Abstract
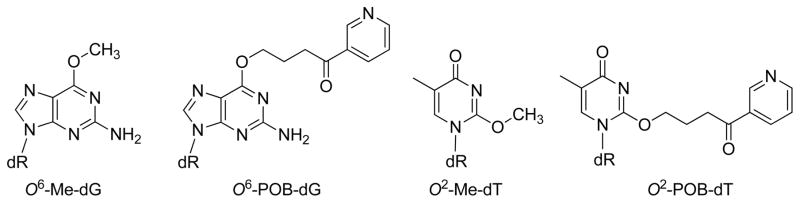
4-(Methylnitrosamino)-1-(3-pyridyl)-1-butanone (NNK) is a potent tobacco carcinogen that forms mutagenic DNA adducts including O6-methyl-2′-deoxyguanosine (O6-Me-dG), O6-[4-(3-pyridyl)-4-oxobut-1-yl]-dG (O6-POB-dG), O2-methylthymidine (O2-Me-dT), and O2-POB-dT. We evaluated the ability of human DNA polymerase ν to bypass this damage to evaluate the structural constraints on substrates for pol ν and to evaluate if there is kinetic evidence suggesting the in vivo activity of pol ν on tobacco-induced DNA damage. Presteady-state kinetic analysis has indicated that O6-Me-dG is a good substrate for pol ν, while O6-POB-dG and the O2-alkyl-dT adducts are poor substrates for pol ν. The reactivity with O6-Me-dG is high with a preference for dCTP > dGTP > dATP > dTTP. The catalytic activity of pol ν toward O6-Me-dG is high and can potentially be involved in its bypass in vivo. In contrast, pol ν is unlikely to bypass O6-POB-dG or the O2-alkyl-dTs in vivo.
Graphical Abstract
INTRODUCTION
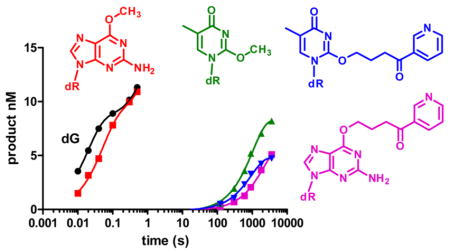
Tobacco and its smoke contains over 4000 compounds and 70 carcinogens.1–4 4-(Methylnitrosamino)-1-(3-pyridyl)-1-buta-none (NNK), one of the more potent carcinogens in tobacco, produces both methyl (Me) and 4-(3-pyridyl)-4-oxobutyl (POB) DNA adducts. Both of these classes of DNA adducts play a role in carcinogenesis.5,6 In particular, O6-Me-dG, O6-POB-dG, and O2-POB-dT (Chart 1) have been implicated in the mutagenicity of NNK.7–9
Chart 1.
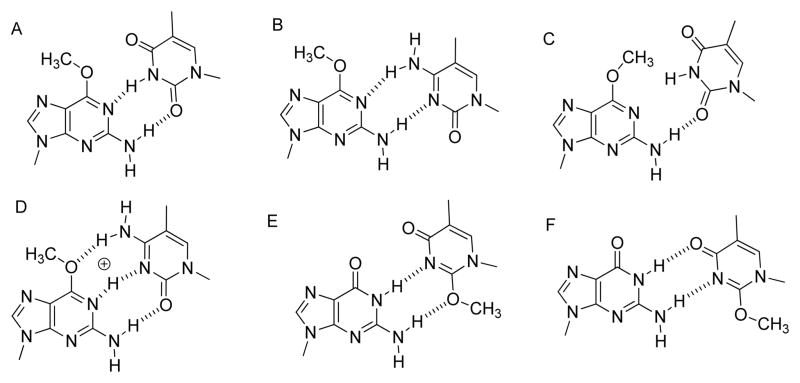
The potent mutagenicity of O6-Me-dG is long known. In vitro studies have shown that many polymerases insert dTTP opposite O6-Me-dG.10,11 When compared with dG in the template, the kcat/Km values for the incorporation of dCTP opposite O6-Me-dG decrease 2–3 orders of magnitude, while that for dTTP increase 1 to 2 orders of magnitude for a variety of polymerases including the Klenow fragment of E. coli DNA polymerase I (Kf) Thermus aquaticus pol I, HIV reverse transcriptase, T7 pol, pol δ/PCNA, pol η, pol κ, and pol ι.10,12–16 Loveless proposed that dTTP is incorporated opposite O6-Me-dG because it is able to form a Watson–Crick-like structure as illustrated in Figure 1A.17 In contrast, dC forms a wobble base pair with O6-Me-dG (Figure 1B). The exact nature of the base pair in the polymerase active site has been elusive. While a function/kinetic study supported a Watson–Crick-like base pair between dT and O6-Me-dG,16 a crystallization study with Bacillus stearothermophilus DNA polymerase I large fragment (BF) found the dT/O6-Me-dG base pair to be that in Figure 1C, while the dC/O6-Me-dG base pair was Watson–Crick like as shown in Figure 1D.18 The question of which polymerase(s) bypass this adduct in vivo still remains. While pol η is the most active eukaryotic Y-family polymerase with O6-Me-dG, the replicative polymerase pol δ is also quite active.19
Figure 1.
Potential base pair structures.
O6-POB-dG is mutagenic in HEK293 cells, causing primarily G to A mutations, consistent with dT pairing with O6-POB-dG.8 In vitro studies have shown that O6-POB-dG is a poorer substrate than O6-Me-dG for pol η, κ, and ι.13 Similar to O6-Me-dG, pol η has the highest reactivity for O6-POB-dG.13
O2-POB-dT may also play a role in NNK mutagenesis, as it is the most abundant POB-adduct in rodents. Bypass of O2-alkyl-dTs requires TLS polymerases in E. coli20,21 as well as human cells.9 In human cells, the bypass of O2-Me-dT and O2-POB-dT requires pol η, ζ, and Rev1.9 In vitro kinetics studies show that pol η is the most active Y-family polymerase in the bypass of O2-alkyl-dTs22–25 However, the activity is low, and there remains a possibility that another polymerase is involved in the bypass of O2-alkyl-dTs.
DNA polymerase ν is an A-family polymerase, with a mutation rate similar to Y-family polymerases.26 The polymer-ase is especially unfaithful with a high propensity for the formation of dG/dT mismatches.26,27 The polymerase has been implicated in the bypass of interstrand cross-links and can also bypass damage in the major groove of the DNA.28,29 Since the dT/dG mispair forms structures similar to the dC/O6-Me-dG and dT/O2-Me-dT base pairs,18,30,31 we hypothesized that pol ν can bypass O6-Me-dG. The ability of pol ν to bypass bulky major groove adducts led us to hypothesize that pol ν can bypass the bulky O6-POB-dG. Because O2-Me-dT can potentially form a wobble or Watson–Crick structures with dA or dG, we also evaluated the reactivity of pol ν with O2-Me-dT and the tobacco-relevant O2-POB-dT.
EXPERIMENTAL SECTION
General
[γ-32P]ATP was purchased from PerkinElmer and T4 polynucleotide kinase from USB/Aflymetrix. The dNTPs (ultrapure grade) were purchased from GE Healthcare, and the concentrations were determined by UV absorbance.32 Oligodeoxynucleotides were synthesized at the Macromolecular Core facility at the PSU College of Medicine. The phosphoramidite for O6-Me-dG was purchased from Glen Research (Sterling, VA). The oligodeoxynucleotides containing O6-POB-dG, O2-Me-dT, and O2-POB-dT were synthesized as described.24,25,33,34 The sequences of the oligodeoxynucleotides are shown in Chart 2 in which X was dG, O6-Me-dG, O6-POB-dG, and Y was dT, O2-Me-dT, and O2-POB-dT. The primer was 32P-labeled with [γ-32P]ATP and annealed with a 20% excess of the template as previously described.35
Chart 2.
Oligodeoxynucleotide sequences
Pol ν was purified by a modification of the method of Takata et al.26 as we previously described.35 The protein has a deletion of a C-terminal poly proline segment, an N-terminal His-tag, and C-terminal FLAG-tag. After lysing the cells, the resulting supernatant was loaded onto a Ni-NTA column (3 mL) and the enzyme eluted with an imidazole gradient. Purity was evaluated by SDS–PAGE and the protein concentration determined by the Qubit protein assay kit (Life Technologies). The active concentration was determined by performing burst kinetics with an excess of DNA.35
Kinetic Analyses
All reactions were performed in 40 mM Tris-HCl (pH 8.0) and 5 mM MgCl2 at 37 °C. The concentrations reported for each species are those that occurred during the reaction. The reactions were initiated by the addition of dNTP containing MgCl2 to preincubated protein and DNA in buffer. Steady-state reactions were performed with 0.1 to 1 nM polymerase and 20–30 nM DNA. The enzyme in excess reactions were conducted with 150 nM polymerase and 15 nM DNA. Slow reactions were quenched with equal volumes of STOP solution containing 10% 0.5 M Na2EDTA, 90% formamide, and 0.025% (w/v) xylene cyanol and bromophenol blue. Fast reactions were carried out with an RQF-3 quench flow apparatus (Kin Tek) and were quenched with 0.3 M Na2EDTA, which was diluted with STOP solution to load on the gel. The progress of the reaction was analyzed by denaturing PAGE, and the radioactivity on the gel was visualized with a Typhoon 9200. The reactions were quantitated by dividing the total radioactivity in the product band(s) by the radioactivity in the product and reactant bands.
Data Analysis
The kinetic constants were fit using GraphPad Prism V5. Computer simulations were performed with DynaFit Version 4 (BioKin Ltd.).36 The steady-state initial rate reactions were fit to the Michelis–Menten equation in eq 1. The time course experiments were fit to the burst equation (eq 2), in which P is the concentration of the product at various times (t), A the amplitude, and k and kss are the rapid and steady-state rate constants. If the burst rate constant demonstrated a dNTP dependence, it was fit to eq 3 in which k was the burst rate constant and KddNTP the apparent dNTP dissociation constant. Similarly, if the burst amplitude exhibited a dNTP concentration dependence, it was fit to the hyperbolic equation to obtain Amax and KA values.
| (1) |
| (2) |
| (3) |
RESULTS
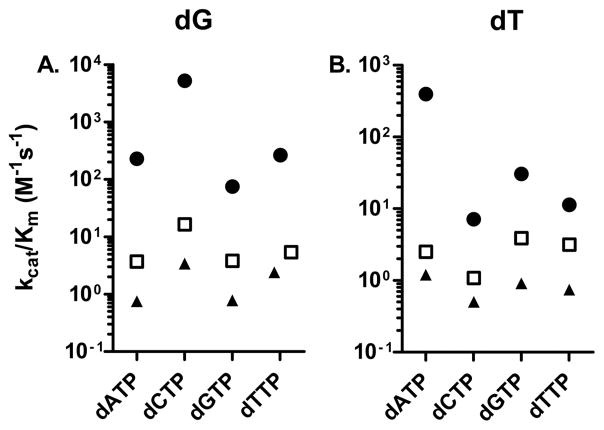
To gain more information about the activity of pol ν, we examined the incorporation of dNTPs opposite four DNA adducts, O6-Me-dG, O6-POB-dG, O2-Me-dT, and O2-POB-dT. The steady state rate constants were determined, and the results are presented in Table 1. The kcat/Km values are summarized in Figure 2. dCTP was inserted opposite dG with a kcat/Km value of 5220 M−1 s−1. The fidelity with undamaged DNA is poor, as expected.27,37 dTTP, is incorporated with a kcat/Km of 264 M−1 s−1, which corresponds to an finc = 0.05. Discrimination against dATP and dGTP were only slightly better, with finc values of 0.011 and 0.014, respectively.
Table 1.
Steady State Kinetic Parameters for the Pol ν Catalyzed Insertion Opposite O6-Alkyl-dG and O2-Alkyl-dTa
| dNTP | template | kcat (s−1 × 10 3) | Km (μM) | kcat/Km (M−1 s−1) | finc |
|---|---|---|---|---|---|
| dATP | dG | 12.5 ± 0.5 | 230 ± 22 | 55 ± 3 | 0.010 |
| dCTP | dG | 30.9 ± 0.7 | 5.9 ± 0.5 | 5220 ± 310 | 1 |
| dGTP | dG | 5.4 ± 0.1 | 76 ± 6 | 75 ± 5 | 0.014 |
| dTTP | dG | 13.7 ± 0.6 | 52 ± 7 | 264 ± 25 | 0.051 |
| dATP | O6-Me-dG | 3.05 ± 0.23 | 820 ± 110 | 3.7 ± 0.2 | 0.23 |
| dCTP | O6-Me-dG | 6.61 ± 0.52 | 400 ± 56 | 16.5 ± 1.1 | 1 |
| dGTP | O6-Me-dG | 3.68 ± 0.38 | 970 ± 160 | 3.8 ± 0.3 | 0.23 |
| dTTP | O6-Me-dG | 4.62 ± 0.55 | 860 ± 180 | 5.4 ± 0.5 | 0.33 |
| dATP | O6-POB-dG | 0.48 ± 0.05 | 636 ± 111 | 0.75 ± 0.07 | 0.22 |
| dCTP | O6-POB-dG | 1.18 ± 0.08 | 350 ± 57 | 3.4 ± 0.3 | 1 |
| dGTP | O6-POB-dG | 0.67 ± 0.07 | 860 ± 150 | 0.78 ± 0.07 | 0.23 |
| dTTP | O6-POB-dG | 0.78 ± 0.04 | 331 ± 46 | 2.4 ± 0.2 | 0.70 |
| dATP | dT | 24.1 ± 1.5 | 61 ± 7 | 396 ± 24 | 1 |
| dCTP | dT | 1.07 ± 0.02 | 150 ± 10 | 7.15 ± 0.35 | 0.018 |
| dGTP | dT | 2.26 ± 0.04 | 74 ± 4 | 30.5 ± 1.5 | 0.077 |
| dTTP | dT | 1.18 ± 0.03 | 104 ± 0 10 | 11.3 ± 0.9 | 0.029 |
| dATP | O2-Me-dT | 1.10 ± 0.05 | 438 ± 43 | 2.51 ± 0.14 | 1 |
| dCTP | O2-Me-dT | 0.71 ± 0.05 | 657 ± 95 | 1.08 ± 0.08 | 0.43 |
| dGTP | O2-Me-dT | 1.48 ± 0.05 | 381 ± 28 | 3.89 ± 0.17 | 1.55 |
| dTTP | O2-Me-dT | 1.23 ± 0.04 | 386 ± 28 | 3.19 ± 0.14 | 1.27 |
| dATP | O2-POB-dT | 0.43 ± 0.02 | 358 ± 34 | 1.20 ± 0.08 | 1 |
| dCTP | O2-POB-dT | 0.55 ± 0.04 | 1100 ± 150 | 0.50 ± 0.03 | 0.42 |
| dGTP | O2-POB-dT | 0.553 ± 0.027 | 605 ± 69 | 0.91 ± 0.06 | 0.76 |
| dTTP | O2-POB-dT | 0.49 ± 0.02 | 660 ± 70 | 0.74 ± 0.05 | 0.6261 |
The kinetic parameters were obtained with 1 nM pol ν and 20 nM DNA. The error is the standard error with three experiments.
Figure 2.
Relative kcat/Km values for the single nucleotide incorporation opposite (A) dG(closed circle), O6-Me-dG (open square), and O6-POB-dG (closed triangle), and (B) dT (closed circle), O2-Me-dT (open square), and O2-POB-dT (closed triangle).
With O6-Me-dG as the template base, the kcat/Km for dCTP decreased 316-fold, while the kcat/Km for dTTP incorporation decreased 49-fold. This effect differs from that observed with Kf(exo-) and Taq polymerase, two high fidelity A-family polymerases. Several studies have shown that the kcat/Km for dCTP incorporation decreases 10,000-fold, while that for dTTP incorporation increases ~100-fold, thereby giving a preference for dTTP incorporation opposite O6-Me-dG.10,16 In contrast to other polymerases, methylation of the O6-position did not increase the kcat/Km of dTTP, and pol ν retains its preference for dCTP incorporation. In addition, the kcat/Km values for the incorporation of dG and dA were very similar to that of dT. Thus, pol ν does not have any preference to insert dTTP opposite O6-Me-dG. We observed a similar pattern with the larger O6-POB-dG, with kcat/Km values that are 2–5-fold lower than those for O6-Me-dG.
With dT as the template base, while the kcat/Km for correct incorporation was less than that for dG as template, but the fidelity was similar. We found the kcat/Km for the incorporation of dGTP opposite dC to be 13-fold greater than the incorporation of dATP opposite dT. Methylation of the O2-position decreases kcat/Km for dATP incorporation 160-fold, a value that is similar to the effect caused by the dG to O6-Me-dG. The O2-methylation, however, essentially abolishes the specificity of the incorporation. The kcat/Km values for dNTP incorporation are all within a factor of 3. Pyridyloxobutylation of the O2-position further reduces the kcat/Km 2–4-fold and similarly abolishes specificity.
The in vitro activities of DNA polymerases are limited by the rate limiting dissociation of the DNA from the polymerase. With respect to pol ν, the reactivity is also limited by a rate limiting conformational change after phosphodiester bond formation. Thus, Michaelis–Menten kinetics may not reflect the phosphoryl-transfer activity of the polymerase. Therefore, we examined the reaction with DNA polymerase in excess over the DNA to examine the rate of phosphodiester bond formation.
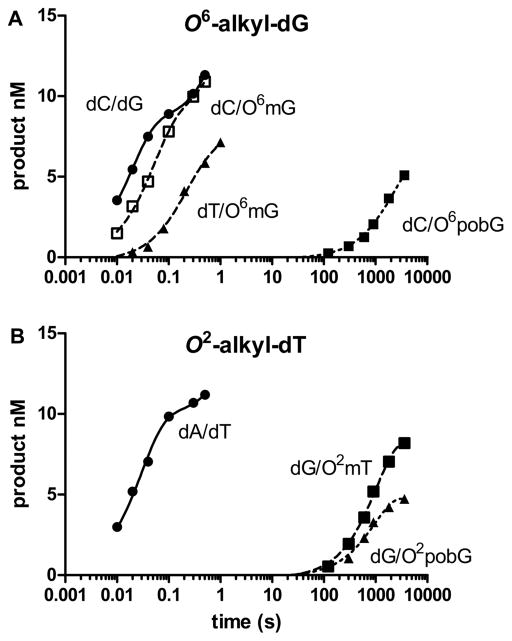
Figure 3 shows the plots with 150 nM pol ν, 15 nM DNA, and 50 μM dNTP. At this concentration of DNA and polymerase, 95% of the DNA should be bound to polymerase, assuming that the modification does not alter the Kd of the pol to undamaged DNA.35 dCTP is incorporated opposite dG with a half-life of 40 ms, while the incorporation opposite O6-Me-dG is only slightly slower with a half-life of 60 ms. The incorporation of dTTP opposite O6-Me-dG is considerably slower with a half-life of 1 s. The incorporation of dCTP opposite O6-POB-dG is extremely slow with a half-life of >1000 s. With dT modification, both O2-Me-dT and O2-POB-dT reacted very slowly. dGTP is the most reactive triphosphate, with t1/2 > 1000 s−1. These results are very different from the steady-state kinetics.
Figure 3.
Incorporation of single dNTPs opposite templates containing (A) dG and (dT) and analogues. The concentration of the DNA was 15 nM, the pol 150 nM, and the dNTP was 50 μM.
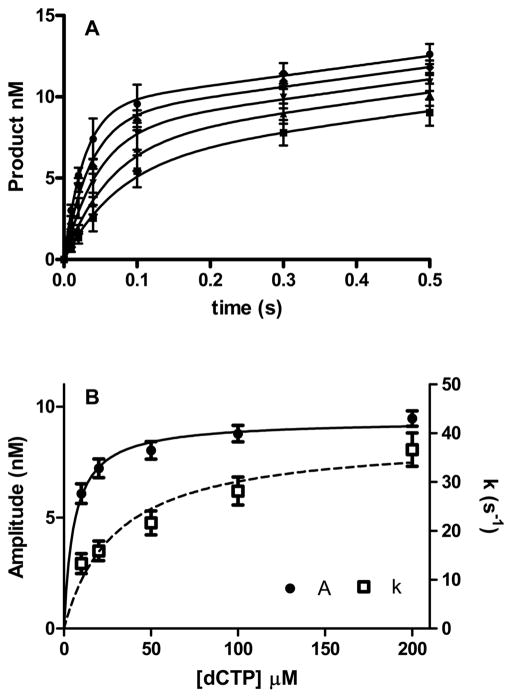
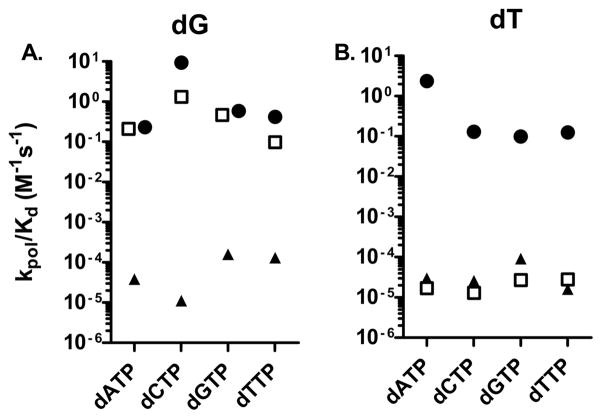
We more fully examined these reactions with the six DNA substrates with pol ν in excess. The method is illustrated in Figure 4 in which 15 nM DNA containing O6-Me-dG and 150 nM pol ν were reacted with various concentrations of dCTP. The time courses (panel A) were fitted to the burst eq (eq 2). Both the burst rates and amplitudes were dependent on the dCTP concentration. Therefore, we fit those parameters to the hyperbolic eq 1 as illustrated in panel B. The results are presented in Table 2, and the kpol/KddNTP values are summarized in Figure 5.
Figure 4.
Analysis of the time course data. (A) Incorporation of pol ν (150 nM) catalyzed dCTP (10, square; 20, triangle; 50, upside down triangle; 100, diamond; and 200, circle μM) incorporation opposite DNA containing O6-Me-dG (15 nM). The lines are the best fit to equation 1. The error bars represent the standard deviation of three determinations. (B) Plot of amplitude (A) and burst rate constant (k) versus dCTP concentration. The lines are the best fit to eq 2, and the error bars are the standard errors.
Table 2.
Kinetic Parameters for the dNTP Dependence of the Pol ν Catalyzed Insertion Opposite O2-Alkyl-dT
| amplitude
|
burst rate constant
|
kss/[pol] s−1 | ||||
|---|---|---|---|---|---|---|
| Amax/[pol] | KA(μM) | kpol(s−1) | KddNTP(μM) | |||
| dATP | dG | 0.49 ± 0.01 | 101 ± 6 | 32 ± 4 | 136 ± 58 | 0.020 ± 0.002 |
| dCTP | dG | 0.65 ± 0.02 | 5.3 ± 1.1 | 59 ± 2 | 6.2 ± 1.1 | 0.038 ± 0.003 |
| dGTP | dG | 0.55 ± 0.01 | 58 ± 7 | 28 ± 2 | 47 ± 15 | 0.020 ± 0.003 |
| dTTP | dG | 0.57 ± 0.02 | 92 ± 12 | 25 ± 3 | 60 ± 28 | 0.017 ± 0.002 |
| dATP | O6-Me-dG | 0.54 ± 0.02 | 106 ± 12 | 20 ± 3 | 93 ± 54 | 0.011 ± 0.003 |
| dCTP | O6-Me-dG | 0.62 ± 0.02 | 5.8 ± 0.9 | 39 ± 4 | 30 ± 11 | 0.041 ± 0.01 |
| dGTP | O6-Me-dG | 0.58 ± 0.02 | 63 ± 9 | 19 ± 1 | 40 ± 16 | 0.021 ± 0.002 |
| dTTP | O6-Me-dG | 0.62 ± 0.0 | 38 ± 5 | 14 ± 2 | 140 ± 50 | 0.011 ± 0.003 |
| dATP | O6-POB-dG | 0.34 ± 0.02 | 88 ± 22 | 0.0014 ± 0.0001 | 38 ± 7 | nd |
| dCTP | O6-POB-dG | 0.73 ± 0.04 | 47 ± 11 | 0.0015 ± 0.0001 | 146 ± 42 | nd |
| dGTP | O6-POB-dG | 0.37 ± 0.01 | 240 ± 31 | 0.0013 ± 0.0002 | nd | |
| dTTP | O6-POB-dG | 0.63 ± 0.03 | 120 ± 18 | 0.00085 ± 0.00005 | nd | |
| dATP | dT | 0.74 ± 0.04 | 5.1 ± 1.8 | 62 ± 5 | 26 ± 7 | 0.027 ± 0.004 |
| dCTP | dT | 0.51 ± 0.02 | 42 ± 9 | 13 ± 1 | 100 ± 21 | 0.008 ± 0.002 |
| dGTP | dT | 0.69 ± 0.01 | 17 ± 2 | 10 ± 1 | 101 ± 41 | 0.007 ± 0.003 |
| dTTP | dT | 0.54 ± 0.02 | 27 ± 6 | 15 ± 1 | 118 ± 27 | 0.012 ± 0.002 |
| dATP | O2-Me-dT | 0.55 ± 0.02 | 39 ± 7 < 50 | 0.00150 ± 0.00014 | 95 ± 32 | nd |
| dCTP | O2-Me-dT | 0.35 ± 0.01 | 16 ± 5 < 50 | 0.00113 ± 0.00002 | 84 ± 7 | nd |
| dGTP | O2-Me-dT | 0.83 ± 0.02 | 24 ± 3 < 50 | 0.00212 ± 0.00016 | 78 ± 22 | nd |
| dTTP | O2-Me-dT | 0.72 ± 0.02 | 22 ± 4 < 50 | 0.00238 ± 0.00009 | 88 ± 12 | nd |
| dATP | O2-POB-dT | 0.37 ± 0.05 | 78 ± 9 | 0.00150 ± 0.00014 | 50 ± 22 | nd |
| dCTP | O2-POB-dT | 0.16 ± 0.01 | 42 ± 11 | 0.00117 ± 0.00002 | 45 ± 4 | nd |
| dGTP | O2-POB-dT | 0.44 ± 0.02 | 19 ± 4 | 0.00122 ± 0.00006 | 13 ± 6 | nd |
| dTTP | O2-POB-dT | 0.21 ± 0.01 | 46 ± 9 | 0.00115 ± 0.00002 | 72 ± 4 | nd |
Figure 5.
Relative kpol/Kd values for the single nucleotide incorporation opposite (A) dG(closed circle), O6-Me-dG (open square), O6-POB-dG (closed triangle), and (B) dT (closed circle), O2-Me-dT (open square), and O2-POB-dT (closed triangle).
The kinetic parameters for the dG template are similar to what we previously observed with a slightly different sequence. In this work, with dT and dG as templates we obtained kpol parameters of 62 and 59 s−1 and apparent KddNTP values of 6.2 and 24 μM, respectively. These values are similar to a kpol of 100 s−1 and a KddNTP of 20 μM found previously.35 Misincorporation occurs with kpol values of 10 to 32 s−1 and apparent KddNTP values of 47 to 136 μM. As we previously observed, the amplitude of the rapid reaction was also dependent on dNTP concentrations. The Amax/[pol] values were similar for all dNTP/template pairings, and ranged from 0.5 to 0.74. The largest differences occurred in the KA values, which were 5 μM for correct incorporation and 17 to 101 μM for misincorporation.
For O6-Me-dG, all dNTPs are reactive with the relative preference of dC > dT and dG > dA. The preference for dCTP is not based upon a single dominant parameter but is due to a combination of effects. dCTP has the largest kpol, 39 s−1 versus 14 s−1 for dTTP. dCTP has the lowest apparent KddNTP of 30 μM and dTTP the largest with 140 μM. While all dNTPs have Amax values from 0.54 to 0.65, and dCTP has the lowest KA at 5.8 μM versus 140 μM for dATP.
In spite of similar kcat/Km values, the other modified nucleotides reacted very slowly in the polymerase in excess experiments. The reduced activities are due to small kpol values. O6-POB-dG is bypassed slowly, with kpol values 10,000-fold lower than those for O6-Me-dG. Similarly, the O2-alkyl-dTs react very slowly with kpol values in the range of 0.001 s−1. The kss values that were calculated were in the range from 0.005 to 0.05 s−1, similar to the kcat values.
Decreased burst amplitudes associated with replication of DNA damage have generally been associated with the formation of inactive complexes.35 For example, the A-family T7 DNA polymerase forms are inactive during the insertion of dCTP and dTTP opposite O6-Me-dG.38 The decreased burst amplitudes associated with the pol ν bypass of the four adducts studied in this article can potentially be due to this phenomena. We modeled the time courses of the insertions of dCTP opposite O6-Me-dG and O6-POB-dG, and dATP opposite O2-Me-dT and O2-POB-dT with the seven-step polymerase mechanism. The results shown in Figures S13–S16 demonstrate that the seven-step polymerase mechanism can explain the reduced burst amplitudes. Thus, there is no reason to invoke a more complicated mechanism to explain the results. However, these data do not exclude the possibility of inactive complexes.
DISCUSSION
Pol ν is a low fidelity polymerase because it is very efficient at catalyzing the incorporation of the wrong dNTP.35 In addition, pol ν produces G/T mispairs at a high frequency, in a sequence-specific manner.27,39 The current paradigm is that high fidelity polymerases use geometric selection to favor the formation of the Watson–Crick base pairs. During the formation of a mispair, BF was found to exist in an “ajar” structure in which the polymerase was in a state intermediate between the open and closed conformations.40 This conformation might allow BF to react with the non-Watson–Crick structures in the active site. Pol ν adopts an ajar structure with a Watson–Crick base pair in binding pocket.41 Therefore, pol ν may naturally exist in a conformation that allows for the rapid formation of mispairs and small damage such as O6-Me-dG and O2-Me-dT. It was this property of pol ν, in addition to the similarities between base-pair structures of the G/T mispair and potential O6-Me-dG and O2-Me-dT base pairs that led us to test whether pol ν was able to rapidly bypass O6-alkyl-dG and O2-alkyl-dT.
Upon the basis of steady-state analysis, the kcat/Km for the incorporation of dCTP decreased 300-fold for O6-Me-dG versus dG, values very similar to those found for other enzymes.10,12–16 However, pol ν differs from other polymerase in the reactivity of dTTP. Typically, methylation of the O6-position of dG increases the kcat/Km for dTTP 100-fold.10,12–16 However, with pol ν, the kcat/Km for the insertion of dTTP opposite O6-Me-dG did not increase but decreased 10-fold. The kcat/Km values for dTTP were similar to those of dATP and dGTP. While high fidelity A-family polymerases select for Watson–Crick geometry, pol ν has less stringent geometrical constraints.
The bypass of various DNA damage with various polymerases have been examined with presteady-state kinetics. The studies show that the reactions are biphasic, with a rapid phase followed by a slower phase. Kinetic modeling of the data indicates the presence of both active and inactive polymerase/DNA/dNTP complexes.24,42,43 In the case of T7 pol, an A-family polymerase, and O6-Me-dG, the rate of the rapid phase is very similar to the rate of correct base pair formation.14,38 The amplitude of the burst was low, 6–12% of the total enzyme concentration. The kinetic data are consistent with a mechanism in which the pol-DNA-dNTP ternary complex exists in both active and inactive conformations. The burst rate is due to the reaction of the active complex, while the slow phase is either reaction of the inactive complex or conversion of the inactive complex to the active complex.
Pol ν catalyzed incorporation opposite O6-Me-dG also exhibits biphasic kinetics. However, in contrast to the results observed with T7 pol, the burst amplitude remains the same for the template O6-Me-dG. The reaction is slightly slower, the kpol for dCTP drops 33%, while the KddNTP rises from 6 to 30 μM. Kinetic modeling was undertaken to determine if unreactive pol/DNA/dNTP complexes are necessary to account for the reduced burst amplitudes (Figure S13–S16). However, just as with the undamaged template,35 a rate limiting step after phosphodiester bond formation can account for the biphasic kinetics. An inactive complex that is off the reaction pathway is not necessary to account for the kinetics. Thus, there is no evidence of active and inactive complexes with pol ν. Whatever the base pair structures in the binding pocket, pol ν rapidly catalyzes phosphodiester bond formation.
Pol ν is able to bypass dA with bulky N6-adducts such as peptides and oligonucleotides that would be derived from protein and DNA cross-links.29 However, in an apparent contradiction, pol ν is inefficient at bypassing the smaller N6-(7,8,9-trihydroxy-7,8,9,10-tetrahydrobenzo[a]pyrene)-d (N6-BP-dA).29 We found that pol ν is very inefficient at bypassing O6-POB-dG. The kpol for the incorporation opposite O6-POB-dG is 5-orders of magnitude lower than that opposite O6-Me-dG. There are no obvious steric clashes in the crystal structure of pol ν 44 that would prevent O6-POB-dG or N6-BP-dA from being a substrate. The mechanism underlying the reduced reactivity of moderately sized DNA adducts in the major groove is unknown.
We evaluated the reactivity of pol ν with O2-Me-dT because O2-Me-dT can potentially form Watson–Crick-like and wobble structures with dG (Figure 1E and F). In this regard, it has similarities with O6-Me-dG. However, we found that O2-Me-dT and O2-POB-dT are very poor substrates for pol ν. The kpol values are ~50,000-fold lower than that for the incorporation of dATP opposite dT. Thus, while pol ν rapidly forms mispairs and can bypass small damage such as O6-Me-dG and thymine glycol, the identity of the damage does influence reactivity. A-family polymerases Kf and T7 pol are sensitive to damage on the minor groove side of the DNA.24,45 This property is conserved with pol ν. Perhaps, as with other A-family polymerases, close contacts with the minor groove of the DNA are involved in providing a high catalytic state.46,47
One aim of this work was to evaluate the possibility that pol ν has the in vitro catalytic activity to potentially bypass these adducts in vivo. The catalytic activity of pol ν toward O6-Me-dG is higher than that reported for any other polymerase.14,19,38 Therefore, pol ν has the potential to contribute to the bypass of O6-Me-dG in vivo. With regard to O6-POB-dG, O2-Me-dT, and O2-POB-dT, the catalytic activity of pol ν is much lower than that for pol η,13,25 and thus, it is highly unlikely to be involved it the bypass of these adducts.
Supplementary Material
Acknowledgments
Funding
This project was funded under NIH grant no. ES021762.
The oligodeoxynucleotide synthesis and MS analysis were performed in the Macromolecular Core facility at the PSU College of medicine. The NMR spectra were recorded in the solution NMR core facility. Core Facility services and instruments used in this project were funded, in part, under a grant with the Pennsylvania Department of Health using Tobacco Settlement Funds.
ABBREVIATIONS
- BF
Bacillus stearothermophilus DNA polymerase I large fragment
- Kf(exo-)
Klenow fragment of proofreading deficient E. coli DNA polymerase I
- NNK
4-(methylnitrosamino)-1-(3-pyridyl)-1-butanone
- O6-Me-dG
O2-methyl-2′-deoxyguano-sine
- O2-Me-dT
O2-methylthymidine
- O6-POB-dG
O6-[4-(3-pyridyl)-4-oxobut-1-yl]-2′-deoxyguanosine
- O2-POB-dT
O2-[4-(3-pyridyl)-4-oxobut-1-yl]thymidine
- POB
4-(3-pyridyl)-4-ox-obut-1-yl
- pol
polymerase
- pol
polymerase
Footnotes
Notes
The Pennsylvania Department of Health specifically disclaims responsibility for any analyses, interpretations or conclusions. The authors declare no competing financial interest.
The Supporting Information is available free of charge on the ACS Publications website at DOI: 10.1021/acs.chemres-tox.6b00318.
Steady-state and polymerase in excess analyses and results from computer simulations (PDF)
Script and data used to perform the computer simulations (ZIP)
References
- 1.Cancer Facts and Figures, 2014. American Cancer Society; Atlanta, GA: 2014. [Google Scholar]
- 2.Hecht SS. Tobacco smoke carcinogens and lung cancer. J Natl Cancer Inst. 1999;91:1194–1210. doi: 10.1093/jnci/91.14.1194. [DOI] [PubMed] [Google Scholar]
- 3.Hecht SS. Tobacco carcinogens, their biomarkers and tobacco-induced cancer. Nat Rev Cancer. 2003;3:733–744. doi: 10.1038/nrc1190. [DOI] [PubMed] [Google Scholar]
- 4.IARC. IARC Monographs on the Evaluation of Carcinogenic Risks to Humans. World Health Organization, International Agency for Research on Cancer; Lyon, France: 2004. Tobacco Smoke and Involuntary Smoking; pp. 53–119. [PMC free article] [PubMed] [Google Scholar]
- 5.Hecht SS, Jordan KG, Choi CI, Trushin N. Effects of deuterium substitution on the tumorigenicity of 4-(metlhylnitrosamino)-1-(3-pyridyl)-1-butanone and 4-(methylnitrosa-mino)-1-(3-pyridyl)-1-butanol in A/J mice. Carcinogenesis. 1990;11:1017–1020. doi: 10.1093/carcin/11.6.1017. [DOI] [PubMed] [Google Scholar]
- 6.Hecht SS, Lin D, Castonguay A, Rivenson A. Effects of alpha-deuterium substitution on the tumorigenicity of 4-(methylnitrosamino)-1-(3-pyridyl)-1-butanone in F344 rats. Carcino-genesis. 1987;8:291–294. doi: 10.1093/carcin/8.2.291. [DOI] [PubMed] [Google Scholar]
- 7.Peterson LA, Hecht SS. O6 -Methylguanine is a critical determinant of 4-(methylnitrosamino)-1-(3-pyridyl)-1-buta-none tumorigenesis in A/J mouse lung. Cancer Res. 1991;51:5557–5564. [PubMed] [Google Scholar]
- 8.Pauly GT, Peterson LA, Moschel RC. Mutagenesis by O6-[4-oxo-4-(3-pyridyl)butyl]guanine in Escherichia coli and human cells. Chem Res Toxicol. 2002;15:165–169. doi: 10.1021/tx0101245. [DOI] [PubMed] [Google Scholar]
- 9.Weerasooriya S, Jasti VP, Bose A, Spratt TE, Basu AK. Roles of translesion synthesis DNA polymerases in the potent mutagenicity of tobacco-specific nitrosamine-derived O-alkylthymidines in human cells. DNA Repair. 2015;35:63–70. doi: 10.1016/j.dnarep.2015.09.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dosanjh MK, Galeros G, Goodman MF, Singer B. Kinetics of extension of O 6 -methylguanine paired with cytosine or thymine in defined oligonucleotide sequences. Biochemistry. 1991;30:11595–11599. doi: 10.1021/bi00113a015. [DOI] [PubMed] [Google Scholar]
- 11.Singh J, Su L, Snow ET. Replication across O6-Methylguanine by Human DNA Polymerase beta in Vitro. J Biol Chem. 1996;271:28391–28398. doi: 10.1074/jbc.271.45.28391. [DOI] [PubMed] [Google Scholar]
- 12.Snow ET, Foote RS, Mitra S. Base-pairing properties of O 6 -methylguanine in template DNA during in vitro DNA replication. J Biol Chem. 1984;259:8095–8100. [PubMed] [Google Scholar]
- 13.Choi JY, Chowdhury G, Zang H, Angel KC, Vu CC, Peterson LA, Guengerich FP. Translesion synthesis across O6-alkylguanine DNA adducts by recombinant human DNA polymerases. J Biol Chem. 2006;281:38244–38256. doi: 10.1074/jbc.M608369200. [DOI] [PubMed] [Google Scholar]
- 14.Woodside AM, Guengerich FP. Effect of the O6-substituent on misincorporation kinetics catalyzed by DNA polymerases at O6-methylguanine and O6-benzylguanine. Biochemistry. 2002;41:1027–1038. doi: 10.1021/bi011495n. [DOI] [PubMed] [Google Scholar]
- 15.Haracska L, Prakash S, Prakash L. Replication past O(6)-methylguanine by yeast and human DNA polymerase eta. Mol Cell Biol. 2000;20:8001–8007. doi: 10.1128/mcb.20.21.8001-8007.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Spratt TE, Levy DE. Structure of the hydrogen bonding complex of O6-methylguanine with cytosine and thymine during DNA replication. Nucleic Acids Res. 1997;25:3354–3361. doi: 10.1093/nar/25.16.3354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Loveless A. Possible relevance or O-6 alkylation of deoxyguanosine to the mutagenicity and carcinogenecity of nitros-amines and nitrosamides. Nature. 1969;223:206–207. doi: 10.1038/223206a0. [DOI] [PubMed] [Google Scholar]
- 18.Warren JJ, Forsberg LJ, Beese LS. The structural basis for the mutagenicity of O6-methyl-guanine lesions. Proc Natl Acad Sci U S A. 2006;103:19701–19706. doi: 10.1073/pnas.0609580103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Washington MT, Johnson RE, Prakash L, Prakash S. Accuracy of lesion bypass by yeast and human DNA polymerase eta. Proc Natl Acad Sci U S A. 2001;98:8355–8226. doi: 10.1073/pnas.121007298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zhai Q, Wang P, Cai Q, Wang Y. Syntheses and characterizations of the in vivo replicative bypass and mutagenic properties of the minor-groove O2-alkylthymidine lesions. Nucleic Acids Res. 2014;42:10529–10537. doi: 10.1093/nar/gku748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jasti VP, Spratt TE, Basu AK. Tobacco-specific nitrosamine-derived O2-alkylthymidines are potent mutagenic lesions in SOS-induced Escherichia coli. Chem Res Toxicol. 2011;24:1833–1835. doi: 10.1021/tx200435d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Andersen N, Wang J, Wang P, Jiang Y, Wang Y. In-vitro replication studies on O2-methylthymidine and O4-methyl-thymidine. Chem Res Toxicol. 2012;25:2523–2531. doi: 10.1021/tx300325q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Andersen N, Wang P, Wang Y. Replication across regioisomeric ethylated thymidine lesions by purified DNA polymerases. Chem Res Toxicol. 2013;26:1730–1738. doi: 10.1021/tx4002995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gowda ASP, Krishnegowda G, Suo Z, Amin S, Spratt TE. Low Fidelity Bypass of O2-(3-Pyridyl)-4-oxobutylth-ymine, the Most Persistent Bulky Adduct Produced by the Tobacco Specific Nitrosamine 4-(Methylnitrosamino)-1-(3-pyridyl)-1-butanone by Model DNA Polymerases. Chem Res Toxicol. 2012;25:1195–1202. doi: 10.1021/tx200483g. [DOI] [PubMed] [Google Scholar]
- 25.Gowda ASP, Spratt TE. DNA Polymerases η and ζ Combine to Bypass O2-[4-(3-Pyridyl)-4-oxobutyl]thymine, a DNA Adduct Formed from Tobacco Carcinogens. Chem Res Toxicol. 2016;29:303–316. doi: 10.1021/acs.chemrestox.5b00468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Takata K-i, Shimizu T, Iwai S, Wood RD. Human DNA polymerase N (POLN) Is a low fidelity enzyme capable of error-free bypass of 5S-thymine glycol. J Biol Chem. 2006;281:23445–23455. doi: 10.1074/jbc.M604317200. [DOI] [PubMed] [Google Scholar]
- 27.Arana ME, Takata K-i, Garcia-Diaz M, Wood RD, Kunkel TA. A unique error signature for human DNA polymerase ν. DNA Repair. 2007;6:213–223. doi: 10.1016/j.dnarep.2006.09.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zietlow L, Smith LA, Bessho M, Bessho T. Evidence for the involvement of human DNA polymerase N in the repair of DNA interstrand cross-links. Biochemistry. 2009;48:11817–11824. doi: 10.1021/bi9015346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yamanaka K, Minko IG, Takata K-i, Kolbanovskiy A, Kozekov ID, Wood RD, Rizzo CJ, Lloyd RS. Novel enzymatic function of DNA polymerase ν in translesion DNA synthesis past major groove DNA–peptide and DNA–DNA cross-links. Chem Res Toxicol. 2010;23:689–695. doi: 10.1021/tx900449u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Johnson SJ, Beese LS. Structures of mismatch replication errors observed in a DNA polymerase. Cell. 2004;116:803–816. doi: 10.1016/s0092-8674(04)00252-1. [DOI] [PubMed] [Google Scholar]
- 31.Wang W, Hellinga HW, Beese LS. Structural evidence for the rare tautomer hypothesis of spontaneous mutagenesis. Proc Natl Acad Sci U S A. 2011;108:17644–17648. doi: 10.1073/pnas.1114496108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Dunn DB, Hall RH. In: Handbook of Biochemistry and Molecular Biology. Fasman GD, editor. CRC Press; Boca Raton, FL: 1986. pp. 65–215. [Google Scholar]
- 33.Krishnegowda G, Sharma AK, Krzeminski J, Gowda ASP, Lin JM, Desai D, Spratt TE, Amin S. Facile syntheses of O2-[4-(3-pyridyl-4-oxobut-1-yl]thymidine, the major adduct formed by tobacco specific nitrosamine 4-methylnitrosamino-1-(3-pyridyl)-1-butanone (NNK) in vivo, and Its site-specifically adducted oligodeoxynucleotides. Chem Res Toxicol. 2011;24:960–967. doi: 10.1021/tx200127j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Coulter R, Blandino M, Tomlinson JM, Pauly GT, Krajewska M, Moschel RC, Peterson LA, Pegg AE, Spratt TE. Differences in the rate of repair of O6-alkylguanines in different sequence contexts by O6-alkylguanine-DNA alkyltransferase. Chem Res Toxicol. 2007;20:1966–1971. doi: 10.1021/tx700271j. [DOI] [PubMed] [Google Scholar]
- 35.Gowda AS, Moldovan GL, Spratt TE. Human DNA Polymerase nu Catalyzes Correct and Incorrect DNA Synthesis with High Catalytic Efficiency. J Biol Chem. 2015;290:16292–16303. doi: 10.1074/jbc.M115.653287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kuzmic P. DynaFit–a software package for enzymology. Methods Enzymol. 2009;467:247–280. doi: 10.1016/S0076-6879(09)67010-5. [DOI] [PubMed] [Google Scholar]
- 37.Takata K-i, Arana ME, Seki M, Kunkel TA, Wood RD. Evolutionary conservation of residues in vertebrate DNA polymerase N conferring low fidelity and bypass activity. Nucleic Acids Res. 2010;38:3233–3244. doi: 10.1093/nar/gkq048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Woodside AM, Guengerich FP. Misincorpora-tion and stalling at O6-methylguanine and O6-benzylguanine: evidence for inactive polymerase complexes. Biochemistry. 2002;41:1039–1050. doi: 10.1021/bi011496f. [DOI] [PubMed] [Google Scholar]
- 39.Arana ME, Potapova O, Kunkel TA, Joyce CM. Kinetic analysis of the unique error signature of human DNA polymerase nu. Biochemistry. 2011;50:10126–10135. doi: 10.1021/bi201197p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wu EY, Beese LS. The Structure of a High Fidelity DNA Polymerase Bound to a Mismatched Nucleotide Reveals an “Ajar” Intermediate Conformation in the Nucleotide Selection Mechanism. J Biol Chem. 2011;286:19758–19767. doi: 10.1074/jbc.M110.191130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lee YS, Gregory MT, Yang W. Human Pol ζ purified with accessory subunits is active in translesion DNA synthesis and complements Pol η in cisplatin bypass. Proc Natl Acad Sci U S A. 2014;111:2954–2959. doi: 10.1073/pnas.1324001111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Vyas R, Efthimiopoulos G, Tokarsky EJ, Malik CK, Basu AK, Suo Z. Mechanistic Basis for the Bypass of a Bulky DNA Adduct Catalyzed by a Y-Family DNA Polymerase. J Am Chem Soc. 2015;137:12131–12142. doi: 10.1021/jacs.5b08027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sherrer SM, Sanman LE, Xia CX, Bolin ER, Malik CK, Efthimiopoulos G, Basu AK, Suo Z. Kinetic Analysis of the Bypass of a Bulky DNA Lesion Catalyzed by Human Y-Family DNA Polymerases. Chem Res Toxicol. 2012;25:730–740. doi: 10.1021/tx200531y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lee YS, Gao Y, Yang W. How a homolog of high-fidelity replicases conducts mutagenic DNA synthesis. Nat Struct Mol Biol. 2015;22:298–303. doi: 10.1038/nsmb.2985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zang H, Harris TM, Guengerich FP. Kinetics of Nucleotide Incorporation Opposite DNA Bulky Guanine N2 Adducts by Processive Bacteriophage T7 DNA Polymerase (Exonuclease-) and HIV-1 Reverse Transcriptase. J Biol Chem. 2005;280:1165–1178. doi: 10.1074/jbc.M405996200. [DOI] [PubMed] [Google Scholar]
- 46.Meyer AS, Blandino M, Spratt TE. E. coli DNA polymerase I (Klenow fragment) uses a hydrogen bonding fork from Arg668 to the primer terminus and incoming deoxynucleotide triphosphate to catalyze DNA replication. J Biol Chem. 2004;279:33043–33046. doi: 10.1074/jbc.C400232200. [DOI] [PubMed] [Google Scholar]
- 47.McCain MD, Meyer AS, Glekas A, Spratt TE. Fidelity of mispair formation and mispair extension is dependent on the interaction between the minor groove of the primer terminus and Arg668 of DNA Polymerase I of E. coli. Biochemistry. 2005;44:5647–5659. doi: 10.1021/bi047460f. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.