Abstract
Aberrations in cholesterol homeostasis are associated with several diseases that can be linked to changes in cellular oxygen usage. Prior biological and physical studies have suggested that membrane cholesterol content can modulate oxygen delivery, but questions of magnitude and biological significance remain open for further investigation. Here, we use molecular dynamics simulations in a first step toward reexamining the rate impact of cholesterol on the permeation of oxygen through phospholipid bilayers. The simulation models are closely compared with published electron paramagnetic resonance (EPR) oximetry measurements. The simulations predict an oxygen permeability reduction due to cholesterol but also suggest that the EPR experiments may have underestimated resistance to oxygen permeation in the phospholipid headgroup region.
1 Introduction
Here, we discuss work implicating membrane cholesterol as a hindrance to oxygen transport on the cellular level and consider related biomedical implications. Further, we offer atomic resolution insight into prior electron paramagnetic resonance (EPR) based estimates of membrane oxygen permeability and resistance to permeation.
Though cholesterol is a normal constituent of higher eukaryotic membranes [1] and lung surfactant [2], its influence on oxygen diffusion is not fully understood. The cholesterol content of normal cells varies widely but often falls in the range 20–40 mol% [1]. Normal red blood cells contain about 50 mol% cholesterol, or a 1:1 ratio of cholesterol with phospholipid [3].
The complexity of biological membranes makes drawing conclusions regarding cholesterol’s influence on permeability particularly difficult, as cholesterol itself is inhomogeneously distributed within the membrane and plays a substantial role in raft formation and stabilization [1]. Lateral inhomogeneity of cholesterol content likely impacts oxygen transport, giving rise to regions of greater and lesser permeability [4–6]. Further, measuring intracellular oxygen, as distinct from extracellular oxygen, presents a substantial methodological challenge [7], not to mention the potential difficulty of tracking oxygen movement and partitioning within cells.
A few cellular studies have successfully characterized transmembrane oxygen diffusion with varying membrane cholesterol content and have suggested that cholesterol reduces the rate of transmembrane oxygen flow [7,8]. Yet, generalization of the findings would require further detail regarding membrane composition and organization for the cells studied. Prior biophysical studies have strongly suggested that membrane cholesterol content directly reduces oxygen permeability [5,6,9]. Subczynski and colleagues have used EPR spin-label oximetry data to estimate permeability coefficients, which describe the rate of diffusional flux across a membrane. However, approximations inherent in the method limit the accuracy of the permeability values to ± 30% [9].
Computer simulations of molecular dynamics provide a particularly valuable means of attaining atomic-resolution insight into biological processes in which molecular structural behavior plays a significant role. The current work represents a first step toward reevaluating the rate impact of membrane cholesterol on oxygen permeability. We present resistance to permeation curves calculated from all-atom molecular dynamics simulations and compare the curves with published data from EPR probe-based oximetry. Then, we provide preliminary evidence indicating that the base-level permeability of membrane phospholipids may be lower than generally assumed.
2 Methods
To examine membrane oxygen permeability with atomic resolution, we used all-atom molecular dynamics simulations, conducted with the GPU-enabled version of Amber 14 biomolecular modeling software [10,11], as reported previously [12]. The simulations used the Amber Lipid14 force field and cholesterol extension [13,14], along with the TIP3P explicit water model [15] and in-house O2 parameters [12]. A tempocholine spin label moiety was modeled using Lipid14 force field atom types. The tempocholine moiety and glycerol backbone were charge-fitted according to the Amber Lipid Framework modular scheme [16].
Data from three simulation systems are reported here, with each system consisting of 128 lipid molecules, total, along with 35 water molecules per lipid. 31 or 35 O2 molecules were included, to reach a concentration of 200 mM for the entire system (lipid and water). This high O2 concentration enables relatively rapid sampling of the configurational space and provides results comparable to those obtained with only one O2 molecule. As a simplified representation of the bulk lipid fraction of animal cell membranes, we used the highly abundant phospholipid 1-palmitoyl,2-oleoylphosphatidylcholine (POPC) for all three simulation systems: 1. pure POPC, 2. POPC mixed in a 1:1 ratio with cholesterol (“POPC/cholesterol”), and 3. POPC with one tempocholine-1-palmitoyl-2-oleoylphosphatidic acid ester (T-PC) molecule per leaflet. All simulations were conducted at a constant pressure of 1 atm and at a fixed temperature of either 37 or 25°C.
From duplicate 300-nanosecond POPC and POPC/cholesterol simulations at 37°C, we estimated resistance to permeation curves for oxygen according to the method of Marrink and Berendsen, using the relative local free energy calculated from the depth-dependent oxygen concentration, along with an estimate of the depth-dependent diffusion coefficient based on the local mean-squared displacement along the bilayer normal [17]. From two POPC simulations incorporating two explicit T-PC molecules and run at 25°C, we observed the probe orientational behavior over 100 nanoseconds. Data are shown for one representative simulation from each set of duplicates.
3 Results and Discussion
Simulation resistance to permeation curves for POPC and POPC/cholesterol (Fig.1A) predict fairly similar permeabilities for both bilayers, given comparable areas under the resistance to permeation curves. This similarity was initially puzzling because prior EPR experiments had estimated POPC/cholesterol to be about three times less permeable to oxygen than pure POPC [9]. Our simulation systems show strong agreement with other experimental measurements, especially electron density profiles and nuclear magnetic resonance (NMR) lipid order parameters (data not shown; as reported in [13,14]). Moreover, we are able to reproduce quite accurately the shape of the EPR resistance to permeation curves used to estimate permeability for POPC and POPC/cholesterol (Fig.1B).
Fig.1. EPR “headgroup” probe may oversample the phospholipid tail region.
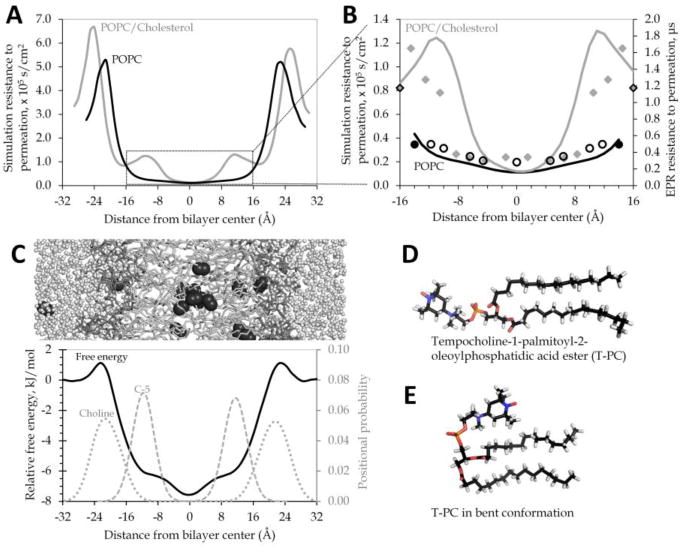
(A) Simulation resistance to permeation gives a similar area under the curve for POPC (black) and POPC/cholesterol (gray). (B) The same simulation resistance to permeation curves (solid lines) differ substantially in contour and area under the curve in the region most thoroughly sampled in published EPR measurements. The secondary axis shows the EPR resistance to permeation values [9], placing the probes according to center of the probability distributions observed in our simulations for the corresponding atoms (oleoyl tail carbons 5, 7, 9, 10, 12, 14, and 16 in open black circles for POPC or solid gray diamonds for POPC/cholesterol; probability distribution data not shown). The T-PC probe positions (solid black circles for POPC or black-outlined diamonds for POPC/cholesterol) have also been estimated from simulations of POPC with one T-PC molecule in each bilayer leaflet. (C) Cropped image of POPC/water/O2 simulation system, positioned above a relative free energy profile for oxygen across a POPC bilayer system at 37°C. Relative local free energy for O2 at 37°C across a model POPC bilayer shows peaks in the headgroup regions and a free energy minimum in the tail region. Positional probability distributions are shown for the POPC headgroup choline (dotted line) and for the fifth carbon of the POPC oleoyl tail (C-5, dashed line). (D–E) Two alternate conformations of the EPR T-PC probe molecule, with the bent conformation favored in preliminary simulations at 25°C.
The EPR curves were calculated from the spin-lattice relaxation times of nitroxide radical spin labels placed at various depths along the membrane. Membrane permeability coefficients were estimated from the respective areas under the resistance to permeation curves [9,17]. The EPR permeability estimates are, thus, sensitive to probe positioning within the membrane. The nitroxide spin label group tempocholine (shown in Fig.1D) was used experimentally for the purpose of detecting oxygen in the headgroup region. Tempocholine is structurally similar to the charged POPC headgroup moiety choline. Yet, it is substantially nonpolar, suggesting that the nitroxide spin label may preferentially associate with the nonpolar hydrocarbon portion of the membrane, rather than with the charged headgroups. Fig.1C juxtaposes a simplified POPC bilayer image with a simulation free energy curve, showing barriers to oxygen permeation at the headgroups. The simulation positional distributions of the headgroup choline moiety and of the fifth carbon atom (C-5) of the oleoyl tail are also shown.
To better understand the orientational behavior of the EPR headgroup probe, we conducted simulations incorporating a preliminary model of the T-PC probe used experimentally (Fig.1D). In our simulations, the nitroxide spin label dwells predominantly within the hydrophobic phospholipid tail region of the bilayer, near C-5 of the oleoyl tail. As such, we find the T-PC molecule predominantly in a bent conformation (Fig.1E). This finding requires experimental confirmation, but it suggests that the permeabilities of POPC and POPC/cholesterol may be more similar than expected from the EPR work. Namely, the EPR T-PC probe may substantially underestimate the resistance to permeation for both bilayers because it may not, in fact, be sampling the headgroup region, where the greatest resistance is seen. The POPC and POPC/cholesterol resistance to permeation curves differ in contour in the region between −16 and 16 Å (Fig.1B), which we suspect to be the portion of the bilayer most sampled by the EPR spin-label probes, including the intended headgroup probe, T-PC. Considering this region alone, we see about a three-fold difference in the area under the resistance to permeation curve, a result that agrees qualitatively with the EPR permeability estimates.
4 Conclusion
The simulation resistance to permeation curves indicate that pure POPC phospholipid and POPC/cholesterol (1:1 mixing ratio) have fairly similar permeabilities to molecular oxygen. Though we predict a less dramatic permeability difference than expected from published EPR estimates, we do see an increase in the overall resistance to permeation for POPC/cholesterol of roughly 20%, compared with POPC alone. This increased resistance should correspond with a permeability reduction of similar magnitude. While modest on a single-membrane level, the rate-reducing effect should be amplified as oxygen is required to cross or circumvent multiple membranes on its path toward mitochondria buried within tissues [18].
Our simulations further predict that the lipid fraction of biological membranes may be less permeable to oxygen than has generally been assumed, regardless of the cholesterol content. This prediction is based on the substantially greater area under the resistance to permeation curve obtained when resistance in the headgroup region is taken fully into account. It should be noted that the current models neglect inhomogeneities in lipid organization within the plane of the membrane, lipid compositional variation within and across leaflets, as well as the presence of membrane proteins. While such factors may moderate cholesterol’s impact in living cells, the models’ simplicity is an advantage for distinguishing the direct influence of cholesterol on membrane permeability. Ongoing work will address other membrane compositional factors that may influence oxygen permeability. Additional biological work is needed to test our predictions and to characterize quantitatively the effect of cholesterol on oxygen permeability in intact, functional cells.
Acknowledgments
We thank Ross Walker and Benjamin Madej for providing advance access to the cholesterol parameters used in this study. James Ryan Bredin developed the O2 parameters used in this work. Daniel Lyons contributed valuable computing expertise. The molecular images were generated using PyMOL software [19], and DataThief software [20] was used to infer the experimental values reported in Figure 1B from published plots. SCP thanks James Kindt and Snežna Rogelj for professional mentoring. This work was supported by the NIH under NIGMS grant P20GM103451 and by a gift from the Glendorn Foundation. The content is solely the responsibility of the authors. We used computing resources of TACC at UT Austin, accessed through XSEDE (funded by NSF grant ACI-1053575), as well as the EXXACT MD SimCluster (“Electra”) at New Mexico Tech.
References
- 1.Mouritsen OG, Zuckermann MJ. What’s so special about cholesterol? Lipids. 2004;39(11):1101–1113. doi: 10.1007/s11745-004-1336-x. [DOI] [PubMed] [Google Scholar]
- 2.Larsson M, Larsson K, Nylander T, Wollmer P. The bilayer melting transition in lung surfactant bilayers: The role of cholesterol. Eur Biophys J. 2003;31(8):633–636. doi: 10.1007/s00249-002-0256-9. [DOI] [PubMed] [Google Scholar]
- 3.Jandl JH. Blood: Textbook of Hematology. 2nd. Little, Brown and Company; Boston, MA: 1996. [Google Scholar]
- 4.Kawasaki K, Yin JJ, Subczynski WK, et al. Pulse EPR detection of lipid exchange between protein-rich raft and bulk domains in the membrane: Methodology development and its application to studies of influenza viral membrane. Biophys J. 2001;80(2):738–748. doi: 10.1016/S0006-3495(01)76053-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Raguz M, Mainali L, Widomska J, Subczynski WK. Using spin-label electron paramagnetic resonance (EPR) to discriminate and characterize the cholesterol bilayer domain. Chem Phys Lipids. 2011;164(8):819–829. doi: 10.1016/j.chemphyslip.2011.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wennberg CL, van der Spoel D, Hub JS. Large influence of cholesterol on solute partitioning into lipid membranes. J Am Chem Soc. 2012;134(11):5351–5361. doi: 10.1021/ja211929h. [DOI] [PubMed] [Google Scholar]
- 7.Khan N, Shen J, Chang TY, et al. Plasma membrane cholesterol: A possible barrier to intracellular oxygen in normal and mutant CHO cells defective in cholesterol metabolism. Biochemistry. 2003;42(1):23–29. doi: 10.1021/bi026039t. [DOI] [PubMed] [Google Scholar]
- 8.Menchaca HJ, Michalek VN, Rohde TD, et al. Decreased blood oxygen diffusion in hypercholesterolemia. Surgery. 1998;124(4):692–698. doi: 10.1067/msy.1998.90944. [DOI] [PubMed] [Google Scholar]
- 9.Widomska J, Raguz M, Subczynski WK. Oxygen permeability of the lipid bilayer membrane made of calf lens lipids. Biochim Biophys Acta. 2007;1768(10):2635–2645. doi: 10.1016/j.bbamem.2007.06.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Case DA, Berryman JT, Betz RM, et al. AMBER 2015. University of California; San Francisco: 2015. [Google Scholar]
- 11.Salomon-Ferrer R, Götz AW, Poole D, et al. Routine microsecond molecular dynamics simulations with AMBER on GPUs. 2. Explicit solvent particle mesh Ewald. J Chem Theory Comput. 2013;9(9):3878–3888. doi: 10.1021/ct400314y. [DOI] [PubMed] [Google Scholar]
- 12.Smith CR, Dotson RJ, Bueche K, et al. Impact of cholesterol on the oxygen permeability of membranes. Submitted. [Google Scholar]
- 13.Dickson CJ, Madej BD, Skjevik ÅA, et al. Lipid14: The Amber lipid force field. J Chem Theory Comput. 2014;10(2):865–879. doi: 10.1021/ct4010307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Madej BD, Gould IR, Walker RC. A parameterization of cholesterol for mixed lipid bilayer simulation within the Amber Lipid14 force field. J Phys Chem B. 2015;119(38):12424–12435. doi: 10.1021/acs.jpcb.5b04924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jorgensen WL, Jenson C. Temperature dependence of TIP3P, SPC, and TIP4P water from NPT Monte Carlo simulations: Seeking temperatures of maximum density. J Comput Chem. 1998;19(10):1179–1186. [Google Scholar]
- 16.Skjevik AA, Madej BD, Walker RC, Teigen K. LIPID11: A modular framework for lipid simulations using Amber. J Phys Chem B. 2012;116(36):11124–11136. doi: 10.1021/jp3059992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Marrink SJ, Berendsen HJC. Simulation of water transport through a lipid membrane. J Phys Chem. 1994;98(15):4155–4168. [Google Scholar]
- 18.Shea R, Smith C, Pias SC. Magnification of cholesterol-induced membrane resistance on the tissue level: Implications for hypoxia. Adv Exp Med Biol. 2016;923:43–50. doi: 10.1007/978-3-319-38810-6_6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Schrödinger LLC. The PyMOL Molecular Graphics System. 2015 Version 1.7.6.5. [Google Scholar]
- 20.Tummers B. DataThief III. 2006 [Google Scholar]


