Abstract
A dynamic hydrogel that sequentially responds to two independent but interrelated physical and biomolecular signals was reported in this work. Once hit by an external light signal, an immobilized internal molecular signal is activated and freed via photoreaction; and subsequently the freed molecular signal works as a self-programming factor of the hydrogel to induce the dissociation of a biomolecular complex to release protein via hybridization reaction. Notably, pulsatile external light input can be converted to periodical protein output from the hydrogel to regulate cell migration. Thus, this hydrogel holds potential as a self-programming platform for biological and biomedical applications such as controlled release of bioactive substances.
INTRODUCTION
Responsive materials are able to change their properties quickly in response to a specific variation of environmental conditions.1–8 These materials have been widely used in drug delivery, cell biology, molecular machine, information technology, and so on.9–15 Using responsive materials to control protein release is one of the most intriguing potential applications,16–17 since a lot of proteins, particularly cytokines and growth factors, can cause severe side effects when delivered systematically.18–19 As hydrogels have good biocompatibility and structural similarities to tissues,20–22 they have been developed as responsive materials for controlled protein release mainly based on the mechanisms of pH-triggered degradation or small molecule-triggered gel-sol transition.23–25 However, it is challenging to apply these responsive hydrogels to realize sequential release of one or multiple proteins as their one-step functional change requires bulk degradation or phase transition.26–30 This problem would be solved if protein release can be decoupled from the bulk change of hydrogels. Recently, photolabile linkers were used to chemically conjugated proteins to the hydrogel for controlled protein release.31 However, chemical conjugation may induce denaturation of proteins owing to the modification of cysteine or lysine in proteins.32–33 Therefore, there is still a need to develop new responsive hydrogels for controlled protein release.
In biological systems, multistep molecular recognition events are used to realize one function. For instance, when light hits a photoreceptor cell, rhodopsin is activated to form metarhodopsin-II, which subsequently recognizes and binds the heterotrimeric G-protein transducin to induce molecular dissociation. Ultimately, this stepwise procedure leads to the conversion of light into an electrophysiological signal for visualization.34–35 Notably, the entire procedure does not need to change other key properties (e.g., cellular integrity) of the photoreceptor cell or involve harsh factors. Inspired by this biological mechanism, we developed a synthetic hydrogel system that responds to two interrelated signals, an external light signal and an internal molecular signal, for regulating the dissociation of a protein-DNA complex to control protein release.
EXPERIMENTAL SECTION
Synthesis of the photolabile linker
Compound 3
To a solution of 1-(4-hydroxy-5-methoxy-2-nitrophenyl)ethan-1-one36 (compound 1, 2.11 g, 10 mmol) in 30 mL acetone was added potassium carbonate (1.65 g, 12 mmol), potassium iodide (0.33 g, 2.0 mmol), 18-crown-6 (0.53 g, 2.0 mmol) and compound 2 (4.25 g, 11 mmol). The mixture was refluxed overnight under argon. After cooling to room temperature, the solvent was evaporated under reduced pressure. The crude reaction mixture was partitioned between EtOAc and H2O. The aqueous layer was extracted twice with EtOAc and the combined organic layers were dried over MgSO4, filtered and concentrated. The crude product was purified by flash chromatography on silica gel to yield 3 (49%). 1H NMR (400 MHz, CDCl3): δ 7.70 (s, 1H), 6.75 (s, 1H), 4.28 (t, J = 4.6 Hz, 2H), 4.20 (d, J = 2.0 Hz, 2H), 3.96 (s, 3H), 3.93 (t, J = 4.8 Hz, 2H), 3.73 (t, J = 2.8 Hz, 2H), 3.69-3.66 (m, 10H), 2.50 (s, 3H), 2.43 (t, J = 2.4 Hz, 1H) ppm. 13C NMR (125 MHz, CDCl3): δ 200.12, 154.38, 148.99, 138.32, 133.00, 108.81, 108.65, 79.67, 70.97, 70.66, 70.63, 70.41, 69.39, 69.25, 69.12, 58.41, 56.62, 30.42 ppm. ESI-MS: [M+H]+ 426.18, [M+NH4]+ 443.21, [M+Na]+ 448.16.
Compound 4
Sodium borohydride (0.11 g, 3.0 mmol) was added to a solution of compound 3 (0.425 g, 1.0 mmol) in 10 mL methanol at 0 °C. The reaction was stirred at 0 °C for 30 min and room temperature for 3 h. The reaction was quenched by addition of 1 mL sat. NH4Cl. The methanol was removed under vacuum and the mixture was dissolved with EtOAc (40 mL) and washed with sat. NaHCO3 and brine, dried over MgSO4, filtered and concentrated under vacuum to afford 4 without purification (96%). 1H NMR (400 MHz, CDCl3): δ 7.64 (s, 1H), 7.30 (s, 1H), 5.56 (q, J = 6.0, 12.4 Hz, 1H), 4.23 (td, J = 1.6, 4.8 Hz, 2H), 4.19 (d, J = 2.4 Hz, 2H), 3.97 (s, 3H), 3.91 (t, J = 4.8 Hz, 2H), 3.73-3.65 (m, 12H), 2.43 (t, J = 2.4 Hz, 1H), 1.54 (d, J = 6.0 Hz, 3H) ppm. 13C NMR (125 MHz, CDCl3): δ 154.23, 147.02, 139.51, 137.18, 109.71, 108.75, 79.66, 74.56, 70.94, 70.66, 70.62, 70.40, 69.49, 69.12, 69.02, 65.75, 58.41, 56.35, 24.29 ppm. ESI-MS: [M-OH]+ 410.17, [M+NH4]+ 445.23, [M+Na]+ 450.18.
Photoresponsive linker 5
N, N′-disuccinimidyl carbonate (0.51 g, 2 mmol) was added to a solution of compound 4 (0.427 g, 1.0 mmol), triethylamine (0.2 mL) and 4-(dimethylamino)pyridine (0.1 g) in 10 mL dry acetonitrile at 0 °C. The reaction was stirred at 0 °C for 30 min and room temperature for 4 h under argon. After concentrating the solution under vacuum, the mixture was diluted by EtOAc (30 mL) and washed with 10% citric acid solution (3 × 20 mL). The organic layer was dried over MgSO4. The crude compound was purified by flash chromatography on silica gel to obtain 5 (86%). 1H NMR (400 MHz, CDCl3): δ 7.70 (s, 1H), 7.05 (s, 1H), 6.47 (q, J = 6.4, 12.8 Hz, 1H), 4.23 (t, J = 4.8 Hz, 2H), 4.17 (d, J = 2.4 Hz, 2H), 4.09 (q, J = 6.4, 14 Hz, 1H), 4.02 (s, 3H), 3.89 (t, J = 4.8 Hz, 2H), 3.72-3.62 (m, 14H), 2.78 (s, 4H), 2.42 (t, J = 2.4 Hz, 1H), 1.74 (d, J = 6.4 Hz, 3H) ppm. 13C NMR (125 MHz, CDCl3): δ 168.50, 154.69, 150.58, 147.78, 139.14, 131.36, 109.74, 107.37, 79.68, 74.55, 70.94, 70.62, 70.39, 69.40, 69.12, 69.06, 60.39, 58.39, 56.51, 25.42, 21.94, 14.21 ppm. ESI-MS: [M+NH4]+ 586.25.
Synthesis of photoresponsive DNA (ON1*)
A 4 μL DMSO-based solution of linker 5 (0.1 M) was added to a solution of the amino-modified oligonucleotide, ON1-NH2, (1 mM, 100 μL) in Tris-HCl buffer (pH 8.0, 100 mM). The mixture was vortexed at room temperature for 4 h. Thereafter, 4 μL of linker 5 solution was further added to the reaction and vortexed for another 4 h at room temperature. The reaction was filtered through a spin filter (MW 5k) and washed with 300 μL of PBS 6 times to remove the excess linker and low molecular weight reaction byproducts. The crude DNA product was collected and resuspended in 100 μL of water, and was used for next step without further purification.
Synthesis of photoresponsive nanogel (NG*)
An azide-functionalized nanogel (NG) was synthesized via a previously reported method with a minor modification.37 The photoresponsive oligonucleotide (ON1*) was covalently conjugated to nanogel via Cu(I)-Catalyzed Azide-Alkyne Cycloaddition (CuAAC).38 In brief, 4.35 mg tris(3-hydroxypropyltriazolylmethyl)amine (THPTA) and 12.5 mg CuSO4·5H2O were added to 1.0 mL water. The mixture was vortexed and formed a clear Cu-THPTA solution (50 mM). Next, 0.55 μL of NaCl solution (5.0 M), 10 μL of ON1* solution (1 mM) and 0.55 μL of Cu-THPTA solution were added to 20 μL of azide-functionalized nanogel (or fluorescent nanogel) solution (7.5 mg·mL−1), followed by the addition of 0.55 μL of sodium ascorbic solution (0.5 M). The solution was first vortexed at room temperature for 2 h. Then, another 0.55 μL of sodium ascorbic solution was added and reacted for another 2 h. The DNA-conjugated photoresponsive NG* was purified via gel electrophoresis, collected by lyophilization and resuspended in water at a final concentration of 2.0 mg·mL−1. This solution was stored at −20 °C until further use. To prepare NG* co-conjugated with ON1* and ON1 (i.e., ON1 with 5′-hexynyl), the two oligonucleotides were used together for the click reaction.
Gel electrophoresis
Gel electrophoresis was used to evaluate the conjugation of ON1* to NG via “click” chemistry. 0.2 μL of the reaction was mixed with ON2 solution (100 pmol) in 10 μL PBS for 1 hour. The samples were loaded into a 10% polyacrylamide gel. The electrophoresis was run in TBE buffer (89 mM tris-borate, 89 mM boric acid, and 2 mM EDTA; pH 8.2) at 80 V for 45 min using a Mini-PROTEAN Tetra Cell system (Bio-Rad, Hercules, CA). The gel was stained in SYBR-Safe™ solution for 30 min and imaged with a green fluorescence filter using a CRI Maestro EX System (Woburn, MA). The images were analyzed using the software provided by the supplier.
Examination of ON1 release from NG*
To perform the photolysis of NG* and monitor the photorelease of ON1, 1 μL of NG* was diluted with 9 μL PBS and the solution was placed under a UV lamp at 365 nm with intensity of 3 mW·cm−2. Upon photolysis for a predetermined period of time, the reaction solution was taken and subject to gel electrophoresis using the method as described above.
Synthesis of the biomimetic system
Superporous hydrogels were prepared according to a reported method.39 The superporous hydrogel was gently blotted with tissue paper to dehydrate. Subsequently, 30 μL of PDGF-BB solution (50 ng) was added and the loaded hydrogel was incubated at room temperature for 4 h. Thereafter, 5.0 μL of NG* was diluted in 10 μL of PBS, which was further loaded into the hydrogel. The hydrogel was incubated at room temperature for another 1 h. The PDGF-BB and NG* loaded hydrogel (i.e., the biomimetic hydrogel system) was directly used for the examination of PDGF-BB retention and signal output without washing treatment.
Examination of NG* distribution in the hydrogel
FITC-doped NG* solution of 10 μL was mixed with 20 μL of TYE665-labeled complementary sequence (TYE665-CS, 100 pmol) in PBS. The mixture was added to the dehydrated hydrogel. After 1 h incubation, the hydrogel was thoroughly washed and subject to observation under a Nikon A1R Spectral Confocal Microscope (Nikon Instruments Inc., Melville, NY) to image the porous network (via TYE665) and the distribution of NG* in the hydrogel. The fluorescein and TYE655 were excited at 488 nm and 645 nm, respectively. Scanning was performed from the surface of the hydrogel to a depth of 100 μm in the z-direction at an optical resolution of 20 μm. Captured images represent a 1280 μm ×1280 μm field.
Ethidium bromide (EB) staining of ON1-ON2 duplexes
EB can be intercalated into the DNA duplexes to produce fluorescence signal. Thus, to evaluate the formation of ON1-ON2 duplexes, superporous hydrogels loaded with NG* were exposed to 365 nm UV light and stained with EB. Hydrogels were incubated in the EB solution (10 μg·mL−1, 1 mL) for 1 h and then incubated in 15 mL of PBS for 2 h washing to remove unbound EB. Hydrogels were imaged (λex/λem = 523/610 nm) with a Maestro In Vivo Imaging System (Woburn, MA).
Examination of protein output
The hydrogels were incubated in 1 mL of release medium (PBS solution of 0.1 wt% BSA, 0.05% v/w Tween 20, and 0.09 wt% sodium azide) at 37 °C. To regulate signal output, the hydrogel was placed under a UV lamp for a predetermined period of UV light exposure (3 mW·cm−2). At predetermined times points, the supernatant was collected and replaced with 1 mL of fresh release medium. The collected supernatant was stored at −20 °C until quantification. PDGF-BB was quantified using a recombinant human PDGF-BB enzyme-linked immunosorbent assay (ELISA) (Peprotech, Rocky Hill, NJ). The absorbance of each sample was measured using a BioTek Synergy™ HT Multi-Mode Microplate Reader (BioTek, Winooski, VT).
Wound closure assay
Smooth muscle cells (SMCs) were cultured in the low serum medium (i.e., basic medium supplemented with 0.5% FBS) overnight in a 24 well plate at a density of 9 × 104 cell/well. Wounds were created by scratching the confluent SMCs with the trimmed pipet tips. After the hydrogels were exposed to UV light (365 nm, 3 mW·cm−2) for 5 min, the hydrogels were put into a transwell insert and co-incubated with the cells in a transwell plate for 6 hours. The cells without any treatment or those co-incubated with the hydrogels without UV exposure were used as controls. Microscopy images of the cells were captured at 0 h and 6 h post treatment for analysis. Statistical analysis was performed in Prisma 5.0 (GraphPad). One-way ANOVA followed by Bonferroni’s Test was used to compare the wound closure distances.
RESULTS AND DISCUSSION
Molecular design of hydrogel system
Figure 1 shows the working principle of the molecular system. Two oligonucleotides are integrated into the system as pendant motifs. The first oligonucleotide (i.e., ON1) is covalently conjugated to a hydrogel nanoparticle (i.e., nanogel) via a photolabile linker; and the second oligonucleotide (i.e., ON2) is covalently conjugated to the bulk hydrogel network with the ability to form a protein-DNA complex. Once the hydrogel system is exposed to an external light signal, ON1 is activated for dissociation from the nanogel. Subsequently, the freed ON1 hybridizes with ON2 and functions as an active intermediate signal to induce the dissociation of the protein-DNA complex and release the bound protein.
Figure 1.
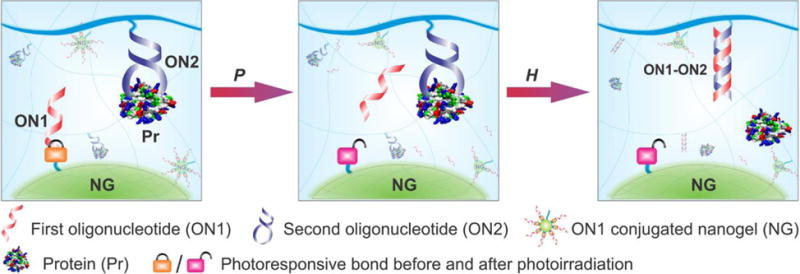
Schematic illustration of the concept. When an external light signal is provided to the biomimetic system, photoreaction (i.e., P) induces the breakage of the light-responsive group of the internal molecular signal (i.e., ON1). The activated ON1 is freed to diffuse to the neighboring DNA-protein complex. ON1 binds to ON2 via hybridization reaction (i.e., H) to induce the conformational change of ON2, which causes the dissociation of the DNA-protein complex for the production of a signal output (i.e., free protein).
The synthesis of hydrogels often involves harsh chemical conditions.26–30 For instance, free radicals were produced in acidic solutions when hydrogels were synthesized in this work. To avoid potential denaturation of proteins by these harsh conditions, we purposely decoupled the synthesis of superporous hydrogels from protein loading. The superporous structure of the hydrogels would allow for easy adsorption of aqueous solutions and loading of proteins into the hydrogels for binding to ON2 after the formation of the bulk hydrogel matrix. An alternative approach to achieving the same purpose of protein loading would be the synthesis of a highly swollen hydrogel, which is not a focus in this current work but worthy of studying in the future. Notably, since the protein is adsorbed into the bulk hydrogel matrix for physical binding to ON2, this system does not involve harsh chemical conditions for protein modification and immobilization. Moreover, ON1 is a pendant motif of the nanogel and the breakage of its light-responsive bond is independent of the bulk properties (e.g., integrity) of the hydrogel. Thus, the current system is different from other elegant stimuli-responsive materials that require bulk change, phase transition or chemical modification of proteins.26–29,31
Preparation of photoresponsive DNA-functionalized nanogel (NG*)
4, 5-Dimethoxy-2-nitrobenzyl alcohol and its derivatives have been studied as photolabile protecting groups for various application, and their photolysis can occur under long wavelength UV light for the caging and highly efficient photorelease of cargoes.40–44 In this work, we synthesized a new photoresponsive linker 5 based on 4, 5-dimethoxy-2-nitrobenzyl alcohol (Figure 2 and Figure S1, Supporting Information). An ethylene glycol oligomer was incorporated into linker 5 for enhanced solubility. Figure 3A shows that linker 5 has three typical absorptions at 246, 305 and 347 nm. The exposure of linker 5 to UV light at 365 nm decreased its absorption at 305 nm and increased absorptions at 261 and 370 nm. These results revealed a fast and clean photoreaction of linker 5 in aqueous solutions.
Figure 2.
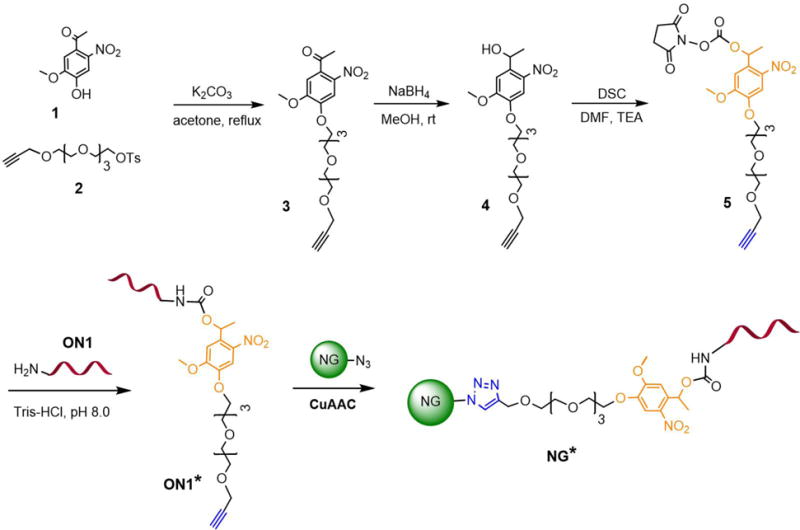
A general outline for the synthesis of photoresponsive NG*. A photoresponsive linker 5 was first synthesized and modified to oligonucleotide ON1, which was further conjugated to NG via CuAAC.
Figure 3.
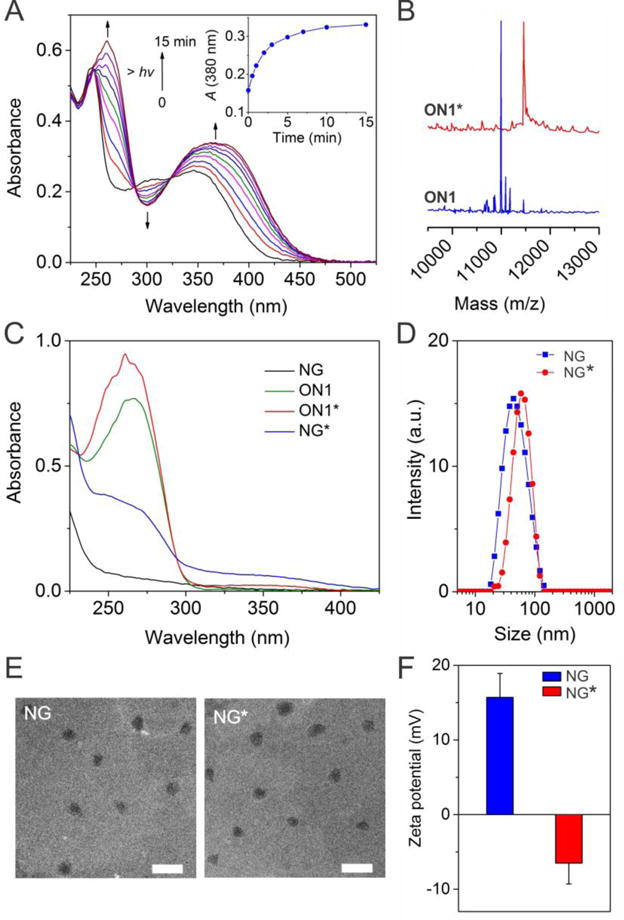
Synthesis and characterizations of NG*. (A) UV-vis absorption spectra monitoring of the photolysis of linker 5 in PBS solution upon UV irradiation (365 nm, 3 mW·cm−2). Inset figure shows the irradiation dependent absorbance change of the solution monitored at 380 nm. (B) Characterization of ON1 before and ON1* using MALDI-TOF analysis. (C) UV-vis absorption spectra of ON1, ON1*, NG and NG* in PBS solution. (D) DLS analysis of NG and NG*. (E) TEM images of NG and NG*. Scale bar: 250 nm. (F) Zeta-potential of NG and NG*. Error bars represent s. e. m (n = 3).
Linker 5 was conjugated with ON1 to synthesize photoresponsive ON1* (Figure 2). MALDI-TOF characterization revealed shift of 454 Da in mass after the conjugation (Figure. 3B), demonstrating the successful conjugation of linker 5 and ON1. As linker 5 has an alkene group, ON1* could be conjugated to the azide-functionalized nanogel (NG) via copper(I)-catalyzed alkyne-azide cycloaddition (CuAAC) to synthesize photoresponsive NG* (Figure 2).
To demonstrate the conjugation of ON1* to NG, we characterized the UV-vis absorption spectrum of NG* (Figure 3C). Since 260 nm is the characteristic absorption peak of DNA, the comparison of the absorption bands of NG and NG* suggested that ON1* was conjugated to NG. It is confirmed by the small absorption of NG* at 358 nm that is the characteristic absorption of the nitrobenzyl derivative. NG* was also characterized with dynamic light scattering (DLS) and transmission electron microscopy (TEM). It is shown in Figure 3D that NG* had an DLS size of 58.8±3.5 nm, which was around 8 nm larger than NG without modification The TEM images show that the diameters of NG* was less than 100 nm (Figure 3E). The evaluation of the surface charge demonstrated that the zeta potential of NG was decreased from 15.7±3.2 to ‐6.5±2.8 mV after the conjugation with ON1* (Figure 3F), further confirming the presence of negatively charged DNA on NG. Taken together, these data confirmed the success of conjugating ON1* to NG via click chemistry. The quantification of DNA suggested that the conjugation efficiency was 40.5 pmol ON1* per μg NG and each NG* had ~40 ON1* (Table S2 and Figure S2).
Photoactivation of ON1* on NG*
We next examined the photoresponsiveness of NG* that was exposed to UV light (Figure 4A). Photoirradiation of NG* induced an increase in absorption at 260 nm and a concomitant redshift of the absorption in the range of 350–400 nm (inset figure) (Figure 4B and Figure S3) whereas photoirradiation of ON1 did not lead to any change in the absorption spectrum (Figure S4). These data suggest that NG* was photoresponsive owing to the photolysis of ON1*.
Figure 4.
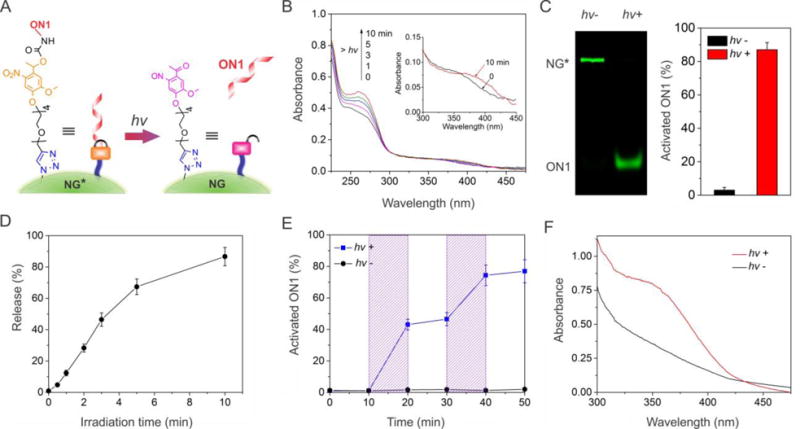
Conversion of the external light signal into the internal molecular signal after the exposure of NG* to the UV light. (A) Illustration of light-induced breakage of the photoresponsive group for ON1 activation. (B) UV-vis absorption spectra of the NG* solution upon photoirradiation. The inset figure shows a red shift in the absorption of NG* after photoirradiation. (C) Gel electrophoresis image and analysis of the NG* solution before and after photoirradiation. The time of irradiation was 10 min. (D) Effects of photoirradiation on the activation of ON1. The release was analyzed via gel electrophoresis and the percentage was calculated by normalization to the theoretical ON1 incorporation. (E) Temporal control of ON1 activation from NG*. The samples were exposed to UV light at 10 and 30 min for 3 min each time. The shaded regions were drawn for the clear legibility of ON1 production. (F) UV-vis absorption spectrum displays the absorption change of the NG*-loaded hydrogel before and after 10 min of photoirradiation. Error bars represent s. e. m (n = 3).
Gel electrophoresis was further performed to verify the states of ON1* of NG*. Without UV irradiation, as shown in lane 1 in Figure 4C, strong fluorescence was observed in the NG* band in contrast to negligible fluorescence in the ON1 band. After UV irradiation, the opposite results were observed (lane 2). Analysis of the band intensities showed that approximately 83% ON1* was lysed to form free ON1 during the irradiation (Figure 4C). Longer exposure time led to more conversion of ON1* to ON1 (Figure 4D and Figure S5). Quantum yield for the photolysis of ON1* (φ) was 0.015 ± 0.002 determined using azobenzene actinometry.45
Since one key advantage of photoirradiation would be the flexibility to regulate a system temporally, we also studied the temporal control of NG* under alternate photoirradiation. The results showed that photoirradiation at two different time points led to the production of free ON1 periodically (Figure 4E), demonstrating that photoirradiation can temporally regulate the function of NG* and the production of free ON1.
As our goal was to use NG* as a self-regulating factor of the bulk hydrogel system, we needed to examine whether the light responsiveness of NG* in aqueous solution could be reproduced in the bulk hydrogel. To this end, we loaded NG* into the superporous hydrogel and treated the NG*-loaded hydrogel with UV irradiation. As revealed by the absorption spectrum (Figure 4F), photolysis of the hydrogel produced a new absorption peak in the range of 350–400 nm, which was attributed to the characteristic absorption of the nitrosobenzyl byproduct. Accordingly, the color of the hydrogel changed from white to pale yellow (Figure S6). These data suggest that NG* could be activated in the bulk hydrogel.
Retention of NG* in ON2-functionalized superporous hydrogel
ON2 is a nucleic acid aptamer. Nucleic acid aptamers are synthetic single-stranded oligonucleotide ligands with the ability to bind target molecules physically with high affinities and specificities.46–47 In this work, we used an aptamer binding to platelet-derived growth factor BB (PDGF-BB) as a model to synthesize the DNA-protein complex. It is important to note that any protein can be developed into the form of DNA-protein complex since aptamers can in principle be selected ex vivo against any target protein. ON2 was conjugated to the bulk hydrogel as a pendant motif via the co-polymerization of methacryl-ON2 with hydrogel monomers. To demonstrate the success of ON2 conjugation, the hydrogel was incubated with a fluorophore-labeled complementary sequence (i.e., TYE665-CS) that is complementary to the ON2. The fluorescent imaging analysis showed that the hydrogels exhibited strong red fluorescence after the TYE665-CS treatment, demonstrating the successful ON2 conjugation to the hydrogel (Figure 5A, a).
Figure 5.
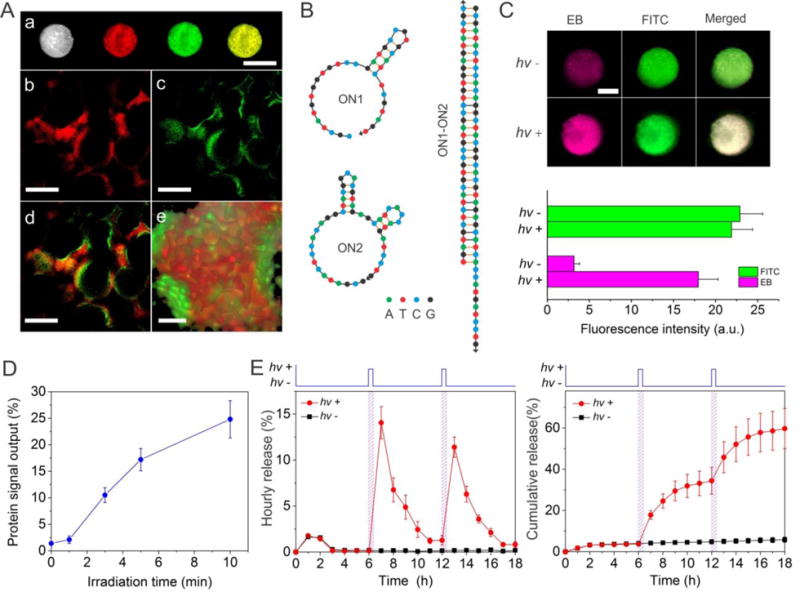
Examination of biomimetic signal output for protein release mediated by sequential photoreaction and hybridization. (A) Images of the hydrogel system. (a): Bright field and fluorescence images of the system loaded with NG* examined at the macroscopic level. Scale bar: 2 mm. (b–e): Confocal fluorescence images. (b): ON2 in the bulk hydrogel was stained with TYE665-CS (red). (c): The nanogel was doped with FITC (green). (d): Merged image of b and c. (e): Z-stack imaging of the system under high magnification. Scale bar: 250 μm for b–d, 50 μm for e. (B) Secondary structures of ON1, ON2 and their duplex as analyzed using the software from IDT (http://www.idtdna.com/calc/analyzer). (C) Fluorescence images of the system before and after photoirradiation. Pink: ethidium bromide (EB); Green: FITC. Scale bar: 2 mm. The values of EB and FITC intensity are also quantitatively shown in the bottom graph for clear comparison. (D) Irradiation time dependent protein signal output from hydrogel system. (E) Measurement of hourly (left) and cumulative (right) protein release when the system was photo-irradiated. The duration of photoirradiation was 5 min. The systems were exposed to UV light at 6 and 12 hours, respectively.
The loading and distribution of NG* within the hydrogel was evaluated using a FITC-labeled NG*. Green fluorescence was observed throughout the bulk hydrogel (Figure 5A). It demonstrates that NG* could be loaded and distributed in the porous bulk hydrogel. As NG* was used to provide ON1 for programmed protein release, it is important to retain NG* in the bulk hydrogel. A straightforward method to achieve this goal is to apply hybridization between ON1 and ON2 to retain NG* in the bulk hydrogel. To satisfy this requirement, ON1 with 5′-hexynyl moiety was directly conjugated to NG via click chemistry (Figure S7). The immobilized ON1 allows for NG* retention as its orientation is favorable for ON1-ON2 hybridization. The result showed that the retention of native NG was less than 42.7% by day 1 and only 1.65% by day 6 (Figure S7B). By contrast, after the initial loss of 4.3% NG*, the majority of NG* was retained in bulk hydrogel and the retention of NG* by day 6 was 91.3%. Since using the ratio of ON1 to NG* at 10:90 led to the retention of the majority of NG* in the ON2-functionalized superporous hydrogel (Figure S7C), this ratio was used in the following studies.
Evaluation of photoreaction and hybridization in hydrogel
After the demonstration of NG* loading in the superporous hydrogel, we studied NG* activation and ON1-ON2 hybridization in the hydrogel system. Once NG* is activated, ON1 would be freed and diffuse to ON2 to form the ON1-ON2 duplex (Figure 5B). To characterize the hybridization, after the treatment of photoirradiation, the hydrogel was stained with ethidium bromide (EB). As EB can be intercalated into the duplexes to produce fluorescence signal, the formation of ON1-ON2 duplexes would increase the fluorescence intensity of the hydrogel system. As expected, the system exhibited weak fluorescence before photoirradiation whereas it exhibited much stronger fluorescence after photoirradiation (Figure 5C, shown in pink color). The intensity of green fluorescence from FITC-doped NG* (used as an internal control) in the system remained constant with or without the treatment of photoirradiation (Figure 5C). Therefore, these data demonstrate that ON1* on NG* can be activated to free ON1 for hybridization with the ON2 in the system.
Examination of protein output of the hydrogel system
To evaluate the outcome of photoirradiation in protein release control, PDGF-BB was loaded into the hydrogel system to form the DNA-protein complex by binding to ON2 (i.e., anti-PDGF-BB aptamer). Since both NG* and PDGF-BB can bind ON2, we examined the effect of NG* loading on PDGF-BB retention. The loading of NG* up to 10 μg led to a negligible effect on PDGF-BB retention (Figure S8). Thus, we used 10 μg NG* per hydrogel in the following experiments.
The conformation of ON2 changes upon hybridization with ON1. Since the conformation of an aptamer is important to its function,48 the ON1-ON2 hybridization would cause the dissociation of the ON2-PDGF-BB complex. Resultantly, PDGF-BB would be released as a signal output. Indeed photoirradiation-mediated ON1 activation and release triggered PDGF-BB output from the system (Figure 5D). The increase of the irradiation time led to more PDGF-BB output. The output was 10.5% after 3 min of photoirradiation and 17.2% after 5 min of photoirradiation. Notably, the hydrogel system did not exhibit a significant change in the elastic modulus after photoirradiation, which suggests that photoirradiation did not change the bulk properties of the system (Figure S9). Since biological systems can often respond to external signals regularly, we further regulated the biomimetic system by light to provide a signal output at different times. Following photoirradiation at 6 and 12 hours, the PDGF-BB release from the system reached peak release levels twice (Figure 5E). The amount of released PDGF-BB was 16.1% and 11.4%, respectively. The signal output decayed to a valley after 5 to 6 hours. By contrast, the signal output of PDGF-BB from the system was as low as 0.2% at each time point if the system was not treated with photoirradiation.
We further evaluated the potential application of this hydrogel system by examining the regulation of cell behavior. Cell migration is important to a variety of physiological processes such as angiogenesis, embryonic development, tissue regeneration and immune surveillance.49 For instance, during the procedure of angiogenesis, smooth muscle cells migrate and grow on the newly formed endothelial cells for the maturation of blood vessels.50 Thus, the regulation of cell migration is important in various applications. Since PDGF-BB is an essential biochemical cue for regulating the migration of smooth muscle cells (SMCs),51 a SMC wound closure assay was used to test the outcome of the signal output. The results showed that the hydrogels with the treatment of photoirradiation could induce the migration of SMCs (Figure 6).
Figure 6.
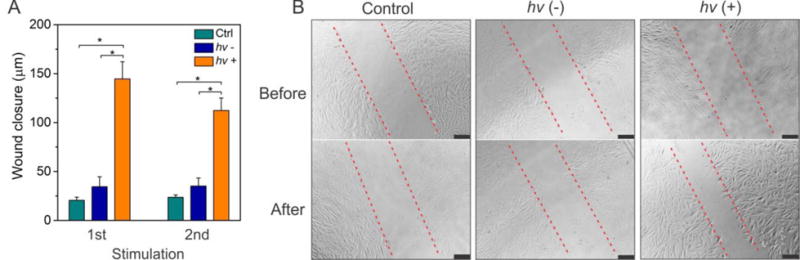
Protein output for stimulation of cell migration. (A) Periodic PDGF-BB output from photoirradiated hydrogels for cell stimulation. After the release experiment started, the hydrogels were photoirradiated twice at 6 and 12 h, respectively. (B) Representative images of SMCs before and after the stimulation with the output PDGF-BB. Cells were cultured for 6h before the imaging. Error bars represent s. e. m (n = 3). *, p < 0.001.
Since the outcome of this sequential photoreaction and hybridization is protein release, one may suggest the direct use of ON1 as an external signal to trigger the protein-DNA complex instead of using sequential photoreaction and hybridization reaction. The concern with using ON1 as an external signal is that the access of the hydrogel to ON1 may be limited in real applications. For instance, ON1 may not have access to the hydrogel system implanted in vivo if ON1 is quickly cleared by the body or if the hydrogel system is not accessible to ON1. Moreover, even if ON1 can approach the vicinity of the hydrogel system, a large amount of ON1 would be needed for sufficient ON1 diffusion into the hydrogel system since a high concentration gradient is a prerequisite for molecular diffusion in a substrate.9 These potential concerns do not exist when ON1 and the DNA-protein complex are integrated into one molecular system as nature does similarly for signal transduction in a cell. Thus, since physical signals such as light irradiation can reach a system remotely, a molecular system regulated by sequential photoreaction and hybridization would be more flexible and feasible for potential applications.
While we demonstrated that an external UV light was converted into an internal biomolecular signal to dissociate a bound protein from a biomolecular complex, it is known that UV irradiation has a limited penetration depth in materials such as living tissues. This potential issue could be addressed by using near infrared (NIR) light responsive photolabile compounds or upconverting nanoparticles due to the deeper tissue penetration of NIR light.52–53 Thus, it is possible that the current system can be optimized in different ways such as the integration of NIR radiation with upconverting nanoparticles or two-photon irradiation in the future work.
CONCLUSION
In summary, this work has demonstrated a hydrogel system that can release proteins via sequential photoreaction and hybridization in a self-programming manner. The procedures of both entrapping and releasing proteins work under mild conditions without the need of changing bulk properties of the system or involving unnecessary chemical conditions. This hydrogel system may be tuned to control the release of other fragile bioactive substances such as vaccines, viruses or cells. We envision that this bioinspired self-programming system is promising for broad biological and biomedical applications.
Supplementary Material
Acknowledgments
This work is supported in part by the U.S. NSF CAREER program under Award No. DMR-1332351 and the National Heart, Lung, and Blood Institute of the NIH under Award No. R01HL122311. We thank the Husk Institute Microscopy Facility (University Park, PA), Prof. Ralph Colby, Dr. Behzad Nazari and Dr. Peng Shi for technical support.
Footnotes
Supporting Information. Experimental methods, synthesis of linkers and additional characterization figures. This material is available free of charge via the Internet at http://pubs.acs.org.
References
- 1.Russew MM, Hecht S. Photoswitches: from molecules to materials. Adv Mater. 2010;22:3348–3360. doi: 10.1002/adma.200904102. [DOI] [PubMed] [Google Scholar]
- 2.Lai JP, Zhang YX, Pasquale N, Lee KB. An upconversion nanoparticle with orthogonal emissions using dual nir excitations for controlled two-way photoswitching. Angew Chem Int Ed. 2014;53:14419–14423. doi: 10.1002/anie.201408219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kumpfer JR, Rowan SJ. Thermo-, photo-, and chemo-responsive shape-memory properties from photo-cross-linked metallo-supramolecular polymers. J Am Chem Soc. 2011;133:12866–12874. doi: 10.1021/ja205332w. [DOI] [PubMed] [Google Scholar]
- 4.Duan QP, Cao Y, Li Y, Hu XY, Xiao TX, Lin C, Pan Y, Wang LY. pH-responsive supramolecular vesicles based on water-soluble pillar[6]arene and ferrocene derivative for drug delivery. J Am Chem Soc. 2013;135:10542–10549. doi: 10.1021/ja405014r. [DOI] [PubMed] [Google Scholar]
- 5.Yan L, Crayton SH, Thawani JP, Amirshaghaghi A, Tsourkas A, Cheng Z. A pH-responsive drug-delivery platform based on glycol chitosan-coated liposomes. Small. 2015;11:4870–4874. doi: 10.1002/smll.201501412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lee MR, Baek KH, Jin HJ, Jung YG, Shin I. Targeted enzyme-responsive drug carriers: Studies on the delivery of a combination of drugs. Angew Chem Int Ed. 2004;43:1675–1678. doi: 10.1002/anie.200353204. [DOI] [PubMed] [Google Scholar]
- 7.Su T, Tang Z, He HJ, Li WJ, Wang X, Liao CN, Sun Y, Wang QG. Glucose oxidase triggers gelation of N-hydroxyimide-heparin conjugates to form enzyme-responsive hydrogels for cell-specific drug delivery. Chem Sci. 2014;5:4204–4209. [Google Scholar]
- 8.Zeng YZ, Lu JQ. Optothermally responsive nanocomposite generating mechanical forces for cells enabled by few-walled carbon nanotubes. ACS Nano. 2014;8:11695–11706. doi: 10.1021/nn505042b. [DOI] [PubMed] [Google Scholar]
- 9.Battig MR, Soontornworajit B, Wang Y. Programmable release of multiple protein drugs from aptamer-functionalized hydrogels via nucleic acid hybridization. J Am Chem Soc. 2012;134:12410–12413. doi: 10.1021/ja305238a. [DOI] [PubMed] [Google Scholar]
- 10.Mura S, Nicolas J, Couvreur P. Stimuli-responsive nanocarriers for drug delivery. Nat Mater. 2013;12:991–1003. doi: 10.1038/nmat3776. [DOI] [PubMed] [Google Scholar]
- 11.Kim J, Yoon J, Hayward RC. Dynamic display of biomolecular patterns through an elastic creasing instability of stimuli-responsive hydrogels. Nat Mater. 2010;9:159–164. doi: 10.1038/nmat2606. [DOI] [PubMed] [Google Scholar]
- 12.DeForest CA, Anseth KS. Cytocompatible click-based hydrogels with dynamically tunable properties through orthogonal photoconjugation and photocleavage reactions. Nat Chem. 2011;3:925–931. doi: 10.1038/nchem.1174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zhang ZY, Chen NC, Li SH, Battig MR, Wang Y. Programmable hydrogels for controlled cell catch and release using hybridized aptamers and complementary sequences. J Am Chem Soc. 2012;134:15716–15719. doi: 10.1021/ja307717w. [DOI] [PubMed] [Google Scholar]
- 14.Abendroth JM, Bushuyev OS, Weiss PS, Barrett CJ. Controlling motion at the nanoscale: rise of the molecular machines. ACS Nano. 2015;9:7746–7768. doi: 10.1021/acsnano.5b03367. [DOI] [PubMed] [Google Scholar]
- 15.Otto S. Dynamic molecular networks: from synthetic receptors to self-replicators. Acc Chem Res. 2012;45:2200–2210. doi: 10.1021/ar200246j. [DOI] [PubMed] [Google Scholar]
- 16.Lu Y, Sun WJ, Gu Z. Stimuli-responsive nanomaterials for therapeutic protein delivery. J Control Release. 2014;194:1–19. doi: 10.1016/j.jconrel.2014.08.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Park TG. Temperature modulated protein release from pH/temperature-sensitive hydrogels. Biomaterials. 1999;20:517–521. doi: 10.1016/s0142-9612(98)00197-5. [DOI] [PubMed] [Google Scholar]
- 18.Lee KY, Peters MC, Anderson KW, Mooney DJ. Controlled growth factor release from synthetic extracellular matrices. Nature. 2000;408:998–1000. doi: 10.1038/35050141. [DOI] [PubMed] [Google Scholar]
- 19.Richardson TP, Peters MC, Ennett AB, Mooney DJ. Polymeric system for dual growth factor delivery. Nat Biotechnol. 2001;19:1029–1034. doi: 10.1038/nbt1101-1029. [DOI] [PubMed] [Google Scholar]
- 20.Hoffman AS. Hydrogels for biomedical applications. Adv Drug Deliver Rev. 2002;54:3–12. doi: 10.1016/s0169-409x(01)00239-3. [DOI] [PubMed] [Google Scholar]
- 21.Yu L, Ding JD. Injectable hydrogels as unique biomedical materials. Chem Soc Rev. 2008;37:1473–1481. doi: 10.1039/b713009k. [DOI] [PubMed] [Google Scholar]
- 22.Li J, Mo LT, Lu CH, Fu T, Yang HH, Tan WH. Functional nucleic acid-based hydrogels for bioanalytical and biomedical applications. Chem Soc Rev. 2016;45:1410–1431. doi: 10.1039/c5cs00586h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Vermonden T, Censi R, Hennink WE. Hydrogels for protein delivery. Chem Rev. 2012;112:2853–2888. doi: 10.1021/cr200157d. [DOI] [PubMed] [Google Scholar]
- 24.Hu YW, Guo WW, Kahn JS, Aleman-Garcia MA, Willner I. A shape-memory DNA-based hydrogel exhibiting two internal memories. Angew Chem Int Edit. 2016;55:4210–4214. doi: 10.1002/anie.201511201. [DOI] [PubMed] [Google Scholar]
- 25.Huang FJ, Liao WC, Sohn YS, Nechushtai R, Lu CH, Willner I. Light-responsive and ph-responsive DNA microcapsules for controlled release of loads. J Am Chem Soc. 2016;138:8936–8945. doi: 10.1021/jacs.6b04773. [DOI] [PubMed] [Google Scholar]
- 26.Tomatsu I, Peng K, Kros A. Photoresponsive hydrogels for biomedical applications. Adv Drug Deliver Rev. 2011;63:1257–1266. doi: 10.1016/j.addr.2011.06.009. [DOI] [PubMed] [Google Scholar]
- 27.Fomina N, Sankaranarayanan J, Almutairi A. Photochemical mechanisms of light-triggered release from nanocarriers. Adv Drug Deliver Rev. 2012;64:1005–1020. doi: 10.1016/j.addr.2012.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Yan B, Boyer JC, Habault D, Branda NR, Zhao Y. Near infrared light triggered release of biomacromolecules from hydrogels loaded with upconversion nanoparticles. J Am Chem Soc. 2012;134:16558–16561. doi: 10.1021/ja308876j. [DOI] [PubMed] [Google Scholar]
- 29.Pasparakis G, Manouras T, Vamvakaki M, Argitis P. Harnessing photochemical internalization with dual degradable nanoparticles for combinatorial photo-chemotherapy. Nat Commun. 2014;5 doi: 10.1038/ncomms4623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Yang HH, Liu HP, Kang HZ, Tan WH. Engineering target-responsive hydrogels based on aptamer-target interactions. J Am Chem Soc. 2008;130:6320–6321. doi: 10.1021/ja801339w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Azagarsamy MA, Anseth KS. Wavelength-controlled photocleavage for the orthogonal and sequential release of multiple proteins. Angew Chem Int Ed. 2013;52:13803–13807. doi: 10.1002/anie.201308174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Spicer CD, Davis BG. Selective chemical protein modification. Nat Commun. 2014;5:4740. doi: 10.1038/ncomms5740. [DOI] [PubMed] [Google Scholar]
- 33.Veronese FM. Peptide and protein pegylation: a review of problems and solutions. Biomaterials. 2001;22:405–417. doi: 10.1016/s0142-9612(00)00193-9. [DOI] [PubMed] [Google Scholar]
- 34.Choe HW, Kim YJ, Park JH, Morizumi T, Pai EF, Krauss N, Hofmann KP, Scheerer P, Ernst OP. Crystal structure of metarhodopsin II. Nature. 2011;471:651–U137. doi: 10.1038/nature09789. [DOI] [PubMed] [Google Scholar]
- 35.Lerea CL, Somers DE, Hurley JB, Klock IB, Buntmilam AH. Identification of specific transducin alpha-subunits in retinal rod and cone photoreceptors. Science. 1986;234:77–80. doi: 10.1126/science.3529395. [DOI] [PubMed] [Google Scholar]
- 36.Azagarsamy MA, Alge DL, Radhakrishnan SJ, Tibbitt MW, Anseth KS. Photocontrolled nanoparticles for on-demand release of proteins. Biomacromolecules. 2012;13:2219–2224. doi: 10.1021/bm300646q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zeng ZY, Hoshino Y, Rodriguez A, Yoo HS, Shea KJ. Synthetic polymer nanoparticles with antibody-like affinity for a hydrophilic peptide. ACS Nano. 2010;4:199–204. doi: 10.1021/nn901256s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kolb HC, Finn MG, Sharpless KB. Click chemistry: diverse chemical function from a few good reactions. Angew Chem Int Ed. 2001;40:2004–2021. doi: 10.1002/1521-3773(20010601)40:11<2004::AID-ANIE2004>3.0.CO;2-5. [DOI] [PubMed] [Google Scholar]
- 39.Gemeinhart RA, Park H, Park K. Pore structure of superporous hydrogels. Polym Advan Technol. 2000;11:617–625. [Google Scholar]
- 40.Zehavi U, Amit B, Patchorn A. Light-sensitive glycosides.1. 6-nitroveratryl β-D-glucopyranoside and 2-nitrobenzyl β-D-glucopyranoside. J Org Chem. 1972;37:2281–285. [Google Scholar]
- 41.Self CH, Thompson S. Light activatable antibodies: models for remotely activatable proteins. Nat Med. 1996;2:817–820. doi: 10.1038/nm0796-817. [DOI] [PubMed] [Google Scholar]
- 42.Heckel A, Mayer G. Light regulation of aptamer activity: an anti-thrombin aptamer with caged thymidine nucleobases. J Am Chem Soc. 2005;127:822–823. doi: 10.1021/ja043285e. [DOI] [PubMed] [Google Scholar]
- 43.Tan XY, Li BB, Lu XG, Jia F, Santori C, Menon P, Li H, Zhang BH, Zhao JJ, Zhang K. Light-triggered, self-immolative nucleic acid-drug nanostructures. J Am Chem Soc. 2015;137:6112–6115. doi: 10.1021/jacs.5b00795. [DOI] [PubMed] [Google Scholar]
- 44.DeForest CA, Tirrell DA. A photoreversible protein-patterning approach for guiding stem cell fate in three-dimensional gels. Nat Mater. 2015;14:523–531. doi: 10.1038/nmat4219. [DOI] [PubMed] [Google Scholar]
- 45.Gauglitz G. Azobenzene as a convenient actinometer for determination of quantum yields of photoreactions. J Photochem. 1976;5:41–47. [Google Scholar]
- 46.Ellington AD, Szostak JW. In vitro selection of rna molecules that bind specific ligands. Nature. 1990;346:818–822. doi: 10.1038/346818a0. [DOI] [PubMed] [Google Scholar]
- 47.Tuerk C, Gold L. Systematic evolution of ligands by exponential enrichment: RNA ligands to bacteriophage T4 DNA polymerase. Science. 1990;249:505–510. doi: 10.1126/science.2200121. [DOI] [PubMed] [Google Scholar]
- 48.Jayasena SD. Aptamers: an emerging class of molecules that rival antibodies in diagnostics. Clin Chem. 1999;45:1628–1650. [PubMed] [Google Scholar]
- 49.Ridley AJ, Schwartz MA, Burridge K, Firtel RA, Ginsberg MH, Borisy G, Parsons JT, Horwitz AR. Cell migration: Integrating signals from front to back. Science. 2003;302:1704–1709. doi: 10.1126/science.1092053. [DOI] [PubMed] [Google Scholar]
- 50.Adams RH, Alitalo K. Molecular regulation of angiogenesis and lymphangiogenesis. Nat Rev Mol Cell Bio. 2007;8:464–478. doi: 10.1038/nrm2183. [DOI] [PubMed] [Google Scholar]
- 51.Abedi H, Dawes KE, Zachary I. Differential effects of platelet-derived growth factor BB on p125 focal adhesion kinase and paxillin tyrosine phosphorylation and on cell migration in rabbit aortic vascular smooth muscle cells and swiss 3T3 fibroblasts. J Biol Chem. 1995;270:11367–11376. doi: 10.1074/jbc.270.19.11367. [DOI] [PubMed] [Google Scholar]
- 52.Smith AM, Mancini MC, Nie SM. Bioimaging second window for in vivo imaging. Nat Nanotechnol. 2009;4:710–711. doi: 10.1038/nnano.2009.326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Idris NM, Gnanasammandhan MK, Zhang J, Ho PC, Mahendran R, Zhang Y. In vivo photodynamic therapy using upconversion nanoparticles as remote-controlled nanotransducers. Nat Med. 2012;18:1580–1585. doi: 10.1038/nm.2933. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


