Abstract
The anthracnose disease caused by Colletotrichum species is well-known as a major plant pathogen that primarily causes fruit rot in pepper and reduces its marketability. Thirty-five isolates representing species of Colletotrichum were obtained from chili fruits showing anthracnose disease symptoms in Chungcheongnam-do and Chungcheongbuk-do, South Korea. These 35 isolates were characterized according to morphological characteristics and nucleotide sequence data of internal transcribed spacer, glyceraldehyde-3-phosphate-dehydrogenase, and β-tubulin. The combined dataset shows that all of these 35 isolates were identified as C. scovillei and morphological characteristics were directly correlated with the nucleotide sequence data. Notably, these isolates were recorded for the first time as the causes of anthracnose caused by C. scovillei on pepper in Korea. Forty cultivars were used to investigate the pathogenicity and to identify the possible source of resistance. The result reveals that all of chili cultivars used in this study are susceptible to C. scovillei.
Keywords: Anthracnose, Colletotrichum scovillei, Pepper
Chili (Capsicum annum L.), also known as pepper, belonging to the Solanaceae family is the fourth major vegetable cultivated globally, first in Asia, and one of the economically important vegetable crops in world given its massive consumption worldwide [1,2]. Chili peppers originated in Mexico and Central America (Mesoamericas) [3]. During the 16th century, chilies were transported to Asia by Portuguese navigators and used in both food and medicine [3]. Regardless of when chili pepper was introduced in Korea, it is one of the most widely used ingredients in Korean cuisine. It is used in sauces, soups, stews, stir fry dishes, marinades, salad as garnish for many dishes, and of course in kimchi [4]. It is supplied as a food ingredient in cuisines and medicinal uses as required and exported to other countries. Interestingly, red chili fruits contain more vitamin A and vitamin C than found in carrots and citrus fruits [5,6]. Moreover, regular consumption of chili fruit is helpful against anorexia, liver congestion, varicose veins, and hemorrhoids [2]. Besides, chili can be used as a natural and organic pesticide, and in the preparation of pepper sprays as a self-defense weapon [7]. In Korea, chili pepper is a widely consumed crop, and was cultivated on approximately 34,514 ha in 2015 [8].
Colletotrichum is a phytopathogenic genus prevalent today all over the world, and is a destructive plant pathogen, causing disease in various crops and fruits. Of the diseases caused by this pathogen, chili anthracnose disease considerably limits chili cultivation, leading to enormous economic losses, with annual losses estimated at more than US $100 million in Korea [1,9]. Fruits lesions are the most economically important aspect of chili anthracnose disease even though the Colletotrichum sp. can cause the anthracnose disease on many other parts of the chili plant. Anthracnose disease is characterized by concentric rings of acervuli within a lesion. The lesions are initially brown, and then turn black because of the formation of setae and sclerotia [10]. To date, only four species of Colletotrichum (C. gloeosporioides, C. coccodes, C. acutatum, and C. dematium) that cause chili anthracnose disease have been reported in Korea [1,11]. Of these species, the major pathogen of chili anthracnose in Korea may be C. acutatum rather than C. gloeosporioides [9]. Currently, tremendous genetic divergence in the population of C. acutatum indicates the taxon should be considered a species complex and should be further separated into distinct species [12,13,14].
Most popular local chili varieties are susceptible to anthracnose disease, and the fungicides must be sprayed at several times during cultivation. Therefore, it is necessary to examine the major causal agent of this disease and the interaction between various popular chili varieties and the pathogen to understand their resistance or susceptibility for further studies. The aim of the present study was to identify and characterize the Colletotrichum species causing chili anthracnose in Korea, to investigate the reaction of four popular local varieties and 36 accessions of Capsicum spp. to isolated C. scovillei, and to identify the possible sources of resistance cultivars for use in plant breeding programs.
MATERIALS AND METHODS
Isolation of the fungus from infected chili fruits
The fruits showing the typical disease symptoms were cut into small fragments (5 mm) and small pieces of disease fruits were surface sterilized by dipping 1% sodium hypochlorite (NaOCl) for 3 min followed by 3 washes with sterilized distilled water and dried on sterilized tissue paper [15]. Then the fruits were placed on Petri dish containing blotter paper and incubated at 25 ± 2℃ with a 12/12-hr dark-light chamber. After 2 days, spore layers were isolated using autoclaved toothpick or glass stick, and were mixed with distilled water and streptomycin (300 ppm for 1 L) in tube. The mixtures were then spread on Water agar media containing streptomycin for 3 days at 25 ± 2℃ [16]. After 2 or 3 days, without bacterial or fungal contaminants, a single isolated spore of emerging fungus was transferred on a potato dextrose agar (PDA; Difco, Sparks, MD, USA) plate to obtain the pure culture. The isolates were identified to the species under light microscopy.
Morphological analysis
To check the detail morphological character of the pathogen, the isolates were cultured on two media, PDA and V8 Juice agar media. Small pieces (5mm) of mycelium plugs were picked up from actively growing area near the edge of a 4–5-day-old culture and placed on PDA and V8 Juice agar media and then incubated at room temperature (25 ± 2℃). After 7 days, colony characters such as colony size, color of the conidial masses and zonation of every culture was recorded from PDA and also V8 Juice media. Conidia were measured using conidia taken from the conidial ooze on PDA media and mounted in a drop of lacto phenol [17], at least 24 conidia were measured for each isolate.
Appressoria were produced using a slide culture [18]. Small pieces (10-mm2) of PDA were placed in vacant Petri dish. The small portion of spore taken from fungal sporulating culture was placed on the edge of this PDA media and immediately covered with a sterilized cover slip. After 7 days, the cover slip (which formed the appressoria across the underside of the cover slip) was removed and mounted in a drop of lacto phenol on a glass slide. At least 20–30 appressoria were measured for each isolate.
Genomic DNA extraction
Genomic DNA was extracted directly from all mycelia of fungal isolates using the method of Cenis (1992) [19]. Aerial mycelia/conidia mat were scraped from 10-day-old cultures and ground with 300 µL of extraction buffer (250 mM NaCl, 200 mM Tris-HCl pH 8.5, 0.5% sodium dodecyl sulfate, 25 mM EDTA) in an Eppendorf tube (1.5 mL). Thereafter, 150 µL of 3M sodium acetate, pH 5.2 are added, and tubes are placed at −20℃ for about ten minutes. Tubes were then centrifuged in a microfuge at 13,000 rpm for 10 min and the supernatant was transferred to another tube. Then, an equal volume of isopropanol was added and placed on ice for 10 min and the tubes were then centrifuged at 13,000 rpm for 20 min. After centrifuged, the pellet was vacuum dried and washed with 500 µL of 70% ethanol and then centrifuged for 3min. After that, the pellet was vacuum dried for some minutes and suspended in 50 µL of distilled water. DNA samples were visualized under ultraviolet illumination by gel electrophoreses and qualities and amounts were compared with standard markers. All samples were stored at 4℃ for future use.
PCR and sequencing
DNA amplification and sequencing were accomplished by PCR. To confirm morphological identification, Glyceraldehyde-3-phostphate dehydrogenase (GAPDH) gene, β-Tublin-2 (TUB2) gene and the internal transcribed spacer (ITS) rDNA regions were amplified using the fungal specific primers GDF/GDR [20], TUBT1 [21]/TUBT2 [22], and ITS1/4 [23] respectively. Condition for amplification were an initial denaturing step of 5 min at 94℃, followed by 33 cycles of 30 sec at 94℃, 30 sec at 54℃ and 1 min at 72℃, and a final denaturing step of 7min at 72℃. PCR products were observed by staining with ethidium bromide (EtBr) on 1% agarose electrophoresis gels in TAE buffer and visualized under UV light. The PCR products were purified and directly sequenced with the same primers. For several strains, sequencing was performed in both directions (forward and reverse) to ensure that there were no misjudge. The sequences data obtained were compared with all fungal sequences and submitted in NCBI GenBank database.
Phylogenetic analysis
All the obtained sequences of GAPDH, TUB2 and ITS were compared with the available sequence data, using BLAST search against the NCBI GenBank database to identify the sequences. Multiple sequences alignment was manually performed with the closely related references sequence of other isolated Colletotrichum species available in NCBI database using ClustalW2 software (http://www.ebi.ac.uk/Tools/phylogenecy/clustalw2-phylogeny/). A neighbor-joining tree with a combined data set ITS, GAPDH and TUB2 were constructed according to maximum likelihood method using the MEGA 7 software ver. 7.0. The reliability of the tree was evaluated with 1,000 bootstrap replication for branch stability. C. truncatum was designated outgroup in all analyses.
Pathogenicity tests
The isolated fungi including CNU151001 (Colletotrichum sp.) were inoculated into healthy fruits to prove the pathogenicity of the fungus. Isolates were cultured on V8 juice agar media at 25℃ under the 12/12-hr dark-light condition because the fungal mycelium can grow rapidly and produce many more conidia on V8 juice media than PDA media. Conidia were harvested from 7-day-old cultures by flushing 5–10 mL of sterilized distilled water onto this culture, which were then filtered through two layers of muslin cloth. The obtaining conidial suspensions were adjusted to get the 1 × 106 conidia/mL concentration using a hemocytometer. For the pathogenicity test, the isolated fungus CNU 151001 was inoculated on healthy fruits by non-wound/drop and wound/drop methods [17,24]. The fruits of four local cultivars (Jumping, PR Jindaegeon, Super Manitta, Cheonnyeon yaksok) and 36 accession of Capsicum sp. from National Agrobiodiversity Center, Rural Development Administration (RDA, Korea) were used to carry out the pathogenicity test. Healthy fruits were rinsed with tap water, sterilized with 1% NaOCl solution for 3 min and rinsed twice with distilled water. Then the fruits were dry and placed on a sterilized paper tissue in moistened clean boxes and inoculated with 10 µL of conidial spore (1 × 106 conidia/mL) suspension using wounding or non-wounding method. To develop the diseases, boxes were incubated for 7 days at 25℃ in the dark. Three fruits per each cultivar and three replicates were carried out for each treatment. All control fruits were treated with 10 µL of sterilized distilled water. To fulfill the Koch's postulates, the symptoms developed on the inoculated fruits were compared with original symptoms and the fungi were re-isolated from inoculated fruits following the above mention procedure.
After inoculation, disease severity of anthracnose on fruit was evaluated based on the modified scale described by Montri et al. [25]. Symptoms were examined 7 days after inoculation and disease severity were observed based on the percentage of disease lesion size dealing with the overall fruit size as the different fruit size and lesion size of individual cultivar where Capsicum chacoense genotypes consisted of small circular fruit while C. baccatum genotype were large, elongated, and wide fruits. A set of disease ratings of point scale, where (−) = no infection and symptom, highly resistant; (+) = 1–5% of the fruit area shows symptom, moderately resistant; (++) ≥ 5–25% of the fruit area shows necrotic lesion, moderately susceptible; (+++) ≥ 25% of the fruit covered with necrotic lesion with acervuli and, lesion often encircling the fruit; abundant acervuli, highly susceptible.
RESULTS
Identification of isolated fungus
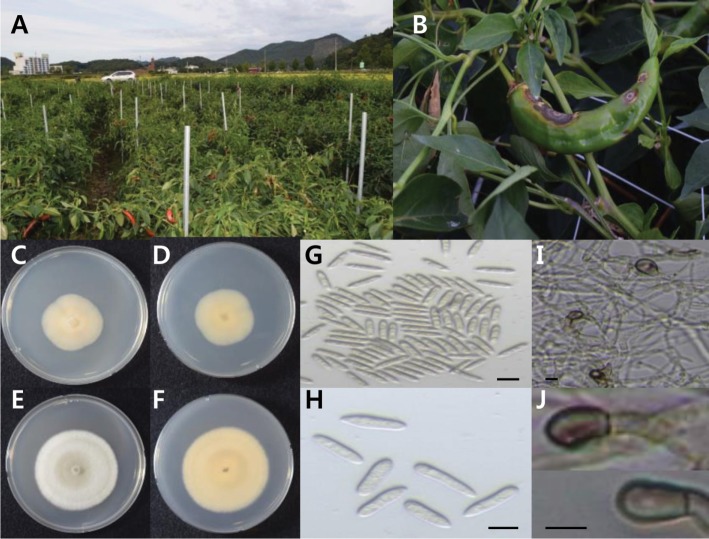
In the summer of 2015, suspected typical anthracnose disease symptoms characterized by a circular, sunken zone with concentric rings of orange conidial masses on fruits of chili were observed in Chungcheongnam-do and Chungcheongbuk-do, South Korea. Fresh specimens were collected from infected plants in the field. Thirty-five isolates of Colletotrichum sp. were obtained from typical anthracnose symptoms of chili fruits (Fig. 1A and 1B). Based on culture features, as well as the shape and size of conidia and appressoria, all of these 35 isolates from infected fruits morphologically aligned and were identified as Colletotrichum scovillei, as proposed by Damm et al. (2012) [26].
Fig. 1. Anthracnose disease caused by Colletotrichum scovillei on chili pepper fruits and morphology characteristics of chili anthracnose. A, An anthracnose-infected field of chili pepper; B, Anthracnose disease symptoms on chili pepper; C, D, Fungal colonies on potato dextrose agar plates (C, upper view; D, reverse view); E, F, Fungal colonies on V8 juice agar plates (E, upper view; F, reverse view); G, H, Morphological characteristics of conidia. One end round and one end (±) acute conidia; I, J, Clavate to irregular shape appressoria (scale bars: G–J = 10 µm).
Morphological characterization
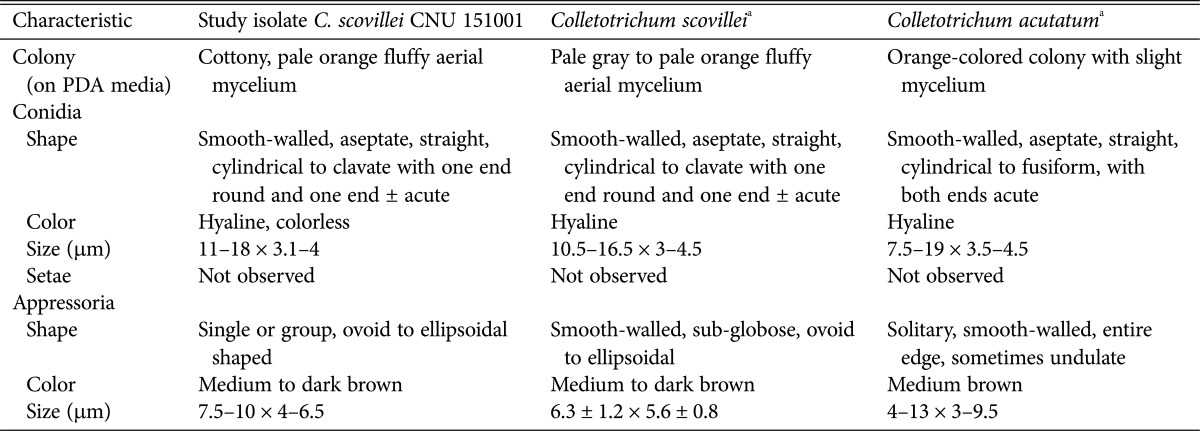
The appearance of colony characters on PDA and V8 juice agar were observed after 7 days of sub-culturing. The mycelia of all of isolated fungi formed cottony, pale orange fluffy aerial mycelial growths on PDA (Fig. 1C and 1D). Colonies on V8 juice agar appeared flat with whole margins, with the surface covered with milky to light olivaceous grey mycelia, olivaceous grey to iron grey in the center (Fig. 1E and 1F). Significant differences were not observed in the growth rate among these 35 isolates and their size ranged from 29–31 mm after 7 days of growth on PDA, and 49–51 mm on V8 juice agar. Conidia in all isolates were smooth-walled, hyaline, straight, aseptate, cylindrical to clavate with one end acute, and other end round, 11 to 18 µm long, and 3.1 to 4 µm wide (Fig. 1G and 1H). Sexual morph was not observed. Acervuli were not developed and chlamydospores were also not observed. No setae were formed. Appressoria were single or grouped, ovoid to ellipsoidal, medium or dark brown color, 7.5–10 × 4–6.5 µm (Fig. 1I and 1J). Significant differences were not observed in the shape and width of appressoria among the 35 isolates. Morphological characteristics typical of C. scovillei [26] are described in Table 1.
Table 1. Comparison of the morphological characteristics of the isolate under study with those of previously reported Colletotrichum scovillei and C. acutatum.
aSource of description and illustration [26].
Phylogenetic analysis
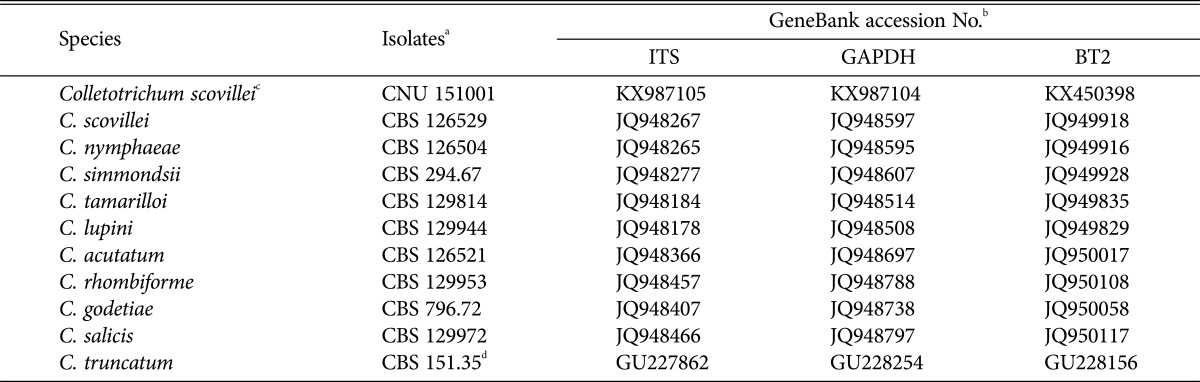
PCR products obtained from ITS, GAPDH and TUB2 gene sequences of isolated fungus CNU151001 were 648 bp, 248 bp, and 563 bp long, respectively. The sequences of all genes revealed a high similarity 99–100% identical to the C. scovillei isolate, but varied from those of other Colletotrichum species. Sequences from all 35 isolates were identical to C. scovillei sequences deposited in GenBank, and no differences were evident between these 35 isolates. The ITS, GAPDH, and TUB2 sequences from a representative isolate (CNU 151001) were deposited in GenBank under the accession Nos. KX987105, KX987104, and KX450398, respectively. The final sequence alignment of a combined ITS, GAPDH and β-tubulin dataset containing 11 taxa (including CNU 151001 and 10 reference sequences from GenBank) was used to compare the tree output. Isolates of Colletotrichum scovillei (Fig. 2) and relevant species from used in this study are described in Table 2. In the current study, similar nucleotide sequences were analyzed among strains belonging to C. scovillei and C. acutatum based on this ITS locus.
Fig. 2. Phylogenetic tree generated by a maximum parsimony analysis of a combined dataset of internal transcribed spacer (ITS), glyceraldehyde 3-phosphate dehydrogenase (GAPDH), and TUB2 gene sequences of Colletotrichum scovillei and those of other related Colletotrichum spp. described by Damm et al. (2012) [26]. Number beside each branch represent bootstrap values obtained after a bootstrap test with 1,000 replications. Bar indicates the number of nucleotide substitutions. The fungal strain analyzed in current study shown in boldface. C. truncatum (CBS 151.35) served as the out-group.
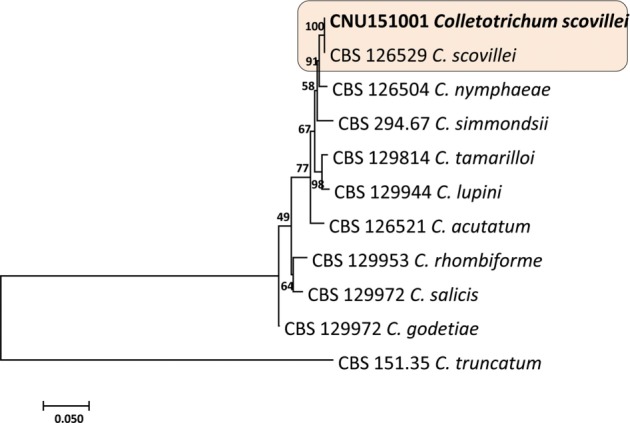
Table 2. Colletotrichum strains used in this study and their GenBank accession numbers.
aIsolates information and sequences retrieved by Damm et al. [26].
bITS, internal transcribed spacer; GAPDH, glyceraldehyde-3-phosphate-dehydrogenase; BT2, β-tubulin.
cIsolated in this study.
dServed as the out-group.
Pathogenicity of CNU151001 to local cultivars and 36 accessions of Capsicum sp
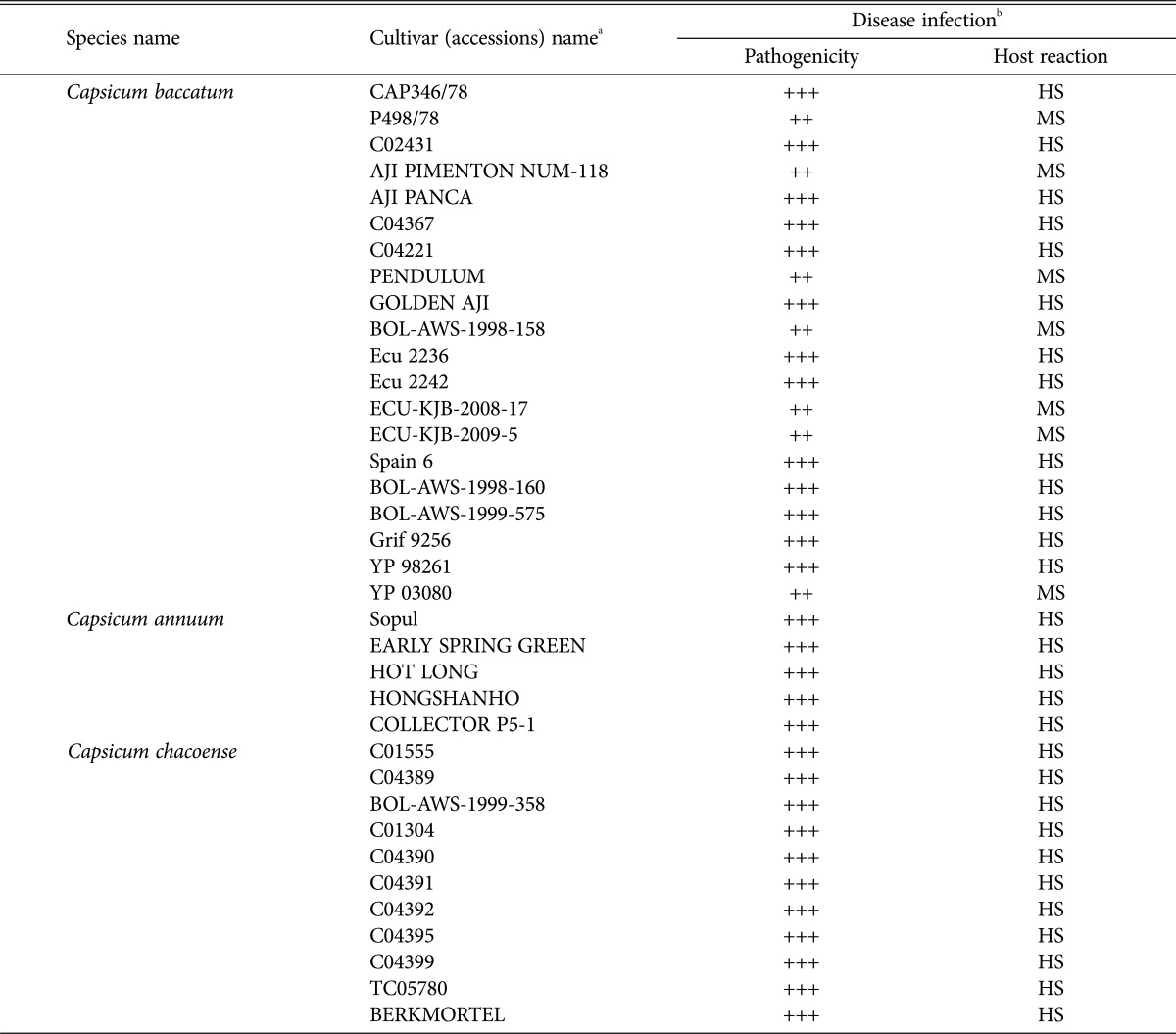
To investigate the responses of chili cultivars to these newly isolated Colletotrichum sp., and to identify the possible source of resistance for use in plant breeding programs, 36 accessions of Capsicum sp. (20 varieties of Capsicum baccatum, 5 varieties of C. annum and 11 C. chaocense) from the RDA of Korea were assessed. To test the pathogenicity of the isolated Colletotrichum, 35 isolates were inoculated onto healthy fruits by wounding methods. All the inoculated pepper fruits showed typical symptoms after 7 days. Of the 35 isolates of Colletotrichum sp., one isolate, CNU151001, showed high susceptibility and was used for pathogen infection of all 36 chili cultivars. Seedlings of 36 cultivars were planted in a plastic house until the fruits were harvested. To assess pathogenicity against these 36 cultivars, the CNU151001 isolate was inoculated on healthy harvested fruits using the wound/drop method, as described. Disease severity was also evaluated as previously described. A list of the chili varieties used in this investigation is presented in Table 3.
Table 3. Pathogenicity of 36 accessions of chilli cultivars to Colletotrichum scovillei 7 days after inoculation with wound/non-wound method.
aThirty-six cultivars of Capsicum sp. provide by the Rural Development Administration, National Agrobiodiversity Center of Korea were used.
b−; 0% (highly resistance), +; 1–5% (moderately resistance), ++; 5–25% (moderately susceptible, MS), +++; > 25% of the fruit covered with necrotic lesion with acervuli (highly susceptible, HS).
All C. scovillei isolates were pathogenic to local cultivars (Jumping, PR Jindaegeon, Super Manitta, and Cheonnyeon yaksok) that served as the original host. Colletotrichum scovillei was pathogenic to all inoculated fruits via both wound/drop and non-wound/drop methods, resulting in typical anthracnose symptoms, and lesions were not significantly different in terms of host response among these four cultivars, with a disease score of “highly susceptible.” Three days after inoculation, typical anthracnose symptoms were observed in terms of the formation of acervuli on the surface of inoculated fruit tissue resulting from the wound/drop inoculation method (Fig. 3A–3E), while in the non-wound/drop inoculation the symptoms appeared 7 days after inoculation (Fig. 3F–3J). Symptoms appeared earlier after inoculation using wound/drop than using the non-wound/drop method. It can therefore be concluded that wounding can accelerate disease progression and injury. Disease symptoms were not visible on control fruits, which were inoculated with sterilized distilled water. The present study indicated that these four local cultivars were highly susceptible to C. scovillei.
Fig. 3. Pathogenicity of anthracnose on pepper fruits after Colletotrichum scovillei CNU151001 inoculation. Symptoms on wound inoculated after 7 days (A–E), and on non-wound (spray) inoculated after 14 days (F–J). Control fruits (A, F), cv. Jumping (B, G), cv. PR Jindaegeon (C, H), cv. Super Manitta (D, I), cv. Cheonnyeon yaksok (E, J).
Typical anthracnose lesions were observed in all 36 accessions of Capsicum sp. infected using the wound/drop inoculation method, with disease scores of “moderately susceptible” and “highly susceptible,” although the degree of pathogenicity differed depending on varieties, and different infection incidences (Table 3). No symptoms were observed on the control fruits. Our study indicates that the most chili varieties might be susceptible to C. scovillei.
DISCUSSION
The high consumption of chili as an important vegetable crop worldwide was the motivation behind this study. Here, we aimed to characterize the etiological agent of anthracnose on chili fruit in Korea, and to explore varieties conferring disease resistance for use in breeding programs. Chili is a widely consumed vegetable in Korea because of its varied use in food and kimchi. In the present study, we identified the Colletotrichum species causing chili anthracnose in Korea by phenotypic and genotypic characterization. Presently, outbreaks of chili anthracnose have considerably affected the chili production area in Korea. Owing to the environment-induced changes in culture and morphological characteristics, C. gloeosporioides and C. acutatum cannot be easily differentiated, and so morphology-based identification has become one of the most challenging issues involved in chili anthracnose disease [1].
In the current study, we used a combined dataset of morphological description and molecular characteristics to confirm the identity of the current isolate. Identification of Colletotrichum species by morphology alone could lead to errors because there are no definite morphotaxonomic characters, and these characters tend to deviate depending on the experimental methods and conditions [27,28]. Therefore, results derived from molecular approaches would be more reliable, stable, and helpful in identifying the Colletotrichum species. A combination of morphological and molecular analyses of ITS-rDNA, GAPDH and TUB2 region, and pathogenicity tests showed that C. scovillei is the major causal agent of anthracnose of chili in Korea.
Colletotrichum scovillei is one of the species that belongs to clade 2 of the C. acutatum species complex [26], and can be differentiated from other species by ACT, TUB2 and GAPDH sequences (with GAPDH being most distinctly differential), while ITS sequences are similar to those of C. acutatum. The sequences data used in this method indicate that the ITS sequence alone cannot clearly differentiate the C. acutatum complex species. Therefore, this locus should not be used alone for species recognition of Colletotrichum sp. isolates. The sequence data in present study indicated that the ITS locus has lower variability than the GAPDH and TUB2 loci, and ITS alone does not have the adequate depth to unambiguously provide characterization results. Moreover, the results of this study confirmed that the combination of a multigene approach with morphology is necessary to accurately identify Colletotrichum species. Morphological characters are variable within the clade of the C. acutatum species complex, and molecular sequence data are needed to reliably differentiate between the constituent taxa.
Most Korean local chili cultivars are susceptible to anthracnose disease and growers therefore normally use fungicides to control this disease. However, continuous use of chemical can cause adversely affect disease resistance and cause environmental pollution. It is therefore important to examine the interactions between various popular and economical chili varieties and pathogen isolates to obtain basic information regarding their resistance and susceptibility for future studies. Information regarding the inheritance of resistance to anthracnose disease in Capsicum is fundamental to successful breeding programs, as it helps the breeder to select the best breeding approach to obtain cultivars. No sources of resistance were identified in any of the 36 accessions of Capsicum species susceptible to Colletotrichum scovillei in chili examined in the present study. Therefore, most of the chili cultivars (C. annum, C. chacoense, and C. baccatum) were highly susceptible to chili anthracnose disease, although some Capsicum baccatum cultivars were moderately susceptible. Although no resistant cultivars were identified in the present study, these results can be the basis of information for further investigation on resistance. According to Potnis et al. (2012) [29], successful crosses have been carried out between C. annum and C. baccatum, which conferred resistance to bacterial spot in sweet pepper. However, resistance in local chili cultivars to anthracnose disease has not been commercialized in Korea [30]. The development of resistant cultivars against chili anthracnose should therefore be emphasized, as this would provide the easiest, safest, cheapest, and most effective method to control chili anthracnose disease [1].
C. scovillei causing chili anthracnose disease was previously reported in Japan, Brazil, and China [31,32,33]. However, to our best knowledge, this is the first report in Korea showing that C. scovillei causes anthracnose disease on pepper fruit. This study updates characterization of C. scovillei. Additional studies with different inoculation test with different cultivars can be conducted as an extension of this finding.
ACKNOWLEDGEMENTS
This work was supported by the Next-Generation BioGreen21 Program (Project No. PJ01118702) of Rural Development Administration, Republic of Korea.
References
- 1.Oo MM, Oh SK. Chilli anthracnose (Colletotrichum spp.) disease and its management approach. Korean J Agric Sci. 2016;43:153–162. [Google Scholar]
- 2.Saxena A, Raghuwanshi R, Gupta VK, Singh HB. Chilli anthracnose: the epidemiology and management. Front Microbiol. 2016;7:1527. doi: 10.3389/fmicb.2016.01527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.MacNeish RS. Ancient mesoamerican civilization. Science. 1964;143:531–537. doi: 10.1126/science.143.3606.531. [DOI] [PubMed] [Google Scholar]
- 4.Park JB. Red pepper and Kimchi in Korea. Chile Pepper Inst Newsl. 1999;8:1–8. [Google Scholar]
- 5.Than PP, Jeewon R, Hyde KD, Pongsupasamit S, Mongkolporn O, Taylor PW. Characterization and pathogenicity of Colletotrichum species associated with anthracnose on chilli (Capsicum spp.) in Thailand. Plant Pathol. 2008;57:562–572. [Google Scholar]
- 6.Osuna-García JA, Wall MM, Waddell CA. Endogenous levels of tocopherols and ascorbic acid during fruit ripening of New Mexican-type chile (Capsicum annuum L.) cultivars. J Agric Food Chem. 1998;46:5093–5096. [Google Scholar]
- 7.Welbaum GE. Vegetable production and practices. Wallingford: CAB International Publisher; 2015. pp. 189–204. [Google Scholar]
- 8.Statistics Korea. Production of chili pepper and sesame in 2016 [Internet] Daejeon: Statistics Korea; 2017. [cited 2017 Sep 1]. Available from: http://kostat.go.kr/portal/eng/pressReleases. [Google Scholar]
- 9.Kim JT, Park SY, Choi W, Lee YH, Kim HT. Characterization of Colletotrichum isolates causing anthracnose of pepper in Korea. Plant Pathol J. 2008;24:17–23. [Google Scholar]
- 10.Roberts PD, Pernezny KL, Kucharek TA Anthracnose caused by Colletotrichum sp. on pepper [Internet] Gainesville (FL): University of Florida; 2001. [cited 2017 Sep 1]. Available from: http://edis.ifas.ufl.edu. [Google Scholar]
- 11.Park KS, Kim CH. Identification, distribution and etiological characteristics of anthracnose fungi of red pepper in Korea. Korean J Plant Pathol. 1992;8:61–69. [Google Scholar]
- 12.Guerber JC, Liu B, Correll JC, Johnston PR. Characterization of diversity in Colletotrichum acutatum sensu lato by sequence analysis of two gene introns, mtDNA and intron RFLPs, and mating compatibility. Mycologia. 2003;95:872–895. [PubMed] [Google Scholar]
- 13.Sreenivasaprasad S, Talhinhas P. Genotypic and phenotypic diversity in Colletotrichum acutatum, a cosmopolitan pathogen causing anthracnose on a wide range of hosts. Mol Plant Pathol. 2005;6:361–378. doi: 10.1111/j.1364-3703.2005.00291.x. [DOI] [PubMed] [Google Scholar]
- 14.Vinnere O, Fatehi J, Wright SA, Gerhardson B. The causal agent of anthracnose of Rhododendron in Sweden and Latvia. Mycol Res. 2002;106:60–69. [Google Scholar]
- 15.Nam MH, Park MS, Lee HD, Yu SH. Taxonomic re-evaluation of Colletotrichum gloeosporioides isolated from strawberry in Korea. Plant Pathol J. 2002;29:317–322. doi: 10.5423/PPJ.NT.12.2012.0188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Prihastuti H, Cai L, Chen H, McKenzie EH, Hyde KD. Characterization of Colletotrichum species associated with coffee berries in northern Thailand. Fungal Divers. 2009;39:89–109. [Google Scholar]
- 17.Cappuccino JG, Sherman N. Microbiology: a laboratory manual. Redwood City (CA): Benjamin-Cummings Publishing Company; 2001. pp. 211–223. [Google Scholar]
- 18.Johnston PR, Jones D. Relationships among Colletotrichum isolates from fruit-rots assessed using rDNA sequences. Mycologia. 1997;89:420–430. [Google Scholar]
- 19.Cenis JL. Rapid extraction of fungal DNA for PCR amplification. Nucleic Acids Res. 1992;20:2380. doi: 10.1093/nar/20.9.2380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Templeton MD, Rikkerink EH, Solon SL, Crowhurst RN. Cloning and molecular characterization of the glyceraldehyde-3-phosphate dehydrogenase-encoding gene and cDNA from the plant pathogenic fungus Glomerella cingulata. Gene. 1992;122:225–230. doi: 10.1016/0378-1119(92)90055-t. [DOI] [PubMed] [Google Scholar]
- 21.O'Donnell K, Cigelnik E. Two divergent intragenomic rDNA ITS2 types within a monophyletic lineage of the fungus Fusarium are nonorthologous. Mol Phylogenet Evol. 1997;7:103–116. doi: 10.1006/mpev.1996.0376. [DOI] [PubMed] [Google Scholar]
- 22.Glass NL, Donaldson GC. Development of primer sets designed for use with the PCR to amplify conserved genes from filamentous ascomycetes. Appl Environ Microbiol. 1995;61:1323–1330. doi: 10.1128/aem.61.4.1323-1330.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.White TJ, Bruns T, Lee S, Taylor J. Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics. In: Innis MA, Gelfand DH, Sninsky JJ, White TJ, editors. PCR protocols: a guide to methods and applications. New York: Academic Press; 1990. pp. 315–322. [Google Scholar]
- 24.Lin Q, Kanchana-udomkan C, Jaunet T, Mongkolporn O. Genetic analysis of resistance to pepper anthracnose caused by Colletotrichum capsici. Thai J Agric Sci. 2002;35:259–264. [Google Scholar]
- 25.Montri P, Taylor PW, Mongkolporn O. Pathotypes of Colletotrichum capsici, the causal agent of chili anthracnose, in Thailand. Plant Dis. 2009;93:17–20. doi: 10.1094/PDIS-93-1-0017. [DOI] [PubMed] [Google Scholar]
- 26.Damm U, Cannon PF, Crous PW. Colletotrichum: complex species or species complexes? Stud Mycol. 2012;73:1–213. doi: 10.3114/sim0010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Cai L, Hyde KD, Taylor PW, Weir BS, Waller JM, Abang MM, Zhang JZ, Yang YL, Phoulivong S, Liu ZY, et al. A polyphasic approach for studying Colletotrichum. Fungal Divers. 2009;39:183–204. [Google Scholar]
- 28.Hyde KD, Cai L, Cannon PF, Crouch JA, Crous PW, Damm U, Goodwin PH, Chen H, Johnston PR, Jones EB, et al. Colletotrichum: names in current use. Fungal Divers. 2009;39:147–182. [Google Scholar]
- 29.Potnis N, Minsavage G, Smith JK, Hurlbert JC, Norman D, Rodrigues R, Stall RE, Jones JB. Avirulence proteins AvrBs7 from Xanthomonas gardneri and AvrBs1.1 from Xanthomonas euvesicatoria contribute to a novel gene-for-gene interaction in pepper. Mol Plant Microbe Interact. 2012;25:307–320. doi: 10.1094/MPMI-08-11-0205. [DOI] [PubMed] [Google Scholar]
- 30.Park HG. Problem of anthracnose in pepper and prospects for its management. In: Oh DG, Kim KT, editors. Abstracts of the first international symposium on chilli anthracnose. Suwon: National Horticultural Research Institute; Rural Development of Administration; 2007. p. 19. [Google Scholar]
- 31.Kanto T, Uematsu S, Tsukamoto T, Moriwaki J, Yamagishi N, Usami T, Sato T. Anthracnose of sweet pepper caused by Colletotrichum scovillei in Japan. J Gen Plant Pathol. 2014;80:73–78. [Google Scholar]
- 32.Caires NP, Pinho DB, Souza JS, Silva MA, Lisboa DO, Pereira OL, Furtado GQ. First report of anthracnose on pepper fruit caused by Colletotrichum scovillei in Brazil. Plant Dis. 2014;98:1437. doi: 10.1094/PDIS-04-14-0426-PDN. [DOI] [PubMed] [Google Scholar]
- 33.Zhao W, Wang T, Chen QQ, Chi YK, Swe TM, Qi RD. First report of Colletotrichum scovillei causing anthracnose fruit rot on pepper in Anhui Province, China. Plant Dis. 2016;100:2168. [Google Scholar]