Abstract
BACKGROUND CONTEXT
Intervertebral disc (IVD) degeneration, a major cause of low back pain, is considered to be induced by daily mechanical loading. Mechanical stress is widely known to affect cell survival and extracellular matrix metabolism in many cell types. Although the involvement of integrin α5β1 transmembrane mechanoreceptor in IVD degeneration has been reported, the precise function of integrin α5β1 remains obscure.
PURPOSE
To reflect IVD tissue response to mechanical stress using a dynamic loading organ culture system and elucidate the functional impact of integrin α5β1 on the pathomechanism of IVD degeneration.
STUDY DESIGN
An ex vivo study using a dynamic loading organ culture system.
METHODS
Ninety-six rat IVD explants were examined. Intervertebral discs were subjected to 1.3 MPa, 1.0 Hz dynamic compressive load in the presence or absence of an Arg-Gly-Asp (RGD) peptide with affinity to the fibronectin binding-site of integrin α5β1. Cell viability and histomorphology were assessed. The localization of integrin α5β1 in the IVD was assessed by immunohistochemistry. Gene expression levels of IVD cells were evaluated using real-time reverse transcription-polymerase chain reaction.
RESULTS
In the nucleus pulposus (NP), cell density and viability were reduced by dynamic compressive load. Histologic degenerative alterations, mainly seen in the NP, were the morphologic changes of NP cells. In both NP and annulus fibrosus (AF), immunohistochemistry revealed localization of integrin α5β1 and that the messenger-RNA expression of integrin α5β1 was increased by dynamic load. Dynamic load induced a catabolic effect, the stimulation of matrix metalloproteinase-3 and -13 gene expressions by NP and AF cells. The RGD peptide partially blocked the histologic alterations and the catabolic effect.
CONCLUSIONS
The dynamic loading organ culture system simulated cellular responses to mechanical loading of the IVD. Our results suggest that IVD cells recognize the mechanical stress through RGD integrins, particularly the α5β1 subtype that is highly expressed in NP and AF cells. Further experiments using this system will provide information about pathomechanisms of IVD degeneration through the mechanotransduction pathways.
Keywords: Intervertebral disc degeneration, Organ culture, Mechanical load, Compression, Mechanoreceptor, Integrin
Introduction
Low back pain causes severe incapacity that impacts the workforce and increases medical expenses, resulting in high socioeconomic costs globally [1]. Intervertebral disc (IVD) degeneration is considered to be a major cause of low back pain [2]. However, the precise pathomechanism of IVD degeneration remains unclear, especially the involvement of mechanical stress in IVD degeneration.
The IVD, an important load-bearing structure exposed to dynamic mechanical loads with daily activity [3], comprises an outer annulus fibrosus (AF) and an enwrapped nucleus pulposus (NP). The undegenerated IVD matrix, which contains an abundance of proteoglycan and collagen, allows movement and elasticity of the IVD tissue. In degenerated IVD tissues, phenotypic changes in NP cells and breakdown of the extracellular matrix (ECM) have been observed [4]. Because NP cells are responsible for tissue homeostasis, they are considered to play a key role in the progression of IVD degeneration [5,6].
The fact that the surrounding ECM affects cell survival, cell differentiation, and cell metabolic activity is widely known [7–9]. Matsumoto et al. [10] reported that lack of mechanical stress induced dedifferentiation of cultured NP cells and suggested that exogenous mechanical stress, such as the physical stress of the daily loading exerted on the spine, was important to maintain the phenotype of NP cells. Lotz and Chin [12] investigated that compressive load induced cell death using a mouse tail compression loading model with 1.3 MPa, which corresponded to the IVD loading produced by lifting a moderate weight in the human lumbar spine [11]. MacLean et al. [13] also reported a dynamic loading of rat tail discs and they concluded that load magnitude (1.0 MPa) and load frequencies (0.01 to 1.0 Hz) had significant effects on the metabolic response of rat tail discs.
To clarify the pathomechanism of IVD degeneration, it is necessary to understand the cell-matrix interactions transducing the mechanical stimuli to biochemical downstream responses, known as “mechanotransduction,” in IVD cells [14,15]. In many cell types, transmembrane receptors, such as integrins and CD44, are considered to transmit mechanical stimuli to the cytoskeleton and lead to subsequent remodeling [16]. A recent study described that “mechanosensing” in NP cells derived from nondegenerated human discs occurs via Arg-Gly-Asp (RGD) integrins, possibly via an integrin α5β1 subtype with an affinity to ligands having RGD amino acid sequences, such as fibronectin [17]. It has been reported that fibronectin binding to integrin α5β1 resulted in elevated expression of Bcl-2 protein, which commonly counteracts apoptosis, and detachment of fibronectin-integrin α5β1 interactions induced apoptosis [18]. Therefore, integrin-mediated events may involve histologic and biochemical alterations of IVD cells induced by dynamic mechanical stimuli.
Consequently, we hypothesized that the mechanoreceptor, integrin α5β1, is involved in the pathomechanism of IVD degeneration induced by mechanical stress. To address our hypothesis, the aims of this study were to develop a dynamic loading organ culture system and elucidate the role of integrin α5β1 in the alteration of cell viability and ECM metabolism in the process of IVD degeneration induced by mechanical stress.
Materials and Methods
All animal procedures were performed under the approval and guidance of the Animal Care and Use Committee at the authors’ institution.
Tissue preparation
After euthanasia using an intraperitoneal injection of 3 mL somnopentyl (Kyorituseiyaku, Tokyo, Japan), two consecutive motion segments, including IVDs and craniocaudal end plates, corresponding to coccygeal 6–7 and 7–8 were aseptically dissected from 48 skeletally mature (14-week-old) male Sprague-Dawley rats (CLEA Japan, Tokyo, Japan). Each IVD explant was flushed with phosphate buffered saline (PBS; Wako Pure Chemical Industries, Tokyo, Japan).
Preculture and experimental groups
Ninety-six IVD explants were precultured at 37°C in 5% CO2 and 95% air for an hour in complete tissue culture media: Dulbecco Modified Eagle Medium (DMEM; Sigma-Aldrich, St. Louis, MO, USA) supplemented with 10% fetal bovine serum (Sigma-Aldrich), 100 U/mL penicillin, and 100 mg/mL streptomycin. Then, IVDs were treated for 2 hours with either 50 µg/mL of an RGD peptide, GRGDSP (Calbiochem, Philadelphia, PA, USA) that binds to integrin ligand fibronectin binding sites or 50 µg/ml of its negative control peptide GRADSP (Calbiochem), or were left untreated according to the culture conditions described below.
Intervertebral disc explants were assigned to one of six culture groups (Table 1) as follows: Group C—unloaded culture with DMEM only (control group); Group T—unloaded culture in the presence of an RGD peptide (GRGDSP); Group N—unloaded culture in the presence of a negative control peptide (GRADSP); Group L—dynamic loaded culture in DMEM with no additive peptide; Group TL—dynamic loaded culture in the presence of GRGDSP; or Group NL—dynamic loaded culture in the presence of GRADSP. Each group consisted of 16 discs. Because the anatomic level of the disc is known to affect cell metabolism and tissue composition [19], equal numbers of each disc level were assigned to each group.
Table 1.
Culture groups
| Medium conditions | |||
|---|---|---|---|
|
|
|||
| Loading status | DMEM only |
DMEM+RGD peptide |
DMEM+negative control peptide |
| Unloaded (without stimulation) | Group C | Group T | Group N |
| Dynamic loaded (1.3 MPa, 1.0 Hz) | Group L | Group TL | Group NL |
DMEM, Dulbecco modified Eagle medium; RGD, Arg-Gly-Asp.
Dynamic loading organ culture system
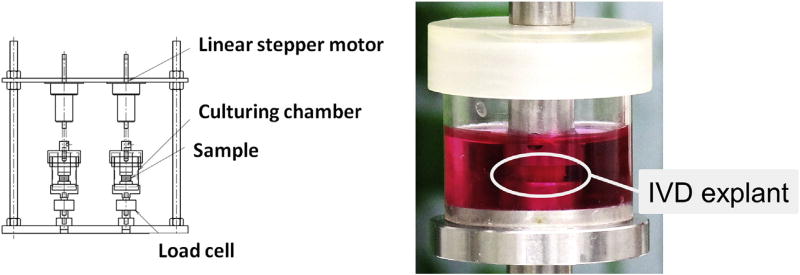
The dynamic loading organ culture system (Fig. 1, Left) comprised two sets in a culture chamber, a linear stepper motor (PFL35T-48Q4; Nippon Pulse Motor, Tokyo, Japan), which gave an axial compressive displacement to the IVD specimen, and a compression load cell (LUX-B-100N-ID; Kyowa Electric Instruments, Tokyo, Japan). These devices were monitored and controlled by the LabVIEW program (National Instrument, Austin, TX, USA).
Fig. 1.
The dynamic loading organ culture system. (Left) The dynamic loading organ culture system and (Right) the close-up view of a culture chamber. IVD explants were cultured with or without dynamic compressive load. IVD, intervertebral disc.
Precultured and treated/untreated IVD explants were sharply cut at their proximal and distal vertebrae in the axial parallel plane close to adjacent end plates. Each IVD specimen was placed on the bottom of a sterilized culture chamber with 7 mL culture media (Fig. 1, Right) corresponding to the culture group and then incubated at 37°C in 5% CO2 for 6 days. Intervertebral disc explants assigned to dynamic loaded groups (Groups L, TL, and NL) were subjected to cyclic uniaxial compressive stress (1.3 MPa, 1.0 Hz at the initial state) throughout the culture period. The applied force for providing 1.3 MPa to the IVD was calculated as 16.0 ± 1.0 N, which was preliminarily adjusted based on the calculated cross-sectional area of the cutting plane (12.2 ± 1.4 mm2 [N = 40]) using a computed tomography scan. Intervertebral disc explants of unloaded groups (Groups C, T, and N) were cultured without mechanical stimuli in the same chamber. The loading displacement was calibrated and culture media were replaced every 2 days.
Vital cell staining
At the end of the culture period, four IVD explants from each culture group were immersed in PBS with 1 µM calcein AM (Sigma-Aldrich) to stain vital cells through the formation of a fluorescent precipitate by active mitochondria and allowed to incubate for 2 hours. Excess dye was removed by placing samples in PBS for 15 minutes.
Paraffin-embedded disc tissue preparation
Ten IVD explants from each group, including the vital cell-stained IVDs, were fixed in 4% paraformaldehyde, decalcified in 10% ethylene-diamine-tetraacetic acid, embedded in paraffin, sectioned at 6 µm thickness in the midsagittal plane through the center of the cross-sections in the pre-cut vertebrae, and a couple of consecutive slides were prepared for each sample for cell viability and histomorphologic and immunohistochemical analyses.
Cell density and cell viability analysis (N = 4)
Four vital cell-stained sections from each group were counterstained with 4′, 6-diamidino-2-phenylindole (DAPI) (Molecular Probes, Eugene, OR, USA). Images were captured under fluorescence spectroscopy for calcein AM (470 nm/495 nm, excitation/emission) and DAPI (360 nm/400 nm) using the BZ-9000 confocal laser scanning microscope (Keyence, Osaka, Japan). To calculate the cell density (total cell number per millimeter2), DAPI-positive nuclei were counted in three random high-power fields (×400) in both the NP and AF regions using the BZ-9000 analysis software (Keyence). Calcein AM-positive cells (vital cells) were counted in the same way, and the percentage of vital cells in the total cell number (% vital cells) was separately calculated in the NP and AF regions.
Histomorphologic analysis (N = 4)
Four sections from each group were stained with Safranin-O fast green and hematoxylin to demonstrate general morphologic structures with proteoglycan distribution. Images were captured using the BZ-9000 microscope, and three researchers, blinded to the study, graded the sections using previously reported histologic degeneration scale [20].
Immunohistochemical analysis (N = 2)
Immunohistochemical staining was performed to determine protein presence and localization of integrin α5 and β1. Sections were incubated at 4°C overnight with one of the following antibodies: 1:100 diluted rabbit-polyclonal anti-integrin α5 antibody (sc-10729; Santa Cruz Biotechnology, Santa Cruz, CA, USA) or 1:100 diluted rabbit-monoclonal anti-integrin β1 antibody (EP1041Y; Epitomics, Burlingame, CA, USA). Negative controls in which rabbit immunoglobulin G (IgG) (Dako, Glostrup, Denmark) replaced the primary antibody (at an equal protein concentration) were included. Subsequently, sections were treated at room temperature for an hour with a peroxidase-labeled anti-rabbit antibody (Nichirei Bioscience, Tokyo, Japan). A brown reaction product was developed using peroxidase substrate 3, 3′-diaminobenzide (Dako). Counterstaining was performed with hematoxylin. Images were immediately captured at ×400 magnification using the BZ-9000 microscope.
RNA extraction and reverse transcription
Six IVD explants from each group were used for gene expression analysis. Each NP and AF tissue were dissected from the explants, pulverized, and total RNA was isolated using the RNeasy Mini Kit (Qiagen, Hilden, Germany) according to the manufacturer’s instructions. Then, 0.1 µg of RNA were reverse-transcribed in the presence of oligo d (T) primer (Applied Biosystems, Foster City, CA, USA).
Quantitative real-time reverse transcription-polymerase chain reaction analysis (N = 6)
Relative messenger-RNA (mRNA) expression levels of integrin-α5, integrin-β1, aggrecan-1, collagen-1a1, collagen-2a1, matrix metalloproteinase (MMP)-3, and MMP-13 were calculated by real-time reverse transcription-polymerase chain reaction using the ABI Prism 7500 sequence detection system (Applied Biosystems). Glyceraldehyde 3-phosphate dehydrogenase mRNA expression was measured as an endogenous control [21]. SYBR green fluorescent dye and predesigned primers (all obtained from Takarabio, Tokyo, Japan) were used according to the manufacturer’s instructions. Primer sequences in the present study are shown in Table 2. Measurements were performed in duplicate. The mRNA expressions of each gene in experimental groups were converted to a relative number representing the amount of mRNA compared with Group C using the 2−ΔΔCt method [22].
Table 2.
Primer sequences for real-time RT-PCR
| Primer sequences | |||
|---|---|---|---|
|
|
|||
| Gene | Sense (5′ → 3′) | Antisense (5′ → 3′) | Accession number |
| GAPDH | GGCACAGTCAAGGCTGAGAATG | TGGTGGTGAAGACGCCAGTA | NM_017008.3 |
| Integrin α5 | ATGAAGCCCTGAAGATGCCCTAC | TAGATGAGCAGACCTAGGAGCAGGA | NM_001108118.1 |
| Integrin β1 | CCTGTGACCCACTGCAAGGA | ACGCCTGCTACAATTGGGATG | NM_017022.2 |
| Aggrecan-1 | TCCGCTGGTCTGATGGACAC | CCAGATCATCACTACGCAGTCCTC | NM_022190.1 |
| Collagen-1a1 | CCAGCAAAGGCAATGCTGAA | TTGTTGCAGAGGCCATGGAG | NM_053304.1 |
| Collagen-2a1 | GAGGGCAACAGCAGGTTCAC | TGTGATCGGTACTCGATGATGG | NM_012929.1 |
| Fibronectin | ACTCATTTATCACGGGCCAGACA | TGGCAGTGGACGAGCTCACTA | NM_001191704.1 |
| MMP-3 | TGGACCAGGGACCAATGGA | GGCCAAGTTCATGAGCAGCA | NM_133523.2 |
| MMP-13 | CCCTGGAATTGGCGACAAAG | GCATGACTCTCACAATGCGATTAC | NM_133530.1 |
GAPDH, glyceraldehyde 3-phosphate dehydrogenase; MMP, matrix metalloproteinase.
Statistical analysis
All values were expressed as mean ± standard deviation. Analysis of variance with Tukey-Kramer post hoc test was used to assess the changes in the effects of experimental group. Statistical significance was assessed with p<.05 using PASW Statistics 18 (SPSS, Chicago, IL, USA).
Results
Analysis of cell density and viability
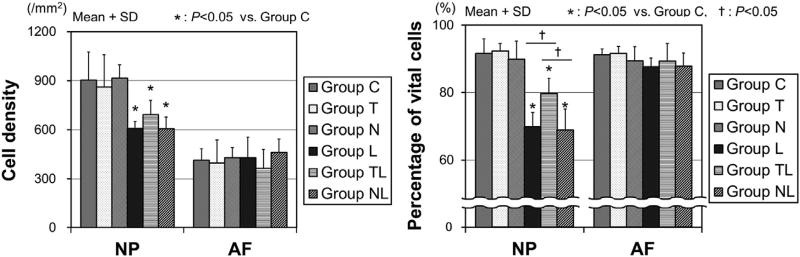
In the NP, cell density significantly decreased in dynamic loaded groups (Group L: 67.4%; Group TL: 76.8%; and Group NL: 67.2%) compared with Group C (Fig. 2, Left). In the AF, cell density was not affected by mechanical loading and RGD peptide. The percentage of vital cells in the NP significantly decreased in the dynamic loaded groups (Group L: 69.9%; Group TL: 79.7%; and Group NL: 68.8%) compared with Group C (91.6%, p<.05) (Fig. 2, Right). However, in the NP, the mean percentage of vital cells of Group TL was significantly higher than Group L and Group NL (p<.05). In the AF, the mean percentage of vital cells was maintained at over 80% in each group, with no statistical differences. Namely in the NP, dynamic load reduced cell viability, and as a result, cell density decreased. The decrease of cell viability in the NP induced by dynamic loading was partially inhibited by the application of the RGD peptide, whereas the decrease of cell density in the NP was not significantly affected by the RGD peptide under the loading condition of this study.
Fig. 2.
Cell density and viability in the NP and AF. (Left) Cell density and (Right) the percentage of vital cells. The data were obtained by N = 4 and expressed as mean+SD. Group C = control. NP, nucleus pulposus; AF, annulus fibrosus; SD, standard deviation.
Histomorphologic analysis
Sections stained with Safranin-O demonstrated that unloaded discs (Groups C, T, and N) had a round-shaped NP comprising half to three-quarters of the disc in midsagittal sections with well-defined borders between the NP and AF regions (Fig. 3, Top). Nucleus pulposus cells had a rounded shape with large vacuoles, like notochordal cells; the proteoglycan matrix was organized in slender bands among the NP cells. The AF showed a pattern of fibrocartilage lamella without a serpentine pattern or ruptured fibers. In dynamic loaded discs (Groups L, TL, and NL), the vacuoles of NP cells were reduced in density and size, the ECM was condensed, and the disc height was decreased. In the AF, the inward bulging of the inner portion (a serpentine pattern) was observed, although the outer AF demonstrated a well-defined lamella pattern without ruptured fibers. Consequently, the histologic degeneration score was significantly increased in dynamic loaded groups: Group L (7.6 ± 1.2); Group TL (6.3 ± 0.5); and Group NL (7.8 ± 1.3), compared with Group C (4.0 ± 0.0) (Fig. 3, Bottom). Group TL showed a lower score than Groups L and NL with a significant difference (p<.05). This histologic alteration, which was mainly observed in the NP, was inhibited using the RGD peptide.
Fig. 3.
Histomorphologic findings and the degeneration score. (Top) Sections stained with Safranin-O. Bars = 300 µm in whole disc photographs and 100 µm in AF and NP photographs. (Bottom) Histologic degeneration scores. The data were obtained by N = 4 and expressed as mean+SD. Group C = control. NP, nucleus pulposus; AF, annulus fibrosus; SD, standard deviation.
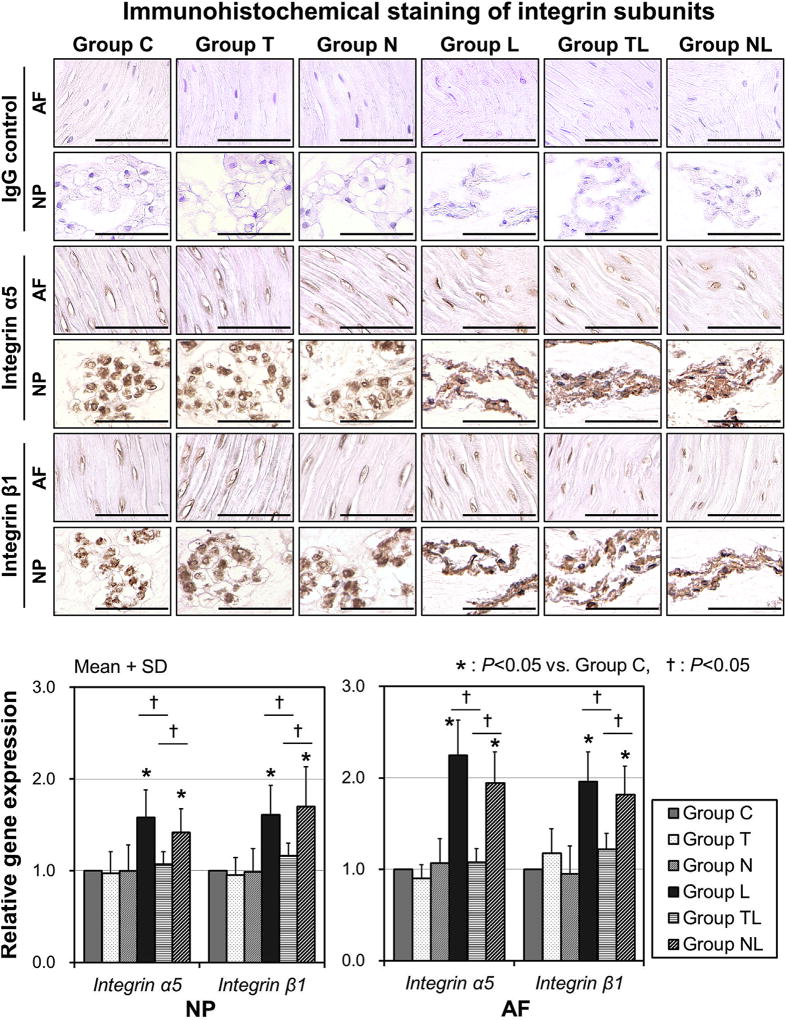
Immunohistochemical staining and mRNA expressions of integrin α5β1
In immunohistochemical staining, IgG controls were all negative and immunoreactivity for integrin α5 and β1 was observed in the NP and AF of all groups. For both integrin α5 and β1, perinuclear patterned staining was particularly prominent in large vacuolated cells in the NP of unloaded groups (Groups C, T, and N), and immunopositivity was observed in flattened NP cells of dynamic loaded groups (Groups L, TL, and NL) (Fig. 4, Top). Among these dynamic loaded groups, immunopositivity and staining patterns were not different in NP cells. Annulus fibrosus cells of all groups showed immunopositivity for integrin α5 and β1, whereas staining patterns did not differ among the six groups.
Fig. 4.
Immunohistochemical staining and messenger-RNA (mRNA) expression of integrin α5β1. (Top) Photomicrographs demonstrating immunohistochemical localization of integrin α5 and β1. Bars = 100 µm. (Bottom) Integrin α5 and β1 mRNA expressions. The data were obtained by N = 6 and expressed as mean+SD. Group C = control. IgG, immunoglobulin G; NP, nucleus pulposus; AF, annulus fibrosus; SD, standard deviation.
The mRNA quantification analysis indicated that integrin α5 and β1 expressions of Group L were both 1.6 ± 0.3-fold upregulated in NP cells (p<.05) and were upregulated 2.2 ± 0.4- and 2.0 ± 0.3-fold, respectively, in AF cells (p<.05), compared with Group C (Fig. 4, Bottom). Upregulation of integrin α5 and β1 by dynamic load was completely inhibited in Group TL using the RGD peptide. These findings suggested that the exogenous RGD peptide blocked an integrin α5β1 upregulation pathway triggered by mechanical load.
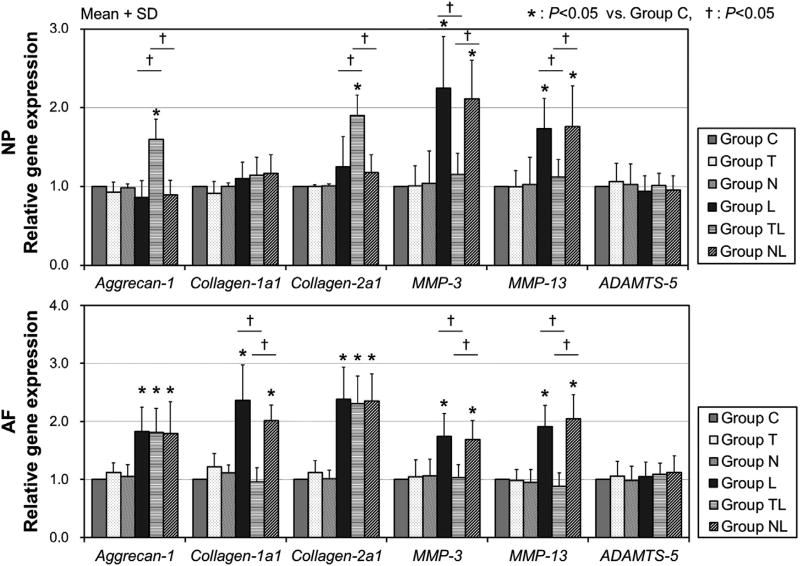
mRNA expressions of extracellular matrix genes
In NP cells, anabolic gene expressions of aggrecan-1 and collagen-2a1 were not affected in Groups L and NL; however, these mRNA expressions were significantly upregulated in Group TL (1.6 ± 0.3- and 1.9 ± 0.3-fold, respectively, p<.05). Messenger-RNA expressions of MMP-3 and -13 in NP cells were 2.2 ± 0.7- and 1.7 ± 0.4-fold, respectively, upregulated in Group L (p<.05) and 2.1 ± 0.5- and 1.8 ± 0.5-fold, respectively, upregulated in Group NL (p<.05) with significant differences, but no upregulation of MMP-3 nor MMP-13 was observed in Group TL (Fig. 5). However, mRNA expression of ADAMTS-5 was not affected by mechanical stress and the RGD peptide in NP cells.
Fig. 5.
Extracellular matrix gene expressions. Anabolic and catabolic gene expressions in NP and AF cells. The data were obtained by N = 6 and expressed as mean+SD. Group C = control. NP, nucleus pulposus; AF, annulus fibrosus; SD, standard deviation; MMP, matrix metalloproteinase.
In AF cells, mRNA expressions of aggrecan-1, collagen-1a1, and collagen-2a1 were significantly upregulated in Groups L and NL (range 1.8–2.4-fold), each compared with Group C (p<.05). The application of an RGD peptide did not affect the upregulation of aggrecan-1 and collagen-2a1 in AF cells, whereas, it inhibited the upregulation of collagen-1a1. The upregulation of collagen-1a1 was only seen in AF cells, not in NP cells. The mRNA expressions of MMP-3 and -13 in AF cells showed similar upregulation pattern of NP cells in Groups L and NL (range 1.7–2.0-fold, p<.05), but not in Group TL. In addition, mRNA expression of ADAMTS-5 was not affected by mechanical stress and the RGD peptide in AF cells.
Discussion
The present study has demonstrated that an ex vivo dynamic culture system using sustained mechanical stimulation with 1.3 MPa, 1.0 Hz cyclic axial compressive displacement induced histologic and genetic alterations in rat IVD tissues.
Elliott and Sarver [23] reported that the compressive mechanical properties of rodent caudal discs were similar to those of the human lumbar spine. Recently, we reported a rat tail temporary static compression model at 1.3 MPa that reproduced different stages of disc degeneration according to loading duration [24]. Although the 1.3 MPa loading corresponded to the human lumbar spinal loading induced by lifting a moderate weight, it seemed to be of higher magnitude for the application of dynamic loading to animal constructs, in comparison with several studies [25,26]. In the present study, the authors preliminarily examined lower magnitude of loading (data not shown). However, the decrease of cell viability induced by the loading was too small to detect the partial block effect of the RGD peptide. Consequently, we decided to use the loading regimen of 1.3 MPa and 1.0 Hz in the present study.
In 1978, Folkman and Moscona [27] reported that most types of normal cells required an attachment to the ECM to be able to proliferate and differentiate. Monolayer culture and three-dimensional matrix culture do not reflect in vivo ECM condition [10,28–30]; therefore, IVD organ culture with native ECM is essential to reveal mechanical stress induced IVD degeneration [31,32]. Because the retention of vertebra and end plates may cause concern about the limitation of culture duration through reduced diffusion, media flow devices were added in some recent studies [26,33]. However, Lim et al. [34] reported a high degree of cell viability (>95%) for up to 14 days in their organ culture system in which a media flow device was not provided. In the present study, cell viability over 90% was maintained in both NP and AF regions of unloaded IVDs on Day 6. From the result of the present study, we consider that our organ culture system does not lose reliability because of decreased cell survival in the IVD tissue through the 6-day culture.
Le Maitre et al. [17] described that human NP and AF cells expressed integrin α5β1 and that this expression was not altered with degree of degeneration; they also referred to the involvement of integrin α5β1 in IVD degeneration. In the present study, rat NP and AF cells highly expressed integrin α5 and β1 subunits compared with the expression level of other subunits (data not shown), and furthermore, high localization of integrin α5 and β1 was confirmed in both unloaded and dynamic loaded IVDs. The integrin family belonged to cell surface receptors that link cells to their surrounding ECM. The signal from integrin family responding to the environmental stimuli regulates cellular homeostasis, including cell survival, differentiation, proliferation, and biosynthetic activity [35,36]. Furthermore, the fact that mechanical stress induced integrin α5β1 mRNA expression and that the RGD peptide inhibited this upregulation suggested that integrin α5β1 was involved in the cellular response of IVDs to mechanical stress.
The authors considered the importance of investigating IVD maturation, specifically the change in NP cell phenotype from notochordal to chondrocyte-like, for a better understanding of IVD degeneration. Notochordal cells maintain nucleus hydration by stimulating surrounding chondrocyte-like NP cells to increase ECM production [37]. In humans, the disappearance of notochordal cells is believed to be among the first signs of disc degeneration [38]. Guehring et al. [39] revealed that notochordal cells are less resistant to mechanical stress than chondrocyte-like cells and that mechanical stress instigates the disappearance of notochordal cells. However, whether this disappearance was caused by the apoptosis or the phenotypic change of notochordal cells remains obscure. In the present study, a major finding of histologic analysis was observed in the morphology of NP cells, in which application of dynamic load affected the morphologic alteration of NP cells from notochordal-like round-shaped large cells to chondrocyte-like mature NP cells. Our results are in accordance with the previous study using a static compression model [39]. Because the present study used an ex vivo organ culture system, other cells were never recruited from different tissues. In addition, the inner layer of the AF did not undergo degenerative changes; therefore, it is reasonable to understand that the disappearance of notochordal cells was mainly caused by phenotypic changes, not by cell death.
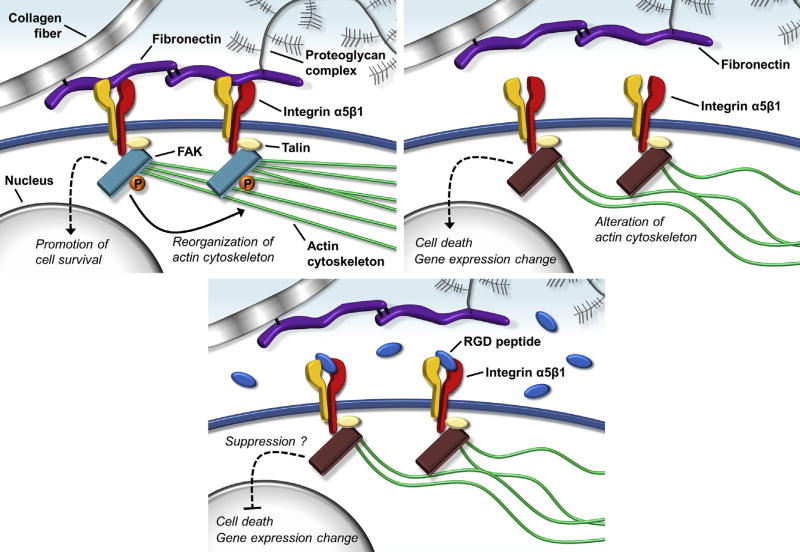
Previous studies revealed that the decrease of NP cell density and viability correlated with the onset of IVD degeneration, especially in the early stage [40,41]. In the present study, the application of dynamic load decreased cell density and viability in the NP region. In addition to these histologic appearances, the results of the mRNA analysis showed that dynamic compressive load enhanced gene expressions of MMP-3 and -13 in NP cells. Matrix metalloproteinase-3 is a major aggrecan-degrading MMP that activates many other MMPs, including MMP-13 [42]. Matrix metalloproteinase-13, one of the collagenases, has a particular affinity for collagen Type II, an ECM substrate that is abundant within the NP region of IVDs [43]. The involvement of MMP-3 and -13 in IVD degeneration has been elucidated by several studies [13,44,45]. Consequently, in the present study, the cellular response of NP tissue to dynamic load may be considered the first signs of disc degeneration (Fig. 6, Top). Furthermore, the RGD peptide suppressed phenotypic changes, decreased cell viability, and upregulated catabolic gene mRNA expressions (Fig. 6, Bottom). Therefore, integrin α5β1 is considered to play an important role in cellular responses to mechanical stress.
Fig. 6.
A functional model of integrin α5β1. (Top Left) Fibronectin-cell interaction via integrin α5β1, (Top Right) fibronectin detachment, and (Bottom) binding of an RGD peptide. FAK, focal adhesion kinase; P, phosphoric acid; RGD, Arg-Gly-Asp.
In the AF, the present study found that cell morphology and ECM structure were not significantly changed by dynamic mechanical load. Alterations in cell density and viability were also not observed. In contrast, we found that NP cells were sensitive to mechanical stress. Therefore, it was considered that spindle-shaped AF cells have stronger resistance to mechanical stress than NP cells. Messenger-RNA expressions of ECM substrates were increased by dynamic load in the AF, although the upregulation of these genes was not observed in the NP. In addition, catabolic genes, MMP-3 and -13, were also upregulated in the AF. However, histologic analysis of the AF did not reveal apparent degenerative findings compared with the NP. The results suggested that both anabolic and catabolic gene upregulations in the AF reflect mechanically induced stimulation of ECM remodeling to adapt to the mechanical perturbations in this study. These metabolic alterations, except for collagen Type I, were inhibited by the RGD peptide; therefore, integrin α5β1 might be involved in these alterations of metabolic activity, leading to ECM remodeling.
From the results of the present study, especially in the early stage of IVD degeneration, mechanical stress transmitted through integrin α5β1 is considered to have caused IVD degeneration. Therefore, at the clinical setting, it would be expected that the preventive application of the integrin α5β1 functional inhibition to the progressing disc degenerative disease will have a greater feasibility than therapeutic treatment of developed IVD degeneration. In concrete terms, a great concern of the authors is the prevention of adjacent IVD disorder after lumbar fusion surgery and the restraint of disc degeneration after removal of disc herniation.
Although the organ culture system of the present study offers improvements by applying a precise mechanical load to IVD explants and maintaining the native ECM components and cell-ECM interactions, it faces some limitations. First, the mechanical load was fixed at 1.3 MPa and 1.0 Hz. Various mechanical loads, especially lower loads, and fewer cycles should be attempted, in which case the key mechanoreceptor may shift from integrin α5β1 to others, CD44 or other integrin subunits. Second, the alteration after mechanical loading was mainly observed in NP cells and the cell phenotype of those NP cells has not been investigated. Namely, the fate of notochordal cells, which are considered to play a major role in IVD degeneration, must be revealed in further studies.
In conclusion, we have demonstrated a novel dynamic organ culture system to investigate IVD cellular responses to mechanical stress under physiologic and pathophysiologic conditions. The mechanotransduction pathway mediated by integrin α5β1 may be involved in IVD maturation, leading to IVD degeneration. Further experiments are now underway to elucidate the pathomechanisms of IVD degeneration through these mechanotransduction pathways.
Acknowledgments
The work was supported by a grant for scientific research from the ministry of the authors’ country.
Footnotes
FDA device/drug status: Not applicable.
Author disclosures: TK: Nothing to disclose. KK: Nothing to disclose. YM: Nothing to disclose. YK: Nothing to disclose. TY: Nothing to disclose. HH: Nothing to disclose. SM: Nothing to disclose. YT: Nothing to disclose. KM: Nothing to disclose. TT: Nothing to disclose. MD: Grant: Grant-in-Aid for Scientific Research from the Ministry of Education, Culture, Sports, Science, and Technology of Japan (Grant number: 24592197) (D, no share, 100%). MK: Nothing to disclose. NI: Nothing to disclose. KM: Nothing to disclose. KN: Nothing to disclose.
The disclosure key can be found on the Table of Contents and at www.TheSpineJournalOnline.com.
The authors declare no conflict of interest regarding this study.
References
- 1.Andersson GB. Epidemiological features of chronic low-back pain. Lancet. 1999;354:581–5. doi: 10.1016/S0140-6736(99)01312-4. [DOI] [PubMed] [Google Scholar]
- 2.Luoma K, Riihimaki H, Luukkonen R, Raininko R, Viikari-Juntura E, Lamminen A. Low back pain in relation to lumbar disc degeneration. Spine. 2000;25:487–92. doi: 10.1097/00007632-200002150-00016. [DOI] [PubMed] [Google Scholar]
- 3.Urban JP, Roberts S. Degeneration of the intervertebral disc. Arthritis Res Ther. 2003;5:120–30. doi: 10.1186/ar629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Roughley PJ. Biology of intervertebral disc aging and degeneration: involvement of the extracellular matrix. Spine. 2004;29:2691–9. doi: 10.1097/01.brs.0000146101.53784.b1. [DOI] [PubMed] [Google Scholar]
- 5.Antoniou J, Steffen T, Nelson F, Winterbottom N, Hollander AP, Poole RA, et al. The human lumbar intervertebral disc: evidence for changes in the biosynthesis and denaturation of the extracellular matrix with growth, maturation, ageing, and degeneration. J Clin Invest. 1996;98:996–1003. doi: 10.1172/JCI118884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Urban JP. The role of the physicochemical environment in determining disc cell behaviour. Biochem Soc Trans. 2002;30(Pt 6):858–64. doi: 10.1042/bst0300858. [DOI] [PubMed] [Google Scholar]
- 7.Gilchrist CL, Chen J, Richardson WJ, Loeser RF, Setton LA. Functional integrin subunits regulating cell-matrix interactions in the intervertebral disc. J Orthop Res. 2007;25:829–40. doi: 10.1002/jor.20343. [DOI] [PubMed] [Google Scholar]
- 8.Hayes AJ, Benjamin M, Ralphs JR. Extracellular matrix in development of the intervertebral disc. Matrix Biol. 2001;20:107–21. doi: 10.1016/s0945-053x(01)00125-1. [DOI] [PubMed] [Google Scholar]
- 9.Labat-Robert J. Cell-matrix interactions in aging: role of receptors and matricryptins. Ageing Res Rev. 2004;3:233–47. doi: 10.1016/j.arr.2003.10.002. [DOI] [PubMed] [Google Scholar]
- 10.Matsumoto T, Kawakami M, Kuribayashi K, Takenaka T, Tamaki T. Cyclic mechanical stretch stress increases the growth rate and collagen synthesis of nucleus pulposus cells in vitro. Spine. 1999;24:315–9. doi: 10.1097/00007632-199902150-00002. [DOI] [PubMed] [Google Scholar]
- 11.Wilke HJ, Neef P, Caimi M, Hoogland T, Claes LE. New in vivo measurements of pressures in the intervertebral disc in daily life. Spine. 1999;24:755–62. doi: 10.1097/00007632-199904150-00005. [DOI] [PubMed] [Google Scholar]
- 12.Lotz JC, Chin JR. Intervertebral disc cell death is dependent on the magnitude and duration of spinal loading. Spine. 2000;25:1477–83. doi: 10.1097/00007632-200006150-00005. [DOI] [PubMed] [Google Scholar]
- 13.Maclean JJ, Lee CR, Alini M, Iatridis JC. Anabolic and catabolic mRNA levels of the intervertebral disc vary with the magnitude and frequency of in vivo dynamic compression. J Orthop Res. 2004;22:1193–200. doi: 10.1016/j.orthres.2004.04.004. [DOI] [PubMed] [Google Scholar]
- 14.Iatridis JC, MacLean JJ, Roughley PJ, Alini M. Effects of mechanical loading on intervertebral disc metabolism in vivo. J Bone Joint Surg Am. 2006;88(Suppl 2):41–6. doi: 10.2106/JBJS.E.01407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hee HT, Zhang J, Wong HK. An in vitro study of dynamic cyclic compressive stress on human inner annulus fibrosus and nucleus pulposus cells. Spine J. 2010;10:795–801. doi: 10.1016/j.spinee.2010.06.009. [DOI] [PubMed] [Google Scholar]
- 16.Setton LA, Chen J. Mechanobiology of the intervertebral disc and relevance to disc degeneration. J Bone Joint Surg Am. 2006;88(Suppl 2):52–7. doi: 10.2106/JBJS.F.00001. [DOI] [PubMed] [Google Scholar]
- 17.Le Maitre CL, Frain J, Millward-Sadler J, Fotheringham AP, Freemont AJ, Hoyland JA. Altered integrin mechanotransduction in human nucleus pulposus cells derived from degenerated discs. Arthritis Rheum. 2009;60:460–9. doi: 10.1002/art.24248. [DOI] [PubMed] [Google Scholar]
- 18.Ruoslahti E, Reed JC. Anchorage dependence, integrins, and apoptosis. Cell. 1994;77:477–8. doi: 10.1016/0092-8674(94)90209-7. [DOI] [PubMed] [Google Scholar]
- 19.Wiseman MA, Birch HL, Akmal M, Goodship AE. Segmental variation in the in vitro cell metabolism of nucleus pulposus cells isolated from a series of bovine caudal intervertebral discs. Spine. 2005;30:505–11. doi: 10.1097/01.brs.0000154615.22311.66. [DOI] [PubMed] [Google Scholar]
- 20.Masuda K, Aota Y, Muehleman C, Imai Y, Okuma M, Thonar EJ, et al. A novel rabbit model of mild, reproducible disc degeneration by an anulus needle puncture: correlation between the degree of disc injury and radiological and histological appearances of disc degeneration. Spine. 2005;30:5–14. doi: 10.1097/01.brs.0000148152.04401.20. [DOI] [PubMed] [Google Scholar]
- 21.Yurube T, Takada T, Hirata H, Kakutani K, Maeno K, Zhang Z, et al. Modified house-keeping gene expression in a rat tail compression loading-induced disc degeneration model. J Orthop Res. 2011;29:1284–90. doi: 10.1002/jor.21406. [DOI] [PubMed] [Google Scholar]
- 22.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(−Delta Delta C(T)) method. Methods. 2001;25:402–8. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 23.Elliott DM, Sarver JJ. Young investigator award winner: validation of the mouse and rat disc as mechanical models of the human lumbar disc. Spine. 2004;29:713–22. doi: 10.1097/01.brs.0000116982.19331.ea. [DOI] [PubMed] [Google Scholar]
- 24.Hirata H, Yurube T, Kakutani K, Maeno K, Takada T, Yamamoto J, et al. A rat tail temporary static compression model reproduces different stages of intervertebral disc degeneration with decreased notochordal cell phenotype. J Orthop Res. 2014;32:455–63. doi: 10.1002/jor.22533. [DOI] [PubMed] [Google Scholar]
- 25.MacLean JJ, Lee CR, Alini M, Iatridis JC. The effects of short-term load duration on anabolic and catabolic gene expression in the rat tail intervertebral disc. J Orthop Res. 2005;23:1120–7. doi: 10.1016/j.orthres.2005.01.020. [DOI] [PubMed] [Google Scholar]
- 26.Hartman RA, Bell KM, Debski RE, Kang JD, Sowa GA. Novel exvivo mechanobiological intervertebral disc culture system. J Biomech. 2012;45:382–5. doi: 10.1016/j.jbiomech.2011.10.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Folkman J, Moscona A. Role of cell shape in growth control. Nature. 1978;273:345–9. doi: 10.1038/273345a0. [DOI] [PubMed] [Google Scholar]
- 28.Le Maitre CL, Frain J, Fotheringham AP, Freemont AJ, Hoyland JA. Human cells derived from degenerate intervertebral discs respond differently to those derived from non-degenerate intervertebral discs following application of dynamic hydrostatic pressure. Biorheology. 2008;45:563–75. [PubMed] [Google Scholar]
- 29.Wang P, Yang L, Hsieh AH. Nucleus pulposus cell response to confined and unconfined compression implicates mechanoregulation by fluid shear stress. Ann Biomed Eng. 2011;39:1101–11. doi: 10.1007/s10439-010-0221-1. [DOI] [PubMed] [Google Scholar]
- 30.Gilchrist CL, Darling EM, Chen J, Setton LA. Extracellular matrix ligand and stiffness modulate immature nucleus pulposus cell-cell interactions. PLoS One. 2011;6:e27170. doi: 10.1371/journal.pone.0027170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lee CR, Iatridis JC, Poveda L, Alini M. In vitro organ culture of the bovine intervertebral disc: effects of vertebral endplate and potential for mechanobiology studies. Spine. 2006;31:515–22. doi: 10.1097/01.brs.0000201302.59050.72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Junger S, Gantenbein-Ritter B, Lezuo P, Alini M, Ferguson SJ, Ito K. Effect of limited nutrition on in situ intervertebral disc cells under simulated-physiological loading. Spine. 2009;34:1264–71. doi: 10.1097/BRS.0b013e3181a0193d. [DOI] [PubMed] [Google Scholar]
- 33.Le Maitre CL, Fotheringham AP, Freemont AJ, Hoyland JA. Development of an in vitro model to test the efficacy of novel therapies for IVD degeneration. J Tissue Eng Regen Med. 2009;3:461–9. doi: 10.1002/term.180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lim TH, Ramakrishnan PS, Kurriger GL, Martin JA, Stevens JW, Kim J, et al. Rat spinal motion segment in organ culture: a cell viability study. Spine. 2006;31:1291–7. doi: 10.1097/01.brs.0000218455.28463.f0. discussion 1298. [DOI] [PubMed] [Google Scholar]
- 35.Giancotti FG, Ruoslahti E. Integrin signaling. Science. 1999;285:1028–32. doi: 10.1126/science.285.5430.1028. [DOI] [PubMed] [Google Scholar]
- 36.Danen EH, Sonnenberg A. Integrins in regulation of tissue development and function. J Pathol. 2003;201:632–41. doi: 10.1002/path.1472. [DOI] [PubMed] [Google Scholar]
- 37.Aguiar DJ, Johnson SL, Oegema TR. Notochordal cells interact with nucleus pulposus cells: regulation of proteoglycan synthesis. Exp Cell Res. 1999;246:129–37. doi: 10.1006/excr.1998.4287. [DOI] [PubMed] [Google Scholar]
- 38.Nerlich AG, Schleicher ED, Boos N. 1997 Volvo Award winner in basic science studies. Immunohistologic markers for age-related changes of human lumbar intervertebral discs. Spine. 1997;22:2781–95. doi: 10.1097/00007632-199712150-00001. [DOI] [PubMed] [Google Scholar]
- 39.Guehring T, Nerlich A, Kroeber M, Richter W, Omlor GW. Sensitivity of notochordal disc cells to mechanical loading: an experimental animal study. Eur Spine J. 2010;19:113–21. doi: 10.1007/s00586-009-1217-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zhao CQ, Wang LM, Jiang LS, Dai LY. The cell biology of intervertebral disc aging and degeneration. Ageing Res Rev. 2007;6:247–61. doi: 10.1016/j.arr.2007.08.001. [DOI] [PubMed] [Google Scholar]
- 41.Buckwalter JA. Aging and degeneration of the human intervertebral disc. Spine. 1995;20:1307–14. doi: 10.1097/00007632-199506000-00022. [DOI] [PubMed] [Google Scholar]
- 42.Visse R, Nagase H. Matrix metalloproteinases and tissue inhibitors of metalloproteinases: structure, function, and biochemistry. Circ Res. 2003;92:827–39. doi: 10.1161/01.RES.0000070112.80711.3D. [DOI] [PubMed] [Google Scholar]
- 43.Millward-Sadler SJ, Costello PW, Freemont AJ, Hoyland JA. Regulation of catabolic gene expression in normal and degenerate human intervertebral disc cells: implications for the pathogenesis of intervertebral disc degeneration. Arthritis Res Ther. 2009;11:R65. doi: 10.1186/ar2693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Guehring T, Omlor GW, Lorenz H, Bertram H, Steck E, Richter W, et al. Stimulation of gene expression and loss of anular architecture caused by experimental disc degeneration—an in vivo animal study. Spine. 2005;30:2510–5. doi: 10.1097/01.brs.0000186591.17114.e9. [DOI] [PubMed] [Google Scholar]
- 45.Yurube T, Nishida K, Suzuki T, Kaneyama S, Zhang Z, Kakutani K, et al. Matrix metalloproteinase (MMP)-3 gene up-regulation in a rat tail compression loading-induced disc degeneration model. J Orthop Res. 2010;28:1026–32. doi: 10.1002/jor.21116. [DOI] [PubMed] [Google Scholar]