Abstract
Interactions between bone marrow stromal cells (BMSCs) and multiple myeloma cells significantly contribute to the progression of multiple myeloma (MM). However, little is known about the molecular mechanisms that regulate these interactions. Connexin-43 (Cx-43) has been implicated in the interplay between BMSCs and MM cells. In this study, we hypothesized that the steroid receptor co-activator-3 (SRC3) expressed in BMSCs regulates the expression of Cx-43 to promote the proliferation and migration of myeloma cells. To address this, we co-cultured a human multiple myeloma cell line, RPMI-8226 transfected with either control BMSCs or sh-SRC3-BMSCs. We found that knocking down SRC3 expression in BMSCs inhibited the proliferation and migration of RPMI-8226 cells. In addition, we found that co-culturing RPMI 8266 cells with BMSCs increased Cx43 expression, while knocking down SRC3 expression in BMSCs decreased Cx43 expression. Moreover, our work revealed that SRC3 in BMSCs regulates Cx43 expression via the mitogen-activated protein kinase (MAPK) pathway. To validate this result in vivo, we knocked down SRC3 expression in BMSCs in nude mice and found that tumor growth and cell apoptosis were significantly decreased. In addition, mice treated with either RPMI 8266 cells overexpressing Cx43 or with a P38 MAPK inhibitor (SB202190) exhibited increased intratumoral leukocyte populations and promoted cell apoptosis in tumor tissue. Our findings demonstrate how SRC3 and Cx43 regulation between BMSCs and myeloma cells mediate cell growth and disease progression, with potential implications for prognosis and therapeutic interventions.
Keywords: bone marrow stromal cells, multiple myeloma, Cx43, SRC3, proliferation, migration, MAPK
Introduction
Multiple myeloma (MM) is a B cell neoplasm characterized by the aberrant clonal expansion of plasma cells (PCs) within the bone marrow (1,2). It is an age-dependent malignancy and leads to osteolytic lesions, immunodeficiency and renal failure. Multiple myeloma accounts for 20% of all deaths from hematological cancers and is the second most common hematological malignancy worldwide (3). Despite advances in novel therapeutics targeting specific myeloma disease-driven pathways, multiple myeloma remains a largely incurable disease with a median survival of 62 months due to relapse and drug resistance (4). Therefore, understanding the mechanisms associated with multiple myeloma pathogenesis is necessary for the development of future therapeutic strategies.
The bone marrow (BM) microenvironment, an intricate and dynamic niche composed of bone marrow stromal cells (BMSCs), fibroblasts, hematopoietic stem cells, progenitor cells, endothelial cells, immune cells, and the extracellular matrix (5), supports myeloma cell growth and contributes to disease progression. Previous studies reported that stromal cells directly promoted the progression of multiple myeloma and drug resistance through cell-to-cell contact and cytokine stimulation (6). Adhesion molecules on the surface of the myeloma cells bind to extracellular matrix proteins and BMSCs (7,8).
Steroid receptor coactivator-3 (SRC3), a multifunctional transcriptional coactivator, is a member of the p160 steroid receptor coactivator family (9,10). SRC3 acts as a bridge between the hormone-activated nuclear receptors (NRs), other coregulators, and the basal transcriptional machinery to regulate target gene transcription and impact multiple growth factor pathways (11,12). SRC3 plays an important role in physiological and pathological functions, and physiological processes including somatic growth, sexual maturation, energy metabolism, female reproductive function, tumorigenesis (9,12). SRC3 is amplified in different cancers and is speculated to be associated with the initiation and/or progression of carcinomas (10–12). In addition, SRC3 influences the radiosensitivity of hematopoietic cells, hematopoietic ability and bone marrow microenvironment (13,14). However, it remains unclear how SRC3 in BMSCs change the bone marrow microenvironment, and promote the proliferation and migration of multiple myeloma.
Connexin 43 (Cx43), a predominant gap junction protein expressed in BMSCs is phosphorylated by Mitogen-activated protein kinase (MAPK) and MAPK is also thought to regulate Cx43 function (15). Gap junctions (GJs) are transmembrane domains that allow direct intercellular communication between neighboring cells. Aberrant function of GJs and reduction in cell-cell coupling via GJs is associated with many pathological conditions, including cancer (16). Cx43 is aberrantly expressed in several cancers, such as liver cancer, prostate cancer, and breast cancer. In breast cancer, therapeutic targeting of Cx43 exhibits strong anticancer effects and few detrimental side effects (17). In addition, Cx43 plays a role in breast cancer cell proliferation, differentiation, and migration (18,19). Therefore, in this study, we tested our hypothesis that SRC3 in BMSCs mediate the bone marrow microenvironment by regulating the expression of Cx43. Our in vitro and in vivo studies suggest that overexpressed SRC3 regulates Cx43 via the MAPK pathway to promote myeloma cell growth.
Materials and methods
Multiple myeloma patients
Patients newly diagnosed (within 6 months) with multiple myeloma (n=20, 14 male and 6 female) were recruited in this study between April 2015 and March 2016 at The Third Affiliated Daping Hospital. All patients had myeloma that was classified as Durie-Salmon stage II or III and/or ISS stage ≥2. The average age of all patients was 65 years. The basic characteristics of multiple myeloma patients were as shown in Table I. This study was approved by the Medical Ethics Committee of the Third Military Medical University. Healthy donors were utilized as control samples. Serum from the patients was collected for the following studies. All the patients signed informed written consents in accordance with the Declaration of Helsinki.
Table I.
Basic characteristics of MM patients.
| Characteristic | No. of patients (%) |
|---|---|
| Total | 20 (100) |
| Male | 14 (70) |
| Female | 6 (30) |
| Age >50 years | 20 (63.2) |
| Durie-Salmon stage ≥2 | 20 (100) |
| ISS stage ≥2 | 20 (100) |
Cell culture
The human multiple myeloma cell lines (RPMI-8226 and U266) were purchased from American Type Culture Collection (Manassas, VA, USA). The human bone marrow-mesenchymal stem cells were obtained from Shanghai Cell Institute (Shanghai, China). The cells were cultured at 37°C in a water-saturated atmosphere of 95% air and 5% CO2 in RPMI-1640 supplemented with 10% heat-inactivated Fetal Bovine Serum (FBS) (Gibco, Carlsbad, CA, USA), 100 U/ml penicillin and 100 µg/ml streptomycin (Amresco, Solon, OH, USA) or human bone marrow mesenchymal stem cell complete medium (Guangzhou, China). For treatment with the P38 inhibitor SB202190, the cells were seeded at 2×105 cells/well in a 24-well plate, and incubated overnight at 37°C. Then, the medium was replaced with fresh medium supplemented with 100 µM of SB202190 (Cell Signaling Technology, Danvers, MA, USA), and the cells were cultured for an additional 4 h. Cells were harvested for further studies.
Co-culture of myeloma cells and BMSCs
The myeloma cells and BMSCs were cultured separately until the fifth passage (50% confluence). Then, myeloma cells and BMSC were co-cultured using a non-contact Transwell system (Corning Inc., Corning, NY, USA). Myeloma cells (4×105 cells/ml) were added to the top of BMSCs and incubated for 24 h. Then, cells were separated and pelleted for the following studies.
Cell Counting Kit-8 (CCK-8) assay
CCK-8 assay was performed by Cell Counting Kit-8 (Dojindo). The RPMI-8226 cells were co-cultured and inoculated into 96-well plates at a density of 5×103 cells/ml, and 100 µl of culture medium was added into each well. After the cells were cultured for 24, 48 and 72 h, 10 µl of CCK-8 reagent was added to each well and incubated for 2 h in 5% CO2 at 37°C. The optical density (OD) was measured by a microplate reader at 450/630 nm.
Lentivirus transfection
To knock down SRC3 expression in BMSCs, cells were transfected with SRC3-specific short hairpin RNA (sh-SRC3) and scrambled shRNA lentiviral vectors. The lentiviral vector green fluorescent protein (GFP) was expressed in all lentiviruses and was used to evaluate the transduction efficiency. The efficiency for SRC3 knockdown was verified by detecting mRNA and protein levels of SRC3. All lentiviral vectors were purchased from GeneChem (Shanghai, China). Multiplicity of infection (MOI) refers to the number of viral particles per cell. Transfections were performed in BMSCs with MOI of 10:1 following the manufacturer's instructions. The medium containing the vector was replaced by complete medium after 10 h.
Cell transfection
The RPMI-8226 cells were transfected with control plasmids (pcDNA3.1) or overexpression plasmids (pcDNA3.1-Cx43). Both the pcDNA3.1 or pcDNA3.1-Cx43 plasmids were purchased from Shanghai GenePharma, Co., Ltd. (Shanghai, China). Before transfection, cells were cultured in 12-well plates till they reached 70% confluency. Opti-MEM medium (Thermo Fisher Scientific, Inc., Waltham, MA, USA) and Lipo 3000 reagent (Thermo Fisher Scientific, Inc.) were used. After transfection for 48 h, cells were collected for subsequent analysis.
Western blotting
Cells were lysed in RIPA buffer (Boston BioProducts, Ashland, MA, USA) containing protease inhibitor cocktail (Roche Diagnostics, Indianapolis, IN, USA). The protein concentration was measured by BCA Protein Assay kit (Pierce, Rockford, IL, USA). Equal amounts of protein samples (50 µg) were separated by 12% SDS-PAGE gel and transferred onto PVDF membranes (Millipore, Billerica, MA, USA). The PVDF membranes were blocked in TBST containing 2% BSA for 1 h, and then incubated with primary antibodies specific for SRC3, Cx43, phosphorylated ERK (pERK), p38 (p-p38) and JNK (p-JNK). The antibodies were all from Cell Signaling Technology. The bands were visualized using ECL Western Blotting Detection Reagents (Kibbutz Beit Haemek, Israel) and subjected to Alpha Innotech Flour Chem-FC2 imaging system (Alpha Innotech).
Quantitative real-time PCR (qPCR)
Total RNA was extracted from cells using TRIzol reagent (Invitrogen, Carlsbad, CA, USA) according to manufacturer's instructions. A total of 1 µg RNA was used as the template for single strand cDNA synthesis utilizing random primers and the Primescript reverse transcriptase (M-MLV, Takara, Shiga, Japan). The PCR amplification conditions were 95°C for 10 sec, 40 cycles of 94°C for 30 sec, 60°C for 30 sec, and 72°C for 30 sec on Applied Biosystems (ABI) step-one plus sequence detection system (Applied Biosystems, Foster City, CA, USA) with SYBR Green PCR Mix (iTAP, Bio-Rad). The qPCR was performed for Cx43, SRC3 and GAPDH. And, the related primer sequence information of Cx43, SRC3 and GAPDH is listed in Table II. Analysis and fold differences were determined using the comparative cycle threshold (CT) method. Fold change was calculated from the ΔΔCT values with the formula 2−ΔΔCT.
Table II.
The specific primer sequences for qRT-PCR.
| ID | Sequence (5′-3′) |
|---|---|
| Cx43 | F: TGGTAAGGTGAAAATGCGAGG |
| R: GCACTCAAGCTGAATCCATAGAT | |
| SRC3 | F: TTGTCTCAACCCACTTCCTT |
| R: CCATACCTAGCTCCACTCATC | |
| GAPDH | F: TGTTCGTCATGGGTGTGAAC |
| R: ATGGCATGGACTGTGGTCAT |
F, forward; R, reverse.
Immunohistochemistry
Immunohistochemistry (IHC) was performed as previously described (20). Tissues were fixed in 4% paraformaldehyde overnight, embedded in paraffin and cut in 5 µm thickness for experiments. Tissue sections were deparaffinized, hydrated, and then heated in a steamer for 2.5 min to retrieve antigen. IHC was carried out with primary monoclonal antibodies (anti-Cx43 or SRC3; Cell Signaling Technology) at 1:500 dilutions for 50 min, followed by incubation with goat anti-rabbit secondary antibody (Thermo Fisher Scientific, Inc., Fremont, CA, USA). Negative control sections were incubated with Phosphate Buffered Saline (PBS) instead of primary antibodies. In this study, we set the negative control, but not the positive control.
Transwell assay
RPMI-8226 cells (2×105) co-cultured with either BMSC or MAPK inhibitor SB202190, were seeded into the Transwell upper chamber (Corning Inc.). The upper chamber was added with serum-free medium, and lower chamber was added with culture medium containing 10% FBS. Moreover, we aseptically coated the outside bottom surface of the inner chamber with Poly-D-lysine polymers so as to promote cell adhesion to the solid substrate. After 48 h, the non-migrated cells were removed from the chamber using a cotton swab and migrated cells were fixed with 2% paraformaldehyde and stained with crystal violet dye. Migrated cells were observed under a phase contrast microscope, and photographed.
Scratch-wound healing assay
Scratch-wound healing assays were performed using RPMI-8226 cells after different co-culture. Cells (5×105) were seeded into 6-well plates containing 5 µg/cm2 of surface area Poly-D-lysine polymers solution and cultured. Scratches were made with a sterile 200 µl pipette tip to create a cell-free zone. Detached cells were washed off with culture medium. Images of the scratch area were captured after 24 h using an inverted phase contrast microscope (Eclipse E400, Nikon Corp., Tokyo, Japan). The remaining wound area was measured using ImageJ software [National Institutes of Health (NIH), Bethesda, MD, USA] and normalized to time 0 wounds. Experiments were performed three times.
Immunofluorescence staining
PMI 8226 cells were stained according to previously described protocols (21). Briefly, cells were fixed in 4% paraformaldehyde and permeabilized with 0.5% Triton X-100 for 15 min. The cells were washed with PBS, blocked with 5% BSA in PBS for 1 h and incubated with primary antibody overnight at 4°C. The primary rabbit polyoclonal anti-Cx43 antibody (dilution 1:1,000; Abcam, Cambridge, MA, USA) and secondary goat anti-mouse IgG H&L (Texas Red®) antibody (dilution 1:1,000; Abcam) were used. The coverslip was subjected to DAPI (1:5,000, Sigma) nuclear counterstain. Cells were visualized by a confocal laser fluorescence microscope (Zeiss LSM 700).
Hoechst staining
Cells were grown on coverslips and then rinsed with PBS 3 times. Then, cells were incubated in the Hoechst labeling solution for 30 min at room temperature. Cells were further rinsed with PBS 3 times. The coverslips were mounted and images were obtained at an excitation wavelength of 353 nm and an emission wavelength of 483 nm.
In vivo tumor growth study
The animal study was approved by the Medical Ethics Committee of the Third Military Medical University in accordance with the Guide for the Care and Use of Laboratory Animals (NIH publication no. 80-23, revised 1996) and animals were treated according to the institutional guidelines. Male nude mice aged between 6–8 weeks were purchased from Shanghai Laboratory Animal Center of China. Each nude mouse was injected with 100 µl cell suspension containing 3×106 RPMI-8226 cells and 3×105 MSC subcutaneously into the right flank for the following groups: A, MM+MSC (RPMI-8226 cells, sh-SRC3-MSC); B, MM+sh-SRC3-MSC+pcDNA3.1 (pcDNA3.1-RPMI-8226 cells, sh-SRC3-MSC); C, MM+sh-SRC3-MSC+Cx43 (Cx43-RPMI-8226 cells, sh-SRC3-MSC); D, MM+sh-SRC3-MSC+inhibitor (pcDNA3.1-RPMI-8226 cells+ SB202190, sh-SRC3-MSC). The tumor size was measured by calipers and the tumor volume calculated with the following formula: V = (length)2 × (width)/2. After 28 days, mice were sacrificed by intraperitoneal administration of an overdose of anaesthetic drug cocktail (240 mg/kg) followed by cervical dislocation. The xenografted tumor tissues were excised and prepared for further analysis.
Terminal deoxynucleotidyl transferase dUTP nick-end labeling (TUNEL) staining
To detect the number of apoptotic nuclei, TUNEL staining was performed. The tumor tissue sections were incubated with permeabilization solution and washed with PBS 2 times. Then, the samples were incubated with TUNEL (Roche Applied Science, Indianapolis, IN, USA) for 60 min at 37°C. The samples were further stained with DAPI (1:5,000, Sigma) nuclear counterstain. Cells were visualized by a confocal laser fluorescence microscope (Zeiss LSM 700).
Statistical analysis
All results are presented as means ± SEM of at least three independent experiments. Student's t-test, Mann-Whitney U test and Log-rank test were used to assess differences between two groups. A P-value of <0.05 was considered to be statistically significant.
Results
Expression of Cx43 in multiple myeloma samples and cell lines
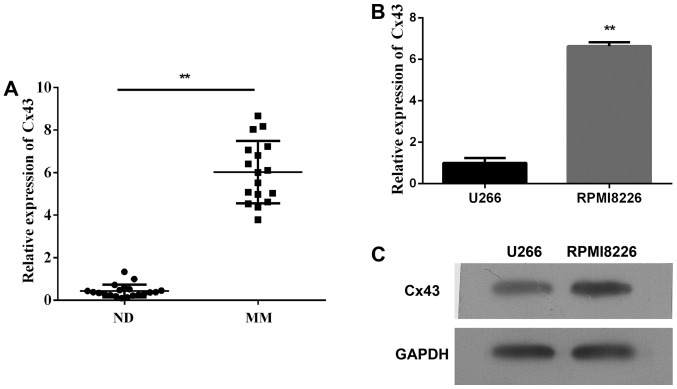
First, we detected the plasma expression levels of Cx43 circulating in patients with multiple myeloma and found that the plasma level of Cx43 significantly increased in patients with newly diagnosed multiple myeloma within 6 months compared with healthy donors (Fig. 1A, P<0.01). Second, we assessed the expression of Cx43 in human multiple myeloma cell lines (RPMI-8226 and U266). As shown in Fig. 1B and C, both MM cell lines expressed Cx43, and the mRNA and protein levels of Cx43 in RPMI-8226 cells were higher than U266 cells. It could be that the expression of Cx43 reduced in the late stages of B cell differentiation. The results are consistent with the data published by Zhang et al (22).
Figure 1.
The expression of Cx43 in multiple myeloma samples and cell lines. (A) Analysis by qPCR measured the expression levels of Cx43 circulating in plasma of patients with multiple myeloma. (B) The mRNA levels of Cx43 in human multiple myeloma cell lines (RPMI-8226 and U266). (C) The protein levels of Cx43 in human multiple myeloma cell lines (RPMI-8226 and U266) by western blots. Data represent three independent experiments (average and SEM of triplicate samples). **P<0.01 vs. control.
SRC3 expressed in BMSCs is involved in the proliferation and migration of multiple myeloma cells
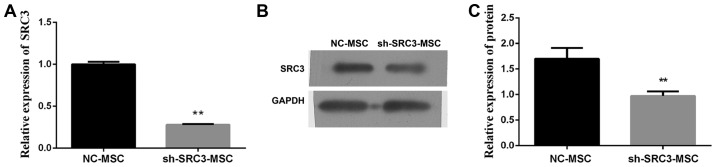
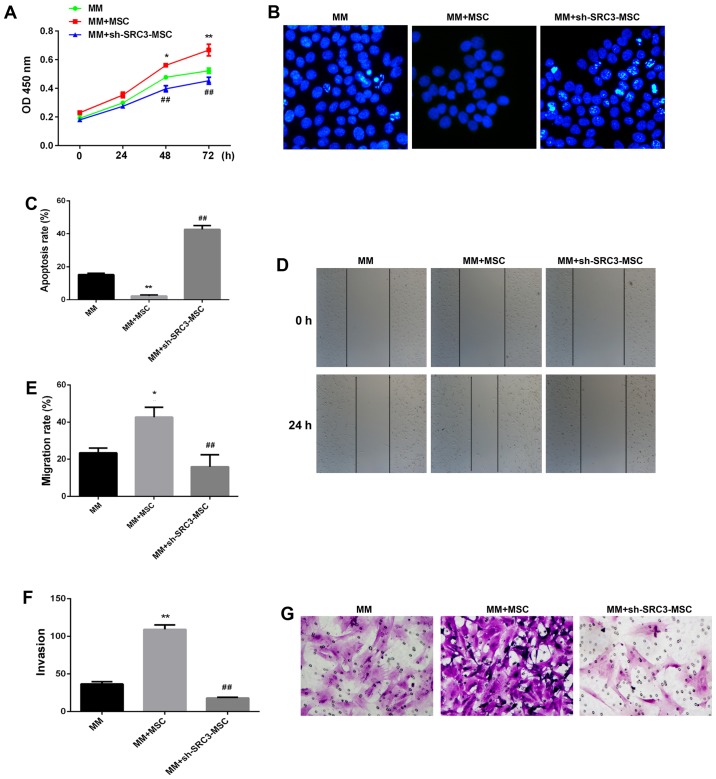
Evidence from the literature suggests that BMSCs promote the proliferation and migration of multiple myeloma cells and contribute to resistance to chemotherapy (23,24). Furthermore, SRC3 influences the radiosensitivity of hematopoietic cells, hematopoietic ability and bone marrow microenvironment (13,14). We wanted to investigate if SRC3 in BMSCs are involved in promoting the proliferation and migration of multiple myeloma cells. We transfected BMSCs with SRC3-specific short hairpin RNA (sh-SRC3) lentiviral vector to knock down the expression of SRC3. We confirmed the efficiency by detecting mRNA and protein levels of SRC3 in BMSCs (Fig. 2A and B). We, next co-cultured the RPMI-8226 cells with either between April 2015 and March 2016 at the third affiliated Daping Hospital control BMSCs or sh-SRC3-BMSCs and evaluated the proliferation and migration ability of RPMI-8226 cells. As shown in Fig. 3A, knocking down SRC3 expression in BMSCs significantly inhibited the proliferation ability (P<0.01) and significantly decreased the rate of apoptosis in RPMI-8226 cells (Fig. 3B and C, P<0.01). In addition, knocking down SRC3 expression in BMSCs inhibited the migration of RPMI-8226 cells assessed by both the wound healing assay (Fig. 3D and E, P<0.01) and Transwell migration assay (Fig. 3F and G, P<0.01).
Figure 2.
Silencing SRC3SRC3 in BMSCs. BMSCs were treated with either sh-SRC3 or sh-NC and the level of SRC3 expression was detected by qPCR (A) and western blots (B). Data represent three independent experiments (average and SEM of triplicate samples). **P<0.01 vs. control; ##P<0.01 vs. MM+sh-SRC3-MSC.
Figure 3.
SRC3 expressed in BMSCs is involved in the proliferation and migration of multiple myeloma cells. The RPMI-8226 cells were co-cultured with either BMSCs or sh-SRC3-BMSCs and their proliferation and migration ability were assessed. (A) Cell proliferation analysis of RPMI-8226 cells after co-culture for 48 h using CCK-8 assay. (B) Hoechst staining of co-cultured RPMI-8226 cells. (C) Cells positive for Hoechst staining were counted. (D and E) Scratch-wound healing assay assessed the migration ability of RPMI-8226 cells after being co-cultured for 48 h. The wound closure was calculated at 24 h under a phase contrast microscope. (F) Transwell migration assay was performed to test the change in migration ability of RPMI-8226 cells after being co-cultured for 48 h. (G) Quantitative assay of migrating cells under a phase contrast microscope. Data represent three independent experiments (average and SEM of triplicate samples). *P<0.05, **P<0.01 vs. control; ##P<0.01 vs. MM+sh-SRC3-MSC.
SRC3 expressed in BMSCs regulates the expression of Cx43 via the MAPK pathway in RPMI-8226 cells
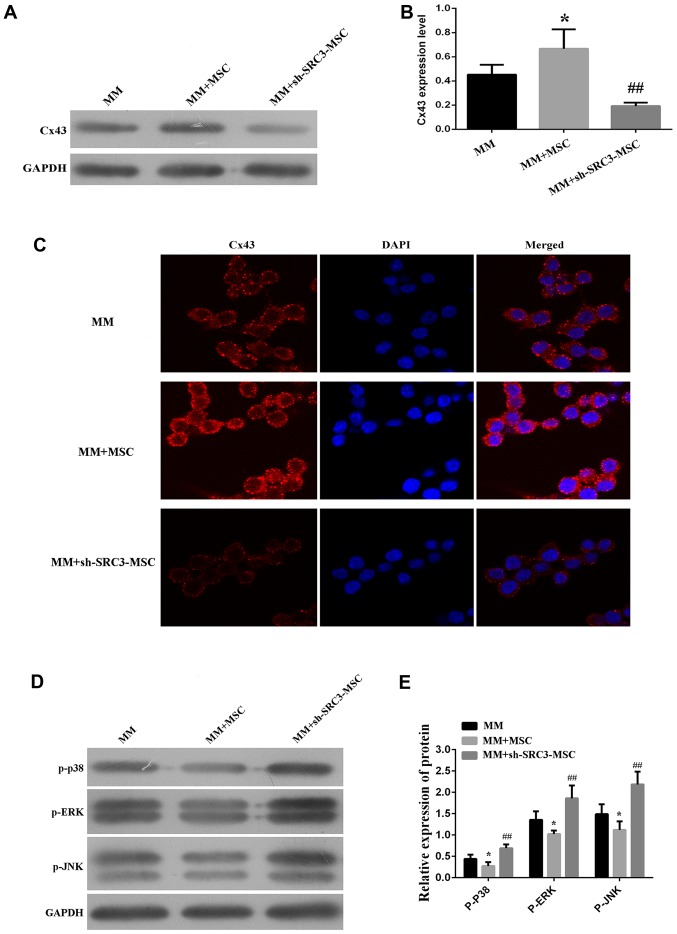
We next asked if SRC3 expression in BMSCs regulated the expression of Cx43. We found that when RPMI-8226 cells were co-cultured with BMSCs, the protein expression of Cx43 was increased (P<0.05). Conversely, when RPMI-8226 cells were co-cultured with BMSCs with knocked down SRC3 expression, the protein level of Cx43 was decreased (Fig. 4A and B, P<0.01). We observed similar results using the immunofluorescence assay (Fig. 4C). The p38 MAPK pathway is implicated in the regulation of cell growth, migration, differentiation, inflammation, survival or apoptosis (25–27). In addition, MAPKs are also involved in the expression of Cx43 in mammary carcinoma and bladder carcinogenesis cells (28–30). We, thus wanted to test if the MAPK pathway mediated SRC3-regulated Cx43 in BMSCs. We co-cultured RPMI-8226 cells transfected with either control BMSCs or sh-SRC3-BMSCs and evaluated the expression of the different signaling molecules in the MAPK pathway, using western blots. As shown in Fig. 4D and E, we found that the protein levels of phosphorylated ERK (pERK), p38 (p-p38) and JNK (p-JNK) were decreased in RPMI-8226 cells co-cultured with BMSCs and elevated in RPMI-8226 cells co-cultured with sh-SRC3-MSC (knockdown of SRC3, P<0.01). These results suggest that SRC3 in BMSCs regulates the expression of Cx43 via the MAPK pathway in RPMI-8226 cells.
Figure 4.
SRC3 in BMSCs may regulate the expression of Cx43 via the MAPK pathway in RPMI-8226 cells. The RPMI-8226 cells were co-cultured with either BMSCs or sh-SRC3-BMSCs and their proliferation and migration ability were assessed. (A) Western blots analyzed the protein levels of Cx43 in RPMI-8226 cells. (B) Densitometry plot of results from (A). The relative expression levels were normalized to GAPDH. (C) Immunofluorescence analyzed the level of Cx43 in RPMI-8226 cells. DAPI was used to stain the cell nucleus. (D) Western blots analyzed the protein levels of phosphorylated ERK (pERK), p38 (p-p38) and JNK (p-JNK) in RPMI-8226 cells. (E) Densitometry plot of results from (D). The relative expression levels were normalized to GAPDH. Data represent three independent experiments (average and SEM of triplicate samples). *P<0.05 vs. control; ##P<0.01 vs. MM+sh-SRC3-MSC.
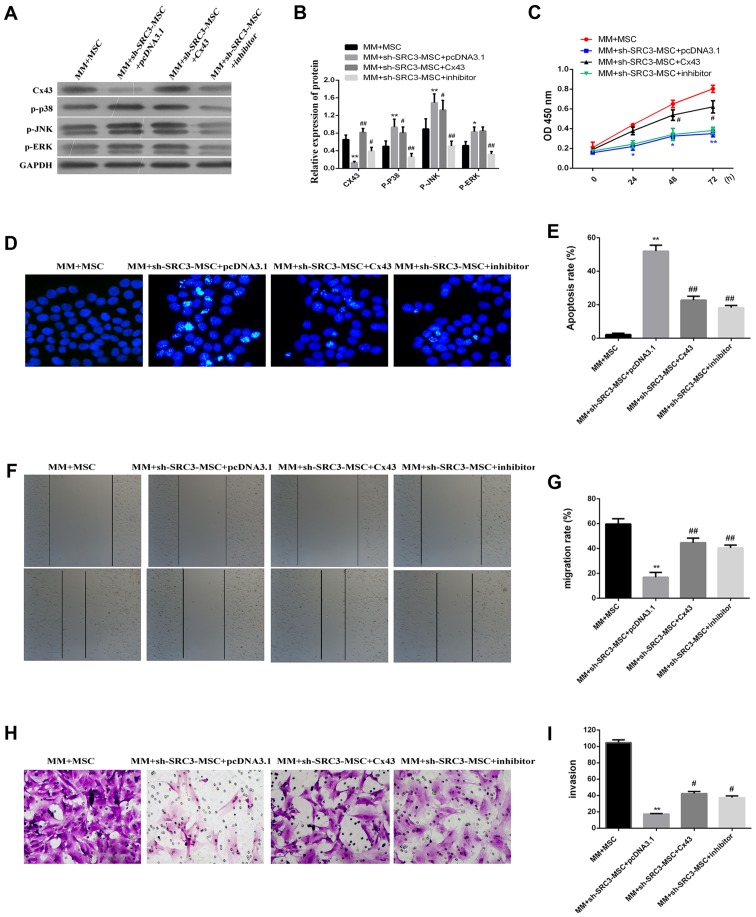
MAPK pathway promotes Cx43-regulated proliferation and migration of RPMI-8226 cells
To gain insight into the possible mechanism associated with the expression of Cx43 regulated by the MAPK pathway, we treated RPMI-8226 cells with the Cx43 overexpression plasmid or with the MAPK inhibitor SB202190. These cells were then co-cultured with either BMSCs or sh-SRC3-MSC. SB202190 inhibits p38 MAP kinase activity by competing with ATP and inhibiting p38 MAPK phosphorylation (31). As shown in Fig. 5A and B, knocking down SRC3 in BMSCs significantly enhanced the protein levels of p-ERK, p-p38 and p-JNK and significantly decreased the protein level of Cx43 level (P<0.01 or P<0.05). We found that the protein levels of p-p38 and p-JNK were down regulated in cells co-cultured with Cx43-MM and sh-SRC3-MSC (P<0.05). We did not observe any significant changes in p-ERK protein levels. SB202190 treatment significantly decreased the expression levels of p-ERK, p-p38 and p-JNK, and signifi-cantly enhanced the expression level of Cx43 in RPMI-8226 cells (Fig. 5A and B, P<0.01 or P<0.05). Knocking down SRC expression and overexpressing Cx43 promoted cell proliferation in RPMI-8226 cells (Fig. 5C, P<0.05) and decreased the proportion of apoptosis in RPMI-8226 cells (Fig. 5D and E, P<0.05). Both the wound healing assay and the Transwell migration assay showed that the migration of RPMI-8226 cells was enhanced by over expressing Cx43 and treating with SB202190 (Fig. 5F and G, P<0.05 and Fig. 5H and I, P<0.05, respectively). Taken together, our data suggest that inactive MAPK pathway enhances the expression of Cx43 and promotes proliferation and migration of RPMI-8226 cells.
Figure 5.
MAPK pathway is involved in promoting the proliferation and migration of RPMI-8226 cells regulated by Cx43. The RPMI-8226 cells were transfected with either pcDNA3.1 or pcDNA3.1-Cx43 for 48 h. These cells were treated with 5 µM MAPK inhibitor SB202190 for 24 h, and then co-cultured with either BMSCs or sh-SRC3-MSC. (A) Western blots analyzed the protein level of Cx43, phosphorylated ERK (pERK), p38 (p-p38) and JNK (p-JNK) in RPMI-8226 cells. (B) Densitometry plot of results from (A). The relative expression levels were normalized to GAPDH. (C) Cell proliferation analysis of RPMI-8226 cells after being co-cultured for 48 h using the CCK-8 assay. (D) Hoechst foci staining for co-cultured RPMI-8226 cells. (E) The cells that stained positive for Hoechst staining were counted. (F and G) Scratch-wound healing assay was used to assess the migration potential of RPMI-8226 cells after being co-cultured for 48 h. The wound closure rate was calculated at 24 h using a phase contrast microscope. (H) Transwell migration assay assessed the change of migration potential of RPMI-8226 cells after being co-cultured for 48 h. Representative images of migrated cells are shown. (I) Relative numbers of migrated cells in the Transwell assay under a phase contrast microscope. Data represent three independent experiments (average and SEM of triplicate samples). *P<0.05, **P<0.01 vs. control; #P<0.05, ##P<0.01 vs. MM+sh-SRC3-MSC+pcDNA3.1.
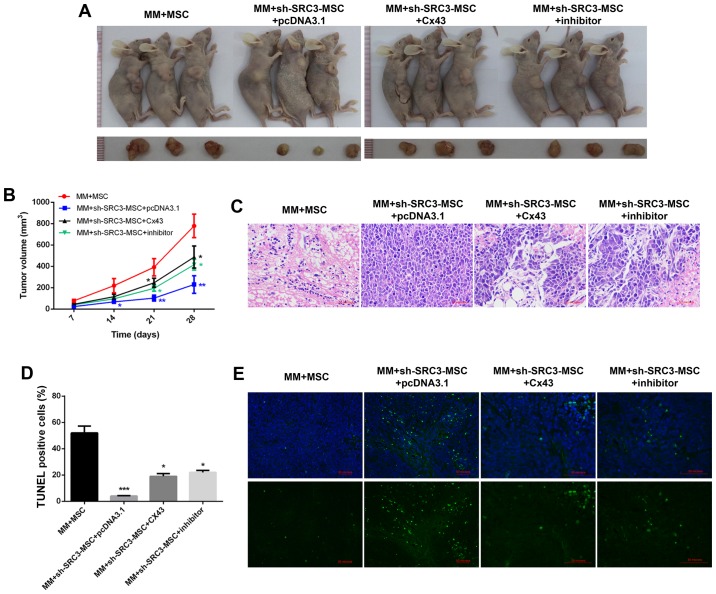
SRC3 expressed in BMSCs promoted tumor growth of multiple myeloma cells by regulating the expression of Cx43
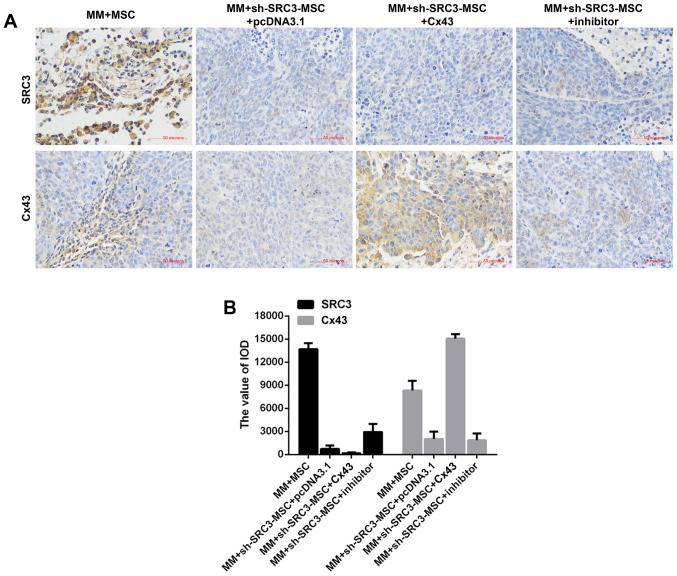
To further validate the molecular mechanism in vivo, we established murine models of multiple myeloma. The BMSCs were treated with sh-SRC3 to knock down the expression of SRC3 and RPMI-8226 cells were overexpressed with either Cx43 or treated with SB202190. The cells were then co-injected into nude mice to establish murine multiple myeloma models. As shown in Fig. 6A and B, compared with the MM+MSCs group, knocking down SRC3 in MSCs (MM+sh-SRC3-MSC+pcDNA3.1) significantly decreased the tumor growth (P<0.01). Both, overexpressing Cx43 and treating with SB202190 promoted the tumor growth (Fig. 6A and B, P<0.05). Histological analysis showed fewer intratumoral leukocyte populations in the MM+sh-SRC3-MSC+pcDNA3.1 group compared with the MM+MSCs group. Intratumoral leukocyte populations increased after overexpressing Cx43 and treating with SB202190 in RPMI-8226 cells (Fig. 6C). Furthermore, TUNEL staining showed that knocking down SRC3 in MSCs (MM+sh-SRC3-MSC+pcDNA3.1) significantly increased cell apoptosis in tumor tissue. Both, overexpressing Cx43 and treating with SB202190 decreased cell apoptosis in tumor tissue (Fig. 6D and E, P<0.05). Moreover, we used immunohis-tochemical staining and found fewer SRC3 positive cells in the MM+sh-SRC3-MSC+pcDNA3.1 group; thus, overexpressing Cx43 and treating with SB202190 influenced the level of SRC3 in tumor tissue. The protein level of Cx43 decreased when SRC3 expression in MSCs was knocked down and increased when cells were overexpressed with Cx43 and treated with SB202190 (Fig. 7).
Figure 6.
SRC3 expressed in BMSCs promoted tumor growth of multiple myeloma cells by regulating the expression of Cx43. The BMSCs were treated with sh-SRC3 to knockdown the expression of SRC3 and RPMI-8226 cells were either overexpressed with Cx43 or treated with MAPK inhibitor SB202190. The cells were co-injected into nude mice to establish murine multiple myeloma models. Each nude mouse was injected with 100 µl of cell suspension containing 3×106 RPMI-8226 cells and 3×105 MSC subcutaneously into the right flank for the following groups. MM+MSC, MM+sh-SRC3-MSC+pcDNA3.1, MM+sh-SRC3-MSC+Cx43, MM+sh-SRC3-MSC+inhibitor. (A) Representative images of tumors from each group. (B) Growth curve of tumors was calculated for each group. (C) Representative images of hematoxylin and eosin staining (x10) of tumors. (D) The proportion of TUNEL positive cells. (E) Representative images of TUNEL staining in tumors. Scale bars, 100 µm. Data represent mean ± SEM. *P<0.05, **P<0.01, ***P<0.001 vs. control; #P<0.05 vs. MM+sh-SRC3-MSC+pcDNA3.1.
Figure 7.
Representative images of immunohistochemical (IHC) staining for SRC3 and Cx43 from each group.
Discussion
The BMSCs in the bone microenvironment promote the proliferation of multiple myeloma cells (5). Interleukin-6 (IL-6), a key inflammatory cytokine required for the growth and survival of multiple myeloma cells, is mostly secreted by BMSCs (32). Adhesion of myeloma cells to BMSCs triggers NF-κB activation, and induces secretion of vascular endothelial growth factor, insulin-like growth factor, and other factors, which support cell growth and chemoresistance of multiple myeloma (33–35). VCAM-1, a cell-surface protein that is highly expressed on BMSCs, promotes the adhesion between BMSCs and multiple myeloma (36). Similarly, in a co-culture system, we found that BMSCs promoted the proliferation and migration of multiple myeloma cells. Knocking down the expression of SRC3 in BMSCs reduced the proliferation and migration of multiple myeloma cells. These results suggested that SRC3 in BMSCs can modify the bone marrow microenvironment, and promote the proliferation and migration of multiple myeloma cells. Moreover, we found that SRC3 in BMSCs promoted the proliferation and migration of multiple myeloma cells by upregulating the expression of Cx43.
We showed that Cx43 is highly expressed in both primary isolated multiple myeloma samples and in multiple myeloma cell lines. Both MM cell lines expressed Cx43, and the mRNA and protein levels of Cx43 in RPMI-8226 cells were higher than U266 cells. Consistently, Fu (37) reported that Cx43 expression was heterogeneous and aberrant in MM cells (RPMI 8266, U266, and XG7 cells). Work from other studies demonstrated that all BMSCs from multiple myeloma patients expressed higher levels of Cx43 than those from normal controls (37), and that Cx43 expressed on BMSCs played an essential role in multiple myeloma cell survival, adhesion, migration and drug resistance (22,38). In our study, we found that Cx43 expressed on multiple myeloma cells promoted the proliferation and migration of multiple myeloma cells, and decreased their cell apoptosis.
In addition, our results revealed that SRC3 in BMSCs regulated the expression of Cx43 via the MAPK pathway in RPMI-8226 cells. The p38 mitogen-activated protein kinase (MAPK) is a subgroup of the MAPK pathway and phosphorylates serine and/or threonine residues of target proteins, subsequently regulating a number of biological processes, including cell growth, differentiation, apoptosis and inflammation (38). p38 MAPK plays a dual role as a regulator of cell death, and can either mediate cell survival or cell death depending on the type of stimulus and the type of the cell (38). In mammary carcinoma cells, over-expressing Cx43 increased the level of the phosphorylated form of p38-MAPK (30). Mechanical stress increased the expression of Cx43 and promoted the osteoblastic differentiation of ligament fibroblasts via ERK1/2 and p38 MAPK pathway (39). In bladder cancer, the highly expressed and cytoplasmic localized Cx43 contributed to oncogenic and aberrant activation of JNK and ERK signaling (28). In our study, knocking down the expression of SRC3 in BMSCs enhanced the protein levels of p-ERK, p-p38 and p-JNK and decreased the protein level of Cx43. The overexpressed Cx43 in RPMI-8226 cells did not influence the protein levels of p-ERK, p-p38 and p-JNK. The inactive MAPK pathway enhanced the expression of Cx43 and promoted cell proliferation and migration of RPMI-8226 cells. In conclusion, SRC3 expressed in BMSCs enhanced the expression of Cx43 via the MAPK pathway and the increased Cx43 promoted cell proliferation and migration of multiple myeloma cells. Our results contribute to a better understanding of the effect of BMSCs in the bone marrow microenvironment and its impact on disease progression.
Acknowledgments
The study was supported by the National Natural Science Foundation of China for Young Scholars (no. 81500175) and the Basic Science and Advanced Technology Research Project of Chongqing City (no. cstc2016jcyjA0381).
References
- 1.Falank C, Fairfield H, Reagan MR. Signaling interplay between bone marrow adipose tissue and multiple myeloma cells. Front Endocrinol (Lausanne) 2016;7(Suppl):67. doi: 10.3389/fendo.2016.00067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Palumbo A, Anderson K. Multiple myeloma. N Engl J Med. 2011;364:1046–1060. doi: 10.1056/NEJMra1011442. [DOI] [PubMed] [Google Scholar]
- 3.Zweegman S, Palumbo A, Bringhen S, Sonneveld P. Age and aging in blood disorders: Multiple myeloma. Haematologica. 2014;99:1133–1137. doi: 10.3324/haematol.2014.110296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.American Cancer Society . Cancer Facts and Figures 2017. American Cancer Society; Atlanta, GA: 2017. https://www.cancer.org/research/cancer-facts-statistics/all-cancer-facts-figures/cancer-facts-figures-2017.html. [Google Scholar]
- 5.Scheideler M, Elabd C, Zaragosi LE, Chiellini C, Hackl H, Sanchez-Cabo F, Yadav S, Duszka K, Friedl G, Papak C, et al. Comparative transcriptomics of human multipotent stem cells during adipogenesis and osteoblastogenesis. BMC Genomics. 2008;9:340. doi: 10.1186/1471-2164-9-340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Mahindra A, Hideshima T, Anderson KC. Multiple myeloma: Biology of the disease. Blood Rev. 2010;24(Suppl 1):S5–S11. doi: 10.1016/S0268-960X(10)70003-5. [DOI] [PubMed] [Google Scholar]
- 7.Podar K, Chauhan D, Anderson KC. Bone marrow microenvironment and the identification of new targets for myeloma therapy. Leukemia. 2009;23:10–24. doi: 10.1038/leu.2008.259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hideshima T, Bergsagel PL, Kuehl WM, Anderson KC. Advances in biology of multiple myeloma: Clinical applications. Blood. 2004;104:607–618. doi: 10.1182/blood-2004-01-0037. [DOI] [PubMed] [Google Scholar]
- 9.Xu J, Li Q. Review of the in vivo functions of the p160 steroid receptor coactivator family. Mol Endocrinol. 2003;17:1681–1692. doi: 10.1210/me.2003-0116. [DOI] [PubMed] [Google Scholar]
- 10.Manier S, Sacco A, Leleu X, Ghobrial IM, Roccaro AM. Bone marrow microenvironment in multiple myeloma progression. J Biomed Biotechnol. 2012;2012:157496. doi: 10.1155/2012/157496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.York B, Yu C, Sagen JV, Liu Z, Nikolai BC, Wu RC, Finegold M, Xu J, O'Malley BW. Reprogramming the posttranslational code of SRC-3 confers a switch in mammalian systems biology. Proc Natl Acad Sci USA. 2010;107:11122–11127. doi: 10.1073/pnas.1005262107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yan J, Tsai SY, Tsai MJ. SRC-3/AIB1: Transcriptional coactivator in oncogenesis. Acta Pharmacol Sin. 2006;27:387–394. doi: 10.1111/j.1745-7254.2006.00315.x. [DOI] [PubMed] [Google Scholar]
- 13.Jin J, Wang Y, Wang J, Xu Y, Chen S, Wang J, Ran X, Su Y. Increased radiosensitivity and radiation-induced apoptosis in SRC-3 knockout mice. J Radiat Res. 2014;55:443–450. doi: 10.1093/jrr/rrt132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jin J, Wang Y, Wang J, Xu Y, Chen SL, Wang JP, Su YP. Impaired hematopoiesis and delayed thrombopoietic recovery following sublethal irradiation in SRC-3 knockout mice. Mol Med Rep. 2014;9:1629–1633. doi: 10.3892/mmr.2014.2043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Warn-Cramer BJ, Cottrell GT, Burt JM, Lau AF. Regulation of connexin-43 gap junctional intercellular communication by mitogen-activated protein kinase. J Biol Chem. 1998;273:9188–9196. doi: 10.1074/jbc.273.15.9188. [DOI] [PubMed] [Google Scholar]
- 16.Nimlamool W, Andrews RMK, Falk MM. Connexin43 phosphorylation by PKC and MAPK signals VEGF-mediated gap junction internalization. Mol Biol Cell. 2015;26:2755–2768. doi: 10.1091/mbc.E14-06-1105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Shishido SN, Delahaye A, Beck A, Nguyen TA. The anticancer effect of PQ1 in the MMTV-PyVT mouse model. Int J Cancer. 2014;134:1474–1483. doi: 10.1002/ijc.28461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kanczuga-Koda L, Sulkowska M, Koda M, Rutkowski R, Sulkowski S. Increased expression of gap junction protein -connexin 32 in lymph node metastases of human ductal breast cancer. Folia Histochem Cytobiol. 2007;45(Suppl 1):S175–S180. [PubMed] [Google Scholar]
- 19.Kanczuga-Koda L, Sulkowski S, Tomaszewski J, Koda M, Sulkowska M, Przystupa W, Golaszewska J, Baltaziak M. Connexins 26 and 43 correlate with Bak, but not with Bcl-2 protein in breast cancer. Oncol Rep. 2005;14:325–329. [PubMed] [Google Scholar]
- 20.Zhou Y, Miao J, Wu H, Tang H, Kuang J, Zhou X, Peng Y, Hu D, Shi D, Deng W, et al. PD-1 and PD-L1 expression in 132 recurrent nasopharyngeal carcinoma: The correlation with anemia and outcomes. Oncotarget. 2017;8:51210–51223. doi: 10.18632/oncotarget.17214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Morsing M, Klitgaard MC, Jafari A, Villadsen R, Kassem M, Petersen OW, Rønnov-Jessen L. Evidence of two distinct functionally specialized fibroblast lineages in breast stroma. Breast Cancer Res. 2016;18:108. doi: 10.1186/s13058-016-0769-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zhang X, Sun Y, Wang Z, Huang Z, Li B, Fu J. Up-regulation of connexin-43 expression in bone marrow mesenchymal stem cells plays a crucial role in adhesion and migration of multiple myeloma cells. Leuk Lymphoma. 2015;56:211–218. doi: 10.3109/10428194.2014.913289. [DOI] [PubMed] [Google Scholar]
- 23.Basak GW, Srivastava AS, Malhotra R, Carrier E. Multiple myeloma bone marrow niche. Curr Pharm Biotechnol. 2009;10:345–346. doi: 10.2174/138920109787847493. [DOI] [PubMed] [Google Scholar]
- 24.Shen X, Guo Y, Yu J, Qi J, Shi W, Wu X, Ni H, Ju S. miRNA-202 in bone marrow stromal cells affects the growth and adhesion of multiple myeloma cells by regulating B cell-activating factor. Clin Exp Med. 2016;16:307–316. doi: 10.1007/s10238-015-0355-4. [DOI] [PubMed] [Google Scholar]
- 25.Khorasanizadeh M, Eskian M, Gelfand EW, Rezaei N. Mitogen-activated protein kinases as therapeutic targets for asthma. Pharmacol Ther. 2017;174:112–126. doi: 10.1016/j.pharmthera.2017.02.024. [DOI] [PubMed] [Google Scholar]
- 26.Mugami S, Dobkin-Bekman M, Rahamim-Ben Navi L, Naor Z. Differential roles of PKC isoforms (PKCs) in GnRH stimulation of MAPK phosphorylation in gonadotrope derived cells. Mol Cell Endocrinol. 2017 Apr 6; doi: 10.1016/j.mce.2017.04.004. Epub ahead of print. [DOI] [PubMed] [Google Scholar]
- 27.Segalés J, Perdiguero E, Muñoz-Cánoves P. Regulation of muscle stem cell functions: A focus on the p38 MAPK signaling pathway. Front Cell Dev Biol. 2016;4:91. doi: 10.3389/fcell.2016.00091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ai XL, Chi Q, Qiu Y, Li HY, Li DJ, Wang JX, Wang ZY. Gap junction protein connexin43 deregulation contributes to bladder carcinogenesis via targeting MAPK pathway. Mol Cell Biochem. 2017;428:109–118. doi: 10.1007/s11010-016-2921-9. [DOI] [PubMed] [Google Scholar]
- 29.Losa D, Köhler T, Bellec J, Dudez T, Crespin S, Bacchetta M, Boulanger P, Hong SS, Morel S, Nguyen TH, et al. Pseudomonas aeruginosa-induced apoptosis in airway epithelial cells is mediated by gap junctional communication in a JNK-dependent manner. J Immunol. 2014;192:4804–4812. doi: 10.4049/jimmunol.1301294. [DOI] [PubMed] [Google Scholar]
- 30.Shishido SN, Nguyen TA. Induction of apoptosis by PQ1 a gap junction enhancer that upregulates connexin 43 and activates the MAPK signaling pathway in mammary carcinoma cells. Int J Mol Sci. 2016;17:178. doi: 10.3390/ijms17020178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Geiger PC, Wright DC, Han DH, Holloszy JO. Activation of p38 MAP kinase enhances sensitivity of muscle glucose transport to insulin. Am J Physiol Endocrinol Metab. 2005;288:E782–E788. doi: 10.1152/ajpendo.00477.2004. [DOI] [PubMed] [Google Scholar]
- 32.Uchiyama H, Barut BA, Mohrbacher AF, Chauhan D, Anderson KC. Adhesion of human myeloma-derived cell lines to bone marrow stromal cells stimulates interleukin-6 secretion. Blood. 1993;82:3712–3720. [PubMed] [Google Scholar]
- 33.Hideshima T, Mitsiades C, Tonon G, Richardson PG, Anderson KC. Understanding multiple myeloma pathogenesis in the bone marrow to identify new therapeutic targets. Nat Rev Cancer. 2007;7:585–598. doi: 10.1038/nrc2189. [DOI] [PubMed] [Google Scholar]
- 34.Kumar S, Witzig TE, Timm M, Haug J, Wellik L, Fonseca R, Greipp PR, Rajkumar SV. Expression of VEGF and its receptors by myeloma cells. Leukemia. 2003;17:2025–2031. doi: 10.1038/sj.leu.2403084. [DOI] [PubMed] [Google Scholar]
- 35.Nefedova Y, Cheng P, Alsina M, Dalton WS, Gabrilovich DI. Involvement of Notch-1 signaling in bone marrow stroma-mediated de novo drug resistance of myeloma and other malignant lymphoid cell lines. Blood. 2004;103:3503–3510. doi: 10.1182/blood-2003-07-2340. [DOI] [PubMed] [Google Scholar]
- 36.Mori Y, Shimizu N, Dallas M, Niewolna M, Story B, Williams PJ, Mundy GR, Yoneda T. Anti-alpha4 integrin antibody suppresses the development of multiple myeloma and associated osteoclastic osteolysis. Blood. 2004;104:2149–2154. doi: 10.1182/blood-2004-01-0236. [DOI] [PubMed] [Google Scholar]
- 37.Fu J. Cx43 expressed on bone marrow stromal cells plays an essential role in multiple myeloma cell survival and drug resistance. Arch Med Sci. 2017;13:236–245. doi: 10.5114/aoms.2017.64722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Koul HK, Pal M, Koul S. Role of p38 MAP kinase signal transduction in solid tumors. Genes Cancer. 2013;4:342–359. doi: 10.1177/1947601913507951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Chen D, Liu Y, Yang H, Chen D, Zhang X, Fermandes JC, Chen Y. Connexin 43 promotes ossification of the posterior longitudinal ligament through activation of the ERK1/2 and p38 MAPK pathways. Cell Tissue Res. 2016;363:765–773. doi: 10.1007/s00441-015-2277-6. [DOI] [PubMed] [Google Scholar]