Abstract
BACKGROUND
Red blood cell (RBC) alloimmunization occurs at a high frequency in sickle cell anemia (SCA) despite serologic matching for Rh (C/c, E/e) and K antigens. RBC minor antigen genotyping allows for prediction of antigens and RH variants that may lead to alloimmunization.
STUDY DESIGN AND METHODS
RBC antigen genotyping was performed on chronically transfused pediatric SCA patients, using PreciseType Human Erythrocyte Antigen (HEA), RHCE, and RHD BeadChip arrays. All patients received C/c, E/e, and K serologically matched units (category 1); patients with prior RBC antibodies were also matched for Fya, Jkb, and any antibodies (category 2). The RBC genotypes of all leukoreduced (LR) units transfused over a 12-month period were determined by the prototype HEA-LR BeadChip assay.
RESULTS
There were 2320 RBC units transfused to 90 patients in 1135 transfusion episodes. Thirty-five (38.9%) patients had homozygous or compound heterozygous RH variants. Seven new alloantibodies were detected, with alloantibody incidence of 0.706/100 units for category 2 transfusions and 0.068/100 units for category 1 (p=0.02). Three patients on category 2 transfusions formed new anti-Jsa and had a higher rate of exposure to Jsa than those who did not form anti-Jsa (20.4 vs. 8.33 exposures/100 units, p=0.02). The most frequent mismatches were S (43.9%), Doa (43.9%), Fya (29.2%), M (28.4%), Jkb (28.1%).
CONCLUSIONS
Alloimmunization incidence was higher in those with prior RBC antibodies, suggesting that past immunologic responders are at higher risk for future alloimmunization and therefore may benefit from more extensive antigen matching beyond C/c, E/e, K, Fya and Jkb.
Keywords: sickle cell disease, alloimmunization, red blood cell genotyping
INTRODUCTION
Chronic transfusion therapy (CTT) is essential for the prevention of severe complications of sickle cell anemia (SCA), particularly stroke prophylaxis. Alloimmunization to red blood cell (RBC) antigens frequently complicates transfusion therapy in SCA, with a prevalence of up to 45%.1–4 Although limited matching of C/c, E/e, and K antigens for all SCA patients has reduced this rate from 27–75% without limited matching to 5–14% with limited matching,1,2,5–11 alloimmunization remains a prevalent and clinically significant problem in SCA.12,13 Alloimmunization may lead to challenges in identifying appropriate antigen-negative blood for patients. Undetected alloantibodies present a risk for patients to receive antigen-positive blood with resultant hemolytic transfusion reactions.14,15 Lastly, patients with a history of one or more alloantibodies are at increased risk of forming new RBC antibodies with subsequent transfusion exposures.16
Extended RBC antigen typing of SCA patients is necessary to provide antigen-matched RBC transfusions and to guide the identification of new RBC antibodies. RBC antigen genotyping allows for DNA-based prediction of minor RBC antigen expression based on single nucleotide polymorphisms (SNPs).17 Additionally, Rh antigen variants are common in SCD and individuals of African descent.4 However altered or partial Rh antigens are not routinely detected by serologic RBC phenotyping, so RHCE and RHD genotyping provides the primary means for detection of potential Rh antigen mismatches that may result in alloimmunization. RBC genotyping offers several advantages over traditional hemagglutination-based antigen typing; these include the ability to type minor antigens accurately in patients for whom anti-sera are not available (such as Jsa/b, Kpa/b, and V/VS), and the ability to accurately type antigens in patients with recent RBC transfusions or patients with RBC allo- or autoantibodies that may interfere with serologic phenotype matching.18
While the incidence of alloantibody formation has been reported in SCA,19 there is little data regarding the frequency of RBC minor antigen mismatches in transfusion therapy for SCA, nor the frequency of exposure to mismatched antigens prior to new RBC alloantibody formation. The main purpose of this study was to determine the frequency of RBC minor antigen mismatches that occur during CTT for SCA patients following standard protocols for limited serologic antigen matching. Additionally, we sought to identify both the incidence of new alloantibody formation as well as identify the frequency of antigen exposures prior to the antibody formation, during CTT.
MATERIALS AND METHODS
A prospective observational study of children ages 3 – 20 years with SCA (HbSS or HbSβ0 thalassemia genotypes) on CTT for at least the past 6 months was conducted at Children’s Healthcare of Atlanta (CHOA) which has 3 hospital-based pediatric hematology infusion centers and blood banks, and at Children’s National Medical Center (CNMC) which has 1 infusion center and blood bank. Written, informed consent and assent were obtained, and this study was approved by the Institutional Review Boards of CHOA and CNMC. Eligible participants received simple transfusions or partial manual exchange (PME) transfusions, receiving 1 – 3 units per transfusion according to institutional weight-based dosing of transfusion volume. Patients receiving chronic exchange transfusions were excluded, as a chronic exchange therapy presents more RBC unit exposures per transfusion episode than simple or PME transfusions do. All transfusion episodes (defined as a single event in which a patient received a prescribed volume of RBC transfusion) over a 12 to 17 month period were recorded, including pre-transfusion antibody screens, RBC unit preservative solution and age of the RBC unit (time from donor collection to transfusion). Segments from the RBC units were collected from all units for donor RBC minor antigen genotyping.
Electronic medical records and the blood bank Laboratory Information Systems of CHOA and CNMC were reviewed to identify patients’ RBC antibody histories including antibody specificities and dates of initial detection at either the current or previous institutions. At CHOA and CNMC, all RBC units for SCD patients are HbS negative and are serologically matched for C/c, E/e, and K antigens (category 1 matching), as consistent with recommendations of the NHLBI Evidence-Based Management of Sickle Cell Disease.20 For SCD patients with ≥1 alloantibody or in cases of persistent detection of warm autoantibodies, RBC units are antigen-negative for the significant antibodies and have extended serologic matching (category 2 matching) for Fya and Jkb (CHOA) or for Fya/b and Jkb antigens (CNMC). All RBC units transfused at CHOA and CNMC are leukoreduced (LR) prior to storage.
All new antibodies identified during the study period were recorded, and previous transfusions during the study period were reviewed to determine the number and frequency of antigen exposures (as determined by HEA-LR) prior to the antibody detection. The incidence of alloantibody formation was calculated for patients on category 1 matched transfusions and category 2 transfusions.
RBC Genotyping (HEA) Methods
RBC minor antigen genotyping of the patients was performed using PreciseType Human Erythrocyte Antigen (HEA) Molecular BeadChip assay (Immucor, Norcross, GA) to identify single nucleotide polymorphisms (SNPs) associated with 35 antigens in 11 blood group systems (Rh, Kell, Kidd, Duffy, MNS, Dombrock, Lutheran, Landsteiner-Wiener, Diego, Colton, and Scianna) and the RHCE, and RHD BeadChip assays to detect >35 RHCE variants and >80 RHD variants (Immucor, Norcross, GA). For each assay, genomic DNA was extracted from whole blood per manufacturer instructions as previously described, for polymerase chain reaction (PCR)-based detection of individual polymorphisms.21–23
For donor unit genotyping, genomic DNA was extracted from 2 – 4 segments of LR RBC units by using the InviGenius Automated DNA Extractor and the InviMag Blood DNA rWBC Kit (Stratec Molecular GmbH, Berlin, Germany). All extracted genomic DNAs were subjected to PCR amplification and analyzed using the prototype HEA -LR BeadChip assay (Immucor, Norcross, GA) which detects the same profile of RBC antigens as the PreciseType HEA assay, as previously reported.24 RH variant testing of LR units was not performed since this testing required a larger quantity of DNA than could be extracted from LR unit segments; therefore units were assumed to express conventional RH haplotypes. However since both the PreciseType and HEA-LR assays are able to detect the 733C>G (RhCE-L245V) and 1006G>T (RhCE-G336C) single nucleotide polymorphisms (SNPs), the frequency of partial e genotypes based on these SNPs was determined for the donor units.
HEA-LR genotype results were compared to available serologic phenotype records for each unit, and in cases of discrepancies, the HEA-LR assay was repeated in duplicate to verify or amend results. In cases where unit genotypes were known from previous donor testing by the blood donor center(s) using the HEA assay, the units were not tested by HEA-LR BeadChip.
HEA and RH Antigen Mismatch Definitions
Each donor unit genotype was compared to the recipient’s genotype to identify antigen mismatches. An antigen mismatch was defined as a recipient exposure to a RBC minor antigen that the recipient does not express. In cases where the HEA-LR result for an antigen was an indeterminate call (IC) or had low signal (LS) intensity, there was assumed to be no mismatch between donor unit and recipient. For recipients with the FY gene promoter region mutation that prevents erythroid-specific GATA-1 transcription factor binding and erythroid Fyb antigen expression, exposure to Fyb+ units was not considered to be a mismatch, since these recipients are not at risk for anti-Fyb alloimmunization.25 For U- (S-s-) patients, exposure to a S+ and/or s+ unit was considered to result in mismatch at the S and/or s antigen (1 or 2 mismatches) and at the U antigen (additional mismatch).
RH genotypes were categorized as either conventional (homozygous for conventional alleles or heterozygous for 1 variant and 1 conventional allele) or variant (homozygous or compound heterozygous for variant alleles, with no conventional allele detected). All units transfused were assumed to have conventional Rh antigen expression unless denoted as homozygous for ce(733G) with/without 1006T. Therefore, for patients with partial e, C, c, or D expression, all antigen-positive units transfused were assumed to be mismatched at the variant antigen, unless specific variant matching had been indicated (e.g. C-, e-, hrB-, D-, etc). In patients with ≥2 SNPs identified in which the BeadChip assay could not distinguish cis vs. trans position of each SNP (e.g. ce48C, 733G), the patient was categorized as not having a variant genotype that could result in an antigen mismatch.
Statistical Methods
Patients’ clinical data and RBC genotyping results were stored in a Research Electronic Data Capture (REDcap) database, and statistical analyses were performed using SAS version 9.4 (SAS Institute, Cary, NC). Comparisons of continuous variables (age and CTT duration) between patients receiving category 1 vs. category 2 matched transfusions were made by student’s t-test or by Wilcoxon Rank Sums test for non-normally distributed variables, with the normality of distribution assessed by the Shapiro-Wilk test. The categorical variable (gender) was compared by chi-squared test. The frequencies of each RBC antigen in SCA patients versus in the donor RBC units were compared by chi-squared or Fisher’s exact test, as appropriate for sample size. Likewise, the frequencies of antigen mismatches among category 1 vs. category 2 matched transfusions were compared by chi-squared or Fisher’s exact test, using the Bonferroni method to account for multiple comparisons. The incidence of new alloantibody detection was calculated as the number of new antibodies detected per number of units transfused during the study period, and incidence rates for category 1 vs. category 2 transfusions. These Poisson incidence rates were compared to a ratio of 1 to test equality. For new alloantibodies detected during the study (e.g. anti-Jsa), the rates of exposure to the antigen in category 1 vs. category 2 transfusions were compared by two-sample test of proportion.
RESULTS
Patient Characteristics
There were 90 patients (82 at CHOA, 8 at CNMC) who were enrolled between June 13, 2013 to May 27, 2015 and had 1,134 transfusion episodes during the 12 month follow-up period. At study entry, 28 (31%) patients were receiving category 2 matched transfusions (25 with RBC alloantibodies, 3 with only RBC autoantibodies). During the study period, 2 patients were switched from category 1 to category 2 matching: 1 patient with a past anti-e antibody who was determined to have the RHCE genotype ce(733G)/ce(733G), and 1 patient who formed a new antibody of undetermined specificity (AUS) during the study. Thus 30 (33%) patients received category 2 matched transfusions during the study. Baseline characteristics of the patients who received category 1 vs. category 2 transfusions are compared in Table 1 showing slightly older age (mean 10.7 vs. 12.5 years, p=0.049).
Table 1.
Patient Characteristics at Study Entry.
| Characteristic | Total N=90 |
Category 1 n=60 |
Category 2 n=30 |
p |
|---|---|---|---|---|
|
| ||||
| Age (years), mean (range) | 11.4 (3.3 – 19.7) | 10.8 (3.3 – 18.0) | 12.5 (4.6 – 19.7) | 0.049 |
|
| ||||
| Gender, male (%) | 44 (48.9%) | 30 (50%) | 14 (46.7%) | 0.77 |
|
| ||||
| Race | 0.11 | |||
| ▪ African-American | 88 (97.8%) | 60 (100%) | 28 (93.3%) | |
| ▪ Hispanic | 2 (2.2%) | 0 (0%) | 2 (6.7%) | |
|
| ||||
| CTT Duration (years), median (range) | 3.9 (0.4 – 13.9) | 3.6 (0.4 – 13.9) | 5.7 (0.7 – 13.7) | 0.087 |
|
| ||||
| No. RBC Alloantibodies | n/a | |||
| ▪ 0 | 64 (71.1%) | 60 (100%) | 4 (13.3%)* | |
| ▪ 1 | 16 (17.8%) | 0 | 16 (53.3%)† | |
| ▪ ≥2 | 10 (11.1%) | 0 | 10 (33.3%) | |
Patients are categorized by the type of transfusion category (1 vs. 2) at study completion. Two patients switched from category 1 to category 2 matched transfusions during the study.
Of the 4 patients in category 2 with no alloantibodies at entry: 3 had autoantibodies, 1 had no antibodies at entry but formed an antibody of undetermined specificity during the study.
Of the 16 category 2 patients with 1 alloantibody at entry: 1 patient was on category 1 matching at entry but was switched to category 2 when it was determined that a past anti-e antibody was likely an alloantibody.
At study entry, there were 41 known RBC alloantibodies in 26 patients. There were 20 (46.5%) antibodies within the Rh blood group (6 anti-C, 4 anti-E, 2 anti-hrB, 1 anti-e, 3 anti-Goa, 1 anti-f, 1 anti-V, 1 anti-VS, 1 anti-Cw), and 10 (23.2%) antibodies within the Kell blood group (2 anti-K, 5 anti-Kpa, 3 anti-Jsa). The specificities of all alloantibodies are in Supplementary Table 1.
Donor RBC Genotyping Results
There were 2,320 RBC units (1,470 category 1, 850 category 2) transfused during the study period (Figure 1). Of these, the donor HEA genotype was obtained in 2,035 (1293 category 1, 1742 category 2): 1,893 by HEA-LR assay and 142 from recorded donor HEA results. Of the 285 units without HEA genotypes, 89 were due to extremely low DNA concentration resulting in HEA-LR assay failure, and 196 were due to lack of unit segments for HEA-LR testing. The frequency of indeterminate calls per antigen is shown in Supplemental Table 2.
Figure 1.
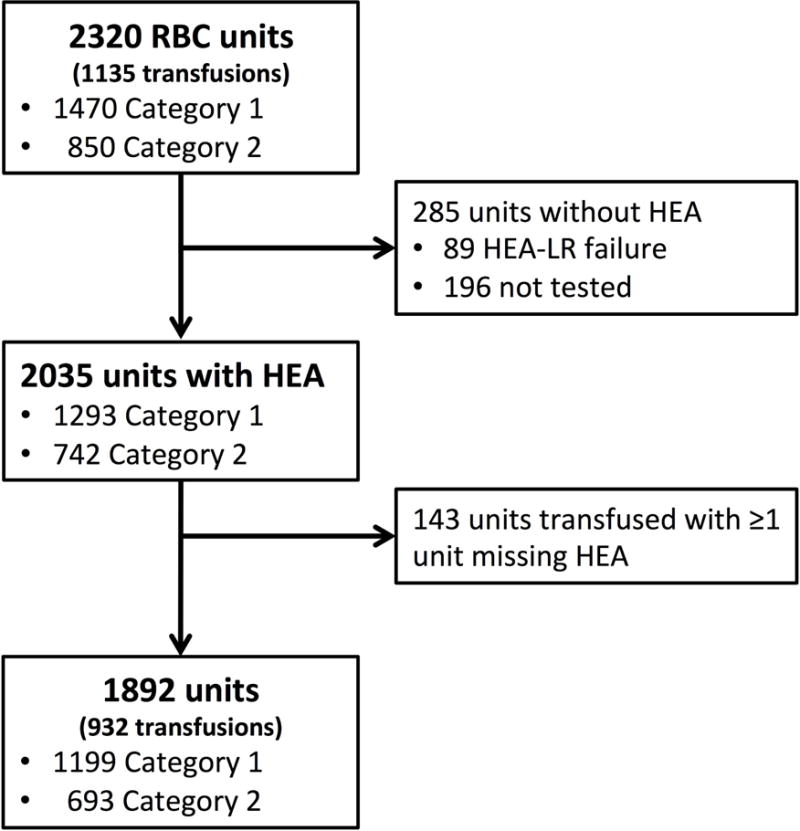
Flow diagram of RBC units and transfusion episodes included in the study.
HEA-LR Assay Concordance
The HEA-LR assay was performed on 1982 units, with assay failure in 89 (4.5%), yielding a success rate similar to previous analyses of HEA-LR.24 The available HEA-LR results (representing 69,370 antigens) were compared to previously known antigen types of 7709 antigens (7219 antigens by serologic typing; 490 antigens by donor whole blood HEA testing). Of the 7709 antigens reviewed for HEA-LR accuracy, 456 (5.9%) were reported indeterminate; 48 (0.62%) were falsely positive, with repeat HEA-LR confirming antigen negative or weak signal. False negative rate could not be assessed in the majority of units because serologic phenotype provided information only on negative antigens; however among the 14 units verified against donor HEA testing, there were 5 (1.0%) false negative antigens. The observed errors were attributed to extremely low DNA concentration in the LR segments.
Patient RBC Genotyping Results
Among the 90 patients, there were 35 (38.9%) patients with homozygous or compound heterozygous variant Rh antigens: 29 (32.2%) with a partial e, 4 (4.4%) with partial C (ce(48C, 733G, 1006T) paired with DIIIa-CE(4–7)-D on RHD, with or without partial e), 1 (1.1%) with partial c (Ce/ceTI), 4 (4.4%) with partial D, and 1 (1.1%) with weak D expression. In 2 patients, RHCE SNPs could not be distinguished as cis (ce/ce(48C, 733G)) vs. trans (ce(48C)/ce(733G)), thus they were categorized as conventional e+ expression, so as to not over-estimate potential e antigen mismatches. Complete lists of patient RHCE and RHD genotypes and frequency of RhCE or RhD antibodies are in Supplementary Tables 3 and 4. Among those with partial e genotypes, 4/29 (13.8%) had anti-e alloantibodies.
Antigen Frequencies of SCA patients vs. donor population
The RBC minor antigen frequencies of the 90 SCA patients were compared to the antigen frequencies found in the 2034 donor units, as shown in Figure 2. The antigen frequency was significantly lower in the patients vs. donors for Fya (11.1% vs. 23.9%, p=0.0049), S (23.3% vs. 38.4%, p=0.0038), and U (95.6% vs. 99.7%, p=0.0005). There was conventional C antigen expression in 21.1% patients vs. 13.4% donor units (p=0.037). There was partial C in 4 (4.4%) patients, but this could not be predicted for units due to lack of RHD genotyping. For the e antigen there were 29 (32.2%) patients with partial e expression (without conventional e), and there were 98 (4.8%) units that were homozygous for the ce(733G) allele which predicts partial e expression.
Figure 2.
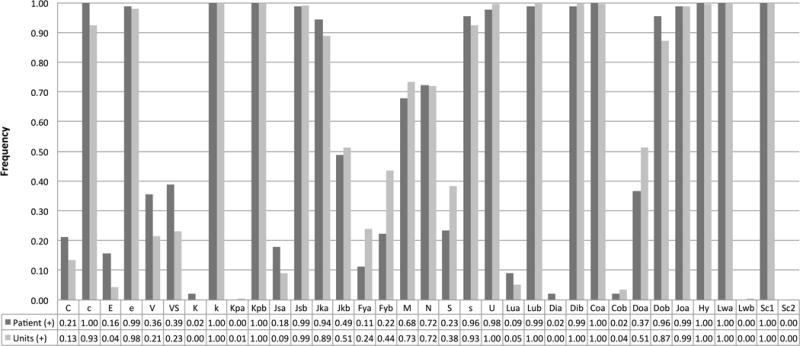
Frequency of each RBC minor antigen among the SCA patients and the donor units. For Rh antigens, conventional and variant antigen expression are shown together. For the e antigen, 68% of patients expressed conventional e, and 31% had partial e (either homozygous or compound heterozygous). Of the e+ units, 4.8% were partial e based on the ce(733G) and ce(1006T) SNPs.
Frequency of antigen mismatch per transfusion
There were 932 transfusion episodes (627 category 1, 305 category 2), comprising 1890 units, in which RBC genotypes were available for all units transfused. Of the 36 minor antigens tested (including RHCE and RHD variants), the mean number of antigen mismatches was 3.5 antigens (median 3.0, range 0 – 9) for category 1 vs. 2.8 antigens (median 3.0, range 0 – 9) for category 2 transfusions; thus the difference in number of antigens mismatched between the two categories is <1 antigen. There were 29 (3.1%) transfusion episodes with 0 antigen mismatches.
The frequency of antigen mismatches per transfusion episode is shown in Figure 3, excluding RhCE and RhD mismatches which could not be fully measured in the units. Category 2 transfusions were intentionally matched for Fya and Jkb, therefore no Fya mismatches were observed in category 2; however 18 (6%) category 2 transfusions were inadvertently mismatched at Jkb. On investigation, these mismatches were attributed to incorrect patient serologic phenotyping of the Jkb antigen in 2 patients. In all transfusions (category 1 and 2), the antigens with a high frequency (>10%) of mismatch were: S (43.9%), Doa (43.9%), Fya (29.2%), M (28.4%), Jkb (28.1%), N (24.0%), V (19.3%), VS (17.9%), and Jsa (13.3%). Most antigens had similar frequencies of mismatch in category 1 vs. 2; however there was a a significantly higher frequency of Dob mismatch in category 2 (2.4% vs. 9.5%, p<0.0001). There was a trend toward higher frequency of V mismatch (21.4% vs. 15.1%, p=0.022) and VS mismatch (20.0% vs. 13.8%, p=0.021); however these were not significant findings when accounting for multiple comparisons.
Figure 3.
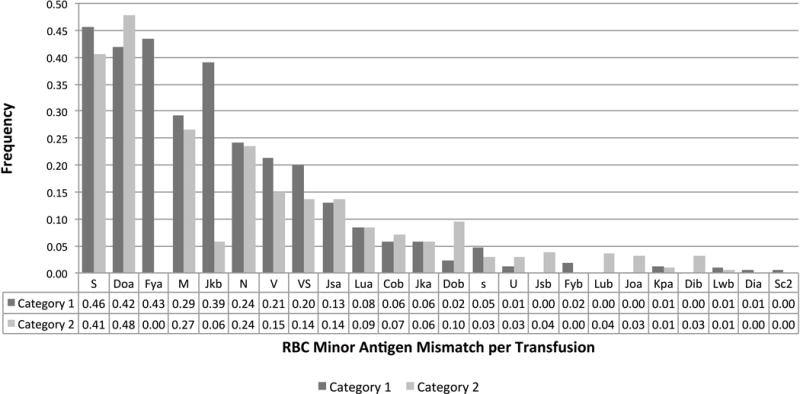
Frequency of RBC minor antigen mismatches per transfusion episode, comparing transfusions that had category 1 level of antigen matching versus category 2. Antigens for which all patients were antigen positive (i.e. k, Kpb, Coa, Hy, LWa, Sc1),antigens that were serologically matched in for all transfusion episodes (i.e. K), and antigens that were not fully characterized in the units (RhD, C/c, E/e) were excluded.
RBC Antibodies
During the study period, there was serologic detection of 7 previously undetected RBC alloantibodies in 5 patients: 3 patients with new anti-Jsa, 1 with anti-e, 1 with anti-Goa, 1 with anti-Wra, and 1 antibody of undetermined specificity (AUS). Additionally, there was new identification of a new warm antibody with e specificity in a patient with RHCE genotype ce(48C)/ceCF; therefore this antibody was determined to represent an anti-e alloantibody. All of the new specific antibodies occurred in patients receiving category 2 matching due to prior alloimmunization; one patient developed a new AUS while receiving category 1 matched transfusions and was changed to category 2 matching. No subsequent antibodies were detected in the patient with AUS. The incidence of alloimmunization was 0.068/100 units for category 1 transfusions and 0.706/100 units for category 2 transfusions (p=0.023).
For Jsa, the frequency of antigen exposures on study prior to the alloantibody detection was assessed. Since donor unit genotyping for the other alloantibodies (anti-Goa, anti-Wra, and e variant determination) was not available, the frequency of these antigen exposures prior to alloantibody detection was not able to be determined. As shown in Figure 4, there were 74 Jsa(−) patients, 3 of whom had anti-Jsa prior to the study thus were receiving Jsa(−) RBC units. Of the remaining 71 patients without prior anti-Jsa, the overall frequency of Jsa antigen exposure was similar for patients on category 1 versus category 2 transfusions (9.96/100 units vs. 9.52/100 units, p=0.87). However, the frequency of Jsa antigen exposure was 20.4/100 units in the 3 patients who developed a new anti-Jsa compared to 8.33/100 units in the 19 patients on category 2-matched transfusions who did not develop an anti-Jsa antibody (p=0.023). For each of the 3 patients who formed anti-Jsa, there were multiple Jsa exposures (mean of 3 RBC units) in the 4 transfusion episodes preceding the antibody detection.
Figure 4.
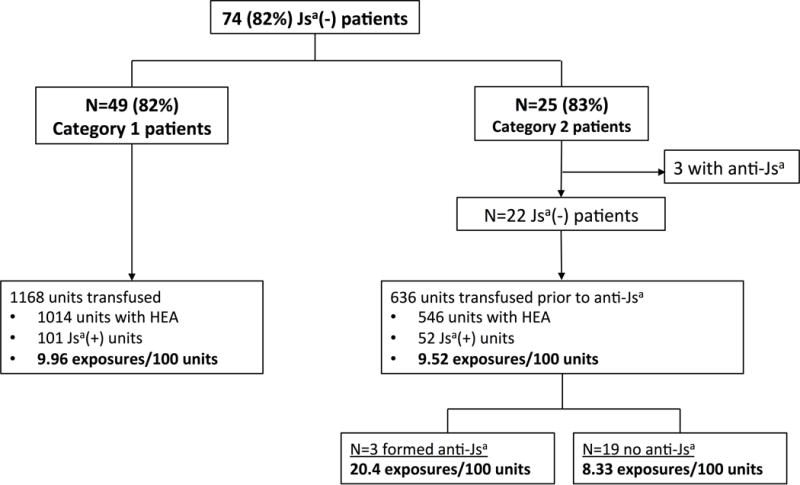
Frequency of Jsa mismatch exposures during transfusions to SCA patients with no prior history of anti-Jsa alloantibodies.
DISCUSSION
RBC alloimmunization is a significant problem among SCA patients, despite minor antigen matching for C/c, E/e, and K as a recommended standard of care for all transfusions.26 While many factors likely contribute to alloimmunization,27–30 exposure to a foreign RBC antigen is the required event to prompt antibody formation. However, it is not known how factors such as frequency or repetition of exposure contribute to the immunologic response. Likewise, the frequency of exposure to minor antigen mismatches during blood transfusion therapy in SCA has not been characterized previously.
This study characterizes the specific RBC minor antigen mismatches that occur during CTT in SCA patients who are receiving minor antigen-matched transfusions. Karafin et al. previously compared antigen frequencies of a cohort of SCA patients receiving C/c, E/e, and K-matched transfusions to those of an African-American targeted donor program, showing that although there were no significant differences in antigen frequencies, there was ongoing development of alloimmunization, demonstrating that antigen mismatches remain problematic.31 Our study uniquely demonstrated the specific antigen matches per transfusion, allowing for comparisons in patients receiving limited vs. extended antigen matching; additionally we were able to demonstrate the frequency of antigen exposure prior to new alloimmunization events in the case of Jsa.
While all transfusions were serologically matched for at least 6 antigens (D, C/c, E/e, K), this study examined 36 antigens, including determination of RHD and RHCE variants among patients that could result in potential Rh mismatches and alloimmunization. For both category 1 and category 2 transfusions, the median number of antigen mismatches per transfusion was low, a finding which is likely influenced by the imposed antigen matching protocols used for SCA. Antigen mismatches were more frequently observed for antigens in which the majority of patients were antigen negative (such as S, Doa, Fya, V, VS) Although antigen-matching protocols (C/c, E/e, K, with or without Fya and Jkb) allow for matching of many immunogenic RBC antigens, this conversely may increase patient exposure to other lower frequency antigens that are more prevalent in African-origin populations than among Caucasian donors, such as V, VS, and Jsa. It should be noted that the number of antigen mismatches may be slightly underrepresented due a 1.6% rate of IC/LS calls per antigen on the HEA-LR results which were not considered donor/recipient mismatches in this study (see supplemental table 2). However, the implicated antigens with the highest IC/LS rates (with the exception of Doa and Dob) were unlikely to be mismatched based on either a high frequency of the IC/LS antigen in the patients (e.g. s antigen), intentional matching for the IC/LS antigen in the units (e.g. C antigen), or the almost universal presence of the Fyb GATA box mutation in SCD patients negating risk of alloimmuniztion (e.g. Fyb antigen).
In our study, there was a high frequency of RHCE homozygous or compound heterozygous variant genotypes lacking expression of conventional antigens. The main limitation of this study was the inability to fully evaluate for RHCE and RHD variant genotypes in the donor units, as the amount of DNA in leukoreduced unit segments is insufficient for PCR-based RH genotyping. Although RH variants were not thoroughly evaluated in the donor units, we presumed that there was a high frequency of Rh antigen mismatches among these patients, particularly with the transfusion of phenotypically e+ units to those with variant e expression. In our cohort, there also was a high frequency of alloimmunization to Rh antigens, similar to the frequency of Rh alloimmunization described by Chou et al. in pediatric SCA patients who had similar transfusion matching,4 including patients with variant e genotypes who had recent development of antibodies with e-like specificity. As some of these Rh antibodies occur in patients with variant RH genotypes and others (such as anti-D) appear transiently among patients with apparent conventional RH genotypes, RH genotyping of both patients and donors is needed to distinguish RBC autoantibodies from Rh alloantibodies and to guide future transfusion antigen matching.
Despite the fact that serologic antigen matching prevents exposure to some of the most immunogenic RBC antigens, there were 7 alloimmunization events in 5 patients during the study period, and the majority of new alloantibodies were directed against low frequency antigens that are not routinely tested by serologic phenotyping. Three of the new alloantibodies were directed against Jsa which occurs in <0.01% of Caucasians compared to 20% of individuals of African descent, and was found to be mismatched in 13% of transfusions in the study period. Therefore, selection of C/c, E/e, and K-matched donors has shifted the pattern of antigen exposure and alloimmunization risk towards an antigen that is more prevalent among African-origin populations. By study completion, 6 SCA patients (6.7% of all patients, 20% of category 2 patients) had alloantibodies to Jsa, demonstrating the importance of identifying Jsa negative donors on a routine basis. Although Jsa negative donors are not rare (as 91% of the donors in the study were Jsa negative), identification of these donor units is a challenge by conventional serologic phenotyping; thus donor RBC genotyping is an important method for locating suitably matched donors for many SCA patients.
Another interesting finding with regards to Jsa alloimmunization in this study was the frequency of exposure to the Jsa antigen among patients who developed new anti-Jsa vs. those who did not. The patients with new Jsa alloimmunization had a significantly higher exposure frequency during the study, and these patients had multiple exposures to Jsa+ units within 3 to 4 sequential transfusions prior to the anti-Jsa detection. It is unknown whether the repetitive nature of the antigen exposure was a causative factor in developing a primary alloimmunization response, or whether the immunologic response only became apparent once the alloantibody titers exceeded the threshold of detection by serologic antibody identification techniques; however these findings do suggest that the frequency or “dosage” of antigen exposure may be a contributing factor to alloimmunization.
Notable differences between chronically transfused SCA patients with prior RBC antibodies versus those without prior RBC antibodies were observed in this study. Although the rates of exposure to individual antigen mismatches, as well as the total number of mismatches per transfusion episode, were overall similar for these two groups of patients, the incidence of new RBC alloantibody detection was markedly higher among patients receiving category 2-matched transfusions. This is in keeping with past observations that alloimmunized patients are a distinct group of immunologic “responders” who have a higher tendency of future alloantibody formation despite increased stringency of antigen matching.27,32–34 Of note, category 2 patients were older and had a longer duration of CTT than category 1 patients, which may confound the tendency towards alloimmunization. Further characterization of the immunologic differences between responder and non-responder patients may help to prevent future alloimmunization.
This study demonstrates that despite serologic RBC minor antigen matching for C/c, E/e, and K, as well as extended matching for Fy, Jkb and any alloantibodies in patients with prior RBC antibodies, there remains a high frequency of individual antigen mismatches for several clinically significant antigens. Yet despite observing mismatch rates approaching nearly 50% of transfusions for some antigens, all of the alloimmunization events that occurred during the study frame were to low frequency antigens. Therefore, although foreign antigen exposure is the requisite event for alloimmunization to occur, there are other factors that must contribute to this event, which may include donor and unit characteristics, patient’s immunologic status, the immunogenicity of individual antigens, the frequency of antigen exposure, or the interaction of all of these factors.27 This study suggests that alloimmunized patients may benefit from more stringent RBC antigen matching, especially to antigens that are more prevalent in donor populations that are ethnically similar to SCA patients, such as Jsa. As further studies continue to explore the causes of RBC alloimmunization in SCA, examining the pattern of foreign antigen exposures prior to sensitization will be important in understanding the triggers for alloimmunization.
Supplementary Material
Acknowledgments
This work was supported by an unrestricted grant from Immucor and by the US Centers for Disease Control and Prevention cooperative agreements DD14-1406, Characterizing the Complications Associated with Therapeutic Blood Transfusions for Hemoglobinopathies. Data management through REDCap was supported by the National Institutes of Health grant UL1 TR000424. The authors would like to thank Christopher Lough, MD of LifeSouth Community Blood Centers and Jose Lima, MD of American Red Cross for their help in providing blood donor RBC genotyping records.
Support: This work was supported by an unrestricted grant from Immucor and by the US Centers for Disease Control and Prevention cooperative agreements DD14-1406
Footnotes
Conflict of Interest: Ross M. Fasano has served as an advisory board member for Immucor; Cassandra D. Josephson has served as a consultant for Immucor. All other authors have no conflicts of interest.
References
- 1.Rosse WF, Gallagher D, Kinney TR, et al. Transfusion and alloimmunization in sickle cell disease. The Cooperative Study of Sickle Cell Disease. Blood. 1990;76:1431–7. [PubMed] [Google Scholar]
- 2.Vichinsky EP, Earles A, Johnson RA, Hoag MS, Williams A, Lubin B. Alloimmunization in sickle cell anemia and transfusion of racially unmatched blood. The New England journal of medicine. 1990;322:1617–21. doi: 10.1056/NEJM199006073222301. [DOI] [PubMed] [Google Scholar]
- 3.Chou ST, Liem RI, Thompson AA. Challenges of alloimmunization in patients with haemoglobinopathies. Br J Haematol. 2012;159:394–404. doi: 10.1111/bjh.12061. [DOI] [PubMed] [Google Scholar]
- 4.Chou ST, Jackson T, Vege S, et al. High prevalence of red blood cell alloimmunization in sickle cell disease despite transfusion from Rh-matched minority donors. Blood. 2013;122:1062–71. doi: 10.1182/blood-2013-03-490623. [DOI] [PubMed] [Google Scholar]
- 5.Castro O, Sandler SG, Houston-Yu P, Rana S. Predicting the effect of transfusing only phenotype-matched RBCs to patients with sickle cell disease: theoretical and practical implications. Transfusion. 2002;42:684–90. doi: 10.1046/j.1537-2995.2002.00126.x. [DOI] [PubMed] [Google Scholar]
- 6.Sins JW, Biemond BJ, van den Bersselaar SM, et al. Early occurrence of red blood cell alloimmunization in patients with sickle cell disease. Am J Hematol. 2016;91:763–9. doi: 10.1002/ajh.24397. [DOI] [PubMed] [Google Scholar]
- 7.Lasalle-Williams M, Nuss R, Le T, et al. Extended red blood cell antigen matching for transfusions in sickle cell disease: a review of a 14-year experience from a single center (CME) Transfusion. 2011;51:1732–9. doi: 10.1111/j.1537-2995.2010.03045.x. [DOI] [PubMed] [Google Scholar]
- 8.Ambruso DR, Githens JH, Alcorn R, et al. Experience with donors matched for minor blood group antigens in patients with sickle cell anemia who are receiving chronic transfusion therapy. Transfusion. 1987;27:94–8. doi: 10.1046/j.1537-2995.1987.27187121485.x. [DOI] [PubMed] [Google Scholar]
- 9.Vichinsky EP, Luban NL, Wright E, et al. Prospective RBC phenotype matching in a stroke-prevention trial in sickle cell anemia: a multicenter transfusion trial. Transfusion. 2001;41:1086–92. doi: 10.1046/j.1537-2995.2001.41091086.x. [DOI] [PubMed] [Google Scholar]
- 10.Sakhalkar VS, Roberts K, Hawthorne LM, et al. Allosensitization in patients receiving multiple blood transfusions. Ann N Y Acad Sci. 2005;1054:495–9. doi: 10.1196/annals.1345.072. [DOI] [PubMed] [Google Scholar]
- 11.O’Suoji C, Liem RI, Mack AK, Kingsberry P, Ramsey G, Thompson AA. Alloimmunization in sickle cell anemia in the era of extended red cell typing. Pediatr Blood Cancer. 2013;60:1487–91. doi: 10.1002/pbc.24530. [DOI] [PubMed] [Google Scholar]
- 12.Telen MJ, Afenyi-Annan A, Garrett ME, Combs MR, Orringer EP, Ashley-Koch AE. Alloimmunization in sickle cell disease: changing antibody specificities and association with chronic pain and decreased survival. Transfusion. 2015;55:1378–87. doi: 10.1111/trf.12940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Nickel RS, Hendrickson JE, Fasano RM, et al. Impact of red blood cell alloimmunization on sickle cell disease mortality: a case series. Transfusion. 2016;56:107–14. doi: 10.1111/trf.13379. [DOI] [PubMed] [Google Scholar]
- 14.Cox JV, Steane E, Cunningham G, Frenkel EP. Risk of alloimmunization and delayed hemolytic transfusion reactions in patients with sickle cell disease. Arch Intern Med. 1988;148:2485–9. [PubMed] [Google Scholar]
- 15.Petz LD, Calhoun L, Shulman IA, Johnson C, Herron RM. The sickle cell hemolytic transfusion reaction syndrome. Transfusion. 1997;37:382–92. doi: 10.1046/j.1537-2995.1997.37497265338.x. [DOI] [PubMed] [Google Scholar]
- 16.Higgins JM, Sloan SR. Stochastic modeling of human RBC alloimmunization: evidence for a distinct population of immunologic responders. Blood. 2008;112:2546–53. doi: 10.1182/blood-2008-03-146415. [DOI] [PubMed] [Google Scholar]
- 17.Casas J, Friedman DF, Jackson T, Vege S, Westhoff CM, Chou ST. Changing practice: red blood cell typing by molecular methods for patients with sickle cell disease. Transfusion. 2015;55:1388–93. doi: 10.1111/trf.12987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fasano RM, Chou ST. Red Blood Cell Antigen Genotyping for Sickle Cell Disease, Thalassemia, and Other Transfusion Complications. Transfus Med Rev. 2016 doi: 10.1016/j.tmrv.2016.05.011. [DOI] [PubMed] [Google Scholar]
- 19.Chou ST, Fasano RM. Management of Patients with Sickle Cell Disease Using Transfusion Therapy: Guidelines and Complications. Hematol Oncol Clin North Am. 2016;30:591–608. doi: 10.1016/j.hoc.2016.01.011. [DOI] [PubMed] [Google Scholar]
- 20.Evidence-Based Management of Sickle Cell Disease: US Department of Health and Human Services. 2014 [Google Scholar]
- 21.Hashmi G. Red blood cell antigen phenotype by DNA analysis. Transfusion. 2007;47:60S–3S. doi: 10.1111/j.1537-2995.2007.01312.x. [DOI] [PubMed] [Google Scholar]
- 22.Hashmi G, Shariff T, Zhang Y, et al. Determination of 24 minor red blood cell antigens for more than 2000 blood donors by high-throughput DNA analysis. Transfusion. 2007;47:736–47. doi: 10.1111/j.1537-2995.2007.01178.x. [DOI] [PubMed] [Google Scholar]
- 23.Hashmi G, Shariff T, Seul M, et al. A flexible array format for large-scale, rapid blood group DNA typing. Transfusion. 2005;45:680–8. doi: 10.1111/j.1537-2995.2005.04362.x. [DOI] [PubMed] [Google Scholar]
- 24.Dang JP, Maurice CB, Orengo L, Stack G. AABB. Anaheim, CA: Transfusion; 2015. Blood Group Genotype Testing on Leukoreduced RBC Segments: Establishing the Feasibility of Hospital-Based Donor Genotyping; p. 142A. [Google Scholar]
- 25.Tournamille C, Colin Y, Cartron JP, Le Van Kim C. Disruption of a GATA motif in the Duffy gene promoter abolishes erythroid gene expression in Duffy-negative individuals. Nat Genet. 1995;10:224–8. doi: 10.1038/ng0695-224. [DOI] [PubMed] [Google Scholar]
- 26.Yawn BP, Buchanan GR, Afenyi-Annan AN, et al. Management of sickle cell disease: summary of the 2014 evidence-based report by expert panel members. JAMA. 2014;312:1033–48. doi: 10.1001/jama.2014.10517. [DOI] [PubMed] [Google Scholar]
- 27.Hendrickson JE, Tormey CA. Understanding red blood cell alloimmunization triggers. Hematology Am Soc Hematol Educ Program. 2016;2016:446–51. doi: 10.1182/asheducation-2016.1.446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Fasano RM, Booth GS, Miles M, et al. Red blood cell alloimmunization is influenced by recipient inflammatory state at time of transfusion in patients with sickle cell disease. Br J Haematol. 2015;168:291–300. doi: 10.1111/bjh.13123. [DOI] [PubMed] [Google Scholar]
- 29.Stowell SR, Girard-Pierce KR, Smith NH, et al. Transfusion of murine red blood cells expressing the human KEL glycoprotein induces clinically significant alloantibodies. Transfusion. 2014;54:179–89. doi: 10.1111/trf.12217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Yazdanbakhsh K, Ware RE, Noizat-Pirenne F. Red blood cell alloimmunization in sickle cell disease: pathophysiology, risk factors, and transfusion management. Blood. 2012;120:528–37. doi: 10.1182/blood-2011-11-327361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Karafin MS, Field JJ, Gottschall JL, Denomme GA. Barriers to using molecularly typed minority red blood cell donors in support of chronically transfused adult patients with sickle cell disease. Transfusion. 2015;55:1399–406. doi: 10.1111/trf.13037. [DOI] [PubMed] [Google Scholar]
- 32.Gehrie EA, Tormey CA. The Influence of Clinical and Biological Factors on Transfusion-Associated Non-ABO Antigen Alloimmunization: Responders, Hyper-Responders, and Non-Responders. Transfus Med Hemother. 2014;41:420–9. doi: 10.1159/000369109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.McPherson ME, Anderson AR, Castillejo MI, et al. HLA alloimmunization is associated with RBC antibodies in multiply transfused patients with sickle cell disease. Pediatr Blood Cancer. 2010;54:552–8. doi: 10.1002/pbc.22327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Nickel RS, Hendrickson JE, Yee MM, et al. Red blood cell transfusions are associated with HLA class I but not H-Y alloantibodies in children with sickle cell disease. Br J Haematol. 2015;170:247–56. doi: 10.1111/bjh.13424. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


