Abstract
Cytochrome P450 2C19 (CYP2C19) is involved in the metabolism of many drugs. Extensive studies have demonstrated that genetic variants and endogenous and environmental factors play important roles in the expression of CYP2C19. However, the role of microRNAs (miRNAs) in controlling CYP2C19 expression has not been investigated completely. In the present study, we performed in silico analysis to rank putative miRNA/CYP2C19 hybrids with regards to the predicted stabilities of their duplexes and then we applied a series of biochemical and molecular assays to elucidate the underlying functional mechanisms for the regulation of CYP2C19 by miRNAs. In silico analysis indicated that hsa-miR-23a-3p and hsa-miR-29a-3p target the coding region of CYP2C19 with hybrid stabilities of −27.5 kcal/mol and −23.3 kcal/mol, respectively. RNA electrophoresis mobility shift assays showed that both hsa-miR-23a-3p and hsa-miR-29a-3p miRNAs were able to bind directly to their cognate targets in the CYP2C19 transcript. Further, a significant inverse correlation was found between chemically-induced up-regulation of hsa-miR-29a-3p and CYP2C19 expression in HepaRG cells. In addition, inverse correlations were also observed in human liver tissue samples between the level of CYP2C19 mRNA expression and both hsa-miR-23a-3p and hsa-miR-29a-3p levels. All these results demonstrated the suppressing role of hsa-miR-29a-3p on CYP2C19 expression.
Keywords: hsa-miR-29a-3p, CYP2C19, Drug metabolizing enzymes, Pharmacogenomics, Inter-individual variability, microRNA
1. Introduction
Human cytochrome P450 2C19 (CYP2C19), an important drug metabolizing enzyme mainly expressed in the liver, plays pivotal roles in the activation or elimination of many therapeutic drugs, endogenous biomolecules, and environmental toxicants, including proton pump inhibitors (e.g., omeprazole, lansoprazole and pantoprazole), antiplatelet drugs (e.g., clopidogrel), anticonvulsants (e.g., phenytoin, methylphenytoin, diazepam, and phenobarbital), antidepressants (e.g., sertraline, citalopram, fluoxetine, and venlafaxine), anti-infective agents (e.g., proguanil, chlorproguanil, and nelfinavir), hormones (e.g., progesterone and testosterone), and pesticides (e.g., chlorpyifos and diazinon) [1–3]. Next to CYP3A4/5 that participates in metabolizing 30% of drugs, CYP2C19 is one of the most important hepatic drug metabolizing enzymes (DMEs), participating in the metabolism of 6–10% of clinically prescribed drugs [4].
Several different factors affect the expression of CYP2C19 and its functional activity in humans. For instance, it has been reported that genetic variations affect CYP2C19 expression and enzyme activity among humans, with more than an 800-fold difference in expression found in a cohort of 427 human liver samples [1,4,5]. Variation in the ability to catalyze 4′-hydroxylation of the CYP2C19 substrate mephenytoin allows individuals in most ethnicities to be categorized as poor metabolizers (PMs) or extensive metabolizers (EMs) and CYP2C19 genetic variants have been associated with PM and EM phenotypes [1]. For example, CYP2C19*1 is an allele found in the majority of people having normal CYP2C19 activity, i.e. the principal EM phenotype. CYP2C19*2 is an allele found in 2–5% Caucasians and 18–23% of Japanese [6] that creates an RNA splicing defect, effectively a null allele, resulting in a PM phenotype. CYP2C19*3 is also a null allele that introduces a premature stop codon in exon 4 [7], accounting for another type of PM. In contrast, CYP2C19*17, a variant involving the 5′-flanking region that provides for enhanced binding to GATA-binding (GATA) proteins [8], results in an elevated expression of CYP2C19 among variant carriers, accounting for ultra-rapid metabolizers. To date, more than 34 individual CYP2C19 variants are listed in its nomenclature system, encompassing a variety of genetic polymorphisms (http://www.cypalleles.ki.se/cyp2c19.htm).
In addition to genetic polymorphisms, endogenous and exogenous stimuli may also affect CYP2C19 expression dramatically through the mediation of transcription factors, such as the estrogen receptor α (ERα) [9], the constitutive androstane receptor (CAR) and the pregnane X receptor (PXR) which utilize the same DNA sequence binding specificity [10], the glucocorticoid receptor (GR) [10], and the GATA-4 transcription factor [11], by binding their response elements located at the CYP2C19 promoter. Drug–drug interactions affecting the normal nuclear receptor-mediated regulation of CYP2C19 have a significant impact on clinical pharmacology. For example, the administration of 17β-estradiol or 17α-ethinylestradiol can down-regulate the expression of CYP2C19 through the interaction of ligand-bound ERα with its cognate estrogen response element (ERE) at position 151/−147 in the CYP2C19 promoter [9], providing a mechanism for impaired CYP2C19 expression associated with the use of oral contraceptives by women. On the other hand, the induction of CYP2C19 by rifampicin or other xenobiotics is due to ligand-dependent activation of transcription through the interaction of PXR/CAR with the CAR response element (CAR-RE) located within the proximal promoter of CYP2C19 [12], providing a mechanistic explanation for increased clearance of some CYP2C19-metabolized drugs, including warfarin [13], mephenytoin [14], and hexobarbital [15], in humans exposed to PXR/CAR agonists.
Epigenetic modifications that influence the expression of various drug metabolizing enzymes provide another mechanism contributing to inter-individual variability in drug metabolism and efficacy. Thus, the epigenetic regulation of DME gene expression has a tremendous impact on the optimization of drug therapy. Although a few CpG islands were detected in the CYP2C19 gene using in silico analysis with the potential to influence gene expression via epigenetic DNA methylation [8], little is known about the actual impact of these sites on CYP2C19 expression [16]. MicroRNAs (miRNAs) provide another epigenetic mechanism for regulating DME gene expression. The miRNAs are ~22 nucleotide small RNA molecules which usually suppress gene expression by targeting partially complementary sequences located in the 3′-untranslated regions (3′-UTR) of mRNA transcripts, resulting in the enhanced degradation of targeted mRNA transcripts or the repression of mRNA translational efficiency [17]. Considerable efforts have been made recently to elucidate the roles of specific miRNA species in regulating DME expression. MiRNAs have been shown to affect the expression of many DMEs, including CYP1B1 [18],CYP2E1 [19], CYP3A4 [20] and SULT1A1 [21]. In the case of CYP2C19, two miRNA binding sites were identified within its 3′-UTR that interacted with miR-103 or miR-107 [22]. Transfection of ectopic miR-103 and miR-107 into human hepatocytes resulted in decreased CYP2C19 expression, suggesting that altered miRNA levels among humans could contribute to the inter-individual variability of CYP2C19 expression.
In the current study, we carried out integrative analyses using in silico, in vivo, and in vitro approaches to investigate the potential interaction and the mechanisms by which miRNAs target CYP2C19. First, we identified a miRNA targeting site in the coding region of CYP2C19 using in silico methods and then we employed a series of biochemical assays to elucidate the interaction between hsa-miR-29a-3p and CYP2C19 mRNA transcripts. Our results demonstrate that hsa-miR-29a-3p can suppress CYP2C19 expression in an Ago1-dependent manner in human liver cells.
2. Materials and methods
2.1. Cell lines and materials
293T human embryonic kidney cells were obtained from the American Type Culture Collection (ATCC, Manassas, VA). HepaRG cells, terminally differentiated hepatic cells retaining many characteristics of primary human hepatocytes, were obtained from Life Technologies (Carlsbad, CA). These cell lines were maintained according to ATCC and Life Technologies recommendations, respectively.
2.2. In silico analyses
Three public databases, microRNA.org (http://www.microrna.org/), PITA (http://genie.weizmann.ac.il/pubs/mir07/mir07_prediction.html) and TargetScan (Release 6.2, http://www.targets-can.org), were screened to identify potential miRNA response elements resident within the CYP2C19 3′-UTR. A fourth database, miRTar.human (http://mirtar.mbc.nctu.edu.tw/human/), was also screened to predict miRNAs that could target sites within the full CYP2C19 transcript, including the 5′-UTR, the protein coding region, and the 3′-UTR. The CellMiner™ database (version 1.5, http://discover.nci.nih.gov/cellminer), which integrates a variety of data for the NCI-60 cell lines including miRNAs levels, gene transcripts levels, and pharmacological data sets, was used to select candidate miRNAs negatively correlated with CYP2C19 expression. This database also was screened to identify chemical compounds that are positively or negatively correlated with hsa-miR-23a-3p or hsa-miR-29a-3p expression. The RNAhybrid program (http://bibiserv2.cebitec.uni-bielefeld.de/rnahybrid) was used to calculate the minimum free energy of hybridization for candidate miRNAs with putative binding sites detected within the CYP2C19 mRNA sequence (NM_000769.1).
2.3. Exogenous CYP2C19 expression assays
The pCMV6-CYP2C19 vector, which expresses the full length cDNA for CYP2C19 (Refseq NM_000769) conjugated with a C-terminal Myc-DDK tag, was purchased from OriGene Technologies (Rockville, MD) and used in the exogenous CYP2C19 expression assays in 293T cells. The CYP2C19-M1 or CYP2C19-M2 constructs, which include single nucleotide synonymous mutations in the putative hsa-miR-29a-3p target sequence in CYP2C19 mRNA, were prepared by site-directed mutagenesis of the pCMV6-CYP2C19 plasmid insert. Briefly, CYP2C19-MUT1-F and CYP2C19-MUT1-R primers, or CYP2C19-MUT2-F and CYP2C19-MUT2-R primers, together with the corresponding CYP2C19-F or CYP2C19-R primer, respectively, were used to amplify mutant DNA fragments (detailed primer sequence information is available pre request). The PCR products were then double-digested with Kpn I and Not I (New England Biolabs, Beverly, MA), and subcloned into Kpn I and Not I-linearized pCMV6-CYP2C19 vectors. The resultant constructs were sequenced to confirm authenticity.
293T Human embryonic kidney cells, cultured in Dulbecco’s Modified Eagle medium with 10% fetal bovine serum (FBS), were seeded at 2 × 105 cells per well in 24-multiwell plates and allowed to grow to 70–80% confluence. Using Lipofectamine 2000 reagent (Life Technologies), 293T cells were transfected with pCMV6-CYP2C19, CYP2C19-Mut1, or CYP2C19-Mut2 plasmids (300 ng) together with 25 nmol/L hsa-miR-29a-3p mimic, hsa-miR-23a-3p mimic, miRNA negative control (all from Thermo Scientific, Tewksbury, MA) or predesigned siRNA oligonucleotide specific for human CYP2C19 (Life Technologies). At least three independent transfection experiments were performed for each combination.
2.4. RNA electrophoretic mobility shift assays
The RNA oligonucleotides corresponding to the hsa-miR-23a-3p and hsa-miR-29a-3p sequences were synthesized and 5′-end labeled with cy5.5™ dye. The 2′-O-methyl-modified RNA oligonucleotides corresponding to the hsa-miR-23a-3p and hsa-miR-29a-3p targeting sequences in CYP2C19 coding region were synthesized and labeled with IRDye®800 dye on their 5′-ends. Additional unlabeled oligonucleotides to be used in competition assays were synthesized, comprising the miRNA negative control, hsa-miR-23a-3p and hsa-miR-29a-3p. All oligonucleotides were synthesized by Integrated DNA Technologies (Coraville, IA). Cytoplasmic extracts were prepared from HepaRG cells using NE-PER Nuclear and Cytoplasmic extraction reagents (Thermo Scientific).
RNA electrophoretic mobility shift assays (EMSAs) were performed according to the LightShift Chemiluminescent RNA EMSA Kit (Thermo Scientific) protocol. Briefly, each 20 μL basic reaction mixture contains 1× REMSA Binding Buffer and an additional 5% glycerol, 200 mM KCl, 100 mM MgCl2, and 200 nmol synthetic miRNA or/and cognate mRNA oligonucleotides. Cytoplasmic extract (2 μg) and tRNA (1 μg), which serves as the nonspecific competitor RNA, were added to reaction mixtures to test the RNA:protein interactions. Antibodies against Ago1 or Ago2 used in the supershift assays were purchased from Abcam (Cambridge, MA). In addition, unlabeled probes at 50-fold molar excess were added to the reaction before the addition of dye-labeled probes in competition assays. The reaction mixtures were incubated at 25 °C for 20 min, separated on a 12% PAGE by electrophoresis at 4 °C, and detected with an Odyssey CLx Infrared Imaging System (LI-COR Biosciences, Lincoln, NE), serially.
2.5. HepaRG cell culture and treatments
Terminally differentiated HepaRG cells were first seeded at 5 × 105 cells per well in 24-multiwell plates and maintained in Williams’ E medium supplemented with the Thaw, Plate, & General Purpose Medium Supplement (Life Technologies) for one day. For additional experiments, these cells were incubated for 7 additional days after adding the Maintenance/Metabolism Medium Supplement (Life Technologies) until hepatocyte-like cells organized in well-delineated trabeculae were noted.
The miRNA mimics, miRNA inhibitors (antagomirs, Thermo Scientific), and CYP2C19-specific siRNA, all with the final concentration of 25 nmol/L, were transiently transfected into cells using Lipofectamine 2000 transfection reagent. The chemical compounds NSC-156306 (N-(4-(9-acridinylamino)-2-methoxyphenyl) methanesulfonamide methanesulfonate; CAS 57,164-86-0) and NSC-642957 (((3aR,4S,9bS)-8-chloro-2,3,3a,4,5,9b-hexahydro-1H-cyclopenta[c]quinolin-4-yl)(phenyl)methanone) were obtained from the Developmental Therapeutics Program (DTP) of the National Cancer Institute (NCI), and suspended in dimethyl sulfoxide (DMSO) to achieve 20 μmol/L concentrations. HepaRG cells were then treated with NSC-156306 or NSC-6642957, at the final concentrations of 0, 10, or 100 nmol/L, respectively. Cells were harvested 48 h after transfection or treatment. Each assay was performed at least three times.
2.6. RNA extraction and quantitative reverse-transcription PCR (qRT-PCR)
Total RNA was extracted from 293T or HepaRG cells using the miRNeasy Mini Kit (Qiagen, Valencia, CA) and first-strand cDNA was synthesized using QuantiTect Reverse Transcription Kit (Qiagen) or NCode™ microRNA First-Strand cDNA Synthesis Kit (Life Technologies), respectively. The qRT-PCR was performed on an ABI Prism7900 Sequence Detection System (Applied Biosystems) based on the SYBR Green method, according to the QuantiFast SYBR® Green RT-PCR Kit (Qiagen) instructions. The RNA expression levels of CYP2C19 or hsa-miR-23a-3p and hsa-miR-29a-3p were calculated relative to expression of GAPDH or U6 small nuclear RNA, respectively.
2.7. Western blotting assay
Protein extracts from 293T or HepaRG cells were prepared using RIPA Lysis buffer (Life Technologies). Antibodies against the DDK tag, CYP2C19, and GAPDH were purchased from Abcam (Cam-bridge, MA). Antibodies against the DDK tag and CYP2C19 were used in western blotting assays to test exogenous and endogenous CYP2C19 expression in 293T and HepaRG cells, respectively. Western blotting was performed according to the Odyssey™ Western Blotting Kit (LI-COR Biosciences) instructions and quantitative analyses were performed based on Odyssey CLx Infrared Imaging System.
2.8. Retrieval of data from online database and statistical analyses
The RNA levels reported for CYP2C19, hsa-miR-23a-3p, and hsa-miR-29a-3p in 96 liver (non-tumor) tissues were obtained from the GSE22058 public dataset. Pearson correlation coefficients were used to test the correlations between CYP2C19 and hsa-miR-23a-3p or hsa-miR-29a-3p expression. For luciferase assays, each assay was carried out using at least 3 independent experiments. Students t-tests were used to compare the differences in CYP2C19, hsa-miR-23a-3p or hsa-miR-29a-3p protein or RNA levels between subgroups, and P < 0.05 was considered significant.
3. Results
3.1. Identification of potential miRNAs modulating CYP2C19
Three public databases, microRNA.org, PITA and TargetScan, were screened initially to identify candidate miRNAs targeting the CYP2C19 3′-UTR, but no potential regulatory miRNA binding sites were detected. We next used the miRTar.Human database to identify possible miRNA binding sites within the full length CYP2C19 transcript (Refseq NM_000769). Possible binding sites for the miRNAs hsa-miR-23a-3p, hsa-miR-29a-3p, hsa-miR-29b-3p and hsa-miR-196a-5p were found in the CYP2C19 mRNA transcript (Table 1). In addition, the expression levels of all four miRNAs correlated negatively with CYP2C19 mRNA levels in the CellMi-ner™ database. We focused on two miRNAs predicted to interact with CYP2C19 mRNA, hsa-miR-23a-3p and hsa-miR-29a-3p, because expected binding energies for the miRNA/CYP2C19 mRNA complexes were the most favorable (ΔG < 20 kcal/mol).
Table 1.
MicroRNAs targeting CYP2C19 coding region predicted by miRTar.Human
| Name | Correlationsa | Free energy (kcal/mol)b |
Location |
|---|---|---|---|
| hsa-miR-23a-3p | −0.276 | −27.5 | 19p13.13 |
| hsa-miR-29a-3p | −0.292 | −23.3 | 7q32.3 |
| hsa-miR-29b-3p | −0.255 | −18.9 | 1q32.2 |
| hsa-miR-196a-5p | −0.267 | −18.6 | 12q13.13 |
Correlations between microRNAs and CYP2C19 expression in CellMiner™ Database.
Calculated by RNAhybrid software.
3.2. hsa-miR-29a-3p suppressed exogenous CYP2C19 expression
To test if hsa-miR-23a-3p and hsa-miR-29a-3p affect CYP2C19 expression, the wild type CYP2C19 expression vector plasmid pCMV6-CYP2C19 was co-transfected into 293T cells together with the hsa-miR-23a-3p mimic, the hsa-miR-29a-3p mimic, CYP2C19-specific siRNA, or the miRNA negative control. The appropriate doses and time points were established by our previous study [23]. As shown in Fig. 1A, compared with the miRNA negative control, co-transfection with hsa-miR-29a-3p or CYP2C19-specific siRNA, but not hsa-miR-23a-3p, efficiently suppressed exogenous CYP2C19 expression in 293T cells (50% for RNA levels and 33% for protein levels, respectively; all P < 0.05). We then constructed the CYP2C19-M1 and CYP2C19-M2 vectors, both containing synonymous mutations in the core hsa-miR-29a-3p targeting sequences and both designed to exhibit less favorable miRNA binding energies than the native sequence (predicted ΔG − 16.7 kcal/mol and −16.8 kcal/mol, respectively). Co-transfection of 293T cells with the either of the mutant CYP2C19-M1 or CYP2C19-M2 expression vectors and with either the hsa-miR-29a-3p mimic or the miRNA negative control oligonucleotide showed that, unlike the case with wild-type CYP2C19 expression, hsa-miR-29a-3p did not affect mutant-CYP2C19 expression (Fig. 1B and C), indicating that the interaction between hsa-miR-29a-3p and wild type CYP2C19 mRNA is specific for the native sequence. Again, co-transfection of CYP2C19-M1 or CYP2C19-M2 plasmids with the CYP2C19-specific siRNA, which recognizes a different portion of the CYP2C19 mRNA transcript, led to inhibition of CYP2C19 mRNA and protein levels. Additionally, we tried to test the suppression effects of hsa-miR-23a-3p or hsa-miR-29a-3p on CYP2C19 expression in HepG2 cells, but neither wild type CYP2C19 nor mutant CYP2C19 was expressed successfully (data not shown).
Fig. 1.
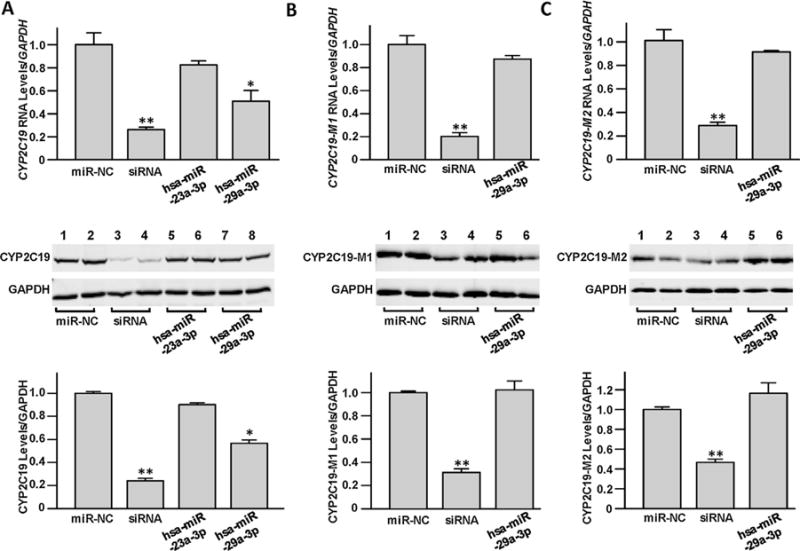
The miRNA hsa-miR-29a-3p inhibited exogenous CYP2C19 expression. The pCMV6-CYP2C19 (A), CYP2C19-M1 (B) or CYP2C19-M2 construct (C), was transiently transfected into 293T cells, together with 50 nmol/L miRNA negative control, CYP2C19-specific siRNA, hsa-miR-23a-3p mimic, or hsa-miR-29a-3p mimic, respectively. Cells were harvested 48 h after transfection, and the relative RNA levels and protein levels of CYP2C19 were tested by quantitative real-time PCR and western blot assays, respectively. Three independent transfection experiments were performed with a triplicate manner. Data were shown as percentage relative to CYP2C19 expression by the pCMV6-CYP2C19 vector transfection together with miRNA negative control. *P < 0.05; **P < 0.001; NC, miRNA negative control.
3.3. hsa-miR-29a-3p suppressed endogenous CYP2C19 expression
HepaRG cells, which express DMEs at levels comparable to those found in primary hepatocytes, were selected to test the regulatory effects of hsa-miR-23a-3p or hsa-miR-29a-3p on endogenous CYP2C19 expression. As shown in Fig. 2A, hsa-miR-23a-3p or hsa-miR-29a-3p levels in HepaRG cells were significantly elevated after transfection with the hsa-miR-23a-3p or hsa-miR-29a-3p mimics. However, only exogenous hsa-miR-29a-3p, but not hsa-miR-23a-3p, was observed to suppress endogenous CYP2C19 expression in HepaRG cells, at both RNA and protein levels (Fig. 2B). In addition, transfection with the hsa-miR-29a-3p inhibitor effectively upregulated CYP2C19 production (Fig. 2C).
Fig. 2.
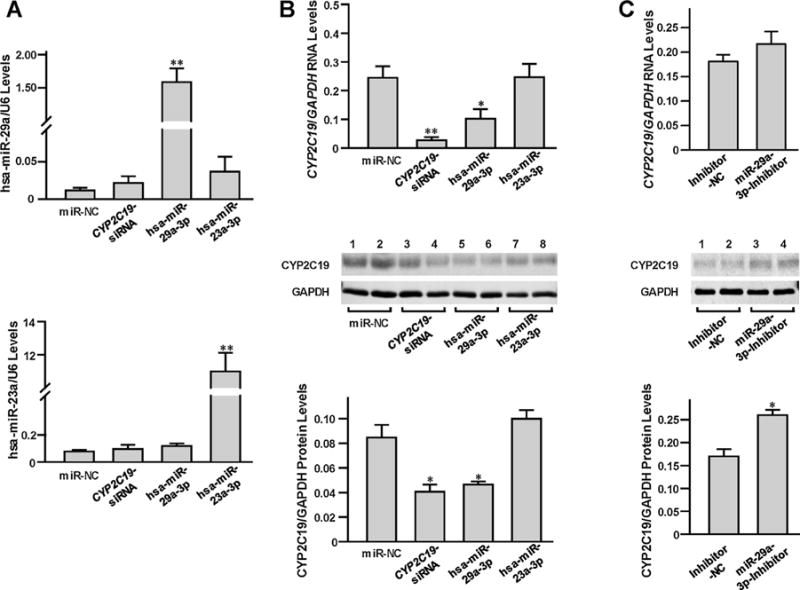
The miRNA hsa-miR-29a-3p inhibited endogenous CYP2C19 expression in HepaRG cells. The 25 nmol/L miRNA negative control, CYP2C19-specific siRNA, hsa-miR-23a-3p mimic, hsa-miR-29a-3p mimic, inhibitor negative control, or hsa-miR-29a-3p inhibitor were transiently transfected into the differentiated HepaRG cells, respectively. Each assay was carried out in at least 3 independent experiments. *P < 0.05; **P < 0.001; miR-NC, miRNA negative control; Inhibitor-NC, inhibitor negative control. (A) Unregulated expression of hsa-miR-29-3p or hsa-miR-23-3p. Data were shown as the relative miRNA levels versus U6 snRNA. (B) Down-regulated CYP2C19 levels resulted from CYP2C19-specific siRNA or hsa-miR-29a-3p mimic transfection. Data were shown as relative CYP2C19 mRNA or protein levels versus GAPDH reference. (C) Up-regulated CYP2C19 levels resulted from hsa-miR-29a-3p inhibitor transfection. Data were shown as relative CYP2C19 mRNA or protein levels versus GAPDH reference.
Further, the chemical compounds NSC-156306 and NSC-642957, which were reported in the CellMiner database™ to induce hsa-miR-29a expression, were used to treat HepaRG cells to investigate whether the chemical induction of hsa-miR-29a levels results in CYP2C19 dysregulation. We observed that the both NSC-156306 and NSC-642957 treatments significantly increased endogenous hsa-miR-29a-3p expression (Fig. 3A, with 2-fold and 3-fold elevations when treated by 10 nmol/L and 100 nmol/L NSC-156306 respectively, or 4-fold elevation when treated with 100 nmol/L NSC-642957 (all P < 0.05). In contrast, Both RNA and protein levels of CYP2C19 were dramatically decreased (Fig. 3B and C). For example, the CYP2C19 RNA levels decreased by 38.9% and 38.7% in cells treated with 10 and 100 nmol/L NSC-156306 or by 46.9% and 56.9% in cells with 10 and 100 nmol/L NSC-642957, respectively (Fig. 3B).
Fig. 3.
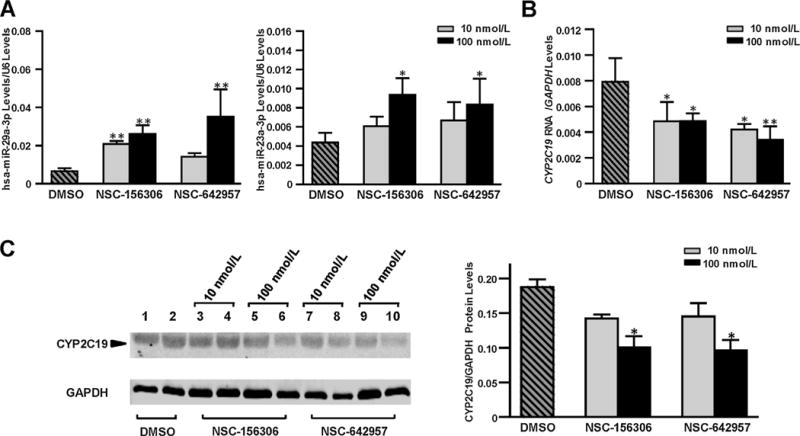
Chemical compounds NSC-156306 and NSC-606170 upregulated endogenous hsa-miR-29a-3p levels and down-regulated CYP2C19 expression in HepaRG cells. Differentiated HepaRG cells were treated with 0, 10 nmol/L, or 100 nmol/L NSC-156306 or NSC-606170 and cells were harvested 48 h after treatments. Each assay was carried out using at least 3 independent experiments. *P <n0.05; **P < 0.001; DMSO, no chemical compound in DMSO reagent. (A) Unregulated expression of hsa-miR-29-3p or hsa-miR-23-3p caused by NSC-156306 or NSC-606170 treatment. Data were shown as the relative miRNA levels versus U6 snRNA. (B) Down-regulated mRNA levels of CYP2C19 caused by NSC-156306 or NSC-606170 treatment. Data were shown as relative CYP2C19 mRNA levels versus GAPDH reference. (C) Down-regulated CYP2C19 protein levels caused by NSC-156306 or NSC-606170 treatment. Data were shown as relative CYP2C19 levels versus GAPDH reference.
3.4. Interactions between hsa-miR-23a-3p or hsa-miR-29a-3p and CYP2C19 coding regions
RNA EMSAs were carried out to determine if either hsa-miR-23a-3p or hsa-miR-29a-3p is able to interact with their cognate CYP2C19 target sequences directly. As shown in Fig. 4A, complexes formed between hsa-miR-23a-3p or hsa-miR-29a-3p and their cognate CYP2C19 mRNA oligonucleotides (Lane 3 or Lane 6) were observed in vitro, and competition assays indicated the binding was sequence-specific (Lane 7 and Lane 8). HepaRG cytoplasmic extract was able to bind the complex formed by hsa-miR-29a-3p and its cognate CYP2C19 mRNA oligonucleotide to form a RNA-protein complex (Fig. 4B, lane 6, complex B). The Ago1-specific antibody was able to interact with the hsa-miR-29a-3p/CYP2C19 RNA duplex-protein complex mentioned above to reform a new supershift complex (RNA-protein-antibody) (Fig. 4B, Lane 7, complex A), suggesting that Ago1 may play a regulatory role in CYP2C19 expression. Additionally, each of these miRNA-mRNA and RNA-protein complexes was eliminated by adding excessive unlabeled hsa-miR-29a-3p oligonucleotides (Fig. 4C, Lane 6 and Lane 9), but not by adding unlabeled nonspecific oligonucleotides (Fig. 4C, Lane 5 and Lane 8), indicating the sequence-specific character of hsa-miR-29a-3p targeting its cognate recognition sequence from the CYP2C19 mRNA transcript. However, RNA EMSAs for hsa-miR-23a-3p revealed neither an RNA-protein complex (Fig. 4B, Lane 2) nor a supershift complex (Fig. 4B, Lane 3 and Lane 4) under our experimental conditions.
Fig. 4.
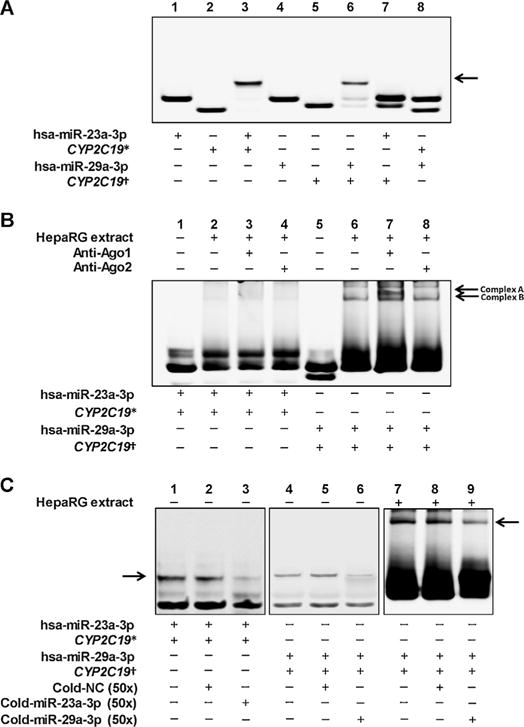
RNA EMSA with mRNA or miRNA oligonucleotides and cytoplasmic extracts from HepaRG cells. * and † indicated the CYP2C19 mRNA oligonucleotides retaining the hsa-miR-23a-3p and hsa-miR-29a-3p targeting sites, respectively. (A) The cy5.5™-labeled hsa-miR-23a-3p or hsa-miR-29a-3p oligonucleotides were incubated with 2′ O-methyl modified and IRDye®800-labeled CYP2C19 mRNA oligonucleotides. Lanes 1, 2, 4, and 5 indicated the mobility of each type of oligonucleotides, respectively; Lanes 3 and 6 indicated the mobility status of the miRNA oligonucleotides incubated with corresponding CYP2C19 mRNA oligonucleotides, respectively; Lanes 7 and 8 indicated the mobility status of the miRNA oligonucleotides incubated with unmatched CYP2C9 mRNA oligonucleotides, respectively. Arrows indicated the oligonucleotide complex formed by miRNA oligonucleotides and mRNA oligonucleotides in Lane 3 and 6. (B) Cytoplasmic extracts from HepaRG cells were incubated with hsa-miR-23a-3p or hsa-miR-29a-3p oligonucleotides, and CYP2C19 mRNA oligonucleotides. Lanes 1 and 5 indicated the mobility status of the miRNA oligonucleotides incubated with corresponding CYP2C19 mRNA oligonucleotides, respectively; Lanes 2 and 6 indicated the mobility status of the miRNA and mRNA oligonucleotides incubated with cell extracts, respectively; Lanes 3, 4, 7, and 8 indicated the mobility status of the oligonucleotide-cytoplasmic extracts complex with Ago1 or Ago2 antibody, respectively. Complex A indicated the supershift complex formed by oligonucleotides, cytoplasmic proteins and antibodies in Lane 6. (C) RNA EMSA in the presence of non-labelled various unlabeled competitors. Arrows indicated the oligonucleotide complexes formed by miRNA oligonucleotides and mRNA oligonucleotides in Lane 7, 8 and 9.
3.5. hsa-miR-23a-3p and hsa-miR-29a-3p levels negatively correlated with CYP2C19 expression in liver tissues
We utilized the RNA levels of CYP2C19 and hsa-miR-29a-3p reported in the GSE22058 public dataset for 96 liver (non-tumor) tissues, and evaluated the correlation between CYP2C19 and hsa-miR-23a-3p or hsa-miR-29a-3p, respectively. We found CYP2C19 expression was significantly and negatively correlated with the expression of hsa-miR-23a-3p or hsa-miR-29a-3p (Pearson r = −0.298 or −0.214, respectively, Fig. 5).
Fig. 5.
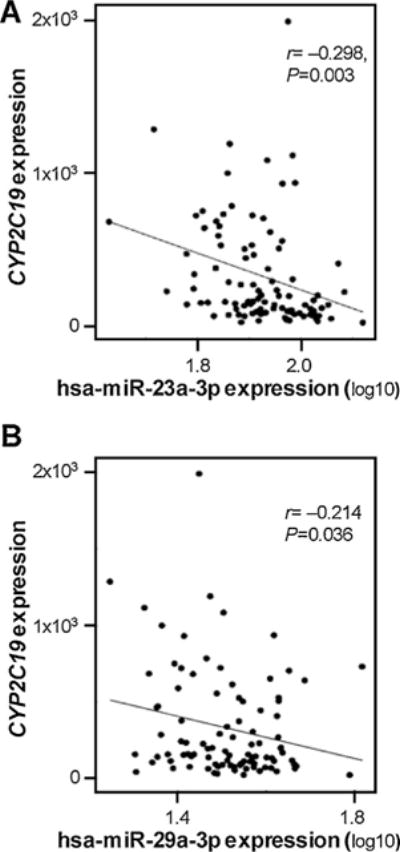
Relationship between CYP2C19 mRNA expression and hsa-miR-23a-3p (A) or hsa-miR-29a-3p levels (B) in 96 non-tumor tissues in GSE22058 public dataset using Spearman correlation analysis.
4. Discussion
In the current study we investigated the regulatory roles of miRNAs in the expression of CYP2C19. From our previous study, we realized that the free energy associated with binding between miRNAs and mRNA targets is a critical parameter for predicting the interactional efficiency and stability of miRNA/mRNA complexes [23]. Here, in silico target prediction and free energy calculations indicated that hsa-miR-29a-3p could potentially target the coding region of CYP2C19 transcripts with a free energy of −23.3 kcal/mol. A series of in vitro and in vivo analyses confirmed the suppressive effect of hsa-miR-29a-3p on CYP2C19 expression. Moreover, we found that the Ago1 protein, but not the Ago2 protein, was detected in the hsa-miR-29a-3p regulatory complex with CYP2C19 mRNA formed using HepaRG cytoplasmic extracts. Our results revealed comprehensively that hsa-miR-29a-3p is involved in negatively regulating the expression of CYP2C19 in liver cells and elucidated the underlying biological mechanism.
Since CYP2C19 is involved in the biotransformation of many clinical drugs and environmental toxicants [1], aberrant CYP2C19 expression and activity might be relevant to drug efficacy or therapeutic failure; it could possibly cause adverse drug reactions in some extreme instances. For example, clopidogrel is one of the most often prescribed antiplatelet drugs used to treat patients with cardiovascular diseases. A meta-analysis consisting of a total of 31 independent studies among 9685 patients revealed that patients even with one allele of reduced/null function CYP2C19 appeared to suffer an increased risk for major adverse cardiovascular events, especially stent thrombosis (odds ratio 2.67; 95% confidence interval 1.69–4.22; P < 0.0001) [24]. Through a genome-wide association study among the Amish population, it was reported that the loss of function CYP2C19*2 variant is a major determinant of clopidogrel response, accounting for 12% of the variation in response. In addition, 13 SNPs on chromosome 10q24 were also associated with decreased clopidogrel response [25]. In a different cohort, it was found that ~30% of the general population harboring CYP2C19*2 have poorer platelet response to clopidogrel and are at a 2.4-fold higher risk of having an ischemic cardiac event or death [25]. It is notable that the substantial variability in the response to clopidogrel among individuals is mostly due to genetic polymorphisms. However, genetic polymorphisms could only explain a portion of the inter-individual variability observed, suggesting that other mechanisms affecting CYP2C19 expression/activity, such as epigenetic mechanisms, may also play an important role in the inter-individual variability in the response to drugs metabolized by CYP2C19. In a practical sense, epigenetic mediators of idiosyncratic drug responses are likely to be most important among individuals who carry the same CYP2C19 genotype.
MiRNAs are a family of small non-coding RNA molecules that can post-transcriptionally regulate gene expression by targeting their cognate mRNA transcripts. Evidence documented that miRNAs play an important role in the modulation of the production of drug metabolizing enzymes and transporters through multi-directional interactions among environmental stimuli, the expression of miRNA molecules and genetic polymorphisms [26–33]. As miRNA expression has been reported to be affected by drugs and since miRNAs themselves may affect drug metabolism and toxicity, miRNA expression possesses a significant potential for affecting drug efficacy and drug safety. In contrast to numerous studies reporting the importance of genetic polymorphisms that influence CYP2C19 gene expression, the role of miRNAs in its regulation has been under-investigated. Zhang et al. [22] reported that miR-103 and miR-107 post-transcriptionally regulate CYP2C8, CYP2C9 and CYP2C19, the only published study available in the literature. Compared to their study, our in silico approach was unable to identify putative miR-103 and miR-107 targeting sites in the 3′-UTR of CYP2C19. However, our strategy did predict that hsa-miR-23a-3p and hsa-miR-29a-3p might efficiently interact with the coding region of CYP2C19. We believe that this minor disparity between the results of these two studies is largely owing to the utilization of different databases and prediction algorithms. Nevertheless, both studies applied in silico, in vitro and in vivo experiments and all validated that specific miRNAs modulate CYP2C19 production in human liver.
Accumulating evidence has shown that miRNAs may also bind within the coding region of mRNA transcripts to cause translation inhibition or mRNA degradation in mammalian cells. For example, miR-602 and miR-608 were reported to target Sonic Hedgehog (SHH) mRNA transcripts within its coding domain sequence [34], while miR-34a showed similar regulatory roles for MDM4 expression [35]. However, in contrast to the well-established roles of miRNAs binding the 3′-UTRs of mRNA transcripts, the regulatory mechanism of miRNAs targeting mRNA coding domain sequences remains to be clarified.
We carried out a series of transfection assays and found that the exogenous and endogenous CYP2C19 levels in liver HepaRG cells and kidney 293T cells (these two cell lines express endogenous CYP2C19) or liver tissue samples exhibited significant negative correlation with hsa-miR-29a-3p levels. In addition, treatments of HepaRG cells with the miRNA-inducing compounds NSC-156306 and NSC-642957 confirmed that the aberrant hsa-miR-29a-3p expression caused by these chemicals consequently leads to dysregulation of CYP2C19 expression. These results considered together prove that hsa-miR-29a-3p acts as negative regulator of CYP2C19 via a specific target site located in the CYP2C19 coding region.
Intriguingly, the small interfering RNAs (siRNAs) and miRNAs have a sorting ability to distinguish specific Ago-RISCs in lower organisms, but appear to have lost this capability in mammals [36]. However, another significant finding in this present study is that hsa-miR-29a-3p has a selective advantage toward binding Ago1 proteins to form the multi-protein RNA-induced silencing complex (RISC). At first, we observed that hsa-miR-29a-3p is able to interact with its cognate CYP2C19 sequence directly in vitro in RNA EMSA. When adding the HepaRG cytoplasmic extracts and Ago1 or Ago2 antibodies, only Ago1 antibodies, but not Ago2 antibodies, interacted with RNA duplex and protein extracts to form the miRNA–mRNA–protein–antibody complex, indicating that Ago1 might be involved in the modulation of CYP2C19 expression. Such a sorting mechanism for Ago family proteins in mammals was previously reported by Yamakawa et al. [37], who found that some novel small RNAs were able to selectively distinguish and load onto human Ago1-RISC. Also, Turchinovich et al. [38] observed that many miRNAs derived from antisense strands of pre-miRNAs (i.e., 3p-miRNAs) were Ago1-biased, while the counterparts derived from the sense strand (i.e., 5p-miRNAs) were Ago2-biased. All the previous studies and our results support the notion that an RNA sequence-specific sorting mechanism remains in mammals, at least for some miRNAs.
In the current study, we tried to understand the extent to which this miRNA molecule contributes to inter-individual variability in the expression of CYP2C19 among a population. However, correlation analyses indicated that only a small portion (less than 5%) was contributed by the modulation of hsa-miR-29a-3p. Notably, all miRNAs and CYP2C19 expression data were obtained using samples described as adjacent liver non-tumor tissues, which are often complicated with cirrhosis. In addition, genetic variants, medication history and environmental exposure status (such as dietary information) for these liver donors were not available. Therefore, the complex and heterogeneous characteristics of these tissue samples might reduce the correlation power. On the other hand, we believe that genetic variation, chemical induction, and epigenetic modulation (including miRNA modulation) can all contribute to the differential expression of CYP2C19; however, genetic variation and chemical induction play a more important role. In the implementation of precision medicine, the miRNA modulation of CYP2C19 should be taken into consideration when the individuals have the same genotype but take a different combination of drugs, by which the increased expression of miRNA through a drug induction could be an important modulator to affect CYP2C9 expression.
In summary, we showed that hsa-miR-29a-3p modulates CYP2C19 expression in liver cells, through an Ago1-dependent manner, and by targeting the coding region directly. Though the precise mechanism is yet to be elucidated, our results supplied important clues to decode the interplay puzzle between miRNAs and target sites in coding region in human drug metabolizing enzymes and transporters.
Acknowledgments
This study was supported and funded by the National Center for Toxicological Research (Project E0752601), U.S. Food and Drug Administration.
Abbreviations
- CYP
cytochrome P450
- miRNA
microRNA
- DMETs
drug metabolizing enzymes and transporters
- PM
poor metabolizer
- EM
extensive metabolizer
- ERα
estrogen receptor α
- CAR
constitutive androstane receptor
- PXR
pregnane X receptor
- RE
response element
- 3′-UTR
3′-untranslated region
- SULT
sulfotransferase
- Ago
argonaute RISC catalytic component
- RISC
RNA-induced silence complex
- FSB
fetal bovine serum
- EMSA
electrophoretic mobility shift assay
Footnotes
Conflict of interest
The authors have no conflict of interest.
Publisher's Disclaimer: Disclaimer
The information in these materials is not a formal dissemination of information by FDA and does not represent agency position or policy.
Author contributions
Participated in study design: Yu, Tolleson, Pogribny, Ning.
Conducted experiments: Yu, Green, Jin, Mei.
Performed data analysis: Yu, Guo, Deng, Ning.
Wrote or contributed to the writing of the manuscript: Yu, Tolleson, Pogribny, Ning.
References
- 1.Desta Z, Zhao X, Shin JG, Flockhart DA. Clinical significance of the cytochrome P450 2C19 genetic polymorphism. Clin Pharmacokinet. 2002;41:913–958. doi: 10.2165/00003088-200241120-00002. [DOI] [PubMed] [Google Scholar]
- 2.Dadson OA, Ellison CA, Singleton ST, Chi LH, McGarrigle BP, Lein PJ, et al. Metabolism of profenofos to 4-bromo-2-chlorophenol, a specific and sensitive exposure biomarker. Toxicology. 2013;306:35–39. doi: 10.1016/j.tox.2013.01.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kappers WA, Edwards RJ, Murray S, Boobis AR. Diazinon is activated by CYP2C19 in human liver. Toxicol Appl Pharmacol. 2001;177:68–76. doi: 10.1006/taap.2001.9294. [DOI] [PubMed] [Google Scholar]
- 4.Zanger UM, Schwab M. Cytochrome P450 enzymes in drug metabolism: regulation of gene expression, enzyme activities, and impact of genetic variation. Pharmacol Ther. 2013;138:103–141. doi: 10.1016/j.pharmthera.2012.12.007. [DOI] [PubMed] [Google Scholar]
- 5.Yang L, Price ET, Chang CW, Li Y, Huang Y, Guo LW, et al. Gene expression variability in human hepatic drug metabolizing enzymes and transporters. PLoS One. 2013;8:e60368. doi: 10.1371/journal.pone.0060368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Goldstein JA, Ishizaki T, Chiba K, De Morais SM, Bell D, Krahn PM, et al. Frequencies of the defective CYP2C19 alleles responsible for the mephenytoin poor metabolizer phenotype in various Oriental, Caucasian, Saudi Arabian and American black populations. Pharmacogenetics. 1997;7:59–64. doi: 10.1097/00008571-199702000-00008. [DOI] [PubMed] [Google Scholar]
- 7.De Morais SM, Wilkinson GR, Blaisdell J, Meyer UA, Nakamura K, Goldstein JA. Identification of a new genetic defect responsible for the polymorphism of (S)-mephenytoin metabolism in Japanese. Mol Pharmacol. 1994;46:594–598. [PubMed] [Google Scholar]
- 8.Helsby NA, Burns KE. Molecular mechanisms of genetic variation and transcriptional regulation of CYP2C19. Front Genet. 2012;3:206. doi: 10.3389/fgene.2012.00206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Mwinyi J, Cavaco I, Pedersen RS, Persson A, Burkhardt S, Mkrtchian S, et al. Regulation of CYP2C19 expression by estrogen receptor alpha: implications for estrogen-dependent inhibition of drug metabolism. Mol Pharmacol. 2010;78:886–894. doi: 10.1124/mol.110.065540. [DOI] [PubMed] [Google Scholar]
- 10.Chen Y, Ferguson SS, Negishi M, Goldstein JA. Identification of constitutive androstane receptor and glucocorticoid receptor binding sites in the CYP2C19 promoter. Mol Pharmacol. 2003;64:316–324. doi: 10.1124/mol.64.2.316. [DOI] [PubMed] [Google Scholar]
- 11.Mwinyi J, Hofmann Y, Pedersen RS, Nekvindova J, Cavaco I, Mkrtchian S, et al. The transcription factor GATA-4 regulates cytochrome P4502C19 gene expression. Life Sci. 2010;86:699–706. doi: 10.1016/j.lfs.2010.02.021. [DOI] [PubMed] [Google Scholar]
- 12.Chen Y, Ferguson SS, Negishi M, Goldstein JA. Induction of human CYP2C9 by rifampicin, hyperforin, and phenobarbital is mediated by the pregnane X receptor. J Pharmacol Exp Ther. 2004;308:495–501. doi: 10.1124/jpet.103.058818. [DOI] [PubMed] [Google Scholar]
- 13.Heimark LD, Gibaldi M, Trager WF, O’Reilly RA, Goulart DA. The mechanism of the warfarin–rifampin drug interaction in humans. Clin Pharmacol Ther. 1987;42:388–394. doi: 10.1038/clpt.1987.168. [DOI] [PubMed] [Google Scholar]
- 14.Feng HJ, Huang SL, Wang W, Zhou HH. The induction effect of rifampicin on activity of mephenytoin 4′-hydroxylase related to M1 mutation of CYP2C19 and gene dose. Br J Clin Pharmacol. 1998;45:27–29. doi: 10.1046/j.1365-2125.1998.00643.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Smith DA, Chandler MH, Shedlofsky SI, Wedlund PJ, Blouin RA. Age-dependent stereoselective increase in the oral clearance of hexobarbitone isomers caused by rifampicin. Br J Clin Pharmacol. 1991;32:735–739. [PMC free article] [PubMed] [Google Scholar]
- 16.Ingelman-Sundberg M, Sim SC, Gomez A, Rodriguez-Antona C. Influence of cytochrome P450 polymorphisms on drug therapies: pharmacogenetic, pharmacoepigenetic and clinical aspects. Pharmacol Ther. 2007;116:496–526. doi: 10.1016/j.pharmthera.2007.09.004. [DOI] [PubMed] [Google Scholar]
- 17.Saito Y, Saito H, Liang G, Friedman JM. Epigenetic alterations and microRNA misexpression in cancer and autoimmune diseases: a critical review. Clin Rev Allergy Immunol. 2014;47:128–135. doi: 10.1007/s12016-013-8401-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Tsuchiya Y, Nakajima M, Takagi S, Taniya T, Yokoi T. MicroRNA regulates the expression of human cytochrome P450 1B1. Cancer Res. 2006;66:9090–9098. doi: 10.1158/0008-5472.CAN-06-1403. [DOI] [PubMed] [Google Scholar]
- 19.Mohri T, Nakajima M, Fukami T, Takamiya M, Aoki Y, Yokoi T. Human CYP2E1 is regulated by miR-378. Biochem Pharmacol. 2010;79:1045–1052. doi: 10.1016/j.bcp.2009.11.015. [DOI] [PubMed] [Google Scholar]
- 20.Pan YZ, Gao W, Yu AM. MicroRNAs regulate CYP3A4 expression via direct and indirect targeting. Drug Metab Dispos. 2009;37:2112–2117. doi: 10.1124/dmd.109.027680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yu X, Dhakal IB, Beggs M, Edavana VK, Williams S, Zhang X, et al. Functional genetic variants in the 3′-untranslated region of sulfotransferase isoform 1A1 (SULT1A1) and their effect on enzymatic activity. Toxicol Sci. 2010:391–403. doi: 10.1093/toxsci/kfq296. doi: http://dx.doi.org/10.1093/toxsci/kfq296. [DOI] [PMC free article] [PubMed]
- 22.Zhang SY, Surapureddi S, Coulter S, Ferguson SS, Goldstein JA. Human CYP2C8 is post-transcriptionally regulated by microRNAs 103 and 107 in human liver. Mol Pharmacol. 2012;82:529–540. doi: 10.1124/mol.112.078386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yu D, Green B, Marrone A, Guo Y, Kadlubar S, Lin D, et al. Suppression of CYP2C9 by microRNA hsa-miR-128-3p in human liver cells and association with hepatocellular carcinoma. Sci Rep. 2015;5:8534. doi: 10.1038/srep08534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mega JL, Simon T, Collet JP, Anderson JL, Antman EM, Bliden K, et al. Reduced-function CYP2C19 genotype and risk of adverse clinical outcomes among patients treated with clopidogrel predominantly for PCI: a meta-analysis. JAMA. 2010;304:1821–1830. doi: 10.1001/jama.2010.1543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Shuldiner AR, O’Connell JR, Bliden KP, Gandhi A, Ryan K, Horenstein RB, et al. Association of cytochrome P450 2C19 genotype with the antiplatelet effect and clinical efficacy of clopidogrel therapy. JAMA. 2009;302:849–857. doi: 10.1001/jama.2009.1232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zhong XB, Leeder JS. Epigenetic regulation of ADME-related genes: focus on drug metabolism and transport. Drug Metab Dispos. 2013;41:1721–1724. doi: 10.1124/dmd.113.053942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Rieger JK, Klein K, Winter S, Zanger UM. Expression variability of absorption, distribution, metabolism, excretion-related microRNAs in human liver: influence of nongenetic factors and association with gene expression. Drug Metab Dispos. 2013;41:1752–1762. doi: 10.1124/dmd.113.052126. [DOI] [PubMed] [Google Scholar]
- 28.Yokoi T, Nakajima M. microRNAs as mediators of drug toxicity. Annu Rev Pharmacol Toxicol. 2013;53:377–400. doi: 10.1146/annurev-pharmtox-011112-140250. [DOI] [PubMed] [Google Scholar]
- 29.Yu AM. Role of microRNAs in the regulation of drug metabolism and disposition. Expert Opin Drug Metab Toxicol. 2009;5:1513–1528. doi: 10.1517/17425250903307448. [DOI] [PubMed] [Google Scholar]
- 30.Rukov JL, Shomron N. MicroRNA pharmacogenomics: post-transcriptional regulation of drug response. Trends Mol Med. 2011;17:412–423. doi: 10.1016/j.molmed.2011.04.003. [DOI] [PubMed] [Google Scholar]
- 31.Kacevska M, Ivanov M, Ingelman-Sundberg M. Epigenetic-dependent regulation of drug transport and metabolism: an update. Pharmacogenomics. 2012;13:1373–1385. doi: 10.2217/pgs.12.119. [DOI] [PubMed] [Google Scholar]
- 32.Kim IW, Han N, Burckart GJ, Oh JM. Epigenetic changes in gene expression for drug-metabolizing enzymes and transporters. Pharmacotherapy. 2014;34:140–150. doi: 10.1002/phar.1362. [DOI] [PubMed] [Google Scholar]
- 33.Ning B, Su Z, Mei N, Hong H, Deng H, Shi L, et al. Toxicogenomics and cancer susceptibility: advances with next-generation sequencing. J Environ Sci Health C Environ Carcinog Ecotoxicol Rev. 2014;32:121–158. doi: 10.1080/10590501.2014.907460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Akhtar N, Makki MS, Haqqi TM. MicroRNA-602 and MicroRNA-608 regulate sonic hedgehog expression via target sites in the coding region in human chondrocytes. Arthritis Rheumatol. 2015;67:423–434. doi: 10.1002/art.38952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Mandke P, Wyatt N, Fraser J, Bates B, Berberich SJ, Markey MP. MicroRNA-34a modulates MDM4 expression via a target site in the open reading frame. PLoS One. 2012;7:e42034. doi: 10.1371/journal.pone.0042034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Su H, Trombly MI, Chen J, Wang X. Essential and overlapping functions for mammalian Argonautes in microRNA silencing. Genes Dev. 2009;23:304–317. doi: 10.1101/gad.1749809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Yamakawa N, Okuyama K, Ogata J, Kanai A, Helwak A, Takamatsu M, et al. Novel functional small RNAs are selectively loaded onto mammalian. Ago1, Nucleic Acids Res. 2014;42:5289–5301. doi: 10.1093/nar/gku137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Turchinovich A, Burwinkel B. Distinct AGO1 and AGO2 associated miRNA profiles in human cells and blood plasma. RNA Biol. 2012;9:1066–1075. doi: 10.4161/rna.21083. [DOI] [PMC free article] [PubMed] [Google Scholar]


