were measured using the Dual-Luciferase Reporter assay system (Promega, Madison, WI, USA). The experiments were carried out in triplicate.
Chromatin immunoprecipitation
Cells were cross-linked with 1% formaldehyde, lysed, and then sheared to an average size of 250–500 bp by sonication. ChIP was performed by incubating chromatin with anti-E2F1 antibody. After that, chromatin-antibody complexes were isolated using Protein A/G Plus-Agarose (Santa Cruz). Finally, the crosslinking was reversed and immunoprecipitated DNA was purified. Enrichment of DNA fragments was analyzed by qRT-PCR. Three independent experiments were performed.
Migration assay
A scratch wound healing assay was used for determining cell migration. Briefly, cell monolayer was scratched with a sterile pipette. The culture was then washed with medium to remove free-floating cells and debris and cultured in serum-free medium for an additional 24 h. The coverage of the scratched area was measured at three different positions. All experiments were performed in triplicates.
Invasion assay
Transfected cells in serum-free media were placed into the upper chamber of an insert coated with Matrigel (BD, San Diego, CA, USA). Media containing 10% FBS were added to the lower chamber. After incubation for 48 h, non-invaded cells on the upper surface of the filter were removed carefully with a cotton swab and the undersurface adherent cells that had invaded through the Matrigel were fixed in methanol and stained with 0.5% crystal violet. The filter membrane was viewed under a microscope and 5 random fields were selected for cell counting. Experiments were independently repeated three times.
Colony formation assay
Stable transfected cells were suspended in medium containing DMEM medium with 10% FBS and 2.5 ml of 0.3% noble agar (Sigma). Cells were plated onto a solidified medium containing 2.5 ml of 0.6% noble agar in a 60-mm dish. The dishes were incubated at 37°C with 5% CO2, and fresh medium was added every 3–4 days. After culture for 25 days, cells were staining with 0.05% crystal violet, and forming colonies (>100 mm in diameter) were counted using microscopy.
Xenografts
Fifteen female BALB/c nude mice were injected with U2OS cells with stable knockdown of E2F1 or DDR1. Mice were randomly chosen for the two experimental groups and one control group, and each received a subcutaneous injection of a viable cell suspension mixture (5×106) containing a 90% U2OS cells. Subcutaneous tumor size was blindly measured twice a week with a caliper. Tumor volumes were calculated using the formula: V = W2 × L × 0.5, where W is tumor width and L is tumor length. All of the mice were sacrificed on the fifth week after injection, and the individual tumors were weighed. The animal experiment was approved by the institutional review board of Zhejiang Rongjun Hospital.
Immunohistochemistry
All procedures performed in studies involving human participants were in accordance with the Ethics Committee of Zhejiang Rongjun Hospital and the Second Hospital of Jiaxing and with the 1964 Helsinki declaration and its later amendments or comparable ethical standards. Osteosarcoma tissues were collected by surgery from 81 patients at Zhejiang Rongjun Hospital (Jiaxing, China) and the Second Hospital of Jiaxing (Jiaxing, China) from 2008 to 2011. All specimens were fixed in 10% neutral formalin, embedded in paraffin and cut into 4-µm sections for immunohistochemical staining. The EnVision™ two-step method was used (Dako, Hamburg, Germany), as well as the following antibodies: antibody against E2F1 and antibody against DDR1. To estimate the score for each slide, at least 10 individual fields at ×200 were chosen, and 100 cancer cells were counted in each field. The immunostaining intensity was divided into four grades: 0, no expression; 1, mildly positive; 2, moderately positive; and 3, markedly positive. The proportion of positive-staining cells was divided into five grades: 0, <10%; 1, 11–25%; 2, 26–50%; 3, 51–75%; and 4, >75%. The staining results were assessed and confirmed by two independent investigators blinded to the clinical data. The percentage of positivity of the tumor cells and the staining intensities were then multiplied in order to generate the IHC score, and graded as 0–6, low expression; 7–12, high expression. Cases with a discrepancy in scores were discussed to obtain a consensus.
Statistical analysis
All statistical analyses were carried out using the GraphPad Prism 5.0 and SPSS 17.0 statistical software. Experimental data are presented as mean ± SD from at least three independent experiments. The χ2 test or Fisher's exact probability test was used to compare clinicopathological features of the patients with protein expression. Kaplan-Meier plots and log-rank tests were used for survival analysis. Spearman test was used in analyzing the correlation. A P-value of ≤0.05 was considered statistically significant.
Results
E2F1 silencing not only reduces osteosarcoma cell proliferation but attenuates migration and invasion
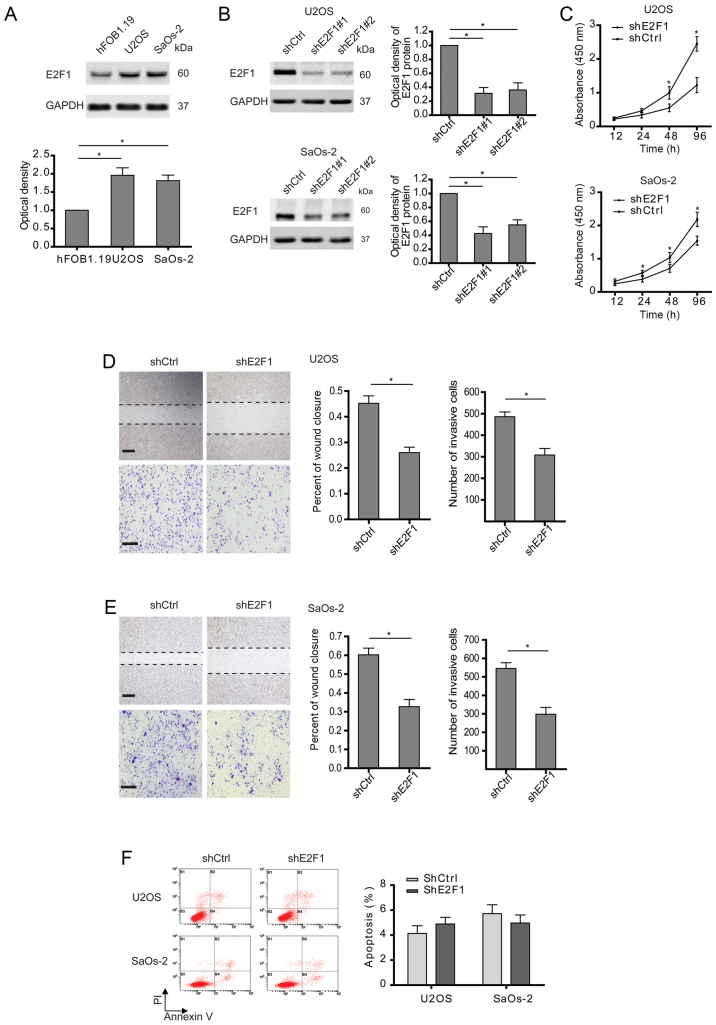
Consistent with previous observations (14), E2F1 was highly expressed in osteosarcoma cells compared with osteoblast (Fig. 1A). To explore the roles of E2F1 in osteosarcoma, two osteosarcoma cell lines (U2OS and SaOS-2) with high E2F1 levels were examined after stable transfection with E2F1 shRNA (Fig. 1B). Two shRNAs targeting E2F1 were checked for their efficacies, and as indicated by western blotting, shE2F1#1 was slightly more effective for inhibiting E2F1 expression than shE2F1#2. So, shE2F1#1 was applied in the following experiments. As expected, cell growth was markedly suppressed in the osteosarcoma cells with E2F1 silence compared to control cells (Fig. 1C). Interestingly, cell migration and invasion determined by wound healing and Transwell assays were also inhibited in the osteosarcoma cells with E2F1 knockdown (Fig. 1D and E). In consideration of the relationship between E2F1 and apoptosis described in osteosarcoma (15), the apoptosis of osteosarcoma cells after abolishing E2F1 was analyzed. As shown in Fig. 1F, knockdown of E2F1 had no significant effect on apoptosis. Therefore, these findings indicate that high expression of E2F1 is essential for the malignancy of osteosarcoma.
Figure 1.
E2F1 silencing not only reduces osteosarcoma cell proliferation but attenuates migration and invasion. (A) The different expressions of E2F1 at the protein level in osteoblast (hFOB1.19) or osteosarcoma (U2OS and SaOs-2) cells were examined. (B) Shscramble and two E2F1 shRNAs were transfected into U2OS or SaOs-2 cells, and the protein levels of E2F1 were measured to indicate their efficacy. (C) Cell viability assay in U2OS or SaOs-2 cells stably transfected with shscramble and shE2F1. (D and E) Cell migration or invasion activity was examined by wound healing or Transwell assay. Representative images at 24 h are shown (left panels), and wound areas or invasive cells were quantified (right graph). Scale bar, 200 µm (wound healing); 100 µm (Transwell). (F) U2OS and SaOs-2 cells stably transfected as indicated, were subjected to cytometric analysis of apoptotic cells after staining with Annexin V/PI. (A-F) Data shown are mean ± SEM, each performed in triplicate. *P<0.05.
Knockdown of E2F1 suppresses the transcription of DDR1 in osteosarcoma cells
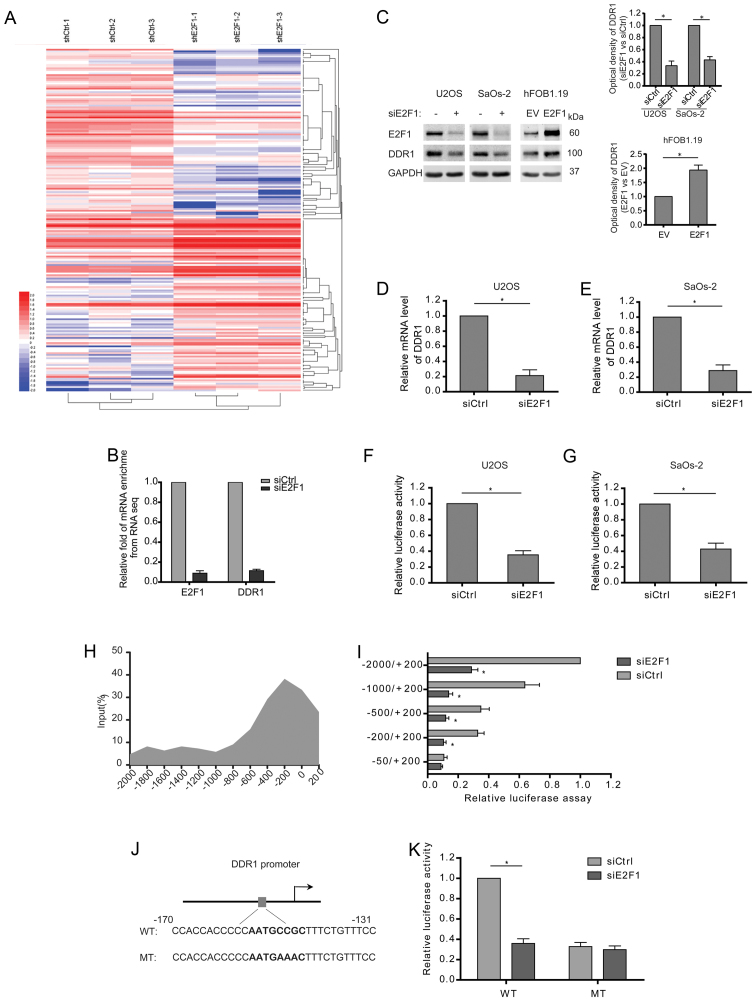
In attempt to understand the potential mechanisms behind E2F1-induced migration and invasion, the expression of mRNA isolated from the osteosarcoma cells with stable knockdown of E2F1 was profiled by RNA-seq analysis. Differentially expressed genes (DEGs) were clustered and displayed in the heat map (Fig. 2A). Especially, the enrichment of DDR1 mRNA was dramatically reduced after E2F1 knockdown (Fig. 2B). Consistently, the expression of DDR1 was decreased in the E2F1-silenced osteosarcoma cells both at the protein and mRNA levels (Fig. 2C–E). To determine whether E2F1 transactivates the transcription of DDR1, 2 kb DDR1 promoter region upstream transcription start site was cloned to a luciferase gene and then co-transfected with E2F1 siRNA. The results showed that DDR1 promoter activity was strongly repressed in response to E2F1 knockdown in osteosarcoma cells (Fig. 2F and G). The direct regulation for DDR1 transcription by E2F1 was further confirmed by chromatin immunoprecipitation (ChIP), which showed a peak binding of endogenous E2F1 to DDR1 promoter at 200 bp upstream the transcription start site (Fig. 2H). Moreover, the truncated DDR1 promoter without the region (−200/−50) failed to respond to E2F1 knockdown (Fig. 2I). Then, we analyzed the promoter sequence around −200 bp by bioinformatic software (JASPAR and Tfsearch), and a putative E2F1 binding site (AATGCCGC) was identified (Fig. 2J). Furthermore, mutation of the site greatly diminished the inhibitory effect of E2F1 silence on the transcription activity of DDR1 promoter (Fig. 2K). Taken together, E2F1 is required for DDR1 transcription by directly binding to the promoter.
Figure 2.
Knockdown of E2F1 suppresses the transcription of DDR1 in osteosarcoma cells. (A) Unsupervised hierarchical clustering and heat map display of genes that were differentially expressed between osteosarcoma cells with and without siE2F1. N=3. (B) Fold enrichment of E2F1 and DDR1 mRNAs from the RNA-seq results. N=3. (C-E) Siscramble and siE2F1 were transfected into U2OS or SaOs-2 cells. Empty vector and E2F1 plasmids were transfected into hFOB1.19 cells. The expression of DDR1 at the mRNA and protein level among the indicated cells is shown. (F and G) Quantification of dual reporter luciferase assay for DDR1 promoter in U2OS or SaOs-2 cells transfected with siscramble and siE2F1. Relative luciferase activities were firefly/Renilla ratios. (H) Chromatin immunoprecipitation (ChIP) assay showing the enrichments of E2F1 at different regions of DDR1 promoter. Relative promoter enrichment was normalized on input material. (I) Quantification of dual reporter luciferase assay of truncated DDR1 promoters in U2OS cells transfected with siscramble and siE2F1. Relative luciferase activities were firefly/Renilla ratios. (J) The graph shows a putative E2F1 binding site on the DDR1 promoter, where E2F1 could potentially bind to and activate DDR1 transcription. (K) Quantification of dual reporter luciferase assay of DDR1 promoter containing the wild or mutant E2F1 binding site in U2OS cells transfected with siscramble and siE2F1. Relative luciferase activities were firefly/Renilla ratios. (D-G, I and K) Data shown are mean ± SEM, each performed in triplicate. *P<0.05.
Silence of DDR1 reduces the levels of p-AKT and p-STAT3 accompanied with an attenuated aggressiveness of osteosarcoma cells
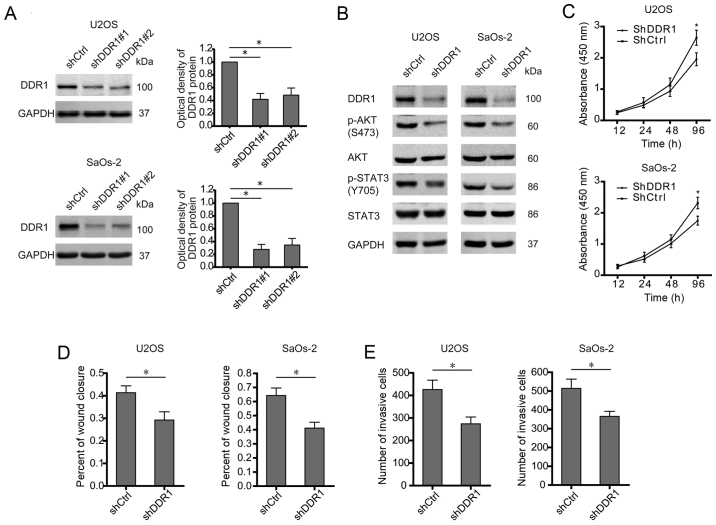
DDR1 serves as docking sites for the assembly of downstream signaling molecules and then activates different target mediators which are involved in several oncogenic processes including survival, metastasis, and chemosensitivity (16,17). In osteosarcoma cells (U2OS and SaOS-2), DDR1 was stably knocked down by two shRNAs (Fig. 3A). Even though no significant difference between shDDR1#1 and shDDR1#2 in knocking down DDR1 was observed, shDDR1#1 showed a more powerful effect than shDDR1#2, especially in SaOs-2 cells. Thus shDDR1#1 was used for silencing DDR1 in this study. As a result, silence of DDR1 abolished the activation of both AKT and STAT3 signaling (Fig. 3B). Similarly with E2F1 knockdown, deprivation of DDR1 undermined the malignant phenotypes of osteosarcoma cells, as shown by decreased proliferation, migration, and invasion (Fig. 3C–E). Therefore, as a target gene of E2F1, DDR1 is also required for triggering oncogenic signaling and aggressive behavior in osteosarcoma.
Figure 3.
Silence of DDR1 reduces the levels of p-AKT and p-STAT3 accompanied with an attenuated aggressiveness of osteosarcoma cells. (A) Shscramble and two DDR1 shRNAs were transfected into U2OS or SaOs-2 cells, and the protein levels of DDR1 were measured to indicate their efficacy. (B) U2OS or SaOs-2 cells were stably transfected with shscramble and shDDR1, and the expression of pAKT, total-AKT, pY705-STAT3, and total-STAT3 was examined. (C) Cell viability assay in U2OS or SaOs-2 cells stably transfected with shscramble and shDDR1. (D and E) Cell migration or invasion activity was examined by wound healing or Transwell assay. Wound areas or invasive cells were quantified. (A-E) Data shown are mean ± SEM, each performed in triplicate. *P<0.05.
DDR1 mediates E2F1-dependent malignant phenotypes of osteosarcoma cells
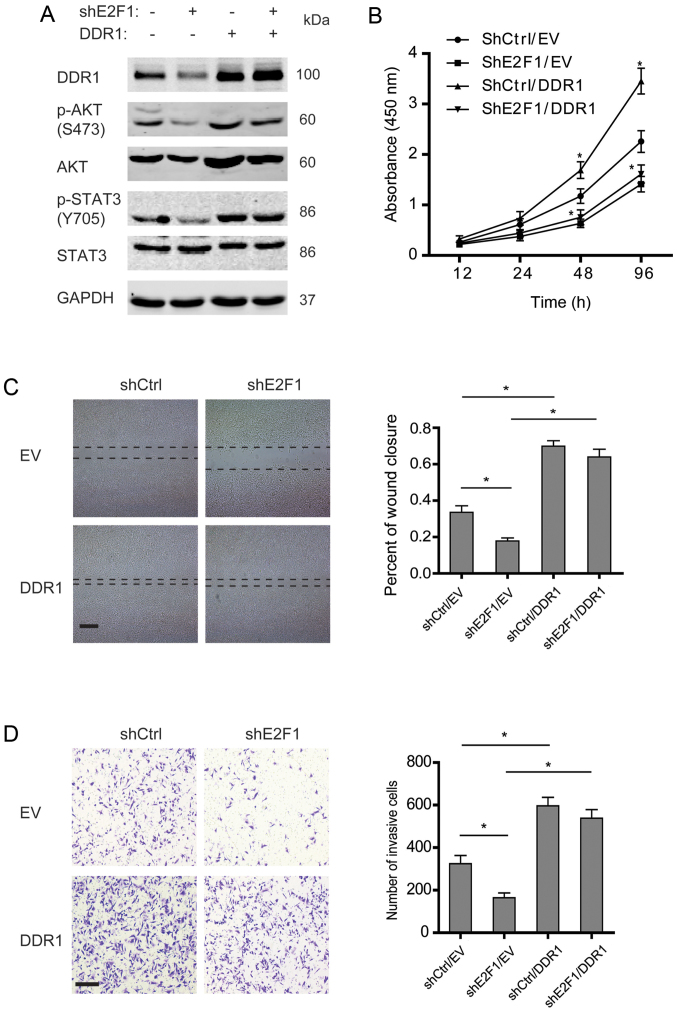
In order to determine the role of DDR1 in E2F1-dependent malignancy of osteosarcoma, DDR1 was overexpressed in the osteosarcoma cells with stable knockdown of E2F1. As shown in Fig. 4A, knockdown of E2F1 decreased phosphorylation of AKT and STAT3, while DDR1 overexpression rescued the reduction. Further studies showed that knockdown of E2F1 impaired the proliferation, migration, and invasion of osteosarcoma cells, and overexpression of DDR1 could enhance the aggressive phenotypes (Fig. 4B–D). Notably, DDR1 overexpression was able to restore the incapability of migration and invasion in the E2F1-silenced osteosarcoma cells but failed to rescue the proliferation, which suggested that DDR1 displayed a crucial role in E2F1-dependent migration and invasion but not proliferation in osteosarcoma.
Figure 4.
DDR1 mediates E2F1-dependent malignant phenotypes of osteosarcoma cells. (A) Shscramble or shE2F1-U2OS cells were transfected with empty vector and DDR1 plasmids, and the expression of pAKT, total-AKT, pY705-STAT3, and total-STAT3 was examined. (B) Cell viability assay in the indicated U2OS cells. (C and D) Cell migration or invasion activity was examined by wound healing or Transwell assay. Representative images at 24 h are shown (left panels), and wound areas or invasive cells were quantified (right graph). Scale bar, 200 µm (wound healing); 100 µm (Transwell). (B-D) Data shown are mean ± SEM, each performed in triplicate. *P<0.05.
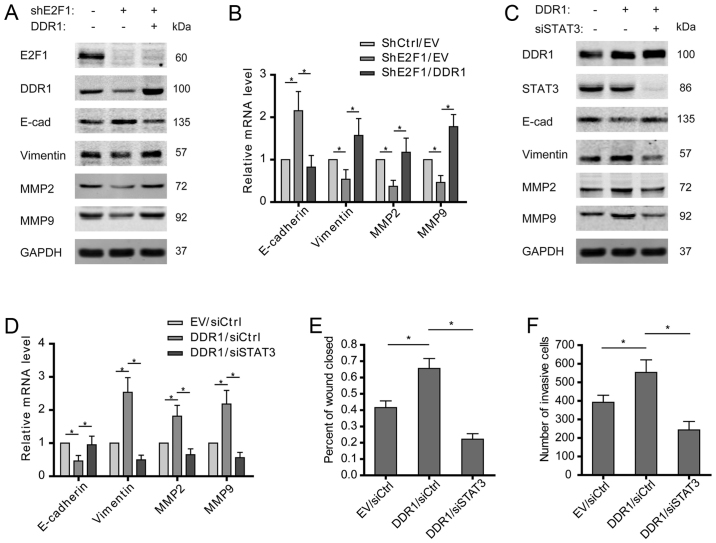
E2F1/DDR1/STAT3 axis drives the EMT of osteosarcoma cells
Activation of epithelial-mesenchymal transition (EMT) is important for driving cancer cell migration and invasion, and STAT3 was reported to be involved in the EMT of various tumors. To clarify whether E2F1/DDR1/STAT3 pathway is indispensable for promoting osteosarcoma EMT, the expression levels of EMT markers in osteosarcoma cells were examined. The results showed that E2F1 knockdown increased E-cadherin but decreased vimentin at both mRNA and protein levels, which was then reversed by overexpression of DDR1 (Fig. 5A and B). Furthermore, overexpressed DDR1 was found to induce the EMT of osteosarcoma cells, as shown by decreased E-cadherin and increased vimentin. As expected, STAT3 depletion abolished the promotive effect of DDR1 on the features of EMT (Fig. 5C and D). Except for the canonical EMT markers expression of MMP2 and MMP9 was also found to rely on an intact E2F1/DDR1/STAT3 signaling in osteosarcoma cells (Fig. 5A–D). Consistently, overexpression of DDR1 promoted the migration and invasion of osteosarcoma cells, which was attenuated by silence of STAT3 (Fig. 5E and F). These findings suggest that E2F1/DDR1/STAT3 pathway may contribute to the migration and invasion of osteosarcoma cells via driving EMT.
Figure 5.
E2F1/DDR1/STAT3 axis drives the EMT of osteosarcoma cells. (A and B) Shscramble or shE2F1-U2OS cells were transfected with empty vector and DDR1 plasmids, and the expression of E-cadherin, vimentin, MMP2, and MMP9 was examined. (C and D) U2OS cells were transfected with empty vector/DDR1 plasmids and siscramble/siSTAT3. The expression of E-cadherin, vimentin, MMP2, and MMP9 was examined. (E and F) Cell migration or invasion activity was examined by wound healing or Transwell assay. Wound areas or invasive cells were quantified. (B and D-F) Data shown are mean ± SEM, each performed in triplicate. *P<0.05.
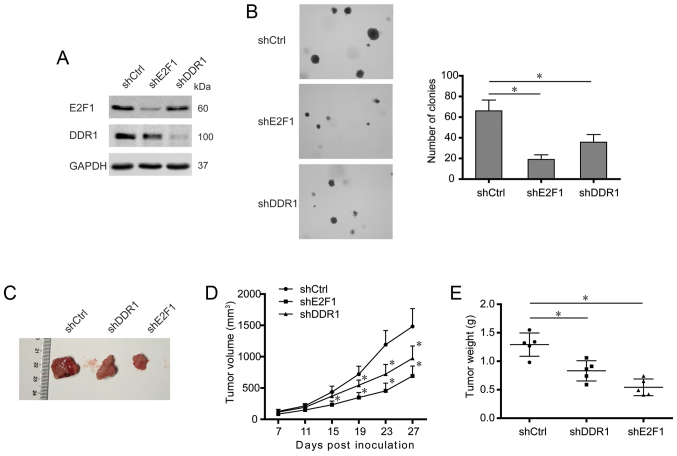
Knockdown of either E2F1 or DDR1 impairs tumorigenesis of osteosarcoma cells
To further elucidate the roles of E2F1 and DDR1 in osteosarcoma development, the osteosarcoma cells with stable knockdown of DDR1 were also constructed (Fig. 6A). Then the colony-formation ability of the osteosarcoma cells was determined by colony number and size in soft-agar colony formation assays. As shown in Fig. 6B, knockdown of either E2F1 or DDR1 significantly decreased the number of colony formation compared with control. Moreover, the constructed osteosarcoma cells were subcutaneously injected into nude mice, and tumor growth was monitored. It was shown that silence of E2F1 or DDR1 apparently reduced the volumes and weights of xenografts (Fig. 6C–E), which suggested an E2F1 and DDR1-dependent tumorigenesis in vivo. Collectively, these data indicate a critical role of E2F1 and DDR1 in tumorigenesis of osteosarcoma cells.
Figure 6.
Knockdown of either E2F1 or DDR1 impairs tumorigenesis of osteosarcoma cells. (A) The expression of E2F1 and DDR1 in U2OS cells stably transfected with shscramble, shE2F1, and shDDR1. (B) Colony formation was assessed by soft-agar colony formation assay and representative colonies are shown (left panels). Quantification of colony number is shown (right graph). (C) Representative images of U2OS xenografts, as indicated. (D and E) Quantification of tumor volumes and weights (n=5 per group). (B) Data shown are mean ± SEM, each performed in triplicate. *P<0.05.
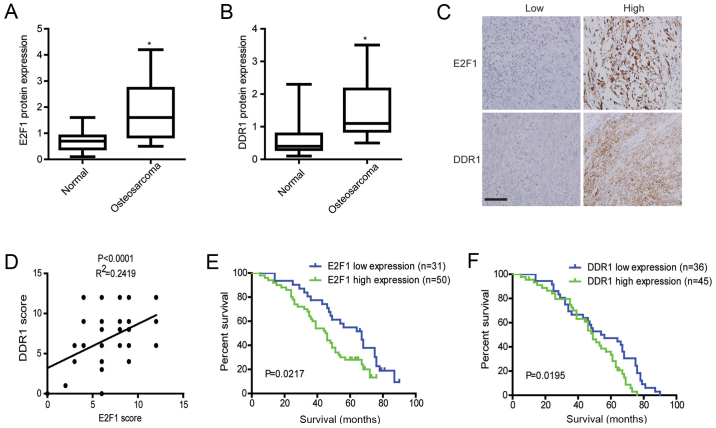
High expression of E2F1 and DDR1 predicts worse survival in osteosarcoma
The results of western blotting showed that the expression levels of E2F1 and DDR1 protein in osteosarcoma tissues were significantly higher than those in adjacent non-cancerous tissues (Fig. 7A and B). Meanwhile, expressions of E2F1 and DDR1 were assessed in osteosarcoma by immunohistochemistry. As demonstrated by E2F1 and DDR1 staining, E2F1 was mainly accumulated in the nucleus and cytoplasm, while DDR1 was diffusely expressed in the cytoplasm and cytoplasmic membrane of osteosarcoma cells (Fig. 7C). Furthermore, E2F1 expression levels paralleled the changes of DDR1 in the osteosarcoma cases as shown by immunohistochemical analyses (Fig. 7D). Also, further analysis showed that E2F1 and DDR1 staining was positively correlated with grades, advanced TNM stages, and metastasis (Table I). Importantly, Kaplan-Meier survival analyses showed that patients with osteosarcoma expressing high E2F1 or DDR1 had a significantly worse overall survival than those expressing low E2F1 or DDR1 (Kaplan-Meier, log-rank p=0.0217 (E2F1); p=0.0195 (DDR1), Fig. 7E and F). These data suggest that abnormally expressed E2F1 and DDR1 promote osteosarcoma development and indicate a poor prognosis for osteosarcoma patients.
Figure 7.
High expression of E2F1 and DDR1 predicts worse survival in osteosarcoma. (A and B) The protein levels of E2F1 and DDR1 in osteosarcoma tissues were determined by western blotting. (C) Immunohistochemical staining of E2F1 and DDR1 in osteosarcoma tissues. Scale bar, 200 µm. (D) The correlation of concurrent immunostaining scores of E2F1 and DDR1 in osteosarcoma tissues. (E) OS curve of patients with osteosarcoma based on E2F1 expression. The cumulative 5-year survival rate of the patients with E2F1 high expressions was 28%, while that of the patients with E2F1 low expressions was 54.84%. (F) OS curve of patients with osteosarcoma based on DDR1 expression. The cumulative 5-year survival rate of the patients with DDR1 high expression was 31.11%, while that of the patients with DDR1 low expression was 47.22%.
Table I.
Correlation of the expression of E2F1 and DDR1 with clinicopathological features in osteosarcoma.
| Characteristics | Cases | E2F1 expression
|
DDR1 expression
|
||||
|---|---|---|---|---|---|---|---|
| Low cases | High cases | P-value | Low cases | High cases | P-value | ||
| 81 | 45 | 36 | 44 | 37 | |||
| Sex | 0.8542 | 0.4856 | |||||
| Male | 40 | 22 | 18 | 19 | 21 | ||
| Female | 41 | 23 | 18 | 25 | 16 | ||
| Age (years) | 0.6425 | 0.2489 | |||||
| <30 | 62 | 35 | 27 | 31 | 31 | ||
| ≥30 | 19 | 10 | 9 | 13 | 6 | ||
| Tumor size (cm) | 0.1462 | 0.0611 | |||||
| <5 | 53 | 30 | 23 | 34 | 19 | ||
| ≥5 | 28 | 15 | 13 | 10 | 18 | ||
| Grade | 0.0157a | 0.0019a | |||||
| Low | 16 | 12 | 4 | 15 | 1 | ||
| High | 65 | 33 | 32 | 29 | 36 | ||
| TNM | 0.0101a | 0.0034a | |||||
| I | 8 | 8 | 0 | 8 | 0 | ||
| II | 42 | 24 | 18 | 28 | 24 | ||
| III | 21 | 12 | 9 | 8 | 13 | ||
| IV | 10 | 1 | 9 | 0 | 10 | ||
| Histological type | 0.1601 | 0.2193 | |||||
| Osteoblastic | 29 | 17 | 12 | 14 | 15 | ||
| Fibroblastic | 6 | 4 | 2 | 5 | 1 | ||
| Chondroblastic | 30 | 16 | 14 | 15 | 15 | ||
| Others | 16 | 8 | 8 | 10 | 6 | ||
| Metastasis | 0.0009a | 0.0002a | |||||
| Yes | 10 | 1 | 9 | 0 | 10 | ||
| No | 71 | 44 | 27 | 44 | 27 | ||
P<0.05.
Discussion
In the past, E2F1 was recognized as a crucial transcription factor that regulated cell cycle and apoptosis. However, the latest studies revealed that E2F1 prompted metastatic and invasive progresses, except for its proliferative and apoptotic functions (18). The diverse behavior of E2F1 rely on different cell context and its various target genes in different cancer types (19). In osteosarcoma, E2F1 was found to be indispensable for proliferation (20), however, overexpression of E2F1 not only exerted a growth-suppressing effect but facilitated the chemotherapeutic drug-induced apoptosis (21,22). Our findings revealed that knockdown of E2F1 had no effect on apoptosis of untreated osteosarcoma cells but raised an apparent growth retardation. More importantly, E2F1 silence significantly suppressed the migration and invasion of osteosarcoma cells, which suggested the necessity of E2F1 for osteosarcoma metastasis. Except for promoting metastasis, high expression of E2F1 sensitizes osteosarcoma to chemotherapy. More and more evidence support that the E2F1-induced apoptosis was dependent on DNA damage which altered the post-translational modifications of E2F1 (23), then phosphorylation and acetylation of E2F1 predominantly strengthened its proapoptotic activity (24).
Gene set enrichment analysis (GSEA) performed by Gao et al indicated that the aberrant expression of E2F1 was a key regulator of the entire metastatic process (25). Increased abundance of E2F1 triggers invasion and metastasis by activating growth receptor signaling pathways, which sheds light on the molecular mechanisms underlying E2F1-induced prometastatic activity. Recent study revealed that E2F1 enhanced metastatic behavior in melanomas by transactivating the VEGFC receptor, VEGFR-3, and epidermal growth factor receptor (EGFR) (6,26). In prostate cancer, E2F1 was found to activate the transcription of androgen receptor which was essential for the initiation and progression of prostate cancer (27). Similarly, we reported that DDR1, a discoidin domain receptor, was directly regulated by E2F1 which was verified as a transcription activator of DDR1 in osteosarcoma. Then, the E2F1-dependent expression of DDR1 facilitated the proliferation, migration, and invasion of osteosarcoma cells. However, overexpression of DDR1 partially restored the impaired migration and invasion caused by E2F1 knockdown, but not for proliferation. So, even though DDR1 could induce the proliferation of osteosarcoma cells, other factors or pathways regulated by E2F1 are necessary for proliferation.
Although the roles of DDR1 in the metastasis of osteosarcoma have rarely been reported, acquisition of a more mesenchymal-like phenotype was associated with expression of DDR1 in other cancer types, such as gastric, colorectal, and pancreatic cancer (13,28,29). In the present study, we found that DDR1 was required for EMT which might contribute to the E2F1-dependent aggressive behavior in osteosarcoma cells. Mechanistically, DDR1 promoted EMT by activating its downstream signaling pathways. Due to the extensive deposition of collagen I characterized in osteosarcoma, activation of highly expressed DDR1 by collagen is believed to activate downstream signaling molecules that display as mediators to drive EMT. In a recent study, a non-canonical DDR1 signaling was found to enable the metastatic reactivation of breast cancer cells by activating JAK2-STAT3 signaling (30). Given the important role of STAT3 as an EMT signaling transducer, we examined the phosphorylated STAT3 (Y705) that was crucial for its dimerization and activation. Also, evidence showed that activation of STAT3 by DDR1 mediated the EMT and malignant phenotypes of osteosarcoma cells. Altogether, we hypothesize that the E2F1/DDR1/STAT3 axis is critical for the metastasis of osteosarcoma.
As another widely reported oncogene regulated by DDR1, Matrix metalloproteinase (MMP) contributes to the matrix components degradation that is required for tumor invasion (31). Several studies reported that DDR1 could play an invasive role in various tumors via inducing MMP2 and MMP9 (32,33). Multiple mechanisms may be employed in the induction of MMPs by DDR1. In breast cancer, type IV collagen increased MMP2 and MMP9 secretion and invasion through activating DDR1-Src signaling (34). Whereas, receptor signaling activated by collagen was not requisite for DDR1 to support invasion in oral squamous cell carcinoma (35). Except for inducing MMPs expression, DDR1 activation by collagen could accelerate the conversion of pro-MMP2 into its active form (36). In the present study, we found that E2F1/DDR1/STAT3 signaling was essential for maintaining the high expression of MMP2 and MMP9 which might enhance an invasive behavior to osteosarcoma.
In conclusion, the present study demonstrates that E2F1 is important for osteosarcoma proliferation, migration and invasion. In addition, our findings identify a novel regulatory pathway, E2F1/DDR1/STAT3 axis, which drives EMT and increases the expressions of MMPs in osteosarcoma cells. As a consequence, overactivation of the pathway results in the aggressive phenotypes of osteosarcoma. Consistently, clinical analysis suggests that high expressions of E2F1 and DDR1 indicate a relative poor prognosis in osteosarcoma patients. Although this study provides a possible molecular mechanism for E2F1-dependent osteosarcoma metastasis, further studies are needed to elucidate the details of the deregulation of E2F1 and its downstream pathways.
Acknowledgments
This study was supported by the Science Technology Department of Zhejiang Province (2014C33122).
References
- 1.Anderson ME. Update on survival in osteosarcoma. Orthop Clin North Am. 2016;47:283–292. doi: 10.1016/j.ocl.2015.08.022. [DOI] [PubMed] [Google Scholar]
- 2.Poletajew S, Fus L, Wasiutyński A. Current concepts on pathogenesis and biology of metastatic osteosarcoma tumors. Ortop Traumatol Rehabil. 2011;13:537–545. doi: 10.5604/15093492.971038. [DOI] [PubMed] [Google Scholar]
- 3.Maximov VV, Aqeilan RI. Genetic factors conferring metastasis in osteosarcoma. Future Oncol. 2016;12:1623–1644. doi: 10.2217/fon-2016-0014. [DOI] [PubMed] [Google Scholar]
- 4.Dyson N. The regulation of E2F by pRB-family proteins. Genes Dev. 1998;12:2245–2262. doi: 10.1101/gad.12.15.2245. [DOI] [PubMed] [Google Scholar]
- 5.Lee JS, Leem SH, Lee SY, Kim SC, Park ES, Kim SB, Kim SK, Kim YJ, Kim WJ, Chu IS. Expression signature of E2F1 and its associated genes predict superficial to invasive progression of bladder tumors. J Clin Oncol. 2010;28:2660–2667. doi: 10.1200/JCO.2009.25.0977. [DOI] [PubMed] [Google Scholar]
- 6.Alla V, Engelmann D, Niemetz A, Pahnke J, Schmidt A, Kunz M, Emmrich S, Steder M, Koczan D, Pützer BM. E2F1 in melanoma progression and metastasis. J Natl Cancer Inst. 2010;102:127–133. doi: 10.1093/jnci/djp458. [DOI] [PubMed] [Google Scholar]
- 7.Liang YX, Lu JM, Mo RJ, He HC, Xie J, Jiang FN, Lin ZY, Chen YR, Wu YD, Luo HW, et al. E2F1 promotes tumor cell invasion and migration through regulating CD147 in prostate cancer. Int J Oncol. 2016;48:1650–1658. doi: 10.3892/ijo.2016.3364. [DOI] [PubMed] [Google Scholar]
- 8.Rammal H, Saby C, Magnien K, Van-Gulick L, Garnotel R, Buache E, El Btaouri H, Jeannesson P, Morjani H. Discoidin domain receptors: Potential actors and targets in cancer. Front Pharmacol. 2016;7:55. doi: 10.3389/fphar.2016.00055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Miao L, Zhu S, Wang Y, Li Y, Ding J, Dai J, Cai H, Zhang D, Song Y. Discoidin domain receptor 1 is associated with poor prognosis of non-small cell lung cancer and promotes cell invasion via epithelial-to-mesenchymal transition. Med Oncol. 2013;30:626. doi: 10.1007/s12032-013-0626-4. [DOI] [PubMed] [Google Scholar]
- 10.Koh M, Woo Y, Valiathan RR, Jung HY, Park SY, Kim YN, Kim HR, Fridman R, Moon A. Discoidin domain receptor 1 is a novel transcriptional target of ZEB1 in breast epithelial cells undergoing H-Ras-induced epithelial to mesenchymal transition. Int J Cancer. 2015;136:E508–E520. doi: 10.1002/ijc.29154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Shimada K, Nakamura M, Ishida E, Higuchi T, Yamamoto H, Tsujikawa K, Konishi N. Prostate cancer antigen-1 contributes to cell survival and invasion though discoidin receptor 1 in human prostate cancer. Cancer Sci. 2008;99:39–45. doi: 10.1111/j.1349-7006.2007.00655.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Huo Y, Yang M, Liu W, Yang J, Fu X, Liu D, Li J, Zhang J, Hua R, Sun Y. High expression of DDR1 is associated with the poor prognosis in Chinese patients with pancreatic ductal adenocarcinoma. J Exp Clin Cancer Res. 2015;34:88. doi: 10.1186/s13046-015-0202-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Xie R, Wang X, Qi G, Wu Z, Wei R, Li P, Zhang D. DDR1 enhances invasion and metastasis of gastric cancer via epithelial-mesenchymal transition. Tumour Biol. 2016;37:12049–12059. doi: 10.1007/s13277-016-5070-6. [DOI] [PubMed] [Google Scholar]
- 14.Pagliarini V, Giglio P, Bernardoni P, De Zio D, Fimia GM, Piacentini M, Corazzari M. Downregulation of E2F1 during ER stress is required to induce apoptosis. J Cell Sci. 2015;128:1166–1179. doi: 10.1242/jcs.164103. [DOI] [PubMed] [Google Scholar]
- 15.Shats I, Gatza ML, Liu B, Angus SP, You L, Nevins JR. FOXO transcription factors control E2F1 transcriptional specificity and apoptotic function. Cancer Res. 2013;73:6056–6067. doi: 10.1158/0008-5472.CAN-13-0453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Summy JM, Gallick GE. Src family kinases in tumor progression and metastasis. Cancer Metastasis Rev. 2003;22:337–358. doi: 10.1023/A:1023772912750. [DOI] [PubMed] [Google Scholar]
- 17.Valencia K, Ormazabal C, Zandueta C, Luis-Ravelo D, Antó I, Pajares MJ, Agorreta J, Montuenga LM, Martínez-Canarias S, Leitinger B, et al. Inhibition of collagen receptor discoidin domain receptor-1 (DDR1) reduces cell survival, homing, and colonization in lung cancer bone metastasis. Clin Cancer Res. 2012;18:969–980. doi: 10.1158/1078-0432.CCR-11-1686. [DOI] [PubMed] [Google Scholar]
- 18.Engelmann D, Pützer BM. The dark side of E2F1: In transit beyond apoptosis. Cancer Res. 2012;72:571–575. doi: 10.1158/0008-5472.CAN-11-2575. [DOI] [PubMed] [Google Scholar]
- 19.Meng P, Ghosh R. Transcription addiction: Can we garner the Yin and Yang functions of E2F1 for cancer therapy? Cell Death Dis. 2014;5:e1360. doi: 10.1038/cddis.2014.326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ahn JD, Kim CH, Magae J, Kim YH, Kim HJ, Park KK, Hong S, Park KG, Lee IK, Chang YC. E2F decoy oligodeoxynucleotides effectively inhibit growth of human tumor cells. Biochem Biophys Res Commun. 2003;310:1048–1053. doi: 10.1016/j.bbrc.2003.09.124. [DOI] [PubMed] [Google Scholar]
- 21.Liontos M, Niforou K, Velimezi G, Vougas K, Evangelou K, Apostolopoulou K, Vrtel R, Damalas A, Kontovazenitis P, Kotsinas A, et al. Modulation of the E2F1-driven cancer cell fate by the DNA damage response machinery and potential novel E2F1 targets in osteosarcomas. Am J Pathol. 2009;175:376–391. doi: 10.2353/ajpath.2009.081160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Russo AJ, Magro PG, Hu Z, Li WW, Peters R, Mandola J, Banerjee D, Bertino JR. E2F-1 overexpression in U2OS cells increases cyclin B1 levels and cdc2 kinase activity and sensitizes cells to antimitotic agents. Cancer Res. 2006;66:7253–7260. doi: 10.1158/0008-5472.CAN-05-3725. [DOI] [PubMed] [Google Scholar]
- 23.Stevens C, Smith L, La Thangue NB. Chk2 activates E2F-1 in response to DNA damage. Nat Cell Biol. 2003;5:401–409. doi: 10.1038/ncb974. [DOI] [PubMed] [Google Scholar]
- 24.Galbiati L, Mendoza-Maldonado R, Gutierrez MI, Giacca M. Regulation of E2F-1 after DNA damage by p300-mediated acetylation and ubiquitination. Cell Cycle. 2005;4:930–939. doi: 10.4161/cc.4.7.1784. [DOI] [PubMed] [Google Scholar]
- 25.Gao Y, Li H, Ma X, Fan Y, Ni D, Zhang Y, Huang Q, Liu K, Li X, Wang L, et al. KLF6 suppresses metastasis of clear cell renal cell carcinoma via transcriptional repression of E2F1. Cancer Res. 2017;77:330–342. doi: 10.1158/0008-5472.CAN-16-0348. [DOI] [PubMed] [Google Scholar]
- 26.Engelmann D, Mayoli-Nüssle D, Mayrhofer C, Fürst K, Alla V, Stoll A, Spitschak A, Abshagen K, Vollmar B, Ran S, et al. E2F1 promotes angiogenesis through the VEGF-C/VEGFR-3 axis in a feedback loop for cooperative induction of PDGF-B. J Mol Cell Biol. 2013;5:391–403. doi: 10.1093/jmcb/mjt035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sharma A, Yeow WS, Ertel A, Coleman I, Clegg N, Thangavel C, Morrissey C, Zhang X, Comstock CE, Witkiewicz AK, et al. The retinoblastoma tumor suppressor controls androgen signaling and human prostate cancer progression. J Clin Invest. 2010;120:4478–4492. doi: 10.1172/JCI44239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hu Y, Liu J, Jiang B, Chen J, Fu Z, Bai F, Jiang J, Tang Z. MiR-199a-5p loss up-regulated DDR1 aggravated colorectal cancer by activating epithelial-to-mesenchymal transition related signaling. Dig Dis Sci. 2014;59:2163–2172. doi: 10.1007/s10620-014-3136-0. [DOI] [PubMed] [Google Scholar]
- 29.Shintani Y, Fukumoto Y, Chaika N, Svoboda R, Wheelock MJ, Johnson KR. Collagen I-mediated up-regulation of N-cadherin requires cooperative signals from integrins and discoidin domain receptor 1. J Cell Biol. 2008;180:1277–1289. doi: 10.1083/jcb.200708137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gao H, Chakraborty G, Zhang Z, Akalay I, Gadiya M, Gao Y, Sinha S, Hu J, Jiang C, Akram M, et al. Multi-organ site metastatic reactivation mediated by non-canonical discoidin domain receptor 1 signaling. Cell. 2016;166:47–62. doi: 10.1016/j.cell.2016.06.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gialeli C, Theocharis AD, Karamanos NK. Roles of matrix metalloproteinases in cancer progression and their pharmacological targeting. FEBS J. 2011;278:16–27. doi: 10.1111/j.1742-4658.2010.07919.x. [DOI] [PubMed] [Google Scholar]
- 32.Juin A, Di Martino J, Leitinger B, Henriet E, Gary AS, Paysan L, Bomo J, Baffet G, Gauthier-Rouvière C, Rosenbaum J, et al. Discoidin domain receptor 1 controls linear invadosome formation via a Cdc42-Tuba pathway. J Cell Biol. 2014;207:517–533. doi: 10.1083/jcb.201404079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Yang SH, Baek HA, Lee HJ, Park HS, Jang KY, Kang MJ, Lee DG, Lee YC, Moon WS, Chung MJ. Discoidin domain receptor 1 is associated with poor prognosis of non-small cell lung carcinomas. Oncol Rep. 2010;24:311–319. doi: 10.3892/or_00000861. [DOI] [PubMed] [Google Scholar]
- 34.Castro-Sanchez L, Soto-Guzman A, Guaderrama-Diaz M, Cortes-Reynosa P, Salazar EP. Role of DDR1 in the gelatinases secretion induced by native type IV collagen in MDA-MB-231 breast cancer cells. Clin Exp Metastasis. 2011;28:463–477. doi: 10.1007/s10585-011-9385-9. [DOI] [PubMed] [Google Scholar]
- 35.Hidalgo-Carcedo C, Hooper S, Chaudhry SI, Williamson P, Harrington K, Leitinger B, Sahai E. Collective cell migration requires suppression of actomyosin at cell-cell contacts mediated by DDR1 and the cell polarity regulators Par3 and Par6. Nat Cell Biol. 2011;13:49–58. doi: 10.1038/ncb2133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ram R, Lorente G, Nikolich K, Urfer R, Foehr E, Nagavarapu U. Discoidin domain receptor-1a (DDR1a) promotes glioma cell invasion and adhesion in association with matrix metalloproteinase-2. J Neurooncol. 2006;76:239–248. doi: 10.1007/s11060-005-6874-1. [DOI] [PubMed] [Google Scholar]