SUMMARY
Zika virus (ZIKV), a mosquito-borne flavivirus, causes devastating congenital birth defects. We isolated a human monoclonal antibody (mAb), ZKA190, that potently cross-neutralizes multi-lineage ZIKV strains. ZKA190 is highly effective in vivo in preventing morbidity and mortality of ZIKV-infected mice. NMR and cryo-electron microscopy show its binding to an exposed epitope on DIII of the E protein. ZKA190 Fab binds all 180 E protein copies, altering the virus quaternary arrangement and surface curvature. However, ZIKV escape mutants emerged in vitro and in vivo in the presence of ZKA190, as well as of other neutralizing mAbs. To counter this problem, we developed a bispecific antibody (FIT-1) comprising ZKA190 and a second mAb specific for DII of E protein. In addition to retaining high in vitro and in vivo potencies, FIT-1 robustly prevented viral escape, warranting its development as a ZIKV immunotherapy.
Graphical Abstract
INTRODUCTION
Effective therapeutics and vaccines against Zika virus (ZIKV) are urgently needed. Although ZIKV generally causes asymptomatic or mild disease, it can lead to neurological complications both in adulthood and during fetal development (Honein et al., 2017), likely due to infection of neural progenitor cells (Frontera and da Silva, 2016; Mlakar et al., 2016). Further, infection of the testes has been shown to cause infertility in mice (Govero et al., 2016).
Cryo-electron microscopy (cryoEM) of ZIKV revealed high structural similarity to other flaviviruses (Kostyuchenko et al., 2016; Sirohi et al., 2016). The smooth-surfaced mature ZIKV particle comprises 180 copies of E protein organized in icosahedral symmetry, each asymmetric unit containing three E proteins. Three E protein dimers lie parallel to each other building a raft, and thirty rafts form a herringbone pattern. The E protein consists of three domains – DI, DII, and DIII (Barba-Spaeth et al., 2016; Kostyuchenko et al., 2016; Rey et al., 1995; Zhao et al., 2016). DIII contains the receptor-binding site and mediates virus attachment (Crill and Roehrig, 2001), whereas the fusion loop in DII facilitates virus-endosomal membrane fusion (Allison et al., 2001). After cell entry, the low endosomal pH triggers dramatic structural rearrangement of E protein dimers into trimers, leading to membrane fusion (Bressanelli et al., 2004; Modis et al., 2004; Stiasny et al., 1996). The post-fusion structure shows DIII rotated ~70° relative to DI, which requires movement of the DI-DIII linker (Bressanelli et al., 2004; Modis et al., 2004).
We recently isolated a panel of anti-ZIKV neutralizing human mAbs and identified a DIII-binding mAb, ZKA190, that showed strong neutralizing activity across ZIKV strains (Stettler et al., 2016). Here, we further characterized ZKA190 in vitro and in vivo. Using a combination of NMR, cryoEM and molecular simulations, we mapped the ZKA190 epitope to an exposed region consisting of the DI-DIII linker and the lateral ridge (LR) region of DIII. Different from other DIII antibodies, ZKA190 Fab binds to all 180 E proteins on the virus surface, altering the E protein quaternary interactions, causing separation of the rafts and flattening their curvature. In vivo studies of ZIKV infection in lethal mouse models demonstrated strong protection of ZKA190 from multiple strains. To minimize the risks of viral escape and antibody-mediated disease enhancement, we engineered a bispecific antibody, which combines ZKA190 with the DII-binding mAb ZKA185 and also includes a mutation in the Fc region (LALA) abolishing binding to Fcγ receptor and complement (Hessell et al., 2017). FIT-1 retains the breadth and potencies of both parental antibodies and was not observed to generate escape mutants, encouraging FIT-1 development as a ZIKV therapeutic.
RESULTS
In vitro characterization of the DIII-specific mAb ZKA190
Recombinant ZKA190 synthesized in the IgG1 wild-type (wt) and IgG Fc-LALA format potently neutralized African, Asian and American ZIKV strains, with IC50 ranging from 0.004 to 0.05 nM (Figure 1A; 0.6 to 8 ng/ml). Although both the ZKA190 IgG and its Fab bound to E protein with similar affinities (Figure 1C; KD values of 0.3 and 0.2 nM, respectively), the Fab neutralized ZIKV infection less potently with a 300-fold higher IC50 (Figure 1B). Since ZIKV has been shown to infect human neural progenitor cells (hNPC) (Garcez et al., 2016; Nowakowski et al., 2016; Onorati et al., 2016; Tang et al., 2016), we tested ZKA190 wt and LALA, and both fully abolished infection and replication of ZIKV in hNPCs (Figure 1D).
Figure 1. Neutralization and Enhancement of ZIKV Infection by ZKA190 mAb.
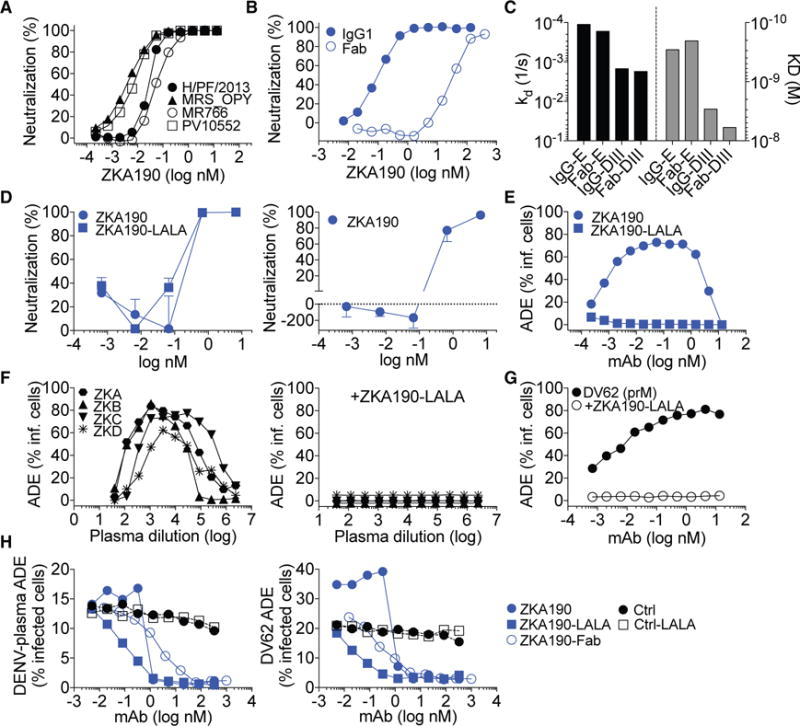
(A) ZKA190 was tested for neutralization of four strains of ZIKV, as determined by the percentage of infected Vero cells in the presence of increasing amounts of mAbs. Data are representative of at least two independent experiments.
(B) Neutralization of ZIKV H/PF/2013 strain by ZKA190 IgG and Fab determined as in (A).
(C) Binding of ZKA190 IgG and Fab to recombinant ZIKV E and DIII proteins as assessed by SPR.
(D) Neutralization of ZIKV strain PRVABC59 infection of hNPCs by ZKA190, ZKA190-LALA and a control mAb as determined by plaque assay on Vero cells (left panel) and indirect immunofluorescence of infected hNPCs using fluorophore-labelled anti-E antibody (right panel).
(E) ADE of ZIKV infection of non-permissive K562 cells by ZKA190 and ZKA190-LALA.
(F) ADE induced in K562 cells when ZIKV is pre-incubated with serial dilutions of plasma or serum from different ZIKV-positive patients (left panel). When ZKA190 LALA is added to the ZIKV-serum complexes, ADE is inhibited (right panel).
(G) ADE induced in K562 cells when ZIKV is pre-incubated with serial dilutions of a prM cross-reactive mAb (DV62) derived from a DENV-immune donor. ZKA190-LALA inhibits ADE of ZIKV when complexed with prM-reactive mAb DV62.
(H) Effect on ADE induced by peak enhancing dilution of a DENV2 plasma sample (left panel) or anti-prM DV62 mAb (right panel) by serial dilutions of indicated mAbs.
Error bars represent the SD of the mean for each determination.
Antibody-dependent enhancement (ADE) of infection may contribute to disease severity in heterologous secondary DENV infections (Halstead, 2014), caused by the binding of non-neutralizing or sub-neutralizing concentrations of E- and prM-specific antibodies to mature, partially mature and fully immature viruses. ZKA190 supports ADE in K562 cells from 0.0001 to 1 nM, while ZKA190-LALA did not show any ADE activity (Figure 1E) and completely inhibited ADE induced by plasma antibodies from four ZIKV-immune donors (Figure 1F).
Anti-prM antibodies are elicited during the human immune response against flaviviruses, and they enhance in vitro infection of immature or partially immature virions (Dejnirattisai et al., 2010). ZKA190-LALA can fully block ADE of ZIKV infection induced by a flavivirus cross-reactive anti-prM mAb, DV62, isolated from a dengue patient (Beltramello et al., 2010) (Figure 1G). Finally, we compared the ability of ZKA190 wt, LALA and Fab to cause or block ADE of ZIKV by human anti-DENV2 plasma or DV62 mAb (Figure 1H). Low concentrations of ZKA190 increased DV62-mediated ADE of ZIKV infection, consistent with its ability to promote the entry of both immature and mature virions, while at concentrations above 1.3 nM (i.e., 200 ng/ml), ZKA190 blocked ADE induced by both anti-DENV plasma and DV62. At concentrations above 0.06 nM, ZKA190 Fab was equally effective as ZKA190-LALA in reducing ADE induced by DV62, indicating that inhibition occurs at a post-attachment step, likely fusion.
ZKA190 binds to a conserved and highly accessible region of DIII
The ZKA190 epitope was determined by solution NMR spectroscopy (Bardelli et al., 2015; Simonelli et al., 2010; 2013). Differences in the NMR spectra of free and mAb-bound DIII identified DIII residues affected by ZKA190 binding, either directly or through allosteric effects (Figure S1A). The ZKA190 epitope consists of the LR of DIII (the BC, DE and FG loops), as well as part of the DI-DIII linker (Figure 2A). These residues are conserved among 217 known ZIKV strains, with the exception of V341I and E393D substitutions in the Uganda 1947 MR766 strains (Figure S1D), which ZKA190 was shown to neutralize (Figure 1A). The ZKA190 epitope is highly accessible on the ZIKV surface, except for the FG loop in the 5-fold vertex (Figure 2B and S1C, molecule A).
Figure 2. ZKA190 Epitope Identified By NMR, CryoEM, and Docking.
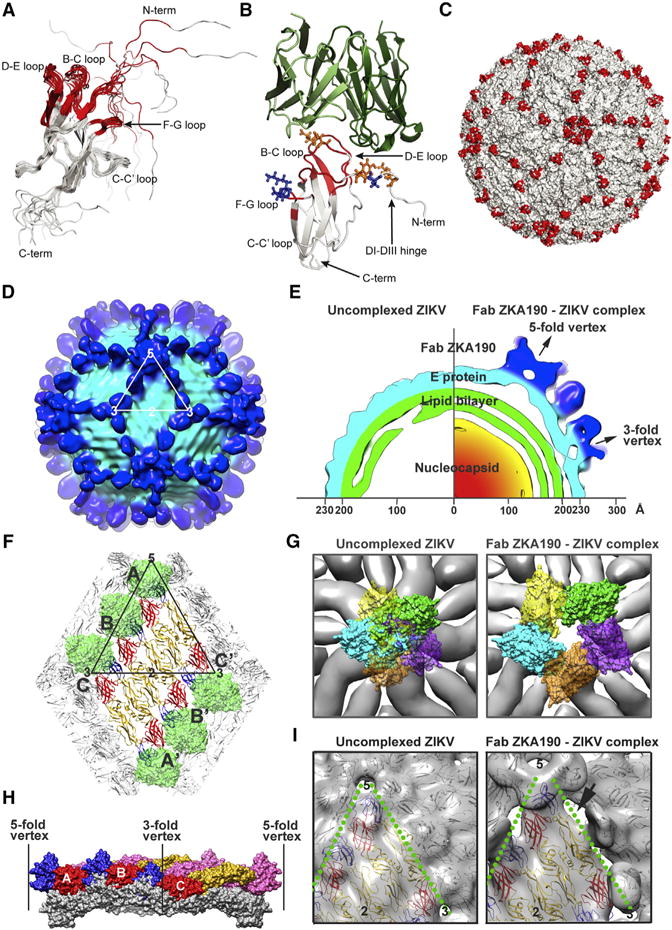
(A) Cartoon representation of the 12 lowest-energy NMR structures of ZIKV DIII, with residues affected by ZKA190 binding in red. Flexibility in the N terminus of the construct is apparent.
(B) Model of the ZKA190:DIII complex derived by computational docking and molecular simulation validated by cryoEM and NMR results. The NMR-identified epitope on DIII (gray) is indicated in red. The ZKA190 heavy and light chains are colored in dark and light green, respectively. DIII residues that affect or do not affect antibody binding when mutated are shown as orange and blue sticks, respectively.
(C) NMR-identified ZKA190 epitope (red) is accessible on the virus surface (white).
(D–I) The ~22Å resolution cryoEM map of the ZIKV:ZKA190 Fab complex at 37°C. (D) Surface of the cryoEM map. One icosahedral asymmetric unit is indicated by a white triangle. Light and dark blue correspond to the E protein shell and the Fab densities, respectively. (E) A quarter of the central section of the uncomplexed (left) and complexed (right) cryoEM maps showing that the E protein layer in the latter moved outward with the highest protrusion at the 5-fold vertex. (F) CryoEM structure of ZIKV:ZKA190 Fab complex. The DIIIs of all E proteins are bound by Fabs (green). E proteins in an asymmetric unit are labelled A, B and C, and the neighboring asymmetric unit within a raft A′, B′ and C′. DI, DII and DIII are colored in red, yellow and blue, respectively. (G) E proteins at the 5-fold vertices move apart upon Fab ZKA190 binding. (Left) Superposition of the Fab ZKA190:E complex onto the uncomplexed ZIKV shows clashes between the Fabs. (Right) In contrast, no clashes are observed in the cryoEM complex structure. E proteins are in gray and Fabs are colored. (H) Superposition of the E protein raft in the free (gray) and ZKA190-complexed (colors) ZIKV; the curvature flattens in the complex structure. E proteins of one asymmetric unit (A, B and C) are in red, yellow and blue; those from another unit are in pink. (I) The rafts separate from each other in the complex (right) compared to the uncomplexed ZIKV structure (left). The inter-raft interface is indicated with a green dotted line, and the gap between rafts is indicated with an arrow.
Computational docking followed by molecular dynamics simulation, guided and validated by both NMR- and cryoEM-derived epitope information as well as DIII mutagenesis, showed that ZKA190 binds through an interface characterized by shape and charge complementarity (Figures 2B and S1E). The computational model fits well to the cryoEM density map of ZIKV-ZKA190 Fab complex (Figure 2C and 3D). Docking suggests that there are no direct contacts between ZKA190 and the FG loop on DIII, thus changes in NMR signals likely derive from allosteric effects. Consistently, mutations of FG loop residues, but not of other NMR-affected residues in the recombinant DIII, did not affect the binding affinity of ZKA190 (Figure 2B and S2).
Figure 3. ZKA190 Aggregates Virus at High Antibody:Virus Molar Ratios and Inhibits a Post-Attachment Stage of Virus Infection.
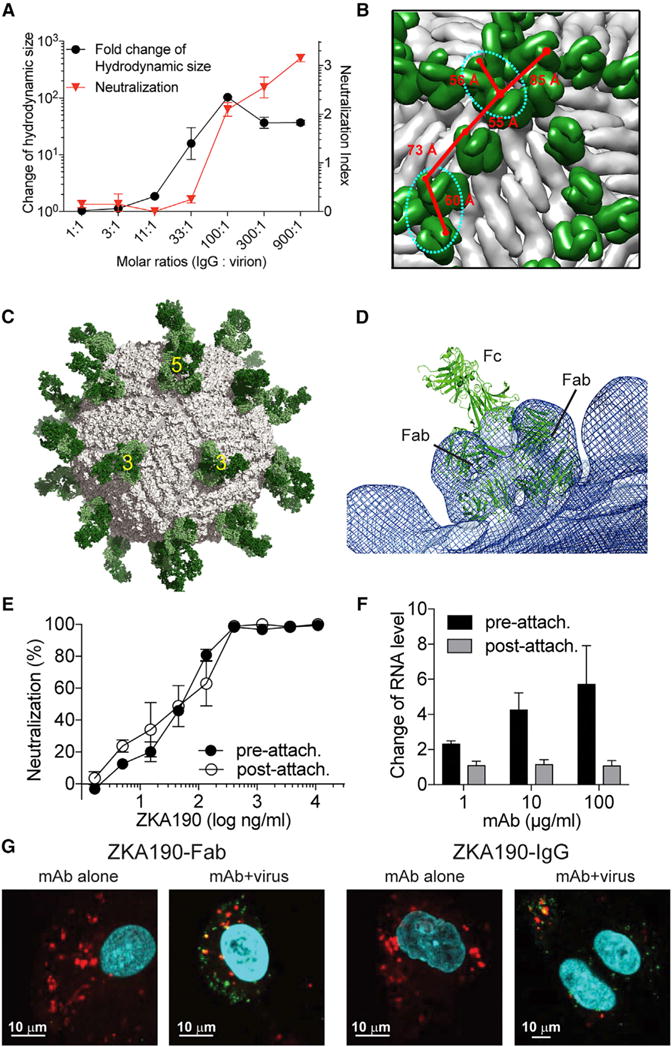
(A) Dynamic light scattering experiments measuring the hydrodynamic size of the virus complexes shows ZKA190 IgG aggregates ZIKV at IgG:virion molar ratio of ≥33:1. The neutralization activity of antibody correlates with the formation of virus-antibody aggregates. The neutralization is expressed as the log10-transformed fold-reduction of the virus titer compared to the virus-only control.
(B) ZKA190 Fab (green) is shown on the viral surface (gray). The close proximity (<86Å) between CH1 domains of two Fabs (red lines) in the cryoEM ZIKV:ZKA190 Fab structure indicates possible bivalent binding of IgG on the same virus particle. However, due to the orientation of the Fabs, only the sites (cyan dotted circles) around the 5-fold and the 3-fold vertices can accommodate bivalent binding of a full IgG.
(C) Molecular Dynamics (MD)-based atomic model of the full ZKA190 IgG (green) bridging across two neighboring E proteins on the viral surface (gray). For clarity, only one IgG binding is shown. Bivalent binding to other sites within these vertices is possible.
(D) Superposition of the MD-based full IgG model (green cartoon) on the cryoEM map (blue mesh) near the 3-fold vertex shows good fitting of the modelled Fabs to the cryoEM density.
(E) Neutralization test of ZIKV when ZKA190 is added pre- or post-attachment of virus to cells that were incubated at 4°C. Results show no difference in the neutralization profile.
(F) RT-PCR detecting the amount of virus remaining on cells when ZKA190 was added pre- and post- virus attachment to cells. When increasing concentrations of mAb were mixed with virus before attaching to cells (“pre-attach.”), there is a gradual increase in the amount of virus on cell surface, likely due to aggregation of virus. Addition of the mAb to virus that had already bound to cells (“post-attach.”) showed that the mAb is unable to strip the virus off the cell surface. Therefore, the mAb can also inhibit a post-attachment stage of the infection cycle.
(G) Confocal microscopy experiments. ZIKV incubated with a concentration exceeding 10,000-fold the IC50 value of either ZKA190 Fab or full IgG were added to Vero cells. The ZIKV:antibody complex is detected inside the cells (green) and co-localizes with endosomes (red, yellow overlay). Endosomes and acidic organelles are marked by Lysotracker red; Alexa 488-conjugated ZKA190 is in green. Nuclei are stained with DAPI (blue).
Data in (A) and (F) are represented as mean ±SD of at least two independent experiments. Data in (E) are represented as mean ± SEM of three independent experiments performed in triplicate.
CryoEM structure of ZKA190 complexed with ZIKV
CryoEM micrographs showed that ZKA190 binds to the virus at 4°C, 37°C and 40°C (Figure S3A). Structures of ZIKV complexed with ZKA190 Fab at 4°C and 37°C were determined to ~22 Å resolution and found to be similar (Figures 2D, S3B and S3C). At this resolution, the Fabs and E protein rafts can only be roughly fitted into the density; the footprint on DIII cannot be accurately evaluated using cryoEM results alone. In contrast to other mAbs against DIII LR of other flaviviruses, Fab ZKA190 bound to all 180 copies of E protein on the viral surface (Figure 2D and S3), achieving full occupancy.
Superposition of the structure of the ZKA190 Fab:E protein complex onto the uncomplexed ZIKV structure showed that Fabs bound to the E protein molecule around the 5-fold vertex clashed with each other (Figure 2G). On the contrary, cryoEM data showed equally strong densities of the variable region of the Fabs at all three individual E proteins in the asymmetric unit (Figures 2F and S4). To accommodate binding of all five Fabs without clashes, the E proteins have to separate from each other in the 5-fold vertex (Figure 2G). Comparison of the cryoEM structures of uncomplexed and ZKA190-bound ZIKV showed that in the latter, the E protein rafts move to a higher radius (Figure 2E) and are separated from each other (Figure 2I); the curvature of the rafts is also flattened (Figure 2H). In the uncomplexed ZIKV cryoEM structure, the E protein ectodomain interacts with the membrane-associated stem regions of E and M proteins on the virus, and this interaction might be disrupted by ZKA190 Fab binding, as suggested by the spatial gap observed between the E protein and the lipid bilayer (Figure 2E). The dissociation may increase the motion of the rafts, explaining the inability to obtain a higher resolution cryoEM map.
Mechanisms of ZKA190 neutralization
The ability of ZKA190 to efficiently neutralize the virus may involve inhibition of either cell attachment or membrane fusion. Another mechanism might involve virus inactivation through aggregation. Virus aggregation due to simultaneously engaging epitopes on different particles by ZKA190 was assessed by dynamic light scattering (DLS). Particles with size similar to single virus particle were detected at low mAb concentration, whereas large viral aggregates appeared at higher concentration (≥ 33 antibodies per virus) and correlated with neutralizing activity (Figure 3A). One caveat is that the contribution of aggregation to neutralization may be different in vivo, as the amount of virus could differ.
IgG ZKA190 may also engage neighboring E proteins on the viral surface. As shown by cryo-EM, the distance between the Fabs’ CH1 regions bound on the viral surface is compatible with that of a full IgG (Figure 3B). However, bivalent binding of a full IgG depends not only on the distance between two Fab molecules, but also on their relative orientation. Full atomistic molecular dynamics simulations of a homology model of ZKA190 IgG showed that a single ZKA190 molecule can bridge adjacent E proteins around the 5- and 3-fold vertices, but not the 2-fold vertex (Figures 3C and 3D). The Fab regions of the modelled ZKA190 IgG correlated well with the cryo-EM structure (Figure 3D). This suggests that ZKA190 cross-linking of neighboring E proteins might prevent the E dimers from rearrangement into trimers that is required for membrane fusion. Both proposed mechanisms imply bivalent binding of ZKA190 IgG. Since ZKA190 Fab also neutralizes ZIKV, albeit less efficiently than the full IgG, additional mechanisms may be involved. ZKA190 might prevent the attachment of ZIKV to target cells or viral fusion to the endosomal membrane.
Neutralization at a post-attachment step is supported by data showing that ZKA190 inhibits in vitro ZIKV infection with similar potency regardless of whether the mAb was added to the virus pre- or post- attachment to cells (Figure 3E). This result is further supported by the qRT-PCR experiments that detect the amount of virus on the cell surface, which does not decrease at increasing antibody concentrations (Figure 3F), but rather increased compared to the virus-only control, likely due to antibody-mediated virus aggregation (Figure 3A). Moreover, confocal microscopy analysis showed that ZIKV-complexed ZKA190 Fab can enter cells at neutralizing concentrations exceeding the IC50 by 10,000-fold (Figure 3G).
In vivo characterization of the DIII-specific mAb ZKA190
ZKA190 and ZKA190-LALA were tested in A129 mice challenged with a lethal dose of ZIKV strain MP1751 (African lineage). Prophylaxis with ZKA190 and ZKA190-LALA one day before virus challenge protected mice from mortality and morbidity at mAb concentrations of 5, 1 or 0.2 mg/kg (Figures 4A–4B). ZKA190-LALA, and to a lesser extent ZKA190, delayed morbidity and mortality as compared to the control at 0.04 mg/kg. Viral titers in blood and organs were reduced significantly, even in the presence of serum antibody levels below 1 μg/ml (Figures S5A–S5A). To determine if low doses of ZKA190 could potentially enhance ZIKV infection, we administered 1 mg/kg, 0.04 mg/kg, 0.02 mg/kg, and 0.01 mg/kg of ZKA190 to AG129 mice and challenged them with a recently isolated ZIKV strain from Nicaragua (Nica 2–16, American lineage). Mice receiving 1 mg/kg of ZKA190 or ZKA190-LALA were completely protected from ZIKV disease (Figures 4C–4D). ZKA190 or ZKA190-LALA at 0.04 mg/kg and 0.02 mg/kg of partially protected mice from mortality and morbidity, and none of the lower antibody doses resulted in enhanced disease or infection. Blood viral titers on day 3 post-infection were reduced compared to control antibody-treated animals in a dose-dependent fashion (Figure S5E).
Figure 4. Prophylactic and Therapeutic Efficacy of ZKA190.
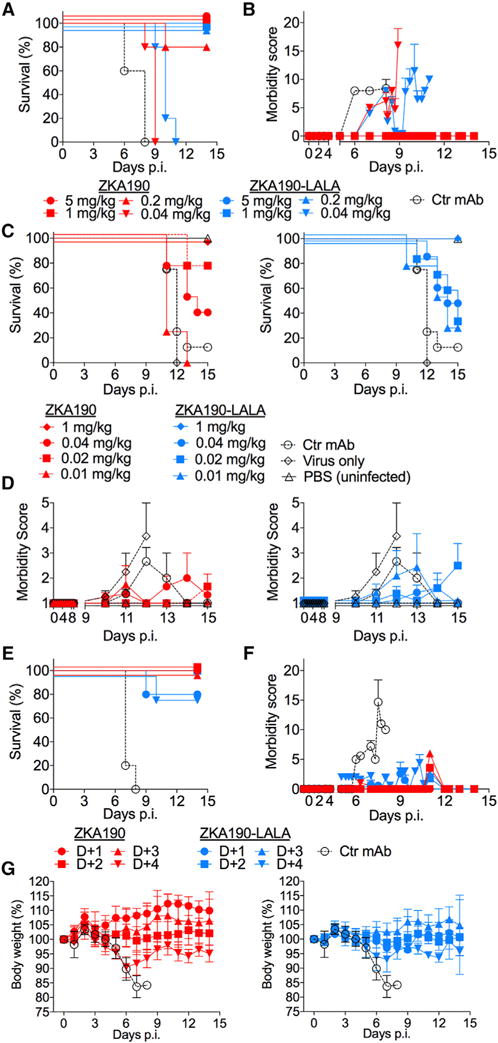
(A to D) ZKA190 is strongly protective against ZIKV infection when administered prophylactically to mice (A129 in (A) and AG129 in (C, D)) challenged with a lethal dose of ZIKV strain MP1745 (A, B) or Nica 2–16 (C, D). Experiments used N=4–8 mice per group. (A, C) Kaplan-Meier survival curves are shown. Significance was determined using the Mantel-Cox log-rank test. Panel A: ZKA190 at 5, 1 and 0.2 mg/kg versus Ctr mAb, P = 0.0031; ZKA190 at 0.04 mg/kg versus Ctr mAb, P = 0.0116; ZKA190-LALA at 5, 1, 0.2 and 0.04 mg/kg versus Ctr mAb, P = 0.0031. Panels C: ZKA190 and ZKA190 LALA at 1 mg/kg versus Ctr mAb, P = 0.0091; ZKA190 at 0.02 mg/kg versus Ctr mAb, P = 0.0035; ZKA190-LALA at 0.04 mg/kg versus Ctr mAb, P = 0.0142. (B, D) Morbidity score of mice monitored over a 14–15 day period (two different scoring methods were used; see (Dowall et al., 2016) for (B) and (Orozco et al., 2012) for panel D). Control mAb is MPE8 mAb (A, specific for RSV F protein (Corti et al., 2013)) and a human IgG polyclonal isotype control (C).
(E – G) ZKA190 or ZKA190-LALA were administered at 15 mg/kg at different time-points after ZIKV infection to A129 mice challenged with a lethal dose of ZIKV strain MP1745. (E) A Kaplan-Meier survival curve is shown. Experiments used N=5 mice per group. Significance was determined by using the Mantel-Cox log-rank test. ZKA190 and ZKA190-LALA administered on day 1, 2, 3 or 4 versus Ctr., P = 0.0016. (F) Morbidity score of mice monitored over a 14-day period according to (Dowall et al., 2016). (G) Mice were monitored over a 14-day period for body weight loss. Control mAb is MPE8 specific for RSV F protein (Corti et al., 2013).
Data in (B), (D) and (F) are represented as mean ± SEM for each determination. Data in (G) are represented as mean ±SD for each determination. See also Figures S5 and S6.
To evaluate therapeutic potential, we administered ZKA190 and ZKA190-LALA to A129 mice at different time-points following ZIKV infection. At a dose of 15 mg/kg, survival rates of 80%–100% were achieved, and morbidity was greatly reduced even when treatment was given four days post-infection (Figure 4E–G). ZKA190 and ZKA190-LALA treatment at all post-infection time-points resulted in significantly reduced viral titers with a clear trend for greater reduction with earlier treatment (Figure S6A–S5C). Of note, ZKA190-LALA showed a reduced antiviral activity on day 5 as compared to ZKA190 when mAbs were given four days post-infection, a result that might relate to the impaired ability of the LALA mAb to facilitate rapid clearance of coated virions.
Of the 16 treated mice, one in vivo escape mutant (Monoclonal Antibody Resistant Mutant 1, MARM1) was isolated containing the amino acid substitution T335R in DIII, which was shown to abrogate ZKA190 binding (Figure S2). Viruses from the other treated mice did not contain any E mutations (Figures S6D–S5E).
In vitro selection of ZIKV escape mutants
To assess the ability of ZKA190 to select for resistant mutants (MARMs) in vitro, ZIKV (H/PF/2013) was passaged in the presence of sub-neutralizing concentrations of ZKA190. MARM2, carrying the DIII E370K mutation, was isolated after three rounds of selection. The mutation abolished neutralization by ZKA190, but not its binding to DIII (Figure S2). The mutations in the in vivo (T335R) and in vitro (E370K) MARMs are located on the BC and DE loops of DIII, respectively, and are consistent with the epitope identified by NMR.
Development of the bispecific FIT-1 antibody
Generation of viral escape mutants can render antibodies useless for treatment. This may be prevented by making a bispecific antibody combining two highly neutralizing mAbs, each specific for a distinct epitope. We selected ZKA185 and ZKA230, which potently neutralize African, Asian, and American strains with IC50 ranging from 0.02 to 0.62 nM (Figure 5A). ZKA185 binds with high affinity to recombinant ZIKV E protein and to Zika virus-like particles (VLP) but not to the isolated DIII (Figure 5B). ZKA230 bound to ZIKV VLPs, but not to recombinant E or DIII, suggesting that it recognizes a quaternary epitope displayed only on the viral surface (Figure 5B). ZKA185 IgG and Fab were shown to bind to E and VLP antigens with similar high affinity by ELISA. Conversely, ZKA230 Fab bound to VLPs with markedly reduced affinity compared to its IgG.
Figure 5. Engineering of ZKA190 into the Bispecific FIT-1 mAb.
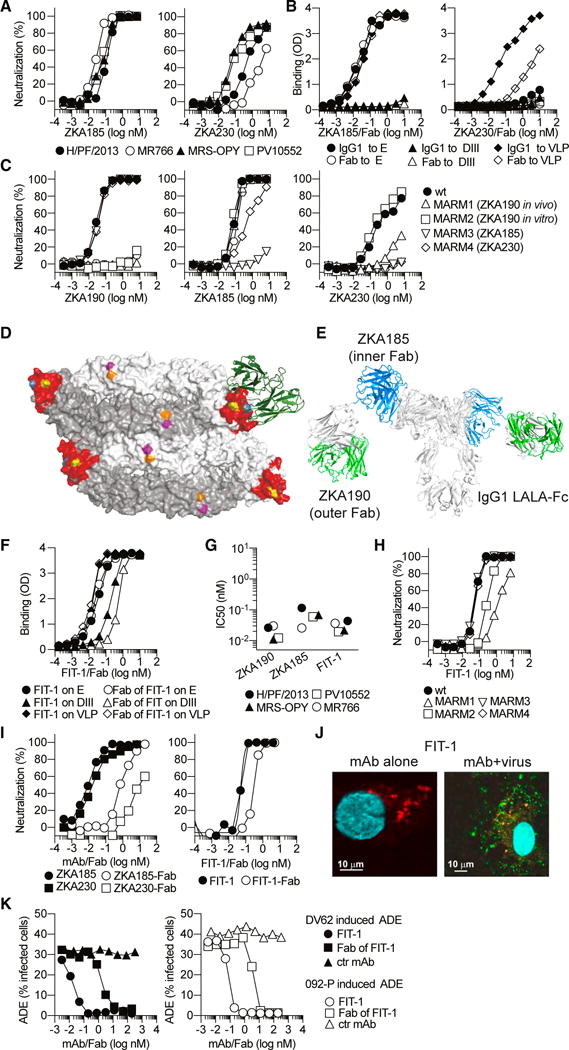
(A) ZKA185 and ZKA230 mAbs were tested for neutralization of four strains of ZIKV, as determined by the percentage of infected Vero cells in the presence of increasing amounts of mAbs. Data are representative of at least two independent experiments.
(B) Binding of ZKA185 and ZKA230 IgG and Fab to recombinant ZIKV VLP, E and DIII antigens as assessed by ELISA.
(C) ZKA190, ZKA185 and ZKA230 were tested for neutralization of H/PF/2013 (wt) and MARMs 1–4.
(D) Surface representation of two E protein dimers bound by ZKA190 (green); the ZKA190 NMR derived epitope is in red; positions mutated in MARMs are indicated in yellow (E370), blue (T335), orange (D67) and magenta (K84).
(E) Model of FIT-1. The natural linkers between inner and outer Fabs allow flexible movement of Fabs in the FIT-1 antibody. The variable regions of ZKA185 and ZKA190 are highlighted in blue and green, respectively.
(F) Binding of FIT-1 IgG and Fab to recombinant ZIKV VLP, E and DIII antigens as assessed by ELISA.
(G and H) ZKA190, ZKA185 and FIT-1 mAbs were tested for neutralization of four strains (IC50 values, G) and four MARMs (H) of ZIKV.
(I) Neutralization of ZIKV H/PF/2013 strain by ZKA185, ZKA230 and FIT-1 IgG and Fab determined as in (A).
(J) Confocal microscopy experiments as shown in Figure 3G.
(K) Effect on ADE induced by peak enhancing dilution of anti-prM DV62 mAb or DENV2 plasma by serial dilutions of FIT-1 IgG and Fab.
We isolated MARMs of ZKA185 (MARM3) and ZKA230 (MARM4) by passaging virus in the presence of sub-neutralizing mAb concentrations. MARM3 contained substitutions at K84E and D67H on DII (Figure 5D). MARM4 showed a mixture of different amino acid substitutions at position 84 (from K to G, E or R). ZKA190 neutralized ZKA185 and ZKA230 MARMs as well as the parental virus (Figure 5C). ZKA185 neutralized both ZKA190 and ZKA230 MARMs. In contrast, ZKA230 neutralized only ZKA190 MARM2.
To gain insight into the development of MARMs capable of escaping from the pressure of multiple mAbs, we serially passaged ZKA190 MARM2 (E370K) in the presence of ZKA185 or ZK230 and found that double MARMs emerged after three to four passages. ZKA230 introduced the K84E mutation while ZKA185 selected for the D76G mutation. These findings indicate that ZIKV can escape multiple mAbs targeting distinct sites when the selection is performed in a stepwise fashion.
MAb ZKA185 was selected to combine with ZKA190 for the development of a bispecific antibody since it potently cross-neutralizes ZIKV strains and does not compete with ZKA190. We produced a bispecific antibody in the tetravalent symmetric format Fabs-in-tandem-Ig (FIT-Ig) (Gong et al., 2017). ZKA190 was placed in the outer and ZKA185 in the inner Fab position (Figure 5E), using the backbone IgG1 antibody in LALA format (here designated FIT-1). FIT-1 bound DIII, E and VLPs in ELISA (Figure 5F) and retained high neutralizing potency against ZIKV strains, with IC50 values largely similar to those of the parental mAbs (Figure 5G). FIT-1 bound to E protein with higher affinity than either of the parental mAbs (KD values: ZKA185 1.8 nM, ZKA190 9.3 nM and FIT-1 KD <1 pM due to slower dissociation rate, presumably through avidity effects) and effectively neutralized all the ZKA190, ZKA185 and ZKA230 MARMs (Figure 5H). In addition, the neutralizing activity of the Fab fragment of FIT-1 (comprising one ZKA190 and one ZKA185) was reduced only by about 6-fold (Figure 5I, right panel), whereas those of the individual ZKA190 and ZKA185 Fabs were reduced by about 100-fold compared to their full-length IgG1 (Figure 1B and Figure 5I). Taken together, these findings suggest the ZKA190 and ZKA185 moieties in FIT-1 are both active.
No MARMs were isolated despite 8 rounds of serial passages in the presence of FIT-1. In contrast, MARMs appeared after 3 to 4 passages with individual mAbs. Simultaneous mutations in both DIII and DII are thus unlikely to occur, suggesting that FIT-1 is a safer therapeutic. Confocal microscopy studies showed that FIT-1, like ZKA190, inhibits virus infection at a post-attachment step, likely fusion (Figure 5J). Finally, FIT-1and its Fab blocked ADE of human anti-DENV2 plasma or DV62 at concentrations above 0.1 and 10 nM, respectively (Figure 5K)
We evaluated the therapeutic potential of FIT-1 by administering three different doses (15, 5 and 1 mg/kg) at three different time-points to mice following ZIKV infection. At a dose of 15 mg/kg, survival rates were 100% without signs of morbidity even when treatment was given three days post-infection (Figure 6); viral titers were abrogated and no escape mutants were detected, indicating high in vivo efficacy. Administration of 5 mg/kg resulted in survival rates of 70–100%; again, no escape mutants were detected on day 5 after infection. The lowest dose tested (1 mg/kg) protected when administered on day 1 or 2, but not day 3, after infection.
Figure 6. Therapeutic efficacy of FIT-1.
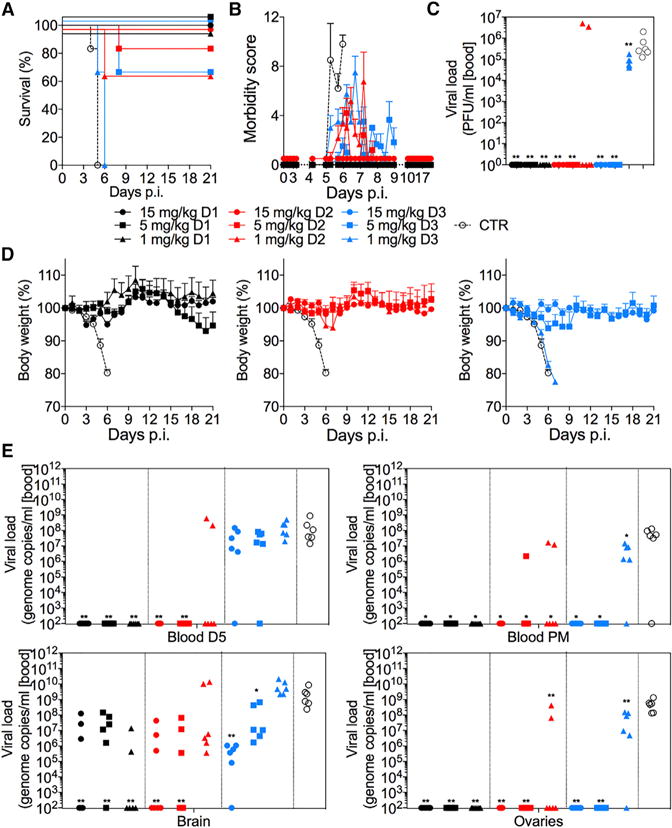
FIT-1 is strongly effective against ZIKV infection when administered therapeutically at different time-points to mice (A129) challenged with a lethal dose of ZIKV strain MP17451. Experiments used N=5–6 mice per group.
(A) Kaplan-Meier survival curves are shown. Significance was determined by using the Mantel-Cox log-rank test. FIT-1 at 15, 5 and 1 mg/kg given either on day 1 or 2 versus Ctr mAb, P = 0.0012; ZKA190 at 15 and 5 mg/kg given on day 3 versus Ctr mAb, P = 0.0012; ZKA190 at 1 mg/kg given on day 3 versus Ctr mAb, P = 0.0170.
(B) Morbidity score of mice monitored over a 21-day period (Dowall et al., 2016).
(C) Viral load was measured as PFUs on day 5 in blood of all animals.
(D) Mice were monitored over a 21-day period for body weight loss. Control mAb in (A) is MPE8 mAb (specific for RSV F protein (Corti et al., 2013)).
(E) Viral load was measured as genomic copies by qPCR on day 5 in blood of all animals and in blood and indicated tissues when animals were culled at the end of the study or when the humane endpoints were met. Significance was determined compared to control antibody treatment by nonparametric unpaired Mann-Whitney U test. *p < 0.05; **p < 0.01.
Data in (B) are represented as mean ± SEM for each determination. Data in (D) are represented as mean ± SD for each determination.
DISCUSSION
The highly potent ZKA190 mAb binds to to the lateral ridge in DIII of the E protein in a unique orientation compared to other flavivirus-neutralizing LR-binding mAbs (Edeling et al., 2014; Nybakken et al., 2005; Zhao et al., 2016) (Figure 7A), resulting in full occupancy of all 180 E proteins. Unlike ZKA190, the cryoEM structures of E16:WNV (Kaufmann et al., 2006) and ZV23:ZIKV (Wang et al., 2016) showed that their Fabs are unable to bind to DIII around the 5-fold vertex. Superposition of the E106:DIII (Edeling et al., 2014), ZV23:DIII and ZV67:DIII (Zhao et al., 2016) structures onto an uncomplexed ZIKV cryoEM structure showed clashes with the neighboring E proteins around the 5-fold vertex (Figure 7B).
Figure 7. Comparison of ZKA190 to other DIII lateral ridge antibodies.
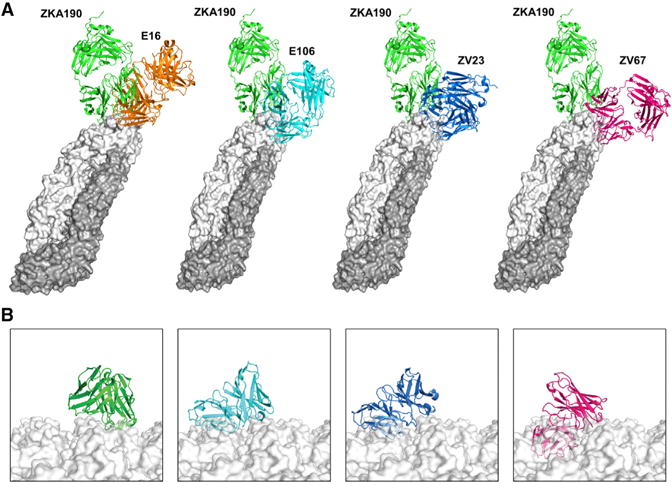
(A) Superposition of the structures of the Fabs E16 (orange, PDB 1ZTX), E106 (cyan, PDB 4L5F), ZV23 (blue, PDB 5GZR) and ZK67 (purple, PDB 5KVG) onto DIII of ZKA190-E protein cryoEM structure.
(B) Fv regions of ZKA190, E106, ZV23 and ZV67 (colors as indicated) over the ZIKV surface, obtained by superimposing their structural complex with DIII to E protein on the viral surface (white surface, the 5-fold vertex is shown). All mAbs, but not ZKA190, clashes with adjacent E proteins when superimposed onto molecule A near the 5-fold vertex.
Our previous structure of uncomplexed ZIKV revealed that the E protein shell is more compact than that of DENV (Kostyuchenko et al., 2016). Binding of ZKA190 disrupts the tight interactions between E proteins in the 5-fold vertices at both 4°C and 37°C, thus allowing full occupancy. ZKA190 may either select a transient viral conformation where the 5-fold vertex is “open” (conformational selection) or cause surface structural changes (allosteric effects). The latter appears more likely since binding occurs even at 4°C, where the thermal motions required for E protein conformational changes are limited. NMR studies and DIII mutants indicated that binding of ZKA190 causes an allosteric effect on the DIII FG loop, which points towards the center of the 5-fold vertex. ZKA190 binding to one DIII near the 5-fold vertex may alter the FG loop conformation, opening the vertex and exposing additional ZKA190 binding sites. Alternatively, the antibody could first bind to the 3- and/or 2-fold vertices, causing allosteric effects throughout the viral surface resulting in opening of the 5-fold vertex, to which other antibodies could then bind; however, experimental evidence for this is lacking.
Multiple mechanisms may contribute to ZKA190 activity. We showed here that high concentrations of ZKA190 induced aggregation of virus particles by crosslinking different particles with the two Fab arms. This correlated with increased neutralization activity, suggesting a role for aggregation in viral neutralization. Molecular dynamics showed that the two Fab arms of ZKA190 can bind within a viral particle, possibly preventing the dramatic rearrangement of the E proteins on the virus surface required for fusion.
RT-PCR and confocal microscopy data indicate that the virus remains attached to cells in the presence of ZKA190 and is able to enter cells in the presence of neutralizing concentrations of ZKA190. Furthermore, similar neutralization profiles of ZKA190 (and FIT-1) in the post- and pre-attachment assays, as well as inhibition by the ZKA190 Fab fragment of DENV plasma and anti-prM antibody-mediated ADE, support the notion that ZKA190 inhibits a post-attachment step, likely fusion. The epitope of ZKA190 encompasses the LR of DIII and the DI-DIII linker region. In a normal infection, the low-pH endosomal environment provokes the E proteins to rearrange from a dimeric structure to a trimeric post-fusion structure. Binding of ZKA190 to the DI–DIII linker and DIII may hinder the movement of DIII, preventing rearrangement and thus membrane fusion, as previously suggested in structural studies of E16:WNV (Kaufmann et al., 2006; Zhang et al., 2015).
Although antibody effector functions can enhance the efficacy of anti-flavivirus antibodies (Chung et al., 2006), comparison of ZKA190 and ZKA190-LALA in vivo efficacy indicates that Fc-dependent mechanisms, such as ADCC or CDC, likely do not play a major role in prophylaxis or therapy. Despite clear evidence of in vitro ADE by ZKA190, and in contrast to a recent report (Bardina et al., 2017), our in vivo results indicate that ZKA190-mediated disease enhancement does not occur even when the mAb is administered at doses expected to provide only partial neutralization.
Numerous flavivirus MARMs are readily isolated in in vitro systems (de Alwis et al., 2012; Zou et al., 2012), similarly to what we observed with ZKA190, ZKA185 and ZKA230 mAbs. However, only a few studies described isolation of MARMs in vivo (Lai et al., 2007). Here, we isolated a ZKA190 MARM from one animal treated therapeutically, although no MARMs were detected in animals treated prophylactically. To reduce the risk of emergence of escape mutants, we produced a bispecific antibody coupling ZKA190 to the anti-DII ZKA185 mAb. The FIT-1 bispecific antibody is highly potent in vitro and in vivo and did not generate escape mutants even after 8 passages.
Flavivirus particles in in vitro preparations represent different maturation states (mature, partially immature, and fully immature) (Lok, 2016). Only partially and fully immature virions have prM on their surfaces, and anti-prM antibodies can confer infectivity to the otherwise non-infectious immature virions (Rodenhuis-Zybert et al., 2010). Since anti-prM antibodies are elicited in anti-flavivirus immune responses (Dejnirattisai et al., 2010), they can contribute significantly to pathogenesis (Dejnirattisai et al., 2010). We showed here that ZKA190 and FIT-1 can block in vitro ADE mediated by the cross-reactive prM mAb DV62, indicating that ZKA190 can effectively block prM antibody-mediated infection by at least partially immature ZIKV particles.
Our current studies in vitro and in lethal adult mouse models showed that the FIT-1 bispecific antibody is a good candidate for development of antibody-based therapy for ZIKV infection. Further studies in pregnant rhesus macaques (Dudley et al., 2016; Nguyen et al., 2017) would enable testing FIT-1 efficacy in blocking fetal infection. The in vivo protective activity of FIT-1 indicates that alternative delivery mechanisms, such as DNA or RNA vectors, or half-life-extending modification of the Fc, might be achievable to make it cost-effective for the prevention of congenital ZIKV disease.
STAR METHODS
KEY RESOURCES TABLE
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| ZKA190 | (Stettler et al., 2016); this study | N/A |
| ZKA190-LALA | This study | N/A |
| ZKA185 | (Stettler et al., 2016); this study | N/A |
| ZKA230 | (Stettler et al., 2016); this study | N/A |
| FIT-1 | This study | N/A |
| 4G2 | Absolute Antibody Ltd | Cat# Ab00230-2.0-BT |
| Bacterial and Virus Strains | ||
| ZIKV H/PF/2013 | European-Virus-Archive | Cat# 001v-EVA1545 |
| ZIKV MR766 | European-Virus-Archive | Cat# 001v-EVA143 |
| ZIKV MRS_OPY_Martinique_PaRi_2015 | European-Virus-Archive | Cat# 001v-EVAg1589 |
| ZIKV PV10552 | Baldanti F., IRCCS, Pavia (IT) (Acc. Number KY003154) | N/A |
| ZIKV PRVABC59 | CDC | N/A |
| ZIKV MP1751 | (Stettler et al., 2016); this study | N/A |
| ZIKV Nica 2-16 | (Tabata et al., 2016) | N/A |
| Chemicals, Peptides, and Recombinant Proteins | ||
| recombinant E protein | Meridian LifeScience | Cat# R01635 |
| ZIKV DIII from ZIKV Brazil 2015 strain (SPH2015) | (Stettler et al., 2016); this study | N/A |
| Critical Commercial Assays | ||
| RNeasy mini kit | QIAGEN | Cat# 74106 |
| qScript cDNA SuperMix | Quantabio | Cat# 95048-100 |
| iQ SYBR Green Supermix | Bio-Rad | Cat# #1708880 |
| Deposited Data | ||
| Fab ZKA190:ZIKV complex incubated at 37°C | This paper | PDB: 5Y0A |
| CryoEM maps of Fab ZKA190:ZIKV complexes incubated at 4°C | This paper | EMD-6794 |
| CryoEM maps of Fab ZKA190:ZIKV complexes incubated at 4°C | This paper | EMD-6793 |
| NMR structure of DIII | This paper | PDB: 5OMZ |
| Chemical shift used for structural calculation and mapping | This paper | BMRB: 34167 |
| Experimental Models: Cell Lines | ||
| EXPI293 | Invitrogen | N/A |
| Adult male fibroblasts | Movement Disorders Bio-Bank | N/A |
| Aedes albopitus C6/36 | ATCC | N/A |
| Vero cells | ATCC | N/A |
| K562 cells | N/A | |
| BHK-21 | ATCC | N/A |
| Experimental Models: Organisms/Strains | ||
| A129 mice, (IFN-alpha/beta receptor−/−) | N/A | |
| AG129 mice | (van den Broek et al., 1995) | N/A |
| Oligonucleotides | ||
| ZIKV_1086_Fwd, 5′- CCGCTGCCCAACACAAG-3′ | This study | N/A |
| ZIKV_1162c_Rev, 5′- CCACTAACGTTCTTTTGCAGACAT-3′ | This study | N/A |
| Probe ZIKV_1107-FAM_UG&Nica 5′- AGCCTACCTTGACA AGCAATCAGACACTCAA-3′ | (Lanciotti et al., 2008) | N/A |
| Zika-E-F1 5′-TGCAAACGCGGTCGCAAACCTGGTTG-3′ | This study | N/A |
| ZIKV-E-R1 5′-CGTGCCAAGGTAATGGAATGTCGTG-3′ | This study | N/A |
| ZIKV-E-f1530 5′-AGCCTAGGACTTGATTGTGAACCGA-3′ | This study | N/A |
| ZIKV-E-R2769 5′-TTACAGATCCCACAACGACCGTCAG-3′ | This study | N/A |
| ZIKV-E-F2 5′-ACTTGGTCATGATACTGCTGATTGC-3′ | This study | N/A |
| ZIKV-E-R2 5′-TCGGTTCACAATCAAGTCCTAGGCT-3′ | This study | N/A |
| ZIKV-E-f2058 5′-GCTAACCCCGTAATCACTGAAAGCA-3′ | This study | N/A |
| ZIKV-E-r2248 5′-AAGACTGCCATTCTCTTGGCACCTC-3′ | This study | N/A |
| ZIKV-NS1-Forward: 5′-TGGAGTTCAACTGACGGTCG-3′ | (Xu et al., 2016) | N/A |
| ZIKV-NS1-Reverse: 5′-TACCCCGAACCCATGATCCT-3′ | (Xu et al., 2016) | N/A |
| Gapdh- Forward: 5′-GGCAAGTTCAAAGGCACAGTC-3′ | (Xu et al., 2016) | N/A |
| Gapdh-Reverse: 5′-CACCAGCATCACCCCATTT-3′ | (Xu et al., 2016) | N/A |
| Software and Algorithms | ||
| PyMol Molecular Graphics Systems | Schrodinger, LLC | RRID: SCR_000305 |
| Prism 7.0 | GraphPad Software | N/A |
| CLC Main Work Bench | CLC bio | N/A |
| UCSF Chimera (v1.11.2) | (Pettersen et al., 2004) | RRID: SCR_004097 |
| Relion | (Scheres, 2012) | http://www2.mrc-lmb.cam.ac.uk/relion/index.php/Main_Page |
| Ctffind3 | (Mindell et al., 2003) | http://grigoriefflab.janelia.org/ctf |
| Leginon | (Carragher et al., 2000) | http://emg.nysbc.org/redmine/projects/leginon/wiki/Leginon_Homepage |
| RosettaDock | (Gray et al., 2003) | RRID: SCR_013393 |
| Assisted Model Building with Energy Refinement (AMBER) | (Case et al., 2017) | RRID: SCR_014230 |
| CARA | (Keller, 2004) | http://cara.nmr-software.org/downloads/3-85600-112-3.pdf |
| CYANA | (Güntert and Buchner, 2015) | RRID: SCR_014229 |
| Zetasizer Nano software version 6.01 | Malvern Instruments | N/A |
CONTACT FOR REAGENTS AND RESOURCE SHARING
Further information and requests for resources and reagents should be directed to and will be fulfilled by the Lead Contact, Davide Corti (davide.corti@humabs.ch)
EXPERIMENTAL MODEL AND SUBJECT DETAILS
Reagents
Cell lines were obtained from American Tissue Culture Collection. Vero and K562 cells were cultured in DMEM or RPMI 1640 medium (Invitrogen) supplemented with 10% FCS (Hyclone), 1% penicillin, 1% streptomycin and L-glutamine (complete DMEM or RPMI; all from Invitrogen)). The ZIKV H/PF/2013, MR766 and MRS_OPY_Martinique_PaRi_2015 (MRS_OPY) strains used in this work were obtained from European-Virus-Archive. The PV10552 strain was isolated in 2016 from the semen of an international traveler in Italy and was kindly provided by Fausto Baldanti from the Policlinico San Matteo, Italy (accession number KY003154). The Puerto Rican 2013 PRVABC59 strain was obtained from the CDC (GenBank Accession #KU501215). The virus had been passaged three times in Vero cells prior to our acquisition. Vero cells were maintained in Eagle Minimal Essential Medium (EMEM) supplemented with 10% fetal bovine serum and 1% penicillin-streptomycin. The virus was passaged two more times in Vero cells to establish a viral stock. Viral stocks were titered by limiting dilution plaque assay on Vero cells. Infections with ZIKV were performed at 37°C for 4 hr (h) before cells were overlaid with methylcellulose. After 6 days, plaques were visualized by crystal violet staining. The murine cross-reactive mAb used to detect ZIKV and DENV infection was produced from the 4G2 hybridoma purchased from ATCC and purified using protein G affinity chromatography (Thermo Scientific). The antigens used in ELISA were: recombinant E protein from ZIKV (Meridian LifeScience, R01635), recombinant DIII synthesized using the sequence from ZIKV Brazil 2015 strain (SPH2015). The DIII of ZIKV was expressed in E. coli Rosetta and Rosetta pLysS cells respectively, with a pET21 vector (Novagen) either in LB or M9 minimal media with aptly labelled ingredients as required for NMR (15N NH4Cl, 13C Glucose, 70% D2O). The cells were induced at OD600 = 0.6 with 0.5 mM of IPTG and harvested after 4 hr post-induction. After sonication and centrifugation, the pellet was washed and centrifuged two times in sodium phosphate buffer pH 7.2, 150 mM NaCl, 1 M urea and 1% Triton-X100. The pellet was resuspended in phosphate buffer pH 7.2 and 8 M urea (buffer A). Following addition of 0.2% of PEI and centrifugation to remove DNA contamination, ammonium sulphate to 60% was added to the supernatant to remove traces of PEI. After centrifugation, the pellet was resuspended in buffer A, and denaturated protein was diluted in refolding buffer (20 mM phosphate buffer pH 7.2, 150 mM NaCl, 200 mM arginine) in order to reach a final protein concentration of 0.2 mg/mL. DIIIs of DENV and ZIKV were purified on a Superdex-75 size exclusion column (GE) in PBS buffer. The elution and dynamic light scattering profiles of DIII were consistent with monomeric species.
METHOD DETAILS
Production of recombinant IgG and LALA variant
Briefly, the VH and VL sequences were cloned into human Igγ1, Igκ and Igλ expression vectors (kindly provided by Michel Nussenzweig, Rockefeller University, New York, NY, USA), essentially as described (47). Recombinant mAbs were produced by transient transfection of EXPI293 cells (Invitrogen), purified by Protein A chromatography (GE Healthcare) and desalted against PBS.
Production of recombinant FIT bispecific antibodies
The three genes encoding for FIT were codon optimized and synthesized by Genscript and cloned as follow: i) the VL of the outer Fab, followed by the full constant region (lambda or kappa), is fused with the VH of the inner and cloned into the Igγ1 expression vector (modified to encode for the LALA mutation). The resulting polypeptide 1 is formed by VL and CL of the outer Fab, VH of the inner Fab fused to IgG1 CH1-hinge-CH2-CH3 domains; ii) the VH gene of the outer Fab (encoding for polypeptide 2 formed by VH and CH1 of the outer Fab) was cloned into the Fab expression vector (Igγ1 expression vector in which a stop codon is introduced after the codon encoding for the CH1 cysteine residue 220); iii) the VL gene of the inner Fab is cloned into the Igκ or Igλ expression vectors (encoding for polypeptide 3 formed by VL and CL of the inner Fab). Recombinant FIT-Ig mAbs were produced by transient transfection of EXPI293 cells (Invitrogen) using a molar ratio of 1:3:3 of the three constructs described above (as described in PCT/US2014/072336), purified by Protein A chromatography (GE Healthcare) and desalted against PBS. The proteins were analyzed by by SDS-PAGE in both reduced and non-reduced conditions and their concentrations determined by BCA (Pierce, Rockford, IL). In non-reduced conditions, FIT-Ig migrated as a major single band of approximately 250 KDa. In reducing conditions, each of the FIT-Ig proteins yielded two bands, one higher MW band is polypeptide 1 of approximately 75 KDa, and one lower MW band corresponds to both polypeptide 2 and 3 overlapped at approximately 25 KDa. To further study the physical properties of FIT-Ig in solution, size exclusion chromatography (SEC) was used to analyze each protein. Purified FIT-Ig, in PBS, was applied on a Superdex 200 Increase 5/150 GL. All proteins were determined using UV detection at 280 nm and 214 nm. FIT-Ig proteins exhibited a single major peak, demonstrating physical homogeneity as monomeric proteins.
Infection of human neural progenitor cells (hNPCs)
Adult male fibroblasts obtained from the Movement Disorders Bio-Bank (Neurogenetics Unit of the Neurological Institute ‘Carlo Besta’, Milan) were reprogrammed using the CytoTune-iPS 2.0 Sendai kit (Life Technologies). hiPSCs were maintained in feeder-free conditions in mTeSR1 (Stem Cell Technologies). To generate embryoid bodies (EBs), dissociated hiPSCs were plated into low-adhesion plates in mTeSR1 supplemented with N2 (0.5×) (ThermoFisher Scientific), human Noggin (0.5 μg/ml, R&D System), SB431542 (5 μM, Sigma), Y27632 (10 μM, Miltenyi Biotec) and penicillin/streptomycin (1%, Sigma) (Marchetto et al., 2010). To obtain rosettes, EBs were plated after 10 days onto matrigel-coated plates (1:100, matrigel growth factor reduced, Corning) in DMEM/F12 (Sigma) with N2 (1×), non-essential amino acids (1%, ThermoFisher Scientific) and penicillin/streptomycin. After 10 days, cells were passaged with Accutase (Sigma) and seeded onto matrigel coated-flasks in NPC media containing DMEM/F12, N2 (0.5%), B27 (0.5%, ThermoFisher Scientific), penicillin/streptomycin and FGF2 (20 ng/ml, ThermoFisher Scientific). hNPCs (3×104) were plated on coverslips in 24-well plates 3 days prior to infection with PRVABC59 strain. Virus stock was incubated with the mAbs 1h prior to addition to hNPCs to obtain an MOI of 0.5. After 4h of virus adsorption, culture supernatant was removed and fresh medium containing the mAbs was re-added. Supernatant was collected 96h post-infection to measure virus titers by plaque assay on Vero cells. Cells were fixed in 4% paraformaldehyde (PFA, Sigma) solution in phosphate-buffered saline (PBS, Euroclone) for 30 min for indirect immunofluorescence. Fixed cells were permeabilized for 30 min in blocking solution, containing 0.2% Triton X-100 (Sigma) and 10% donkey serum (Sigma), and incubated overnight at 4°C with the primary antibodies in blocking solution. The following antibody was used for detection: anti-envelope (1:200, Millipore, MAB10216). Then, cells were washed with PBS and incubated for 1h with Hoechst and anti-mouse Alexa Fluor-488 secondary antibodies (1:1,000 in blocking solution, ThermoFisher Scientific). After PBS washes, cells were washed again and mounted.
Virus neutralization and antibody-dependent enhancement
Neutralization of ZIKV infection by mAbs was measured using a micro-neutralization flow cytometry-based assay. Different dilutions of mAbs were mixed with ZIKV (MOI of 0.35) for 1 hr at 37°C and added to 5000 Vero cells/well in 96-well flat-bottom plates. After four days for ZIKV, the cells were fixed with 2% formaldehyde, permeabilized in PBS containing 1% fetal claf serum (Hyclone) and 0.5% saponin, and stained with the mouse mAb 4G2. The cells were incubated with a goat anti-mouse IgG conjugated to Alexa Fluor488 (Jackson Immuno- Research, 115485164) and analyzed by flow cytometry. The neutralization titer (50% inhibitory concentration [IC50]) is expressed as the antibody concentration that reduced the infection by 50% compared to virus-only control wells. ADE was measured by a flow cytometry-based assay using K562 cells. mAbs and ZIKV (MOI 0.175) were mixed for 1 hr at 37°C and added to 5000 K562 cells/well. After four days, cells were fixed, permeabilized, and stained with mAb m4G2 The number of infected cells was determined by flow cytometry, as described above.
Size measurement of ZKA190 IgG — ZIKV complex by dynamic light scattering
ZIKV was purified as described above and diluted in NTE buffer (corresponding to ~0.2 mg/ml E protein). Immune complexes were formed by mixing ZIKV with serial dilutions of ZKA190 IgG in IgG/virion molar ratios ranging from 900:1 to 1:1 (concentration of virion was determined using SDS-PAGE gel by comparing the virus E proteins with BSA standards). After one hour incubation at 4°C, the diameter of the immune complexes were measured using dynamic light scattering (DLS) and the infectivity was determined using standard plaque assay in BHK-21 cells. Neutralization is represented as the fold reduction of the remaining virus infectivity in each condition compared to the virus only control (log10 (virus titer (PFU/ml) in the absence of mAb)/(virus titer (PFU/ml) in the presence of mAb). For the size measurement, 15 μl of the sample was loaded into a quartz cuvette and measured at 25°C by the Zetasizer Nano S machine (Malvern). Data were analyzed using Zetasizer Nano software version 6.01. The deconvolution of the measured correlation curve was done using a non-negative least-squares algorithm. Each sample is measured 10 times and each data point is a repeat of two independent experiments. Size of the immune complex is shown as intensity weighted mean hydrodynamic size of the ensemble collection of particles in diameter. Our purified virus alone sample has a polydispersity index of 0.2, indicating that the sample is monodisperse and acceptable for DLS size measurement. Aggregation of immune complex has very broad size distribution, resulting in a polydispersity index higher than 0.6. Therefore, the measured size is not very accurate for the aggregations. However, DLS was only used to probe the formation of aggregation in this experiment and the exact size of the aggregation is less important.
Pre- and postattachment neutralization assays
The pre- and post-attachment neutralization assays were conducted as previously described (Fibriansah et al., 2015). For the pre-attachment neutralization assay, BHK-21 cells were grown in 24-well plates. Different dilutions of ZKA190 IgG were incubated with 100 plaque forming units (PFU) of ZIKV for 1 hr at 4°C and subsequently added to the cells and further incubated for 1 hr at 4°C. For the post-attachment neutralization assays, the cell monolayer was pre-chilled at 4°C for 15 min before 100 PFU of ZIKV was added. The cells was co-incubated with the virus for 1 hr at 4°C and then washed twice with ice-cold serum-free medium to get rid of the unbound virus. Subsequently, different dilutions of ZKA190 IgG were added to the cells and further incubated for 1 hr at 4°C. The cells were finally washed twice with ice-cold serum-free medium and overlaid with 1% aquacide in RPMI medium supplemented with 2% fetal bovine serum. The cell monolayer was fixed and stained after 5-days incubation at 37°C in order to visualize the plaque formed by un-neutralized ZIKV.
RT–PCR to quantitate virus on cell surface
The number of ZIKV remaining on the surface of BHK-21 cells after ZKA190 treatment was measured by quantitative RT–PCR as previously described (Fibriansah et al., 2015). The experiment procedure is similar to the pre- and post-attachment neutralization assay described above but the quantification of the amount of virus particles attached on the cell surface were conducted immediately after the final wash of the cells. In pre-attachment condition, 105 PFU of ZIKV was incubated with neutralizing concentrations of ZKA190 IgG for 1 hr at 4°C and then added to the cells pre-chilled at 4°C. After one hour of incubation at 4°C, the cells were washed twice and total RNA was purified using RNeasy mini kit (Qiagen). In post-attachment condition, the cells were first incubated with the same amount of ZIKV at 4°C for 1 hr and then washed twice to get rid of the unbound virus. Neutralizing concentrations of ZKA190 IgG were then added to the cell and incubated at 4°C for 1 hr. Finally, the cells were washed twice and total RNA was purified. Complementary DNA was synthesized using qScript cDNA SuperMix (Quantabio). Realtime quantitative PCR (qPCR) was performed using iQ™ SYBR Green supermix kit (Bio-Rad) in a CFX 96 Real-Time System (Bio-Rad). Glyceraldehyde-3-phosphate dehydrogenase was used as the housekeeping gene to normalize samples. The analysis of relative levels of ZIKV RNA in different samples was performed by comparative 2-ΔΔCT method (Livak and Schmittgen, 2001). Gene specific primers (ZIKV-NS1-Forward: 5′-TGGAGTTCAACTGACGGTCG-3′; ZIKV-NS1-Reverse: 5′-TACCCCGAACCCATGATCCT-3′; Gapdh- Forward: 5′-GGCAAGTTCAAAGGCACAGTC-3′; Gapdh-Reverse: 5′-CACCAGCATCACCCCATTT-3′) were used for the real-time qPCR experiment.
Effect of LALA antibodies on ADE
ZIKV (MOI 0.175) was mixed with plasma from primary ZIKV-infected donors for 30 min at 37°C. ZKA190-LALA was added at 50 μg/ml, mixed with 5000 K562 cells/well and incubated for three days. Cells were then stained with 4G2 and analyzed by flow cytometry, as described above.
ELISA
A standard ELISA was used to determine binding of mAbs to ZIKV DIII or E proteins. ELISA plates were coated overnight at 4°C with 3 μg/ml of antigens. Plates were blocked with a 1% w/v solution of Bovine Serum Albumin (BSA; Sigma) in PBS, incubated with serial dilutions of mAbs for 1.5 hr at room temperature. After washing, antibody binding was revealed using goat antihuman IgG coupled to alkaline phosphatase (Jackson Immunoresearch). Plates were then washed, substrate (p-NPP, Sigma) was added and plates were read at 405 nm. Serum levels of human IgG in blood samples of infected mice was determined using a sandwhich ELISA specific for human IgG antibodies. ELISA plates were coated overnight at 4°C 5 μg/ml of goat anti-human IgG Fc-specific, mouse-adsorbed, antibodies (Southern Biotech). A standard based on the spiked-in mAb used in the challenge study diluted in mouse serum was used to interpolate the concentration of human mAbs in mouse sera samples. Plates were blocked with a 1% w/v solution of Bovine Serum Albumin (BSA; Sigma) in PBS, incubated with serial dilutions of sera samples for 1.5 hr at room temperature. After washing, human IgG levels were revealed using goat anti-human IgG Fc-specific (mouse-adsorbed) antibodies coupled to alkaline phosphatase (Southern Biotech). Plates were then washed, substrate (p-NPP, Sigma) was added and plates were read at 405 nm.
Surface Plasmon Resonance
Binding properties of the complexes between mAbs IgG or Fab and DIII (wild-type and mutants) or E protein were determined at 25°C with a ProteOn XPR-36 instrument (Bio-Rad). The antibodies were immobilized on the surface of a GLM sensor chip through standard amine coupling. Serial dilution of DIII and E-protein in the nanomolar range were injected at a flow rate of 100 μl/min (contact time 4 minutes); dissociation was followed for 10 min. Analyte responses were corrected for responses from buffer-only injection both on a channel with antibody immobilized and on a channel with no antibody immobilized. Curve fitting and data analysis were performed with Bio-Rad ProteOn Manager software (version 3.1.0.6).
NMR spectroscopy
Spectra were recorded on a Bruker Avance 700 MHz NMR spectrometer at 300 K. For assignments of backbone resonances standard triple resonance experiments (HNCO, HN(CA)CO, HN(CO)CACB, HNCACB were used, while sidechains were annotated using HCCH-TOCSY and HBHA(CO)NH experiments. All NMR experiments were processed using Topspin 2.1 (Bruker Biospin) and analysed with CARA. NOESY cross peaks were automatically assigned using the CYANA “noeassign” macro based on the manually assigned chemical shifts. Upper-distance restraints used for the structure calculations in CYANA using the standard simulated annealing protocol were derived from 70 ms 15N- and 13C-resolved NOESY spectra.
Backbone dynamics of ZIKV DIII were derived from 15N relaxation measurements recorded on 600 and 700 MHz spectrometers. Proton-detected versions of the CPMG (R2), inversion-recovery (R1) and 15N{1H}-steady-state NOE were utilized.
Delay settings for the T2 series were in the range of 0 to 0.25 sec and for the T1 series between 0.02 to 2 sec. The 15N{1H}-NOE experiment used a relaxation delay of 5 s. The R1 and R2 relaxation rates were derived from least-squares fits of corresponding exponential functions to the measured data using home-written scripts. The relaxation data were analyzed in a model-free approach using the software package DYNAMICS. The program ROTDIF was used to calculate the overall correlation time from the relaxation data (8.5 ns). NMR epitope mapping was performed as previously described (Bardelli et al., 2015; Simonelli et al., 2010; 2013). Briefly, overlay of N-HSQC spectra of labelled DIII free or bound to ZKA190 Fab allowed identification of DIII residues whose NMR signal changed upon complex formation, indicating that they were affected by ZKA190 binding. Changes were identified by manual inspection and by the Chemical Shift Perturbation (CSP), CSP=((ΔδH)2 +(ΔδN/10)2)1/2. NMR samples were typically 800μM of [15N, 13C]-labeled DIII in 20 mM sodium phosphate, 50 mM NaCl, pH 6.0. Perdeuterated (nominally 70%) 2H, 15N DIII samples were used for NMR epitope mapping with a DIII:ZKA190 Fab ratio of 1:1.1; DIII concentration was typically 0.4 mM.
Antibody modelling, computational docking and Molecular Dynamics simulations
The ZKA190, ZKA185 and ZKA230 variable fragments were modeled according to the canonical structure method with the program RosettaAntibody (Sircar et al., 2009) as previously described (Pedotti et al., 2011). Ten models, mainly differing in the H3 loop conformation, were generated and independently used for docking. For ZKA190, either the NMR calculated or X-ray derived structure of DIII (PDB 5JHM) was used for docking. Docking was performed using RosettaDock 2.3 as previously described (Simonelli et al., 2010). In summary, each antibody model was independently docked to DIII. Among the thousands of computationally generated complexes, those in better agreement with NMR and cryoEM experimental data were selected and further refined by computational docking. Models were selected to maximize fit to the cryoEM density map and presence of NMR-defined epitope residues at the computational interface in an iterative process. For ZKA185 and ZKA230, four adjacent E protein monomers extract from the CryoEM structure of ZIKV were used for docking. The best solutions according to the algorithm were evaluated in terms of their agreement with the experimental data (MARMs) After refinement, selected models of all the three antibodies were subjected to a 100ns molecular dynamics (MD) simulation to adjust the local geometry and verify that the structure was energetically stable. MD was performed with the AMBER package proteins were centered in a cubic box filled with TIP3P water model and 0.15M Na+Cl− ions using the ff14SB force field. After energy minimization and temperature and pressure equilibration of 100ps, 100ns MD simulations were performed. The trajectory files generated were analyzed after removal of periodic boundary conditions.
Modelling of ZKA190 full IgG bound to the viral surface
A homology model of ZKA190 full IgG was generated using the x-ray structure of a full IgG1 (PDB 1HZH) as template. Analysis of the cryo-EM, NMR and docking data showed that adjacent ZKA190 Fab binding sites on the viral surface are likely compatible with distances and orientation of the two arms of a full IgG; a pair on the 3-fold (~60 Å) and one on the 5-fold vertex (~56Å) (Figure 3B). Models of the full IgG were aligned with the Fabs on these binding sites and restrained molecular dynamics simulations were used to generate atomistic models of the full antibody bridging across two E proteins on the viral surface. A 1ns simulation with restraints on the distance between the two Fabs of the full IgG was first performed to allow movement of the two arms into an appropriate orientation. Another 1 ns simulation of 16641 Cα distance restraints moved the arms in their final position. Finally, 10’000 steps of energy minimizations were performed. The final result shows good agreement to both cryo-EM and NMR data.
Confocal Microscopy
Vero cells were plated at 7,500 cells/well on 12 mm-diameter coverslips in 24-well plates and incubated overnight. Cells were infected with ZIKV H/PF/2013 (MOI of 100) in the presence or absence of neutralizing concentrations of Alexa 488 conjugated mAbs (0.7 μM) at 37°C for 3 h, washed with PBS, and fixed with 2% paraformaldehyde in PBS for 30 min at room temperature. Acidified endosome were identified with Lysotracker red (Invitrogen) by adding the dye (50 nM) to the cells for the last 30 min of the incubation prior to fixation. Fixation was followed by extensive washes in PBS and 50 mM glycine and finally the coverslips were prepared for microscopy analysis using Vectashield mounting medium for fluorescence with DAPI (Vector Laboratories). Samples were analyzed by confocal microscopy using a Leica TCS SP5 microscope with a 63×/1.4 N.A. objective. Image analysis and processing was performed with FIJI software.
Sequence analysis from ex vivo ZIKV samples
Envelop protein (E) sequences were obtained from National Center for Biotechnology Information (NCBI) Zika Virus Variation Resource (Brister et al., 2014) as of November 24th, 2016. After aligning the protein sequences with Clustal Ω (Sievers et al., 2011) and purging the sequences that did not cover the DIII domain, 217 sequences were obtained. An amino acid distribution analysis was performed for each residue of the DIII protein sequences. The related 217 nucleotide sequences of the E protein were retrieved and aligned using MAFFT (Katoh and Standley, 2013). The identical nucleotide sequences were collapsed into unique sequences (138 in total) and a phylogenic tree was derived by maximum-likelihood method (bootstrapped 1,000 times) using MEGA 7 software (Kumar et al., 2016).
MARMs selection
Two-thousands TCID50 of H/PF/2013 ZIKV in 500 μl were incubated with 250 μl containing varying concentrations of mAb (8 different concentrations, starting with a final concentration of 200 μg/ml and performing serial 1:4 dilutions). The mixture was incubated for 45 minutes at 37°C, followed by the addition of 250 μl of a Vero cells suspension (3.2 × 106 cells) and an incubation in a 24 well plate for three-four days at 37°C to allow virus propagation to occur. After each step of selection, 500 μl of supernatants from three conditions were selected: the lowest concentration of mAb at which full protection of the monolayer was observed, one concentration at which we observed a partial cytophatic effect (CPE) effect on the cell monolayer and one concentration at which 100% of the cell monolayer was destroyed by the ZIKV CPE. The tube was spun down for 5 min at 1000 × g, aliquoted and stored at −80°C. Half of the volume was again pre-mixed with varying concentrations of mAb to repeat the selection and propagation process. The remaining supernatant was used for micro-neutralization assays and subsequent sequencing of the virus.
Sequencing of MARMs virus
To identify the escape mutations of the selected MARMs virus, a genomic RNA extraction was done followed by a one-step-PCR to amplify and sequence the ZIKV E protein amplicon. Cell supernatant (140 μl) from the MARMs selection was used for RNA extraction with the QIAamp Viral RNA mini kit (Qiagen). cDNA synthesis and PCR amplification were performed together using the SuperScript III One-Step RT-PCR with Platinium Taq (Invitrogen). For one reaction 25 μl reaction mix, 8 μl sterile water, 2 μM of each primer, 1 μl RNAse out (Life Technologies), 2 μl Superscript III RT/Platinum TaqMix and 12 μl RNA were used giving a final reaction volume of 50 μl. For the E protein N-terminal part, the primer pair Zika-E-F1 5′-TGCAAACGCGGTCGCAAACCTGGTTG-3′ and ZIKV-E-R1 5′-CGTGCCAAGGTAATGGAATGTCGTG-3′ and for the C-terminal part the primer pair ZIKV-E- f1530 5′-AGCCTAGGACTTGATTGTGAACCGA-3′ and ZIKV-E-R2769 5′-TTACAGATCCCACAACGACCGTCAG-3′ were used. The cycling conditions were 54°C for 40 min, 94°C for 2 min followed by 45 cycles of 94°C for 45 s, 50°C for 45 s and 68°C for 1.5 min with a final elongation step at 68°C for 5 min and a final cooling step at 4°C. The PCR products were analyzed and extracted from a 1.5 % agarose gel and further purified with the GFX PCR DNA and Gel Band Purification kit (GE Healthcare). For the sequencing reaction 8 μl of purified PCR product was mixed with 2 μM primer in a final volume of 10 μl and sent for sequencing (Microsynth). E protein N-terminal products were sequenced with ZIKV-E-F2 5′-ACTTGGTCATGATACTGCTGATTGC-3′ and ZIKV-E-R2 5′-TCGGTTCACAATCAAGTCCTAGGCT-3′, C-terminal PCR products with ZIKV-E-f2058 5′-GCTAACCCCGTAATCACTGAAAGCA-3′ and ZIKV-E-r2248 5′-AAGACTGCCATTCTCTTGGCACCTC-3′. Sequences were assembled and analyzed using CLC Main Workbench software (CLC Bio, version 5).
ZIKV infection of A129 mice
ZIKV infection studies were conducted in the laboratories of Public Health England (PHE), UK. Female A129 mice (IFN-alpha/beta receptor −/−) aged 5–8 weeks were supplied from a UK approved breeding colony (B&K Universal Ltd, UK). All procedures with animals were undertaken according to the United Kingdom Animals (Scientific Procedures) Act 1986. These studies were approved by the ethical review process of Public Health England, Porton Down, UK and the Home Office, UK via an Establishment Licence (PEL PCD 70/1707) and project Licence (30/2906). PHE has Animal Welfare Assurance that has been reviewed and approved by the Office of Laboratory Animal Welfare (OLAW) — ref #A5560-01. Humane endpoints were defined as 20% weight loss, neurological signs or immobility. When any of these endpoints were reached, animals were culled to prevent unnecessary suffering.
Assessment of prophylactic and therapeutic activity of ZKA190 and FIT-1 in A129 mice
Mice were administered mAbs (ZKA190, ZKA190-LALA and MPE8) diluted in PBS at different doses via the intraperitoneal (i.p.) route in a volume of 500 μl. MAbs were administered either 1 day before or 1, 2, 3 or 4 days after virus challenge. Animals were challenged subcutaneously with 10 pfu ZIKV (strain MP 1751) and followed for 14 days. Weights and temperatures were monitored daily and clinical observations were recorded at least twice per day. On day 5 post-challenge, 50 μl of blood was collected from each animal into a RNAprotect tube (QIAGEN,UK) and frozen at −80°C. At the end of the study (14 days post-challenge) or when animals met human endpoints, necropsies were undertaken, and blood and sections of brain, spleen, liver, kidney and ovary were collected for virological analysis.
Assessment of prophylactic activity of ZKA190 in AG129 mice
To test whether ZKA190 was capable of inducing ADE of ZIKV infection in vivo, 4- to 6-week-old AG129 mice lacking the interferon α/β and γ receptors (van den Broek et al., 1995) were used to test low doses of mAb prior to challenge. Animals were bred at the University of California, Berkeley, and all experimental procedures involving the use of AG129 mice were pre-approved and performed according to the guidelines of the Institutional Animal Care and Use Committee of University of California, Berkeley. Mice were injected intraperitoneally (i.p.) with antibodies or phosphate buffered saline (PBS) in a total volume of 100 μl and then infected 20–24 hr later with 103 focus-forming units (FFU) of ZIKV strain Nica 2–16 (Tabata et al., 2016) in a 50-μl volume by subcutaneous (s.c.) foot pad injection. Mice were monitored over a 15-day period post-infection; body weight was recorded daily, and morbidity was scored using a standardized 5-point system (Orozco et al., 2012). Mice were euthanized if they lost more than 20% of their initial weight and/or if they progressed to stage 5 on the morbidity scale (moribund, consistent with partial paralysis and neurological symptoms).
Viral load quantification from AG129 ZIKV infected mice
Blood samples from ZIKV-infected mice or controls were collected from the facial (submandibular) vein of anesthetized mice on days 3 and 6 post-infection. Blood specimens were allowed to clot at room temperature, and serum was separated by centrifugation. ZIKV RNA was extracted from serum samples using the QIAamp Viral RNA Mini Kit (Qiagen), and RNA levels were determined by TaqMan one-step quantitative reverse transcriptase PCR (qRT-PCR) using a standard curve with 10-fold dilutions of ZIKV Nica 2–16 RNA. Primer sets used for this assay: ZIKV_1086_Fwd, 5′-CCGCTGCCCAACACAAG-3′, ZIKV_1162c_Rev, 5′- CCACTAACGTTCTTTTGCAGACAT-3′; Probe ZIKV_1107-FAM_UG&Nica 5′- AGCCTACCTTGACAAGCAATCAGACACTCAA-3′ (Lanciotti et al., 2008).
Viral load quantification from A129 ZIKV infected mice
Tissue samples from A129 mice were weighed and homogenized into PBS using ceramic beads and an automated homogenizer (Precellys, UK) using six 5 s cycles of 6500 rpm with a 30 s gap. Two hundred μl of tissue homogenate or blood solution was transferred into 600 μL RLT buffer (Qiagen, UK) for RNA extraction using the RNeasy Mini extraction kit (Qiagen, UK); samples were passed through a QIAshredder (Qiagen, UK) as an initial step. A ZIKV specific realtime RT-PCR assay was utilized for the detection of viral RNA from subject animals. The primer and probe sequences were adopted from a published method (52) with in-house optimization and validation performed to provide optimal mastermix and cycling conditions. Real-time RT-PCR was performed using the SuperScript III Platinum One-step qRT-PCR kit (Life Technologies, UK). The final mastermix (15 μl) was comprised of 10 μl of 2× Reaction Mix, 1.2 μl of PCR-grade water, 0.2 μl of 50 mM MgSO4, 1 μl of each primer (ZIKV 1086 and ZIKV 1162c both at 18 μM working concentration), 0.8 μl of probe (ZIKV 1107-FAM at 25 μM working concentration) and 0.8 μl of SSIII enzyme mix. Five μl of template RNA was added to the mastermix, yielding a final reaction volume of 20 μl. The cycling conditions used were 50°C for 10 min, 95°C for 2 min, followed by 45 cycles of 95°C for 10 s and 60°C for 40 s, plus a final cooling step of 40°C for 30 s. Quantification analysis using fluorescence was performed at the end of each 60°C step. Reactions were run and analyzed on the 7500 Fast platform (Life Technologies, UK) using 7500 software version 2.0.6. Quantification of viral load in samples was performed using a dilution series of quantified RNA oligonucleotide (Integrated DNA Technologies). The oligonucleotide comprised the 77 bases of ZIKV RNA targeted by the assay, based on GenBank accession AY632535.2 and was synthesized to a scale of 250 nmole with HPLC purification.
DIII coding region sequencing from biological samples
For each sample, an attempt was made to reverse-transcribe and amplify the whole genome using the v2 amplicon sequencing protocol kindly supplied by J. Quick and N. Loman of the Zibra project (Faria et al., 2016) (www.zibraproject.org/data/amplicon_sequencing_protocol_v2.pdf). The two amplicon pools produced for each sample were quantified using the dsDNA HS assay (Life Technologies) and combined, before 1 ug of DNA was used as input to the EXP-NBD002 barcoding kit with barcodes NB01–NB09. Samples 1–9 and 10–18 were pooled separately and two sequencing libraries produced using the SQK-NSK007 kit. Each library was sequenced using a FLO-MIN105 Flow cell on a MinION Mk1B device and base calling was performed using the Metrichor workflow “2D Basecalling plus Barcoding for FLO-MIN105 250bps” (Oxford Nanopore Technologies). Between 3989 and 12801 2D reads were generated per sample. FastQ data from 2D reads was extracted using poretools (Loman and Quinlan, 2014) and mapped using BWA-MEM (Li and Durbin, 2009) to the sequence of the original MP1741 stock used in this study (Genbank KY288905). Coverage across the genome varied due to differences in amplification efficiency between amplicons, but mean coverage of the DIII coding region was greater than 60-fold (range 64-fold to 410-fold) for all samples other than 14 and 16, which were not included for further analysis. The align_trim.py script from the Zibraproject.org pipeline (https://github.com/zibraproject/zika-pipeline, branch master, commit ID a4364fa8901b06d6d8bd058c19acd2c5e780179e) was used with an MP1741 adapted BED file of primer binding sites to trim primer sequences from the mapped reads. Samtools (Li et al., 2009) was used to covert the trimmed SAM file to a sorted and indexed BAM which was used as input along with FAST5 files for nanopolish v0.5 variants module (Loman et al., 2015) (Quick et al., 2016) (https://github.com/jts/nanopolish, branch master, commit ID 6406f78c01d05f968bc841c03e4ccb4ea208677a) to identify SNPs that met minimum criteria of read depth 20, support fraction 0.75 and minimum quality value of 200.
Virus sample preparation for cryoEM studies
Aedes albopitus C6/36 cells (ATCC) were propagated in RPMI 1640 media supplemented with 10% fetal bovine serum at 29°C. The cells were inoculated with ZIKV strain H/PF/2013 (Baronti et al., 2014) at a MOI of 0.5 and incubated at 29°C with 2% of fetal bovine serum. The virus containing media were harvested 4 days post-infection and clarified from cell debris by centrifugation. The supernatant was then precipitated by 8% of polyethylene glycol 8000 in NTE buffer (12mM Tris, 120 mM NaCl, 1 mM EDTA, pH 8.0) overnight at 4°C and pelleted by centrifugation at 14,000 g for 1 h. The resulting pellet was then resuspended in NTE buffer and further purified through a 24% (w/v) sucrose cushion followed by a 10–30% (w/v) potassium tartrate density gradient. The virus band in the gradient was then extracted and buffer exchanged into NTE buffer and concentrated using a concentrator with 100-kDa molecular weight cut-off filter. The concentration of the virus was estimated by comparing the viral E protein band with bovine serum albumin at known concentrations as standard in a Coomassie blue-stained SDS-PAGE.
CryoEM sample preparation
For preparation of the antibody-virus complex for cryoEM imaging, an excessive amount of recombinant Fab fragment of ZKA190 was added to the purified ZIKV to achieve a molar ratio of three Fab molecules for two E proteins. The complexes were incubated at 4, 37 and 40°C for 30 min, respectively. To prepare the cryoEM grids, 2.3 μl of sample was applied to a lacey copper grid with a thin carbon film, blotted for 1 s with an FEI Vitrobot in 100% humidity and then plunged into liquid ethane. The frozen grids were kept in liquid nitrogen before imaging.
CryoEM data collection and image processing
CryoEM images of the frozen ZKA190 Fab:ZIKV complexes were collected on a Titan Krios (FEI) microscope equipped with 300 kV field emission gun. Leginon was used for automated data collection (Carragher et al., 2000). The magnification was 81,871, giving a pixel size of 1.71 Å. The images were recorded in single image mode on Falcon II direct electron detector (FEI) with a total dose of 20 e−Å−2. The images were taken at underfocus range between 1.0 and 3.5 μm. A total of 902 and 738 micrographs were collected for ZKA190 Fab:ZIKV complexes at 4°C and 37°C, respectively. The astigmatic defocus parameters were estimated using ctffind3 (Mindell and Grigorieff, 2003) and used in orientation search and 3D reconstruction in Relion (Scheres, 2012). Particles were picked using Relion template-based autopicking program, visually examined to discard nonviral particles, and subsequently subjected to Relion 2D classification to produce 2D class averages. Classes containing junks and broken particles were excluded from further processing. In total, 10,641 and 6,728 particles in the ZKA190Fab:ZIKV complex samples that were incubated at 4°C and 37°C were selected for further processing. The 3D orientation search and reconstruction was done with icosahedral symmetry using Relion auto-refine procedure, where two models are refined independently for two random halves of the data to prevent over-fitting (Scheres, 2012). The mature ZIKV map (EMDB: EMD-8139) was filtered to 60 Å and used as the starting model. The 3D refinement produced structures with resolutions of 22 Å for both complexes at Fourier shell correlation (FSC) cut-off of 0.5.
Model fitting
As the E protein raft was intact (Figure S4A), the cryoEM map was first interpreted by fitting in the E protein dimers using the “fit-in-map” in Chimera (Pettersen et al., 2004). We tried fitting the E protein densities of both ZKA190 Fab:ZIKV complex cryoEM maps at 4°C and 37°C by using either the uncomplexed ZIKV (PDB 5IZ7) or the crystal structure of the ZIKV E protein dimers (PDB 5JHM). The crystal structure of the ZIKV E protein dimer fits better to the raft density than that of the cryoEM uncomplexed ZIKV. The difference between the cryoEM ZIKV E protein dimer structure and the crystal structure of ZIKV E ectodomain is in the DI–DII hinge angle. In the uncomplexed ZIKV cryoEM structure, the DI–DII hinge is fixed at a certain angle because its ectodomain interacts with the membrane associated stem regions of E and M proteins on the virus. However, in the ZKA190 Fab:ZIKV structure, the E protein raft is lifted up, as observed by the separation of the E protein shell density from the viral lipid bilayer membrane (Figure 2E). This may explain why the crystal structure of recombinant E protein fits better to the cryoEM density map of the complex. We then fitted the variable and constant domains of the Fab ZKA190 homology model into their corresponding densities above the DIII of E protein molecule C around the 3-fold vertex, as this density shows the best shape for the Fab (Figure S4A). The fitted molecule C and ZKA190 Fab were then locked and this E protein-ZKA190 Fab complex was superimposed as a rigid body to E molecules A and B, respectively. These Fab molecules correlated well with their corresponding densities. Slight adjustment of the positions of the Fab above molecules A and B was done to further optimize the fit (Figure S4A). No crashes between the symmetry related molecules were detected.
Structure analysis
Electrostatic potentials of protein surfaces were calculated using Coulombic Surface Coloring in Chimera (Pettersen et al., 2004).
QUANTIFICATION AND STATISTICAL ANALYSIS
Statistical analysis
Mortality data were expressed using Kaplan-Meier survival curves, and log-rank (Mantel-Cox) tests were used to determine statistical significance using Prism 7.0 software (GraphPad).
DATA AND SOFTWARE AVAILABILITY
Coordinates of Fab ZKA190:ZIKV complex incubated at 37°C was deposited in the Protein Data Bank under accession code 5Y0A and the NMR structure of DIII under PDB:5OMZ and chemical shift used for structural calculation and mapping under BMRB code 34167. The cryoEM maps of Fab ZKA190:ZIKV complexes incubated at 4°C and 37°C were deposited in the Electron Microscopy Database under accession number EMD:6794 and EMD:6793, respectively.
Supplementary Material
Highlights.
ZKA190 is a potent ZIKV neutralizing mAb targeting DIII of the E protein
ZKA190 binds to all 180 E proteins on ZIKV virion distorting quaternary structure
Zika virus escapes from ZKA190 as well as DII-specific neutralizing antibodies
A ZKA190-based DII–DIII bispecific antibody prevents ZIKV escape and protects animals
Acknowledgments
We thank Victor A. Kostyuchenko and P. Robert Beatty for scientific discussion and Kuiama Lewandowski at PHE for the technical support in viral sequencing and Rocco D’Antuono for microscope assistance. We thank Fabio Benigni at Humabs for critical reading of the manuscript and Antonio Piralla from San Matteo Hospital in Pavia for helping in the sequencing of MARMs. We thank the European Virus Archive for consenting to the use of the ZIKV H/PF/2013 strain. The work was supported by Singapore Ministry of Education Tier 3 grant (MOE2012-T3-1-008), National Research Foundation Investigatorship award (NRF-NRFI2016-01), National Research Foundation Competitive Research Project grant (NRF2016NRF-CRP001-063) and the Duke-NUS Signature Research Programme funded by the Ministry of Health, Singapore, awarded to S-M.L, and grant 5U19AI109761 from the US National Institutes of Health to E.H. L.V. is grateful for support by G. Noseda, SNF grant 310030_166445, KFS-3728-08-2015 and Lions Club Monteceneri. A.L. is the scientific founder of Humabs BioMed SA. A.L. holds shares in Humabs BioMed. K.S., F.Z., S.J., E.C., M.B. and D.C. are employees of Humabs Biomed.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
SUPPLEMENTAL INFORMATION
Supplemental Information includes six figures and can be found with this article online at http://dx.doi.org/10.1016/j.cell.2017.09.002.
AUTHOR CONTRIBUTIONS
F.Z. and S. Jaconi produced and purified antibodies. M.F. produced figures and carried out bioinformatic analysis. S.B. performed in vitro cellular neutralization, ADE assays and MARMs selection. D.A.E., T.B., V.G., S.P., S.D. and R.H. designed and carried out in vivo studies. J.W., T.-S.N., E.X.Y.L., G.F., J.S. designed and performed structural cryoEM research. M.B., M.P., L.S. designed and performedNMR structural research, docking, molecular dynamics, mutagenesis, binding and immunofluorescence studies; A.C. designed and performed molecular dynamics simulations; S. Jurt and O.Z. performed NMR studies. I.P., A.R., V.B. and E.V. designed and performed in vitro studies on hNPCs infection models. K.S. carried out binding studies, in vitro cellular neutralization, MARMs isolation and sequencing, analyzed the data and coordinated in vitro characterization activities. E.C. and M.B. provided supervision and contributed analyzing data. F.S. and A.L. edited the paper and provided supervision. J.W., E.H., S.-M.L., L.V., and D.C. designed research, analyzed data, and wrote the paper.
References
- Allison SL, Schalich J, Stiasny K, Mandl CW, Heinz FX. Mutational evidence for an internal fusion peptide in flavivirus envelope protein E. J Virol. 2001;75:4268–4275. doi: 10.1128/JVI.75.9.4268-4275.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barba-Spaeth G, Dejnirattisai W, Rouvinski A, Vaney MC, Medits I, Sharma A, Simon-Lorière E, Sakuntabhai A, Cao-Lormeau VM, Haouz A, et al. Structural basis of potent Zika—dengue virus antibody cross-neutralization. Nature. 2016:1–23. doi: 10.1038/nature18938. [DOI] [PubMed] [Google Scholar]
- Bardelli M, Livoti E, Simonelli L, Pedotti M, Moraes A, Valente AP, Varani L. Epitope mapping by solution NMR spectroscopy. J Mol Recognit. 2015;28:393–400. doi: 10.1002/jmr.2454. [DOI] [PubMed] [Google Scholar]
- Bardina SV, Bunduc P, Tripathi S, Duehr J, Frere JJ, Brown JA, Nachbagauer R, Foster GA, Krysztof D, Tortorella D, et al. Enhancement of Zika virus pathogenesis by preexisting antiflavivirus immunity. Science. 2017;356:175–180. doi: 10.1126/science.aal4365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baronti C, Piorkowski G, Charrel RN, Boubis L, Leparc-Goffart I, de Lamballerie X. Complete coding sequence of zika virus from a French polynesia outbreak in 2013. Genome Announc. 2014;2:e00500-14–e00500-14. doi: 10.1128/genomeA.00500-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beltramello M, Williams KL, Simmons CP, Macagno A, Simonelli L, Quyen NTH, Sukupolvi-Petty S, Navarro-Sanchez E, Young PR, de Silva AM, et al. The human immune response to Dengue virus is dominated by highly cross-reactive antibodies endowed with neutralizing and enhancing activity. Cell Host Microbe. 2010;8:271–283. doi: 10.1016/j.chom.2010.08.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bressanelli S, Stiasny K, Allison SL, Stura EA, Duquerroy S, Lescar J, Heinz FX, Rey FA. Structure of a flavivirus envelope glycoprotein in its low-pH-induced membrane fusion conformation. Embo J. 2004;23:728–738. doi: 10.1038/sj.emboj.7600064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brister JR, Bao Y, Zhdanov SA, Ostapchuck Y, Chetvernin V, Kiryutin B, Zaslavsky L, Kimelman M, Tatusova TA. Virus Variation Resource–recent updates and future directions. Nucleic Acids Res. 2014;42:D660–D665. doi: 10.1093/nar/gkt1268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carragher B, Kisseberth N, Kriegman D, Milligan RA, Potter CS, Pulokas J, Reilein A. Leginon: an automated system for acquisition of images from vitreous ice specimens. Journal of Structural Biology. 2000;132:33–45. doi: 10.1006/jsbi.2000.4314. [DOI] [PubMed] [Google Scholar]
- Case DA, Cerutti DS, Cheatham TE, Darden TA, Duke RE, Giese TJ, Gohlke H, Goetz AW, Greene D, Homeyer N, et al. AMBER 2017. University of California; San Francisco: 2017. [Google Scholar]
- Chung KM, Nybakken GE, Thompson BS, Engle MJ, Marri A, Fremont DH, Diamond MS. Antibodies against West Nile Virus nonstructural protein NS1 prevent lethal infection through Fc gamma receptor-dependent and -independent mechanisms. J Virol. 2006;80:1340–1351. doi: 10.1128/JVI.80.3.1340-1351.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corti D, Bianchi S, Vanzetta F, Minola A, Perez L, Agatic G, Guarino B, Silacci C, Marcandalli J, Marsland BJ, et al. Cross-neutralization of four paramyxoviruses by a human monoclonal antibody. Nature. 2013;501:439–443. doi: 10.1038/nature12442. [DOI] [PubMed] [Google Scholar]
- Crill WD, Roehrig JT. Monoclonal antibodies that bind to domain III of dengue virus E glycoprotein are the most efficient blockers of virus adsorption to Vero cells. J Virol. 2001;75:7769–7773. doi: 10.1128/JVI.75.16.7769-7773.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Alwis R, Smith SA, Olivarez NP, Messer WB, Huynh JP, Wahala WMPB, White LJ, Diamond MS, Baric RS, Crowe JE, et al. Identification of human neutralizing antibodies that bind to complex epitopes on dengue virions. Proc Natl Acad Sci USA. 2012 doi: 10.1073/pnas.1200566109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dejnirattisai W, Jumnainsong A, Onsirisakul N, Fitton P, Vasanawathana S, Limpitikul W, Puttikhunt C, Edwards C, Duangchinda T, Supasa S, et al. Cross-reacting antibodies enhance dengue virus infection in humans. Science. 2010;328:745–748. doi: 10.1126/science.1185181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dowall SD, Graham VA, Rayner E, Atkinson B, Hall G, Watson RJ, Bosworth A, Bonney LC, Kitchen S, Hewson R. A Susceptible Mouse Model for Zika Virus Infection. PLoS Negl Trop Dis. 2016;10:e0004658–13. doi: 10.1371/journal.pntd.0004658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dudley DM, Aliota MT, Mohr EL, Weiler AM, Lehrer-Brey G, Weisgrau KL, Mohns MS, Breitbach ME, Rasheed MN, Newman CM, et al. A rhesus macaque model of Asian-lineage Zika virus infection. Nat Commun. 2016;7:12204. doi: 10.1038/ncomms12204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edeling MA, Austin SK, Shrestha B, Dowd KA, Mukherjee S, Nelson CA, Johnson S, Mabila MN, Christian EA, Rucker J, et al. Potent dengue virus neutralization by a therapeutic antibody with low monovalent affinity requires bivalent engagement. PLoS Pathog. 2014;10:e1004072. doi: 10.1371/journal.ppat.1004072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faria NR, Sabino EC, Nunes MRT, Alcantara LCJ, Loman NJ, Pybus OG. Mobile real-time surveillance of Zika virus in Brazil. Genome Med. 2016;8:97. doi: 10.1186/s13073-016-0356-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fibriansah G, Tan JL, Smith SA, de Alwis R, Ng TS, Kostyuchenko VA, Jadi RS, Kukkaro P, de Silva AM, Crowe JE, et al. A highly potent human antibody neutralizes dengue virus serotype 3 by binding across three surface proteins. Nat Commun. 2015;6:6341. doi: 10.1038/ncomms7341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frontera JA, da Silva IRF. Zika Getting on Your Nerves? The Association with the Guillain—Barré Syndrome. N Engl J Med. 2016 doi: 10.1056/NEJMe1611840. NEJMe1611840–NEJMe1611842. [DOI] [PubMed] [Google Scholar]
- Garcez PP, Loiola EC, Madeiro da Costa R, Higa LM, Trindade P, Delvecchio R, Nascimento JM, Brindeiro R, Tanuri A, Rehen SK. Zika virus impairs growth in human neurospheres and brain organoids. Science. 2016:1–7. doi: 10.1126/science.aaf6116. [DOI] [PubMed] [Google Scholar]
- Gong S, Ren F, Wu D, Wu X, Wu C. Fabs-in-tandem immunoglobulin is a novel and versatile bispecific design for engaging multiple therapeutic targets. MAbs. 2017;165:0–00. doi: 10.1080/19420862.2017.1345401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Govero J, Esakky P, Scheaffer SM, Fernandez E, Drury A, Platt DJ, Gorman MJ, Richner JM, Caine EA, Salazar V, et al. Zika virus infection damages the testes in mice. Nature. 2016 doi: 10.1038/nature20556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gray JJ, Moughon S, Wang C, Schueler-Furman O, Kuhlman B, Rohl CA, Baker D. Proteinprotein docking with simultaneous optimization of rigid-body displacement and side-chain conformations. J Mol Biol. 2003;331:281–299. doi: 10.1016/s0022-2836(03)00670-3. [DOI] [PubMed] [Google Scholar]
- Güntert P, Buchner L. Combined automated NOE assignment and structure calculation with CYANA. J Biomol NMR. 2015;62:453–471. doi: 10.1007/s10858-015-9924-9. [DOI] [PubMed] [Google Scholar]
- Halstead SB. Dengue Antibody-Dependent Enhancement: Knowns and Unknowns. Microbiol Spectr. 2014;2:249–271. doi: 10.1128/microbiolspec.AID-0022-2014. [DOI] [PubMed] [Google Scholar]
- Hessell AJ, Hangartner L, Hunter M, Havenith CE, Beurskens FJ, Bakker JM, Lanigan CM, Landucci G, Forthal DN, Parren PW, et al. Fc receptor but not complement binding is important in antibody protection against HIV. Nature. 2007;449:101–104. doi: 10.1038/nature06106. [DOI] [PubMed] [Google Scholar]
- Honein MA, Dawson AL, Petersen EE, Jones AM, Lee EH, Yazdy MM, Ahmad N, Macdonald J, Evert N, Bingham A, et al. Birth Defects Among Fetuses and Infants of US Women With Evidence of Possible Zika Virus Infection During Pregnancy. Jama. 2017 doi: 10.1001/jama.2016.19006. [DOI] [PubMed] [Google Scholar]
- Katoh K, Standley DM. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol. 2013;30:772–780. doi: 10.1093/molbev/mst010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaufmann B, Nybakken GE, Chipman PR, Zhang W, Diamond MS, Fremont DH, Kuhn RJ, Rossmann MG. West Nile virus in complex with the Fab fragment of a neutralizing monoclonal antibody. Proc Natl Acad Sci USA. 2006;103:12400–12404. doi: 10.1073/pnas.0603488103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keller R. The Computer Aided Resonance Assignment Tutorial. Cantina Verlag; 2004. [Google Scholar]
- Kostyuchenko VA, Lim EXY, Zhang S, Fibriansah G, Ng TS, Ooi JSG, Shi J, Lok SM. Structure of the thermally stable Zika virus. Nature. 2016;533:425–428. doi: 10.1038/nature17994. [DOI] [PubMed] [Google Scholar]
- Kumar S, Stecher G, Tamura K. MEGA7: Molecular Evolutionary Genetics Analysis version 7.0 for bigger datasets. Mol Biol Evol. 2016:msw054. doi: 10.1093/molbev/msw054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lai CJ, Goncalvez AP, Men R, Wernly C, Donau O, Engle RE, Purcell RH. Epitope determinants of a chimpanzee dengue virus type 4 (DENV-4)-neutralizing antibody and protection against DENV-4 challenge in mice and rhesus monkeys by passively transferred humanized antibody. J Virol. 2007;81:12766–12774. doi: 10.1128/JVI.01420-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lanciotti RS, Kosoy OL, Laven JJ, Velez JO, Lambert AJ, Johnson AJ, Stanfield SM, Duffy MR. Genetic and serologic properties of Zika virus associated with an epidemic, Yap State, Micronesia, 2007. Emerg Infect Dis. 2008;14:1232–1239. doi: 10.3201/eid1408.080287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Durbin R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics. 2009;25:1754–1760. doi: 10.1093/bioinformatics/btp324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, Marth G, Abecasis G, Durbin R, 1000 Genome Project Data Processing Subgroup The Sequence Alignment/Map format and SAMtools. Bioinformatics. 2009;25:2078–2079. doi: 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- Lok SM. The Interplay of Dengue Virus Morphological Diversity and Human Antibodies. Trends Microbiol. 2016;24:284–293. doi: 10.1016/j.tim.2015.12.004. [DOI] [PubMed] [Google Scholar]
- Loman NJ, Quinlan AR. Poretools: a toolkit for analyzing nanopore sequence data. Bioinformatics. 2014;30:3399–3401. doi: 10.1093/bioinformatics/btu555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loman NJ, Quick J, Simpson JT. A complete bacterial genome assembled de novo using only nanopore sequencing data. Nat Methods. 2015;12:733–735. doi: 10.1038/nmeth.3444. [DOI] [PubMed] [Google Scholar]
- Marchetto MCN, Carromeu C, Acab A, Yu D, Yeo GW, Mu Y, Chen G, Gage FH, Muotri AR. A model for neural development and treatment of Rett syndrome using human induced pluripotent stem cells. Cell. 2010;143:527–539. doi: 10.1016/j.cell.2010.10.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mindell JA, Grigorieff N. Accurate determination of local defocus and specimen tilt in electron microscopy. Journal of Structural Biology. 2003;142:334–347. doi: 10.1016/s1047-8477(03)00069-8. [DOI] [PubMed] [Google Scholar]
- Mlakar J, Korva M, Tul N, Popović M, Poljšak-Prijatelj M, Mraz J, Kolenc M, Resman Rus K, Vesnaver Vipotnik T, Fabjan Vodušek V, et al. Zika Virus Associated with Microcephaly. N Engl J Med. 2016;374:951–958. doi: 10.1056/NEJMoa1600651. [DOI] [PubMed] [Google Scholar]
- Modis Y, Ogata S, Clements D, Harrison SC. Structure of the dengue virus envelope protein after membrane fusion. Nature. 2004;427:313–319. doi: 10.1038/nature02165. [DOI] [PubMed] [Google Scholar]
- Nguyen SM, Antony KM, Dudley DM, Kohn S, Simmons HA, Wolfe B, Salamat MS, Teixeira LBC, Wiepz GJ, Thoong TH, et al. Highly efficient maternal-fetal Zika virus transmission in pregnant rhesus macaques. PLoS Pathog. 2017;13:e1006378–22. doi: 10.1371/journal.ppat.1006378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nowakowski TJ, Pollen AA, Di Lullo E, Sandoval-Espinosa C, Bershteyn M, Kriegstein AR. Expression Analysis Highlights AXL as a Candidate Zika Virus Entry Receptor in Neural Stem Cells. Cell Stem Cell. 2016;18:591–596. doi: 10.1016/j.stem.2016.03.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nybakken GE, Oliphant T, Johnson S, Burke S, Diamond MS, Fremont DH. Structural basis of West Nile virus neutralization by a therapeutic antibody. Nature. 2005;437:764–769. doi: 10.1038/nature03956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Onorati M, Li Z, Liu F, Sousa AMM, Nakagawa N, Li M, Dell’Anno MT, Gulden FO, Pochareddy S, Tebbenkamp ATN, et al. Zika Virus Disrupts Phospho-TBK1 Localization and Mitosis in Human Neuroepithelial Stem Cells and Radial Glia. Cell Rep. 2016;16:2576–2592. doi: 10.1016/j.celrep.2016.08.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orozco S, Schmid MA, Parameswaran P, Lachica R, Henn MR, Beatty R, Harris E. Characterization of a model of lethal dengue virus 2 infection in C57BL/6 mice deficient in the alpha/beta interferon receptor. Journal of General Virology. 2012;93:2152–2157. doi: 10.1099/vir.0.045088-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pedotti M, Simonelli L, Livoti E, Varani L. Computational docking of antibody-antigen complexes, opportunities and pitfalls illustrated by influenza hemagglutinin. Int J Mol Sci. 2011;12:226–251. doi: 10.3390/ijms12010226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pettersen EF, Goddard TD, Huang CC, Couch GS, Greenblatt DM, Meng EC, Ferrin TE. UCSF Chimera–a visualization system for exploratory research and analysis. J Comput Chem. 2004;25:1605–1612. doi: 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
- Quick J, Loman NJ, Duraffour S, Simpson JT, Severi E, Cowley L, Bore JA, Koundouno R, Dudas G, Mikhail A, et al. Real-time, portable genome sequencing for Ebola surveillance. Nature. 2016;530:228–232. doi: 10.1038/nature16996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rey FA, Heinz FX, Mandl C, Kunz C, Harrison SC. The envelope glycoprotein from tick-borne encephalitis virus at 2 A resolution. Nature. 1995;375:291–298. doi: 10.1038/375291a0. [DOI] [PubMed] [Google Scholar]
- Rodenhuis-Zybert IA, van der Schaar HM, da Silva Voorham JM, van der Ende-Metselaar H, Lei HY, Wilschut J, Smit JM. Immature dengue virus: a veiled pathogen? PLoS Pathog. 2010;6:e1000718. doi: 10.1371/journal.ppat.1000718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scheres SHW. RELION: implementation of a Bayesian approach to cryo-EM structure determination. Journal of Structural Biology. 2012;180:519–530. doi: 10.1016/j.jsb.2012.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sievers F, Wilm A, Dineen D, Gibson TJ, Karplus K, Li W, Lopez R, McWilliam H, Remmert M, Söding J, et al. Fast, scalable generation of high-quality protein multiple sequence alignments using Clustal Omega. Mol Syst Biol. 2011;7:539–539. doi: 10.1038/msb.2011.75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simonelli L, Beltramello M, Yudina Z, Macagno A, Calzolai L, Varani L. Rapid structural characterization of human antibody-antigen complexes through experimentally validated computational docking. J Mol Biol. 2010;396:1491–1507. doi: 10.1016/j.jmb.2009.12.053. [DOI] [PubMed] [Google Scholar]
- Simonelli L, Pedotti M, Beltramello M, Livoti E, Calzolai L, Sallusto F, Lanzavecchia A, Varani L. Rational Engineering of a Human Anti-Dengue Antibody through Experimentally Validated Computational Docking. PLoS ONE. 2013;8:e55561. doi: 10.1371/journal.pone.0055561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sircar A, Kim ET, Gray JJ. RosettaAntibody: antibody variable region homology modeling server. Nucleic Acids Res. 2009;37:W474–W479. doi: 10.1093/nar/gkp387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sirohi D, Chen Z, Sun L, Klose T, Pierson TC, Rossmann MG, Kuhn RJ. The 3.8 Å resolution cryo-EM structure of Zika virus. Science. 2016:aaf5316. doi: 10.1126/science.aaf5316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stettler K, Beltramello M, Espinosa DA, Graham V, Cassotta A, Bianchi S, Vanzetta F, Minola A, Jaconi S, Mele F, et al. Specificity, cross-reactivity, and function of antibodies elicited by Zika virus infection. Science. 2016;353:823–826. doi: 10.1126/science.aaf8505. [DOI] [PubMed] [Google Scholar]
- Stiasny K, Allison SL, Marchler-Bauer A, Kunz C, Heinz FX. Structural requirements for low-pH-induced rearrangements in the envelope glycoprotein of tick-borne encephalitis virus. J Virol. 1996;70:8142–8147. doi: 10.1128/jvi.70.11.8142-8147.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tabata T, Petitt M, Puerta-Guardo H, Michlmayr D, Wang C, Fang-Hoover J, Harris E, Pereira L. Zika Virus Targets Different Primary Human Placental Cells, Suggesting Two Routes for Vertical Transmission. Cell Host Microbe. 2016 doi: 10.1016/j.chom.2016.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang H, Hammack C, Ogden SC, Wen Z, Qian X, Li Y, Yao B, Shin J, Zhang F, Lee EM, et al. Zika Virus Infects Human Cortical Neural Progenitors and Attenuates Their Growth. Stem Cell. 2016:1–5. doi: 10.1016/j.stem.2016.02.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van den Broek MF, Müller U, Huang S, Aguet M, Zinkernagel RM. Antiviral defense in mice lacking both alpha/beta and gamma interferon receptors. J Virol. 1995;69:4792–4796. doi: 10.1128/jvi.69.8.4792-4796.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Q, Yang H, Liu X, Dai L, Ma T, Qi J, Wong G, Peng R, Liu S, Li J, et al. Molecular determinants of human neutralizing antibodies isolated from a patient infected with Zika virus. Sci Transl Med. 2016;8:369ra179–369ra179. doi: 10.1126/scitranslmed.aai8336. [DOI] [PubMed] [Google Scholar]
- Xu M, Lee EM, Wen Z, Cheng Y, Huang WK, Qian X, Tcw J, Kouznetsova J, Ogden SC, Hammack C, et al. Identification of small-molecule inhibitors of Zika virus infection and induced neural cell death via a drug repurposing screen. Nat Med. 2016;22:1101–1107. doi: 10.1038/nm.4184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X, Sheng J, Austin SK, Hoornweg TE, Smit JM, Kuhn RJ, Diamond MS, Rossmann MG. Structure of acidic pH dengue virus showing the fusogenic glycoprotein trimers. J Virol. 2015;89:743–750. doi: 10.1128/JVI.02411-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao H, Fernandez E, Dowd KA, Speer SD, Platt DJ, Gorman MJ, Govero J, Nelson CA, Pierson TC, Diamond MS, et al. Structural Basis of Zika Virus-Specific Antibody Protection. Cell. 2016:1–26. doi: 10.1016/j.cell.2016.07.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zou G, Kukkaro P, Lok SM, Ng JKW, Tan GK, Hanson BJ, Alonso S, MacAry PA, Shi PY. Resistance analysis of an antibody that selectively inhibits dengue virus serotype-1. Antiviral Res. 2012;95:216–223. doi: 10.1016/j.antiviral.2012.06.010. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


