Abstract
At the core of the Saccharomyces Genome Database (SGD) are chromosomal features that encode a product. These include protein-coding genes and major noncoding RNA genes, such as tRNA and rRNA genes. The basic entry point into SGD is a gene or open-reading frame name that leads directly to the locus summary information page. A keyword describing function, phenotype, selective condition, or text from abstracts will also provide a door into the SGD. A DNA or protein sequence can be used to identify a gene or a chromosomal region using BLAST. Protein and DNA sequence identifiers, PubMed and NCBI IDs, author names, and function terms are also valid entry points. The information in SGD has been gathered and is maintained by a group of scientific biocurators and software developers who are devoted to providing researchers with up-to-date information from the published literature, connections to all the major research resources, and tools that allow the data to be explored. All the collected information cannot be represented or summarized for every possible question; therefore, it is necessary to be able to search the structured data in the database. This protocol describes the YeastMine tool, which provides an advanced search capability via an interactive tool. The SGD also archives results from microarray expression experiments, and a strategy designed to explore these data using the SPELL (Serial Pattern of Expression Levels Locator) tool is provided.
MATERIALS
Equipment
Internet-connected computer with web browser
METHOD
At any step in this protocol, support is available via the comprehensive Help documents maintained by the SGD. These can be accessed via the help button at the top of each page or, for specific features, a small red button with a question mark in its center is provided and will link to the help pages specific for that feature.
The SGD is continually updated; therefore, specific items presented in this protocol may not appear on the SGD website exactly as described.
Using YeastMine
This series of steps presents a method to identify all uncharacterized yeast genes that are essential for life (lethal when mutated) and that have a human homolog associated with a disease phenotype. The set of yeast genes that fulfill these criteria can be found using YeastMine (Balakrishnan et al. 2012). YeastMine is a query tool for all the data that are contained in the SGD and downloads site. Video tutorials are available for the basic features of YeastMine and can be accessed from links in the help section, www.yeastgenome.org/help/video-tutorials. These videos provide an effective way to learn the interface and operations that are provided by YeastMine. The procedure described here will use Templates, Lists, and List Operations. There are many situations where an advanced query of the data is the only way to obtain the desired information. In general, this occurs when a very specific answer is sought from a complex set of criteria or when large amounts of data are desired. YeastMine is a specialized data warehouse containing everything that is included within the main SGD database in addition to other large data sets. YeastMine is built using an open source tool called InterMine (Kalderimis et al. 2014).
An effective strategy to address a complex query is to break down the query into smaller questions and to then combine the results of these smaller questions. The answer to a complex question is obtained using basic list operations. There are four basic operations: union—the combination of two lists; intersection—the elements that are common between two lists; subtraction—the elements that are not common between two lists; and asymmetric difference—the elements that are not common in one of the two lists. In the procedure below, three simple queries are used to define a list of genes that are essential, a list of all genes that have a human homolog, and a list of genes that are associated with human disease. The union of the first two lists gives all essential yeast genes that have a human homolog, making a fourth list. The fourth list is compared to the third to provide the answer to the question: What are all the essential yeast genes that have a human homolog associated with a disease phenotype?
This procedure describes how to identify the set of uncharacterized essential yeast genes that have a human homolog associated with a disease phenotype annotation defined by OMIM (Online Mendelian Inheritance in Man; www.omim.org; Amberger et al. 2008). This is an easy query for YeastMine and is not something you would typically expect to find precomputed. An open-reading frame (ORF) defined as uncharacterized is believed to be a protein-coding gene; however, it does not yet have a function assigned (Fisk et al. 2006). OMIM is a detailed encyclopedia of human genetic diseases. OMIM includes details on genes and their variants and uses phenotype ontologies to define the characteristics of the disease.
-
1
Open the URL www.yeastgenome.org in any modern web browser (e.g., Chrome, Firefox, or Safari).
-
2
Open the “Help” page and under “Video Tutorials” open the “YeastMine” page. Watch the YeastMine video tutorials to become familiar with its basic features.
-
3
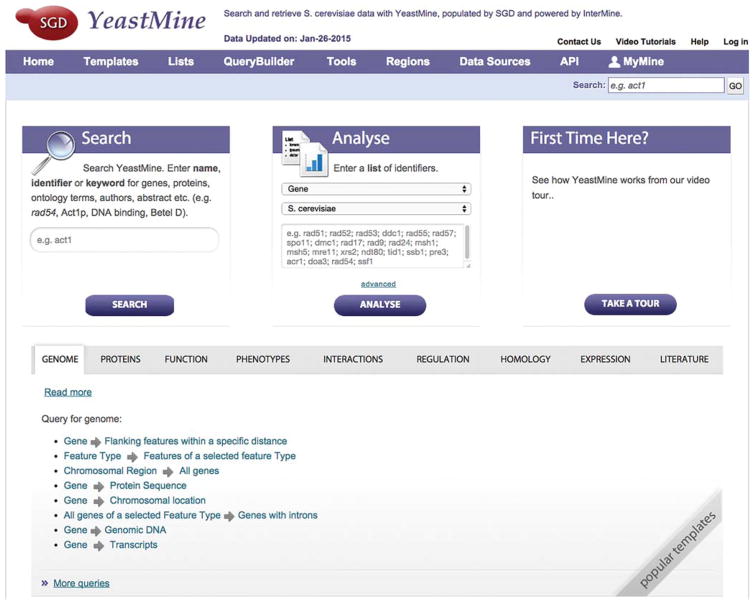
Open YeastMine (Fig. 1) by selecting “Advanced Search” at the top right of any SGD page (just below the Search box) and set up a myMine account so you can save the lists you create and any custom templates you define.
Custom templates can also be created using instructions from the video tutorials (Step 2). -
4
Make a list of all essential yeast genes. From the YeastMine homepage, select the Templates tab and then select the Phenotypes filter, followed by the Phenotype –> Genes template and locate “inviable” from the long list of observables. Start the search by clicking Show Results; there will be over a thousand rows in the resulting table. Now click on “Create/Add to list,” select “All 1284 Genes” (listed in columns 1, 2, 3, 4, 5), and name this list “essential genes.”
-
5
Create a list of all yeast genes with a human homolog that have a disease phenotype defined in OMIM. From the YeastMine homepage, select the “Templates” tab and then select the “Homology” filter, followed by the “Yeast gene –> OMIM human homolog(s) –> OMIM Disease Phenotype(s).” Select “constrain to be” and the predefined gene list “Uncharacterized_ORFs,” then “Show Results.” This results in 562 rows. Create another list of the 197 genes found in columns 1, 2, and 3; the columns used are indicated as you mouse over the options in the “Create New List” menu. Name this list “uncharacterized_with_human_disease_phenotype.”
-
6
Select “Lists” from the purple bar and then “View”. Locate the two lists you have just created, “essential genes” and “uncharacterized_with_human_disease_phenotype.” Select your two lists and click “Intersect” to find the genes in common. Name this list “yeast-gene-human-phenotype.” Click on the list name to begin exploring more information about these genes.
There are three uncharacterized yeast genes that fit the criteria of having an inviable phenotype annotation, have a human homolog, and the human homolog has a disease phenotype annotation defined by OMIM. The three genes are FMP27/YLR454W, FSF1/YOR271C, and YJR141W. It is possible that fewer genes may qualify for this list if any of these three are reclassified to the Verified set (i.e., genes that have been experimentally shown to be expressed and to have a function in the cell). SGD is continually defining new templates as new data types are being made available via YeastMine. New templates are also added at the request of users. In the near future more regulation, protein complex, protein modification, transcription, and cellular pathway details will be provided by YeastMine.
FIGURE 1.
Home page for YeastMine. All information available from the SGD database and downloads site is available via a powerful search interface. Predefined templates are provided for many common searches.
Exploring Microarray Data
The SGD provides access to a complete set of microarray expression data sets via a tool called SPELL (Hibbs et al. 2007). This tool allows a gene or set of genes to be used as a query to more than 250 data sets (studies) that include more than 400 experimental conditions (arrays). There are several links from the SGD Locus pages to the expression array tool.
-
7
In SGD, search for AIM17 and open the AIM17 Locus Overview page. Click on the Expression tab link at the top of the page. Then click on “SPELL” under the Expression Overview graph to go to the site spell.yeastgenome.org.
This provides standard red/green visualization of the microarray expression ratios. The columns are experimental conditions, and the rows are genes. With a search for AIM17 the rows represent those genes that are most similar to AIM17 based on the Pearson correlation statistic comparing the expression ratios. The second row is TPS2 indicating it has the best-adjusted correlation score across all arrays presented. The rows below these two are genes in decreasing order that are less correlated with the observed expression of AIM17. This tool allows selection of a group of genes (query set) and then observation of the genes that are most similar to the query set. -
8
Select the check box for TPS2 (keeping the AIM17 box checked) and then the “Update” icon.
The resulting view changes to provide the correlation of AIM17 and TPS2 with the other genes. The most correlated genes are TPS1 and TSL1. The contribution of each experiment to the correlation is provided along the top of the graphic. Scrolling to the right illustrates that data sets from Orlando et al. (2008) are the top ranking experiments but they only contribute 1.4% of the correlation within the queried data sets. -
9
Select the plus sign above the graphic labeled “Options for Filtering Results by Dataset Tags.” This reveals topic tags (keywords) that can be queried for each dataset. A complete list of all tags and their definitions are found by clicking on “Dataset Tags.” For this example with AIM17 and TPS2, select only respiration and then Update.
The top two correlated genes remain TPS1 and TSL1. Below the correlation graphic, the GO Term Enrichment results are shown. In this case, the set of genes shown are enriched for “trehalose metabolic process.” This is not surprising as TPS1 and TPS2 are components of the alpha, alpha-trehalose-phosphate synthase complex.The YeastMine tool explored above (Steps 1–7) also has the numerical values for these expression experiments. You can use templates from the Expression tab to identify expression scores for genes, or list out all genes associated with an expression dataset.
Acknowledgments
I am grateful to all the present and past staff of the Saccharomyces Genome Database project for their dedication to accuracy and service to life science educators and researchers. I also want to thank the yeast research community for their support and suggestions. This work was supported by the National Human Genome Research Institute (grant number U41 HG001315), and Funding for open access charge was provided by the National Institutes of Health. The content is solely the responsibility of the author and does not necessarily represent the official views of the National Human Genome Research Institute or the National Institutes of Health.
References
- Amberger J, Bocchini CA, Scott AF, Hamosh A. McKusick’s Online Mendelian Inheritance in Man (OMIM) Nucleic Acids Res. 2008;37:D793–D796. doi: 10.1093/nar/gkn665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balakrishnan R, Park J, Karra K, Hitz BC, Binkley G, Hong EL, Sullivan J, Micklem G, Cherry JM. YeastMine—An integrated data warehouse for Saccharomyces cerevisiae data as a multipurpose tool-kit. Database (Oxford) 2012 doi: 10.1093/database/bar062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fisk DG, Ball CA, Dolinski K, Engel SR, Hong EL, Issel-Tarver L, Schwartz K, Sethuraman A, Botstein D, Cherry JM. Saccharomyces Genome Database Project. Saccharomyces cerevisiae S288C genome annotation: A working hypothesis. Yeast. 2006;23:857–865. doi: 10.1002/yea.1400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hibbs MA, Hess DC, Myers CL, Huttenhower C, Li K, Troyanskaya OG. Exploring the functional landscape of gene expression: Directed search of large microarray compendia. Bioinformatics. 2007;23:2692–2699. doi: 10.1093/bioinformatics/btm403. [DOI] [PubMed] [Google Scholar]
- Kalderimis A, Lyne R, Butano D, Contrino S, Lyne M, Heimbach J, Hu F, Smith R, Stepan R, Sullivan J, et al. InterMine: Extensive web services for modern biology. Nucleic Acids Res 2014. 2014;42:W468–W472. doi: 10.1093/nar/gku301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orlando DA, Lin CY, Bernard A, Wang JY, Socolar JE, Iversen ES, Hartemink AJ, Haase SB. Global control of cell-cycle transcription by coupled CDK and network oscillators. Nature. 2008;453:944–947. doi: 10.1038/nature06955. [DOI] [PMC free article] [PubMed] [Google Scholar]