Abstract
In our previous study, three bacterial strains, Bacillus megaterium KU143, Microbacterium testaceum KU313, and Pseudomonas protegens AS15, were selected as effective biocontrol agents against Aspergillus flavus on stored rice grains. In this study, we evaluated the inhibitory effects of the volatiles produced by the strains on A. flavus growth and aflatoxin production on stored rice grains. The three strains significantly reduced mycelial growth of A. flavus in dual-culture assays compared with the negative control strain, Sphingomonas aquatilis KU408, and an untreated control. Of these tested strains, volatiles produced by B. megaterium KU143 and P. protegens AS15 markedly inhibited mycelial growth, sporulation, and conidial germination of A. flavus on agar medium and suppressed the fungal populations in rice grains. Moreover, volatiles produced by these two strains significantly reduced aflatoxin production in the rice grains by A. flavus. To our knowledge, this is the first report of the suppression of A. flavus aflatoxin production in rice grains using B. megaterium and P. protegens volatiles.
Keywords: Aflatoxin, Aspergillus flavus, Bacillus megaterium, Bacterial volatiles, Pseudomonas protegens
Rice (Oryza sativa L.) is one of the most important food crops worldwide. Stored rice grains are often subjected to contamination by a wide range of fungi, causing significant economic losses and poor grain quality [1]. Fungi belonging to the genera Aspergillus and Penicillium are the most detected in stored rice grains [2,3]. Of these fungi, Aspergillus flavus is one of the most significant, since it is prevalent on stored food products and produces the harmful aflatoxins. It usually exists as a saprophyte in the soil, where it is associated with decaying organic matters [4]. The xerophilic nature of A. flavus allows it to tolerate the dry conditions found in storage facilities and to dominate the fungal populations found on rice grains [5,6].
Aflatoxins are a group of secondary metabolites, primarily produced in nature by a few species of the genus Aspergillus, and are considered one of the most dangerous mycotoxins, based on their high toxicity to humans and animals that have directly or indirectly consumed contaminated food [7]. Geographical areas with high level of dietary intake of aflatoxins have increased levels of liver disease and cancer [8]. Consumption of food highly contaminated with aflatoxins has caused several outbreaks of aflatoxicosis, including those in western India and Kenya where several hundred individuals were affected and many lost their lives [9,10]. Hence, management of levels of aflatoxins in stored rice grains is critically important.
One of the most effective methods for controlling aflatoxin levels on stored rice grains is to avoid contamination by eliminating or reducing A. flavus populations [3]. Several chemical and physical methods have been applied for the control of A. flavus and aflatoxins on stored grains. A number of fungicides, including benomyl, iprodione, and thiabendazole, are used for controlling fungi on grains in storage [11,12]; however, because of their adverse effects on grain quality and health concerns associated with the residues from chemical application, there is an urgent need to identify alternative, non-toxic approaches to control fungal and aflatoxin contamination of grains [13]. Biological control is therefore emerging as a promising alternative to chemical control methods, as it offers an effective, environmentally sound approach to the management of fungal and aflatoxin contamination of stored grains. Several biocontrol agents have been tested against A. flavus and aflatoxin production on stored grains, including strains of Bacillus subtilisRhodococcus erythropolis, and Pseudomonas fluorescens; these microorganisms were reported to inhibit the growth and aflatoxin production of A. flavus on grains [14,15]. Further, several volatile compounds from plants were reported to inhibit aflatoxin production by Aspergillus spp. [16,17].
In our previous study, three potential biocontrol bacterial strains (Bacillus megaterium KU143, Microbacterium testaceum KU313 and Pseudomonas protegens AS15) were selected from a total of 460 bacterial strains [18,19], isolated from stored rice grains collected from 11 different rice-processing complexes in Korea [2]. These three selected bacterial strains exhibited biocontrol activity against A. flavus and aflatoxin production on stored rice grains; in particular, P. protegens AS15 could degrade aflatoxin B1 and utilize it in liquid nutrient-deficient media [19]. The objectives of this study was to evaluate the inhibitory effects of the volatiles produced by these antagonistic bacterial strains on A. flavus growth and aflatoxin production on stored rice grains.
To prepare bacterial suspensions, the three bacterial strains, B. megaterium KU143, M. testaceum KU313, and P. protegens AS15, and a negative control strain Sphingomonas aquatilis KU408 [19] were streaked on nutrient agar (NA) and incubated at 28℃ for 2 days; then, single colonies from each strain culture were transferred to nutrient broth and incubated in a rotary shaker (160rpm) in the dark at 28℃ for 2 days. After incubation, bacterial cells were harvested by centrifugation at 5,000×g at for 15min, suspended in a 10-mM MgSO4 solution, and washed twice with the same solution by centrifugation. Bacterial cells were adjusted to 108 cells/mL (OD600 =0.5) in 10-mM MgSO4 solution using a spectrophotometer. In addition, A. flavus AF57 isolated from stored rice grains [2] and A. flavus KCCM 60330 (an aflatoxigenic strain), obtained from the Korean Culture Center of Microorganisms (KCCM; Seoul, Korea) were used in this study. The latter strain was used for assessment of aflatoxin production. Conidial suspensions were prepared by culturing fungal isolates on potato dextrose agar (PDA) for 5 days at 28℃. Conidia were harvested in sterile distilled water (SDW) containing 0.03% Tween 20 and adjusted to 107conidia/mL using a hemocytometer. Stored unhulled rice grains (cv. Ilpum) from Korea University Farm (Namyangju, Korea) were used in this study and surface-sterilized using 1% sodium hypochlorite for 3min, followed by 70% ethanol for 5min [20]; then, washed three times in SDW and blotted dry on filter papers (Whatman No. 1) before fungal inoculation.
To test the antifungal activities of the bacterial strains against A. flavus AF57, a dual-culture assay was conducted on PDA as previously described [19]. Briefly, bacterial strains or SDW (control) were line-streaked in the center of PDA plates, which were inoculated on the opposite edges with 2 µL of the prepared conidial suspensions. When the mycelia of A. flavus AF57 in the control plates almost reached the streaked lines, mycelial inhibition (mm) was measured; at the same time, mycelial growth area (cm2) was assessed using ImageJ software after photography [21].
To examine the antifungal effects of the volatiles produced by the three bacterial strains on mycelial growth, sporulation, and conidial germination of A. flavus AF57, in vitro tests were conducted using I-plates (Petri plates 90mm in diameter, separated in the middle with a partition that allowed only volatiles to pass from one side to the other, without any direct contact between the cultured organisms on either side) (Fisher Scientific, Pittsburgh, PA, USA). For the in vitro tests, 100 µL of bacterial suspensions of the test and negative control strains and 10-mM MgSO4 solution (untreated control) were smeared on the NA side of the I-plates and incubated at 28℃. After 24hr of incubation, 2 µL of the conidial suspension (107 conidia/mL) of A. flavus AF57 were drop-inoculated onto the opposite side of the I-plates, which contained PDA. Plates were then sealed with Parafilm M (Sigma-Aldrich, St. Louis, MO, USA) and incubated in the dark at 28℃. When mycelia almost reached the center of the control plates, the mycelial growth area (cm2) of A. flavus AF57 on the PDA side of the I-plates was measured, as described above. To assess fungal sporulation, fungal conidia in each I-plate were harvested in SDW containing 0.03% Tween 20 and vortexed for 1min. Then, numbers of conidia were determined using a hemocytometer; the total numbers of the conidia harvested from each I-plate were divided by the mycelial growth area (cm2) of the plate. Sporulation was expressed as the log number of conidia per mycelial growth area (cm2) of each sample. For evaluation of A. flavus AF57 conidial germination, conidia from the PDA side of the I-plates were harvested in SDW containing 0.03% Tween 20. The harvested conidia were filtered through two layers of sterile cheesecloth and transferred to potato dextrose broth at a final concentration of 105 conidia/mL. After stationary incubation of each sample (three sub-replicates) at 28℃ in the dark for 10hr, the numbers of germinated conidia among a total of 100 conidia per sample were determined.
To examine the effect of the volatiles produced by the tested bacterial strains against the population of A. flavus AF57 on unhulled rice grains, in vivo tests were conducted using I-plates as follows. Aliquots (100 µL) of the prepared treatments described above were smeared on I-plates containing NA and incubated at 28℃. After 24hr, surfacesterilized unhulled rice grains (2g) were inoculated with the 200 µL of the A. flavus AF57 conidial suspension (equivalent to 106/g rice grains) and placed on the other side of the I-plates. The plates were then sealed with Parafilm and incubated for 7 days at 28℃. The populations of A. flavus AF57 on rice grains were assessed as previously described [19]. Briefly, rice grains were finely ground using an analytical mill (IKA A11 basic, IKA Works, Wilmington, DE, USA), 1g of grains were suspended in 10mL of SDW, and were incubated in a rotary shaker (160rpm) at 28℃ for 30min. After incubation, 10-fold serial dilutions of rice suspensions were prepared and 200 µL of the diluted suspensions were smeared on 18% glycerol agar (DG18; Fluka 40587, Sigma-Aldrich) [22]. These smeared DG18 plates were incubated for 4 days at 28℃ for assessment of the fungal population. The fungal population was expressed as the log number of colony-forming units/g dry weight of rice grains.
To examine the effects of bacterial volatiles on aflatoxin production, the aflatoxigenic A. flavus strain KCCM 60330 was used in this test rather than A. flavus AF57, which produces low levels of aflatoxins. This experiment was conducted using I-plates as described above. Treatments and assessment of fungal population were conducted as shown above. Total aflatoxins, including aflatoxin B1 and B2 produced by A. flavus KCCM 60330 were analyzed using a competitive direct enzyme-linked immunosorbent assay with Veratox HS (Neogen Co., Lansing, MI, USA), following the procedure described by Mannaa et al. [19].
Statistical analysis of the data was conducted using Statistical Analysis Systems (SAS Institute, Cary, NC, USA). All experiments were performed twice with three replications per treatment. Combined data from repeated experiments were analyzed after confirmation of homogeneity of variances, using Levene's test [23]. For analysis of fungal populations, data were analyzed after logarithmic transformation. Analysis of variance was determined using the general linear model procedure and means were compared using the least significant difference at p<0.05.
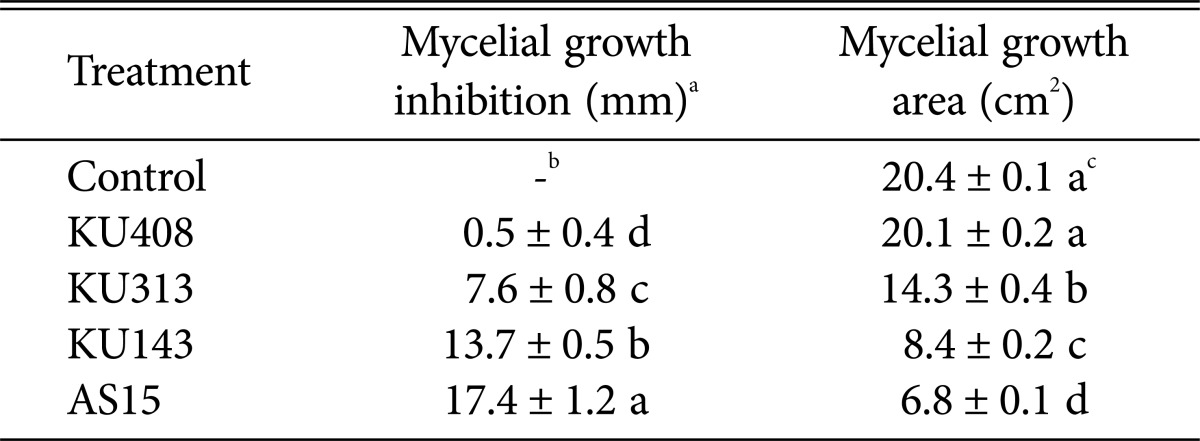
The results of dual-culture assays exhibited that the three tested antagonistic strains, B. megaterium KU143, M. testaceum KU313, and P. protegens AS15, caused significant (p<0.05) reduction in mycelial growth compared with the negative control strain, S. aquatilis KU408, and SDW control plates (Table 1, Fig. 1A). Among the tested bacterial strains, P. protegens AS15 caused the most marked reduction in mycelial growth (Table 1). However, the negative control bacterial strain did not inhibit mycelial growth, producing results similar to those of the SDW control (Table 1).
Table 1. Antifungal activities of the bacterial strains, Microbacterium testaceum KU313, Bacillus megaterium KU143, Pseudomonas protegens AS15, and Sphingomonas aquatilis KU408 (negative control), on mycelial growth of Aspergillus flavus AF57 using dual-culture assays on PDA.
aMycelial growth inhibition (mm) was determined using a dualculture assay on potato dextrose agar (PDA) plates, which were streaked with the bacterial strains or sterile distilled water (control) in the center. The plates were then inoculated with conidial suspension of A. flavus AF57 on the opposite edges from the bacteria. Mycelial inhibition was evaluated when mycelia reached the centers of the control plates and mycelial growth area (cm2) was determined using ImageJ software.
bNot detected.
cValues are presented as the means±standard errors of six replicates from repeated experiments. Different letters in each column indicate significant (p<0.05) differences according to the least significant difference test.
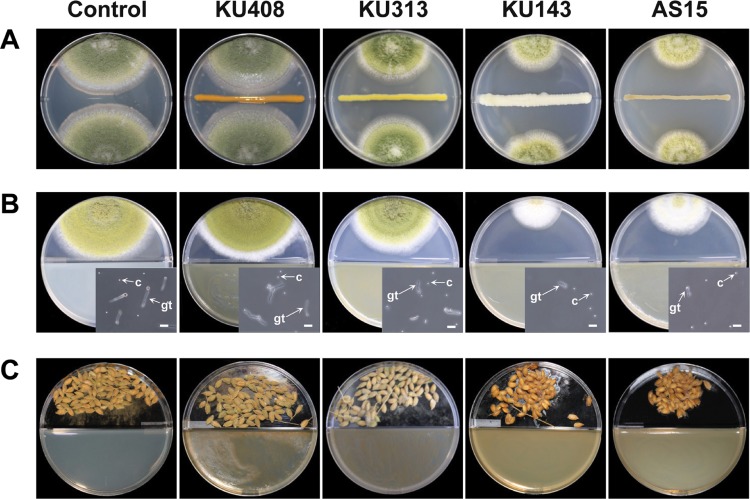
Fig. 1. Effects of the antifungal or volatiles produced by the bacterial strains, Microbacterium testaceum KU313, Bacillus megaterium KU143, Pseudomonas protegens AS15, and Sphingomonas aquatilis KU408 (negative control), on Aspergillus flavus AF57 mycelial growth determined using dual-culture assays on potato dextrose agar (PDA) (A) and PDA on I-plates (B), and unhulled rice grains on I-plates inoculated with A. flavus AF57 (C). In the dual-culture assays, bacterial strains or sterile distilled water (control) were line-streaked in the center of the plates, which were then inoculated with conidial suspension of A. flavus AF57 on the opposite edges from the bacteria. In I-plate tests, bacterial strains or 10-mM MgSO4 solution (untreated control) were smeared on one side (nutrient agar) of the I-plates and the other side (PDA or rice grains) was inoculated with conidial suspension of A. flavus AF57 24hr after the bacterial treatment. Insets of Fig. 1B are photographs of the conidial germination of A. flavus AF57 affected by the bacterial volatiles and untreated control. c, conidia; gt, germ tube (scale bars: B inset=20 µm).
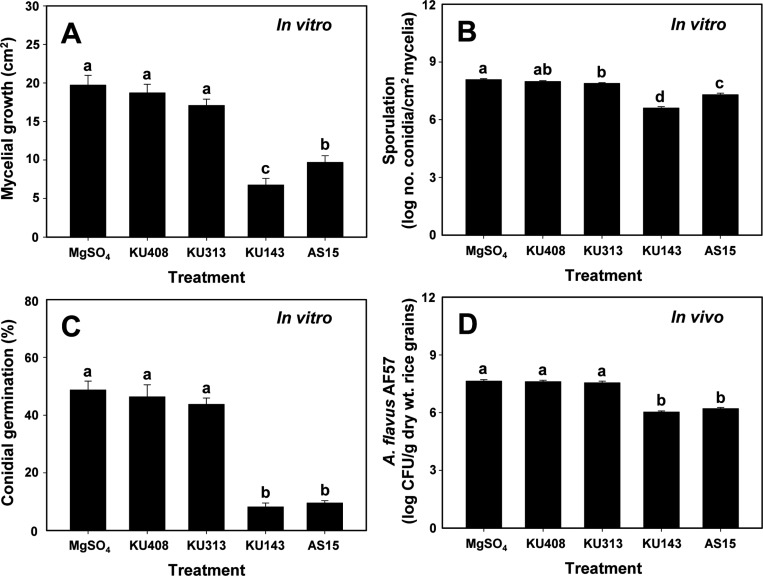
For the effects of bacterial volatiles against mycelial growth, sporulation, and conidial germination of A. flavus AF57 on I-plates (Fig. 1B), volatiles produced by B. megaterium KU143 and P. protegens AS15 significantly (p<0.05) reduced the mycelial growth area compared with S. aquatilis KU408 (bacterial negative control) and MgSO4 solution (untreated control) (Fig. 2A). Volatiles produced by B. megaterium KU143 and P. protegens AS15 significantly (p<0.05) inhibited sporulation of A. flavus AF57 compared with negative and untreated controls (Fig. 2B). In addition, volatiles generated by B. megaterium KU143 and P. protegens AS15 significantly (p<0.05) inhibited conidial germination (%) of A. flavus AF57 compared with negative and untreated controls (Fig. 2C). In contrast, volatiles produced by M. testaceum KU313 did not significantly (p>0.05) inhibit mycelial growth, sporulation, or conidial germination of A. flavus AF57 compared with the bacterial negative control or untreated control (Fig. 2A-C). Testing of the effects of bacterial volatiles on the population of A. flavus AF57 on unhulled rice grains (Fig. 1C) showed that those produced by B. megaterium KU143 and P. protegens AS15 significantly (p<0.05) reduced fungal populations compared with bacterial negative and untreated controls (Fig. 2D). However, volatiles produced by M. testaceum KU313 failed to inhibit the fungal growth in rice grains (Fig. 2D).
Fig. 2. Antifungal activities of the volatiles produced by the bacterial strains, Microbacterium testaceum KU313, Bacillus megaterium KU143, Pseudomonas protegens AS15, and Sphingomonas aquatilis KU408 (bacterial negative control), on mycelial growth (A), sporulation (B), and conidial germination on potato dextrose agar (C), and fungal population on unhulled rice grains inoculated with Aspergillus flavus AF57 (D). Bacterial strains were smeared on one side of the I-plates and the other side was inoculated with conidial suspension of A. flavus AF57 24hr after bacterial treatment. Mycelial growth area (cm2) was measured using ImageJ software after the fungal mycelia reached the center of 10-mM MgSO4 solution-treated plates (untreated control). Sporulation was determined as total harvested spores divided by the mycelial growth area. For conidial germination, the numbers of germinated conidia among a total of 100 conidia were determined 10hr after incubation in potato dextrose broth at 28℃ in the dark. At 7 days after inoculation, fungal populations on rice grains were determined by assessment of colony-forming units (CFU) on DG18 plates, which were smeared with the diluted rice suspensions and incubated for 4 days at 28℃ in the dark. Different letters on vertical bars with error bars (standard errors, n=6) indicate significant (p<0.05) differences between treatments according to the least significant difference test.
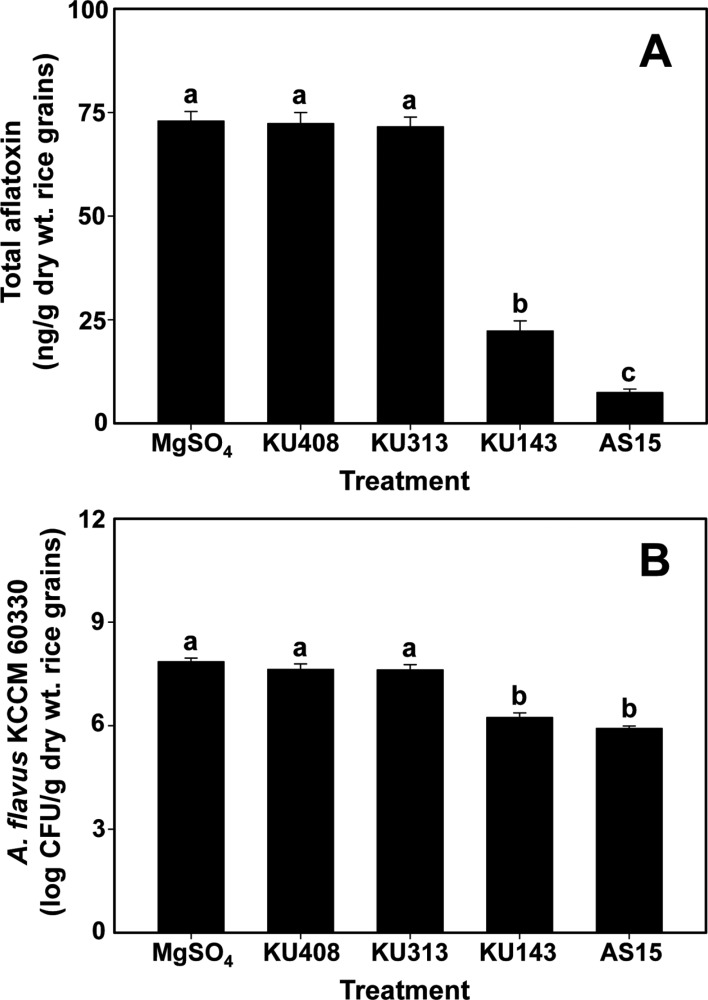
When the effect of bacterial volatiles on aflatoxin production was examined on unhulled rice grains using the aflatoxigenic A. flavus KCCM 60330, volatiles produced by B. megaterium KU143 and P. protegens AS15 significantly (p<0.05) reduced total aflatoxin production compared with the bacterial negative control strain, S. aquatilis KU408, and untreated MgSO4 control (Fig. 3A). In particular, volatiles generated by P. protegens AS15 and B. megaterium KU143 markedly inhibited total aflatoxin production by A. flavus KCCM 60330 to 7.4 and 22.3ng/g dry weight rice grains (percent total aflatoxin reduction relative to untreated control, 90.4% and 69.9%) compared with 72.9ng/g dry weight rice grains for the untreated MgSO4 control, respectively (Fig. 3A). In contrast, volatiles produced by M. testaceum KU313 did not inhibit total aflatoxin production in rice grains (Fig. 3A). In addition, the bacterial volatiles generated by B. megaterium KU143 and P. protegens AS15 significantly (p<0.05) inhibited the fungal growth of A. flavus KCCM 60330 compared with bacterial negative and untreated controls (Fig. 3B), similar to the observations in the experiment using A. flavus AF57 (Fig. 2D).
Fig. 3. Biocontrol activities of the volatiles produced from bacterial strains, Microbacterium testaceum KU313, Bacillus megaterium KU143, and Pseudomonas protegens AS15, and Sphingomonas aquatilis KU408 (bacterial negative control), on total aflatoxin production (A) and the population of the aflatoxigenic Aspergillus flavus KCCM 60330 in stored rice grains (B). In I-plate tests, bacterial strains or 10-mM MgSO4 solution (untreated control) were smeared on one side (nutrient agar) of the I-plates and the other side (rice grains) was inoculated with conidial suspension of A. flavus KCCM 60330 24hr after the bacterial treatment. Total aflatoxins in rice grains were quantified using a Veratox HS ELISA kit. Fungal populations were determined 7 days after inoculation by assessment of colony-forming units (CFU) on DG18 plates smeared with the diluted rice suspensions and incubated for 4 days at 28℃ in the dark. Different letters on the vertical bars with error bars (standard errors, n=6) indicate significant (p<0.05) differences between treatments according to the least significant difference test.
Production of antimicrobial or systemic resistance-inducing volatile compounds by antagonistic bacterial strains is one of the biocontrol mechanisms against plant pathogens [24,25]. Volatiles from antagonistic rhizobacteria were reported to stimulate pepper fruit ripening and to inhibit mycelial growth, sporulation, and spore germination of Colletotrichum acutatum and Phytophthora capsici [25,26]. There were also reports demonstrating the influence of volatiles of plant or microbial origin on the growth of fungal species, including A. flavus [27,28]. In our previous study [19], the antagonistic bacterial strains, B. megaterium KU143, M. testaceum KU313, and P. protegens AS15, were selected as effective biocontrol agents against A. flavus. The results of the current study demonstrate that, among these strains, B. megaterium KU143 and P. protegens AS15 produce antifungal volatile organic compounds (VOCs), which can inhibit the mycelial growth, sporulation, and conidial germination of A. flavus AF57 on artificial medium and fungal populations on unhulled rice grains inoculated with A. flavus AF57 (rice-originated isolate) or KCCM 60330 (aflatoxigenic isolate). In contrast, our results do not provide evidence that M. testaceum KU313 can produce antifungal volatiles, although it was found to be an effective antagonistic strain against A. flavus on rice grains [19]. The biocontrol activity of M. testaceum KU313 may be attributable to the production of non-volatile antifungal compounds, as indicated by the results of the dual-culture assay in this study and evaluation of effective colonization ability on rice grains in our previous study [19]. Further, in our previous study, the cell-free culture filtrates from these bacterial strains were proven to inhibit conidial germination, germ-tube lengths, and mycelial dry weight of A. flavus, indicating that these strains can produce extracellular antifungal compounds [19].
Since aflatoxins were discovered as early as 1960, various chemical compounds, natural products, and microbial extracts have been screened as potential inhibitors of aflatoxin biosynthesis [29]. Extensive research efforts have been directed toward selection of microorganisms with aflatoxin inhibition ability in co-cultivation with aflatoxigenic Aspergillus spp. [30]. Previously, specific plant volatiles, including alcohols, aldehydes, and ketones have been reported to inhibit fungal growth and aflatoxin production by A. flavus [16,17,31,32]. Roze et al. [32] demonstrated that volatiles, including formic acid pentyl ester, hexanoic acid ethyl ester, pentanoic acid methyl ester, 3,5-octadien-2-one, and 2-pentenal from willow bark inhibited aflatoxin production in Aspergillus parasiticus grown on a minimal medium; aflatoxin inhibition was correlated with a two-fold reduction in expression of the ver-1 gene, which encodes an enzyme to the aflatoxin biosynthesis pathway. In another study, soybean volatile compounds, including aldehydes, (E)-2-hexenal, and (E)-2-heptenal effectively inhibited A. flavus growth and aflatoxin production [31]. The biocontrol yeast, Pichia anomala, was reported to produce 2-phenylethanol, which inhibited aflatoxin production by down regulation of expression of aflatoxin biosynthesis genes in A. flavus [33]. In this study, volatiles produced by B. megaterium KU143 and P. protegens AS15 significantly reduced aflatoxin production by A. flavus KCCM 60330 in rice grains. Considering the suppression of A. flavus growth and toxin production by these bacterial volatiles, P. protegens AS15 inhibited aflatoxin production more than fungal population, indicating the possible degradation of the aflatoxin or suppression of toxin production by A. flavus. However, reduction of the aflatoxins by the treatment with B. megaterium KU143 may result from the inhibitory effect on the fungal growth. Further study to identify the VOCs responsible for the suppression of aflatoxin production will be needed to facilitate understanding of the related processes and to develop an effective and applicable method to control aflatoxins in stored crop grains.
Taken together, the results of the present study suggest that the antagonistic bacterial strains, B. megaterium KU143 and P. protegens AS15, produce effective antifungal volatile compounds that inhibit A. flavus growth on artificial medium and unhulled rice grains. Furthermore, the volatiles from these two bacterial strains reduced aflatoxin production by A. flavus in stored rice grains. To our knowledge, this is the first report of the inhibition of A. flavus aflatoxin production in rice grains using the volatiles produced by B. megaterium and P. protegens.
ACKNOWLEDGEMENTS
This work was supported by a Korea University Grant. We thank anonymous reviewers for valuable comments on this manuscript.
References
- 1.Shotwell OL, Hesseltine CW, Stubblefield RD, Sorenson WG. Production of aflatoxin on rice. Appl Microbiol. 1966;14:425–428. doi: 10.1128/am.14.3.425-428.1966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Oh JY, Jee SN, Nam Y, Lee H, Ryoo MI, Kim KD. Populations of fungi and bacteria associated with samples of stored rice in Korea. Mycobiology. 2007;35:36–38. doi: 10.4489/MYCO.2007.35.1.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Mannaa M, Kim KD. Microbe-mediated control of mycotoxigenic grain fungi in stored rice with focus on aflatoxin biodegradation and biosynthesis inhibition. Mycobiology. 2016;44:67–78. doi: 10.5941/MYCO.2016.44.2.67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Dvořáčková I. Aflatoxins and human health. Boca Raton (FL): CRC Press; 1989. [Google Scholar]
- 5.Park JW, Choi SY, Hwang HJ, Kim YB. Fungal mycoflora and mycotoxins in Korean polished rice destined for humans. Int J Food Microbiol. 2005;103:305–314. doi: 10.1016/j.ijfoodmicro.2005.02.001. [DOI] [PubMed] [Google Scholar]
- 6.Klich MA. Environmental and developmental factors influencing aflatoxin production by Aspergillus flavus and Aspergillus parasiticus. Mycoscience. 2007;48:71–80. [Google Scholar]
- 7.Squire RA. Ranking animal carcinogens: a proposed regulatory approach. Science. 1981;214:877–880. doi: 10.1126/science.7302565. [DOI] [PubMed] [Google Scholar]
- 8.Yeh FS, Yu MC, Mo CC, Luo S, Tong MJ, Henderson BE. Hepatitis B virus, aflatoxins, and hepatocellular carcinoma in southern Guangxi, China. Cancer Res. 1989;49:2506–2509. [PubMed] [Google Scholar]
- 9.Azziz-Baumgartner E, Lindblade K, Gieseker K, Rogers HS, Kieszak S, Njapau H, Schleicher R, McCoy LF, Misore A, DeCock K, et al. Case-control study of an acute aflatoxicosis outbreak, Kenya, 2004. Environ Health Perspect. 2005;113:1779–1783. doi: 10.1289/ehp.8384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Krishnamachari KA, Nagarajan V, Bhar R, Tilak TBG. Hepatitis due to aflatoxicosis: an outbreak in western India. Lancet. 1975;1:1061–1063. doi: 10.1016/s0140-6736(75)91829-2. [DOI] [PubMed] [Google Scholar]
- 11.White DG, Toman J, Burnette DC, Jacobsen BJ. The effect of postharvest fungicide application on storage fungi of corn during ambient air drying and storage. Plant Dis. 1993;77:562–568. [Google Scholar]
- 12.White DG, Toman J., Jr Effects of postharvest oil and fungicide application on storage fungi in corn following high-temperature grain drying. Plant Dis. 1994;78:38–43. [Google Scholar]
- 13.Bluma R, Amaiden MR, Daghero J, Etcheverry M. Control of Aspergillus section Flavi growth and aflatoxin accumulation by plant essential oils. J Appl Microbiol. 2008;105:203–214. doi: 10.1111/j.1365-2672.2008.03741.x. [DOI] [PubMed] [Google Scholar]
- 14.Kimura N, Hirano S. Inhibitory strains of Bacillus subtilis for growth and aflatoxin-production of aflatoxigenic fungi. Agric Biol Chem. 1988;52:1173–1179. [Google Scholar]
- 15.Reddy KR, Reddy CS, Muralidharan K. Potential of botanicals and biocontrol agents on growth and aflatoxin production by Aspergillus flavus infecting rice grains. Food Control. 2009;20:173–178. [Google Scholar]
- 16.Zeringue HJ, Jr, McCormick SP. Aflatoxin production in cultures of Aspergillus flavus incubated in atmospheres containing selected cotton leaf-derived volatiles. Toxicon. 1990;28:445–448. doi: 10.1016/0041-0101(90)90083-j. [DOI] [PubMed] [Google Scholar]
- 17.Wright MS, Greene-McDowelle DM, Zeringue HJ, Bhatnagar D, Cleveland TE. Effects of volatile aldehydes from Aspergillus-resistant varieties of corn on Aspergillus parasiticus growth and aflatoxin biosynthesis. Toxicon. 2000;38:1215–1223. doi: 10.1016/s0041-0101(99)00221-4. [DOI] [PubMed] [Google Scholar]
- 18.Lee SY, Oh JY, Ryoo MI, Kim KD. Biological control of the rice storage fungi Aspergillus and Penicillium species by antagonistic bacteria originated from rice. Plant Pathol J. 2007;23:328. [Google Scholar]
- 19.Mannaa M, Oh JY, Kim KD. Microbe-mediated control of Aspergillus flavus in stored rice grains with a focus on aflatoxin inhibition and biodegradation. Ann Appl Biol. 2017 Aug 14; doi: 10.1111/aab.12381. [Epub] [DOI] [Google Scholar]
- 20.Gu Q, Han N, Liu J, Zhu M. Expression of Helicobacter pylori urease subunit B gene in transgenic rice. Biotechnol Lett. 2006;28:1661–1666. doi: 10.1007/s10529-006-9141-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Schneider CA, Rasband WS, Eliceiri KW. NIH Image to ImageJ: 25 years of image analysis. Nat Methods. 2012;9:671–675. doi: 10.1038/nmeth.2089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hocking AD, Pitt JI. Dichloran-glycerol medium for enumeration of xerophilic fungi from low-moisture foods. Appl Environ Microbiol. 1980;39:488–492. doi: 10.1128/aem.39.3.488-492.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Levene H. Robust tests for equality of variances. In: Olkin I, Ghurye SG, Hoeffding W, Madow WG, Mann HB, editors. Contributions to probability and statistics: essays in honor of Harold Hotelling. Stanford (CA): Stanford University Press; 1960. pp. 278–292. [Google Scholar]
- 24.Ryu CM, Farag MA, Hu CH, Reddy MS, Kloepper JW, Paré PW. Bacterial volatiles induce systemic resistance in Arabidopsis. Plant Physiol. 2004;134:1017–1026. doi: 10.1104/pp.103.026583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sang MK, Kim JD, Kim BS, Kim KD. Root treatment with rhizobacteria antagonistic to Phytophthora blight affects anthracnose occurrence, ripening, and yield of pepper fruit in the plastic house and field. Phytopathology. 2011;101:666–678. doi: 10.1094/PHYTO-08-10-0224. [DOI] [PubMed] [Google Scholar]
- 26.Sang MK, Kim KD. The volatile-producing Flavobacterium johnsoniae strain GSE09 shows biocontrol activity against Phytophthora capsici in pepper. J Appl Microbiol. 2012;113:383–398. doi: 10.1111/j.1365-2672.2012.05330.x. [DOI] [PubMed] [Google Scholar]
- 27.Gueldner RC, Wilson DM, Heidt AR. Volatile compounds inhibiting Aspergillus flavus. J Agric Food Chem. 1985;33:411–413. [Google Scholar]
- 28.Utama IM, Wills RB, Ben-Yehoshua S, Kuek C. In vitro efficacy of plant volatiles for inhibiting the growth of fruit and vegetable decay microorganisms. J Agric Food Chem. 2002;50:6371–6377. doi: 10.1021/jf020484d. [DOI] [PubMed] [Google Scholar]
- 29.Holmes RA, Boston RS, Payne GA. Diverse inhibitors of aflatoxin biosynthesis. Appl Microbiol Biotechnol. 2008;78:559–572. doi: 10.1007/s00253-008-1362-0. [DOI] [PubMed] [Google Scholar]
- 30.Mishra HN, Das C. A review on biological control and metabolism of aflatoxin. Crit Rev Food Sci Nutr. 2003;43:245–264. doi: 10.1080/10408690390826518. [DOI] [PubMed] [Google Scholar]
- 31.Cleveland TE, Carter-Wientjes CH, De Lucca AJ, Boué SM. Effect of soybean volatile compounds on Aspergillus flavus growth and aflatoxin production. J Food Sci. 2009;74:H83–H87. doi: 10.1111/j.1750-3841.2009.01078.x. [DOI] [PubMed] [Google Scholar]
- 32.Roze LV, Koptina AV, Laivenieks M, Beaudry RM, Jones DA, Kanarsky AV, Linz JE. Willow volatiles influence growth, development, and secondary metabolism in Aspergillus parasiticus. Appl Microbiol Biotechnol. 2011;92:359–370. doi: 10.1007/s00253-011-3339-7. [DOI] [PubMed] [Google Scholar]
- 33.Hua SS, Beck JJ, Sarreal SB, Gee W. The major volatile compound 2-phenylethanol from the biocontrol yeast, Pichia anomala, inhibits growth and expression of aflatoxin biosynthetic genes of Aspergillus flavus. Mycotoxin Res. 2014;30:71–78. doi: 10.1007/s12550-014-0189-z. [DOI] [PubMed] [Google Scholar]