Abstract
Holoprosencephaly (HPE), a common developmental defect of the forebrain and midface, has a complex etiology. Heterozygous, loss-of-function mutations in the Sonic hedgehog (SHH) pathway are associated with HPE. However, mutation carriers display highly variable clinical presentation, leading to an “autosomal dominant with modifier” model, in which the penetrance and expressivity of a predisposing mutation is graded by genetic or environmental modifiers. Such modifiers have not been identified. Boc encodes a SHH co-receptor and is a silent HPE modifier gene in mice. Here, we report identification of missense BOC variants in HPE patients. Consistent with these alleles functioning as HPE modifiers, individual variant BOC proteins had either loss- or gain-of-function properties in cell-based SHH signaling assays. Therefore, in addition to heterozygous loss-of-function mutations in specific SHH pathway genes and an ill-defined environmental component, our findings identify a third variable in HPE: low frequency modifier genes, BOC being the first identified.
Keywords: Holoprosencephaly, Sonic hedgehog, BOC, Modifier gene, Gene variant, Birth defect
Many structural birth defects are thought to arise from a complex combination of genetic and environmental risk factors (Krauss and Hong, 2016). Holoprosencephaly (HPE; MIM# 236100), a common and often devastating defect in midline patterning of the forebrain and midface, is a prototypical example. HPE encompasses a phenotypic spectrum that ranges from failure to partition the forebrain into hemispheres and cyclopia, to mild midfacial anomalies that occur without forebrain involvement (Geng and Oliver, 2009). Heterozygous, loss-of-function mutations in components of the Sonic hedgehog (SHH; MIM# 600725) signaling pathway are frequently associated with HPE (Roessler and Muenke, 2010). However, highly variable clinical presentation is seen in mutation carriers, even within pedigrees. Furthermore, in many “sporadic” cases, mutations in SHH are inherited from a parent with little or no clinical manifestation (Solomon et al., 2012). Statistical analyses have led to an “autosomal dominant with modifier” model, in which the penetrance and expressivity of a predisposing heterozygous mutation is graded by modifiers (Roessler et al., 2012). Such modifiers may be genetic or environmental in nature. While bona fide pathogenic mutations in SHH and other genes associated with HPE continue to be catalogued, identification of potential HPE modifiers is in its infancy.
The binding of SHH to its primary cell surface receptor PTCH1 (MIM# 601309) initiates a signaling cascade to modulate GLI transcription factors, which induce expression of pathway target genes (Lee et al., 2016). Among the direct target genes is GLI1 (MIM# 165220) itself. SHH and PTCH1 both bind to several co-receptors, including CDON (MIM# 608707), BOC (MIM# 608708), and GAS1 (MIM# 139185) (Bae et al., 2011; Izzi et al., 2011; Tenzen et al., 2006). Studies with knockout mice revealed that these three co-receptors have overlapping and partially redundant function in supporting SHH signaling during development; embryos lacking all three have a nearly complete loss of pathway activity (Allen et al., 2011). Heterozygous, loss-of-function mutations in CDON and GAS1 have been identified in HPE patients (Bae et al., 2011; Pineda-Alvarez et al., 2012; Ribeiro et al., 2010). Consistent with these observations, mice with targeted mutations in either Cdon or Gas1 display a range of HPE phenotypes (Allen et al., 2007; Hong and Krauss, 2012; Martinelli and Fan, 2007; Seppala et al., 2007; Zhang et al., 2006). In contrast, mice lacking Boc do not have HPE (Zhang et al., 2011). However, genetic removal of Boc from Cdon−/− or Gas1−/− mice exacerbates their HPE phenotype (Seppala et al., 2014; Zhang et al., 2011). Therefore, Boc functions as a silent HPE modifier gene in mice. Here we address the potential role of BOC in HPE pathogenesis in humans.
We performed high-throughput screening of BOC (NM_033254.3) for 360 HPE patients using High Resolution Melting, as described (Kauvar et al., 2011). Additionally, 384 unrelated individuals were screened as controls. HPE and control samples with melting profiles that deviated from wild type melting curves were directly sequenced for variant confirmation. A total of eight different amino acid substitution variants in BOC were identified in HPE patients, including one that was found in two unrelated patients and two that were present in a single patient (Table 1). There are over 400 BOC missense variants reported in ExAC. We note that most of the variants we identified are rare (Table 1), including p.Gly556Glu, which has not been previously reported. An exception is p.Pro828Arg, which has a minor allele frequency of 0.0017, more common than the ~1:10,000 birth frequency of HPE (Leoncini et al., 2008), and consistent with possible function as a modifier allele. All individuals were also studied for the four genes most commonly screened in HPE (SHH, ZIC2 (MIM# 603073), SIX3 (MIM# 603714), and TGIF (MIM# 602630)). One patient had a truncating mutation in ZIC2, and another patient had a deletion of TGIF (Table 1).
Table 1.
Sequence variations in BOC
| Singleton patient |
Four gene screena |
BOC gDNA [hg19] |
BOC cDNA [NM_033254.2]b |
BOC Protein [NP_150279.1] |
Protein domain |
Consensus Predictiond |
Experiment | ExAC MAF |
ACMGe | ACMG predictione |
|---|---|---|---|---|---|---|---|---|---|---|
| BL9321f | ZIC2 c.1206C>G; p.Y402* | chr3:112969488 C/T chr3:112997000 G/T (phase not known) |
c.184C>T c.1598G>T |
p.Pro62Ser p.Arg533Leu |
Ig FN1 |
Benign Benign |
N.T. Suppressor of gain of function |
0.000008238 0.000008237 |
BP4/BP5 BP4/BP5 |
Likely benign Likely benign |
| LCL100 g | Normal | chr3:112997069 G/A | c.1667G>A | p.Gly556Glu | FN1 | Damaging | Gain of function | None | PM2;PP3 | VOUS |
| FB604 | Normal | chr3:112998740 G/C | c.2090G>C | p.Arg697Pro | linker FN2 to FN3 | Damaging | Benign | 0.0005543 | PP3;BS1 | VOUS |
| BL1062 | Normal | chr3:112998761 A/C | c.2111A>C | p.Tyr704Ser | linker FN2 to FN3 | Benign | N.T. | 0.0003142 | BS1;BP4 | Likely benign |
| BL6788 | Normal | chr3:113002309 C/G | c.2483C>G | p.Pro828Argc | linker FN3 to TM | Benign | Suppressor of gain of function | 0.001668 | BS1;BP4 | Likely benign |
| BL7348 h | del TGIF/dup 18p | chr3:113002309 C/G | c.2483C>G | p.Pro828Argc | linker FN3 to TM | Benign | Suppressor of gain of function | 0.001668 | BS1;BP4/BP5 | Benign |
| BL6768 i | Normal | chr3:113002401 G/A | c.2575G>A | p.Gly859Arg | TM | Damaging | N.T. | 0.00002471 | PP3 | VOUS |
| BL9282 j | Normal | chr3:113005589 T/A | c.3225T>A | p.Ser1075Arg | intracellular | Benign | N.T. | 0.0001979 | BS1;BP4 | Likely benign |
Pineda-Alvarez et al. (2010) by Sanger sequencing or cytogenetic analysis.
Variant screening of BOC (MIM#608708) was essentially identical to that described in Kauvar et al. 2010 with Sanger sequencing as confirmation. Minor Allele Frequency (MAF) as determined by ExAC [http://exac.broadinstitute.org].
Also detected in a healthy parent of an HPE child, a product of a first cousin marriage.
dbNSFP v.3.3a consensus (http://annovar.openbioinformatics.org/en/latest/): determined by concurrence >50% of [SIFT, PolyPhen2HDIV, PolyPhenHVAR, LRT, MutationTaster, MutationAssessor, FATHMM, PROVEAN, FATHMM-MKL, MetaSVM, MetaLR] as damaging or <50% as benign.
Justifications for assertions of pathogenicity incorporate the principal accepted guidelines in Richards, S. et al. (2015) for a simple autosomal dominant disorder with high penetrance. Variants with an allele frequency greater than 1;10,000 (the live birth incidence of HPE in the newborn nursery) are either likely benign or, if shown to be abnormal in function, can act as modifiers.
Semilobar HPE, microcephaly, elevated palate, hypotelorism, global developmental delay, spasticity, diabetes insipidus.
Semilobar HPE, hypoplastic corpus callosum, asymmetric hydrocephalous, generalized atrophy, brachycephaly.
Hypotelorism, microcephaly, cebocephaly, cleft lip.
Lobar HPE, midline cleft lip and palate.
Lobar HPE.
To explore the functional consequences of HPE-associated BOC variants, we developed a cell-based assay for BOC activity. Exogenous expression of BOC had little ability to enhance SHH signaling in wild type mouse embryo fibroblasts (MEFs) or even Cdon−/−;Boc−/− MEFs (not shown). To work in as sensitive a cell system as possible, we used Cdon−/−;Boc−/−;Gas1−/− MEFs (TKO cells) (Mathew et al., 2014). When treated with escalating doses of recombinant SHH N-terminus protein (referred to simply as SHH), TKO cells expressed the direct target gene Gli1 very inefficiently, relative to wild type MEFs, as determined by qRT-PCR (Supp. Fig. S1A). To determine whether exogenous expression of CDON or BOC could restore SHH responsiveness to TKO cells, we transfected the cells with expression vectors encoding one or the other of these co-receptors, treated the cultures with SHH, and quantified Gli1 expression. CDON effectively rescued SHH-dependent Gli1 induction, whereas BOC expression had little effect (Supp. Fig. S1B). These results suggest that BOC may require a factor not present in MEFs to act as a productive SHH co-receptor. CDON and GAS1 may play some role in this phenomenon. However, as BOC had little activity in wild type MEFs, and Cdon−/−;Gas1−/− embryos have BOC-dependent SHH activity (Allen et al., 2011; Allen et al., 2007), a need for additional factors for BOC function seems likely.
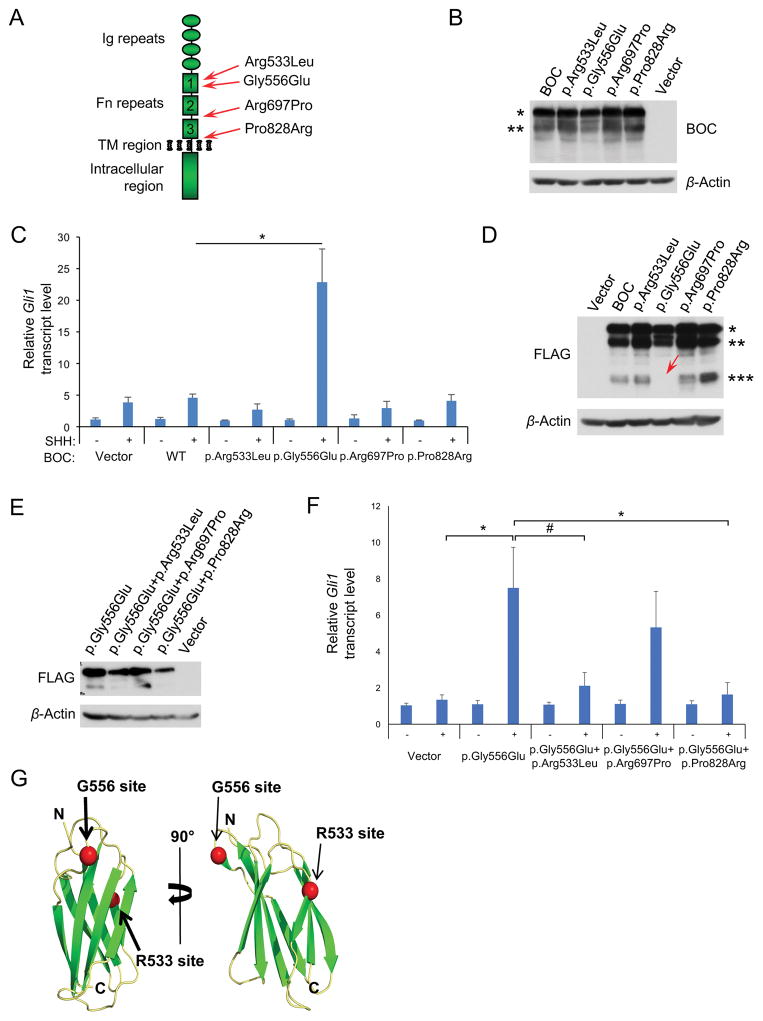
BOC and CDON have Ig repeats followed by fibronectin-type III (Fn3) repeats in their ectodomains (Fig. 1A). The third Fn3 repeat of BOC and CDON bind to SHH, whereas Fn3 repeats 1 and 2 each associate with PTCH1 (Bae et al., 2011; Izzi et al., 2011; Kavran et al., 2010). We constructed expression vectors encoding four of the HPE-associated BOC variants: p.Arg533Leu, p.Gly556Glu, p.Arg697Pro, and p.Pro828Arg. The p.Arg533Leu and p.Gly556Glu substitutions are in Fn3 repeat 1, p.Arg697Pro is in the linker region between Fn3 repeats 1 and 2, and p.Pro828Arg is in the linker region between Fn3 repeat 3 and the transmembrane region (TM). These variants were selected because: 1) they were located in the Fn repeat region of BOC, which is critical for Shh signaling (Song et al., 2015); and 2) they resemble similarly located, HPE-associated, loss-of-function, missense mutations in CDON in that they were highly non-conservative substitutions in evolutionarily conserved residues (Bae et al., 2011). Prioritization of these variants was not based on either retrospective allele frequency or Annovar annotation criteria, which were not available at the time of study design.
FIGURE 1. HPE-associated BOC variants display gain- and loss-of-function activities.
A: Schematic diagram of BOC protein and the location of HPE-associated variants. B: Western blot analysis of BOC and BOC variant expression. HEK293T cells were transiently transfected with empty vector (vector), wild type BOC (BOC), and the indicated variants. Forty-eight hours later cells were lysed and prepared for Western blots with antibodies to human BOC and β-actin as a loading control. C: Expression of p.Gly556Glu, but not other BOC variants, restored the response to SHH by TKO cells. Cultures were treated with 3 ng/ml SHH and analysis of Gli1 expression was performed and quantified qRT-PCR. WT, wild type BOC. D: BOC p.Gly556Glu displayed altered product formation upon limited proteolysis. Wild type BOC and BOC variants with a C-terminal FLAG tag were expressed in 293T cells as in B, and incubated at 4° C prior to Western blot analysis with anti-FLAG antibodies. * indicates full-length BOC, ** indicates a product likely formed by incomplete N-linked glycosylation (Kang et al., 2002), and *** indicates a proteolytic degradation product. The red arrow indicates the absence of the proteo-lytic degradation product with the p.Gly556Glu variant. This was seen reproducibly. E: Western blot analysis of BOC double mutant expression. 293T cells were transiently transfected with empty vector (vector), or vectors encoding the indicated variants. F: The Pro828Arg substitution, but not the Arg697Pro substution, prevents p.Gly556Glu from conferring SHH responsiveness to TKO cells. The Arg533Leu substitution also diminished p.Gly556Glu activity but fell short of statistical significance. Cultures were treated with 3 ng/ml SHH, and analysis of Gli1 expression was performed by qRT-PCR. *p<0.05; #p=0.06 with Student’s t-test. G: Sites of BOC variations mapped on Fn3 domain from 4U3H. Beta strand and loop topology are indicated in green and yellow, respectively. Sites of R553 and G556 are indicated by the red spheres.
Each variant was expressed at easily detectable levels when transiently transfected in HEK293T cells (Fig. 1B). When expressed in TKO cells subsequently treated with SHH, the p.Arg533Leu, p.Arg697Pro, and p.Pro828Arg variants were similar to wild type BOC in having little ability to promote ligand-dependent Gli1 expression (Fig. 1C). Surprisingly, the p.Gly556Glu variant had substantial activity in this assay, conferring on BOC the ability to promote SHH signaling. Therefore, p.Gly556Glu is a ligand-dependent, gain-of-function mutant. When lysates of cells expressing wild type BOC or the HPE-associated variants were analyzed by Western blotting after conditions of limited proteolysis, p.Gly556Glu did not produce a product formed by the others (Fig. 1D). This result suggests that p.Gly556Glu may have an altered conformation that contributes to its gain of function. A similar observation was made for some of the HPE-associated CDON variants that displayed a loss of function (Bae et al., 2011).
The ability of p.Gly556Glu to confer SHH-dependent Gli1 expression to TKO cells offered an opportunity to assess whether the other three HPE-associated variants might have the anticipated loss of function property by constructing BOC expression vectors that harbor the p.Gly556Glu change along with, independently, each of the other three. We reasoned that if these other variations crippled BOC’s ability to function as a SHH co-receptor, they would block Gli1 expression stimulated by p.Gly556Glu in the TKO cell assay. Each double variant (p.Gly556Glu/Arg533Leu, p.Gly556Glu/Arg697Pro, and p.Gly556Glu/Pro828Arg) was expressed at a similar level as p.Gly556Glu itself in transient transfectants (Fig. 1E). When assayed for the ability to promote SHH-dependent Gli1 expression, the p.Gly556Glu/Pro828Arg double variant lost the ability to do so, while p.Gly556Glu/Arg697Pro was still nearly as active as p.Gly556Glu (Fig. 1F). The p.Gly556Glu/Arg533Leu double variant resembled p.Gly556Glu/Pro828Arg in having diminished activity relative to p.Gly556Glu, but this fell just short of statistical significance (p = 0.06). We conclude that p.Pro828Arg is a loss-of-function variant in this context and p.Arg533Leu is likely to be one. Furthermore, we anticipate that this is so even in the context of otherwise wild type BOC, as would be the case in the individuals with HPE who harbored these variants. In contrast, p.Arg697Pro may be a benign variant, or it may affect BOC function in a way not revealed by this assay.
Arg533 and Gly556 are both within BOC Fn3 repeat 1 [Fn3(1)]. Although Fn3 repeats have considerable sequence variability, they adopt a similar three-dimensional fold. We aligned the sequence of Fn3(1) with the closest hit in the Protein Data Bank (http://www.rcsb.org/pdb/home/home.do), an engineered Fn3 domain (RCSB code 4U3H; 32% sequence identity) (Porebski et al., 2015). The sequence of Fn3(1) was then aligned with this structure to gain information on the locations of Arg533 and Gly556 (Fig. 1G). The Arg533 homolog in 4U3H is a surface exposed serine. Substituting a hydrophobic leucine for the larger, charged arginine at such a position could therefore potentially disrupt interactions with binding partners mediated by that surface. The Gly556 equivalent maps to a position just after a conserved beta hairpin connecting two beta strands. This glycine is conserved in many Fn repeats, and its backbone phi is 138°, which is unfavorable for non-glycine residues. Substituting the larger, charged glutamate in this position is likely to cause local disruptions of this loop. We note that this loop (called the F-G loop) is where the RGD sequence of the Fn3(10) repeat in fibronectin resides and that this sequence is essential for fibronectin to bind integrin receptors (Dickinson et al., 1994). Mapping the sites of the p.Arg533Leu and p.Gly556Glu variants on additional closely related Fn3 domain structures gave very similar results as for the Fn3 domain from the PDB entry 4U3H (data not shown). Importantly, the p.Arg533Leu and p.Gly556Glu variants are distant from one another, on opposite sides of the molecule. It therefore seems unlikely that they are disrupting the same interaction. This is consistent with the notion that p.Arg533Leu would be a function-perturbing variant for BOC generally, not only in the context of the gain-of-function p.Gly556Glu variant.
The identification of silent modifier genes that can influence the severity of developmental defects is uniquely challenging (Kousi and Katsanis, 2015). In our consideration(s) of such potential modifiers, we are referring to variant molecules that can occur at any allele frequency and whose biological effects are contingent on a specific genetic driver or environmental insult with which they interact. Occasionally, variants with the ability to synergize in a biological process may co-occur and functionally interact. Such instances would qualify as direct gene:gene interaction partners (Hong et al., 2016). It is these context-dependent interactions that are the most difficult to predict using current bioinformatic analysis tools. In fact, our retrospective analysis of currently used bioinformatics tools demonstrated that the ACMG guidelines are unhelpful in the autosomal dominant with modifier model that we think best explains HPE; none of the three BOC variants with perturbed function were predicted to be so by ACMG (Table 1). While these annotations are generally useful in identifying strong genetic drivers, the modifier category is not a consideration. For example, the BP5 category is allowed for variants that co-occur with a strong driver mutation as an indication that the variant should be considered as benign (Richards et al., 2015). Similarly, although most genetic drivers for HPE (or similar malformation syndromes) are both novel and clearly damaging, the effects of genetic variation at additional loci cannot be disregarded (Mouden et al., 2016). Given the complexity of SHH signal reception and transduction, it should not be surprising that bioinformatics predictions that relate to only one molecule at a time, to the exclusion of other co-morbid variants, may be unsatisfactory. In some important respects, all of the bioinformatics predictions were either imprecise, ambiguous, or misleading when applied to the variants tested in this study (Table 1).
The incomplete penetrance and wide phenotypic spectrum that characterize HPE can be commonly seen even in pedigrees in which bona fide deleterious mutations in SHH or SIX3 are inherited (Solomon et al., 2012; Solomon et al., 2009). This strongly suggests that modifiers, genetic and/or environmental, play a role in the clinical outcome (Roessler et al., 2012). Our findings provide strong support for BOC as a modifier gene in HPE. First, studies with mice support this role. Loss of Boc in mice does not result in HPE, but its genetic removal enhances the phenotypic severity of the HPE seen in Cdon and Gas1 mutant mice (Seppala et al., 2014; Zhang et al., 2011). Furthermore, in the case of Cdon mutants, Boc’s effects are gene-dosage sensitive (Zhang et al., 2011). Mice have proven to be a good model for HPE. Mice with targeted mutations in virtually all the genes thought to be drivers of human HPE also have HPE, including Shh, Six3, Zic2, TGIF, Gli2, Cdon, and Gas1 mutants (Geng and Oliver, 2009). Therefore, HPE-associated variants in BOC are likely to act on an essential substrate, either genetic or teratogenic, in the causation of HPE.
Second, functional analyses of HPE-associated BOC variants are most consistent with a modifier role. In model systems, loss of SHH signaling during early midline patterning is causally associated with HPE (Chen, 2016; Geng et al., 2016; Hong and Krauss, 2013; Roessler and Muenke, 2010). Furthermore, human HPE-associated heterozygous mutations in SHH pathway genes virtually always show a loss of function in cell- or model organism-based assays (Geng and Oliver, 2009; Roessler and Muenke, 2010). Therefore, in HPE patients where these mutations are found, they are extremely likely to be the genetic drivers on which modifiers act. In contrast, modifier alleles can, by definition, encode phenotypic suppressors or enhancers (Kousi and Katsanis, 2015). Strikingly, the p.Gly556Glu BOC variant identified here displayed a gain-of-function in SHH signaling. This is the opposite of what would be expected in causation of HPE, but is consistent with the notion that it may have served as a protective modifier allele in the patient carrying it. Approximately 97% of HPE cases succumb in utero and are lost to follow-up analysis. We speculate that this patient may have survived long enough for sequence analysis due to p.Gly556Glu acting as a phenotypic suppressor of an unidentified genetic or environmental substrate that caused a loss-of-function defect in SHH signaling. Similar effects have been demonstrated in mouse models of HPE, in which genetic removal of one copy of the negative SHH pathway regulator, Ptch1, largely restored normal rostroventral midline patterning (Geng et al., 2016; Hong and Krauss, 2013). Two other BOC variants identified in HPE patients displayed a loss-of-function property that is consistent with their acting more conventionally, i.e., as phenotypic enhancers of a primary defect that acted, directly or indirectly, to depress SHH signaling. Consistent with this notion, the patient with the p.Arg533Leu variant also had a mutation in ZIC2, and one patient with a p.Pro828Arg variant also had a deletion of TGIF (Table 1). Therefore, as predicted for a modifier gene, we have identified BOC variants that show either gains or losses of function in patients with HPE. As discussed above, the algorithms for predicting perturbation of function remain imperfect, but even as they improve, it will be extremely difficult to predict whether a specific variant may have an unexpected type of perturbation in function. The p.Gly556Glu variant is a point in case, as its gain-of-function phenotype could only be identified through functional analysis.
As with many birth defects, understanding the multifactorial etiology of HPE has been difficult. Along with heterozygous loss-of-function mutations in a select group of genes and a poorly illuminated environmental component, our findings with BOC variants argue for a third category of variable: low frequency modifier genes, BOC being the first such factor identified.
Supplementary Material
Acknowledgments
Grant sponsors: This work was supported by grants NIH DE024748, GM118751, CA198074; Cancer Prevention and Research Institute of Texas (CPRIT) award RR160023; and by the Division of Intramural Research, NHGRI, NIH.
Footnotes
DISCLOSURE STATEMENT
No authors of this manuscript have a conflict of interest to declare.
References
- Allen BL, Song JY, Izzi L, Althaus IW, Kang JS, Charron F, Krauss RS, McMahon AP. Overlapping roles and collective requirement for the co-receptors Gas1, Cdo and Boc in Shh pathway function. Dev Cell. 2011;20:775–787. doi: 10.1016/j.devcel.2011.04.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allen BL, Tenzen T, McMahon AP. The Hedgehog-binding proteins Gas1 and Cdo cooperate to positively regulate Shh signaling during mouse development. Genes Dev. 2007;21:1244–1257. doi: 10.1101/gad.1543607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bae GU, Domené S, Roessler E, Schachter K, Kang JS, Muenke M, Krauss RS. Mutations in CDON, Encoding a Hedgehog Receptor, Result in Holoprosencephaly and Defective Interactions with Other Hedgehog Receptors. Am J Hum Genet. 2011;89:231–240. doi: 10.1016/j.ajhg.2011.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen JK. I only have eye for ewe: the discovery of cyclopamine and development of Hedgehog pathway-targeting drugs. Nat Prod Rep. 2016;33:595–601. doi: 10.1039/c5np00153f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dickinson CD, Veerapandian B, Dai XP, Hamlin RC, Xuong NH, Ruoslahti E, Ely KR. Crystal structure of the tenth type III cell adhesion module of human fibronectin. J Mol Biol. 1994;236:1079–1092. doi: 10.1016/0022-2836(94)90013-2. [DOI] [PubMed] [Google Scholar]
- Geng X, Acosta S, Lagutin O, Gil HJ, Oliver G. Six3 dosage mediates the pathogenesis of holoprosencephaly. Development. 2016;143:4462–4473. doi: 10.1242/dev.132142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geng X, Oliver G. Pathogenesis of holoprosencephaly. J Clin Invest. 2009;119:1403–1413. doi: 10.1172/JCI38937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hong M, Krauss RS. Cdon mutation and fetal ethanol exposure synergize to produce midline signaling defects and holoprosencephaly spectrum disorders in mice. PLOS Genet. 2012;8:e1002999. doi: 10.1371/journal.pgen.1002999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hong M, Krauss RS. Rescue of Holoprosencephaly in Fetal Alcohol-Exposed Cdon Mutant Mice by Reduced Gene Dosage of Ptch1. PLOS ONE. 2013;8:e79269. doi: 10.1371/journal.pone.0079269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hong S, Hu P, Marino J, Hufnagel SB, Hopkin RJ, Toromanovic A, Richieri-Costa A, Ribeiro-Bicudo LA, Kruszka P, Roessler E, Muenke M. Dominant-negative kinase domain mutations in FGFR1 can explain the clinical severity of Hartsfield syndrome. Hum Mol Genet. 2016;25:1912–1922. doi: 10.1093/hmg/ddw064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Izzi L, Lévesque M, Morin S, Laniel D, Wilkes BC, Mille F, Krauss RS, McMahon AP, Allen BL, Charron F. Boc and Gas1 each form distinct Shh receptor complexes with Ptch1 and are required for Shh-mediated cell proliferation. Dev Cell. 2011;20:788–801. doi: 10.1016/j.devcel.2011.04.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang JS, Mulieri PJ, Hu Y, Taliana L, Krauss RS. BOC, an Ig superfamily member, associates with CDO to positively regulate myogenic differentiation. EMBO J. 2002;21:114–124. doi: 10.1093/emboj/21.1.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kauvar EF, Hu P, Pineda-Alvarez DE, Solomon BD, Dutra A, Pak E, Blessing B, Proud V, Shanske AL, Stevens CA, Rosenfeld JA, Shaffer LG, Roessler E, Muenke M. Minimal evidence for a direct involvement of twisted gastrulation homolog 1 (TWSG1) gene in human holoprosencephaly. Mol Genet Metab. 2011;102:470–480. doi: 10.1016/j.ymgme.2010.12.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kavran JM, Ward MD, Oladosu OO, Mulepati S, Leahy DJ. All Mammalian Hedgehog Proteins Interact with Cell Adhesion Molecule, Down-regulated by Oncogenes (CDO) and Brother of CDO (BOC) in a Conserved Manner. J Biol Chem. 2010;285:24584–24590. doi: 10.1074/jbc.M110.131680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kousi M, Katsanis N. Genetic Modifiers and Oligogenic Inheritance. Cold Spring Harb Perspect Med. 2015;5:a017145. doi: 10.1101/cshperspect.a017145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krauss RS, Hong M. Gene–Environment Interactions and the Etiology of Birth Defects. Curr Top Dev Biol. 2016;116:569–580. doi: 10.1016/bs.ctdb.2015.12.010. [DOI] [PubMed] [Google Scholar]
- Lee RTH, Zhao Z, Ingham PW. Hedgehog signalling. Development. 2016;143:367–372. doi: 10.1242/dev.120154. [DOI] [PubMed] [Google Scholar]
- Leoncini E, Baranello G, Orioli IM, Annerén G, Bakker M, Bianchi F, Bower C, Canfield MA, Castilla EE, Cocchi G, Correa A, De Vigan C, Doray B, Feldkamp ML, Gatt M, Irgens LM, Lowry RB, Maraschini A, McDonnell R, Morgan M, Mutchinick O, Poetzsch S, Riley M, Ritvanen A, Gnansia ER, Scarano G, Sipek A, Tenconi R, Mastroiacovo P. Frequency of holoprosencephaly in the International Clearinghouse Birth Defects Surveillance Systems: searching for population variations. Birth Defects Res A Clin Mol Teratol. 2008;82:585–591. doi: 10.1002/bdra.20479. [DOI] [PubMed] [Google Scholar]
- Martinelli DC, Fan CM. Gas1 extends the range of Hedgehog action by facilitating its signaling. Genes Dev. 2007;21:1231–1243. doi: 10.1101/gad.1546307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mathew E, Zhang Y, Holtz Alexander M, Kane Kevin T, Song Jane Y, Allen Benjamin L, Pasca di Magliano M. Dosage-Dependent Regulation of Pancreatic Cancer Growth and Angiogenesis by Hedgehog Signaling. Cell Reports. 2014;9:484–494. doi: 10.1016/j.celrep.2014.09.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mouden C, Dubourg C, Carre W, Rose S, Quelin C, Akloul L, Hamdi-Roze H, Viot G, Salhi H, Darnault P, Odent S, Dupe V, David V. Complex mode of inheritance in holoprosencephaly revealed by whole exome sequencing. Clin Genet. 2016;89:659–668. doi: 10.1111/cge.12722. [DOI] [PubMed] [Google Scholar]
- Pineda-Alvarez DE, Roessler E, Hu P, Srivastava K, Solomon BD, Siple CE, Fan CM, Muenke M. Missense substitutions in the GAS1 protein present in holoprosencephaly patients reduce the affinity for its ligand, SHH. Hum Genet. 2012;131:301–310. doi: 10.1007/s00439-011-1078-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Porebski BT, Nickson AA, Hoke DE, Hunter MR, Zhu L, McGowan S, Webb GI, Buckle AM. Structural and dynamic properties that govern the stability of an engineered fibronectin type III domain. Protein Eng Des Sel. 2015;28:67–78. doi: 10.1093/protein/gzv002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ribeiro LA, Quiezi RG, Nascimento A, Bertolacini CP, Richieri-Costa A. Holoprosencephaly and holoprosencephaly-like phenotype and GAS1 DNA sequence changes: Report of four Brazilian patients. Am J Med Genet A. 2010;152A:1688–1694. doi: 10.1002/ajmg.a.33466. [DOI] [PubMed] [Google Scholar]
- Richards S, Aziz N, Bale S, Bick D, Das S, Gastier-Foster J, Grody WW, Hegde M, Lyon E, Spector E, Voelkerding K, Rehm HL ACMG Laboratory Quality Assurance Committee. Standards and guidelines for the interpretation of sequence variants: a joint consensus recommendation of the American College of Medical Genetics and Genomics and the Association for Molecular Pathology. Genet Med. 2015;17:405–424. doi: 10.1038/gim.2015.30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roessler E, Muenke M. The molecular genetics of holoprosencephaly. Am J Med Genet C Semin Med Genet. 2010;154C:52–61. doi: 10.1002/ajmg.c.30236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roessler E, Vélez JI, Zhou N, Muenke M. Utilizing prospective sequence analysis of SHH, ZIC2, SIX3 and TGIF in holoprosencephaly probands to describe the parameters limiting the observed frequency of mutant gene×gene interactions. Mol Genet Metab. 2012;105:658–664. doi: 10.1016/j.ymgme.2012.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seppala M, Depew MJ, Martinelli DC, Fan CM, Sharpe PT, Cobourne MT. Gas1 is a modifier for holoprosencephaly and genetically interacts with sonic hedgehog. J Clin Invest. 2007;117:1575–1584. doi: 10.1172/JCI32032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seppala M, Xavier GM, Fan CM, Cobourne MT. Boc modifies the spectrum of holoprosencephaly in the absence of Gas1 function. Biol Open. 2014;3:728–740. doi: 10.1242/bio.20147989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Solomon B, Bear K, Wyllie A, Keaton A, Dubourg C, David V, Mercier S, Odent S, Hehr U, Paulussen A, Clegg NJ, Delgado MR, Bale SJ, Lacbawan F, Ardinger HH, Aylsworth AS, Bhengu NL, Braddock S, Brookhyser K, Burton B, Gaspar H, Grix A, Horovitz D, Kanetzke E, Kayserili H, Lev D, Nikkel SM, Norton M, Roberts R, Saal H, Schaefer GB, Schneider A, Smith EK, Sowry E, Spence MA, Shalev SA, Steiner CE, Thompson EM, Winder TL, Balog JZ, Hadley DW, Zhou N, Pineda-Alvarez DE, Roessler E, Muenke M. Genotypic and phenotypic analysis of 396 individuals with mutations in Sonic Hedgehog. Journal of Medical Genetics. 2012;49:473–479. doi: 10.1136/jmedgenet-2012-101008. [DOI] [PubMed] [Google Scholar]
- Solomon BD, Lacbawan F, Jain M, Domene S, Roessler E, Moore C, Dobyns WB, Muenke M. A novel SIX3 mutation segregates with holoprosencephaly in a large family. Am J Med Genet A. 2009;149A:919–925. doi: 10.1002/ajmg.a.32813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song JY, Holtz AM, Pinskey JM, Allen BL. Distinct structural requirements for CDON and BOC in the promotion of Hedgehog signaling. Dev Bio. 2015;402:239–252. doi: 10.1016/j.ydbio.2015.03.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tenzen T, Allen BL, Cole F, Kang JS, Krauss RS, McMahon AP. The cell surface membrane proteins Cdo and Boc are components and targets of the hedgehog signaling pathway and feedback network in mice. Dev Cell. 2006;10:647–656. doi: 10.1016/j.devcel.2006.04.004. [DOI] [PubMed] [Google Scholar]
- Zhang W, Hong M, Bae GU, Kang JS, Krauss RS. Boc modifies the holoprosencephaly spectrum of Cdo mutant mice. Dis Model Mech. 2011;4:368–380. doi: 10.1242/dmm.005744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang W, Kang JS, Cole F, Yi MJ, Krauss RS. Cdo functions at multiple points in the Sonic Hedgehog pathway, and Cdo-deficient mice accurately model human holoprosencephaly. Dev Cell. 2006;10:657–665. doi: 10.1016/j.devcel.2006.04.005. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.