Abstract
Background
The hypermethylated in cancer 1/Sirtuin 1 (HIC1/SIRT1) axis plays an important role in regulating the nucleotide excision repair pathway, which is the main oxaliplatin-induced damage repair system. Based on prior evidence that the variable number of tandem repeat (VNTR) sequence located near the promoter lesion of HIC1 is associated with HIC1 gene expression, we tested the hypothesis that this VNTR is associated with clinical outcome in metastatic colorectal cancer (mCRC) patients receiving oxaliplatin-based chemotherapy.
Methods
Four independent cohorts were tested. Patients treated with oxaliplatin-based chemotherapy served as the training cohort (n=218), and those treated without oxaliplatin served as the control cohort (n=215). Two cohorts of patients receiving oxaliplatin-based chemotherapy, were used for validation studies (n=176 and n=73). The VNTR near HIC1 was analyzed by PCR and gel electrophoresis, and was tested for associations with RR, PFS, and OS.
Results
In the training cohort, patients harboring at least five tandem repeats (TRs) in both alleles showed a significantly shorter PFS compared with those with fewer than four TRs in at least one allele (9.5 vs. 11.6 months, HR 1.93, P=0.012); these findings remained statistically significance after multivariate analysis (HR 2.00, 95% CI: 1.13–3.54, P=0.018). This preliminary association was confirmed in the validation cohort, and patients with at least five TRs in both alleles showed a worse PFS compared with the other cohort (7.9 vs. 9.8 months, HR 1.85, P=0.044).
Conclusions
Our findings suggest that the VNTR near HIC1 could be a predictive marker for oxaliplatin-based chemotherapy in mCRC patients.
Keywords: HIC1, VNTR polymorphism, predictive marker, metastatic colorectal cancer, oxaliplatin
Introduction
Acquired resistance to chemotherapy and molecularly targeted therapies used to treat human cancers is mediated by molecular alterations. Thus, a better understanding of these alterations could lead to the discovery and identification of potential biomarkers to predict treatment responses, as well as reduce acquired resistance to such therapies.
Oxaliplatin is a third-generation platinum drug that is used as a chemotherapy backbone to treat patients with metastatic colorectal cancer (mCRC) in combination with infused 5-fluorouracil/leucovorin (FOLFOX), capecitabine (CapeOX), and infused 5-fluorouracil/leucovorin plus irinotecan (FOLFOXIRI)1. The antitumor effect of oxaliplatin is considered to be the disruption of DNA replication and transcription, primarily by cross-linking the same strand of DNA2. Oxaliplatin-induced DNA damage is primarily repaired by the nucleotide excision repair (NER) pathway, and activated NER may contribute to oxaliplatin resistance3. Therefore, genes involved in the NER pathway, such as ERCC1 (excision repair cross-complementation group 1), have been extensively studied over the past decade as predictive markers for both oxaliplatin efficacy and resistance4–6. However, no predictive markers for oxaliplatin have been prospectively validated.
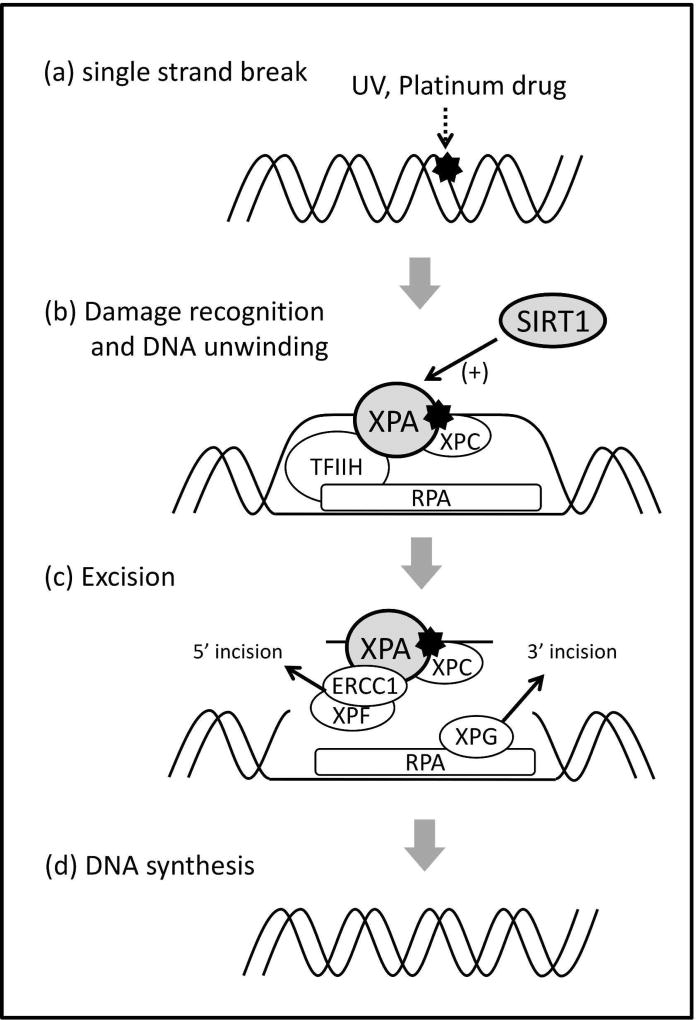
The hypermethylated in cancer 1/Sirtuin 1 (HIC1/SIRT1) axis plays an important role in the regulation of DNA repair pathways7. SIRT1 is a deacetylase that regulates several types of substrates or proteins to maintain genomic instability. Among the targets of SIRT1, XPA (xeroderma pigmentosum complementation group A), which is involved in the NER pathway, is deacetylated by SIRT1; this modification leads to the promotion of the NER pathway8 (Figure 1). HIC1, a sequence-specific transcriptional repressor, directly binds to the SIRT1 promoter and suppresses SIRT1 transcription9. Thus, the inactivation of HIC1 results in the upregulation of SIRT1 expression and activates the NER pathway, which may contribute to oxaliplatin resistance.
Figure 1. The NER pathway.
The single-strand break induced by UV or platinum drugs (a) is recognized by damage-sensor molecules, such as XPC, followed by the binding of several other proteins, such as TFIIH, XPA, and RPA. TFIIH unwinds the DNA duplex, whereas XPA verifies the DNA damage by binding to the chemically altered nucleotides (b). Then, XPA allows for the binding of a complex of nucleases (ERCC1/XPF), which is directed to the damaged strand by RPA to create an incision 5’ to the lesion. After the 5’ incision, RPA recruits a nuclease (XPG), which makes an incision on the 3’ side of the damaged lesion (c). Following this process, DNA polymerase synthesizes new DNA to replace the damage (d). XPA is deacetylated by SIRT1, leading to NER promotion by facilitating its interaction with RPA.
Given prior evidence that the variable number of tandem repeat (VNTR) sequences located near the HIC1 promoter, denoted as D17S5, is associated with HIC1 gene expression (Supplementary Figure S1), we hypothesized that the VNTR near HIC1 may cause interindividual differences in clinical outcome in metastatic colorectal cancer (mCRC) patients treated with oxaliplatin-based chemotherapy. This study presents the potential value of the VNTR near HIC1 as a polymorphic genetic marker in predicting oxaliplatin efficacy in different patient cohorts.
Methods
Patients and samples
This study consists of four independent cohorts: a control cohort, a training cohort, and two validation cohorts. Patients in each cohort are mCRC patients enrolled in clinical trials who received chemotherapy, with or without oxaliplatin (Table 1). Of the patients enrolled in the randomized phase III TRIBE study10, those treated with oxaliplatin-based chemotherapy (FOLFOXIRI + bevacizumab) served as the training set (TRIBE-B cohort, n = 218), and those treated without oxaliplatin (FOLFIRI + bevacizumab) served as the control set (TRIBE-A cohort, n = 215). Two cohorts of patients receiving oxaliplatin-based chemotherapy were used for validation studies (MOMA cohort, n = 176; USA cohort, n = 73). All patients provided an informed consent before entering the randomized trials as well as information regarding the use of their tumor tissue to explore relevant molecular parameters. This study was conducted adhering to the REporting recommendations for tumor MARKer prognostic studies (REMARK). The tissue analysis was approved by the University of Southern California (USC) Institutional Review Board of Medical Sciences and conducted at the USC/Norris Comprehensive Cancer Center in accordance with the Declaration of Helsinki and Good Clinical Practice Guidelines.
Table 1.
Summary of cohorts used in this study
| Control cohort | Training cohort | Validation cohort | ||
|---|---|---|---|---|
| Cohort name | TRIBE-A | TRIBE-B | MOMA | USA |
| Clinical trial | TRIBE study (arm A) randomized phase III (2008–2011, Italy) | TRIBE study (arm B) randomized phase III (2008–2011, Italy) | MOMA study(a) randomized phase II (2012–2016, Italy) | 3C-04-10 study(b) phase II (2005–2009, USA) |
| Ethnic background | Caucasian | Caucasian | Caucasian | Caucasian 32.9% Hispanic 35.6% Asian 26.0% African American 5.5% |
| Number of patients(c) | 215 | 218 | 176 | 73 |
| Chemotherapy | FOLFIRI(d)+ bevacizumab | FOLFOXIRI + bevacizumab | FOLFOXIRI + bevacizumab | FOLFOX or CapeOx + bevacizumab |
| Oxaliplatin-based | (−) | (+) | (+) | (+) |
ClinicalTrials.gov Identifier: NCT02271464
ClinicalTrials.gov Identifier: NCT00159432
patients without available samples were excluded from this study.
combination of infused 5-fluorouracil/leucovorin plus irinotecan
Genotyping of the VNTR near HIC1
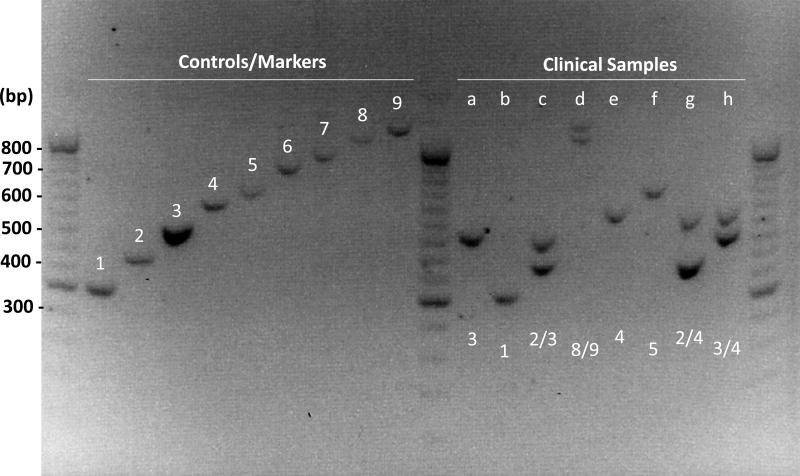
Tandem repeats (TRs) at the D17S5 loci are found approximately 1000 base pairs upstream of the HIC1 transcription start site, and a 70-bp-long sequence is tandemly repeated one to more than ten times11. Genomic DNA was extracted from the blood using the QIAamp DNAeasy Kit (Qiagen), according to the manufacturer’s instructions. To determine the genotypes for TR length, a DNA fragment containing the TR region was amplified by PCR using forward primer 5'-GTTGGGAGCGGGGAAATGCA-3' and reverse primer 5'-TGCATTGCTAACCTGCCCCA-3'. The PCR protocol is consisted of: 1 cycle for 5 min at 95 °C, 30 cycles for 30 sec at 95 °C, 30 sec at 60 °C, 90 sec at 72 °C, and 1 cycle for 7 min at 72 °C. PCR products were separated on 1.2% agarose gels containing biotin and detected by UV fluorescence (Figure 2). Investigators involved in VNTR analyses were blinded to patient clinical data.
Figure 2. Genotype identification of the VNTR near HIC1.
The size of the PCR product size was calculated by the following formula: 260 + 70 × N (N: number of TRs). Control samples showing one to nine TRs were used as markers to identify the genotype for clinical samples. For example, sample (a) depicts a genotype of homozygous 3/3 TRs, and sample (d) shows a genotype of heterozygous 8/9 TRs.
Statistical analysis
To explore the predictive value of the VNTR near HIC1 for oxaliplatin-based chemotherapy, the primary outcome measure was defined as progression-free survival (PFS), which was calculated from the start date of the first-line chemotherapy until the first observation of disease progression or death. If progression or death was not observed, PFS was the day of the last computed tomography scan. We also evaluated the association with response rate (RR) and overall survival (OS), which are defined as the percentage of patients experiencing complete responses and partial responses, according to the Response Evaluation Criteria in Solid Tumors (RECIST) guidelines, and the period from the start date of therapy to the date of death or the date of last contact if alive, respectively. The differences in baseline patient characteristics between the four cohorts were examined using the Fisher’s exact test or the Kruskal-Wallis test where appropriate. The power to detect an association between a VNTR in the recessive model and PFS was 80% when the minimum hazard ratio (HR) varied from 1.84 to 3.19 in 20% to 40% of patients with two copies of the minor allele using a two-sided log-rank test at a significant level of 0.05 in the training cohort (n = 218, 155 PFS events). The power was greater than 70% and 46%, respectively, using the same test to detect the same range of HRs with the same allele frequencies in the recessive model in the validation cohorts (MOMA, n = 176, 119 PFS events; USA, n = 73, 62 PFS events). The associations between polymorphisms and PFS and OS were investigated using Kaplan-Meier curves, the log-rank test, and the Fisher’s exact test. A Cox proportional hazards regression model with stratification factors was fitted to re-evaluate the association between the VNTR and PFS and OS, considering imbalances in the distributions of baseline characteristics among cohorts. The baseline demographics and clinical characteristics that remained significantly associated with endpoints after the multivariate analysis (P < 0.1) were included in the final model. All analyses were performed with two-sided tests at a significance level of 0.05 by using the SAS 9.4 version (SAS Institute, Cary, North Carolina, USA).
Results
Compared with the TRIBE-B training cohort, no differences in characteristics were observed in the TRIBE-A control and the MOMA validation cohorts. The USA validation cohort was composed of younger patients (Table 2). The median PFS, OS, and median follow-up period were 9.7, 25.6, and 49.9 months, respectively, in the TRIBE-A cohort; 11.4, 28.6, and 48.0 months, respectively, in the TRIBE-B cohort; 9.5, 25.4, and 25.3 months, respectively, in the MOMA cohort; and 13.0, 31.1, and 43.3 months, respectively, in the USA cohort.
Table 2.
Baseline clinical characteristics of TRIBE-A, TRIBE-B, MOMA, and USA cohorts
|
TRIBE-A (n = 215) [n (%)] |
TRIBE-B (n = 218) [n (%)] |
MOMA (n = 176) [n (%)] |
USA (n = 73) [n (%)] |
P-value (a) | |
|---|---|---|---|---|---|
| Gender | |||||
| Male | 128 (59.5) | 130 (59.6) | 100 (56.8) | 45 (61.6) | 0.89 |
| Female | 87 (40.5) | 88 (40.4) | 76 (43.2) | 28 (38.4) | |
| Age | |||||
| Median (range) | 60 (29–75) | 60 (29–75) | 61 (23–74) | 55 (28–74) | 0.002 |
| ≤65 | 155 (72.1) | 150 (68.8) | 125 (71.0) | 61 (83.6) | 0.11 |
| >65 | 60 (27.9) | 68 (31.2) | 51 (29.0) | 12 (16.4) | |
| Performance Status | |||||
| ECOG 0 | 177 (82.3) | 195 (89.4) | 153 (86.9) | - | 0.12 |
| ECOG 1 | 37 (17.2) | 23 (10.6) | 23 (13.1) | - | |
| unknown (b) | 1 (0.5) | ||||
| Primary Tumor Site | |||||
| Right | 53 (24.7) | 73 (33.5) | 63 (35.8) | 19 (26.0) | 0.11 |
| Left | 147 (68.4) | 134 (61.5) | 113 (64.2) | 53 (72.6) | |
| unknown (b) | 15 (7.0) | 11 (5.0) | 1 (1.3) | ||
| Time to Metastasis | |||||
| Synchronous | 177 (82.3) | 172 (78.9) | - | 62 (84.9) | 0.45 |
| Metachronous | 38 (17.7) | 46 (21.1) | - | 11 (15.1) | |
| liver metastasis | |||||
| Yes | 66 (30.7) | 76 (34.9) | 52 (29.5) | 33 (45.2) | 0.083 |
| No | 149 (69.3) | 142 (65.1) | 124 (70.5) | 40 (54.8) | |
| Number of metastatic sites | |||||
| 1 | 92 (42.8) | 96 (44.0) | - | 41 (56.2) | 0.12 |
| ≥2 | 123 (57.2) | 122 (56.0) | - | 32 (43.8) | |
| Adjuvant chemotherapy | |||||
| Yes | 26 (12.1) | 27 (12.4) | - | 4 (5.5) | 0.24 |
| No | 189 (87.9) | 191 (87.6) | - | 69 (94.5) | |
| RAS wild type | |||||
| Yes | 53 (24.7) | 58 (26.6) | 43 (24.4) | - | 0.24 |
| No | 110 (51.2) | 108 (49.5) | 119 (67.6) | - | |
| unknown (b) | 52 (24.2) | 52 (23.9) | 14 (8.0) | - |
Based on the Chi-square test or the Kruskal-Wallis test whenever appropriate.
Not included in the test.
Based on our previous findings that the VNTR near HIC1 has different expression patterns for HIC1 between fewer than four TRs and at least five TRs (Supplementary Figure S1), we applied a cut-off value for the number of TRs between four and five. Allele with fewer than four TRs and those with at least five TRs were defined as small (S) and large (L), respectively. We compared clinical outcomes between the patients harboring fewer than four TRs in both alleles (S/S), those with fewer than four TRs in one allele (S/L), and those carrying at least five TRs in both alleles (L/L).
Association of TRs with clinical outcomes in the training and control cohorts
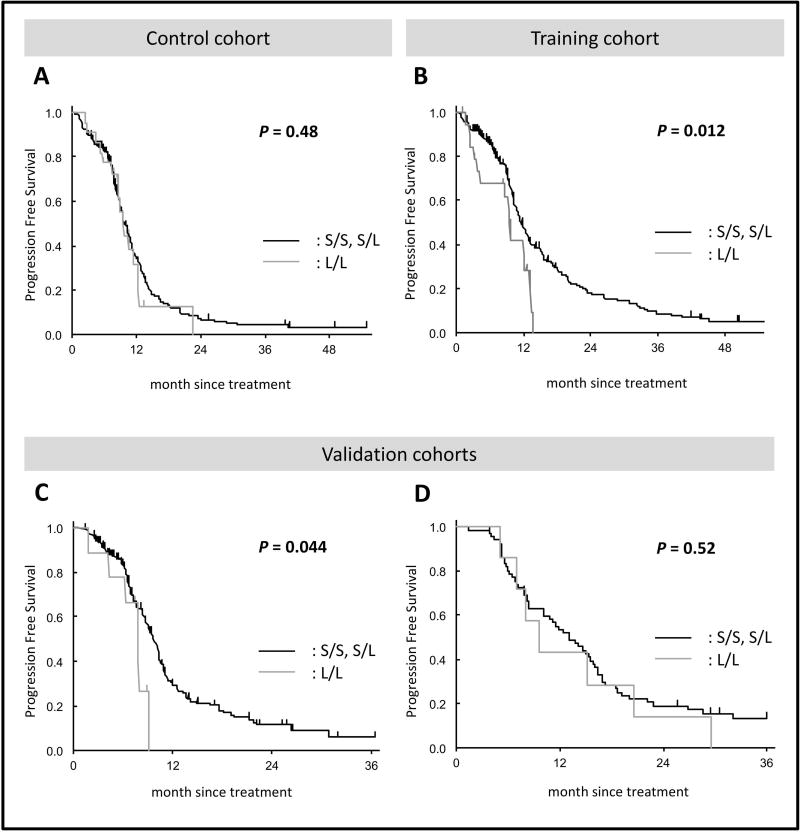
Table 3 shows the association between the VNTR near HIC1 and clinical outcome. The frequency of genotype of the S/S, S/L, and L/L were 77%, 13%, and 10%, respectively, in the TRIBE-A cohort, and 82%, 9%, and 9%, respectively, in the TRIBE-B cohort. In the TRIBE-B training cohort, patients with the L/L genotype had a significantly shorter PFS compared with those with either the S/L or L/L genotype. The median PFS was 9.5 months for patients with the L/L genotype compared with 11.6 months for those with the other genotypes [HR 1.93, 95% confidence interval (CI): 1.11–3.35, P = 0.012] (Figure 3B). This difference remained statistically significant after multivariate analysis (HR 2.00, 95% CI: 1.13–3.54, P = 0.018). Conversely, these correlations were not observed in the TRIBE-A control cohort (Figure 3A). These data suggest that the VNTR near HIC1 may associate with oxaliplatin efficacy in mCRC patients.
Table 3.
Association of the VNTR near HIC1 with clinical outcome in four cohorts
| Genotype | n | RR | PFS | OS | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
|
|
|
|
|||||||||||
| (%) | P-value | univariate (a) | multivariate (b) | univariate (a) | multivariate (b) | ||||||||
|
|
|
|
|
||||||||||
| median month (95% CI) |
HR (95%CI) | P-value | P-value | median month (95% CI) |
HR (95%CI) | P-value | P-value | ||||||
| Training cohort (TRIBE-B) | |||||||||||||
| S/S | 179 | 68% | 0.58 | 11.7 (10.4, 13.1) | 1 (Reference) | 0.041 | 1 (Reference) | 0.049 | 28.5 (25.0, 34.2) | 1 (Reference) | 0.61 | 1 (Reference) | 0.78 |
| S/L | 19 | 76% | 11.0 (8.9, 14.0) | 1.11 (0.63, 1.96) | 0.80 (0.41, 1.56) | 25.8 (10.4, 51.8) | 1.16 (0.67, 2.02) | 0.94 (0.50, 1.76) | |||||
| L/L | 20 | 61% | 9.5 (3.7, 13.2) | 1.95 (1.12, 3.39) | 1.96 (1.10, 3.48) | 30.8 (8.4, 42.0) | 1.27 (0.75, 2.13) | 1.19 (0.70, 2.03) | |||||
| S/S+S/L | 198 | 69% | 0.60 | 11.6 (10.4, 13.0) | 1 (Reference) | 0.012 | 1 (Reference) | 0.018 | 28.5 (25.0, 34.2) | 1 (Reference) | 0.40 | 1 (Reference) | 0.50 |
| L/L | 20 | 61% | 9.5 (3.7, 13.2) | 1.93 (1.11, 3.35) | 2.00 (1.13, 3.54) | 30.8 (8.4, 42.0) | 1.25 (0.74, 2.10) | 1.20 (0.71, 2.04) | |||||
|
| |||||||||||||
| Validation cohort 1 (MOMA) | |||||||||||||
| S/S+S/L | 167 | 65% | 0.99 | 9.8 (8.8, 10.4) | 1 (Reference) | 0.044 | 1 (Reference) | 0.105 | 25.4 (19.8, 29.1) | 1 (Reference) | 0.104 | 1 (Reference) | 0.217 |
| L/L | 9 | 67% | 7.9 (1.9, 9.1) | 2.13 (0.96, 4.71) | 1.92 (0.87, 4.24) | 14.4 (6.9, 41.2) | 2.07 (0.83, 5.17) | 1.79 (0.71, 4.48) | |||||
|
| |||||||||||||
| Validation cohort 2 (USA) | |||||||||||||
| S/S+S/L | 66 | 60% | 0.44 | 13.0 (10.1, 15.8) | 1 (Reference) | 0.52 | 1 (Reference) | 0.68 | 31.1 (24.2, 42.7) | 1 (Reference) | 0.70 | 1 (Reference) | 0.71 |
| L/L | 7 | 43% | 9.6 (7.0, 20.7) | 1.29 (0.59, 2.84) | 0.83 (0.34, 2.02) | 32.3 (9.8, 37.7) | 1.22 (0.43, 3.45) | 1.23 (0.42, 3.61) | |||||
|
| |||||||||||||
| Control cohort (TRIBE-A) | |||||||||||||
| S/S+S/L | 193 | 56% | 0.24 | 9.7 (9.0, 11.0) | 1 (Reference) | 0.484 | 1 (Reference) | 0.079 | 25.6 (21.5, 29.1) | 1 (Reference) | 0.851 | 1 (Reference) | 0.240 |
| L/L | 22 | 71% | 9.5 (7.5, 12.2) | 1.19 (0.72, 1.98) | 1.61 (0.95, 2.73) | 25.5 (15.1, 48.4) | 0.95 (0.57, 1.59) | 1.38 (0.81, 2.34) | |||||
The P value was based on the Fisher’s exact test for tumor response, the log-rank test for PFS and OS in the univariate analysis (a) and the Wald test for PFS and OS in the multivariable Cox regression model, adjusted for age, ECOG performance status, primary tumor site, number of metastatic sites, resection of the primary tumors, RAS mutation status, adjuvant chemotherapy in the TRIBE-A and TRIBE-B cohorts; adjusted for ECOG performance status, liver limited metastasis in the MOMA cohort; adjusted for number of metastasis, treatment type, and sex in the USA cohort (b). Correlations with p < 0.05 are marked with bold text style.
Figure 3. Progression-free survival in four cohorts.
Kaplan-Meier estimates of progression-free survival in patients with fewer than four TRs in at least one allele (S/S and S/L) and those with at least five TRs in both alleles.
Validation studies
In the MOMA validation cohort (S/S: 81%, S/L: 14%, and L/L: 5%), the preliminary association observed in the training cohort was confirmed. Patients with the L/L genotype showed a significantly worse PFS compared with those with either S/L or S/S genotype after univariate analysis (median 7.9 vs. 9.8 months, HR 2.13, P = 0.044) (Figure 3C), yet this correlation was not statistically significant after multivariate analysis (HR 1.92, 95% CI: 0.87–4.24, P = 0.105).
Furthermore, to exclude the impact of irinotecan on clinical outcome, we examined the association of the VNTR in patients treated with FOLFOX or CapeOx backbone in the USA cohort. There was no evidence for an association with PFS between patients with the L/L and those with the S/L or S/S genotype (Figure 3D). However, patients with more than five TRs in both alleles (n = 4) showed a borderline significant trend toward shorter PFS compared with the other cohort (n = 69) after univariate analysis (8.1 vs. 13.0 months, HR 2.28, 95% CI: 0.80–6.47, P = 0.098) (Supplementary Figure S2).
Subgroup analysis
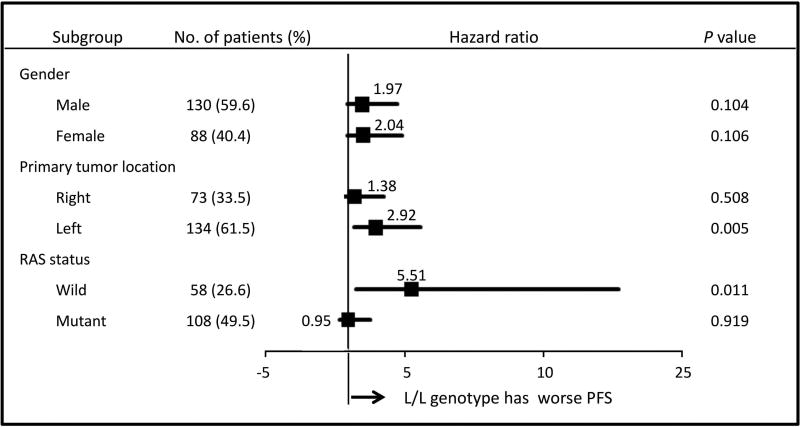
Given the biological features of CRC previously reported12–14, subgroup analyses by gender, primary tumor location, and RAS status were performed. Primary tumor locations were grouped into right-side (cecum, ascending colon, and transverse colon) and left-side (descending colon, sigmoid colon, and rectum). RAS status was divided into two groups: all RAS wild-type and other types. Figure 4 shows the associations of the VNTR near HIC1 with PFS in each subgroup of the TRIBE-B training cohort. Significant associations were observed, particularly in patients with mCRC tumors derived from the left-side (univariate: 8.6 vs. 12.4 months, HR 2.68, P = 0.003, multivariate: HR 2.92, 95% CI: 1.38–6.15, P = 0.005) or with RAS wild-type tumors (univariate: 9.3 vs. 11.3 months, HR 2.78, P = 0.042, multivariate: HR 5.51, 95% CI: 1.49–20.4, P = 0.011) after both univariate and multivariate analyses. However, these associations were not confirmed in the MOMA validation cohort (data not shown).
Figure 4. Subgroup analysis of progression-free survival in the TRIBE-B cohort.
The P value was based on the Wald test for PFS in the multivariable Cox regression model, adjusted for age, ECOG performance status, primary tumor site, number of metastatic sites, resection of the primary tumor, RAS mutation status, and adjuvant chemotherapy.
Discussion
Although many drug combinations have emerged as possible options for the first-line chemotherapy for mCRC patients, several controversial issues remain, including the choice of an optimal chemotherapy backbone. In the clinic, either FOLFIRI or FOLFOX (or CapeOx) is selected as the first-line chemotherapy backbone based on practical issues, patient preference, and toxicity profiles, as no substantial differences in efficacy have been shown between these regimens15, 16. FOLFOXIRI has been proposed as a feasible option with a high antitumor activity and acceptable toxicity17, 18, yet the eligibility criteria have some limitations due to the high intensity of the treatment. Therefore, biomarkers predicting drug efficacy, resistance, and/or toxicity could help the selection of the most favorable first-line chemotherapy regimen for mCRC patients.
In this study, we found that the VNTR near HIC1 was significantly associated with PFS in patients who received oxaliplatin-based chemotherapy but not in those who received irinotecan-based chemotherapy, suggesting that this VNTR may be a predictive marker for oxaliplatin efficacy.
TRs, stretches of DNA composed of two or more contiguous copies of the same genomic sequence, exhibit a length polymorphism that could be potentially used as a genetic marker. Although the functional role of TRs is unclear, growing evidence has revealed that certain TRs located in the 5'-untranslated region exert functional effects on gene expression by modulating the binding of transcription factors, the distance between promoter elements, splicing efficiency, or DNA structure19. We previously found that the VNTR near HIC1 was significantly associated with HIC1 expression: Once the smaller allele exceeds four repeats, higher levels of HIC1 expression were no longer observed, and a trend of decreasing expression was evident (Supplementary Figure S1). In the present study, we found a clear difference in PFS between patients with fewer than four TRs and those with at least five TRs. Although this observation was not confirmed in the USA cohort, potentially due to the small sample size, the result that patients with at least 6 TRs in both alleles showed a trend toward worse PFS is consistent with the inverse correlation of a larger number of TRs and lower levels of gene expression.
Downregulation of HIC1 associated with large numbers of TRs may lead to the overexpression of SIRT1 due to the transcriptional suppression by HIC19. The biological rationale between HIC1/SIRT1 expression and oxaliplatin efficacy, however, remains currently unclear. NER is a repair pathway for single-strand breaks: DNA damage induced by UV and platinum drugs (e.g., cisplatin, carboplatin, and oxaliplatin) is repaired by the NER system20–22. In the NER pathway, XPA, a target of SIRT1, is thought to be the core factor responsible for platinum drug resistance8, as several studies have demonstrated that XPA is overexpressed in cisplatin-resistant cancers23–25. Among the multi-step process of DNA repair, XPA plays a role during the damage recognition or verification, interacting with RPA (replication protein A) (Figure 1). While the acetylated status of XPA reduces the NER activity, deacetylation of XPA by SIRT1 could promote NER activity by facilitating its interaction with RPA, in turn leading to platinum drug resistance8. In a recent study by Asaka et al.26, a cell line overexpressing SIRT1 showed higher viability after treatment with cisplatin than a control cell line. In a mouse xenograft model, tumors overexpressing SIRT1 exhibited stronger resistance to cisplatin compared with control tumors. Taken together, HIC1/SIRT1 expression levels affect NER activity via XPA modulation, resulting in interindividual differences in oxaliplatin efficacy.
In mCRC, distinct molecular profiles based on gender, primary tumor site, and RAS status have been reported12–14. DNA repair systems may differ by primary tumor location, as represented by differences in microsatellite instability27. In addition, RAS status reportedly influences DNA damage responses28. A previous study identified a distinct ERCC1 expression profile based on primary tumor location and RAS status, suggesting the existence of regulatory mechanisms between the NER pathway, tumor location, and RAS status29. Interestingly, we observed herein a strong association between the VNTR near HIC1 and PFS, particularly in patients with metastatic tumors derived from the left-side of the colon or with RAS wild-type tumors. However, these associations were not confirmed in the validation study, potentially due to the small numbers of patients with the minor L/L genotype. Future studies with a larger sample size are warranted to confirm these associations and underlying biological mechanisms.
In conclusion, this study showed for the first time the potential value of the VNTR polymorphism near HIC1 as a predictive marker of outcome after oxaliplatin-based chemotherapy in mCRC patients. If our findings are prospectively validated, since the TR polymorphism can be easily examined by PCR of a blood sample, the VNTR near HIC1 would be a promising biomarker to individualize the use of oxaliplatin in the clinic.
Supplementary Material
Acknowledgments
Funding:
This work was supported by the National Institutes of Health (P30CA014089-27S1), the Gloria Borges Wunderglo Project, the Dhont Family Foundation, the Dave Butler Research Fund, and the Call to Cure Research Fund.
Martin D. Berger received a grant from the Swiss Cancer League (BIL KLS-3334-02-2014) and the Werner and Hedy Berger-Janser Foundation for cancer research.
Y. Miyamoto received a grant from Japan Society for the Promotion of Science (S2606).
Disclosure of Potential Conflicts of Interest:
A. Falcone has received compensation for participation to Advisory Boards and Grants to my Institution from Amgen, Bayer, Roche, Sanofi, Servier, Lilly, and Merck.
Footnotes
Author Contributions:
Conceptualization: S. Okazaki, T. Helentjaris, H-J. Lenz
Methodology: S. Okazaki, W. Zhang, Y. Ning
Validation: S.Okazaki
Formal analysis: S. Cao, D. Yang
Investigation: S. Okazaki, M. Schirripa, M. Berger, Y. Miyamoto, M. Suenaga
Resources: F. Loupakis, S. Iqubal, A. Barzi, C. Cremolini, A. Falcone, F. Battaglin, L. Salvatore, B. Borelli
Writing – original draft: S. Okazaki
Writing – review and editing: H-J. Lenz
Supervision: H-J. Lenz
Funding acquisition: H-J. Lenz
References
- 1.Cremolini C, Schirripa M, Antoniotti C, et al. First-line chemotherapy for mCRC—a review and evidence-based algorithm. Nat Rev Clin Oncol. 2015;12:607–619. doi: 10.1038/nrclinonc.2015.129. [DOI] [PubMed] [Google Scholar]
- 2.Culy CR, Clemett D, Wiseman LR. Oxaliplatin. A review of its pharmacological properties and clinical efficacy in metastatic colorectal cancer and its potential in other malignancies. Drugs. 2000;60:895–924. doi: 10.2165/00003495-200060040-00005. [DOI] [PubMed] [Google Scholar]
- 3.Martinez-Balibrea E, Martínez-Cardús A, Ginés A, et al. Tumor-Related Molecular Mechanisms of Oxaliplatin Resistance. Mol Cancer Ther. 2015;14:1767–1776. doi: 10.1158/1535-7163.MCT-14-0636. [DOI] [PubMed] [Google Scholar]
- 4.Shirota Y, Stoehlmacher J, Brabender J, et al. ERCC1 and thymidylate synthase mRNA levels predict survival for colorectal cancer patients receiving combination oxaliplatin and fluorouracil chemotherapy. J Clin Oncol. 2001;19:4298–4304. doi: 10.1200/JCO.2001.19.23.4298. [DOI] [PubMed] [Google Scholar]
- 5.Yin M, Yan J, Martinez-Balibrea E, et al. ERCC1 and ERCC2 polymorphisms predict clinical outcomes of oxaliplatin-based chemotherapies in gastric and colorectal cancer: a systemic review and meta-analysis. Clin Cancer Res. 2011;17:1632–1640. doi: 10.1158/1078-0432.CCR-10-2169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hatch SB, Swift LP, Caporali S, et al. XPF protein levels determine sensitivity of malignant melanoma cells to oxaliplatin chemotherapy: suitability as a biomarker for patient selection. Int J Cancer. 2014;134:1495–1503. doi: 10.1002/ijc.28454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chen WY, Wang DH, Yen RC, Luo J, Gu W, Baylin SB. Tumor suppressor HIC1 directly regulates SIRT1 to modulate p53-dependent DNA-damage responses. Cell. 2005;123:437–448. doi: 10.1016/j.cell.2005.08.011. [DOI] [PubMed] [Google Scholar]
- 8.Fan W, Luo J. SIRT1 regulates UV-induced DNA repair through deacetylating XPA. Mol Cell. 2010;39:247–258. doi: 10.1016/j.molcel.2010.07.006. [DOI] [PubMed] [Google Scholar]
- 9.Fleuriel C, Touka M, Boulay G, Guérardel C, Rood BR, Leprince D. HIC1 (Hypermethylated in Cancer 1) epigenetic silencing in tumors. Int J Biochem Cell Biol. 2009;41:26–33. doi: 10.1016/j.biocel.2008.05.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Loupakis F, Cremolini C, Masi G, et al. Initial therapy with FOLFOXIRI and bevacizumab for metastatic colorectal cancer. N Engl J Med. 2014;371:1609–1618. doi: 10.1056/NEJMoa1403108. [DOI] [PubMed] [Google Scholar]
- 11.Odelberg SJ, Plaetke R, Eldridge JR, et al. Characterization of eight VNTR loci by agarose gel electrophoresis. Genomics. 1989;5:915–924. doi: 10.1016/0888-7543(89)90134-1. [DOI] [PubMed] [Google Scholar]
- 12.Ali RH, Marafie MJ, Bitar MS, et al. Gender-associated genomic differences in colorectal cancer: clinical insight from feminization of male cancer cells. Int J Mol Sci. 2014;15:17344–17365. doi: 10.3390/ijms151017344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Glebov OK, Rodriguez LM, Nakahara K, et al. Distinguishing right from left colon by the pattern of gene expression. Cancer Epidemiol Biomarkers Prev. 2003;12:755–762. [PubMed] [Google Scholar]
- 14.Network CGA. Comprehensive molecular characterization of human colon and rectal cancer. Nature. 2012;487:330–337. doi: 10.1038/nature11252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tournigand C, André T, Achille E, et al. FOLFIRI followed by FOLFOX6 or the reverse sequence in advanced colorectal cancer: a randomized GERCOR study. J Clin Oncol. 2004;22:229–237. doi: 10.1200/JCO.2004.05.113. [DOI] [PubMed] [Google Scholar]
- 16.Colucci G, Gebbia V, Paoletti G, et al. Phase III randomized trial of FOLFIRI versus FOLFOX4 in the treatment of advanced colorectal cancer: a multicenter study of the Gruppo Oncologico Dell'Italia Meridionale. J Clin Oncol. 2005;23:4866–4875. doi: 10.1200/JCO.2005.07.113. [DOI] [PubMed] [Google Scholar]
- 17.Souglakos J, Mavroudis D, Kakolyris S, et al. Triplet combination with irinotecan plus oxaliplatin plus continuous-infusion fluorouracil and leucovorin as first-line treatment in metastatic colorectal cancer: a multicenter phase II trial. J Clin Oncol. 2002;20:2651–2657. doi: 10.1200/JCO.2002.08.015. [DOI] [PubMed] [Google Scholar]
- 18.Falcone A, Ricci S, Brunetti I, et al. Phase III trial of infusional fluorouracil, leucovorin, oxaliplatin, and irinotecan (FOLFOXIRI) compared with infusional fluorouracil, leucovorin, and irinotecan (FOLFIRI) as first-line treatment for metastatic colorectal cancer: the Gruppo Oncologico Nord Ovest. J Clin Oncol. 2007;25:1670–1676. doi: 10.1200/JCO.2006.09.0928. [DOI] [PubMed] [Google Scholar]
- 19.Gymrek M, Willems T, Guilmatre A, et al. Abundant contribution of short tandem repeats to gene expression variation in humans. Nat Genet. 2016;48:22–29. doi: 10.1038/ng.3461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hoeijmakers JH. Genome maintenance mechanisms for preventing cancer. Nature. 2001;411:366–374. doi: 10.1038/35077232. [DOI] [PubMed] [Google Scholar]
- 21.Marrache S, Pathak RK, Dhar S. Detouring of cisplatin to access mitochondrial genome for overcoming resistance. Proc Natl Acad Sci U S A. 2014;111:10444–10449. doi: 10.1073/pnas.1405244111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ahmad S. Platinum-DNA interactions and subsequent cellular processes controlling sensitivity to anticancer platinum complexes. Chem Biodivers. 2010;7:543–566. doi: 10.1002/cbdv.200800340. [DOI] [PubMed] [Google Scholar]
- 23.Weaver DA, Crawford EL, Warner KA, Elkhairi F, Khuder SA, Willey JC. ABCC5, ERCC2, XPA and XRCC1 transcript abundance levels correlate with cisplatin chemoresistance in non-small cell lung cancer cell lines. Mol Cancer. 2005;4:18. doi: 10.1186/1476-4598-4-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Honecker F, Mayer F, Stoop H, et al. Xeroderma pigmentosum group a protein and chemotherapy resistance in human germ cell tumors. Lab Invest. 2003;83:1489–1495. doi: 10.1097/01.lab.0000090221.95883.41. [DOI] [PubMed] [Google Scholar]
- 25.Stevens EV, Raffeld M, Espina V, et al. Expression of xeroderma pigmentosum A protein predicts improved outcome in metastatic ovarian carcinoma. Cancer. 2005;103:2313–2319. doi: 10.1002/cncr.21031. [DOI] [PubMed] [Google Scholar]
- 26.Asaka R, Miyamoto T, Yamada Y, et al. Sirtuin 1 promotes the growth and cisplatin resistance of endometrial carcinoma cells: a novel therapeutic target. Lab Invest. 2015;95:1363–1373. doi: 10.1038/labinvest.2015.119. [DOI] [PubMed] [Google Scholar]
- 27.Thibodeau SN, Bren G, Schaid D. Microsatellite instability in cancer of the proximal colon. Science. 1993;260:816–819. doi: 10.1126/science.8484122. [DOI] [PubMed] [Google Scholar]
- 28.Grabocka E, Commisso C, Bar-Sagi D. Molecular pathways: targeting the dependence of mutant RAS cancers on the DNA damage response. Clin Cancer Res. 2015;21:1243–1247. doi: 10.1158/1078-0432.CCR-14-0650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Maus MK, Hanna DL, Stephens CL, et al. Distinct gene expression profiles of proximal and distal colorectal cancer: implications for cytotoxic and targeted therapy. Pharmacogenomics J. 2015;15:354–362. doi: 10.1038/tpj.2014.73. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.