Abstract
Background
Cutaneous leishmaniasis (CL) is an important health problem in the New World affecting civilian and military populations that are frequently exposed in endemic settings. The Peruvian region of Madre de Dios located near the border with Brazil is one of the most endemic CL regions in South America with more than 4,451 reported cases between 2010 and 2015 according to the Peruvian epidemiology directorate. However, little is known regarding the diversity and distribution of sand fly vectors in this region. In this study, we aimed to characterize the sand fly fauna in this endemic setting and identify sand fly species naturally infected with Leishmania possibly involved in pathogen transmission.
Methods
Sand fly collections were carried out during 2014 and 2015 in the communities of Flor de Acre, Villa Primavera, Mavila and Arca Pacahuara using CDC light traps and Shannon traps. Collected specimens were identified and non-blood-fed females were selected for Leishmania infection screening using kinetoplastid DNA-PCR (kDNA-PCR) and nested Real time PCR for species identification.
Results
A total of 10,897 phlebotomines belonging to the genus Lutzomyia (58 species) and Brumptomyia (2 species) were collected. Our study confirmed the widespread distribution and abundance of Lutzomyia (Trichophoromyia) spp. (24%), Lu. whitmani (19.4%) and Lu. yucumensis (15.8%) in the region. Analysis of Shannon diversity index indicates variability in sand fly composition across sites with Villa Primavera presenting the highest sand fly diversity and abundance. Leishmania screening by kDNA-PCR resulted in 45 positive pools collected from Flor de Acre (34 pools), Mavila (10 pools) and Arca Pacahuara (1 pool) and included 14 species: Lu. yucumensis, Lu. aragoi, Lu. sallesi, Lu. sherlocki, Lu. shawi, Lu. walkeri, Lu nevesi, Lu. migonei, Lu. davisi, Lu. carrerai, Lu. hirsuta, Lu. (Trichophoromyia) spp., Lu. llanosmartinsi and Lu. whitmani. Lutzomyia sherlocki, Lu. walkeri and Lu. llanosmartinsi had the highest infection rates (8%, 7% and 6%, respectively). We identified Leishmania guyanensis in two Lu. whitmani pools, and L. braziliensis in two Lu. llanosmartinsi pools and one Lu. davisi pool.
Conclusions
Based on our collections there is high sand fly diversity in Madre de Dios, with differences in sand fly abundance and species composition across sites. We identified 14 sand fly species naturally infected with Leishmania spp., having detected natural infection with L. (V.) guyanensis and L. (V.) braziliensis in three sand fly species. These results suggest the presence of several potential vectors that vary in their spatial and geographical distribution, which could explain the high prevalence of CL cases in this region.
Author summary
Leishmaniasis is a neglected disease that affects more than 2 million people worldwide. The identification of putative Leishmania vectors is an important step towards the design of better control strategies and estimating the risk of transmission in endemic areas. In this paper the authors explored the distribution of sand flies and identified potential vectors in a largely unexplored setting in the Southeastern Peruvian Amazon Basin. Three new sand fly species Lutzomyia naiffi, Lu. dereuri and Lu. flabellata are reported for Peru. In addition, they found fourteen sand fly species naturally infected with Leishmania that comprised seven new reports for Peru and one for the Americas. This information will serve as a baseline for future surveillance and intervention studies in this highly endemic area.
Introduction
Leishmaniasis is a group of neglected tropical diseases caused by digenic protozoan of the genus Leishmania. This disease affects over 12 million people in more than 98 countries worldwide and causes more than 1.5 million cutaneous leishmaniasis (CL) new cases per year [1]. Leishmania parasites cause a wide spectrum of clinical manifestations that are divided into cutaneous, mucosal (ML) and visceral leishmaniasis (VL) [2, 3]. In the New World, CL is the most common clinical form of disease [1] leading to disfigurement, functional impairment and stigma in affected patients. CL is mainly caused by species of the Viannia subgenus, including L. (Viannia) braziliensis, L. (V.) peruviana, L. (V.) guyanensis and L. (V.) panamensis [4, 5] that are widely distributed in the Amazonian region.
The transmission cycle of leishmaniasis is highly dependent on the interaction of the sand fly vector and the mammalian host. In the New World, transmission occurs by the bite of infected phlebotomine sand flies of the genus Lutzomyia. Although, more than 500 sand fly species have been reported in the Americas, only 30 are known vectors of leishmaniasis [6, 7]. This evidence underscores the need to study the distribution and identification of possible Leishmania vectors.
Peru is among the ten countries that hold more than 75% of all CL infections worldwide [1] with 5,955 cases reported in 2015. The Amazonian region of Madre de Dios located near the border with Brazil is a highly endemic leishmaniasis area with an incidence rate of 9 cases per 10,000 person-years and contributing with up to 13% of all leishmaniasis cases in the country.
This high incidence in leishmaniasis cases in this and other Amazonian regions could be a result of the rich diversity of sand flies, leishmaniasis reservoirs and human driven activities like illegal mining, logging, and chestnut harvesting [8–10].
The presence of Leishmania infection in humans has been extensively documented in Madre de Dios with reports of L. (V.) braziliensis, L. (V.) lainsoni and L. (V.) guyanensis [5, 11–13]. However, information about Leishmania vectors, reservoirs, their role in disease transmission and the variables influencing their distribution is still limited [14–16]. For instance, a surveillance study using molecular methods for parasite identification, failed to detect Leishmania on more than 80 wild native rodents [16].
We conducted vector surveillance during 2014 and 2015 in different sites located in the region of Madre de Dios near the border with Brazil and employed molecular methods to identify natural Leishmania infections. Our results allowed us to characterize the dynamics of the sand fly populations and contributed to the understanding of pathogen transmission in the Southeastern Peruvian Amazon.
Methods
Ethics statement
This study (NAMRU6.2014.0007) was exempt from NAMRU-6 IRB review as this project did not involve humans as the subject of the study evaluation.
Therefore, this study did not meet the definition of research involving human subjects, and 32 CFR 219 does not apply. Sand fly collections were performed under approval from the General Directorate of Forestry and Wild Fauna from the Ministry of Agriculture and Irrigation of Peru (Resolución Directoral No. 0406-2013-MINAGRI-DGFFS/DGEFFS)
Study sites for 2014
In 2014, sand flies were collected in February, May and September in the community of Villa Primavera (11° 02' 33.5"S, 69° 34' 24.6"W, 295 m.a.s.l.), and in May and September in the community of Flor de Acre (11° 19' 54.3"S, 69° 36' 20.6"W, 292 m.a.s.l) (Fig 1).
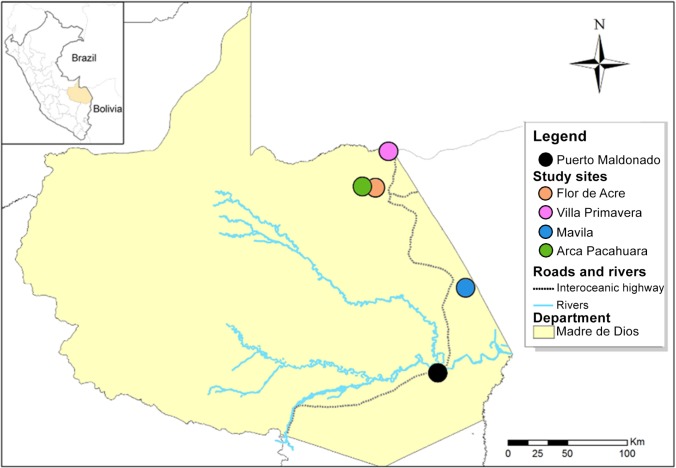
Fig 1. Map of the study area.
This figure illustrates the Peruvian region of Madre de Dios and its capital city (Puerto Maldonado) crossed by the interoceanic highway. Collection sites are showed as points in the map as colored circles.
Flor de Acre was selected as a study site based on previous evidence of Leishmania vectors [17] whereas Villa Primavera was chosen due to its location at 13 km from the Brazilian border constituting a stopover.
Study sites for 2015
In 2015, sand flies were collected in June and August in the communities of Flor de Acre, Arca Pacahuara (11° 19’ 57.6”S, 69° 36’ 53.8”W, 272 m.a.s.l.) and Mavila (11° 57’ 53.1”S, 69° 09’ 46.2”W, 206 m.a.s.l.) (Fig 1). The change in study sites was based on preliminary results from 2014 and reports of human CL in Arca Pacahuara and Mavila by the Peruvian Ministry of Health.
All study sites have a humid sub-tropical climate with annual temperatures between 19°C to 36°C with occasional low temperature periods that can reach 13°C. These sites have an annual precipitation of more than 3,000 mm that occurs mostly between January to April.
The sites are experiencing a rapid change of land use and deforestation due to illegal mining, agriculture, Brazil nut extraction, and livestock farming that are the major economic activities in the area.
Sand fly collections and morphological identification
At each site, sand flies were collected inside, immediately outside and in the surrounding area of four households using six CDC light traps per site. Collections were conducted for 12 hours per night (18:00–06:00) during five consecutive nights. In addition, Shannon traps were used for collections outside and in the areas surround the houses from 18:00 to 21:00.
Sand flies were transported in 70% ethanol to the Entomology Department at NAMRU-6 in Lima where they were identified using keys developed by Young and Duncan [18] and Galati [19]. Female specimens were processed using a modified protocol to allow molecular analysis [17]. Briefly, the head and the two last abdominal segments were separated and placed in lactophenol for two hours at room temperature. These regions contain key taxonomic structures (cibarium, palpomeres, flagellomeres and spermathecae) which are used for species identification. The remaining parts of the sand fly were preserved in absolute ethanol at -20°C for molecular biology.
Male specimens were placed in 20% KOH for 12 to 24 hours at room temperature. Then, specimens were clarified with lactophenol for two hours at room temperature. Male and female specimens were mounted permanently on Euparal.
DNA extraction and molecular detection of Leishmania
We selected non-blood-fed female sand flies that were pooled in sets of 1–10 specimens according to species, study site, household, collection date and trap type.
DNA from non-blood fed female sand flies was isolated using the DNeasy Blood & Tissue kit (QIAGEN) following the standard manufacturer’s protocol for isolation of insect genomic DNA.
In order to detect the presence of Lutzomyia DNA, we employed a PCR targeting the Lutzomyia 12S ribosomal DNA using primers T1B 5′-AAA CTA GGA TTA GAT ACC CT-3′ and T2A 5′-AAT GAG AGC GAC GGG CGA TGT-3′ as previously described [17, 20]. The reactions were prepared in 25 μL that contained1X Taq polymerase buffer (Invitrogen, Carlsbad, CA), 1.5 mM MgCl2, 125 μM dNTPs, 0.5 μM of each primer, 1 unit of Taq DNA polymerase (Invitrogen), and 5 μL of DNA sample. The PCR was run on a thermocycler under the following cycling conditions: initial denaturation at 94°C for 5 minutes followed by 35 cycles of denaturation 94°C for 20 sec, annealing at 56°C for 30 sec, and extension at 72°C for 25 sec; and a final extension step at 72°C for 5 min. This reaction generates a 360 bp product in the presence of sand fly DNA that serves to rule out the presence of PCR inhibitors in extracted DNA and as a positive control.
The presence of Leishmania DNA was detected by a PCR that targets the Leishmania minicircle [21–23]. This region is a high copy number DNA sequence present in the kinetoplast of Leishmania and other related protozoa that has been shown to be highly sensitive and specific [21, 23]. Reactions were carried out in 20 μL of PCR mixture containing 1X Taq polymerase buffer (Invitrogen, Carlsbad, CA), 1.5 mM MgCl2, 125 μM dNTPs, 0.5 μM of each primer, 1 unit of Taq DNA polymerase (Invitrogen), and 4 μL of DNA sample. The thermal cycling conditions consisted of an initial denaturation at 94°C for 5 min followed by 35 cycles of denaturation at 94°C for 45 sec, annealing at 58°C for 45 sec, and extension at 72°C for 60 sec; and a final extension step at 72°C for 5 min. This reaction generates a 120 bp amplification band that is considered positive for the Leishmania genus.
In order to detect the infecting Leishmania species on the kDNA positive samples we employed a FRET based nested Real Time PCR [24]. This method detects mutations on the 6PGD and MPI genes yielding different melting peaks according to the Leishmania species. For the first round of amplification, we prepared a 50μL reaction containing 1X Taq polymerase buffer (Invitrogen), 1.5 mM MgCl2, 200 μM dNTPs (Invitrogen), 0.8 μM or 1 μM of each primer (6PGD and MPI, respectively), 1.5 units of Taq DNA polymerase (Invitrogen), and 5 μL of DNA sample. The amplification setting consisted of an initial denaturation at 94°C for 5min followed by 35 cycles of denaturation at 94°C for 45 sec, annealing at 57°C (for MPI) or 62°C (for 6PGD) for 45 sec, and extension at 72°C for 90 sec; and a final extension at 72°C for 7 min for MPI or 5 min for 6PGD.
The second amplification round was performed on a 20μL reaction for each gene containing 1X LightCycler 480 Genotyping Master (Roche, Indianapolis, IN), 1.25 μM of forward primer, 0.25 μM of reverse primer, 0.75 μM of anchor and sensor probes, and 5 μL of PCR product from the first reaction.
The amplification setting was performed on a LightCycler 480 and consisted of an initial denaturation at 95°C for 5 min followed by 45 cycles of denaturation at 95°C for 10 sec, annealing at 60°C for 20 sec under a single acquisition step) and extension at 72°C for 20 sec. A melting curve analysis was performed at the end of the amplification cycles by heating the amplicons at 95°C for 10 sec, cooling at 50°C for 59 sec and then gradually increasing the temperature to 80°C with one acquisition step each °C.
Melting curves were analyzed using the LightCycler 480 Software Version 1.0 as previously described [24].
Data analysis
We calculated the Shannon-Wiener (H) diversity index in PAST v3.12[25] for each study site using the equations , where “p” represents the proportion in which each species “n” was collected (∑p = 1). The Hutcheson t-test was employed to assess the statistical significance of differences in Shannon diversity indexes between study sites [26].
Species abundance was calculated in Excel 2010 (Microsoft) using the Index of Species Abundance (ISA) [27] using the formula ISA = (a + Rj)/k. Briefly, for each site we established a rank of abundance from 1 (the species with the highest value) to the number of species collected (leaving in blank species not represented in the site or using the average for ties between 2 or more species). Then, we calculated “a” as the number of zero observations for each species in all sites multiplied “c” which is the single largest rank in all the data set plus 1. The value of Rj corresponds to the sum of ranks for a given species in all the sites whereas “k” corresponds to the number of sites. The resulting ISA values were converted into the Standardized Index of Species Abundance (SISA) using the formula SISA = (c − ISA)/(c − 1).
Results
During the two years of the study, we collected 10,897 sand flies belonging to the genus Lutzomyia (10,800 specimens, 99.1%) and Brumptomyia (97 specimens, 0.9%). The majority of specimens were collected in the areas surrounding houses (76.57%), followed by primary forest patches (14.44%) and inside of the houses (8.99%).
Overall, we identified 58 Lutzomyia species and two Brumptomyia species with three new sand fly reports for Peru; Lu. naiffi, Lu. dereuri and Lu. flabellata that were collected in Flor de Acre (S1 and S2 Tables).
We were not able to identify the female specimens of the subgenus Trichophoromyia up to the species level due to the high similarity among females of this subgenus and the fact that we detected five distinct Lutzomyia (Trichophoromyia) species based on male morphology (Lu. auraensis, Lu. loretonensis, Lu. clitella, Lu. nemorosa and Lu. ubiquitalis). The same situation occurs for the subgenus Pressatia with three distinct species (Lu. choti, Lu. calcarata and Lu. triacantha).
Lutzomyia (Trichophoromyia) auraensis (male specimens), Lutzomyia (Trichophoromyia) spp and Lu. davisi were the most abundant species in the study sites (SISA = 0.98, 0.95, 0.93 for 2014 and 0.99, 0.97 and 0.95 for 2015, respectively). These species are suspected vectors of leishmaniasis and may have an important role in its transmission in the area.
Sand fly collections of 2014
In February 2014, we collected 311 specimens in Villa Primavera with Lu. (Trichophoromyia) spp. (n = 239, 76.8%), Lu. davisi (n = 20, 6.43%) and Lu. shawi (n = 10, 3.22%) as the most abundant species out of 24 recorded (S1 Table). Collections were not performed in Flor de Acre during this month due to extreme weather conditions in the area.
In May 2014, we collected 4,629 specimens, 4.99% in Villa Primavera and 95.01% in Flor de Acre (S2 Table). The most abundant species on this collection were Lu. (Trichophoromyia) spp (n = 156, 67.5%) and Lu. davisi (n = 33, 14.2%) out of 24 species recorded in Villa Primavera; and Lu. yucumensis (n = 1,564; 35.6%) and Lu. (Trichophoromyia) spp (n = 2,158; 49.1%) out of 18 species recorded in Flor de Acre.
In September 2014, the number of collected specimens was lower than May with 2,643 specimens, 2.72% in Villa Primavera and 97.28% in Flor de Acre. The most abundant species were Lu. aragoi (n = 17, 23.6%) in Villa Primavera and Lu. whitmani (n = 2,033; 79.07%) in Flor de Acre, out of 16 and 34 species recorded, respectively.
Flor de Acre had a higher phlebotomine species richness (44 species in 5,999 specimens) than Villa Primavera (36 species in 410 specimens). However, Villa Primavera presented a significantly (p<0.05) higher Shannon diversity index (H = 2.15±0.16) than Flor de Acre (H = 1.72±0.31) due differences in the number of collected specimens.
Sand fly collections of 2015
In June 2015, we collected 2,948 specimens; 18.69% in Arca Pacahuara, 16.82% in Flor de Acre and 64.48% in Mavila. Out of 9 species recorded, Lu. (Trichophoromyia) spp stood out as the most predominant species on the three sites (S2 Table).
In August 2015 we collected 366 specimens, 18.58% in Arca Pacahuara (9 species), 18.03% in Flor de Acre (16 species) and 63.39% in Mavila (21 species). This low number of specimens could be the result of a drop in rainfall from June towards September.
Out of all species recorded, the most predominant species were Lu. (Trichophoromyia) spp in Arca Pacahuara (n = 53, 77.9%), Lu. whitmani in Flor de Acre (n = 16, 24.24%) and Lu. davisi (n = 56, 24.14%) in Mavila (S2 Table).
Mavila presented a higher species richness (33 species in 1,240 specimens) than Flor de Acre (22 species in 461 specimens) and Arca Pacahuara (13 species in 346 specimens). However, Flor de Acre had a significantly (p<0.05) higher Shannon diversity index (H = 1.79±0.12) than Arca Pacahuara (H = 0.69±0.14) and Mavila (H = 1.62±0.08).
Sand flies naturally infected with Leishmania
Female sand fly specimens were grouped into 850 pools based on species; trap type, month of collection and site. The 12S ribosomal DNA PCR confirmed the presence of sand fly DNA on all samples and served as an internal control for DNA quality by ruling out the presence of PCR inhibitors.
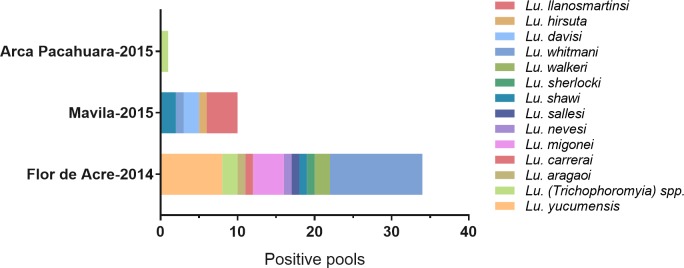
Leishmania specific kinetoplast PCR detected parasite DNA on 45 pools from 14 different Lutzomyia species (S3 Table, Fig 2). Pools collected in Flor de Acre in 2014 and Mavila in 2015 accounted for 75.6% (n = 34) and 22.2% (n = 10) of all positives.
Fig 2. Positive pools detected by kDNA PCR.
The figure shows that Mavila and Flor de Acre presented the highest number of positive pools. These pools belonged to up to 14 different Lutzomyia species.
The group of sand fly species where we detected natural Leishmania infections in our study accounted for nearly 70% of all phlebotomines collected, and within this group Lu. (Trichophoromyia) spp, Lu. whitmani, Lu. yucumensis and Lu. davisi stood out as the predominant species (Fig 2).
The estimated minimum infection rates in 2014 for Lu. whitmani, Lu. yucumensis, Lu. (Trichophoromyia) spp. and Lu. carrerai carrerai were 1.09, 0.97%, 0.22% and 0.81%, respectively. In 2015, the minimum infection rates Lu. (Trichophoromyia) spp and Lu. davisi were 0.21% and 0.29%, respectively (S4 Table).
FRET-based Real-Time PCR was employed to identify the species of Leishmania present in the kDNA-positive pools. This assay confirmed the presence of Leishmania (Viannia) guyanensis in two pools of Lu. whitmani collected from Flor de Acre (S3 Table). Additionally, we detected L. braziliensis in two pools of Lu. llanosmartinsi and on one pool of Lu. davisi collected from Mavila in 2015 (S3 Table). We could not identify the Leishmania species in the remaining kDNA-positives due to lack of detectable amplification product on the Real-Time PCR assay.
To assess the relation of potential vector versus non-vector species we estimated their ratio for each locality. Our results indicate that Flor de Acre presented the highest potential vector versus non-vector ratio (2.3:1) (Table 1).
Table 1. Ratio of vector versus non-vector species per site (V:N).
The table shows the number of different vector species collected at each study location, the ratio versus non-vector species and the predominant vector found at each site.
| Study site | Year | Ratio (V:N) | Vector species | Collected vectors (%) | Predominant vector species (♀>100) |
|---|---|---|---|---|---|
| Flor de Acre | 2014 | 2.3:1 | 14 | 69.9% | Lu. whitmani, Lu. yucumensis, Lu. auraensis, Lu. (Trichophoromyia) spp, Lu. davisi, Lu. carrerai, Lu. migonei |
| 2015 | 1.9:1 | 12 | 66.2% | Lu. auraensis, Lu. davisi | |
| Arca Pacahuara | 2015 | 0.62:1 | 8 | 38.3% | Lu. auraensis, Lu. (Trichophoromyia) spp. |
| Mavila | 2015 | 0.7:1 | 10 | 40.5% | Lu. auraensis, Lu. (Trichophoromyia) spp, Lu. davisi |
| Villa Primavera | 2014 | 0.7:1 | 10 | 39.8% | Lu. (Trichophoromyia) spp, Lu. davisi |
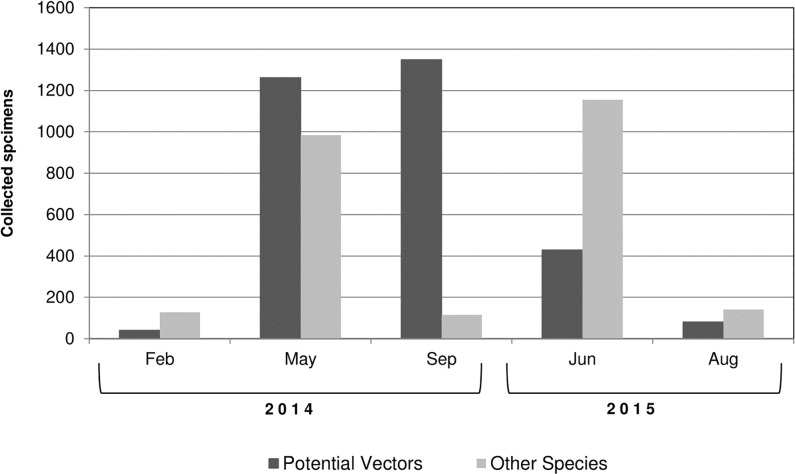
The majority of phlebotomines were collected in May, September of 2014 and June 2015 (42.48%, 24.25% and 27.05%, respectively). In the other hand, a lower sand fly density was found during February and August (2.85% and 3.36% of the total, respectively). Putative Leishmania vectors were overrepresented in May, September and June (Fig 3).
Fig 3. Relative abundance of sand fly specimens.
This figure shows changes in sand fly abundance of putative vectors and non-vectors by month of collection.
Discussion
The Peruvian region of Madre de Dios is an important focus of CL in Peru with multiple Leishmania species reported in humans [5]. This area has been experiencing a dramatic change in land use due to the increase of agriculture, logging and economic activities that are related to the trans-oceanic highway that connects Peru, Brazil and Bolivia. This economic boost has led to the appearance of new communities along this highway such as Arca Pacahura and Mavila that are mainly formed by immigrants from nearby Peruvian regions (Cusco, Puno and Arequipa) [28].
It is known that leishmaniasis is highest among people living near to forest edges or working in forested areas [29]. In this sense, the colonization of previously forested areas has resulted in the rapid emergence and spread of CL cases in Madre de Dios placing this disease as an important public health problem.
The study sites have environmental variables that favored a similar sand fly biodiversity to those found in low latitude areas of Central America and South America where 1 ha of forest can contain up to 50 different species [30]. The number and recorded species collected in our study support this high biodiversity which is similar to previous reports from the region of Acre on the Brazilian side of the Peru-Brazil border [9, 10, 31].
Our results show that there is important variation in the number of species across sites and time of collection (S1 Table). However, members of the Trichophoromyia and Psychodopygus subgenus are consistently abundant in all study sites according to the SISA estimates (S1 and S2 Tables).
Differences in abundance and diversity between study sites could result in different risks of Leishmania transmission. In this regard, our results of 2014 showed the extent of this variation with up to 7,000 sand flies collected in Flor de Acre versus only 600 in Villa Primavera. On the same year, Lu. whitmani was the predominant species on Flor de Acre accounting with 30% of all collected individuals whereas in Villa Primavera it represented only 0.8% of all collected sand flies.
This difference in collected specimens could be a result of the different degrees of deforestation in the two sites. Villa Primavera is a small village on a highly deforested area near the transoceanic highway while Flor de Acre is located far from the highway and surrounded by a primary forest.
Interestingly, the sites studied in 2015 presented a different sand fly composition from the ones in 2014. In these areas, Lu. (Trichophoromyia) spp. accounted for the majority of specimens collected (90% in Arca Pacahuara, 73% in Mavila and 46% in Flor de Acre). Among these sites, Mavila accounted for the majority of collected sand flies (2,133 versus 619 in Arca Pacahuara and 562 in Flor de Acre). The presence of primary forested areas appeared to be related to these differences given the location of Mavila in the deep jungle.
Our study has also shown that abundance and sand fly diversity can vary in the same area, potentially complicating control activities due to differences in sand fly behavior. In this regard, Lu. yucumensis and Lu. (Trichophoromyia) spp were the most abundant species in Flor de Acre in May 2014. However, their abundance decreased towards September 2014 and Lu. whitmani replaced them as the dominant species.
Regardless of these variations, the most abundant species at each site are suspected putative Leishmania vectors with confirmed PCR infection (Lu. (Trichophoromyia) spp., Lu. whitmani, Lu. yucumensis, and Lu. davisi). However, less prevalent species presented higher minimum infection rates such as Lu. sallesi/cortelezzi (14.29%), Lu. walkeri (12.5%) and Lu. sherlocki (9%) (S4 Table).
The abundance of the subgenera Trichophoromyia, Psychodopygus and Nyssomyia is consistent with previous studies conducted in Peru and Brazil [10, 17, 31] and indicates that species from these subgenera are predominant in the Peruvian and Brazilian Amazon Basins and could play and important role in leishmaniasis transmission in the region.
In terms of natural Leishmania infection, our study has shown that the proportion of infected sand flies without considering Lutzomyia species is not statistically different across sites according to the Fisher exact test. This suggests that variations in Leishmania transmission at each site will likely depend on the behavior and vector competence of the predominant species rather than differences in the prevalence of Leishmania.
Infection with Leishmania (V.) guyanensis in Lu. whitmani and L. (V.) braziliensis in Lu. llanosmartinsi and Lu. davisi suggests a role for these species in leishmaniasis transmission in Peru and Brazil [31]. This finding is further supported by the isolation of these two Leishmania species from tissue biopsies from patients with CL in Madre de Dios [5, 12, 13].
Our minimum infection rates for Lu. davisi and Lu. (Trichophoromyia) spp. are similar to the ones obtained in a previous study conducted in 2010 on Flor de Acre [17] suggesting that the composition of potential vectors has remained constant in this area. However, our infection rates differ from other studies conducted in the neighboring Brazilian state of Acre. In this state, Lu. davisi and Lu. (Trichophoromyia) spp. presented higher infection rates whereas Lu. whitmani presented a lower infection rate than in our study (1.84, 2.05 and 0.5%, respectively) [10, 31].
It is important to note that Lu. whitmani has been suggested as one of the most important vectors of CL in various regions of Brazil [32]. This species is highly anthropophilic and has been frequently found in areas undergoing deforestation. Previous epidemiological assessments from deforested areas indicate that transmission of leishmaniasis relies mainly on this species [32–34].
This is the first report of natural Leishmania infection in Lu. sherlocki and the first report of seven sand fly species naturally infected with Leishmania in Peru: Lu. whitmani, Lu. sherlocki, Lu. llanosmartinsi, Lu. shawi, Lu. yucumensis, Lu. nevesi and Lu. walkeri. These species are widely distributed in the Amazon Basin underscoring the need to assess their vectorial competency [9, 17, 31]. In this regard, future studies will be oriented towards the identification of the infecting Leishmania species in the kDNA positive/ Real Time PCR negative samples employing alternative methods.
Although we did not identify Lu. longipalpis during the study execution, we collected specimens of Lu. migonei in Flor de Acre and Mavila. This species has been reported as a confirmed vector of L. (L.) infantum [35] highlighting a potential risk for the introduction of visceral leishmaniasis into this region.
An important limitation to consider for this study is that the finding of Leishmania DNA in a sand fly species is not a conclusive evidence of its role as vector. Vectorial role is confirmed by a series of criteria that include the capacity of the sand fly to maintain and transmit the parasite and the isolation of the same parasite strain from the human and the mammalian reservoir in the focus [6, 36]. Further studies should be oriented towards assessing the vectorial competency of putative vectors identified on this study.
Leishmaniasis control is highly dependent on the understanding of the ecology and disease epidemiology in endemic settings. In this sense, sand fly surveillance and identification of natural Leishmania infection are critical to assess the role of each species and design better and efficient control strategies [6, 36].
Our results underscore the need for increased control efforts against the sand fly vectors in the area and shed light into the potential effects of human activities in the epidemiology of the leishmaniasis in the Peruvian Amazon Basin.
Supporting information
(XLSX)
(XLSX)
(XLSX)
(XLSX)
Acknowledgments
We thank Micaela Arrunategui, Edwin Mamani, Jose Villavicencio and Antonio Silva from (DIRESA-Madre de Dios); Luis Angel Rosales (NAMRU-6 Puerto Maldonado); Dr. Carlos Magallanes (Centro de Salud de Iñapari) and Dr. Andrew Taniguchi (DUKE University) for all their support in fieldwork activities. We also appreciate the support in sand fly collections of Michael Tapscott, Grace Perrotta and Megan Murphy. We thank Dr. Nils Valencia (Laboratorio de Ecología, Museo de Historia Natural UNMSM) for the input and suggestions for entomological data analysis. We are grateful to the Ministerio de Agricultura y Riego de Perú, Dirección General Forestal y de Fauna Silvestre for permission to conduct these studies under the auspices of Resolución Directoral No. 0406-2013-MINAGRI-DGFFS/DGEFFS.
Disclaimer
The views expressed in this article are those of the author and do not necessarily reflect the official policy or position of the Department of the Navy, Department of Defense, nor the U.S. Government.
Copyright statement
Some authors of this manuscript are military service members and employees of the U.S. Government. This work was prepared as part of their official duties. Title 17 U.S.C. §105 provides that “Copyright protection under this Title is not available for any work of the United States Government”. Title 17 U.S.C. §101 defines a U.S. Government work as a work prepared by a military service member or employee of the U.S. Government as part of that person’s official duties.
Data Availability
All relevant data are within the paper and its Supporting Information files.
Funding Statement
This work was supported by an award to HOV from the Armed Forces Health Surveillance Branch (AFHSB) and its Global Emerging Infections Surveillance and Response (GEIS) Section, PROMIS ID P0372_14_N6 for FY2014. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Alvar J, Velez ID, Bern C, Herrero M, Desjeux P, Cano J, et al. Leishmaniasis Worldwide and Global Estimates of Its Incidence. Plos One. 2012;7(5). doi: ARTN e35671 doi: 10.1371/journal.pone.0035671 PubMed PMID: ISI:000305338500009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.David CV, Craft N. Cutaneous and mucocutaneous leishmaniasis. Dermatologic therapy. 2009;22(6):491–502. Epub 2009/11/06. doi: 10.1111/j.1529-8019.2009.01272.x . [DOI] [PubMed] [Google Scholar]
- 3.Murray HW, Berman JD, Davies CR, Saravia NG. Advances in leishmaniasis. Lancet. 2005;366(9496):1561–77. Epub 2005/11/01. doi: 10.1016/S0140-6736(05)67629-5 . [DOI] [PubMed] [Google Scholar]
- 4.Banuls AL, Hide M, Prugnolle F. Leishmania and the leishmaniases: a parasite genetic update and advances in taxonomy, epidemiology and pathogenicity in humans. Advances in parasitology. 2007;64:1–109. Epub 2007/05/15. doi: 10.1016/S0065-308X(06)64001-3 . [DOI] [PubMed] [Google Scholar]
- 5.Lucas CM, Franke ED, Cachay MI, Tejada A, Cruz ME, Kreutzer RD, et al. Geographic distribution and clinical description of leishmaniasis cases in Peru. The American journal of tropical medicine and hygiene. 1998;59(2):312–7. Epub 1998/08/26. . [DOI] [PubMed] [Google Scholar]
- 6.Killick-Kendrick R. Phlebotomine vectors of the leishmaniases: a review. Medical and veterinary entomology. 1990;4(1):1–24. Epub 1990/01/01. . [DOI] [PubMed] [Google Scholar]
- 7.Kato H, Gomez EA, Caceres AG, Uezato H, Mimori T, Hashiguchi Y. Molecular epidemiology for vector research on leishmaniasis. International journal of environmental research and public health. 2010;7(3):814–26. Epub 2010/07/10. doi: 10.3390/ijerph7030814 ; PubMed Central PMCID: PMC2872317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Grimaldi G Jr., Tesh RB, McMahon-Pratt D. A review of the geographic distribution and epidemiology of leishmaniasis in the New World. The American journal of tropical medicine and hygiene. 1989;41(6):687–725. Epub 1989/12/01. . [DOI] [PubMed] [Google Scholar]
- 9.Araujo-Pereira T, Fuzari AA, Andrade Filho JD, Pita-Pereira D, Britto C, Brazil RP. Sand fly fauna (Diptera: Psychodidae: Phlebotominae) in an area of leishmaniasis transmission in the municipality of Rio Branco, state of Acre, Brazil. Parasites & vectors. 2014;7:360 Epub 2014/08/12. doi: 10.1186/1756-3305-7-360 ; PubMed Central PMCID: PMC4141082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Azevedo AC, Costa SM, Pinto MC, Souza JL, Cruz HC, Vidal J, et al. Studies on the sandfly fauna (Diptera: Psychodidae: Phlebotominae) from transmission areas of American Cutaneous Leishmaniasis in state of Acre, Brazil. Mem Inst Oswaldo Cruz. 2008;103(8):760–7. . [DOI] [PubMed] [Google Scholar]
- 11.Kato H, Caceres AG, Mimori T, Ishimaru Y, Sayed AS, Fujita M, et al. Use of FTA cards for direct sampling of patients' lesions in the ecological study of cutaneous leishmaniasis. Journal of clinical microbiology. 2010;48(10):3661–5. Epub 2010/08/20. doi: 10.1128/JCM.00498-10 ; PubMed Central PMCID: PMC2953078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Arevalo J, Ramirez L, Adaui V, Zimic M, Tulliano G, Miranda-Verastegui C, et al. Influence of Leishmania (Viannia) species on the response to antimonial treatment in patients with American tegumentary leishmaniasis. The Journal of infectious diseases. 2007;195(12):1846–51. Epub 2007/05/12. doi: 10.1086/518041 . [DOI] [PubMed] [Google Scholar]
- 13.Yardley V, Croft SL, De Doncker S, Dujardin JC, Koirala S, Rijal S, et al. The sensitivity of clinical isolates of Leishmania from Peru and Nepal to miltefosine. The American journal of tropical medicine and hygiene. 2005;73(2):272–5. Epub 2005/08/17. . [PubMed] [Google Scholar]
- 14.Cáceres AG, Galati EA. Lista de Phlebotominae (Diptera: Psychodidae) para el Perú y especies consideradas como vectores naturales e incriminadas en la transmisión de patógenos de la leishmaniosis tegumentaria y la enfermedad de Carrión (verruga peruana). Revista Peruana de Medicina Experimental y Salud Pública. 2001;18(3–4):100–6. [Google Scholar]
- 15.Kato H, Caceres AG, Gomez EA, Mimori T, Uezato H, Marco JD, et al. Molecular mass screening to incriminate sand fly vectors of Andean-type cutaneous leishmaniasis in Ecuador and Peru. The American journal of tropical medicine and hygiene. 2008;79(5):719–21. . [PubMed] [Google Scholar]
- 16.Shender LA, De Los Santos M, Montgomery JM, Conrad PA, Ghersi BM, Razuri H, et al. Native rodent species are unlikely sources of infection for Leishmania (Viannia) braziliensis along the Transoceanic Highway in Madre de Dios, Peru. Plos One. 2014;9(7):e103358 doi: 10.1371/journal.pone.0103358 ; PubMed Central PMCID: PMCPMC4111550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Valdivia HO, De Los Santos MB, Fernandez R, Baldeviano GC, Zorrilla VO, Vera H, et al. Natural Leishmania infection of Lutzomyia auraensis in Madre de Dios, Peru, detected by a fluorescence resonance energy transfer-based real-time polymerase chain reaction. The American journal of tropical medicine and hygiene. 2012;87(3):511–7. Epub 2012/07/18. doi: 10.4269/ajtmh.2012.11-0708 ; PubMed Central PMCID: PMC3435357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Young DG, Duncan MA. Guide to the identification and geographic distribution of Lutzomyia sand flies in Mexico, the West Indies, Central and South America (Diptera: Psychodidae): Associated Publishers; 1994. [Google Scholar]
- 19.Rangel EF, Lainson R. Classificação de Phlebotominae Flebotomíneos do Brasil: Fiocruz; 2003. p. 23–51. [Google Scholar]
- 20.Beati L, Caceres AG, Lee JA, Munstermann LE. Systematic relationships among Lutzomyia sand flies (Diptera: Psychodidae) of Peru and Colombia based on the analysis of 12S and 28S ribosomal DNA sequences. International journal for parasitology. 2004;34(2):225–34. Epub 2004/03/24. doi: 10.1016/j.ijpara.2003.10.012 . [DOI] [PubMed] [Google Scholar]
- 21.Passos VM, Lasmar EB, Gontijo CM, Fernandes O, Degrave W. Natural infection of a domestic cat (Felis domesticus) with Leishmania (Viannia) in the metropolitan region of Belo Horizonte, State of Minas Gerais, Brazil. Mem Inst Oswaldo Cruz. 1996;91(1):19–20. . [DOI] [PubMed] [Google Scholar]
- 22.Degrave W, Fernandes O, Campbell D, Bozza M, Lopes U. Use of molecular probes and PCR for detection and typing of Leishmania—a mini-review. Mem Inst Oswaldo Cruz. 1994;89(3):463–9. . [DOI] [PubMed] [Google Scholar]
- 23.Rodgers MR, Popper SJ, Wirth DF. Amplification of kinetoplast DNA as a tool in the detection and diagnosis of Leishmania. Exp Parasitol. 1990;71(3):267–75. . [DOI] [PubMed] [Google Scholar]
- 24.Tsukayama P, Nunez JH, De Los Santos M, Soberon V, Lucas CM, Matlashewski G, et al. A FRET-based real-time PCR assay to identify the main causal agents of New World tegumentary leishmaniasis. PLoS Negl Trop Dis. 2013;7(1):e1956 doi: 10.1371/journal.pntd.0001956 ; PubMed Central PMCID: PMCPMC3536805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hammer Ø, Harper D, Ryan P. Paleontological Statistics Software: Package for Education and Data Analysis. Palaeontologia Electronica. 2001. [Google Scholar]
- 26.Hutcheson K. A test for comparing diversities based on the Shannon formula. J Theor Biol. 1970;29(1):151–4. . [DOI] [PubMed] [Google Scholar]
- 27.Roberts D, Hsi B. An Index of Species Abundance for Use with Mosquito Surveillance Data 1 2. Environmental Entomology. 1979;8(6):1007–13. [Google Scholar]
- 28.Sanchez A. Migraciones internas en el Peru 2015. Available from: http://www.oimperu.org/sitehome/sites/default/files/Documentos/03-03-2015_Publicacion%20Migraciones%20Internas_OIM.PDF.
- 29.Yadon ZE, Rodrigues LC, Davies CR, Quigley MA. Indoor and peridomestic transmission of American cutaneous leishmaniasis in northwestern Argentina: a retrospective case-control study. The American journal of tropical medicine and hygiene. 2003;68(5):519–26. . [DOI] [PubMed] [Google Scholar]
- 30.Young D, Arias J. Flebotomíneos vectores de leishmaniasis en las Americas. Organização Pan-Americana da Saúde, caderno técnico (3) 1992. [Google Scholar]
- 31.Teles CB, Santos AP, Freitas RA, Oliveira AF, Ogawa GM, Rodrigues MS, et al. Phlebotomine sandfly (Diptera: Psychodidae) diversity and their Leishmania DNA in a hot spot of American Cutaneous Leishmaniasis human cases along the Brazilian border with Peru and Bolivia. Mem Inst Oswaldo Cruz. 2016;0:0 doi: 10.1590/0074-02760160054 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Souza NA, Andrade-Coelho CA, Vilela ML, Peixoto AA, Rangel EF. Seasonality of Lutzomyia intermedia and Lutzomyia whitmani (Diptera: Psychodidae: Phlebotominae), occurring sympatrically in area of cutaneous leishmaniasis in the State of Rio de Janeiro, Brazil. Mem Inst Oswaldo Cruz. 2002;97(6):759–65. . [DOI] [PubMed] [Google Scholar]
- 33.Stolf HO, Marques A, Marques ME, Yoshida EL, Dillon NL. Surto de leishmaniose tegumentar americana em Itaporanga, São Paulo (Brasil). Rev Inst Med Trop São Paulo. 1993;35:437–42. [PubMed] [Google Scholar]
- 34.FORATTINI OP. Novas observações sobre a biologia de flebótomos em condições naturais (Diptera, Psychodidae). Arq Hig. 1960:209–15. [Google Scholar]
- 35.Guimaraes VC, Pruzinova K, Sadlova J, Volfova V, Myskova J, Filho SP, et al. Lutzomyia migonei is a permissive vector competent for Leishmania infantum. Parasites & vectors. 2016;9:159 doi: 10.1186/s13071-016-1444-2 ; PubMed Central PMCID: PMCPMC4797322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ready PD. Biology of phlebotomine sand flies as vectors of disease agents. Annu Rev Entomol. 2013;58:227–50. doi: 10.1146/annurev-ento-120811-153557 . [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(XLSX)
(XLSX)
(XLSX)
(XLSX)
Data Availability Statement
All relevant data are within the paper and its Supporting Information files.