Abstract
Mitochondria are involved in the generation of energy, cell growth and differentiation, cellular signaling, cell cycle control, and cell death. To date, the mitochondrial basis of cancer disparities is unknown. The goal of this review is to provide an understanding and a framework of mitochondrial determinants that may contribute to cancer disparities in racially different populations.
Mitochondria, which are multi-functional, have been implicated in the initiation and progression of cancers in relation to metabolic alterations in transformed cells. Due to ethnic-based diversity, the mitochondrial genome (mtDNA) could be a basis for inherited racial disparities and for acquired somatic mutations during tumorigenesis. In African Americans, several germline, population-specific haplotype variants in mtDNA as well as depletion of mtDNA have been linked to cancer predisposition and cancer disparities. Indeed, depletion of mtDNA and mutations in mtDNA or nuclear genome (nDNA)-encoded mitochondrial proteins lead to mitochondrial dysfunction and promote resistance to apoptosis, the epithelial-to-mesenchymal transition, and metastatic disease, which in turn can contribute to cancer disparity and tumor aggressiveness related to racial disparities. Ethnic differences at the level of expression or genetic variations in nDNA encoding the mitochondrial proteome, including mitochondria-localized mtDNA replication and repair proteins, miRNA, transcription factors, kinases and phosphatases, and tumor suppressors and oncogenes may underlie susceptibility to high-risk and aggressive cancers found in African Americans and other ethnicities.
The mitochondrial retrograde signaling that alters the expression profile of nuclear genes in response to dysfunctional mitochondria is a mechanism for tumorigenesis. In ethnic populations, differences in mitochondrial function may alter the cross talk between mitochondria and the nucleus at epigenetic and genetic levels, which can also contribute to cancer health disparities. Targeting mitochondrial determinants and mitochondrial retrograde signaling could provide a promising strategy for the development of selective anticancer therapy for dealing with cancer disparities. Further, agents that restore mitochondrial function to optimal levels should permit sensitivity to anticancer agents for the treatment of aggressive tumors that occur in racially diverse populations and hence help in reducing racial disparities.
Keywords: Mitochondria, racial, cancer diversity, disparity, African American, Caucasian, mitochondrial DNA, mipigenetics, numtogenesis, genomic instability, retrograde, mitochondria-to-nucleus, anterograde, nuclear mitochondria, cancer prevention, cancer therapy, epigenetic, exosome
1. INTRODUCTION
Mitochondria, often called the powerhouse of the cell, are dynamic organelles found in nearly all eukaryotic cells. Mitochondria are the site of the citric acid cycle (TCA) and oxidative phosphorylation (OXPHOS), metabolic processes that convert pyruvate molecules into ATP, the energy currency of the cell. Apart from producing ATP, mitochondria are also involved in cellular activities such as maintenance of redox potential, apoptosis, fatty acid and heme biosynthesis, and regulation of oxidative stress. The role of mitochondria in cancer was described in 1931 as the discovery of a metabolic rearrangement in cancer cells, a phenomenon known as the Warburg effect. Otto Warburg discovered that cancer cells, due to a defect in mitochondrial oxidative phosphorylation (OXPHOS), produced abnormally high levels of lactate from glucose, even under aerobic conditions. Consistently, in human cancers, the mitochondrial genes that encode the components of the electron transport chain (ETC) of OXPHOS are frequently mutated and/or repressed. Furthermore, mitochondria are also the primary site of generation of reactive oxygen species (ROS). The ROS, upon exceeding a threshold, lead to extensive damage to mitochondria and changes in mitochondrial membrane potential. The resulting mitochondrial dysfunction can cause pro-cancer changes in expression of nuclear genes. This phenomenon is known as mitochondria-to-nucleus retrograde signaling.
Recent clinical advances in cancer prevention, diagnosis, and treatment have helped reduce cancer mortality rates in the Caucasian population. For this disease, however, there is a distinct disparity between African Americans (AAs) and other ethnic populations. Past studies have attributed racial disparities to differences in socioeconomic, educational, cultural and envirnonmental factors. However, it is increasingly recognized that these disparities may also be due to biological differences including differences in mitochondrial biology, genetics and function. Unfortunately, to date a comprehensive mitochondrial basis of cancer disparity is lacking. The present review provides an understanding of mitochondrial determinants involved in tumorigenesis and highlights, where available, the underlying mitochondrial basis for cancer disparities in AAs and other ethnic populations. Restoring mitochondrial function and targeting mitochondrial determinants and mitochondria-to-nucleus retrograde signaling could provide an effective strategy for the development of the selective anticancer therapy that would reduce cancer disparities between diverse populations.
2. DISPARITIES AT THE MITOCHONDRIAL GENOME LEVEL AND THEIR ROLE IN CANCER
The mitochondrial genome is a 16.6 kb, closed-circular, double-helical molecule that is inherited only through the mother. The mtDNA encodes 13 proteins of the OXPHOS system, two rRNAs, and 22 tRNAs [344]. The mtDNA-encoded proteins include 13 subunits that make up four OXPHOS complexes (Complex I, III, IV, and V; see Figure 1b.) Complexes I to IV are electron transport proteins, and Complex V is an ATP synthase [344, 370]. Nuclear DNA encodes the remaining OXPHOS complexes (73 subunits). Nuclear DNA-encoded subunits (38 for Complex I, 10 each for protein Complexes III and IV, and 15 for Complex V) are translated in the cytosol and imported into the mitochondrial compartment [344]. All four subunits constituting complex II are encoded by nuclear DNA (Figure 1).
Figure 1. mtDNA and nDNA encode subunits comprising the OXPHOS complexes.
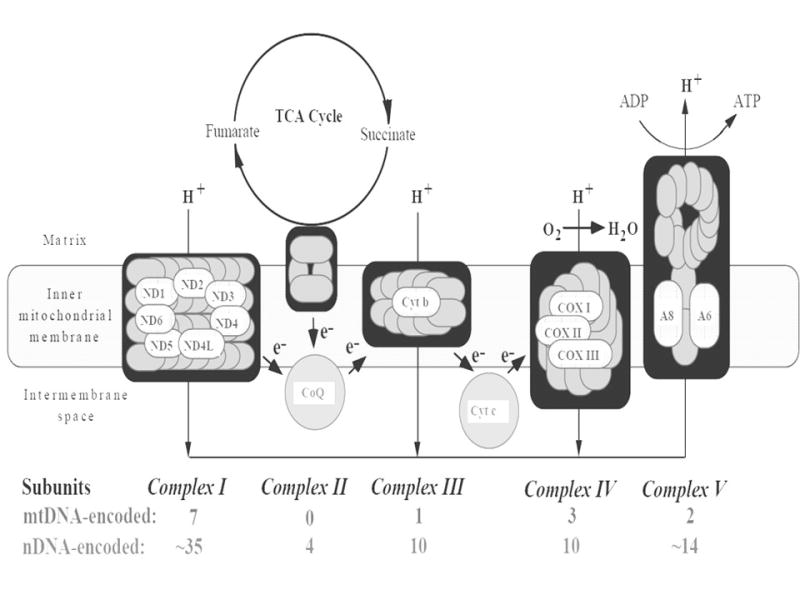
mtDNA encodes 13 protein subunits involved in OXPHOS complexes; the other OXPHOS subunits are encoded by nDNA.
The remaining approximately two thousand mitochondrial proteins, including those involved in the replication, transcription, and translation of mtDNA, are encoded by nuclear genes and are translocated into the mitochondria [324]. In various aspects, mtDNA differs from nuclear DNA: a) although constituting <1% of the total cellular DNA content, mtDNA is polyploid or present as hundreds to thousands of copies; b) mtDNA lacks introns, and only 10% of the mitochondrial genome consists of non-coding sequences; c) mtDNA also lacks the histone bound nucleo-protein structure; d) transcription of mtDNA produces a polycistronic precursor RNA that is processed further into mRNAs, rRNAs, and tRNAs; and e) mtDNA replication occurs independently of the cell cycle [369]. Owing to proximity to the OXPHOS machinery, mtDNA is constantly exposed to the harmful effects of the ROS generated by the ‘leakage’ of electrons from the respiration complexes. This, combined with the absence of any histone-mediated protection and the natural error-prone replication of mtDNA polymerase γ [344], results in a tenfold higher accumulation of mutations in mtDNA compared to nuclear DNA [121]. Furthermore, due to the high density of coding sequences in mtDNA, mutations in mtDNA have more functional and epidemiological consequences than those in nuclear DNA. mtDNA mutations affect mitochondrial respiratory enzyme complexes and can promote oncogenesis through a) increased production of ROS [98], b) further damage to the mtDNA [95, 341], and c) resistance to apoptosis [190, 342].
2.1 Germline mitochondrial genome variations
Persistence of some mutations in certain populations creates haplogroups; a particular mtDNA haplogroup within a population will carry a unique mutation. A haplogroup can be further divided into haplotypes, generally based on restriction fragment-length polymorphisms [233]. Among human populations, there are more than 25 mtDNA haplogroups, and many of these correlate with a predisposition to cancer.
For a Japanese group, Tanaka et al. [359] retrospectively classified 30 haplotypes based on 149 polymorphisms in the coding region. The individual haplotypes were as follows: F, B5, B4a, B4b, B4c, A, N9a, N9b, Y, M10+M11+M12, M7a, M7b2, M7c, M8+Z+C, G1, G2, M9, D5, D4a, D4b, D4d, D4e, D4g, D4h, D4j, D4k, D4k, D4l, D4m, and D4n. The haplogroup M7b2 was associated with a higher risk of leukemia [372]. Booker et al. classified nine main European haplotypes (H, I, J, K, T, U, V, W, X) in patients with prostate and renal cancers and reported a higher risk of renal cancer for haplogroup U [38]. In a cohort study, they demonstrated an association for four germline mutations in cytochrome oxidase subunit I (T6253C, C6340T, G6261A, and A6663G).
A variety of studies have associated breast cancer risk with certain mtDNA single nucleotide polymorphisms (SNPs) in various populations. In European-American, Polish, Malay, North Indian and AA populations, the G10398A substitution in the N haplogroup affecting the ND3 locus is linked to a higher susceptibility to breast cancer [12, 50, 79, 80, 83, 191, 361]. For AA women, there is a cumulative increase in risk when the T4216C substitution in the ND1 locus is present along with G10398A. Although the A10398G and T16519C substitutions increase breast cancer risk, the T3197C and G13708A SNPs decrease the risk [12] (Figure 2).
Figure 2. mtDNA alterations associated with cancer disparity.
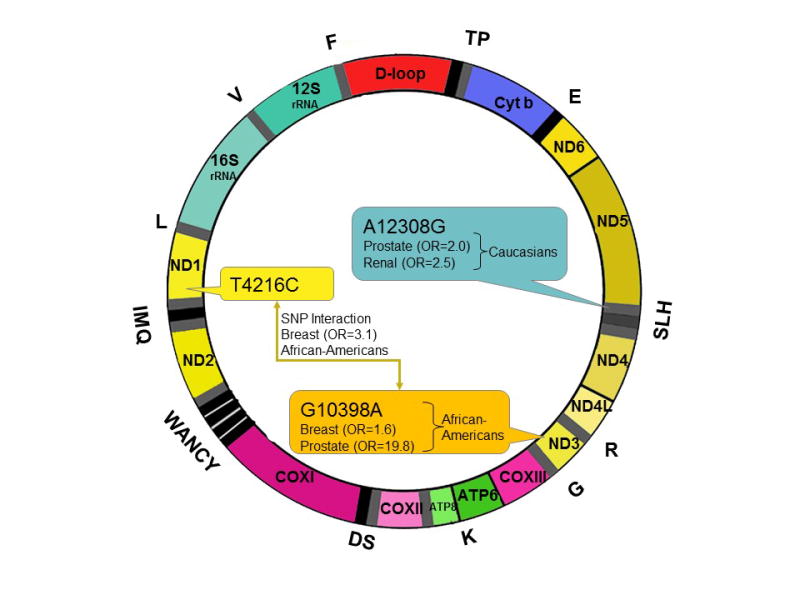
Three non-synonymous and tRNA substitutions are identified in epidemiologic studies as being associated with an increased risk of cancer in specific populations. G10398A in ND3 is associated with increased risk of breast (OR=1.6) and prostate cancer (OR=19.8) in AAs. A12308G in tRNALeu2 is a marker of the mtDNA haplogroup U. In Caucasians, this haplogroup is associated with increased risk of prostate (OR=1.95) and renal cancer (OR=2.52).* In isolation, the T4216C substitution in ND1 confers no increased risk. However, when the T4216C substitution is present with G10398A, the risk of breast cancer is increased (OR=3.1) in AAs. Further, mtDNA depletion is reported to be associated with cancer disparities for AAs.
Other cancers that exhibit a higher prevalence in the presence of germline mtDNA mutations include esophageal and pancreatic cancers [369]. The 10398A polymorphism, for instance, confers a higher risk for esophageal cancer for Indian populations [83]. In Chinese, there is an association between mtDNA haplogroups D4a and D5a and increased risk of esophageal cancer [212]. In European haplotypes, the heritable SNP rs2857285 in the ND4 gene is associated with a more invasive form of ovarian cancer [104]. For patients with pancreatic cancer, germline mutation 16519T in the displacement loop (D-loop) worsened the prognosis [274]. Also, there is an association between a G5460A SNP (in haplogroup H) encoding an A331T substitution in the ND2 gene with pancreatic cancer [196]. Wang et al. [380] and Halfdanarson et al. [132], however, did not find any significant correlation between mtDNA polymorphisms and pancreatic cancer in Caucasian populations. In a multi-ethnic cohort study, the missense 4917 SNP in the ND2 gene was associated with a risk of colorectal cancer in European-Americans but not Africans, Asians, or Latinos. The same study also found that, for the T haplotype, the risk of colorectal cancer was elevated [206].
Although there are many studies correlating mtDNA germline polymorphisms and haplogroups to various cancer types, correlating a specific mutation to a specific cancer type is difficult. Since the mutation and polymorphism rates in mtDNA are high, the same mutation may arise in various populations and lead to different risks in those populations.
2.2 Germline mitochondrial genome depletion
Our previous study found that normal prostate and prostate tumors from AA men contain lower mtDNA copy numbers [185]. In this context, it is noteworthy that lower mtDNA content leads to the acquisition of aggressive, androgen-independent development of prostate cancers and to the epithelial-to-mesenchymal transition involved in metastasis [143, 273]. Our results demonstrate that depletion of mtDNA promotes apoptotic resistance and tumor aggressiveness [190, 281, 282]. In a prostate cancer cell line derived from an AA tumor, mtDNA depletion results in apoptotic resistance [63]. mtDNA depletion promotes features of stem cell-like properties such as over-expression of Oct3/3, Nanog, and CD44 [125, 223]. This feature is reported for prostate [223], ovarian [149], and breast [125] cancers. A low mtDNA content can be caused by mutations in the p53 gene, the POLG gene, and the gene for the mitochondrial transcription factor, TFAM [189, 338–340]. In a yeast screen designed to identify nuclear genes involved in the maintenance of mtDNA, Zhang and Singh identified more than 50 human homologs whose inactivation caused depletion of mtDNA [414]. It is likely that germline variants or somatic mutations in these genes alter mtDNA content and may promote apoptotic resistance and risk of cancers. The reduced mtDNA invokes mitochondria-to-nucleus cross talk, thus providing a molecular basis for risk of cancer and an underlying mechanism contributing to tumor aggressiveness. In addition to mtDNA depletion, mtDNA variants associated with different races also lead to mitochondria-to-nucleus cross-talk [191]. Such constitutive mito-nuclear cross talk may contribute to high risk, tumor aggressiveness, and cancer disparities in AAs and other ethnic populations.
2.3 Somatic mitochondrial genome mutations
Mutations in mtDNA have been reported in many cancers [263]. An exhaustive list of somatic mtDNA mutations and their frequencies has been prepared by Lee [202]. Although these mutations occur throughout the mitochondrial genome, in human cancers, the displacement loop (or D-loop) region is a mutational “hot spot.” The D-loop is a non-coding control region (np 16024-516) that houses cis-regulatory elements required for replication and transcription of mtDNA. Mutations in this region may affect copy numbers and expression of mitochondrial genes, thus making D-loop instabilities a driver of oncogenesis. Maurya et al., who analyzed the D-loop regions in 14 urothelial cell carcinomas, found 28 somatic mutations, which included nine insertion/deletion changes and two single-base substitutions [251]. In oral squamous cell carcinomas, nine mutations, including one point mutation, two base deletions, three insertion mutations, and three heterozygous mutations, were detected in the D-loop region [410]. In another study, somatic mutations in the D-loop were identified in a cohort of Chinese squamous cell carcinomas, but they did not correlate with prognosis or survival [217]. There are also D-Loop mutations in acute lymphoblastic leukemia (ALL) cases, with 89 G insertions, 95 G insertions, 182 C/T substitutions, 308 C insertions, and 311 C insertions, making a total of 132 mutations at 25 locations [397]. In a comprehensive study involving 54 hepatocellular carcinomas, 31 gastric cancers, 31 lung cancers, and 25 colorectal cancers, the incidence of somatic D-loop mutations in cancers of later stages was higher than that of early-stage cancers [203].
In addition to D-loop disruptions, deletions, point mutations, insertions, and duplications in other parts of the mitochondrial genome are known. Somatic mutations in mtDNA genes have been noted in human ovarian, thyroid, salivary, kidney, liver lung, colon, gastric, brain, bladder, head and neck, prostate, and breast cancers, and in leukemia [369, 398]. For example, a 40-bp insertion localized in the COX I gene appears to be specific for renal cell oncocytoma [386], and a deletion mutation is resulting in the loss of mtDNA within NADH dehydrogenase subunit III is associated with renal carcinoma [147, 186]. In cancers of AA women, two variants of the cytochrome b gene, 13G and I2-992T, are more frequent in breast cancer patients compared to healthy controls. As determined in a multi-ethnic study, there is also a positive correlation between the SNP T4216C in the ND1 gene and colorectal cancer [4]. In a population-based study involving prostate cancer patients of European and AA descent, the frequency of COX I missense mutations was higher in cancer patients compared to healthy controls, with some of these sequence variants possibly representing germ line mutations [300]. Specifically, the authors associated two SNPs, T6221C and T7389C, with a higher incidence of prostate cancer. In renal cell carcinomas, the co-occurrence of somatic and germ-line mtDNA mutations has been reported [321]. In contrast, somatic variants of the ATP6 and ND3 genes in the Mexican Mestizo populations could not be linked to a higher risk of prostate cancer [54].
Brandon et al. [42] suggested that mtDNA mutations in tumors can be divided into two main groups: (1) severe mutations that inhibit OXPHOS, increase ROS production, and promote tumor cell proliferation and (2) milder mutations that permit tumors to adapt to new environments. The severely deleterious tumorigenic mutations that inhibit mitochondrial respiration could be advantageous in the initial phases of tumor growth when the tumor requires mitochondrial ROS to drive cell proliferation. However, the adaptive mtDNA mutations may permit the tumor cells to flourish in new environments as they metastasize. Since the migratory tumor cells could be exposed to similar environmental challenges as the humans who migrated out of Africa, the same mtDNA mutations might be adaptive in both tumors and people [42].
2.4 Somatic mtDNA copy number variations
Both an increase and a decrease in mtDNA copy number are associated with an increased risk for tumorigenesis [68, 220, 244, 244,164, 235, 377, 381, 382]. Depletion in mtDNA has been reported in breast [235, 367], colorectal [202], gastric [393, 202] hepatocellular [202], kidney [423, 424], lung [202], ovarian [381], and prostate [185] cancers. An increase in mtDNA content has been reported in Burkitt’s lymphoma, chronic lymphocytic leukemia, head & neck cancer, non-Hodgkin’s lymphoma, small lymphocytic lymphoma [425], and papillary thyroid carcinoma [235]. Depletion of mtDNA may result in disruption of mitochondrial respiration, and the ensuing dysfunction in mito-nuclear signaling can drive oncogenesis. An increase in mtDNA copy number might occur as a compensatory response to mitochondrial dysfunction, resulting in increased ROS production, altered mito-nuclear cross talk, and tumorigenesis.
Mutations in or depletion of mtDNA can have a causative effect in carcinogenesis through disruption of the OXPHOS enzyme complexes and the ensuing oxidative stress and retrograde signaling. Conversely, mtDNA dysfunction may not directly generate the cancer phenotype but influence tumor progression and maintenance by causing a metabolic shift from respiration to aerobic glycolysis [369]. To a great extent, the physiological role of mtDNA in initiating and maintaining tumorigenesis has been elucidated through the use of trans-mitochondrial cybrids –hybrid cells that combine nuclear genes from one cell and mitochondrial genes from another. This technique is useful in dissociating the function of mtDNA genes from those of the nuclear genes. Prostate cell (PC3) cybrids harboring the T8993G mtDNA mutation generate tumors that are seven times larger than wild-type cybrids, which barely grew in mice [38]. Additionally, after transplantation into nude mice, cybrids constructed using a common HeLa nucleus and mitochondria containing a point mutation in ATP synthase subunit 6 conferred a growth advantage in early tumor stages, possibly through the prevention of apoptosis [137].
2.5 Contribution of numtogenesis
Intact mitochondria containing mtDNA, mitochondrial RNA (mtRNA), and mitochondrial proteins localize into the nucleus [13, 33, 41]. Indeed, the nuclear copies of mtDNA described as NUMTs (nuclear mtDNA) are present in at least 85 sequenced eukaryotic genomes [139]. We have named this phenomenon leading to the presence of nuclear mitochondria as numtogenesis [422]. In humans, numtogenesis is estimated to occur at a rate of ~5 × 10−6 per germ cell per generation [89].
Evolutionary studies suggest that, in humans and other mammals, the insertion sites of germline NUMTs are distributed non-randomly [379]. Germline NUMTs tend not to originate from the mtDNA displacement loop (D-loop); they tend to be located in damage-prone regions of the nuclear genome, such as open chromatin and fragile sites [379]. These results implicate NUMTs in the repair of double-strand breaks [138]. The mechanism for NUMT accumulation is not well understood. The most parsimonious mechanism explaining NUMT accumulation involves de novo transposition from the mitochondria to the nucleus; however, NUMTs also accumulate via segmental duplication (sometimes within repetitive elements), and possibly by RNA retro-transposition [64, 130, 270, 271]. The human genome contains between 755 and 1,105 germline NUMTs, with mtDNA identities ranging from 64–100% [270]. Our preliminary data suggest the existence of NUMT diversity between AAs and Caucasian Americans and among other populations.
3. DISPARITIES AT THE NUCLEAR GENOME LEVEL AND THEIR ROLE IN CANCER
3.1 Mitochondrial oxidative phosphorylation
Except for 13 proteins, all other mitochondrial proteins, as well as the regulatory apparatus of the mitochondria, are encoded by nuclear genes (Figure 1). In human cancers, there are mutations in genes encoding OXPHOS proteins, TCA cycle enzymes, mtDNA regulatory elements, and mitochondrial biogenesis.
Succinate dehydrogenase (SDH), or complex II of the ETC, serves as a link between the TCA cycle and the ETC, and the gene encoding it is frequently mutated in paraganglioma, breast cancer, gastric cancer, and renal carcinoma. The SDH complex consists of four subunits, A, B, C, and D, encoded respectively by the SDHA, SDHB, SDHC, and SDHD genes. Patients with hereditary paraganglioma carry germline mutations in the SDHD gene [22], whereas mutations in SDHC cause an autosomal dominant form of paraganglioma [275]. The main mechanism of SDH mutation-derived tumorigenesis is disruption of the ETC, leading to increases in the levels of cell-damaging ROS [153]. Also, murine fibroblasts deficient in SDHA cause succinate accumulation and translocation of HIF-1α into the nucleus. Mutations in SDHA are seen in pituitary adenoma [104], and SDHB mutations are associated with an early onset of familial renal carcinoma [305]. Patients suffering from gastrointestinal stromal tumors [159] and breast cancers [176] also show SDH inactivation in the tumor tissues. Germline mutations in the gene encoding fumarate dehydrogenase (FDH), the TCA cycle enzyme that reversibly hydrates fumarate to malate, is seen in skin leiomyomata, renal cell cancers, and uterine leiomyomas [204, 205]. Bennedbaek et al. identified, in Danish patients with pheochromocytoma or paraganglioma, eight germline variants in the SDHB, SDHC, and SDHD genes [24].
Heterozygous missense mutations in the isocitrate dehydrogenase (IDH) gene are frequently seen in acute myelocytic leukemia (AML), gliomas, and astrocytomas [401]. Both the cytosolic (IDH1) and mitochondrial (IDH2) isoforms display mutated versions and are implicated in tumorigenesis. In AML, IDH2-R140 is the most common mutation [241, 284], whereas the IDH2-R172 mutation is seen in gliomas [400]. Under physiological conditions, IDH de-carboxylates isocitrate to α-ketoglutarate and reduces NADP+ to NADPH. Mutated forms of IDH convert isocitrate to 2-hydroxyglutarate, an oncometabolite that alters global gene expression patterns through DNA methylation and chromatin remodeling [71, 229]. In some recent studies, the relevance of germline variants of IDH genes has been investigated. In a cohort of AML patients, the G-allele of the IDH1105 SNP was associated with shorter survival and a poorer prognosis compared to the T-allele [107]. In another study, the SNPs rs12478635 in the IDH1 gene and rs11632348 in the IDH2 gene exhibited associations with death risk for patients with hepatocellular carcinoma [413].
3.2 Mitochondrial antioxidant system
The reverse electron transport from complex II to complex I in the ETC leads to the formation of reactive oxygen species (ROS). Although low levels of ROS serve as signal transducers and promote normal cellular functions, excessive levels induce mtDNA damage and destabilize the mitochondrial membrane potential, ultimately initiating cancer [347]. Mitochondria, therefore, house antioxidant enzymes that quench these free radicals and prevent cellular damage. Deregulation/mutations in manganese superoxide dismutase (MnSOD/SOD2), glutathione peroxidase (Gpx), and thioredoxin-2 (Trx2) are seen in several human cancers [69]. SOD2 catalyzes the conversion of the superoxide anion (O2−) to hydrogen peroxide (H2O2). The role of SOD2 in cancer initiation is concentration-dependent and akin to that of a “two-edged sword.” Under physiological conditions, SOD2 scavenges ROS and acts primarily as a tumor suppressor. This activity of SOD2 is aided by Sirt2 deacetylase, and loss of the latter promotes oncogenesis. Under high ROS conditions, however, the scavenging properties of SOD2 prevent ROS-induced apoptosis and thereby allow malignant cells to survive [151]. Likewise, in breast cancers and skin carcinomas, SOD2 is associated with a protective effect [209], but it has a pro-tumorigenic function in ovarian clear cell carcinoma [141].
Variants of the SOD2 gene are linked to increased susceptibility to various cancers. Five SNPs are currently identified: rs7855, rs5746151, rs5746136, rs2758331, and rs4880. The rs4880 SNP in the mitochondrial targeting sequence involves a T47C substitution that results in an Ala to Val change in the mitochondrial targeting sequence. The Ala allele is present at a higher frequency in colorectal cancers of Hispanic patients compared to non-Hispanic white patients, even though this SNP is not particularly associated with higher rates of colorectal cancer [348]. In a recent study of pediatric medulloblastoma patients, the T47C SNP was linked to a higher incidence of the disease [43]. In a Chinese population, T47C is associated with a poorer prognosis for gastric cancer [395] and for squamous cell carcinoma [224]. In a Korean cohort, the allelic frequency of the T5482C SNP was higher in patients compared to healthy controls [134]. In the Hispanic population, this substitution is linked to higher incidence of colorectal cancer [348].
Gpx is involved in the removal of peroxides from the cytosol and mitochondria. The mitochondrial isoforms, Gpx1 and Gpx4, act as tumor suppressors by reducing ROS [165, 210]. The Gpx1 Pro198Leu SNP is associated with a higher risk of bladder cancer [55]. Gpx4 expression is downregulated in pancreatic and breast cancer cells [59, 405], and there is a Gpx4 deletion in large B-cell lymphomas and renal cancers [403].
The mitochondrial thioredoxin (Trx) superfamily of thiol-disulfide oxidoreductases comprises Trx2, Trx2 reductase (TrxR2), and Trx-dependent peroxidase (Prx3) [303]. Try2 transfers reducing equivalents from cysteine residues to disulphide bonds and maintains proteins in a reduced state. Trx2 modulates the activities of transcription factors and apoptosis signaling factors. On the other hand, Prx3 reduces H2O2 generated during mitochondrial respiration [145]. The Trx superfamily is overexpressed in human cancers; Trx2 and Prx3 are elevated in multiple myeloma and thymoma, respectively, and protect cancer cells from apoptosis.
3.3 Mitochondrial DNA replication
The replication of mtDNA is regulated by several nuclear-encoded proteins, including DNA polymerase γ (POLG1 and POLG2) [88], TFAM [298], and mitochondrial RNA polymerase (POLMRT) [40]. POLG and TFAM are essential for maintaining mtDNA integrity and copy number. Mutations in either can lead to depletion of mtDNA, which in turn initiates mitochondrial dysfunction. In POLG1, there are numerous somatic and germ-line mutations that are associated with various cancers. Singh et al. detected POLG1 mutations in 63% of breast tumors [340]. In malignant breast tissue, there were 17 mutations spanning over the exonuclease domain and linker region of POLG associated with a complete loss of mtDNA. Mutations in the POLG exonuclease domain correlated with a higher frequency of rare point mutations in the mtDNA control region [93]. A recent study revealed that POLG1 expression is upregulated in myelomas, melanomas, and prostate cancers and is downregulated in lung, head and neck, brain, bladder, and esophageal cancer and in leukemia [338]. In addition, the expression levels of POLG1 correlates with the copy numbers of POLG1. In primary tumors, there is a higher frequency of POLG1 mutations, mostly missense mutations and substitutions. At the 1143 amino acid position in the polymerase domain, there is a missense variation that changes glutamic acid to glycine. The European-American population shows a six-fold higher allele frequency of E1143G compared to the AA population. It remains to be determined if E1143G, along with the POLG1 variants, T251I and P587L, are involved in the predisposition to cancer. As determined with a German cohort of patients, an SNP in the promoter region of POLG (rs2856268, A>G) is associated with higher risk of breast cancer [290]. In the AA population, variants in the CAG repeat sequence length of the POLG gene are associated with increased risk of breast cancer [10] and testicular cancer [32]. As determined for an Indian population, the SNPs, rs41553913 at POLRMT and rs9905016 at POLG2, increase the risk of oral leukoplakia and cancer [85]. The authors speculate that, in cancer tissues, these polymorphisms are associated with increased mtDNA replication, although the mechanism remains elusive.
In colorectal cancers [126] and breast cancers [15], truncating mutations in TFAM are associated with mtDNA depletion and oxidative stress. In murine models, complete knockout of TFAM leads to mtDNA loss, OXPHOS breakdown, and embryonic lethality [373].
3.4 Mitochondrial DNA repair
There are primarily six kinds of lesions that affect mtDNA: 1) Alkylation of bases that is usually induced by chemotherapeutic agents [317]. Alkylation damage can also be caused by the endogenous pool of S-adenosylmethionine that exists in the mitochondria [146]. 2) Hydrolytic deamination of bases and formation of abasic sites [90, 219]. 3) Adduction of DNA bases with carcinogens such as acetaldehyde, cisplatin, and DMBA, resulting in crosslinking of bases [195]. 4) Mismatched bases that occur due to replication errors [170] or incorporation of damaged bases during replication [35]. 5) DNA single- and double-strand breaks that are either induced by carcinogens or occur as a result of inefficient repair of other lesions. 6) Oxidative damage to bases and the sugar-phosphate backbone from ROS.
Due to its proximity to the ETC chain and sites of ROS generation, mtDNA is particularly susceptible to oxidative damage. In human fibroblasts, an H2O2-producing system of glucose and glucose oxidase leads to a greater accumulation of strand breaks, oxidized bases, and abasic sites in mtDNA compared to nDNA [31]. In fact, strand breaks induced by H2O2 occur at a tenfold higher frequency in mtDNA, with the sugar-phosphate backbone as the primary target [337]. Oxidative damage to DNA bases results in two main products: thymine glycol, the main modified pyrimidine [383], and 7,8-dihydro-8oxo-20-deoxyguanosine (8-oxodG), the main modified purine [90]. Although both oxidized bases block DNA polymerase, 8-oxodG is more mutagenic and is responsible for the characteristic G→T substitutions [135]. Other oxidative lesions include 8-hydroxyguanine, FAPy-adenine, 8-hydroxyadenine, 5,6-dihydroxyuracil, 5-hdroxyuracil, 5-hydroxycytosine, and 5-hydroxymethyluracil [5,8].
Previously, the consensus was that the DNA repair pathways in mitochondria were less developed than in the nucleus since it was thought that mitochondria were unable to repair UV-induced dimerization [74] and alkylation damage [259]. Recently, improvements in sub-cellular localization techniques using fluorescent tagging of proteins and immunogold labeling have provided evidence that the major DNA repair pathways also exist in mitochondria [35].
During DNA replication, base mismatches are often introduced by ‘slippage’ errors of DNA polymerase or randomly by alkylation, deamination, or oxidation of the bases. The DNA mismatch repair (MMR) pathway operates in the nucleus to correct these errors. Briefly, the MSH proteins (MutS homolog family) recognize and bind to the mismatched base sites [194], followed by recruitment of the MLH (MutL homolog) proteins [264], downstream excision, and re-synthesis of the DNA strand [252]. MMR was first identified in rat mitochondrial lysates wherein the G→T mismatch was corrected in a bi-directional, ATP-dependent manner [245]. In another study, the mechanism for mitochondrial MMR was somewhat elucidated with the discovery of the MLH1 protein in mammalian mitochondria [332]. However, mitochondrial MMR appears to be largely independent of the MLH proteins, as the Y-box binding protein (YB-1) is the major MMR initiator in mitochondria of HeLa cells [100]. Loss of YB-1 in the HeLa cells lead to higher rates of mtDNA mutagenesis.
The best-understood pathway for DNA repair in mitochondria is base excision repair (BER). Since BER is the preferred pathway for the repair of oxidative DNA base lesions [352], mitochondria use it because of the susceptibility mtDNA to ROS. Indeed, the most common oxidative base lesion, 8-oxodG, is more efficiently repaired in the mitochondria than in the nucleus. The first indication of mitochondrial BER was the discovery of mitochondrial uracil-DNA glycosylase (UDG) by Anderson and Friedberg in 1980 [7]. Later studies provided direct evidence of BER in the mitochondria [103, 201].
Mitochondrial BER starts with a) lesion recognition and strand scission, followed by b) gap tailoring, and converging at the c) DNA ligation step [352]. The initial step of recognizing the DNA lesion is mediated by DNA glycosylases. Mono-functional DNA glycosylases like UDG1 recognize the damaged base and hydrolyze the N-glycosidic bond, leaving the DNA in an abasic state. Following removal of the damaged base, the resulting apurinic/apyrimidinic (AP) site is incised by AP endonuclease. The mitochondrial isoform of APE1 is generated by truncation of the nuclear localization signal [62]. In contrast, bi-functional DNA glycolysis involves intrinsic AP-lyase activity. In mammalian mitochondria, there are at least four bi-functional DNA glycosylases: 8-oxoguanine DNA glycosylase (OGG1) [99], nth-like 1 (NTHL1) [172], Nei-like 1 (NEIL1), and Nei-like 2 (NEIL2) [237].
DNA glycosylases produce single-strand breaks that have a blocking 5′ or 3′ end that must be removed before the correct base can be inserted, and DNA ligation can occur. The 5′-deoxyribose phosphate (dRP) generated by the action of UNG1 and APE1 is removed by the dRP lyase activity of DNA polymerase γ, leaving a single strand gap with 5′-P and 3′-OH ends [226]. Similarly, the 3′-phospho-α,β-unsaturated aldehyde left by OGG1 is removed by the phosphodiesterase activity of APE1 [352], leaving a single-strand gap with 5′-P and 3′-OH ends. The final step is DNA synthesis and ligation by DNA polymerase γ and DNA ligase IIIα, respectively [352].
Variations in OGG1 correlate with various cancers. The C1245G SNP has a Ser326Cys change in the catalytic subunit of OGG1, which has an impaired capacity to excise 8-oxoguanine [160]. In a Brazilian group, this variant of OGG1 was present in 42% of the non-small cell lung carcinoma (NSCLC) patients and in 34% of healthy controls [78]. In a meta-analysis of Asian patients, Ser326Cys was associated higher risk of lung cancer [207]. A higher susceptibility to head and neck cancer was linked to the C1245G SNP [345]. Breast cancer risk is increased in individuals with the OGGI C >G variant [178, 268]. In certain North Indian ethnic groups, the SNP C1245G at exon seven was detected in about a quarter of the population, although its relevance in cancer risk remains to be evaluated [236].
3.5 Transcription factors in the mitochondria
The regulation of mtDNA genes depends on various nuclear-encoded transcription factors. The mitochondria-localized transcription factors, called mitoTFs, mediate respiration, apoptosis, and mtDNA transcription. The mitoTFs are broadly classified into two groups: a) the hormone and steroid receptors, such as those for estrogen, progesterone, androgen, and glucocorticoids and b) downstream factors activated upon binding of hormones and cytokines to their respective receptors (e.g., p53 HER1, and HER2).
3.5.1 Estrogen receptor in the mitochondria
First described by Jenson [162], the estrogen receptor (ER) is present as two isoforms, ERα and ERβ, each encoded by separate genes, ESR1 and ESR2 [187]. ERα and ERβ bind to estrogens, resulting in receptor dimerization and recruitment to the promoter region of target genes at their estrogen response elements (EREs) [278]. ERβ is predominant in mitochondria, whereas ERα is concentrated in the cytosol [49, 66]. Chen et al. [66] identified, in ERβ, a mitochondrial-targeting peptide signal (mTP) that is essential for proteins to be translocated into the mitochondria. Furthermore, the presence of EREs in the mtDNA points to the mitochondrial localization of ERs. The binding of ERβ to mtDNA was confirmed in MCF-7 breast cancer cells [65]. Apart from directly regulating the genes of mtDNA, ER also influences mitochondrial functions indirectly through the transcriptional regulation of nuclear genes encoding mitochondrial proteins. For instance, in MCF-7 cells, ERα activates respiratory factor-1 (NRF-1), which in turn activates TFAM and promotes mitochondrial biogenesis [250].
In breast cancer cells, binding of the estrogen, 17β estradiol (E2), to ERα and ERβ results in an increase in proliferation, survival, and invasion [183]. Also, in MCF-7 cells, exposure to E2 increases the binding of ERα and ERβ to mtDNA and increases the expression of genes encoding cytochrome c oxidase subunits I and II [65]. In lung cancer cells, there is a pro-survival, anti-apoptotic role for ERβ [415]. Tamoxifen, an anti-cancer drug that induces oxidative stress and apoptosis via a mitochondria-dependent pathway also acts through binding with the ERs as both agonist and antagonist [301]. SNPs in the estrogen receptor gene are involved in breast cancer susceptibility. In a meta-analysis, three SNPs (rs2077647:T>C, rs2228480:G>A, and rs3798577:T>C) correlated with breast cancer risk in various ethnic populations [211]. Amongst Caucasians, the rs2228480 AA genotype was associated with a lower risk compared to the GG genotype, whereas the rs3798577TT genotype correlated with increased risk in an Asian population. In all the ethnic groups, rs2077647:T>C was associated with a higher risk of breast cancer, albeit non-significantly. In a recent study, the 13950T/C SNP of ERβ was associated with higher risk of uterine leiomyomas [374].
3.5.2 Progesterone receptor in the mitochondria
The response to progesterone is mediated by two nuclear isoforms of the progesterone receptor (PR), PRA and PRB [16]. The nuclear PRs (nPRs) contain an N-terminal transcriptional regulatory domain, a DNA-binding domain, a D-domain or hinge region, and a hormone binding domain [157]. Although, under normal conditions, the two isoforms are co-expressed in progesterone-targeted tissues (breast, endometrium, cervix, and ovaries), their ratios vary widely during malignant transformation. For instance, PRB is overexpressed in highly invasive forms of cervical, endometrial, and ovarian cancers [112]. In contrast, in breast cancers, an excess of PRA correlates with a poor prognosis, and a greater proportion of PRB predicts better survival [16].
MCF-10A breast epithelial cells lacking nPRs proliferate rapidly in the presence of progestin [23]. This phenomenon is attributed to stimulation of mitochondrial respiration and a concomitant inhibition of apoptosis by progestin. The authors hypothesize two scenarios: an indirect paracrine action of progesterone on mitochondrial function and/or the direct action of progesterone through ligand binding with a mitochondrial PR. In fact, a truncated PR (PR-M) has been cloned from human adipose and aortic cDNA libraries [320]; the encoded protein lacks the DNA-binding and regulatory domains but retains the hormone-binding domain. Subsequently, Dai et al. showed that the PR-M localizes to the mitochondria of T47D breast epithelial cells and HeLa cells [81]. The functionality of PR-M was established by an increase in mitochondrial membranes and increased cellular respiration upon exposure to progestin. Progesterone action via PR-M may be a mechanism for adapting to the increased energy demands of invasive cancer cells. This hypothesis was strengthened by results of a study that correlated increased PR-M expression in leiomyomas with increased mitochondrial membrane potential [108].
Three polymorphisms of the PR (Alu insertion, 331G/Aand, and Val660Leu) have been investigated in the context of cancer susceptibility. In a Chinese cohort, the Alu insertion and Val660Leu polymorphisms were associated with a risk of ovarian cancer, whereas the 331G/A SNP could not be correlated with any cancer risk [213]. In a Caucasian cohort of endometrial cancer patients, the 331G/A SNP could not be correlated with a cancer risk [279], but, in another study, the Alu insertion variant was associated with a higher breast cancer incidence in Mexican women [113].
3.5.3 Androgen receptor in the mitochondria
The androgen receptor (AR), which is activated by ligand-specific binding of the androgenic receptors, testosterone, and dihydrotestosterone [313, 387], is involved in the transcriptional regulation of genes that maintain the male sexual phenotype. Binding of androgens to AR also induces proliferation of prostate cells and initiates prostate cancer; in most prostate cancers, AR is constitutively activated and overexpressed [402]. Solakidis et al. reported the mitochondrial localization of AR in the midsection of spermatozoa (346), and Chaudhary et al. [70] linked the regulation of mitochondrial fission to AR. Mitochondrial fission is initiated upon translocation of the cytoplasmic dynamin-related protein 1 (Drp1) to the mitochondrial outer membrane [408]; impairment of Drp1 results in fragmented mitochondria, which lead to apoptosis [60]. Co-ordination between mitochondrial fission and fusion is essential for optimal cellular proliferation and turnover [19]. In LNCap cells, Drp1 is regulated in an androgen-dependent manner at both the transcriptional and translational levels. The transcript levels of AR and Drp1 are positively correlated, and androgen-mediated Ser-616 phosphorylation of Drp1 reduces mitochondrial fission and promotes cell proliferation [70]. This may be a mechanism utilized by invasive prostate cancer cells to escape apoptosis. It remains to be determined if localization of AR in the mitochondria can be related to mitochondria-mediated survival of cancer cells.
Inherited variations in the AR gene are associated with higher incidences of several types of cancers. The non-coding CAG repeat sequence has been investigated in the context of cancer risk. Generally, repeat lengths <22 are classified as short, whereas those >22 are classified as long. For Mexican men, a lower number of CAG repeats correlates to earlier and more aggressive forms of prostate cancer [118]. For Asian women, there is a link between shorter CAG repeats and a higher risk of developing epithelial ovarian cancer [254]. In contrast, longer CAG repeats are present at a higher frequency in breast cancer patients of the Han Chinese population [82]. Although the CAG repeat length does not affect prostate cancer risk amongst European men, carriers of the SNP rs1204038A allele are more likely to develop this cancer [21]. Koocheckpour et al. performed a comprehensive analysis of the prevalence of germline and somatic AR mutations in AA men suffering from prostate cancer. Compared to Caucasian males, somatic missense AR mutations were more frequent in the AAs. Furthermore, the A-allele of the E213 G/A SNP was more frequent amongst the AA men, although this polymorphism could not be correlated to a higher cancer risk.
3.5.4 Glucocorticoid receptor in the mitochondria
The glucocorticoid receptor (GR), which binds cortisol and other glucocorticoids and is expressed in almost all eukaryotic cells, regulates expression of genes related to inflammation, immunity, metabolism, and development [304]. The mitochondrial localization of GR regulates the expression of mtRNA encoding for the OXPHOS elements [325]. Analysis of human and rodent mtDNA revealed the presence of glucocorticoid-responsive elements (GREs) within the D-loop and within the coding regions of genes such as those for cytochrome oxidase I, ND I, and 12S RNA [152]. As determined by ChiP analysis of mitochondria isolated from hepatocarcinoma cells, the binding of GR was shown in the D-loop and in the ND1 and 12S rRNA sites [294], thus proving a role of GR in transcription of mitochondrial genes.
In the context of tumor physiology, GR induces apoptosis at the mitochondrial level by indirectly upregulating (by repressing miRNA 17-92) the expression of the pro-apoptotic Bcl-2 family member, BIM [266]. BIM, along with other pro-apoptotic proteins such as BAD and BID, causes the release of cytochrome c into the cytosol and triggers caspase-mediated apoptosis [243]. For this reason, glucocorticoids are included in the chemotherapeutic regimens for non-Hodgkin’s leukemia, multiple myeloma, chronic lymphoblastic leukemia (CLL), and ALL [11, 326]. Heideri et al. investigated the GR-mediated apoptotic pathways in the mitochondria, placing them downstream of the p38-MAPK and RUNX1/c-Jun signaling pathways and identifying therapeutic targets in glucocorticoid-resistant leukemias [140]. For Han Chinese, SNPs in the non-coding region of GR were linked to a higher incidence of pediatric ALL [396]. Five SNPs were analyzed, of which the rs41423247 and rs7701443 polymorphisms were significantly associated with a poorer prognosis.
3.5.5 Tumor suppressors in the mitochondria
The tumor suppressor p53 is involved in various cellular functions, including proliferation, senescence, apoptosis, autophagy, metabolism, and differentiation. This is accomplished through transcriptional regulation of an array of target genes and integration of various signaling pathways. In human cancers, this master regulator is the most frequently mutated gene [178]. The regulatory properties of p53 in the cytoplasm have been studied extensively; there is rapid translocation of p53 to the mitochondria (outer membrane) in response to hypoxia, oxidative damage, and DNA damage [9, 238]. Moreover, in p53−/− cancer cells, a p53 fusion protein with a specific mitochondrial targeting sequence induces apoptosis and cell cycle arrest, bypassing the nuclear circuit [238, 257]. There is the translocation of p53 to the mitochondrial matrix [189] wherein p53 forms complexes with the mitochondrial chaperone proteins, Hsp60 and Hsp70 [238]. Of note, p53 is present in mitochondria even in the absence of cellular stress [91], suggesting that endogenous p53 is involved in normal mitochondrial functions. Despite evidence of mitochondrial localization of p53, the mechanism of its entry, in the absence of a mitochondrial targeting sequence [239], is a matter of debate. One hypothesis is that stress-induced binding of Mdm2 to p53 leads to the ubiquitination of p53 and provides a targeting signal to the mitochondria; upon entry, the p53 is deubiquitinated by a mitochondrial ubiquitin-specific protease [239].
There is considerable evidence for various functions of p53 in mitochondria, namely mtDNA maintenance and repair, apoptosis, and respiration. For HCT116 cells, there is an association between the mitochondrial DNA polymerase γ (POLG) and p53, with p53 also binding to mtDNA and enhancing the function of POLG [2]. Association of p53 with POLG also improves the latter’s proofreading capacity [14] during mtDNA replication. Similarly, the gap-filling activity of POLG is impaired in mitochondria derived from the livers of p53−/− mice and is restored by introducing a recombinant p53 [2]. Binding of the mitochondrial transcription factor TFAM to oxidized DNA bases is enhanced upon physical interaction with p53 [409]. The transcriptional regulation of mtDNA genes by p53 has, however, not been confirmed, despite evidence of p53 binding sites in the human mitochondrial genome [142]. The rapid, stress-induced translocation of p53 to the outer mitochondrial membrane is a prelude to the action of p53 in mitochondrial outer membrane permeabilization (MOMP), which releases cytochrome c into the cytosol and triggers the apoptotic machinery [323]. Also, the binding of p53 to pro-apoptotic elements such as Bax [417] and pro-caspase-3 [111] has been reported.
In malignant cells, the tumor suppressive actions of p53 are often directed towards subverting the glycolytic pathway and counteracting the Warburg effect. In leukemia cells, p53 induces the expression of TIGAR (Tp53 induced glycolysis and apoptosis regulator), inhibits glycolysis, and increases the production of ROS, ultimately clearing damaged cells via apoptosis [25]. p53 also prevents the uptake of glucose, the substrate for glycolysis, by blocking the expression of the glucose transporters, GLUT1 and GLUT4 [328]. In cancer cells, p53 hinders glycolysis through the use of alternate substrates. Vousden et al. identified mitochondrial phosphate-activated glutaminase (GLS2) as a transcriptional target of p53 [378]. Activation of GLS2 shifts the cellular metabolism from glycolysis to aerobic respiration and glutaminolysis. Consistent with these findings, a dominant-negative mutant of p53 up-regulates the expression of hexokinase II, which, in AS-30D hepatoma cells, docks at the mitochondrial outer membrane and increases glycolytic activity [246, 247]. Similarly, deletion of p53 in human colon cancer cells leads to decreased expression of cytochrome c oxidase and a disruption in mitochondrial activity and structure [420]. For both murine and human cancer cell lines, Matoba et al. reported the synthesis of cytochrome c oxidase (SCO2), an effector necessary for respiratory chain function, as the downstream target of p53 in the regulation of mitochondrial respiration [248]. Disruption of the SCO2 gene in p53+/+ cells also directed the cellular metabolism towards glycolysis in the same way as a loss of p53. Conversely, disruption of the mitochondrial ETC increases the accumulation of p53 and leads to activation of apoptotic pathways. Both loss of complex I [76] and impairment of complex III function [175] lead to the accumulation of p53.
Although the somatic loss of p53 usually accompanies the development of most human cancers, there are germline mutations/SNPs in the familial Li-Fraumeni syndrome, encompassing cancers such as premenopausal breast cancer, bone and soft-tissue sarcomas, adrenal cortical carcinomas, and brain tumors [255]. The hotspot of polymorphisms in p53 resides in codon 172, which encodes its protein binding and interaction domain. The SNP rs2602141, resulting in an arginine-to-leucine substitution, is associated with leukemia, prostate cancer, esophageal cancer, and lung cancer in Asians and Caucasians but not in other ethnic groups [174].
3.5.6 FOXO3 transcription factor in the mitochondria
The forkhead box (FOX) family of proteins consists of 19 sub-families of transcription factors that share a highly conserved DNA-binding domain of approximately 110 amino acids, the forkhead box domain (also known as the winged-helix domain). Within this family, the O subgroup contains four members: FOXO1 (FKHR), FOXO3 (FKHRL1), FOXO4 (AFX), and FOXO6 [77]. The first three are ubiquitously expressed but at various levels, depending on the tissue [77]. FOXO3 localizes to mitochondria [46], which may contribute to the various physiological and pathophysiological functions of this transcription factor. In tumors of AAs, the expression of FOXO3a is lower by 65% as compared to adjacent normal tissues [206].
3.5.7 Aryl hydrocarbon receptor in the mitochondria
The cellular mechanisms that respond to various toxic compounds involve the aryl hydrocarbon receptor (AHR). The AHR is a ligand-activated transcription factor within the Per-Arnt-Sim (PAS) domain superfamily. Exposure to the most potent AHR ligand, 2,3,7,8-tetrachlorodibenzo-p-dioxin (TCDD), is associated with various pathological effects. A portion of the cellular pool of AHR is found in the mitochondria [150], localized to the inter-membrane space of the organelle [150]. The AHR interacts with ATP5α1, a subunit of the ATP synthase complex, and modulates mitochondrial function [360]. An ethnic variability in the allelic distribution of AHR receptor codon 554 and an assessment of variant receptor function have been described [391].
3.6 miRNAs in mitochondria and their role in cancer
MicroRNAs (miRNAs) are small, non-coding RNAs, with 18–22 nucleotides, which regulate expression of mRNA. The miRNAs bind to 3′ untranslated regions (UTRs) in a sequence-specific manner and either inhibit translation or degrade the mRNA. Post-transcriptional regulation by miRNAs is implicated in proliferation, apoptosis, and differentiation; in several human cancers, miRNAs are de-regulated.
One of the first studies to link miRNA-mediated mitochondrial dysfunction with cancer showed that mir-15a induces an efflux of cytochrome c and disrupts the mitochondrial membrane potential [116]. In CLL, mir-15a is frequently deleted [48]. In various human cancers, mir-143, which targets the pro-proliferative factor ERK5 and induces apoptosis through mitochondrial dysfunction, is downregulated [3]. miRNAs are linked to the control of mitochondrial dynamics and mtDNA integrity, factors associated with cancer initiation. The mir-30 family regulates mitochondrial fission and apoptosis through dynamin-related protein 1 (DRP1) and the p53 axis [208]. In some tumors, miR-21 levels are increased and downregulate the expression of PTEN, deactivate PINK1, and prevent the clearing of damaged mitochondria [253, 415]. In hepatocellular carcinoma cells, mir-200a, which targets the mitochondrial transcription factor TFAM [404] and acts as a tumor suppressor [416], is downregulated in breast cancers [404].
miRNAs are implicated in the regulation of the mitochondrial metabolic apparatus. They modulate several enzymes of the Krebs cycle at the transcriptional level. In glioma cells, miR-183 downregulates the levels of isocitrate dehydrogenase-2 (IDH-2) [358]. Succinate dehydrogenase (SDH) is targeted by miR-210, and, in response to oxidative stress, malate dehydrogenase levels in mouse neuronal cells are elevated through reduction in miR-743a levels [106]. Proteins of the ETC are also targets of miRNAs. Over-expression of miR-338 inhibits the expression of the cytochrome oxidase-IV subunit [208], whereas over-expression of miR-181c leads to down-regulation of the cytochrome oxidase-I subunit [84]. In various cancers, including malignant mesothelioma (MM) [365], gastric cancer [109], and breast cancer [421], miR-126 is frequently deleted. In an MM cell line, mir-216 has a tumor suppressor role [363, 364], and its over-expression down-regulates ATP-citrate lyase and increases production of ATP and citrate. Although cancer cells switch to aerobic glycolysis and divert the metabolites into anabolic pathways to support malignant growth, this proliferative program is suspended during metastasis. Invasive cancer cells instead favor OXPHOS and increased ATP production. In breast cancer cells, this is mediated by the transcription co-activator, PGC-1α [200]. In breast cancer tissues, miR-485-3p and miR-485-5p inhibit PGC-1α to regulate mitochondrial respiration and are consequently down-regulated [227]. An indirect route for inducing cancer is through ROS generation and subsequent disruption of the ETC. By inhibiting the antioxidant enzymes, Mn superoxide dismutase (MnSOD), glutathione peroxidase, and theorductase-2, miR-17* suppresses tumorigenicity of prostate cancer by inducing cancer cell death [394].
Although miRNAs are largely encoded by the nuclear DNA and perform their activities in the cytosol, pre-miRNAs and mature miRNAs are present in human mitochondria [18]. After predicting 33 pre-miRNA and 25 miRNA candidates in the mitochondrial genome by in silico analysis, miR-365, pre-miR302a, and pre-let-7b were co-localized in the mitochondria. Forty-six miRNAs that were at significantly higher levels in the mitochondrial RNA fraction were isolated. Furthermore, putative target sites for these miRNAs – referred to as mitomiRs – were detected in mtDNA. Although the presence of miRNAs in mitochondria is not indicative of their origin, the pre-miRNAs suggest the possibility of post-transcriptional processing of miRNAs in the mitochondria. The functional significance of mitomiRs was underscored by the co-localization of argonaute 2 (Ago2), part of the RNA-induced signaling complex (RISC), with mtDNA-encoded Cox3 mRNA [17]. Further, in the mitochondria of Hela cells, there is an enrichment of 13 nuclear-encoded miRNAs. Regardless of their origin, the mitomiRs contribute to the regulation of genes involved in mitochondrial functions. Thus, in several human cancers, there is de-regulation of mitomiRs, which is associated with mitochondrial dysfunction or mitochondria-associated mechanisms.
3.7 Kinases in mitochondria and their role in cancer
The signaling pathways that regulate and integrate cellular functions are largely dependent on concerted phosphorylation and de-phosphorylation of the pathway mediators and targets. Phosphorylation of proteins occurs via the kinases, and de-phosphorylation depends on the action of phosphatases. Although the regulatory activities of these signaling proteins have been documented for the cytoplasm and the nucleus, especially in the context of steady-state and pathological conditions such as cancer, their specific functions in the mitochondria have only recently been uncovered [216, 309].
3.7.1 Activated protein kinase B (Akt) in mitochondria
Akt, a serine-threonine kinase, is implicated in cellular processes such as apoptosis, metabolism, and proliferation. An early indication of the mitochondrial functions of Akt was the discovery that Akt phosphorylates and inactivates the apoptotic protein BAD, thereby promoting cell survival [86]. A later study implicated Raf-1 in the anti-apoptotic action of Akt, as the expression of a dominant-negative version of the mitochondrial Raf-1 in Akt-expressing cells rendered them susceptible to apoptosis [232]. Subsequently, in response to mitochondrial dysfunction, Akt was detected in the mitochondrial outer and inner membranes [27, 322].
Akt is part of the PI3K-Akt signaling pathway that is active in cancer cells and promotes cell survival by modifying cellular metabolism and inhibiting apoptosis [20]. The functional significance of Akt in mitochondria is highlighted by its role in regulating both glycolysis and OXPHOS. In cancer cells, Akt activates hexokinase II at the outer mitochondrial membrane and promotes glycolysis [120]. However, over-expression of Akt increases mitochondrial respiration in PTEN−/− murine fibroblasts relative to that in wild-type cells [119]. The authors hypothesize that the Akt-mediated increase in OXPHOS could be part of a metabolic adaptation in response to inactivation of the tumor suppressive PTEN. The role of the PI3K-Akt pathway in apoptosis is complex. In a recent study, the PI3K-Akt-mTOR pathway was found to be involved in the chemo-resistance of human seminoma cells through negative regulation of the mitochondrial apoptotic pathway [114]. Furthermore, the activation of HK-II by mitochondrial Akt had an anti-apoptotic effect on the cells [308]. In contrast, a pro-apoptotic role of Akt was seen in HCT11 cells expressing a mutant k-ras oncogene; Akt rapidly translocated into the mitochondria and increased ROS production, leading to cell death [155].
Recently, Buroker reviewed the genetic variants of Akt and their involvement in various cancers [44]. Three rSNPS (rs10157763, rs10927067 and rs2125230) in Akt codon one are associated with an aggressive form of prostate cancer. Other intron one rSNPs (rs4132509, rs12031994, rs2345994) are associated with risk of renal cell carcinoma. All these SNPs reside in the transcription factor binding site and consequently affect downstream signaling pathways.
3.7.2 Pyruvate dehydrogenase kinase 1 in mitochondria
Pyruvate dehydrogenase kinase 1 (PDK1) is involved in the phosphorylation and inactivation of pyruvate dehydrogenase (PDH). PDK1, along with the pyruvate dehydrogenase complex (PDC), resides in the mitochondrial matrix and is involved in balancing the metabolic output of mitochondria between glycolysis, the citric acid cycle, and OXPHOS.
In a recent study, the ‘gatekeeper’ role of PDK1 (and thus cellular metabolism) was elucidated in oncogene-induced senescence [171]. Senescence is commonly defined as a block in cellular proliferation with no cessation in the metabolic activities of cells. Oncogene-induced senescence (OIS) is hypothesized to be a protective mechanism that withdraws the cells from the proliferative pool and thus averts malignant transformation [58]. In human melanoma cells, forced expression of the oncogene BRAF leads to a senescent phenotype with the suppression of PDK1 and simultaneous induction of the PDH-activating enzyme, pyruvate dehydrogenase phosphatase 2 (PDP2). The resulting activation of PDH increases respiration and production of ROS. Upon forced expression of either PDK1 or PDP2, OIS is abolished, leading to the development of BRAF melanomas. Consistent with these results, the RNAi-mediated attenuation of PDK1 and the EGF receptor in glioblastoma cells reversed the Warburg effect towards OXPHOS and inhibited the growth of glioblastomas [371].
3.6.3 HER1 and HER2 kinases in mitochondria
The epidermal growth factor receptor (EGFR) family of receptor tyrosine kinases consists of four members: EGFR/ErbB-1/HER-1, ErbB-2/HER-2/neu, ErbB-3/HER-3, and ErbB-4/HER-4; all except ErbB-3 are associated with tyrosine kinase activity. Ligand-bound, activated EGFR family receptors trigger downstream signaling pathways, including PI3K, MAPK, STAT, phospholipase C, and Ca2+/calceneurin [73]. The HER1 and HER2 kinases, which are involved in cancer initiation, progression, invasiveness, and metastasis, are expressed and/or constitutively activated in various human malignancies, especially in breast cancer [225], lung cancer [117], and glioblastoma multiforme [148].
The mitochondrial localization of HER1 was established by use of immunofluorescence and immune-electron microscopy [412]. The translocation of HER1 into mitochondria is facilitated by EGF-dependent binding to the cytochrome c subunit II (COX2); the resulting phosphorylation of COX2 leads to a decline in CoxII activity, with a resultant drop in cellular ATP levels [96]. The same study also reported a putative mitochondrial localization sequence in HER1. A later study showed that the mitochondrial uptake of HER1 was increased upon exposure to apoptosis inducers and that, in cancer cells, mitochondrial targeting of HER1 was responsible for drug resistance [115, 116]. Together, these findings indicate that mitochondrial HER is involved in modulating metabolism, apoptosis, and cell survival. Further studies are needed to clarify the connection between the mitochondrial localization of HER1 and oncogenesis. The presence of HER2 in mitochondria was reported by Ding et al., who linked HER2 with the regulation of metabolism and resistance to treatment [102]. HER2 possibly interacts with complex IV of the ETC [349].
A dinucleotide CA repeat polymorphism in intron 1 of HER1, ranging from 14 to 21 repeats, has been implicated in the regulation of EGFR expression. Liu et al. evaluated the allelic distribution of this polymorphism in various ethnic groups [222]. The longer allele 20 is higher in Asian individuals; the shorter allele 16 is the most common allele in Caucasians and AAs. In a retrospective study by Nomura et al., both the longer and shorter CA allele was found to be present in East Asian individuals with non-small cell lung carcinoma, with the latter present at a higher frequency [276]. In the same study, two SNPs, G216T and C191A, were present at higher frequencies in lung cancer tissues of patients of Northern European and AA descent compared to East Asians. Another functional SNP, rs4444903, is associated with a higher risk of hepatocellular carcinoma [1]. In Austrian women, a polymorphism in the HER2 codon 655, rs1136201, corresponds to a more aggressive form of breast cancer [384].
Also localized in the mitochondria are the kinases, Abl, ATM, Src, JNK, ERK1/2, P38MAPK, GSK3B, PKA, PKC, and PINK1 [216]. However, their role in cancer disparities is unknown.
3.8 Phosphatases in mitochondria
Both protein and lipid phosphatases localize in the mitochondria [216]. Protein phosphatases include MAP kinase phosphatase (MKP1), Src homology two domain-containing phosphatase 2 (Shp2), and protein tyrosine phosphatases (PTPs). A lipid phosphatase, PTEN, also localizes to the mitochondria. To date, a role for these mitochondrial phosphatases in cancer disparities is not described.
3.9 Oncogenes in mitochondria
The Ras family of proteins is small GTPases that function as binary molecular ‘switches,’ depending on their GTP/GDP binding status [375]. There are four proteins in the Ras family: HRAS, NRAS, KRASA, and KRASB, the latter two being splice variants of KRAS [295]. The Ras proto-oncogenes are expressed in all cell lineages and are mutated in 20–30% of human tumors [39]. Ras-mediated oncogenesis involves the downstream PI3K-Akt pathway and leads to increased cell proliferation, survival, and invasiveness [128]. The localization of all Ras isoforms in the mitochondria has been confirmed [302,306, 310, 390].
Mitochondrial Ras affects metabolism, apoptosis, and mitochondrial biogenesis, which can often be correlated with tumor initiation and/or maintenance. In a murine model of acute myeloid leukemia, NRAS was associated with the anti-apoptotic Bcl-2 protein and protected the leukemic cells from apoptosis [277]. Similarly, mouse fibroblasts lacking NRAS displayed mitochondrial dysfunction and increased apoptosis that could be alleviated upon mitochondrially targeted delivery of NRAS [390]. Cells transformed with the K-ras gene, however, were susceptible to apoptosis and were involved in protein kinase C (PKC)-mediated translocation of KRAS to the outer mitochondrial membrane in association with the Bcl-XL anti-apoptotic protein 30]. In the mitochondria, mutant KRAS also increase ROS via the PI3K-Akt pathway, leading to cell death. In HEK293 cells, translocation of HRAS and KRAS into the mitochondria leads to a reduction of mitochondrial membrane potential and respiration, thus causing a metabolic shift to glycolysis [406]. In glioblastoma cells, RAS-mediated signaling is implicated in the inactivation of pyruvate dehydrogenase, the enzyme that feeds acetyl-CoA into the citric acid cycle for cellular respiration [291]. For human hepatocellular carcinoma, KRAS-induced mitophagy (clearance of diseased mitochondria) and the resulting loss of mitochondria has been linked to early tumorigenesis [180].
A germline SNP, rs61764370, located in the 3′UTR of the KRAS oncogene alters the binding capacity of the let-7 miRNA. Uvirova et al. found, in a Czech cohort, an association of this somatic variant across breast cancers, colorectal cancers, non-small cell lung cancers, and brain tumors [368]. In another study, the rs61764370 SNP was associated with a higher risk of chronic myeloid leukemia in Mexican-Mestizo women [129]. An indication of the role of HRAS polymorphisms in cancer susceptibility came from a study of an Italian gastric cancer cohort. Thirteen rare inheritable variants were detected in the tandem-repeat sequence downstream of the structural part of the HRAS gene and were correlated with a higher incidence of cancer [297]. In a meta-analysis of the prevalence of genetic polymorphisms in breast cancer, rare alleles of HRAS were associated with higher risk [288]. Regarding NRAS, the only relevant genetic variant discovered is the G138R SNP, which is associated with higher colorectal cancer risk among Taiwanese patients [61].
4. DISPARITIES AT THE NUCLEAR MITOCHONDRIAL PROTEIN LEVEL AND THEIR ROLE IN CANCER
Although most mitochondrial proteins are synthesized in the cytosol by cytoplasmic ribosomes and then translocated into the mitochondria with the help of the mitochondrial translocase complex, several mitochondria-targeted proteins are not exclusively mitochondrial. This dual targeting could involve other organelles, including the endoplasmic reticulum, the nucleus, and the cytosol. The shuttling of proteins between the mitochondria and nucleus can be passive and, especially if the proteins are smaller than 25–30 kDa or selective require the presence of nuclear or mitochondrial localization signals [407]. Examples of these dual-localized proteins are the transcription factors and kinases that have been discussed in previous sections. A small number of primarily mitochondrial proteins can also be found in the nucleus; the functions of the proteins could be different in the two organelles.
Fumarate hydratase (FDH), the enzyme that catalyzes a reversible conversion of fumarate to malate, is a TCA cycle enzyme that is localized and active in the mitochondrial matrix. To elucidate the role of a cytosolic version of FDH, Yogev et al. generated a nuclear fum−/− yeast strain in which a copy of the gene was inserted into the mitochondria genome [407]. Although the mutant cells had a normal TCA cycle, they displayed a higher sensitivity to radiation-induced DNA damage. This effect was reversed when a cytosolic version of FDH (lacking the mitochondrial targeting signal) was introduced into these cells. Furthermore, FDH was translocated into the nucleus of HeLa cells exposed to radiation or hydroxyurea. These findings point to a role of FDH in DNA damage in the nucleus. This hypothesis is strengthened by the increased predisposition to renal cancer and leiomyomatosis following bi-allelic loss of the FDH gene.
The pyruvate dehydrogenase complex (PDC), a large multi-protein complex residing in the mitochondrial matrix, generates acetyl-CoA and feeds it into the TCA cycle. PDC, localized in the nucleus, is involved in the acetyl-CoA synthesis and histone acetylation [351]. Upon mutagenic stress, the nuclear levels of PDC increased, and the mitochondrial levels concomitantly decreased, prompting the authors to postulate that the PDC is translocated from the mitochondria to the nucleus as an intact complex. Further studies are needed to correlate this function of PDC to cancer initiation.
The mitochondrial coenzyme Q biosynthesis protein 7 (COQ7) is a monooxygenase catalyzing the hydroxylation of 5-demethoxyubiquinone to 5-hydroxyubiquinone. Although the presence of a mitochondrial localization signal marks COQ7 as an exclusively mitochondrial protein, Monaghan et al. have recently reported its localization in the nuclei of HeLa cells [267]. This was attributed to a nuclear localization signal that could be cleaved before transfer into the mitochondria. Accumulation of COQ7 in the nucleus is stimulated by ROS and lowered by antioxidants. Moreover, in human cells, deletion of COQ7 increases the levels of ROS and the ROS-responsive genes, SOD2 and NRF2. COQ7 is associated with chromatin and implicated in the transcriptional regulation of TIMM22 (mitochondrial import inner membrane translocase subunit Tim22) [327].
5. DISPARITY AT THE LEVEL OF MITOCHONDRIA-TO-NUCLEUS CROSS TALK AND ITS ROLE IN CANCER
In human cells, mitochondrial dysfunctions lead to mitochondria-to-nucleus retrograde responses. Mitochondria are multi-tasking organelles that integrate pathways of cellular respiration, biosynthesis, apoptosis, and redox potential. Due to the paucity of genes in the mitochondrial genome, most proteins and regulatory RNAs needed for mitochondrial structure and function are encoded by the nuclear genome. Thus, to maintain functioning mitochondria and a healthy cell, constant communication between the nucleus and mitochondria is required. Deregulated expression of nuclear genes encoding the mitochondrial proteins can result in various pathologies. Further, mitochondrial dysfunction arising from cellular/environmental imbalances can signal changes in nuclear gene expression, leading to cellular adaptation. The mito-nuclear cross talk can, therefore, be split into two broad categories: 1) anterograde signaling that involves the regulation of mitochondrial biogenesis and functions through nuclear-encoded transcription factors and other proteins and 2) retrograde signaling that relays metabolic and oxidative changes in the mitochondria to the nucleus and leads to reconfiguration of the nuclear transcriptome.
5.1 Mitochondrial retrograde signaling
Mitochondrial retrograde signaling, first observed in Saccharomyces cerevisiae [214], has been reported in mammalian cell. The main ‘sensors’ that trigger a retrograde response include mtDNA mutations and/or loss in copy numbers, disruption of the OXPHOS machinery, an increase in oxidative stress, and loss of mitochondrial membrane potential. Retrograde signaling causes changes in nuclear gene expression that can lead to metabolic rearrangement (relevant in the context of cancer cells), apoptosis, mitophagy, and DNA repair [28, 45, 122, 161]. Mitochondrial retrograde signaling is linked to various pathological conditions. Shoffner et al. showed that, in the MERRF (myoclonic epilepsy with ragged red fibers) condition, disruption of the ETC and the resulting low production of ATP-triggered retrograde signaling [336]. To compensate for impaired muscle activity, the proliferation of the damaged mitochondria was increased in the muscle cells. A direct association between mitochondrial dysfunction and retrograde signaling and aging has also been found by use of DNA polymerase γ knock-in (Polg−/−) mice. A knock-in D257A mutation in the exonuclease domain of POLG abolishes its DNA-proofreading capacity and increases the frequency of mtDNA mutations, with the mice displaying signs of premature aging [188, 366]. Mitochondrial retrograde signaling has also been implicated as a causal factor in tumorigenesis [29, 36, 37, 56, 94, 95, 193]. In particular, studies involving murine models and cybrids have established a causal link between mtDNA defects and cancer [169]. Mice heterozygous for TFAM, the regulator of mtDNA replication and transcription, were crossed with mice heterozygous for adenomatous polyposis coli multiple intestinal neoplasias (APCMin+/−). The double-heterozygote offspring showed an increase in the rates of tumor incidence and growth [392].
Even though various nuclear targets of stress-initiated retrograde signaling have been identified, the complete genome-wide rearrangement remains to be characterized. The transcriptional profiles of cells responding to mitochondrial retrograde signaling have been established and compared that to those of the ‘resting’ state. To determine changes mediated by retrograde signaling, most of these investigations involved use of Rho0 or mtDNA-depleted versions of the parental cells [28, 29, 94, 95, 97, 122–125, 231, 258]. The consensus is that the genes affected by retrograde signaling control diverse functions, in particular, mitochondrial biogenesis, metabolism, cell adhesion, metastasis, and apoptosis. In Rho0 cells, Guha et al. found higher expression of the general transcription factor TFIIH and the hematopoietic-specific AML1 (RUNX1). Delsite et al. observed that Rho0 cells display reduced expression of cytochrome p450 (metabolism), CDK inhibitor p19 (cell differentiation), and signaling molecules (PKCγ and protein tyrosine phosphatase C) [94]. For Rho0 breast epithelial cells, Kulawiec et al. reported an alternate regulation of the p53-mediated signaling, leading to increases in DNA breaks and chromosomal rearrangements [192]. In osteosarcoma cells, there is up-regulation of HIF1α, which confers metabolic advantages to malignant cells. The retrograde signaling activates the JNK and PGC1α transcription factors that initiate pro-survival and mitochondrial biogenesis pathways [156].
In tumor cells, the metabolic switch and retrograde signaling often re-program the genetic components of cellular respiration. In several tumor cell lines, there is activation of the Akt transcription factor in response to mitochondrial respiratory stress [122, 286]. ROS-mediated disruption of complex I of the ETC chain is linked with Akt activation [330]. Moreover, Akt phosphorylates a transcriptional co-activator, hnRNAP2 (heterogeneous ribonucleoprotein A), which is overexpressed during retrograde signaling. Implicated in the post-transcriptional processing of mRNA [242, 385], hnRNPA2 has been considered, in several clinical studies, as an early biomarker for lung epithelial carcinoma and metastasis [362, 419]. There is over-expression of hnRNAP2 in cancers of the brain, colon, breast, cervix, and ovary [285]. Guha et al. confirmed that hnRNPA2 is up-regulated in response to low mtDNA and mitochondrial stress signaling; it acts as a proto-oncogene by increasing the proliferation, survival, and invasiveness of cancer cells. Silencing of hnRNPA2 leads to apoptosis [123].
The regulatory action of hnRNPA2 in enabling cells to adapt to a malignant condition (such as through a metabolic shift) occurs by two mechanisms. It acts as a co-activator of transcription factors such as Akt and IGHR1 [122, 123], and it introduces alternative splicing as a means of regulation. In breast cancer cells, the hnRNPA 1 and two proteins modulate expression of the pyruvate kinase gene by alternative splicing, an action that leads to expression of pyruvate kinase isoform M (PKM) [67, 86]. This embryonic isoform, which is re-expressed in cancers, promotes aerobic glycolysis, as opposed to the adult isoform, PKM1, which promotes OXPHOS [72]. In breast and colorectal epithelial cells, alternative splicing of the GTPase Rac1b by hnRNPA1 correlates with cancerous transformation [287]. Furthermore, in hepatocellular cancer cells, hnRNPA2 binds to the telomeric repeat TTAGGGn sequence and human telomerase (hTERT), probably to maintain telomere length and indirectly affect the checkpoints for cell cycle arrest [262]. This is an indication of an epigenetic link between mitochondrial dysfunction (and retrograde signaling) and nuclear gene expression.
There are several mechanisms of mitochondrial retrograde signaling. The most widely studied are a) ROS-mediated, b) the mitochondrial unfolding protein response, c) Ca2+ gradient and calcineurin, d) NAD+/NADH-mediated, e) ATP-mediated, AMP-activated protein kinase signaling, and f) inter-genomic mipigenetic mechanisms. These may respond to different stimuli, but they often share various signaling molecules and converge on the common goal of nuclear genome regulation.
5.2 Mitochondrial ROS-mediated signaling
Inefficient electron transport through the ETC complexes leads to formation of free radicals or ROS, which, upon exceeding the capacity of the antioxidant enzymes, leads to damage of the ETC and respiratory stress [181, 399]. The production of ROS in mitochondria is enhanced by various stresses, including hypoxia, chemotherapy, radiation, and other pathophysiological injuries [110]. Neurological disorders such as the NARP (neuropathy, ataxia, and retinitis pigmentosa) syndrome are ascribed to the action of ROS on mitochondrial biogenesis [389]. Mitochondrial ROS production and ROS-mediated signaling are the main causes of lung fibrosis following alveolar injury [418]. Apart from inducing retrograde signaling, ROS also acts indirectly by oxidatively damaging mtDNA, as the proximity of mtDNA to the ETC makes it particularly susceptible to mutations.
ROS-mediated signaling is implicated in tumor initiation and progression. Formentini et al. demonstrated that human colorectal cancer cells overexpress ATPase inhibitory factor 1 (IFI1), which depolarizes the mitochondrial membrane and increases ROS production, which, in turn, activates NFkB and pro-survival genes [110]. The IFI1-ROS nexus is also responsible for the metabolic switch of cancer cells to aerobic glycolysis [319]. Increased production of ROS activates HIF1α and HIF2α, which, in melanomas, turn on expression of the Src oncogene and promote metastasis [136]. With cybrid technology, Ishikawa et al. showed that ROS-induced mutations in the mtDNA-encoded NADH dehydrogenase subunit 6 (ND6) increased the metastatic potential of tumor cells engrafted in mice [154]. In cancer cells, ROS-mediated deregulation of mitochondrial redox leads to activation of the Akt proliferation and survival pathways [286]. Singh’s group has demonstrated that retrograde mitochondria-to-nucleus signaling involves regulation of NADPH oxidase (Nox1) and that this enzyme is over-expressed in most breast and ovarian tumors [58].
5.3 Mitochondrial unfolded protein in signaling
The mitochondrial unfolded protein response (UPR) is a type of retrograde signaling that is initiated upon the efflux of peptides from the damaged mitochondrial matrix proteins to the cytosol [182, 228]. These peptides can then translocate into the nucleus and activate nuclear transcription factors. By maintaining an equilibrium between folded and unfolded proteins, the UPR is responsible for mitochondrial integrity and biogenesis. Mitochondria have molecular chaperones to facilitate protein folding and proteases that degrade misfolded proteins. Increased accumulation of unfolded, misfolded, and unassembled proteins due to damage to the protein-folding machinery can lead to organelle dysfunction. Due to the potential sensory function of damaged mitochondrial peptides, the mtUPR is recognized for its role in synchronizing expression of mitochondrial and nuclear genes [168]. Currently, two pathways of mtUPR are postulated in mammalian cells. In the first model [316], accumulation of unfolded proteins in the mitochondrial matrix with the help of heat-shock protein (HSP) chaperones leads to their cleavage by Clp proteases and subsequent efflux of the peptides. The damaged peptides then activate JNK-mediated stress signaling. In the second model, accumulation of unfolded proteins in the inter-membrane space, mtUPR is mediated by the estrogen receptor. ERα-mediated mtUPR upregulates the transcriptional activators, NRF1 and NRF2, which control mitochondrial biogenesis [280].
In various patho-physiological conditions, such as neurodegenerative disorders and aging, the mtUPR acts as a rapid stress response. The accumulation of amyloid beta protein in the mitochondria is a characteristic of Alzheimer’s disease (AD) [57]. Although amyloid β protein is cleaved by the mitochondrial HTRA2 chaperone/Omi serine protease complex, a direct role of mtUPR in the pathology of AD has not been demonstrated [283]. There is an accumulation of α-synuclein in the brain mitochondria of Parkinson’s disease patients (PD) [101]. The added presence of HSP60 in the brains of these patients is an indication of the involvement of mtUPR in PD [100, 330]. mtUPR is implicated in the regulation of the aging of hematopoietic stem cells (HSCs) [265]. Activation of mtUPR in aged HSCs results in an induction of the NAD+-dependent deacetylase, SIRT7. SIRT7 binds to the transcription factor, NRF1, which attenuates mitochondrial translation, leads to HSC quiescence and prevents differentiation.
5.4 Mitochondrial Ca2+ signaling
Mitochondria maintain a Ca2+ gradient across the inner membrane by use of gated Ca2+ channels and Na+/H+-Ca2+ exchangers [295]. The uptake and release of Ca2+ regulate various physiological processes, including ATP production. Disruption of mitochondrial Ca2+ signaling triggers the opening of permeability transition pores (mPTPs) and initiates the apoptotic cascade [131, 354]. Since the uptake of Ca2+ depends on the mitochondrial membrane potential, any disruption in the latter due to low mtDNA copy numbers, the breakdown of mitochondrial respiration, or treatment with mitochondrial ionophores disrupts the uptake of Ca2+. The leakage of Ca2+ into the cytosol leads to activation of the phosphatase, calcineurin (Cn) [6, 124]. Ca2+/Cn-mediated retrograde signaling culminates in changes in the expression of an array of kinases, phosphatases, and nuclear transcription factors that affect Ca2+ storage, mitochondrial biogenesis, mtDNA transcription, cell survival, and apoptosis [6, 29, 124].
The signaling pathways activated by the Ca2+/Cn relay in mouse skeletal myoblast C2C12 cells have been studied by Guha et al. Increased expression of Cn leads to activation of calmodulin-dependent protein kinase (CAMK), which activates the downstream targets, Akt Ca2+/Cn-dependent transcription factor (NFAT), cAMP response element binding protein (CREB), CCAAT enhancer binding protein delta (CEBPδ), and NFκB, which are then translocated into the nucleus.
5.5 Mitochondrial signaling mediated by NAD+/NADH
Nicotinamide adenine dinucleotide (NAD) in its oxidized (NAD+, electron acceptor) and reduced (NADH, electron donor) forms is a metabolite that is localized to mitochondria, cytoplasm, and in the nucleus. As a coenzyme, NAD regulates electron transport and the TCA cycle in the mitochondria. It also regulates glycolysis [53, 318, 406]. NAD+ serves as a substrate for two families of proteins involved in response to the DNA damage. These proteins include the PARP family and the sirtuin family of NAD+-dependent deacetylases (SIRTs) [278, 318].
Of the 17 members of PARP family, PARP1, 2, and 3 functions in DNA repair. PARP1 and PARP2 are involved in base-excision repair (BER). However, PARP3 is involved in signaling double-strand breaks (DSB). The PARPs, when sensing DSBs in the nuclear DNA, synthesize poly(ADP-ribose). PARPs utilize NAD+ as a substrate and transfer the ADP-ribose moiety of NAD+ to acceptor proteins. PARP1 and PARP2 are then auto-PARylated, and further recruit and PARylate other repair enzymes [34, 47, 315, 318]. PARP1, which localize to the mitochondria [311], PARylate POLG and EXOG and, in contrast to its function in the nucleus, inhibit BER [355].
The sirtuin (SIRT) proteins also use NAD+ as a substrate. SIRTs deacetylate proteins by the removal of acetyl groups from lysine residues [256]. The SIRT family contains seven members. SIRT1, SIRT6, and SIRT7 localize in the nucleus; SIRT2 in the cytoplasm; and SIRT3, SIRT4, and SIRT5 in the mitochondria [256, 278]. SIRT1 regulates mitochondrial biogenesis. It deacetylates peroxisome proliferator-activated receptor-γ co-activator 1α (PGC-1α). An increase in Ca2+ levels in the mitochondria influences the NAD+/NADH ratio in the cytoplasm by causing an efflux of NADH from the mitochondria to the cytosol [240]. This change influences the NAD+-consuming SIRT enzymes and provides a link between Ca2+ fluctuations, NAD+ levels, DNA damage and repair, and mitochondrial dysfunction [240].
5.6 ATP-mediated signaling
AMP-activated protein kinase (AMPK), a sensor of intracellular energy (ATP:AMP ratio) levels [334], is a serine/threonine kinase that triggers orchestrated responses to restore ATP levels and maintain energy balance [51, 334]. p53 is a target of AMPK phosphorylation, and this phosphorylation triggers the accumulation of mitochondrial p53 that promotes apoptosis via the Bak-Bcl-xL complex [166]. In response to oxidative stress, the ataxia telangiectasia mutated (ATM) kinase phosphorylates AMPK [333]. Phosphorylated AMPK, in turn, phosphorylates SIRT1 and PGC-1α, which are involved in mitochondrial nuclear signaling responses [144, 158, 314]. Mitochondrial SIRT4 also contributes to mito-nuclear crosstalk via AMPK-mediated signaling [181].
5.7 Mito-nuclear mechanisms underlying tumorigenesis
The mipigenetic mechanism is defined as mito-nuclear intergenomic cross-talk at epigenetic and genetic levels involving gene expression and genomic instability [339]. Epigenetic and genetic mechanisms regulate gene expression. Our studies have identified an effect on the epigenetic landscape of the nuclear genome as a consequence of mitochondrial dysfunction. In particular, we demonstrated reversible and irreversible changes in genomic DNA methylation profiles of the nuclear genome [258].
5.7.1 Epigenetic changes in nuclear genome
Perturbed DNA methylation profiles of certain loci within the human nuclear genome are associated with depleted mtDNA. Depletion of the mitochondrial genome in two different cell types resulted in the aberrant methylation of promoter CpG islands (high CG-rich regions) that were unmethylated in the parental cell line [258]. Repletion of mtDNA resulted in the partial re-establishment of methylation profiles back to their original parental state. These results provide an insight into the role of mitochondria in establishing or maintaining nuclear DNA methylation at the genomic level [258].
There is an interdependent relationship between mitochondria and the nuclear genome, including DNA methylation in both the nucleus and the mitochondria [258]. Mammalian mitochondria have mitochondrial DNA methyltransferase 1 (mtDNMT1) activity, 5-methylcytosine (5mC), and 5-hydroxymethylcytosine (5hmC). Shock et al. showed that translocation of nuclear DNMT1 to the mitochondrial matrix is regulated by expression of a conserved mitochondria-targeting sequence, upstream of the gene’s transcription start site within the nuclear-encoded gene [335]. Alterations in mtDNMT1 affect transcription from the light and heavy strands of mtDNA, suggesting a correlation between 5hmC- and 5mC-mediated transcriptional regulation of mtDNA by a nuclear-encoded gene. These findings provide evidence implicating epigenetic regulation of the mitochondrial genome by nuclear-encoded, translocated mtDNMT1 relative to mitochondrial dysfunction. Takasugi et al. described epigenetic regulatory mechanisms affecting the transcriptional control of nuclear DNA-encoded mitochondrial proteins [356]. They used a systematic approach to identifying the presence of differentially methylated regions within nuclear-encoded mitochondrial proteins in a tissue-dependent manner and demonstrated the presence of differentially methylated regions in 636 of 899 nuclear-encoded mitochondrial genes that have functional roles related to the mitochondria.
5.7.2 Genetic changes in the nuclear genome
We have demonstrated that mtDNA dysfunction in human cells to leads a reduced rate of repair of oxidative DNA damage and to increased frequencies of mutation within the nuclear genome [95]. The mitochondria-mediated mutator phenotype is suppressed by inactivating subunits of error-prone DNA repair. Error-prone repair polymerases are involved in the bypass of several types of DNA lesions that have the potential to inhibit chromosome replication [299]. Thus, mitochondrial dysfunction limits or decreases nuclear DNA repair, resulting in unrepaired DNA lesions, which are subsequently converted into mutations by error-prone repair. Increased error-prone bypass of DNA lesions may be characteristic of imbalanced pools of deoxyribose nucleoside triphosphates (dNTPs). Mitochondrial dysfunction induces an imbalanced pool of dNTPs [97]. nDNA mutations may activate proto-oncogenes and/or inactivate tumor suppressor genes, leading to genomic instability, which is involved in the development of cancers.
The functional state of mitochondria in human cells is monitored by the mitochondria damage checkpoint (mitocheckpoint) [258, 343]. The mitocheckpoint coordinates and maintains the balance between apoptotic and anti-apoptotic signals. When mitochondrial dysfunction occurs, the mitocheckpoint is activated to restore normal mitochondrial function and avoid production of mitochondria-defective cells. This response induces changes to the nuclear epigenome (Figure 3) [258].
Figure 3. Differences in mipigenetic mechanisms may contribute to cancer disparity.
Schematic depiction of mechanisms probably involved in cancers induced by mitochondrial dysfunction. Mitochondrial dysfunction or suboptimal function can arise due to mutations or variants in genes involved in nuclear genes encoding the mitochondrial proteome or mitochondrial genes encoding the OXPHOS. See text and Figure 2 for variants described in AAs, Caucasian Americans, and other ethnic populations. A transient mitochondrial dysfunction turns on the mitocheckpoint control to restore the normal mitochondrial function. Restoration involves intergenomic mipigenetic cross talk at epigenetic (e.g., DNA methylation) and genetic levels (gene expression). A severe mitochondrial dysfunction may result in cellular senescence, leading to apoptosis and mitochondrial diseases. A persistent mitochondrial dysfunction arising due to mutations in either nuclear or mitochondrial genes can cause senescence and result in genetic instability in the nuclear genome as well as resistance to apoptosis, underlying tumorigenic cell transformation, and development of cancer.
If mitochondria are severely dysfunctional, they can trigger senescence resulting in apoptosis and mitochondrial diseases (Figure 3). If mitochondrial dysfunction is persistent and defective, mitochondria accumulate in the cell, leading to instability of the nuclear genome [97, 299]. Nuclear genome instability causes cells to acquire new functions, such as resistance to apoptosis and cellular transformation [281, 282], migration, and invasive characteristics involved in tumorigenesis (Figure 3) [191, 258]. Cellular senescence provides a barrier to tumorigenesis [293]. Our studies suggest that a mitochondrial defect leads to cellular senescence and that senescence bypass is induced due to the instability of the nuclear genome [282]. We propose that mitochondrial dysfunction-induced cellular senescence acts as an additional mitocheckpoint mechanism that suppresses tumor development (Figure 3).
6. CONCLUSIONS AND FUTURE PERSPECTIVE
We have presented a list of genetic factors that relate to mitochondrial function. We suggest that variants or mutations in either mitochondrial or nuclear genome-encoded mitochondrial proteins contribute to mitochondrial dysfunction or to suboptimal mitochondrial function, which contributes to cancer diversity and tumor aggressiveness related to racial disparities. Ethnic differences, either at the level of expression or genetic variations in mitochondria-localized transcription factors, kinases and phosphatases, and tumor suppressors and oncogenes may underlie susceptibility to cancer and aggressiveness of cancers frequently observed in AAs and other populations. Differences in mitochondrial function may alter the mipigenetic cross talk between mitochondria and the nucleus and contribute to cancer disparities. Agents that restore mitochondrial function to optimal levels should permit sensitivity to anticancer agents, including radiation treatment of aggressive tumors, thereby reducing racial disparities.
Acknowledgments
This study was supported by NIH grant R01 CA204430 and VA 5I0I BX001716. We apologize to colleagues whose work we were not able to cite due to space limitations or might have missed inadvertently.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
8. CONFLICT OF INTEREST
Authors declare that they have no conflict of interest.
References
- 1.Abu Dayyeh BK, Yang M, Fuchs BC, Karl DL, Yamada S, Sninsky JJ, O’Brien TR, Dienstag JL, Tanabe KK, Chung RT, Morishima C, Gretch DR, Apodaca MC, Shankar R, Antonov N, Snow KK, Stoddard AM, Curto TM, Goodman ZD, David GL, Garcia-Tsao G, Kutner M, Lemon SM, Perrillo RP. A functional polymorphism in the epidermal growth factor gene is associated with risk for hepatocellular carcinoma. Gastroenterology. 2001;141:141–49. doi: 10.1053/j.gastro.2011.03.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Achanta G, Sasaki R, Feng L, Carew JS, Lu W, Pelicano H, Keating MJ, Huang P. Novel role of p53 in maintaining mitochondrial genetic stability through interaction with DNA Pol gamma. EMBO J. 2005;24:3482–92. doi: 10.1038/sj.emboj.7600819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Akao Y, Nakagawa Y, Naoe T. MicroRNA-143 and -145 in colon cancer. DNA Cell Biol. 2007;26:3113–20. doi: 10.1089/dna.2006.0550. [DOI] [PubMed] [Google Scholar]
- 4.Akouchekian M, Houshmand M, Akbari MH, Kamalidehghan B, Dehghan M. Analysis of mitochondrial ND1 gene in human colorectal cancer. J Res Med Sci. 2001;16:50–55. [PMC free article] [PubMed] [Google Scholar]
- 5.Alexeyev MF. Is there more to aging than mitochondrial DNA and reactive oxygen species? FEBS J. 2009;276:5768–87. doi: 10.1111/j.1742-4658.2009.07269.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Amuthan G, Biswas G, Ananadatheerthavarada HK, Vijayasarathy C, Shephard HM, Avadhani NG. Mitochondrial stress-induced calcium signaling, phenotypic changes and invasive behavior in human lung carcinoma A549 cells. Oncogene. 2002;21:7839–49. doi: 10.1038/sj.onc.1205983. [DOI] [PubMed] [Google Scholar]
- 7.Anderson CT, Friedberg EC. The presence of nuclear and mitochondrial uracil-DNA glycosylase in extracts of human KB cells. Nucleic Acids Res. 1908;8:875–88. [PMC free article] [PubMed] [Google Scholar]
- 8.Anson RM, Bohr VA. Mitochondria, oxidative DNA damage, and aging. J Am Aging Assoc. 2000;23:199–218. doi: 10.1007/s11357-000-0020-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Arima Y, Nitta M, Kuninaka S, Zhang D, Fujiwara T, Taya Y, Nakao M, Saya H. Transcriptional blockade induces p53-dependent apoptosis associated with translocation of p53 to mitochondria. J Biol Chem. 2005;280:19166–76. doi: 10.1074/jbc.M410691200. [DOI] [PubMed] [Google Scholar]
- 10.Azrak S, Ayyasamy V, Zirpoli G, Ambrosone C, Bandera EV, Bovbjerg DH, Jandorf L, Ciupak G, Davis W, Pawlish KS, Liang P, Singh K. CAG repeat variants in the POLG1 gene encoding mtDNA polymerase-gamma and risk of breast cancer in African-American women. PLoS One. 2012;7:e29548. doi: 10.1371/journal.pone.0029548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bachmann PS, Gorman R, Papa RA, Bardell JE, Ford J, Kees UR, Marshall GM, Lock RB. Divergent mechanisms of glucocorticoid resistance in experimental models of pediatric acute lymphoblastic leukemia. Cancer Res. 2007;67:4482–90. doi: 10.1158/0008-5472.CAN-06-4244. [DOI] [PubMed] [Google Scholar]
- 12.Bai RK, Leal SM, Covarrubias D, Liu A, Wong LJ. Mitochondrial genetic background modifies breast cancer risk. Cancer Res. 2007;67:4687–94. doi: 10.1158/0008-5472.CAN-06-3554. [DOI] [PubMed] [Google Scholar]
- 13.Bakeeva LE, Skulachev VP, Sudarikova YV, Tsyplenkova VG. Mitochondria enter the nucleus (one further problem in chronic alcoholism) Biochemistry (Mosc) 2001;66:1335–41. doi: 10.1023/a:1013374410540. [DOI] [PubMed] [Google Scholar]
- 14.Bakhanashvili M, Grinberg S, Bonda E, Simon AJ, Moshitch-Moshkovitz S, Rahav G. p53 in mitochondria enhances the accuracy of DNA synthesis. Cell Death Differ. 2008;15:1865–74. doi: 10.1038/cdd.2008.122. [DOI] [PubMed] [Google Scholar]
- 15.Balliet RM, Capparelli C, Guido C, Pestell TG, Martinez-Outschoorn UE, Lin Z, Whitaker-Menezes D, Chiavarina B, Pestell RG, Howell A, Sotgia F, Lisanti MP. Mitochondrial oxidative stress in cancer-associated fibroblasts drives lactate production, promoting breast cancer tumor growth: understanding the aging and cancer connection. Cell Cycle. 2011;10:4065–73. doi: 10.4161/cc.10.23.18254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bamberger AM, Milde-Langosch K, Schulte HM, Löning T. Progesterone receptor isoforms, PR-B and PR-A, in breast cancer: correlations with clinico-pathologic tumor parameters and expression of AP-1 factors. Horm Res. 2000;54:32–37. doi: 10.1159/000063434. [DOI] [PubMed] [Google Scholar]
- 17.Bandiera S, Hanein S, Lyonnet S, Henrion-Caude A. Mitochondria as novel players of the cellular RNA interference. J Biol Chem. 2011;286 doi: 10.1074/jbc.L111.240259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Barrey E, Saint-Auret G, Bonnamy B, Damas D, Boyer O, Gidrol X. Pre-microRNA and mature microRNA in human mitochondria. PLoS One. 2001;6 doi: 10.1371/journal.pone.0020220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Barsoum MJ, Yuan H, Gerencser AA, Liot G, Kushnareva Y, Gräber S, Kovacs I, Lee WD, Waggoner J, Cui J, White AD, Bossy B, Martinou JC, Youle RJ, Lipton SA, Ellisman MH, Perkins GA, Bossy-Wetzel E. Nitric oxide-induced mitochondrial fission is regulated by dynamin-related GTPases in neurons. EMBO J. 2006;25:3900–11. doi: 10.1038/sj.emboj.7601253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Barthel A, Okino ST, Liao J, Nakatani K, Li J, Whitlock JP, Jr, Roth RA. Regulation of GLUT1 gene transcription by the serine/threonine kinase Akt1. J Biol Chem. 1999;274:20281–86. doi: 10.1074/jbc.274.29.20281. [DOI] [PubMed] [Google Scholar]
- 21.Bauer MF, Hofmann S, Neupert W, Brunner M. Protein translocation into mitochondria: the role of TIM complexes. Trends Cell Biol. 2000;10:25–31. doi: 10.1016/s0962-8924(99)01684-0. [DOI] [PubMed] [Google Scholar]
- 22.Baysal BE, Ferrell RE, Willett-Brozick JE, Lawrence EC, Myssiorek D, Bosch A, van der Mey A, Taschner PE, Rubinstein WS, Myers EN, Richard CW, 3rd, Cornelisse CJ, Devilee P, Devlin B. Mutations in SDHD, a mitochondrial complex II gene, in hereditary paraganglioma. Science. 2000;287:848–51. doi: 10.1126/science.287.5454.848. [DOI] [PubMed] [Google Scholar]
- 23.Behera MA, 1, Dai Q, Garde R, Saner C, Jungheim E, Price TM. Progesterone stimulates mitochondrial activity with subsequent inhibition of apoptosis in MCF-10A benign breast epithelial cells. Am J Physiol Endocrinol Metab. 2009;297:1089–96. doi: 10.1152/ajpendo.00209.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bennedbæk M, Rossing M, Rasmussen ÅK, Gerdes AM, Skytte AB, Jensen UB, Nielsen FC, Hansen TV. Identification of eight novel SDHB, SDHC, SDHD germline variants in Danish pheochromocytoma/paraganglioma patients. Hered Cancer Clin Pract. 2016;14:13. doi: 10.1186/s13053-016-0053-6. 2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bensaad K, Tsuruta A, Selak MA, Vidal MN, Nakano K, Bartrons R, Gottlieb E, Vousden KH. TIGAR, a p53-inducible regulator of glycolysis and apoptosis. Cell. 2006;126:107–20. doi: 10.1016/j.cell.2006.05.036. [DOI] [PubMed] [Google Scholar]
- 26.Bentmar Holgersson M, Giwercman A, Bjartell A, Wu FC, Huhtaniemi IT, O’Neill TW, Pendleton N, Vanderschueren D, Lean ME, Han TS, Finn JD, Kula K, Forti G, Casanueva FF, Bartfai G, Punab M, Lundberg Giwercman Y. Androgen receptor polymorphism-dependent variation in prostate-specific antigen concentrations of European men. Cancer Epidemiol Biomarkers Prev. 2014;23:2048–56. doi: 10.1158/1055-9965.EPI-14-0376. [DOI] [PubMed] [Google Scholar]
- 27.Bijur GN, Jope RS. Rapid accumulation of Akt in mitochondria following phosphatidylinositol 3-kinase activation. J Neurochem. 2003;87:1427–35. doi: 10.1046/j.1471-4159.2003.02113.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Biswas G, Adebanjo OA, Freedman BD, Anandatheerthavarada HK, Vijayasarathy C, Zaidi M, Kotlikoff M, Avadhani NG. Retrograde Ca2+ signaling in C2C12 skeletal myocytes in response to mitochondrial genetic and metabolic stress: a novel mode of inter-organelle crosstalk. EMBO J. 1999;18:522–33. doi: 10.1093/emboj/18.3.522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Biswas G, Tang W, Sondheimer N, Guha M, Bansal S, Avadhani NG. A distinctive physiological role for IkappaBbeta in the propagation of mitochondrial respiratory stress signaling. J Biol Chem. 2008;283:12586–94. doi: 10.1074/jbc.M710481200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bivona TG, Quatela SE, Bodemann BO, Ahearn IM, Soskis MJ, Mor A, Miura J, Wiener HH, Wright L, Saba SG, Yim D, Fein A, Pérez de Castro I, Li C, Thompson CB, Cox AD, Philips MR. PKC regulates a farnesyl-electrostatic switch on K-Ras that promotes its association with Bcl-XL on mitochondria and induces apoptosis. Mol Cell. 2006;21:481–93. doi: 10.1016/j.molcel.2006.01.012. [DOI] [PubMed] [Google Scholar]
- 31.Blair IA. DNA adducts with lipid peroxidation products. J Biol Chem. 2008;283:15545–49. doi: 10.1074/jbc.R700051200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Blomberg Jensen M, Leffers H, Petersen JH, Daugaard G, Skakkebaek NE, Rajpert-De Meyts E. Association of the polymorphism of the CAG repeat in the mitochondrial DNA polymerase gamma gene (POLG) with testicular germ-cell cancer. Ann Oncol. 2008;19:1910–14. doi: 10.1093/annonc/mdn407. [DOI] [PubMed] [Google Scholar]
- 33.Bloom GD. A nucleus with cytoplasmic features. J Cell Biol. 1967;35:266–8. doi: 10.1083/jcb.35.1.266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Boehler C, Gauthier LR, Mortusewicz O, Biard DS, Saliou JM, Bresson A, Sanglier-Cianferani S, Smith S, Schreiber V, Boussin F, Dantzer F. Poly(ADP-ribose) polymerase 3 (PARP3), a newcomer in cellular response to DNA damage and mitotic progression. Proc Natl Acad Sci. 2011;108:2783–8. doi: 10.1073/pnas.1016574108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Boesch P, Weber-Lotfi F, Ibrahim N, Tarasenko V, Cosset A, Paulus F, Lightowlers RN, Dietrich A. DNA repair in organelles: Pathways, organization, regulation, relevance in disease and aging. Biochim Biophys Acta. 2011;1813:186–200. doi: 10.1016/j.bbamcr.2010.10.002. [DOI] [PubMed] [Google Scholar]
- 36.Bonora E, Evangelisti C, Bonichon F, Tallini G, Romeo G. Novel germline variants identified in the inner mitochondrial membrane transporter TIMM44 and their role in predisposition to oncocytic thyroid carcinomas. Br J Cancer. 2006;95:1529–36. doi: 10.1038/sj.bjc.6603455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Bonora E, Porcelli AM, Gasparre G, Biondi A, Ghelli A, Carelli V, Baracca A, Tallini G, Martinuzzi A, Lenaz G, Rugolo M, Romeo G. Defective oxidative phosphorylation in thyroid oncocytic carcinoma is associated with pathogenic mitochondrial DNA mutations affecting complexes I and III. Cancer Res. 2006;66:6087–96. doi: 10.1158/0008-5472.CAN-06-0171. [DOI] [PubMed] [Google Scholar]
- 38.Booker LM, Habermacher GM, Jessie BC, Sun QC, Baumann AK, Amin M, Lim SD, Fernandez-Golarz C, Lyles RH, Brown MD, Marshall FF, Petros JA. North American white mitochondrial haplogroups in prostate and renal cancer. J Urol. 2006;175:468–72. doi: 10.1016/S0022-5347(05)00163-1. [DOI] [PubMed] [Google Scholar]
- 39.Bos JL. ras oncogenes in human cancer: a review. Cancer Res. 1989;49:4682–4689. [PubMed] [Google Scholar]
- 40.Boultwood J, Fidler C, Mills KI, Frodsham PM, Kusec R, Gaiger A, Gale RE, Linch DC, Littlewood TJ, Moss PA, Wainscoat JS. Amplification of mitochondrial DNA in acute myeloid leukemia. Br J Haematol. 1996;95:426–31. doi: 10.1046/j.1365-2141.1996.d01-1922.x. [DOI] [PubMed] [Google Scholar]
- 41.Brandes D, Schofield BH, Anton E. Nuclear mitochondria? Science. 1965;149:1373–4. doi: 10.1126/science.149.3690.1373. [DOI] [PubMed] [Google Scholar]
- 42.Brandon M, Baldi P, Wallace DC. Mitochondrial mutations in cancer. Oncogene. 2006;25:4647–62. doi: 10.1038/sj.onc.1209607. [DOI] [PubMed] [Google Scholar]
- 43.Brown AL, Lupo PJ, Okcu MF, Lau CC, Rednam S, Scheurer ME. SOD2 genetic variant associated with treatment-related ototoxicity in cisplatin-treated pediatric medulloblastoma. Cancer Med. 2015;4:1679–86. doi: 10.1002/cam4.516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Buroker NE. Regulatory SNPs and transcriptional factor binding sites in ADRBK1, AKT3, ATF3, DIO2, TBXA2R and VEGFA. Transcription. 2014;5:e964559. doi: 10.4161/21541264.2014.964559. 2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Butow RA, Avadhani NG. Mitochondrial signaling: the retrograde response. Mol Cell. 2004;14:1–15. doi: 10.1016/s1097-2765(04)00179-0. [DOI] [PubMed] [Google Scholar]
- 46.Caballero-Caballero A, Engel T, Martinez-Villarreal J, Sanz-Rodriguez A, Chang P, Dunleavy M, Mooney CM, Jimenez-Mateos EM, Schindler CK, Henshall DC. Mitochondrial localization of the forkhead box class O transcription factor FOXO3a in brain. J Neurochem. 2013;124:749–56. doi: 10.1111/jnc.12133. [DOI] [PubMed] [Google Scholar]
- 47.Caldecott KW. Single-strand break repair and genetic disease. Nat Rev Genet. 2008;9:619–31. doi: 10.1038/nrg2380. [DOI] [PubMed] [Google Scholar]
- 48.Calin GA, Garzon R, Cimmino A, Fabbri M, Croce CM. MicroRNAs and leukemias: how strong is the connection? Leuk Res. 2006;30:653–55. doi: 10.1016/j.leukres.2005.10.017. [DOI] [PubMed] [Google Scholar]
- 49.Cammarata PR, Chu S, Moor A, Wang Z, Yang SH, Simpkins JW. Subcellular distribution of native estrogen receptor alpha and beta subtypes in cultured human lens epithelial cells. Exp Eye Res. 2004;78:861–71. doi: 10.1016/j.exer.2003.09.027. [DOI] [PubMed] [Google Scholar]
- 50.Canter JA, Kallianpur AR, Parl FF, Millikan RC. Mitochondrial DNA G10398A polymorphism and invasive breast cancer in African-American women. Cancer Res. 2005;65:8028–33. doi: 10.1158/0008-5472.CAN-05-1428. [DOI] [PubMed] [Google Scholar]
- 51.Canto C, Auwerx J. AMP-activated protein kinase and its downstream transcriptional pathways. Cell Mol Life Sci. 2010;67:3407–23. doi: 10.1007/s00018-010-0454-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Canto C, Auwerx J. Calorie restriction: is AMPK a key sensor and effector? Physiology (Bethesda) 2011;26:214–24. doi: 10.1152/physiol.00010.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Canto C, Auwerx J. PGC-1alpha, SIRT1 and AMPK, an energy sensing network that controls energy expenditure. Current opinion in lipidology. 2009;20:98–105. doi: 10.1097/MOL.0b013e328328d0a4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Canto P, Benítez Granados J, Martínez Ramírez MA, Reyes E, Feria-Bernal G, García-García E, Tejeda ME, Zavala E, Tapia A, Rojano-Mejía D, Méndez JP. Genetic variants in ATP6 and ND3 mitochondrial genes are not associated with aggressive prostate cancer in Mexican-Mestizo men with overweight or obesity. Aging Male. 2016;17:1–5. doi: 10.1080/13685538.2016.1185409. [DOI] [PubMed] [Google Scholar]
- 55.Cao M, Mu X, Jiang C, Yang G, Chen H, Xue W. Single-nucleotide polymorphisms of GPX1 and MnSOD and susceptibility to bladder cancer: a systematic review and meta-analysis. Tumor Biol. 2014;35:759–64. doi: 10.1007/s13277-013-1103-6. [DOI] [PubMed] [Google Scholar]
- 56.Carew JS, Huang P. Mitochondrial defects in cancer. Mol Cancer. 2002;1:9. doi: 10.1186/1476-4598-1-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Caspersen C, Wang N, Yao J, Sosunov A, Chen X, Lustbader JW, Xu HW, Stern D, McKhann G, Yan SD. Mitochondrial Aβ: A potential focal point for neuronal metabolic dysfunction in Alzheimer’s disease. FASEB J. 2005;19:2040–41. doi: 10.1096/fj.05-3735fje. [DOI] [PubMed] [Google Scholar]
- 58.Cavalli G, Biavasco R, Borgiani B, Dagna L. Oncogene-induced senescence as a new mechanism of disease: the paradigm of erdheim-chester disease. Front Immunol. 2014;5:281. doi: 10.3389/fimmu.2014.00281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Cejas P, García-Cabezas MA, Casado E, Belda-Iniesta C, De Castro J, Fresno JA, Sereno M, Barriuso J, Espinosa E, Zamora P, Feliu J, Redondo A, Hardisson DA, Renart J, González-Barón M. Phospholipid hydroperoxide glutathione peroxidase (PHGPx) expression is downregulated in poorly differentiated breast invasive ductal carcinoma. Free Radic Res. 2007;4:681–87. doi: 10.1080/10715760701286167. [DOI] [PubMed] [Google Scholar]
- 60.Chang CR, Blackstone C. Dynamic regulation of mitochondrial fission through modification of the dynamin-related protein Drp1. Ann N Y Acad Sci. 2010;1201:34–39. doi: 10.1111/j.1749-6632.2010.05629.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Chang PY, Chen JS, Chang NC, Chang SC, Wang MC, Tsai SH, Wen YH, Tsai WS, Chan EC, Lu JJ. NRAS germline variant G138R and multiple rare somatic mutations on APC in colorectal cancer patients in Taiwan by next generation sequencing. Oncotarget. 2016 doi: 10.18632/oncotarget.8885. epub ahead of print. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Chattopadhyay R, Wiederhold L, Szczesny B, Boldogh I, Hazra TK, Izumi T, Mitra S. Identification and characterization of mitochondrial abasic (AP)-endonuclease in mammalian cells. Nucleic Acids Res. 2006;34:2067–76. doi: 10.1093/nar/gkl177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Chaudhary AK, Bhat TA, Kumar S, Kumar A, Kumar R, Underwood W, Koochekpour S, Shourideh M, Yadav N, Dhar S, Chandra D. Mitochondrial dysfunction-mediated apoptosis resistance associates with defective heat shock protein response in African-American men with prostate cancer. Br J Cancer. 2016;114:1090–100. doi: 10.1038/bjc.2016.88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Chen D, Xue W, Xiang J. The intra-nucleus integration of mitochondrial DNA (mtDNA)in cervical mucosa cells and its relation with c-myc expression. J Exp Clin Cancer Res. 2008;27:36. doi: 10.1186/1756-9966-27-36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Chen JQ, Delannoy M, Cooke C, Yager JD. Mitochondrial localization of ERalpha and ERbeta in human MCF7 cells. Am J Physiol Endocrinol Metab. 2004;286:1011–22. doi: 10.1152/ajpendo.00508.2003. [DOI] [PubMed] [Google Scholar]
- 66.Chen JQ, Russo PA, Cooke C, Russo IH, Russo J. ERbeta shifts from mitochondria to nucleus during estrogen-induced neoplastic transformation of human breast epithelial cells and is involved in estrogen-induced synthesis of mitochondrial respiratory chain proteins. Biochim Biophys Acta. 2007;1773:1732–46. doi: 10.1016/j.bbamcr.2007.05.008. [DOI] [PubMed] [Google Scholar]
- 67.Chen M, David CJ, Manley JL. Concentration-dependent control of pyruvate kinase M mutually exclusive splicing by hnRNP proteins. Nat Struct Mol Biol. 2012;19:346–54. doi: 10.1038/nsmb.2219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Chen T, He J, Shen L, Fang H, Nie H, Jin T, Wei X, Xin Y, Jiang Y, Li H, Chen G, Lu J, Bai Y. The mitochondrial DNA 4,977-bp deletion and its implication in copy number alteration in colorectal cancer. BMC Med Genet. 2011;12:8. doi: 10.1186/1471-2350-12-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Chen Y, Zhang H, Zhou HJ, Ji W, Min W. Mitochondrial Redox Signaling and Tumor Progression. Cancers (Basel) 2016:8. doi: 10.3390/cancers8040040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Choudhary V, Kaddour-Djebbar I, Lakshmikanthan V, Ghazaly T, Thangjam GS, Sreekumar A, Lewis RW, Mills IG, Bollag WB, Kumar MV. Novel role of androgens in mitochondrial fission and apoptosis. Mol Cancer Res. 2011;9:1067–77. doi: 10.1158/1541-7786.MCR-10-0445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Chowdhury R, Yeoh KK, Tian YM, Hillringhaus L, Bagg EA, Rose NR, Leung IK, Li XS, Woon EC, Yang M, McDonough MA, King ON, Clifton IJ, Klose RJ, Claridge TD, Ratcliffe PJ, Schofield CJ, Kawamura A. The oncometabolite 2-hydroxyglutarate inhibits histone lysine demethylases. EMBO Rep. 2011;12:463–69. doi: 10.1038/embor.2011.43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Christofk HR, Vander Heiden MG, Harris MH, Ramanathan A, Gerszten RE, Wei R, Fleming MD, Schreiber SL, Cantley LC. The M2 splice isoform of pyruvate kinase is important for cancer metabolism and tumor growth. Nature. 2008;452:230–33. doi: 10.1038/nature06734. [DOI] [PubMed] [Google Scholar]
- 73.Citri A, Yarden Y. EGF-ERBB signalling: towards the systems level. Nat Rev Mol Cell Biol. 2006;7:505–16. doi: 10.1038/nrm1962. [DOI] [PubMed] [Google Scholar]
- 74.Clayton DA, Doda JN, Friedberg EC. The absence of a pyrimidine dimer repair mechanism in mammalian mitochondria. PNAS. 1974;71:2777–81. doi: 10.1073/pnas.71.7.2777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Cohen HY, Miller C, Bitterman KY, Wall NR, Hekking B, Kessler B, Howitz KT, Gorospe M, de Cabo R, Sinclair DA. Calorie restriction promotes mammalian cell survival by inducing the SIRT1 deacetylase. Science. 2004;305:390–2. doi: 10.1126/science.1099196. [DOI] [PubMed] [Google Scholar]
- 76.Compton S, Kim C, Griner NB, Potluri P, Scheffler IE, Sen S, Jerry DJ, Schneider S, Yadava N. Mitochondrial dysfunction impairs tumor suppressor p53 expression/function. J Biol Chem. 2011;286:20297–312. doi: 10.1074/jbc.M110.163063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Coomans de Brachène A, Demoulin JB. FOXO development factors in cancer and development. Cell Mol Life Sci. 2016;73:1159–72. doi: 10.1007/s00018-015-2112-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Couto PG, Bastos-Rodrigues L, Carneiro JG, Guieiro F, Bicalho MA, Leidenz FB, Bicalho AJ, Friedman E, De Marco L. DNA Base-Excision Repair Genes OGG1 and NTH1 in Brazilian Lung Cancer Patients. Mol Diagn Ther. 2015;19:389–95. doi: 10.1007/s40291-015-0164-1. [DOI] [PubMed] [Google Scholar]
- 79.Covarrubias D, Bai RK, Wong LJ, Leal SM. Mitochondrial DNA variant interactions modify breast cancer risk. J Hum Genet. 2008;53:924–28. doi: 10.1007/s10038-008-0331-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Czarnecka AM, Krawczyk T, Plak K, Klemba A, Zdrozny M, Arnold RS, Kofler B, Golik P, Szybinska A, Lubinski J, Mossakowska M, Bartnik E, Petros JA. Mitochondrial genotype and breast cancer predisposition. Oncol Rep. 2010;24:1521–34. doi: 10.3892/or_00001014. [DOI] [PubMed] [Google Scholar]
- 81.Dai Q, Shah AA, Garde RV, Yonish BA, Zhang L, Medvitz NA, Miller SE, Hansen EL, Dunn CN, Price TM. A truncated progesterone receptor (PR-M) localizes to the mitochondrion and controls cellular respiration. Mol Endocrinol. 2013;27:741–53. doi: 10.1210/me.2012-1292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Dang J, Peng L, Zhong HJ, Huo ZH. Androgen receptor (CAG)n polymorphisms and breast cancer risk in a Han Chinese population. Genet Mol Res. 2015;14:10258–66. doi: 10.4238/2015.August.28.10. [DOI] [PubMed] [Google Scholar]
- 83.Darvishi K, Sharma S, Bhat AK, Rai E, Bamezai RN. Mitochondrial DNA G10398A polymorphism imparts maternal haplogroup N a risk for breast and esophageal cancer. Cancer Lett. 2007;249:249–55. doi: 10.1016/j.canlet.2006.09.005. [DOI] [PubMed] [Google Scholar]
- 84.Das S, Bedja D, Campbell N, Dunkerly B, Chenna V, Maitra A, Steenbergen C. miR-181c regulates the mitochondrial genome, bioenergetics, and propensity for heart failure in vivo. PLoS One. 2014;9:e96820. doi: 10.1371/journal.pone.0096820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Datta S, Ray A, Roy R, Roy B. Association of DNA sequence variation in mitochondrial DNA polymerase with mitochondrial DNA synthesis and risk of oral cancer. Gene. 2016;575:650–54. doi: 10.1016/j.gene.2015.09.039. [DOI] [PubMed] [Google Scholar]
- 86.Datta SR, Dudek H, Tao X, Masters S, Fu H, Gotoh Y, Greenberg ME. Akt phosphorylation of BAD couples survival signals to the cell-intrinsic death machinery. Cell. 1997;91:231–41. doi: 10.1016/s0092-8674(00)80405-5. [DOI] [PubMed] [Google Scholar]
- 87.David CJ, Chen M, Assanah M, Canoll P, Manley JL. HnRNP proteins controlled by c-Myc deregulate pyruvate kinase mRNA splicing in cancer. Nature. 2010;463:364–68. doi: 10.1038/nature08697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Davis AF, Ropp PA, Clayton DA, Copeland WC. Mitochondrial DNA polymerase gamma is expressed and translated in the absence of mitochondrial DNA maintenance and replication. Nucleic Acids Res. 1996;24:2753–59. doi: 10.1093/nar/24.14.2753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Srinivasainagendra V, Sandel MW, Singh B, Sundaresan A, Mooga VP, Bajpai P, Tiwari HK, Singh KK. Migration of mitochondrial DNA in the nuclear genome of colorectal adenocarcinoma. Genome Med. 2017;9(1):31. doi: 10.1186/s13073-017-0420-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.De Bont R, van Larebeke N. Endogenous DNA damage in humans: a review of quantitative data. Mutagenesis. 2004;19:169–85. doi: 10.1093/mutage/geh025. [DOI] [PubMed] [Google Scholar]
- 91.De S, Kumari J, Mudgal R, Modi P, Gupta S, Futami K, Goto H, Lindor NM, Furuichi Y, Mohanty D, Sengupta S. RECQL4 is essential for the transport of p53 to mitochondria in normal human cells in the absence of exogenous stress. J Cell Sci. 2012;125:2509–22. doi: 10.1242/jcs.101501. [DOI] [PubMed] [Google Scholar]
- 92.De Vos WH, Houben F, Kamps M, Malhas A, Verheyen F, Cox J, et al. Repetitive disruptions of the nuclear envelope invoke temporary loss of cellular compartmentalization in laminopathies. Hum Mol Genet. 2011;20:4175–86. doi: 10.1093/hmg/ddr344. [DOI] [PubMed] [Google Scholar]
- 93.Del Bo R, Bordoni A, Sciacco M, Di Fonzo A, Galbiati S, Crimi M, Bresolin N, Comi GP. Remarkable infidelity of polymerase gammaA associated with mutations in POLG1 exonuclease domain. Neurology. 2003;61:903–08. doi: 10.1212/01.wnl.0000092303.13864.be. [DOI] [PubMed] [Google Scholar]
- 94.Delsite R, Kachhap S, Anbazhagan R, Gabrielson E, Singh KK. Nuclear genes involved in mitochondria-to-nucleus communication in breast cancer cells. Mol Cancer. 2002;1:6. doi: 10.1186/1476-4598-1-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Delsite RL, Rasmussen LJ, Rasmussen AK, Kalen A, Goswami PC, Singh KK. Mitochondrial impairment is accompanied by impaired oxidative DNA repair in the nucleus. Mutagenesis. 2003;18:497–503. doi: 10.1093/mutage/geg027. [DOI] [PubMed] [Google Scholar]
- 96.Demory ML, Boerner JL, Davidson R, Faust W, Miyake T, Lee I, Hüttemann M, Douglas R, Haddad G, Parsons SJ. Epidermal growth factor receptor translocation to the mitochondria: regulation and effect. J Biol Chem. 2009;284:36592–604. doi: 10.1074/jbc.M109.000760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Desler C, Marcker ML, Singh KK, Rasmussen LJ. The importance of mitochondrial DNA in aging and cancer. J Aging Res. 2011;2011:407536. doi: 10.4061/2011/407536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Desouki MM, Kulawiec M, Bansal S, Das GM, Singh KK. Cross talk between mitochondria and superoxide generating NADPH oxidase in breast and ovarian tumors. Cancer Biol Ther. 2005;4:1367–73. doi: 10.4161/cbt.4.12.2233. [DOI] [PubMed] [Google Scholar]
- 99.deSouza-Pinto NC, Eide L, Hogue BA, Thybo T, Stevnsner T, Seeberg E, Klungland A, Bohr VA. Repair of 8-oxodeoxyguanosine lesions in mitochondrial DNA depends on the oxoguanine DNA glycosylase (OGG1) gene and 8-oxoguanine accumulates in the mitochondrial DNA of OGG1-defective mice. Cancer Res. 2001;61:5378–81. [PubMed] [Google Scholar]
- 100.deSouza-Pinto NC, Mason PA, Hashiguchi K, Weissman L, Tian J, Guay D, Lebel M, Stevnsner TV, Rasmussen LJ, Bohr VA. Novel DNA mismatch-repairactivity involving YB-1 in human mitochondria. DNA Repair (Amst) 2009;8:704–19. doi: 10.1016/j.dnarep.2009.01.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Devi L, Raghavendran V, Prabhu BM, Avadhani NG, Anandatheerthavarada HK. Mitochondrial import and accumulation of alpha-synuclein impair complex I in human dopaminergic neuronal cultures and Parkinson disease brain. J Biol Chem. 2008;283:9089–100. doi: 10.1074/jbc.M710012200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Ding Y, Liu Z, Desai S, Zhao Y, Liu H, Pannell LK, Yi H, Wright ER, Owen LB, Dean-Colomb W, Fodstad O, Lu J, LeDoux SP, Wilson GL, Tan M. Receptor tyrosine kinase ErbB2 translocates into mitochondria and regulates cellular metabolism. Nat Commun. 2012;3:1271. doi: 10.1038/ncomms2236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Driggers WJ, LeDoux SP, Wilson GL. Repair of oxidative damage within the mitochondrial DNA of RINr 38 cells. J Biol Chem. 1993;268:22042–45. [PubMed] [Google Scholar]
- 104.Dwight T, Mann K, Benn DE, Robinson BG, McKelvie P, Gill AJ, Winship I, Clifton-Bligh RJ. Familial SDHA mutation associated with pituitary adenoma and pheochromocytoma/paraganglioma. J Clin Endocrinol Metab. 2013;98:1103–08. doi: 10.1210/jc.2013-1400. [DOI] [PubMed] [Google Scholar]
- 105.Earp MA, Brooks-Wilson A, Cook L, Le N. Inherited common variants in mitochondrial DNA and invasive serous epithelial ovarian cancer risk. BMC Res Notes. 2013;6:425. doi: 10.1186/1756-0500-6-425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Eichner LJ, Perry MC, Dufour CR, Bertos N, Park M, St-Pierre J, Giguère V. miR-378(∗) mediates metabolic shift in breast cancer cells via the PGC-1β/ERRγ transcriptional pathway. Cell Metab. 2010;12:352–61. doi: 10.1016/j.cmet.2010.09.002. [DOI] [PubMed] [Google Scholar]
- 107.Fasan A, Haferlach C, Eder C, Alpermann T, Quante A, Peters A, Kern W, Haferlach T, Schnittger S. Evaluation of IDH1G105 polymorphism as prognostic marker in intermediate-risk AML. Ann Hematol. 2015;94:1991–2001. doi: 10.1007/s00277-015-2488-7. [DOI] [PubMed] [Google Scholar]
- 108.Feng Q, Crochet JR, Dai Q, Leppert PC, Price TM. Expression of a mitochondrial progesterone receptor (PR-M) in leiomyomata and association with increased mitochondrial membrane potential. J Clin Endocrinol Metab. 2014;99:390–99. doi: 10.1210/jc.2013-2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Feng R, Chen X, Yu Y, Su L, Yu B, Li J, Cai Q, Yan M, Liu B, Zhu Z. miR-126 functions as a tumor suppressor in human gastric cancer. Cancer Lett. 2010;298:50–63. doi: 10.1016/j.canlet.2010.06.004. [DOI] [PubMed] [Google Scholar]
- 110.Formentini L, Sánchez-Aragó M, Sánchez-Cenizo L, Cuezva JM. The mitochondrial ATPase inhibitory factor 1 triggers a ROS-mediated retrograde pro-survival and proliferative response. Mol Cell. 2012;45:731–42. doi: 10.1016/j.molcel.2012.01.008. [DOI] [PubMed] [Google Scholar]
- 111.Frank AK, Pietsch EC, Dumont P, Tao J, Murphy ME. Wild-type and mutant p53 proteins interact with mitochondrial caspase-3. Cancer Biol Ther. 2011;11:740–45. doi: 10.4161/cbt.11.8.14906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Fujimoto J, Ichigo S, Hirose R, Sakaguchi H, Tamaya T. Clinical implication of expression of progesterone receptor form A and B mRNAs in secondary spreading of gynecologic cancers. J Steroid Biochem Mol Biol. 1997;62:449–54. doi: 10.1016/s0960-0760(97)00057-5. [DOI] [PubMed] [Google Scholar]
- 113.Gallegos-Arreola MP, Figuera LE, Flores-Ramos LG, Puebla-Pérez AM, Zúñiga-González GM. Association of the Alu insertion polymorphism in the progesterone receptor gene with breast cancer in a Mexican population. Arch Med Sci. 2015;11:551–60. doi: 10.5114/aoms.2015.52357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Gan Y, Wang Y, Tan Z, Zhou J, Kitazawa R, Jiang X, Tang Y, Yang J. TDRG1 regulates chemosensitivity of seminoma TCam-2 cells to cisplatin via PI3K/Akt/mTOR signaling pathway and mitochondria-mediated apoptotic pathway. Cancer Biol Ther. 2016;22:1–10. doi: 10.1080/15384047.2016.1178425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Gao H, Ding X, Wei D, Cheng P, Su X, Liu H, Aziz F, Wang D, Zhang T. Erlotinib in patients with advanced non-small-cell lung cancer: A meta-analysis. Transl Lung Cancer Res. 2012;1:129–44. doi: 10.3978/j.issn.2218-6751.2012.06.01. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Gao SM, Chen C, Wu J, Tan Y, Yu K, Xing CY, Ye A, Yin L, Jiang L. Synergistic apoptosis induction in leukemic cells by miR-15a/16-1 and arsenic trioxide. Biochem Biophys Res Commun. 2010;403:203–08. doi: 10.1016/j.bbrc.2010.10.137. [DOI] [PubMed] [Google Scholar]
- 117.Gazdar AF. Activating and resistance mutations of EGFR in non-small-cell lung cancer: role in clinical response to EGFR tyrosine kinase inhibitors. Oncogene. 2009;(Suppl 1):S24–S31. doi: 10.1038/onc.2009.198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Gómez R, Torres-Sánchez L, Camacho-Mejorado R, Burguete-García AI, Vázquez-Salas RA, Martínez-Nava GA, Santana C, Noris G. Androgen receptor CAG polymorphism and sporadic and early-onset prostate cancer among Mexican men. J Hum Genet. 2016 doi: 10.1038/jhg.2016.49. epub ahead of print. [DOI] [PubMed] [Google Scholar]
- 119.Goo CK, Lim HY, Ho QS, Too HP, Clement MV, Wong KP. PTEN/Akt signaling controls mitochondrial respiratory capacity through 4E-BP1. PLoS One. 2012;7:e45806. doi: 10.1371/journal.pone.0045806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Gottlob K, Majewski N, Kennedy S, Kandel E, Robey RB, Hay N. Inhibition of early apoptotic events by Akt/PKB is dependent on the first committed step of glycolysis and mitochondrial hexokinase. Genes Dev. 2001;15:1406–18. doi: 10.1101/gad.889901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Grossman LI, Shoubridge EA. Mitochondrial genetics and human disease. Bioessays. 1996;18:983–91. doi: 10.1002/bies.950181208. [DOI] [PubMed] [Google Scholar]
- 122.Guha M, Fang JK, Monks R, Birnbaum MJ, Avadhani NG. Activation of Akt is essential for the propagation of mitochondrial respiratory stress signaling and activation of the transcriptional coactivator heterogeneous ribonucleoprotein A2. Mol Biol Cell. 2010;21:3578–89. doi: 10.1091/mbc.E10-03-0192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Guha M, Pan H, Fang JK, Avadhani NG. Heterogeneous nuclear ribonucleoprotein A2 is a common transcriptional coactivator in the nuclear transcription response to mitochondrial respiratory stress. Mol Biol Cell. 2009;20:4107–19. doi: 10.1091/mbc.E09-04-0296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Guha M, Srinivasan S, Biswas G, Avadhani NG. Activation of a novel calcineurin-mediated insulin-like growth factor-1 receptor pathway, altered metabolism, and tumor cell invasion in cells subjected to mitochondrial respiratory stress. J Biol Chem. 2007;282:14536–46. doi: 10.1074/jbc.M611693200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Guha M, Srinivasan S, Ruthel G, Kashina AK, Carstens RP, Mendoza A, Khanna C, Van Winkle T, Avadhani NG. Mitochondrial retrograde signaling induces epithelial-mesenchymal transition and generates breast cancer stem cells. Oncogene. 2014;33:5238–50. doi: 10.1038/onc.2013.467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Guo J, Zheng L, Liu W, Wang X, Wang Z, Wang Z, French AJ, Kang D, Chen L, Thibodeau SN, Liu W. Frequent truncating mutation of TFAM induces mitochondrial DNA depletion and apoptotic resistance in microsatellite-unstable colorectal cancer. Cancer Res. 2011;71:2978–87. doi: 10.1158/0008-5472.CAN-10-3482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Guo Z, Deshpande R, Paull TT. ATM activation in the presence of oxidative stress. Cell Cycle. 2010;9:4805–11. doi: 10.4161/cc.9.24.14323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Gupta S, Ramjaun AR, Haiko P, Wang Y, Warne PH, Nicke B, Nye E, Stamp G, Alitalo K, Downward J. Binding of ras to phosphoinositide 3-kinase p110alpha is required for ras-driven tumorigenesis in mice. Cell. 2007;129:957–68. doi: 10.1016/j.cell.2007.03.051. [DOI] [PubMed] [Google Scholar]
- 129.Gutiérrez-Malacatt H, Ayala-Sanchez M, Aquino-Ortega X, Dominguez-Rodriguez J, Martinez-Tovar A, Olarte-Carrillo I, Martinez-Hernandez A, C CC, Orozco L, Cordova EJ. The rs61764370 Functional Variant in the KRAS Oncogene is Associated with Chronic Myeloid Leukemia Risk in Women. Asian Pac J Cancer Prev. 2016;17:2265–70. doi: 10.7314/apjcp.2016.17.4.2265. [DOI] [PubMed] [Google Scholar]
- 130.Hadler HI, Devadas K, Mahalingam R. Selected nuclear LINE elements with mitochondrial DNA-like inserts are more plentiful and mobile in tumor than in normal tissue of mouse and rat. J Cell Biochem. 1998;68:100–9. doi: 10.1002/(sici)1097-4644(19980101)68:1<100::aid-jcb10>3.0.co;2-l. [DOI] [PubMed] [Google Scholar]
- 131.Hajnóczky G, Csordás G. Calcium signaling: fishing out molecules of mitochondrial calcium transport. Curr Biol. 2010;20:888–91. doi: 10.1016/j.cub.2010.09.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Halfdanarson TR, Wang L, Bamlet WR, de Andrade M, McWilliams RR, Cunningham JM, Petersen GM. Mitochondrial genetic polymorphisms do not predict survival in patients with pancreatic cancer. Cancer Epidemiol Biomarkers Prev. 2008;17:2512–13. doi: 10.1158/1055-9965.EPI-08-0460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Hallmann A, Milczarek R, Lipinski M, Kossowska E, Spodnik JH, Wozniak M, et al. Fast perinuclear clustering of mitochondria in oxidatively stressed human choriocarcinoma cells. Folia Morphol (Warsz) 2004;63:407–12. [PubMed] [Google Scholar]
- 134.Han L, Lee SW, Yoon JH, Park YG, Choi YJ, Nam SW, Lee JY, Wang YP, Park WS. Association of SOD1 and SOD2 single nucleotide polymorphisms with susceptibility to gastric cancer in a Korean population. APMIS. 2013;121:246–56. doi: 10.1111/j.1600-0463.2012.02963.x. [DOI] [PubMed] [Google Scholar]
- 135.Hanes JW, Thal DM, Johnson KA. Incorporation and replication of 8-oxodeoxyguanosine by the human mitochondrial DNA polymerase. J Biol Chem. 2006;281:36241–48. doi: 10.1074/jbc.M607965200. [DOI] [PubMed] [Google Scholar]
- 136.Hanna SC, Krishnan B, Bailey ST, Moschos SJ, Kuan PF, Shimamura T, Osborne LD, Siegel MB, Duncan LM, O’Brien ET, 3rd, Superfine R, Miller CR, Simon MC, Wong KK, Kim WY. HIF1α and HIF2α independently activate SRC to promote melanoma metastases. Clin Invest. 2013;123:2078–93. doi: 10.1172/JCI66715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Hayashi J, Ohta S, Kikuchi A, Takemitsu M, Goto Y, Nonaka I. Introduction of disease-related mitochondrial DNA deletions into HeLa cells lacking mitochondrial DNA results in mitochondrial dysfunction. PNAS. 1991;88:10614–18. doi: 10.1073/pnas.88.23.10614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Hazkani-Covo E, Covo S. Numt-mediated double-strand break repair mitigates deletions during primate genome evolution. PLoS Genet. 2008;4:e1000237. doi: 10.1371/journal.pgen.1000237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Hazkani-Covo E, Zeller RM, Martin W. Molecular poltergeists: mitochondrial DNA copies (numts) in sequenced nuclear genomes. PLoS Genet. 2010;6:e1000834. doi: 10.1371/journal.pgen.1000834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Heidari N, Miller AV, Hicks MA, Marking CB, Harada H. Glucocorticoid-mediated BIM induction and apoptosis are regulated by Runx2 and c-Jun in leukemia cells. Cell Death Dis. 2012;3:e349. doi: 10.1038/cddis.2012.89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Hemachandra LP, Shin DH, Dier U, Iuliano JN, Engelberth SA, Uusitalo LM, Murphy SK, Hempel N. Mitochondrial Superoxide Dismutase Has a Protumorigenic Role in Ovarian Clear Cell Carcinoma. Cancer Res. 2015;75:4973–84. doi: 10.1158/0008-5472.CAN-14-3799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Heyne K, Mannebach S, Wuertz E, Knaup KX, Mahyar-Roemer M, Roemer K. Identification of a putative p53 binding sequence within the human mitochondrial genome. FEBS Lett. 2004;578:198–202. doi: 10.1016/j.febslet.2004.10.099. [DOI] [PubMed] [Google Scholar]
- 143.Higuchi M, Kudo T, Suzuki S, Evans TT, Sasaki R, Wada Y, Shirakawa T, Sawyer JR, Gotoh A. Mitochondrial DNA determines androgen dependence in prostate cancer cell lines. Oncogene. 2006;25:1437–45. doi: 10.1038/sj.onc.1209190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Ho L, Titus AS, Banerjee KK, George S, Lin W, Deota S, Saha AK, Nakamura K, Gut P, Verdin E, Kolthur-Seetharam U. SIRT4 regulates ATP 47 homeostasis and mediates a retrograde signaling via AMPK. Aging (Albany NY) 2013;5:835–49. doi: 10.18632/aging.100616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Holmgren A. Antioxidant function of thioredoxin and glutaredoxin systems. Antioxid Redox Signal. 2000;2:811–20. doi: 10.1089/ars.2000.2.4-811. [DOI] [PubMed] [Google Scholar]
- 146.Horne DW, Holloway RS, Wagner C. Transport of Sadenosylmethionine in isolated rat liver mitochondria. Arch Biochem Biophys. 1997;343:201–06. doi: 10.1006/abbi.1997.0167. [DOI] [PubMed] [Google Scholar]
- 147.Horton TM, Petros JA, Heddi A, Shoffner J, Kaufman AE, Graham SD, Jr, Gramlich T, Wallace DC. Novel mitochondrial DNA deletion found in a renal cell carcinoma. Genes Chromosomes Cancer. 1996;15:95–101. doi: 10.1002/(SICI)1098-2264(199602)15:2<95::AID-GCC3>3.0.CO;2-Z. [DOI] [PubMed] [Google Scholar]
- 148.Huang PH, Xu AM, White FM. Oncogenic EGFR signaling networks in glioma. Sci Signal. 2009;2 doi: 10.1126/scisignal.287re6. [DOI] [PubMed] [Google Scholar]
- 149.Huang R, Wang J, Zhong Y, Liu Y, Stokke T, Trope CG, Nesland JM, Suo Z. Mitochondrial DNA Deficiency in Ovarian Cancer Cells and Cancer Stem Cell-like Properties. Anticancer Res. 2015;35:3743–53. [PubMed] [Google Scholar]
- 150.Hwang HJ, Dornbos P, Steidemann M, Dunivin TK, Rizzo M, LaPres JJ. Mitochondrial-targeted aryl hydrocarbon receptor and the impact of 2, 3, 7, 8-tetrachlorodibenzo-p-dioxin on cellular respiration and the mitochondrial proteome. Toxicol Appl Pharmacol. 2016;304:121–32. doi: 10.1016/j.taap.2016.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Inoue T, Suzuki-Karasaki Y. Mitochondrial superoxide mediates mitochondrial and endoplasmic reticulum dysfunctions in TRAIL-induced apoptosis in Jurkat cells. Free Radic Biol Med. 2013;61:273–84. doi: 10.1016/j.freeradbiomed.2013.04.020. [DOI] [PubMed] [Google Scholar]
- 152.Ioannou IM, Tsawdaroglou N, Sekeris CE. Presence of glucocorticoid responsive elements in the mitochondrial genome. Anticancer Res. 1988;8:1405–09. [PubMed] [Google Scholar]
- 153.Ishii T, Yasuda K, Akatsuka A, Hino O, Hartman PS, Ishii N. A mutation in the SDHC gene of complex II increases oxidative stress, resulting in apoptosis and tumorigenesis. Cancer Res. 2005;65:203–09. [PubMed] [Google Scholar]
- 154.Ishikawa K, Takenaga K, Akimoto M, Koshikawa N, Yamaguchi A, Imanishi H, Nakada K, Honma Y, Hayashi J. ROS-generating mitochondrial DNA mutations can regulate tumor cell metastasis. Science. 2008;320:661–64. doi: 10.1126/science.1156906. [DOI] [PubMed] [Google Scholar]
- 155.Iskandar K, Rezlan M, Yadav SK, Foo CH, Sethi G, Qiang Y, Bellot GL, Pervaiz S. Synthetic Lethality of a Novel Small Molecule against Mutant KRAS-Expressing Cancer Cells Involves AKT-Dependent ROS Production. Antioxid Redox Signal. 2016;24:781–94. doi: 10.1089/ars.2015.6362. [DOI] [PubMed] [Google Scholar]
- 156.Ivanov VN, Ghandhi SA, Zhou H, Huang SX, Chai Y, Amundson SA, Hei TK. Radiation response and regulation of apoptosis induced by a combination of TRAIL and CHX in cells lacking mitochondrial DNA: a role for NF-κB-STAT3-directed gene expression. Exp Cell Res. 2011;317:1548–66. doi: 10.1016/j.yexcr.2011.03.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Jacobsen BM, Richer JK, Sartorius CA, Horwitz KB. Expression profiling of human breast cancers and gene regulation by progesterone receptors. J Mammary Gland Biol Neoplasia. 2003;8:257–68. doi: 10.1023/b:jomg.0000010028.48159.84. [DOI] [PubMed] [Google Scholar]
- 158.Jager S, Handschin C, St-Pierre J, Spiegelman BM. AMP-activated protein kinase (AMPK) action in skeletal muscle via direct phosphorylation of PGC-1alpha. Proc Natl Acad Sci. 2007;104:12017–22. doi: 10.1073/pnas.0705070104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Janeway KA, Kim SY, Lodish M, Nosé V, Rustin P, Gaal J, Dahia PL, Liegl B, Ball ER, Raygada M, Lai AH, Kelly L, Hornick JL, NIH Pediatric and Wild-Type GIST Clinic. O’Sullivan M, de Krijger RR, Dinjens WN, Demetri GD, Antonescu CR, Fletcher JA, Helman L, Stratakis CA. Defects in succinate dehydrogenase in gastrointestinal stromal tumors lacking KIT and PDGFRA mutations. PNAS. 2011;108:314–18. doi: 10.1073/pnas.1009199108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Janik J, Swoboda M, Janowska B, Cieśla JM, Gackowski D, Kowalewski J, Olinski R, Tudek B, Speina E. 8-Oxoguanine incision activity is impaired in lung tissues of NSCLC patients with the polymorphism of OGG1 and XRCC1 genes. Mutat Res. 2011:709–710. 21–31. doi: 10.1016/j.mrfmmm.2011.02.009. [DOI] [PubMed] [Google Scholar]
- 161.Jazwinski SM, Kriete A. The yeast retrograde response as a model of intracellular signaling of mitochondrial dysfunction. Front Physiol. 2012;3:39. doi: 10.3389/fphys.2012.00139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Jensen EV, Jacobson HI. Basic guides to the mechanism of estrogen action. Recent Prog Horm Res. 1962;18:318–414. [Google Scholar]
- 163.Jeong J, Juhn K, Lee H, Kim SH, Min BH, Lee KM, Cho MH, Park GH, Lee KH. SIRT1 promotes DNA repair activity and deacetylation of Ku70. Experimental &molecular medicine. 2007;39:8–13. doi: 10.1038/emm.2007.2. [DOI] [PubMed] [Google Scholar]
- 164.Jiang WW, Masayesva B, Zahurak M, Carvalho AL, Rosenbaum E, Mambo E, Zhou S, Minhas K, Benoit N, Westra WH, Alberg A, Sidransky D, Koch W, Califano J. Increased mitochondrial DNA content in saliva associated with head and neck cancer. Clin Cancer Res. 2005;11:2486–91. doi: 10.1158/1078-0432.CCR-04-2147. [DOI] [PubMed] [Google Scholar]
- 165.Jin L, Li D, Alesi GN, Fan J, Kang HB, Lu Z, Boggon TJ, Jin P, Yi H, Wright ER, Duong D, Seyfried NT, Egnatchik R, DeBerardinis RJ, Magliocca KR, He C, Arellano ML, Khoury HJ, Shin DM, Khuri FR, Kang S. Glutamate dehydrogenase 1 signals through antioxidant glutathione peroxidase 1 to regulate redox homeostasis and tumor growth. Cancer Cell. 2015;27:257–70. doi: 10.1016/j.ccell.2014.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Jones RG, Plas DR, Kubek S, Buzzai M, Mu J, Xu Y, Birnbaum MJ, Thompson CB. AMP-activated protein kinase induces a p53-dependent metabolic checkpoint. Mol Cell. 2005;18:283–93. doi: 10.1016/j.molcel.2005.03.027. [DOI] [PubMed] [Google Scholar]
- 167.Joo HY, Yun M, Jeong J, Park ER, Shin HJ, Woo SR, Jung JK, Kim YM, Park JJ, Kim J, Lee KH. SIRT1 deacetylates and stabilizes hypoxia-inducible factor-1alpha (HIF-1alpha) via direct interactions during hypoxia. Biochem Biophys Res Commun. 2015;462:294–300. doi: 10.1016/j.bbrc.2015.04.119. [DOI] [PubMed] [Google Scholar]
- 168.Jovaisaite V, Auwerx J. The mitochondrial unfolded protein response—synchronizing genomes. Curr Opin Cell Biol. 2015;33:74–81. doi: 10.1016/j.ceb.2014.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Kaipparettu BA, Ma Y, Park JH, Lee TL, Zhang Y, Yotnda P, Creighton CJ, Chan WY, Wong LJ. Crosstalk from non-cancerous mitochondria can inhibit tumor properties of metastatic cells by suppressing oncogenic pathways. PLoS One. 2013;8:61747. doi: 10.1371/journal.pone.0061747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Kamiya H. Mutagenic potentials of damaged nucleic acids produced by reactive oxygen/nitrogen species: approaches using synthetic oligonucleotides and nucleotides: survey and summary. Nucleic Acids Res. 2003;31:517–31. doi: 10.1093/nar/gkg137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Kaplon J, Zheng L, Meissl K, Chaneton B, Selivanov VA, Mackay G, van der Burg SH, Verdegaal EM, Cascante M, Shlomi T, Gottlieb E, Peeper DS. A key role for mitochondrial gatekeeper pyruvate dehydrogenase in oncogene-induced senescence. Nature. 2013;498:109–12. doi: 10.1038/nature12154. [DOI] [PubMed] [Google Scholar]
- 172.Karahalil B, de Souza-Pinto NC, Parsons JL, Elder RH, Bohr VA. Compromised incision of oxidized pyrimidines in liver mitochondria of mice deficient in NTH1 and OGG1 glycosylases. J Biol Chem. 2003;278:33701–07. doi: 10.1074/jbc.M301617200. [DOI] [PubMed] [Google Scholar]
- 173.Kennedy SG, Kandel ES, Cross TK, Hay N. Akt/Protein kinase B inhibits cell death by preventing the release of cytochrome c from mitochondria. Mol Cell Biol. 1999;19:5800–10. doi: 10.1128/mcb.19.8.5800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Khan MH, Khalil A, Rashid H. Evaluation of the p53 Arg72Pro polymorphism and its association with cancer risk: a HuGE review and meta-analysis. Genet Res (Camb) 2015;97:e7. doi: 10.1017/S0016672315000075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Khutornenko AA, Roudko VV, Chernyak BV, Vartapetian AB, Chumakov PM, Evstafieva AG. Pyrimidine biosynthesis links mitochondrial respiration to the p53 pathway. PNAS. 107:12828–33. doi: 10.1073/pnas.0910885107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Kim HJ, Winge DR. Emerging concepts in the flavinylation of succinate dehydrogenase. Biochim Biophys Acta. 2013;1827:627–36. doi: 10.1016/j.bbabio.2013.01.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Kim HS, Kim BH, Jung JE, Lee CS, Lee HG, Lee JW, Lee KH, You HJ, Chung MH, Ye SK. Potential role of 8-oxoguanine DNA glycosylase 1 as a STAT1 coactivator in endotoxin-induced inflammatory response. Free Radic Biol Med. 2016;93:12–22. doi: 10.1016/j.freeradbiomed.2015.10.415. [DOI] [PubMed] [Google Scholar]
- 178.Kim MP, Zhang Y, Lozano G. Mutant p53: Multiple Mechanisms Define Biologic Activity in Cancer. Front Oncol. 2015;5:249. doi: 10.3389/fonc.2015.00249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Kim R, Kubal T. To give or not to give anti-epidermal growth factor receptor (EGFR) monoclonal antibodies to patients with KRAS G13D mutation in advanced colorectal cancer. Clin Colorectal Cancer. 2012;11:85–87. doi: 10.1016/j.clcc.2011.10.005. [DOI] [PubMed] [Google Scholar]
- 180.Kim SJ, Syed GH, Siddiqui A. Hepatitis C virus induces the mitochondrial translocation of Parkin and subsequent mitophagy. PLoS Pathog. 2013;9:e1003285. doi: 10.1371/journal.ppat.1003285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.Kirkinezos IG, Moraes CT. Reactive oxygen species and mitochondrial diseases. Semin Cell Dev Biol. 2001;12:449–57. doi: 10.1006/scdb.2001.0282. [DOI] [PubMed] [Google Scholar]
- 182.Kirstein-Miles J, Morimoto RI. Peptides signal mitochondrial stress. Cell Metab. 2010;11:177–78. doi: 10.1016/j.cmet.2010.02.011. [DOI] [PubMed] [Google Scholar]
- 183.Ko BH, Paik JY, Jung KH, Lee KH. 17beta-estradiol augments 18F-FDG uptake and glycolysis of T47D breast cancer cells via membrane-initiated rapid PI3K-Akt activation. J Nucl Med. 2010;51:1740–47. doi: 10.2967/jnumed.110.074708. [DOI] [PubMed] [Google Scholar]
- 184.Koochekpour S, Buckles E, Shourideh M, Hu S, Chandra D, Zabaleta J, Attwood K. Androgen receptor mutations and polymorphisms in African American prostate cancer. Int J Biol Sci. 2014;10:643–51. doi: 10.7150/ijbs.8974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185.Koochekpour S, Marlowe T, Singh KK, Attwood K, Chandra D. Reduced mitochondrial DNA content associates with poor prognosis of prostate cancer in African American men. PLoS One. 2013;8:e74688. doi: 10.1371/journal.pone.0074688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186.Kovacs A, Storkel S, Thoenes W, Kovacs G. Mitochondrial and chromosomal DNA alterations in human chromophobe renal cell carcinomas. J Pathol. 1992;167:273–77. doi: 10.1002/path.1711670303. [DOI] [PubMed] [Google Scholar]
- 187.Kuiper GG, Gustafsson JA. The novel estrogen receptor-beta subtype: potential role in the cell- and promoter-specific actions of estrogens and anti-estrogens. FEBS Lett. 1997;410:87–90. doi: 10.1016/s0014-5793(97)00413-4. [DOI] [PubMed] [Google Scholar]
- 188.Kujoth GC, Hiona A, Pugh TD, Someya S, Panzer K, Wohlgemuth SE, Hofer T, Seo AY, Sullivan R, Jobling WA, Morrow JD, Van Remmen H, Sedivy JM, Yamasoba T, Tanokura M, Weindruch R, Leeuwenburgh C, Prolla TA. Mitochondrial DNA mutations, oxidative stress, and apoptosis in mammalian aging. Science. 2005;309:481–84. doi: 10.1126/science.1112125. [DOI] [PubMed] [Google Scholar]
- 189.Kulawiec M, Ayyasamy V, Singh KK. p53 regulates mtDNA copy number and mitocheckpoint pathway. J Carcinog. 2009;8:8. doi: 10.4103/1477-3163.50893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 190.Kulawiec M, Owens KM, Singh KK. Cancer cell mitochondria confer apoptosis resistance and promote metastasis. Cancer Biol Ther. 2009;8:1378–85. doi: 10.4161/cbt.8.14.8751. [DOI] [PubMed] [Google Scholar]
- 191.Kulawiec M, Owens KM, Singh KK. mtDNA G10398A variant in African-American women with breast cancer provides resistance to apoptosis and promotes metastasis in mice. J Hum Genet. 2009;54:647–54. doi: 10.1038/jhg.2009.89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 192.Kulawiec M, Safina A, Desouki MM, Still I, Matsui S, Bakin A, Singh KK. Tumorigenic transformation of human breast epithelial cells induced by mitochondrial DNA depletion. Cancer Biol Ther. 2008;7:1732–43. doi: 10.4161/cbt.7.11.6729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 193.Kulawiec M, Arnouk H, Desouki MM, Kazim L, Still I, Singh KK. Proteomic anlysis of proteins involved in mitochondria-to-nucleus retrograde response in human cancer cells. Cancer Biol Ther. 2006;5 doi: 10.4161/cbt.5.8.2880. [DOI] [PubMed] [Google Scholar]
- 194.Kunkel TA, Erie DA. DNA mismatch repair. Annu Rev Biochem. 2005;74:681–710. doi: 10.1146/annurev.biochem.74.082803.133243. [DOI] [PubMed] [Google Scholar]
- 195.La DK, Swenberg JA. DNA adducts: biological markers of exposure and potential applications to risk assessment. Mutat Res. 1996;365:129–46. doi: 10.1016/s0165-1110(96)90017-2. [DOI] [PubMed] [Google Scholar]
- 196.Lam ET, Bracci PM, Holly EA, Chu C, Poon A, Wan E, White K, Kwok PY, Pawlikowska L, Tranah GJ. Mitochondrial DNA sequence variation and risk of pancreatic cancer. Cancer Res. 2012;72:686–95. doi: 10.1158/0008-5472.CAN-11-1682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 197.Landerer E, Villegas J, Burzio VA, Oliveira L, Villota C, Lopez C, et al. Nuclear localization of the mitochondrial ncRNAs in normal and cancer cells. Cell Oncol (Dordr) 2011;34:297–305. doi: 10.1007/s13402-011-0018-8. [DOI] [PubMed] [Google Scholar]
- 198.Lapucci A, Pittelli M, Rapizzi E, Felici R, Moroni F, Chiarugi A. Poly(ADPribose) polymerase-1 is a nuclear epigenetic regulator of mitochondrial DNA repair and transcription. Molecular pharmacology. 2011;79:932–40. doi: 10.1124/mol.110.070110. [DOI] [PubMed] [Google Scholar]
- 199.Lascorz J, Bevier M, Schönfels WV, Kalthoff H, Aselmann H, Beckmann J, Egberts J, Buch S, Becker T, Schreiber S, Hampe J, Hemminki K, Försti A, Schafmayer C. Polymorphisms in the mitochondrial oxidative phosphorylation chain genes as prognostic markers for colorectal cancer. BMC Med Genet. 2012;13:31. doi: 10.1186/1471-2350-13-31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 200.LeBleu VS, O’Connell JT, Gonzalez Herrera KN, Wikman H, Pantel K, Haigis MC, de Carvalho FM, Damascena A, Domingos Chinen LT, Rocha RM, Asara JM, Kalluri R. PGC-1α mediates mitochondrial biogenesis and oxidative phosphorylation in cancer cells to promote metastasis. Nat Cell Biol. 2014;16:992–1003. doi: 10.1038/ncb3039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 201.LeDoux SP, Wilson GL, Beecham EJ, Stevnsner T, Wassermann K, Bohr VA. Repair of mitochondrial DNA after various types of DNA damage in Chinese hamster ovary cells. Carcinogenesis. 1992;13:1967–73. doi: 10.1093/carcin/13.11.1967. [DOI] [PubMed] [Google Scholar]
- 202.Wu HC, Huang KH, Yeh TS, Chi CW. Somatic alterations in mitochondrial DNA and mitochondrial dysfunction in gastric cancer progression. World J Gastroenterol. 2015;20:3950–59. doi: 10.3748/wjg.v20.i14.3950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 203.Lee HC, Yin PH, Lin JC, Wu CC, Chen CY, Wu CW, Chi CW, Tam TN, Wei YH. Mitochondrial genome instability and mtDNA depletion in human cancers. Ann N Y Acad Sci. 2005;1042:109–22. doi: 10.1196/annals.1338.011. [DOI] [PubMed] [Google Scholar]
- 204.Lehtonen HJ, Blanco I, Piulats JM, Herva R, Launonen V, Aaltonen LA. Conventional renal cancer in a patient with fumarate hydratase mutation. Hum Pathol. 2007;38:793–96. doi: 10.1016/j.humpath.2006.10.011. [DOI] [PubMed] [Google Scholar]
- 205.Lehtonen R, Kiuru M, Vanharanta S, Sjöberg J, Aaltonen LM, Aittomäki K, Arola J, Butzow R, Eng C, Husgafvel-Pursiainen K, Isola J, Järvinen H, Koivisto P, Mecklin JP, Peltomäki P, Salovaara R, Wasenius VM, Karhu A, Launonen V, Nupponen NN, Aaltonen LA. Biallelic inactivation of fumarate hydratase (FH) occurs in nonsyndromic uterine leiomyomas but is rare in other tumors. Am J Pathol. 2004;164:17–22. doi: 10.1016/S0002-9440(10)63091-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 206.Li E, Ji P, Ouyang N, Zhang Y, Wang XY, Rubin DC, Davidson NO, Bergamaschi R, Shroyer KR, Burke S, Zhu W, Williams JL. Differential expression of miRNAs in colon cancer between African and Caucasian Americans: implications for cancer racial health disparities. Int J Oncol. 2014;45:587–94. doi: 10.3892/ijo.2014.2469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 207.Li H, Hao X, Zhang W, Wei Q, Chen K. The hOGG1 Ser326Cys polymorphism and lung cancer risk: a meta-analysis. Cancer Epidemiol Biomarkers Prev. 2008;17:1739–45. doi: 10.1158/1055-9965.EPI-08-0001. [DOI] [PubMed] [Google Scholar]
- 208.Li J, Donath S, Li Y, Qin D, Prabhakar BS, Li P. miR-30 regulates mitochondrial fission through targeting p53 and the dynamin-related protein-1 pathway. PLoS Genet. 2010;6:e1000795. doi: 10.1371/journal.pgen.1000795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 209.Li JJ, Oberley LW, St Clair DK, Ridnour LA, Oberley TD. Phenotypic changes induced in human breast cancer cells by overexpression of manganese-containing superoxide dismutase. Oncogene. 1995;10:1989–2000. [PubMed] [Google Scholar]
- 210.Li S, Yan T, Yang JQ, Oberley TD, Oberley LW. The role of cellular glutathione peroxidase redox regulation in the suppression of tumor cell growth by manganese superoxide dismutase. Cancer Res. 2000;60:3927–39. [PubMed] [Google Scholar]
- 211.Li X, Zhong Y, Lu J, Axcrona K, Eide L, Syljuåsen RG, Peng Q, Wang J, Zhang H, Goscinski MA, Kvalheim G, Nesland JM, Suo Z. MtDNA depleted PC3 cells exhibit Warburg effect and cancer stem cell features. Oncotarget. 2016;7:40297–313. doi: 10.18632/oncotarget.9610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 212.Li XY, Su M, Huang HH, Li H, Tian DP, Gao YX. mtDNA evidence: genetic background associated with related populations at high risk for esophageal cancer between Chaoshan and Taihang Mountain areas in China. Genomics. 2007;90:474–81. doi: 10.1016/j.ygeno.2007.06.006. [DOI] [PubMed] [Google Scholar]
- 213.Liao J, Ding D, Sun C, Weng D, Meng L, Chen G, Ma D. Polymorphisms of progesterone receptor and ovarian cancer risk: a systemic review and meta-analysis. J Obstet Gynaecol Res. 2015;41:178–87. doi: 10.1111/jog.12519. [DOI] [PubMed] [Google Scholar]
- 214.Liao X, Butow RA. RTG1 and RTG2: two yeast genes required for a novel path of communication from mitochondria to the nucleus. Cell. 1993;72:61–71. doi: 10.1016/0092-8674(93)90050-z. [DOI] [PubMed] [Google Scholar]
- 215.Lim JH, Lee YM, Chun YS, Chen J, Kim JE, Park JW. Sirtuin 1 modulates cellular responses to hypoxia by deacetylating hypoxia-inducible factor 1alpha. Mol Cell. 2010;38:864–78. doi: 10.1016/j.molcel.2010.05.023. [DOI] [PubMed] [Google Scholar]
- 216.Lim S, Smith KR, Lim SS, Tian R, Lu J, Tan M. Regulation of mitochondrial function by protein phosphorylation and de-phosphorylation. Cell Biosci. 2016;6:25–39. doi: 10.1186/s13578-016-0089-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 217.Lin JC, Wang CC, Jiang RS, Wang WY, Liu SA. Impact of somatic mutations in the D-loop of mitochondrial DNA on the survival of oral squamous cell carcinoma patients. PLoS One. 2015;10:e0124322. doi: 10.1371/journal.pone.0124322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 218.Lin RJ, Xiao DW, Liao LD, Chen T, Xie ZF, Huang WZ, Wang WS, Jiang TF, Wu BL, Li EM, Xu LY. MiR-142-3p as a potential prognostic biomarker for esophageal squamous cell carcinoma. J Surg Oncol. 2012;105:175–82. doi: 10.1002/jso.22066. [DOI] [PubMed] [Google Scholar]
- 219.Lindahl T. Instability and decay of the primary structure of DNA. Nature. 1993;362:709–15. doi: 10.1038/362709a0. [DOI] [PubMed] [Google Scholar]
- 220.Liou CW, Lin TK, Chen JB, Tiao MM, Weng SW, Chen SD, Chuang YC, Chuang JH, Wang PW. Association between a common mitochondrial DNA D-loop polycytosine variant and alteration of mitochondrial copy number in human peripheral blood cells. J Med Genet. 2010;47:723–28. doi: 10.1136/jmg.2010.077552. [DOI] [PubMed] [Google Scholar]
- 221.Liu T, Chen L, Sun X, Wang Y, Li S, Yin X, Wang X, Ding C, Li H, Di W. Progesterone receptor PROGINS and +331G/A polymorphisms confer susceptibility to ovarian cancer: a meta-analysis based on 17 studies. Tumor Biol. 2014;35:2427–36. doi: 10.1007/s13277-013-1322-x. [DOI] [PubMed] [Google Scholar]
- 222.Liu W, Innocenti F, Chen P, Das S, Cook EH, Jr, Ratain MJ. Interethnic difference in the allelic distribution of human epidermal growth factor receptor intron 1 polymorphism. Clin Cancer Res. 2003;9:1009–12. [PubMed] [Google Scholar]
- 223.Liu Y, Liu L, Ma X, Yin Y, Tang B, Li Z. Characteristics and neural-like differentiation of mesenchymal stem cells derived from foetal porcine bone marrow. Biosci Rep. 2013;33:e00032. doi: 10.1042/BSR20120023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 224.Liu Y, Zha L, Li B, Zhang L, Yu T, Li L. Correlation between superoxide dismutase 1 and 2 polymorphisms and susceptibility to oral squamous cell carcinoma. Exp Ther Med. 2014;7:171–78. doi: 10.3892/etm.2013.1375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 225.Lo HW, Hsu SC, Hung MC. EGFR signaling pathway in breast cancers: from traditional signal transduction to direct nuclear translocalization. Breast Cancer Res Treat. 2006;95:211–18. doi: 10.1007/s10549-005-9011-0. [DOI] [PubMed] [Google Scholar]
- 226.Longley MJ, Prasad R, Srivastava DK, Wilson SH, Copeland WC. Identification of 5′-deoxyribose phosphate lyase activity in human DNA polymerase gamma and its role in mitochondrial base excision repair in vitro. NAS. 1998;95:12244–48. doi: 10.1073/pnas.95.21.12244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 227.Lou C, Xiao M, Cheng S, Lu X, Jia S, Ren Y, Li Z. MiR-485-3p and miR-485-5p suppress breast cancer cell metastasis by inhibiting PGC-1α expression. Cell Death Dis. 2016;7:2159. doi: 10.1038/cddis.2016.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 228.Loveland B, Wang CR, Yonekawa H, Hermel E, Lindahl KF. Maternally transmitted histocompatibility antigen of mice: a hydrophobic peptide of a mitochondrially encoded protein. Cell. 1990;60:971–80. doi: 10.1016/0092-8674(90)90345-f. [DOI] [PubMed] [Google Scholar]
- 229.Lu C, Ward PS, Kapoor GS, Rohle D, Turcan S, Abdel-Wahab O, Edwards CR, Khanin R, Figueroa ME, Melnick A, Wellen KE, O’Rourke DM, Berger SL, Chan TA, Levine RL, Mellinghoff IK, Thompson CB. IDH mutation impairs histone demethylation and results in a block to cell differentiation. Nature. 2012;483:474–78. doi: 10.1038/nature10860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 230.Luna A, Aladjem MI, Kohn KW. SIRT1/PARP1 crosstalk: connecting DNA damage and metabolism. Genome integrity. 2013;4:6. doi: 10.1186/2041-9414-4-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 231.Magda D, Lecane P, Prescott J, Thiemann P, Ma X, Dranchak PK, Toleno DM, Ramaswamy K, Siegmund KD, Hacia JG. mtDNA depletion confers specific gene expression profiles in human cells grown in culture and in xenograft. BMC Genomics. 2008;9:521. doi: 10.1186/1471-2164-9-521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 232.Majewski M, Nieborowska-Skorska M, Salomoni P, Slupianek A, Reiss K, Trotta R, Calabretta B, Skorski T. Activation of mitochondrial Raf-1 is involved in the anti-apoptotic effects of Akt. Cancer Res. 1999;59:2815–19. [PubMed] [Google Scholar]
- 233.Malhi RS, Mortensen HM, Eshleman JA, Kemp BM, Lorenz JG, Kaestle FA, Johnson JR, Gorodezky C, Smith DG. Native American mtDNA prehistory in the American Southwest. Am J Phys Anthropol. 2003;120:108–24. doi: 10.1002/ajpa.10138. [DOI] [PubMed] [Google Scholar]
- 234.Mambo E, Chatterjee A, de Souza-Pinto NC, Mayard S, Hogue BA, Hoque MO, Dizdaroglu M, Bohr VA, Sidransky D. Oxidized guanine lesions and hOgg1 activity in lung cancer. Oncogene. 2005;24:4496–508. doi: 10.1038/sj.onc.1208669. [DOI] [PubMed] [Google Scholar]
- 235.Mambo E, Chatterjee A, Xing M, Tallini G, Haugen BR, Yeung SC, Sukumar S, Sidransky D. Tumor-specific changes in mtDNA content in human cancer. Int J Cancer. 2005;116:920–24. doi: 10.1002/ijc.21110. [DOI] [PubMed] [Google Scholar]
- 236.Mandal RK, Mittal T, Kapoor R, Mittal RD. NER and BER repair gene polymorphisms in a healthy north Indian cohort and comparison with different ethnic groups worldwide. Asian Pac J Cancer Prev. 2010;11:1601–04. [PubMed] [Google Scholar]
- 237.Mandal SM, Hegde ML, Chatterjee A, Hegde PM, Szczesny B, Banerjee D, Boldogh I, Gao R, Falkenberg M, Gustafsson CM, Sarkar PS, Hazra TK. Role of human DNA glycosylase Nei-like 2 (NEIL2) and single strand break repair protein polynucleotide kinase 3′-phosphatase in maintenance of mitochondrial genome. J Biol Chem. 2012;287:2819–29. doi: 10.1074/jbc.M111.272179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 238.Marchenko ND, Zaika A, Moll UM. Death signal-induced localization of p53 protein to mitochondria. A potential role in apoptotic signaling. J Biol Chem. 2000;275:16202–12. doi: 10.1074/jbc.275.21.16202. [DOI] [PubMed] [Google Scholar]
- 239.Marchenko ND, Moll UM. The role of ubiquitination in the direct mitochondrial death program of p53. Cell Cycle. 2007;6:1718–23. doi: 10.4161/cc.6.14.4503. [DOI] [PubMed] [Google Scholar]
- 240.Marcu R, Wiczer BM, Neeley CK, Hawkins BJ. Mitochondrial matrix Ca(2)(+) accumulation regulates cytosolic NAD(+)/NADH metabolism, protein acetylation, and sirtuin expression. Mol Cell Biol. 2014;34:2890–902. doi: 10.1128/MCB.00068-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 241.Mardis ER, Wilson RK. Cancer genome sequencing: a review. Hum Mol Genet. 2009;18:163–68. doi: 10.1093/hmg/ddp396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 242.Martinez-Contreras R, Cloutier P, Shkreta L, Fisette JF, Revil T, Chabot B. hnRNP proteins and splicing control. Adv Exp Med Biol. 2007;623:123–47. doi: 10.1007/978-0-387-77374-2_8. [DOI] [PubMed] [Google Scholar]
- 243.Martinou JC, Youle RJ. Mitochondria in apoptosis: Bcl-2 family members and mitochondrial dynamics. Dev Cell. 2011;21:92–101. doi: 10.1016/j.devcel.2011.06.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 244.Marucci G, Maresca A, Caporali L, Farnedi A, Betts CM, Morandi L, de Biase D, Cerasoli S, Foschini MP, Bonora E, Vidone M, Romeo G, Perli E, Giordano C, d’Amati G, Gasparre G, Baruzzi A, Carelli V, Eusebi V. Oncocytic glioblastoma: a glioblastoma showing oncocytic changes and increased mitochondrial DNA copy number. Hum Pathol. 2013;44:1867–76. doi: 10.1016/j.humpath.2013.02.014. [DOI] [PubMed] [Google Scholar]
- 245.Mason PA, Matheson EC, Hall AG, Lightowlers RN. Mismatch repair activity in mammalian mitochondria. Nucleic Acids Res. 2003;31:1052–58. doi: 10.1093/nar/gkg167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 246.Mathupala SP, Ko YH, Pedersen PL. Hexokinase II: cancer’s double-edged sword acting as both facilitator and gatekeeper of malignancy when bound to mitochondria. Oncogene. 2006;25:4777–86. doi: 10.1038/sj.onc.1209603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 247.Mathupala SP, Heese C, Pedersen PL. Glucose catabolism in cancer cells. The type II hexokinase promoter contains functionally active response elements for the tumor suppressor p53. J Biol Chem. 1997;272:22776–80. doi: 10.1074/jbc.272.36.22776. [DOI] [PubMed] [Google Scholar]
- 248.Matoba S, Kang JG, Patino WD, Wragg A, Boehm M, Gavrilova O, Hurley PJ, Bunz F, Hwang PM. p53 regulates mitochondrial respiration. Science. 2006;312:1650–53. doi: 10.1126/science.1126863. [DOI] [PubMed] [Google Scholar]
- 249.Matsuyama M, Suzuki H. Seizing mechanism and fate of intranuclear mitochondria. Experientia. 1972;28:1347–8. doi: 10.1007/BF01965337. [DOI] [PubMed] [Google Scholar]
- 250.Mattingly KA, Ivanova MM, Riggs KA, Wickramasinghe NS, Barch MJ, Klinge CM. Estradiol stimulates transcription of nuclear respiratory factor-1 and increases mitochondrial biogenesis. Mol Endocrinol. 2008;22:609–22. doi: 10.1210/me.2007-0029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 251.Maurya PK, Noto C, Rizzo LB, Rios AC, Nunes SO, Barbosa DS, Sethi S, Zeni M, Mansur RB, Maes M, Brietzke E. The role of oxidative and nitrosative stress in accelerated aging and major depressive disorder. Prog Neuropsychopharmacol Biol Psychiatry. 2016;65:134–44. doi: 10.1016/j.pnpbp.2015.08.016. [DOI] [PubMed] [Google Scholar]
- 252.Mendillo ML, Hargreaves VV, Jamison JW, Mo AO, Li S, Putnam CD, Woods VL, Jr, Kolodner RD. A conserved MutS homolog connector domain interface interacts with MutL homologs. PNAS. 2009;106:22223–28. doi: 10.1073/pnas.0912250106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 253.Meng F, Henson R, Wehbe-Janek H, Ghoshal K, Jacob ST, Patel T. MicroRNA-21 regulates expression of the PTEN tumor suppressor gene in human hepatocellular cancer. Gastroenterology. 2007;133:647–58. doi: 10.1053/j.gastro.2007.05.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 254.Meng X, Lu P, Chu Z, Fan Q. The androgen receptor cytosine-adenine-guanine repeat length contributes to the development of epithelial ovarian cancer. Oncotarget. 2016;7:2105–12. doi: 10.18632/oncotarget.6012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 255.Merino D, Malkin D. p53 and hereditary cancer. Subcell Biochem. 2014;85:1–16. doi: 10.1007/978-94-017-9211-0_1. [DOI] [PubMed] [Google Scholar]
- 256.Michishita E, Park JY, Burneskis JM, Barrett JC, Horikawa I. Evolutionarily conserved and nonconserved cellular localizations and functions of human SIRT proteins. Molecular biology of the cell. 2005;16:4623–35. doi: 10.1091/mbc.E05-01-0033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 257.Mihara M, Erster S, Zaika A, Petrenko O, Chittenden T, Pancoska P, Moll UM. p53 has a direct apoptogenic role at the mitochondria. Mol Cell. 2003;11:577–90. doi: 10.1016/s1097-2765(03)00050-9. [DOI] [PubMed] [Google Scholar]
- 258.Minocherhomji S, Tollefsbol TO, Singh KK. Mitochondrial regulation of epigenetics and its role in human diseases. Epigenetics. 2012;7:326–34. doi: 10.4161/epi.19547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 259.Miyaki M, Yatagai K, Ono T. Strand breaks of mammalian mitochondrial DNA induced by carcinogens. Chem Biol Interact. 1977;17:321–29. doi: 10.1016/0009-2797(77)90095-3. [DOI] [PubMed] [Google Scholar]
- 260.Mizumachi T, Muskhelishvili L, Naito A, Furusawa J, Fan CY, Siegel ER, Kadlubar FF, Kumar U, Higuchi M. Increased distributional variance of mitochondrial DNA content associated with prostate cancer cells as compared with normal prostate cells. Prostate. 2008;68:408–17. doi: 10.1002/pros.20697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 261.Mizumachi T, Suzuki S, Naito A, Carcel-Trullols J, Evans TT, Spring PM, Oridate N, Furuta Y, Fukuda S, Higuchi M. Increased mitochondrial DNA induces acquired docetaxel resistance in head and neck cancer cells. Oncogene. 2008;27:831–38. doi: 10.1038/sj.onc.1210681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 262.Mizuno H, Honda M, Shirasaki T, Yamashita T, Yamashita T, Mizukoshi E, Kaneko S. Heterogeneous nuclear ribonucleoprotein A2/B1 in association with hTERT is a potential biomarker for hepatocellular carcinoma. Liver Int. 2012;32:1146–55. doi: 10.1111/j.1478-3231.2012.02778.x. [DOI] [PubMed] [Google Scholar]
- 263.Modica-Napolitano JS, Singh KK. Mitochondrial dysfunction in cancer. Mitochondrion. 2004;4:755–62. doi: 10.1016/j.mito.2004.07.027. [DOI] [PubMed] [Google Scholar]
- 264.Modrich P. Mechanisms in eukaryotic mismatch repair. J Biol Chem. 2006;281:30305–09. doi: 10.1074/jbc.R600022200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 265.Mohrin M, Shin J, Liu Y, Brown K, Luo H, Xi Y, Haynes CM, Chen D. Stem cell aging. A mitochondrial UPR-mediated metabolic checkpoint regulates hematopoietic stem cell aging. Science. 2015;347:1374–77. doi: 10.1126/science.aaa2361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 266.Molitoris JK, McColl KS, Distelhorst CW. Glucocorticoid-mediated repression of the oncogenic microRNA cluster miR-17~92 contributes to the induction of Bim and initiation of apoptosis. Mol Endocrinol. 2011;20:409–20. doi: 10.1210/me.2010-0402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 267.Monaghan RM, Whitmarsh AJ. Mitochondrial Proteins Moonlighting in the Nucleus. Trends Biochem Sci. 2015;40:728–35. doi: 10.1016/j.tibs.2015.10.003. [DOI] [PubMed] [Google Scholar]
- 268.Mordukhovich I, Beyea J, Herring AH, Hatch M, Stellman SD, Teitelbaum SL, Richardson DB, Millikan RC, Engel LS, Shantakumar S, Steck SE, Neugut AI, Rossner P, Jr, Santella RM, Gammon MD. Polymorphisms in DNA repair genes, traffic-related polycyclic aromatic hydrocarbon exposure and breast cancer incidence. Int J Cancer. 2016;139:310–21. doi: 10.1002/ijc.30079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 269.Mosquera-Miguel A, Alvarez-Iglesias V, Carracedo A, Salas A, Vega A, Carracedo A, Milne R, de León AC, Benitez J, Carracedo A, Salas A. Is mitochondrial DNA variation associated with sporadic breast cancer risk? Cancer Res. 2008;68:623–25. doi: 10.1158/0008-5472.CAN-07-2385. [DOI] [PubMed] [Google Scholar]
- 270.Mourier T, Hansen AJ, Willerslev E, Arctander P. The Human Genome Project reveals a continuous transfer of large mitochondrial fragments to the nucleus. Mol Biol Evol. 2001;18:1833–7. doi: 10.1093/oxfordjournals.molbev.a003971. [DOI] [PubMed] [Google Scholar]
- 271.Mourier T. Reverse transcription in genome evolution. Cytogenet Genome Res. 2005;110:56. doi: 10.1159/000084938. [DOI] [PubMed] [Google Scholar]
- 272.Naito A, Carcel-Trullols J, Xie CH, Evans TT, Mizumachi T, Higuchi M. Induction of acquired resistance to antiestrogen by reversible mitochondrial DNA depletion in breast cancer cell line. Int J Cancer. 2008;122:1506–11. doi: 10.1002/ijc.23235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 273.Naito A, Cook CC, Mizumachi T, Wang M, Xie CH, Evans TT, Kelly T, Higuchi M. Progressive tumor features accompany epithelial-mesenchymal transition induced in mitochondrial DNA-depleted cells. Cancer Sci. 2008;99:1584–8. doi: 10.1111/j.1349-7006.2008.00879.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 274.Navaglia F, Basso D, Fogar P, Sperti C, Greco E, Zambon CF, Stranges A, Falda A, Pizzi S, Parenti A, Pedrazzoli S, Plebani M. Mitochondrial DNA D-loop in pancreatic cancer: somatic mutations are epiphenomena while the germline 16519 T variant worsens metabolism and outcome. Am J Clin Pathol. 2006;126:593–601. doi: 10.1309/GQFCCJMH5KHNVX73. [DOI] [PubMed] [Google Scholar]
- 275.Niemann S, Müller U. Mutations in SDHC cause autosomal dominant paraganglioma, type 3. Nat Genet. 2000;26:268–70. doi: 10.1038/81551. [DOI] [PubMed] [Google Scholar]
- 276.Nomura M, Shigematsu H, Li L, Suzuki M, Takahashi T, Estess P, Siegelman M, Feng Z, Kato H, Marchetti A, Shay JW, Spitz MR, Wistuba II, Minna JD, Gazdar AF. Polymorphisms, mutations, and amplification of the EGFR gene in non-small cell lung cancers. PLoS Med. 2007;4:125. doi: 10.1371/journal.pmed.0040125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 277.Omidvar N, Kogan S, Beurlet S, le Pogam C, Janin A, West R, Noguera ME, Reboul M, Soulie A, Leboeuf C, Setterblad N, Felsher D, Lagasse E, Mohamedali A, Thomas NS, Fenaux P, Fontenay M, Pla M, Mufti GJ, Weissman I, Chomienne C, Padua RA. BCL-2 and mutant NRAS interact physically and functionally in a mouse model of progressive myelodysplasia. Cancer Res. 2007;67:11657–67. doi: 10.1158/0008-5472.CAN-07-0196. [DOI] [PubMed] [Google Scholar]
- 278.Osborne CK, Schiff R. Estrogen-receptor biology: continuing progress and therapeutic implications. J Clin Oncol. 2005;23:1616–22. doi: 10.1200/JCO.2005.10.036. [DOI] [PubMed] [Google Scholar]
- 279.Pabalan N, Pineda MR, Jarjanazi H, Christofolini DM, Barbosa CP, Bianco B. Association of the +331G/A progesterone receptor gene (PgR) polymorphism with risk of endometrial cancer in Caucasian women: a meta-analysis. Arch Gynecol Obstet. 2015;291:115–22. doi: 10.1007/s00404-014-3344-z. [DOI] [PubMed] [Google Scholar]
- 280.Papa L, Germain D. Estrogen receptor mediates a distinct mitochondrial unfolded protein response. J Cell Sci. 2011;124:1396–1402. doi: 10.1242/jcs.078220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 281.Park SY, Chang I, Kim JY, Kang SW, Park SH, Singh K, Lee MS. Resistance of mitochondrial DNA-depleted cells against cell death: role of mitochondrial superoxide dismutase. J Biol Chem. 2004;279:7512–20. doi: 10.1074/jbc.M307677200. [DOI] [PubMed] [Google Scholar]
- 282.Park SY, Choi B, Cheon H, Pak YK, Kulawiec M, Singh KK, Lee MS. Cellular aging of mitochondrial DNA-depleted cells. Biochem Biophys Res Commun. 2004;325:1399–405. doi: 10.1016/j.bbrc.2004.10.182. [DOI] [PubMed] [Google Scholar]
- 283.Parks JK, Smith TS, Trimmer PA, Bennett JP, Jr, Parker WD., Jr Neurotoxic Abeta peptides increase oxidative stress in vivo through NMDA-receptor and nitric-oxide-synthase mechanisms, and inhibit complex IV activity and induce a mitochondrial permeability transition in vitro. J Neurochem. 2001;76:1050–56. doi: 10.1046/j.1471-4159.2001.00112.x. [DOI] [PubMed] [Google Scholar]
- 284.Paschka P, Schlenk RF, Gaidzik VI, Habdank M, Krönke J, Bullinger L, Späth D, Kayser S, Zucknick M, Götze K, Horst HA, Germing U, Döhner H, Döhner K. IDH1 and IDH2 mutations are frequent genetic alterations in acute myeloid leukemia and confer adverse prognosis in cytogenetically normal acute myeloid leukemia with NPM1 mutation without FLT3 internal tandem duplication. J Clin Oncol. 2010;28:3636–43. doi: 10.1200/JCO.2010.28.3762. [DOI] [PubMed] [Google Scholar]
- 285.Patry C, Bouchard L, Labrecque P, Gendron D, Lemieux B, Toutant J, Lapointe E, Wellinger R, Chabot B. Small interfering RNA-mediated reduction in heterogeneous nuclear ribonucleoparticule A1/A2 proteins induces apoptosis in human cancer cells but not in normal mortal cell lines. Cancer Res. 2003;63:7679–88. [PubMed] [Google Scholar]
- 286.Pelicano H, Xu RH, Du M, Feng L, Sasaki R, Carew JS, Hu Y, Ramdas L, Hu L, Keating MJ, Zhang W, Plunkett W, Huang P. Mitochondrial respiration defects in cancer cells cause activation of Akt survival pathway through a redox-mediated mechanism. J Cell Biol. 2006;175:913–23. doi: 10.1083/jcb.200512100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 287.Pelisch F, Pozzi B, Risso G, Muñoz MJ, Srebrow A. DNA damage-induced heterogeneous nuclear ribonucleoprotein K sumoylation regulates p53 transcriptional activation. J Biol Chem. 2012;287:30789–99. doi: 10.1074/jbc.M112.390120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 288.Peng S, Lü B, Ruan W, Zhu Y, Sheng H, Lai M. Genetic polymorphisms and breast cancer risk: evidence from meta-analyses, pooled analyses, and genome-wide association studies. Breast Cancer Res Treat. 2011;127:309–24. doi: 10.1007/s10549-011-1459-5. [DOI] [PubMed] [Google Scholar]
- 289.Polyak K, Li Y, Zhu H, Lengauer C, Willson JK, Markowitz SD, Trush MA, Kinzler KW, Vogelstein B. Somatic mutations of the mitochondrial genome in human colorectal tumors. Nat Genet. 1998;20:291–93. doi: 10.1038/3108. [DOI] [PubMed] [Google Scholar]
- 290.Popanda O, Seibold P, Nikolov I, Oakes CC, Burwinkel B, Hausmann S, Flesch-Janys D, Plass C, Chang-Claude J, Schmezer P. Germline variants of base excision repair genes and breast cancer: A polymorphism in DNA polymerase gamma modifies gene expression and breast cancer risk. Int J Cancer. 2013;132:55–62. doi: 10.1002/ijc.27665. [DOI] [PubMed] [Google Scholar]
- 291.Prabhu A, Sarcar B, Miller CR, Kim SH, Nakano I, Forsyth P, Chinnaiyan P. Ras-mediated modulation of pyruvate dehydrogenase activity regulates mitochondrial reserve capacity and contributes to glioblastoma tumorigenesis. Neuro Oncol. 2015;17:1220–30. doi: 10.1093/neuonc/nou369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 292.Price AL, Patterson NJ, Plenge RM, Weinblatt ME, Shadick NA, Reich D. Principal components analysis corrects for stratification in genome-wide association studies. Nat Genet. 2006;38:904–09. doi: 10.1038/ng1847. [DOI] [PubMed] [Google Scholar]
- 293.Prieur A, Peeper DS. Cellular senescence in vivo: a barrier to tumorigenesis. Curr Opin Cell Biol. 2008;20:150–5. doi: 10.1016/j.ceb.2008.01.007. [DOI] [PubMed] [Google Scholar]
- 294.Psarra AM, Sekeris CE. Glucocorticoids induce mitochondrial gene transcription in HepG2 cells: role of the mitochondrial glucocorticoid receptor. Biochim Biophys Acta. 2011;1813:1814–21. doi: 10.1016/j.bbamcr.2011.05.014. [DOI] [PubMed] [Google Scholar]
- 295.Pylayeva-Gupta Y, Grabocka E, Bar-Sagi D. RAS oncogenes: weaving a tumorigenic web. Nat Rev Cancer. 2011;11:761–64. doi: 10.1038/nrc3106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 296.Raffaello A, De Stefani D, Rizzuto R. The mitochondrial Ca(2+) uniporter. Cell Calcium. 2012;52:16–21. doi: 10.1016/j.ceca.2012.04.006. [DOI] [PubMed] [Google Scholar]
- 297.Ranzani GN, Salerno-Mele P, Maltoni M, Talarico D, Della Valle G, Amadori D. Study of the c-Ha-ras-1 locus polymorphism in an Italian population with high incidence of gastric cancer. Mol Biol Med. 1988;5:145–53. [PubMed] [Google Scholar]
- 298.Rao JU, Engelke UF, Sweep FC, Pacak K, Kusters B, Goudswaard AG, Hermus AR, Mensenkamp AR, Eisenhofer G, Qin N, Richter S, Kunst HP, Timmers HJ, Wevers RA. Genotype-specific differences in the tumor metabolite profile of pheochromocytoma and paraganglioma using untargeted and targeted metabolomics. J Clin Endocrinol Metab. 2015;100:214–22. doi: 10.1210/jc.2014-2138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 299.Rasmussen AK, Chatterjee A, Rasmussen LJ, Singh KK. Mitochondria-mediated nuclear mutator phenotype in Saccharomyces cerevisiae. Nucleic Acids Res. 2003;31:3909–17. doi: 10.1093/nar/gkg446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 300.Ray AM, Zuhlke KA, Levin AM, Douglas JA, Cooney KA, Petros JA. Sequence variation in the mitochondrial gene cytochrome c oxidase subunit I and prostate cancer in African American men. Prostate. 2009;69:956–60. doi: 10.1002/pros.20943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 301.Razandi M, Pedram A, Jordan VC, Fuqua S, Levin ER. Tamoxifen regulates cell fate through mitochondrial estrogen receptor beta in breast cancer. Oncogene. 2013;232:3274–85. doi: 10.1038/onc.2012.335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 302.Rebollo A, Pérez-Sala D, Martínez-A C. Bcl-2 differentially targets K-, N-, and H-Ras to mitochondria in IL-2 supplemented or deprived cells: implications in prevention of apoptosis. Oncogene. 1999;18:4930–39. doi: 10.1038/sj.onc.1202875. [DOI] [PubMed] [Google Scholar]
- 303.Rhee SG, Chae HZ, Kim K. Peroxiredoxins: a historical overview and speculative preview of novel mechanisms and emerging concepts in cell signaling. Free Radic Biol Med. 2005;38:1543–52. doi: 10.1016/j.freeradbiomed.2005.02.026. [DOI] [PubMed] [Google Scholar]
- 304.Rhen T, Cidlowski JA. Anti-inflammatory action of glucocorticoids–new mechanisms for old drugs. N Engl J Med. 2005;353:1711–23. doi: 10.1056/NEJMra050541. [DOI] [PubMed] [Google Scholar]
- 305.Ricketts C, Woodward ER, Killick P, Morris MR, Astuti D, Latif F, Maher ER. Germline SDHB mutations and familial renal cell carcinoma. J Natl Cancer Inst. 2008;100:1260–62. doi: 10.1093/jnci/djn254. [DOI] [PubMed] [Google Scholar]
- 306.Rimessi A, Marchi S, Patergnani S, Pinton P. H-Ras-driven tumoral maintenance is sustained through caveolin-1-dependent alterations in calcium signaling. Oncogene. 2014;33:2329–40. doi: 10.1038/onc.2013.192. [DOI] [PubMed] [Google Scholar]
- 307.Rizzuto R, De Stefani D, Raffaello A, Mammucari C. Mitochondria as sensors and regulators of calcium signaling. Nat Rev Mol Cell Biol. 2012;13:566–78. doi: 10.1038/nrm3412. [DOI] [PubMed] [Google Scholar]
- 308.Roberts DJ, Tan-Sah VP, Smith JM, Miyamoto S. Akt phosphorylates HK-II at Thr-473 and increases mitochondrial HK-II association to protect cardiomyocytes. J Biol Chem. 2013;288:23798–806. doi: 10.1074/jbc.M113.482026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 309.Rohlenova K, Neuzil J, Rohlena J. The role of Her2 and other oncogenes of the PI3K/Akt pathway in mitochondria. Biol Chem. 2016;397:607–15. doi: 10.1515/hsz-2016-0130. [DOI] [PubMed] [Google Scholar]
- 310.Romano D, Maccario H, Doherty C, Quinn NP, Kolch W, Matallanas D. The differential effects of wild-type and mutated K-Ras on MST2 signaling are determined by K-Ras activation kinetics. Mol Cell Biol. 2013;33:1859–68. doi: 10.1128/MCB.01414-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 311.Rongvaux A, Andris F, Van Gool F, Leo O. Reconstructing eukaryotic NAD metabolism. BioEssays: news and reviews in molecular, cellular and developmental biology. 2003;25:683–90. doi: 10.1002/bies.10297. [DOI] [PubMed] [Google Scholar]
- 312.Rossi MN, Carbone M, Mostocotto C, Mancone C, Tripodi M, Maione R, Amati P. Mitochondrial localization of PARP-1 requires interaction with mitofilin and is involved in the maintenance of mitochondrial DNA integrity. J Biol Chem. 2009;284:31616–24. doi: 10.1074/jbc.M109.025882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 313.Roy AK, Lavrovsky Y, Song CS, Chen S, Jung MH, Velu NK, Bi BY, Chatterjee B. Regulation of androgen action. Vitam Horm. 1999;55:309–52. doi: 10.1016/s0083-6729(08)60938-3. [DOI] [PubMed] [Google Scholar]
- 314.Ruderman NB, Xu XJ, Nelson L, Cacicedo JM, Saha AK, Lan F, Ido Y. AMPK and SIRT1: a long-standing partnership? Am J Physiol Endocrinol Metab. 2010;298:E751–60. doi: 10.1152/ajpendo.00745.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 315.Rulten SL, Fisher AR, Robert I, Zuma MC, Rouleau M, Ju L, Poirier G, Reina-San-Martin B, Caldecott KW. PARP-3 and APLF function together to accelerate non-homologous end-joining. Mol Cell. 2011;41:33–45. doi: 10.1016/j.molcel.2010.12.006. [DOI] [PubMed] [Google Scholar]
- 316.Ryan MT, Hoogenraad NJ. Mitochondrial-nuclear communications. Annu Rev Biochem. 2007;6:701–22. doi: 10.1146/annurev.biochem.76.052305.091720. [DOI] [PubMed] [Google Scholar]
- 317.Rydberg B, Lindahl T. Nonenzymatic methylation of DNA by the intracellular methyl group donor S-adenosyl-L-methionine is a potentially mutagenic reaction. EMBO. 1982;1:211–16. doi: 10.1002/j.1460-2075.1982.tb01149.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 318.Saki M, Prakash A. DNA damage related crosstalk between the nucleus and mitochondria. Free Radic Biol Med. 2016;S0891–5849:31086–93. doi: 10.1016/j.freeradbiomed.2016.11.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 319.Sánchez-Aragó M, Formentini L, Cuezva JM. Mitochondria-mediated energy adaption in cancer: the H(+)-ATP synthase-geared switch of metabolism in human tumors. Antioxid Redox Signal. 2013;19:285–98. doi: 10.1089/ars.2012.4883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 320.Saner KJ, Welter BH, Zhang F, Hansen E, Dupont B, Wei Y, Price TM. Cloning and expression of a novel, truncated, progesterone receptor. Mol Cell Endocrinol. 2003;200:155–63. doi: 10.1016/s0303-7207(02)00380-5. [DOI] [PubMed] [Google Scholar]
- 321.Sangkhathat S, Kusafuka T, Yoneda A, Kuroda S, Tanaka Y, Sakai N, Fukuzawa M. Renal cell carcinoma in a pediatric patient with an inherited mitochondrial mutation. Pediatr Surg Int. 2005;21:745–48. doi: 10.1007/s00383-005-1471-0. [DOI] [PubMed] [Google Scholar]
- 322.Sarbassov DD, Guertin DA, Ali SM, Sabatini DM. Phosphorylation and regulation of Akt/PKB by the rictor-mTOR complex. Science. 2005;307:1098–101. doi: 10.1126/science.1106148. [DOI] [PubMed] [Google Scholar]
- 323.Sayan BS, Sayan AE, Knight RA, Melino G, Cohen GM. p53 is cleaved by caspases generating fragments localizing to mitochondria. J Biol Chem. 2006;281:13566–73. doi: 10.1074/jbc.M512467200. [DOI] [PubMed] [Google Scholar]
- 324.Schatz G. The protein import system of mitochondria. J Biol Chem. 1996;13:31763–66. doi: 10.1074/jbc.271.50.31763. [DOI] [PubMed] [Google Scholar]
- 325.Scheller K, Seibel P, Sekeris CE. Glucocorticoid and thyroid hormone receptors in mitochondria of animal cells. Int Rev Cytol. 2003;222:1–61. doi: 10.1016/s0074-7696(02)22011-2. [DOI] [PubMed] [Google Scholar]
- 326.Schmidt S, Rainer J, Ploner C, Presul E, Riml S, Kofler R. Glucocorticoid-induced apoptosis and glucocorticoid resistance: molecular mechanisms and clinical relevance. Cell Death Differ. 2004;11:S45–S55. doi: 10.1038/sj.cdd.4401456. [DOI] [PubMed] [Google Scholar]
- 327.Schulz AM, Haynes CM. UPR(mt)-mediated cytoprotection and organismal aging. Biochim Biophys Acta. 2015;1847:1448–56. doi: 10.1016/j.bbabio.2015.03.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 328.Schwartzenberg-Bar-Yoseph F, Armoni M, Karnieli E. The tumor suppressor p53 down-regulates glucose transporters GLUT1 and GLUT4 gene expression. Cancer Res. 2004;64:2627–33. doi: 10.1158/0008-5472.can-03-0846. [DOI] [PubMed] [Google Scholar]
- 329.Setiawan VW, Chu LH, John EM, Ding YC, Ingles SA, Bernstein L, Press MF, Ursin G, Haiman CA, Neuhausen SL. Mitochondrial DNA G10398A variant is not associated with breast cancer in African-American women. Cancer Genet Cytogenet. 2008;181:16–19. doi: 10.1016/j.cancergencyto.2007.10.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 330.Sharma LK, Fang H, Liu J, Vartak R, Deng J, Bai Y. Mitochondrial respiratory complex I dysfunction promotes tumorigenesis through ROS alteration and AKT activation. Hum Mol Genet. 2011;20:4605–16. doi: 10.1093/hmg/ddr395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 331.Shavali S, Brown-Borg HM, Ebadi M, Porter J. Mitochondrial localization of alpha-synuclein protein in alpha-synuclein overexpressing cells. Neurosci Lett. 2008;439:125–28. doi: 10.1016/j.neulet.2008.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 332.Sheridan C, Martin SJ. Mitochondrial fission/fusion dynamics and apoptosis. Mitochondrion. 2010;10:640–48. doi: 10.1016/j.mito.2010.08.005. [DOI] [PubMed] [Google Scholar]
- 333.Shiloh Y, Ziv Y. The ATM protein kinase: regulating the cellular response to genotoxic stress, and more. Nat Rev Mol Cell Biol. 2013;14:197–210. [PubMed] [Google Scholar]
- 334.Shirwany NA, Zou MH. AMPK: a cellular metabolic and redox sensor. A minireview Front Biosci (Landmark Ed) 2014;19:447–74. doi: 10.2741/4218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 335.Shock LS, Thakkar PV, Peterson EJ, Moran RG, Taylor SM. DNA methyltransferase, cytosine methylation, and cytosine hydroxymethylation in mammalian mitochondria. Proc Natl Acad Sci U S A. 2011;108:3630–5. doi: 10.1073/pnas.1012311108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 336.Shoffner JM, Lott MT, Lezza AM, Seibel P, Ballinger SW, Wallace DC. Myoclonic epilepsy and ragged-red fiber disease (MERRF) is associated with a mitochondrial DNA tRNA(Lys) mutation. Cell. 1990;61:931–37. doi: 10.1016/0092-8674(90)90059-n. [DOI] [PubMed] [Google Scholar]
- 337.Shokolenko I, Venediktova N, Bochkareva A, Wilson GL, Alexeyev MF. Oxidative stress induces degradation of mitochondrial DNA. Nucleic Acids Res. 2009;37:2539–48. doi: 10.1093/nar/gkp100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 338.Singh B, Owens KM, Bajpai P, Desouki MM, Srinivasasainagendra V, Tiwari HK, Singh KK. Mitochondrial DNA polymerase POLG1 Disease Mutations and Germline Variants Promote Tumorigenic Properties. PLoS One. 2015;10:e0139846. doi: 10.1371/journal.pone.0139846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 339.Singh KK. MIPIGENETICS and MIPIGENOMICS: Integrating mitochondria-induced mayhem contributing to mystondria. 2015 http://dx.doi.org/10.1016/j.mito.2015.07.024.
- 340.Singh KK, Ayyasamy V, Owens KM, Koul MS, Vujcic M. Mutations in mitochondrial DNA polymerase-gamma promote breast tumorigenesis. J Hum Genet. 2009;54:516–24. doi: 10.1038/jhg.2009.71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 341.Singh KK, Kulawiec M, Still I, Desouki MM, Geradts J, Matsui S. Intergenomic cross talk between mitochondria and the nucleus plays an important role in tumorigenesis. Gene. 2005;354:140–46. doi: 10.1016/j.gene.2005.03.027. [DOI] [PubMed] [Google Scholar]
- 342.Singh KK. Mitochondria damage checkpoint in apoptosis and genome stability. FEMS Yeast Res. 2004;5:127–32. doi: 10.1016/j.femsyr.2004.04.008. [DOI] [PubMed] [Google Scholar]
- 343.Singh KK. Mitochondria damage checkpoint, aging, and cancer. Ann N Y Acad Sci. 2006;1067:182–90. doi: 10.1196/annals.1354.022. [DOI] [PubMed] [Google Scholar]
- 344.Singh KK, editor. Mitochondrial DNA Mutations in Aging, Disease, and Cancer. Springer; New York, USA: 1998. [Google Scholar]
- 345.Sliwinski T, Przybylowska K, Markiewicz L, Rusin P, Pietruszewska W, Zelinska-Blizniewska H, Olszewski J, Morawiec-Sztandera A, Mlynarski W, Majsterek I. MUTYH Tyr165Cys, OGG1 Ser326Cys and XPD Lys751Gln polymorphisms and head neck cancer susceptibility: a case control study. Mol Biol Rep. 2011;38:1251–61. doi: 10.1007/s11033-010-0224-x. [DOI] [PubMed] [Google Scholar]
- 346.Solakidi S, Psarra AM, Nikolaropoulos S, Sekeris CE. Estrogen receptors alpha and beta (ERalpha and ERbeta) and androgen receptor (AR) in human sperm: localization of ERbeta and AR in mitochondria of the midpiece. Hum Reprod. 2005;20:3481–87. doi: 10.1093/humrep/dei267. [DOI] [PubMed] [Google Scholar]
- 347.Spitz DR, Azzam EI, Li JJ, Gius D. Metabolic oxidation/reduction reactions and cellular responses to ionizing radiation: a unifying concept in stress response biology. Cancer Metastasis Rev. 2004;23:311–22. doi: 10.1023/B:CANC.0000031769.14728.bc. [DOI] [PubMed] [Google Scholar]
- 348.Stoehlmacher J, Ingles SA, Park DJ, Zhang W, Lenz HJ. The -9Ala/-9Val polymorphism in the mitochondrial targeting sequence of the manganese superoxide dismutase gene (MnSOD) is associated with age among Hispanics with colorectal carcinoma. Oncol Rep. 2002;9:235–38. [PubMed] [Google Scholar]
- 349.Sun Y, Lin H, Zhu Y, Ma C, Ye J, Luo J. Induction or suppression of expression of cytochrome C oxidase subunit II by heregulin beta 1 in human mammary epithelial cells is dependent on the levels of ErbB2 expression. J Cell Physiol. 2002;192:225–33. doi: 10.1002/jcp.10132. [DOI] [PubMed] [Google Scholar]
- 350.Sunba MS, Rahi AH, Morgan G. Tumors of the anterior uvea. II. Intranuclear cytoplasmic inclusions in malignant melanoma of the iris. Br J Ophthalmol. 1980;64:453–6. doi: 10.1136/bjo.64.6.453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 351.Sutendra G, Kinnaird A, Dromparis P, Paulin R, Stenson TH, Haromy A, Hashimoto K, Zhang N, Flaim E, Michelakis ED. A nuclear pyruvate dehydrogenase complex is important for the generation of acetyl-CoA and histone acetylation. Cell. 2014;158:84–97. doi: 10.1016/j.cell.2014.04.046. [DOI] [PubMed] [Google Scholar]
- 352.Svilar D, Goellner EM, Almeida KH, Sobol RW. Base excision repair and lesion-dependent sub-pathways for repair of oxidative DNA damage. Antioxid Redox Signal. 2011;14:2491–507. doi: 10.1089/ars.2010.3466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 353.Sykora P, Croteau DL, Bohr VA, Wilson DM. Aprataxin localizes to mitochondria and preserves mitochondrial function. Proc Natl Acad Sci. 2011;108:7437–42. doi: 10.1073/pnas.1100084108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 354.Szabadkai G, Simoni AM, Bianchi K, De Stefani D, Leo S, Wieckowski MR, Rizzuto R. Mitochondrial dynamics and Ca2+ signaling. Biochim Biophys Acta. 2006;1763:442–49. doi: 10.1016/j.bbamcr.2006.04.002. [DOI] [PubMed] [Google Scholar]
- 355.Szczesny B, Brunyanszki A, Olah G, Mitra S, Szabo C. Opposing roles of mitochondrial and nuclear PARP1 in the regulation of mitochondrial and nuclear DNA integrity: implications for the regulation of mitochondrial function. Nucleic Acids Res. 2014;42:13161–73. doi: 10.1093/nar/gku1089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 356.Takasugi M, Yagi S, Hirabayashi K, Shiota K. DNA methylation status of nuclear-encoded mitochondrial genes underlies the tissue-dependent mitochondrial functions. BMC Genomics. 2010;11:481. doi: 10.1186/1471-2164-11-481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 357.Takemura G, Takatsu Y, Sakaguchi H, Fujiwara H. Intranuclear mitochondria in human myocardial cells. Pathol Res Pract. 1997;193:305–11. doi: 10.1016/s0344-0338(97)80008-8. [DOI] [PubMed] [Google Scholar]
- 358.Tanaka H, Sasayama T, Tanaka K, Nakamizo S, Nishihara M, Mizukawa K, Kohta M, Koyama J, Miyake S, Taniguchi M, Hosoda K, Kohmura E. MicroRNA-183 upregulates HIF-1α by targeting isocitrate dehydrogenase 2 (IDH2) in glioma cells. J Neurooncol. 2013;111:273–83. doi: 10.1007/s11060-012-1027-9. [DOI] [PubMed] [Google Scholar]
- 359.Tanaka M, Takeyasu T, Fuku N, Li-Jun G, Kurata M. Mitochondrial genome single nucleotide polymorphisms and their phenotypes in the Japanese. Ann N Y Acad Sci. 2004;1011:7–20. doi: 10.1007/978-3-662-41088-2_2. [DOI] [PubMed] [Google Scholar]
- 360.Tappenden DM, Lynn SG, Crawford RB, Lee K, Vengellur A, Kaminski NE, Thomas RS, LaPres JJ. The aryl hydrocarbon receptor interacts with ATP5α1, a subunit of the ATP synthase complex, and modulates mitochondrial function. Toxicol Appl Pharmacol. 2011;254:299–310. doi: 10.1016/j.taap.2011.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 361.Tengku Baharudin N, Jaafar H, Zainuddin Z. Association of mitochondrial DNA 10398 polymorphism in invasive breast cancer in malay population of peninsular malaysia. Malays J Med Sci. 2012;19:36–42. [PMC free article] [PubMed] [Google Scholar]
- 362.Tockman MS, Gupta PK, Pressman NJ, Mulshine JL. Cytometric validation of immunocytochemical observations in developing lung cancer. Diagn Cytopathol. 1993;9:615–22. doi: 10.1002/dc.2840090604. [DOI] [PubMed] [Google Scholar]
- 363.Tomasetti M, Nocchi L, Staffolani S, Manzella N, Amati M, Goodwin J, Kluckova K, Nguyen M, Strafella E, Bajzikova M, Peterka M, Lettlova S, Truksa J, Lee W, Dong LF, Santarelli L, Neuzil J. MicroRNA-126 suppresses mesothelioma malignancy by targeting IRS1 and interfering with the mitochondrial function. Antioxid Redox Signal. 2014;21:2109–25. doi: 10.1089/ars.2013.5215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 364.Tomasetti M, Santarelli L, Neuzil J, Dong L. MicroRNA regulation of cancer metabolism: role in tumor suppression. Mitochondrion. 2014;19:29–38. doi: 10.1016/j.mito.2014.06.004. [DOI] [PubMed] [Google Scholar]
- 365.Tomasetti M, Staffolani S, Nocchi L, Neuzil J, Strafella E, Manzella N, Mariotti L, Bracci M, Valentino M, Amati M, Santarelli L. Clinical significance of circulating miR-126 quantification in malignant mesothelioma patients. Clin Biochem. 2012;45:575–81. doi: 10.1016/j.clinbiochem.2012.02.009. [DOI] [PubMed] [Google Scholar]
- 366.Trifunovic A, Wredenberg A, Falkenberg M, Spelbrink JN, Rovio AT, Bruder CE, Bohlooly-Y M, Gidlöf S, Oldfors A, Wibom R, Törnell J, Jacobs HT, Larsson NG. Premature ageing in mice expressing defective mitochondrial DNA polymerase. Nature. 2004;429:417–23. doi: 10.1038/nature02517. [DOI] [PubMed] [Google Scholar]
- 367.Tseng LM, Yin PH, Chi CW, Hsu CY, Wu CW, Lee LM, Wei YH, Lee HC. Mitochondrial DNA mutations and mitochondrial DNA depletion in breast cancer. Genes Chromosomes Cancer. 2006;45:629–38. doi: 10.1002/gcc.20326. [DOI] [PubMed] [Google Scholar]
- 368.Uvirova M, Simova J, Kubova B, Dvorackova N, Tomaskova H, Sedivcova M, Dite P. Comparison of the prevalence of KRAS-LCS6 polymorphism (rs61764370) within different tumor types (colorectal, breast, non-small cell lung cancer and brain tumors). A study of the Czech population. Biomed Pap Med Fac Univ Palacky Olomouc Czech Repub. 2015;159:466–71. doi: 10.5507/bp.2015.029. [DOI] [PubMed] [Google Scholar]
- 369.van Gisbergen MW, Voets AM, Starmans MH, de Coo IF, Yadak R, Hoffmann RF, Boutros PC, Smeets HJ, Dubois L. Lambin P How do changes in the mtDNA and mitochondrial dysfunction influence cancer and cancer therapy? Challenges, opportunities and models. Mutat Res Rev. 2015;764:16–30. doi: 10.1016/j.mrrev.2015.01.001. [DOI] [PubMed] [Google Scholar]
- 370.Van Waveren C, Moraes CT. Transcriptional co-expression and co-regulation of genes coding for components of the oxidative phosphorylation system. BMC Genomics. 2008;9:18. doi: 10.1186/1471-2164-9-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 371.Velpula KK, Bhasin A, Asuthkar S, Tsung AJ. Combined targeting of PDK1 and EGFR triggers regression of glioblastoma by reversing the Warburg effect. Cancer Res. 2013;73:7277–89. doi: 10.1158/0008-5472.CAN-13-1868. [DOI] [PubMed] [Google Scholar]
- 372.Verma M, Kumar D. Application of mitochondrial genome information in cancer epidemiology. Clin Chim Acta. 2007;383:41–50. doi: 10.1016/j.cca.2007.04.018. [DOI] [PubMed] [Google Scholar]
- 373.Vermulst M, Bielas JH, Loeb LA. Quantification of random mutations in the mitochondrial genome. Methods. 2008;46:263–68. doi: 10.1016/j.ymeth.2008.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 374.Veronica M, Ali A, Venkateshwari A, Mamata D, Nallari P. Association of estrogen and progesterone receptor gene polymorphisms and their respective hormones in uterine leiomyomas. Tumor Biol. 2016;37:8067–74. doi: 10.1007/s13277-015-4711-5. [DOI] [PubMed] [Google Scholar]
- 375.Vetter IR, Wittinghofer A. The guanine nucleotide-binding switch in three dimensions. Science. 2001;294:1299–304. doi: 10.1126/science.1062023. [DOI] [PubMed] [Google Scholar]
- 376.Villa AM, Doglia SM. Mitochondria in tumor cells studied by laser scanning confocal microscopy. J Biomed Opt. 2004;9:385–94. doi: 10.1117/1.1646414. [DOI] [PubMed] [Google Scholar]
- 377.Vivekanandan P, Daniel H, Yeh MM, Torbenson M. Mitochondrial mutations in hepatocellular carcinomas and fibrolamellar carcinomas. Mod Pathol. 2010;23:790–798. doi: 10.1038/modpathol.2010.51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 378.Vousden KH. Alternative fuel–another role for p53 in the regulation of metabolism. PNAS. 2010;107:7117–78. doi: 10.1073/pnas.1002656107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 379.Wang D, Timmis JN. Cytoplasmic organelle DNA preferentially inserts into open chromatin. Genome Biol Evol. 2013;5:1060–4. doi: 10.1093/gbe/evt070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 380.Wang L, Bamlet WR, de Andrade M, Boardman LA, Cunningham JM, Thibodeau SN, Petersen GM. Mitochondrial genetic polymorphisms and pancreatic cancer risk. Cancer Epidemiol Biomarkers Prev. 2007;16:1455–59. doi: 10.1158/1055-9965.EPI-07-0119. [DOI] [PubMed] [Google Scholar]
- 381.Wang Y, Liu VW, Xue WC, Cheung AN, Ngan HY. Association of decreased mitochondrial DNA content with ovarian cancer progression. Br J Cancer. 2006;95:1087–91. doi: 10.1038/sj.bjc.6603377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 382.Wang Y, Liu VW, Xue WC, Tsang PC, Cheung AN, Ngan HY. The increase of mitochondrial DNA content in endometrial adenocarcinoma cells: a quantitative study using laser-captured micro-dissected tissues. Gynecol Oncol. 2005;98:104–10. doi: 10.1016/j.ygyno.2005.04.015. [DOI] [PubMed] [Google Scholar]
- 383.Wang ZH, Samuels S, Gama Sosa MA, Kolodny EH. 5-Fluorocytosine-mediated apoptosis and DNA damage in glioma cells engineered to express cytosine deaminase and their enhancement with interferon. J Neurooncol. 1998;36:219–29. doi: 10.1023/a:1005883128175. [DOI] [PubMed] [Google Scholar]
- 384.Watrowski R, Castillo-Tong DC, Wolf A, Schuster E, Fischer MB, Speiser P, Zeillinger R. HER2 Codon 655 (Ile/Val) Polymorphism and Breast Cancer in Austrian Women. Anticancer Res. 2015;35:5901–04. [PubMed] [Google Scholar]
- 385.Weighardt F, Biamonti G, Riva S. The roles of heterogeneous nuclear ribonucleoproteins (hnRNP) in RNA metabolism. Bioessays. 1996;18:747–56. doi: 10.1002/bies.950180910. [DOI] [PubMed] [Google Scholar]
- 386.Welter C, Kovacs G, Seitz G, Blin N. Alteration of mitochondrial DNA in human oncocytomas. Genes Chromosomes Cancer. 1989;1:79–82. doi: 10.1002/gcc.2870010112. [DOI] [PubMed] [Google Scholar]
- 387.Wilson CM, McPhaul MJ. A and B forms of the androgen receptor are present in human genital skin fibroblasts. PNAS. 1994;91:1234–38. doi: 10.1073/pnas.91.4.1234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 388.Woischnik M, Moraes CT. Pattern of organization of human mitochondrial pseudogenes in the nuclear genome. Genome Res. 2002;12:885–93. doi: 10.1101/gr.227202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 389.Wojewoda M, Duszyński J, Szczepanowska J. NARP mutation and mtDNA depletion trigger mitochondrial biogenesis which can be modulated by selenite supplementation. Int J Biochem Cell Biol. 2011;43:1178–86. doi: 10.1016/j.biocel.2011.04.011. [DOI] [PubMed] [Google Scholar]
- 390.Wolfman JC, Planchon SM, Liao J, Wolfman A. Structural and functional consequences of c-N-Ras constitutively associated with intact mitochondria. Biochim Biophys Acta. 2006;1763:1108–24. doi: 10.1016/j.bbamcr.2006.07.015. [DOI] [PubMed] [Google Scholar]
- 391.Wong JM, Harper PA, Meyer UA, Bock KW, Morike K, Lagueux J, Ayotte P, Tyndale RF, Sellers EM, Manchester DK, Okey AB. Ethnic variability in the allelic distribution of human aryl hydrocarbon receptor codon 554 and assessment of variant receptor function in vitro. Pharmacogenetics. 2001;11:85–94. doi: 10.1097/00008571-200102000-00010. [DOI] [PubMed] [Google Scholar]
- 392.Woo DK, Green PD, Santos JH, D’Souza AD, Walther Z, Martin WD, Christian BE, Chandel NS, Shadel GS. Mitochondrial genome instability and ROS enhance intestinal tumorigenesis in APC (Min/+) mice. Am J Pathol. 2012;180:24–31. doi: 10.1016/j.ajpath.2011.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 393.Wu CW, Yin PH, Hung WY, Li AF, Li SH, Chi CW, Wei YH, Lee HC. Mitochondrial DNA mutations and mitochondrial DNA depletion in gastric cancer. Genes Chromosomes Cancer. 2005;44:19–28. doi: 10.1002/gcc.20213. [DOI] [PubMed] [Google Scholar]
- 394.Xu Y, Fang F, Zhang J, Josson S, St Clair WH, St Clair DK. miR-17* suppresses tumorigenicity of prostate cancer by inhibiting mitochondrial antioxidant enzymes. PLoS One. 2010;5:e14356. doi: 10.1371/journal.pone.0014356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 395.Xu Z, Chen Y, Gu D, Lee NP, Sun S, Gong W, Tan Y, Luk JM, Chen J. SOD2 rs4880 CT/CC genotype predicts poor survival for Chinese gastric cancer patients received platinum and fluorouracil based adjuvant chemotherapy. Am J Transl Res. 2015;7:4014–10. [PMC free article] [PubMed] [Google Scholar]
- 396.Xue L, Li C, Wang Y, Sun W, Ma C, He Y, Yu Y, Cai L, Wang L. Single nucleotide polymorphisms in non-coding region of the glucocorticoid receptor gene and prednisone response in childhood acute lymphoblastic leukemia. Leuk Lymphoma. 2015;56:1704–09. doi: 10.3109/10428194.2014.951848. [DOI] [PubMed] [Google Scholar]
- 397.Yacoub HA, Mahmoud WM, El-Baz HA, Eid OM, El-Fayoumi RI, Mahmoud MM, Harakeh S, Abuzinadah OH. New haplotypes of the ATP synthase subunit 6 gene of mitochondrial DNA are associated with acute lymphoblastic leukemia in Saudi Arabia. Asian Pac J Cancer Prev. 2014;15:10433–38. doi: 10.7314/apjcp.2014.15.23.10433. [DOI] [PubMed] [Google Scholar]
- 398.Yadav SS, Seth S, Khan AJ, Maurya SS, Dhawan A, Pant S, Pant MC, Parmar D. Association of polymorphism in cytochrome P450 2C9 with susceptibility to head and neck cancer and treatment outcome. Appl Transl Genom. 2013;3:8–13. doi: 10.1016/j.atg.2013.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 399.Yakes FM, Van Houten B. Mitochondrial DNA damage is more extensive and persists longer than nuclear DNA damage in human cells following oxidative stress. PNAS. 1997;94:514–19. doi: 10.1073/pnas.94.2.514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 400.Yan H, Parsons DW, Jin G, McLendon R, Rasheed BA, Yuan W, Kos I, Batinic-Haberle I, Jones S, Riggins GJ, Friedman H, Friedman A, Reardon D, Herndon J, Kinzler KW, Velculescu VE, Vogelstein B, Bigner DD. IDH1 and IDH2 mutations in gliomas. N Engl J Med. 2009;360:765–73. doi: 10.1056/NEJMoa0808710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 401.Yang H, Ye D, Guan KL, Xiong Y. IDH1 and IDH2 mutations in tumorigenesis: mechanistic insights and clinical perspectives. Clin Cancer Res. 2012;18:5562–71. doi: 10.1158/1078-0432.CCR-12-1773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 402.Yang JC, Ok JH, Busby JE, Borowsky AD, Kung HJ, Evans CP. Aberrant activation of androgen receptor in a new neuropeptide-autocrine model of androgen-insensitive prostate cancer. Cancer Res. 2009;69:151–60. doi: 10.1158/0008-5472.CAN-08-0442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 403.Yang WS, SriRamaratnam R, Welsch ME, Shimada K, Skouta R, Viswanathan VS, Cheah JH, Clemons PA, Shamji AF, Clish CB, Brown LM, Girotti AW, Cornish VW, Schreiber SL, Stockwell BR. Regulation of ferroptotic cancer cell death by GPX4. Cell. 2014;156:317–31. doi: 10.1016/j.cell.2013.12.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 404.Yao J, Zhou E, Wang Y, Xu F, Zhang D, Zhong D. microRNA-200a inhibits cell proliferation by targeting mitochondrial transcription factor A in breast cancer. DNA Cell Biol. 2014;33:291–300. doi: 10.1089/dna.2013.2132. [DOI] [PubMed] [Google Scholar]
- 405.Yeh CC, Hou MF, Wu SH, Tsai SM, Lin SK, Hou LA, Ma H, Tsai LY. A study of glutathione status in the blood and tissues of patients with breast cancer. Cell Biochem Funct. 2006;24:555–59. doi: 10.1002/cbf.1275. [DOI] [PubMed] [Google Scholar]
- 406.Ying H, Kimmelman AC, Lyssiotis CA, Hua S, Chu GC, Fletcher-Sananikone E, Locasale JW, Son J, Zhang H, Coloff JL, Yan H, Wang W, Chen S, Viale A, Zheng H, Paik JH, Lim C, Guimaraes AR, Martin ES, Chang J, Hezel AF, Perry SR, Hu J, Gan B, Xiao Y, Asara JM, Weissleder R, Wang YA, Chin L, Cantley LC, DePinho RA. Oncogenic Kras maintains pancreatic tumors through regulation of anabolic glucose metabolism. Cell. 2012;149:656–70. doi: 10.1016/j.cell.2012.01.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 407.Yogev O, Yogev O, Singer E, Shaulian E, Goldberg M, Fox TD, Pines O. Fumarase: a mitochondrial metabolic enzyme and a cytosolic/nuclear component of the DNA damage response. PLoS Biol. 2010;8 doi: 10.1371/journal.pbio.1000328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 408.Yoon Y, McNiven MA. Mitochondrial division: New partners in membrane pinching. Curr Biol. 2001;11:R67–R70. doi: 10.1016/s0960-9822(01)00011-2. [DOI] [PubMed] [Google Scholar]
- 409.Yoshida Y, Izumi H, Torigoe T, Ishiguchi H, Itoh H, Kang D, Kohno K. P53 physically interacts with mitochondrial transcription factor A and differentially regulates binding to damaged DNA. Cancer Res. 2003;63:3729–34. [PubMed] [Google Scholar]
- 410.Yuan RT, Sun Y, Bu LX, Jia MY. Gene mutations in the D-loop region of mitochondrial DNA in oral squamous cell carcinoma. Mol Med Rep. 2015;11:4496–500. doi: 10.3892/mmr.2015.3240. [DOI] [PubMed] [Google Scholar]
- 411.Yuan S, Wang F, Chen G, Zhang H, Feng L, Wang L, Colman H, Keating MJ, Li X, Xu RH, Wang J, Huang P. Effective elimination of cancer stem cells by a novel drug combination strategy. Stem Cells. 2013;31:23–34. doi: 10.1002/stem.1273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 412.Yue X, Song W, Zhang W, Chen L, Xi Z, Xin Z, Jiang X. Mitochondrially localized EGFR is subjected to autophagic regulation and implicated in cell survival. Autophagy. 2008;4:641–49. [Google Scholar]
- 413.Zhang H, Guo X, Dai J, Wu Y, Ge N, Yang Y, Ji J, Zhang H. Genetic variations in IDH gene as prognosis predictors in TACE-treated hepatocellular carcinoma patients. Med Oncol. 2014;31:278. doi: 10.1007/s12032-014-0278-z. [DOI] [PubMed] [Google Scholar]
- 414.Zhang H, Singh KK. Global determinants of mitochondrial copy number. PLoS One. 2014;9:e105242. doi: 10.1371/journal.pone.0105242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 415.Zhang JG, Wang JJ, Zhao F, Liu Q, Jiang K, Yang GH. MicroRNA-21 (miR-21) represses tumor suppressor PTEN and promotes growth and invasion in non-small cell lung cancer (NSCLC) Clin Chim Acta. 2010;411:846–52. doi: 10.1016/j.cca.2010.02.074. [DOI] [PubMed] [Google Scholar]
- 416.Zhang L, Yang F, Yuan JH, Yuan SX, Zhou WP, Huo XS, Xu D, Bi HS, Wang F, Sun SH. Epigenetic activation of the MiR-200 family contributes to H19-mediated metastasis suppression in hepatocellular carcinoma. Carcinogenesis. 2013;34:577–86. doi: 10.1093/carcin/bgs381. [DOI] [PubMed] [Google Scholar]
- 417.Zhao Y, Chaiswing L, Velez JM, Batinic-Haberle I, Colburn NH, Oberley TD, St Clair DK. p53 translocation to mitochondria precedes its nuclear translocation and targets mitochondrial oxidative defense protein-manganese superoxide dismutase. Cancer Res. 2005;65:3745–50. doi: 10.1158/0008-5472.CAN-04-3835. [DOI] [PubMed] [Google Scholar]
- 418.Zhou G, Dada LA, Wu M, Kelly A, Trejo H, Zhou Q, Varga J, Sznajder JI. Hypoxia-induced alveolar epithelial-mesenchymal transition requires mitochondrial ROS and hypoxia-inducible factor 1. Am J Physiol Lung Cell Mol Physiol. 2009;297:L1120–30. doi: 10.1152/ajplung.00007.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 419.Zhou J, Mulshine JL, Unsworth EJ, Scott FM, Avis IM, Vos MD, Treston AM. Purification and characterization of a protein that permits early detection of lung cancer. Identification of heterogeneous nuclear ribonucleoprotein-A2/B1 as the antigen for monoclonal antibody 703D4. J Biol Chem. 1996;271:10760–66. doi: 10.1074/jbc.271.18.10760. [DOI] [PubMed] [Google Scholar]
- 420.Zhou S, Kachhap S, Singh KK. Mitochondrial impairment in p53-deficient human cancer cells. Mutagenesis. 2003;18:287–92. doi: 10.1093/mutage/18.3.287. [DOI] [PubMed] [Google Scholar]
- 421.Zhu M, Yi M, Kim CH, Deng C, Li Y, Medina D, Stephens RM, Green JE. Integrated miRNA and mRNA expression profiling of mouse mammary tumor models identifies miRNA signatures associated with mammary tumor lineage. Genome Biol. 2011;12:R77. doi: 10.1186/gb-2011-12-8-r77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 422.Srinivasainagendra V, Sandel MW, Singh B, Sundaresan A, Mooga VP, Bajpai P, Tiwari HK, Singh KK. Migration of mitochondrial DNA in the nuclear genome of colorectal adenocarcinoma. Genome Med. 2017;9:31. doi: 10.1186/s13073-017-0420-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 423.Melkonian SC, Wang X, Gu J, Matin SF, Tannir NM, Wood CG, Wu X. Mitochondrial DNA copy number in peripheral blood leukocytes and the risk of clear cell renal cell carcinoma. Carcinogenesis. 2015;36:249–255. doi: 10.1093/carcin/bgu248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 424.Xing J, Chen M, Wood CG, Lin J, Spitz MR, Ma J, Amos CI, Shields PG, Benowitz NL, Gu J, de Andrade M, Swan GE, Wu X. Mitochondrial DNA content: its genetic heritability and association with renal cell carcinoma. J Natl Cancer Inst. 2008;100:1104–1112. doi: 10.1093/jnci/djn213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 425.Lan Q, Lim U, Liu CS, Weinstein SJ, Chanock S, Bonner MR, Virtamo J, Albanes D, Rothman N. A prospective study of mitochondrial DNA copy number and risk of non-Hodgkin lymphoma. Blood. 2008;112:4247–4249. doi: 10.1182/blood-2008-05-157974. [DOI] [PMC free article] [PubMed] [Google Scholar]


